Abstract
Early neural fate commitment is a key process in neural development and establishment of the central nervous system, and this process is tightly controlled by extrinsic signals, intrinsic factors, and epigenetic regulation. Here, we summarize the main findings regarding the regulatory network of epigenetic mechanisms that play important roles during early neural fate determination and embryonic development, including histone modifications, chromatin remodeling, DNA modifications, and RNA-level regulation. These regulatory mechanisms coordinate to play essential roles in silencing of pluripotency genes and activating key neurodevelopmental genes during cell fate commitment at DNA, histone, chromatin, and RNA levels. Moreover, we discuss the relationship between epigenetic regulation, signaling pathways, and intrinsic factors during early neural fate specification.
Keywords: Early neural fate commitment, Epigenetic regulation, Histone modification, DNA methylation, Chromatin remodeling
Introduction
During gastrulation of vertebrate embryonic development, pluripotent epiblast cells undergo massive reorganization and fate specification into three distinct germ layers: ectoderm, mesoderm, and endoderm. The ectodermal cells are then patterned under the action of inductive signals and intrinsic factors to differentiate into neural or epidermal cells [1, 2]. Early neural fate commitment requires sequential activation of neural lineage regulators that are cooperatively modulated by epigenetic mechanisms [3].
In recent years, the capacity of pluripotent cells to differentiate into the neural lineage has been extensively studied from temporal and spatial perspectives in vivo and in vitro [4, 5]. Epigenetic control of key neurodevelopmental genes is a heritable process that occurs through cell proliferation and differentiation, and acts as an imperative mechanism to determine the cell fates of descendants of pluripotent cells [6]. Developmental genes are retained in a ‘poised state’ that is dynamically modulated by epigenetic markers, which are activated through cooperation with differentiation-inducing signals and intrinsic differentiation programs. Consequently, the permanent activation or repression of distinct developmental genes restricts cell fates concomitantly [7].
The specific epigenetic modulation of histones, DNA, and RNA regulate the chromatin state, chromatin accessibility, DNA binding activity, and/or transcriptional activity [8]. Numerous studies have demonstrated that epigenetic regulation plays essential roles in neural fate commitment. During this process, pluripotency genes are switched off and neurodevelopmental genes become accessible to transcriptional activators, and in turn are transcriptionally activated [3]. Here, we review the epigenetic mechanisms, such as histone methylation/acetylation, DNA methylation/hydroxylation, and RNA modifications, which underlie the regulation of differentiation capacity during early embryonic development.
Early neural fate commitment
The mammalian central nervous system consists of several neuronal subtypes, which are derived from a common pool of neural progenitor cells (NPCs) [9]. Hence, the differentiation program from pluripotent stem cells in the inner cell mass (ICM) of the embryo into NPCs, referred to as early neural fate commitment, is the biological basis for establishment of the central nervous system and brain architecture [6]. Theoretically, cells in the ICMs have the potential to differentiate into all of the cell types for embryonic development, during which these cells undergo sequential cell fate restriction, accompanied by the concurrent loss of the potent state [10]. During neural fate commitment, ectodermal cells that do not ingress through the primitive streak acquire a neural fate but not a mesendoderm lineage. This differentiation process involves an intricate transcriptional program switching between activated and silenced genes to determine the differentiation potential of each cell [11]. Knowledge of how the differentiation capacity of pluripotent cells is spatially and temporally controlled during early neural development is fundamental for the understanding of neural fate commitment and neurodevelopmental disorders.
Evidence from different species supports the notion that the mechanisms underlying the neural fate commitment are evolutionarily conserved [12]. During the early neural induction process, ectoderm cells autonomously differentiate into the neural lineage, which is inhibited by bone morphogenetic proteins (BMPs) [13–15]. Fibroblast growth factors (FGFs), wingless/int class proteins (WNTs) and Nodal are also involved in neural fate commitment [16–18]. These extracellular signaling pathways are integrated to orchestrate establishment of an appropriate neural fate [19].
In addition to extrinsic cues, the transcriptional activity of developmental genes is also regulated by transcriptional and epigenetic factors. In undifferentiated embryonic stem cells (ESCs), pluripotency genes, such as Oct4, Nanog, and Sox2, are transcriptionally activated, whereas key neurodevelopmental genes are silenced or expressed at basal levels under the control of epigenetic markers or cell cycle regulators to maintain the pluripotent state of ESCs [11, 20]. Simultaneously, many neural regulators, the associated chromatin of which is maintained in a reversible silenced state, are poised to accept the stimulating developmental cues; this poised state confers the differentiation potential of ESCs [6]. Sox2, Zfp521, SIP1, Pou3f1, and other transcription factors coordinate with each other to initiate and/or reinforce the appropriate neural fate commitment process [3]. During ESC neural differentiation, Pou3f1 functions as an upstream regulator of Sox2 and Zfp521 to promote neural differentiation. Other intrinsic factors, such as Zic2/3, Otx2, and Sox2, may participate in the repression of non-neural lineages by repressing the expression of Sox17 and Eomes [3]. Along with the inactivation of pluripotency genes, neurodevelopmental genes are sequentially activated in an ordered regulatory cascade to initiate the neural differentiation process [21]. How the transition of epigenetic states associated with key genes during early neural fate commitment is accurately controlled by numerous types of epigenetic regulators has become a topic of intense interest in recent years and will be reviewed and discussed here.
Research model for neural differentiation
To study early neurodevelopment, Xenopus, zebrafish, chick, and mouse embryos are extensively employed for embryo manipulation and functional assays [9]. By using these animal models in the past decades, it has been established that BMP, Nodal, FGF, and WNT signaling pathways play crucial roles in early embryonic patterning [16–18, 22, 23]. We have also demonstrated that BMP signaling acts through activation of distinct downstream targets, such as AP2gamma and Ovol2, to regulate ectoderm patterning and mesendoderm specification [24, 25]. Although the general rules for neural fate commitment are conserved across various species, inconsistent results regarding the details of their execution have been obtained from different species, probably due to the divergence of species and the molecular details of their germ-layer interactions [17]. This indicates that different species likely employ differential regulatory mechanisms for early neural fate commitment. Therefore, amenable in vitro models are urgently required to recapitulate embryonic neurodevelopment for mechanistic studies.
