Abstract
During organism development, a diversity of cell types emerges with disparate, yet stable profiles of gene expression with distinctive cellular functions. In addition to gene promoters, the genome contains enhancer regulatory sequences, which are implicated in cellular specialization by facilitating cell-type and tissue-specific gene expression. Enhancers are DNA binding elements characterized by highly sophisticated and various mechanisms of action allowing for the specific interaction of general and tissue-specific transcription factors (TFs). However, eukaryotic organisms package their genetic material into chromatin, generating a physical barrier for TFs to interact with their cognate sequences. The ability of TFs to bind DNA regulatory elements is also modulated by changes in the chromatin structure, including histone modifications, histone variants, ATP-dependent chromatin remodeling, and the methylation status of DNA. Furthermore, it has recently been revealed that enhancer sequences are also transcribed into a set of enhancer RNAs with regulatory potential. These interdependent processes act in the context of a complex network of chromatin interactions, which together contributes to a renewed vision of how gene activation is coordinated in a cell-type-dependent manner. In this review, we describe the interplay between genetic and epigenetic aspects associated with enhancers and discuss their possible roles on enhancer function.
Keywords: Enhancer, Promoter, Histone modifications, Chromatin, CTCF, Topological associating domains, Non-coding RNAs, Enhancer RNA
Introduction
It was recognized almost 35 years ago that enhancers are important DNA regulatory elements that eukaryotic cells use to generate specificity by controlling differential programs of gene expression. Next-generation sequencing methodologies such as ChIP-seq and RNA-seq have enabled us to integrate chromatin and transcriptional states and explore the dynamic regulation of gene expression during cellular differentiation and development. In particular, enhancers have emerged as critical players in distinguishing transcriptional states that show a high degree of variation between different cell types. Enhancer DNA regulatory elements were first described in monkey tumor virus studies [1, 2]. The simian virus SV40 enhancer consists of a 72 base pairs (bp)-long repeated sequence whose deletion reduces the viral protein levels expressed in early stages of infection, and as a result, abolishing virus viability [1]. The existence of enhancers with similar stimulatory characteristics was subsequently reported in eukaryotic genomes [3–7]. The first cellular enhancer was found within an intron of the mouse immunoglobulin heavy chain gene whose stimulatory activity depended on a subset of nuclear factors with high cell-type specificity [3, 4, 8]. In this review, we will cover the most relevant chromatin-associated aspects of enhancer functions, including recent discoveries involving long non-coding RNAs (lncRNAs) and enhancer RNAs (eRNAs).
How do enhancers affect transcription?
The way in which enhancers stimulate transcription is a central question that remains poorly understood. As enhancers were characterized based on their property to increase the transcriptional levels of target genes, the amount of gene product or the number of cells that activate transcription must be important. Therefore, two models have been suggested to explain their function: the binary model and the progressive or rheostatic model [9]. The binary model for enhancer function proposes that enhancers increase the probability that a higher portion of cells activate transcription at a given locus within a cell population [9–13]. In the progressive model, enhancers increase the number of RNA molecules transcribed from the target gene, but not the number of cells that initiate transcription [14]. It is currently unsolved if these models of enhancer function can be generalized or even if alternative models exist.
Transcription factors as mediators of enhancer activity
A central characteristic of enhancers is to function as TF-binding platforms that affect transcription by direct stimulation of their target promoters, located a few kilobases (kb) up to megabases away [15–18]. Although many enhancers are found in intergenic regions, they can also be localized within protein coding genes, in particular introns [19, 20]. Enhancer sequences are commonly considered to be 200–500 bp in length where DNA-binding sites for multiple TFs are clustered [18]. Many lineage-specific TFs and their recruitment to binding sites within enhancers govern lineage-specific gene transcription [21–23]. Two models for TF binding at enhancers have been proposed [24]. In the “enhanceosome” model, the DNA sequence acts as a scaffold for the ordered and cooperative binding of TFs to form a protein complex that can activate transcription. In this scenario, enhancer activity emerges from a network of interactions and its action is lost just by the absence of one protein [25, 26]. In contrast, in the “billboard” model, TF binding is independent from each other and TFs do not act as a single unit [24].
TFs typically have short recognition sequences (4–10 nucleotides), often with a high degree of degeneracy, which increases the probability to find their target sequences in the genome. For example, the predicted estrogen receptor recognition sequence occurs more than one million times in the human genome, but only 10,000–16,000 binding sites are occupied in a human cell line [20, 27, 28]. It is unclear why a major proportion of binding sites are not occupied. For example, DNA packaging into nucleosomes could block the accessibility to their binding sites [29]. However, clustering of binding sites contributes to overcome the chromatin barrier by increasing the cooperativity between TFs like in the “enhanceosome” model [18, 21, 25, 28, 30]. Cooperative binding was observed using reconstituted nucleosome cores and DNA harboring different combinations of binding sites for different and unrelated TFs including GAL4, USF, and NF-κB. The binding of the first TF stimulates the binding of the second by up to two orders of magnitude [31]. This evidence supports the idea that cooperative TF binding to chromatinized DNA could significantly increase the affinity of each factor for nucleosomal DNA at regulatory elements in the genome and thereby overcoming the natural chromatin barrier (Fig. 1) [21–23].
