Abstract
Exosomes are vesicles released from cells by fusion of multivesicular bodies (MVBs) with the plasma membrane. This study aimed to investigate whether the phosphoinositide kinase PIKfyve affects this process. Our results show that in PC-3 cells inhibition of PIKfyve by apilimod or depletion by siRNA increased the secretion of the exosomal fraction. Moreover, quantitative electron microscopy analysis showed that cells treated with apilimod contained more MVBs per cell and more intraluminal vesicles per MVB. Interestingly, mass spectrometry analysis revealed a considerable enrichment of autophagy-related proteins (NBR1, p62, LC3, WIPI2) in exosomal fractions released by apilimod-treated cells, a result that was confirmed by immunoblotting. When the exosome preparations were investigated by electron microscopy a small population of p62-labelled electron dense structures was observed together with CD63-containing exosomes. The p62-positive structures were found in less dense fractions than exosomes in density gradients. Inside the cells, p62 and CD63 were found in the same MVB-like organelles. Finally, both the degradation of EGF and long-lived proteins were shown to be reduced by apilimod. In conclusion, inhibition of PIKfyve increases secretion of exosomes and induces secretory autophagy, showing that these pathways are closely linked. We suggest this is due to impaired fusion of lysosomes with both MVBs and autophagosomes, and possibly increased fusion of MVBs with autophagosomes, and that the cells respond by secreting the content of these organelles to maintain cellular homeostasis.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2309-8) contains supplementary material, which is available to authorized users.
Keywords: Exosomes, Extracellular vesicles, PIKfyve, Secretory autophagy, Exophagy, Phosphoinositides
Introduction
Exosomes are nanovesicles (30–150 nm in diameter) released from cells by fusion of multivesicular bodies (MVBs) with the plasma membrane [1–3]. Initially exosomes were proposed to represent waste from cells [2], and have recently been shown to be a route to cellular disposal of tumor-suppressor miRNAs, hence increasing the metastatic properties of the parental cell [4]. In addition, these vesicles are suggested to play a role in cell-to-cell communication [3, 5, 6] and have been implicated in numerous physiological and pathological functions. Exosomes contain proteins [7], lipids and other metabolites [8, 9], mRNAs [10], small RNAs such as miRNAs [10–12] and probably DNA fragments [13, 14], which can be delivered to recipient cells.
Although several proteins and lipids have been reported to regulate exosome release, the molecular mechanism is not completely understood. Endosomes mature into late endosomes/MVBs and during this process the limiting membrane buds and pinches off into the lumen generating intraluminal vesicles (ILVs). ILVs can have different fates. They can be degraded after fusion of MVBs with lysosomes or escape degradation through back-fusion with the MVB membrane [15]. In addition, ILVs can be secreted to the cellular environment as exosomes after fusion of MVBs with the plasma membrane. Still, it is not known how MVBs are marked for fusion with either the plasma membrane or with lysosomes. Some endosomal sorting complexes required for transport (ESCRTs) proteins such as Hrs [16], Tsg101 [17] and VPS4 [17], as well as the ESCRT-associated protein Alix [18], have been reported to affect exosome release, probably through an effect on MVB biogenesis. Exosome biogenesis can also occur through ESCRT-independent mechanisms. The tetraspanins CD9 [19], CD82 [19] and Tspan8 [20] have been reported to affect exosome release, and neutral sphingomyelinase 2 has been shown to regulate exosome biogenesis in some [21], but not all cell lines [22]. Several Rab proteins have also been shown to regulate exosome release, probably through affecting transport of MVBs to the plasma membrane [23–25].
It has been speculated that exosome secretion and autophagy are coordinated processes that contribute to maintenance of cellular homeostasis [26]. Autophagy is a protective catabolic process that provides nutrients during starvation and eliminates intracellular waste products, such as damaged organelles, aggregated proteins or invading pathogens. The cytoplasmic cargo is sequestered within double-membrane vesicles termed autophagosomes, which can fuse with either MVBs to form amphisomes, or directly with lysosomes for cargo degradation [27]. Beside its role in degradation, the autophagic machinery is also involved in a process termed secretory autophagy [28]. Secretory autophagy is part of unconventional secretion and releases numerous cytoplasmic substrates from the cell [28]. Since secretory autophagy has been observed in cells with lysosomal dysfunction [28], the secretion process might be an alternative way of eliminating waste products from the cell. This phenomenon is observed in certain neurodegenerative diseases, which are associated with deposition of aggregation-prone proteins as well as autophagy dysfunction [29, 30].
Phosphoinositides are membrane lipids that recruit cytosolic proteins containing specific recognition domains to membranes. These lipids modify signal transduction, help specifying organelle identity and regulate membrane transport [31, 32]. Seven different phosphoinositides have been identified, and they can be converted into each other by phosphoinositide kinases and phosphatases [31]. Phosphatidylinositol-3,5-bisphosphate (PI(3,5)P2) is a low abundance phosphoinositide mainly localized to late endosomes/MVBs and lysosomes [33–35]. PIKfyve (Fab1 in yeast and flies) is the kinase that generates PI(3,5)P2 from PI(3)P. In addition, PIKfyve generates PI(5)P, either directly from PI [36] or indirectly through production of PI(3,5)P2, which subsequently can be dephosphorylated by phosphatases [37].
PIKfyve has been implicated in several membrane transport events, both in lower and higher eukaryotes. Deletion of Fab1 in yeast results in swollen vacuoles [38], and treatment with siRNA against PIKfyve or addition of PIKfyve inhibitors leads to formation of swollen endosomal/lysosomal structures in various mammalian cell lines [39–41]. Inhibition or mutation of PIKfyve (and Fab1 in Drosophila melanogaster) also results in accumulation of amphisomes, probably due to impaired fusion with lysosomes [42, 43]. The substrate for PIKfyve, PI(3)P, is well known to play a critical role both for initiation of autophagy and autophagosome maturation [32]. Recently, also PI(5)P was shown to be a regulator of autophagosome biogenesis [44]. Pharmacological inhibition and knockdown of PIKfyve using siRNA also lead to a defect in endosome to trans-Golgi-network (TGN) retrograde trafficking, shown by increased degradation of cation-independent mannose 6-phosphate receptor (CI-MPR) as well as delayed transport of the Shiga toxin B-chain, CD8-CI-MPR and CD8-furin from the cell surface to the TGN [39, 45]. Interestingly, PIKfyve is shown to negatively regulate exocytosis of secretory granules from neuroendocrine cells [46]. Recently, PIKfyve inhibition with YM-201636 and apilimod was found to enhance exocytosis of amylase from pancreatic acinar cells [47].
Since PIKfyve has been implicated in both endosomal morphology and various membrane trafficking events we have investigated whether PIKfyve is involved in the release of exosomes. Our study shows that both inhibition and depletion of PIKfyve by apilimod or specific siRNAs, respectively, increase release of materials recovered by ultracentrifugation at 100,000×g (called here exosomal fractions), from PC-3 cells. Both small extracellular vesicles and other particulate materials are present in these fractions, and we show that p62-positive protein structures are present in the exosomal fractions after PIKfyve inhibition, indicating that secretory autophagy is induced. The observed effects might be a consequence of impaired fusion of lysosomes with both MVBs and autophagosomes, and facilitated fusion between MVBs and autophagosomes, generating amphisomes.
Materials and methods
Materials
Ham’s F-12/DMEM (1:1 mixture) with Glutamax, DMEM with Glutamax, RPMI and EBSS were from Gibco Invitrogen (Invitrogen, Carlsbad, CA, USA). OptiPrep, concanamycin A and 3-methyladenine were purchased from Sigma-Aldrich (St. Louis, MO, USA). EDTA-free protease inhibitor cocktail was from Roche Applied Science (Mannheim, Germany). Bicinchoninic acid (BCA) protein assay kit was from Pierce (Thermo Scientific, Rockford, IL, USA) and Cathepsin D activity assay was from Abcam (Cambridge, UK). Mini-protean TGX gels and Tranfer-Blot Turbo Transfer Pack were from Bio-Rad (Hercules, CA, USA). PVDF membranes and 0.02 µM Anotop 25 filters were from Millipore (Billerica, MA, USA) and Whatman (Dassel, Germany), respectively. The antibodies used for Western blotting were: rabbit anti-p62 (MBL, Woburn, MA, USA), rabbit anti-LC3 (Cell Signaling), mouse anti-NBR1 (Abnova), rabbit anti-caveolin-1 and mouse anti-TSG101 (both from BD Biosciences, Heidelberg, Germany), rabbit anti-PIKfyve (Sigma-Aldrich, St. Louis, MO, USA), rabbit anti-Alix and mouse anti-CD29 (both from Merck Millipore, Billerica, MA, USA). HRP-conjugated secondary antibodies were from Jackson Immunoresearch (West Grove, PA, USA) and Li-Cor infrared dye secondary antibodies were from Li-Cor Biosciences (Lincoln, NE, USA). Apilimod was purchased from MedChem Express (NJ, USA). L-[3,4,5-3H(N)]leucine and 125I-EGF were from PerkinElmer (Waltham, MA, USA).The mCherry-GFP-LC3 plasmid was a kind gift from Harald Stenmark.
Cell culture and treatment
The human prostate cancer epithelial cell line PC-3 was obtained from ATCC (Manassas, VA, USA). PC-3 cells were cultured in Ham’s F-12/DMEM (1:1 mixture) with Glutamax supplemented with 7 % foetal calf serum (FCS), 100 units/ml penicillin and 100 units/ml streptomycin, in a humidified 5 % CO2 atmosphere at 37 °C. Cells were preincubated with apilimod or control (DMSO) in complete medium for 2 h, washed twice, and then incubated for 18–19 h in serum-free medium with the same concentration of the compounds.
Knockdown of PIKfyve
ON-TARGET plus siRNA oligonucleotides against PIKfyve and ON-TARGET plus non-targeting siRNA (control) were from Dharmacon RNAi Technologies (IL, USA). To knock down PIKfyve, two different siRNA oligonucleotides against the enzyme were used: J-005058-13 (PIKfyve-1): GGCACAAGCUAUAGCAAUU and J-005058-14 (PIKfyve-2): GAGAUGAGUAUGCGCUGUA. siRNA oligonucleotides against PIKfyve (25 nM) and non-targeting siRNA (control, 25 nM) were delivered to the cells using Lipofectamine RNAiMAX transfection reagent (Life Technologies), according to the manufacturer’s protocol. Two days after transfection, cells were washed twice and incubated for 18–19 h in serum-free medium to collect exosomes. Cells were lysed 2 or 3 days after transfection for measurement of knockdown efficiency. Knockdown efficiency after two days was measured to ensure that the level of PIKfyve was reduced before serum-free medium was added to the cells for exosome collection.