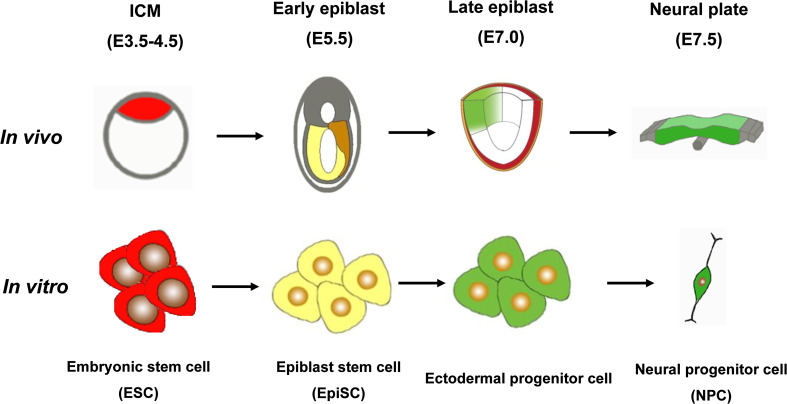
Since mouse ESCs (mESCs) were successfully established in vitro [26, 27], numerous studies have reported various methods to direct mESCs differentiation into neuroectoderm cells under appropriate culture conditions [5]. These in vitro differentiation models have been subsequently used for exploring functional roles of signaling pathways and their underlying mechanisms during pluripotent cell fate specification. Another type of pluripotent stem cells, named epiblast stem cells (EpiSCs), are derived from epiblast tissues of early mouse embryos or differentiating mESCs and are equivalent to late epiblasts [22, 28, 29]. The neural differentiation of EpiSCs can also recapitulate the neural differentiation program [22, 30], thereby offering a novel system to study the mechanisms of early embryonic development at specific developmental stages. Combined with our recently reported ectodermal progenitor cells [23, 31], in vitro cultured cells can recapitulate nearly every stage of early mouse embryonic development (Fig. 1).
Fig. 1.
In vitro differentiation models recapitulate in vivo early neurodevelopment. Established cell lines cultured in vitro correspond to the different states of mouse embryos at specific developmental stages in vivo
Previously, knowledge of human neural development was mainly restricted to the morphological architecture of neural tissues and structural brain development. With the development of high-throughout RNA-sequencing technology, the precise spatiotemporal regulation of the human neural transcriptome and epigenome has recently been reported [32–34], which will enhance our understanding of human neurodevelopment. However, in view of the material and ethical limitations, it is difficult to investigate the molecular events of early human neural fate commitment. Therefore, human ESCs (hESCs), which are capable of differentiating into all cell types [35], allow the functional evaluation of human neurodevelopment under highly reproducible conditions, because NPCs can be efficiently derived from hESCs and patterned into regionalized neural progenitors [36, 37]. Moreover, the early stages of hESC neural differentiation are highly similar to early anatomical brain development and can recapitulate early human neurodevelopment [38]. Together, in vitro neural differentiation systems from ESCs allow investigation of the differentiation potential at various developmental stages in the presence or absence of environmental cues.
Epigenetics
Epigenetic mechanisms, which involve DNA, RNA, and histone modifications, result in mitotically heritable modulation of gene expression without changing gene coding sequences [39]. The epigenetic code composed of histone and DNA modifications establishes a specific chromatin state at specific genes to maintain the poised nature of specific cell types. Emerging evidence has demonstrated that cell-intrinsic programs, in addition to extracellular signals, play an imperative role in developmental fate restriction of stem cells. Particularly, the long-term repression of developmental genes that define specific cell lineages contributes to the fate restriction. DNA or histone modifications regulate target gene transcription through influencing the chromatin state and accessibility of DNA to transcription factors [6].
During neural fate commitment of pluripotent stem cells, neurodevelopmental genes are activated while pluripotency genes are silenced. The expression of these regulated genes is modulated by epigenetic modifications, which are achieved by a temporary bivalent state of chromatin that eventually proceeds to permanent long-term silencing. The comprehensive epigenetic mechanisms cooperatively ensure proper neurodevelopment and avoid precocious differentiation. Recent findings suggest that cell fates can be reset by the alteration of epigenetic marks, highlighting the theoretical and practical importance of epigenetic regulation for regenerative medicine [39].
Histone modifications
Various types of histone modifications, including methylation, acetylation, sumoylation, phosphorylation, and ubiquitination, regulate gene expression by controlling histone protein stability, chromatin structure, and DNA accessibility. These histone modifications compose the histone code, the readout of which directly affects target gene transcription [40]. The histones—H2A, H2B, H3, and H4—are post-translationally modified at many sites on their N- and C-terminal tails, and the regulatory outcomes of some specific histone modifications have been decoded. For example, acetylation of histone H3 lysines 9 and 14 (H3K9/14) represents a transcriptionally active state, whereas trimethylated histone H3K9 represents inactive heterochromatin. The functional implications of specific histone modifications have been summarized in many reviews [41, 42], and will not be reviewed here. Histone modifications are reversibly modulated by various types of modification-specific enzymes, including histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), histone lysine demethylases (KDMs), the E1–E3 ubiquitinating enzyme complex, de-ubiquitinating enzymes, and SUMO-activating and -conjugating enzymes.
Accumulating evidence has demonstrated that histone modifications are involved in temporally and spatially precise gene expression patterns during neurodevelopment [6]. The enzymes responsible for histone modifications function coordinately over short time-scales and respond quickly to the changing environments of early embryonic development. The global histone modification landscape suggests that histone modifications are dynamic and are enriched in different sets of developmental genes at different neurodevelopmental stages, linked to cell identity and transcriptional consistency [21, 43, 44].