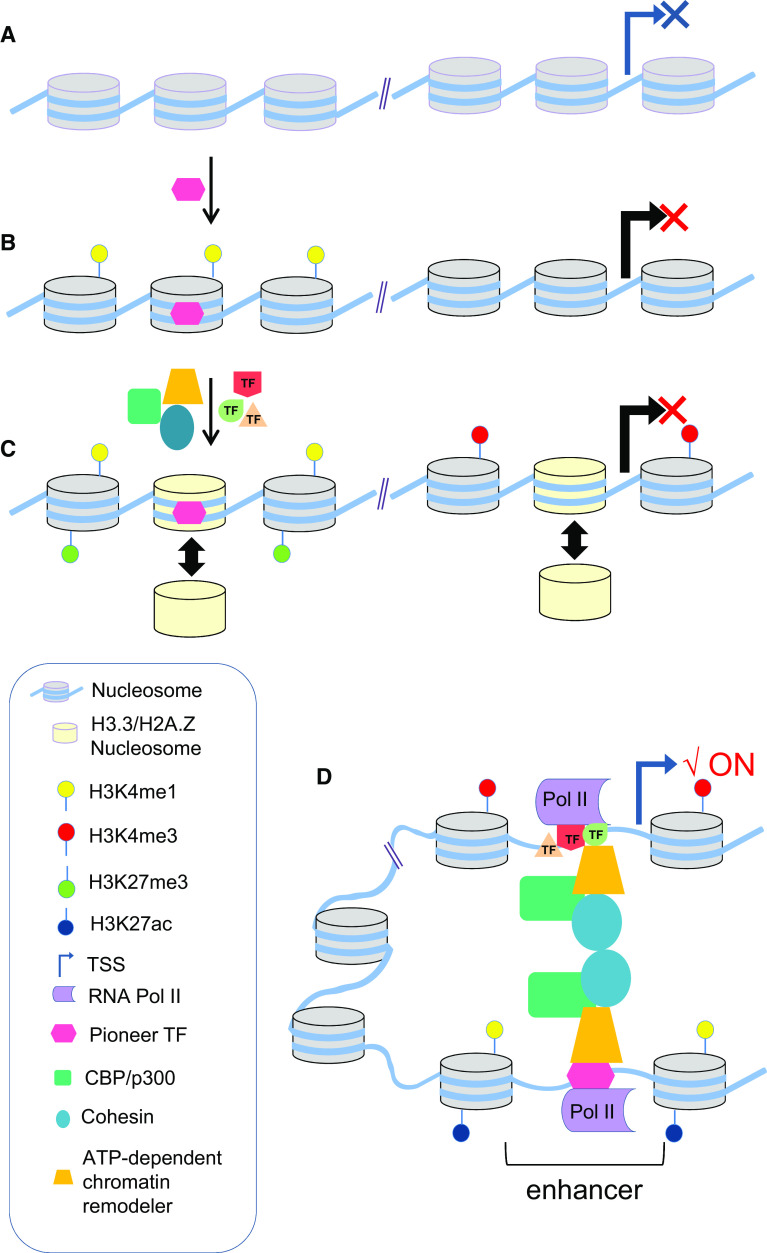
Fig. 1.
Chromatin dynamics at enhancers and promoters during gene activation. a Silent chromatin characterized by a regular pattern of nucleosomes positioned over regulatory elements including the enhancer and its target promoter. b Initial binding of a Pioneer TF within nucleosomes first to the enhancer facilitates the binding of additional TFs and co-factors to open the chromatin through histone modifications. c Additional TFs and the action of ATP-dependent chromatin remodeling complexes in response to hormones or other differentiation signals facilitate the recruitment of the transcription machinery to both the promoter and the enhancer. Histone variant exchange at regulatory elements may contribute to chromatin relaxation. d Physical contacts between enhancer and its target promoter occurs when the gene is transcribed. Under this condition, the enhancer is characterized by its association with high levels of the H3K27ac histone modification
DNA accessibility differs between cell types and changes dynamically during organism development, cell differentiation and in response to external and internal cell stimuli [32, 33]. Such changes in DNA accessibility are believed to be mediated by regulatory proteins, including a special class of TFs called pioneer factors (Fig. 1). Pioneer or nucleation factors, such as FOXA, PU.1, OCT4, and GATA-1, initially bind their consensus sequence within the context of the chromatin, at the level of the solenoid or 30 nm fiber facilitating the binding of additional TFs and co-factors to further open the chromatin in response to hormones or other differentiation signals [23, 29].
The induction of pluripotent stem cells (iPS) from somatic cells through the overexpression of pluripotency TFs contributed to the finding that some TFs are able to recognize their DNA binding sites within silent chromatin [21, 23, 34, 35]. Pluripotency factors including OCT4, SOX2, and KLF4 preferentially recognize partial motifs displayed on the nucleosome surface [23]. The majority (70 %) of pluripotency TF binding initiates gene activation by first binding enhancer sequences of genes that promote reprogramming [23, 35–38]. Pioneer factors are hypothesized to direct lineage specification by changing chromatin states at their binding sites. One well-studied example is OCT4, a TF involved in establishing and maintaining the pluripotent state. In one study, OCT4 was shown to change the chromatin state at the MYOD1 locus. Overexpression of OCT4 resulted in its binding to the enhancer of MYOD1, and a switch from H3K4me1 to H3K4me3 at the MYOD1 promoter. Interestingly, the H3K27me3 mark, which was present before OCT4 overexpression, remained [39]. This bivalent promoter state has been described in embryonic stem cells and could be mediated in part by OCT4 [36].
It is important to consider that TF binding does not always imply direct stimulation of gene expression. For example, in the case of pioneer factors that can access DNA where other factors cannot, binding is primarily required to initiate additional chromatin remodeling events that will prepare the chromatin template for the binding of other cell-type-specific TFs (Fig. 1) [39]. The ability of TFs to activate transcription on chromatin templates is also dependent on the recruitment of coactivator proteins [21, 30, 40, 41]. By definition, coactivators lack sequence-specific DNA binding competency, but they function as histone modifiers or by recruiting complexes with histone modifier capabilities (i.e., histone acetyltransferases), ATP-dependent chromatin remodelers, or mediators of long-range chromatin contacts [42–44]. Therefore, pioneer TFs and their regulatory partners can selectively bind to the site where an enhancer will be remodeled in order to be fully activated in a regulated and highly specific manner.
Histone modifications at enhancers
The histone H3 lysine 4 mono-methylation is present in nucleosomes linked to enhancer elements
Systematic genome-wide studies of histone post-translational modifications have revealed new insights into transcriptional regulation [33, 45, 46]. In particular, H3K4me1 was the first histone modification globally linked to distal regulatory regions [19] in a tissue-specific manner [33]. Comparably, histone H3K4me3 mark was predominantly enriched at gene promoter regions [19]. Nevertheless, a clear-cut discrimination between enhancers and promoters cannot be made based on their histone H3K4 methylation status as histone H3K4me2 or H3K4me3 marks have been also detected at active enhancers bound by the RNA Pol II [47–51].
