Abstract
Osteoblasts and chondrocytes produce a large number of extracellular matrix proteins to generate and maintain the skeletal system. To cope with their functions as secretory cells, these cells must acquire a considerable capacity for protein synthesis and also the machinery for the quality-control and transport of newly synthesized secreted proteins. The unfolded protein response (UPR) plays a crucial role during the differentiation of these cells to achieve this goal. Unexpectedly, however, studies in the past several years have revealed that the UPR has more extensive functions in skeletal development than was initially assumed, and the UPR critically orchestrates many facets of skeletal development and homeostasis. This review focuses on recent findings on the functions of the UPR in the differentiation of osteoblasts, chondrocytes, and osteoclasts. These findings may have a substantial impact on our understanding of bone metabolism and also on establishing treatments for congenital and acquired skeletal disorders.
Keywords: Unfolded protein response, ER stress, Osteoblast, Chondrocyte, Osteoclast, IRE1α, PERK, ATF6
Introduction
The skeletal system is primarily composed of extracellular matrix proteins and calcium phosphate. To develop and maintain this system, osteoblasts and chondrocytes must produce a large number of extracellular matrix proteins, including collagens, noncollagenous proteins, proteoglycans, and glycoproteins. Therefore, these cells must constantly cope with the burden of protein synthesis overload and efficiently transport and secrete newly synthesized proteins. Secreted proteins are synthesized and acquire their three-dimensional conformation in the endoplasmic reticulum (ER). To maintain the integrity of the skeletal system, these proteins must be quality controlled, and only properly folded proteins are secreted extracellularly. The proteins that fail to acquire a proper conformation accumulate in the ER lumen and ultimately lead to a condition called ER stress [1–5]. In addition to protein synthesis overload, various cellular stresses, such as hypoxia, low glucose, intracellular calcium fluctuation, and ultraviolet exposure, can damage the microenvironment of the ER, resulting in an increased number of unfolded proteins in the ER lumen and ER stress. Because prolonged ER stress is harmful to cellular homeostasis, cells are equipped with machinery called the unfolded protein response (UPR) to comply with this undesirable condition. The UPR is a highly conserved cellular mechanism, and found in species from yeasts to humans [6]. In mammals, there are three major branches of the UPR that function as sensors for ER stress and transmit signals to downstream signaling molecules and to the nucleus. Upon ER stress, the UPR suppresses general protein synthesis, promotes the degradation of unfolded proteins, and increases the capacity of the ER to alleviate ER stress and recover the functions of the ER. However, when the cellular stress becomes unmanageable and irreversible, the UPR induces apoptosis to circumvent the ensuing damage to the surrounding milieu and cells [7].
The three major UPR branches are mediated by three different classes of transmembrane proteins in the ER: inositol-requiring protein-1α (IRE1α), activating transcription factor 6 (ATF6), and pancreatic ER kinase (PERK). The functions of these molecules in the UPR have been a subject of intensive study over the past decade [1–5]. In accordance with the fundamental contributions of the UPR in maintaining the ER and the quality control of secreted proteins, studies over the past decade have identified critical roles for the UPR in secretory cells, particularly plasma cells [8, 9] and pancreatic β-cells [10, 11]. However, more recent studies have revealed unexpected roles of the UPR that are apparently not related to the conventional function of the UPR in cells and tissues that are not specialized in producing secretory proteins, including angiogenesis [12], lipogenesis [13], the pathology of Huntington’s disease [14], and the regulation of innate immunity [15], to name a few examples.
In a similar vein, in addition to its function as an adaptation mechanism against ER stress, the UPR in the skeletal system was shown to be an essential regulator of osteoblast and chondrocyte differentiation [16–19]. Furthermore, recent studies have also suggested that the UPR is important for the development of osteoclasts, which are multinucleated cells responsible for bone resorption [20, 21]. As both osteoblasts and chondrocytes have a highly developed ER system, it is conceivable that the UPR works to expand the protein synthesis capacity of these cells. However, studies over the past decade have radically changed our understanding of the functions of the UPR in skeletal development and homeostasis. This review focuses on current data on the molecular function of the UPR in the regulation of the skeletal system and its impact on skeletal disorders.
The three major branches of the UPR
The IRE1α axis
The branch of UPR mediated by IRE1α (encoded by Ern1) is the most highly conserved and is present in species from yeasts to mammals. IRE1α is a type-1 transmembrane protein that harbors both a kinase domain and an endoribonuclease domain [2, 22]. Upon ER stress, the molecular chaperone immunoglobulin heavy chain-binding protein (BiP, also referred to as GRP78) dissociates from the luminal domain of IRE1α and facilitates its homodimerization and autophosphorylation. Autophosphorylation of IRE1α leads to conformational changes and the activation of the endoribonuclease domain. The activated endonuclease targets its substrate, Xbp1 mRNA, and splices out 26 nucleotides, resulting in a translational frame shift that yields the transcription factor XBP1s. The protein product derived from the unspliced Xbp1 mRNA (XBP1u) is highly unstable, and its potential functions are not fully clarified [23–25]. The kinase domain of IRE1α is involved in recruiting TNF receptor-associated factor 2 (TRAF2), which in turn activates JUN N-terminal kinase (JNK) [26]. The consequence of IRE1α-mediated JNK activation is not fully understood; however, past studies have suggested that it may be involved in the regulation of autophagy [27, 28] and the promotion of apoptosis [29]. Although Xbp1 mRNA is the preferred substrate for the IRE1α endoribonuclease, under certain conditions, various other mRNAs are also targeted and degraded by the IRE1α endoribonuclease, a mechanism termed regulated IRE1-dependent decay of mRNA (RIDD) [30, 31]. RIDD was originally reported to be involved in the degradation of mRNAs encoding membrane-bound and secreted proteins, thereby reducing the protein synthesis overload [32, 33]. Furthermore, recent studies have shown that RIDD has roles in the regulation of lipid metabolism [13, 34], protection against liver damage [35], and adaptive and innate immunity [36–38]. Whereas, under overt ER stress conditions, RIDD promotes apoptosis by degrading mRNAs encoding pro-survival factors and micro-RNAs targeting pro-apoptotic factors [39, 40]. How the activity of RIDD is regulated is not fully understood and will be one of the focuses of research in this field [30].
XBP1s, which is derived from the spliced version of the Xbp1 transcript, is a member of the CREB/ATF basic leucine-zipper (bZIP) family of transcription factors. The conventional transcriptional targets of XBP1s include luminal ER protein chaperones, disulfide isomerases, glycosylases, and components of the ER to Golgi network and the ER-associated degradation (ERAD) pathway [41, 42]. Collectively, the IRE1α-XBP1 pathway promotes the removal and degradation of unfolded proteins and the expansion of ER capacity, thereby maintaining the homeostasis of the functions and microenvironment of the ER [9, 43]. However, recent studies have revealed that the functions of the IRE1α-XBP1 pathway are not limited to the maintenance of the ER system but also include hepatic lipogenesis [44], the innate immune response [45], hexosamine biosynthesis [46], the HIF-1α-mediated hypoxia pathway [47], and cell differentiation [42, 48–50], suggesting that the IRE1α-XBP1 pathway has diverse functions that are not necessarily related to those in the classical UPR. Corroborating the importance of this pathway in vivo, mice lacking IRE1α or XBP1 are embryonic lethal and die at approximately 9.5–11.5 dpc and 11.5–14.5 dpc, respectively [26, 51].
The PERK axis
PERK (encoded by the Eif2ak3 gene) is a type I transmembrane protein that is localized in the ER and contains a protein kinase domain in its cytoplasmic tail. The dissociation of BiP from the luminal domain promotes dimerization and autophosphorylation. Activated PERK directly phosphorylates the α-subunit of eukaryotic translation initiation factor-2 (eIF2α), which results in an attenuation of global protein synthesis by interfering with 5′-cap assembly [52]. In contrast to this general suppression of protein synthesis, the translation of activating transcription factor 4 (ATF4), a transcription factor belonging to the cAMP response element-binding protein (CREB)/ATF family, is selectively promoted by the phosphorylation of eIF2α. The transcriptional targets of ATF4 include the genes that are involved in amino acid import, glutathione biosynthesis, and resistance to oxidative stress [53]. Collectively, the UPR mediated by PERK has two distinct roles; one is to alleviate the protein synthesis overload under ER stress conditions, and the other is to protect the cell from various endogenous and exogenous stresses by increasing the production of ATF4 [54]. In addition to these cytoprotective functions of PERK, ATF4 also induces the transcription of Ddit3 (encodes the CCAAT/enhancer-binding protein homologous protein, CHOP) [55], which is associated with ER stress-mediated apoptosis [56]. Therefore, PERK not only promotes cell survival, but also mediates programed cell death under certain conditions. Of interest, CHOP induces the transcription of Ppp1r15a, which encodes an eIF2α-directed phosphatase GADD34. In turn, GADD34, along with another eIF2α-directed phosphatase CReP (encoded by Ppp1r15b), dephosphorylates eIF2α and thereby forms a negative feedback loop in regulating the level of eIF2α phosphorylation [57].
Mice lacking PERK are not embryonically lethal, indicating that PERK is not necessarily essential for embryonic development [11, 58]. However, these mice progressively develop diabetes mellitus due to a loss of insulin-producing pancreatic β-cells and exhibit growth retardation, indicative of a potential role for PERK in skeletal development.
The ATF6 axis and the OASIS/ATF6 family genes
ATF6 is a type II transmembrane protein that contains a bZIP transcription factor domain in its cytoplasmic domain. Upon dissociation of BiP from its luminal domain, ATF6 translocates from the ER to the Golgi and is cleaved by site-1 and site-2 proteases (S1P and S2P, respectively) [59, 60]. The cytoplasmic domain is released from the Golgi membrane by S2P and moves into the nucleus where it functions as a transcription factor to promote the expression of genes that are involved in ER quality control, including Hspa5 (encodes BiP) and Xbp1. Mice lacking ATF6 develop normally and exhibit no apparent defects, at least under unchallenged conditions [61, 62], indicating that the contribution of ATF6 to the maintenance of the ER homeostasis is relatively minor and can possibly be compensated by XBP1.
Five transcription factors share structural homology with ATF6: old astrocyte specifically induced substance (OASIS, encoded by Creb3l1) [63], BBF2 human homolog on chromosome 7 (BBF2H7, encoded by Creb3l3) [64], Luman (encoded by Creb3) [65], CREBH (encoded by Creb3l3) [66], and CREB4 (encoded by Creb3l4) [67]. All are transmembrane proteins that are localized in the ER and contain a bZIP transcription factor domain in their cytoplasmic domain. Similar to ATF6, these proteins translocate from the ER to the Golgi upon ER stress and are processed by S1P and S2P. The released cytoplasmic domain enters the nucleus to function as a transcription factor. However, unlike ATF6, each of the family members has a rather distinct and specific expression pattern, indicating that each gene has tissue- or cell-specific functions. As discussed in the present review, OASIS, BBF2H7, and Luman are preferentially expressed in osteoblasts, chondrocytes, and immune cells, respectively, and exhibit highly specialized functions in skeletal development [17, 21, 64].
