Abstract
Methionine biosynthesis relies on the sequential catalysis of multiple enzymes. Escherichia coli, the main bacteria used in research and industry for protein production and engineering, utilizes the three-step trans-sulfurylation pathway catalyzed by L-homoserine O-succinyl transferase, cystathionine gamma synthase and cystathionine beta lyase to convert L-homoserine to L-homocysteine. However, most bacteria employ the two-step direct-sulfurylation pathway involving L-homoserine O-acetyltransferases and O-acetyl homoserine sulfhydrylase. We previously showed that a methionine-auxotroph Escherichiacoli strain (MG1655) with deletion of metA, encoding for L-homoserine O-succinyl transferase, and metB, encoding for cystathionine gamma synthase, could be complemented by introducing the genes metX, encoding for L-homoserine O-acetyltransferases and metY, encoding for O-acetyl homoserine sulfhydrylase, from various sources, thus altering the Escherichia coli methionine biosynthesis metabolic pathway to direct-sulfurylation. However, introducing metX and metY from Corynebacterium glutamicum failed to complement methionine auxotrophy. Herein, we generated a randomized genetic library based on the metX and metY of Corynebacterium glutamicum and transformed it into a methionine-auxotrophic Escherichia coli strain lacking the metA and metB genes. Through multiple enrichment cycles, we successfully isolated active clones capable of growing in M9 minimal media. The dominant metX mutations in the evolved methionine-autotrophs Escherichia coli were L315P and H46R. Interestingly, we found that a metY gene encoding only the N-terminus 106 out of 438 amino acids of the wild-type MetY enzyme is functional and supports the growth of the methionine auxotroph. Recloning the new genes into the original plasmid and transforming them to methionine auxotroph Escherichia coli validated their functionality. These results show that directed enzyme-evolution enables fast and simultaneous engineering of new active variants within the Escherichia coli methionine direct-sulfurylation pathway, leading to efficient complementation.
Keywords: Methionine biosynthesis, Directed enzyme evolution, Direct-sulfurylation, L-homoserine O-Acetyltransferases, O-acetyl homoserine sulfhydrylase, Escherichia coli
Highlights
-
•
Altering the methionine biosynthesis pathway in E. coli by enzyme directed-evolution.
-
•
Identify novel mutations in the enzyme MetX.
-
•
Identify a large deletion in MetY that retains the enzyme functionality.
-
•
Parallel simultaneous evolution of MetX and MetY enzymes.
-
•
Exploring the potential of E. coli for bioproduction of methionine.
1. Introduction
Methionine biosynthesis is an important metabolic pathway in microorganisms and plants, encompassing a series of consecutive enzymatic steps (Ravanel et al., 1998; Valley et al., 2012; Wirtz and Droux, 2005). As an essential amino acid for vertebrates, methionine is also highly sought for biotechnological applications, including in the food, feed, and pharmaceutical industries (Neubauer and Landecker, 2021). Within the cellular environment, methionine is vital not only for initiating protein biosynthesis but also for protein folding and stability (Aledo, 2019). Moreover, it plays a significant role as the starting point for various metabolic processes, including S-adenosylmethionine (SAM), a prominent methyl group donor in cell methylation reactions (Aledo, 2019; Bennett et al., 2009; Brosnan et al., 2007).
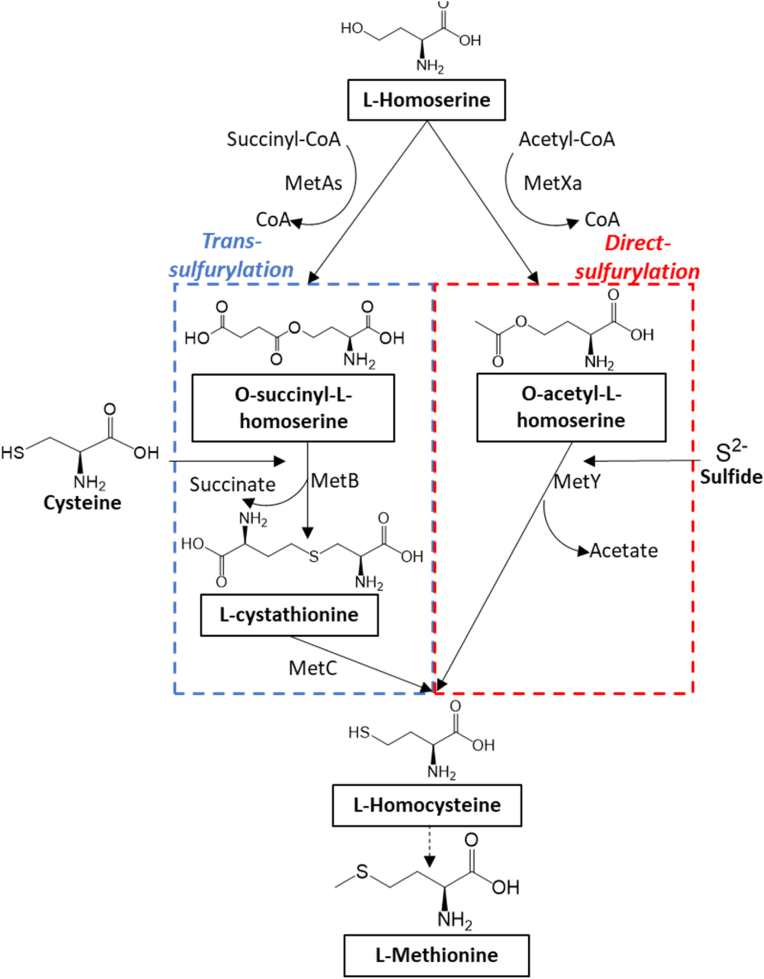
Bacterial methionine biosynthesis relies on two distinct pathways, direct-sulfurylation and trans-sulfurylation (Fig. 1). Both pathways convert L-homoserine to L-homocysteine, which is further converted by the enzyme methionine synthase (metE/H) to L-methionine (Ferla and Patrick, 2014; Hacham et al., 2003; Vermeij and Kertesz, 1999). However, the two pathways differ in the sulfur source used and the number of steps required to synthesize L-homocysteine (Foglino et al., 1995; Ito et al., 2008; Vermeij and Kertesz, 1999). In the trans-sulfurylation pathway, three steps are catalyzed by the enzymes L-homoserine O-succinyl transferase (HST; EC 2.3.1.46, MetA), cystathionine gamma synthase (CgS; EC 2.5.1.48, MetB) and cystathionine beta lyase (CbL; EC 4.4.1.13, MetC) (Aitken et al., 2011). MetA synthesizes O-succinyl L-homoserine from homoserine and succinyl CoA. MetB synthesizes cystathionine from cysteine and O-succinyl L-homoserine. MetC converts cystathionine into L-homocysteine. The direct-sulfurylation is a more efficient two-step pathway catalyzed by the enzymes L-homoserine O-acetyltransferases (HAT; EC 2.3.1.31, MetX) synthesizes O-acetyl L-homoserine from homoserine and acetyl-CoA and the O-acetyl homoserine sulfhydrylase (OAHS; EC 2.5.1.49, MetY), synthesizes L-homocysteine from O-acetyl L-homoserine and an inorganic sulfur source (Ferla and Patrick, 2014).
