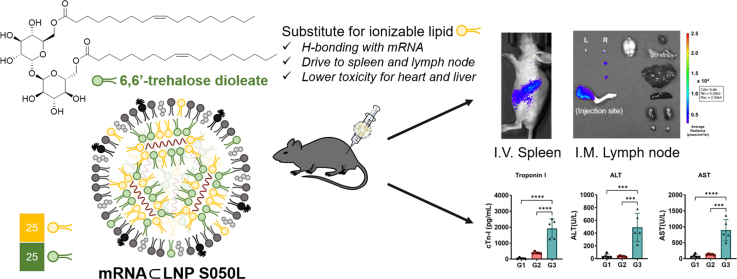
Abstract
The rapid development of messenger RNA (mRNA) vaccines formulated with lipid nanoparticles (LNPs) has contributed to control of the COVID-19 pandemic. However, mRNA vaccines have raised concerns about their potential toxicity and clinical safety, including side effects, such as myocarditis, anaphylaxis, and pericarditis. In this study, we investigated the potential of trehalose glycolipids-containing LNP (LNP S050L) to reduce the risks associated with ionizable lipids. Trehalose glycolipids can form hydrogen bonds with polar biomolecules, allowing the formation of a stable LNP structure by replacing half of the ionizable lipids. The efficacy and safety of LNP S050L were evaluated by encapsulating the mRNA encoding the luciferase reporter gene and measuring gene expression and organ toxicity, respectively. Furthermore, mice immunized with an LNP S050L-formulated mRNA vaccine expressing influenza hemagglutinin exhibited a significant reduction in organ toxicity, including in the heart, spleen, and liver, while sustaining gene expression and immune efficiency, compared to conventional LNPs (Con-LNPs). Our findings suggest that LNP S050L, a trehalose glycolipid-based LNP, could facilitate the development of safe mRNA vaccines with improved clinical safety.
Keywords: Trehalose glycolipid, Lipid nanoparticle, mRNA vaccine, Toxicity, Immunogenicity
Graphical abstract
Highlights
-
•
Trehalose glycolipids can replace half of the ionizable lipids in the LNP formulation.
-
•
LNP S050L, the optimized LNP incorporating trehalose glycolipids, effectively migrates to the lymph nodes and spleen.
-
•
The mRNA vaccine formulated with LNP S050L demonstrates robust gene expression and immune efficiency comparable to the control LNP.
-
•
Mice inoculated with LNP S050L-formulated mRNA vaccines exhibited reduced heart and liver toxicity compared to the control LNP.
1. Introduction
Messenger RNA (mRNA) vaccines have been in the spotlight since the onset of the COVID-19 pandemic. The success of the Pfizer-BioNTech and Moderna COVID-19 vaccines, which have demonstrated high potency in clinical trials, has propelled recent advancements in mRNA vaccine technologies [1]. This success has enabled the rapid development and production of mRNA vaccines, accelerating research and development [2]. Finally, research on mRNA vaccines has earned the honor of being awarded the Nobel Prize in Physiology or Medicine. As COVID-19 mRNA vaccines are being administered worldwide, the advantages and necessity of mRNA vaccine technology have been highlighted, and some concerns have been revealed. Adverse reactions to the COVID-19 mRNA vaccine have been reported, ranging from mild symptoms to serious adverse events, such as myocarditis, pericarditis, thrombosis with thrombocytopenia syndrome, and anaphylaxis [[3], [4], [5]].
Lipid nanoparticles (LNPs) are a classic delivery system for nucleic acid therapeutics and mRNA vaccines. Currently, LNPs are composed of four components: ionizable lipids, phospholipids, cholesterol and polyethylene glycol (PEG)-conjugated lipids, and Onpattro® for siRNA therapeutics and mRNA-1273 and BNT162b2 for COVID-19 mRNA vaccines are commercially available [6]. Among these four components, ionizable lipids interact with RNA through electrostatic interactions and are key components that determine the delivery efficacy of mRNA. At the same time, they have an adjuvant effect, helping to produce appropriate neutralizing antibodies against antigens produced from mRNA. Because cationic lipids initially carrying nucleic acids are known to be toxic [7], the ionizable heads were replaced with cationic heads to reduce toxicity. Subsequently, cone-shaped lipids with branched carbon chains and ester bonds introduced into the hydrophobic lipid increase biodegradability and delivery efficiency and promote escape from endosomes [8]. Although remarkable results have been achieved in terms of delivery efficacy and reduction in toxicity, COVID-19 mRNA vaccines have been reported to cause various adverse effects. Among the components of LNPs, ionizable lipids are the main synthetic chemicals that induce acute immune responses and toxicity [9].
Focus on the toxicity of LNPs has led to the idea of replacing ionizable lipids with nontoxic materials. The substitute should interact with the mRNA to form a stable nanoparticle and take over the adjuvanticity of an ionizable lipid [10,11]. Herein, we selected trehalose glycolipids as ionizable lipid substitutes. Trehalose is a natural disaccharide comprising two glucose molecules. Some bacteria, fungi, plants, and invertebrates synthesize trehalose as an energy source and use it to survive freezing temperatures and water shortages. An aqueous trehalose solution exhibited a concentration-dependent clustering tendency [12]. Because of their ability to form hydrogen bonds, highly concentrated trehalose self-bonds in water form clusters of various sizes. Trehalose also interacts directly with nucleic acids to promote the dissolution of double-stranded DNA and stabilizes single-stranded nucleic acids [13]. Combining this hydrogen-bonding characteristic with other cationic functional groups, backbone and side chain trehalose polymers have been applied to deliver plasmid DNA into cells [14].
Trehalose is present in the trehalose glycolipid form in bacteria and is believed to play a protective role against harsh environmental conditions. 6,6′-trehalose dimycolate (TDM), discovered in Mycobacterium tuberculosis, is a well-known naturally occurring trehalose glycolipid (Fig. S1) [15]. Both Synthetic and naturally occurring trehalose glycolipids protected the supported phospholipid monolayer from dehydration as a minor component of the overall membrane, up to a concentration of at least 20 mol%. Dependence of dry protection on the synthetic trehalose glycolipid fraction was almost identical to that of naturally occurring TDM. This was not significantly related to the type of hydrophobic tail structure, suggesting that the interaction between the trehalose head groups and the surrounding molecules is a determinant of dehydration protection.
Trehalose glycolipids are known immunomodulators because they act as ligands of macrophage-inducible C-type lectin (Mincle) [16]. As a simplified synthetic analog of TDM, 6,6′-trehalose dibehenate (TDB) has been used in liposomal subunit vaccines as an adjuvant acting as a Mincle ligand (Fig. S1) [17,18]. Sulfolipids in the form of sulfate esters activate bacterial phagocytosis [19]. 6,6′-Aryl trehalose derivatives and trehalose-based amide and sulfonamide are also reported as a Mincle ligand [[20], [21], [22]]. Other biological functions of trehalose and trehalose glycolipids include anti-inflammatory, antitumor, adjuvant, and antibacterial activities, as well as the ability to induce granulomatosis and angiogenesis [[23], [24], [25]]. Additionally, the cardioprotective effects of trehalose have recently been reported, with one possible mechanism being the reduction of inflammation associated with the development of heart disease [26,27].
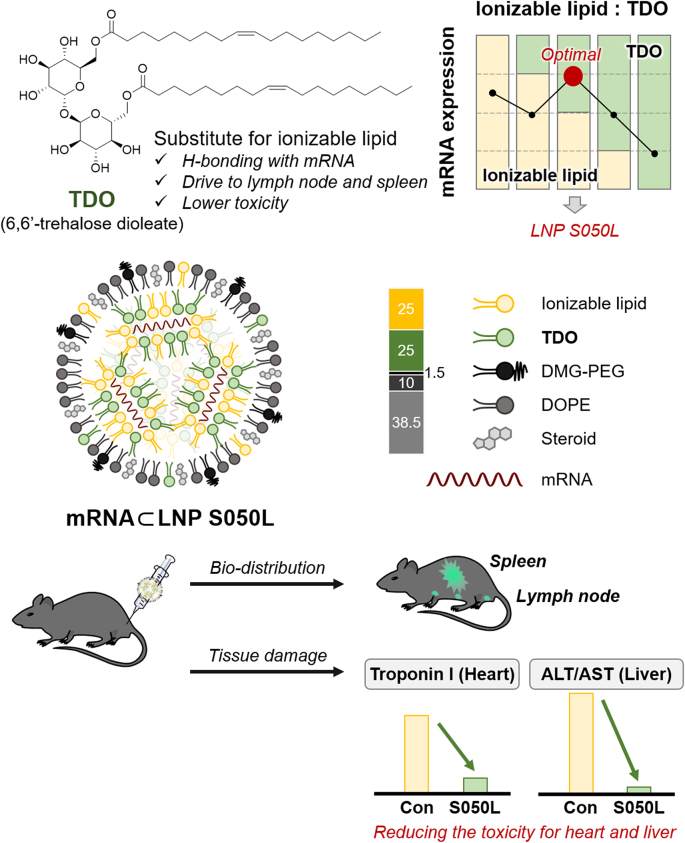
In this study, we devised a novel LNP platform that replaced a portion of ionizable lipids with trehalose glycolipids to enhance the safety of mRNA vaccine delivery. These trehalose glycolipids can strongly interact with nucleic acids and surrounding molecules through hydrogen bonding. Trehalose glycolipid analogs were intended to promote a stable formulation of particles and reduce the toxicity caused by ionizable lipids, maintaining their delivery efficacy for mRNA vaccines. Fig. 1 schematically illustrates the discovery of an optimal lipid nanoparticle platform based on trehalose glycolipids. The LNPs for optimization were formulated with Renilla luciferase (R/L) mRNA. Each combination of LNPs showed varying mRNA translation in vivo depending on the ratio of trehalose glycolipids replacing ionizable lipids, the type of steroid lipids or ionizable lipids used, and the N/P ratio. The best combination was selected based on the luminescence signal from R/L, which indicated the delivery efficiency of the mRNA. The top-performing LNP (LNP S050L) underwent structural, physicochemical, and biological analyses in comparison to Con-LNP (Conventional LNP featuring the same lipid combination as mRNA-1273). Additionally, we compared the toxicity of the LNP S050L with that of Con-LNP via organ expression, immunogenicity, and toxicology studies. The initial findings suggest that LNP S050L could potentially be safer than Con-LNPs. We showed that the trehalose glycolipid-containing LNP has potential cardioprotective and anti-inflammatory properties, which could be a promising approach for the development of mRNA vaccines for clinical applications.
