Abstract
The CRISPR RNA-guided Cas9 nuclease gene-targeting system has been extensively used to edit the genome of several organisms. However, most mutations reported to date have been are indels, resulting in multiple mutations and numerous alleles in targeted genes. In the present study, a large deletion of 105 kb in the TYR (tyrosinase) gene was generated in rabbit via a dual sgRNA-directed CRISPR/Cas9 system. The typical symptoms of albinism accompanied significantly decreased expression of TYR in the TYR knockout rabbits. Furthermore, the same genotype and albinism phenotype were found in the F1 generation, suggesting that large-fragment deletions can be efficiently transmitted to the germline and stably inherited in offspring. Taken together, our data demonstrate that mono and biallelic large deletions can be achieved using the dual sgRNA-directed CRISPR/Cas9 system. This system produces no mosaic mutations or off-target effects, making it an efficient tool for large-fragment deletions in rabbit and other organisms.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2143-z) contains supplementary material, which is available to authorized users.
Keywords: Rabbit, CRISPR/Cas9, Tyrosinase, Albinism
Introduction
Rabbits are classic model animals and have more similarities to humans than mice and rats in terms of physiology, anatomy, and genetics. They also require less maintenance and have a shorter gestation period than pigs and monkeys. Currently, rabbits are extensively used as an appropriate animal model for cardiovascular/metabolic diseases and ophthalmic studies [1].
The Cas9/gRNA system was developed to take advantage of the RNA-guided Cas9 protein; when combined with a short, guide RNA (sgRNA), it can be used to target and cleave DNA sequences [1–5]. It has been successfully used to create gene-targeted in Drosophila [6, 7], Caenorhabditis elegans [8], rat [9, 10], monkey [11], zebrafish [12–14], mouse [15] and rabbit [16] with high efficiency. All of these genetically modified animals were generated via double-strand break and non-homologous end joining mediated repair, leading to the introduction of small insertions or deletions in the open reading frame (ORF) of the target locus. Further breeding and genetic testing of the homozygous offspring were therefore necessary. Recently, the use of a dual sgRNA-directed system to create large gene deletions has provided an efficient tool for the deletion of gene clusters, removal of long non-coding RNAs (lncRNAs), and elimination of gene regulatory sequences [17–19]. Co-injection of a dual sgRNA-directed CRISPR/Cas9 system was able to induce the complete deletion of a 65-kb fragment including the entire Dip2a gene in a mouse zygote [21]. However, there has been no report of large deletions via the CRISPR/Cas9 system in rabbit.
Here, we aimed to investigate the feasibility of generating large-scale genomic deletions via the dual sgRNA system with cytoplasm microinjection pronuclear-stage embryos. In the present study, we targeted the tyrosinase gene TYR, a major gene linked to albinism [20, 21], and generated a large-fragment genome deletion of TYR (105-kb) in rabbit. We demonstrate that a dual sgRNA-guided CRISPR/Cas9 system is an efficient method for large-fragment gene deletions in rabbit.
Materials and methods
Ethics statement
The rabbits used in this study were New Zealand white and Lianshan black rabbits. All animal studies were conducted according to experimental practices and standards approved by the Animal Welfare and Research Ethics Committee at Jilin University.
DNA constructs and in vitro transcription
The Cas9 expression construct, 3xFLAG-NLS-SpCas9-NLS, was synthesized and cloned into the pCS2+ vector. The construct was linearized with NotI and transcribed in vitro using the mMessage mMachine SP6 Kit (Ambion, USA) and the RNeasy Mini Kit (Qiagen).
To create a gRNA expression vector, the T7 promoter followed by two BbsI sites was synthesized upstream of the gRNA scaffold and cloned into the pUC57-Simple vector (Addgene ID 51306). A pair of complementary oligonucleotides encoding the 20-nt guide sequences were annealed at 95 °C for 5 min and ramped down to 25 °C to generate the dsDNA fragment, which was then cloned into the BbsI-digested gRNA expression vector.
PCR products for in vitro transcription of gRNAs were amplified using T7-F: (5′-GAAATTAATACGACTCACTATA-3′) and T7-R: (5′-AAAAAAAGCACCGA CTCGGTGCCAC-3′) primers. The sgRNAs were transcribed using the T7 RNA Synthesis Kit (Ambion) and purified by miRNeasy Mini Kit (Qiagen) according to the manufacturer. The concentration and quality of synthesized mRNAs were determined by Nanodrop 2000 and agarose gel electrophoresis, respectively.
