Abstract
Ubiquitin-related modifier 1 (Urm1) is a ubiquitin-like molecule (UBL) with the dual capacity to act both as a sulphur carrier and posttranslational protein modifier. Here we characterize the Drosophila melanogaster homologues of Urm1 (CG33276) and its E1 activating enzyme Uba4 (CG13090), and show that they function together to induce protein urmylation in vivo. Urm1 conjugation to target proteins in general, and to the evolutionary conserved substrate Peroxiredoxin 5 (Prx5) specifically, is dependent on Uba4. A complete loss of Urm1 is lethal in flies, although a small number of adult zygotic Urm1 n123 mutant escapers can be recovered. These escapers display a decreased general fitness and shortened lifespan, but in contrast to their S. cerevisiae counterparts, they are resistant to oxidative stress. Providing a molecular explanation, we demonstrate that cytoprotective JNK signaling is increased in Urm1 deficient animals. In agreement, molecular and genetic evidence suggest that elevated activity of the JNK downstream target genes Jafrac1 and gstD1 strongly contributes to the tolerance against oxidative stress displayed by Urm1 n123 null mutants. In conclusion, Urm1 is a UBL that is involved in the regulation of JNK signaling and the response against oxidative stress in the fruit fly.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-2121-x) contains supplementary material, which is available to authorized users.
Keywords: Urm1, Drosophila, Oxidative stress, UBL, JNK pathway
Introduction
The utilization of posttranslational modifications (PTMs) has in the past decades been acknowledged as an ingenious strategy of cells to multiply and regulate the functions of most proteins encoded by the genome of eukaryotic organisms. The ubiquitin-like molecules (UBLs) form a family of evolutionary conserved PTMs, critical for the formation of reversible, short-lived signaling centers, and functions by dynamically modulating target protein properties such as enzymatic activity, subcellular localization, half-life and inter-/intra-molecular interactions [1, 2]. Similar to ubiquitin, the founding and most studied member of the UBL family, all UBLs adopt a compact globular fold, the β-grasp superfold, and commonly rely on the sequential activity of E1 activating, E2 conjugating and E3 ligating enzymes to covalently attach to their target proteins via the highly conserved C-terminal di-glycine (GG) motif [1, 2]. In many cases the proper interpretation of UBL modifications in a cellular context relies on specialized ubiquitin/UBL-binding domains (UBDs) that recognize specific UBL motifs and further transmit their messages inside the cell [3].
Several of the UBLs have been found to respond to various cellular or environmental stimuli, such as increased sumoylation induced as a result of irradiation and DNA damage [4] and the enhanced ISGylation that follows stimulation of the immune system [5]. Similarly, conjugation of the Ubiquitin-related modifier 1 (Urm1) has been reported to be specifically induced in both yeast and mammalian cells in response to oxidative stress, a process known as urmylation [6–8]. Urm1 is an evolutionary conserved UBL of 11 kDa which phylogenetically appears to represent an evolutionary breakpoint where UBLs first started to be employed as PTMs in eukaryotic cells [9, 10]. It shares homologies with the prokaryotic UBL ancestors MoaD and ThiS, two sulphur carriers that function in molybdopterin and thiamine biosynthesis, respectively, as well as with eukaryotic UBLs that orchestrate cellular behavior by conjugating to target proteins [11]. Indeed, Urm1 appears to be an ancient remnant, which in higher species has developed dual capacity both to act as a sulphur carrier, specifically functioning in the modification of tRNA bases by thiolation [12–15], and as a posttranslational modifier that conjugates to lysine residues in multiple target proteins following oxidative stress [8, 16]. Urmylation has been shown to rely on the Urm1-specific E1 enzyme Ubiquitin-like modifier activating enzyme 4 (Uba4) [6], in mammals known as Molybdenum cofactor synthesis 3 (MOCS3) [17], but to date there have been no reports describing any E2, E3 or deurmylating enzymes in the Urm1 conjugation machinery. Among the identified targets of urmylation, which include proteins in the urmylation and ubiquitylation pathways, nuclear import, RNA biology and oxidative stress [8], the S. cerevisiae Alkyl hydroperoxide reductase 1 (Ahp1) and the Urm1 E1 activating enzyme Uba4 are the only targets which have been studied in more detail [8, 16, 18]. A report from investigations performed in the archaeon Sulfolobus acidocaldarius has recently identified 29 additional novel target proteins conjugated by Urm1 at lysine residues, and interestingly pointed towards a role of Urm1 in directing target protein degradation by the 20S proteasome [19].
In agreement with urmylation being triggered in response to oxidative stress, yeast strains deficient in urm1 or uba4 are sensitive to oxidizing agents [7], suggesting an important role of the Urm1/Uba4 conjugation machinery in the response against oxidative stress. To protect cells against dangerous reactive oxygen species (ROS), which arise naturally as a result of cellular respiration, but also as a consequence of environmental pollutants, UV radiation and therapeutic oxygen treatment [20, 21], multiple response systems have evolved. The cellular defense systems against oxidative stress include the immediate activity of antioxidant enzymes, such as the family of peroxiredoxins that degrade hydroperoxides to water, but also a transcriptional upregulation of cytoprotective genes including the gluthathione S-transferases (GSTs), aldehyde dehydrogenases (ALDH) and NAD(P)H:quinone oxidoreductases (NQO1) [20, 22, 23]. In response to oxidative and genotoxic insults, multiple signaling pathways come together to trigger the necessary reprogramming of the cell required to withstand the stress, among which JNK, p38 MAPK, p53, NK-κB and Nrf2 have been recognized as the most important [24]. An insufficient response to oxidizing agents results in increased levels of ROS, which can cause severe damage to cellular proteins, nucleic acids and lipids and in extension contribute to the initiation and progression of many human diseases including cancer, diabetes and cardiovascular disease, as well as premature ageing [23, 25, 26].
Despite being described as a UBL modifier more than a decade ago [6], the understanding of the role of Urm1 conjugation to target proteins has remained poor. Whereas the sulphur carrier activity of Urm1 has been fairly well described, studies regarding the conjugation machinery as well as characterization of putative targets and functions have been few [6–8, 16, 19]. To learn more about urmylation and its functions in a multicellular organism, we have utilized the Drosophila melanogaster model system to study Urm1 and its E1 activating enzyme Uba4. We have identified and characterized the annotated genes CG33276 and CG13090 as the homologues of Urm1 and Uba4/MOCS3 in Drosophila melanogaster, respectively. Drosophila Urm1 and Uba4 interact endogenously in fly tissues and are able to induce urmylation of multiple target proteins, including the evolutionary conserved substrate Peroxiredoxin 5 (Prx5), upon overexpression. Furthermore, employing Urm1 null mutants generated in our laboratory, we show that Urm1 is essential for viability and that flies lacking Urm1 protein display a shortened lifespan and an increased resistance to oxidative stress. Loss of Urm1 leads to an upregulation of the JNK pathway and increased levels of the cytoprotective JNK-regulated genes Jafrac1 and gstD1, providing a molecular explanation for the observed tolerance of Urm1 null flies to paraquat.
Results
Characterization of the Urm1 modifier in Drosophila melanogaster
The Drosophila genome contains a clear Urm1 fly homolog, encoded by the annotated gene CG33276. Sequence alignment analysis with Urm1 homologues in different species reveals that CG33276 displays all the characteristic features of a typical UBL, including a small size of 11 kDa, a predicted β-grasp structural fold and a conserved C-terminal di-glycine (GG) motif (Fig. 1a). Based on these findings, together with a shared sequence identity of 66 and 37 % with the Urm1 counterparts in human and S. cerevisiae, respectively, we hereafter refer to CG33276 as Urm1. Phylogenetic analysis places Drosophila Urm1 together with other eukaryotic Urm1 genes and an evolutionary ancestor shared with the sulphur carriers MoaD and ThiS in E. coli (Fig. 1b).
Fig. 1.
