Abstract
Genomic instability drives cancer progression by promoting genetic abnormalities that allow for the multi-step clonal selection of cells with growth advantages. We previously reported that the IL-9-dependent TS1 cell line sequentially acquired activating substitutions in JAK1 and JAK3 upon successive selections for growth factor independent and JAK inhibitor-resistant cells, suggestive of a defect in mutation avoidance mechanisms. In the first part of this paper, we discovered that the gene encoding mutL homolog-1 (MLH1), a key component of the DNA mismatch repair system, is silenced by promoter methylation in TS1 cells. By means of stable ectopic expression and RNA interference methods, we showed that the high frequencies of growth factor-independent and inhibitor-resistant cells with activating JAK mutations can be attributed to the absence of MLH1 expression. In the second part of this paper, we confirm the clinical relevance of our findings by showing that chronic myeloid leukemia relapses upon ABL-targeted therapy correlated with a lower expression of MLH1 messenger RNA. Interestingly, the mutational profile observed in our TS1 model, characterized by a strong predominance of T:A>C:G transitions, was identical to the one described in the literature for primitive cells derived from chronic myeloid leukemia patients. Taken together, our observations demonstrate for the first time a causal relationship between MLH1-deficiency and incidence of oncogenic point mutations in tyrosine kinases driving cell transformation and acquired resistance to kinase-targeted cancer therapies.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2310-2) contains supplementary material, which is available to authorized users.
Keywords: MLH1, DNA mismatch repair, Janus kinase (JAK), Oncogenic mutations, Tyrosine kinase inhibitor, Drug resistance
Introduction
Malignant cell transformation requires the acquisition of various aberrant phenotypic hallmarks through the accumulation of advantageous genetic abnormalities, defined as driver mutations [1]. The notion of “mutator phenotype” has been formulated to reconcile the disparity between the low spontaneous mutation rate of normal cells, and the thousands of mutations present in a single cancer genome [2]. This concept states that malignant cells exhibit a higher mutation rate that increases the likelihood of acquiring advantageous alterations needed to overcome each selective bottleneck in cancer evolution [2]. As a result, across multiple rounds of selections for driver mutants, there is a simultaneous progressive enrichment of inactivating mutations in caretaker genes that will further enhance random mutagenesis and fuel tumorigenesis.
The mutator hypothesis also has major implications, concerning the response to targeted therapies. Indeed, it predicts that cancers constitute a highly heterogeneous population of stochastically mutated malignant cells within which drug-resistant variants pre-exist and are selected upon treatment causing rapid disease relapse [3]. The importance of such pre-existing mutants is best exemplified in chronic myeloid leukemia patients treated with imatinib by the emergence of tumors variants with point mutations in the kinase domain of BCR-ABL, which could be detected at low frequency in the bone marrow before treatment [4].
Whether all cancers invariably acquire a mutator phenotype as a driving force for progression is still controversial [5]. However, it is well accepted that certain tumors exhibit a marked genetic instability under the form of chromosomal rearrangements or small deletions/insertions within regions of short tandem repeated nucleotide tracks called microsatellites, a phenotype referred to as microsatellite instability (MSI).
The MMR system guarantees genome fidelity by recognizing and correcting post-replicative errors that escape proofreading activity of DNA polymerases. MMR process is initiated by the binding of a heterodimeric mismatch recognition complex composed of MutS homolog-2 and -6 (MSH2 and MSH6) to mispaired or misinserted bases in the newly synthesized DNA strand. Subsequent recruitment of a second heterodimer of MutL homolog-1 (MLH1) and post-meiotic segregation protein-2 (PMS2) at the site of the mismatch triggers downstream events that eventually lead to exonuclease-1-mediated removal of the error and resynthesis by DNA polymerases δ or ε [6]. Inefficient MMR confers a strong mutator phenotype characterized by a mutation rate 100- to 1000-fold greater in MMR-deficient cells as compared with normal cells [7–9].
In addition to resolving post-replicative mismatches, MMR proteins also participate to DNA damage signaling [10]. Consequently, cells lacking a functional MMR system tolerate the presence of injured DNA resulting in primary refractoriness to clinically used genotoxic chemotherapeutics, such as 6-thioguanine (6-TG) [11].
The link between MMR deficiency and tumorigenesis is well established in HNPCC patients who inherit a heterozygous germ-line mutation in one of the MMR components (mainly MSH2 and MLH1) and are prone to cancer development further to somatic inactivation of the remaining wild-type allele [9]. Besides familial cancers, MMR defects are present in a significant proportion (12–20 %) of sporadic counterparts of the HNPCC-spectrum tumors (gastro-intestinal, urologic, and gynecologic tracts) [12–15]. More recently, growing evidence for involvement of MMR defects in sporadic hematological malignancies has also been provided [16–22]. In contrast to HNPCC, MMR deficiency in sporadic cancers is commonly caused by silencing of MLH1 owing to promoter methylation.
We previously described a murine lymphoid cell line, called TS1, which was initially dependent on interleukin-9 (IL-9) for proliferation, but sequentially gained growth factor (GF)-independence and resistance to JAK inhibitors at high frequencies (1/20,000 for GF-independent clones and 1 in 2 millions for JAK inhibitor-resistant subclones) [23]. Each of the two successive selection steps was accompanied by newly acquired activating point mutations in one of the kinases associated with the IL-9 receptor, namely JAK1 and JAK3. Our data showed that the relative expansion of the mutated cells occurred in culture prior to the selective pressure exerted by cytokine withdrawal or by the inhibitor. The high phenotypic plasticity of the TS1 cell line is somewhat reminiscent of the mutator concept, where selection from a large spectrum of randomly pre-existing variants facilitates rapid adaptation to environmental pressures. In this study, we took advantage of the TS1 model to investigate the molecular mechanisms underlying the acquisition of oncogenic and inhibitor-resistant mutations in JAK kinases.
Materials and methods
Cell culture
TS1 and TS2 cells were cultured in the presence of murine IL-9 (100 U/ml) in Iscove-Dulbecco’s medium supplemented with 10 % fetal bovine serum, 50 µM β-mercaptoethanol, 0.55 mM l-arginine, 0.24 mM l-asparagine, and 1.25 mM l-glutamine. Genome-wide demethylation was achieved by supplementation of cell medium with 0.5–2 µM 5-AZA-2′deoxycytidine (Sigma-Aldrich, cat #A3656) during 48 h.
Plasmids construction, cell electroporation, and selection of stable transfectants
Murine MLH1 cDNA was inserted into the pEF6V5HisTOPO plasmid (Invitrogen). 107 cells were electroporated with 50 µg of DNA (280 V, 1500 µF, 75 Ω) and selected in the presence of blasticidin 40 µg/ml (Invivogen, ANT-BL-1) 24 h after electroporation.
Retroviral infection with shRNA and selection of stable transfectants
Retroviruses were obtained by transient transfection of HEK293 cells with pLKO.1 lentiviral vector coding for shRNA against murine MLH1 (TRCN00000042721, Thermo Scientifics) or anti-GFP (SHC004, Sigma), packaging vector psPAX2, and VSV-G protein vector pMD2-G following the RNAi Consortium recommendations. HEK293 supernatants were harvested at 24 and 48 h post-transfection and used to spin-infect 0.5 × 106 cells. Three days after infection, stable transfectants were selected with puromycin 5 µg/ml (MP biomedicals, cat #194539).