Microinjection and embryo transfer
The protocol for microinjection of pronuclear-stage embryos has been described in detail by our published protocols [22]. In brief, zygotes were collected from sexually mature Lianshan black rabbits, that had undergone superovulation by six times and a 12-h interval of intravenous injection of follicle-stimulating hormone (FSH). The rabbits were mated 18 h after intravenous injection of 100 IU human chorionic gonadotrophin (hCG). The oviducts were then flushed with 5 mL DPBS-BSA for collection of pronuclear-stage embryos, which were transferred to embryo culture medium for microinjection.
Mixtures of in vitro-transcribed mRNA derived from the gRNAs (25 ng/μL) and Cas9 (100 ng/μL) were injected into the cytoplasm of pronuclear stage embryos. The injected embryos were transferred to embryo culture medium for 30–60 min, followed by transfer of approximately 30–50 injected embryos into the oviduct of the recipient mother.
Mutation detection in embryos and pups by PCR
Each injected zygote was collected at the blastocyst stage and incubated in embryo lysis buffer at 50 °C for 20 min and 90 °C for 5 min in a BIO-RAD PCR machine. Genomic DNA from TYR knockout and WT rabbits was isolated using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. PCR primers used for mutation detection are listed in Supplementary Table S1. PCR products were gel purified and cloned into pGM-T (Tiangen, Beijing, China). Ten positive plasmid clones were sequenced and DNAMAN was used for sequence analysis.
Off-target assay
Potential off-target sites (POTS) of the sgRNAs were predicted using the CRISPR online design tool (http://tools.genomeengineering.org). The top five POTS that were most likely to produce off-target mutations were selected and subjected to PCR and sequence analysis. Vector NTI and DNAMAN were used for sequence analysis. Primers are shown in Supplementary Table S3.
T7 endonuclease I (T7EI) assay
A T7 endonuclease I (T7EI) assay was performed as described previously [23]. Briefly, the genomic DNA of each Cas9/gRNA-injected blastocyst and its pups was extracted as mentioned above. The regions containing the off-target sites were amplified by PCR with gene-specific primers (Supplementary Table S2), then the PCR products were denatured and annealed under the following conditions: 95 °C for 5 min, 95 °C for 5 min, 95–85 °C at −2 °C/s, 85–25 °C at −0.1 °C/s, hold at 4 °C. The annealed samples were digested with T7EI (NEB M0302L), separated and measured on an ethidium bromide-stained 10 % polyacrylamide TAE gel.
Histology and western blotting
Skin and eye tissues from TYR knockout and WT rabbits were fixed with 4 % paraformaldehyde for 48 h, embedded in paraffin wax, and slide sectioned. Skin and eye sections were stained with hematoxylin and eosin and analyzed by microscope (Nikon ts100).
For western blotting, the eyes from TYR knockout and WT rabbits were homogenized in 150 µL of lysis buffer and protein concentrations were measured using the BCA Protein Assay Kit (Beyotime). Anti-TYR polyclonal antibody (1:2000; abcam) was used as the primary antibody, and anti-GAPDH monoclonal antibody (1:2000; Beyotime) was used as an internal control. The image was quantified using ImageJ software (NIH) and all the data are expressed as mean ± SEM.
Real-time quantitative PCR (qRT-PCR)
Total RNA was isolated from skin tissue using TRNzol reagent (TIANGEN, Beijing, China) according to the manufacturer’s instructions. RNA was first treated with DNase I (Fermentas) and reverse transcribed to cDNA using the BioRT cDNA First-Strand Synthesis Kit (Bioer Technology, Hangzhou, China). The primers used in this study are listed in Supplementary Table S1. qRT-PCR was performed using the BIO-RAD iQ5 Multicolor Real-Time PCR Detection System with the BioEasy SYBR Green I Real Time PCR Kit (Bioer Technology, Hangzhou, China). The formula was used to determine relative gene expression, which was normalized to the amount of GAPDH mRNA. All experiments were repeated three times for each gene. All the data are expressed as mean ± SEM.
Statistical analyses
Data from qRT-PCR and western blotting were analyzed with t tests using Graphpad Prism software. A P value of <0.05 was considered statistically significant.
