Abstract
All organs consisting of single cells are consistently maintaining homeostasis in response to stimuli such as free oxygen, DNA damage, inflammation, and microorganisms. The cell cycle of all mammalian cells is regulated by protein expression in the right phase to respond to proliferation and apoptosis signals. Post-translational modifications (PTMs) of proteins by several protein-editing enzymes are associated with cell cycle regulation by their enzymatic functions. Ubiquitination, one of the PTMs, is also strongly related to cell cycle regulation by protein degradation or signal transduction. The importance of deubiquitinating enzymes (DUBs), which have a reversible function for ubiquitination, has recently suggested that the function of DUBs is also important for determining the fate of proteins during cell cycle processing. This article reviews and summarizes the diverse roles of DUBs, including DNA damage, cell cycle processing, and regulation of histone proteins, and also suggests the possibility for therapeutic targets.
Keywords: Cell cycle, Deubiquitinating enzyme, DNA damage, Ubiquitination
Introduction
Cellular response to maintaining genomic stability from the several genotoxic stresses is an important reaction in cancer cell homeostasis. The expression and activation of the cell-cycle checkpoint proteins begin with diverse proteins such as transcription factors and post-translational modifying enzymes in each phase by diverse stimulations in both prokaryotic and eukaryotic cells. In addition, the damaged cells undergo cell cycle arrest and programmed cell death, and these processes are regulated by the orchestration of various proteins. For example, upstream checkpoint kinases such as ataxia telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) play an important role in the initiation of DNA damage response (DDR) [1]. Besides, p53, a well-characterized cell-cycle checkpoint regulator, contributes to cell cycle arrest and/or apoptosis by the DDR signaling cascade [2]. Checkpoint proteins can control cell cycle progression, and they monitor whether the previous phase has been suitably completed in order to progress to the next phase. The way proteins ensure cell cycle arrest, proliferation, and apoptosis in cells through signal transduction is one of important questions in the field of cell biology.
In mammalian cells, most proteins are subjected to post-translational modifications (PTMs). Ubiquitination is one of these PTMs, and it regulates the proteins’ fate by a series of enzymatic cascade reactions [3]. In the course of these enzymatic reactions, ubiquitin (Ub) as a 76-residue polypeptide is activated by Ub-activating enzyme (E1) with high-energy thioester. Activated ubiquitin is subsequently transferred to a cysteine residue of Ub-conjugating enzymes (E2). The E3 Ub-ligase enzyme catalyzes between the carboxyl terminus as a glycine (Gly or G) residue of Ub and a lysine (Lys or K) residue of a targeted protein, and this interaction finally makes an isopeptide bond (Fig. 1a) [4]. Ubiquitination is involved in the regulation of diverse functions in the cells, including protein degradation by the 26S proteasome, immune response, protein transport, transcription, and cell cycle progression [5]. Ubiquitin can mark substrates by its monomeric (monoUb) or polymeric (polyUb) form as a chain. In addition, Ub moieties can make diverse branches through their Lys residues by utilizing the editing function of E3 Ub-ligases (K6, K11, K27, K29, K33, K48, and K63) in cells [6]. In general, the protein marked by K48-linked polyUb chains is recognized by the 26S proteasome, and it leads to proteolysis [7]. On the other hand, K63-linked polyUb chains serve in intracellular signaling as a monoUb [8]. A proteomic approach to quantification for Ub–Ub links with seven Lys sites showed a relative abundant order of K48 > K63 and K11 ≫ K33, K27, and K6 in yeast [6].
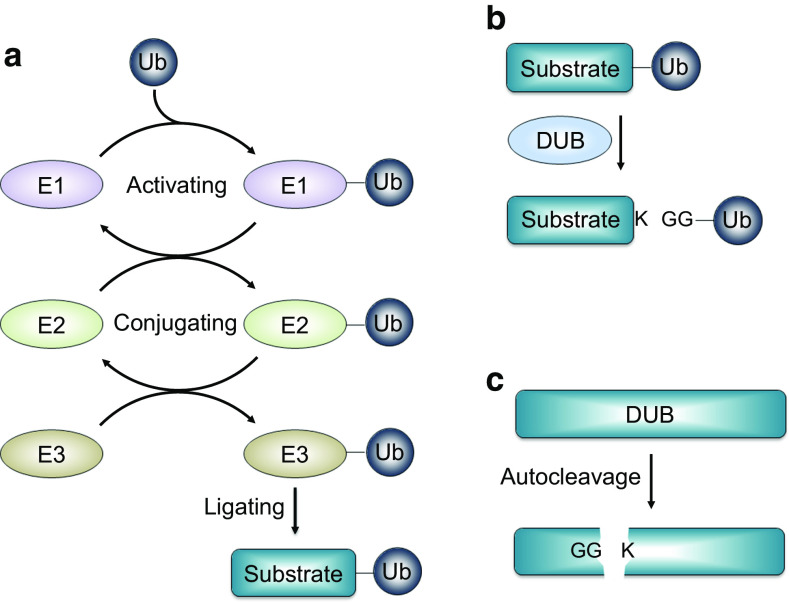
Fig. 1.
The role of DUBs within the ubiquitin–proteasome system. a Ubiquitin is composed of 76 amino acids, and it undergoes a series of enzyme reactions, such as E1 (activating enzyme), E2 (conjugating enzyme), and E3 (ligase enzyme), to attach to target proteins. b Most proteins are ubiquitinated on one or more multiple-lysine sites, and DUBs dissociate the ubiquitin-substrate bond through recognizing and cleaving at the diGly site. c A scheme of autocleavage for DUBs
DUBs belong to the cysteine protease subfamily and negatively regulate ubiquitination by disassembling the Ub chains between diGly (GG) peptides on the ubiquitin proximal site and Lys (K) peptide on targeted proteins, and accordingly have a crucial role in the regulation of the ubiquitination process and its subsequent physiological functions in cells [9] (Fig. 1b). Several DUBs also cleave themselves on their own diGly sites [10, 11] (Fig. 1c). Therefore, DUBs control numerous bioactivities in cell cycle regulation, signal transduction, membrane trafficking, DNA damage response, immune response, and programmed cell death [12]. The human genome encodes approximately 100 DUB enzymes, and these can be classified into at least six families according to their functional and structural properties: ubiquitin-specific proteases (USPs), ubiquitin carboxy-terminal hydrolases (UCHs), ovarian-tumor proteases (OTUs), Machado–Joseph diseases (MJDs), JAB1/MPN/MOV34 metalloenzymes (JAMMs), and monocyte chemotactic protein-induced proteases (MCPIPs) [13]. DUBs have specific conserved domains, including Cys, Asp/Asn, and His domains, which determine their catalytic activity. However, JAMMs lacking in these three domains are zinc metalloproteases [9]. Each class of DUB enzymes differs in the size and arrangement of conserved sequences. The structural analysis performed based on their conserved sequences revealed that there are various conserved motifs throughout these DUBs [9], and an elegant review also has described and compared the structures of DUBs recently [14].
Cell-cycle checkpoint proteins are synthesized and produced to assist in the progression of the cell cycle. It is well known that various cancer cells have mutations or overexpression of checkpoint proteins. However, the regulatory mechanism of PTM in the ubiquitination and deubiquitination that regulate the fate of cell-cycle checkpoint proteins in cancer cells is not completely understood. Here, we focus on how DUBs regulate cell cycle checkpoints, and the potentially utilizing DUB inhibitors in cancer therapy process by understanding the enzymatic mechanisms of DUBs.
DNA damage and DUBs
Cells exposed to DNA damage agents activate DDR pathways to counteract DNA lesions. Commonly, base excision repair (BER), direct repair (DR), homologous recombination (HR), nucleotide excision repair (NER), non-homologous end joining (NHEJ), and mismatch repair (MMR) are classified as types of DNA lesion progression [15]. Cell cycle checkpoint is coordinated with DNA repair as part of DDR. We have summarized DUBs according to the type of DNA lesions upon DDR (Fig. 2). The DNA damage checkpoint halts the progression through cell cycle in the G1, S, or G2 phase until the recovery of genomic damages by sensor proteins occurs. The initiation of signal transduction to respond to DNA damage is conducted by upstream kinases such as ATM and ATR. In general, these two extremely huge protein kinase complexes directly phosphorylate several substrates to accomplish their signal transduction for DDR [16]. In addition, ATM and ATR target CHK1 and/or CHK2 to regulate cyclin-dependent kinases (CDK) activity through various pathways and phosphorylate and activate p53 [17, 18].
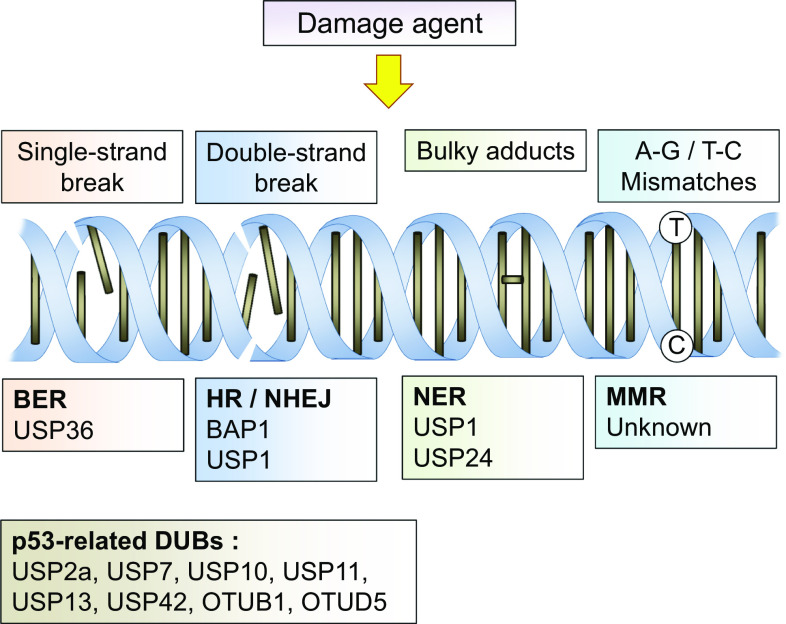
Fig. 2.