Fig. 1.
Direct-sulfurylation and trans-sulfurylation methionine biosynthesis pathways in bacteria. The first step in methionine biosynthesis involves the activation of homoserine through an acylation step catalyzed by two enzymes encoded by metAs and metXa genes. In the trans-sulfurylation pathway (blue box), the enzyme homoserine succinyl transferase (HST, MetAs) converts homoserine and succinyl-CoA into O-succinyl-L-homoserine (OSH). Cysteine and O-succinyl-L-homoserine (OSH) are converted into cystathionine by cystathionine-γ-synthase (CgS, MetB). Cystathionine is converted into homocysteine by cystathionine-β-lyase (CbL, MetC). In the direct-sulfurylation pathway, the enzyme homoserine acetyl transferase (HAT, MetXa) converts homoserine and acetyl-CoA into O-acetyl-L-homoserine (OAH) which in a single step is further converted into homocysteine by O-acetylhomoserine sulfhydrylase (OAHS, MetY). In both pathways, the enzyme methionine synthase (MetE/H, not shown in the figure) converts homocysteine to methionine.
Most bacteria predominantly use the direct-sulfurylation pathway for methionine biosynthesis. However, E. coli is among the group of bacteria that employs the trans-sulfurylation pathway (Bastard et al., 2017; Brewster et al., 2021; Hwang et al., 2007; Weissbach and Brot, 1991). This pathway in E. coli is well characterized and was found to be tightly regulated due to the high energetic cost associated with methionine production (Kaleta et al., 2013), and with feedback inhibition from methionine and SAM (Ferla and Patrick, 2014; Hacham et al., 2003; Sbodio et al., 2019). Recent studies have demonstrated that introducing direct-sulfurylation genes from various organisms into E. coli can significantly enhance its methionine biosynthesis levels, bypassing the strict regulation of the natural trans-sulfurylation pathway (Belfaiza et al., 1998; Bourhy et al., 1997; Ochrombel et al., 2021; Schipp et al., 2020). Moreover, we recently showed that by completely removing the trans-sulfurylation enzymes encoded by metA and metB from E. coli and substituting them with the direct-sulfurylation genes metX and metY, E. coli could successfully synthesize methionine via the direct-sulfurylation pathway (Gruzdev et al., 2023). Interestingly, while MetX and MetY enzymes from bacteria such as Deinococcus geothermalis (DG) and Cyclobacterium marinum (CM) effectively restored methionine production in methionine-auxotrophic E. coli, the insertion of this pair of genes from Corynebacterium glutamicum (CG), a bacteria known to utilize both of the trans and direct-sulfurylation pathways, failed to do so (Gruzdev et al., 2023; Hwang et al., 2002). To understand the molecular basis of the complementation and restore methionine biosynthesis in auxotrophic E. coli, we evolved the non-complementing metX and metY genes from CG to produce a functionally active enzyme pair. The concept of directed enzyme evolution involves creating a diverse genetic library and applying appropriate selection pressure, enriching and identifying variants with the desired activity profile (Badran and Liu, 2015; Wang et al., 2021; Xu et al., 2018). The process relies on two central steps repeated iteratively: introducing genetic diversity, either randomly or through rational design, and systematically selecting variants that display the targeted phenotypes (Cherry and Fidantsef, 2003; Wang et al., 2021). In this study, we employed error-prone PCR to create a genetic library with random mutations in the MetX and MetY enzymes (Wilson and Keefe, 2001). Following several selection rounds, we successfully identified and further characterized promising new variants of the MetX and MetY enzymes that enabled the recovery of methionine-auxotroph E. coli on a methionine-depleted minimal medium.
2. Materials and methods
2.1. Generation of auxotroph E. coli
The generation of methionine auxotroph E. coli was described elsewhere (Gruzdev et al., 2023). Briefly, genes in E. coli MG1655 were deleted by the lambda red recombinase procedure (Datsenko and Wanner, 2000), with the pKD4 plasmid carrying the KnR cassette as a PCR reaction template. Following each knockout cycle, the cassette was removed, and the triple knockout ΔmetABJ was further generated.
2.2. M9 minimal media
The M9 minimal media was prepared by dissolving the following components in 1 L H2O, 6.0 g Na2HPO4, 3.0 g KH2PO4, 0.5 g NaCl, 1.5 g NH4Cl, 4.0 g Glucose, 0.3 g MgSO4, 0.02 g CaCl2, 250 μL trace metal mix and 250 μL Vitamin mix (Tables S2 and S3, respectively).
2.3. Complementation of ΔmetABJ with a plasmid carrying the metX/Y genes
Electrocompetent ΔmetABJ mutants were transformed with pTrcHis plasmid carrying the CG metXY synthetic operon. Following transformation, several colonies growing on LB agar plates supplemented with 50 μg/ml ampicillin and kanamycin were tested for the presence of the correct plasmid by colony PCR (primers are listed in Table S4). Positive clones were inoculated in a 5 ml M9 and incubated overnight at 37 °C (constant orbital shaking 250 rpm).
2.4. Growth evaluation of the bacteria with and without methionine
For the growth evaluation assays, bacteria were inoculated into a 96-well plate filled with M9 media with or without the addition of 20 μM methionine. Each well was supplemented with 100 μg/mL ampicillin and 50 μg/mL kanamycin. Growth was monitored at 37 °C with orbital shaking and absorbance at 600 nm was measured every 10 min in a plate reader (Synergy H1 microplate reader, BioTek).
2.5. Construction of a randomized genetic library of metX and metY and enrichment cycles
A genetic library was constructed based on the metX and metY genes with the GeneMorph II Random Mutagenesis Kit (Agilent, Santa Clara, CA, USA), adjusted to produce an average of 4 nonsynonymous mutations per gene. Following mutagenesis PCR, libraries were cloned back into the original vector and transformed into ΔmetABJ E. coli cells and then plated on an LB plate supplemented with 100 μg/mL amp and 50 μg/mL kana. All colonies were collected and grown for enrichment in 200 ml M9 minimal media supplemented with 100 μg/mL amp, 50 μg/mL kan, and 0.1 mM IPTG at 37 °C. For enrichment, at OD = 0.8, 1 ml of the culture was rediluted in M9 and regrown. Primers used for error-prone PCR and sequencing of the construct are listed in Table S4. The full sequence of the metX/Y construct is listed in Table S5.
2.6. Structural modeling and visualization
Protein structures were predicted by AlphaFold2 with MMseqs2. Structural visualization was performed using UCSF ChimeraX (Pettersen et al., 2021). Calculations of RMSDs were done in ChimeraX using the Needleman-Wunsch alignment algorithm. The Similarity matrix was produced by BLOSUM-62 and the sequence similarity score was calculated by Clustal 2.1 (Madeira et al., 2022).