Selection of growth factor-independent clones and JAK inhibitor-resistant subclones
For the selection of GF-independent clones, IL-9-dependent TS1 or TS2 cells were washed three times in PBS and seeded in 96-well plates at a concentration of 20,000–200 cells/well in the absence of cytokines. After 1–2 weeks, GF-independent clones were picked up from plates, where less than 20 % of wells were positive for proliferation, corresponding to a probability of clonality superior to 0.9 according to the Poisson distribution. For the selection of JAK inhibitor-resistant subclones, GF-independent TS1 clones were seeded in 96-well plates at a density of 30,000 cells/well in the presence of 300–600 nM of pan-JAK inhibitor CMP6 (Calbiochem, cat #420097). After 2–3 weeks, proliferation-positive wells from plates meeting clonality conditions were counted.
Microarray
Whole transcripts were amplified from 250 ng total RNA and converted into single-stranded DNA using the Ambion® WT expression kit. 5.5 µg DNA was fragmented, labeled (Genechip® WT terminal labeling kit, Affymetrix), and hybridized on Genechip® Mouse Genome 1.0 ST array (Affymetrix). Data are accessible on the GEO platform (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70395).
RNA extraction, cDNA synthesis, and qRT-PCR
Total RNA was isolated from 106 cells using TriPure Isolation Reagent (Roche). Reverse transcription was performed on 1 µg of total RNA using an oligo-dT primer and M-MULV Reverse transcriptase (RevertAid Reverse Transcriptase, Thermo Scientific). qRT-PCR was performed using the qPCR core-kit for Sybrgreen I (Eurogentec) and primers sets annealing to murine MLH1 (forward: 5′-AGATGCTCCGTAACCATTCCTTTGTG-3′ and reverse: 5′-TAGAACAGCTCTTCACTGAGCTTGGTA-3′), murine β-actin (forward: 5′-TCCTGAGCGCAAGTACTCTGT-3′ and reverse: 5′-CTGATCCACATCTGCTGGAAG-3′), human MLH1 (forward: 5′-ACAGTAGCTGATGTTAGGACACTACC-3′ and reverse: 5′-CTATCAGTTCTCGACTAACAGCATTTCC-3′), and human EF1 (forward: 5′-GCTTCACTGCTCAGGTGAT-3′ and reverse: 5′-GCCGTGTGGCAATCCAAT-3′).
Bisulfite treatment of genomic DNA and sequencing
Bisulfite treatment of genomic DNA was performed as previously described [24]. PCR amplification was performed with primers annealing to the bisulfite-modified sequence of the murine promoter of MLH1 (forward: 5′-GTTTTTAGAAATGAGTTAATAGGAAGAG-3′ and reverse: 5′-TACCAATTTTCTATCATCTCTTTAATAACATTAACC-3′). PCR products were cloned into the PCR2.1TOPO vector (invitrogen) and transformed into DH5α bacteria. Plasmid DNA was extracted from seven colonies for each PCR amplification (Nucleospin plasmid-8, Macherey-Nagel), and inserts were sequenced using the DYEnamic Big Dye Terminator Kit (Amersham).
Western blot
Western blot analysis was performed as previously described [23]. Blots were probed with anti-MLH1 (Cell Signaling Technology, #4256) and anti-β-actin (Sigma, #A5441) antibodies.
CML patient samples
Peripheral blood leucocyte RNA was extracted from eight patients at diagnosis, and eight patients relapsing after one or more TKI were obtained from the Cliniques universitaires Saint-Luc, Brussels. Reverse transcription was performed on 1 µg of total RNA using an oligo-dT primer and M-MULV Reverse transcriptase (RevertAid Reverse Transcriptase, Thermo Scientific). An independent cohort of 17 diagnostic and 10 relapse samples obtained from the University of Bordeaux was used as a validation cohort. In all samples, the BCR-ABL/ABL transcript levels were above 10 % (IS). Written informed consent was obtained in accordance with the Declaration of Helsinki from all patients and from parents or guardians on behalf of children who participated in this study. The study was approved by the local Ethics Committee: Comité d’Ethique Hospitalo-Facultaire Saint-Luc, UCL, and Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale (CCPRB) de Bordeaux at the University of Bordeaux.
Results
MLH1 expression is repressed by promoter methylation in TS1 cells
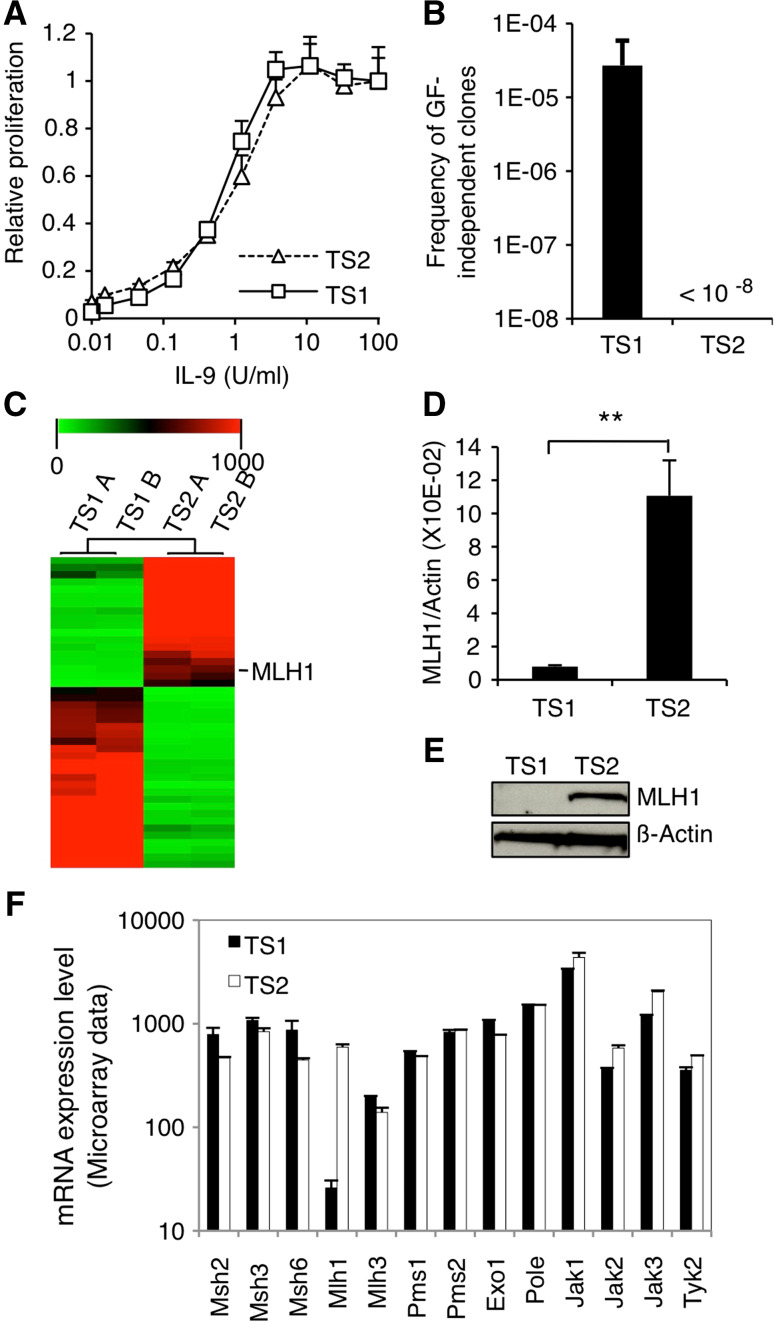
TS1 and TS2 are T-lymphocyte-derived cell lines that strictly depend on the presence of IL-9 for survival [25, 26] (Fig. 1a). IL-9 withdrawal results in rapid cell death in both cell lines, though few TS1 cells occasionally succeed to proliferate independently of growth factor at a frequency of 1/20,000 cells. We previously showed that 100 % of those GF-independent TS1 clones acquired activating point mutations in JAK1 and/or JAK3 [23] suggestive of a high rate of spontaneous mutagenesis. Interestingly, IL-9-starved TS2 cells failed to give rise to GF-independent clones (108 cells tested), indicating that this phenotype was specific for the TS1 cell line (Fig. 1b).