In ESCs, pluripotency genes that are highly transcriptionally active are associated with active histone modification marks, such as H3K9 acetylation (H3K9ac) and histone H3 lysine 27 acetylation (H3K27ac) [21, 43]. Moreover, the Polycomb group (PcG) class of histone modification enzymes plays an essential role in establishment of the bivalent chromatin state that represses key developmental gene expression and maintains pluripotency [11]. Key developmental regulators of ESC differentiation are trimethylated at H3K27 (H3K27me3) and/or H3K4 (H3K4me3) by Polycomb repressive complexes (PRCs) and Trithorax group (TrxG) protein mixed-lineage leukemia (MLL), respectively [7, 45]. The combination of these inhibitory and activating marks constitutes ‘bivalent’ chromatin. These genes are poised for activation upon appropriate differentiation cues. For instance, the chromatin state of several key neural genes, such as Pax6, Sox1, Ngn2, and Ascl1, is ‘bivalent’ in pluripotent ESCs, but becomes activated upon neural differentiation [46].
As ESCs commitment to neural fates, they gradually lose their pluripotency, and non-neural genes become inaccessible to transcription factors due to the loss of associated transcriptionally active histone modifications (such as H3K9 acetylation [21]). These genes are therefore silenced in fate-committed NPCs. In contrast, the poised state of neural genes is rewritten by specific histone modifiers for transcriptional activator accessibility. For instance, several neural genes lose the H3K27me3 modification but retain the H3K4me3 mark, resulting in the upregulation of these neural regulators [4], and these genes coordinately promote the ESC neural differentiation program. Histone demethylases (i.e., KDM6B, KDM7A, LSD1) [6, 47] and methyltransferases [the enhancer of Zeste homolog 2 (Ezh2) and G9a] [48, 49] are essential for early embryogenesis and neural fate commitment, and disruption of these histone-modifying proteins leads to deregulated neurodevelopmental timing and neuronal output. Moreover, a group of genes responsible for terminal neuronal differentiation also possess ‘bivalent’ marks but remain silenced in NPCs. Upon neurogenesis, their marks are removed, indicating that NPCs have cell identity-specific poised genes required for their terminal differentiation potential [6]. The mechanism underlying this interesting observation has yet to be elucidated.
Additionally, HDAC-mediated histone deacetylation is tightly linked to transcriptional repression during early neural fate commitment. Inhibition of HDAC activity by the inhibitors valproic acid and Trichostatin A retains ESC pluripotency and promotes the generation of pluripotent stem cells (iPSCs) [50, 51]. Furthermore, disruption of histone deacetylation during ESC neural differentiation impacts neural fate commitment with complex results depending on the timing of HDAC inhibition [21, 52, 53], indicating that HDAC activity is essential for neural determination during a specific temporal window. Moreover, different HAT (p300 and CBP) and HDAC (HDAC1–11 and Sirt1–7) members are involved in maintaining ESC pluripotency and sequential neural differentiation [16, 21, 54–56], which are possibly coordinated by specific cofactors [57].
DNA modifications
DNA modifications play essential roles in genomic imprinting, X-chromosome inactivation, gene regulation, and the maintenance of epigenetic memory. DNA methylation (5mC) is the major epigenetic mechanism for long-term gene silencing, whereas other DNA modifications, such as 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), are the intermediate products of DNA demethylation catalyzed by Ten-eleven translocation (Tet) and TDG proteins. DNA bearing these forms may represent transcriptionally active states [58–60]. The genome-wide dynamic changes in DNA methylation patterns as well as a group of epigenetic enzymes are involved in various biological processes, including early fate commitment and embryogenesis.
DNA methylation mainly occurs at CpG sites of approximately half of all tissue-specific promoter regions for maintaining gene silencing. At present, three types of DNA methyltransferases have been identified: Dnmt1 maintains DNA methylation patterns across mitotic divisions; Dnmt2 has weak methyltransferase activity; and Dnmt3a and Dnmt3b are involved in de novo methylation by adding methyl groups onto unmethylated DNA. Dnmt3l that is a close homolog of Dnmt3a and Dnm3b but lacks the catalytic methyltransferase domain interacts with unmethylated H3K4. MeCP2, a member of the MBD family of proteins, binds to methylated DNA near promoter regions of specific genes, maintaining silencing through recruitment of the Sin3a-HDAC co-repressor complex. Deletion of Dnmt1 or Dnmt3a/3b results in embryonic lethality or postnatal death, indicating the essential role of DNA methyltransferases during development. After the establishment of DNA methylation patterns during zygote formation, DNA methyltransferases start to re-build the DNA methylation patterns during implantation, germ layer, and cell type differentiation. Dnmt3a/3b are required for ESC pluripotency by establishing stem-cell-specific DNA methylation patterns [7, 61]. Although previous reports suggested that Dnmt-mediated DNA methylation was mainly involved in neuronal differentiation processes, DNA methylation patterns change dynamically during early neural commitment [43], indicating that DNA methylation is involved this process. Upon ESC neural differentiation, Dnmt3a/3b participates in silencing of pluripotency genes by establishing DNA methylation patterns [62].
DNA demethylation, which is mainly catalyzed by the Tet proteins (Tet1, Tet2, and Tet3) to promote 5mC-to-5hmC conversion as the first step, is an essential process for neural fate commitment from pluripotent cells. Genome-wide analysis of Tet1 and 5hmC distribution in ESCs has revealed that Tet proteins are involved in the regulation of dynamic changes during ESC differentiation, and 5hmC and Tet1 are enriched at bivalently marked genes in ESCs [63, 64]. Several studies have reported the convergent role of Tet proteins in neurodevelopment. Xenopus Tet3 directly activates master neural genes (rx, pax6, and ngn2) involved in eye and neural development [65], and Tet1 regulates neural lineage specification and promotes active DNA demethylation in the adult brain and mESCs [64, 66, 67]. DNA methylation dynamics are correlated with brain development and maturation [68, 69]. Furthermore, a global epigenomic transition from 5mC enrichment in hESCs to 5hmC enrichment in NPCs has been reported during hESC neural differentiation [70], and TET protein-mediated 5hmC generation is an important epigenetic mechanism for master neural gene activation [65, 70]. Three members of the Tet family of proteins have been identified to catalyze DNA hydroxylation across the entire genome, each of which are differentially expressed in different tissues [71]. It is now widely accepted that the Tet proteins need to interact with specific partners to specifically regulate target gene expression in particular biological processes [65, 72, 73]. For instance, AF9 physically interacts with TET2 to direct 5hmC generation and neurodevelopmental gene activation during hESC neural differentiation [70].