The UPR in osteoblast differentiation and functions
Osteoblasts are derived from mesenchymal progenitor cells that are also capable of differentiating into various cell lineages, including chondrocytes, tenocytes, and adipocytes. The cell fate decision into the osteoblast lineage is determined by the expression of RUNX2, a member of the Runt-containing family of transcription factors, followed by that of Osterix, a zinc-finger-containing transcription factor [68–73]. These two osteoblast-specific transcription factors are indispensable for osteoblast differentiation, as evidenced by a nearly complete loss of osteoblasts and calcified tissues in mutant mice lacking either of these transcription factors [74–76]. In more mature osteoblasts, ATF4, one of the downstream targets of the PERK axis of the UPR, plays critical roles in maintaining osteoblast functions [77]. Mature osteoblasts produce a large number of extracellular matrix proteins, such as type I collagen and various noncollagenous proteins, to form osteoids. Osteoids are the organic matrix of bone and serve as a biological scaffold for calcium phosphate mineral (hydroxyapatite) deposition. Approximately 70 % of the bone volume consists of hydroxyapatite, whereas the rest consists of extracellular matrix proteins produced by osteoblasts. To handle the burden of matrix protein production, osteoblasts have a highly developed ER system, similar to that observed in plasma cells and pancreatic β cells.
In humans, mutations in the COL1A1 and COL1A2 genes often result in a failure to assemble the proper triple-helix formation of type I procollagen and lead to dominantly inherited forms of osteogenesis imperfecta (OI) [78–82]. Although the possible involvement of the UPR in the development of OI is not fully understood, the associated ER stress in fibroblasts derived from patients with OI and OI mice models suggest that the over-activated UPR is causally related to the pathogenesis of OI [78, 83].
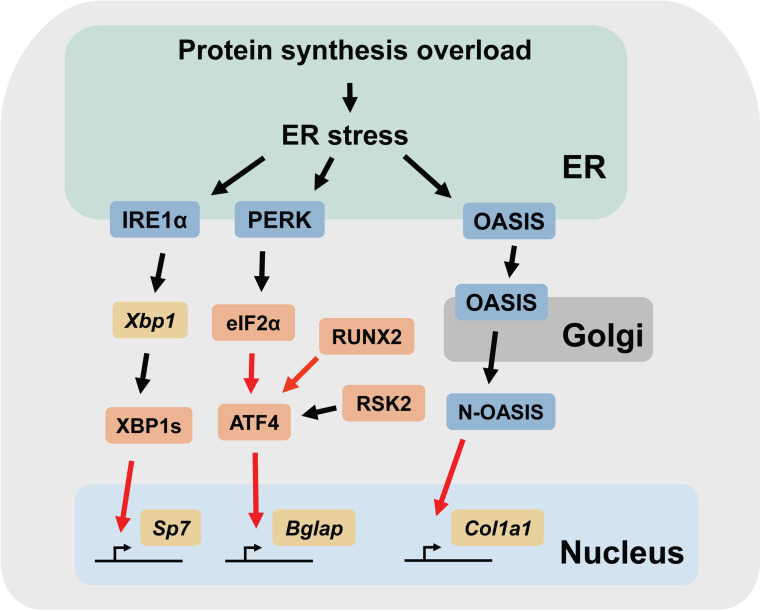
This section mainly describes three mediators of the UPR (IRE1α, PERK, and OASIS, respectively) and their potential roles in osteoblast differentiation, placing emphasis on their functions that are not directly related to the conventional functions of the UPR (Fig. 1).
Fig. 1.
Schematic representation of the UPR signaling pathways involved in osteoblast differentiation. The black arrows indicate post-translational modifications (e.g., phosphorylation, splicing, and translocation). The red arrows indicate transcriptional regulation. Note that other cellular stresses other than protein synthesis overload may also be involved in triggering ER stress. N-OASIS N-terminal fragment of OASIS
The IRE1α-XBP1 pathway promotes Osterix expression
Because the null mutation of Ern1 (encodes IRE1α) leads to embryonic lethality in mice (between 9.5 and 11.5 dpc) [26], the initial study was performed using immortalized mouse embryonic fibroblasts (mEFs) derived from Ern1 −/− embryos and BMP2-induced osteoblastic differentiation assays in vitro [18, 84]. Recombinant BMP2 is widely used to promote osteoblastic differentiation in primary osteoblasts, mEFs, and stromal and osteoblastic cell lines in vitro. BMP2-treated cells express osteoblast-specific transcription factors and osteoblastic extracellular proteins and ultimately form calcified matrix. In wild-type mEFs, the expression of RUNX2 increases approximately 24 h after BMP2 treatment, followed by the induction of the transcripts for type I collagen, an early marker of osteoblast differentiation. Subsequently, the expression of the Osterix transcripts increases and peaks at approximately 72 h after BMP2 treatment. The levels of the total Xbp1, Xbp1s, and Hspa5 transcripts drastically increase at approximately 24 h and reach their plateaus by 48 h after the treatment [18]. These observations indicate that the expression of RUNX2 is induced in BMP-treated mEFs prior to the onset of ER stress, whereas the expression of Osterix transcripts is promoted after ER stress is triggered by the increase in matrix protein synthesis.
The target genes of the transcription factor XBP1s include Dnajc3, Dnajb9, Dnajb11, Edem1, Pdia6, and Serp1 [41], all of which encode proteins that reside in the ER and have functions related to the maintenance of ER functions. Given that mature osteoblasts must have a highly developed ER system to sustain the production of extracellular matrix proteins and that the processing of the transcripts for Xbp1 (and thus the activation of IRE1α) occurs in BMP2-treated mEFs, the potential involvement of the IRE1α-XBP1 pathway was investigated. mEFs derived from IRE1α-deficient embryos (henceforth referred to as Ern1 −/− mEFs) grew normally and did not show any apparent defects under unchallenged conditions. However, they failed to differentiate into the osteoblast lineage when they were incubated with BMP2. Consistently, the levels of the RUNX2, Osterix, type 1 collagen, and osteocalcin transcripts were all significantly reduced in the Ern1 −/− mEFs compared to those in the wild-type mEFs. BMP2-treated wild-type mEFs transfected with siRNAs against either the XBP1 or IRE1α transcripts showed a significant decrease in the expression of the osteoblastic genes, similar to the Ern1 −/− mEFs. However, gene silencing of the transcripts for TRAF2, a target molecule of IRE1α kinase [26], showed no effects on the expression of these genes. These data indicate that XBP1 and not TRAF2 is the molecule downstream of IRE1α that is involved in the defective osteoblastic differentiation in the Ern1 −/− mEFs.
The failure of the Ern1 −/− mEFs to differentiate into the osteoblastic lineage can theoretically be explained by a defect in expanding the capacity of the ER to meet the protein synthesis demands during osteoblast differentiation. However, quite unexpectedly, additional analyses revealed that XBP1s functions as a transcription factor for one of the master regulators of osteoblast differentiation, Osterix [75]. The promoter region of the Sp7 gene (encodes Osterix) has two potential binding sites for XBP1s that are highly conserved across species. XBP1s binds to these sites and promotes Osterix transcription, whereas it has no effects on RUNX2 transcription. The forced expression of Osterix in Ern1 −/− mEFs effectively rescued the expression of alkaline phosphatase, a marker for osteoblasts, in the BMP2-treated cells, suggesting that the defective osteoblastic differentiation of Ern1 −/− mEFs is, at least in part, derived from the reduced expression of Osterix. An analysis of the gene expression patterns in BMP2-treated mEFs also fits with this idea, where the expression of the XBP1s transcripts precedes those of Osterix. Notably, treatment with thapsigargin, a potent chemical inducer of ER stress, alone was sufficient to induce the transcription of the Osterix mRNA in wild-type mEFs that were preincubated with BMP2 for 16 h [18]. This observation indicates that epigenetic factors may also be involved in the transcriptional regulation of Osterix by XBP1s. Due to the embryonic lethality of Ern1 −/− animals, the physiological relevance and the contribution of the IRE1α-XBP1 pathway to skeletal development remains to be addressed.
PERK is required for the late-phase differentiation of osteoblasts
PERK is highly expressed in the pancreas and skeletal tissues in adult mice, indicating that PERK is functionally involved in the regulation of the secretory cells in these tissues. Mutations in the EIF2AK3 gene (encodes PERK) cause a rare autosomal recessive disease, Wolcott–Rallison syndrome [85], which is characterized by early onset diabetes, skeletal dysplasia, and growth retardation. A recent study revealed an association between a haplotype in the EIF2AK3 gene and a lower bone mineral density in humans, suggesting a potential role for PERK in skeletal homeostasis [86].
In mice, the systemic abrogation of the Eif2ak3 gene is not embryonic lethal but leads to the progressive loss of pancreatic β-cells (which secrete insulin) and, consequently, diabetes mellitus [11, 58], as observed in patients with Wolcott–Rallison syndrome. Eif2ak3 −/− mice also exhibit growth retardation and severe osteopenia, which are the other hallmarks of Wolcott–Rallison syndrome. Detailed analyses of the skeletal defects showed that Eif2ak3 −/− mice exhibit markedly reduced bone formation activity, highlighted by lower bone mass, fewer osteoblasts, and reduced mineral deposition compared to the wild-type animals [16]. The expression levels of the transcripts for the markers of mature osteoblasts, including alkaline phosphatase, type I collagen, osteocalcin, and bone sialoprotein, were all reduced in the skeletal tissues collected from Eif2ak3 −/− mice compared to those from wild-type mice. In contrast, there was no significant difference in the transcript levels of osteopontin, an early marker of osteoblast differentiation, in the skeletal tissues of Eif2ak3 −/− and wild-type mice, indicating that PERK is required for the maturation or the late phase differentiation of osteoblasts [16, 87]. These defects were also observed in mutant mice in which PERK is specifically inactivated in osteoblasts [16], indicating that the osteopenia in Eif2ak3 −/− mice was not secondary to the defects in other tissues and that PERK functions in osteoblasts in a cell-autonomous manner. The studies also showed that osteoblasts derived from Eif2ak3 −/− mice have less mature type I collagen and more procollagen than those from the wild-type mice, presumably due to an impairment in procollagen processing and/or in the transport system in the ER-Golgi secretory pathway. In support of the latter hypothesis, electron microscopic analyses revealed grossly enlarged ER in Eif2ak3 −/− mice, most likely due to the retention of newly synthesized proteins in the ER lumen [58, 87].
Because PERK induces the transcription of Atf4 through the phosphorylation of eIF2α [88] and Atf4 −/− mice develop severe osteopenia [77], it is theoretically possible that ATF4 is involved in the skeletal defects observed in Eif2ak3 −/− mice. ATF4 is a transcription factor that belongs to the CREB protein family. ATF4 regulates the transcription of osteoblastic genes, most importantly osteocalcin, through its interaction with RUNX2 [89, 90] and promotes amino acid transport and synthesis of type I collagen in osteoblasts [89, 91]. ATF4 is also a substrate for a growth factor-related kinase, RSK2, in osteoblasts. RSK2 is encoded by the RPS6KA3 gene, mutations in which are causally related to Coffin–Lowry Syndrome, an X-linked mental retardation associated with various skeletal anomalies in humans [92]. Mutant mice lacking either RSK2 or ATF4 exhibit similar skeletal defects, including reduced bone mass and bone formation rate, due to impaired osteoblast differentiation and type I collagen synthesis [77]. Taken together, these observations suggest an intriguing idea that the skeletal phenotypes in both Wolcott–Rallison syndrome and Coffin–Lowry Syndrome may be the result of a failure to induce ATF4 activity, where transcriptional induction is defective in Wolcott–Rallison syndrome and the phosphorylation of ATF4 is defective in Coffin–Lowry Syndrome. However, the potential involvement of ATF4 in the skeletal phenotype of Eif2ak3 −/− mice remains somewhat controversial. Saito et al. demonstrated that the PERK-eIF2α-ATF4 pathway was indeed activated in BMP2-treated osteoblasts and that the expression of ATF4 was significantly reduced in Eif2ak3 −/− osteoblasts compared to wild-type osteoblasts [87]. In a similar vein, BMP-induced ATF4 expression was significantly enhanced by salubrinal, a specific inhibitor of eIF2α phosphatases (which, in turn, enhances the activity of eIF2α) [93, 94]. However, Wei et al. analyzed the expression of ATF4 in calvarial osteoblasts collected from wild-type and Eif2ak3 −/− mice and did not observe changes in the expression or phosphorylation of ATF4 between the two [16]. In support of this view, it has also been shown that the expression of ATF4 in osteoblasts is dependent on RUNX2 [77]. To make the issue even more complicated, a recent study suggests that ATF4 expression in osteoblasts may not be as critical for skeletal development as these studies had indicated (discussed in a later section) [95]. The reasons for these apparent discrepancies are currently unclear; nevertheless, additional studies are warranted to clarify the roles of the PERK-eIF2α-ATF4 pathway in skeletal development in vivo and to define the degree to which the expression of ATF4 in osteoblasts is dependent on the UPR mediated by the PERK axis.