Isolation of exosomes
Exosomes were isolated from the conditioned media of cells after 18–19 h incubation as previously described [48]. Briefly, the medium was centrifuged at 300×g for 10 min, then the supernatant was centrifuged at 1000×g for 10 min, and thereafter at 10,000×g for 30 min to remove dead cells and cell debris. The supernatant was then ultracentrifuged at 100,000×g for 70 min. The exosome pellet was washed with PBS, and centrifuged again at 100,000×g for 70 min. The exosome pellets were resuspended in equal volumes of PBS or lysis buffer before further analyses. All centrifugation steps were carried out at 4 °C. Exosomes were isolated from cells grown on 10 cm plates, using half the amount of exosomes from one plate for either nanoparticle tracking analysis, immunoblot or total protein measurement (BCA assay). For proteomics, density gradients and electron microscopy (EM) more material was needed. Therefore, the conditioned media from eight and four 10 cm plates were combined for density gradients and EM, respectively. The details for proteomics are described below. We have previously characterized the exosomes isolated by this method using electron microscopy and Western blotting with several exosome associated proteins [12, 49, 50].
OptiPrep density gradient separation
The continuous gradient was prepared diluting a stock solution of OptiPrep (60 % w/v aqueous iodixanol) in 0.25 M sucrose/10 mM Tris, pH 7.5 to 30 and 5 %. To prepare a sedimentation gradient, the 5 % Optiprep dilution was layered on top of the 30 % dilution and a continuous gradient was made by use of Gradient Master (80°, 2 min, 20 rpm). Exosomes were isolated by ultracentrifugation as described above and then layered on top of the OptiPrep density gradient and centrifuged at 100,000×g for 18 h at 4 °C. After centrifugation, starting from the top of the tube, 12 individual 1 ml gradient fractions of increasing density were collected and diluted with 5 ml PBS, followed by centrifugation at 100,000×g for 70 min at 4 °C. The pellets were resuspended in equal volumes of PBS or lysis buffer and used for further analyses.
Nanoparticle tracking analysis (NTA)
The concentration and the size distribution of exosomal fractions were measured by NTA. Exosome pellets were resuspended in the amount of PBS (filtered through a 0.02 µM Anotop 25 filter) needed to obtain a concentration within the recommended range (2 ×108–1 × 109 particles per ml) and vortexed for 1 min. The samples were then loaded into a NS500 instrument (Malvern Instruments Ltd, Worcestershire, UK). Five videos, each of 60 s, were acquired for every sample. Videos were subsequently analyzed with the NTA 2.3 software, which identifies and tracks the centre of each particle under Brownian motion to measure the average distance the particles move on a frame-by-frame basis.
SDS-PAGE and immunoblotting
Similar volumes of exosomal fractions and similar volumes of cell lysates were mixed with loading buffer, and run on 4–20 % gradient TGX gels. The proteins were transferred to PVDF membranes using a Tranfer-Blot Turbo Transfer Pack. Membranes were incubated with the specified primary and secondary antibodies. Blots were visualized with the Amersham ECL Prime Western blot detection (GE Healthcare, Little Chalfont, UK) on the Universal Hood II Bio-Rad scanner (Bio-Rad, Hercules, CA, USA) or the Odyssey imaging system (Li-Cor Biosciences).
Protein synthesis
After incubation with the PIKfyve inhibitor apilimod, cells were washed once in leucine-free Hepes medium, and incubated for 30 min in leucine-free Hepes medium supplemented with 2 µCi/ml [3H]leucine to allow incorporation of the radioactive isotope into newly synthesized proteins. The proteins were precipitated by adding 5 % trichloroacetic acid, the precipitate was washed once in the same solution and then dissolved in 0.1 M KOH. The radioactivity was measured using a β-counter (Packard, Meriden, CT, USA).
Endocytosis and degradation of 125I-EGF
Cells were seeded in four-well plates and treated with apilimod (0.5 µM) or control (DMSO) for 21 h before they were incubated for 10 min at 37 °C with 125I-EGF (0.75 μCi/ml) in HEPES and then washed three times with cold PBS. For 125I-EGF degradation, cells were thereafter incubated for 1 h at 37 °C in the presence or absence of apilimod. Then, the media was removed, BSA (0.5 mg/ml) and TCA (5 %) were added and centrifuged for 3 min at 6000×g. Cells were lysed in 0.1 M KOH. Degradation of 125I-EGF was calculated as percentage of radioactivity in the TCA-soluble fraction relative to the total radioactivity in the cells, the TCA-soluble and nonsoluble fractions. For 125I-EGF endocytosis, cells were incubated for 6 min on ice with low pH buffer (0.5 M NaCl and 0.2 M CH3COOH, pH 2.5). Then, the buffer was transferred and counted, the cells washed once with the same low pH buffer (which was thereafter counted), and lysed in 0.1 M KOH. Endocytosis of EGF was calculated as percentage of radioactivity in the cells relative to the sum of radioactivity removed by the low pH buffer and remaining in the cells.
Cathepsin D activity assay
Cells were seeded in 6-well plates and treated with apilimod (0.5 µM) or control (DMSO) for 21 h. Then the cells were lysed and cathepsin D activity was measured using a Cathepsin D activity assay kit following the manufacturer’s protocol.
Degradation of long-lived proteins
Cells were seeded in 24-well plates and radiolabeled with [14C]valine (0.27 µCi/ml) in complete RPMI 1640 (with 10 % FCS) for 24 h. Subsequently, cells were washed with medium, and chased in complete RPMI supplemented with 10 mM cold valine (to prevent reincorporation of radiolabeled valine) in the presence or absence of apilimod for 17 h. Thereafter, cells were washed to remove degraded short-lived proteins, before incubation with complete RPMI or Earle’s Balanced Salt Solution (EBSS, to induce starvation) in the presence or absence of apilimod for 4 h. To precipitate non-degraded proteins, BSA (3 mg/ml) and TCA (10 %) were added to the cells and incubated at 4 °C overnight. All liquid was transferred to tubes and centrifuged at 6000×g for 10 min at 4 °C, and 0.2 M KOH was added to the wells to solubilize the precipitate. After 6000×g centrifugation, the supernatant was counted using a β-counter. To dissolve the pellet, 0.2 M KOH was added, and it was then combined with the re-solubilized precipitate from the wells and counted using a β-counter. The degradation of long-lived proteins was calculated as percentage of radioactivity in the TCA-soluble fraction relative to the total radioactivity in the TCA-soluble and nonsoluble fractions.
Electron microscopy
Cells for conventional electron microscopy were fixed in 2 % glutaraldehyde in 0.1 M PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA and 2 mM MgCl2 at pH 6.9), postfixed with 1 % OsO4 and 1.5 % KFeCN, en bloc stained in 4 % uranyl acetate, and Epon embedded after dehydration in graded ethanol series. Ultrathin sections were cut on a Leica UFC6 ultramicrotome and collected on formvar/carbon coated grids and poststained with 2 % lead citrate. Cells for immuno-EM were fixed with 4 % paraformaldehyde and 0.1 % glutaraldehyde in 0.1 M PHEM buffer, pelleted and embedded in 10 % gelatin. After solidification on ice small blocks were prepared and infused with 2.3 M sucrose before freezing in liquid N2. Ultrathin Sects. (70–100 nm) were cut on a Leica UFC microtome with an attached cryochamber at −110 °C and collected with a 50:50 mixture of 2 % methyl cellulose and 2.3 M sucrose. Double labeling was performed essentially as described earlier [51] using primary and secondary antibodies followed by different sized protein A gold (University Medical Center, Utrecht, Netherlands). Exosomal fractions for negative staining were dropped directly onto and left on formvar/carbon coated grids for 20 min, washed with PBS and then stained/embedded with 0.4 % uranyl acetate/1.8 % methyl cellulose. Single and double labeling was performed as described above. Samples were observed on a JEOL-JEM 1230 at 80 kV and images were recorded with a Morada digital camera. Further image processing was performed with Adobe Photoshop.
Confocal fluorescence microscopy
PC-3 cells were grown on coverslips and transfected with mCherry-GFP-LC3 plasmid [52] using Fugene6 (Roche) transfection reagent according to the manufacturer’s instructions. The cells were thereafter treated in the presence or absence of apilimod (0.5 µM) for 21 h and concanamycin A (50 nM) during the last 4 h of incubation, before fixation in 3 % paraformaldehyde and mounting with ProLong Gold antifade mounting medium with DAPI (Molecular Probes). The cells were imaged using a Zeiss LSM780 laser scanning confocal microscope (Carl Zeiss MicroImaging) equipped with an Ar-Laser multiline (458/488/514 nm), a DPSS-561 10 (561 nm), and a Laser diode 405–30 CW (405 nm). The objective used was a Zeiss Plan-Apochromat 63×/1.40 Oil DIC M27. Images were acquired using the ZEN 2010 software (Carl Zeiss MicroImaging). Image processing and analysis was done with LSM780 software, ImageJ and Adobe Illustrator. A total of 39–52 cells were analyzed per condition, and the experiment was performed three times. The number of autophagosomes (yellow puncta) and autolysosomes (red-only puncta) per cell were quantified.
In-solution digest
In three independent experiments, exosomes were isolated by ultracentrifugation from PC-3 cells grown on 15 cm plates, using five plates per condition. In each experiment, the conditional media from five plates were combined, giving 3 µg protein of isolated exosomal fractions (estimated by the BCA assay) in one volume of PBS which was mixed with four volumes of cold acetone (with 1 M HCl) and methanol at –20 °C, followed by centrifugation at 15,000×g for 15 min. The subsequent pellets were dried in a Speed-Vac. The pellets were dissolved in 50 µl of a fresh solution of 100 mM ammonium bicarbonate with 6 M urea, and reduced with 10 mM dithiothreitol at 30 °C for 30 min. The samples were then incubated with 25 mM iodoacetamide to alkylate exposed side chains for 1 h at room temperature protected from light. The enzymatic digestion was initiated by adding 1 µg Lys-C to the samples and incubating them at 37 °C for 2 h. Finally, 240 µl 50 mM ammonium bicarbonate with 10 µg of trypsin was added and the samples were first incubated for 1 h at 37 °C, followed by 15 h at 30 °C. Peptides were purified by C18 Zip Tips (Millipore, Billerica, MA, USA). Prior to LC–MS analysis, 5 µl formic acid was added to the digested exosomes.