Fig. 1.
MLH1 expression is lost in TS1 cells. a TS1 and TS2 cells were seeded in 96-well plates at a density of 1000 cells/well in the presence of increasing concentrations of murine IL-9. After 48 h of culture, methyl-H3 thymidine was added to the cells for 4 h and thymidine incorporation was measured. Thymidine incorporation values were standardized relative to the value in the presence of IL-9 100 U/ml. b TS1 and TS2 cells were plated in the absence of IL-9 on 96-well plates at different densities ranging from 200 to 40,000 cells/well. 1–2 weeks later, wells positive for proliferation were visually screened. For TS1 cells, histogram represents mean ± SEM of seven independent selection experiments. Concerning TS2 cells, no GF-independent clones arose from a total of 108 tested cells. c Microarray data are represented as hierarchical clustering of transcripts ×20 differentially expressed in TS1 versus TS2 corresponding to a list of 44 genes (biological duplicates). The image was obtained with the Multiple Experiment Viewer program (MeV). The line corresponding to MLH1 transcript is indicated. d MLH1 mRNA levels were assessed by quantitative RT-PCR in TS1 and TS2 cells. Histograms represent mean ± SD of biological triplicates. Student’s t test was performed to determine p value. e MLH1 protein levels were assessed by western blot in TS1 and TS2 cells. The membrane was probed with anti-β-actin antibody as loading control. f mRNA expression levels of the different components of the MMR system and JAK kinases in TS1 and TS2 cell lines based on the microarray transcriptome analysis. Histograms represent mean ± SD of biological duplicates
Based on this observation, we hypothesized that TS1 cells have lost the expression of crucial genome maintenance protein(s). Using a microarray technique, we compared the transcriptomes of TS1 and TS2 cells and obtained a list of 44 genes showing at least a 20-fold differential expression (Fig. 1c; Supplementary Table 1). Among these genes, we identified MLH1 as a relevant candidate, because of its pivotal role in MMR, namely coordination of mismatch recognition with effective repair. Expression values for MLH1 transcript were 25-fold lower in TS1 compared with TS2 cells. All other components of the MMR system showed similar expression profiles between TS1 and TS2 cells (Fig. 1f). Loss of MLH1 expression in TS1 versus TS2 cells was confirmed both at the mRNA level by quantitative RT-PCR and at the protein level by the western blot analysis (Fig. 1d, e).
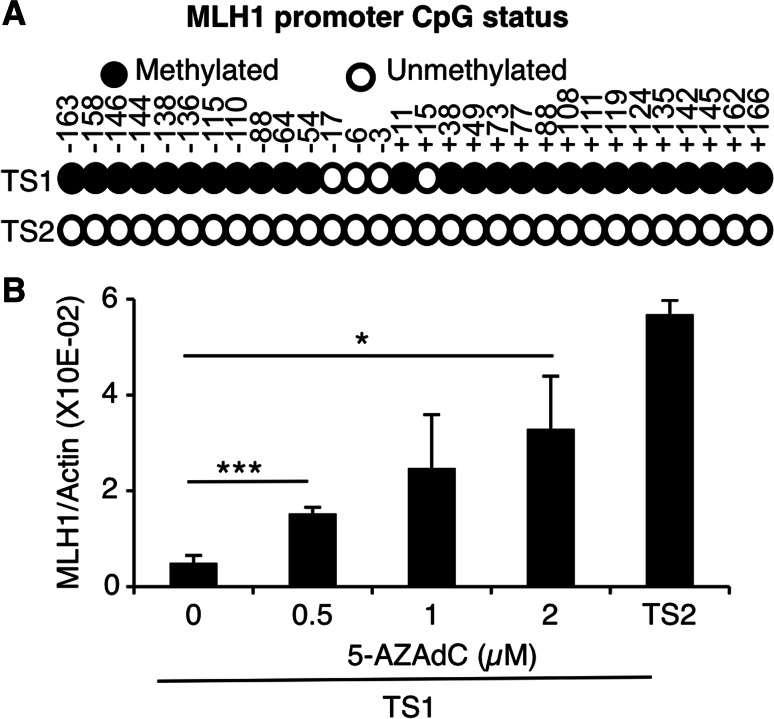
Given that the promoter of MLH1 is frequently hypermethylated in sporadic human tumors, we determined its cytosine methylation status through sequencing of bisulfite-treated genomic DNA. As shown in Fig. 2a, most CpG sites present in the MLH1 promoter region appeared to be methylated in TS1 cells, while this promoter was totally unmethylated in TS2. In line with these data, TS1 cells treated with demethylating agent 5-AZA-2′deoxycytidine (5-AZAdC) partially recovered MLH1 expression in a dose-dependent manner (Fig. 2b). No mutations were observed in the coding sequence of MLH1, MSH2, MSH6, and PMS2 in TS1 and TS2 cell lines, excluding inactivating mutations of MMR system components as a cause of the mutator phenotype. Considering those results, we postulated that MMR deficiency caused by silencing of MLH1 locus accounts for the high rate of spontaneous mutagenesis in TS1 cells.
Fig. 2.