OASIS/CREB3L1, a homologue of ATF6, promotes the transcription of Col1a1
ATF6 is induced by BMP2 in an osteoblastic cell line, MC3T3-E1, in a RUNX2-dependent manner and promotes the transcription of osteocalcin and the calcification of the extracellular matrix [96]. However, mice lacking ATF6 did not display any apparent developmental defects and grew normally, at least under unchallenged conditions [61]. ATF6 plays an important role in optimizing ER functions by inducing genes that protect against chronic ER stress. Accordingly, mice lacking ATF6 are more sensitive to chemically induced ER stress than wild-type mice, due to an impaired adaptation to chronic ER stress [61]. Therefore, additional analyses of Atf6 −/− mice under both unchallenged and challenged conditions may be necessary to uncover the potential role of ATF6 in skeletal development and homeostasis.
OASIS (encoded by Creb3l1) is a membrane-bound transcription factor that shares structural similarities with ATF6. OASIS was originally identified as a molecule that is highly upregulated in astrocytes after long passages in culture (hence the name OASIS, old astrocyte specifically induced substance). In mice, high levels of OASIS expression are present in astrocytes, the digestive tract, salivary glands, and skeletal tissues. In bone, OASIS is specifically expressed in osteoblasts, but not in osteocytes or osteoclasts, suggesting that OASIS has an osteoblast-specific role in skeletal development. Mice lacking OASIS (Creb3l1 −/− mice) were not embryonic lethal, but exhibited growth retardation and often developed spontaneous fractures in the long bones [17]. X-rays and micro-CT analyses revealed that the Creb3l1 −/− mice exhibit severe osteopenia. Consistently, the mutant mice had markedly lower bone mineral density, lower bone volume, and a reduced bone formation rate compared to the wild-type controls. A gene expression analysis revealed that the expression levels of the transcripts for type I collagen (Col1a1 and Col1a2) were significantly lower in the bone tissue from the Creb3l1 −/− mice compared to the wild-type mice, whereas there were no significant changes in the expression levels of the transcripts for RUNX2 or Osterix. Similarly, the loss of OASIS did not affect the expression of other UPR-related genes, including Hspa5, Ddit3, Atf4, Xbp1, P4hb (encodes PDI), Dnajb9 (encodes ERdj4), or Edem1, in the calvaria. These observations indicate that OASIS is not involved in the regulation of osteoblast differentiation and does not affect the other branches of the UPR. The diminished expression of type I collagen in skeletal tissues in the Creb3l1 −/− mice led the authors to ask whether OASIS was involved in the transcriptional regulation of type I collagen in osteoblasts [17]. A nucleotide sequence analysis of the promoter region of the Col1a1 gene uncovered an unfolded protein response element (TGACGTGG)-like sequence (CGACGTGG), which resembles the conventional binding motif of OASIS, in the osteoblast-specific regulatory region (−2.3 kbp) [97]. Luciferase reporter assays and chromatin immunoprecipitation experiments confirmed that OASIS binds to this putative binding motif and promotes the transcription of Col1a1. These results indicate that OASIS functions as a major transcription factor for type I collagen in osteoblasts and that the loss of OASIS in osteoblasts results in severe osteopenia due to impaired collagen production. Notably, a recent study identified siblings with a homozygous deletion in the CREB3L1 gene who exhibited severe osteopenia, thus linking CREB3L1 to a previously unknown form of a recessive OI in humans [98, 99]. This finding underscores that OASIS is an essential and indispensable regulator of the skeletal system in humans.
As observed in Eif2ak3 −/− mice, osteoblasts from Creb3l1 −/− mice also exhibited grossly enlarged ERs, which were not identified in osteocytes or chondrocytes [17]. Because the reduced production of type I collagen (due to reduced transcription) in Creb3l1 −/− mice does not theoretically result in the expansion of the ER lumen in osteoblasts, this observation indicates that OASIS has other functions in addition to regulating collagen production and could potentially regulate transport in the ER-Golgi secretory pathway. In agreement with this idea, a study showed that the introduction of human OASIS (CREB3L1) to Drosophila and human cells promoted the expression of secretory pathway-related genes in these cells [100]. This observation indicates that the functions of OASIS are highly conserved across species and that mouse OASIS is also involved in the transcriptional regulation of the genes that regulate the secretory pathway. In addition, it is also worth noting that the introduction of a transgene consisting of a 2.3 kbp osteoblast-specific type I collagen promoter and the coding sequence for the Creb3l1 gene into Creb3l1 −/− mice was able to fully rescue the osteopenia and also the impaired production of type I collagen and the dilation of the ER [101]. This result strongly suggests that OASIS functions in a cell-autonomous manner to maintain osteoblast functions. However, quite unexpectedly, the growth retardation (lower body weight and shorter body length) observed in the Creb3l1 −/− mice was not rescued by the introduction of the transgene. Notably, the serum levels of growth hormone and insulin-like growth factor-1 were both decreased in the Creb3l1 −/− mice compared to the wild-type mice, and the levels of these hormones were not recovered by the forced expression of OASIS in osteoblasts using the transgene. These results pose an intriguing hypothesis that the OASIS expressed in cells outside the bone, most likely the pituitary gland, where these hormones are produced, indirectly regulates skeletal growth by producing secretory factors. At any rate, these observations suggest that OASIS has diverse and critical functions in regulating skeletal development and postnatal growth.
The UPR in cartilage development and chondrocytes
Most of the long bones are formed through endochondral ossification, a process by which calcified cartilage molds are replaced with bone [69, 72, 102, 103]. The contribution of membranous ossification, by which mesenchymal cells directly differentiate into osteoblasts and form bone, is comparably minor, at least during embryonic skeletal development. Endochondral ossification is responsible for forming the templates for future bones and also for the longitudinal growth of long bones during embryogenesis and postnatal development. Endochondral bone formation commences with the condensation of mesenchymal cells, which subsequently form the cartilage. At the early stage of endochondral ossification, SOX9, a transcription factor of the family of SRY-related high mobility group box of proteins, plays an indispensable role in directing these undifferentiated cells into the chondrocyte lineage [104, 105]. At a later stage, SOX9, along with transcription factors in its family, SOX5 and SOX6, regulates the production of chondrocyte-specific extracellular matrix proteins, including type II collagen and the proteoglycan aggrecan [105]. The primitive cartilage enlarges as the chondrocytes proliferate and deposit extracellular matrix proteins. The chondrocytes in the center of the cartilage further differentiate into hypertrophic chondrocytes and promote the mineralization of their surrounding matrix. The calcified cartilage matrix is subsequently invaded by blood vessels, osteoclasts and osteoblasts and is finally replaced by bone matrix and hydroxyapatite. Not surprisingly, these processes are spatially and temporarily regulated by highly complex signaling pathways mediated by various secreted factors, including PTHrP, Ihh, BMPs/GDFs, and FGF [69, 102, 103].
Recent studies have shown that ER stress may be involved in the pathology of chondrodysplasia and osteoarthritis in humans [106, 107]. There are more than 300 different types of chondrodysplasia, which clearly demonstrates its highly heterogeneous nature [107]. Mutations in the genes encoding extracellular matrix proteins can also lead to chondrodysplasia, and there are several studies showing a causal relationship between ER stress and the pathogenesis of chondrodysplasia caused by mutations in the extracellular matrix proteins [80, 108–111]. Studies have also shown that the factors that are involved in the development of osteoarthritis, such as biomechanical stimuli, IL-1β, and nitric oxide, can also induce the UPR in chondrocytes in vitro [112, 113] and that the UPR is activated in the cartilage tissues of osteoarthritis patients [112, 114]. Accordingly, mice lacking CHOP exhibited less severe cartilage degradation compared to wild-type animals in a knee osteoarthritis mouse model [115], indicating that the UPR mediated by CHOP exacerbates osteoarthritis. However, the precise roles of the UPR in the development of osteoarthritis in humans remain to be elucidated.
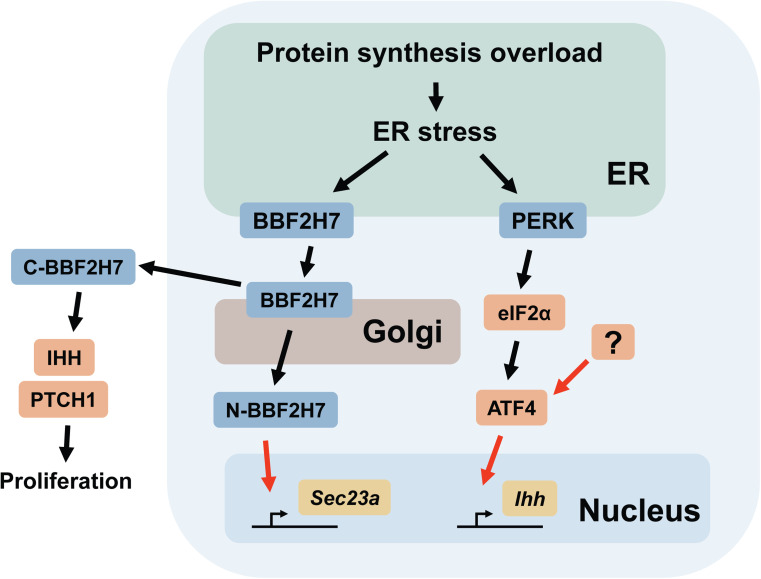
This section mainly describes two branches of the UPR (mediated by IRE1α and BBF2H7) and ATF4, as well as their roles in chondrocyte differentiation (Fig. 2). Although it is not yet clear to what extent the activity of ATF4 is dependent on the PERK-eIF2α pathway in chondrocytes, a discussion on ATF4 is included in this section because of its important functions in skeletal development.
Fig. 2.