Mass spectrometric analyses
The digested exosomal fractions were divided in three, and two parts were separately injected into the HPLC, giving two technical replicates per sample. For each replicate, one-third of the volume (1 µg) was injected into an Ultimate 3000 nano-UHPLC system (Dionex, Sunnyvale CA, USA) connected to a linear quadrupole ion trap-orbitrap (LTQ-Orbitrap XL) mass spectrometer (ThermoScientific, Bremen, Germany) equipped with a nanoelectrospray ion source. An Acclaim PepMap 100 column (C18, 3 μm, 100 Å) (Dionex) with a capillary of 25 cm bed length was used for separation by liquid chromatography. Solvent A was 0.1 % formic acid, and solvent B contained 90 % aqueous acetonitrile in 0.1 % formic acid. A flow rate of 300 nl/min was employed with a solvent gradient of 4 % B to 60 % B in 230 min. The mass spectrometer was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Survey full scan MS spectra (from m/z 300 to 2000) were acquired in the Orbitrap with resolution R = 60,000 at m/z 400 (after accumulation to a target of 500,000 charges in the LTQ). The method used allowed sequential isolation of the most intense ions, up to six, depending on signal intensity, for fragmentation on the linear ion trap using collision induced dissociation at a target value of 10,000 charges.
For liquid chromatography separation, an Acclaim PepMap 100 column (C18, 3 µM beads, 100 Å, 75 μm inner diameter) (Dionex, Sunnyvale CA, USA) capillary of 50 cm bed length was used. A flow rate of 300 nl/min was employed with a solvent gradient of 4–35 % B in 207 min, to 50 % B in 20 min and then to 80 % B in 2 min. Solvent A was 0.1 % formic acid and solvent B was 0.1 % formic acid/90 % acetonitrile.
Data processing
Quantification of Western blots was performed using ImageJ (Fiji) software. All experiments were carried out using duplicates and run in at least three independent experiments. Statistical analysis was carried out using a paired two-tailed t test. A P value <0.05 was considered significant.
Data processing for proteomic analysis
Data were acquired using Xcalibur v2.5.5 and raw files were processed to generate peak list in Mascot generic format (*.mgf) using ProteoWizard release version 3.0.331. Database searches were performed using Mascot in-house version 2.4. to search from Swiss-Prot selected for Homo sapiens (11.2013, 20,252 entries) assuming the digestion enzyme trypsin with at maximum one missed cleavage, fragment ion mass tolerance of 0.60 Da, and a parent ion tolerance of 10 ppm. Carbamidomethyl of cysteine was specified in Mascot as a fixed modification. Oxidation of methionine, acetylation of the N-terminus and phosphorylation of serine, threonine and tyrosine were specified in Mascot as variable modifications. Scaffold (version Scaffold_4.3.2, Proteome Software Inc., Portland, OR, USA) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0 % probability by the Peptide Prophet algorithm [53] with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99.0 % probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm [54]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. MS/MS spectra from protein hits identified with only 1 peptide were investigated manually.
For comparing data sets Fisher’s exact test (CI 95 %) was used to determine significant changes between the subproteomes of exosomal fractions from apilimod-treated PC-3 cells and control cells. The label-free quantitative measurement of individual samples used top 3 precursor intensities from total ion chromatogram (TOP3TIC), and only protein hits significantly altered (heteroscedastic two-sided t test, P < 0.05) for both were considered.
Results
PIKfyve inhibition and knockdown increase release to the exosomal fraction
To investigate whether PIKfyve is involved in exosome release, PC-3 cells were treated with the PIKfyve inhibitor apilimod [41]. Treatment with apilimod did not affect cell proliferation or protein synthesis (Supplementary Fig. 1), showing that the inhibitor was well tolerated by the cells. Massive induction of vacuoles, a phenotypic hallmark of PIKfyve inhibition [38–40], was clearly observed by light microscopy in cells treated with apilimod (Supplementary Fig. 2).
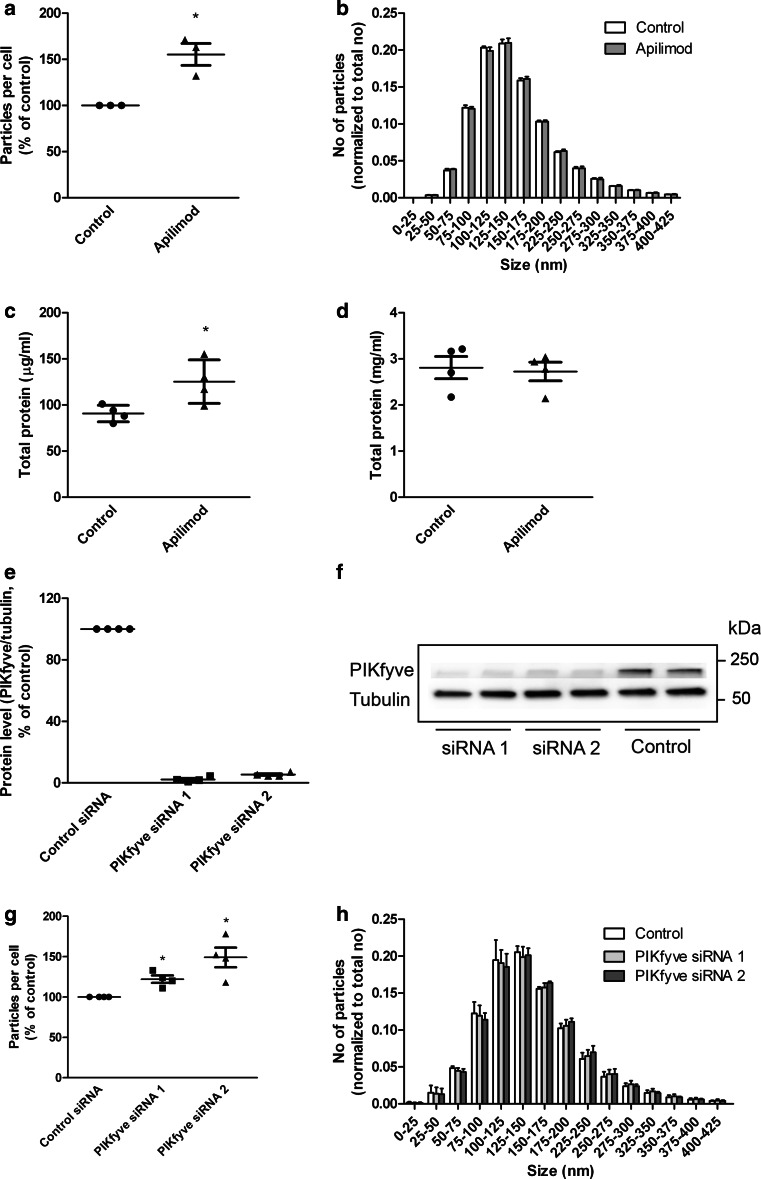
To quantify release of exosomes, PC-3 cells were pretreated with or without apilimod for 2 h, and exosomes released during the next 18–19 h in the presence or absence of inhibitor were then isolated from the conditioned media by ultracentrifugation. Other small extracellular vesicles might co-pellet with exosomes at 100,000×g, but the pellet should be enriched with exosomes, therefore, we will refer to this pellet as exosomal fraction or exosome preparation. As quantified by nanoparticle tracking analysis (NTA) treatment with apilimod increased the number of released particles per cell compared to control (Fig. 1a), whereas the size distribution of the exosome preparation was not affected (Fig. 1b). The increased release to the exosomal fraction was verified by measuring the total protein amount in the exosomal fractions from control or apilimod treated cells (Fig. 1c). Control experiments showed that the total protein level in the cell lysates was not affected by apilimod treatment (Fig. 1d).
Fig. 1.
Apilimod treatment or knockdown of PIKfyve increases the release of particles to the exosomal fraction. a PC-3 cells were treated in the presence or absence of apilimod (0.5 µM) before exosomes were isolated and concentration of the exosomal fractions was measured by NTA. b Size distribution of exosomal fractions shown as number of particles normalized by total number of particles for each treatment. c Total protein content in exosomal fractions. d Total protein content in cells after apilimod treatment. e and f Knockdown efficiency 3 days after transfection with siRNA against PIKfyve (25 nM). g The concentration of the exosomal fractions was quantified by NTA 3 days after transfection with siRNA. h Size distribution of exosomal fractions shown as number of particles normalized by total number of particles for each treatment. The results are plotted as dot plots showing the value obtained for each individual experiment as a single dot + mean values + standard error of the mean from 3–4 independent experiments. *; P < 0.05 versus control
To verify that the increased release to the exosomal fraction induced by apilimod is due to PIKfyve inhibition, PC-3 cells were treated with two different siRNA oligos to knock down PIKfyve. The knockdown efficiency 3 days after addition of siRNA was more than 95 % for both oligos (Fig. 1e, f). We, therefore, decided to collect exosomes during the third day of knockdown. To ensure that the level of PIKfyve was reduced at the beginning of exosome collection, knockdown efficiency was also measured after 2 days. Approximately 80 % knockdown of PIKfyve was observed for both oligos. Cell proliferation after 3 days was inhibited by 26–27 % by both oligos compared to non-targeting control. Exosomes were isolated and quantified by NTA as above, and in accordance with the apilimod data, the number of released particles per cell was significantly increased by both siRNA oligos (Fig. 1g). The size distribution of the exosomal fraction was unchanged after PIKfyve knockdown (Fig. 1h). In conclusion, these experiments indicate that PIKfyve inhibition or depletion increases release to the exosomal fraction.
PIKfyve inhibition by apilimod increases the number of ILVs per MVB
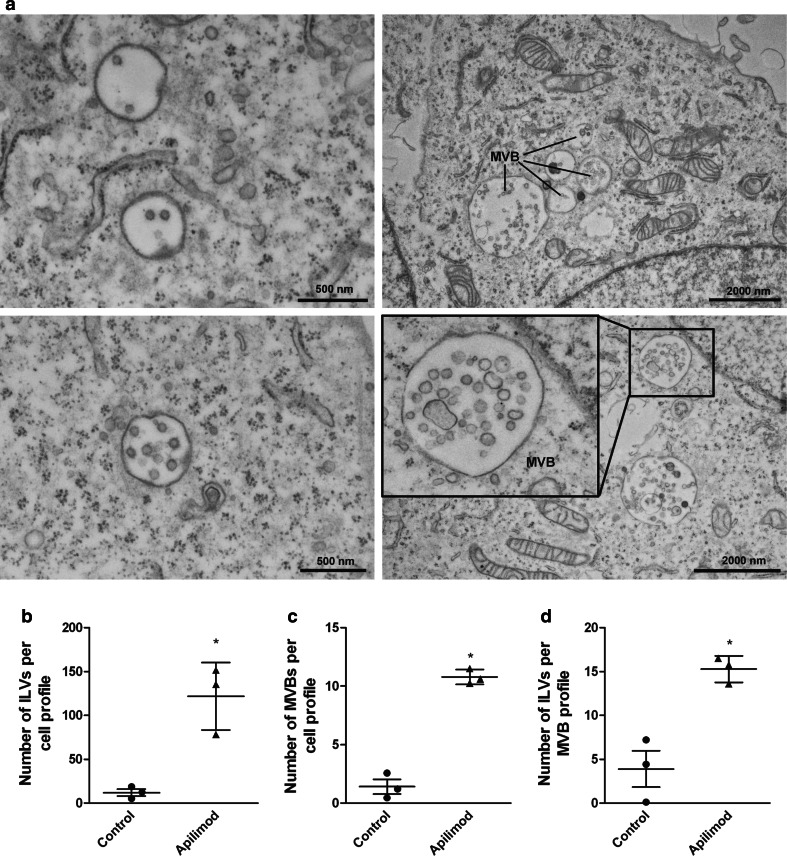
Exosome release can be divided into three main steps; MVB biogenesis, transport of MVBs to the plasma membrane and finally fusion of MVB with the plasma membrane. To obtain information about which step is affected by PIKfyve, we performed quantitative electron microscopy (EM) on PC-3 cells treated with apilimod. Apilimod treatment dramatically increased the size of the MVBs (Fig. 2a), whereas the ILVs were still morphologically homogenous. In addition, apilimod treatment considerably increased the number of ILVs per cell profile (Fig. 2b). This increase was due to both significantly increased number of MVBs per cell profile (Fig. 2c) and significantly increased number of ILVs per MVB profile (Fig. 2d).