Fig. 1.
Schematic illustration of novel LNP S050L based on trehalose glycolipids. A partial substitution of ionizable lipids with trehalose glycolipids maintained high efficacy in mRNA expression and immune response. The toxicity to the heart and liver was reduced compared to Con-LNPs and the mRNAs encapsulated within LNP S050L were traced in the lymph node and spleen.
2. Result and discussion
2.1. Rational design and optimization of LNP platform incorporating trehalose glycolipid
As a component of LNP, 6,6′-trehalose dioleate (TDO) was synthesized as previously reported [28,29]. Both TDO and TDB were initially tested for formulation with mRNAs by replacing a portion of the ionizable lipids. However, LNPs containing TDB were excluded because the formulated LNP containing TDB precipitated during centrifugal filtration for ethanol removal, whereas TDO resulted in stable LNPs. The length of the carbon chains in trehalose glycolipids affects the particle stability [30]. Generally, helper lipids in LNPs have a C14–C18 carbon chain. Thus, TDO with a C18 chain is more favorable than TDB with a C22 chain, leading to stable particle formation through appropriate hydrophobic packing.
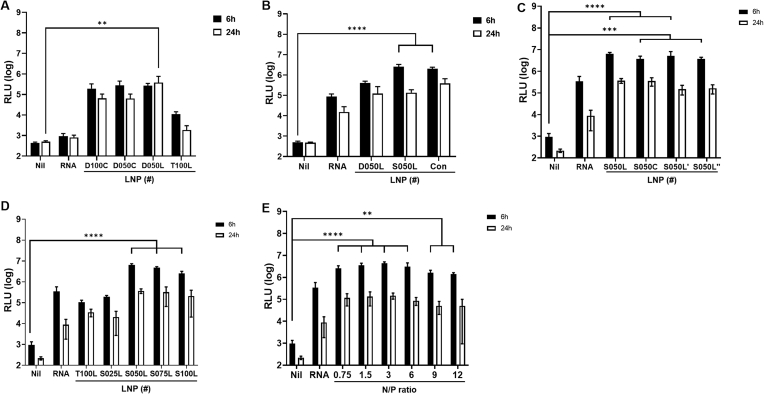
The composition of the LNPs incorporating TDO is presented in Table 1. We designated the LNPs based on the type (D for DLin-MC3-DMA; S for SM-102; T for TDO) and relative content (25–100%) of used ionizable lipids and their substitutes, as well as the type of cholesterol and its alternatives (C for cholesterol; L, L′, L″ for alternatives). The LNPs incorporating TDO were formulated with R/L mRNA and the resulting R/L mRNA-encapsulated LNPs (R/L mRNA⊂LNPs) were screened and selected by measuring R/L expression level in vivo after 6 and 24 h of intradermal (I.D.) injection. Although the hydrogen bonding properties of TDO did not completely replace the electrostatic interactions of the ionizable lipids (LNP T100L vs. LNP D050L in Fig. 2A), half of the ionizable lipids were completely replaced by TDO, improving the efficacy of R/L expression in vivo (LNP D050C vs LNP D100C in Fig. 2A).
Table 1.
LNP composition for optimization.
| LNP | Ionizable lipid (mol%) | Trehalose glycolipid (mol%) | Steroid (mol%) | Helper lipid (mol%) | PEG-conjugated lipid (mol%) |
|---|---|---|---|---|---|
| Con-LNP | SM-102 (50) | – | Cholesterol (38.5) | DSPC (10) | DMG-PEG (1.5) |
| D100C | DLin-MC3-DMA (50) | – | Cholesterol (38.5) | DOPE (10) | DMG-PEG (1.5) |
| D050C | DLin-MC3-DMA (25) | TDO (25) | Cholesterol (38.5) | DOPE (10) | DMG-PEG (1.5) |
| D050L | DLin-MC3-DMA (25) | TDO (25) | n-Butyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
| T100L | – | TDO (50) | n-Butyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
| S025L | SM-102 (12.5) | TDO (37.5) | n-Butyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
| S050L | SM-102 (25) | TDO (25) | n-Butyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
| S075L | SM-102 (37.5) | TDO (12.5) | n-Butyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
| S100L | SM-102 (50) | – | n-Butyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
| S050C | SM-102 (25) | TDO (25) | Cholesterol (38.5) | DOPE (10) | DMG-PEG (1.5) |
| S050L′ | SM-102 (25) | TDO (25) | 3-methylpentyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
| S050L″ | SM-102 (25) | TDO (25) | iso-Pentyl lithocholate (38.5) | DOPE (10) | DMG-PEG (1.5) |
Fig. 2.
In vivo optimization of trehalose glycolipids-containing LNPs. The optimal LNPs exhibiting superior efficiency was subsequently cherry-picked, considering the interplay between helper lipids and the molar ratio of ionizable lipids. The luminescence signal of R/L at 6 and 24 h after injection of R/L mRNA⊂LNPs was expressed on a logarithmic scale. (A) A proof of concept was conducted with a primary optimizing group (LNP D100C, D050C, D050L, and T100L). (B) A cone-shaped SM-102 (LNP S050L) was replaced DLin-MC3-DMA (LNP D050L). (C) Steroid lipid structure was optimized by changing alkyl groups in steroids (LNP S050L, S050C, S050L′, and S050L″). (D) Optimization of the substitution ratio of TDO to ionizable lipid (SM-102) was investigated by changing the ratio (LNP T100L, S025L, S050L, S075L, and S100L). (E) Optimization of N/P ratio of LNP S050L. N/P ratio: moles of cationizable nitrogen in ionizable lipids divided by moles of phosphodiesters in mRNAs. Nil: the group injected with saline. RNA: the group injected with only mRNA. Data are represented as the mean ± standard deviation (SD). Statistical significance was analyzed using two-way ANOVA. Statistically significant differences were defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
LNP D050C was optimized by modifying the steroid lipid components. Cholesterol, which is primarily used as a steroid lipid, facilitates membrane fusion and the structural stability of LNPs [[31], [32], [33]]. It is characterized by a small and polar hydroxyl group at one terminus, while the opposite end presents a series of large, rigid nonpolar hydrocarbon rings coupled with a flexible carbon chain. This structural configuration significantly impacts the dynamics of lipid bilayer and is reported to induce phase separation in the present of a multicomponent lipid mixture. The unique properties are also facilitating the fusion between LNPs and the endosomal membrane, as well as initial uptake, thus playing a crucial role in enhancing the efficiency of endosomal escape. Recently, an effective formulation of mRNA⊂LNPs was developed by replacing cholesterol with various steroid-based compounds [34]. In the present study, we focused on lithocholic acid, which is used in various delivery systems, with the aim to mimic the structure of cholesterol, including its small polar group, rigid steroid backbone, and flexible carbon chain [[35], [36], [37]]. Lithocholic acid is a bile acid naturally synthesized from cholesterol and has been reported to exert anti-inflammatory and anticancer effects under controlled conditions. In addition, the strong binding capacity of lithocholic acid to albumin suggests that LNPs composed of lithocholic acid derivatives may exhibit a distinct delivery pattern from conventional LNPs, which are primarily distributed in the liver through an apolipoprotein E-dependent mechanism [38]. Three lithocholic acid derivatives (n-butyl lithocholate, 3-methylpentyl lithocholate, and iso-pentyl lithocholate) were synthesized by esterification of carboxylic acid with short-chain alcohols [29]. The R/L mRNA⊂LNP composed of n-buthyl lithocholate instead of cholesterol (LNP D050L) showed similar R/L expression efficacy to that of the LNP containing cholesterol (LNP D050C) at 6 h after injection. However, when examining the R/L expression level at 24 h after injection, it was evident that n-butyl lithocholate showed a significant improvement compared to cholesterol (LNP D050L vs. LNP D050C in Fig. 2A).
DLin-MC3-DMA, the ionizable lipids of the LNP D050L, were substituted with SM-102, a lipid used in mRNA-1273 (Fig. 2B). Upon replacing the ionizable lipid with SM-102, which possesses a cone-shaped structure, LNP S050L was observed that the expression level of R/L increased more than 10-fold compared to the case with DLin-MC3-DMA at 6 h after injection (LNP S050L vs. LNP D050L in Fig. 2B). The mRNA delivery efficiency was then compared to the Con-LNP platform based on the R/L expression level. LNP S050L demonstrates comparable delivery efficiency to Con-LNP (LNP S050L vs. Con-LNP in Fig. 2B).