Schematic representation of the UPR signaling pathways involved in chondrocyte differentiation. The black arrows indicate post-translational modifications (e.g., phosphorylation, translocation, and proteolytic cleavage). C-BBF2H7 is released extracellularly and forms a complex with IHH and PTCH1. The red arrows indicate transcriptional regulation. The mediator(s) of ATF4 in chondrocytes remain to be elucidated and are indicated as “?”. Note that other cellular stresses other than protein synthesis overload may also be involved in triggering ER stress. N-BBF2H7 N-terminal fragment of BBF2H7, C-BBF2H7 C-terminal fragment of BBF2H7
XBP1 expression in chondrocytes has a minor impact on skeletal development
The potential roles of the IRE1α-XBP1 pathway in cartilage development were studied using mutant mice in which XBP1 was specifically deleted in cartilage tissues under the control of a Col2a1 promoter [116]. The mutant mice did not exhibit any obvious developmental or growth defects compared with their littermate control mice; however, they had shorter long bones (a decrease of approximately 13 %) at 2 weeks of age compared to wild-type mice. The difference in the length of the long bones was transient and resolved by 7 weeks of age. A histological analysis revealed that the mutant mice had a shorter hypertrophic zone and a delay in the formation of the secondary ossification center at 2 weeks of age. The lack of XBP1 in the growth plate also resulted in a moderate decrease in the proliferation of chondrocytes in the proliferating zone, as evaluated by bromodeoxyuridine labeling. In contrast, there were no significant differences in the expression levels of the transcripts for chondrocyte proliferation markers or UPR-related genes. Furthermore, the abrogation of XBP1 in a mouse model of metaphyseal chondrodysplasia type Schmid, a skeletal disorder caused by autosomal dominant mutations in COL10A1 in humans (which results in ER stress in chondrocytes), did not aggravate the skeletal defects observed in these mice [108, 117]. These observations suggest that the expression of XBP1 in chondrocytes has a relatively minor contribution to cartilage development, even when ER stress is enhanced with a mutated collagen gene [118]. Given that the IRE1α-XBP1 pathway is implicated in the pathology of osteoarthritis [114, 119], it will be of interest to evaluate the roles of XBP1 in joint destruction and arthritis using animal models.
BBF2H7/CREB3L2 is critical for transport vesicle formation and chondrocyte proliferation
BBF2H7 (encoded by Creb3l2I) is a membrane-bound transcription factor that shares structural homology with ATF6 and OASIS [64, 120]. The gene was originally identified as a novel gene that fuses with the FUS gene in low-grade fibromyxoid sarcoma, a rare malignant soft tissue tumor [120]. The chimeric FUS/CREB3L2 gene is specific for this tumor and may be involved in its pathogenesis [121]. In mice, the transcripts for BBF2H7 are expressed in a relatively wide variety of tissues, including the heart, lung, liver, kidney, adrenal gland, bladder, cerebrum, ovary, spleen, testis, and prostate; however, they are most highly expressed in the proliferating zone of the growth plate [19, 64]. Accordingly, the cartilage-specific expression of BBF2H7 is mediated by one of the master regulators of chondrogenesis, the SOX9 transcription factor [122].
Mice lacking BBF2H7 (Creb3l2 −/− mice) were not embryonic lethal, but they were significantly smaller than their wild-type littermates, and they died of dyspnea shortly after birth [19]. Whole-mount skeletal staining using alizarin red and alcian blue showed that the Creb3l2 −/− mice had highly under-developed skeletal systems, which were highlighted by short limbs and a severely reduced cartilage matrix. Histologically, the Creb3l2 −/− mice exhibited a structurally disorganized growth plate characterized by a lack of columnar structure and a decrease in the size of the proliferating zone. Most importantly, the chondrocytes in the proliferating zone of the Creb3l2 −/− mice displayed a grossly enlarged ER caused by the accumulation of aggregates of type II collagen and cartilage oligomeric matrix protein, a noncollagenous extracellular matrix protein. These observations indicate that the chondrodysplasia in the Creb3l2 −/− mice was not due to the reduced production of cartilage matrix proteins, but was derived from a defect in the export of the matrix proteins from the ER to the Golgi apparatus. A differential gene expression analysis revealed that the expression of various genes that are related to the secretory pathway was decreased in the chondrocytes collected from the Creb3l2 −/− mice compared to those from the wild-type controls. Among these genes, the expression of Sec23a, which encodes one of the components of the coat protein II (COPII) complex, was the most significantly reduced in chondrocytes lacking BBF2H7. The COPII complex, which consists of four protein subunits (SEC13, SEC31, SEC24, and SEC23A), mediates vesicle budding from the ER and forms COPII vesicles that transport the proteins synthesized in the ER to the Golgi apparatus. A nucleotide sequence analysis revealed that the promoter region of the Sec23a gene contains a cAMP response element-like sequence (TAACGTAA). BBF2H7 specifically binds to this motif and promotes the transcription of Sec23a, indicating that BBF2H7 regulates COPII vesicle formation by promoting Sec23a transcription. An in vitro chondrocyte differentiation model showed that the forced expression of SEC23A effectively recovered the defective cartilage matrix formation in the Creb3l2 −/− mesenchymal cells, similar to reintroducing BBF2H7. The result strongly suggests that the BBF2H7-SEC23A pathway is critically involved in cartilage development through the formation of the COPII vesicles.
SEC23A-deficient mice have recently been documented [123]; however, because the mutant mice were embryonic lethal at midgestation (11.5–12.5 dpc), the skeletal defects observed in Creb3l2 −/− mice were not reproduced in this study. Nevertheless, an analysis of the SEC23A-deficient cells and embryos showed that loss of SEC23A results in impaired collagen secretion and cartilage matrix formation, similar to that observed in BBF2H7-deficient mice and cells. Furthermore, a single amino acid substitution in the SEC23A gene is associated with Craniolenticulosutural dysplasia in humans, an autosomal recessive syndrome characterized by late-closing fontanels, sutural cataracts, facial dysmorphisms, and skeletal defects [124]. Skin fibroblasts collected from patients with this disorder showed a highly dilated ER lumen, similar to that observed in BBF2H7-deficient cells [19]. Taken together, these observations further endorse the idea that the BBF2H7-SEC23A pathway functions as one of the master regulators of cartilage matrix secretion during skeletal development.
An analysis of the Creb3l2 −/− mice demonstrated that BBF2H7 plays an indispensable role in cartilage matrix production by promoting the transcription of Sec23a and, thereby, COPII vesicle formation. However, an additional study unexpectedly revealed an unconventional role for BBF2H7 in regulating chondrocyte proliferation [125]. As with other members of the ATF6/OASIS family genes, BBF2H7 is transported from the ER to Golgi upon ER stress. BBF2H7 is subsequently cleaved in the Golgi, and in a manner similar to that of the other members of the ATF6/OASIS family of proteins, the N-terminal fragment (which contains the bZIP domain) translocates into the nucleus and functions as a transcription factor. The fate and the potential biological functions of the C-terminal fragment, however, remained overlooked and unaddressed. Surprisingly, Saito et al. found that the C-terminal fragment of BBF2H7 is released to the extracellular space and directly binds to Indian hedgehog (encoded by Ihh) and its receptor Patched-1 (encoded by Ptch1) [125]. IHH is synthesized by early hypertrophic chondrocytes and stimulates chondrocyte proliferation and the production of parathyroid hormone-related peptide (PTHrP). PTHrP, in turn, acts on proliferating chondrocytes to keep them from differentiating into hypertrophic chondrocytes and, thereby, indirectly suppresses the production of IHH. Thus, the IHH-PTHrP pathway forms a negative-feedback mechanism that is crucial for determining the length of the long bones [69, 72, 102, 103]. Notably, the binding of the C-terminal fragment of BBF2H7 to IHH and PTCH1 stimulates hedgehog signaling by facilitating the interaction between IHH and PTCH1 and enhances cell proliferation. In fact, the number of cells in the proliferating zone is significantly lower in Creb3l2 −/− mice than in wild-type mice, which is mainly due to decreased chondrocyte proliferation in the proliferating zone [125]. Taken together, these two studies elegantly demonstrate that the N-terminal and C-terminal ends of the cleaved products from BBF2H7 have indispensable and unique roles in skeletal development; the former is to promote the formation of transport vesicles and the latter is to promote chondrocyte proliferation.
Interestingly, the aforementioned low-grade fibromyxoid sarcoma also exhibits a fusion gene of FUS and CREB3L1 (encodes OASIS), although this presentation is much rarer than cases with the typical FUS-CREB3L2 (encodes BBF2H7) fusion gene [126]. This observation may indicate that, while the functions of CREB3L1 and CREB3L2 are widely different from one another during skeletal development, they may contribute in a similar fashion to the tumorigenesis of this rare form of soft-tissue tumor.
ATF4 regulates chondrocyte proliferation and differentiation
As described in the previous section, ATF4 has been implicated as a crucial regulator of osteoblast differentiation by promoting the expression of osteoblast-specific genes and collagen synthesis [77, 89]. Interestingly, ATF4 is also expressed in proliferative and prehypertrophic chondrocytes, and, as indicated by its expression pattern, an analysis of Atf4 −/− mice showed that ATF4 is also critically involved in the regulation of chondrocyte differentiation and proliferation [127]. Atf4 −/− mice exhibit delayed ossification and low bone mass as well as dwarfism and short limbs [77], which suggests that endochondral ossification is defective in these mice. The study by Wang et al. showed that the Atf4 −/− mice have reduced chondrocyte proliferation and delayed hypertrophic mineralization and that ATF4 functions as an essential transcription factor for Ihh in chondrocytes [127]. Reactivation of hedgehog signaling by purmorphamine, an agonist for Smoothened, a key component of the hedgehog signaling pathway [128], was able to rescue the limb defects in the Atf4 −/− embryos, suggesting that the dwarfism in the Atf4 −/− mice was derived from suppressed Ihh expression due to the lack of ATF4 in chondrocytes. Surprisingly, an additional study using a mutant mouse model in which ATF4 is specifically expressed in chondrocytes under the control of a Col2a1 promoter in Atf4 −/− mice (Atf4 −/−; Col2a1-Atf4 mice) showed that the expression of ATF4 in chondrocytes is sufficient to rescue the developmental and skeletal growth defects in Atf4 −/− mice [95]. Although this observation does not necessarily negate the autonomous functions of ATF4 in osteoblasts, the data suggest that the activity of ATF4 in chondrocytes can indirectly regulate osteoblast activity and bone mass through the production of IHH. A study showing that chondrocyte-specific abrogation of the Ihh gene resulted in a continuous loss of trabecular bone further supports the idea that chondrocyte-derived IHH is critically involved in the maintenance of bone mass by promoting osteoblast activity [129].
As UPR-related genes are induced during chondrogenesis [19], it is likely that PERK is also activated during that process and enhances the transcription of Atf4, at least to some degree. However, little is currently known about the mechanism by which the transcription of Atf4 is regulated in chondrocytes and whether it is dependent on or independent of PERK activity. Although Eif2ak3 −/− mice are defective in both membranous and endochondral ossification and exhibit growth retardation [58, 87], the potential contribution of PERK to chondrocyte differentiation has not been extensively investigated. Further study is warranted to elucidate the contribution of the UPR branch mediated by the PERK axis, or lack of it, to the transcriptional activation of Atf4 during chondrocyte differentiation.