Fig. 2.
Apilimod treatment increases the number of ILVs per cell section. a EM on PC-3 cells after apilimod (right panels) and control (left panels) treatment. Scale bars are as indicated. b The number of ILVs per cell profile. c The number of MVBs per cell profile. d The number of ILVs per MVB profile. The ILVs and MVBs in 20–21 cells were counted per treatment in each experiment. Only MVBs containing typical ILV structures were counted. The results are plotted dot plots showing the value obtained for each individual experiment as a single dot + mean values + standard error of the mean from 3 independent experiments, *; P < 0.05 versus control
Autophagy-related proteins appear in the exosomal fractions after apilimod treatment
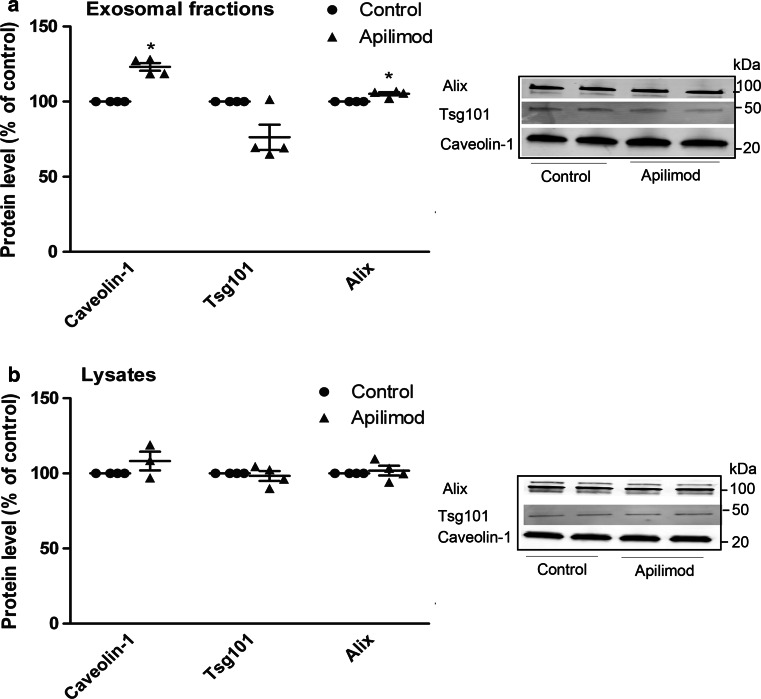
Since PIKfyve inhibition seemed to increase the release of particles without altering the particle size distribution or changing the size of ILVs in the MVBs, it was of interest to see whether PIKfyve inhibition simply increases the rate of secretion, or whether both the rate and the nature of the particles are altered. Interestingly, although apilimod treatment increase the number of released particles, the levels of known exosomal markers were not increased to the same extent. Apilimod treatment only modestly increased the levels of Alix and caveolin-1, whereas the level of Tsg101 was unchanged in the exosomal fraction (Fig. 3a). This was not due to reduced cellular expression of these proteins (Fig. 3b). Rather, this might indicate that PIKfyve inhibition not only increases the rate of exosome secretion, but also alters the composition of the exosomal fraction.
Fig. 3.
Apilimod treatment modestly increases the level of well-known exosome-associated proteins in exosomal fractions. a PC-3 cells were seeded on 10 cm plates and pretreated with apilimod (0.5 µM) or DMSO (0.1 %, as control) for 2 h before collection of exosomes for 18–19 h in the presence of inhibitors. Exosomes were isolated by ultracentrifugation and lysed in total 80 µl (lysis buffer plus sample buffer), before 25 µl of this mixture was loaded per well and run on SDS-PAGE gel. Each treatment was run in duplicates per experiment. Representative Western blot and quantification of proteins from exosomal fractions from several independent experiments. b Representative Western blot and quantification of cellular proteins from several independent experiments. The results are plotted as dot plots showing the value obtained for each individual experiment as a single dot + mean values + standard error of the mean. *; P < 0.05 versus control, #; P < 0.05 versus apilimod, n = 3–4
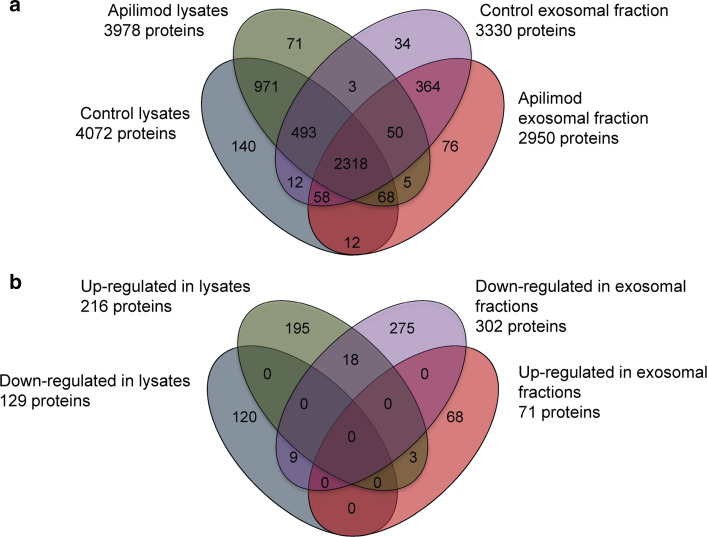
To further address the effect of PIKfyve-inhibition on exosomal protein composition a proteomic analysis was performed. For this purpose we chose to inhibit PIKfyve with apilimod rather than to deplete PIKfyve by siRNAs, since this gives a rapid and probably more complete inhibition of the enzyme. Exosomes isolated from PC-3 cells in the presence or absence of apilimod as described above were collected from three independent experiments and subjected to nano-LC-high resolution mass spectrometry after tryptic digestion. The corresponding cell lysates were also analyzed in the same manner. In total 4675 proteins (3493 proteins in exosomes and 4201 proteins in lysates) were reliably (protein FDR 1 %, peptide FDR 0.1 %) identified and present in 2 of 3 experiments. Among the proteins characterized, 971 were solely found in lysates (treated and untreated), whilst 364 were only detected in exosomal preparations (treated and untreated) (Fig. 4a). Our main focus was to investigate the fraction of proteins significantly changed by PIKfyve inhibition in the exosomal fractions, but also in the lysates. Interestingly, in exosomal fractions, the level of 71 proteins (Supplementary Table 1) was found to be significantly increased, and 302 proteins (Supplementary Table 2) were found to be down-regulated by apilimod treatment (Fig. 4b). The proteins that were affected by apilimod treatment showed low overlap between exosomal fractions and lysates, indicating that the changes in the exosomal fraction induced by apilimod are not directly reflected by the cellular protein composition.
Fig. 4.
Proteomic analysis of exosomal fractions and cell lysates after apilimod treatment revealed 4675 unique proteins. a Venn diagram showing the overlap of Uniprot protein entries (www.uniprot.org) for all confidently identified proteins in exosomal fractions and cells, after treatment with apilimod or control. b Venn diagram showing significantly changed proteins in exosomal fractions and cells after apilimod treatment. Changes between subproteomes were tested by Fisher’s Exact test (confidence interval 95 %)
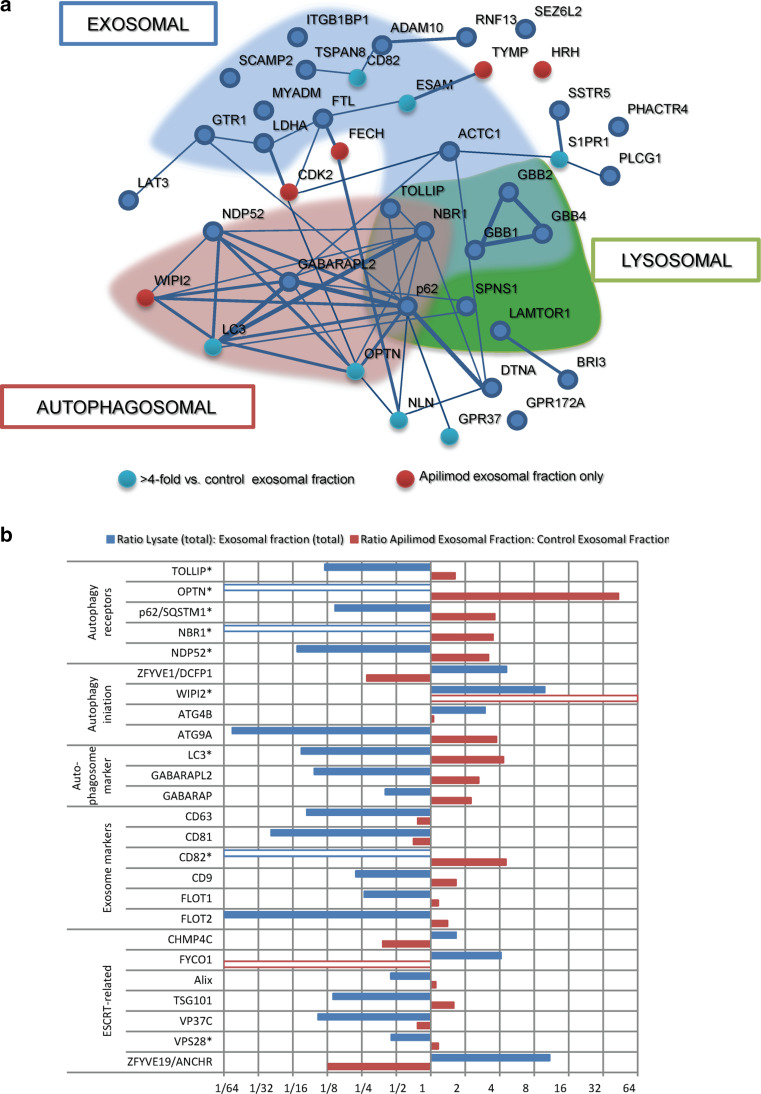
A further investigation of the proteins enriched in exosomal fractions after apilimod treatment through interaction analysis revealed the particular presence of autophagy-related proteins (Fig. 5a). Several LC3-interacting autophagy-receptor-type proteins, such as p62, NBR1, CACO2 and OPTN [55], were enriched in exosomal fractions after treatment of cells with apilimod, while proteins involved in earlier stages of autophagosome formation (Atg2a, Atg16L and Atg5) were not found (Fig. 5b; Supplementary Table 1). The autophagosomal marker proteins LC3, GABARAP and GABARAPL2 [55, 56] were strongly enriched in exosomal fractions after apilimod treatment of cells (Fig. 5b; Supplementary Table 1). Interestingly, a protein important for phagophore assembly, the initial stage of autophagosomal biosynthesis, the phosphoinositide-binding and WD40-containing protein WIPI2 [57, 58], was uniquely detected in exosome preparations from cells treated with apilimod. In contrast, the double-FYVE-containing protein 1, ZFYVE1, also a phagophore/omegasome-initiator protein [59], was down-regulated in exosome preparations after treatment with apilimod. Among the detected autophagosomal proteins only p62, TOLLIP and NBR1 have previously been described in exosomes [60, 61].