The expression efficiency of R/L mRNA⊂LNPs in which cholesterol was replaced with 3-methylpentyl lithocholate and iso-pentyl lithocholate (LNP S050L′ and LNP S050L'', respectively) was also verified, and results showed that 3-methylpentyl lithocholate was as effective as n-butyl lithocholate (Fig. 2C). However, considering both the expression efficiency at 6 h and the persistence of expression at 24 h after injection, the cholesterol substitute was optimized for n-butyl lithocholate. This phenomenon was observed in a similar pattern in LNPs utilizing DLin-MC3-DMA (Fig. S2A)
Then, to confirm the ratio of trehalose glycolipid and ionizable lipid, R/L mRNA⊂LNPs of the selected ratio were formulated and the R/L expression efficiency was observed (Fig. 2D and Fig. S2B). As the substitution ratio of TDO to ionizable lipids increased, the luminescence signal also increased. It reached its peak at a 50% substitution ratio, but further increases caused the signal to decrease again. In other words, LNP S050L with a TDO: ionizable lipid ratio of 1:1 exhibited the highest expression efficiency and was selected as the optimal LNP. Finally, the amount of lipid mixture relative to mRNA was also optimized (Fig. 2E). Upon analyzing the R/L expression efficiencies as influenced by the N/P ratio, which is defined as the ratio of the potential positive charge of the ionizable lipids to the negative charge of the mRNA phosphate groups, a pattern of gradual enhancement in delivery efficiency was observed. This improvement continued up to an N/P ratio of 3, after which a further increase in the N/P ratio resulted in a diminishing efficiency. Importantly, this trend remained consistent across different types of ionizable lipids (Fig. S2C), leading to the identification of 3 as the optimal N/P ratio for efficient delivery.
Ultimately, we determined the optimal conditions for formulating LNP S050L platform with mRNA at an N/P ratio of 3. Under optimal conditions, the complete replacement of TDO with the helper lipid (DOPE) led to a significant reduction in mRNA encapsulation efficiency, dropping to around 50% (LNP S050L vs. LNP P050L in Fig. S2D). This observation highlights the critical importance of TDO in the mRNA encapsulation process, serving as an effective substitute for ionizable lipids. Parallel studies are underway to verify the efficacy of this LNP platform for delivering ssRNA adjuvants or mRNA vaccines against influenza and human papillomavirus [39,40].
2.2. Characterization of LNP S050L
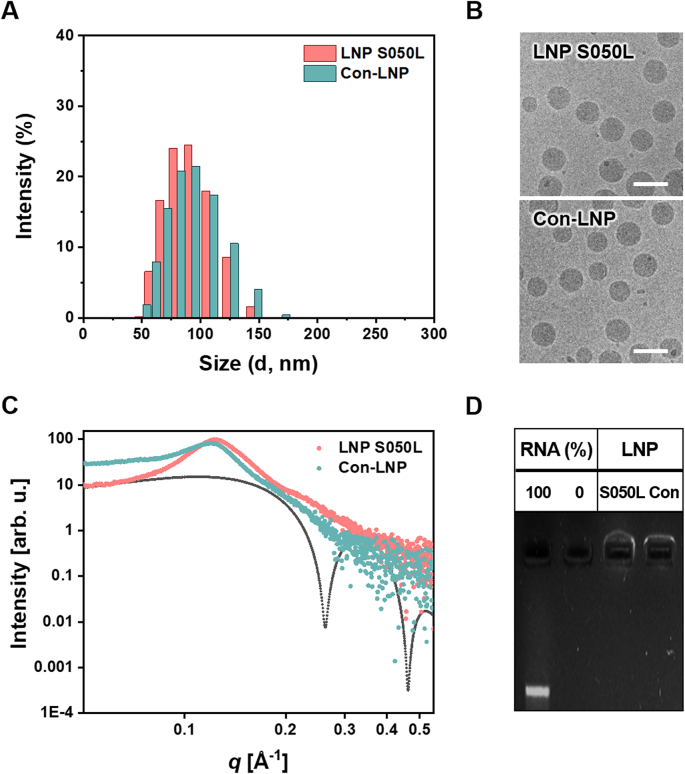
The structural and physicochemical properties of the top-performing LNP S050L were analyzed (Table 2, Fig. 3). When compared to R/L mRNA⊂Con-LNP, R/L mRNA⊂LNP S050L exhibited similar size, polydispersity index (PDI), zeta (ζ) potential, encapsulation efficiency (EE), and pKa value (Table 2). The structures of the LNPs were determined using dynamic light scattering (DLS), cryogenic transmission electron microscopy (cryo-TEM), and small-angle X-ray scattering (SAXS). The size distribution of LNP S050L almost overlapped with that of the Con-LNPs (Fig. 3A). According to a report by Moderna Inc., the size of LNPs in the best-performing formulations ranged from 75 to 95 nm, and LNP S050L met the reported size criteria [41]. Both LNP S050L and Con-LNP exhibited typical cryo-TEM images of lipid nanoparticles with an electrodense core, as previously reported (Fig. 3B) [42,43]. Both types of LNPs had uniform sizes and demonstrated a strong correlation with the average size and distribution determined using DLS. To evaluate the structural differences between the LNPs, Synchrotron SAXS scans were performed (Fig. 3C). Scattering from both LNPs showed neither a lipid bilayer form factor (black dotted line) nor an inverted hexagonal structure. Instead, scattering from both LNPs showed a single broad characteristic peak at q ∼0.1 Å−1, presumably from packing structure of mRNA inside of LNP. This type of scattering curve indicates the typical glassy and short-range ordered internal structure of LNPs [43]. Con-LNP, however, showed a larger average characteristic distance of mRNA-LNP complex (52.8 Å) compared to that of LNP S050L (50.8 Å). Approximate domain size of such complex in Con-LNP (110.2 Å) was also smaller than that of LNP S050L (128.62 Å), indicating that LNP S050L had more compact and long-range ordered structure than Con-LNP.
Table 2.
Physical and chemical properties of LNP S050L compared to Con-LNP.
| LNP | Z-average (nm) | PDI | ζ (mV) | EE (%) | pKa |
|---|---|---|---|---|---|
| LNP S050L | 87.3 ± 0.6 | 0.05 | 10.7 ± 0.4 | 90.9 | 6.75 ± 0.01 |
| Con-LNP | 86.3 ± 1.4 | 0.07 | 9.2 ± 1.5 | 91.8 | 6.73 ± 0.02 |
Fig. 3.
Characterization of LNP S050L compared to Con-LNP. (A) Size distributions, (B) TEM images, (C) SAXS scans, and (D) mRNA encapsulation efficiencies of LNP S050L and Con-LNP.
The EEs of LNP S050L and Con-LNPs were determined to be greater than 90% based on the ribogreen assay. The gel electrophoresis results, as shown in Fig. 3D, mirrored the EE results, with both LNPs efficiently encapsulating the cargo mRNA, resulting in no free RNA bands being observed in gel electrophoresis after formulation.
2.3. Monitoring in vivo bio-distribution and residence of mRNA formulated with LNP S050L
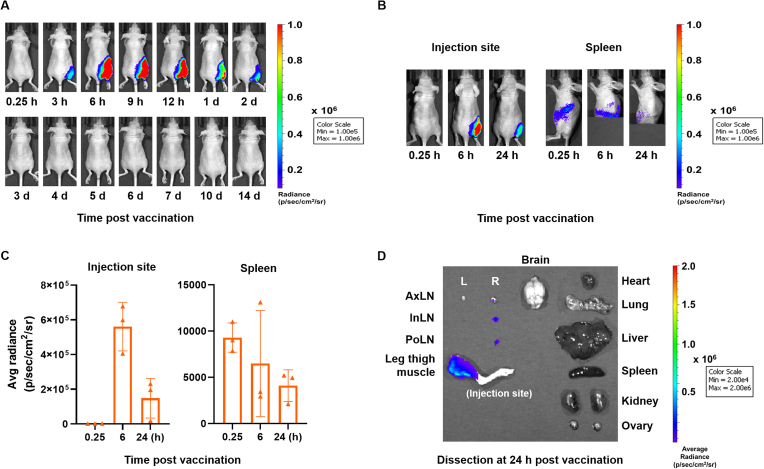
To evaluate the expression level of the mRNA to monitor in vivo bio-distribution and residence, mRNA (5 μg) encoding firefly luciferase reporter gene (F/L mRNA) formulated with LNP S050L or Con-LNP was injected into mice via intramuscular (I.M.) injection, and bioluminescence signals were monitored using an in vivo imaging system (IVIS). Luminescence signals from F/L mRNA were observed at the highest levels 6 h after injection and lasted until 48 h at the injection site. Interestingly, the bioluminescence signal from the luciferase persisted longer than that induced via I.M. injection (Fig. 4A). In vivo bio-distribution of F/L mRNA⊂LNP S050L was rapidly spread from the injection site to other organs. Most of the luciferases from F/L mRNA⊂LNP S050L were located in the injection site, and some of luciferases migrated to the spleen and inguinal lymph node (LN) (Fig. 4B and C). To evaluate ex vivo bio-distribution of luciferases from injected F/L mRNA⊂LNP S050L, mice were euthanized and dissected 24 h after administration. Luciferase remained at the injection site and was observed on the right side of the draining LNs, which were localized near the injection site (Fig. 4D). The bio-distribution F/L mRNA⊂LNPs was also investigated through intravenous (I.V.) injection (Fig. S3). In mice receiving I.V. injections, the luciferase signal was expressed more rapidly than that in the group receiving I.M. injection. Luminescence from the F/L mRNA⊂LNP S050L, administered I.V. injection and circulated through the blood vessels, were primarily detected in the liver and spleen from at an early stage. They were mainly observed in the spleen after 24 h, with trace amounts found in the lungs. These results suggest that LNP S050L can act as an mRNA vaccine delivery platform for both I.M. and I.V. injections.
Fig. 4.
Bio-distribution of LNP S050L. Real-time whole-body imaging of LNP S050L at different time points post-injection. Mice were injected with 5 μg of mRNA. (A) In vivo distribution patterns of F/L mRNA⊂LNP S050L were evaluated after I.M. injections. The maximum F/L mRNA expression was observed at 6 h. (B, C) In vivo bio-distribution analysis of the I.M. injection revealed F/L bioluminescence signal in injection site and spleen. (D) Ex vivo bio-distribution in the I.M. injection group. F/L bioluminescence signal remained at the injection site and migrated toward to the right side of the draining lymph node. AxLN: Axillary lymph node, InLN: Inguinal lymph node, PoLN: Popliteal Lymph node. All data are presented as the mean ± SD of values from independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by a two-tailed Student's t-test.