The UPR in osteoclastogenesis
Osteoclasts are the only cells that are capable of bone resorption in vivo and, therefore, exhibit highly specialized functions [130–132]. The progenitor cells of osteoclasts are derived from monocytes and macrophage lineage cells. It is conceivable that bone-forming osteoblasts and cartilage-forming chondrocytes undergo ER stress during skeletal formation, and the roles of the UPR in these cells have been widely investigated; however, there are only a few studies in the literature that explore the potential contribution of the UPR to osteoclastogenesis. Osteoclast progenitor cells express the receptor activator of NFκB (RANK) on the cell surface, which functions as a receptor for RANK ligand (RANKL), a membrane-bound ligand expressed on osteoblasts and osteocytes. Therefore, cell–cell interactions between osteoclast precursors and osteoblasts/osteocytes is deemed essential for the binding of RANK and RANKL. Upon the binding of the ligand, RANK recruits adapter proteins, such as TRAF6, and activates various signaling molecules, including MAP kinases and NFκB. The intracellular signaling elicited by RANK ultimately induces the expression of NFATc1, the master transcriptional regulator of osteoclastogenesis, through the signaling cascades mediated by NFκB and c-Fos [133]. To function as a transcription factor, NFATc1 must be dephosphorylated by the serine/threonine phosphatase Calcineurin, which is a downstream target of the Ca2+-calmodulin pathway. NFATc1 dephosphorylation reveals its nuclear-localization signal and allows it to translocate to the nucleus. Activated RANK also induces Ca2+ oscillations, which are mediated by phospholipase Cγ (PLCγ) and inositol trisphosphate receptors (IP3Rs), Ca2+-channels located in the membrane of the ER [134]. The activation of Calcineurin, which is dependent on Ca2+-calmodulin signaling, is elicited by the outflow of Ca2+ from the ER. NFATc1 binds to the promotor of the Nfatc1 gene and stimulates its own transcription in a positive-feedback manner. The robust induction of NFATc1 induces the expression of osteoclast-specific genes in osteoclast precursors, resulting in the formation of mature, multinucleated osteoclasts [135, 136].
Mature, activated osteoclasts release hydrogen ions to absorb the mineralized bone matrix and secrete proteases to degrade the bone matrix proteins. Because the production of increased amounts of these enzymes could potentially lead to ER stress, it was initially assumed that the UPR was activated towards the end of and not during osteoclast differentiation. However, in contrast to this assumption, the UPR is transiently induced during osteoclast differentiation and subsides as osteoclastogenesis proceeds [20, 21]. An in vitro osteoclast formation assay showed that all three major branches of the UPR are activated in BMMs and in an osteoclast precursor-like cell line, RAW264.7 cells, 2–3 days after RANKL stimulation, suggesting that ER stress occurs during osteoclastogenesis and that the UPR is involved in the regulation of osteoclastogenesis.
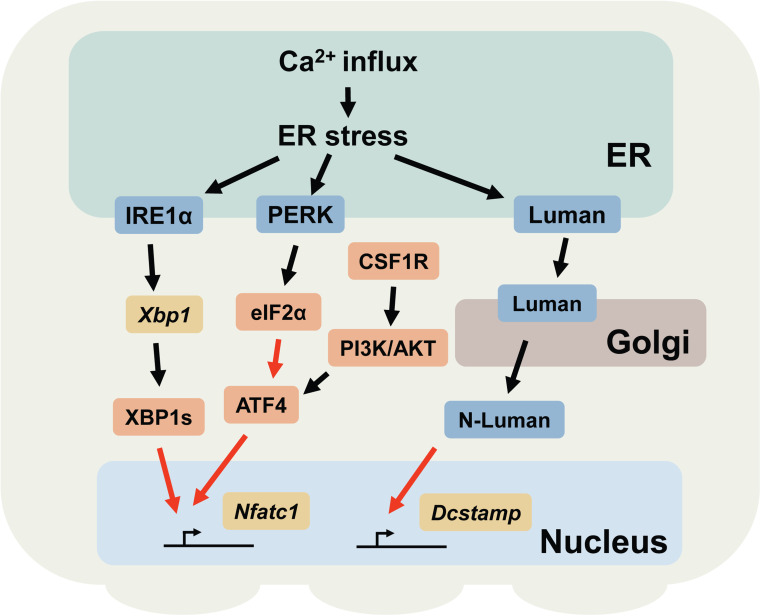
This section mainly describes two branches of the UPR (mediated by IRE1α and Luman, respectively) and ATF4, as well as their roles in osteoclast differentiation (Fig. 3). As is the case for chondrocytes, to what extent the activity of ATF4 is mediated by the PERK-eIF2α pathway in osteoclasts is not fully understood. Nevertheless, a discussion on ATF4 is included in this section because the PERK axis of the UPR is also activated during osteoclastogenesis [20].
Fig. 3.
Schematic representation of the UPR signaling pathways involved in osteoclast differentiation. The black arrows indicate post-translational modifications (e.g., phosphorylation, splicing, and translocation). The red arrows indicate transcriptional regulation. Note that other cellular stresses other than Ca2+ influx may also be involved in triggering ER stress. N-Luman N-terminal fragment of Luman
The IRE1α-XBP1 pathway mediates NFATc1 expression
The crucial roles of the IRE1α-XBP1 pathway in osteoclastogenesis have recently been reported [20]. The splicing of Xbp1 transcripts and translocation of XBP1s into the nucleus are transiently induced in BMMs 2–3 days after RANKL stimulation. Pharmacological inhibition of the IRE1α endonuclease or gene silencing of the transcripts for Ern1 or Xbp1 markedly suppressed osteoclastogenesis. Accordingly, the mutant mice in which bone marrow cells are devoid of IRE1α showed an increase in bone mass due to decreased osteoclastic bone resorption. Notably, two UPR element-like sequences (GGAAG) [42] flank the tandem NFATc1-binding motifs in the promoter region of the Nfatc1 gene. Chromatin immunoprecipitation and luciferase reporter experiments have confirmed that XBP1s binds to these UPR element-like sequences and enhances the transcription of Nfatc1. These data suggest that the IRE1α-XBP1 pathway functions as a positive regulator of osteoclastogenesis by enhancing the transcription of Nfatc1 in osteoclast precursors. In contrast to osteoblasts and chondrocytes, osteoclasts do not produce significant amounts of secreted proteins during differentiation; therefore, it was assumed that ER stress is triggered by a stimulus other than the over-production of secreted proteins. It is known that the ER is a major store for Ca2+ within cells, and fluctuations in ER Ca2+ levels can disturb the microenvironment of the ER lumen and result in ER stress [137, 138]. As mentioned above, activated RANK induces Ca2+ outflow from the ER and induces Ca2+ oscillations during osteoclastogenesis. It is thus conceivable that ER stress is induced by RANK signaling-mediated Ca2+ oscillations. Our observations suggested that this is in fact the case [20]. The induction of Ca2+ outflow from the ER by ATP, an agonist for the phospholipase C-coupled P2Y receptor [139, 140], concomitantly induced ER stress in an osteoclast precursor-like cell line, RAW264.7 cells. Furthermore, the processing of Xbp1 transcripts, which reflects the degree of ER stress, was significantly reduced in osteoclast precursors lacking the calcium channels IP3R1 and two compared to that in wild-type cells. These observations indicate that the Ca2+ outflow from ER triggered by RANK signaling is, at least in part, responsible for triggering ER stress during osteoclastogenesis. Taken together, these data suggest that Ca2+ outflow from the ER during osteoclastogenesis has a dual role in activating NFATc1; one as an activator of NFATc1 through the Ca2+-calmodulin pathway and the other as a transcriptional activator of the Nfatc1 gene through the IRE1α-XBP1 pathway.
ATF4 functions as a transcription factor for Nfatc1
Among the other UPR-related molecules, ATF4 is also critically involved in the regulation of osteoclastogenesis. ATF4 is a transcription factor that is critically involved in the development of osteoblasts and chondrocytes, as highlighted by the severely hampered skeletal formation in Atf4 −/− mice [77, 127] (and as discussed in former sections). Interestingly, ATF4 induces the expression of NFATc1 by directly enhancing the transcription of the Nfatc1 gene. Accordingly, osteoclast precursors lacking ATF4 are defective in forming multinucleated osteoclasts, even when they are cultured with a high concentration of RANKL in vitro (up to 200 ng/ml) [141]. A histological analysis revealed that Atf4 −/− mice have fewer tartrate-resistant acid phosphatase (TRACP)-positive osteoclasts than wild-type controls. Furthermore, a transgenic mouse line that expresses ATF4 under the control of an Acp5 (encodes TRACP) promoter exhibits severe bone loss due to increased osteoclastic bone resorption. Although the translation of Atf4 is efficiently activated by the PERK-eIF2α pathway, it was shown that the expression of ATF4 is mainly regulated by the CSF1R (the receptor for CSF1, also known as c-fms)-PI3K-AKT signaling pathway in osteoclast precursors. However, given that the transcription of Atf4 is enhanced in a manner similar to that of other UPR-related genes [20] and that osteoclast activity is reduced in mice lacking PERK [16], it is possible that the expression of ATF4 is, at least in part, regulated by ER stress during osteoclast differentiation.
In addition to the pro-osteoclastogenic effects of ATF4 [141], pharmacological inhibition of eIF2α phosphatase by Salubrinal and Guanabenz has been shown to suppress osteoclastogenesis through reducing NFATc1 expression [142–144]. Because increased phosphorylation of eIF2α theoretically leads to enhanced ATF4 activity, these studies implicate that the PERK-eIF2α pathway exhibits opposing effects on NFATc1 expression and osteoclastogenesis. Further studies are required to resolve the apparent discrepancies and to elucidate to what degree the PERK-mediated UPR affects the activities of eIF2α and ATF4 in osteoclast precursors following RANKL stimulation.
Luman/CREB3 regulates the cell–cell fusion of osteoclast precursors
Luman (encoded by Creb3) is a transmembrane transcription factor that belongs to the ATF6/OASIS family of proteins. It is highly expressed in trigeminal ganglion neurons and monocytes. Like other members of this gene family, Luman resides in the ER and is transferred to the Golgi apparatus upon ER stress. Luman is proteolytically cleaved in the Golgi apparatus to release an N-terminal fragment, which subsequently translocates into the nucleus and functions as a transcription factor [62, 145]. The first line of evidence of the potential involvement of Luman in osteoclastogenesis came from a study to identify the binding partners for Luman using a yeast-two-hybrid analysis [146]. In this study, DC-STAMP, a molecule implicated in cell-to-cell fusion during osteoclast and giant cell development [147], was found to form a complex with Luman [146]. The formation of the Lumen-DC-STAMP complex renders Luman resistant to proteolytic cleavage (and thereby inhibits the release of the cytoplasmic domain of Lumen) and presumably suppresses the transcription mediated by Luman [21, 146]. Similar to the splicing of Xbp1 transcripts, Luman is induced and activated 24–48 h after RANKL stimulation, further corroborating the idea that the UPR is triggered in the early stage of osteoclastogenesis [20]. The promoter region of the Dcstamp gene contains two cAMP response element-like binding sites, which are potential binding motifs for Luman. A luciferase reporter assay and gel shift assay showed that Luman binds to these sites and promotes the transcription of Dcstamp. shRNA-mediated gene silencing of the Luman transcripts in BMMs significantly suppressed the expression of the Dcstamp transcripts, but not the Nfatc1 transcripts. Accordingly, the BMMs transfected with shRNAs against the Luman transcripts failed to form multinucleated osteoclasts but were able to differentiate into TRACP-positive mononucleated osteoclasts when they were incubated in the presence of CSF1 and RANKL. These data show that Luman has a dual function in regulating DC-STAMP activity: one is to function as a transcription factor to induce DC-STAMP expression and the other is to stabilize DC-STAMP proteins by forming a complex [21]. In contrast, Luman does not appear to participate in the regulation of osteoclast differentiation per se, as evidenced by the unaltered expression levels of Nfatc1 transcripts in Dcstamp-suppressed BMMs. While it is strongly indicated that Luman is involved in the fusion of osteoclast precursors, it is not clear to what extent Luman contributes to the transcriptional regulation of Dcstamp under physiological conditions, given that c-Fos and NFATc1 are also shown to drive the transcription of Dcstamp [148]. Nevertheless, because the defect in forming multinucleated osteoclasts does not have a profound impact on overall osteoclastic bone resorption in vivo [147], it is possible that a lack of Luman has a minor impact on skeletal development and bone homeostasis. The generation of Luman-deficient mice will help to address these issues.