Involvement of DUBs in DNA repair mechanisms. DNA damage reagents cause several types of genomic instability including single-strand breaks (SSBs) to base mismatches. DNA repair is triggered by several repair systems, such as base excision repair (BER), nucleotide excision repair (NER), homologous recombination (HR), non-homologous end-joining (NHEJ), and mismatch repair (MMR). Several DUBs are involved in and associated with DNA repair mechanisms and regulation of p53 in DNA damage responses
In base excision repair (BER)
Cells exposed to chemical toxins, ionizing radiation (IR), ultraviolet radiation (UV), and reactive oxygen species (ROS) can cause DNA lesions and produce DNA single-strand breaks (SSBs) and double-strand breaks (DSBs) [19]. SSBs brought about by ROS and IR are repaired by BER. In BER, damaged bases are recognized by DNA glycosylase and removed from the double helix, and then polymerases and ligases excise the damaged section [20]. The most proliferous cells with unrepaired SSBs from the exposure of physically damaging agents result in the blockage of cell cycle progression and collapse of DNA replication forks in the S phase [21]. Moreover, these harmful situations possibly accompany DSBs [22]. Poly(ADP-ribose) polymerase (PARP) enzymes are considered to be monitoring proteins in an abundance of SSBs, and they induce cell death through the release of apoptosis-inducing factors (AIFs) from mitochondria-related stimulation [23]. PARP1 and PARP2, as the PARP superfamily members, are primarily detected and activated in SSBs, and these proteins subsequently synthesize poly (ADP-ribose) (PAR) chains at DNA breaks through a zinc finger motif [21]. Activated PARP1 is detected in BER, and it interacts with the XRCC1-DNA ligase III complex to regulate DNA ligation [24]. PARP modifies not only its targeting proteins but also itself with PAR modification (PARsylation) [25], and a recent in vitro study showed that only PARsylated PARP1 is recognized by Iduna as a PAR-dependent E3 ligase, and this protein–protein interaction leads to PARP1 polyubiquitination [26]. A further study identified a PARP1 regulating an E3 ligase such as checkpoint with fork-head associated and ring finger (CHFR) in the early stage of DDR [27]. The functional interaction between PARP1 and CHFR was also shown in mitosis, and we will discuss this issue in the next section. Even though PARP1-regulating DUBs have not been identified as of yet, USP36, one of the DUBs, was identified as a PARP1 binding partner by global proteomic analysis [28]. A genome-wide analysis showed that Ubp10 (known as USP36) is induced by oxidative stress in budding yeast [29]. Our biochemical analyses showed that USP36 stabilizes and regulates mitochondrial superoxide dismutase SOD2 in mammalian cells [30, 31]. Moreover, a study on Drosophila revealed that the mutation of USP36 leads to nuclear protein aggregates such as H2B, and selective autophagy induced by deficiency of USP36 [32]. This finding supports the role of USP36 on genomic functions through the regulation of histone protein. Previous studies have demonstrated that USP36 is localized into rDNA locus, and it catalyzes the ubiquitin on H2B with deubiquitinating activity [33–36]. The mechanistic details of USP36 in DNA damage have been studied. For example, a recent study revealed that high oxidative stress induced Ubp10 inactivation, and it leads to an increase of protein carbonylation in Ubp10-depleted cells [37]. Ubiquitination and carbonylation can be a marker of ROS [38], which also strongly supports this study.
In nucleotide excision repair (NER)
Whereas BER is concerned with small base adduction, NER recognizes and processes in the bulkier distorting base lesions such as cyclobutane pyrimidine dimers (CPD) and pyrimidine(6-4)pyrimidone photoproducts (6-4PP) in DNA helix by UV. Generally, NER is classified into two subpathways as global genomic NER (GG-NER), which repairs DNA lesions of the entire genome, and transcription coupled with NER (TC-NER), which is removal of transcription-blocking lesions [39]. GG-NER in DNA distorting is recognized by two types of protein complexes as a UV-damaged DNA binding protein (UV-DDB), which is known as DDB1-DDB2-containing the E3-ubiquitin ligase CUL4A complex and XPC-RAD23B protein complex [40]. UV-DDB is associated with UV-damage response to regulate genome stability [41]. DDB1 (known as p127) was found to be an adaptor protein that allosterically regulates CUL4 E3 ligase activity [42], and this complex is recruited by DDB2 to the site of damaged DNA in regulating the initiation of GG-NER [43, 44]. PCNA has a role in the processing of DNA replication through interactions between several binding proteins and DNA, and its ubiquitination is tightly regulated during DDR to orchestrate the function of binding partners by PTMs [45]. The PTMs of PCNA with ubiquitin or small ubiquitin-like modifier (SUMO) in the DNA damage condition have been studied over the past decade [46]. A recent study added a phenomenon for the PTMs of PCNA with ISG15 ubiquitin-like modifier (ISG15) in triggering translesion DNA synthesis (TLS) [47]. The coordinators for the PTMs of PCNA have been well summarized and reviewed in several outstanding papers [45, 48, 49], and we will discuss PCNA-associated DUBs and their binding proteins in this review. A part of the DDB2 protein degradation in UV-exposed cells is shown in that DDB2 requires PCNA to degrade itself by ubiquitination [50]. USP1-UAF1 makes a complex by UV irradiation; this complex regulates PCNA monoubiquitination, and this interaction leads to the regulation of DNA synthesis. The role of the USP1-UAF1 complex will be discussed in detail. In addition, a recent study showed that USP24 stabilizes DDB2 through its deubiquitinating activity [51]. XPC also undergoes the proteasomal degradation by ubiquitination in cells, and USP7 has deubiquitinating activity in XPC regulation under the UV damage condition [52]. In addition, the UV-irradiated cell that is depleted with USP7 decreased the processing of 6-4PP [52]. Thus, this kind of evidence suggests that several DUBs are directly or indirectly associated with the DDB2 regulation pathway in NER.
In homologous recombination (HR), nucleotide and non-homologous end joining (NHEJ)
The HR and NHEJ pathways have functions for dealing with chromosome repair for DSBs [19]. HR mostly acts in the S and G2 phases during the cell cycle, and it can be separated into three stages as pre-synapsis, synapsis, and post-synapsis to recover DNA sequences in the damaged location [53]. The nucleotides around the DSB ends resected to 3′-OH by the Mre11-Rad50-Xrs2 (MRX) complex during pre-synapsis. In synapsis, Rad51 that binds with ssDNA forms a D-loop through DNA strand invasion and the sister chromatid is utilized as a template for DNA synthesis. RNAi-based screening at IR-induced foci for identification of the regulator protein for HR protein assembly found BRAC1-associated protein 1 (BAP1) [54]. BAP1 belongs to the UCH subfamily (UCH-L1, UCH-L3, UCH37, and BAP1) and is known to be a tumor suppressor protein that is mutated in various melanomas [55]. Experimental evidence with BAP1 knock-out cells showed the novel function of BAP1 in HR and the BAP1-deficient cells sensitized to DSBs and the deubiquitinating activity of BAP1 or its phosphorylation is needed for DNA repair [54]. USP1 is a well-defined DUB for both the regulation of HR repair and the Fanconi anemia pathway through the interaction of its binding partner, UAF1 [56, 57]. During the cell cycle process, USP1 eliminates monoUb on PCNA to regulate error-prone TLS polymerase. Moreover, USP1 has a di-Gly (GG-K) site on its His domain, and it leads to autocleavage by its protease activity upon DNA damage condition [11]. Cleaved USP1 undergoes proteasomal degradation, and it leads to an increase in the level of monoubiquitinated PCNA [11]. UAF1 has been identified as an USP1 interacting protein, and this interaction was increased in DNA damage condition [58]. Further studies have suggested that USP1 has to interact with UAF1 to have a function in deubiquitinating activity [59–61]. A mouse model for USP1 in DDR showed that USP1−/− mouse embryonic fibroblast cells (MEFs) increased UV sensitivity and monoUb of both PCNA and FANCD2 [62]. Further studies have supported this phenomenon. Murai and colleagues employed USP1−/−, UAF1−/−, and USP1−/−UAF1−/− DT40 cells to investigate the USP1/UAF1 complex in DDR, and these cells also increased the level of mono-Ub for both PCNA and FANCD2 [63]. In addition, USP1- and/or UAF1-depleted DT40 cells sensitized the treatment of several DNA damage reagents, such as camptothecin and PARP inhibitors, and resulted in reduced HR repair [63]. A recent study showed the importance of the UAF1-USP1 regulatory mechanism in HR repair in UAF1-deficient mice, which can be observed in embryonic lethality in E7.5, and its embryonic stem cells (ESCs) showed hypersensitivity to several DNA damage reagents and chromosomal abnormality [64]. BRCA1, known as a tumor suppressor, is also well characterized in cancer and is defined as a DNA damage regulator to respond to HR [65]. The molecular events and pathways of BRCA1 in HR have been reviewed in an outstanding article [66]. However, a DUB as an essential regulator for BRCA1 has not been identified yet.