2.7. Molecular dynamics simulation
The coordinates of the wild-type protein and the L315P mutant were modeled using AlphaFold2 with MMseqs2 and Amber relaxation (Mirdita et al., 2022). The lowest energy model was then implemented into CHARMM GUI (Brooks et al., 2009; Lee et al., 2016) for proper formulation of the syntax and ionization states. Molecular dynamics (MD) simulations were done using GROMACS software version 2020.1 using charmm27 force field. The protein was embedded in a cubic water box, with a distance of 1.0 nm of solvent on all sides of the protein. Water molecules were described using the TIP3P model. Counter ions (Cl− and Na+) were inserted to achieve a neutral simulation cell. Energy minimization was carried out using the steepest descent algorithm, followed by the conjugate gradient algorithm. The system was equilibrated by 50 ps MD simulation in the canonical (NVT) ensemble using the modified Berendsen thermostat (Bussi et al., 2007) for fixing the temperature of the system at 310 K, followed by 50 ps MD simulation in the isothermal–isobaric (NPT) ensemble, using the Parrinello–Rahman pressure coupling method (Akya et al., 2019; Bussi et al., 2007) for maintaining the pressure at fixed 1 bar. The MD simulation, starting from the previous NPT simulation, was equilibrated for another 2 ns with no position restraints, followed by an additional 8 ns of dynamics for averaging H bonds.
3. Results
3.1. E. coli ΔmetABJ transformed with metX/Y from CG is a methionine-auxotroph strain
To test the ability of E. coli to produce sufficient amounts of methionine to support growth, we used a strain auxotrophic for methionine (ΔmetABJ) expressing the CG wild-type metX and metY genes (NCBI OQ291222) from a plasmid (ΔmetABJ-CG) (Gruzdev et al., 2023). We then evaluated the ability of this strain to grow on M9 minimal media, which contained glucose, ammonium chloride, and MgSO4 as the carbon, nitrogen, and sole sulfate sources, respectively. As depicted in Fig. 2, the ΔmetABJ-CG strain showed growth in the presence of methionine (red curve) but failed to grow without it (blue curve). This indicates that methionine limits bacterial growth and that the metX and metY genes from CG are insufficient for autonomous methionine production in E. coli, thus necessitating external supplementation of methionine.
Fig. 2.
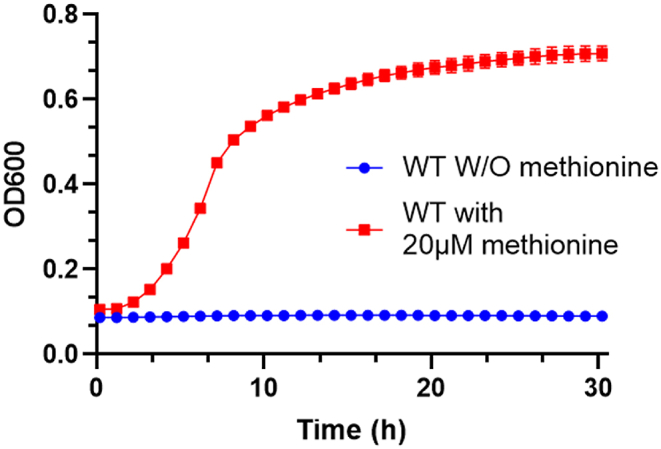
Methionine Dependency of the ΔmetABJ-CG Strain. The graph shows the growth dependency of the ΔmetABJ-CG strain on methionine in M9 minimal media, comparing the growth curve of the strain supplemented with 20 μM methionine (red curve) against its growth without methionine supplementation (blue curve). Data points represent the mean ± standard deviation (SD) from three independent replicates.
3.2. Identifying active MetX/Y variants in a methionine auxotrophic E. coli
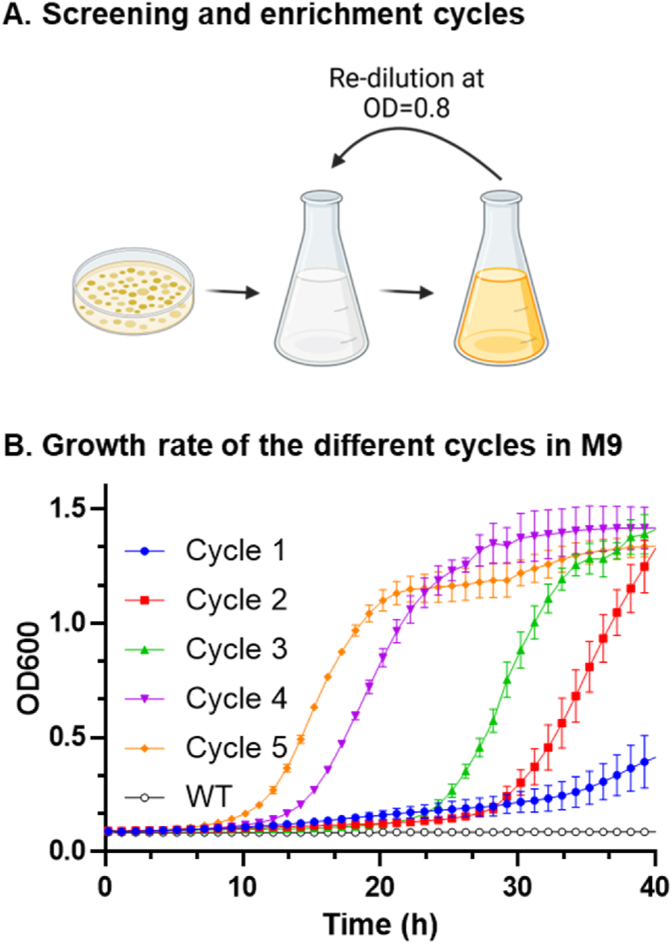
To identify metX and metYvariants from CG capable of supporting methionine synthesis in the E. coli ΔmetABJ, we generated a randomized genetic library from these genes. We calibrated the error rate of the PCR such that approximately four non-synonymous mutations will be inserted per 1000 base pairs. The actual library size, resulting from the transformation efficiency, was ∼1.5 × 105. This library was then introduced into ΔmetABJ E. coli, initiating the selection process to identify active variants of the MetX/Y enzymes. We initiated this process by transforming these cells with our randomized library and cultivating them in the M9 minimal medium. Fig. 3A illustrates the enrichment process used to select variants that effectively complement ΔmetABJ E. coli cells, which was performed by diluting the primary culture by a factor of 1000 upon reaching an OD600 = 0.8, allowing for regrowth in the M9 medium. The dilution and regrowth step was repeated for five cycles. After each cycle, we plated aliquots from the culture on M9 agar plates containing ampicillin and kanamycin for selection and confirmed their ability to grow in the absence of methionine. Moreover, the presence of metY and metX genes was verified using colony PCR. Fig. 3B illustrates the growth curves of these enrichment cycles. Notably, advanced cycles exhibited an accelerated growth rate, indicating the successful enrichment of methionine-producing bacterial variants.
Fig. 3.
Enrichment of Active MetX/Y Variants. (A) Schematic of the enrichment process: The panel illustrates the sequential steps to select active genetic variants from the library. (B) The growth patterns of the wild-type strain and enrichment cycles following the generation of the genetic library demonstrate the enhancement in growth rates with each cycle.