Results
Dual sgRNA-directed large deletion of the TYR gene in zygotes
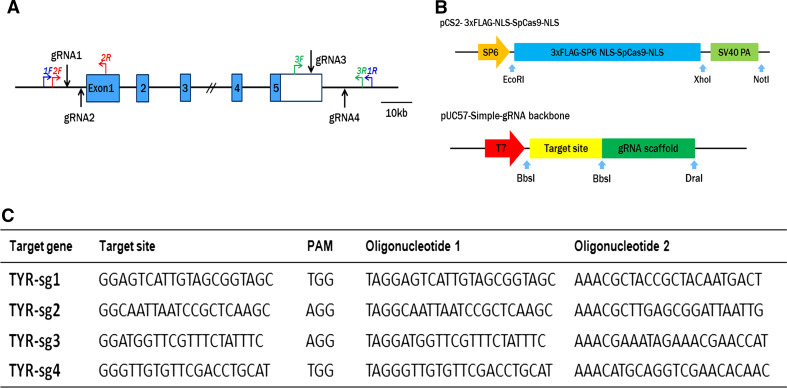
For targeting the rabbit TYR gene, four sgRNAs were designed based on the website tool. Two sgRNAs targeted sites 43 and 83 bp upstream of the TYR locus (sgRNA1 and sgRNA2, respectively), and the other two sites (sgRNA3 and sgRNA4) were located in the 3′ UTR of the last exon (Fig. 1a, c). Three pairs of primers were designed and used for the detection of mutations via PCR (Fig. 1a). The pCS2-3xFLAG-NLS-SpCas9- NLS vector and the pUC57-Simple-gRNA backbone vector (Addgene ID 51306), including the sgRNA sequences were transcribed in vitro and used for cytoplasmic microinjection (Fig. 1b).
Fig. 1.
Genome editing of TYR gene via the Cas9/gRNA system. a Schematic diagram of the four sgRNA target sites at the tyrosinase (TYR) gene locus. The CDS region is indicated by blue rectangles. SgRNA target sites are indicated by black arrow. SgRNA1and sgRNA2 are located upstream of the TYR locus. SgRNA3 and sgRNA4 are located in the 3′ UTR. 1/2/3 F and 1/2/3 R represent the PCR primer pairs used for mutation detection. The scale bar represents 10 kb. b Constructs and schematic illustration of the Cas9/gRNA system used in this study. c Target sequence of the four sgRNA used in this study
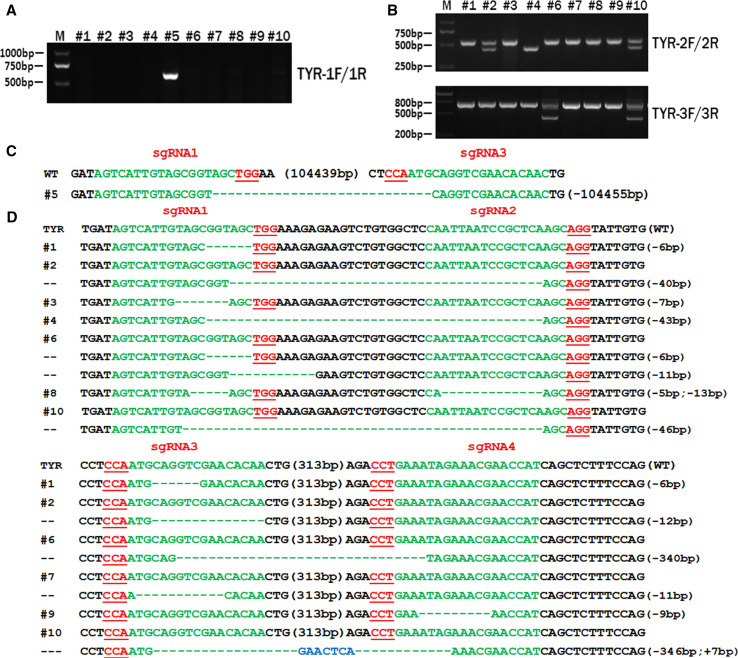
To determine the efficiency of dual sgRNA-directed large deletion of the TYR gene in zygotes, in vitro-transcribed mRNA of Cas9 and the four sgRNAs were mixed and microinjected into the cytoplasm of pronuclear-stage embryos at a concentration of 100 ng/μL (Cas9) and 25 ng/μL (sgRNA). Out of 18 injected embryos, 14 developed to the blastocyst stage; PCR products derived from 10 blastocysts were then sequenced to determine mutation efficiency in the zygotes. As shown in Fig. 2a and confirmed by sequence analysis, the desired large fragment deletion of TYR was found in blastocyst #5, demonstrating that a large deletion of 105 kb can be achieved via the dual sgRNA system and cytoplasmic microinjection of pronuclear-stage embryos (Fig. 2c).
Fig. 2.