2.4. Comparison study of LNP S050L with Con-LNP on their toxicity
Before conducting in vivo toxicity testing, cytotoxicity was confirmed in three cell types (HepG2, HEK293, and fibroblasts) using empty LNPs without mRNA (Fig. S4). Both LNP S050L and Con-LNPs maintained a cell viability of over 80%, even at high lipid concentrations, with no observed cytotoxicity. For in vivo toxicity, a different approach was used compared to standard toxicity protocols. Instead of following the general toxicity verification protocol, which typically results in most indicators returning to normal levels and no serious toxicity observed for Con-LNPs, we performed an autopsy on the first day after the final administration to confirm acute side effects during vaccination. This approach allowed us to detect any increase in specific indicators of Con-LNP, which may reflect the acute adverse effects of vaccination.
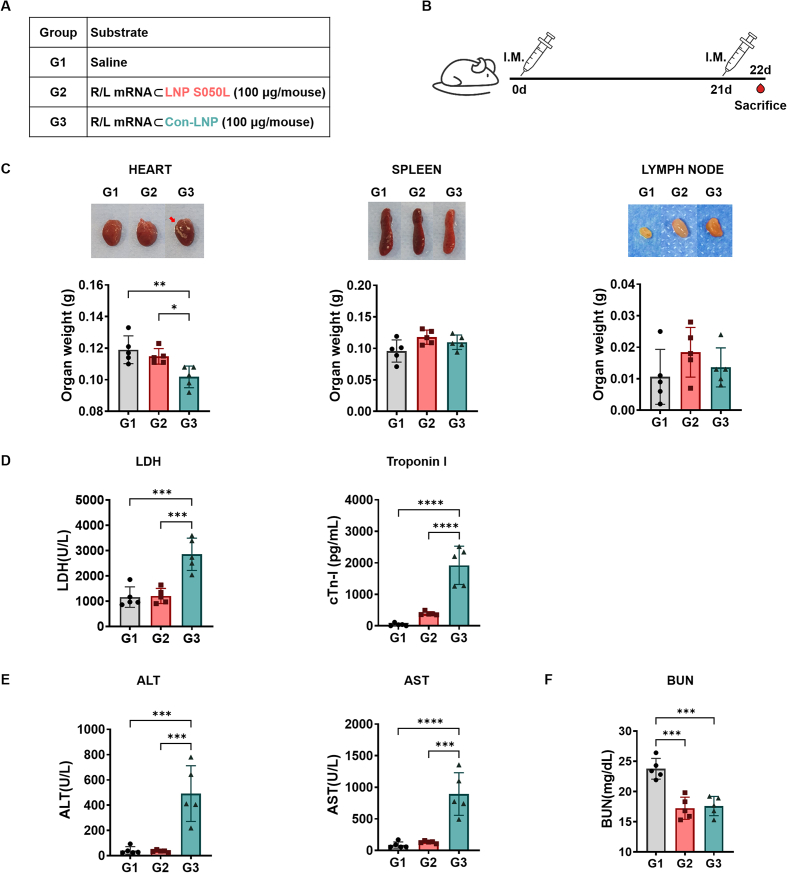
To assess the toxicity of LNP S050L compared to Con-LNP in vivo, we vaccinated 6-week-old mice via I.M. injection with the R/L mRNA (100 μg/mice, within pathological ranges) formulated with LNP S050L (G2) and Con-LNP (G3) in a prime/boost schedule and collected their blood samples, as shown in Fig. 5A and B. Calcification and weight loss were observed in the hearts of the Con-LNP group; however, no morphological changes were observed in the saline (G1) and LNP S050L groups. The weight of immune organs, including the spleen and LNs, was slightly increased in both LNP S050L (G2) and Con-LNP (G3) groups compared to that in the saline group (G1), suggesting that the R/L mRNA⊂LNPs might stimulate immune responses [1,2,44]. Moreover, the spleens of the Con-LNP group exhibited a vague red color and looser tissue structure than those of the other groups, indicating that LNP S050L reduced tissue damage compared to the Con-LNP injection (Fig. 5C). Similarly, although there were no significant differences between the saline and LNP S050L groups, the levels of tissue damage markers, including lactate dehydrogenase (LDH), troponin-I (Tn-I), aspartate aminotransferase (AST), and alanine aminotransferase (ALT), were significantly increased in the Con-LNP group (Fig. 5D and E). Furthermore, the blood urea nitrogen (BUN) level was decreased in the groups injected with mRNA⊂LNP compared to that in the saline group (Fig. 5F). A comparative evaluation of LNP S050L and Con-LNP revealed distinct differences in the toxic effects associated with these two formulations, especially in the types and contents of ionizable lipids. Consistent with our results, previous studies have supported the idea that trehalose glycolipids, which are used in LNP S050L instead of SM-102, can have beneficial effects on cardiac remodeling and cardiovascular health [26,27]. Additionally, it was recently reported that LNP containing ALC-0315, which is structurally similar to SM-102 used in Con-LNPs, rather increased liver toxicity at a high dose (5 mg/kg) [45].
Fig. 5.
In vivo toxicity of LNP S050L and Con-LNP. Mice were immunized with saline or R/L mRNA⊂LNPs to evaluate the state of the major organs and the toxicity of the LNPs. (A, B) Overview of the experimental groups and immunization schedules. (C) Representative images of the heart, spleen, and lymph nodes from immunized mice captured during euthanization. Pericardial calcification was observed in the group immunized with con-LNPs (G3). (D) Hematological safety profiles of serum samples collected from immunized mice during euthanization. The results showed an increase in alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), and troponin I activity and a decrease in blood urea nitrogen (BUN) levels in the serum. Data are represented as the mean ± SD. Statistical significance was analyzed using one-way ANOVA. Statistically significant differences were defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
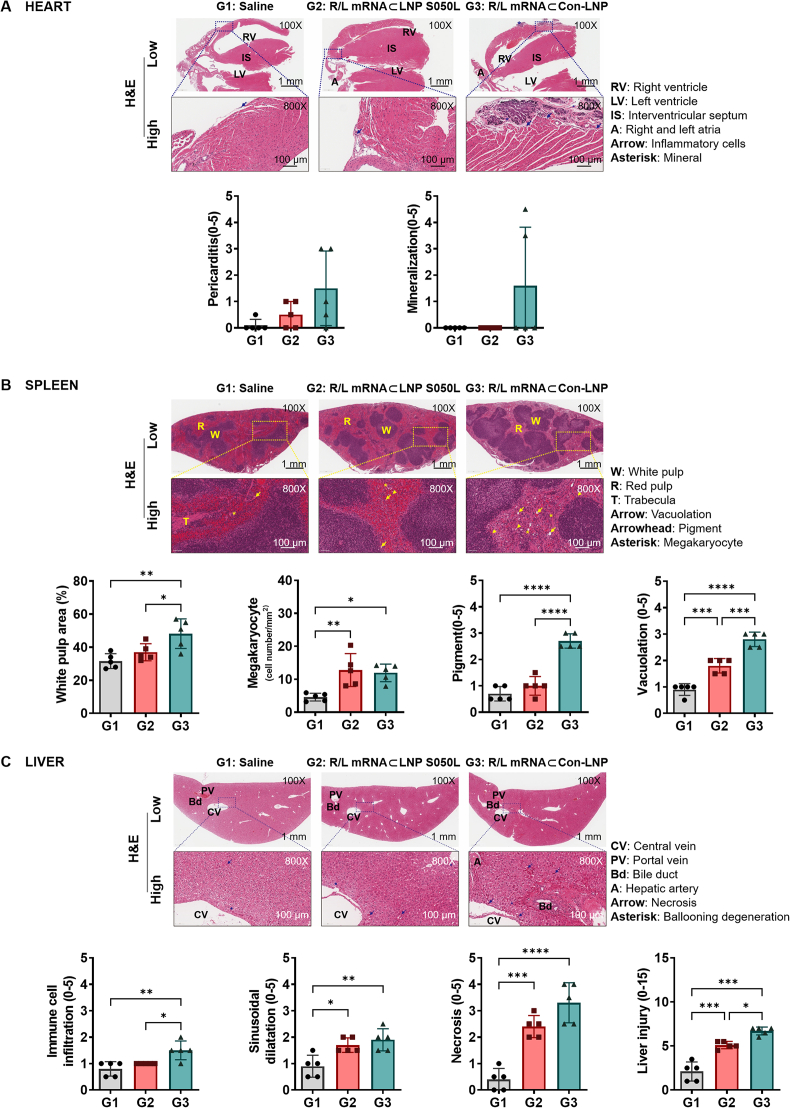
2.5. Effects of LNP S050L on histopathological toxicity of mRNA vaccine
The potential toxicities of LNP S050L and Con-LNP were further assessed via histological analysis of various organs commonly affected by mRNA vaccines. In the heart tissue analysis, pericarditis was observed in the LNP S050L group, and a relative pathological trend of pericarditis and mineralization was observed in the Con-LNP group. However, this difference was not statistically significant because of high intragroup variation (Fig. 6A). The Con-LNP group exhibited more severe histopathological changes in the spleen tissue than the LNP S050L group, with noticeable increases in white pulp area, pigment, and vacuolation. However, no significant differences were observed in the total area of the spleen or the number of megakaryocytes between the two groups. This indicated that the histopathological changes observed in the Con-LNP group were mitigated in the LNP S050L group (Fig. 6B). Furthermore, in the liver tissue, immune cell infiltration, sinusoidal dilatation, and necrosis were significantly increased in both LNP S050L and Con-LNP groups compared with those in the saline group. However, the LNP S050L group showed significantly lower immune cell infiltration, especially necrosis, than the Con-LNP group (Fig. 6C). In a recent study of a novel library of ionizable lipids synthesized through modifications of the lipid tail chain and linker, a microfluidic mixing technique was employed to create stable LNP complexes in conjunction with other helper lipids [46]. They developed novel LNPs using this library, which exhibited greater specificity toward specific cell types and lower toxicity in a comparative analysis with the conventional LNP. This study underscores the potential of tail and linker structural variations to facilitate novel functionalities within LNPs for enhanced mRNA delivery. This further emphasized the significance of tail and linker modifications in the design of efficient LNPs for drug delivery. Likewise, the utilization of trehalose glycolipid-based LNPs, incorporating trehalose glycolipids modified with an additional lipid tail chain, showed reduced toxicity compared to Con-LNPs, while maintaining equivalent immunogenicity.