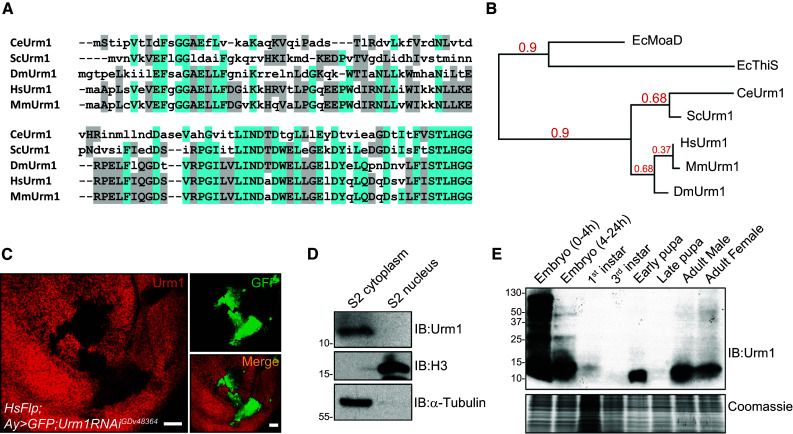
The Drosophila Urm1 homologue, CG33276, is an evolutionary conserved UBL with a highly dynamic pattern of expression. a Sequence alignment of Drosophila Urm1 (CG33276) with homologous counterparts in C. elegans, S. cerevisiae, H. sapiens and M. musculus, demonstrates a high homology between Drosophila Urm1 and mammalian Urm1 orthologues, as well as a conservation of the C-terminal di-glycine (GG) motif used for protein conjugation [66, 67]. b Phylogenetic representation of the evolutionary conservation of Urm1, generated by utilizing the Phylogeny.fr platform, indicates that Drosophila Urm1 shows a close relationship with its mammalian counterparts, and that all Urm1 genes in higher species share a common ancestry in the prokaryotic sulphur carriers MoaD and ThiS in E. coli [66, 67]. c Clonal expression of an RNAi transgene targeting Urm1 (marked by GFP) in third instar wing discs using FLP/FRT-mediated recombination, both verifies the specificity of the anti-Urm1 antibody and indicates a cytoplasmic localization of Urm1 in the surrounding wild type tissue. Scale bar 20 μm. d Urm1 protein is restricted to the cytoplasmic fraction of Drosophila embryonic S2 cells. e Urm1 expression and conjugation to target proteins shows a highly dynamic profile during development, displaying high levels in early embryonic stages which declines in larval stages to later peak again during larval/pupal transition. In adults Urm1 expression remains at fairly constant levels both in males and females. Western blot on whole cell lysates from control w 1118 flies at the developmental stages indicated
To initiate our studies of Urm1, we first generated antibodies specifically recognizing fly Urm1. Clonal expression of an RNAi transgene targeting Urm1 in third instar wing discs verified the specificity of the antibody and in addition indicated Urm1 as a protein primarily residing in the cytoplasm (Fig. 1c). This finding was further supported by immunoblotting cytoplasmic and nuclear extracts prepared from Drosophila S2 cells (Fig. 1d, Suppl. Fig. 1a), as well as the detection of Flag-tagged Urm1 in the cytoplasm of wing imaginal disc cells, expressed under control of the GAL4/UAS system (Suppl. Fig. 1b). In fly tissues, Urm1 appears to be ubiquitously expressed during all embryonic stages (Suppl. Fig. 1c), as well as in third instar larval imaginal discs, as indicated by a primarily cytoplasmic staining of imaginal disc cells, which is lost in clones where Urm1 has been knocked down by RNAi (Suppl. Fig. 1d). In agreement with these results, Western blot analysis of Drosophila lysates at different developmental stages showed a strong enrichment of both Urm1 and Urm1 conjugates during embryogenesis, in particular at very early stages (Fig. 1e), implicating important functions of Urm1 in early developmental processes. After embryogenesis, Urm1 decrease to barely detectable levels, but increase again to later peak at early pupal stages. In late pupae Urm1 protein expression is again strongly reduced, but reappears once more in adult flies where it appears to remain at stable levels. Interestingly, when analyzing the mRNA levels of monomeric Urm1, Urm1 appears to be transcribed at considerably higher levels in pupal and adult stages, possibly reflecting a need of the cells to rapidly being able to post-transcriptionally increase Urm1 levels in response to environmental stress, or an active degradation of Urm1 mRNA or protein at certain developmental stages (Suppl. Fig. 1e). Moreover, the excessive urmylation in Drosophila embryos is likely to be dependent on the high levels of maternally contributed Urm1 protein, deposited in the egg during oogenesis.
Drosophila Uba4 interacts with Urm1 and potentiates urmylation in vivo
Similar to all UBLs, Urm1 depends on a conjugation machinery in order to posttranslationally modify target proteins. However, in contrast to other UBLs, Urm1 does not appear to be regulated by any E2 or E3 enzymes, but is merely controlled by its E1-activating enzyme Uba4, known as MOCS3 in mammals. To identify the fly Uba4/MOCS3 homologue, we screened the Drosophila genome for genes containing a Rhodanese homology domain (RHOD) and found eight candidates of which only one, CG13090, displayed the conserved structural configuration shared by Uba4 in yeast and mammals—a RHOD domain in combination with an E1-like domain. Upon sequence alignment (Suppl. Fig. 2) we found that CG13090 exhibits an overall sequence identity with human and yeast MOCS3/Uba4 of 51 and 44 %, respectively (Fig. 2a). Together with a close phylogenetic relationship with orthologues in other species (Fig. 2b), this led us to conclude that the fly Uba4 homologue is encoded by the annotated gene CG13090.
Fig. 2.
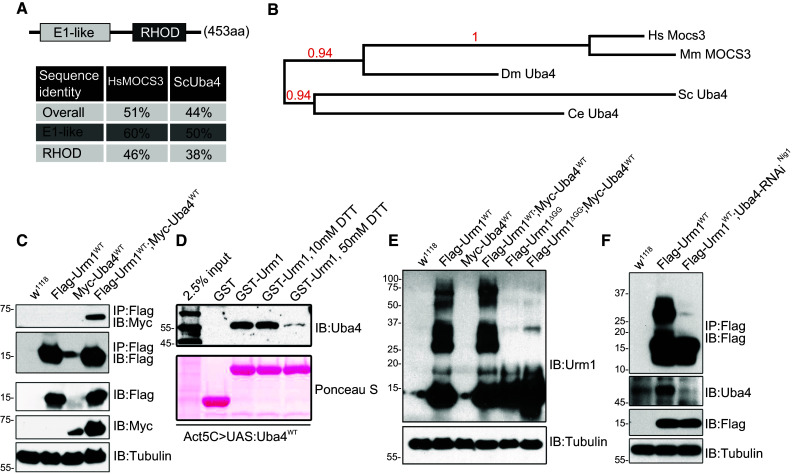
Drosophila Uba4 interacts with Urm1 and potentiates urmylation in vivo. a Schematic representation and evolutionary conservation of the Drosophila Uba4 homolog, CG13090. The Drosophila Uba4 is composed of an N-terminal E1-like domain and C-terminal Rhodanese homology domain (RHOD), similar to its human and yeast counterparts. Percent identity matrix was calculated using the online ClustalW2 tool developed by EMBL-EBI [68, 69]. b Phylogenetic analysis of the evolutionary conservation of Uba4, generated by utilizing the Phylogeny.fr platform [66, 67]. c Urm1 and Uba4 interacts in vivo. Immunoprecipitation of Flag-tagged Urm1 followed by immunoblotting with anti-Myc antibodies reveals that Flag-tagged Urm1 interacts with Myc-tagged Uba4. Proteins were overexpressed ubiquitously in adult flies using the GAL4/UAS system. d The interaction between Drosophila Urm1 and Uba4 is sensitive to reducing agents. A recombinant GST-tagged Urm1 protein was employed to pull down Uba4 from lysates of flies in which Uba4 is overexpressed under control of the Actin5C promotor (GAL4/UAS system). The addition of increasing concentrations of DTT abolishes the ability of Urm1 to bind Uba4. e Ectopic expression of Urm1 induces urmylation of multiple target proteins in adult flies, a process which is not observed when overexpressing a truncated version of Urm1 lacking the C-terminal GG motif used for conjugation (Urm1 ΔGG). f Urm1 conjugation to target proteins in adult flies is strongly reduced by RNAi-mediated knockdown of Uba4 expression
Consistent with the formation of a conjugation machinery complex, Drosophila Urm1 and Uba4 interact in lysates prepared from flies expressing tagged versions of the proteins (Fig. 2c). In contrast to other UBLs, Uba4 has recently been reported to interact with Urm1 via an acyl-disulphide bond, leading to the formation of thiocarboxylated Urm1 [17]. Consistent with these findings, we observe that the interaction between Drosophila Urm1 and Uba4 is sensitive to the presence of reducing agents such as DTT (Fig. 2d). Despite intensive efforts, we have failed to detect anything else than a weak pattern of protein urmylation in adult wild type flies, either under normal conditions or following various stress situations. However, upon tagged Urm1 overexpression, the appearance of multiple high-molecular weight bands in Western blots indicates that urmylation indeed occurs in living flies (Fig. 2e). Removal of the C-terminal GG motif of Urm1 (Urm1 ΔGG) prevents tagged Urm1 from conjugating to target proteins (Fig. 2e), a finding that reinforces that the utilization of the GG motif for protein conjugation is conserved in evolution. Consistent with the assumption that CG13090 represents the Drosophila Uba4 gene, RNAi-mediated knockdown of Uba4 strongly reduces urmylation in the fly (Fig. 2f). As noted previously, we observe the highest levels of endogenous urmylation in early embryos (Fig. 1e).
Loss of Urm1 causes lethality and decreased longevity
To further investigate the function of Urm1, we generated a mutant in which the entire Urm1 locus is deleted by imprecise immobilization of the KG{SuPor-P} KG08138 element, inserted in the immediate vicinity of the Urm1 translation-initiation ATG site [27] (Fig. 3a). The Urm1 n123 null allele and control revertant line Urm1 rv164 obtained by precise excision of KG{SuPor-P} KG08138, were both identified by PCR and verified by sequence analysis (Fig. 3b), as well as by the presence or absence of Urm1 protein expression in protein lysates from adult flies (Fig. 3c).
Fig. 3.