3.3. Sequence analysis of the evolved MetX and MetY variants
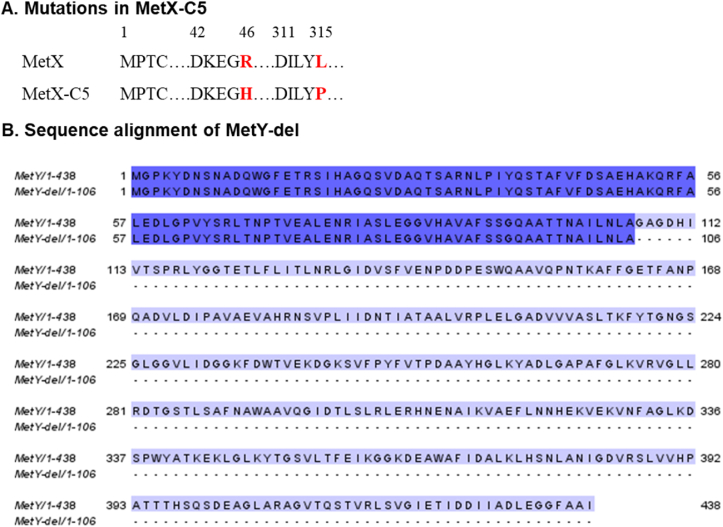
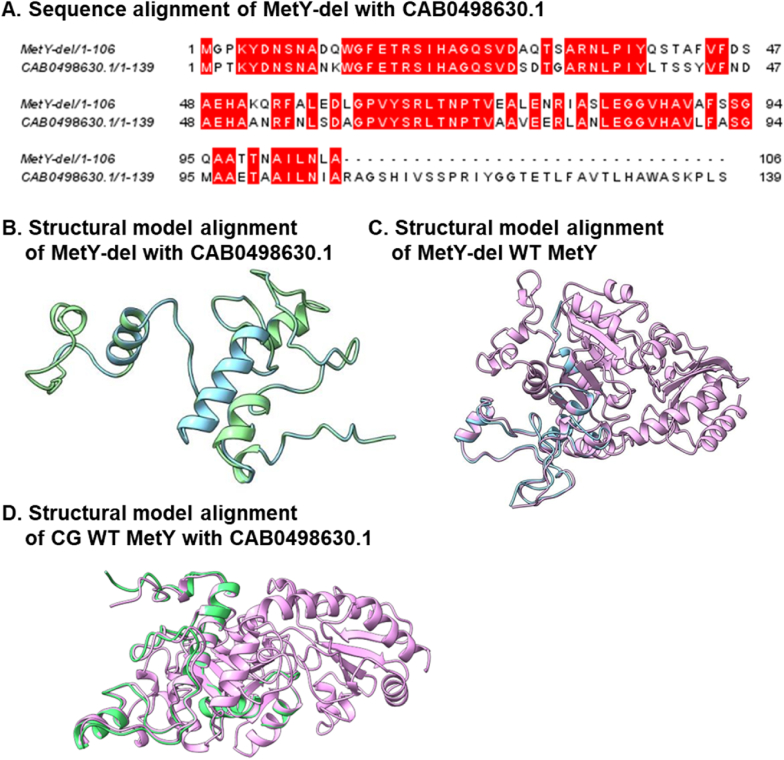
Following multiple rounds of enrichment, we sequenced six colonies from the fifth cycle (C5) and characterized the mutations. The sequencing results of all colonies showed identical two specific mutations in metX (MetX-C5) and a significant deletion in metY in part of the colonies. The enzyme amino acid sequences and sequencing results of both the wild-type and variants are presented in Table S1. Fig. 4A highlights the mutations in MetX-C5, an arginine to histidine substitution at position 46 (R46H), and a leucine to proline substitution at position 315 (L315P). Fig. 4B shows a sequence alignment, comparing the wild-type MetY gene with its truncated variant (MetY-del). This shortened variant comprises only the first 106 amino acids, a substantial reduction from the 438 amino acids of the full-length wild-type enzyme.
Fig. 4.
Sequence and mutations in the evolved MetX-C5 and MetY-del Variants. (A) Specific single-point mutations of R46H and L315P were discovered in the MetX enzyme and are highlighted in red. (B) Alignment of the sequence of MetY-del with the wild-type enzyme. The 106 amino acids retained in the MetY-del variant are highlighted in dark purple.
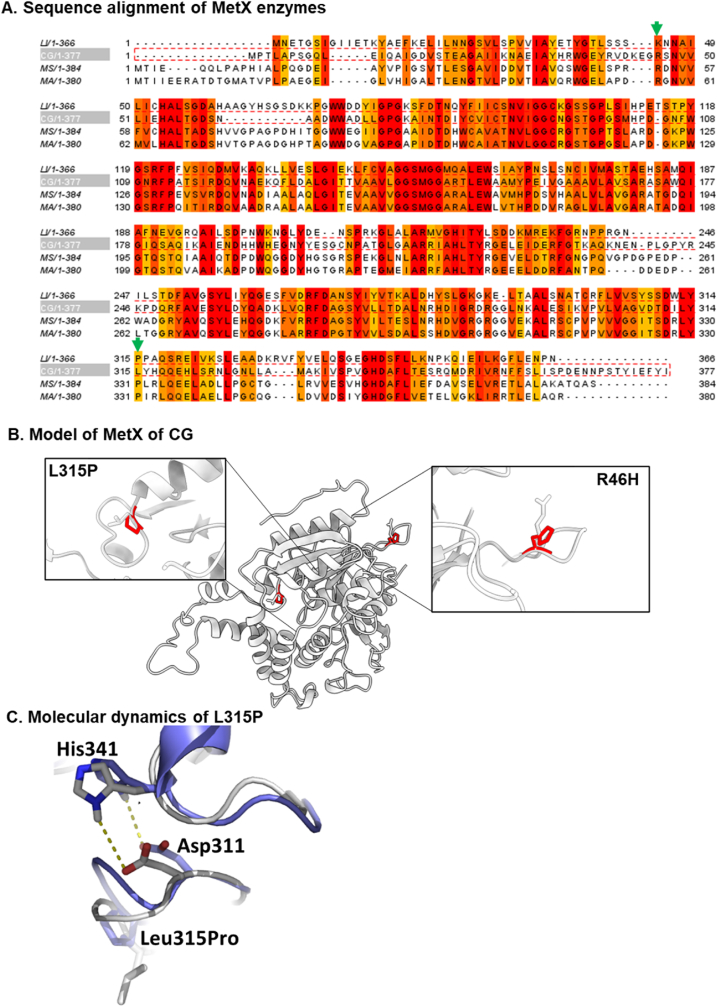
3.4. Sequence alignment and structural analysis of MetX
Fig. 5A shows the alignment of MetX of CG with additional MetX enzymes. The arrows point to the position in which the two mutations occurred. Interestingly, the L315P mutation is highly conserved in other MetX enzymes. This segment is central to the catalytic site. Fig. 5B shows the 3D model of MetX CG enzyme, with the mutated residues His and Pro highlighted in red on top of the wild-type enzyme (gray). As shown in other MetX enzyme structures, the acquisition of the conserved proline residue supports the correct structural formation of the catalytic site tunnel and architecture of the conserved catalytic triad (Chaton et al., 2019). In MetX-C5, the Pro residue is located at the N-terminus of an alpha helix, which is composed of residues Y316 to N325. This follows L313 and Y314, both of which are part of the acetyl-moiety binding site (Sagong et al., 2019). The single rotatable bond of proline and its ability to break the continuation of the helix make the L315P at the beginning of the Y316 – N325 α-helix contribute to the overall stabilization of the protein. Therefore, the evolution of the native LYL to the conserved LYP motif enables the activation of the catalytic site within the E. coli. This is consistent with the functional role of Leu317 and Tyr318 of MetX in Mycobacterium smegmatis, which is hindered by Leu and restored by the Pro residue. Fig S1 shows multiple sequence alignments of additional bacterial strains that harbor the conserved Pro residue, predominantly within the LYP sequence. Specific interactions involving the pyrrolidine ring atoms could provide an additional contribution to the catalytic activity (Bhattacharyya and Chakrabarti, 2003). The P315 ring could potentially bind with the adjacent backbone oxygen of Asp311, one of the three amino acids comprising the catalytic triad. This stabilization could enhance the enzyme's catalytic activity within the cell. To further study the effect of L315P on protein structure, we performed MD simulation. The results revealed that the hydrogen bond formed between His341 and Asp311, two conserved residues within the catalytic site, is indeed shorter in the L315P variant compared to the wild-type. Specifically, the average bond length was 3.8 Å in the wild type and 3.4 Å in the L315P variant (Fig. 5C). This reduction in length facilitates the correct structural rearrangement necessary for catalysis activation. The specific orientation of the R46H mutation most likely did not change the enzyme activity. Considering that the Arg is buried within a positive pocket formed by Glu 44 and Asp 42, a mutation to another positive yet less flexible residue as His is not expected to induce a crucial stabilizing effect. However, both mutations impart the bacteria with an overall positive contribution, enabling it to grow faster in minimal media containing no methionine supplementation.