In p53 regulation upon DNA damage
The kinetics of p53 upon DDR has also been widely evidenced in several biochemical and mice studies. These investigations revealed that many residues on p53 protein are coordinated by diverse kinases to respond to DDR. Upon DNA damage, seven serine residues (S6, S9, S15, S20, S33, S36, and S36) and two tyrosine residues (T18 and T81) on p53 are phosphorylated, and six lysine residues (K370, K372, K373, K381, K382, and K386) are acetylated [67], and these modifications induce transactivation to several p53 transcriptional targets [68]. In addition, six lysine residues for p53 acetylation can be shared with ubiquitination [67]. So far, several studies have identified approximately 27 E3 ubiquitin ligases for p53, and MDM2 is the most studied negative regulator for p53 stability [69]. In contrast, eight DUBs (USP2a, USP7, USP10, USP11, USP13, USP42, OTUB1, and OTUD5) have been identified for deubiquitination of p53 [70–77]. These findings were expected in that the regulatory mechanism for DDR via p53 is tightly regulated by the ubiquitination or deubiquitination, and another unknown DUB can also be a regulator for p53 in cell cycle processing.
DUBs and cell cycle processing
In mammalian cells, cell cycle processing can be divided into four phases as G1, S, G2, and mitosis. The cell cycle progression is determined by the expression and activation of several relative regulators such as CDKs, cyclin-dependent kinase inhibitors (CDKIs), cyclins, and aurora kinases. Several studies have reported on the direct or indirect function of DUBs in the cell cycle processing (Fig. 3).
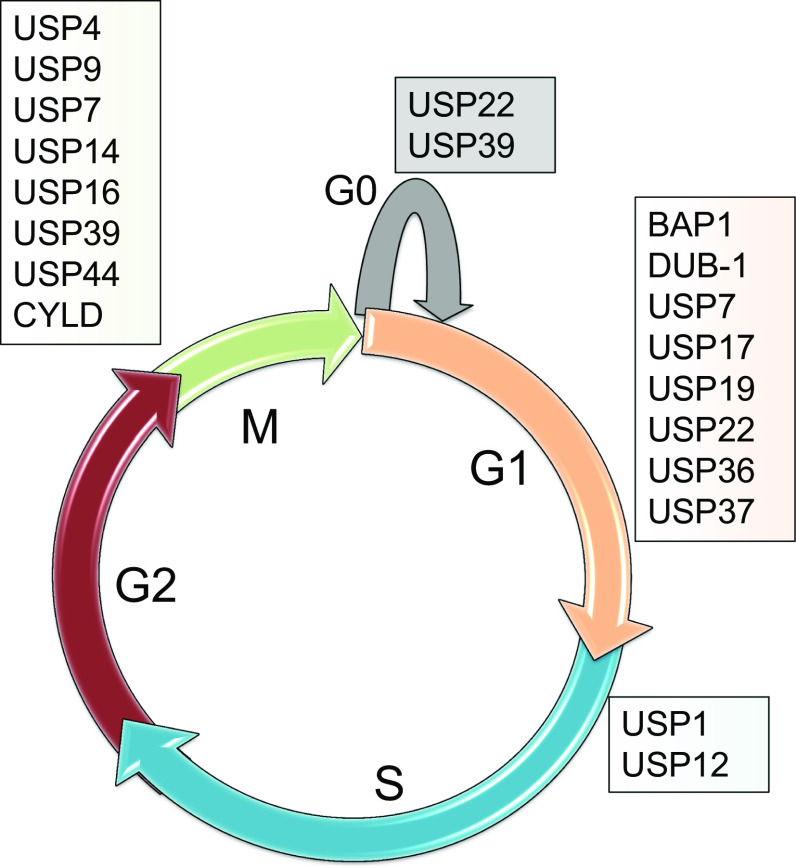
Fig. 3.
DUBs in cell cycle regulation. Living cells process the cell cycle in four sequential phases: the first gap phase (G1); the DNA synthesis phase (S) for DNA replication; the second gap phase for preparing mitosis (G2); and finally mitosis. Withdrawal of cells after many cellular divisions leads to the quiescent phase (G0). Each distinct phase is controlled by several DUBs
Interphase (G1, S and G2 phase)
Synthesis of RNA and proteins occurs in the G1 phase, which prepares for the onset of DNA replication. USP17 was identified as a cytokine inducible immediate-early DUB, and it can modulate cell proliferation by blocking Ras-converting enzyme 1 (RCE1) activity [78, 79]. The ability of USP17 for regulation of cell proliferation is elucidated by Ras-mediated CDKIs activation [79, 80]. Lack of USP17 leads to an accumulation of CDKIs such as p21Cip1 and p27Kip1, resulting in the inhibition of the G1-S transition. USP17 can also regulate apoptosis through the interaction of several apoptosis-associated factors, and detailed properties have been well reviewed by a recent article [81]. DUB-1 is also known as a cytokine inducible DUB in murine and cells are arrested in the G1 phase by the overexpression of DUB-1 [82]. Further studies revealed that DUB-1 is regulated by the JAK/STAT pathway and interacts with dynein heavy chain during cell cycle processing [3, 83], but the essential binding partner of DUB-1 for regulation of the G1-S transition has not yet been identified yet. Lu and colleagues found that USP19 is associated with the regulation of stability for p27Kip1 during the cell cycle [84]. p27Kip1, one of the CDKIs, is ubiquitinated by two E3 ligases, KPC1/KPC2 and Skp1-Cul1-F-box (SCF)Skp2 in the G1-S phase. Lu and colleagues have observed that p27Kip1 was accumulated from G1 to early S phases by depletion of USP19. In addition, USP19 can stabilize KPC1 by its deubiquitinating activity, and their interaction leads to the degradation of p27Kip1. However, most cells are not controlled by the absence or presence of USP19 by p27Kip1 regulation during G1-S transition [85]. NCI-H226 non-small lung cancer cells, as BAP1-deficient cells, are used to study BAP1 function. Overexpression of BAP1 accelerates G1-S transition, and it leads to cell death [86]. The human herpes simplex virus-associated host cell factor 1 (HCF-1) was identified as a BAP1 interacting protein [87]. During G1-S transition, E2F proteins (E2Fs) promote transcription of cell cycle-associated proteins, and HCF-1 has the function of an E2Fs coactivator [88]. BAP1 has deubiquitinating activity for inhibition of HCF-1 ubiquitination, specifically dissociating Lys48-linked ubiquitin chains from HCF-1 [89]. Therefore, deubiquitination of HCF-1 by BAP1 regulates G1-S processing. Similarly, overexpression of USP37 also accelerates G1-S transition [90]. The E3 ubiquitin ligase anaphase-promoting complex (APC/C) controls the overall cell cycle. Because APC/C consists of multimeric proteins, the activity of APC/C depends on its substrates during cell cycle processing [91]. Cyclin A is a substrate of APC/C, and APC/C subsequently ubiquitinates and degrades cyclin A from the G1-S phase to mitosis [91]. A recent study showed the role of USP37 for G1-S transition, including the regulation of cyclin A [90]. The transcription factors E2Fs induce USP37 in the G1 phase, and CDK2 as a kinase phosphorylates USP37 to confer deubiquitinating activity [90]. These events lead to the stabilization of cyclin A before mitotic entry, and the expression of USP37 subsequently decreases by E3 ligase SCFβTrCP in the G2 phase [90, 92]. Interestingly, p27 was destroyed by the KPC1-USP19 complex as we mentioned previously [84], but USP37 rescued p27 expression by its deubiquitinating activity [93].
Mitosis
Mitosis is the most strictly regulated phase in the cell cycle. Although it is the shortest phase in the entire cell cycle, many dynamic cellular events rapidly compared to other periods [94]. In comparison with interphase, which is the period including cell growth and replication of DNA, the key point of mitosis is cell division (also known as cytokinesis). There are six well known steps involved in mitosis–prophase, prometaphase, metaphase, anaphase, telophase, and cytokinesis. Through this entire cycle flow, five main significant events occur—disassociation of the nuclear envelope, chromatid condensation, attachment of microtubules, moving to opposite pole and cytokinesis—and must occur successively and without mistakes [95]. Otherwise, cells undergoing mitosis may acquire metabolically related disorders [95]. Some modulators or regulators are required for repairing a damaged cell cycle from abnormal progressions [96]. One of the vital events for making progression in mitosis is spindle assembly. APC/C has shown their functions and regulations in the attachment of microtubules. In mitosis, APC/C mainly functions in triggering the metaphase-anaphase transition via degradation of two main proteins; securin and cyclin B. Ubiquitination of the separase inhibitor securin by the E3-ubiquitin ligase activity of APC results in proteolytic degradation [97, 98]. Separase is activated and its protease activity cleaves cohesin rings so that they detach from each sister chromatid, resulting in the transition from metaphase to anaphase. A recent biochemical study in yeast screened both securin- and cyclin B-associated DUBs [99]. Ubp1, Ubp2, Ubp3, Ubp10, and Ubp12 were screened for both polyubiquitinated securin and cyclin B regulating DUBs [99]. In addition, Ubp6 (known as human USP14) and Ubp14 (known as human isopeptidase T) have DUB activity for the monoubiquitination of both securin and cyclin B [99]. Regulations by Aurora B during spindle assembly are noticeable, and a recent study has shown that mRNA levels of Aurora B are intimately associated with USP39 [100]. USP39 is required for maintenance of the mitotic spindle checkpoint and its function for cytokinesis as a stout assistant [100]. Besides, not only USP39 but also USP44 and USP4 are significantly involved in the regulation of spindle assembly checkpoint. Spindle assembly checkpoint monitors whether attachment between all paired-sister chromatids at their kinetochore by microtubules is normally stable. USP44 plays a role in the regulation of mitotic checkpoint interacting with cell-division cycle protein 20 (CDC20). USP44 participates in the cytokinesis promoting activity of APC/C by stabilizing CDC20-MAD2 as a result of the deubiquitinating activity to CDC20 [101]. In addition, USP44-null mouse has the defects of regulating mitotic cell cycle checkpoint and in chromosome lagging [102]. This observation indicates that USP44 takes part in the functional role of centrosome and the role of the natural formation of the mitotic spindle. Further, USP44 functions as a tumor suppressor by protecting against the missegregation of chromosomes [102]. Other observations of deubiquitinating activity with USP7 in mitosis have suggested that USP7 could be essential for mitosis regulation. Claspin synthesized in the S phase functions as an adaptor protein for Chk1 signaling, and it is degraded by both E3 ligases SCFβTrCP and APC/C in mitosis and G1 phase processing, respectively [103]. USP7 counteracts with SCFβTrCP but not APC/C in claspin stability during mitosis. Oppositely, USP7 does have deubiquitinating activity for claspin degradation that is ubiquitinated by SCFβTrCP during the G1 phase [103]. Depletion of USP7 increases the genomic stability, and the cells that completely depleted USP7 lead to a blockage of mitosis and the G1 phase [104, 105].