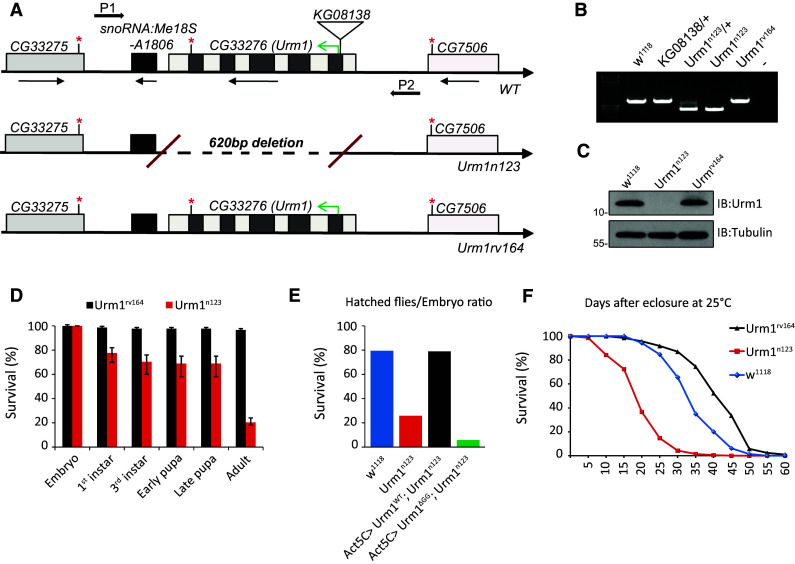
Flies lacking Urm1 are characterized by a reduced survival and longevity. a Schematic representation of the Urm1 n123 null mutant allele and the Urm1 rv164 revertant control line, obtained by imprecise and precise excision of the P{SUPor-P} element CG33276 KG08138 inserted in the 5′flanking region of the CG33276 coding region, respectively. b PCR results for the Urm1 n123 and Urm1 rv164 alleles, employing the primers P1 and P2 indicated in a. The reduced size of the PCR fragment in Urm1 n123 flies indicates a deletion in the Urm1 genomic locus, whereas the Urm1 rv164 flies contains a PCR band identical to w 1118 controls. c Urm1 n123 null mutant flies do not express any Urm1 protein. Total cell lysates of flies of the indicated genotypes, blotted with anti-Urm1 antibodies. d Loss of Urm1 causes lethality in Drosophila. Zygotic homozygous Urm1 n123 flies show an increased lethality primarily at embryonic and pupal stages, as compared with control Urm1 rv164 flies. n = 400 for both genotypes. e Ubiquitous overexpression of UAS:Urm1 WT fully rescues the lethality of the Urm1 n123 mutants, whereas the expression of an UAS:Urm1 ΔGG transgene contributes to an increased lethality in the Urm1 n123 mutant background (n > 1000 collected embryos for all genotypes). f Homozygous Urm1 n123 adult escapers display a significantly reduced lifespan, as compared with revertant Urm1 rv164 and w 1118 controls. Only female flies were tested, n = 200 for all genotypes
Loss of Urm1 is in most cases lethal, causing a small fraction of zygotic homozygous flies to die during embryogenesis (20 %) and the majority at late pupal stages (60 %) (Fig. 3d). This lethality is fully rescued by the ubiquitous expression of a UAS:Urm1 WT transgene, but not an Urm1 variant lacking the C-terminal GG motif (UAS:Urm1 ΔGG) (Fig. 3e). The survival of zygotic Urm1 n123 flies (20 %) is most likely explained by high levels of maternally contributed Urm1 (Fig. 1e and data not shown), a hypothesis which is further supported by the fully penetrant embryonic lethality displayed by the progeny of homozygous zygotic Urm1 n123 mutant males and females. A variable number of homozygous Urm1 n123 mutants, primarily of the female sex, eclose as adult escapers after a small, but significant, delay in development. The amount of escapers (ranging between 3 and 20 %) appears to be highly dependent on food quality and degree of competition from heterozygous siblings, indicating a decreased general fitness in the absence of Urm1 expression. In agreement, homozygous adult Urm1 n123 flies are smaller, less active and display a significantly reduced fecundity and shortened lifespan, as compared with control Urm1 rv164 and w 1118 flies (Fig. 3f). In keeping with a genetic interaction and position of Urm1 and Uba4 in the same cellular pathway, the survival of homozygous Urm1 n123 mutants is strongly decreased by the simultaneous removal of one copy of Uba4 (i.e. flies homozygous for Urm1 n123 and heterozygous for a deficiency uncovering Uba4) (Suppl. Fig. 3a). Further emphasizing the connection between Urm1 and Uba4, misexpression of Urm1 or Uba4 in the developing wing disc cause a similar loss of wing vein material, a phenotype which gains higher penetrance upon simultaneous co-expression of both Urm1 and Uba4 (Suppl. Fig. 3c). However, Urm1 and Uba4 are not interdependent, since Uba4 is not affected on the protein level in Urm1 n123 mutants (Suppl. Fig. 3b).
Urm1n123 mutant flies are resistant to oxidative stress
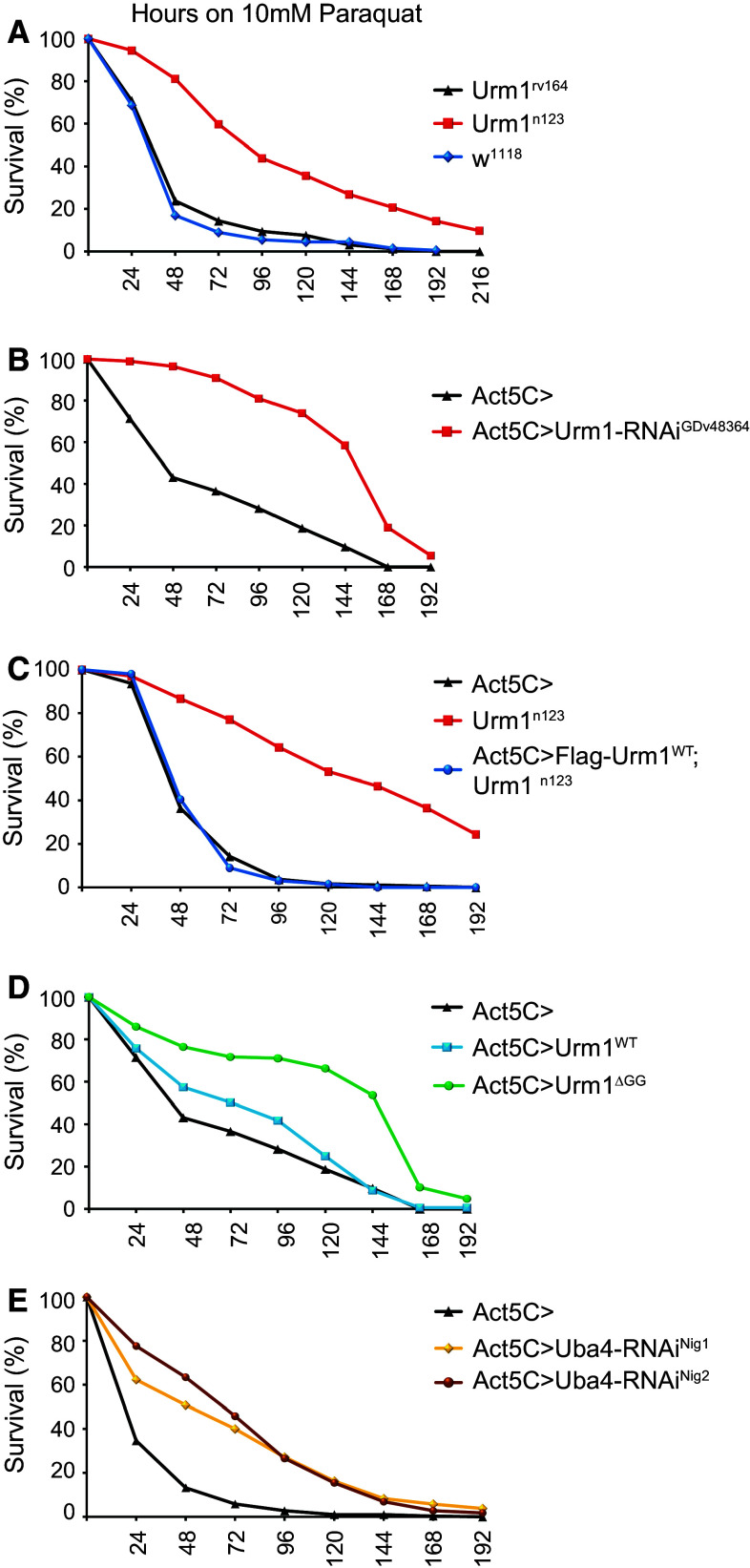
Given the reported sensitivity of yeast strains deficient for urm1 and/or uba4 to oxidizing agents [16], we were interested to investigate whether flies lacking Urm1 show a similar phenotype. In contrast, we found that loss of Urm1 in Drosophila melanogaster results in increased tolerance to oxidative stress (Fig. 4a). Urm1 n123 mutant flies display a median survival of 88 h when kept on 10 mM paraquat (Methyl viologen dichloride hydrate) diluted in 5 % sucrose, as compared with 36 and 35 h in control Urm1 rv164 and w 1118 flies, respectively. In support of this finding, flies in which Urm1 has been ubiquitously knocked down by RNAi also show a high tolerance to paraquat treatment (Fig. 4b), as well as to oxidative stress induced by hydrogen peroxide (Suppl. Fig. 4a). Importantly, the lethality and oxidative stress resistance caused by loss of Urm1 is rescued by a GAL4/UAS-mediated reintroduction of a wild type Urm1 transgene (UAS:Flag-Urm1 WT), but not an Urm1 transgene lacking the C-terminal GG-motif required for conjugation (UAS:Urm1 ΔGG) (Figs. 3e, 4c), suggesting that Urm1 conjugation is essential for Urm1 functionality in Drosophila. Consistent with this hypothesis, overexpression of the Urm1ΔGG protein appears to act in a dominant negative fashion, blocking the activity of endogenous Urm1 and thus resulting in a similar phenotype as loss of Urm1 per se (Fig. 4d). In agreement, whereas the Urm1 WT transgene fully rescues the lethality of the Urm1 n123 mutants, expression of Urm1 ΔGG in fact increases the lethality of zygotic Urm1 n123 flies (Fig. 3e). Overexpression of Urm1WT does not appear to have any major influence on sensitivity to oxidative stress when expressed on its own (Fig. 4d).
Fig. 4.