Fig. 5.
Analysis of MetX and MetX-C5. (A) Sequence alignment of MetX CG with MetX of Leptospira interrogans (LI), Mycolicibacterium smegmatis (MS), and Mycobacteroides abscessus (MA). Green arrows show the position of the R46H and L315P mutations. Sequence alignment was done with JalView version 2.11.3.2. (B) The structure of wild-type MetX CG and the MetX-C5 with the two mutations are shown in red color. (C) Superposition of wild-type (gray) and L315P MetX mutant (light blue). The hydrogen bond in the L315P variant between His341 and Asp311 has a shorter length (3.4 Å) relative to the wild-type protein (3.8 Å).
3.5. Analysis of MetY-del variant
To decipher the functionality of the truncated MetY-del, we initially utilized BLAST to identify similar protein sequences. This revealed a protein from Corynebacterium diphtheriae, annotated as O-acetylhomoserine amino carboxypropyl transferase (E.C. 2.5.1.49, NCBI CAB0498630.1). This protein exhibited considerable similarity with MetY-del in molecular size and sequence (Fig. 6A). Table 1 shows the calculated sequence percent identity matrix for MetY WT, MetY-del and the CAB0498630.1 enzyme. Notably, there is a high identity of 83.94% between the identified variant MetY-del and the CAB0498630 enzyme. This finding suggests a potential functional similarity between these proteins.
Fig. 6.
Analysis of MetY-del and O-Acetylhomoserine amino carboxypropyl transferase from C. diphtheriae. (A) Sequence alignment of the two enzymes (MetY-del in the top row). Sequence alignment was done with JalView version 2.11.3.2. (B) Structure alignment of MetY-del (light blue) with CAB0498630.1. (C) Structure alignment of MetY-del (light blue) with CG WT MetY (pink). (D) Structure alignment of CG WT MetY of CG (pink) with CAB0498630.1.
Table 1.
Percent identity matrix.
| (%) | CAB0498630.1 | WT | MetY-del |
|---|---|---|---|
| CAB0498630.1 | 100 | 72.58 | 66.94 |
| WT | 72.58 | 100 | 83.94 |
| MetY-del | 66.94 | 83.94 | 100 |
Complementing the sequence analysis, we utilized AlphaFold to model the structures of both proteins. Fig. 6B displays the structural alignment of MetY-del (in light blue) and the O-acetylhomoserine amino carboxypropyl transferase (in green). Fig. 6C displays the structural alignment of MetY-del (in light blue) and MetY WT from CG. In agreement with the sequence similarity, the alignment revealed considerable structural similarity, except for differences in the α-helix regions. Fig. 6D shows the structure alignment of MetY WT (pink) with CAB0498630.1 (green). Table 2 summarizes the RMSD values in Å of the three proteins. This supports the notion that deleting the large amino acids segment resulting in MetY-del can indeed generate a functional enzyme.
Table 2.
RMSDs of WT, MetY-del and CAB0498630.1 enzymes.
| Å | CAB0498630.1 | WT | MetY-del |
|---|---|---|---|
| CAB0498630.1 | 100 | 0.787 | 0.347 |
| WT | 0.787 | 100 | 0.795 |
| MetY-del | 0.347 | 0.795 | 100 |
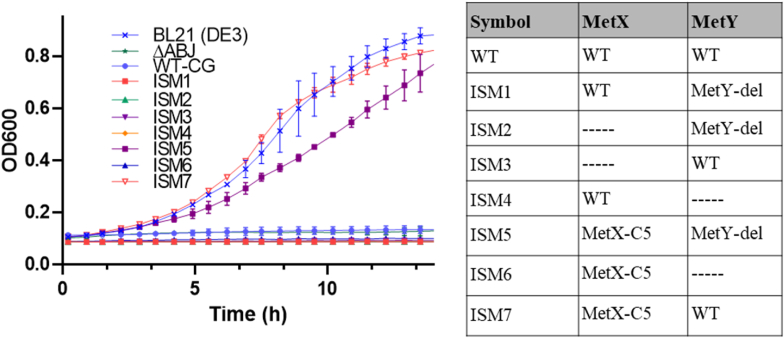
3.6. Validating the activity of the new MetX and MetY variants
To confirm the functionality of the new MetX and MetY variants, each variant gene was re-cloned into the original empty plasmid and then used to transform the original E. coli ΔmetABJ strain. This approach ensures that unintended mutations elsewhere in the plasmid or in the bacterial genome do not influence any observed activity. It also imparts uniform regulatory conditions for all genetic configurations. Fig. 7 displays the growth curves in M9 minimal media of E. coli ΔmetABJ harboring each modified plasmid. In addition, the figure shows the growth of E. coli BL21 (DE3), a common bacteria utilized in the lab. Notably, the plasmids ISM5 and ISM7, carrying wild-type MetY and MetY-del, respectively, along with the MetX-C5 variant, showed successful bacterial proliferation. The strain with wild-type MetY exhibited a higher growth rate, suggesting that the activity of the full-length protein is better than that of the shorter variant MetY-del. Indeed, the growth curve of the latter is comparable to that of the BL21 strain. In contrast, strains with other genetic configurations failed to grow, thereby validating the functionality of these specific enzyme variants and underscoring the necessity of both enzymes for bacterial growth.