Dual sgRNA-directed large deletion of TYR in zygotes. a Determination of 105-kb deletions of TYR gene in rabbit embryos by PCR. A clear band demonstrates the large-fragment deletion of TYR in #5 embryos; primers 1F and 1R were used for large-fragment determination. M DNA ladder. b Mutation detection for each sgRNA in zygotes. Primers TYR-2F/2R were used for mutation determination for sgRNA1 and sgRNA2, and TYR-3F/3R were used for sgRNA3 and sgRNA 4. M DNA marker. c T-cloning sequencing of deletion of TYR in #5 blastocyst. PAM sites are underlined and highlighted in red; target sequences are green; deletions (−) and insertions (+) are shown. WT wild-type control. d T-cloning sequencing of the target site for each sgRNA in injected embryos. PAM sites are underlined and highlighted in red; target sequences are green; deletions (−) and insertions (+) are shown. WT wild-type control
The mutation efficiency of the CRISPR/Cas9 system for each sgRNA was also determined by T-cloning and Sanger sequencing. PCR primers 2F and 2R were used for detection of mutations with sgRNA1 and sgRNA2, while 3F and 3R were used for sgRNA3 and sgRNA4 (Fig. 1a). As shown in Fig. 2b, the desired deletion of TYR was found in samples #2, #4, #6, and #10. The T-cloning and Sanger sequencing results demonstrate that the mutation efficiency of the four sgRNAs was as high as 100 % in all tested embryos, with mutation efficiencies of 82.6, 39.1, 52.1, and 43.5 % for sgRNA1, sgRNA2, sgRNA3, and sgRNA4, respectively (Fig. 2d; Table S2). All these results indicate that the CRISPR/Cas9 system is an efficient tool for gene mutation and precise genomic deletion of large fragments in zygotes.
Generation of the whole gene deletion of TYR in rabbit by dual sgRNAs
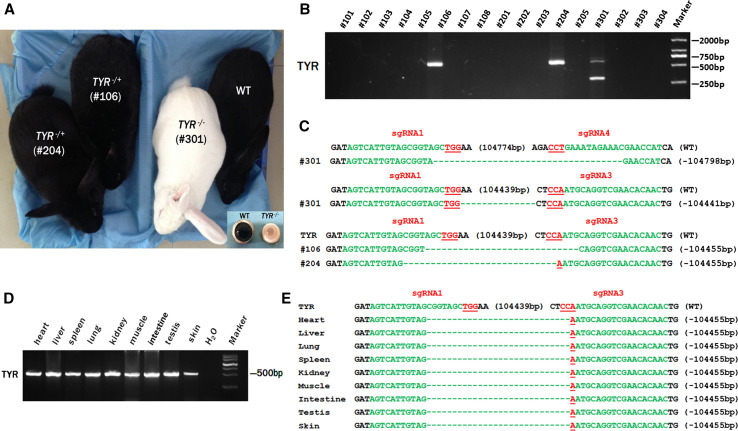
To generate the whole gene deletions of TYR in rabbit, 169 injected embryos were transferred to 5 pseudo-pregnant recipient rabbits. Three of these recipient mothers were pregnant to term and gave birth to 17 live pups (Table 1). The TYR biallelic gene deletion rabbit (#301) showed the typical albino phenotype, with a complete loss of dark pigment in the skin and eyes, while the TYR monoallelic gene deletion rabbits (#106 and #204) and WT littermates had black skin and eyes (Fig. 3a). The T-cloning and PCR-sequencing results showed that a large-fragment deletion of TYR was detected in rabbits #106, #204 and #301 (Fig. 3b, c), while the other rabbits carried other deletions or mutations. In addition, TYR-E3-F/R primers were used for heterozygous determination, revealing that samples #106 and #204 were heterozygous, with one allele carrying a 105-kb deletion and the other being the WT allele (Fig. 3c). Of interest, although #106 and #204 carried a large-fragment deletion of TYR, the typical symptoms of albinism were not observed in these two rabbits. Sequence analysis confirmed that the homozygous genotype (A/A) was associated with a non-pigmented phenotype (Fig. S2), as has been demonstrated in previous studies [10, 24].
Table 1.
Generation of genetically targeted rabbits using CRISPR/Cas9
| Recipients | gRNA/Cas9 mRNA (ng/uL) | Embryos injected | Embryos transferred (% microinjected) | Pregnancy | Pups obtained (% transferred) | Pups with mutations (% pups) | Pups with large fragment deletion (% pups) | Pups with color change |
|---|---|---|---|---|---|---|---|---|
| 1 | 25/100 | 40 | 30 (75 %) | No | ||||
| 2 | 25/100 | 45 | 40 (89 %) | Yes | 8 (20 %) | 6 (75 %) | 1 (16.7 %) | 1 |
| 3 | 25/100 | 40 | 34 (85 %) | No | ||||
| 4 | 25/100 | 37 | 30 (81 %) | Yes | 5 (16.7 %) | 5 (100 %) | 1 (20 %) | 1 |
| 5 | 25/100 | 40 | 35 (88 %) | Yes | 4 (11.4 %) | 4 (100 %) | 1 (25 %) | 1 |
Fig. 3.