Loss of Urm1 results in an increased resistance to oxidative stress. a Urm1 deficient flies show an increased survival when exposed to oxidative stress induced by 10 mM paraquat. Urm1 n123 mutants displayed a median survival of 88 h on 10 mM paraquat, as compared with 36 h in the Urm1 rv164 revertant and 35 h in w 1118 controls, respectively. b Ubiquitous downregulation of Urm1 by RNA interference results in an increased survival under conditions of induced oxidative stress (10 mM paraquat). c The increased resistance of Urm1 n123 flies to oxidative stress can be rescued by ubiquitous overexpression of UAS:Flag-Urm1 WT, induced as a UAS transgene driven by the Actin5C-GAL4 driver in a homozygous Urm1 n123 background. d Overexpression of an Urm1 variant lacking the C-terminal GG-motif (Urm1 ΔGG) used for conjugation also generates flies with an increased tolerance to oxidative stress, indicating a dominant negative mode of action for Urm1ΔGG. e Ubiquitous downregulation of Uba4 by RNA interference, employing two independent Uba4-RNAi transgenic fly strains, cause an increased tolerance to oxidative stress induced by 10 mM paraquat. All graphs in a–e are representatives of at least three independent replicates, testing age-matched female flies, n > 300 for Urm1 n123, Urm1 rv164 and w 1118, n = 200 for all other genotypes tested. All flies used employing the GAL4/UAS system, were heterozygous for Actin5C-GAL4, as well as for the indicated UAS transgene, control Act5C > flies represent the progeny of Actin5C-GAL4 flies crossed to w 1118 controls
Similar to the results obtained for Urm1 n123 mutant flies, flies in which Uba4 has been ubiquitously knocked down, employing transgenically expressed constructs targeting Uba4, also show an increased resistance to oxidative stress (Fig. 4e), reinforcing that Drosophila Urm1 and Uba4 are acting in the same pathway.
Prx5 is targeted by urmylation in response to oxidative stress
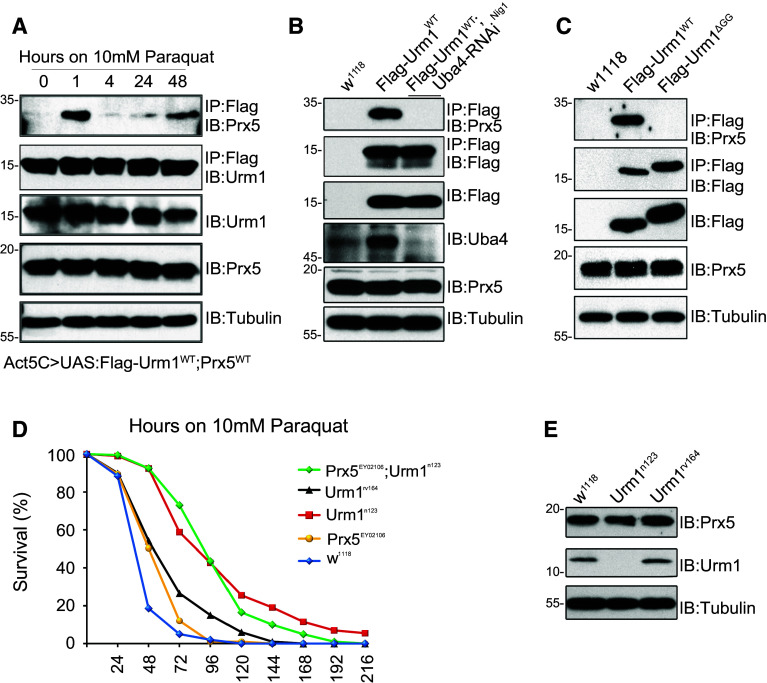
The best characterized target of Urm1 conjugation is the Alkyl hydroperoxide reductase (Ahp1) in S. cerevisiae [16], which is encoded by Peroxiredoxin 5 (Prx5) in Drosophila [28, 29]. We therefore wished to analyze if Prx5 is a target of urmylation also in flies and whether the identification of Prx5 as an urmylation target could contribute to the understanding of Urm1 function during oxidative stress. In order to determine if Urm1 is conjugated to Prx5, we utilized a Flag-tagged Urm1WT protein to immunoprecipitate either ectopically expressed or endogenous Prx5. In non-stimulated cells Prx5 could not be detected in Urm1 immunoprecipitates, but rapidly appeared as a prominent binding partner of Urm1 following paraquat treatment (Fig. 5a). Urmylated Prx5 showed a size shift from ~17 to ~30 kDa (Suppl. Fig. 4b), which is in agreement with conjugation of one Flag-tagged Urm1 moiety. Similar to findings in yeast [18], urmylation of Prx5 was found to depend on Uba4, as Prx5-Urm1 conjugates were lost in flies where Uba4 levels have been reduced by RNAi (Fig. 5b). A Flag-tagged Urm1ΔGG protein failed to interact with Prx5 in response to oxidative stress, indicating that Urm1 indeed is covalently conjugated to Prx5 via its C-terminal GG-motif and suggesting that the targeting of Prx5 by urmylation is evolutionary conserved (Fig. 5c). Consistent with reports in yeast [16, 30], Drosophila Prx5 EY02106 null mutants are sensitive to oxidizing agents [29] and in addition, a forced overexpression of Prx5 increases the survival of flies during conditions of oxidative stress [29]. However, while Prx5 is clearly urmylated following paraquat treatment, we have found no evidence that would support an upregulation, activation or stabilization of Prx5 as a plausible explanation for the tolerance observed in Urm1 n123 flies. A concurrent loss of Prx5 does not affect the survival of Urm1 mutant flies (Fig. 5d), nor is the protein level of Prx5 affected in Urm1 n123 mutant lysates (Fig. 5e). Interestingly, Prx5 EY02106 mutant flies appear to benefit from a simultaneous loss of Urm1, indicating a dominant cytoprotective effect of Urm1 deficiency which is unrelated to the activity of Prx5 (Fig. 5d).
Fig. 5.
The Drosophila homolog of Ahp1, Peroxiredoxin 5 (Prx5), is a target of urmylation in vivo. a Prx5 is urmylated in response to oxidative stress. Flies with an induced ubiquitous expression of UAS:Flag-Urm1 WT and UAS:Prx5 WT was exposed to 10 mM paraquat for the time points indicated. Immunoprecipitation using anti-Flag antibodies followed by immunoblotting with anti-Prx5 antibodies indicates a strong conjugation of Urm1 to Prx5 after 1 h of stimulation. b Urm1 conjugation to Prx5 is abolished in flies where the level of Uba4 has been reduced by expression of an Uba4-RNAi transgenic construct under control of Actin5C-GAL4. c The urmylation of Prx5 in response to paraquat treatment is dependent on the C-terminal GG-motif in Urm1. Flag-tagged Urm1 lacking the GG-motif (UAS:Urm1 ΔGG) fails to interact with endogenous Prx5 in fly lysates after 1 h exposure to 10 mM paraquat. d Prx5 is not promoting the oxidative stress tolerance in Urm1 n123 mutant flies. A simultaneous loss of Prx5 in an Urm1 n123 mutant background does not alter the survival rates of Urm1 deficient flies exposed to oxidative stress (compare homozygous Urm1 n123 mutants with Urm1 n123; Prx5 EY02106 double mutants). In contrast, Prx5 EY02106 null flies benefit from a simultaneous loss of Urm1, suggesting that Urm1 operates in a cytoprotective pathway acting in parallel to Prx5. The graph is a representative of three independent replicates, only testing female flies, n = 200 for all genotypes tested. e Prx5 is not affected on the protein level by loss of Urm1. Total cell lysates from Urm1 n123 and control flies were immunoblotted using anti-Prx5 antibodies
Urm1 is a novel negative regulator of the JNK signaling pathway
In order to explain the oxidative stress tolerance observed in Urm1 deficient flies at the molecular level, we investigated a number of the major pathways known to be involved in oxidative stress. We found that one key pathway, the JNK pathway, is clearly upregulated in Urm1 n123 mutant flies, as indicated by increased JNK phosphorylation (Fig. 6a, b). The c-Jun NH(2)-terminal kinase (JNK), which is a member of a subfamily of the mitogen-activated protein (MAP) kinases, has been ascribed a prominent role in the response against various environmental stressors, as well as in inflammatory responses, cell survival and development [31, 32]. Increased JNK phosphorylation was observed when immunoblotting Urm1 n123 fly lysates with anti-pJNK antibodies and further confirmed by immunohistochemical analysis of somatic Urm1 n123 null mutant clones in imaginal discs (Fig. 6a, panel 1 and 2). Hyperactivation of JNK signaling at the level of the downstream transcription factor dJun, observed as an increased nuclear accumulation of dJun [33], was also observed (Fig. 6a, panel 3). In Drosophila, the JNK pathway has shown to induce a battery of genes involved in the cytoprotective response evoked by oxidative stress [34] and consistent with our results, some of these genes are also upregulated in the absence of Urm1. Firstly, the peroxiredoxin Jafrac1, previously reported to be upregulated downstream of stress-induced JNK, as well as Keap1/Nrf2, signaling and to confer resistance to oxidative stress [35, 36], is upregulated both in lysates prepared from homozygous Urm1 n123 flies (Fig. 6b), and in clones lacking Urm1 (Fig. 6a, panel 4). Secondly, we also found that the prototypical oxidative stress response protein Glutathione S-transferase D1, GstD1 [37], is strongly induced in Urm1 n123 mutants (Fig. 6c), using a transgenic gstD-GFP reporter fly strain, generated by Sykiotis and Bohmann [38].
Fig. 6.