In mouse and human embryonic stem cells, enhancers directing promoter activity of genes linked to developmental processes are pre-labeled by H3K4me1. This leads to the hypothesis that this modification participates in enhancer priming [18, 52–54]. Histone H3K4me1, unlike H3K27ac or RNA Pol II, appears to persist at enhancers even after their disengagement from promoters [55–57]. Once incorporated, H3K4me1 can be read by chromodomain-containing acetyltransferases like TIP60, which catalyzes acetylation of H2A at Lys5 [58, 59]. In addition, histone H3K4me1 contributes to maintain a permissive chromatin state by repelling the interaction of proteins that recognize unmethylated H3 lysine 4, BHC80 (CoREST-LSD1 complex) and de novo DNA methylation complex [60, 61]. Thus, H3K4 methylation may protect distal regulatory elements from being targeted by DNA methylation and keep them in a poised state, until ready for activation with the proper signal. Nevertheless, the presence of H3K4me1 at enhancers in Drosophila melanogaster, an organism lacking Dnmt3 homolog and low levels of DNA methylation [62], suggests that other mechanisms may be at play, potentially involving direct recognition of H3K4me1.
Histone acetyltransferases occupancy at enhancers
In line with the positive correlation between histone acetylation and gene expression [48, 63], genome wide binding of enzymes with histone acetyltransferase activity represents a central aspect in enhancer function [52, 64]. CBP/p300 proteins are the most studied co-factors with intrinsic histone acetyltransferase (HATs) activity [65]. Genomic occupancy of p300/CBP during development in humans shows that 95 % of p300 in vivo binding is found at promoter distal regions [52, 65]. Consistent with these data, p300 binding is an accurate predictor of in vivo enhancers in the developing mouse embryo. Additionally, most p300 bound regions coincide with DNase I hypersensitive sites (DHS) and active gene expression during development [64]. CBP and p300 HATs acetylate over 70 proteins, including themselves, however, histone 3 lysine 18 (H3K18) and H3K27 are their major in vivo histone targets [66–68]. One role for CBP/p300 enhancer binding may be to integrate metabolic information of the levels of acetyl coenzyme A (which acts as donor of acetyl groups) [69]. For example, in yeast where the Gcn5p/SAGA complex catalyzes the acetylation of histones at promoters of genes important for growth according to increased levels of acetyl coenzyme A [70]. Alternatively, CBP/p300 may recruit RNA Pol II to enhancers marked with H3K4me1 to transcribe the enhancer itself and generate a special class of transcripts, collectively called eRNAs, which have been implicated in the mechanism of action of enhancers (Fig. 2) [71, 72].
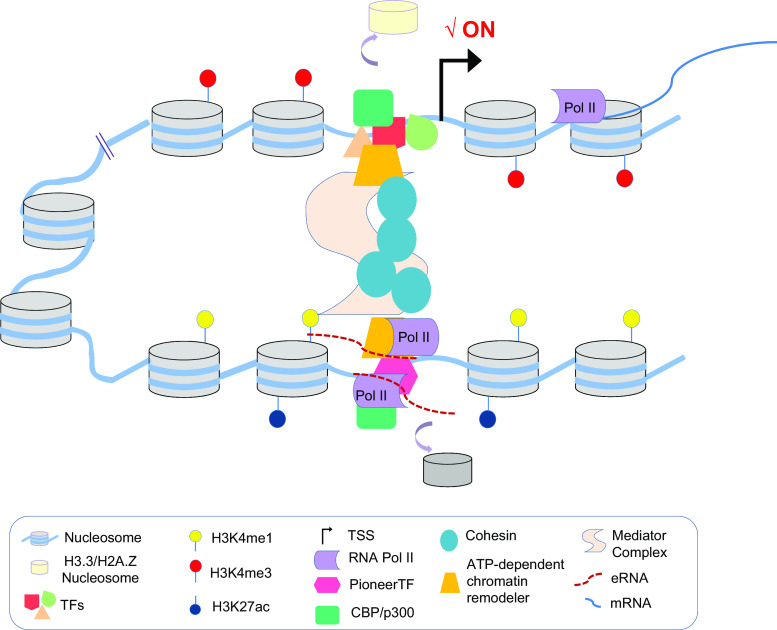
Fig. 2.
Gene transcription is coupled to non-coding transcription at enhancers. Active enhancers are transcribed in a cell-type-specific manner producing bidirectional eRNAs. eRNA transcription coincides with transcription of target gene, high levels of H3K27ac, and a common architecture of transcriptional initiation characterized by nucleosome depleted region formation. eRNAs can exert their regulatory action through the stabilization of chromatin loops
Apart from CBP/p300, other HATs have been found to interact with enhancers [72]. A possible reason for binding of different HATs to enhancer regions could be a differential recruitment of cofactors or TFs. Moreover, TFs could be potentially modified by HATs at target enhancers affecting their activity or protein interactions [73]. In conclusion, histone acetyltransferases are widely associated with enhancer elements, which argue in favor of a functional role for acetylation in enhancer function.
Histone H3K27ac and H3K27me3 marks demarcate active and poised enhancers
Despite a cell-type-specific genome-wide correlation between enhancers and the presence of H3K4me1 histone modification or the p300 coactivator complex [19, 45], the use of these parameters does not accurately predict enhancer activity on target promoters. Therefore, histone chromatin analysis at enhancer sequences during ESC differentiation revealed a clear correlation between the H3K27ac histone mark and an increased gene activity of proximal genes; in contrast, the presence of H3K27me3 showed the opposite trend [52–54, 56]. To some extent, the presence of acetylation at many enhancers may attenuate nucleosome stability [25] or improve chromatin accessibility. Since acetylation reduces the affinity of histones for negatively charged DNA, it may help TFs to access their binding sites more efficiently [74].
The different combinations of histone marks at enhancers on a genome-wide scale in different cell types permitted the classification of poised enhancers containing H3K4me1 and H3K27me3 histone marks and active enhancers associated with H3K4me1 and H3K27ac histone marks [52, 53]. Detailed analysis revealed that poised enhancers are lineage-specific enhancers that become activated upon differentiation [52]. Nucleosome remodeling accompanies changes in the H3K27ac and H3K4me1 histone marks at active enhancers (Fig. 1). At the time of TF binding, active enhancers exhibit single peak of RNA Pol II enrichment at the center. On the contrary, poised enhancers show a single peak of H3K4me1 and H3K27me3 histone marks without RNA Pol II occupancy [53, 56]. As previously mentioned, poised enhancers contain the H3K27me3 mark instead of H3K27ac. Remarkably, in D. melanogaster, the H3K27me3 histone demethylase UTX and the chromatin remodeler Brm directly bind to CBP/p300 and both brm mutations and UTX knockdown reduce H3K27ac levels at enhancer regions [75]. This provides a mechanism that couples H3K27 demethylation and the presence/action of a histone acetyltransferase (CBP) in order to switch from the inactive to the active enhancer status.