MLH1 expression is repressed by promoter methylation in TS1 cells. a Methylation status of the MLH1 promoter was determined by bisulfite sequencing of genomic DNA in TS1 and TS2 cells. Circles represent CpG sites (methylated in black, unmethylated in white). Data represent sequencing results of seven clones from the PCR product for each cell line. Numbers indicate the nucleotide position relative to the origin of transcription. b MLH1 mRNA levels were assessed by quantitative RT-PCR in TS1 cells treated with 0/0.5/1/2 µM of 5-AZA-2′deoxycytidine for 48 h. Histograms represent mean ± SEM of three independent experiments. One-way ANOVA test was performed to determine p values (*p < 0.05, ***p < 0.001)
Ectopic MLH1 expression in TS1 cells prevents oncogenic and inhibitor-resistant point mutations in JAK kinases
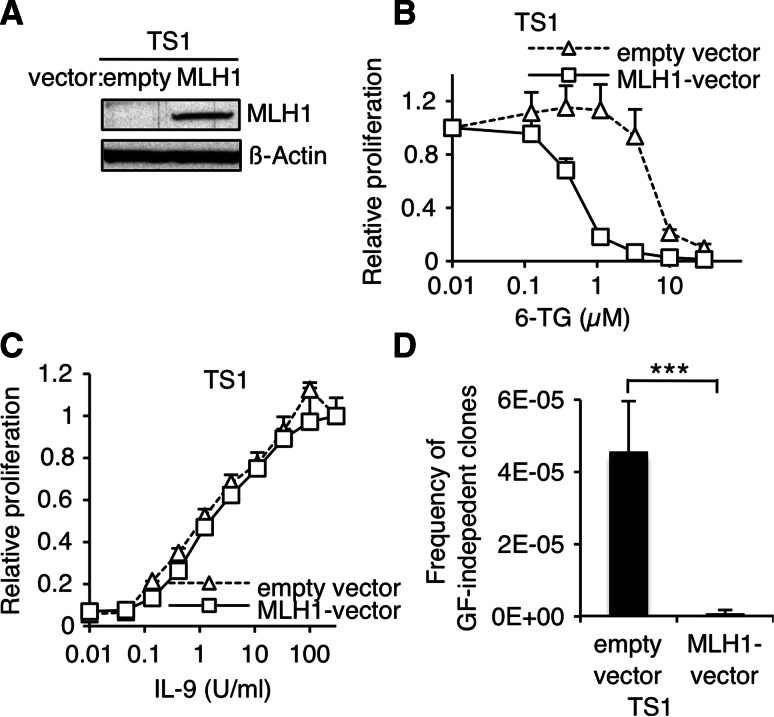
To demonstrate that loss of MLH1 is responsible for the acquisition of oncogenic mutations, we electroporated IL-9-dependent TS1 cells with an expression vector coding for MLH1 or the empty vector as control. Doing so, we obtained three independent sets of MLH1-expressing cells and paired control cells coming from distinct electroporation experiments. Effective restoration of MLH1 protein expression was confirmed by western blot on a representative set of cells (Fig. 3a). Moreover, reconstitution of DNA damage-signaling function of MMR system in MLH1-electroporated TS1 cells was demonstrated by their increased sensitivity to genotoxic agent 6-TG compared to control cells (Fig. 3b). Exogenous MLH1 expression did not modify the proliferation of TS1 cells in response to IL-9 (Fig. 3c). By contrast, when MLH1-expressing cells were plated in the absence of IL-9 to select GF-independent clones, the frequency of GF-independent clones displays a 50-fold decrease compared to control cells (Fig. 3d). Because in TS1 cells, GF-independence is systematically associated with acquisition of point mutations [23], this difference reflects a reduced mutation rate in MLH1-expressing cells.
Fig. 3.
Ectopic MLH1 expression in TS1 cells decreases the frequency of GF-independent clones. a MLH1 protein levels were assessed by western blot in TS1 cells electroporated with MLH1 or empty vector. The membrane was probed with anti-β-actin antibody as loading control. b TS1 cells electroporated with MLH1 or empty vector were seeded in 96-well plates at a density of 1000 cells/well in the presence of IL-9 100 U/ml and with increasing concentrations of 6-TG. After 48 h of culture, methyl-H3 thymidine was added to the cells for 4 h and thymidine incorporation was measured. Thymidine incorporation values were standardized relative to the value in the absence of 6-TG. c TS1 cells electroporated with MLH1 or empty vector were seeded in 96-well plates at a density of 1000 cells/well in the presence of increasing concentrations IL-9. After 48 h of culture, methyl-H3 thymidine was added to the cells for 4 h and thymidine incorporation was measured. Thymidine incorporation values were standardized relative to the value in the presence of IL-9 100 U/ml. d TS1 cells were plated in the absence of IL-9 on 96-well plates at different densities ranging from 200 to 20,000 cells/well. 1–2 weeks later, wells positive for proliferation were visually screened. Histograms represent mean ± SEM of six independent selection experiments (3 independent electroporated bulks, each bulk was tested twice). Student’s t test was performed to determine the p value (***p < 0.001)
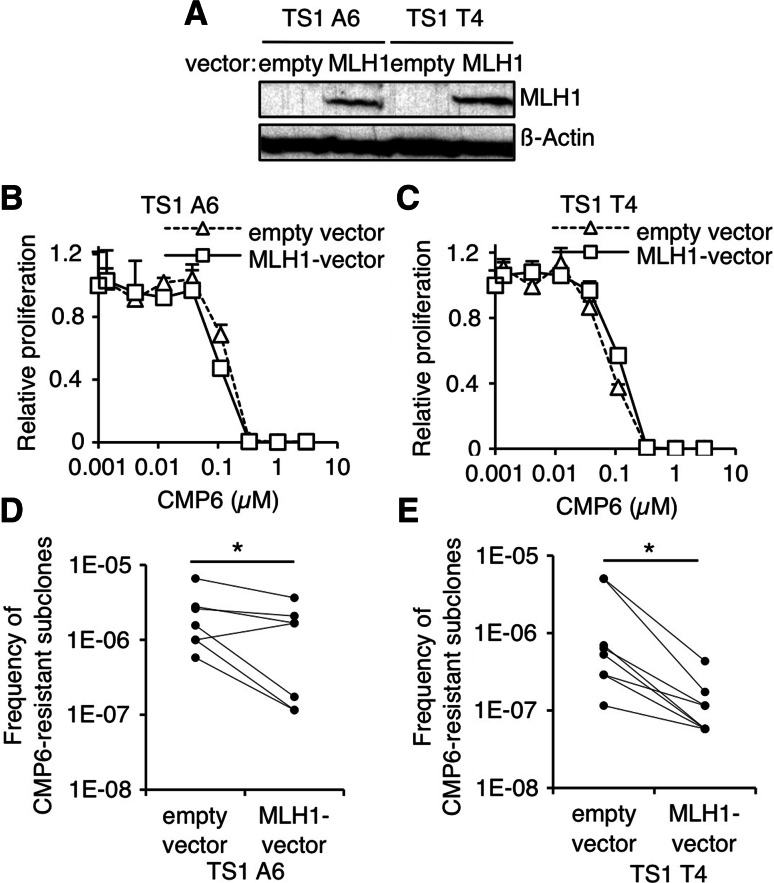
As previously shown, GF-independent clones were sensitive to JAK inhibitors, such as CMP6, in short-term assays. However, resistant subclones arose upon long-term culture in the presence of inhibitor as a result of secondary mutations in JAK1 or JAK3 [23]. Hence, we asked whether restoration of MLH1 in TS1 GF-independent clones could prevent the occurrence of secondary mutations and thereby the emergence of CMP6-resistant subclones. Two different GF-independent clones (TS1 A6 and T4) were electroporated with MLH1 vector (Fig. 4a). In short-term assays, ectopic MLH1 expression had no impact on the sensitivity of clones to CMP6 (Fig. 4b, c). However, when cells were seeded in the presence of the inhibitor for 3 weeks, significantly less CMP6-resistant subclones emerged from cells that express MLH1 as compared to control cells (Fig. 4d, e).
Fig. 4.