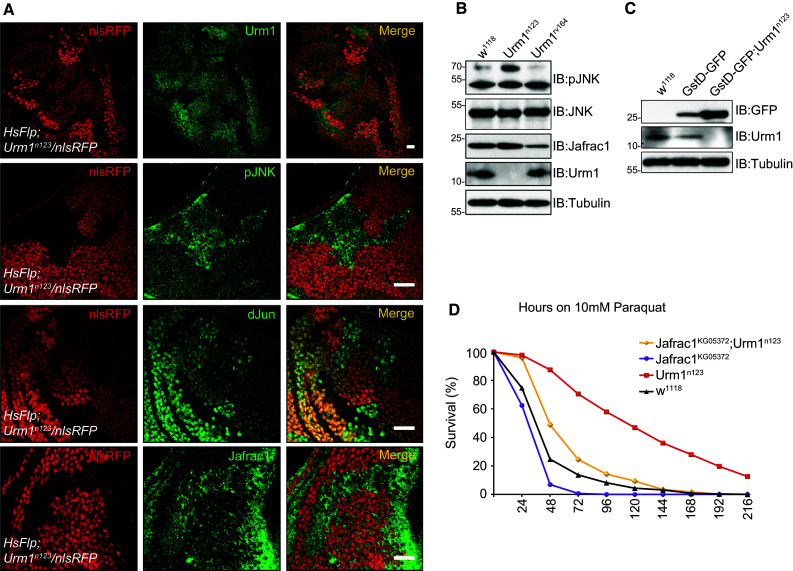
Loss of Urm1 results in an upregulation of the JNK signaling pathway and induction of cytoprotective genes such as Jafrac1 and gstD1. a Clones deficient of Urm1 expression shows increased levels of JNK phosphorylation, nuclear accumulation of dJun and expression of Jafrac1. Urm1 loss-of-function clones (P{w[+mW.hs] = FRT(w[hs])2A; Urm1 n123), induced by heat shock and marked by loss of nlsRFP expression were stained with the antibodies indicated. Note that the nlsRFP positive cells surrounding the clones serve as control with a w 1118 genetic background. Scale bars = 20 μm. b Urm1 n123 mutant flies exhibit a constitutive upregulation of the JNK signaling pathway and Jafrac1 expression. Protein lysates from Urm1 n123 flies display significantly higher levels of JNK phosphorylation, as well as an increased level of Jafrac1 protein expression, as compared with control flies (white 1118 and Urm1 rv164). Importantly, the level of total JNK is not affected in Urm1 n123 mutants. c Transcription from the gstD1 promoter is strongly upregulated in the absence of Urm1. The transgenic gstD-GFP reporter generated by Sykiotis and co-workers [38] was monitored by anti-GFP immunoblotting of total cell lysates of flies homozygous for the Urm1 n123 mutation. d Urm1 interacts genetically with Jafrac1. The removal of Jafrac1 in a homozygous Urm1 n123 background (Jafrac1 KG05372/KG05372 ; Urm1 n123/n123) reduce the resistance of Urm1 mutants to oxidative stress. n = 200 for all genotypes tested
Having established that loss of Urm1 results in a hyperactivation of the JNK pathway, we wanted to address whether JNK-mediated upregulation of oxidative stress response genes could explain the oxidative stress tolerance phenotype of Urm1 n123 mutants. We therefore generated double mutants lacking expression of both Urm1 and Jafrac1 (Jafrac1 KG05372; Urm1 n123) and tested their sensitivity to paraquat. In agreement with our hypothesis, a simultaneous loss of Jafrac1 clearly reduced the ability of Urm1 n123 mutant flies to survive in the presence of 10 mM paraquat, suggesting that JNK-mediated upregulation of Jafrac1 at least partially explains the oxidative stress resistance displayed by Urm1 deficient animals (Fig. 6d).
Discussion
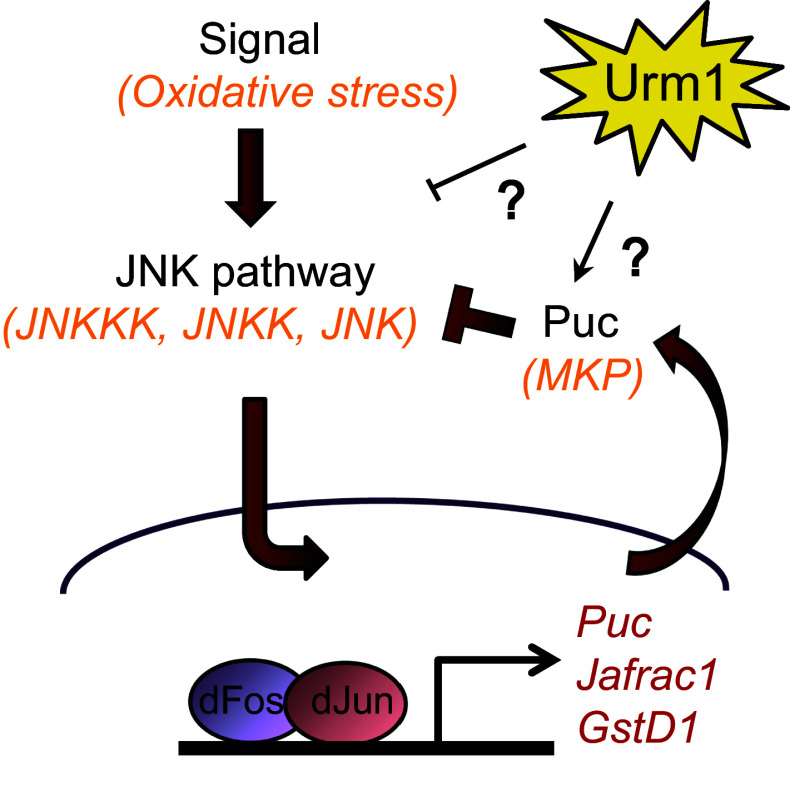
In conclusion, we have identified the UBL Urm1 and its E1 activating enzyme Uba4 in Drosophila melanogaster and shown that they form a conjugation machinery complex with the capacity to induce urmylation of multiple target proteins in vivo, of which we provide molecular proof for one, the Ahp1 homologue Prx5. We demonstrate that complete loss of Urm1 causes embryonic lethality, whereas high levels of maternal contribution allows the survival of a small percentage of adult zygotic Urm1 n123 mutant escaper flies, which display a reduced lifespan but increased resistance to oxidative stress. Importantly, similar phenotypes are induced by RNAi-mediated reduction of Uba4 expression, a finding that together with genetic interaction experiments clearly places Urm1 and Uba4 in the same cellular pathway. One hypothesis for explaining the tolerance of Urm1 deficient flies to oxidative stress, is that loss of Urm1 directly or indirectly results in an activation of the JNK pathway and by extension elevated expression levels of multiple oxidative stress response genes, including Jafrac1 and gstD1 (model in Fig. 7).
Fig. 7.
Model describing the oxidative stress tolerance displayed by Urm1 deficient flies. On a yet unidentified level, loss of Urm1 negatively affects signaling in the JNK pathway. In the absence of Urm1, the JNK pathway is hyperactivated, resulting in a nuclear accumulation of the dJun transcription factor and subsequent upregulation of multiple cytoprotective genes, including Jafrac1 and gstD1. As a consequence, zygotic homozygous Urm1 n123 mutant flies display an increased tolerance to oxidative stress
Oxidative stress has in both yeast and mammalian cells been described as a strong inducer of Urm1 conjugation to target proteins and in agreement, yeast strains deficient of urm1 or uba4 are sensitive to multiple environmental stressors including the oxidizing agents diamide and tert-butyl hydroperoxide (t-BOOH) [8, 16]. The contradictory tolerance of Urm1 null (Urm1 n123) flies to oxidative stress initially struck us as an artefact, but the congruent phenotypes induced by RNAi-mediated knockdown of Urm1, as well as misexpression of a putative Urm1 dominant negative transgene (Urm1 ΔGG) strongly support these results. Although we have not been able to demonstrate a stable urmylation response following oxidative stress, we can clearly show that the bona fide Urm1 conjugation target protein Prx5 is rapidly urmylated in flies exposed to paraquat, and thus that Urm1 acts as a UBL that posttranslationally modifies target proteins in higher organisms. This is further emphasized by the appearance of multiple high-molecular weight bands recognized by anti-Urm1 antibodies on a Western blot of lysates prepared from flies ectopically expressing tagged Urm1 and Uba4 proteins.
Despite being urmylated in response to oxidative stress, we have found no proof that a stabilization or activation of Prx5 would contribute to the paraquat resistance observed in Urm1 n123 flies. Loss of Prx5 has no effect in an Urm1 mutant background, and alterations of the Urm1 cellular landscape (by transgenic expression of either Urm1 WT, Urm1 ΔGG or Urm1-RNAi) does not interfere with the increased tolerance to oxidative stress that have been associated with elevated levels of Prx5 (data not shown and [29]). In contrast, loss of Urm1 appears to be beneficial in Prx5 EY02106 null mutants, suggesting that an Urm1 deficiency positively regulates a cytoprotective pathway which acts in parallel to Prx5. However, the Uba4-dependent Urm1 conjugation to Prx5 is nevertheless an important finding, which strongly reinforces the evolutionary conservation of Prx5/Ahp1 urmylation in peroxide modification, previously reported in yeast and human cells [8, 16, 18]. Moreover, the rapid and transient nature of Prx5 urmylation is interesting and may point towards a role of urmylation in the cellular machinery that immediately senses and responds to oxidative stress, the suggested switch mechanism that determines the destiny of the cell—to stay and fight or to die [39–41].