Resting phase (G0 phase)
Three DUBs—Rpn11, Uch37, and USP14—are incorporated in the 26S proteasome [106–108]. Rnp11 has deubiquitinating activity to target proteins ATP-dependently and positively promotes substrate degradation [109]. On the contrary, Rpn11, Uch37, and USP14 deubiquitinate target proteins, and negatively promote substrate degradation in the proteasome through an Ub-chain trimming activity; namely, Uch37 and USP14 have a role as a proteasome inhibitor [109]. The biological function of USP14 has shown that depletion of USP14 increases G0/G1 and reduces S phase population in the cells [110]. In addition, clinical analysis has revealed that the survival ratio of lung adenocarcinoma patients depends on USP14 expression level [110].
Commonly, the canonical function of DUBs is involved in dissociation of ubiquitin from target proteins, and this reaction is related to several cellular processes. However, the non-canonical function of DUBs also contributes cellular regulation through protein–protein interaction. UCH-L1 can be a good example of the non-canonical function of DUBs. UCH-L1 is a well-characterized DUB in Parkinson’s disease and several cancer progressions [111]. A recent study has shown that cell cycle and proliferation were enhanced by interaction between UCH-L1 and CDKs (CDK1, CDK4, and CDK5) [111]. However, the role of their interaction in the cell cycle and proliferation is not dependent on UCH-L1 deubiquitinating activity [111].
Histones and DUBs in the cell cycle
Some studies have demonstrated and proved that PTMs of histone proteins from DUBs are charged with many considerable tasks in cell cycle, including chromatin formation and structure. During the cell cycle, histones are covered by DNA, and linked histones between each nucleosome are, in total, divided into mainly five kinds of histone protein: H1/H5, H2A, H2B, H3, and H4; however, only H2A, H2B, H3, and H4 are considered as core histones [112]. Several decades ago, Richmond et al. investigated the crystal structure of a histone octamer wrapped in DNA [113]. Although each of the four core histones seems not to be similar in sequence, each has a histone-fold motif which is called helix-loop-helix [114]. Further, it was estimated that all four core histone proteins commonly contain 20–24 % of arginine and lysine and have a highly positive charge [115].
H2A and DUBs
Histone ubiquitination is fundamentally required for the orchestration of nucleus events including chromatin stability, transcriptional regulation, DNA repair, X chromosome inactivation, cell cycle progression, and gene silencing. Despite the fact that chromosomal histone proteins are in a stable state, extra histones not attached to chromatin undergo proteasomal degradation [116]. The first identification of ubiquitination on histone protein (H2A) was studied several decades ago [117]. Further research discovered that histone ubiquitination including cross-talk between ubiquitination and methylation was identified [118]. In addition, trans-histones have provided insights into research on PTMs and cellular mechanisms [119]. The ubiquitination ratio of chromosomal H2A and H2B has shown that more ubiquitins tend to be conjugated at H2A (about 5–15 %) than at H2B (1–2 %) [120–122]. In general, H2A and H2B are conjugated to ubiquitin at their Lys-119 and Lys-120, respectively, and the counteraction of monoUb-histone proteins occurs by several DUBs [123, 124]. The consequence of H2A monoUb on K119 residue tends to be accompanied by gene silencing [125]. Therefore, segregation of ubiquitin from this site by DUBs is consistent with gene activation and cell cycle progression [126]. In addition to the bulk of the monoUb pattern on H2A and H2B, the polyUb form of H2A was also found in many cell types [127]. A recent study has demonstrated other ubiquitin-binding sites of H2A located at the N-terminal sequence, K13, and K15, which are targets for E3 ligase RNF8 and RNF168 upon DDR [128].
Lots of specific binding substrates of H2A are involved in the dissociation of H1 (also known as a linker histone), which links each nucleosome to histone proteins [129, 130]. It has been suggested that ubiquitins bound to H2A are removed for chromatin condensation in mitosis and are related to apoptotic response [131, 132]. Therefore, The H2A deubiquitination is closely related to chromatin stabilization, gene expression, cell cycle progression, and DDR [126, 133]. Several DUBs for the deubiquitination of H2A have been found; USP3 (most homologous to Ubp8 in yeast), USP16 (homologous to Ubp-M in yeast), USP21, USP22 (homologous to Ubp-8 in yeast), USP29, USP44, and Dub-2A [129, 131, 134–137].
Not only is H2B deubiquitinated by USP3, but H2A is also the enzymatic target of USP3, which is the most homologous to Ubp-8 in yeast [138]. H2B deubiquitination by USP3 will also be discussed later. Recent studies have examined and demonstrated that USP3 deubiquitinates ubiquitin conjugated at K13 on H2A, and it plays a vital role in genome stability and cell cycle progression, especially in the S phase transition [137]. Cesare and colleagues observed the tumor development and an unstable pattern of chromosomal integrity in USP3-depeleted animals [139]. Further, USP3 was used to demonstrate the necessity of H2A ubiquitination in response to IR-induced DSB. Overexpressed USP3 led to an interruption in the recruitment of RNF168 at the DSB sites, indicating that the ubiquitinated H2A is indispensable to RNF168-included complex formation [140]. Recently, Hu and colleagues found that a decreased H2A ubiquitination level is also related to USP7 [141]. However, whether USP7 directly deubiquitinates H2A is still unknown. Instead, they demonstrated that the interaction between USP7 and HSCARG (also known as NmrA-like family domain containing 1, NMRAL1) results in reduced PRC1 complex-mediated H2A ubiquitination [141]. HSCARG is a newly identified protein that interacts with USP7 and is included in the dehydrogenase family but does not show dehydrogenase activity [142]. Several functions and substrates of USP16 have been revealed and found in the cell cycle. A recent study identified that the H2A lysine sites for deubiquitination by USP16 were not only K119 but also K15, and that the detachment of ubiquitin from K15 on H2A is responsible for DNA damage response [143]. It has been shown that one of the vital functions of USP16 in the cell cycle was identified by a knockdown experiment [126]. Deficiency of USP16 leads to defects in mitosis, resulting in decreased growth rate and G2/M transition, indicating the importance of USP16 for cell growth and cell cycle progression [126]. Further studies have also suggested that the deubiquitinating activity of USP16 and its important role in cell cycle regulation are prerequisites for S10 phosphorylation on H3, which plays a vital role in genome stability and acts as a marker for chromatin condensation in the cell cycle, especially in mitosis [126, 144]. Based on microarray expression from the regenerating hepatocytes, Nakagawa and colleagues revealed that USP21 conducts a hydrolyase activity on histone H2A proteins, resulting in the detachment of conjugated ubiquitin [136]. Deubiquitinated H2A by USP21 could exert influence on di- or tri-methylation at H3K4, which is required for transcription initiation, via a ‘trans-histone’ pathway [136]. USP21 has been relatively less understood for its histone-related functions in the cell cycle. A further study on the influence of H2A-USP21 on cell cycle regulation is needed. USP22 also deubiquitinates both H2A and H2B by acting as a component of the TFTC/STAGA complexes that function in transcriptional activation in vitro and that have drawn much attention as an important factor involved in either transcription activation or G1/S transition [121, 122, 135]. Zhang et al. demonstrated that the remarkable effect of USP22 depletion by RNA interference (RNAi) is the cell cycle arrest in the G1 phase [135]. USP29 was also found to mediate the deubiquitination of the H2A-conjugated ubiquitin, and its activity results in cell cycle regulation [134]. Both USP29 and USP44 were identified as participating in H2A deubiquitination and playing roles including suppression of IR-induced 53BP1 formation and its recruitment into damaged chromatin sites for DSB [134]. The deubiquitinating activity of USP44 toward H2B was recently investigated with ESCs [145]. So, further studies on USP44-H2B interactions and their following influences on the cell cycle are needed. Recent works have shown that USP46 and USP12 also target and detach the ubiquitin from both H2A and H2B in vitro and in vivo, by interacting with the WD40 repeat-containing protein 48 (WDR48), which is required for these two DUB’s activities [146, 147]. DUB-2A and its deubiquitinating activity toward H2A were also investigated [129]. DUB-2A targets K119 on H2A to counteract the H2A E3 ligase activity and is involved in transcriptional regulation, including transcription initiation and elongation [129]. γH2AX is a member of H2A family and has been elucidated for its PTMs because it is also ubiquitinated by an ubiquitin E3 ligase, RNF168, on K13 and K15 [148]. In addition to the discovery of ubiquitination factors toward γH2AX, the DUB of γH2AX, USP3, was also recently found. Ectopic expression results in deubiquitination of γH2Ax under the UV-induced DNA damage condition [149].
H2B and DUBs
H2B ubiquitination as well as H2A play many important roles in cell cycle regulation and in DNA damage response even though the ubiquitination on H2B does not frequently occur compared to H2A. It was revealed that the ubiquitination at the H2BK123 (K120 in humans) site by the Rad6-Bre1 complex in yeast influences on H3K4 and H3K79 methylation, resulting in Rad53 kinase activation, which is involved in DNA damage response and cell cycle arrest [119, 150]. Therefore, DUBs that antagonize these effects modifying histone proteins might play a role in cell cycle progression. Collectively, less than 10 DUBs involved in the detachment of ubiquitin on H2B have been investigated so far: USP3, USP7, USP12, USP15 (indirectly regulates the ubiquitination of H2B), USP22, USP42, USP44, USP46, and USP49.