Fig. 6.
Histological analysis. Representative H&E-stained images of the hearts, spleens, and livers of immunized mice during euthanization. Data are represented as the mean ± SD. Statistical significance was analyzed using one-way ANOVA. Statistically significant differences were defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
2.6. Effects of LNP S050L on immune response of mRNA vaccine
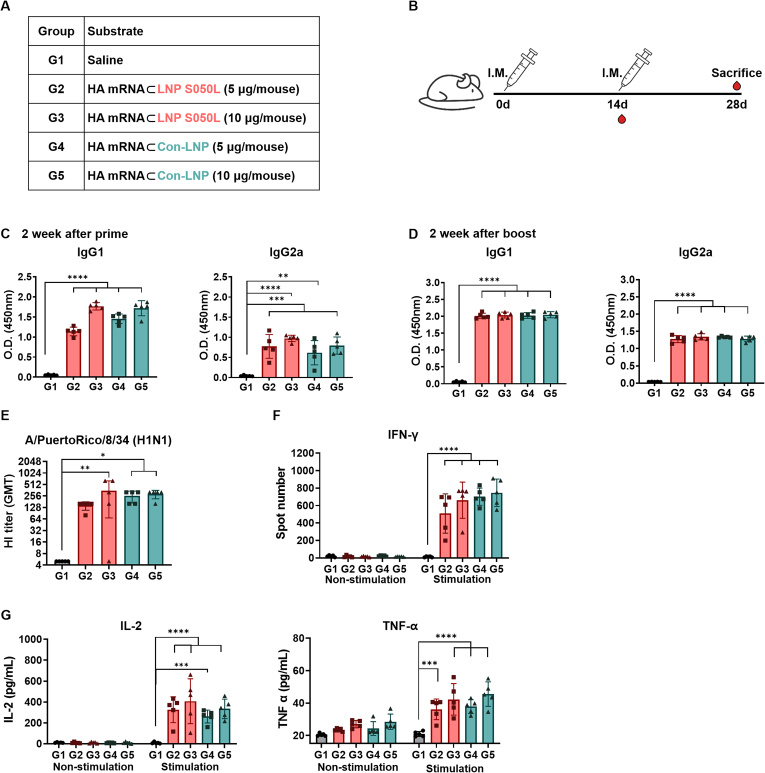
To assess the impact of different LNPs on humoral responses, 6-week-old mice were intramuscularly vaccinated with hemagglutinin (HA) mRNA (5 or 10 μg/mouse), encoding the HA of the influenza virus, encapsulated with either LNP S050L or Con-LNP. Blood samples were collected following a prime/boost schedule outlined in Fig. 7A and B. Mice immunized with HA mRNA⊂LNP S050L (G2 and G3) or HA mRNA⊂Con-LNP (G4 and G5) exhibited a significant increase in IgG1 and IgG2a antibodies two weeks after priming, with antibody titers gradually increasing in a concentration-dependent manner (Fig. 7C). Similarly, two weeks after boosting, IgG1 and IgG2a antibodies were significantly elevated in both groups, although no concentration-dependent pattern was observed (Fig. 7D). These data indicate comparable increases in antibody responses in mice injected with either HA mRNA⊂LNP S050L or HA mRNA⊂Con-LNP. Furthermore, to evaluate vaccine effectiveness, neutralizing antibodies were assessed through a hemagglutination inhibition (HI) assay using serum from immunized mice (Fig. 7E). The HI titer tended to increase remarkably in a dose-dependent manner in both mRNA vaccine groups compared to the saline-injected group. Additionally, we assessed the cell-mediated immune responses using ELISPOT for measuring the number of splenocytes secreting IFN-γ to the antigen stimulation with HA-specific peptides. As shown in Fig. 7F, the number of IFN-γ secreting cells was significantly increased in all groups in a dose-dependent manner in consistent with that of antibodies indicating activation of cellular response and antiviral property [47]. Moreover, the levels of antigen-specific cytokine in splenocyte supernatants after stimulation with HA-specific peptides. The levels of TNF-α and IL-2 were comparably increased in both groups with statistical significance (Fig. 7G). Simultaneously, the toxicity of the mRNA vaccines was also compared in the same dosage (Fig. S5). The results demonstrated that cell-mediated immune response was appropriately induced by both HA mRNA⊂LNP S050L and HA mRNA⊂Con-LNP indicating the HA mRNA⊂LNP S050L elicited a robust immune response comparable to that elicited by Con-LNPs.
Fig. 7.
In vivo immune response of the LNP S050L and Con-LNP. Mice were immunized with saline and HA mRNA⊂LNPs to compare the immune responses to the two distinct LNPs. (A, B) Overview of the experimental groups and immunization schedules. (C) Serum samples were collected from mice 2 weeks after the first immunization with mRNA-LNPs, and the levels of IgG1 and IgG2a were measured using ELISA. (D) Serum samples were collected from mice 2 weeks after the second immunization with mRNA⊂LNPs, and the levels of IgG1 and IgG2a were measured using ELISA. (E). HI titer against vaccine strains measured by HI assay using serum collected 2weeks after boost. (F) The numbers of HA peptide-Specific IFN-γ cells in splenocytes were measured using ELISPOT. (G) The concentration of TNF-α, IL-2, and IFN-γ was measured with ELISA in splenocyte culture supernatants. Data are represented as the mean ± SD. Statistical significance was analyzed using one-way ANOVA. Statistically significant differences were defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
These findings indicated that LNPs containing trehalose glycolipids can confer protection against viral challenges by eliciting a robust immune response comparable to that elicited by Con-LNPs in preclinical models. The inclusion of trehalose glycolipids and the modification of lipid tail chains can contribute to the development of safe LNPs that are both effective in mRNA delivery and in inducing an appropriate immune response. Further research into the optimization of these modifications may lead to the development of improved LNPs for therapeutic applications.
The immunomodulatory effects of trehalose glycolipid-based LNPs enhanced humoral responses and may offer protection against pathogens. However, this raises intriguing questions about the underlying mechanisms that contribute to their immune efficacy compared to that of LNPs containing ionizable lipids. A previous study developed a synthetic vaccine targeting the Mincle receptor by conjugating TDB to protein carriers [17,18]. Mincle receptors on immune cells specifically recognize certain bacterial and fungal infections, making them promising targets for vaccine development. Consequently, the integration of trehalose glycolipids into LNPs may facilitate the engagement of these specific immune receptors and trigger distinct signaling pathways, which could explain the observed immune efficacy. Additional research is needed to enhance the design of trehalose glycolipid-based LNPs to ensure efficient vaccine delivery and effective immune system modulation, with a comprehensive understanding of the associated mechanisms.
3. Conclusion
In this study, we designed and optimized a novel LNP platform based on trehalose glycolipids for the delivery of mRNA vaccines. Our study revealed efficient intracellular delivery of mRNA using this LNP platform, leading to a robust immune response. Importantly, the use of trehalose glycolipid-based LNPs offers distinct advantages over conventional LNPs, effectively reducing the risk of potential side effects. These findings have significant scientific implications, as they offer a promising alternative approach for enhancing the safety and efficacy of mRNA vaccines. Incorporating trehalose glycolipids as partial substitutes for ionizable lipids presents a promising strategy not only for improving the safety and efficacy of mRNA vaccines in infectious diseases but also in fields, such as cancer immunotherapy and personalized medicine. This study marks a significant advancement in the pursuit of safe and effective mRNA vaccines with the potential to revolutionize vaccine development and facilitate the widespread adoption of mRNA-based therapies in various clinical settings.
4. Materials and methods
4.1. Materials
The DLin-MC3-DMA was purchased from MedChemExpress (Monmouth Junction, NJ, USA). SM102 was purchased from Hanmi Fine Chemical Co. Ltd. (Siheung, South Korea). 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2‐distearoyl‐sn‐glycero‐3‐phosphocholine (DSPC), and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Cholesterol and 6,6′-trehalose dibehenate (TDB) were purchased from Sigma-Aldrich (Burlington, Massachusetts, USA). 6,6′-trehalose dioleate (TDO) and alkyl lithocholate were synthesized as reported [29]. Other reagents for the synthesis were purchased from Sigma–Aldrich and Tokyo Chemical Industry (Tokyo, Japan) and were used without further purification.
4.2. Preparation of mRNA
The DNA template for mRNA platform was designed using the intergenic region of the internal ribosome entry site of encephalomyocarditis virus. The DNA template was also designed to incorporate four restriction enzyme sequences, creating multiple cloning sites between the untranslated regions to allow the insertion of the R/L and F/L for examining in vivo expression and toxicity with bioluminescence imaging [48]. For assessing in vivo immune response, the antigen sequence for the mRNA vaccine was a DNA fragment encoding the HA protein of the influenza A virus (A/Puerto Rico/8/1934) and was cloned into a plasmid vector with backbone sequence elements as described in our previous study [40]. We used the EZTM T7 High-Yield In vitro Transcription Kit (Enzynomics, Daejeon, Korea) to generate mRNA from DNA templates through in vitro transcription. Plasmids containing the desired sequences were linearized using the Not1 restriction enzyme. The reactions involving the mRNA platforms were incubated overnight at 37 °C to allow for efficient transcription. Subsequently, DNase 1 treatment was performed at the same temperature for 30 min to remove remaining template DNA. The resulting transcripts were precipitated by the addition of lithium chloride and incubated at −20 °C for 30 min. After centrifugation (13,000 rpm for 15 min), the supernatant was removed and then pellets were washed with 70% ethanol (700 μL). The mRNA pellets were then resuspended in sterile distilled water. To minimize double-stranded RNA (dsRNA) contamination, a second purification step involving cellulose purification was performed. The concentration of the synthesized mRNA was determined using a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, USA). Finally, the resuspended RNA was aliquoted and stored at −80 °C for subsequent experimental procedures. The final mRNAs containing the R/L, F-Luc2, and HA sequences in the open reading frames were named R/L, F/L, and HA mRNA, respectively.