Ectopic MLH1 expression in GF-independent TS1 clones decreases the frequency of CMP6-resistant subclones. a MLH1 protein levels were assessed by western blot in TS1 A6 and T4 GF-independent clones electroporated with MLH1 or empty vector. The membrane was probed with anti-β-actin antibody as loading control. b, c GF-independent TS1 A6 and T4 clones electroporated with MLH1 or empty vector were seeded in 96-well plates at a density of 1000 cells/wells in the presence of increasing concentrations of CMP6. After 48 h of culture, methyl-H3 thymidine was added to the cells for 4 h and thymidine incorporation was measured. Thymidine incorporation values were standardized relative to the value in the absence of CMP6. d, e GF-independent TS1 A6 and T4 clones electroporated with MLH1 or empty vector were plated in the presence of CMP6 (300–600 nM) on 96-well plates at a density of 30,000 cells/well. 2–3 weeks later, wells positive for proliferation were visually screened. Graphs represent paired frequency values from seven and eight independent selection experiments (each of the four electroporated set was at least tested once). Paired Wilcoxon test was performed to determine p values (*p < 0.05)
Altogether, our data demonstrate that ectopic expression of MLH1 in TS1 cells significantly diminishes the incidences of GF-independent clones and CMP6-resistant subclones.
MLH1 knock-down in TS2 cells promotes oncogenic point mutations in JAK3
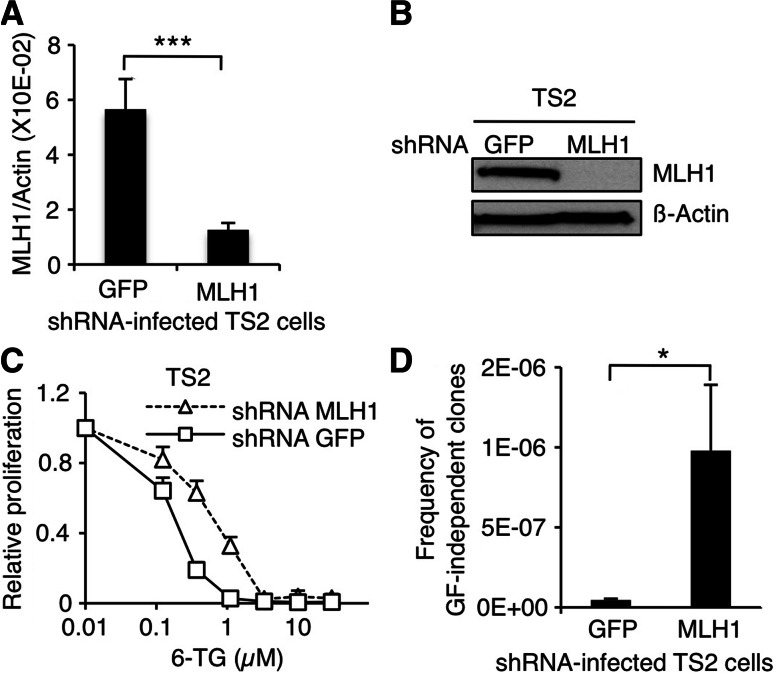
We next sought to determine if the TS2 cell line could give rise to GF-independent clones with acquired mutations in JAK1 and JAK3 upon MLH1 knock-down (KD). To that end, TS2 cells were infected with retroviruses coding for shRNAs against MLH1 or against GFP as a control. In doing so, we obtained seven paired TS2 bulks coming from seven independent infection experiments. As presented in Fig. 5a, shRNA-targeting MLH1 successfully silenced 78 % of the mRNA expression. Subsequent reduction in MLH1 protein amount was corroborated by western blot on a representative set of cells (Fig. 5b). MLH1-KD TS2 cells showed a decreased sensitivity to 6-TG compared to control cells, indicating that effective MMR-dependent DNA damage signaling was impaired (Fig. 5c).
Fig. 5.
shRNA-mediated MLH1 knock-down in TS2 cells increases the frequency of GF-independent clones. a MLH1 mRNA levels assessed by quantitative RT-PCR in TS2 cells infected with retroviruses coding for shRNA against MLH1 or GFP. Histograms represent mean ± SEM of seven independent infection experiments. Student’s t test was performed to determine p value. b MLH1 protein levels assessed by western blot in shRNA-infected TS2. The membrane was probed with anti-β-actin antibody as loading control. c TS2 cells infected with retroviruses coding for shRNA against MLH1 or GFP were seeded in 96-well plates at a density of 1000 cells/well in the presence of IL-9 100 U/ml and with increasing concentrations of 6-TG. After 48 h of culture, methyl-H3 thymidine was added to the cells for 4 h and thymidine incorporation was measured. Thymidine incorporation values were standardized relative to the value in the absence of 6-TG. d TS2 cells infected with retroviruses coding for shRNA against MLH1 or GFP were plated in the absence of IL-9 on 96-well plates at a density of 40,000 cells/well. 1–2 weeks later, wells positive for proliferation were visually screened. Histograms represent mean ± SEM of seven independent selection experiments (each set of infected cells was tested once). Student’s t test was performed to determine p values (*p < 0.05, ***p < 0.001)
Altogether, 162 × 106 shRNA-infected TS2 cells were plated in the absence of IL-9 with the hope to select GF-independent clones from MLH1-KD cells. In line with our expectations, a series of GF-independent clones arose at an average frequency of 1 in a million MLH1-KD TS2 cells (Fig. 5d). By contrast, only rare (three) GF-independent clones emerged from the control TS2 cells, at an extremely low frequency (<2 × 10−8). Fourty-five GF-independent TS2 clones (including the three clones from the control cells) were picked up and screened for mutations in JAK1 and JAK3 (Table 1). 89 % (40/45) of them had acquired a mutation in JAK3, whereas none did in JAK1. Altogether, we identified nine different substitutions affecting six conserved residues between mouse and human JAK3. All residues were previously shown to be mutated in GF-independent TS1 clones [23] and for three of them (A572, R657, and V674), mutations have been also described in human leukemia [27, 28]. The remaining five clones displayed no mutation indicating that acquiring a mutation in JAK3 is not the only way for TS2 cells to proliferate independently of growth factor. This contrasts with the TS1 cell line, where 100 % of GF-independent clones gained a mutation in JAK1 and/or JAK3.
Table 1.
List of the JAK3 mutations identified in growth factor-independent TS2 clones
| shRNA | JAK1 | JAK3 | Number of sequenced clones (n = 45) |
|---|---|---|---|
| Anti-MLH1 | WT | A572V | 9 |
| WT | L586S | 6 | |
| WT | R657H | 3 | |
| WT | R657C | 6 | |
| WT | F666C | 2 | |
| WT | V674A | 10 | |
| WT | V674G | 1 | |
| WT | T848A | 1 | |
| WT | WT | 4 | |
| Anti-GFP | WT | R657H | 1 |
| WT | V674F | 1 |
The table shows the 9 different identified substitutions and their respective absolute frequencies for in a total of 45 characterized TS2 clones. All six affected amino acids are conserved in mouse and human JAKs (amino acid numbers correspond to human sequence)
Thus, these data show that selection of GF-independent clones with activating mutations in JAK3 from the MLH1-expressing TS2 cell line is a rare event, but its incidence is strongly increased upon MLH1 silencing.