To identify cellular machineries affected by Urm1 we have utilized available tools and antibodies to measure oxidative stress levels and evaluate the activity of known players in the oxidative stress response, including the Nrf2 [42] and PI3 Kinase pathways [43], p38 and pERK [44], the HIF-1 oxygen sensing system [45], expression of oxidative stress protective genes such as Heme oxygenase [46] and multiple peroxiredoxin genes [47], as well as general apoptotic markers. By using this strategy, we detected an ectopic activation of the JNK signaling pathway in the absence of Urm1, both in fly lysates and somatic clones lacking Urm1 expression in imaginal discs. For this we provide molecular evidence on multiple levels of the JNK pathway, including increased phosphorylation of JNK, enhanced nuclear accumulation of the downstream transcription factor dJun [33, 48], as well as upregulation of the JNK-controlled cytoprotective genes Jafrac1 and gstD1 in tissues lacking Urm1 [34, 36]. Drosophila Jafrac1, which is the fly equivalent of human Peroxiredoxin II [49], has interestingly been reported to reduce ROS levels and contribute to an enhanced stress resistance, as well as extended lifespan in flies [36]. Indeed, the increased levels of Jafrac1 in Urm1 mutant flies clearly contributes to the oxidative stress tolerance of Urm1 mutant flies, given that a concurrent loss of Urm1 and Jafrac1 at least partially reverts the Urm1 n123 phenotype.
In most animals, an increased tolerance to various stressors, including oxidative stress, is strongly related to a prolonged longevity [26], as is the case for several components of the JNK pathway [34, 50]. However, in at least one case in the literature, one mutant organism has been described to, analogous to Urm1 n123 mutants, be resistant to oxidative stress and simultaneously display a decreased lifespan. Interestingly, this mutant is a loss-of-function allele of the 2-Cys type peroxiredoxin PRDX-2 in C. elegans [51], which is strongly related to Drosophila Jafrac1. PRDX-2 has been suggested to perform functions as divergent as (1) thioredoxin peroxidase activity, protective against oxidative stress, (2) chaperone activity, activated during heat stress and (3) to act as a signaling molecule, inhibiting the transcription of phase-II detoxificating enzymes, of which the last has been suggested as the main cause of the oxidative stress tolerance displayed by PRDX-2 deficient animals [51]. However, in flies Jafrac1 follow the common rule of acting protective against stress and to prolong longevity [36]. So, the question of how loss of Urm1 can cause stress resistance together with a decreased lifespan still remains. A plausible hypothesis may lie in the dual nature of JNK signaling. As being one of the most ancient conserved signaling molecules, JNK has shown to be essential during development and display beneficial functions during multiple stress responses in many species, but can like a double-edge sword switch function and contribute to cell death and disease [31, 32]. How JNK signaling can alternate between valuable and detrimental activities is not fully understood, but is believed to be influenced by multiple criteria, including mode and duration of activation, cross-talk with other pathways, tissue specificity and the age of animals [52–55]. In agreement with such studies, loss of Urm1 could in young animals contribute to a beneficial JNK signaling activity and oxidative stress tolerance, which in older animals could turn into pro-apoptotic activities and premature aging. The dynamic functionality of JNK signaling, together with the apparent evolutionary transition of JNK-like proteins, from primarily acting in osmotic to oxidative stress in yeast versus mammals, respectively [56, 57], may explain the reported divergence in Urm1 activity during oxidative stress in different species. It is not unlikely that JNK in Drosophila has gained other cellular functions and levels of regulation, as compared with yeast. However, since increased JNK has been reported to prolong longevity in flies [34, 50], the shortened lifespan we observe in Urm1 Δ123 mutants may also be caused by interference with other signaling routes.
Another interesting question is how Urm1 influences the JNK pathway, whether the effect is directly caused by urmylation of one of the components in the pathway or indirect in nature. Despite intense efforts, we have not been able to verify an Urm1 conjugation to any of the major components in the fly JNK pathway (Hemipterous, Basket, Puckered, Kayak or dJun) in either embryos or adult flies. However, in light of the recently described role of S. acidocaldarius Urm1 in protein degradation [19], it is plausible that this investigation should be performed in the presence of proteasomal inhibitors. Hence, at this point we do not know whether Urm1 has the capacity to conjugate to any of the components of the JNK cascade, but in light of the sulphur carrier activity of Urm1, it is interesting to note that thiolation has been reported to influence JNK activation. Firstly, the JNKKK protein apoptosis-signal regulated kinase 1 (ASK1) is inhibited by binding to the reduced form of the redox protein Thioredoxin (Trx) [58] and secondly, the JNK inhibitory phosphatases Puckered and MKP4 are known to be strongly influenced by redox status, given that they both belong to the class I cysteine-based dual-specificity PTPs, which are regulated by a reversible oxidation of their catalytic-site cysteine [52, 59–61]. To pinpoint where and how Urm1 interferes with the JNK pathway will require further experimental analysis, but will be an important issue to address in future studies. Moreover, in light of the involvement of the JNK pathway in a large variety of cellular processes, it will be interesting to study whether Urm1 also plays a role in other JNK-regulated events.
Since its discovery the role of Urm1 as a combined sulphur carrier and UBL modifier has puzzled the scientific community. Here we provide evidence from a multicellular organism that urmylation indeed occurs in vivo, and that Urm1 displays multiple biologically relevant functions at different time points of the Drosophila life cycle. The high evolutionary conservation of Urm1 is consistent with our finding that Urm1 is an essential gene, which strongly influences cellular signaling in eukaryotic cells. This initial characterization of Urm1 and Uba4 in Drosophila and the generation of new tools can now lay the ground for further investigations of Urm1 at the cellular, as well as physiological and developmental levels. In conclusion, Urm1 is a UBL modifier essential for life and longevity in Drosophila, which negatively influences the JNK pathway, resulting in an upregulation of JNK-dependent cytoprotective genes and resistance to oxidative stress in Urm1 deficient animals.
Materials and methods
Drosophila stocks
Standard Drosophila husbandry procedures were employed. Flies were raised and crossed at room temperature (RT) unless otherwise stated. white 1118 was used as wild type control. Actin5C-GAL4, MS1096-GAL4, P{SUPor-P}CG33276 KG08138 (#14938), P{EPgy2}Prx5 EY02106 CG7215 EY02106 (#15852), P{SUPor-P}Jafrac1 KG05372 (#14440), P{ry[+t7.2] = Delta2-3}99B (#1610), Df(2L)BSC215/CyO (#9643), Df(2L)Exel7038/CyO (#7809) and P{w[+mW.hs] = FRT(w[hs])}2A (#1997) was obtained from Bloomington Drosophila Stock Center (BDSC), Indiana, USA. Urm1 RNAi lines P{GD15862}v48364 and P{GD15862}v483643/TM3 was from Vienna Drosophila Resource Center, Vienna, Austria [62]. Uba4 RNAi lines CG13090R-1 and CG13090R-2 were obtained from NIG-Fly Stock Center (Kyoto, Japan). Clones were induced using hsFlp (#1929) in combination with and P{AyGAL4}25 P{UAS-GFP.S65T}Myo31DF T2 (#4411) for overexpression clones and P{His2Av-mRFP1}III.1 P{FRT(w hs )}2A (#34498) for somatic clones, respectively (BDSC). Clones were induced by heat shocking first instar larvae in a 37 °C water bath for 75 min. The gstD-GFP reporter has been previously described [38], as well as UAS:Prx5 flies [29].
Generation of Urm1 mutants
The Drosophila Urm1 n123 null allele was generated by imprecise excision of the P{SUPor-P} element CG33276 KG08138 (#14938), inserted in the 5′flanking region of the CG33276 coding region, and identified by PCR using the primers 5′-GAGGTTCCCGGCATGTTCACC-3′ and 5′-CTTCGAATTACCCGCCAAACCG-3′. The revertant line Urm1 rv164 is a precise deletion of the CG33276 KG08138 element. Both the Urm1 n123 and Urm1 rv164 lines was cleaned from an unidentified background mutation on the CG33276 KG08138 chromosome by back-crossing to white 1118 six times. For clonal analysis the Urm1 n123 mutation was recombined onto the P{w[+mW.hs] = FRT(w[hs])}2A chromosome.
Generation of Urm1 and Uba4 transgenic flies
pUAST:Urm1WT was generated by amplification of the entire Urm1 cDNA (IP20063) by PCR and subsequent subcloning into pUAST using XhoI/XbaI. pUAST:Urm1ΔGG was generated in a similar manner. pUAST:Flag-Urm1WT was generated by inserting a 3xFlag tag in the N-terminus of Urm1 using BglII/XhoI. pUAST:Flag-Urm1ΔGG was generated by first making a pEntr™/D-TOPO vector containing Urm1ΔGG and subsequently moving the Urm1ΔGG fragment into the pTFW vector (DGRC) by standard Gateway LR Clonase methodology (Life Technologies). The size difference between Flag-Urm1WT and Flag-Urm1ΔGG is caused by different linker lengths resulting from different strategies of plasmid preparation (Fig. 5b). Uba4 was amplified by PCR and ligated into pUAST (XhoI/XbaI) to generate pUAST:Uba4WT. In pUAST:Myc-Uba4WT, a 6x-Myc tag was inserted in the N-terminus of Uba4 using BglII/XhoI.
Generation of Urm1 and Uba4 antibodies
The entire Urm1 or Uba4 cDNA was subcloned into the pETM-11 vector using NcoI/XhoI and transformed into E. coli BL21(DE3). Protein expression was induced at 37 °C using 1 mM IPTG and the resulting 6xHIS-Urm1 and 6xHIS-Uba4 fusion proteins was purified using Ni-NTA agarose according to the manufacturer’s instructions (QIAGEN). Two rabbits were immunized with SDS-PAGE gel pieces containing the 6xHIS-Urm1 protein (Agrisera), of which one showed a good response against Urm1. One rat was similarly injected with 6xHIS-Uba4 (Agrisera), as well as one rabbit. The rabbit polyclonal Urm1 and Uba4antibodies were further refined by Protein A IgG purification (Thermo Scientific), followed by affinity purification against GST-Urm1 or GST-Uba4, and was subsequently used at 1:250–1:500 for both Western blot and immunohistochemistry (IHC). Rat anti-Uba4 was used at 1:500 for Western Blot.
