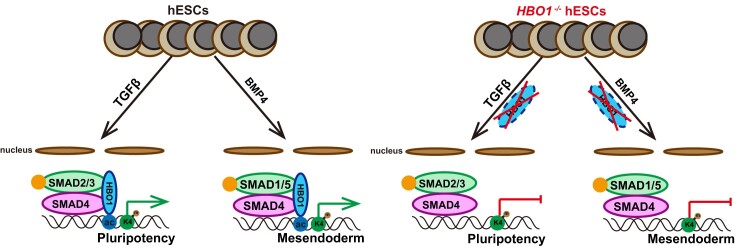
Abstract
TGF-β signaling family plays an essential role to regulate fate decisions in pluripotency and lineage specification. How the action of TGF-β family signaling is intrinsically executed remains not fully elucidated. Here, we show that HBO1, a MYST histone acetyltransferase (HAT) is an essential cell intrinsic determinant for TGF-β signaling in human embryonic stem cells (hESCs). HBO1−/− hESCs fail to response to TGF-β signaling to maintain pluripotency and spontaneously differentiate into neuroectoderm. Moreover, HBO1 deficient hESCs show complete defect in mesendoderm specification in BMP4-triggered gastruloids or teratomas. Molecularly, HBO1 interacts with SMAD4 and co-binds the open chromatin labeled by H3K14ac and H3K4me3 in undifferentiated hESCs. Upon differentiation, HBO1/SMAD4 co-bind and maintain the mesoderm genes in BMP4-triggered mesoderm cells while lose chromatin occupancy in neural cells induced by dual-SMAD inhibition. Our data reveal an essential role of HBO1, a chromatin factor to determine the action of SMAD in both human pluripotency and mesendoderm specification.
Graphical Abstract
Graphical Abstract.
Introduction
During early mammalian embryonic development, the epiblast cells undergo pluripotency exit and lineage specification to differentiate into three germ layer fates (the neuroectoderm, mesoderm and endoderm) instructed by the external signaling morphogens (1,2). TGF-β signaling is an evolutionarily conserved morphogen signaling that regulates cell fate decision in both embryonic development and various stages of adult life (3–5). Specifically, TGF-β signaling plays important roles in regulating stem cell maintenance, cell proliferation and early embryonic lineage decision and is also involved in tumorigenesis (3–5). TGF-β signaling family contains two subfamily branches, the TGF-β/Nodal/Activin branch and the bone morphogenic protein (BMP) branch. During early development, two branches differentially regulate fate decision in early germ layer specification (6,7). BMP signaling serves as an essential morphogen to specify mesoderm and primitive streak patterning, while neural fate specification usually requires dual suppression of TGF-β/BMP branch signaling (8–11). In addition, TGF-β branch signaling is essential to maintain the pluripotency and self-renewal of human embryonic stem cells (ESCs), while BMP4 signaling was reported to trigger human ESC differentiation (12–14).
The majority effects of TGF-β family signaling are mediated by SMAD transcription factors that become phosphorylated and activated upon ligand-receptor binding (15). The phosphorylated SMAD2/3 induced by TGF-β branch signaling or SMAD1/5/8 by BMP signaling form a complex with SMAD4, the co-SMAD and enter nuclei to recruit other factors such as chromatin related factors (CRFs) to mediate gene regulations (15–17). SMAD2/3 was reported to bind CBP/p300, the histone acetyltransferase (HAT) catalyzing histone acetylation for loosening chromatin and promoting transcription (18). Other CRFs were also reported to associate with SMADs and potentialize SMAD functions, such as TRIM33 (19), GCN5 (20), SMIF (21), etc. However, most of these reported SMAD associated CRFs are not known as lineage specific factors that specify a particular lineage fate. At the meantime, the cell intrinsic determinants for TGF-β signaling action in early embryonic development remain less elucidated.
CRFs and chromatin modification play important roles to regulate normal development and stem cell differentiation (22–24). For example, polycomb repressive complex proteins (PRC) maintain the developmental genes in a repressive but poised state in undifferentiated ESCs through histone H3K27 trimethylations (H3K27me3) (25–27). Recently, some CRFs were shown to play a role to regulate specific lineage fate choice in pluripotent stem cell differentiation. We and others have shown that PRC specifically determines neuroectoderm fate specification in human ESC differentiation (28,29). hESCs deficient in EZH1 or EZH2, the PRC members failed to generate the whole neuroectoderm lineage but maintained mesendoderm in teratoma formation (28). Other CRFs involved in specific lineage decision were also reported, such as Whsc1 in mesendoderm (30), Setd2 in endoderm (31), Zrf1 in ectoderm (32), etc.
Histone acetylation is a canonical epigenetic mechanism to maintain the open chromatin structure and active transcription in eukaryotic cells (33). HBO1 (also known as KAT7, MYST2) is a HAT of the MYST family that belongs to three HAT families in eukaryotic cells, and mainly catalyzes acetylation of histone H4 and H3K14 (34,35). HBO1 is highly conserved from yeast to human and widely expressed in various cell types (36). However, HBO1 functions are far from elucidated, particularly in early embryonic fate decision. Here in this study, we reveal that HBO1 is an essential cell intrinsic determinant for TGF-β signaling/SMAD action to regulate human pluripotency as well as mesendoderm specification.