Generation of large deletion of TYR in rabbits by dual sgRNAs. a Phenotype of TYR mutant rabbit; #301 is the TYR biallelic mutant (homozygote), which exhibited the typical albinism phenotype; #204 and #106 are the TYR monoalleic mutation (heterozygote) and the wild-type rabbit. The box shows the light pink-tinted iris and pupil of the #301 rabbit compared with the dark iris of wild-type rabbits. WT wild-type control. b The mutation determination of 105 kb deletions of TYR gene in founder rabbits by PCR. Results showed that three (#106, #204 and #301) of the 17 founders were large deletion of TYR rabbit. Primers used were TYR-F1 and R1 (Table S1). M DNA marker. c T-cloning sequences of mutant alleles in the large deletion of TYR rabbit (#106, #204 and #301). The PAM sites are underlined and highlighted in red; the target sequences are green; deletions (−) and insertions (+) are shown. WT wild-type control. d Chimera analysis of different tissues from #204 by PCR-sequencing. All detected tissues showed the same PCR band. Primers used were F1 and R1 (Table S1). M DNA marker, WT wild-type control. e T-cloning sequences of mutant alleles in different tissues from #204. PAM sites are underlined and highlighted in red; target sequences are green; deletions (−) and insertions (+) are shown. WT wild-type control
To test if the rabbits were chimeras in different tissues or not, the genomic DNA of the heart, liver, spleen, lung, kidney, muscle, intestine, testis and skin from sample #204 was isolated for PCR and Sanger sequencing analysis. As demonstrated in Fig. 3d, e, all tissues from #204 exhibited the same mutations and cleavage bands in all tested tissues, suggesting that no chimeric mutations were present in the gene-targeted rabbits.
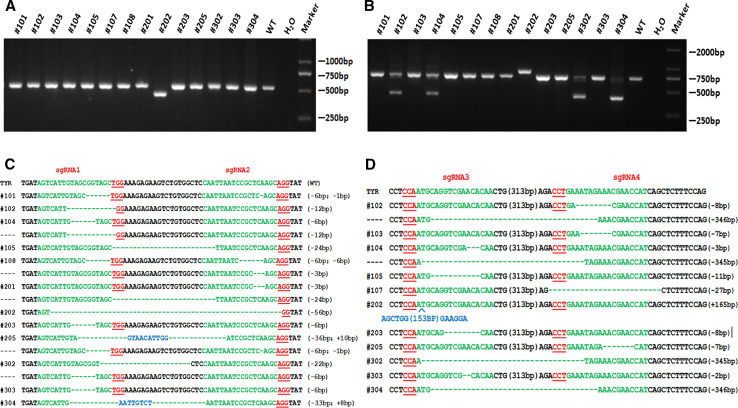
The mutation efficiency of each sgRNA was also determined by PCR-sequencing and T-cloning analysis as described above. As shown in Fig. 4, at least one TYR target site was destroyed by CRISPR/Cas9 in the rabbits, with indels in the founders ranging from 1 to 346 bp. The results demonstrated the dual sgRNAs directed CRISPR/Cas9 system is efficiently in mutations and large gene deletions of the rabbit TYR gene in this study.
Fig. 4.
PCR and T-cloning sequencing of the target site for each sgRNA in founder rabbit. a Mutation detection of sgRNA1 and sgRNA2 in founder rabbit by PCR. Primers TYR-2F/2R were used for mutation determination. M DNA marker, WT wild-type control. b Mutation detection of sgRNA3 and sgRNA4 in founder rabbit by PCR. Primers TYR-3F/3R were used for mutation determination. M DNA marker, WT wild-type control. c T-cloning sequencing of the target sites of sgRNA1 and sgRNA2 in founder rabbits. PAM sites are underlined and highlighted in red; target sequences are green; deletions (−) and insertions (+) are shown. WT wild-type control. d T-cloning sequencing of the target sites of sgRNA3 and sgRNA4 in founder rabbits. PAM sites are underlined and highlighted in red; target sequences are green; deletions (−) and insertions (+) are shown. WT wild-type control
Phenotype identification of TYR gene deletion in rabbits
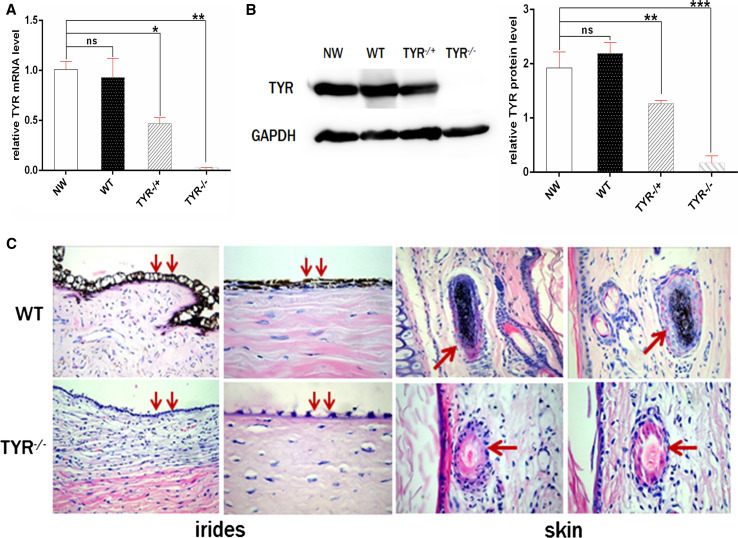
We further examined whether the gene mutations caused a reduction in gene expression or related phenotypes. As shown in Fig. 5a, TYR mRNA was reduced significantly, by about 50 %, in TYR +/− rabbits, with virtually no expression in TYR −/− rabbits compared to WT rabbits. These results were also confirmed by western blot and gray-scale analysis at the protein level (Fig. 5b), indicating that both TYR protein and mRNA were completely eliminated in TYR −/− rabbits and had decreased expression in TYR +/− rabbits. In addition, histological HE staining showed the absence of melanin in hair follicles and irises of TYR −/− rabbits but not in the WT littermates (Fig. 5c). These results indicate that typical symptoms of albinism, such as white skin color and red eyes, were observed in TYR −/− rabbits.