Genomic Drosophila DNA extraction, PCR and sequencing
Genomic fly DNA was extracted by manual homogenization of single frozen candidate flies in 50 µl squishing buffer (10 mM Tris-HCl pH 8.2, 1 mM EDTA, 25 mM NaCl) freshly supplemented with 200 µg/ml Proteinase K and incubated for 40 min at 37 °C. After heat inactivation of Proteinase K for 10 min at 85 °C, the DNA samples were cleared by centrifugation at top speed for 2 min and either used immediately for PCR analysis or stored at −20 °C. PCR was performed according to standard protocols using DreamTaq polymerase (Thermo Scientific, Waltham, USA). PCR fragments were purified using the QIAEX II gel extraction kit (QIAGEN) and sequence information was obtained by employing the BigDye™ Terminator Signaling Kit v3.1 (Applied Biosystems).
Immunostaining of Drosophila imaginal discs
Imaginal discs from third instar larvae were dissected in PBS, fixed in 4 % formaldehyde for 25 min, washed 4 × 15 min in PBST (1 × PBS with 0.1% Triton X-100) and blocked at least 1 h in 5 % Normal Goat Serum (NGS) before incubation with primary antibodies overnight at 4 °C. Antibodies were diluted in 1 x PBS with 1 % NGS at the following concentrations; Rabbit anti-Urm1 at 1:500, rabbit anti-Active-JNK (pTPpY) at 1:1000 (Promega), rabbit anti-Jafrac1 at 1:1000 (kind gift from J. Santaren [63]), rabbit anti-dJun at 1:1000 (kind gift from D. Bohmann [64]). The next day the discs were washed in PBST, incubated with fluorescently labelled secondary antibodies (Jackson ImmunoResearch), washed again and mounted on Poly-l-lysine coated slides prior to visualization by Nikon C1 confocal microscope, magnifications 40× Plan Apo NA 0,95, 60× Plan Apo VC NA 1,40 oil and 100× Plan Apo VC NA 1,40 oil and EZ-C1 software.
Drosophila protein lysate preparation, immunoprecipitation and western blot
Protein lysate preparation Adult Drosophila flies were homogenized in lysis buffer (50 mM HEPES pH7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 % Triton X-100, 10% Glycerol, 25 mM NAF,10 µM ZnCl2) supplemented with Complete Protease Inhibitor Cocktail (Roche), 1 mM PMSF and 10 mM N-Ethylmaleimide (Sigma-Aldrich). The lysates were cleared by centrifugation at top speed for 30 min at 4°C and the total protein concentration was determined using standard Bradford protein assay (Bio-Rad). 1–2 mg of total protein were used for IP and GST-pull down, unless otherwise stated.
GST pull-down: GST-Urm1 fusion protein was purified according to standard protocols from E. coli BL21(DE3) transformed with pGEX-4T1:Urm1 and linked to Gluthathione Sepharose 4B (GE Healthcare). Equivalent amounts of GST or GST-Urm1 (corresponding to 2–5 µl of coupled Sepharose beads) were incubated with 1 mg total protein fly lysate in the absence or presence of DTT on a rotator at 4 °C overnight. The pull-downs were washed 4 times with lysis buffer and finally resolved by boiling in 2× Sample buffer (63 mM Tris–HCl pH 6.8, 10 % Glycerol, 2 % SDS, 0.0025 % Bromophenol Blue) at 95 °C for 5 min.
Immunoprecipitation: Immunoprecipitations were performed using Anti-FLAG® M2 Magnetic Beads (Sigma-Aldrich) according to instructions provided by the manufacturer.
Western Blot: GST pull-downs, immunoprecipitations and protein lysates were separated by SDS-PAGE and transferred to a 0.2 μm PVDF membrane (Merck Millipore) using a semi-dry Trans-Blot Turbo transfer system (Bio-Rad). GST pull-downs were analysed by Ponceau S staining. All membranes were blocked in 5 % BSA in 1× TBS (50 mM Tris−HCl, pH 7.5, 150 mM NaCl) for 1 h at RT and incubated with primary antibodies overnight at 4 °C. Antibodies were diluted in 1x TBS with 5 % BSA at the following dilutions; rabbit anti-Urm1 at 1:500, rat anti-Uba4 at 1:500, rabbit anti-Uba4 at 1:250, mouse anti-Tubulin 1:5000 (Sigma-Aldrich), mouse anti-Flag M2 at 1:1000 (Sigma-Aldrich), mouse anti-Myc 9E10 at 1:500 (Abcam), mouse anti-GFP JL-8 at 1:2000 (Clontech), rabbit anti-Histone H3 at 1:10,000 (Abcam), rabbit anti-Active-JNK (pTPpY) at 1:1000 (Promega), rabbit anti-JNK(FL) at 1:250 (sc-571, Santa Cruz Biotechnology), rabbit anti-Jafrac1 at 1:1000 (kind gift from R. Lehmann [65]) and rabbit anti-Prx5 at 1:1000 (kind gift from S. Radyuk [28]). The membranes were washed 5 × for 10 min in TBST (1 × TBS with 0.075 % Tween-20), incubated with HRP-linked secondary antibodies (GE Healthcare) for 1 h and washed again 5 × 10 min, all at RT, before detection by ECL (Thermo Scientific or GE Healthcare) followed by autoradiography.
S2 cell nuclear/cytoplasmic lysate preparation
S2 Cells were harvested by centrifugation at 1500×g and homogenized in lysis buffer (10 % Sucrose, 10 mM Tris-HCl pH 8.0, 10 mM NaCl, 3 mM MgCl2, 2 mM DTT, 0.2% Triton X-100, 1 mM PMSF). After 3 min centrifugation at 1500×g, the supernatant (containing cytoplasmic fraction) was collected and the pellet (containing the nuclear fraction) was resolved in the nuclear buffer (10 mM Tris-HCL pH 8.0, 1 mM EDTA pH 8.0, 0.14 M NaCl, 2 mM DTT and 1 mM PMSF).
Lifespan analysis
Flies of the indicated genotypes were collected 0–24 h after eclosure and sorted into females and males after 3 days. 40 flies were kept in each vial, which were monitored for survival and transferred to fresh food every 2–3 days until no flies remained. Flies were kept at 25 °C with a humidity of 60 % and exposed to an artificial light–dark cycle of 12:12 h.
ROS experiments
Age-matched flies of the indicated number and genotypes were starved for 4 h in empty vials, before exposure to 10 mM paraquat (Methyl viologen dichloride hydrate, Sigma-Aldrich), or 2 % H2O2 (Sigma-Aldrich), dissolved in 5 % sucrose and soaked into Whatman GB005 filter papers (GE Healthcare). The flies were scored for survival every 24 h and transferred to a fresh vial with newly prepared paraquat every 48 h.
Alignments and phylogenetic analysis
Sequence alignments were made using the multiple sequence alignment method (MUSCLE) server provided by the online Phylogeny.fr platform [66, 67]. The protein sequence accession numbers used are for D. melanogaster Urm1 NP_996018.2, S. cerevisiae Urm1 NP_012258.3, C. elegans Urm1 NP_001255080.1, M. musculus Urm1 NP_080891.1, H. sapiens Urm1 NP_112176.1, E. coli MoaD NP_415305.1, E. coli ThiS YP_491469.1, D. melanogaster Uba4 NP_001260256.1, S. cerevisiae Uba4p NP_011979.1, C. elegans MOCS3 NP_501359.1, M. musculus MOCS3 NP_001153802.1 and H. sapiens MOCS3 NP_055299.1. Phylogenetic trees were generated using the PhyML tool provided by the online Phylogeny.fr platform [66, 67].
Preparation and mounting of adult Drosophila wings
Wings of the indicated genotypes were washed in Methanol for 5 min and mounted on glass slides in a drop of pre-warmed Canada balsam (Sigma). The slides were heated to 65 °C for 20 min to allow bubbles to dissipate before imaging was performed using a Nikon SMZ1500 stereo microscope with NIS-E F software.
Quantitative PCR
Total RNA of the indicated developmental stages of the w 1118 control strain, extracted using TRIzol® (Thermo Fischer Scientific), was template for cDNA synthesis with Random Hexamers and SuperScript®II Reverse Transcriptase (Thermo Fischer Scientific). The cDNA was purified with the DNA Clean and Concentrator™ kit (Zymo Research), before qPCR was performed employing the KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems), employing the primers GGGCGGAGTTACTATTTGGT and TCATAACCGATTTCACTCAAGTTT for Urm1, together with TGGGCGATCTCGCCGCAGTA and CAGAGTGCGTCGCCGCTTCA for RpL32.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We would like to thank members of the scientific community who generously shared reagents critical to this work. In addition, we thank Ruth H. Palmer, Mattias Alenius and Yasuo Yamazaki for critical reading of the manuscript and for scientific discussions. We also acknowledge the Drosophila Genomics Resource Center (DGRC), supported by NIH grant 2P40OD010949-10A1, for providing cDNA clones and vectors and the Bloomington Drosophila Stock Center (NIH P40OD018537) for fly stocks. This work was supported by the Swedish Foundation for Strategic research (SSF), The Swedish Research Council, Jeanssons Stiftelser and Insamlingsstiftelsen för medicinsk forskning vid Umeå Universitet.