It has been studied that USP3 deubiquitinates not only H2A but also H2B-attached ubiquitin [137]. Similarly, not only H2A, but USP7 also is involved in H2B deubiquitination [151]. It interacts with 5′-monophosphate synthetase (GMPS), and its interaction could trigger the H2B ubiquitin cleavage by enzymatic activity for deubiquitination [151]. USP12 and USP46 were co-studied as putative substrates of H2A and H2B, and it was revealed that both have a role in deubiquitinating each histone protein [146]. The activity of these two DUBs was identified from their developmental functions in the Xenopus model [146]. However, cell cycle-related influences from USP12 and USP46 activities toward both H2A and H2B are not clearly understood. Recently, it has been suggested that USP15 indirectly participates in the ubiquitin detachment of non-nucleosomal free H2Bub via direct interaction with the squamous cell carcinoma antigen recognized by T-cells 3 (SART3, also known as TIP110 or p110), but does not have direct interaction with H2B E3 ligase [152]. The deubiquitinating activity of USP22 on H2B has been revealed and is required for SAGA-related gene activation [153]. A further study on the association between USP22 and H2B suggested that USP22 is involved in cell cycle progression [135, 153, 154]. Meanwhile, rather than the relatively well understood effects of USP42 on p53 regulation, a recent study found that USP42 also cleaves the conjugated ubiquitin on H2B through its deubiquitinating activity [75, 145]. In addition, a further study demonstrated that a main function of USP42 on H2B is accompanied by transcriptional regulation [155]. An increased H2Bub level was found under the USP44-knockdown condition, indicating that USP44 negatively regulates H2B ubiquitination [145]. In contrast with the USP44-H2A interaction, during the differentiation of stem cells, USP44-mediated H2B deubiquitination is involved in the regulation of gene expression which is closely related to the cell cycle event [145]. The newly discovered H2B-DUB, USP49, counteracts the ubiquitination of H2B and influences the processing of pre-mRNA both in vitro and in vivo [151].
Research upon histone H2B has not been active in comparison to that of H2A. So, the cell cycle-involved H2B DUBs are not much studied. Further studies on H2B and its DUBs in the cell cycle are needed. The other two core histone proteins, H3 and H4, are poorly understood and their ubiquitin sites have not been found. However, the PTMs of their residues play many critical roles in cell cycle events, and, the ubiquitination or deubiquitination of the previous two core histones (H2A and H2B) could exert profound influences on the PTMs of histones H3 and H4 in many cases.
DUBs for therapeutic targets
The ubiquitin–proteasome system for anticancer drugs has been targeted, and the proteasome inhibitor bortezomib (known as Velcade) was approved by the FDA in 2003 for the treatment of multiple myeloma (MM) and mantle cell lymphoma [156]. However, the ubiquitin–proteasome system is highly incorporated in intracellular regulation, and side effects in treatment with the proteasome inhibiter have arisen [157]. Therefore, the development of specific drugs to minimize toxicity in the ubiquitin–proteasome system is required. Since Sowa et al. mapped the DUBs and their interacting protein networks [28], DUBs are considered as potential therapeutic targets for generation of anticancer drugs. As we described above, DUBs are defined by their ability to dissociate the ubiquitin from substrates that usually contain Lys residue, and all DUBs have conserved motifs, which are important in regulating DUB activity. It is presumed that the active site of each class of DUB has subtle difference, and blocking of specific DUB active site or its substrate binding site by small molecules can be a good target for development of anti-cancer drugs. Understanding of the cell-based assays to identify antagonists and agonists for DUBs is required for drug development. Recently, an excellent review paper has addressed the development of DUB inhibitors by activity-based probes with the cell-based assay [158]. In addition, the possibility of whether DUBs can be used in a chemical drug for cancer treatment has been suggested [9, 159], and the development of anticancer drugs with DUBs based on small-molecule screening is still being investigated by many world-wide research groups. It is feasible that the identified and developed DUB inhibitors have affected and targeted only one DUB [9]. Because the identified DUB inhibitors predominantly have functions for targeting different cell cycle phases, the side effects of DUB inhibitors need to be evaluated and considered from various angles to apply clinical therapy.
Fanconi anemia (FA) is mainly caused by the dysfunction of several FA proteins, and the mutation of 15 FA genes has been identified in FA patients so far [160]. Because most FA is related to a genetic disorder, it has been considered as an important model for the study of several genetic diseases and cancers [66]. The FA pathway is impaired in terms of the repair of DNA interstrand crosslinks (ICLs), and several DNA repair-related proteins are orchestrated in ICLs to complete DNA repair [66]. For example, deubiquitination of both FANCD2 and FANCI terminates the final step of ICLs by the USP1-UAF1 complex [57, 58]. Pimozide and GW7647 were firstly identified as USP1-UAF1 inhibitors, and the treatment of these drugs increases FANCD2 monoubiquitination [161]. However, pimozide and GW7647 have no effect on H2A ubiquitination [161]. Further, the USP1 catalytic activity inhibitor, C527, has been identified [162]. ID1 is an USP1-binding substrate, and it regulates the cell proliferation and cell cycle processes [163]. Functionally, USP1 promotes ID1 stability by its deubiquitinating activity. Analysis for the treatment of C527 has shown that C527 inhibits leukemic cell growth, as shown with pimozide [162]. Lately, ML323 as an USP1-UAF1 inhibitor was screened [164], and the drug was shown to prevent the deubiquitination of PCNA and FANCD2 [164]. The treatment of ML323 had no effect on cell cycle regulation. However, a combination treatment with cisplatin as a DNA damage agent and ML323 increases S1 phase arrest and apoptosis compared to cisplatin along in cancer cells [164].
USP7 is the most important DUB for the p53-mediated cell cycle regulation pathway in several kinds of cancer cells [165], and inhibitors targeting USP7 are currently being considered for preclinical trials [166]. HBX 19,818, P05091, and analogues such as P045204 and P22077, and HBX 41,108, have recently been identified as USP7 inhibitors, and these have shown to be effective as anti-cancer compounds. HBX 19,818 covalently binds USP7 and blocks deubiquitinating activity for USP7, inducing p53-mediated apoptosis in cancer cells [167]. A further study revealed that P05091 has a great anti-proliferative effect on reducing multiple myeloma when combined with other drugs such as dexamethasone, lenalidomide, and/or suberoylanilide hydroxamic acid [168]. A recent study revealed the effect of the USP7 inhibitor P22077 for neuroblastoma (NB), and treatment with P22077 led to p53-mediated cell growth inhibition and apoptosis in an NB mouse model [169]. Because USP7 is highly expressed in NB patients, P022077 can be an NB-specific drug [169]. A natural compound, Spongiacidin C, from the marine sponge Stylissa massa was screened as a USP7 inhibitor recently, and it has shown inhibition of deubiquitinating activity for USP7, but the specific in vivo function remains to be determined [170].
IU1 was identified as a small molecule inhibitor of USP14 [171]. IU1 enhanced the proteasome activity via inhibition of USP14 and stimulated target protein degradation [171]. Further studies have suggested the role of IU1 in cells such as the inhibition of dengue virus replication [172]. In contrast to the role of bortezomib for targeting the inhibition of whole proteasome activity, targeting a specific component of proteasome can suggest a valuable aspect of MM treatment for IU1.
USP30 was first identified as a mitochondrial-associated DUB, and it regulates mitochondria morphology through localization of the outer mitochondrial membrane depending on deubiquitinating activity [173]. A further study added a role of USP30 in mitochondria, revealing that it has an opposite function with parkin as an E3 ligase for mitochondrial degradation [174]. Recently, a diterpenoid derivative 15-oxospiramilactone (S3) was identified as an USP30 inhibitor, which promotes mitochondrial fusion [175]. The effect of S3 remains unevaluated in Parkinson’s disease (PD), but it may prove to be a valuable drug for PD.
Most DUBs share similar domains such as Cys and His and these domains are closed to the regulation of deubiquitinating activity. This has indicated an advantage for developing inhibitors targeting multiple DUBs. Several inhibitors targeting not only one specific DUB, but two or more DUBs have been screened and studied. The role of WP1130, as a JAK-STAT-signaling inhibitor, was characterized for its anti-proliferative and apoptotic effect in chronic myeloid leukemia (CML) through down-regulation of the Bcr/Abl protein [176–178]. A further study identified the ability of WP1130 in that several DUBs, such as USP5, USP9X, USP14, and UCH37, inhibited their deubiquitinating activity through treatment with WP1130 without restriction of the 26S proteasome activity [179]. The observation that the cells form aggresome and undergo apoptosis through treatment with WP1130 has suggested that WP1130 has an effect on the modification of Ub-K63 chains in Bcr/Abl in CML [180]. In addition, apoptosis through down-regulation of the proapoptotic protein Mcl-1 through the inhibition of USP9X through treatment with WP1130 was detected not only in CML but also in several cancers [181–184]. Similar to WP1130, the small molecule b-AP15 inhibits proteasome-associated DUBs such as UCH37 and USP14 without affecting proteasome activity [185, 186]. Upon targeting UCH37 and USP14 by b-AP15, cancer cells undergo apoptosis regardless of the expression of TP53 and anti-apoptotic protein B cell lymphoma 2 (Bcl-2) [185]. Moreover, a recent study identified the effect of b-AP15 on the cell cycle, in which it induces cell cycle arrest through down-regulation of cell-cycle checkpoint proteins, including cdc25c, cdc2, and cyclin B1 in MM [186].