Fig. 5.
Phenotype identification of TYR mutated rabbits. a Expression of TYR gene was determined by qRT-PCR. WT wild-type control, NW New Zealand white rabbit. The data were analyzed by t tests using Graphpad Prism software. A probability of P < 0.05 was considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.005. ns not significant, WT wild-type control. b Western blot and gray-scale analysis of the expression of TYR protein. The image was quantified using ImageJ software (NIH) and all the data are expressed as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005. WT wild-type control. c H&E staining of the irides and skin from the WT and TYR mutated rabbits. The arrows indicate the melanin in the iris and the basal layer of the epidermis. WT wild-type control
Off-target analysis in TYR knockout rabbits
To test whether off-target effects occurred in these genetically modified rabbits, we screened the rabbit genome and predicted five POTS for each sgRNA, Primers and mismatch sites are listed in Table S3. Genomic DNA from the modified mutant rabbits was amplified by PCR and T7E1 analysis (Fig. S1). The sequence results showed that none of the sequencing reads exhibited mutation, suggesting that no off-target effects occurred at the 20 POTS in the rabbits derived from the dual sgRNA system with cytoplasmic microinjection in pronuclear-stage embryos.
Heritability of large fragment deletions
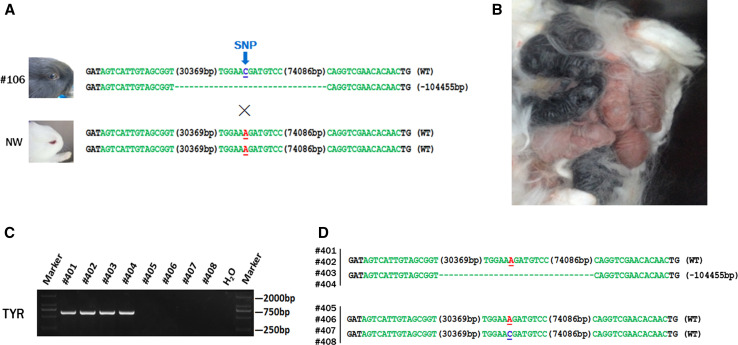
To study whether the large fragment deletions were heritable or not, the female founder #106 was mated with New Zealand white (NW) rabbits (Fig. 6a). Rabbit #106 was a heterozygote with a large-fragment deletion of TYR, though the typical symptom of albinism was not observed. Sequence analysis demonstrated that the NW rabbits are homozygous (A/A) at nucleotide 1118, while the Lianshan black rabbit has a common C/C or A/C, indicating the homozygous A/A genotype is associated with a non-pigmented phenotype (Fig. S2; Fig. 6a). Results demonstrated that four of eight F1 rabbits showed the typical albino phenotype, with a complete loss of dark pigment in the skin and eyes. Furthermore, T-cloning and PCR sequencing results confirmed that a large-fragment deletion of TYR was detected in pups of rabbits #401, #402, #403, and #404, but not in those of rabbits #405, #406, #407, and #408, demonstrating that large-fragment deletions are heritable and that genome modifications induced by CRISPR/Cas9 are transmitted to the germ-line (Fig. 6c, d).
Fig. 6.