A limited set of modifications including H3K4me1/3, H3K27ac, and H3K27me3 are ubiquitously utilized to map and demarcate active and poised enhancers and promoters. Yet, these modifications represent only a fraction of the full repertoire of histone modifications at enhancers and promoters. Evidence is still lacking as to whether histone marks such as H3K4me1 or H3K27ac are sufficient, necessary, or even mechanistically involved in the activity of these regulatory elements [76, 77]. Both marks have fairly high turnover rates, and it is thus likely that neither is inherited across mitosis nor is instructive for future cellular generations [78].
Remodeling the chromatin structure to access enhancer sequences
Chromatin remodeling by changing nucleosome density or position at enhancers is critical to generate a regulatory environment typically “devoid” of nucleosomes [also known as nucleosome depleted regions (NDR)] such that the DNA is accessible to be bound by TFs. In fact, chromatin remodeling has been widely associated with cis-regulatory elements in the genome such as promoters and enhancers [79–81]. At least one-third of the identified DHSs in 125 cell types were tissue-specific, located distally from annotated promoters [81], highly enriched for the H3K27ac histone mark, and associated with active enhancer sequences [33].
Nucleosome positioning and eviction at enhancers through chromatin remodeling complexes
For years, an intense debate has developed in terms of the role of DNA sequence in nucleosomal positioning. Sequence analysis of enhancers responsive to androgen stimulation show enrichment for an AT-rich motif (AA/TT/TA/AT) at NDRs that is absent at nucleosome-enriched regions. In contrast, GC-rich motifs show the opposite tendency [82]. Similar observations of nucleotide composition at NDR of regulatory elements like enhancers have been reported in yeast and D. melanogaster [83–85]. Despite these observations, there is no clear consensus about the role of those sequences in nucleosome positioning and chromatin remodeling.
In contrast, ATP-dependent chromatin remodeling complexes directly contribute to nucleosomal positioning. Unlike TFs, these complexes do not bind directly to the DNA but contain protein domains that recognize other proteins or covalent histone modifications that assist them to associate to specific regions of the chromatin [86]. Importantly, in humans there is evidence that functional enhancers are associated with nucleosome remodeling [87]. SATB1 (special AT-rich sequence binding 1) is a protein found predominantly in thymocytes that recruits the CHRAC and ACF nucleosome mobilizing complexes within the interleukin-2 receptor gene (IL-2Rα), in particular, toward the exon 1 and the SBS700 region of the gene, repressing its expression through changes in chromatin accessibility [88]. Local alteration in the pattern of cleavage by micrococcal nuclease is evident between wild-type and SATB1 null thymocyte nuclei possibly due to changes in nucleosome distribution over the enhancer [88].
Furthermore, the recruitment of the ATP-dependent chromatin remodeling SWI/SNF (Smarca4/BRG1) complex to enhancers is critical to initiate and establish the transcriptional program that promotes oligodendrocyte differentiation and subsequent myelination of the central nervous system (CNS) in humans [89]. The oligodendrocyte-lineage determination factor (Olig2) recruits the chromatin remodeling factor Brg1 to enhancer elements that exhibit enrichment in H3K27ac and that control oligodendrocyte differentiation [52, 89]. A Brg1 ATPase mutant domain inhibits myelin gene expression, suggesting that ATPase activity is essential for Brg1 function to promote myelination programs by chromatin remodeling of Olig2-enhancers [89].
Another ATP-dependent chromatin remodeling enzyme implicated in the regulation of enhancer activity in mammals is CHD7, a member of the Chromodomain Helicase DNA-binding protein family. Genetic mutations of CHD7 are a cause of CHARGE syndrome [86, 90]. In mouse embryonic stem cells (mESCs), CHD7 co-localizes on a genome-wide level with loci exhibiting high levels of p300, H3K4me1, DHSs, and binding of master regulators like OCT4, SOX2, and NANOG, suggesting its association with enhancer sequences [91]. Significant correlation between CHD7 occupancy and reduced gene expression suggests that CHD7 functions to limit the expression of a subset of ESC-specific genes [91]. However, whether CHD7 changes the chromatin structure associated with enhancers remains to be explored.
Nucleosome positioning changes according to gene activity, and is an important element of transcriptional regulation [45]. Recent genome-wide data confirm that DNA accessibility is highly dynamic and changes during lineage specification, and that particular patterns of nucleosome positioning along enhancer sequences emerge during gene activation [33, 45, 82, 92].
In prostate cancer cells, the androgen receptor primarily binds to enhancer sequences [82]. Before androgen receptor activation, androgen receptor binding loci are already marked with two well-positioned H3K4me2-containing nucleosomes that flank the binding site, along with a well-positioned H2A.Z/H3K4me2 containing nucleosome occluding the binding site. After androgen receptor activation, the central H3K4me2-modified nucleosome is destabilized and the region between the flanking nucleosome is increased probably reflecting the binding of the androgen receptor that could act as boundary that direct the positioning of nearby nucleosomes [82]. However, it is unknown if the eviction of the central nucleosome is a consequence of active chromatin remodeling or if rather the presence of the H2A.Z histone variant causes nucleosome instability.
Nucleosome positioning is also important at the 3′ chicken α-globin enhancer. When the enhancer reaches its highest activity, two flanking nucleosomes are well positioned and two additional nucleosomes, shielding the functional part, are evicted concomitant with the appearance of two micrococcal hypersensitive sites (MHSs). Introduction of unrelated lambda DNA increasing the distance between this two the MHSs affects enhancer activity and nucleosome organization [92, 93]. The local chromatin configuration of the enhancer is dependent on the binding of the erythroid-specific factor GATA-1, probably by recruiting Brg1, since it physically interacts with GATA-1 at the α-globin locus of murine fetal liver cells [94, 95]. The artificial expression of the pioneer factor GATA-1 in HeLa cell line resulted in the formation of DHSs and hyperacetylation of histones within GATA-1 bound regulatory elements. Functional analysis of GATA-1 binding at interaction sites in an erythroid context revealed that GATA-1 plays a major role in defining both chromatin structure and enhancer activity of the 3′ chicken α-globin enhancer since point mutations of GATA-1 interaction sites abolish both the DHSs and the enhancer capacity to trans-activate a reporter gene [92]. In an attempt to distinguish between the two models of α-globin enhancer activity (binary vs. progressive), we identified a novel regulatory element which modulates the activity of the chicken α-globin enhancer [92, 93]. This element, named upstream enhancer element (UEE), increases the activity of this enhancer but not the numbers of cells in which the enhancer is active in agreement with the progressive model [93]. We propose that this mode of action is in part facilitated by the fixed positioning of nucleosomes located upstream and downstream of the core enhancer since the UEE is important for positioning the nucleosome located upstream of the enhancer’s core [92, 93].