Fig. 7.
Comparative growth analysis of E. coli ΔmetABJ with various MetX/Y variants. Each growth curve of E. coli ΔmetABJ represents a different combination of MetX and MetY variants within the ISM plasmids. ΔABJ represents the non-transformed auxotroph bacteria, and BL21 (DE3) is the commonly used lab strain.
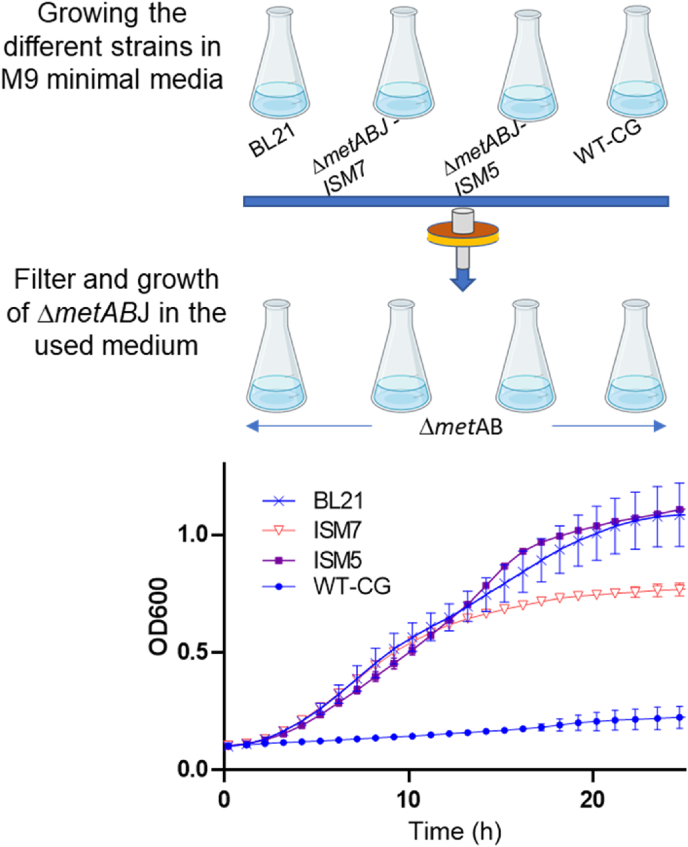
3.7. Bioavailability of extracellular methionine secreted from the different strains
To assess the levels and bioavailability of methionine secreted by each strain, we collected the spent medium at the end of growth phase for each strain. This medium was filtered through a 0.22 μm membrane and added to a fresh M9 minimal medium, which contained no methionine, at a 1:1 ratio. Consequently, the only source of methionine was the filtered spent medium (Fig. 8). Auxotroph bacteria ΔmetABJ in the medium originated from BL21, ISM5, and ISM7 strains were able to grow, suggesting the bioavailability of methionine. Although the growth rates resulting from the addition of all three media were comparable, the bacterial growth following supplementation with the medium originating from BL21 and ISM5 achieved a higher cell density. This occurred despite the superior growth of bacteria transformed with ISM7 compared to ISM5 (Fig. 7). These results align with the ability of the evolved enzymes and the transformed bacteria to produce methionine.
Fig. 8.
Evaluating methionine bioavailability through growth curves of methionine-auxotroph ΔmetABJ E. coli in the spent medium of each indicated strain. (Top) Illustration of the experimental scheme employed to assess the bioavailability of methionine in the medium post-growth of each strain. (Bottom) Growth curves of the auxotroph E. coli ΔmetABJ in M9 minimal medium. The medium was supplemented with spent and filtered medium following the growth of the indicated strains.
4. Discussion
Methionine is a critical amino acid in food, feed, and pharmaceutical industries, and its efficient biosynthesis is of significant public and commercial interest. The potential of engineering E. coli, a workhorse in industrial biotechnology, to enhance methionine production is particularly promising. By altering its natural methionine biosynthesis pathway through the incorporation of evolved MetX and MetY enzymes, one can potentially boost the production of this essential amino acid. This will not only address the need for methionine but will also open avenues for more sustainable and cost-effective production methods. The ability to engineer E. coli for enhanced methionine biosynthesis is thus a critical step forward for such biotechnological innovations. This study focused on the evolution of the direct-sulfurylation enzymes MetX and MetY from C. glutamicum in a methionine-auxotroph E. coli. Although E. coli is a common bacteria for the production of amino acids, its methionine biosynthesis pathway relies on the less abundant and heavily regulated trans-sulfurylation pathway, a challenge for efficient methionine production (Li et al., 2017; Veeravalli and Laird, 2015). Previous studies have shown that clearance of negative regulation of the methionine biosynthesis routes has enhanced the ability to produce methionine (Li et al., 2017; Nakamori et al., 1999; Sagong et al., 2022; Usuda and Kurahashi, 2005). Herein, we explored the ability to harness laboratory evolution of MetX and MetY, to transform an auxotroph-methionine E. coli into a prototroph. The evolution of the CG enzymes MetX and MetY led to the identification of specific mutations that conferred functionality in the introduced direct-sulfurylation enzymes in E. coli. The mutations identified in MetX and MetY, particularly those in the active site of MetX, likely improve the enzymes' catalytic efficiency or substrate specificity to fit to the E. coli cellular environment or evade the cellular regulation mechanism within the cell. Indeed, the deletion found in MetY points towards a potential structural simplification that may be more compatible with E. coli's cellular machinery. Despite the deletion of a large segment, MetY-del retained the ability to support the complementation of the methionine auxotroph. This result is congruent with the evolutionary notion that proteins can undergo truncation while maintaining essential functional characteristics (Bartha et al., 2015; Higdon et al., 2023), often leading to increased efficiency or altered regulatory control. Interestingly, the classification of the protein domains by InterPro algorithm (Paysan-Lafosse et al., 2023) reveals that MetY-del lacks the PLP binding domain, a cofactor of the MetY enzyme, implying on the lower activity of MetY-del. Additionally, some of the other domains have become shorter, which could potentially diminish their activity compared to WT MetY. This raises a question about the evolutionary enrichment of the MetY-del enzyme. Despite its lower activity, it is possible that the selection of MetY-del was directed by transcription/translation or other energetic costs ultimately leading to enhanced strain fitness. Moreover, although the strain harboring ISM7 plasmid has an accelerated growth rate in comparison to ISM5 and comparable to wild-type E.coli bacteria (Fig. 7), the auxotroph ΔABJ strain grew better on spent medium originated from ISM5 (Fig. 8). This could suggest that the latter exported higher levels of extracellular methionine, which may have resulted in a reduced growth rate.