4.3. Formulation of mRNA⊂LNP
All lipid components, such as ionizable lipids (DLin-MC3-DMA and SM102), trehalose glycolipids (TDO and TDB), phospholipids (DOPE and DSPC), lithocholic acid derivatives (n-butyl lithocholate, 3-methylpentyl lithocholate, iso-pentyl lithocholate), cholesterol, and DMG-PEG, were dissolved in a chloroform/methanol mixture solvent (1/1, v/v) at a concentration of 50 μg/μL. The lipid components were mixed in the molar ratios listed in Table 1 and Table S1 and concentrated under reduced pressure. Lipid mixtures and mRNA were dissolved in ethanol and citrate buffers (pH 4.0, 50 mM), respectively. mRNA⊂LNPs were formulated using NanoAssemblr® Ignite™ (Precision Nanosystems Inc., Canada) at a total flow rate of 10 mL/min with the mixing volume ratio (1:3) of lipid and mRNA solutions. In Table 1 and Table S1, the N/P ratio is directly proportional to the mol% of ionizable lipids, and the N/P ratio at 25 mol% is 3. The final mRNA⊂LNP were washed twice with 1X DPBS and concentrated using 624R Centrifuge (LABOGENE Co., Ltd., South Korea) with Amicon® Ultra-15 Centrifugal Filter (Merck Millipore, Germany).
4.4. Measurement of size and zeta potential for mRNA⊂LNP
The mRNA⊂LNP solutions were prepared by diluting 100 times with DPBS (1X) for size and PDI measurements or deionized water (DIW) for zeta potential measurements. The size, PDI, and zeta potential of the LNPs were measured using a PANalytical Zetasizer Ultra (Malvern, USA) with 12 mm Square Polystyrene Cuvettes (DTS0012, for size and PDI) and folded capillary zeta cells (DTS1070, for zeta potential).
4.5. EE and gel retardation assay for mRNA⊂LNP
The mRNA EE of LNP was evaluated using fluorescent Ribogreen and gel retardation assays. mRNA standard solutions were prepared by serial dilution of the mRNA stock solution with 1X TE buffer (for the fluorescence assay) or RNase-free water (for the gel electrophoresis).
For the fluorescent ribogreen assay, the mRNA⊂LNP solutions were diluted 500 times with 1X TE buffer, allowing the measurement of any remaining free mRNA outside the LNP. To determine the total amount of RNA both inside and outside the LNP, the LNP solutions were diluted 500 times and lysed in TE buffer containing 0.5% Triton X. After adding Quant-it Ribogreen dye (Invitrogen™, USA) to all samples in the 96-well plate, fluorescence intensities were recorded at 520 nm using the Cytation 5 cell imaging multi-mode reader (BioTek, USA) with an excitation wavelength of 485 nm. The amount of mRNA in the sample was calculated using a linear standard curve based on the mRNA concentration, enabling determination of the mRNA EE of the LNPs.
For the gel retardation assay, a 1% agarose gel containing 1X MOPS buffer (20 mM 3-(N-morpholino) propanesulfonic acid, 5 mM sodium acetate, 1 mM EDTA, pH 7.0), formaldehyde (7%), and Midory green dye (NIPPON Genetics EUROPE, Germany) was prepared. The mRNA⊂ LNP solutions and mRNA standard solutions were mixed with loading buffer (6% glycerol and 0.2X blue juice) and loaded into the wells of a 1% agarose gel. After the sample-loaded agarose gel was placed in an electrophoresis system filled with 1X MOPS buffer, the system was run at full voltage for 5 min. The fluorescent gel images were measured using the GelDoc Go Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) in gel green mode.
4.6. Measurement of pKa for mRNA⊂LNP
The pKa of the mRNA⊂LNP was evaluated using a fluorescent 6-(p-Toluidino)-2-naphthalene sulfonic acid sodium salt (TNS) assay. LNP solutions (1 mM for total lipids) in 1X DPBS and TNS solution (300 μM) in DIW were prepared. The buffer solutions (consisting of 20 mM sodium phosphate monobasic, 20 mM ammonium acetate, 25 mM sodium citrate, and 150 mM NaCl) were also prepared at 0.5 intervals from pH 2.5 to pH 11.0. Then, 2 μL of LNP solutions and 2 μL of TNS solution were added to the 96 μL of various buffer solutions in the 96-well plate. The TNS fluorescence intensities were measured using the Cytation 5 cell imaging multi-mode reader (BioTek, USA) (λex = 322 nm and λem = 431 nm) at 25 °C. Normalized intensities were calculated using the following equation: , where F.I. = TNS fluorescence intensity of the sample solution measured at each pH, F·I.Max = maximum fluorescence intensity among the measured values, and F·I.min = minimum fluorescence intensity among the measured values. The pKa value for each LNP was defined as the pH corresponding to 50% of the (F·I.Max - F·I.min) value on the sigmoidal curve obtained using the KaleidaGraph program (Synergy Software, PA, USA).
4.7. Cryo-TEM for mRNA⊂LNP
The cryo-TEM images for R/L mRNA⊂LNPs (LNP S050L and Con-LNP) were measured using an FEI Tecnai™ G2 F20 scanning TEM (FEI Company, USA) from the Advanced Analysis Center at the Korea Institute of Science and Technology (KIST, Seoul headquarters, South Korea).
4.8. SAXS analysis for mRNA⊂LNP
Synchrotron SAXS was used for studying the R/L mRNA⊂LNP structures. R/L mRNA⊂LNPs were concentrated to 20 mM based on the total lipid concentration in saline. The measurements were conducted using beamline 4C at the Pohang Accelerator Laboratory (Pohang, South Korea). The sample-to-detector distance was approximately 1 m and the average photon energy was 16.9 keV. The LNP samples were transferred to a borosilicate capillary (Hilgenberg, Germany) at the beamline. The prepared capillaries were analyzed at an exposure time of 30 s. Scattered photons were recorded using a Rayonix 2D SX 165 CCD detector (Rayonix, USA) and radially averaged using custom software provided by the beamline. The obtained 1D scattering plot was analyzed using SASfit software.
4.9. Animals
Female ICR mice (6-week-old) for the ear expression and toxicity test and BALB/c mice for the immune response test were procured from Dae-Han Biolink (Eumseong, Chungcheongbukdo, South Korea) and acclimatized for 1 week. All mice were housed under specific-pathogen-free conditions at a controlled temperature of 23 ± 2 °C, with a 12 h light/dark cycle. Animal experiments were conducted following the ethical guidelines and approved by the Institutional Animal Care and Use Committee of the Catholic University of Korea (Approval No. CUK-IACUC-2022-025) and Seoul National University Hospital (Approval No. SNUIACUC-220401-2-2).
4.10. Luciferase activity assay to optimize LNP formulation for expression in vivo
R/L expression was evaluated in vivo after ear injection in mice. Mice were anesthetized using 5% isoflurane and then the synthesized R/L mRNA (5 μg/20 μL)-formulated LNPs was intradermally injected into mouse ear skin using a 30G insulin syringe (BD, NJ, USA). After a period of 6 h post-injection, the mice were euthanized, and their ears were harvested. The harvested ears were collected in Renilla lysis buffer (300 μL) and then chopped, followed by homogenization. Following a brief centrifugation step of 1 min, the R/L Assay system (Promega Corp., WI, USA) was used to measure luciferase activity. The analysis was performed in accordance with the manufacturer's instructions. Luminescence levels were quantified using the GloMax® instrument from Promega Corp. (WI, USA).
4.11. Bioluminescence imaging (BLI) of mRNA expression in vivo to monitor bio-distribution and persistence
BLI was performed using the IVIS Spectrum system (PerkinElmer, Waltham, MA, USA) to monitor in vivo reporter expression. The mice were anesthetized using a combination of isoflurane (1.5%) and oxygen during the IVIS scans. A codon optimized F/L mRNA was engineered to enhance its sensitivity for in vivo observation [49]. The F/L mRNA⊂LNP S050L at a dose of 5 μg F/L mRNA per mouse were injected via I.M. injection into the right side of the thigh leg or I.V. injection into the lateral tail vein in 10–12 weeks BALB/c nude mice. D-luciferin (Promega Corp., WI, USA) was administered via intraperitoneal injection at a dose of 3 mg per mouse as the luciferase substrate. Bioluminescence images of the F/L mRNA expression were captured at specific time points. All images were sequentially acquired at 5 min intervals, and the maximum signals were used for quantitative analysis. BLI signals were acquired using an IVIS spectrum (PerkinElmer, MA, USA) and analyzed using Living Imaging software (PerkinElmer, ver. 4.7.4).
4.12. In vitro cytotoxicity test
The cytotoxicity of LNPs (LNP S050L and Con-LNP) against HepG2, HEK293, and fibroblasts was evaluated using a cell counting kit-8 (CCK-8) (CK04, DOJINDO Laboratories, Japan). The LNPs were formulated without mRNA. The LNP solutions were prepared by diluting to 2 mM, 1.5 mM, 1 mM, 0.5 mM, 0.1 mM, 50 μM, 10 μM, and 5 μM of total lipids concentration with DMEM cell growth media containing 10% DPBS. HepG2 ( cells/well), HEK293 ( cells/well), and fibroblast cells ( cells/well) were seeded into 96-well cell culture plates and incubated overnight in a CO2 incubator (5% CO2 and 37 °C). After the cells were treated with various concentrations of LNP solution for 24 h, the solutions were removed. 10% CCK-8 solution in DMEM was added to each well and incubated for 2 h in a CO2 incubator. The optical densities (O.D.) at 450 nm were measured using the Cytation 5 cell imaging multi-mode reader (BioTek, USA) at 25 °C.