Non-cell-autonomous contribution of the UPR to the regulation of osteoclast differentiation
In addition to the cell-autonomous participation of the UPR in osteoclastogenesis through the IRE1α-XBP1 pathway, Luman, and ATF4, there are only a few studies that suggest a non-cell-autonomous contribution of the UPR to osteoclast differentiation [84, 149]. Comparative gene expression analyses using the transcripts from BMP2-treated wild-type and Ern1 −/− mEFs (which lack IRE1α) identified Pth1r (encodes parathyroid hormone 1 receptor, PTH1R) as a candidate target gene for XBP1s [84]. The expression of Pth1r transcripts is induced in wild-type mEFs after BMP2 treatment; however, the increase in Pth1r transcripts was markedly suppressed in Ern1 −/− mEFs. Chromatin immunoprecipitation experiments and luciferase reporter assays showed that XBP1s binds to the promoter of the Pth1r gene and promotes the transcription of Pth1r. BMP2-treated mEFs transfected with an siRNA against Xbp1 transcripts are less sensitive to parathyroid hormone (PTH) and, thus, expressed fewer RANKL transcripts upon stimulation with PTH, a potent inducer of RANKL in osteoblasts and stromal cells [150], compared to wild-type mEFs. Consequently, mEFs transfected with siRNAs against either Ern1 or Xbp1 transcripts were incapable of supporting PTH-mediated osteoclastogenesis due to the impaired expression of RANKL. Similarly, a different study showed that multiple myeloma cells enhance the expression of RANKL in bone marrow stromal cells in an XPP1-dependent manner, thus supporting osteoclast formation in multiple myeloma bone lesions [149]. The study did not identify the pathway that triggers XBP1s expression in stromal cells or the mechanism by which XBP1s regulates the expression of RANKL. Nevertheless, these studies show that osteoclastogenesis can be indirectly regulated by the UPR in a non-cell-autonomous manner.
Conclusions and future directions
Studies over the past decade have clearly shown that the UPR is a critical regulator of skeletal development and homeostasis (some of the most important findings are summarized in Table 1). From an evolutionary biology perspective, the UPR must have developed to handle ER stress and recover cellular functions as the need for producing extracellular matrix proteins and secreted proteins increase. Therefore, it is quite intriguing that the UPR machinery in mammals (and possibly in other vertebrates) has acquired functions that are apparently unrelated to the conventional roles of the UPR. In the case of the skeletal system, osteoblasts, chondrocytes, and osteoclasts all have adopted the UPR in such a way that it not only functions passively to normalize the ER homeostasis upon ER stress but also actively regulates cell differentiation and maturation. It is thus tempting to speculate that these cells use ER stress as a temporal and spatial cue to determine when to promote the expression of a certain set of genes during differentiation.
Table 1.
UPR-related molecules and their suggested functions in skeletal development
| Functions | |
|---|---|
| XBP1 |
Promotes the transcription of Sp7 (encodes Osterix) [18], Nfatc1 [20], and Pth1r (encodes parathyroid hormone 1 receptor) [84] Directly or indirectly regulates the expression of RANKL [149] Potentially regulates longitudinal bone growth [116] |
| ATF4 |
Interacts with RUNX2 and promotes the transcription of osteoblastic genes, including Bglap (encodes osteocalcin) [89–91] Regulates chondrocyte proliferation through promoting the transcription of Ihh [127] Promotes the transcription of Nfatc1 [141] |
| OASIS/CREBL1 |
Promotes the transcription of Col1a1 [17] Potentially regulates the production of growth hormone and insulin-like growth factor-1 [101] |
| BBF2H7/CREB3L2 |
Promotes the transcription of Sec23a [19] Regulates chondrocyte proliferation through activating hedgehog signaling [125] |
| Luman/CREB3 |
Forms a complex with DC-STAMP [21, 146] Promotes the transcription of Dcstamp [21] |
| eIF2α | Suppresses NFATc1 expression [142–144] |
In addition to the physiological roles of the UPR in skeletal development and homeostasis, we still do not fully understand how the UPR contributes to the pathogenesis of skeletal diseases, including osteoarthritis, osteoporosis, congenital skeletal disorders, and bone metastasis, or whether the UPR could be a molecular target for these disorders. Nevertheless, because over-activated UPR signaling is often associated with certain types of skeletal disorders, such as osteoarthritis [112, 114] and OI [78, 83], pharmacological suppression of UPR signaling could potentially alleviate the symptoms of these disorders. Furthermore, it would also be interesting to investigate whether the UPR is involved in the pathology of bone tumors, such as osteosarcoma and chondrosarcoma. Because these tumors often produce an unusually large amount of extracellular matrix, it is likely that these tumors are equipped with a secretory system that is highly resistant to protein synthesis overload and various cellular stresses. Therefore, the branches of the UPR could be a therapeutic target in these tumors, as is the case for immunoglobulin-producing myeloma [151–153]. Deepening our understanding of the network between skeletal homeostasis and the UPR will provide important biological insights into skeletal metabolism, as well as the basis for therapeutic interventions for skeletal disorders.
Acknowledgments
We would like to thank Yoshiaki Toyama, Masaya Nakamura, Morio Matsumoto, and the members of the Molecular Bone Biology Laboratory at the Department of Orthopedic Surgery, Keio University School of Medicine for their comments and suggestions for the present review. This work was supported in part by MEXT KAKENHI (24390358).
Abbreviations
- ATF6
Activating transcription factor 6
- BBF2H7
BBF2 human homolog on chromosome 7
- BiP
Immunoglobulin heavy chain-binding protein
- bZIP
Basic leucine-zipper
- CHOP
CCAAT/enhancer-binding protein homologous protein
- CREB
cAMP response element-binding protein
- eIF2α
Eukaryotic translation initiation factor-2
- ERAD
ER-associated degradation
- ER
Endoplasmic reticulum
- IRE1α
Inositol-requiring protein-1α
- mEFs
Mouse embryonic fibroblasts
- OASIS
Old astrocyte specifically induced substance
- OI
Osteogenesis imperfecta
- PERK
Pancreatic ER kinase
- RIDD
Regulated IRE1-dependent decay of mRNA
- TRACP
Tartrate-resistant acid phosphatase
- TRAF2
TNF receptor-associated factor 2
- UPR
Unfolded protein response
- XBP1
X-box binding protein 1
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14(1):20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35(5):551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115(10):2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 5.Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146(6):743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 6.Hollien J. Evolution of the unfolded protein response. Biochim Biophys Acta. 2013;1833(11):2458–2463. doi: 10.1016/j.bbamcr.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7(9):880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4(4):321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 9.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Lipson KL, Fonseca SG, Ishigaki S, Nguyen LX, Foss E, Bortell R, Rossini AA, Urano F. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4(3):245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. doi: 10.1016/S1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 12.Binet F, Sapieha P. ER stress and angiogenesis. Cell Metab. 2015;22(4):560–575. doi: 10.1016/j.cmet.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 13.So JS, Hur KY, Tarrio M, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Lichtman AH, Iwawaki T, Glimcher LH, Lee AH. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16(4):487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal RL, Figueroa A, Court FA, Thielen P, Molina C, Wirth C, Caballero B, Kiffin R, Segura-Aguilar J, Cuervo AM, Glimcher LH, Hetz C. Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum Mol Genet. 2012;21(10):2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinon F, Glimcher LH. Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr Opin Immunol. 2011;23(1):35–40. doi: 10.1016/j.coi.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei J, Sheng X, Feng D, McGrath B, Cavener DR. PERK is essential for neonatal skeletal development to regulate osteoblast proliferation and differentiation. J Cell Physiol. 2008;217(3):693–707. doi: 10.1002/jcp.21543. [DOI] [PubMed] [Google Scholar]
- 17.Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Wanaka A, Imaizumi K. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009;11(10):1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- 18.Tohmonda T, Miyauchi Y, Ghosh R, Yoda M, Uchikawa S, Takito J, Morioka H, Nakamura M, Iwawaki T, Chiba K, Toyama Y, Urano F, Horiuchi K. The IRE1alpha-XBP1 pathway is essential for osteoblast differentiation through promoting transcription of Osterix. EMBO Rep. 2011;12(5):451–457. doi: 10.1038/embor.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito A, Hino S, Murakami T, Kanemoto S, Kondo S, Saitoh M, Nishimura R, Yoneda T, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Imaizumi K. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol. 2009;11(10):1197–1204. doi: 10.1038/ncb1962. [DOI] [PubMed] [Google Scholar]
- 20.Tohmonda T, Yoda M, Iwawaki T, Matsumoto M, Nakamura M, Mikoshiba K, Toyama Y, Horiuchi K. IRE1alpha/XBP1-mediated branch of the unfolded protein response regulates osteoclastogenesis. J Clin Invest. 2015;125(8):3269–3279. doi: 10.1172/JCI76765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanemoto S, Kobayashi Y, Yamashita T, Miyamoto T, Cui M, Asada R, Cui X, Hino K, Kaneko M, Takai T, Matsuhisa K, Takahashi N, Imaizumi K. Luman is involved in osteoclastogenesis through the regulation of DC-STAMP expression, stability and localization. J Cell Sci. 2015;128(23):4353–4365. doi: 10.1242/jcs.176057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91(4):1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 23.Vidal RL, Hetz C. Unspliced XBP1 controls autophagy through FoxO1. Cell Res. 2013;23(4):463–464. doi: 10.1038/cr.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172(4):565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, Lemberg MK. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014;33(21):2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 27.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Li X, Cai MY, Ma K, Yang J, Zhou J, Fu W, Wei FZ, Wang L, Xie D, Zhu WG. XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res. 2013;23(4):491–507. doi: 10.1038/cr.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001;276(17):13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- 30.Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39(5):245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Coelho DS, Domingos PM. Physiological roles of regulated Ire1 dependent decay. Front Genet. 2014;5:76. doi: 10.3389/fgene.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313(5783):104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 33.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186(3):323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, Ron D, Tabas I, Hussain MM. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7(5):445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hur KY, So JS, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Iwawaki T, Glimcher LH, Lee AH. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209(2):307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho JA, Lee AH, Platzer B, Cross BC, Gardner BM, De Luca H, Luong P, Harding HP, Glimcher LH, Walter P, Fiebiger E, Ron D, Kagan JC, Lencer WI. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe. 2013;13(5):558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Osorio F, Tavernier SJ, Hoffmann E, Saeys Y, Martens L, Vetters J, Delrue I, De Rycke R, Parthoens E, Pouliot P, Iwawaki T, Janssens S, Lambrecht BN. The unfolded-protein-response sensor IRE-1alpha regulates the function of CD8alpha + dendritic cells. Nat Immunol. 2014;15(3):248–257. doi: 10.1038/ni.2808. [DOI] [PubMed] [Google Scholar]