Footnotes
B. Khoshnood and I. Dacklin have contributed equally to this work.
References
- 1.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Ann Rev Biochem. 2012;81(81):323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 3.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399(3):361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D, Zhang DE. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine. 2011;31(1):119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275(11):7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 7.Goehring AS, Rivers DM, Sprague GF., Jr Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol Biol Cell. 2003;14(11):4329–4341. doi: 10.1091/mbc.E03-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci USA. 2011;108(5):1763–1770. doi: 10.1073/pnas.1014402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedrioli PG, Leidel S, Hofmann K. Urm1 at the crossroad of modifications. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9(12):1196–1202. doi: 10.1038/embor.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Liu M, Qiu R, Ji C. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell. 2011;2(8):612–619. doi: 10.1007/s13238-011-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2008 doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B, Lu J, Bystrom AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae . RNA. 2008;14(10):2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009 doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 14.Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem. 2008;283(41):27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 15.Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci USA. 2008;105(47):18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goehring AS, Rivers DM, Sprague GF., Jr Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003;2(5):930–936. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz J, Chowdhury MM, Hanzelmann P, Nimtz M, Lee EY, Schindelin H, Leimkuhler S. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47(24):6479–6489. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 18.Judes A, Ebert F, Bar C, Thuring KL, Harrer A, Klassen R, Helm M, Stark MJ, Schaffrath R. Urmylation and tRNA thiolation functions of ubiquitin-like Uba4.Urm1 systems are conserved from yeast to man. FEBS Lett. 2015;589(8):904–909. doi: 10.1016/j.febslet.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 19.Anjum RS, Bray SM, Blackwood JK, Kilkenny ML, Coelho MA, Foster BM, Li S, Howard JA, Pellegrini L, Albers SV, Deery MJ, Robinson NP. Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius . Nat Commun. 2015;6:8163. doi: 10.1038/ncomms9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor CT, Pouyssegur J. Oxygen, hypoxia, and stress. Ann N Y Acad Sci. 2007;1113:87–94. doi: 10.1196/annals.1391.004. [DOI] [PubMed] [Google Scholar]
- 21.Zaher TE, Miller EJ, Morrow DM, Javdan M, Mantell LL. Hyperoxia-induced signal transduction pathways in pulmonary epithelial cells. Free Radic Biol Med. 2007;42(7):897–908. doi: 10.1016/j.freeradbiomed.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295(4):C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Wang Y, Su Y. Peroxiredoxins, a novel target in cancer radiotherapy. Cancer Lett. 2009;286(2):154–160. doi: 10.1016/j.canlet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- 24.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 25.Le Lay S, Simard G, Martinez MC, Andriantsitohaina R. Oxidative stress and metabolic pathologies: from an adipocentric point of view. Oxid Med Cell Longev. 2014;2014:908539. doi: 10.1155/2014/908539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43(4):477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 27.St Pierre SE, Ponting L, Stefancsik R, McQuilton P, FlyBase C. FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42((Database issue)):D780–D788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalak K, Orr WC, Radyuk SN. Drosophila peroxiredoxin 5 is the second gene in a dicistronic operon. Biochem Biophys Res Commun. 2008;368(2):273–278. doi: 10.1016/j.bbrc.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radyuk SN, Michalak K, Klichko VI, Benes J, Rebrin I, Sohal RS, Orr WC. Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila . Biochem J. 2009;419(2):437–445. doi: 10.1042/BJ20082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Spector D, Godon C, Labarre J, Toledano MB. A new antioxidant with alkyl hydroperoxide defense properties in yeast. J Biol Chem. 1999;274(8):4537–4544. doi: 10.1074/jbc.274.8.4537. [DOI] [PubMed] [Google Scholar]
- 31.Johnson GL, Nakamura K. The c-jun kinase/stress-activated pathway: regulation, function and role in human disease. Biochim Biophys Acta. 2007;1773(8):1341–1348. doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19(2):142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Musti AM, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275(5298):400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 34.Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila . Dev Cell. 2003;5(5):811–816. doi: 10.1016/S1534-5807(03)00323-X. [DOI] [PubMed] [Google Scholar]
- 35.Hochmuth CE, Biteau B, Bohmann D, Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila . Cell Stem Cell. 2011;8(2):188–199. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee KS, Iijima-Ando K, Iijima K, Lee WJ, Lee JH, Yu K, Lee DS. JNK/FOXO-mediated neuronal expression of fly homologue of peroxiredoxin II reduces oxidative stress and extends life span. J Biol Chem. 2009;284(43):29454–29461. doi: 10.1074/jbc.M109.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawicki R, Singh SP, Mondal AK, Benes H, Zimniak P. Cloning, expression and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem J. 2003;370(Pt 2):661–669. doi: 10.1042/bj20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila . Dev Cell. 2008;14(1):76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox AG, Winterbourn CC, Hampton MB. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem J. 2010;425(2):313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 40.Park J, Lee S, Lee S, Kang SW. 2-cys peroxiredoxins: emerging hubs determining redox dependency of Mammalian signaling networks. Int J Cell Biol. 2014;2014:715867. doi: 10.1155/2014/715867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poynton RA, Hampton MB. Peroxiredoxins as biomarkers of oxidative stress. Biochim Biophys Acta. 1840;2:906–912. doi: 10.1016/j.bbagen.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee N, Bohmann D. A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One. 2012;7(4):e34063. doi: 10.1371/journal.pone.0034063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodson M, Darley-Usmar V, Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCubrey JA, Lahair MM, Franklin RA. Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid Redox Signal. 2006;8(9–10):1775–1789. doi: 10.1089/ars.2006.8.1775. [DOI] [PubMed] [Google Scholar]
- 45.Movafagh S, Crook S, Vo K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: new developments in an old debate. J Cell Biochem. 2015;116(5):696–703. doi: 10.1002/jcb.25074. [DOI] [PubMed] [Google Scholar]
- 46.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86(2):583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 47.Radyuk SN, Klichko VI, Spinola B, Sohal RS, Orr WC. The peroxiredoxin gene family in Drosophila melanogaster . Free Radic Biol Med. 2001;31(9):1090–1100. doi: 10.1016/S0891-5849(01)00692-X. [DOI] [PubMed] [Google Scholar]
- 48.Schreck I, Al-Rawi M, Mingot JM, Scholl C, Diefenbacher ME, O’Donnell P, Bohmann D, Weiss C. c-Jun localizes to the nucleus independent of its phosphorylation by and interaction with JNK and vice versa promotes nuclear accumulation of JNK. Biochem Biophys Res Commun. 2011;407(4):735–740. doi: 10.1016/j.bbrc.2011.03.092. [DOI] [PubMed] [Google Scholar]
- 49.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300(5619):650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 50.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121(1):115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 51.Olahova M, Taylor SR, Khazaipoul S, Wang J, Morgan BA, Matsumoto K, Blackwell TK, Veal EA. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc Natl Acad Sci USA. 2008;105(50):19839–19844. doi: 10.1073/pnas.0805507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120(5):649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 53.Lamb JA, Ventura JJ, Hess P, Flavell RA, Davis RJ. JunD mediates survival signaling by the JNK signal transduction pathway. Mol Cell. 2003;11(6):1479–1489. doi: 10.1016/S1097-2765(03)00203-X. [DOI] [PubMed] [Google Scholar]
- 54.Sabio G, Kennedy NJ, Cavanagh-Kyros J, Jung DY, Ko HJ, Ong H, Barrett T, Kim JK, Davis RJ. Role of muscle c-Jun NH2-terminal kinase 1 in obesity-induced insulin resistance. Mol Cell Biol. 2010;30(1):106–115. doi: 10.1128/MCB.01162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Twumasi-Boateng K, Wang TW, Tsai L, Lee KH, Salehpour A, Bhat S, Tan MW, Shapira M. An age-dependent reversal in the protective capacities of JNK signaling shortens Caenorhabditis elegans lifespan. Aging Cell. 2012;11(4):659–667. doi: 10.1111/j.1474-9726.2012.00829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae . Microbiol Mol Biol Rev. 1998;62(4):1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol. 2010;13(6):677–683. doi: 10.1016/j.mib.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17(9):2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cavigelli M, Li WW, Lin A, Su B, Yoshioka K, Karin M. The tumor promoter arsenite stimulates AP-1 activity by inhibiting a JNK phosphatase. EMBO J. 1996;15(22):6269–6279. [PMC free article] [PubMed] [Google Scholar]
- 60.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24(5):981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun L, Yu MC, Kong L, Zhuang ZH, Hu JH, Ge BX. Molecular identification and functional characterization of a Drosophila dual-specificity phosphatase DMKP-4 which is involved in PGN-induced activation of the JNK pathway. Cell Signal. 2008;20(7):1329–1337. doi: 10.1016/j.cellsig.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez J, Agudo M, Van Damme J, Vandekerckhove J, Santaren JF. Polypeptides differentially expressed in imaginal discs define the peroxiredoxin family of genes in Drosophila . Eur J Biochem. 2000;267(2):487–497. doi: 10.1046/j.1432-1327.2000.01022.x. [DOI] [PubMed] [Google Scholar]
- 64.Bohmann D, Ellis MC, Staszewski LM, Mlodzik M. Drosophila Jun mediates Ras-dependent photoreceptor determination. Cell. 1994;78(6):973–986. doi: 10.1016/0092-8674(94)90273-9. [DOI] [PubMed] [Google Scholar]
- 65.DeGennaro M, Hurd TR, Siekhaus DE, Biteau B, Jasper H, Lehmann R. Peroxiredoxin stabilization of DE-cadherin promotes primordial germ cell adhesion. Dev Cell. 2011;20(2):233–243. doi: 10.1016/j.devcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36((Web Server issue)):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38((Web Server issue)):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.