Generation of F1s carrying large deletions of TYR. a Sequence analysis of rabbit containing a large deletion of TYR (#106) and New Zealand white rabbit. SNP is indicated by the blue arrows. Deletions (−) and insertions (+) are shown. WT wild-type allele, NW New Zealand white rabbit. b Picture of the F1 TYR mutant rabbit; four of the F1 rabbits exhibited the typical albinism phenotype; others are wild-type rabbits. c Determination of large deletions of TYR in F1 rabbits by PCR. A clear band demonstrates a large-fragment deletion of TYR is present in #401, #402, #403, and #404. Primers 1F and 1R were used for mutant determination. M DNA ladder. d T-cloning sequence analysis of the F1 rabbits. SNP is indicated by the blue arrows. Deletions (−) and insertions (+) are shown. WT wild-type allele
Discussion
In recent studies, CRISPR/Cas9-directed gene insertions or deletions have been successfully introduced to create genetic modification in many organisms [5–16, 25]. Mutations to the gene TYR mediated by the CRISPR/Cas9 system have been reported in mice, resulting in the typical albino phenotype [26, 27]. In this report, we described the feasibility of applying the Cas9/gRNA system to produce a large deletion (105 kb)of the TYR gene in rabbits. To our knowledge, this is the first description of a large biallelic gene deletion in rabbit by the dual sgRNA system and cytoplasmic microinjection, demonstrating that this system can be used efficiently not only in cells and mice, but also in rabbit.
Large-fragment gene deletions were first reported using TALENs, which produced deletions of approximately 1 Mb in zebrafish, but the efficiency was very low [28]. Zhang et al. showed that the efficiency of a 65-kb deletion was about 6.0 % in injected mouse embryos and about 21.4 % in live pups using the CRISPR/Cas9 system [29]. According to a previous study, the use of multiple sgRNAs increased the targeting efficiency of a 23-kb deletion in the Rian locus from 16 to 33.3 % in human cells [30]. Therefore, for improving efficiency of gene targeting, four sgRNAs and Cas9 mRNA were mixed together and used for cytoplasm microinjection in this study. Our data showed that the efficiency of the large 105-kb deletion was 10 and 17.67 % in rabbit embryos and live pups, respectively. These results are consistent with previous studies, suggesting that the CRISPR/Cas9 system is better than TALENs, ZFNs, and other traditional gene editing tools [30–32]. In addition, we found that the mutation efficiency of a single sgRNA was as high as 100 % in both injected embryos and pups, and the efficiency for large-fragment gene deletions was approximately 70 % (Table S2). We hypothesize that the efficiency of large gene deletions is dependent on gene length, though more research on the mechanisms of CRISPR/Cas9-directed deletions is needed in future studies.
In this study, we found that a TYR −/− rabbit (#301) exhibited the typical symptoms of albinism, while TYR +/− rabbits (#106 and #204) did not. Furthermore sequencing demonstrated that there is a common C nucleotide at position 1118 in the TYR WT allele of rabbit #106. This is consistent with a previous study showed that a C to A mutation at position 1118 resulted in an amino acid replacement (Thr373 to Lys373) in the last N-glycosylation site, which was associated with a non-pigmented phenotype [33]. In addition, the results of qRT-PCR and western blotting showed that the expression of TYR was completely eliminated by the large-fragment deletion in the TYR −/− rabbit, suggesting that it is possible to knock out whole gene fragments when constructing gene knockout models with the CRISPR/Cas9 system.
Off-target effects are a major concern in the Cas9-mediated gene editing system [7, 15]. However, such effects were not found in the present study. We suspected that a low concentration of sgRNA and Cas9 mRNA would reduce off-target effects, as they would immediately degrade after targeting the aimed gene. Furthermore, it is particularly important to avoid mismatches in the seed sequences (8–12 bases closest to PAM), which are critical for site-specific cleavage in the CRISPR/Cas9 system. In all, the protocol for mRNA injection into zygotes used in this study should be a valuable and efficient strategy for gene targeting, not only in rabbit but also in other mammalian species.
In summary, this the first report of a large-fragment gene deletion in rabbit, indicating that the dual sgRNA-directed CAS9/gRNA system may provide a simple and fast method for the large deletion of genes in mammalian genomes. This may also contribute to the functional study of gene clusters, lncRNAs, and regulatory sequences in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. Off-target analysis of the 4 sgRNAs in KO founders. PCR and T7EI assays of the PCR products of candidate off-target sites for 4 sgRNAs in founder #301 (primer sequences listed in Table S3). No fragment was found in T7EI assays (JPEG 1693 kb)
Figure S2. Schematic diagram of the SNP located in exon3 of TYR gene. Schematic diagram of exon3 of TYR in TYR KO rabbits and WT rabbits. The CDS region is indicated by blue rectangles; the SNP is located in a yellow circle and indicated by the arrows. WT, wild-type control; NW, New Zealand white rabbit; #301 and #106 are TYR KO rabbits (JPEG 1932 kb)
Figure S3. Sequence diagram of POTS in TYR KO founders. Sequence diagram of 20 potential off-target sites for sgRNA1, sgRNA2, sgRNA3, and sgRNA4 showing no double curve in any sequencing diagrams. Blue area represents sequencing of the POTS (JPEG 5527 kb)
Acknowledgments
We thank Peiran Hu at the Embryo Engineering Center for the critical technical assistance. This work was financially supported by the National Natural Science Foundation of China (Grant No. 31201080 and 31272394).