The elegant studies with anti-cancer drugs to define its functions in several cancers solve their uncovered function. For example, paclitaxel has been known as an anticancer drug targeting microtubule assembly [187]. USP4 has drawn attentions for its functions in cell signaling pathways and binding partners during mitosis, and a recent study demonstrated that the down-regulation of USP4 by treating with paclitaxel results in spindle assembly checkpoint bypass [188]. Metformin has been used for diabetes treatment, cancers, and polycystic ovary syndrome (PCOS) [189, 190]. Increasing evidence suggests that metformin affects the inhibition of cell proliferation and tumor growth by induction of cell cycle arrest in the G0-G1 phase [191–193]. In a variety of metformin-induced intracellular signaling, USP7 is positively targeted by treatment with metformin [194]. Recently, 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as a potential inhibitor of USP8 has been identified [195], but their biological functions should be elucidated for pre-clinical trial.
Conclusions
A number of in vivo and in vitro studies on the activity, function, and turnover of proteins have disclosed the complicated molecular cascades that contribute to cell cycle regulation. However, these molecular cascades regulate cell cycle progression and are associated with several PTMs, such as acetylation, methylation, phosphorylation, and ubiquitination. For maintenance of cellular homeostasis, in part, protein turnover needs to be eliminated and synthesized in cells. Most proteins are targeted by either lysosome or the 26S proteasome degradation pathway. In addition, approximately 80 % of intra cellular proteins are followed by proteasomal degradation through the attachment of ubiquitins. Several hundreds of E3 ligases lead to substrate degradation, whereas relatively far fewer numbers of DUBs are responsible for detaching or trimming ubiquitins from ubiquitinated proteins. Therefore, each single DUB might have the responsibility for many substrates that participate in many different types of cellular processes. Although many challenges have arisen, now several international research groups have identified DUB inhibitors at the cell, pre-clinical, and clinical levels to improve or identify the targeting of several diseases. As numerous studies have been validated with DUBs so far, DUBs may be one of the most ideal and attractive targets for several diseases and cancers.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2013-0141).
Abbreviations
- ATM
Ataxia telangiectasia mutated
- ATR
ATM and Rad3-related
- DDR
DNA damage response
- DUB
Deubiquitinating enzyme
- UPP
Ubiquitin proteasomal pathway
- USP
Ubiquitin-specific protease
- UCH
Ubiquitin carboxy terminal hydrolases
- OTU
Ovarian tumor domain
- MJD
Machado–Joseph disease
- JAMM
Jab1/MPN domain-associated metalloisopeptidase
References
- 1.Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013 doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458(7242):1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim KH, Ramakrishna S, Baek KH. Molecular mechanisms and functions of cytokine-inducible deubiquitinating enzymes. Cytokine Growth Factor Rev. 2013;24(5):427–431. doi: 10.1016/j.cytogfr.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover A. Stanhill A (2014) The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim Biophys Acta. 1843;1:86–96. doi: 10.1016/j.bbamcr.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 7.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 8.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15(3):1265–1273. doi: 10.1128/MCB.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim KH, Baek KH. Deubiquitinating enzymes as therapeutic targets in cancer. Curr Pharm Des. 2013;19(22):4039–4052. doi: 10.2174/1381612811319220013. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura N, Harada K, Kato M, Hirose S. Ubiquitin-specific protease 19 regulates the stability of the E3 ubiquitin ligase MARCH6. Exp Cell Res. 2014;328(1):207–216. doi: 10.1016/j.yexcr.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8(4):339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 12.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya S, Ghosh MK. Cell death and deubiquitinases: perspectives in cancer. Biomed Res Int. 2014;2014:435197. doi: 10.1155/2014/435197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahtoe DD, Sixma TK. Layers of DUB regulation. Trends Biochem Sci. 2015;40(8):456–467. doi: 10.1016/j.tibs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol. 2012;9(3):144–155. doi: 10.1038/nrclinonc.2012.3. [DOI] [PubMed] [Google Scholar]
- 16.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 17.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 18.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24(23):3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 20.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 22.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci USA. 2001;98(15):8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol. 2007;11(6):644–653. doi: 10.1016/j.cbpa.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 24.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31(19):5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7(7):517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 26.Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, Lee Y, Ko HS, Lee BD, Poirier GG, Dawson VL, Dawson TM. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci USA. 2011;108(34):14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Wu J, Paudyal SC, You Z, Yu X. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 2013;41(3):1698–1710. doi: 10.1093/nar/gks1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlandi I, Bettiga M, Alberghina L, Vai M. Transcriptional profiling of ubp10 null mutant reveals altered subtelomeric gene expression and insurgence of oxidative stress response. J Biol Chem. 2004;279(8):6414–6425. doi: 10.1074/jbc.M306464200. [DOI] [PubMed] [Google Scholar]
- 30.Kim MS, Yoo KJ, Kang I, Chung HM, Baek KH. A novel cysteine protease HeLa DUB-1 responsible for cleaving the ubiquitin in human ovarian cancer cells. Int J Oncol. 2004;25(2):373–379. [PubMed] [Google Scholar]
- 31.Kim MS, Ramakrishna S, Lim KH, Kim JH, Baek KH. Protein stability of mitochondrial superoxide dismutase SOD2 is regulated by USP36. J Cell Biochem. 2011;112(2):498–508. doi: 10.1002/jcb.22940. [DOI] [PubMed] [Google Scholar]
- 32.Taillebourg E, Gregoire I, Viargues P, Jacomin AC, Thevenon D, Faure M, Fauvarque MO. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy. 2012;8(5):767–779. doi: 10.4161/auto.19381. [DOI] [PubMed] [Google Scholar]
- 33.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, Berger SL. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17(4):585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Emre NC, Berger SL. Histone H2B ubiquitylation and deubiquitylation in genomic regulation. Cold Spring Harb Symp Quant Biol. 2004;69:289–299. doi: 10.1101/sqb.2004.69.289. [DOI] [PubMed] [Google Scholar]
- 35.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25(14):6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calzari L, Orlandi I, Alberghina L, Vai M. The histone deubiquitinating enzyme Ubp10 is involved in rDNA locus control in Saccharomyces cerevisiae by affecting Sir2p association. Genetics. 2006;174(4):2249–2254. doi: 10.1534/genetics.106.063099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orlandi I, Bettiga M, Alberghina L, Nystrom T. Vai M (2010) Sir2-dependent asymmetric segregation of damaged proteins in ubp10 null mutants is independent of genomic silencing. Biochim Biophys Acta. 1803;5:630–638. doi: 10.1016/j.bbamcr.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Chora S, McDonagh B, Sheehan D, Starita-Geribaldi M, Romeo M, Bebianno MJ. Ubiquitination and carbonylation as markers of oxidative-stress in Ruditapes decussatus . Mar Environ Res. 2008;66(1):95–97. doi: 10.1016/j.marenvres.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 39.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18(1):73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 40.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18(1):64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443(7111):590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 42.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6(10):1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 43.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA. 2006;103(8):2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luijsterburg MS, Goedhart J, Moser J, Kool H, Geverts B, Houtsmuller AB, Mullenders LH, Vermeulen W, van Driel R. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J Cell Sci. 2007;120(Pt 15):2706–2716. doi: 10.1242/jcs.008367. [DOI] [PubMed] [Google Scholar]
- 45.Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol. 2013;14(5):269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- 46.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 47.Park JM, Yang SW, Yu KR, Ka SH, Lee SW, Seol JH, Jeon YJ, Chung CH. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol Cell. 2014;54(4):626–638. doi: 10.1016/j.molcel.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 48.Matunis MJ. On the road to repair: PCNA encounters SUMO and ubiquitin modifications. Mol Cell. 2002;10(3):441–442. doi: 10.1016/S1097-2765(02)00653-6. [DOI] [PubMed] [Google Scholar]
- 49.Kirchmaier AL. Ub-family modifications at the replication fork: regulating PCNA-interacting components. FEBS Lett. 2011;585(18):2920–2928. doi: 10.1016/j.febslet.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Cazzalini O, Perucca P, Mocchi R, Sommatis S, Prosperi E, Stivala LA. DDB2 association with PCNA is required for its degradation after UV-induced DNA damage. Cell Cycle. 2014;13(2):240–248. doi: 10.4161/cc.26987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Lubin A, Chen H, Sun Z, Gong F. The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability. Cell Cycle. 2012;11(23):4378–4384. doi: 10.4161/cc.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang Q, Wani AA. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquinating XPC and preventing XPC from UV-induced and VCP/p97-regulated proteolysis. J Biol Chem. 2014 doi: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18(1):99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, Barbour H, Corbeil L, Hebert J, Drobetsky E, Masson JY, Di Noia JM, el Affar B. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014;111(1):285–290. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13(3):153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Santisteban I, Peters GJ, Giovannetti E, Rodriguez JA. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol Cancer. 2013;12:91. doi: 10.1186/1476-4598-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17(3):331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D’Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28(5):786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 59.Villamil MA, Chen J, Liang Q, Zhuang Z. A noncanonical cysteine protease USP1 is activated through active site modulation by USP1-associated factor 1. Biochemistry. 2012;51(13):2829–2839. doi: 10.1021/bi3000512. [DOI] [PubMed] [Google Scholar]