4.13. Toxicological analysis
Female ICR mice were randomly assigned to three groups (n = 5) and received I.M. injections of a effective and high dose of mRNA (5, 10, 100 μg/40 μL) to maximize potential toxicity exposure. The injections were administered twice, with a 3-week interval between injections. One day after the second round of injections, the mice were euthanized and serum samples were collected. The major tissues including the heart, spleen, LNs, and liver were carefully dissected and collected for further analysis. The collected serum samples were subjected to comprehensive toxicology panel analysis conducted by the Korea Pathology Technical Center (KP&T). This panel aimed to assess the systemic effects of mRNA encapsulated in LNPs by measuring various parameters, such as LDH, Tn-I, AST, ALT, and BUN in the serum. The collected tissues were processed for histological analysis.
4.14. Histological analysis
The heart, spleen, LNs, and liver were dissected according to established protocols. Before further processing, the excised heart, spleen, and LNs were gently blotted to remove excess moisture and weighed for accurate measurements. For histological analysis, the heart, spleen, and liver specimens were fixed in a 4% paraformaldehyde solution, embedded in paraffin, and cut into 4-μm thick sections using a microtome. These sections were stained with hematoxylin and eosin to visualize tissue morphology and cellular structures. The stained sections were observed under a light microscope for imaging purposes. Histopathological changes were evaluated using a standardized five-point scoring system as follows: 0, no abnormality, 1 = minimal, 2 = mild, 3 = moderate, 4 = moderately severe, and 5 = severe.
4.15. Immunization
Female BALB/c mice were randomly divided into five groups (n = 5) for mRNA delivery. The mice were immunized intramuscularly with 5 or 10 μg of HA mRNA vaccines encoding the HA sequence of influenza strain A/Puerto Rico/8/1934H1N. The immunization schedule consisted of two injections: an initial prime injection followed by a boost injection, with a 2-week interval between injections. Once the immunization protocol was completed, the mice were euthanized and whole blood samples were collected. The collected blood samples were processed to separate serum components. Serum was obtained by allowing the blood samples to clot for 2 h, followed by centrifugation to separate the liquid portion.
4.16. Enzyme-linked immunosorbent assay (ELISA)
An ELISA was performed to assess the levels of antigen-specific IgG1 and IgG2a in mouse serum. In brief, a 96-well plate was coated with HA at a concentration of 100 ng per well and left to incubate overnight at 4 °C. Subsequently, the wells were blocked with 100 μL of blocking buffer (1% BSA in PBS) for 1 h at room temperature. The diluted serum samples were added to the wells and incubated for 2 h at room temperature. After the incubation period, the wells were washed thrice with 200 μL of PBS-T (PBS containing Tween 20). Horseradish peroxidase-conjugated anti-mouse IgG1 (Bethyl Laboratories, Montgomery, TX, USA) and IgG2a (Novus Biologicals, Centennial, CO, USA) antibodies were then added to the wells and incubated at room temperature for 1 h. The antibodies were appropriately diluted in PBS (1:1000–1:10,000). Following three washes with PBS-T, tetramethylbenzidine substrate was added to the wells and the plates were incubated for 15 min. The reaction was stopped by the addition of 2 N H2SO4. Finally, the optical density was measured at 450 nm using a microplate reader (GloMax Explorer, Promega, Seoul, Republic of Korea).
To quantify cytokine levels in splenocyte culture supernatants, splenocytes were harvested from immunized mice and seeded at a density of 5 × 10^5 cells per well in a 96-well plate. Subsequently, they were stimulated with a 5 μg mixture of HA-specific T cell epitope peptides (IYSTVASSL, LYEKVKSQL, DYEELREQL, SFERFEIFPKE, HNTNGVTAACSH, KLKNSYVNKKGK, NAYVSVVTSNYNRRF, and CPKYVRSAKLRM) per well for 72 h at 37 °C. The concentrations of TNF-α and IL-2 were assessed using ELISA kits from Invitrogen (Thermo Fisher Scientific Inc.), following the manufacturer's protocols. Quantification of these cytokines was achieved through the utilization of standard curves, with results expressed as picograms (pg) per milliliter of supernatant.
4.17. Enzyme-linked immunospot (ELISPOT)
Splenocytes were stimulated with a 5 μg/well mixture of HA-specific T cell epitope peptides (IYSTVASSL, LYEKVKSQL, DYEELREQL, SFERFEIFPKE, HNTNGVTAACSH, KLKNSYVNKKGK, NAYVSVVTSNYNRRF, and CPKYVRSAKLRM) for 48 h at 37 °C. These HA peptides were synthesized by Peptron (Daejeon, Korea). The detection of IFN-γ secreting T cells was performed through ELISPOT assays utilizing the mouse IFN-γ ELISpotBASIC kit from Mabtech (Stockholm, Sweden), following the manufacturer's instructions.
4.18. Hemagglutination inhibition (HI) assay
The HI assay was conducted according to the guidelines provided by the World Health Organization in their “Manual for the laboratory diagnosis and virological surveillance of influenza.” In summary, mouse sera were subjected to treatment with receptor-destroying enzyme (Denka Seiken, Tokyo, Japan) at 37 °C overnight, followed by inactivation at 56 °C for 30 min. To minimize nonspecific responses, the sera were adsorbed onto chicken red blood cells. Serial dilutions of the sera were prepared in 25 μL of PBS using V-bottom 96-well microtiter plates and incubated with standardized viral suspensions (4 HA U/25 μL) for 1 h at a temperature ranging from 18 to 25 °C. Subsequently, 50 μL of 0.5% chicken red blood cells was added, and the plates were further incubated for 45 min at 18–25 °C. Antibody titers, expressed as geometric mean titers, were calculated by determining the reciprocal of the highest serum dilution that completely inhibited agglutination in duplicate experiments. The detectable antibody titer had a lower limit of 1:10, and titers below 1:10 were considered 1:5 for data analysis.
4.19. Statistical analysis
Statistical analyses were performed using Prism 8 software (GraphPad Software Inc., CA, USA). Data are presented as mean ± standard deviation (SD). An unpaired Student's t-test was used to compare the two groups. Multiple comparisons between experimental groups were performed using one-way or two-way ANOVA, followed by the Tukey-Kramer multiple comparison test. Statistically significant differences were defined as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Ethics approval and consent to participate
This research did not involve the use of human subjects. As such, ethical approval was not required. The study solely focused on the validation of the efficacy and toxicity of a novel lipid nanoparticle in mice, following the ethical guidelines. The study was approved by the Institutional Animal Care and Use Committee of the Catholic University of Korea (Approval No. CUK-IACUC-2022-025) and Seoul National University Hospital (Approval No. SNUIACUC-220401-2-2). All data used in this research were obtained and analyzed in accordance with relevant ethical standards and legal regulations.
Funding
This research received financial support from the Ministry of Science and ICT, the Ministry of Food and Drug Safety, and institutional funding of KIST in South Korea. The funding source had no role in the design, conduct, analysis, or reporting of the study.
CRediT authorship contribution statement
Seo-Hyeon Bae: Writing – original draft, Validation, Formal analysis, Data curation. Soyeon Yoo: Writing – review & editing, Writing – original draft, Validation, Formal analysis, Data curation. Jisun Lee: Writing – review & editing, Writing – original draft, Validation, Formal analysis, Data curation. Hyo-Jung Park: Writing – original draft, Validation, Formal analysis, Data curation. Sung Pil Kwon: Validation, Methodology, Data curation. Harin Jin: Validation. Sang-In Park: Formal analysis, Data curation. Yu-Sun Lee: Validation. Yoo-Jin Bang: Validation. Gahyun Roh: Validation. Seonghyun Lee: Validation. Sue Bean Youn: Validation. In Woo Kim: Writing – original draft, Validation, Formal analysis. Ho Rim Oh: Validation, Formal analysis. Ashraf K. El-Damasy: Validation, Methodology. Gyochang Keum: Validation, Supervision, Investigation, Data curation. Hojun Kim: Writing – original draft, Validation, Supervision, Formal analysis, Data curation, Conceptualization. Hyewon Youn: Writing – review & editing, Writing – original draft, Supervision, Project administration. Jae-Hwan Nam: Writing – review & editing, Supervision, Project administration, Investigation. Eun-Kyoung Bang: Writing – review & editing, Writing – original draft, Supervision, Project administration, Investigation, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following personal relationships which may be considered as potential competing interests: Sang-In Park is currently employed by SML Biopharm.The authors declare the following personal relationships which may be considered as potential competing interests: is currently employed by Jamia Millia Islamia.
All authors have seen and approved the final version of the manuscript being submitted. They warrant that the article is the authors' original work, hasn't received prior publication and isn't under consideration for publication elsewhere.
Acknowledgements
E.-K. Bang was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. NRF-2021M3E5E3080563, RS-2023-00229101), the Ministry of Food and Drug Safety (No. 22213MFDS421), and the Korea Institute of Science and Technology (KIST) Institutional Program (No. 2E32852).
H. Kim was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. RS-2023-00209955) and the Korea Institute of Science and Technology (KIST) Institutional Program (No. 2E33111).
J. H. Nam was supported by grants from the Ministry of Food and Drug Safety (grant number 22213MFDS421) and partially supported by the Brain Korea 21 Four Program.
H. Youn was supported by a grant from the Ministry of Food and Drug Safety (RS-2023-00217026).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.05.012.
Contributor Information
Hyewon Youn, Email: hwyoun@snu.ac.kr.
Jae-Hwan Nam, Email: jhnam@catholic.ac.kr.
Eun-Kyoung Bang, Email: eunkbang@kist.re.kr.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supporting Information is available: chemical structure of trehalose glycolipids, LNP composition for optimization, in vivo optimization of LNPs, in vivo bio-distribution by I.V. injection, in vitro cytotoxicity, and in vivo toxicity study.