Footnotes
Y. Song and L. Yuan contributed equally to this work.
Contributor Information
Zhanjun Li, Phone: (86)431-87836176, Email: lizj_1998@jlu.edu.cn.
Liangxue Lai, Phone: (86)431-87836176, Email: lai_liangxue@gibh.ac.cn.
References
- 1.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 11.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 12.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Q, Zhang Q, Yang H, Zou Q, Tang C, Fan N, Lai L. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen. 2014;3:12. doi: 10.1186/2045-9769-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans . Genetics. 2014;198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oetting WS, King RA. Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat. 1999;13:99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 21.Oetting WS. The tyrosinase gene and oculocutaneous albinism type 1 (OCA1): a model for understanding the molecular biology of melanin formation. Pigment Cell Res. 2000;13:320–325. doi: 10.1034/j.1600-0749.2000.130503.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Fan N, Song J, Zhong J, Guo X, Tian W, Zhang Q, Cui F, Li L, Newsome PN, Frampton J, Esteban MA, Lai L. Generation of knockout rabbits using transcription activator-like effector nucleases. Cell Regen. 2014;3:3. doi: 10.1186/2045-9769-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 24.Yen ST, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, Swinton PG, Martin JF, Behringer RR. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol. 2014;393:3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, Zhang N, Zhu K, Xu J, Hu B, Leng Q, Huang X. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol. 2014;46:49–55. doi: 10.1016/j.biocel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Mizuno S, Dinh TT, Kato K, Mizuno-Iijima S, Tanimoto Y, Daitoku Y, Hoshino Y, Ikawa M, Takahashi S, Sugiyama F, Yagami K. Simple generation of albino C57BL/6J mice with G291T mutation in the tyrosinase gene by the CRISPR/Cas9 system. Mamm Genome. 2014;25:327–334. doi: 10.1007/s00335-014-9524-0. [DOI] [PubMed] [Google Scholar]
- 27.Seruggia D, Fernandez A, Cantero M, Pelczar P, Montoliu L. Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis. Nucleic Acids Res. 2015;43:4855–4867. doi: 10.1093/nar/gkv375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, Zhu Z, Lin S, Zhang B. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Jia R, Palange NJ, Satheka AC, Togo J, An Y, Humphrey M, Ban L, Ji Y, Jin H, Feng X, Zheng Y. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One. 2015;10:e0120396. doi: 10.1371/journal.pone.0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han J, Zhang J, Chen L, Shen B, Zhou J, Hu B, Du Y, Tate PH, Huang X, Zhang W. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014;11:829–835. doi: 10.4161/rna.29624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Wang J, Shen B, Chen L, Su Y, Yang J, Zhang W, Tian X, Huang X. Dual sgRNAs facilitate CRISPR/Cas9-mediated mouse genome targeting. FEBS J. 2014;281:1717–1725. doi: 10.1111/febs.12735. [DOI] [PubMed] [Google Scholar]
- 32.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, Chernomorsky R, Boucher M, Elsasser AL, Esau L, Zheng J, Griffiths JA, Wang X, Su H, Xue Y, Dominguez MG, Noguera I, Torres R, Macdonald LE, Stewart AF, DeChiara TM, Yancopoulos GD. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 33.Aigner B, Besenfelder U, Muller M, Brem G. Tyrosinase gene variants in different rabbit strains. Mamm Genome. 2000;11:700–702. doi: 10.1007/s003350010120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Off-target analysis of the 4 sgRNAs in KO founders. PCR and T7EI assays of the PCR products of candidate off-target sites for 4 sgRNAs in founder #301 (primer sequences listed in Table S3). No fragment was found in T7EI assays (JPEG 1693 kb)
Figure S2. Schematic diagram of the SNP located in exon3 of TYR gene. Schematic diagram of exon3 of TYR in TYR KO rabbits and WT rabbits. The CDS region is indicated by blue rectangles; the SNP is located in a yellow circle and indicated by the arrows. WT, wild-type control; NW, New Zealand white rabbit; #301 and #106 are TYR KO rabbits (JPEG 1932 kb)
Figure S3. Sequence diagram of POTS in TYR KO founders. Sequence diagram of 20 potential off-target sites for sgRNA1, sgRNA2, sgRNA3, and sgRNA4 showing no double curve in any sequencing diagrams. Blue area represents sequencing of the POTS (JPEG 5527 kb)