Nucleosome eviction at promoters has also been described, for example upon transcriptional activation of the yeast PHO5 gene promoter and for the mammalian MMTV LTR promoter regulated by steroid hormones [96, 97]. Therefore, nucleosome eviction is a phenomena that is shared by both enhancers and promoters sequences, which further suggests, along with enrichment of shared histone marks (like H3K4me1), that both regulatory sequences have common mechanisms of action (see later “Similarities between enhancers and promoters”).
Together, these results suggest that the coordinated repositioning and/or eviction of nucleosomes through chromatin remodeling complexes represent a critical step to render functional enhancers.
DNA hydroxymethylation in enhancer function
An alternative proposed mechanism involved in the regulation of enhancer activity is related to DNA methylation and hydroxymethylation. It has been well established that cytosine DNA methylation (5mC) is an essential epigenetic modification catalyzed by DNA methyltransferases [98, 99]. Genome-wide studies show a negative correlation between DNA methylation and chromatin accessibility [81]. Similarly, TFs and co-activator binding are inversely correlated with DNA methylation [85, 100, 101]. Concordantly, active enhancers are normally depleted of DNA methylation [100, 102].
5mC can be further oxidized by TET proteins through the active demethylation pathway generating 5hmC, 5fC, and 5caC, which can regenerate unmodified cytosines [103, 104]. Importantly, cytosine 5-hydroxymethylation (5hmC), which is the initial intermediate in the enzymatic cytosine demethylation cascade [103, 104], is a novel epigenetic modification that overlaps with histone H3K4me1 and H3K27ac at enhancers during differentiation [105, 106]. A single base resolution map of 5hmC in mouse and human ESCs, revealed that 5hmC is most abundant at both poised and active enhancers [107], rather than at CpG-rich promoters, as was previously suggested [108, 109]. Importantly, 5hmC enrichment at enhancers is characterized by a bimodal distribution flanking TF-binding sites [107].
The role of 5hmC on enhancer function remains to be determined, but one possibility is that a protein directly recognizes this DNA modification, which then can affect transcriptional activation. Proteins have been identified that preferentially recognize the 5hmC mark. For instance, the peptide product of the Uhrf2 gene whose expression increases upon differentiation of neural progenitor cells (NPC) [104, 110, 111]. Alternatively, new candidate molecules that can shape the DNA methylation landscape by recruiting both DNMT and TET enzymes to chromatin are long non-coding RNAs [112, 113]. However, further studies are needed to demonstrate that these mechanisms can occur at enhancer regions.
Long non-coding RNA with enhancer-like function
Most of the mammalian genome (70–80 %) has the potential to be transcribed into non-coding RNAs (ncRNAs) [114–116]. Recent experimental data support a role for lncRNAs in transcriptional regulation [117, 118]. LncRNAs have been implicated in cellular processes such as X chromosome inactivation, genomic imprinting, development, cell differentiation, and several pathologies, among many other functions [119, 120]. Using a model of primary human keratinocytes, long non-coding RNAs that respond to cell differentiation signals were found to be upregulated [118]. These non-coding transcripts act by stimulating transcription and behave similarly to classical enhancers. It has been observed that this kind of transcript interacts with the Mediator complex, thereby favoring long-range chromatin interactions between two loci [121]. As an alternative molecular mechanism to stimulate gene expression, lncRNAs might produce a more permissive chromatin environment at regulatory regions by direct recruitment of chromatin remodeling complexes [122]. Therefore, particular attention needs to be paid to define whether lncRNAs themselves possess or contribute to enhancer function and if the Mediator complex as a critical player in these interdependent interactions.
Non-coding transcription at enhancers, a novel regulatory mechanism
Detailed genome-wide analysis by RNA-seq and Global Run-On sequencing (GRO-seq) assays at high resolution, together with the data accumulated by the ENCODE Project reveals that a majority of active enhancers are transcribed in a cell-type-specific manner and produce relatively short transcripts (200-300 nucleotides), known as enhancer RNAs or eRNAs [45, 49, 51, 71, 114, 123–126]. This type of transcript has been observed in different cellular contexts including primary neuronal cultures, myogenic cells, mouse macrophages, and breast cancer cells among many others [71, 124–127]. eRNAs exhibit a 5′ cap and are generally not spliced or polyadenylated. Interestingly, the majority of enhancer transcription is bidirectional (Fig. 2) [128]. Apparently, there is a tight correlation between eRNA expression and the transcription of nearby genes, even though eRNA relative abundance is frequently low [128]. In line with this, transcription of eRNAs is positively correlated with an acquisition of active histone marks at enhancers, particularly H3K27ac, and the absence of the repressive histone H3K27me3 mark (Fig. 2) [129, 130].
An important question is how eRNAs become activated as specifically regulated transcription units and, of course, which mechanisms define the precise initiation site of enhancer transcription. Additionally, an aspect that remains unanswered is if eRNA represent a general feature of enhancer function, and if so, which is the hierarchy of eRNA transcription during enhancer action.
eRNAs have been implicated in different processes. For example, they participate in the formation of enhancer-promoter contacts in collaboration with the cohesin complex (RAD21 and SMC3) (Fig. 2) [125] and they can interact with the negative elongation factor (NELF) complex to facilitate the transition of paused RNA polymerase II into productive elongation by a decoy mechanism upon induction of immediate early genes in neurons [44, 131]. Alternatively, in macrophages, co-repressor/histone deacetylase complex NCoR-HDAC3 can be recruited by the nuclear receptors Rev-Erbs at the response elements in enhancers and promoters of target genes to establish a macrophage-specific program of repression. The repressive function of Rev-Erbs is dependent of their ability to inhibit eRNAs transcription, therefore suggesting eRNAs transcription is essential for gene expression of enhancer target genes [124].
In summary, there is increasing evidence that implicates eRNAs in the activity of enhancers. Their mechanism of action remains to be further investigated but there is no doubt that they have a relevant role in transcription regulation. One unexplored function of eRNA may have to do with multiple long-distance interactions that can be mediated by CTCF or even the Mediator complex. Importantly, they may be linked to different diseases, in particular, because a significant number of SNPs are found in enhancer regions perturbing chromatin remodeling activities and/or TFs binding [132].