Materials and methods
Cell lines and culture conditions
The human embryonic stem cell lines H1 (Wi Cell) and knockout cell lines H1-HBO1−/−−1/2# and hESCs-HBO1−/−/OE were grown on Matrigel (Corning) coated plates in the mTeSR1 (STEMCELL Technologies) medium. Culture medium was changed every day. All cell lines were passaged every 3 days and maintained at 5% CO2.
Establishing the gene knockout or knock-in cell lines in human ESCs
Cas9 protein and guide RNA were expressed through pX330 (Addgene). Guide RNAs (gRNAs) for the knockout and knock-in of HBO1 were designed on the website (CCTop, http://crispr.cos.uni-heidelberg.de). The knockout donor plasmid contained a left homology arm, a LoxP-flanked PGK-puromycin cassette and a right homology arm. In addition, the left homology arm of the knock-in donor plasmid contained a 3 × FLAG tag. These donor plasmids were used for targeting.
After all the plasmids were constructed, then 4 μg of each pX330 plasmid and donor plasmid were electroporated into 1 × 106 H1 hESCs by Nucleofector 2b Device (Lonza). These hES cells were cultured in mTeSR1 medium with Thiazovivin (0.1 μM, Selleck) for 1 day, then screened by puromycin (1 μg/ml, Gibco).
To identify gene knockout or knock-in cell lines, Genomic DNA extracted by the TIANamp Genomic DNA Kit (Tiangen) was used in PCR experiments. For gene knockout, the KO-F/R primers were used to amplify a ∼2 kb product of the targeted integration. For gene knock-in, the KI-F/R primers were used to amplify a ∼2 kb product of the targeted integration. All gRNA sequences and primer sequences are listed in Supplementary Table S1.
Establishing inducible over-expression or gene knockout cell lines in human ESCs
Inducible gene over-expression was through the Tet-on system. Firstly, typical coding protein sequences of HBO1 were cloned in the inducible over-expression plasmid FUW-HBO1. FUW-HBO1 and rtTA plasmids were in company with pMD2G and psPAX2 respectively co-transfection in 293T for producing viral particles, which were concentrated by ultracentrifugation. These cells were selected with doxycycline (2 μg/ml, DOX) and puromycin, known as hESCs-HBO1-OE.
Endogenous HBO1 was deleted in the inducible over-expression cell line. For targeting, the same method as gene knockout in hESCs. Positive clones were selected by puromycin and picked and cultured in mTeSR1 with DOX, known as hESCs-HBO1−/−-OE. We got a total of three clones of hESCs-HBO1−/−/OE, and these clones showed consistent phenotype. All primer sequences are listed in Supplementary Table S1.
Overexpression of HBO1 mutants in the hESCs-HBO1−/−-OE
High fidelity polymerase (Vazyme) was used in PCR to produce the wild type and all mutant cDNAs of human HBO1. Using homologous recombination technology, these PCR products were put into the pSIN-3 × FLAG vector, respectively. After that, 293T cells were used to package these plasmids into lentiviruses. hESCs-HBO1−/−-OE were cultured in mTeSR1 medium on Matrigel-coated 6-well plates without DOX for 1 day and infected with these lentivirus containing these above cDNAs for 1 day. Then, we used puromycin (1 mg/ml, Gibco) to screen positive cells. These primer sequences are listed in the Supplementary Table S2.
RNA extraction and quantitative real-time PCR
Total RNA was extracted with Trizol (Invitrogen), and complementary DNA was generated using oligo dT (Takara) and RT ACE (Toyobo). mRNA levels were quantified by real-time PCR using SYBR Green (Vazyme) and CFX96 machine (BIO-RAD) and were represented relative to GAPDH expression. All the data were measured in three duplicates. All primer sequences are listed in Supplementary Table S2.
Western blot analysis
Total proteins of hES cells were extracted using RIPA buffer (Beyotime) loaded on 4–20% SDS-PAGE gel and transferred to PVDF membranes (Millipore), and incubated with primary antibodies at room temperature for 2 h. The membranes were washed in TBST, and incubated with HRP-conjugated secondary antibodies. HRP was detected by ECL (Trans), and visualized by GelView 6000Plus (BLT). The information for antibodies is listed in Supplementary Table S3.
Flow cytometry analysis
The cells were dissociated into single cells by Accutase (Sigma) treatment, and fixed in solution buffer (BD, 554655) for about 20 min at room temperature, washed twice with 2% fetal bovine serum (FBS, Vistech) in PBS. Cell suspensions were stained with antibodies, incubated at 37°C for 30 min, then further stained with secondary antibodies for 30 min, and analyzed with a CytoFLEX S (BECKMAN) using FlowJo software. The information for antibodies is listed in Supplementary Table S3.