Fig. 5.
Autophagy-related proteins are up-regulated in exosomal fractions after PIKfyve inhibition. a Interactions between proteins in exosomal fractions found to be more than twofold up-regulated after apilimod treatment were analyzed through STRING (string.db.org). Proteins included in GO-annotation autophagosomal (GO:0005776), exosomal (GO:0070062) or lysosomal (GO:0005764) are emphasized. Strength of interaction is indicated by thickness of interconnecting line. b Selected proteins in exosomal fractions that were affected by apilimod treatment. Red bars ratio of protein level in exosomal fractions after apilimod treatment versus protein level in control exosomal fractions. Blue bars protein level in total lysates (control and apilimod) versus protein level in total exosomal fractions (control and apilimod). *; P < 0.05 versus control exosomal fractions. Empty bars not found in lysates/exosomal fractions
The interaction analysis also showed that treatment with apilimod increased the level of several proteins of lysosomal origin in exosomal fractions, including the WD40-motif containing members of the G-protein beta protein-class (GBB1, 2 and 4, Fig. 5a), which also were among the most abundant proteins in these fractions from PC-3 cells. A known binding partner for GBB1, GNAI2, was also found to be enriched after apilimod. GBB1 also interacts with sphingosine-1-phosphate receptor 2, which is among the most highly enriched proteins (69-fold enrichment in exosomal fractions after apilimod treatment compared to control, Supplementary Table 1). Finally, the phospholipase C gamma 1 (PLCG1), which also has been indicated to interact with GBB1 [62, 63], was enriched in exosomal fractions after apilimod treatment. Among the binding partners of GBB2, CDK2 was found to be uniquely present in exosomal fractions after apilimod treatment and not in control condition.
Enrichment of exosomal proteins after apilimod treatment also appeared in the interaction analysis (Fig. 5a). Selected proteins involved in autophagosome/exosome biogenesis and/or MVB formation are shown in Fig. 5b. CD82 and caveolin-1 were only detected in exosomal fractions by MS (though caveolin-1 was detected in lysates by Western blot, Fig. 3b), and both were enriched in exosomal fractions after apilimod treatment.
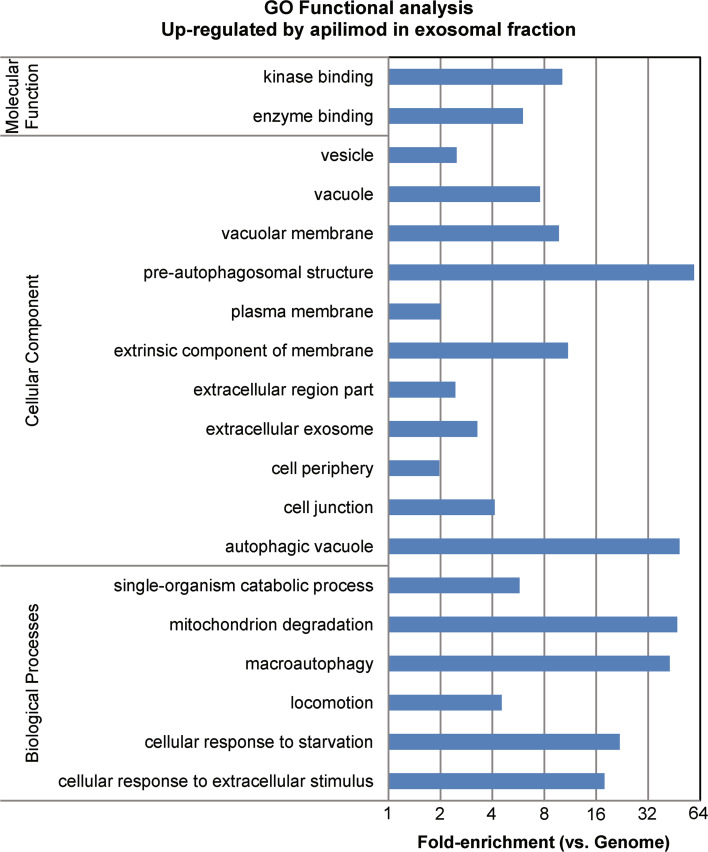
In line with the interaction analysis, Gene Ontology functional analysis showed that macroautophagy, as well as other autophagy-related functions, were highly enriched biological processes (macroautophagy; 43-fold, P = 3.3 × 10−9) in exosomal fractions from apilimod treated cells (Fig. 6). In contrast, pathways involved in RNA processing, splicing and translation, as well as pathways involved in focal adhesion and cell cycle regulation, were down-regulated in exosomal fractions after apilimod treatment (Supplementary Fig. 3).
Fig. 6.
Pathways up-regulated in exosomal fractions after apilimod treatment of cells. Functional analysis was performed on proteins in exosomal fractions significantly changed by apilimod. Analysis performed with DAVID-algorithm (http://david.abcc.ncifcrf.gov/) and fold-enrichment of GO-entries compared to genome
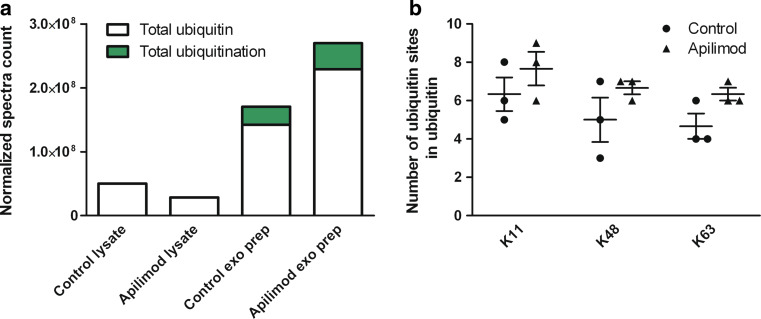
Ubiquitin, a signaling protein moiety involved in degradation and sorting among others, was enriched in exosome preparations compared to lysates, and apilimod treatment significantly increased the level of ubiquitin in the exosome preparations (Fig. 7a). In addition, ubiquitination analysis on the proteome showed increased ubiquitination and increased number of ubiquitination sites on ubiquitin (polyubiquitin) in the exosome preparation after apilimod treatment (Fig. 7a, b). Intriguingly, ubiquitination was completely absent in lysates (Fig. 7a). Polyubiquitination of ubiquitin at sites K11, K48 and K63 were occurring with increased frequency in exosomal fractions after apilimod treatment (Fig. 7b). Increased level of ubiquitin in the exosome preparation after apilimod was confirmed by Western blot (Supplementary Fig. 4).
Fig. 7.