- 38.Benhamron S, Hadar R, Iwawaky T, So JS, Lee AH, Tirosh B. Regulated IRE1-dependent decay participates in curtailing immunoglobulin secretion from plasma cells. Eur J Immunol. 2014;44(3):867–876. doi: 10.1002/eji.201343953. [DOI] [PubMed] [Google Scholar]
- 39.Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, Papa FR, Oakes SA. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338(6108):818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138(3):562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27(1):53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24(24):4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320(5882):1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang ZV, Deng Y, Gao N, Pedrozo Z, Li DL, Morales CR, Criollo A, Luo X, Tan W, Jiang N, Lehrman MA, Rothermel BA, Lee AH, Lavandero S, Mammen PP, Ferdous A, Gillette TG, Scherer PE, Hill JA. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014;156(6):1179–1192. doi: 10.1016/j.cell.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, Mai J, Shen H, Hu DZ, Adoro S, Hu B, Song M, Tan C, Landis MD, Ferrari M, Shin SJ, Brown M, Chang JC, Liu XS, Glimcher LH. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508(7494):103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwakoshi NN, Pypaert M, Glimcher LH. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J Exp Med. 2007;204(10):2267–2275. doi: 10.1084/jem.20070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Todd DJ, McHeyzer-Williams LJ, Kowal C, Lee AH, Volpe BT, Diamond B, McHeyzer-Williams MG, Glimcher LH. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206(10):2151–2159. doi: 10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bettigole SE, Lis R, Adoro S, Lee AH, Spencer LA, Weller PF, Glimcher LH. The transcription factor XBP1 is selectively required for eosinophil differentiation. Nat Immunol. 2015;16(8):829–837. doi: 10.1038/ni.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14(2):152–157. [PMC free article] [PubMed] [Google Scholar]
- 52.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 53.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 54.Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23(1):169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318(5):1351–1365. doi: 10.1016/S0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 56.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12(7):982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ron D, Harding HP (2012) Protein-folding homeostasis in the endoplasmic reticulum and nutritional regulation. Cold Spring Harb Perspect Biol 4(12). doi:10.1101/cshperspect.a013177 [DOI] [PMC free article] [PubMed]
- 58.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22(11):3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 60.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 61.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Liang G, Audas TE, Li Y, Cockram GP, Dean JD, Martyn AC, Kokame K, Lu R. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Mol Cell Biol. 2006;26(21):7999–8010. doi: 10.1128/MCB.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, Iseki K, Wanaka A, Imaizumi K. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol. 2005;7(2):186–194. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- 64.Kondo S, Saito A, Hino S, Murakami T, Ogata M, Kanemoto S, Nara S, Yamashita A, Yoshinaga K, Hara H, Imaizumi K. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol. 2007;27(5):1716–1729. doi: 10.1128/MCB.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DenBoer LM, Hardy-Smith PW, Hogan MR, Cockram GP, Audas TE, Lu R. Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem Biophys Res Commun. 2005;331(1):113–119. doi: 10.1016/j.bbrc.2005.03.141. [DOI] [PubMed] [Google Scholar]
- 66.Omori Y, Imai J, Watanabe M, Komatsu T, Suzuki Y, Kataoka K, Watanabe S, Tanigami A, Sugano S. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 2001;29(10):2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagamori I, Yabuta N, Fujii T, Tanaka H, Yomogida K, Nishimune Y, Nojima H. Tisp40, a spermatid specific bZip transcription factor, functions by binding to the unfolded protein response element via the Rip pathway. Genes Cells. 2005;10(6):575–594. doi: 10.1111/j.1365-2443.2005.00860.x. [DOI] [PubMed] [Google Scholar]
- 68.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13(1):27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 69.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 70.Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99(5):1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 71.Karsenty G. Bone formation and factors affecting this process. Matrix Biol. 2000;19(2):85–89. doi: 10.1016/S0945-053X(00)00053-6. [DOI] [PubMed] [Google Scholar]
- 72.Hartmann C. Transcriptional networks controlling skeletal development. Curr Opin Genet Dev. 2009;19(5):437–443. doi: 10.1016/j.gde.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 73.Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13(7):791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 74.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 75.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 76.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 77.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin–Lowry Syndrome. Cell. 2004;117(3):387–398. doi: 10.1016/S0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 78.Chessler SD, Wallis GA, Byers PH. Mutations in the carboxyl-terminal propeptide of the pro alpha 1(I) chain of type I collagen result in defective chain association and produce lethal osteogenesis imperfecta. J Biol Chem. 1993;268(24):18218–18225. [PubMed] [Google Scholar]
- 79.Pace JM, Wiese M, Drenguis AS, Kuznetsova N, Leikin S, Schwarze U, Chen D, Mooney SH, Unger S, Byers PH. Defective C-propeptides of the proalpha2(I) chain of type I procollagen impede molecular assembly and result in osteogenesis imperfecta. J Biol Chem. 2008;283(23):16061–16067. doi: 10.1074/jbc.M801982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boot-Handford RP, Briggs MD. The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res. 2010;339(1):197–211. doi: 10.1007/s00441-009-0877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dalgleish R. The human type I collagen mutation database. Nucleic Acids Res. 1997;25(1):181–187. doi: 10.1093/nar/25.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dalgleish R. The human collagen mutation database 1998. Nucleic Acids Res. 1998;26(1):253–255. doi: 10.1093/nar/26.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lisse TS, Thiele F, Fuchs H, Hans W, Przemeck GK, Abe K, Rathkolb B, Quintanilla-Martinez L, Hoelzlwimmer G, Helfrich M, Wolf E, Ralston SH, Hrabe de Angelis M. ER stress-mediated apoptosis in a new mouse model of osteogenesis imperfecta. PLoS Genet. 2008;4(2):e7. doi: 10.1371/journal.pgen.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tohmonda T, Yoda M, Mizuochi H, Morioka H, Matsumoto M, Urano F, Toyama Y, Horiuchi K. The IRE1alpha-XBP1 pathway positively regulates parathyroid hormone (PTH)/PTH-related peptide receptor expression and is involved in pth-induced osteoclastogenesis. J Biol Chem. 2013;288(3):1691–1695. doi: 10.1074/jbc.C112.424606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Julier C, Nicolino M. Wolcott–Rallison syndrome. Orphanet J Rare Dis. 2010;5:29. doi: 10.1186/1750-1172-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J, Hoppman N, O’Connell JR, Wang H, Streeten EA, McLenithan JC, Mitchell BD, Shuldiner AR. A functional haplotype in EIF2AK3, an ER stress sensor, is associated with lower bone mineral density. J Bone Miner Res. 2012;27(2):331–341. doi: 10.1002/jbmr.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, Imaizumi K. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem. 2011;286(6):4809–4818. doi: 10.1074/jbc.M110.152900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 89.Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem. 2005;280(35):30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- 90.Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol Biol Cell. 2008;19(12):5373–5386. doi: 10.1091/mbc.E08-03-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem. 2004;279(45):47109–47114. doi: 10.1074/jbc.M410010200. [DOI] [PubMed] [Google Scholar]
- 92.Pereira PM, Schneider A, Pannetier S, Heron D, Hanauer A. Coffin–Lowry syndrome. Eur J Hum Genet. 2010;18(6):627–633. doi: 10.1038/ejhg.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tanaka K, Kaji H, Yamaguchi T, Kanazawa I, Canaff L, Hendy GN, Sugimoto T. Involvement of the osteoinductive factors, Tmem119 and BMP-2, and the ER stress response PERK-eIF2alpha-ATF4 pathway in the commitment of myoblastic into osteoblastic cells. Calcif Tissue Int. 2014;94(4):454–464. doi: 10.1007/s00223-013-9828-1. [DOI] [PubMed] [Google Scholar]
- 94.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307(5711):935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 95.Wang W, Lian N, Ma Y, Li L, Gallant RC, Elefteriou F, Yang X. Chondrocytic Atf4 regulates osteoblast differentiation and function via Ihh. Development. 2012;139(3):601–611. doi: 10.1242/dev.069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jang WG, Kim EJ, Kim DK, Ryoo HM, Lee KB, Kim SH, Choi HS, Koh JT. BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J Biol Chem. 2012;287(2):905–915. doi: 10.1074/jbc.M111.253187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossert J, Eberspaecher H, de Crombrugghe B. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129(5):1421–1432. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Symoens S, Malfait F, D’Hondt S, Callewaert B, Dheedene A, Steyaert W, Bachinger HP, De Paepe A, Kayserili H, Coucke PJ. Deficiency for the ER-stress transducer OASIS causes severe recessive osteogenesis imperfecta in humans. Orphanet J Rare Dis. 2013;8:154. doi: 10.1186/1750-1172-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shaker JL, Albert C, Fritz J, Harris G (2015) Recent developments in osteogenesis imperfecta. F1000Res 4 (F1000 Faculty Rev):681 [DOI] [PMC free article] [PubMed]
- 100.Fox RM, Hanlon CD, Andrew DJ. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J Cell Biol. 2010;191(3):479–492. doi: 10.1083/jcb.201004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murakami T, Hino S, Nishimura R, Yoneda T, Wanaka A, Imaizumi K. Distinct mechanisms are responsible for osteopenia and growth retardation in OASIS-deficient mice. Bone. 2011;48(3):514–523. doi: 10.1016/j.bone.2010.10.176. [DOI] [PubMed] [Google Scholar]