Another form of methylated DNA, N 6-methyladenine (6 mA), is a covalent modification of DNA to protect against restriction enzymes in bacteria. Very recently, three studies reported dynamic deposition of 6 mA with spatiotemporal patterns during development in three different eukaryotic genomes: Chlamydomonas reinhardtii, Caenorhabditis elegans, and Drosophila melanogaster [74–77]. Moreover, 6 mA is associated with gene regulatory events and is highly abundant in early staged embryos of Drosophila melanogaster [76], supporting the hypothesis that 6 mA very possibly acts as an epigenetic mark with restricted functions in pluripotent cells and early neural fate commitment.
DNA and histone modifications, which correlate with the spatiotemporal changes of epigenetic landscapes, have cross talk to regulate the stem cell differentiation along the neural lineage. In undifferentiated ESCs and non-neural lineages, some neural genes are suppressed by both PRC2 and repressor element RE1 binding transcription factor (REST, also known as neuron-restrictive silencer factor (NRSF)). In ESCs and NPCs, REST recruits a corepressor complex that consists of CoREST, HDAC, mSin3A, and MeCP2 to the RE1 site to control chromatin states [78]. Upon neural differentiation, the REST complexes associate with G9a, SUV39H1, TET proteins, and other epigenetic modifiers to induce permanent silencing of non-neural genes and activation of neural genes via H3K9 methylation, DNA methylation, H3K4 demethylation, and DNA hydroxylation [73, 79, 80]. Thus, DNA methylation may interact with histone modifications at the level of chromatin, and both modifications may influence each other through unknown mechanisms.
Chromatin remodeling in early neural development
In addition to the epigenetic regulation described above, chromatin remodeling is another imperative mechanism for controlling gene expression and cell fate commitment. In mammals, chromatin-remodeling complexes are classified into four groups based on the ATPase subunit present: SWI (switching or SNF); imitation switch (ISWI); Mi-2/NuRD; and others [81]. Each complex is composed of approximately nine proteins that include both conserved and non-conserved components, which define the characteristics of the specific chromatin complex. The ATPase activity of remodeling complexes is correlated with the disruption of nucleosome-DNA contacts, nucleosome movement along the DNA, and nucleosome changes. Similar to transcription factors, chromatin-remodeling complexes are differentially expressed in cell- and tissue-specific patterns and interact with other transcriptional regulators or complexes to modulate target gene transcription. The interacting proteins important for neural development mainly include REST and some co-repressors (CoREST, MeCP2, and Sin3A), which act by influencing the activity of chromatin-remodeling complexes.
Growing evidence demonstrates that chromatin-remodeling complexes are involved in vertebrate neural development. Several complexes or their components, such as ISWI complex and Brg1 (a SWI/SNF protein), are specifically expressed in the neural plate, brain, and spinal cord. Depletion of Brg1 maintains NPC proliferation and prevents neuronal differentiation by mediating transactivation of Ngn and NeuroD [82]. Moreover, the ISWI complex is essential for eye development and cataract formation through the induction of BMP4 and the inhibition of Sonic hedgehog [83]. In contrast, pluripotent ESCs contain another specialized form of the BAF complex termed esBAF that is distinguished by the presence of a BAF155 homodimer and Brg1 [84]. These findings suggest that chromatin-remodeling complexes play crucial roles in neural development and ESC maintenance. Chromatin-remodeling complexes appear to primarily participate in neuronal differentiation processes [85]. However, it is unclear whether chromatin-remodeling complexes are involved in the early fate transition from ESCs to NPCs. Based on our RNA-seq analysis of gene expression patterns during ESC neural differentiation (unpublished data), we believe that chromatin-remodeling complexes very likely participate in the early neural conversion from ESCs into NPCs.
RNA-level regulation
RNA-level regulation, including non-coding RNAs (different classes of small RNAs and long non-coding RNAs (lncRNAs)) and RNA modifications, have emerged as key regulators of gene expression and genome stability in various biological processes. The formation of RNA scaffolds serves as a unifying mechanism for silencing gene transcription and modifying chromatin structures by small RNAs and lncRNAs. In past decades, different kinds of non-coding RNAs have been observed in the brain with divergent roles through mediating epigenetic modifications and chromatin remodeling [86, 87], demonstrating that RNA-level regulation is an imperative mechanism for neurodevelopment.
RNA interference (RNAi) is an important pathway by which small RNAs modify chromatin and target gene expression. RNAi was first discovered in Caenorhabditis elegans as a response to exogenous double-stranded RNA (dsRNA), which results in the sequence-specific gene silencing [88]. The endogenous RNAi pathway is initiated by Dicer, which binds and cleaves dsRNAs to produce double-stranded fragments of 20–25 base pairs, called small interfering RNAs (siRNAs) [89]. These siRNAs are incorporated into the RNA-induced silencing complex (RISC), which together binds to the target RNAs, thereby causing the sequence-specific destruction of mRNAs [90]. Considering the function of siRNAs in preventing mRNA translation, the RNAi pathway has been used as an effective tool to knock down target genes in vitro to investigate specific gene functions in biological processes, such as neural fate commitment [91, 92].
Another large family of small noncoding RNAs is microRNAs (miRNAs), which act as key post-transcriptional regulators in stem cell maintenance and differentiation. miRNAs are noncoding transcripts of 18–25 nucleotides that are derived from long primary transcripts [93]. Much like siRNAs, mature miRNAs are incorporated into RISC, where they are used to target RISC to specific mRNAs for degradation or inhibition of translation [94]. miRNAs are involved in many biological processes, such as developmental timing, cell proliferation, cell death, and nervous system patterning [95]. Many miRNAs, such as let-7, miR-124, miR-9, and miR-125, are highly enriched in the brain or nervous system [96–99], indicating a possible role of miRNAs in neural fate determination and neural development. let-7b has been reported to regulate neural stem cell proliferation and differentiation by targeting TLX and the cell cycle regulator-Cyclin D1 [99]. Overexpression or knockdown of miRNAs in an ESC-derived neural differentiation system demonstrated that miR-124a and miR-9 participate in regulation of the STAT3 pathway [100].