Ubiquitin and ubiquitination is increased in exosomal fractions after apilimod treatment. a Total ubiquitin and total ubiquitination in lysates and exosome preparations after apilimod and control treatment. b Polyubiquitination of ubiquitin at sites K11, K48 and K63 in exosome preparations after apilimod and control treatment. exo prepr; exosome preparation
Secretory autophagy is induced by apilimod
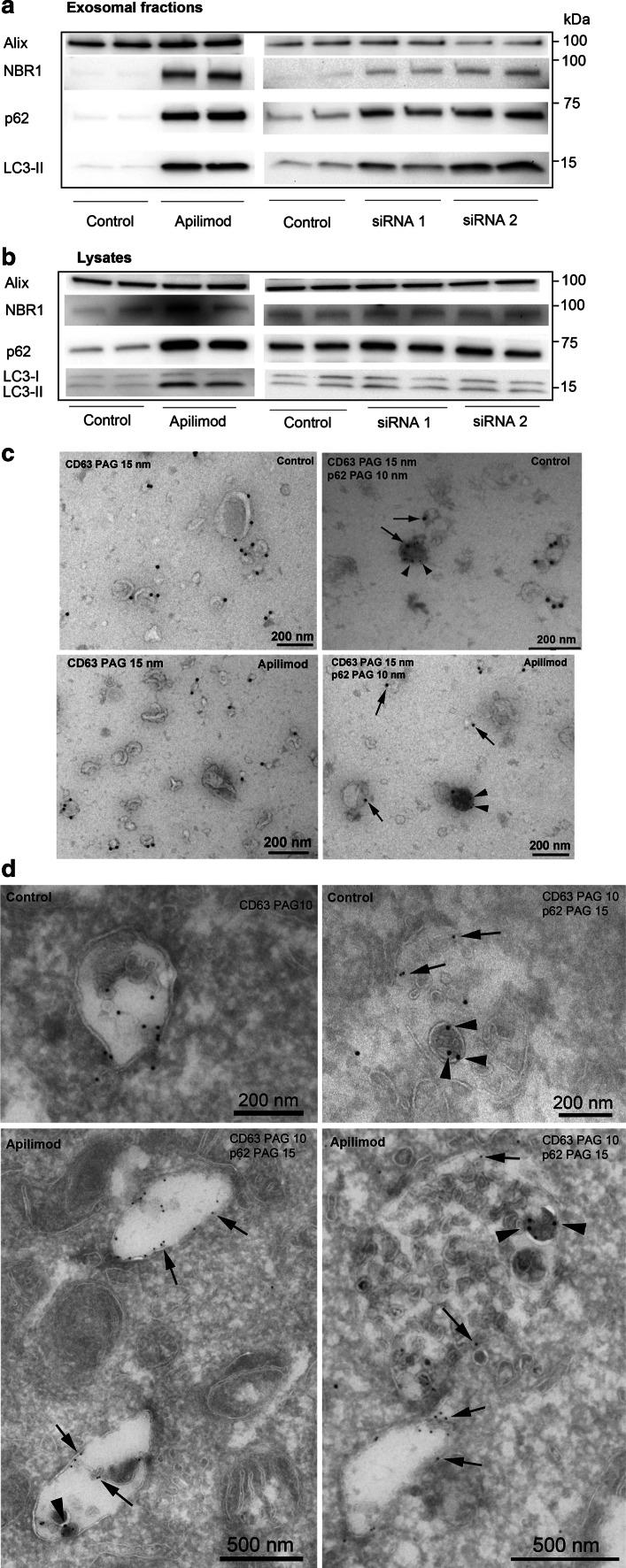
To verify the results from the proteomic study, Western blotting was performed to identify autophagy-related proteins in the exosomal preparation. Exosomes were isolated after treatment with apilimod or siRNA against PIKfyve as previously described, before Western blotting. In line with the proteomics data, a strong up-regulation of the autophagy-related proteins LC3, p62 and NBR1 in exosomal fractions after apilimod treatment of cells was observed (Fig. 8a). As expected, only the lipidated, membrane-associated form of LC3 (LC3-II) was detected in the exosomal fractions (Supplementary Fig. 5). Depletion of PIKfyve by siRNA also increased the level of LC3-II, p62 and NBR1 in the exosome preparations, but to a lesser degree than apilimod (Fig. 8a). In the cell lysates, treatment with apilimod increased the level of LC3-II, p62 and NBR1, but the induction was not as strong as in exosomal fractions (Fig. 8b). Knockdown of PIKfyve resulted in a small increase in p62 in the lysates (Fig. 8b). To further confirm the presence of autophagy markers in the exosomal fractions, EM was performed, showing p62-positive structures in the exosome preparation after apilimod treatment, whereas p62 only rarely was detected in exosomal fraction from control cells. Strikingly, the material marked with p62 was electron dense and had a different morphology than exosomes (Fig. 8c). However, most of the secreted material in the exosome preparation still had the characteristic exosomal shape and was positive for CD63, a well-known exosomal marker [64], indicating that the p62-positive structures only represent a sub-population of the exosomal preparation. This was confirmed by immuno-EM on whole PC-3 cells, which rarely showed labeling of p62 in MVBs in control cells (upper panel of Fig. 8d). In apilimod treated cells we observed p62-positive material, which colocalized with CD63-positive ILVs in MVBs, and importantly that each MVB contained much more ILVs than p62-positive material (Fig. 8d). This further suggests that the p62-positive structures constitute a minor part of the exosomal preparation. It should be noted that CD63 was often also found along the limiting membrane of endosomes with fewer ILVs (lower left panel in Fig. 8d) and that not all ILVs in MVBs are positive for CD63 (lower right panel in Fig. 8d).
Fig. 8.
p62 and CD63 are located in the same MVB-like structures after apilimod treatment. a PC-3 cells were seeded on 10 cm plates and pretreated with apilimod (0.5 µM) or DMSO (0.1 %, as control) for 2 h, before collection of exosomes for 18–19 h in the presence of inhibitor. Exosomes were also isolated 3 days after addition of siRNA against PIKfyve (siRNA1 and siRNA2, 25 nM) and non-targeting control (25 nM). Exosomes were isolated by ultracentrifugation and lysed in total 80 µl (lysis buffer plus sample buffer), before 25 µl of this mixture was loaded per well and run on SDS-PAGE gel. Each treatment was run in duplicates per experiment. Representative Western blot of proteins from exosomal fractions. b Representative Western blot of cellular proteins. c Immuno-EM on exosomal fractions after control (upper panels) and apilimod treatment (lower panels) from PC-3 cells that were double-labeled with CD63 (15 nm, arrows) and p62 (10 nm, arrowheads). d Immuno-EM on control PC-3 cells (upper panels) and apilimod treated PC-3 cells (lower panels) that were single labeled with CD63 (10 nm, arrows, upper left panel) or double-labeled with CD63 (10 nm, arrows) and p62 (15 nm, arrowheads) (other panels). Scale bars as indicated
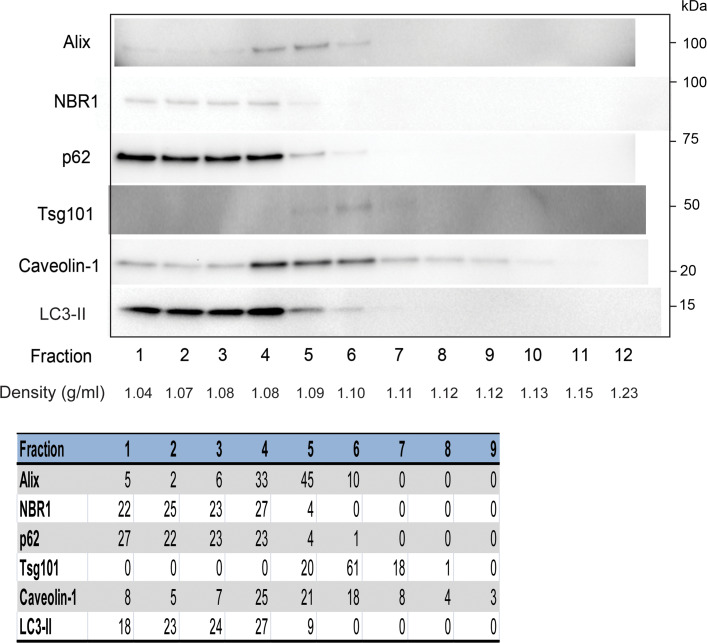
To quantify the contribution of p62-positive structures to the exosomal fraction after apilimod treatment, exosomes were separated from p62-positive structures by OptiPrep density gradient (5–30 %) centrifugation. p62, LC3-II and NBR1 appeared in less dense fractions than the exosomal markers (Fig. 9). The fractions positive for p62, LC3-II and NBR1 (fraction 1–3) were combined and the fractions positive for caveolin-1, Tsg101 and Alix (fraction 4–9) were combined, before they were analyzed by NTA. This showed that the p62-positive structures represent approximately 10–20 % of the total exosomal fraction (Supplementary Fig. 6a). Thus, the increased number of particles in the exosomal fraction after apilimod treatment seems to be caused by both increased exosome release and released p62-positive structures. EM on fraction 1–3 and fraction 4–9 was also performed (Supplementary Fig. 6b–c).
Fig. 9.
Partial separation of p62-positive structures from exosomes by OptiPrep density gradient. Exosomes were separated from p62-positive structures by OptiPrep density gradient (5–30 %) centrifugation at 100,000×g for 18 h. Twelve fractions were collected and Western blotting was used to detect which fractions contained exosomal proteins and autophagy-related proteins. Representative Western blot showing the 12 fractions. The bands were quantified and calculated as % of total signal per protein for fraction 1–9
PIKfyve inhibition reduces both autophagic flux and EGF degradation
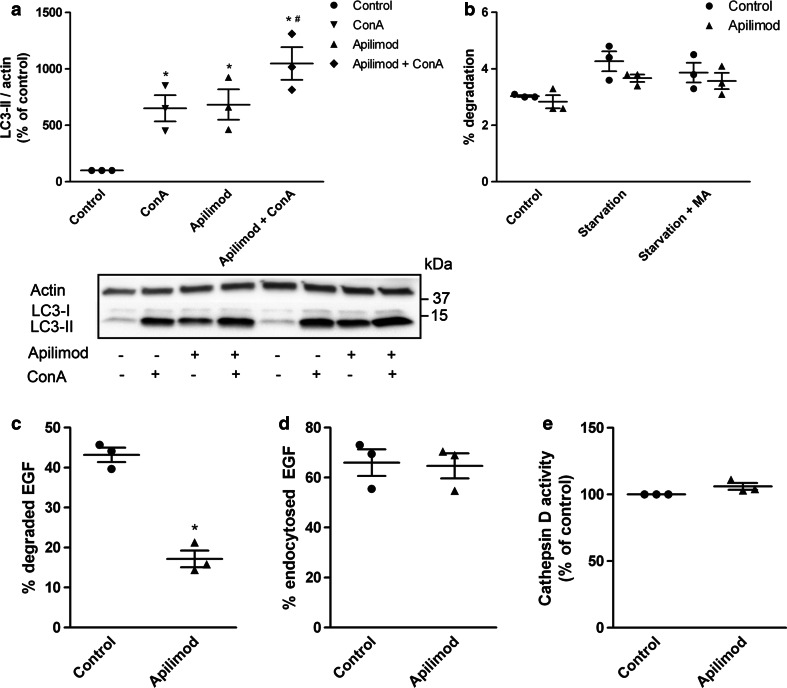
Since autophagy was found to be a highly enriched biological process in the exosome preparation from apilimod treated cells, we wanted to know whether autophagic activity was affected in these cells. First, we measured the autophagic flux by immunoblot of LC3. The level of basal autophagic turnover in these cells was determined by a four hour treatment with the lysosomal inhibitor concanamycin A (Con A) (Fig. 10a), and surprisingly, apilimod treatment for 21 h increased the level of LC3-II to a level comparable to Con A alone (Fig. 10a). Such high levels of LC3-II could either be due to a massive increase in the generation of LC3-II-positive autophagosomes, or an inhibited degradation of autophagosomes. Since the combined treatment of apilimod and Con A only led to a modest further increase in LC3-II levels, the former possibility can be ruled out. This indicates that although apilimod slightly induces the on-rate of autophagy, the major effect of apilimod seems to be inhibition of autophagic degradation, at least during the final hours of the incubation. To verify the effect of apilimod on autophagic turnover, a pulse-chase assay for degradation of long-lived proteins was performed. Treatment with apilimod tended to decrease the starvation-induced autophagic degradation of long-lived proteins (Fig. 10b), suggesting that autophagic flux is inhibited by apilimod.
Fig. 10.