An additional point within the context of the functional validation of the evolved enzymes in the ΔmetABJ E. coli is the adaptability of metabolic pathways. Herein, we simultaneously evolved two enzymes within the same pathway on the same plasmid. An interesting point to consider is whether such parallel evolution is important. On the one hand, the mutational space is dramatically increased due to the larger operon. However, if cooperative activity of the two enzymes is beneficial, this enables the screening for sequence combinations that otherwise could be neglected.
A question arises regarding the most efficient evolutionary path to generate functional enzymes. We relied on introducing random mutations throughout the gene, mimicking the stochastic nature of evolutionary changes. This method is useful when multiple enzymes catalyze a series of chemical reactions, as it allows for exploring a vast and often unexplored sequence space. Rational design, on the other hand, involves introducing specific mutations based on known structures, mechanisms, interactions, and known metabolic flux. While this method can be more targeted, it is limited by the number of calculated mutations and potential combinations and may miss beneficial mutations that are not intuitively obvious. An additional important factor is the evolutionary starting point. Starting with a non-active enzyme, as in the case of the CG-derived MetX and MetY in E. coli, can be challenging due to the initial lack of function. However, it offers the opportunity to discover novel adaptations and functionalities that may not be present in the native enzyme. This approach can yield enzymes better suited to new environments or substrates. On the other hand, beginning with an active enzyme allows for a more incremental improvement approach, focusing on enhancing or altering specific properties such as substrate specificity, stability, or catalytic efficiency. While this approach may offer a quicker path to improved function, it may not cover the large space of functional diversity achievable when starting from a non-active baseline. Of note, we sequenced representative colonies of the final enrichment cycles and found a dominant mutation in Metx-C5 and the single variant MetY-del. However, other variants in earlier enrichment cycles could exhibit additional variants capable of producing methionine in the auxotroph bacteria. Although less efficient than MetX-C5, such variants could set a starting point for additional genetic libraries, leading to new and unexplored sequences. The search for alternative sequences, together with structure-function studies of current variants, is the focus of our following studies.
5. Conclusions
Our study shows that through the substitution of the native trans-sulfurylation E. coli enzymes with direct-sulfurylation and by the application of directed enzyme evolution, we engineered E. coli capable of producing a sufficient amount of methionine to support growth. Together with improving regulation and metabolic flux, enzyme optimization could be a promising approach to support methionine biosynthesis and thus pose an important avenue for a broad range of biotechnological applications.
CRediT authorship contribution statement
Matan Gabay: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Inbar Stern: Methodology, Investigation, Formal analysis, Data curation. Nadya Gruzdev: Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Adi Cohen: Methodology, Investigation. Lucia Adriana-Lifshits: Visualization, Software, Methodology, Formal analysis. Tamar Ansbacher: Writing – original draft, Visualization, Validation, Software, Investigation, Formal analysis. Itamar Yadid: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Project administration, Funding acquisition, Formal analysis, Data curation, Conceptualization. Maayan Gal: Writing – review & editing, Writing – original draft, Validation, Supervision, Project administration, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Acknowledgements
This research was supported by the Israeli Ministry of Agriculture (grant# 21-36-0003), the Gertner Institute for Medical Nano System at Tel Aviv University and the Israeli Ministry of Science and Technology (grant# 0005749). L.A.L is grateful for Ph.D. scholarship financial support from the ADAMA Center for Novel Delivery Systems and the Gertner Institute for the Medical Nano System in Tel Aviv University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2024.e00236.
Contributor Information
Itamar Yadid, Email: itamarya@migal.org.il.
Maayan Gal, Email: mayyanga@tauex.tau.ac.il.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Aitken S.M., Lodha P.H., Morneau D.J.K. The enzymes of the transsulfuration pathways: active-site characterizations. Biochim. Biophys. Acta. 2011;1814:1511–1517. doi: 10.1016/j.bbapap.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Akya A., Farasat A., Ghadiri K., Rostamian M. Identification of HLA-I restricted epitopes in six vaccine candidates of Leishmania tropica using immunoinformatics and molecular dynamics simulation approaches. Infect. Genet. Evol. 2019;75 doi: 10.1016/j.meegid.2019.103953. [DOI] [PubMed] [Google Scholar]
- Aledo J.C. Methionine in proteins: The Cinderella of the proteinogenic amino acids. Protein Sci. 2019;28:1785–1796. doi: 10.1002/pro.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran A.H., Liu D.R. In vivo continuous directed evolution. Curr. Opin. Chem. Biol. 2015;24:1–10. doi: 10.1016/j.cbpa.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartha I., Rausell A., McLaren P.J., Mohammadi P., Tardaguila M., Chaturvedi N., Fellay J., Telenti A. The characteristics of Heterozygous protein truncating variants in the Human genome. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard K., Perret A., Mariage A., Bessonnet T., Pinet-Turpault A., Petit J.-L., Darii E., Bazire P., Vergne-Vaxelaire C., Brewee C., Debard A., Pellouin V., Besnard-Gonnet M., Artiguenave F., Médigue C., Vallenet D., Danchin A., Zaparucha A., Weissenbach J., Salanoubat M., de Berardinis V. Parallel evolution of non-homologous isofunctional enzymes in methionine biosynthesis. Nat. Chem. Biol. 2017;13:858–866. doi: 10.1038/nchembio.2397. [DOI] [PubMed] [Google Scholar]
- Belfaiza J., Martel A., Margarita D., Saint Girons I. Direct sulfhydrylation for methionine biosynthesis in Leptospira meyeri. J. Bacteriol. 1998;180:250–255. doi: 10.1128/jb.180.2.250-255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R., Chakrabarti P. Stereospecific interactions of proline residues in protein structures and complexes. J. Mol. Biol. 2003;331:925–940. doi: 10.1016/s0022-2836(03)00759-9. [DOI] [PubMed] [Google Scholar]