LncRNAs are non-protein coding transcripts longer than 200 nucleotides, and more than 35,000 human non-coding transcripts have been identified [101]. These lncRNAs may be located within the nucleus or cytoplasm, and are often transcribed from either strand of a protein-coding locus, typically without an open reading frame [102]. Intriguingly, many lncRNAs are expressed in the brain and central nervous system and the expression of many lncRNAs is restricted to specific developmental stages (i.e., ESC differentiation) and cell types (i.e., central nervous system) [103–105]. The potential roles of lncRNAs are mainly associated with gene transcription regulation and epigenetic regulation [106–108]. Therefore, numerous studies have begun to investigate the specific functions of lncRNAs in specific developmental processes, such as early neural fate commitment. For instance, the lncRNA TUNA is essential for mESC neural differentiation by forming a complex with three RNA-binding complexes [109]. A group of 35 lncRNAs including RMST, lncRNA-N1, lncRNA-N2, and lncRNA-N3 are associated with the neural differentiation of hESCs [110]. Additionally, other lncRNAs, such as Nkx2.2AS and Evf2, are also involved in neural fate commitment [111, 112]. Considering the large number of lncRNAs and the cooperation between lncRNAs and histone modifiers [110], deciphering the lncRNA world will require considerable effort.
Recent findings concerning N 6-methyladenosine (m 6 A) modifications, which widely exist on messenger RNAs or long non-coding RNAs in mammalian cells, have provided insights into the roles that RNA modifications play in regulating developmental events [113, 114]. More than 100 chemical RNA modifications have been found since the 1970s [115, 116]; however, the functions of RNA modifications remain unclear. RNA methylation is catalyzed by Mettl3, Mettl14, and WTAP while demethylation is catalyzed FTO and ALKBH5 [117]. Recent studies have shown that the m 6 A pathway is important to regulate the stability of specific target RNAs that regulate stemness maintenance and differentiation [118–121]. Reduction of m 6 A levels by the depletion of Mettl3 or Mettl14 in mESCs promotes the self-renewal of mESCs and inhibits their neural differentiation capacity [121]. However, the function of m 6 A in pluripotency maintenance and differentiation capacity of ESCs remains controversial [122], indicating the need to discern the exact functions of m 6 A in stage-specific neural differentiation of ESCs and in vivo neurodevelopment.
As a key player in gene transcriptional regulation, RNA-level regulation plays important roles in the occurrence and development of neurodegenerative diseases such as Parkinson’s, Alzheimer’s, and Huntington’s diseases [123, 124]. Clinical evidence shows that deregulated miRNAs disrupt the expression levels of key developmental proteins and cell cycle regulators, leading to the occurrence of sporadic neurodegenerative disorders [124–126]. Moreover, lncRNAs (for example, the antisense transcriptional lncRNAs of PINK1 and HAR1) also play important roles in the development of neurodegenerative diseases through effects on the cell cycle and apoptosis [127–129]. Despite these recent discoveries, more work is needed to uncover the relationship between RNA-level regulation and neurodegenerative diseases.
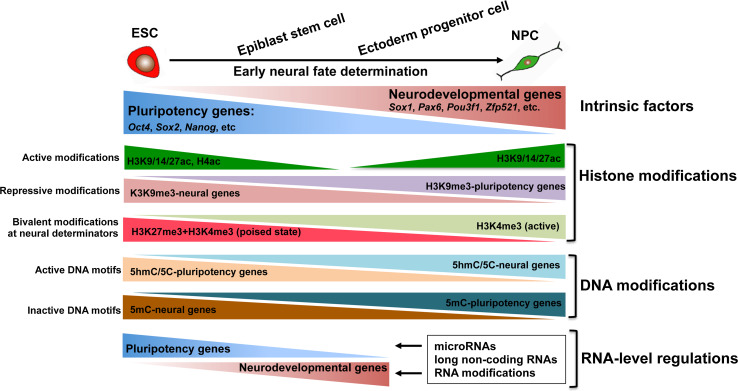
In summary, early neural fate commitment is accompanied by silencing of pluripotency genes and activation of neurodevelopmental genes. This is accomplished by the coordination of epigenetic mechanisms, including histone modifications, DNA modifications, chromatin remodeling, and RNA-level regulation (Fig. 2). In pluripotent ESCs, pluripotency genes are associated with active epigenetic marks (i.e., H3K9/14/27ac, H4ac, 5C, and 5hmC) to permit transcriptional activation; these active marks are removed upon neural differentiation. Some key neural fate determinants are repressed and maintained in a poised state, marked by H3K27me3 and H3K4me3. During the process of neural fate commitment, H3K27me3 is removed, leaving only H3K4me3, which permits transcription of these neural regulators. Additionally, chromatin-remodeling complexes and RNA-level regulation (miRNAs, lncRNAs, and RNA modifications) participate in the stepwise fate commitment from ESCs into NPCs, thereby ensuring sequential neural lineage determination.
Fig. 2.
Epigenetic regulation during fate commitment from pluripotent embryonic stem cells into neural progenitor cells. Histone modifications, DNA modifications, and RNA-mediated regulation are involved in the multi-step process of neural fate commitment by silencing pluripotency genes and activating neurodevelopmental genes
The interplay between intrinsic factors, signaling pathways, and epigenetic regulations
Neural fate commitment from pluripotent stem cells occurs in a cell-autonomous (intrinsic) manner but is also affected by epigenetic regulations and extrinsic signals. During early neurodevelopment, embryonic cells undergo symmetric or asymmetric divisions in a micro-niche, and this process is accompanied by the gradual loss of differentiation potential. Although the molecular steps of neural fate determination have been primarily defined, the functional heterogeneity of differentiating cells may be linked to multiple factors, such as asymmetrical division, epigenetic regulation, the cell cycle, and the signaling activity of various pathways. Intrinsic factors, signaling pathways, and epigenetic regulation are known to be individually involved in early neural fate commitment [3]. In recent years, scientists have realized that functional interaction between different regulatory mechanisms drives various biological processes. For instance, we previously found that the intrinsic factor Pou3f1 promotes neural fate commitment via inhibition of the BMP and WNT signaling pathways [130] and that histone deacetylation promotes neural induction by restricting the Nodal-dependent mesendoderm fate [16]. This demonstrates that epigenetic regulations, intrinsic factors, and signaling pathways can influence each other during ESC neural differentiation. The interaction between signaling pathways and epigenetic regulations is of special interest for understanding the mechanism underlying early neural fate commitment.