Similarities between enhancers and promoters
Evidence for enhancer transcription has led the field to compare the nucleosome and chromatin architecture of enhancers and TSSs [51]. Examination of nascent RNA identified common architecture of transcriptional initiation characterized by NDR formation and the symmetry of two nucleosomes flanking divergent transcription pairs at enhancers suggesting that the same principle applies to both enhancers and promoters (Fig. 2). However, to what extent the chromatin architecture associated with promoters and enhancers accounts for transcription initiation will require further analysis since NDR formation has been also observed at the 3′ end of genes [83, 133–135]. Whether a universal architecture for promoters and enhancers is applicable to all loci is still controversially debated [82, 136].
Since the line distinguishing transcription start sites from enhancers has become blurred by the finding that transcription originates from enhancers and promoters, histone modifications seem to not be responsible for differences in transcription levels [51], therefore it is possible that chromatin features cannot distinguish between them (Figs. 1, 2). However, at the functional level, both elements work differently as the excision of the proximal promoter region does not elicit transcription despite the presence of the enhancer [19, 71].
In any case, different regulatory mechanisms remain to be studied. For example, and based in genome-wide scale studies, in particular, with antibodies against a sub-set of histone modification signatures, active enhancer for some cell types and tissues have been found, with active promoters having different signatures. Based on such difference it has been proposed that this is a group of cis-regulatory elements with dynamic signatures also named cREDS [137]. This type of regulatory element shows histone marks associated with active enhancers like H3K27ac and H3K4me1 in a particular tissue, but in other cell types, histone modifications shift towards H3K4me3 [137]. In addition, intragenic enhancers can function as promoters of the embedded gene generating abundant, spliced and multi-exonic poly(A)+ RNAs but with low protein coding potential and thus far with unknown function [138]. In conclusion, there are many convergent features between promoters and enhancers that deserve a new visualization and strategies to study them.
Super-enhancers coordinate the expression of tissue-specific genes
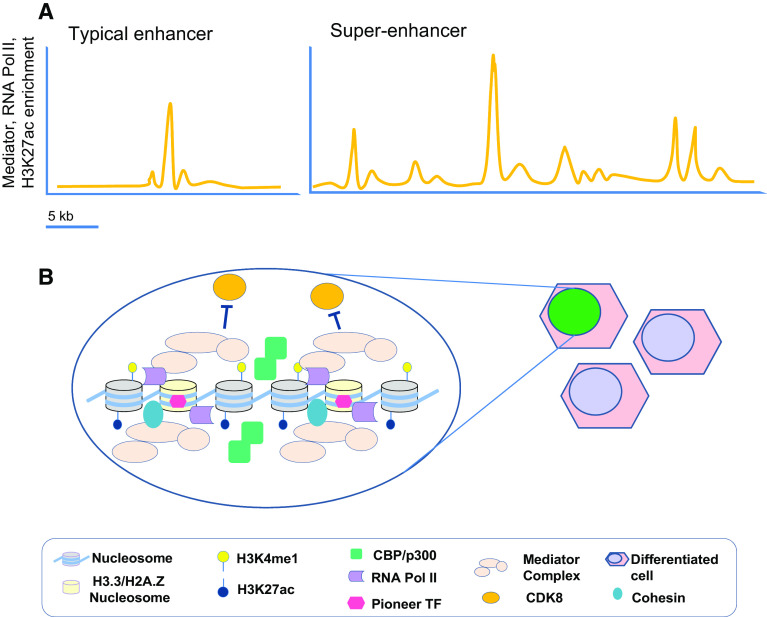
Several studies have implicated the Mediator complex, which is essential for the maintenance of stem cells and their differentiation [139–141] as a facilitator of the RNA Pol II recruitment to enhancers via TFs (Fig. 3) [142, 143]. Recently, large areas of up to 50 kb showing a great enrichment of Mediator complex components were termed super-enhancers [144]. Despite the difficulty to functionally define super-enhancers, they frequently harbor one or multiple (up to six) DHSs, each one with enhancer features including RNA Pol II association, high density of TF binding, and the highest enrichment of H3K27ac histone mark (Fig. 3) [144, 145]. Intriguingly, Mediator subunits seem to have different roles in diverse cell types, and they can interact with structural proteins such as cohesin in a tissue-specific manner [139, 146]. Future work is necessary to unravel the possible mechanisms driving cell fates regulated by the Mediator complex.
Fig. 3.
a Comparison of genomic features associated with typical vs. super-enhancers. Super-enhancers are not functionally defined but they harbor chromatin enhancer features like very high enrichment of Mediator complex, H3K27ac, RNA Pol II and TFs, they span large genomic areas, and regulate genes that specify cell identity. b Mediator complex binds genomic regions in a tissue-specific manner and may be directly involved in integrating external and internal metabolic signals to modulate gene expression. The dynamic structure and composition of the Mediator complex may be a key feature in regulating the process of transcription since its interaction with the CDK8 subunit prevents its interaction with RNA Pol II
Three-dimensional circuitry of enhancer sequences
Regulatory elements like enhancers and promoters interact in space through loop formation and this has important consequences for gene expression [15]. Loop formation is thought to increase the local concentration of TFs or cofactors involved in transcriptional regulation including cohesin or Mediator [139, 147, 148]. Most of the loops occur within topologically associating domains (TADs) in metazoans like D. melanogaster and mammals. TADs are megabase-sized chromosomal regions often invariant between tissues. Sequences inside TADs interact more frequently with each other than with sequences outside the TADs [149–152]. These structures have been proposed to create a microenvironment where genes and regulatory sequences can interact with high frequency, and share histone modifications and patterns of gene expression [149, 152]. For example, Shh gene expression depends on an enhancer located 1 Mb away from its target promoter. Both enhancer and promoter are inside the same TAD and contact each other through loop formation [17].