- Bourhy P., Martel A., Margarita D., Saint Girons I., Belfaiza J. Homoserine O-acetyltransferase, involved in the Leptospira meyeri methionine biosynthetic pathway, is not feedback inhibited. J. Bacteriol. 1997;179:4396–4398. doi: 10.1128/jb.179.13.4396-4398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster J.L., Pachl P., McKellar J.L.O., Selmer M., Squire C.J., Patrick W.M. Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan J.T., Brosnan M.E., Bertolo R.F.P., Brunton J.A. Methionine: a metabolically unique amino acid. Livest. Sci. 2007;112:2–7. [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Chaton C.T., Rodriguez E.S., Reed R.W., Li J., Kenner C.W., Korotkov K.V. Structural analysis of mycobacterial homoserine transacetylases central to methionine biosynthesis reveals druggable active site. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-56722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J.R., Fidantsef A.L. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 2003;14:438–443. doi: 10.1016/s0958-1669(03)00099-5. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferla M.P., Patrick W.M. Bacterial methionine biosynthesis. Microbiology. 2014;160:1571–1584. doi: 10.1099/mic.0.077826-0. [DOI] [PubMed] [Google Scholar]
- Foglino M., Borne F., Bally M., Ball G., Patte J.C. A direct sulfhydrylation pathway is used for methionine biosynthesis in Pseudomonas aeruginosa. Microbiology. 1995;141(Pt 2):431–439. doi: 10.1099/13500872-141-2-431. [DOI] [PubMed] [Google Scholar]
- Gruzdev N., Hacham Y., Haviv H., Stern I., Gabay M., Bloch I., Amir R., Gal M., Yadid I. Conversion of methionine biosynthesis in Escherichia coli from trans- to direct-sulfurylation enhances extracellular methionine levels. Microb. Cell Fact. 2023;22:151. doi: 10.1186/s12934-023-02150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Gophna U., Amir R. In vivo analysis of various substrates utilized by cystathionine gamma-synthase and O-acetylhomoserine sulfhydrylase in methionine biosynthesis. Mol. Biol. Evol. 2003;20:1513–1520. doi: 10.1093/molbev/msg169. [DOI] [PubMed] [Google Scholar]
- Higdon A.L., Won N.H., Brar G.A. bioRxiv; 2023. Truncated Protein Isoforms Generate Diversity of Protein Localization and Function in Yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B.-J., Park S.-D., Kim Y., Kim P., Lee H.-S. Biochemical analysis on the parallel pathways of methionine biosynthesis in Corynebacterium glutamicum. J. Microbiol. Biotechnol. 2007;17:1010–1017. [PubMed] [Google Scholar]
- Hwang B.-J., Yeom H.-J., Kim Y., Lee H.-S. Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. J. Bacteriol. 2002;184:1277–1286. doi: 10.1128/JB.184.5.1277-1286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Nagamune H., Tamura H., Yoshida Y. Identification and molecular analysis of betaC-S lyase producing hydrogen sulfide in Streptococcus intermedius. J. Med. Microbiol. 2008;57:1411–1419. doi: 10.1099/jmm.0.2008/001677-0. [DOI] [PubMed] [Google Scholar]
- Kaleta C., Schäuble S., Rinas U., Schuster S. Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol. J. 2013;8:1105–1114. doi: 10.1002/biot.201200267. [DOI] [PubMed] [Google Scholar]
- Li H., Wang B.S., Li Y.R., Zhang L., Ding Z.Y., Gu Z.H., Shi G.Y. Metabolic engineering of Escherichia coli W3110 for the production of L-methionine. J. Ind. Microbiol. Biotechnol. 2017;44:75–88. doi: 10.1007/s10295-016-1870-3. [DOI] [PubMed] [Google Scholar]
- Madeira F., Pearce M., Tivey A.R.N., Basutkar P., Lee J., Edbali O., Madhusoodanan N., Kolesnikov A., Lopez R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022;50:W276–W279. doi: 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamori S., Kobayashi S., Nishimura T., Takagi H. Mechanism of L-methionine overproduction by Escherichia coli: the replacement of Ser-54 by Asn in the MetJ protein causes the derepression of L-methionine biosynthetic enzymes. Appl. Microbiol. Biotechnol. 1999;52:179–185. doi: 10.1007/s002530051506. [DOI] [PubMed] [Google Scholar]
- Neubauer C., Landecker H. A planetary health perspective on synthetic methionine. Lancet Planet. Health. 2021;5:e560–e569. doi: 10.1016/S2542-5196(21)00138-8. [DOI] [PubMed] [Google Scholar]
- Ochrombel I., Fischer D., Bathe B., Hasselmeyer M., Hampel M., Pedall J. US Patent; 2021. Method for Producing L-Methionine. [Google Scholar]
- Paysan-Lafosse T., Blum M., Chuguransky S., Grego T., Pinto B.L., Salazar G.A., Bileschi M.L., Bork P., Bridge A., Colwell L., Gough J., Haft D.H., Letunić I., Marchler-Bauer A., Mi H., Natale D.A., Orengo C.A., Pandurangan A.P., Rivoire C., Sigrist C.J.A., Sillitoe I., Thanki N., Thomas P.D., Tosatto S.C.E., Wu C.H., Bateman A. InterPro in 2022. Nucleic Acids Res. 2023;51:D418–D427. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S., Gakière B., Job D., Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagong H.-Y., Hong J., Kim K.-J. Crystal structure and biochemical characterization of O-acetylhomoserine acetyltransferase from Mycobacterium smegmatis ATCC 19420. Biochem. Biophys. Res. Commun. 2019;517:399–406. doi: 10.1016/j.bbrc.2019.07.117. [DOI] [PubMed] [Google Scholar]
- Sagong H.-Y., Lee D., Kim I.-K., Kim K.-J. Rational engineering of homoserine O-Succinyltransferase from Escherichia coli for reduced feedback inhibition by methionine. J. Agric. Food Chem. 2022;70:1571–1578. doi: 10.1021/acs.jafc.1c07211. [DOI] [PubMed] [Google Scholar]
- Sbodio J.I., Snyder S.H., Paul B.D. Regulators of the transsulfuration pathway. Br. J. Pharmacol. 2019;176:583–593. doi: 10.1111/bph.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipp C.J., Ma Y., Al-Shameri A., D'Alessio F., Neubauer P., Contestabile R., Budisa N., di Salvo M.L. An engineered Escherichia coli strain with synthetic metabolism for in-cell production of Translationally active methionine Derivatives. Chembiochem. 2020;21:3525–3538. doi: 10.1002/cbic.202000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda Y., Kurahashi O. Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl. Environ. Microbiol. 2005;71:3228–3234. doi: 10.1128/AEM.71.6.3228-3234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley C.C., Cembran A., Perlmutter J.D., Lewis A.K., Labello N.P., Gao J., Sachs J.N. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J. Biol. Chem. 2012;287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeravalli K., Laird M.W. Toward an era of utilizing methionine overproducing hosts for recombinant protein production in Escherichia coli. Bioengineered. 2015;6:132–135. doi: 10.1080/21655979.2015.1030544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij P., Kertesz M.A. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J. Bacteriol. 1999;181:5833–5837. doi: 10.1128/jb.181.18.5833-5837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xue P., Cao M., Yu T., Lane S.T., Zhao H. Directed evolution: Methodologies and applications. Chem. Rev. 2021;121:12384–12444. doi: 10.1021/acs.chemrev.1c00260. [DOI] [PubMed] [Google Scholar]
- Weissbach H., Brot N. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 1991;5:1593–1597. doi: 10.1111/j.1365-2958.1991.tb01905.x. [DOI] [PubMed] [Google Scholar]
- Wilson D.S., Keefe A.D. Random mutagenesis by PCR. Curr. Protoc. Mol. Biol. Chapter. 2001;8(Unit8.3) doi: 10.1002/0471142727.mb0803s51. [DOI] [PubMed] [Google Scholar]
- Wirtz M., Droux M. Synthesis of the sulfur amino acids: cysteine and methionine. Photosynth. Res. 2005;86:345–362. doi: 10.1007/s11120-005-8810-9. [DOI] [PubMed] [Google Scholar]
- Xu G., Zhao Q., Huang B., Zhou J., Cao F. Directed evolution of a penicillin V acylase from Bacillus sphaericus to improve its catalytic efficiency for 6-APA production. Enzym. Microb. Technol. 2018;119:65–70. doi: 10.1016/j.enzmictec.2018.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.