The transcriptional activation of some neural regulators (e.g., Sox1, Sox2, Pax6, Nestin, Znf521, and Pou3f1) is regulated by epigenetic marks. During neural differentiation of ESCs, the promoters of many neural genes lose the H3K27me3 modification of the bivalent state but retain the H3K4me3 mark, resulting in the activation of these genes [4, 49]. During neural fate conversion, the expression of the H3K27 demethylase Kdm6b is upregulated to remove the H3K27me3 modification from the promoters of some neural genes (Pax6 and Nestin). Depletion of Kdm6b severely impairs the differentiation of ESCs into neural cells, suggesting that Kdm6b is required for resolution of the bivalent state [131]. In our previous study, we revealed that transcriptional activation of key neural regulators (e.g., ZNF521, ZIC2, HESX1, and POU3F1) is modulated by histone H3K9 acetylation [132]. Moreover, the activation of SOX5, MASH1, ZNF521, and MAP2 also rely on AF9-TET2 complex-mediated DNA hydroxylation and subsequent demethylation [70]. These findings suggest that the regulatory network between intrinsic factors and epigenetic regulation plays a key role in cell fate determination. However, how the sequential activation of key neural regulators is controlled by multiple kinds of epigenetic regulations remains unclear. Furthermore, whether the epigenetic regulation can be modulated by intrinsic factors also remains largely unknown. We believe that answering these questions will be essential for elucidating the key mechanism involved in neural fate specification.
Chromatin is a dynamic and plastic structure that integrates extracellular signals to generate a coordinated transcriptional response in specific cell types under particular conditions. It is widely accepted that cell fate and developmental marks can be reset on genomes by epigenetic alterations. The epigenetic basis of neural differentiation arises from the need to maintain separate gene expression patterns in both pluripotent ESCs and NPCs. Chromatin modifiers with opposing activities play key roles in the dynamic regulation of epigenetic marks, re-defining the related signaling pathways and activating downstream transcription factors. Furthermore, non-coding RNAs provide additional layers for the regulation of gene expression during cell fate specification. The multiple layers of developmental gene regulation guarantee the rapid transition of pluripotent ESCs into neural lineages.
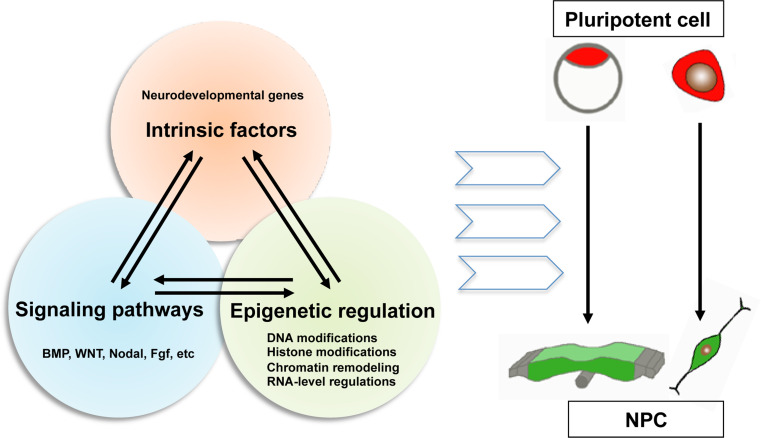
As summarized in Fig. 3, early neural fate commitment may be regulated by the interplay between neural regulators, signaling pathways, and epigenetic mechanisms. Cooperation between distinct regulatory pathways is likely at the core of the transition between the short- and long-term epigenetic alterations required for neural development. Generally, key transcriptional factors or neurodevelopmental genes (especially pioneer factors, such as Zfp521, Pou3f1, and Pax6) are the core machinery for neural fate conversion [3]. Expression of these core factors is induced or controlled by extracellular signals and epigenetic regulations. Meanwhile, the signaling by extrinsic cues is modulated by transcription factors and epigenetic modifiers, and epigenetic machinery is also regulated by signaling pathways and developmental genes. Decoding the complex regulatory network emerging from the interplay of these three aspects is crucial for understanding the molecular basis of cell fate specification in early embryos. Post-translational modification is not restricted to histone proteins, which adds another layer to the intricate regulatory network underlying cell fate specification [133, 134]. These findings blur the boundary between epigenetic regulation and intrinsic factors, and will drive future research into a new world in lineage commitment.
Fig. 3.
The interplay between intrinsic factors, signaling pathways, and epigenetic regulation. The neural fate transition from pluripotent embryonic stem cells is cooperatively modulated by the interaction between intrinsic factors, signaling pathways, and epigenetic regulation
Perspectives
Epigenetic modulation by histone and DNA modifications as well as RNA-level regulation is crucial for early embryonic development and neural fate commitment. It drives critical processes ranging from cell identity maintenance, cell proliferation, and differentiation. The acquisition of distinct epigenetic profiles at pluripotency and neurodevelopmental genes determines a developmental stage-specific identity. Deregulation of the epigenome at the DNA, RNA, or histone levels could lead to various neurodevelopmental disorders in response to environmental changes. Epigenetic mechanisms provide a dynamic regulation pathway over the relatively static genome in a reversible and rapid manner without genetic alterations. Although great advances have been made recently, many challenging questions regarding epigenetic regulation during cell lineage commitment still remain unanswered.