- 60.Garcia-Santisteban I, Zorroza K, Rodriguez JA. Two nuclear localization signals in USP1 mediate nuclear import of the USP1/UAF1 complex. PLoS One. 2012;7(6):e38570. doi: 10.1371/journal.pone.0038570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villamil MA, Liang Q, Chen J, Choi YS, Hou S, Lee KH, Zhuang Z. Serine phosphorylation is critical for the activation of ubiquitin-specific protease 1 and its interaction with WD40-repeat protein UAF1. Biochemistry. 2012;51(45):9112–9123. doi: 10.1021/bi300845s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D’Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16(2):314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D’Andrea AD. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol. 2011;31(12):2462–2469. doi: 10.1128/MCB.05058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park E, Kim JM, Primack B, Weinstock DM, Moreau LA, Parmar K, D’Andrea AD. Inactivation of Uaf1 causes defective homologous recombination and early embryonic lethality in mice. Mol Cell Biol. 2013;33(22):4360–4370. doi: 10.1128/MCB.00870-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–518. doi: 10.1016/S1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 66.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26(13):1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JT, Gu W. SIRT1: regulator of p53 deacetylation. Genes Cancer. 2013;4(3–4):112–117. doi: 10.1177/1947601913484496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hock AK, Vousden KH. The role of ubiquitin modification in the regulation of p53. Biochim Biophys Acta. 2014;2014(1):137–149. doi: 10.1016/j.bbamcr.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 70.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26(4):976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416(6881):648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 72.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140(3):384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ke JY, Dai CJ, Wu WL, Gao JH, Xia AJ, Liu GP, Lv KS, Wu CL. USP11 regulates p53 stability by deubiquitinating p53. J Zhejiang Univ Sci B. 2014;15(12):1032–1038. doi: 10.1631/jzus.B1400180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Li Y, Hao Y, Choi A, Ke H, Ma D, Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J. 2011;30(24):4921–4930. doi: 10.1038/emboj.2011.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31(3):576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo J, Lu Z, Lu X, Chen L, Cao J, Zhang S, Ling Y, Zhou X. OTUD5 regulates p53 stability by deubiquitinating p53. PLoS One. 2013;8(10):e77682. doi: 10.1371/journal.pone.0077682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burrows JF, McGrattan MJ, Johnston JA. The DUB/USP17 deubiquitinating enzymes, a multigene family within a tandemly repeated sequence. Genomics. 2005;85(4):524–529. doi: 10.1016/j.ygeno.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 79.Burrows JF, Kelvin AA, McFarlane C, Burden RE, McGrattan MJ, De la Vega M, Govender U, Quinn DJ, Dib K, Gadina M, Scott CJ, Johnston JA. USP17 regulates Ras activation and cell proliferation by blocking RCE1 activity. J Biol Chem. 2009;284(14):9587–9595. doi: 10.1074/jbc.M807216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de la Vega M, Burrows JF, McFarlane C, Govender U, Scott CJ, Johnston JA. The deubiquitinating enzyme USP17 blocks N-Ras membrane trafficking and activation but leaves K-Ras unaffected. J Biol Chem. 2010;285(16):12028–12036. doi: 10.1074/jbc.M109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramakrishna S, Suresh B, Baek KH. Biological functions of hyaluronan and cytokine-inducible deubiquitinating enzymes. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbcan.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 82.Zhu Y, Carroll M, Papa FR, Hochstrasser M, D’Andrea AD. DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc Natl Acad Sci USA. 1996;93(8):3275–3279. doi: 10.1073/pnas.93.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee MY, Ajjappala BS, Kim MS, Oh YK, Baek KH. DUB-1, a fate determinant of dynein heavy chain in B-lymphocytes, is regulated by the ubiquitin-proteasome pathway. J Cell Biochem. 2008;105(6):1420–1429. doi: 10.1002/jcb.21961. [DOI] [PubMed] [Google Scholar]
- 84.Lu Y, Adegoke OA, Nepveu A, Nakayama KI, Bedard N, Cheng D, Peng J, Wing SS. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol Cell Biol. 2009;29(2):547–558. doi: 10.1128/MCB.00329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu Y, Bedard N, Chevalier S, Wing SS. Identification of distinctive patterns of USP19-mediated growth regulation in normal and malignant cells. PLoS One. 2011;6(1):e15936. doi: 10.1371/journal.pone.0015936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, Wilkinson KD. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68(17):6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, Liu J, O’Rourke K, Dixit VM, Wilson AC. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29(8):2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27(1):107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 89.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284(49):34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, Kirkpatrick DS, Jackson PK, Fang G, Dixit VM. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell. 2011;42(4):511–523. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 91.Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burrows AC, Prokop J, Summers MK. Skp1-Cul1-F-box ubiquitin ligase (SCF(betaTrCP))-mediated destruction of the ubiquitin-specific protease USP37 during G2-phase promotes mitotic entry. J Biol Chem. 2012;287(46):39021–39029. doi: 10.1074/jbc.M112.390328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Das CM, Taylor P, Gireud M, Singh A, Lee D, Fuller G, Ji L, Fangusaro J, Rajaram V, Goldman S, Eberhart C, Gopalakrishnan V. The deubiquitylase USP37 links REST to the control of p27 stability and cell proliferation. Oncogene. 2013;32(13):1691–1701. doi: 10.1038/onc.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duro E, Marston AL. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes Dev. 2015;29(2):109–122. doi: 10.1101/gad.255554.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Topham CH, Taylor SS. Mitosis and apoptosis: how is the balance set? Curr Opin Cell Biol. 2013;25(6):780–785. doi: 10.1016/j.ceb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 96.Janssen A, Medema RH. Mitosis as an anti-cancer target. Oncogene. 2011;30(25):2799–2809. doi: 10.1038/onc.2011.30. [DOI] [PubMed] [Google Scholar]
- 97.Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339(6222):280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 98.Zur A, Brandeis M. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001;20(4):792–801. doi: 10.1093/emboj/20.4.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schaefer JB, Morgan DO. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem. 2011;286(52):45186–45196. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Leuken RJ, Luna-Vargas MP, Sixma TK, Wolthuis RM, Medema RH. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle. 2008;7(17):2710–2719. doi: 10.4161/cc.7.17.6553. [DOI] [PubMed] [Google Scholar]
- 101.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446(7138):876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Y, Foreman O, Wigle DA, Kosari F, Vasmatzis G, Salisbury JL, van Deursen J, Galardy PJ. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest. 2012;122(12):4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J Cell Biol. 2009;184(1):13–19. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3(6):689–692. doi: 10.4161/cc.3.6.924. [DOI] [PubMed] [Google Scholar]
- 105.Giovinazzi S, Sirleto P, Aksenova V, Morozov VM, Zori R, Reinhold WC, Ishov AM. Usp7 protects genomic stability by regulating Bub3. Oncotarget. 2014;5(11):3728–3742. doi: 10.18632/oncotarget.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 107.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25(24):5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20(18):5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu N, Liu C, Bai C, Han YP, Cho WC, Li Q. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int J Mol Sci. 2013;14(6):10749–10760. doi: 10.3390/ijms140610749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kabuta T, Mitsui T, Takahashi M, Fujiwara Y, Kabuta C, Konya C, Tsuchiya Y, Hatanaka Y, Uchida K, Hohjoh H, Wada K. Ubiquitin C-terminal hydrolase L1 (UCH-L1) acts as a novel potentiator of cyclin-dependent kinases to enhance cell proliferation independently of its hydrolase activity. J Biol Chem. 2013;288(18):12615–12626. doi: 10.1074/jbc.M112.435701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15(11):703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 113.Ferreri AJ, Illerhaus G, Zucca E, Cavalli F, International Extranodal Lymphoma Study G Flows and flaws in primary central nervous system lymphoma. Nat Rev Clin Oncol. 2010 doi: 10.1038/nrclinonc.2010.9-c1. [DOI] [PubMed] [Google Scholar]
- 114.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88(22):10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hadnagy A, Beaulieu R, Balicki D. Histone tail modifications and noncanonical functions of histones: perspectives in cancer epigenetics. Mol Cancer Ther. 2008;7(4):740–748. doi: 10.1158/1535-7163.MCT-07-2284. [DOI] [PubMed] [Google Scholar]
- 116.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11(8):925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J Biol Chem. 1975;250(18):7182–7187. [PubMed] [Google Scholar]
- 118.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277(32):28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 119.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418(6893):104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17(22):2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 121.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29(1):92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 122.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8(4):284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 123.Osley MA. Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomic Proteomic. 2006;5(3):179–189. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- 124.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 125.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 126.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449(7165):1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 127.Nickel BE, Allis CD, Davie JR. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28(3):958–963. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- 128.Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 2012;11(13):2538–2544. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27(4):609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29(1):69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mimnaugh EG, Kayastha G, McGovern NB, Hwang SG, Marcu MG, Trepel J, Cai SY, Marchesi VT, Neckers L. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 2001;8(12):1182–1196. doi: 10.1038/sj.cdd.4400924. [DOI] [PubMed] [Google Scholar]