PIKfyve inhibition affects the autophagy pathway. a Cellular LC3-II levels were measured by Western blot after cells were treated with or without concanamycin A (50 nM) during the last 4 h of the preincubation step with apilimod. b Degradation of long-lived proteins. c Degradation of 125I-EGF. d Endocytosis of 125I-EGF. e Cathepsin D activity as measured by Cathepsin D activity assay. MA 3-methyladenine. The results are plotted as dot plots showing the value obtained for each individual experiment as a single dot + mean values + standard error of the mean from 3 independent experiments. *; P < 0.05 versus control, #; P < 0.05 versus apilimod
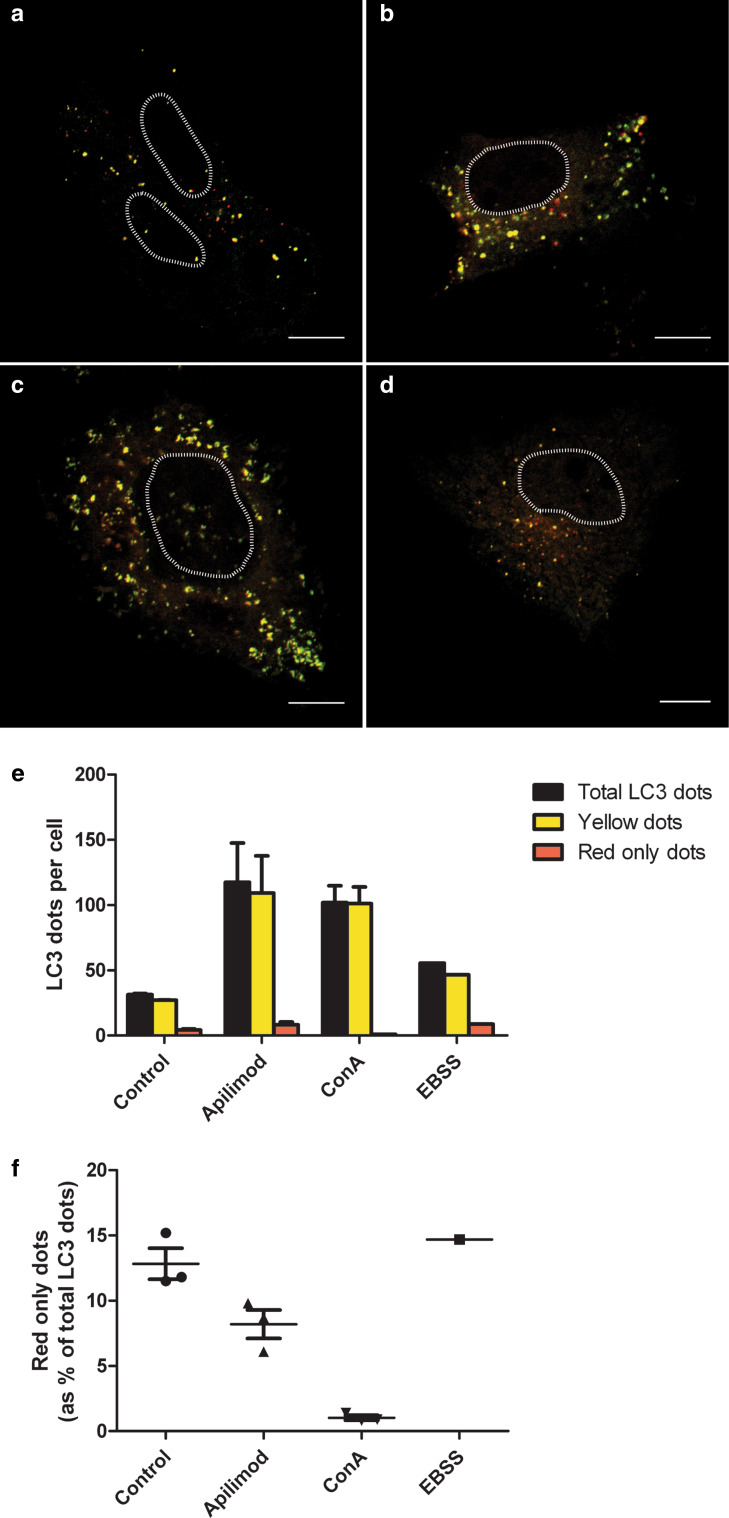
Since our results indicated a decrease in autophagic turnover after apilimod treatment, we wondered whether the degradation through the MVB pathway would also be affected by apilimod. To address this, endocytosis and degradation of 125I-EGF was measured. Indeed, EGF degradation was reduced by approximately 60 % after apilimod treatment (Fig. 10c), whereas endocytosis of EGF was not affected (Fig. 10d). As degradation of both EGF and long-lived proteins was reduced, we investigated the lysosomal function by measuring cathepsin D activity. Treatment with apilimod did not change the cathepsin D activity (Fig. 10e), indicating that the lysosomes are still functioning. To further characterize the fusion of autophagosomes with lysosomes after apilimod treatment, PC-3 cells were transfected with a double-tagged LC3 (mCherry-GFP-LC3), which will appear yellow (green merged with red fluorescence) in the non-acidic autophagosomes and emit red only fluorescence in the autolysosomes due to quenching of GFP in these acidic structures. As expected, activation of autophagy by starvation increased the total number of autophagosomes, whereas blocking the acidification of the autolysosomes by Con A led to accumulation of autophagosomes and abolished the appearance of red-only puncta (Fig. 11). In line with the immunoblot data (Fig. 10a), treatment with apilimod increased the total number of LC3-positive puncta, however, the fraction of red-only puncta was reduced compared to control or EBSS-treated cells, suggesting that the generation of proper autolysosomes is partly inhibited. Altogether, these results suggested that the fusion of lysosomes with both MVBs and autophagosomes is partly inhibited by apilimod.
Fig. 11.
PIKfyve inhibition partly inhibits the fusion of autophagosomes with lysosomes. PC-3 cells were transfected with a double-tagged LC3 protein (mCherry-GFP-LC3), which will emit yellow (green merged with red) fluorescence in non-acidic structures and appear as red only in the autolysosomes due to quenching of GFP in these acidic structures. a Control cells, b cells treated with apilimod (0.5 µM) for 21 h, c cells treated with concanamycin A (50 nM) for 4 h and d cells grown in EBSS for 4 h. Nuclei are marked with a dashed line. Scale bar is 10 µM. e The number of yellow LC3 dots and red only LC3 dots per cell for each condition was quantified. Total LC3 dots are the sum of yellow and red only LC3 dots. f Percentage of red only LC3 dots of the total LC3 dots. From each experiment 12–22 cells were counted per condition and data are presented as mean values + standard error of the mean from 3 independent experiments. EBSS was included as a control, but only in one experiment
Discussion
This study shows that inhibition of PIKfyve by apilimod or depletion by siRNA increases the release of particles to the exosomal fraction from PC-3 cells. Interestingly, the exosome preparation after PIKfyve inhibition is strongly enriched in autophagy-related proteins. Moreover, p62-positive structures are found in the exosome preparation after apilimod treatment, indicating that not only exosomes are secreted, but also the content of amphisomes through induction of secretory autophagy.
NTA measurements do not distinguish between exosomes and protein aggregates. Since the exosome preparation after apilimod treatment contains a mixture of exosomes and p62-positive structures (possibly protein aggregates), it is not clear from this analysis whether the increased secretion of particles is due to increased secretion of exosomes or p62-positive structures, or both. However, EM data showed that p62-positive structures only represent a small sub-population, and that most of the material in the exosome preparation is exosomes. Supporting this, each MVB in apilimod-treated cells contains mainly ILVs and just a few, or no, p62-positive structures. In addition, quantitative EM showed that the number of ILVs per MVB is dramatically increased by apilimod. Using OptiPrep density gradient centrifugation separation of exosomes from p62-positive structures was obtained. Counting by NTA further confirmed that the majority of the exosomal fraction is exosomes, rather than p62-positive structures. This means that the apilimod-induced increase in number of secreted particles cannot solely be due to secretion of p62-positive structures, but also increased secretion of exosomes. Our results emphasize that investigations on exosomes under conditions where autophagy is affected should be interpreted with care.
The additional increase in LC3-II level when cells are treated with the combination of apilimod and concanamycin A indicates that autophagy is slightly induced by apilimod. Induction of autophagy can lead to secretion of substances through secretory autophagy [65], a route for unconventional secretion of cytoplasmic substrates [66]. Secretion of autophagy receptors such as p62 and NBR1 through this pathway has to our knowledge not been previously shown in mammalian cells. In Dictyostelium p62 has been shown to be involved in the secretion of ejectosome [67], otherwise little is known about the role of the autophagy receptors in secretory autophagy. In degradative selective autophagy, the process by which specific organelles, protein aggregates or pathogens are degraded, cargo selection is mediated by autophagic cargo receptors such as p62, NBR1, NDP52 (CALCOCO2) and NIX, which contain an LC3-interacting region (LIR) and, therefore, can bind directly to LC3 [55]. In addition to LC3, p62 and NBR1 bind to ubiquitinated protein aggregates and thereby recruit phagophores to these aggregates. How cargo selection is mediated in secretory autophagy is, however, not known.
We detected increased level of ubiquitin, as well as increased number of ubiquitination sites on ubiquitin (polyubiquitin) in the exosome preparation after apilimod treatment. Previously, exosomes have been shown to contain ubiquitinated proteins and many of the ubiquitinated proteins are polyubiquitinated [68]. In addition, p62-positive protein aggregates are polyubiquitinated, which could contribute to the increased level of ubiquitin and polyubiquitin in the exosome preparation after apilimod treatment.
The observed increased number of ILVs per MVB and increased number of MVBs per cell after apilimod treatment, indicate that either degradation of the MVBs is inhibited or MVB biogenesis and ILV formation are upregulated. In addition, apilimod treatment reduces EGF degradation, indicating that the degradation pathway is affected. Decreased EGFR degradation has previously been reported after treatment with another PIKfyve inhibitor (MF4) [45]. As apilimod does not affect cathepsin D activity, the lysosomes are most likely still functional, whereas the fusion between MVBs and lysosomes seems to be inhibited. The increased cellular level of LC3-II after apilimod treatment in combination with reduced starvation-induced degradation of long-lived proteins, indicates that apilimod also inhibits the autophagic flux. This is in agreement with previous studies showing increased LC3-II levels after treatment with other PIKfyve inhibitors [42, 45]. In accordance, immunofluorescence microscopy showed that apilimod treatment increased the number of autophagosomes and reduced the relative amount of autolysosomes, indicating a partial inhibition of fusion of lysosomes with autophagosomes. Thus, our results suggest that PIKfyve inhibition reduces the fusion of lysosomes with both MVBs and autophagosomes, or with amphisomes. In line with our data, Li et al. showed that an elevation in PI(3,5)P2 occurs immediately before endolysosomal fusion [35], and inhibition of PIKfyve has been suggested to inhibit fusion of lysosomes with both MVBs and autophagosomes [35, 45]. However, inhibition of fusion with lysosomes cannot alone explain the increased size of the MVB-like structures, since the growing membrane needs a lipid source. Since p62 and CD63 positive structures are found in the same MVB-like structures, apilimod treatment might also facilitate fusion of MVBs with autophagosomes, generating amphisomes. Alternatively, increased fusion of early endosomes, as described by Ikonomov et al. [69] could contribute to enlargement of MVB-like structures. Another possibility is that increased level of PI(3)P could generate more phagophores and thereby contribute with more membrane to the amphisomes.