TS1 cells and primitive CML cells share a common mutational profile dominated by T:A>C:G transitions
Next, we investigated if the loss of MLH1 would be translated into a mutational signature present in our collection of TS1-derived JAK1 and JAK3 mutations. As shown in Fig. 6, nucleotide exchanges in TS1 cells are strongly biased towards T:A>C:G transitions in comparison with the expected pattern of mutations arising spontaneously in normal human cells [29]. Thus, in TS1 cells, the accumulation of T:A>C:G substitutions is favored, potentially as a reflect of MLH1-deficiency or MMR-defect, in general. Our data are consistent with studies in bacteria demonstrating that the spectrum of base substitutions is dominated by T:A>C:G mutations in the absence of efficient MMR [30, 31].
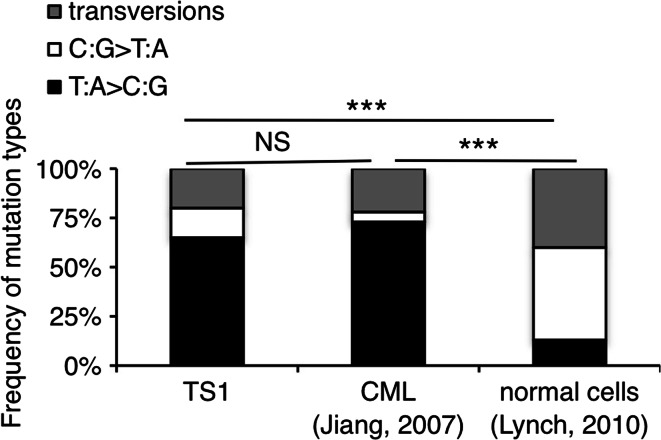
Fig. 6.
TS1 and CML cells share a common mutational profile distinct from normal cells. The diagrams present the percentages of T:A>C:G and C:G>T:A transitions as well as transversions (substitutions of a purine by a pyrimidine base and conversely) occurring in JAK1 and JAK3 genes for the TS1 cells, in BCR-ABL for CML cells [32, 33] and in unselected regions of the genome for the normal cells [29]. χ 2 test was performed to determine the p values (***p < 0.001; NS non-significant)
As already mentioned in the introduction, a fraction of CML patients treated with imatinib experience disease relapse owing to the emergence of pre-existing tumors variants with point mutations in the kinase domain of BCR-ABL. This is somewhat reminiscent of the selection of JAK inhibitor-resistant subclones with acquired mutations in our MLH1-deficient TS1 model. Interestingly, Grant et al. showed that point mutations in BCR-ABL arising in primitive CML cells from Imatinib-naive and Imatinib-resistant patients are characterized by a predominance of T:A>C:G transitions and an under-representation of C:G>T:A transitions, as a result of an unknown mutator process [32, 33]. Knowing this, we compared the profile of nucleotide exchanges in our TS1 model with the one described in CML cells. As shown in Fig. 6, TS1 cells and primitive CML cells display an identical mutational pattern, which differs significantly from the standard profile. This analysis indicates that a defect in MLH1 is a potential suspect for the mutator mechanism operative in CML cells that renders them prone to acquire new mutations in BCR-ABL.
MLH1 expression is down-regulated in BCR-ABL inhibitor-resistant versus newly diagnosed CML patients
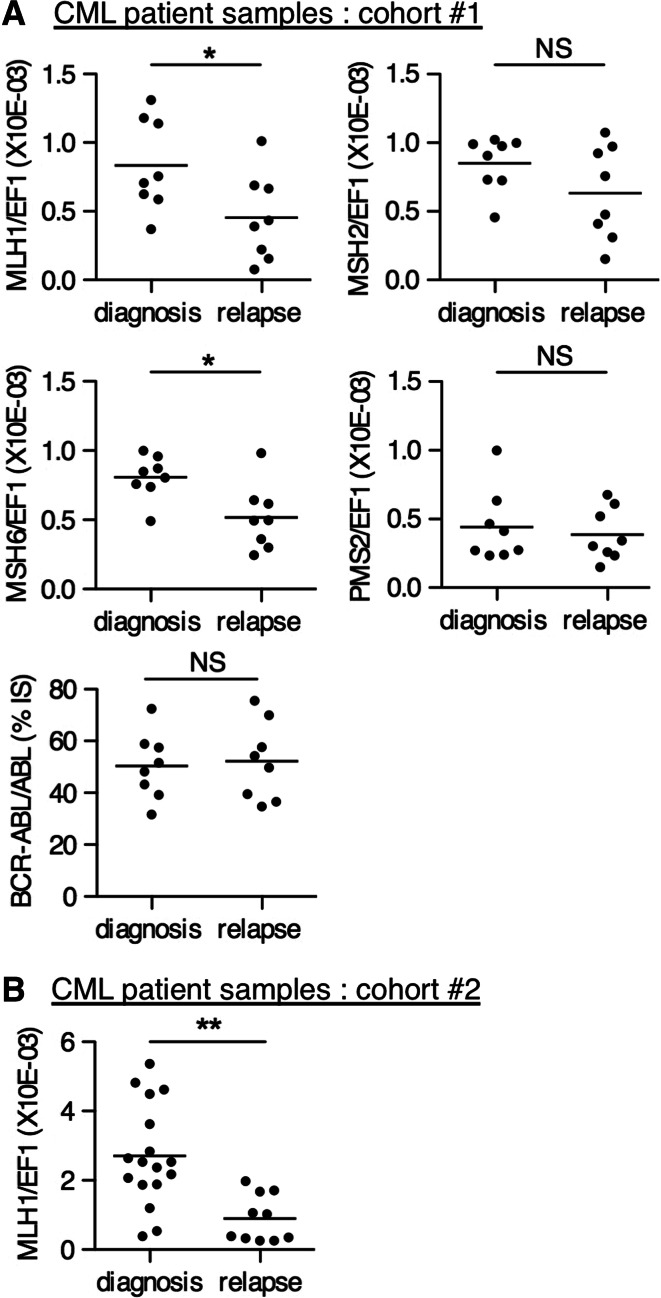
Given the mutational bias described above, we sought to compare the expression of MLH1 in peripheral blood samples coming from BCR-ABL inhibitor-resistant patients and newly diagnosed inhibitor-naïve ones. First, we assessed the mRNA expression of MLH1 in a test cohort, including 16 non-paired CML samples (8 at diagnosis and 8 at relapse), from patients referred to the Saint-Luc Hospital in Brussels. For six out of the eight relapsed samples, acquired mutations in BCR-ABL could be detected. In this cohort, MLH1 expression was significantly down-regulated in the samples at relapse as compared to samples at diagnosis (Fig. 7a, left panel). BCR-ABL/ABL ratios were similar between the two groups (Fig. 7a, right panel). For the other components of the MMR system, we observed no difference in the expression of MSH2 and PMS2, whereas MSH6 mRNA levels displayed a reduction in relapsed patients versus patients at diagnosis (Fig. 7a), but this was only tested in the Saint-Luc Hospital cohort representing a limited set of data.
Fig. 7.