References
- 1.Polack F.P., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yasmin F., et al. Adverse events following COVID-19 mRNA vaccines: a systematic review of cardiovascular complication, thrombosis, and thrombocytopenia, Immun. Inflamm. Dis. 2023;11(3):e807. doi: 10.1002/iid3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Husby A., Køber L. COVID-19 mRNA vaccination and myocarditis or pericarditis. Lancet. 2022;399(10342):2168–2169. doi: 10.1016/S0140-6736(22)00842-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weintraub E.S., Oster M.E., Klein N.P. Myocarditis or pericarditis following mRNA COVID-19 vaccination. JAMA Netw. Open. 2022;5(6) doi: 10.1001/jamanetworkopen.2022.18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Contr. Release. 2006;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Buschmann M.D., Carrasco M.J., Alishetty R., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9(1):65. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moghimi S.M., Simberg D. Pro-inflammatory concerns with lipid nanoparticles. Mol. Ther. 2022;30(6):2109–2110. doi: 10.1016/j.ymthe.2022.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobiyama K., Ishii K.J. Making innate sense of mRNA vaccine adjuvanticity. Nat. Immunol. 2022;23:474–476. doi: 10.1038/s41590-022-01168-4. [DOI] [PubMed] [Google Scholar]
- 11.Han X., et al. Adjuvant lipidoid-substituted lipid nanoparticles augment the immunogenicity of SARS-CoV-2 mRNA vaccines. Nat. Nanotechnol. 2023;18:1105–1114. doi: 10.1038/s41565-023-01404-4. [DOI] [PubMed] [Google Scholar]
- 12.Sapir L., Harries D. Linking trehalose self-association with binary aqueous solution equation of state. J. Phys. Chem. B. 2011;115(4):624–634. doi: 10.1021/jp109780n. [DOI] [PubMed] [Google Scholar]
- 13.Bezrukavnikov S., et al. Trehalose facilitates DNA melting: a single-molecule optical tweezers study. Soft Matter. 2014;10(37):7269–7277. doi: 10.1039/c4sm01532k. [DOI] [PubMed] [Google Scholar]
- 14.Vinciguerra D., Gelb M.B., Maynard H.D. Synthesis and application of trehalose materials. JACS Au. 2022;2(7):1561–1587. doi: 10.1021/jacsau.2c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaire G., Tenu J.-P., Petit J.-F., Lederer E. Natural and synthetic trehalose diesters as immunomodulators. Med. Res. Rev. 1986;6(3):243–274. doi: 10.1002/med.2610060302. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa E., et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J. Exp. Med. 2009;206(13):2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desel C., et al. The Mincle-activating adjuvant TDB induces MyD88-dependent Th1 and Th17 responses through IL-1R signaling. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0053531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanna C.C., et al. Synthetic vaccines targeting Mincle through conjugation of trehalose dibehenate. Chem. Commun. 2022;58(49):6890–6893. doi: 10.1039/d2cc02100e. [DOI] [PubMed] [Google Scholar]
- 19.Middlebrook G., Coleman C.M., Schaefer W.B. Sulfolipid from virulent tubercle bacilli. Proc. Natl. Acad. Sci. U.S.A. 1959;45(12):1801–1804. doi: 10.1073/pnas.45.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasheed O.K., et al. 6,6'-Aryl trehalose analogs as potential Mincle ligands. Bioorg. Med. Chem. 2020;28(14) doi: 10.1016/j.bmc.2020.115564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasheed O.K., Buhl C., Evans J.T., Ryter K.T. Design of trehalose-based amide/sulfonamide C-type lectin receptor signaling compounds. ChemMedChem. 2021;16(8):1246–1251. doi: 10.1002/cmdc.202000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch A.T., et al. Trehalose diamide glycolipids augment antigen-specific antibody responses in a Mincle-dependent manner. Bioorg. Chem. 2021;110 doi: 10.1016/j.bioorg.2021.104747. [DOI] [PubMed] [Google Scholar]
- 23.Khan A.A., Stocker B.L., Timmer M.S.M. Trehalose glycolipids-synthesis and biological activities. Carbohydr. Res. 2012;356:25–36. doi: 10.1016/j.carres.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Echigo R., et al. Trehalose treatment suppresses inflammation, oxidative stress, and vasospasm induced by experimental subarachnoid hemorrhage. J. Transl. Med. 2012;10:80. doi: 10.1186/1479-5876-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalifeh M., Barreto G.E., Sahebkar A. Trehalose as a promising therapeutic candidate for the treatment of Parkinson's disease. Br. J. Pharmacol. 2019;176(9):1173–1189. doi: 10.1111/bph.14623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frati G., Vecchione C., Sciarretta S. Novel beneficial cardiovascular effects of natural activators of autophagy. Circ. Res. 2018;123(8):947–949. doi: 10.1161/CIRCRESAHA.118.313530. [DOI] [PubMed] [Google Scholar]
- 27.Sciarretta S., et al. Trehalose-induced activation of autophagy improves cardiac remodeling after myocardial infarction. J. Am. Coll. Cardiol. 2018;71(18):1999–2010. doi: 10.1016/j.jacc.2018.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul N.K., Twibanire J.-d’A.K., Grindley T.B. Direct synthesis of maradolipids and other trehalose 6-monoesters and 6,6′-diesters. J. Org. Chem. 2013;78(2):363–369. doi: 10.1021/jo302231v. [DOI] [PubMed] [Google Scholar]
- 29.Bang E.-K., Keum G., Kang T., Jeon B.S., Lee A.S., Nam J.H., Bae S.H. Pharmaceutical composition of lipid nanoparticles for delivering nucleic acid medicines containing trehalose derivative and novel structure-maintaining compound. PCT Application No. PCT/KR2022/002372. 2022 [Google Scholar]
- 30.Kallerup R.S., et al. Influence of trehalose 6,6′-diester (TDX) chain length on the physicochemical and immunopotentiating properties of DDA/TDX liposomes. Eur. J. Pharm. Biopharm. 2015;90:80–89. doi: 10.1016/j.ejpb.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Yang S.-T., Kreutzberger A.J.B., Lee J., Kiessling V., Tamm L.K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L., Anchordoquy T.J. Effect of cholesterol nanodomains on the targeting of lipid-based gene delivery in cultured cells. Mol. Pharm. 2010;7(4):1311–1317. doi: 10.1021/mp100097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue L., et al. Responsive biomaterials: optimizing control of cancer immunotherapy. Nat. Rev. Mater. 2024;9:100–118. doi: 10.1038/s41578-023-00617-2. [DOI] [Google Scholar]
- 34.Patel S., et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11(1):983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janout V., Lanier M., Regen S.L. Molecular umbrellas. J. Am. Chem. Soc. 1996;118(6):1573–1574. doi: 10.1021/ja953261v. [DOI] [Google Scholar]
- 36.Kim S.J., Bang E.-K., Kwon H.J., Shim J.S., Kim B.H. Modified oligonucleotides containing lithocholic acid in their backbones: their enhanced cellular uptake and their mimicking of hairpin structures. Chembiochem. 2004;5(11):1517–1522. doi: 10.1002/cbic.200400150. [DOI] [PubMed] [Google Scholar]
- 37.Han J., et al. Lithocholic acid-based peptide delivery system for an enhanced pharmacological and pharmacokinetic profile of xenopus GLP-1 analogs. Mol. Pharm. 2018;15(7):2840–2856. doi: 10.1021/acs.molpharmaceut.8b00336. [DOI] [PubMed] [Google Scholar]
- 38.Spada A., Emami J., Tuszynski J.A., Lavasanifar A. The uniqueness of albumin as a carrier in nanodrug delivery. Mol. Pharm. 2021;18(5):1862–1894. doi: 10.1021/acs.molpharmaceut.1c00046. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y.-S., et al. Analysis of the immunostimulatory effects of cytokine-expressing internal ribosome entry site–based RNA adjuvants and their applications. J. Infect. Dis. 2023 doi: 10.1093/infdis/jiad392. [DOI] [PubMed] [Google Scholar]
- 40.Park H.-J., et al. Analyzing immune responses to varied mRNA and protein vaccine sequences. NPJ Vaccines. 2023;8:84. doi: 10.1038/s41541-023-00684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassett K.J., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung A.K.K., et al. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C Nanomater. Interfaces. 2012;116(34):18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arteta M.Y., Kjellman T., Bartesaghi S., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2018;115(15):E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carragher D.M., Rangel-Moreno J., Randall T.D. Ectopic lymphoid tissues and local immunity. Semin. Immunol. 2008;20(1):26–42. doi: 10.1016/j.smim.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferraresso F., Strilchuk A.W., Juang L.L., Poole L.G., Luyendyk J.P., Kastrup C.J. Comparison of DLin-MC3-DMA and ALC-0315 for siRNA delivery to hepatocytes and hepatic stellate cells. Mol. Pharm. 2022;19(7):2175–2182. doi: 10.1021/acs.molpharmaceut.2c00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naidu G.S., et al. A combinatorial library of lipid nanoparticles for cell type-specific mRNA delivery. Adv. Sci. 2023;10(19) doi: 10.1002/advs.202301929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivashkiv L.B. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018;18:545–558. doi: 10.1038/s41577-018-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko H.L., Park H.-J., Kim J., Kim H., Youn H., Nam J.-H. Development of an RNA expression platform controlled by viral internal ribosome entry sites. J. Microbiol. Biotechnol. 2019;29(1):127–140. doi: 10.4014/jmb.1811.11019. [DOI] [PubMed] [Google Scholar]
- 49.Song M.G., et al. In vivo imaging of differences in early donor cell proliferation in graft-versus-host disease hosts with different pre-conditioning doses. Mol. Cells. 2012;33(1):79–86. doi: 10.1007/s10059-012-2228-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.