It has been speculated that exosomes play a role in alleviating intracellular stress, and that autophagy and the exosome pathway are linked [26]. Fader et al. showed that induction of autophagy can inhibit the secretion of exosomes [70]. This was suggested to be due to fusion of MVB with autophagosomes and thereby directing MVBs to a degradative pathway instead. In line with this, Sahu et al. showed that inhibition of autophagy increased exosomal secretion of GAPDH [71]. Our study supports this idea. The apilimod-induced exosome secretion was associated with reduced autophagic degradation. When a cell is no longer able to degrade material in the lysosomes, secretion of the content of MVBs and amphisomes could be a way to rescue the cell. This supports the idea of exosomes as waste disposal, the first function ascribed to these vesicles [2]. A question then rises how this affects the microenvironment and the neighboring cells, and whether this is beneficial or not for a multicellular organism.
In conclusion, PIKfyve inhibition increases exosome secretion and induces secretory autophagy. This might be caused by impaired fusion of lysosomes with both MVBs and autophagosomes, and increased fusion of MVBs with autophagosomes. The altered fusion affinities generate enlarged MVB-like structures or amphisomes, which contain more ILVs per organelle explaining the increased exosome release and the secretion of p62-positive structures.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was funded by The South-Eastern Norwegian Regional Health Authority, The Norwegian Cancer Society and The Research Council of Norway, and supported by The Research Council of Norway through its Centers of Excellence funding scheme, project number 179571. We thank Anne Engen, Anne Grethe Myrann and Marianne Smestad for their excellent technical assistance and Tore Skotland for valuable and interesting discussions. We also thank the proteomics core facility, especially Bernd Thiede and Christian Koehler, at the University of Oslo.
Abbreviations
- CI-MPR
Cation-independent mannose 6-phosphate receptor
- EM
Electron microscopy
- ESCRTs
Endosomal sorting complexes required for transport proteins
- ILV
Intraluminal vesicle
- MS
Mass spectrometry
- MVB
Multivesicular body
- NTA
Nanoparticle tracking analysis
- PI(3)P
Phosphatidylinositol-3-phosphate
- PI(3,5)P2
Phosphatidylinositol-3,5-bisphosphate
- PI(5)P
Phosphatidylinositol-5-phosphate
- TGN
Trans-golgi network
References
- 1.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101(3):942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- 3.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostenfeld MS, Jeppesen DK, Laurberg JR, Boysen AT, Bramsen JB, Primdal-Bengtson B, Hendrix A, Lamy P, Dagnaes-Hansen F, Rasmussen MH, Bui KH, Fristrup N, Christensen EI, Nordentoft I, Morth JP, Jensen JB, Pedersen JS, Beck M, Theodorescu D, Borre M, Howard KA, Dyrskjot L, Orntoft TF. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014;74(20):5758–5771. doi: 10.1158/0008-5472.CAN-13-3512. [DOI] [PubMed] [Google Scholar]
- 5.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841(1):108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166(12):7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 8.Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C, Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J. 2004;380(Pt 1):161–171. doi: 10.1042/bj20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorente A, Skotland T, Sylvanne T, Kauhanen D, Rog T, Orlowski A, Vattulainen I, Ekroos K, Sandvig K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831(7):1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hessvik NP, Phuyal S, Brech A, Sandvig K, Llorente A. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim Biophys Acta. 2012;1819(11–12):1154–1163. doi: 10.1016/j.bbagrm.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Ronquist KG, Ronquist G, Carlsson L, Larsson A. Human prostasomes contain chromosomal DNA. Prostate. 2009;69(7):737–743. doi: 10.1002/pros.20921. [DOI] [PubMed] [Google Scholar]
- 14.Lazaro-Ibanez E, Sanz-Garcia A, Visakorpi T, Escobedo-Lucea C, Siljander P, Ayuso-Sacido A, Yliperttula M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate. 2014;74(14):1379–1390. doi: 10.1002/pros.22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Goot FG, Gruenberg J. Intra-endosomal membrane traffic. Trends Cell Biol. 2006;16(10):514–521. doi: 10.1016/j.tcb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun. 2010;399(3):384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 17.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 18.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 19.Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190(6):1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, Lochnit G, Preissner KT, Zoller M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70(4):1668–1678. doi: 10.1158/0008-5472.CAN-09-2470. [DOI] [PubMed] [Google Scholar]
- 21.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 22.Phuyal S, Hessvik NP, Skotland T, Sandvig K, Llorente A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014;281(9):2214–2227. doi: 10.1111/febs.12775. [DOI] [PubMed] [Google Scholar]
- 23.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 24.Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005;6(2):131–143. doi: 10.1111/j.1600-0854.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 25.Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Gronborg M, Mobius W, Rhee J, Barr FA, Simons M. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189(2):223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baixauli F, Lopez-Otin C, Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–116. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ejlerskov P, Rasmussen I, Nielsen TT, Bergstrom AL, Tohyama Y, Jensen PH, Vilhardt F. Tubulin polymerization-promoting protein (TPPP/p25alpha) promotes unconventional secretion of alpha-synuclein through exophagy by impairing autophagosome-lysosome fusion. J Biol Chem. 2013;288(24):17313–17335. doi: 10.1074/jbc.M112.401174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsson P, Loganathan K, Sekiguchi M, Matsuba Y, Hui K, Tsubuki S, Tanaka M, Iwata N, Saito T, Saido TC. Abeta secretion and plaque formation depend on autophagy. Cell reports. 2013;5(1):61–69. doi: 10.1016/j.celrep.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 31.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 32.Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584(7):1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390(6656):187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 34.Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323(Pt 3):597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RG, Weisman LS, Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci USA. 2013;110(52):21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shisheva A, Sbrissa D, Ikonomov O. Plentiful PtdIns5P from scanty PtdIns(3,5)P or from ample PtdIns? PIKfyve-dependent models: evidence and speculation (response to: DOI 10.1002/bies.201300012) Bioessays. 2015;37(3):267–277. doi: 10.1002/bies.201400129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCartney AJ, Zhang Y, Weisman LS. Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. BioEssays. 2014;36(1):52–64. doi: 10.1002/bies.201300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6(5):525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, Stenmark H, Cullen PJ. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci. 2006;119(Pt 19):3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jefferies HB, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, Kaizawa H, Ohishi T, Workman P, Waterfield MD, Parker PJ. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9(2):164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai X, Xu Y, Cheung AK, Tomlinson RC, Alcazar-Roman A, Murphy L, Billich A, Zhang B, Feng Y, Klumpp M, Rondeau JM, Fazal AN, Wilson CJ, Myer V, Joberty G, Bouwmeester T, Labow MA, Finan PM, Porter JA, Ploegh HL, Baird D, De Camilli P, Tallarico JA, Huang Q. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem Biol. 2013;20(7):912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin S, Harper CB, May LM, Coulson EJ, Meunier FA, Osborne SL. Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PLoS One. 2013;8(3):e60152. doi: 10.1371/journal.pone.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Current Biol: CB. 2007;17(20):1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 44.Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC. PI(5)P regulates autophagosome biogenesis. Mol Cell. 2015;57(2):219–234. doi: 10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10(7):883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osborne SL, Wen PJ, Boucheron C, Nguyen HN, Hayakawa M, Kaizawa H, Parker PJ, Vitale N, Meunier FA. PIKfyve negatively regulates exocytosis in neurosecretory cells. J Biol Chem. 2008;283(5):2804–2813. doi: 10.1074/jbc.M704856200. [DOI] [PubMed] [Google Scholar]
- 47.Messenger SW, Thomas DD, Cooley MM, Jones EK, Falkowski MA, August BK, Fernandez LA, Gorelick FS, Groblewski GE. Early to late endosome trafficking controls secretion and zymogen activation in rodent and human pancreatic acinar cells. Cell Mol Gastroenterol Hepatol. 2015;1(6):695–709. doi: 10.1016/j.jcmgh.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llorente A, de Marco MC, Alonso MA. Caveolin-1 and MAL are located on prostasomes secreted by the prostate cancer PC-3 cell line. J Cell Sci. 2004;117(22):5343–5351. doi: 10.1242/jcs.01420. [DOI] [PubMed] [Google Scholar]
- 49.Llorente A, van Deurs B, Sandvig K. Cholesterol regulates prostasome release from secretory lysosomes in PC-3 human prostate cancer cells. Eur J Cell Biol. 2007;86(7):405–415. doi: 10.1016/j.ejcb.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 50.Phuyal S, Skotland T, Hessvik NP, Simolin H, Overbye A, Brech A, Parton RG, Ekroos K, Sandvig K, Llorente A. The ether lipid precursor hexadecylglycerol stimulates the release and changes the composition of exosomes derived from PC-3 cells. J Biol Chem. 2015;13(290):4225–4237. doi: 10.1074/jbc.M114.593962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113(1):123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179(3):485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 54.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 55.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117(Pt 13):2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 57.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6(4):506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 58.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55(2):238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nanao T, Koike M, Yamaguchi J, Sasaki M, Uchiyama Y. Cellular localization and tissue distribution of endogenous DFCP1 protein. Biomed Res (Tokyo, Japan) 2015;36(2):121–133. doi: 10.2220/biomedres.36.121. [DOI] [PubMed] [Google Scholar]
- 60.Kharaziha P, Chioureas D, Rutishauser D, Baltatzis G, Lennartsson L, Fonseca P, Azimi A, Hultenby K, Zubarev R, Ullen A, Yachnin J, Nilsson S, Panaretakis T. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget. 2015;6(25):21740–21754. doi: 10.18632/oncotarget.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteom: MCP. 2010;9(6):1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruceanu C, Alda M, Turecki G. Lithium: a key to the genetics of bipolar disorder. Genome medicine. 2009;1(8):79. doi: 10.1186/gm79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu J, Wong C. A computational screen for mouse signaling pathways targeted by microRNA clusters. RNA (New York, NY) 2008;14(7):1276–1283. doi: 10.1261/rna.997708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 65.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30(23):4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol. 2010;188(4):527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerstenmaier L, Pilla R, Herrmann L, Herrmann H, Prado M, Villafano GJ, Kolonko M, Reimer R, Soldati T, King JS, Hagedorn M. The autophagic machinery ensures nonlytic transmission of mycobacteria. Proc Natl Acad Sci USA. 2015;112(7):E687–E692. doi: 10.1073/pnas.1423318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes contain ubiquitinated proteins. Blood Cells Mol Dis. 2005;35(3):398–403. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Ikonomov OC, Sbrissa D, Shisheva A. Localized PtdIns 3,5-P2 synthesis to regulate early endosome dynamics and fusion. Am J Physiol Cell Physiol. 2006;291(2):C393–C404. doi: 10.1152/ajpcell.00019.2006. [DOI] [PubMed] [Google Scholar]
- 70.Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9(2):230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 71.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20(1):131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.