MLH1 mRNA expression is down-regulated in imatinib-resistant versus newly diagnosed CML patient samples. MLH1 mRNA levels assessed by quantitative RT-PCR in CML blood samples taken at diagnosis or at relapse after imatinib treatment from two independent sets of patients, a samples of our test cohort (cohort #1) from Saint-Luc Hospital, Brussels (n = 8 for both diagnosis and relapse) and b samples of the validation cohort (cohort #2) from the University of Bordeaux (n = 17 for diagnosis, n = 10 for relapse). MSH2, MSH6, PMS2 levels, as well as BCR-ABL/ABL ratios of our test cohort from Saint-Luc Hospital are also shown. Horizontal lines represent the mean values. Non-parametric Mann–Whitney one-tailed test was performed to calculate the p values (*p < 0.05, **p < 0.01, NS non-significant
The difference in MLH1 expression was further confirmed on a separate validation cohort coming from the University of Bordeaux comprising 17 diagnosed and 10 relapsed samples from which 6 acquired a mutation in BCR-ABL (Fig. 7b). Clinical details of the patients are described in Supplementary Table 2. Thus, selection of BCR-ABL inhibitor-resistant tumor subclones in CML patients is correlated with the selection of cells with a reduced MLH1 expression, corroborating the clinical significance of our previous in vitro findings.
Discussion
In this paper, we made the clinically relevant observation that loss of MLH1 expression continuously promotes the appearance of point mutations in kinases leading to genetically distinct cell variants within a population that are selected by their fitness to adapt upon cytokine deprivation or therapeutic pressure. This finding stems from experiments carried out on the MLH1-deficient TS1 model, in which sequential acquisition of activating mutations in JAK1 and JAK3 are required to overcome successively IL-9 withdrawal and exposure to JAK inhibitor. Restoration of MLH1 expression at both stages, IL-9-dependent TS1 cells or GF-independent TS1 clones, significantly reduces the frequencies of further transformed cells with newly acquired JAK mutations, whereas conversely, down-regulation in MLH1-proficient TS2 cells enhances the incidence of GF-independent clones harboring mutations in JAK3.
At the moment, ruxolitinib is the only JAK inhibitor that achieved FDA approval. Although no clinical resistance mediated by acquired mutations has been reported yet upon treatment with ruxolitinib, we believe that our model system would be also relevant in other cancer therapies for which patient-derived drug-resistant mutations are described, as illustrated here for BCR-ABL+ CML. Indeed, it seems very likely that mutations in MMR-deficient cells are not restricted to JAK1 and JAK3 genes, but rather occur randomly across the genome. Therefore, extrapolation of our findings to other oncogenes strongly suggests that MLH1-silenced tumors are predisposed to develop secondary resistance against any clinically used targeted therapy.
Imatinib, an ABL1 inhibitor, represents an appropriate clinical illustration of targeted therapy whose effectiveness is limited by the emergence of pre-existing tumor subclones carrying drug-resistant mutations in the kinase domain of BCR-ABL resulting in CML relapse [34]. It has been assumed that the appearance of CML cells harboring such tyrosine kinase inhibitor (TKI)-resistant mutations is fostered by the increased genomic instability in the subset of cells from which the leukemic cells are derived, the so-called leukemia stem cells (LSC) [35]. This genomic instability in LSC, manifested as chromosomal aberrations and point mutations (including BCR-ABL mutations) [32], is believed to stem from a combination of increased DNA damages and inefficient repair [36, 37]. BCR-ABL itself promotes genetic instability through excessive ROS production [38–40] and inhibition of MLH1-PMS2 dimers assembly, but does directly inhibit the expression of MMR proteins [41]. The cause of reduced MLH1 expression at relapse remains unknown. Promoter hypermethylation was not be detected by bisulfite sequencing and Methylation-Specific MLPA, and no haploinsufficiency could be detected in our validation cohort (data not shown).
Here, we provide arguments in favor of a down-regulation of MLH1 expression playing a role in genetic instability of LSC. We show that the pattern of BCR-ABL arising in vivo and in vitro in primitive CD34+ cells from Imatinib-naive and Imatinib-resistant CML patients strikingly resembles the JAK1 and JAK3 mutations occurring spontaneously in our MLH1-deficient TS1 cells [32]. Indeed, both mutational profiles are characterized by a marked prevalence of T:A>C:G transitions, which is distinct from the natural predominance of C:G>T:A mutations due to spontaneous cytosine deamination [29].
A link between genomic instability and high risk of relapse upon TKI therapy was already evoked by the observation that the overall frequency of BCR-ABL mutations increases with sequential TKI use [42]. In particular, patients who harbor BCR-ABL mutations conferring resistance to first-line inhibitor are more likely to develop additional mutations on second-line or third-line TKI therapy than patients without such mutations [43, 44]. This correlation suggests that, in this specific subset of patients, a higher genetic instability boosts spontaneous mutagenesis and fosters the rapid emergence of resistant clones upon treatment. In our paper, we demonstrate that MLH1 expression is significantly down-regulated in leukemic cells from BCR-ABL inhibitor-resistant CML patients compared to newly diagnosed ones. This indicates that therapy relapse might be due to selection of a subset of leukemic progenitors with lower MLH1 expression that consequently possess a higher likelihood to acquire additional mutations in BCR-ABL.
In conclusion, our results indicate that MLH1-deficient tumors are associated with a high risk of relapse upon oncogene inhibitor as monotherapy. Exploiting the Achilles heel of MMR-deficient tumors by making use of synthetic lethal interactions or boosting anti-tumor immune surveillance represents attractive tumor-specific therapeutic avenues. Therefore, combining oncogene-targeted drugs with synthetic lethal approaches or immunotherapy could specifically and efficiently ablate the MMR-defective cancer cells while avoiding the selection of tumor variants harboring drug-resistant mutations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported in part by the Belgian Program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming (Grant IAP-P7/39 to J.C.R.), by the Actions de Recherche Concertées of the Communauté Française de Belgique (Grant ARC09/14-021 to J.C.R.), by the Fondation contre le Cancer, by the Foundation Salus Sanguinis, Belgium, and by the Fonds pour la formation à la Recherche dans l’Industrie et dans l’Agriculture (F.R.I.A). E.L. is the recipient of a fellowship from FRS-FNRS Télévie. L.K. is a MD fellow of the Fonds National de la Recherche Scientifique (F.N.R.S.), Belgium.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Footnotes
L. Knoops and J.-C. Renauld shared senior co-authorship.
Contributor Information
Laurent Knoops, Phone: + 32 2 764 74 41, Email: laurent.knoops@uclouvain.be.