- 132.Raboy B, Parag HA, Kulka RG. Conjugation of [125I]ubiquitin to cellular proteins in permeabilized mammalian cells: comparison of mitotic and interphase cells. EMBO J. 1986;5(5):863–869. doi: 10.1002/j.1460-2075.1986.tb04296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Belle JI, Nijnik A. H2A-DUBbing the mammalian epigenome: expanding frontiers for histone H2A deubiquitinating enzymes in cell biology and physiology. Int J Biochem Cell Biol. 2014;50:161–174. doi: 10.1016/j.biocel.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 134.Mosbech A, Lukas C, Bekker-Jensen S, Mailand N. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J Biol Chem. 2013;288(23):16579–16587. doi: 10.1074/jbc.M113.459917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29(1):102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22(1):37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17(22):1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 138.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17(21):2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lancini C, van den Berk PC, Vissers JH, Gargiulo G, Song JY, Hulsman D, Serresi M, Tanger E, Blom M, Vens C, van Lohuizen M, Jacobs H, Citterio E. Tight regulation of ubiquitin-mediated DNA damage response by USP3 preserves the functional integrity of hematopoietic stem cells. J Exp Med. 2014;211(9):1759–1777. doi: 10.1084/jem.20131436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 141.Hu B, Li S, Zhang X, Zheng X. HSCARG, a novel regulator of H2A ubiquitination by downregulating PRC1 ubiquitin E3 ligase activity, is essential for cell proliferation. Nucleic Acids Res. 2014;42(9):5582–5593. doi: 10.1093/nar/gku230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zheng X, Dai X, Zhao Y, Chen Q, Lu F, Yao D, Yu Q, Liu X, Zhang C, Gu X, Luo M. Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein. Proc Natl Acad Sci USA. 2007;104(21):8809–8814. doi: 10.1073/pnas.0700480104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Z, Yang H, Wang H. The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage. J Biol Chem. 2014;289(47):32883–32894. doi: 10.1074/jbc.M114.599605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, Garcia-Muse T, Aguilera A. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52(4):583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 145.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, Feldmesser E, Brik A, Yu X, Hanna J, Aberdam D, Domany E, Oren M. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46(5):662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Joo HY, Jones A, Yang C, Zhai L, Smith ADt, Zhang Z, Chandrasekharan MB, Sun ZW, Renfrow MB, Wang Y, Chang C, Wang H. Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J Biol Chem. 2011;286(9):7190–7201. doi: 10.1074/jbc.M110.158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cohn MA, Kee Y, Haas W, Gygi SP, D’Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem. 2009;284(8):5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150(6):1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 149.Sharma N, Zhu Q, Wani G, He J, Wang QE, Wani AA. USP3 counteracts RNF168 via deubiquitinating H2A and gammaH2AX at lysine 13 and 15. Cell Cycle. 2014;13(1):106–114. doi: 10.4161/cc.26814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280(11):9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 151.Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, Erdjument-Bromage H, Renfrow M, He H, Tempst P, Townes TM, Giles KE, Ma L, Wang H. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013;27(14):1581–1595. doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29(6):653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 153.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115(6):1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 155.Hock AK, Vigneron AM, Vousden KH. Ubiquitin-specific peptidase 42 (USP42) functions to deubiquitylate histones and regulate transcriptional activity. J Biol Chem. 2014;289(50):34862–34870. doi: 10.1074/jbc.M114.589267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 157.Palumbo A, Mateos MV, Bringhen S, San Miguel JF. Practical management of adverse events in multiple myeloma: can therapy be attenuated in older patients? Blood Rev. 2011;25(4):181–191. doi: 10.1016/j.blre.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 158.Kramer HB, Nicholson B, Kessler BM, Altun M. Detection of ubiquitin-proteasome enzymatic activities in cells: application of activity-based probes to inhibitor development. Biochim Biophys Acta. 2012;1823(11):2029–2037. doi: 10.1016/j.bbamcr.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wing SS. Deubiquitinating enzymes—the importance of driving in reverse along the ubiquitin-proteasome pathway. Int J Biochem Cell Biol. 2003;35(5):590–605. doi: 10.1016/S1357-2725(02)00392-8. [DOI] [PubMed] [Google Scholar]
- 160.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493(7432):356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A, Zhuang Z. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol. 2011;18(11):1390–1400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mistry H, Hsieh G, Buhrlage SJ, Huang M, Park E, Cuny GD, Galinsky I, Stone RM, Gray NS, D’Andrea AD, Parmar K. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther. 2013;12(12):2651–2662. doi: 10.1158/1535-7163.MCT-13-0103-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146(6):918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 164.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, Luci DK, Yuan B, Simeonov A, Jadhav A, Xiao H, Wang Y, Maloney DJ, Zhuang Z. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10(4):298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cheon KW, Baek KH. HAUSP as a therapeutic target for hematopoietic tumors (review) Int J Oncol. 2006;28(5):1209–1215. [PubMed] [Google Scholar]
- 166.Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys. 2011;60(1–2):61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 167.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, Colland F. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19(4):467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 168.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, Weinstock J, Kingsbury WD, Hideshima T, Shah PK, Minvielle S, Altun M, Kessler BM, Orlowski R, Richardson P, Munshi N, Anderson KC. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH, Ma IT, Rojas Y, Zhao Y, Yu Y, Zhang H, Shohet JM, Nuchtern JG, Kim ES, Yang J. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013;4:e867. doi: 10.1038/cddis.2013.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yamaguchi M, Miyazaki M, Kodrasov MP, Rotinsulu H, Losung F, Mangindaan RE, de Voogd NJ, Yokosawa H, Nicholson B, Tsukamoto S. Spongiacidin C, a pyrrole alkaloid from the marine sponge Stylissa massa, functions as a USP7 inhibitor. Bioorg Med Chem Lett. 2013;23(13):3884–3886. doi: 10.1016/j.bmcl.2013.04.066. [DOI] [PubMed] [Google Scholar]
- 171.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nag DK, Finley D. A small-molecule inhibitor of deubiquitinating enzyme USP14 inhibits dengue virus replication. Virus Res. 2012;165(1):103–106. doi: 10.1016/j.virusres.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 173.Nakamura N, Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol Biol Cell. 2008;19(5):1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510(7505):370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 175.Yue W, Chen Z, Liu H, Yan C, Chen M, Feng D, Yan C, Wu H, Du L, Wang Y, Liu J, Huang X, Xia L, Liu L, Wang X, Jin H, Wang J, Song Z, Hao X, Chen Q. A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Res. 2014;24(4):482–496. doi: 10.1038/cr.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bartholomeusz GA, Talpaz M, Kapuria V, Kong LY, Wang S, Estrov Z, Priebe W, Wu J, Donato NJ. Activation of a novel Bcr/Abl destruction pathway by WP1130 induces apoptosis of chronic myelogenous leukemia cells. Blood. 2007;109(8):3470–3478. doi: 10.1182/blood-2006-02-005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bartholomeusz G, Talpaz M, Bornmann W, Kong LY, Donato NJ. Degrasyn activates proteasomal-dependent degradation of c-Myc. Cancer Res. 2007;67(8):3912–3918. doi: 10.1158/0008-5472.CAN-06-4464. [DOI] [PubMed] [Google Scholar]
- 178.Pham LV, Tamayo AT, Li C, Bornmann W, Priebe W, Ford RJ. Degrasyn potentiates the antitumor effects of bortezomib in mantle cell lymphoma cells in vitro and in vivo: therapeutic implications. Mol Cancer Ther. 2010;9(7):2026–2036. doi: 10.1158/1535-7163.MCT-10-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70(22):9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 180.Sun H, Kapuria V, Peterson LF, Fang D, Bornmann WG, Bartholomeusz G, Talpaz M, Donato NJ. Bcr-Abl ubiquitination and Usp9x inhibition block kinase signaling and promote CML cell apoptosis. Blood. 2011;117(11):3151–3162. doi: 10.1182/blood-2010-03-276477. [DOI] [PubMed] [Google Scholar]
- 181.Peddaboina C, Jupiter D, Fletcher S, Yap JL, Rai A, Tobin RP, Jiang W, Rascoe P, Rogers MK, Smythe WR, Cao X. The downregulation of Mcl-1 via USP9X inhibition sensitizes solid tumors to Bcl-xl inhibition. BMC Cancer. 2012;12:541. doi: 10.1186/1471-2407-12-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang S, Kollipara RK, Srivastava N, Li R, Ravindranathan P, Hernandez E, Freeman E, Humphries CG, Kapur P, Lotan Y, Fazli L, Gleave ME, Plymate SR, Raj GV, Hsieh JT, Kittler R. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proc Natl Acad Sci USA. 2014;111(11):4251–4256. doi: 10.1073/pnas.1322198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cox JL, Wilder PJ, Wuebben EL, Ouellette MM, Hollingsworth MA, Rizzino A. Context-dependent function of the deubiquitinating enzyme USP9X in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2014;15(8):1042–1052. doi: 10.4161/cbt.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Thrane S, Pedersen AM, Thomsen MB, Kirkegaard T, Rasmussen BB, Duun-Henriksen AK, Laenkholm AV, Bak M, Lykkesfeldt AE, Yde CW. A kinase inhibitor screen identifies Mcl-1 and Aurora kinase A as novel treatment targets in antiestrogen-resistant breast cancer cells. Oncogene. 2014 doi: 10.1038/onc.2014.351. [DOI] [PubMed] [Google Scholar]
- 185.D’Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17(12):1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 186.Tian Z, D’Arcy P, Wang X, Ray A, Tai YT, Hu Y, Carrasco RD, Richardson P, Linder S, Chauhan D, Anderson KC. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123(5):706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Le XF, Bast RC., Jr Src family kinases and paclitaxel sensitivity. Cancer Biol Ther. 2011;12(4):260–269. doi: 10.4161/cbt.12.4.16430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24(13):1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Johnson NP. Metformin use in women with polycystic ovary syndrome. Ann Transl Med. 2014;2(6):56. doi: 10.3978/j.issn.2305-5839.2014.04.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kasznicki J, Sliwinska A, Drzewoski J. Metformin in cancer prevention and therapy. Ann Transl Med. 2014;2(6):57. doi: 10.3978/j.issn.2305-5839.2014.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hirsch HA, Iliopoulos D, Struhl K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc Natl Acad Sci USA. 2013;110(3):972–977. doi: 10.1073/pnas.1221055110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67(22):10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- 193.Kisfalvi K, Eibl G, Sinnett-Smith J, Rozengurt E. Metformin disrupts crosstalk between G protein-coupled receptor and insulin receptor signaling systems and inhibits pancreatic cancer growth. Cancer Res. 2009;69(16):6539–6545. doi: 10.1158/0008-5472.CAN-09-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xu Y, Lu S. Metformin inhibits esophagus cancer proliferation through upregulation of USP7. Cell Physiol Biochem. 2013;32(5):1178–1186. doi: 10.1159/000354517. [DOI] [PubMed] [Google Scholar]
- 195.Colombo M, Vallese S, Peretto I, Jacq X, Rain JC, Colland F, Guedat P. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. Chem Med Chem. 2010;5(4):552–558. doi: 10.1002/cmdc.200900409. [DOI] [PubMed] [Google Scholar]