Despite extensive annotation of enhancer sequences through mapping of histone modifications like H3K4me1, H3K27ac, or cofactors like p300 [19, 52, 64, 153, 154] enhancers looping dynamics have not been fully investigated. Using chromosome conformation capture-based (Hi-C) method to map long-range chromatin interactions maps for specific regulatory sequences elucidated a complex network of interactions between all annotated promoters and enhancers was recently elucidated in mouse embryonic stem cells (mESCs) and fetal liver cells (FLC) [154]. Less than half of the promoter-enhancer interactions are conserved between mESCs and FLCs. This also holds for human embryonic stem cells (hESCs) and differentiated human cells suggesting that enhancer looping interactions are heavily rewired during cells differentiation in mouse and human [154–156]. In contrast, in D. melanogaster, a small set of developmental enhancers active in mesodermal embryonic cells (6–8 h after egg laying) show no significant change of looping interactions with their target promoters despite a clear induction of gene expression for their associated genes when compared with multipotent cells (3–4 h after egg laying) where mesodermal genes are not expressed [157]. Authors suggest that those enhancer looping interactions are pre-established very early in development and possibly primed for rapid gene expression since paused RNA Pol II was found at the sites of loop interaction. Thus, in human and mouse cells, differentiation is accompanied by an extensive change of enhancer-promoter looping interactions while in D. melanogaster enhancer-promoter looping interactions may be set very early during embryonic development. Whether these differences originate from the small number of enhancer sequences analyzed in D. melanogaster or it reflects a biological difference in the dynamics of enhancer-promoter looping interactions, possibly due to differences in early development between vertebrates and invertebrates (D. melanogaster embryonic development lasts just 24 h) remains to be determined.
Looping interactions seem to be very different between cell types but little is known about their dynamics as a response to different stimuli in differentiated cells. In humans, a high-resolution genome-wide map of chromatin contacts (Hi-C) in primary fibroblast cells treated and untreated with TNF alpha suggest that despite a clear activation of TNF alpha response genes, the vast majority of enhancer chromatin contacts with promoters of TNF alpha response genes are unchanged [155]. This trend was also evident in other cell types and under different stimuli. This suggests that at least in human differentiated cells, pre-existing looping interactions between enhancer and promoters of genes responsive to signaling inducible TFs are a feature of the 3D regulatory landscape of the genome. Even though, looping interactions seem to be pre-established at enhancers and promoters in differentiated cells the connectivity landscape of these regulatory sequences is very different between cell types [154–156].
The molecular mechanisms by which enhancer and promoter interactions are established are just becoming clear but the CCCTC-binding factor (CTCF) seems to be directly involved. CTCF is a DNA-binding protein best characterized as an insulator associated protein and that has been suggested to be a critical player for genome organization through formation of chromatin loops with cohesin (Figs. 1, 2). Remarkably, 92 % of chromatin loops identified in human cells have CTCF binding sites (CBS) in a convergent orientation [155]. Most of the loci at the anchors of the loops were enhancers and promoters, which suggest that CTCF binding in a convergent orientation, could bring together regulatory elements though loop formation. At the protocadherin (Pcdh) gene cluster, the inversion of CBS with a reversed orientation at the Pcdh enhancer using the CRISPR-Cas9 system abolished loop formation between enhancer and target promoters in human and mouse neurons [158]. This data strongly support that the convergent orientation of CBS is critical for loop formation.
The use of the CRISPR-Cas9 system for genome editing has opened a whole new era of research and will enable us to test the function of enhancers endogenously by direct removal or disruption of their sequence in vivo. Moreover, this new technology will allow to understand the mechanistic consequences of mutations within enhancer sequences that are associated with certain diseases, e.g., by creating them in animal models [159, 160]. For instance, analysis of human patient-derived cells demonstrated that mutations identified in human limb malformation syndromes affected proper interactions between enhancers and promoters of the WNT6/IHH/EPHA4/PAX3 region mainly by disrupting TAD borders. This highlights the importance of insulation of genes and their regulatory elements, like enhancers, by TADs to ensure a proper program of gene expression, protecting against non-canonical interactions with other regulatory elements [160].
The emerging picture suggests that enhancers act as complex networks that favor three-dimensional contacts with regulatory regions in the genome where context-dependent signaling outcomes may determine cell fate choices.
Conclusions and prospects
Functional specialization of cells and tissue types is vital for all multicellular organisms. This requires cells to respond to developmental and environmental cues by generating specific patterns of gene expression on the basis of an identical set of genetic material. Enhancers are the main regulators that enable cell-type-specific gene expression. To gain such accuracy, enhancers are bound by specific TFs and 3D chromatin architecture-mediating proteins, marked by specific post-translational histone modifications, and generate non-coding transcripts called eRNAs. In addition, high-resolution maps of promoter–enhancer interactions are providing new insights into enhancer function, generating hypotheses which can now be directly tested using genome editing techniques. With these new tools it will be possible in the future to explore, in great detail, how a cell adopts its unique transcriptional identity.
Acknowledgments
We acknowledge Karin Meier for critical reading of the manuscript. This work was supported by the DGAPA-PAPIIT, UNAM (IN209403, IN203811 and IN201114), CONACyT (42653-Q, 128464 and 220503) and Fronteras de la Ciencia-2015 (Grant 290) to FR-T, and by a PhD fellowship from CONACyT and Programa de Apoyo a los Estudios del Posgrado (PAEP), UNAM to EG-G and RA-M. Additional support was provided by the Ph.D. Graduate Program, “Doctorado en Ciencias Bioquímicas y Biomédicas”, to the Instituto de Fisiología Celular, Universidad Nacional Autónoma de México.