Jean-Christophe Renauld, Phone: + 32 2 764 74 64, Email: jean-christophe.renauld@bru.licr.org.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61(8):3230–3239. [PubMed] [Google Scholar]
- 3.Fox EJ, Prindle MJ, Loeb LA. Do mutator mutations fuel tumorigenesis? Cancer Metastasis Rev. 2013;32(3–4):353–361. doi: 10.1007/s10555-013-9426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100(3):1014–1018. doi: 10.1182/blood.V100.3.1014. [DOI] [PubMed] [Google Scholar]
- 5.Bodmer W, Bielas JH, Beckman RA. Genetic instability is not a requirement for tumor development. Cancer Res. 2008;68(10):3558–3560. doi: 10.1158/0008-5472.CAN-07-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 7.Glaab WE, Tindall KR. Mutation rate at the hprt locus in human cancer cell lines with specific mismatch repair-gene defects. Carcinogenesis. 1997;18(1):1–8. doi: 10.1093/carcin/18.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Spampinato CP, Gomez RL, Galles C, et al. From bacteria to plants: a compendium of mismatch repair assays. Mutat Res. 2009;682(2–3):110–128. doi: 10.1016/j.mrrev.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Lynch HT, Snyder CL, Shaw TG, et al. Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer. 2015;15(3):181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 10.O’Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27(4):682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- 11.Glaab WE, Risinger JI, Umar A, et al. Resistance to 6-thioguanine in mismatch repair-deficient human cancer cell lines correlates with an increase in induced mutations at the HPRT locus. Carcinogenesis. 1998;19(11):1931–1937. doi: 10.1093/carcin/19.11.1931. [DOI] [PubMed] [Google Scholar]
- 12.Ionov Y, Peinado MA, Malkhosyan S, et al. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363(6429):558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 13.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95(12):6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpkins SB, Bocker T, Swisher EM, et al. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet. 1999;8(4):661–666. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann A, Zanardo L, Bocker-Edmonston T, et al. Frequent microsatellite instability in sporadic tumors of the upper urinary tract. Cancer Res. 2002;62(23):6796–6802. [PubMed] [Google Scholar]
- 16.Gu L, Cline-Brown B, Zhang F, et al. Mismatch repair deficiency in hematological malignancies with microsatellite instability. Oncogene. 2002;21(37):5758–5764. doi: 10.1038/sj.onc.1205695. [DOI] [PubMed] [Google Scholar]
- 17.Siu LL, Chan JK, Wong KF, et al. Specific patterns of gene methylation in natural killer cell lymphomas: p73 is consistently involved. Am J Pathol. 2002;160(1):59–66. doi: 10.1016/S0002-9440(10)64349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matheson EC, Hall AG. Assessment of mismatch repair function in leukaemic cell lines and blasts from children with acute lymphoblastic leukaemia. Carcinogenesis. 2003;24(1):31–38. doi: 10.1093/carcin/24.1.31. [DOI] [PubMed] [Google Scholar]
- 19.Mao G, Yuan F, Absher K, et al. Preferential loss of mismatch repair function in refractory and relapsed acute myeloid leukemia: potential contribution to AML progression. Cell Res. 2008;18(2):281–289. doi: 10.1038/cr.2008.14. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths EA, Gore SD, Hooker CM, et al. Epigenetic differences in cytogenetically normal versus abnormal acute myeloid leukemia. Epigenetics. 2010;5(7):590–600. doi: 10.4161/epi.5.7.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hangaishi A, Ogawa S, Mitani K, et al. Mutations and loss of expression of a mismatch repair gene, hMLH1, in leukemia and lymphoma cell lines. Blood. 1997;89(5):1740–1747. [PubMed] [Google Scholar]
- 22.Morimoto H, Tsukada J, Kominato Y, et al. Reduced expression of human mismatch repair genes in adult T-cell leukemia. Am J Hematol. 2005;78(2):100–107. doi: 10.1002/ajh.20259. [DOI] [PubMed] [Google Scholar]
- 23.Springuel L, Hornakova T, Losdyck E, et al. Cooperating JAK1 and JAK3 mutants increase resistance to JAK inhibitors. Blood. 2014;124(26):3924–3931. doi: 10.1182/blood-2014-05-576652. [DOI] [PubMed] [Google Scholar]
- 24.De Smet C, Lurquin C, Lethe B, et al. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19(11):7327–7335. doi: 10.1128/MCB.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uyttenhove C, Simpson RJ, Van Snick J. Functional and structural characterization of P40, a mouse glycoprotein with T-cell growth factor activity. Proc Natl Acad Sci USA. 1988;85(18):6934–6938. doi: 10.1073/pnas.85.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louahed J, Kermouni A, Van Snick J, et al. IL-9 induces expression of granzymes and high-affinity IgE receptor in murine T helper clones. J Immunol. 1995;154(10):5061–5070. [PubMed] [Google Scholar]
- 27.Bergmann AK, Schneppenheim S, Seifert M, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer. 2014;53(4):309–316. doi: 10.1002/gcc.22141. [DOI] [PubMed] [Google Scholar]
- 28.Kiel MJ, Velusamy T, Rolland D, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124(9):1460–1472. doi: 10.1182/blood-2014-03-559542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci USA. 2010;107(3):961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H, Popodi E, Tang H, et al. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci USA. 2012;109(41):E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long H, Sung W, Miller SF, et al. Mutation rate, spectrum, topology, and context-dependency in the DNA mismatch repair-deficient Pseudomonas fluorescens ATCC948. Genome Biol Evol. 2014;7(1):262–271. doi: 10.1093/gbe/evu284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, Saw KM, Eaves A, et al. Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst. 2007;99(9):680–693. doi: 10.1093/jnci/djk150. [DOI] [PubMed] [Google Scholar]
- 33.Grant H, Jiang X, Stebbing J, et al. Analysis of BCR-ABL1 tyrosine kinase domain mutational spectra in primitive chronic myeloid leukemia cells suggests a unique mutator phenotype. Leukemia. 2010;24(10):1817–1821. doi: 10.1038/leu.2010.179. [DOI] [PubMed] [Google Scholar]
- 34.Iqbal Z, Aleem A, Iqbal M, et al. Sensitive detection of pre-existing BCR-ABL kinase domain mutations in CD34+ cells of newly diagnosed chronic-phase chronic myeloid leukemia patients is associated with imatinib resistance: implications in the post-imatinib era. PLoS One. 2013;8(2):e55717. doi: 10.1371/journal.pone.0055717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Bolton-Gillespie E, Schemionek M, Klein HU, et al. Genomic instability may originate from imatinib-refractory chronic myeloid leukemia stem cells. Blood. 2013;121(20):4175–4183. doi: 10.1182/blood-2012-11-466938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrotti D, Jamieson C, Goldman J, et al. Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest. 2010;120(7):2254–2264. doi: 10.1172/JCI41246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burke BA, Carroll M. BCR-ABL: a multi-faceted promoter of DNA mutation in chronic myelogeneous leukemia. Leukemia. 2010;24(6):1105–1112. doi: 10.1038/leu.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JH, Chu SC, Gramlich JL, et al. Activation of the PI3K/mTOR pathway by BCR-ABL contributes to increased production of reactive oxygen species. Blood. 2005;105(4):1717–1723. doi: 10.1182/blood-2004-03-0849. [DOI] [PubMed] [Google Scholar]
- 39.Koptyra M, Falinski R, Nowicki MO, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108(1):319–327. doi: 10.1182/blood-2005-07-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sattler M, Verma S, Shrikhande G, et al. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. J Biol Chem. 2000;275(32):24273–24278. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- 41.Stoklosa T, Poplawski T, Koptyra M, et al. BCR/ABL inhibits mismatch repair to protect from apoptosis and induce point mutations. Cancer Res. 2008;68(8):2576–2580. doi: 10.1158/0008-5472.CAN-07-6858. [DOI] [PubMed] [Google Scholar]
- 42.Cortes J, Jabbour E, Kantarjian H, et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood. 2007;110(12):4005–4011. doi: 10.1182/blood-2007-03-080838. [DOI] [PubMed] [Google Scholar]
- 43.Shah NP, Skaggs BJ, Branford S, et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest. 2007;117(9):2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soverini S, Gnani A, Colarossi S, et al. Philadelphia-positive patients who already harbor imatinib-resistant Bcr-Abl kinase domain mutations have a higher likelihood of developing additional mutations associated with resistance to second- or third-line tyrosine kinase inhibitors. Blood. 2009;114(10):2168–2171. doi: 10.1182/blood-2009-01-197186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.