Our current understanding of epigenetics has mainly been achieved using genome-wide analysis at cell population levels. The function of epigenetic marks and their correlation with transcription has thus far been explored by disrupting epigenetic modifiers. However, the function of epigenetic factors may not perfectly equate with the identified epigenetic marks they establish because epigenetic factors may possess additional functions or act on other histone or DNA sites. CRISPR/Cas9-mediated genome editing [135] to engineer precise site mutations in regulatory DNA loci or sites of histone modification will permit a detailed investigation of the functions of specific epigenetic modifications. Additionally, heterogeneity in cell populations indicates that the results obtained from cell populations may not represent the real state in each single cell. Together with the development of single cell-based assays (e.g., RNA-seq and DNA methylome [136, 137]), determining the landscape of epigenetic modifications at the single-cell level may enhance our understanding of epigenetic regulation during neural fate determination and other lineage commitment processes.
Because of the complexity of epigenetic regulations, extensive genomic analysis to data has failed to identify consensus epigenetic marks in pluripotent ESCs and NPCs, and individual researchers have each mainly focused on a single type of modification. Actually, different epigenetic modifications in DNA, histone, and RNA may compose an epigenetic network where each modification affects the state and activity of other modifications. We envision that decoding the diverse epigenetic code by combining the DNA, RNA, and histone modifications underlying neural development will be an exciting endeavor in the post-genome era.
The spatial–temporal control of neural fate commitment is achieved by the cooperation of multiple developmental genes, extracellular signals, intrinsic factors, and epigenetic marks. Various means of sequentially activating gene expression during an appropriate time window are employed in neural commitment, suggesting a diversification in the modes of regulating gene transcription. Determining how epigenetic regulations, intrinsic factors, and extracellular signals together orchestrate the neural fate program is a future challenge, and dissecting the entire regulatory network during early embryogenesis will aid in preventing and treating neurodevelopmental disorders.
Acknowledgments
This work was supported in part by the “Strategic Priority Research Program” of the Chinese Academy of Sciences, Grant No. XDA01010201, National Key Basic Research and Development Program of China (2014CB964804, 2015CB964500), National Natural Science Foundation of China (31430058, 31571513, 31501178, and 91519314), and Chinese Postdoctoral Science Foundation (2013M541560).
References
- 1.Wilson SI, Edlund T. Neural induction: toward a unifying mechanism. Nat Neurosci. 2001;4(Suppl):1161–1168. doi: 10.1038/nn747. [DOI] [PubMed] [Google Scholar]
- 2.Streit A, et al. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406(6791):74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- 3.Tang K, et al. Intrinsic regulations in neural fate commitment. Dev Growth Differ. 2015;57(2):109–120. doi: 10.1111/dgd.12204. [DOI] [PubMed] [Google Scholar]
- 4.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stavridis MP, Smith AG. Neural differentiation of mouse embryonic stem cells. Biochem Soc Trans. 2003;31(Pt 1):45–49. doi: 10.1042/bst0310045. [DOI] [PubMed] [Google Scholar]
- 6.Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11(6):377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- 7.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Ramirez MA, Nicoli S. Role of miRNAs and epigenetics in neural stem cell fate determination. Epigenetics. 2014;9(1):90–100. doi: 10.4161/epi.27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temple S. The development of neural stem cells. Nature. 2001;414(6859):112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 10.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coskun V, Tsoa R, Sun YE. Epigenetic regulation of stem cells differentiating along the neural lineage. Curr Opin Neurobiol. 2012;22(5):762–767. doi: 10.1016/j.conb.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pera EM, et al. Active signals, gradient formation and regional specificity in neural induction. Exp Cell Res. 2014;321(1):25–31. doi: 10.1016/j.yexcr.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 13.Hawley SH, et al. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes Dev. 1995;9(23):2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- 14.Sasai Y, et al. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus . Nature. 1995;376(6538):333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- 15.Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376(6538):331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- 16.Liu P, et al. Histone deacetylation promotes mouse neural induction by restricting Nodal-dependent mesendoderm fate. Nat Commun. 2015;6:6830. doi: 10.1038/ncomms7830. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Jing N. Pluripotent stem cell studies elucidate the underlying mechanisms of early embryonic development. Genes (Basel) 2011;2(2):298–312. doi: 10.3390/genes2020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linker C, Stern CD. Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development. 2004;131(22):5671–5681. doi: 10.1242/dev.01445. [DOI] [PubMed] [Google Scholar]
- 19.Pera EM, et al. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17(24):3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li VC, Kirschner MW. Molecular ties between the cell cycle and differentiation in embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(26):9503–9508. doi: 10.1073/pnas.1408638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao Y, et al. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J Biol Chem. 2015;290(16):9949. doi: 10.1074/jbc.A114.603761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K, et al. Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development. 2010;137(13):2095–2105. doi: 10.1242/dev.049494. [DOI] [PubMed] [Google Scholar]
- 23.Li L, et al. Location of transient ectodermal progenitor potential in mouse development. Development. 2013;140(22):4533–4543. doi: 10.1242/dev.092866. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, et al. The zinc finger transcription factor Ovol2 acts downstream of the bone morphogenetic protein pathway to regulate the cell fate decision between neuroectoderm and mesendoderm. J Biol Chem. 2013;288(9):6166–6177. doi: 10.1074/jbc.M112.418376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao Y, et al. AP2gamma regulates neural and epidermal development downstream of the BMP pathway at early stages of ectodermal patterning. Cell Res. 2012;22(11):1546–1561. doi: 10.1038/cr.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 27.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 29.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 30.Vallier L, et al. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4(6):e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, et al. Ectodermal progenitors derived from epiblast stem cells by inhibition of Nodal signaling. J Mol Cell Biol. 2015;7(5):455–465. doi: 10.1093/jmcb/mjv030. [DOI] [PubMed] [Google Scholar]
- 32.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523(7559):212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 36.Zhang SC, et al. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19(12):1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 37.Yan Y, et al. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23(6):781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang SC. Neural subtype specification from embryonic stem cells. Brain Pathol. 2006;16(2):132–142. doi: 10.1111/j.1750-3639.2006.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egger G, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 40.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 41.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15(2):163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 43.Ziller MJ, et al. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature. 2015;518(7539):355–359. doi: 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benayoun BA, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158(3):673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 46.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 47.Huang C, et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 2010;20(2):154–165. doi: 10.1038/cr.2010.5. [DOI] [PubMed] [Google Scholar]
- 48.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee ER, Murdoch FE, Fritsch MK. High histone acetylation and decreased Polycomb repressive complex 2 member levels regulate gene specific transcriptional changes during early embryonic stem cell differentiation induced by retinoic acid. Stem Cells. 2007;25(9):2191–2199. doi: 10.1634/stemcells.2007-0203. [DOI] [PubMed] [Google Scholar]