References
- 1.Khoury G, Gruss P. Enhancer elements. Cell. 1983;33:313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- 2.Banerji J, Rusconi S, Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-X. [DOI] [PubMed] [Google Scholar]
- 3.Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 4.Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 5.Walker MD, Edlund T, Boulet AM, Rutter WJ. Cell-specific expression controlled by the 5′-flanking region of insulin and chymotrypsin genes. Nature. 1983;306:557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- 6.Edlund T, Walker MD, Barr PJ, Rutter WJ. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science. 1985;230:912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- 7.Goodbourn S, Zinn K, Maniatis T. Human β-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985;41:509–520. doi: 10.1016/S0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- 8.Schöler HR, Gruss P. Cell type-specific transcriptional enhancement in vitro requires the presence of trans-acting factors. EMBO J. 1985;4:3305–3313. doi: 10.1002/j.1460-2075.1985.tb04036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko MSH, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DIK. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutherland HGE, Martin DIK, Whitelaw E. A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol Cell Biol. 1997;3:1607–1614. doi: 10.1128/MCB.17.3.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yie J, Senger K, Thanos D. Mechanism by which the IFN-β enhanceosome activates transcription. Proc Natl Acad Sci USA. 1999;96:13108–13113. doi: 10.1073/pnas.96.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandaltzopoulos R, Becker PB. Heat shock increases the reinitiation rate from potentiated chromatin templates. Mol Cell Biol. 1998;18:361–367. doi: 10.1128/MCB.18.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 18.Calo E, Wysocka J. Modification of enhancer chromatin: what, how and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 20.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 23.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnosti DN, Kulkarni MM. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 25.Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/S1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 26.Panne D. The enhaceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Vega VB, Lin CY, Lai KS, Kong SL, Xie M, Su X, et al. Multiplatform genome-wide identification and modeling of functional human estrogen receptor binding sites. Genome Biol. 2006;7:R82. doi: 10.1186/gb-2006-7-9-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol Syst Biol. 2010;6:456. doi: 10.1038/msb.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siersbæk R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 31.Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/MCB.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell O, Tiwari VK, Thomä NH, Schübeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 33.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A et al (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330 [DOI] [PMC free article] [PubMed]
- 34.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 35.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci USA. 2011;108:14497–14502. doi: 10.1073/pnas.1111309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sammons MA, Zhu J, Drake AM, Berger SL. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2014;25:179–188. doi: 10.1101/gr.181883.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- 39.Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, et al. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik S, Roeder RG. Dynamic regulation of Pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 42.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin remodeling complexes. Mol Cell Biol. 2000;5:1899–1910. doi: 10.1128/MCB.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Näär AM, Lemon BD, Tijan R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 44.Becker PB, Hörz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 45.ENCODE Project Consortium et al Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 47.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koch F, Andrau JC. Initiating RNA polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription. 2011;2:263–268. doi: 10.4161/trns.2.6.18747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Core LJ, Martins AL, Dank CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bogdanovic O, Fernandez-Miñán A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, et al. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 2012;22:2043–2053. doi: 10.1101/gr.134833.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, et al. Tissue specific analysis of chromatin states identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 57.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeong KW, Kim K, Situ AJ, Ulmer TS, An W, Stallcup MR. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol. 2011;18:1358–1365. doi: 10.1038/nsmb.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;488:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lyko F, Ramsahoye BA, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 63.Cho H, Orphanides G, Sun X, Yang X-J, Ogryzko V, Lees E, et al. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/MCB.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/S0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 66.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CB/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holmqvist P-H, Mannervik M. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription. 2013;4:18–23. doi: 10.4161/trns.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metab. 2014;20:26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daitoku H, Sakamaki J, Fukamizu A. Regulation of FoxO transcription factors by acetylation and protein–protein interactions. Biochem Biophys Acta. 2001;1813:1954–1960. doi: 10.1016/j.bbamcr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 74.Speranzini V, Pilotto S, Sixma TK, Mattevi A. Touch, act and go: landing and operating on nucleosomes. EMBO J. 2016;35:376–388. doi: 10.15252/embj.201593377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol. 2012;32:2323–2334. doi: 10.1128/MCB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hödl M, Basler K. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol. 2012;22:2253–2257. doi: 10.1016/j.cub.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 77.Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 78.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, et al. Genome-scale mapping of DNase I sensitivity in vivo using DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 81.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–364. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson JD, Widom J. Poly (dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 87.Shones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 89.Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 91.Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Escamilla-Del-Arenal M, Recillas-Targa F. GATA-1 modulates the chromatin structure and activity of the chicken α-globin 3′ enhancer. Mol Cell Biol. 2008;28:575–586. doi: 10.1128/MCB.00943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.García-González E, Recillas-Targa F. A regulatory element affects the activity and chromatin structure of the chicken α-globin 3′ enhancer. Biochim Biophys Acta. 2014;1839:1233–1241. doi: 10.1016/j.bbagrm.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 94.Xu Z, Meng X, Cai Y, Koury MJ, Brandt SJ. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem J. 2006;399:297–304. doi: 10.1042/BJ20060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim SI, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 2009;37:6019–6027. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction release additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Richard-Foy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 99.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 101.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bock C, Beerman I, Lien W-H, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, et al. Genome-wide analysis reveals TET-and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 105.Sérandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, et al. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Arab K, Park YT, Lindroth AM, Schäfer A, Oakes C, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 114.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 116.Mousavi K, Zare H, Koulnis M, Sartorelli V. The emerging roles of eRNAs in transcriptional regulatory networks. RNA Biol. 2014;11:106–110. doi: 10.4161/rna.27950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 120.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, et al. A rapid, extensive and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lam MTY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mousavi K, Zare H, Dell’Orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 128.Lam MTY, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schaukowitch K, Joo JY, Liu X, et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–41. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang C, Pugh F. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jansen A, Verstrepen KJ. Nucleosome positioning in Saccharomyces cerevisiae . Microbiol Mol Biol Rev. 2011;75:301–320. doi: 10.1128/MMBR.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang H, Liu H, Sun X. Nucleosome distribution near the 3′ ends of genes in the human genome. Biosci Biotechnol Biochem. 2013;77:2051–2055. doi: 10.1271/bbb.130399. [DOI] [PubMed] [Google Scholar]
- 136.Duttke SH, Lacadie SA, Ibrahaim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U. Human promoters are intrinsically directional. Mol Cell. 2015;57:674–684. doi: 10.1016/j.molcel.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 139.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Risley MD, Clowes C, Yu M, Mitchell K, Hentges KE. The Mediator complex protein Med31 is required for embryonic growth and cell proliferation during mammalian development. Dev Biol. 2010;342:146–156. doi: 10.1016/j.ydbio.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 141.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M. p300-mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 2000;14:1899–1907. [PMC free article] [PubMed] [Google Scholar]
- 144.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 145.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fiering S, Whitelaw E, Martin DI. To be or not to be active: the stochastic nature of enhancer action. BioEssays. 2000;4:381–387. doi: 10.1002/(SICI)1521-1878(200004)22:4<381::AID-BIES8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 147.Plank JL, Dean A. Enhancer function: mechanistic and genome-wide insights come together. Mol Cell. 2014;55:5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 152.Bouwman BA, de Laat W. Getting the genome in shape: the formation of loops, domain and compartments. Genome Biol. 2015;16:154. doi: 10.1186/s13059-015-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo . Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 154.Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 155.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares-Cadete F, Sugar R, Javierre BM, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ghavi-Helm Y, Klein FA, Pakodzi T, Ciglar L, Noordermeer D, Huber W, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 158.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. CRISPR inversion of CTCF sites alters genome topology an enhancer/promoter function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]