- 102.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 103.Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013;5(1):a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 105.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu-Bryan R, Terkeltaub R. Emerging regulators of the inflammatory process in osteoarthritis. Nat Rev Rheumatol. 2015;11(1):35–44. doi: 10.1038/nrrheum.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, LeMerrer M, Mortier G, Mundlos S, Nishimura G, Rimoin DL, Robertson S, Savarirayan R, Sillence D, Spranger J, Unger S, Zabel B, Superti-Furga A. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A(5):943–968. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rajpar MH, McDermott B, Kung L, Eardley R, Knowles L, Heeran M, Thornton DJ, Wilson R, Bateman JF, Poulsom R, Arvan P, Kadler KE, Briggs MD, Boot-Handford RP. Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet. 2009;5(10):e1000691. doi: 10.1371/journal.pgen.1000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hartley CL, Edwards S, Mullan L, Bell PA, Fresquet M, Boot-Handford RP, Briggs MD. Armet/Manf and Creld2 are components of a specialized ER stress response provoked by inappropriate formation of disulphide bonds: implications for genetic skeletal diseases. Hum Mol Genet. 2013;22(25):5262–5275. doi: 10.1093/hmg/ddt383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kung LH, Rajpar MH, Briggs MD, Boot-Handford RP. Hypertrophic chondrocytes have a limited capacity to cope with increases in endoplasmic reticulum stress without triggering the unfolded protein response. J Histochem Cytochem. 2012;60(10):734–748. doi: 10.1369/0022155412458436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kung LH, Rajpar MH, Preziosi R, Briggs MD, Boot-Handford RP. Increased classical endoplasmic reticulum stress is sufficient to reduce chondrocyte proliferation rate in the growth plate and decrease bone growth. PLoS One. 2015;10(2):e0117016. doi: 10.1371/journal.pone.0117016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Husa M, Petursson F, Lotz M, Terkeltaub R, Liu-Bryan R. C/EBP homologous protein drives pro-catabolic responses in chondrocytes. Arthritis Res Ther. 2013;15(6):R218. doi: 10.1186/ar4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oliver BL, Cronin CG, Zhang-Benoit Y, Goldring MB, Tanzer ML. Divergent stress responses to IL-1beta, nitric oxide, and tunicamycin by chondrocytes. J Cell Physiol. 2005;204(1):45–50. doi: 10.1002/jcp.20261. [DOI] [PubMed] [Google Scholar]
- 114.Takada K, Hirose J, Senba K, Yamabe S, Oike Y, Gotoh T, Mizuta H. Enhanced apoptotic and reduced protective response in chondrocytes following endoplasmic reticulum stress in osteoarthritic cartilage. Int J Exp Pathol. 2011;92(4):232–242. doi: 10.1111/j.1365-2613.2010.00758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Uehara Y, Hirose J, Yamabe S, Okamoto N, Okada T, Oyadomari S, Mizuta H. Endoplasmic reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthr Cartil. 2014;22(7):1007–1017. doi: 10.1016/j.joca.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 116.Cameron TL, Gresshoff IL, Bell KM, Pirog KA, Sampurno L, Hartley CL, Sanford EM, Wilson R, Ermann J, Boot-Handford RP, Glimcher LH, Briggs MD, Bateman JF. Cartilage-specific ablation of XBP1 signaling in mouse results in a chondrodysplasia characterized by reduced chondrocyte proliferation and delayed cartilage maturation and mineralization. Osteoarthr Cartil. 2015;23(4):661–670. doi: 10.1016/j.joca.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 117.Cameron TL, Bell KM, Gresshoff IL, Sampurno L, Mullan L, Ermann J, Glimcher LH, Boot-Handford RP, Bateman JF. XBP1-independent UPR pathways suppress C/EBP-beta mediated chondrocyte differentiation in ER-stress related skeletal disease. PLoS Genet. 2015;11(9):e1005505. doi: 10.1371/journal.pgen.1005505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cameron TL, Bell KM, Tatarczuch L, Mackie EJ, Rajpar MH, McDermott BT, Boot-Handford RP, Bateman JF. Transcriptional profiling of chondrodysplasia growth plate cartilage reveals adaptive ER-stress networks that allow survival but disrupt hypertrophy. PLoS One. 2011;6(9):e24600. doi: 10.1371/journal.pone.0024600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bateman JF, Rowley L, Belluoccio D, Chan B, Bell K, Fosang AJ, Little CB. Transcriptomics of wild-type mice and mice lacking ADAMTS-5 activity identifies genes involved in osteoarthritis initiation and cartilage destruction. Arthritis Rheum. 2013;65(6):1547–1560. doi: 10.1002/art.37900. [DOI] [PubMed] [Google Scholar]
- 120.Storlazzi CT, Mertens F, Nascimento A, Isaksson M, Wejde J, Brosjo O, Mandahl N, Panagopoulos I. Fusion of the FUS and BBF2H7 genes in low grade fibromyxoid sarcoma. Hum Mol Genet. 2003;12(18):2349–2358. doi: 10.1093/hmg/ddg237. [DOI] [PubMed] [Google Scholar]
- 121.Panagopoulos I, Storlazzi CT, Fletcher CD, Fletcher JA, Nascimento A, Domanski HA, Wejde J, Brosjo O, Rydholm A, Isaksson M, Mandahl N, Mertens F. The chimeric FUS/CREB3l2 gene is specific for low-grade fibromyxoid sarcoma. Genes Chromosom Cancer. 2004;40(3):218–228. doi: 10.1002/gcc.20037. [DOI] [PubMed] [Google Scholar]
- 122.Hino K, Saito A, Kido M, Kanemoto S, Asada R, Takai T, Cui M, Cui X, Imaizumi K. Master regulator for chondrogenesis, Sox9, regulates transcriptional activation of the endoplasmic reticulum stress transducer BBF2H7/CREB3L2 in chondrocytes. J Biol Chem. 2014;289(20):13810–13820. doi: 10.1074/jbc.M113.543322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu M, Tao J, Vasievich MP, Wei W, Zhu G, Khoriaty RN, Zhang B. Neural tube opening and abnormal extraembryonic membrane development in SEC23A deficient mice. Sci Rep. 2015;5:15471. doi: 10.1038/srep15471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boyadjiev SA, Fromme JC, Ben J, Chong SS, Nauta C, Hur DJ, Zhang G, Hamamoto S, Schekman R, Ravazzola M, Orci L, Eyaid W. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38(10):1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 125.Saito A, Kanemoto S, Zhang Y, Asada R, Hino K, Imaizumi K. Chondrocyte proliferation regulated by secreted luminal domain of ER stress transducer BBF2H7/CREB3L2. Mol Cell. 2014;53(1):127–139. doi: 10.1016/j.molcel.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 126.Mertens F, Fletcher CD, Antonescu CR, Coindre JM, Colecchia M, Domanski HA, Downs-Kelly E, Fisher C, Goldblum JR, Guillou L, Reid R, Rosai J, Sciot R, Mandahl N, Panagopoulos I. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest. 2005;85(3):408–415. doi: 10.1038/labinvest.3700230. [DOI] [PubMed] [Google Scholar]
- 127.Wang W, Lian N, Li L, Moss HE, Wang W, Perrien DS, Elefteriou F, Yang X. Atf4 regulates chondrocyte proliferation and differentiation during endochondral ossification by activating Ihh transcription. Development. 2009;136(24):4143–4153. doi: 10.1242/dev.043281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sinha S, Chen JK. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem Biol. 2006;2(1):29–30. doi: 10.1038/nchembio753. [DOI] [PubMed] [Google Scholar]
- 129.Maeda Y, Nakamura E, Nguyen MT, Suva LJ, Swain FL, Razzaque MS, Mackem S, Lanske B. Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proc Natl Acad Sci USA. 2007;104(15):6382–6387. doi: 10.1073/pnas.0608449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Edwards JR, Mundy GR. Advances in osteoclast biology: old findings and new insights from mouse models. Nat Rev Rheumatol. 2011;7(4):235–243. doi: 10.1038/nrrheum.2011.23. [DOI] [PubMed] [Google Scholar]
- 131.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4(8):638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 132.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 133.Xu F, Teitelbaum SL. Osteoclasts: new insights. Bone Res. 2013;1(1):11–26. doi: 10.4248/BR201301003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kuroda Y, Hisatsune C, Nakamura T, Matsuo K, Mikoshiba K. Osteoblasts induce Ca2+ oscillation-independent NFATc1 activation during osteoclastogenesis. Proc Natl Acad Sci USA. 2008;105(25):8643–8648. doi: 10.1073/pnas.0800642105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakashima T, Hayashi M, Takayanagi H. New insights into osteoclastogenic signaling mechanisms. Trends Endocrinol Metab. 2012;23(11):582–590. doi: 10.1016/j.tem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 136.Negishi-Koga T, Takayanagi H. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol Rev. 2009;231(1):241–256. doi: 10.1111/j.1600-065X.2009.00821.x. [DOI] [PubMed] [Google Scholar]
- 137.Ruiz A, Matute C, Alberdi E. Endoplasmic reticulum Ca(2+) release through ryanodine and IP(3) receptors contributes to neuronal excitotoxicity. Cell Calcium. 2009;46(4):273–281. doi: 10.1016/j.ceca.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 138.Kiviluoto S, Vervliet T, Ivanova H, Decuypere JP, De Smedt H, Missiaen L, Bultynck G, Parys JB. Regulation of inositol 1,4,5-trisphosphate receptors during endoplasmic reticulum stress. Biochim Biophys Acta. 2013;1833(7):1612–1624. doi: 10.1016/j.bbamcr.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 139.Lazarowski ER, Boucher RC, Harden TK. Mechanisms of release of nucleotides and integration of their action as P2X- and P2Y-receptor activating molecules. Mol Pharmacol. 2003;64(4):785–795. doi: 10.1124/mol.64.4.785. [DOI] [PubMed] [Google Scholar]
- 140.Wagner LE, 2nd, Yule DI. Differential regulation of the InsP(3) receptor type-1 and -2 single channel properties by InsP(3), Ca(2)(+) and ATP. J Physiol. 2012;590(14):3245–3259. doi: 10.1113/jphysiol.2012.228320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cao H, Yu S, Yao Z, Galson DL, Jiang Y, Zhang X, Fan J, Lu B, Guan Y, Luo M, Lai Y, Zhu Y, Kurihara N, Patrene K, Roodman GD, Xiao G. Activating transcription factor 4 regulates osteoclast differentiation in mice. J Clin Invest. 2010;120(8):2755–2766. doi: 10.1172/JCI42106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yokota H, Hamamura K, Chen A, Dodge TR, Tanjung N, Abedinpoor A, Zhang P. Effects of salubrinal on development of osteoclasts and osteoblasts from bone marrow-derived cells. BMC Musculoskelet Disord. 2013;14:197. doi: 10.1186/1471-2474-14-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hamamura K, Tanjung N, Yokota H. Suppression of osteoclastogenesis through phosphorylation of eukaryotic translation initiation factor 2 alpha. J Bone Miner Metab. 2013;31(6):618–628. doi: 10.1007/s00774-013-0450-0. [DOI] [PubMed] [Google Scholar]
- 144.He L, Lee J, Jang JH, Sakchaisri K, Hwang J, Cha-Molstad HJ, Kim KA, Ryoo IJ, Lee HG, Kim SO, Soung NK, Lee KS, Kwon YT, Erikson RL, Ahn JS, Kim BY. Osteoporosis regulation by salubrinal through eIF2alpha mediated differentiation of osteoclast and osteoblast. Cell Signal. 2013;25(2):552–560. doi: 10.1016/j.cellsig.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Raggo C, Rapin N, Stirling J, Gobeil P, Smith-Windsor E, O’Hare P, Misra V. Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol Cell Biol. 2002;22(16):5639–5649. doi: 10.1128/MCB.22.16.5639-5649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Eleveld-Trancikova D, Sanecka A, van Hout-Kuijer MA, Looman MW, Hendriks IA, Jansen BJ, Adema GJ. DC-STAMP interacts with ER-resident transcription factor LUMAN which becomes activated during DC maturation. Mol Immunol. 2010;47(11–12):1963–1973. doi: 10.1016/j.molimm.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 147.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K, Oike Y, Takeya M, Toyama Y, Suda T. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202(3):345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yagi M, Ninomiya K, Fujita N, Suzuki T, Iwasaki R, Morita K, Hosogane N, Matsuo K, Toyama Y, Suda T, Miyamoto T. Induction of DC-STAMP by alternative activation and downstream signaling mechanisms. J Bone Miner Res. 2007;22(7):992–1001. doi: 10.1359/jbmr.070401. [DOI] [PubMed] [Google Scholar]
- 149.Xu G, Liu K, Anderson J, Patrene K, Lentzsch S, Roodman GD, Ouyang H. Expression of XBP1s in bone marrow stromal cells is critical for myeloma cell growth and osteoclast formation. Blood. 2012;119(18):4205–4214. doi: 10.1182/blood-2011-05-353300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Datta NS, Abou-Samra AB. PTH and PTHrP signaling in osteoblasts. Cell Signal. 2009;21(8):1245–1254. doi: 10.1016/j.cellsig.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, Munshi N, Horner J, Ivanova EV, Protopopov A, Anderson KC, Tonon G, DePinho RA. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11(4):349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, Hu Y, Fabre C, Minami J, Ohguchi H, Kiziltepe T, Ikeda H, Kawano Y, French M, Blumenthal M, Tam V, Kertesz NL, Malyankar UM, Hokenson M, Pham T, Zeng Q, Patterson JB, Richardson PG, Munshi NC, Anderson KC. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119(24):5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, Koong AC. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]