- 50.Mali P, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ware CB, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4(4):359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balmer NV, et al. Epigenetic changes and disturbed neural development in a human embryonic stem cell-based model relating to the fetal valproate syndrome. Hum Mol Genet. 2012;21(18):4104–4114. doi: 10.1093/hmg/dds239. [DOI] [PubMed] [Google Scholar]
- 53.Hezroni H, Sailaja BS, Meshorer E. Pluripotency-related, valproic acid (VPA)-induced genome-wide histone H3 lysine 9 (H3K9) acetylation patterns in embryonic stem cells. J Biol Chem. 2011;286(41):35977–35988. doi: 10.1074/jbc.M111.266254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107(18):8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang F, et al. Coactivators p300 and CBP maintain the identity of mouse embryonic stem cells by mediating long-range chromatin structure. Stem Cells. 2014;32(7):1805–1816. doi: 10.1002/stem.1705. [DOI] [PubMed] [Google Scholar]
- 57.Gallinari P, et al. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res. 2007;17(3):195–211. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- 58.Li Z, et al. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 2015;43(8):3986–3997. doi: 10.1093/nar/gkv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo F, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 60.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayashi H, Kikyo N. Epigenetic regulation of open chromatin in pluripotent stem cells. Transl Res. 2015;165(1):18–27. doi: 10.1016/j.trsl.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li JY, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27(24):8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Y, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151(6):1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo JU, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Numata S, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90(2):260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiao Y, et al. AF9 promotes hESC neural differentiation through recruiting TET2 to neurodevelopmental gene loci for methylcytosine hydroxylation. Cell Discov. 2015;1:15017. doi: 10.1038/celldisc.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langemeijer SM, Aslanyan MG, Jansen JH. TET proteins in malignant hematopoiesis. Cell Cycle. 2009;8(24):4044–4048. doi: 10.4161/cc.8.24.10239. [DOI] [PubMed] [Google Scholar]
- 72.Yu C, et al. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science. 2013;342(6165):1518–1521. doi: 10.1126/science.1244587. [DOI] [PubMed] [Google Scholar]
- 73.Perera A, et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11(2):283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 74.Heyn H, Esteller M. An adenine code for DNA: a second life for N6-methyladenine. Cell. 2015;161(4):710–713. doi: 10.1016/j.cell.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 75.Fu Y, et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas . Cell. 2015;161(4):879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang G, et al. N6-methyladenine DNA modification in Drosophila . Cell. 2015;161(4):893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 77.Greer EL, et al. DNA methylation on N6-adenine in C. elegans . Cell. 2015;161(4):868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ballas N, et al. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121(4):645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 79.Singh SK, et al. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453(7192):223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jorgensen HF, et al. REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development. 2009;136(5):715–721. doi: 10.1242/dev.028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cho KS, Elizondo LI, Boerkoel CF. Advances in chromatin remodeling and human disease. Curr Opin Genet Dev. 2004;14(3):308–315. doi: 10.1016/j.gde.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 82.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132(1):105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 83.Dirscherl SS, Henry JJ, Krebs JE. Neural and eye-specific defects associated with loss of the imitation switch (ISWI) chromatin remodeler in Xenopus laevis . Mech Dev. 2005;122(11):1157–1170. doi: 10.1016/j.mod.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 84.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009;106(13):5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309(5740):1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- 87.Mattick JS, Makunin IV (2005) Small regulatory RNAs in mammals. Hum Mol Genet 14 Spec No 1: R121–R132 [DOI] [PubMed]
- 88.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 89.Zamore PD, et al. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 90.Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296(5571):1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 91.Jepsen K, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 92.Wang L, et al. Neurogenin 1 mediates erythropoietin enhanced differentiation of adult neural progenitor cells. J Cereb Blood Flow Metab. 2006;26(4):556–564. doi: 10.1038/sj.jcbfm.9600215. [DOI] [PubMed] [Google Scholar]
- 93.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 94.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 95.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 96.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smirnova L, et al. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21(6):1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 98.Le MT, et al. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29(19):5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao C, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci. 2010;107(5):1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krichevsky AM, et al. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24(4):857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 102.Birney E, et al. Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19(7–8):454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 104.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7(8):612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 107.Sanchez-Elsner T, et al. Noncoding RNAs of Trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311(5764):1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 108.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 109.Lin N, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53(6):1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tochitani S, Hayashizaki Y. Nkx2. 2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun. 2008;372(4):691–696. doi: 10.1016/j.bbrc.2008.05.127. [DOI] [PubMed] [Google Scholar]
- 112.Bond AM, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12(8):1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 114.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2(5):611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 116.Grosjean H. RNA modification: the Golden Period 1995–2015. RNA. 2015;21(4):625–626. doi: 10.1261/rna.049866.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fu Y, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 118.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clancy MJ, et al. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30(20):4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Geula S, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 121.Batista PJ, et al. m 6 A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jalkanen AL, Wilusz J. Stem cell RNA epigenetics: M 6 arking your territory. Cell Stem Cell. 2014;15(6):669–670. doi: 10.1016/j.stem.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 123.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13(8):528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32(4):199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 125.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6(17):2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 126.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20(12):617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 127.Morais VA, et al. Parkinson’s disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1(2):99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scheele C, et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genom. 2007;8(1):74. doi: 10.1186/1471-2164-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 130.Zhu Q, et al. The transcription factor Pou3f1 promotes neural fate commitment via activation of neural lineage genes and inhibition of external signaling pathways. Elife. 2014;3:e02224. doi: 10.7554/eLife.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Burgold T, et al. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008;3(8):e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Qiao Y, et al. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J Biol Chem. 2015;290(4):2508–2520. doi: 10.1074/jbc.M114.603761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Glozak MA, et al. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 134.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16(1):5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 135.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 137.Guo H, et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23(12):2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]