Immunofluorescence analysis
The cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 (sigma) and 10% FBS (Vistech) in PBS, and stained with primary antibodies overnight at 4°C. After washing thrice in PBS for 5 min each time, the cells were stained with secondary antibodies for 1.5 h at room temperature. Images were captured with an LSM 800 microscope (Zeiss). The information for antibodies is listed in Supplementary Table S3.
Co-immunoprecipitation analysis
Briefly, Co-immunoprecipitation experiments were performed in 293T cells based on double overexpression of both proteins. 293T cells were seeded in a 6-well plate and co-transfected with the over-expression plasmid pSIN-FLAG-HBO1 and pSIN-HA-SMAD4. In addition, Co-IP experiments werer performed in KI-3 × FALG hESCs in the presence or not of Benzonase (Beyotime). Proteins were extracted from these cells using an immunoprecipitation kit (Biolinkedin) following the manufacturer's instructions. The beads-protein complex was washed three times in wash buffer, and boiled with 30 μl 1 × SDS-PAGE sample loading buffer for 10 min and subjected to western blot analysis. The information for antibodies is listed in Supplementary Table S3.
Teratoma formation analysis
Teratoma formation assays were performed as described previously (28). Briefly, knockout and WT hES cells (1 × 106) were resuspended in 30% matrigel (Corning) in DMEM/F12 (Hyclone) and injected subcutaneously into severe combined immunodeficient mice. Four weeks later, mice with tumors were euthanized, and tumors were fixed in 4% paraformaldehyde for several days, embedded in paraffin, sectioned, and stained with hematoxylin and eosin for histological analysis. We had replicates for teratoma formation for each cell lines and there were two teratomas generated from each WT hESC line or HBO1−/− hESC line. Mice were used according to animal care standards and all experimental procedures were approved by the local ethical committee.
hESC lineage specification
Neuroectoderm differentiation was performed as described previously (37). Briefly, hESCs were passaged at a 2:1 ratio on new six-well plates in mTeSR1 medium and changed into neural induction medium with 5 μM SB431542 (Sigma) and 5 μM Dorsomorphin (Sigma) the next day. After 8 days of induction, cells were passaged on a new Matrigel plate at a 1:2 ratio and changed into N2B27 medium. After 16 days, canonical neural rosettes appeared and were selected. These neural rosettes were named Neural progenitor cells (NPC).
For primitive streak differentiation as described previously (38), hESCs were dissociated by Accutase (Sigma) and on new six-well plates at a 1:3 ratio in mTeSR1 medium with thiazovivin (0.1 μM, Selleck) and changed into induction medium the next day. The medium contained 40 ng/ml BMP4 (Peprotech), 30 ng/ml ACTIVIN A (Sino Biological Inc.), 20 ng/ml bFGF (Sino Biological Inc.), 6 μM CHIR99021 (Selleck), 10 μM LY294002 (Selleck) with 5 μM SB431542 (Sigma) and 5 μM Dorsomorphin (Sigma). After 2 days, the cells were named T+ cells.
For gastrulation-like specification as described previously (39), prior to differentiation, the ∼80% confluent hESCs were dissociated by Dispase (Sigma) at 37°C for 5 min. After washing cells three times in DMEM/F12 (Hyclone), the cells were scraped off on the plates and passaged at a 1:4 ratio on new six-well plates in mTeSR1 medium with thiazovivin (0.1 μM, Selleck). After overnight culture (designated as day 0), the hESCs were induced for gastrulation-like specification in mTeSR1 medium with BMP4 (50 ng/ml, R&D) for 3 days.
Gene expression RNA-seq analysis
Total RNA was extracted using Trizol (Invitrogen), and sequencing libraries for RNA-seq analysis were carried out using the VAHTS Universal V8 RNA-seq Library Prep Kit for Illumina (Vazyme #NR605) following the manufacturer's instructions. The samples were run on the sequencing platform of Illumina NovaSeq 6000.
Raw reads were filtered by Trimmomatic (v0.35) and then mapped to the human reference genome (hg38) using Hisat2 (v2.0.4). Gene expression was calculated using SAMtools (v1.3.1) and htseq-count (v0.6.0), then filtered by a threshold of at least 20 average raw read counts among samples, and normalized Using EDASeq (v2.12.0). Differential expression was analyzed Using DESeq (v 1.18.1). Differences in gene expression with a P-value <5% and a fold-change >2 were considered significant differences. A heatmap was performed using pheatmap (v1.0.10) and Gene ontology analysis was prepared using clusterProfiler (v3.6.0).
CUT&tag-seq analysis
The sequencing libraries for CUT&Tag-seq analysis were carried out using the Hyperactive Universal CUT&Tag Assay Kit for Illumina Pro (Vazyme, #TD904) following the manufacturer's instructions. The samples were run on the sequencing platform of Illumina NovaSeq 6000. Two biological replicates were performed for each sample.
Cutadapt (v1.13) was used to cut adaptors of raw fastq files with parameters -m 35 -e 0.1. Processed sequences were then aligned to reference human genome (hg38) by bowtie2 (v2.4.1), and parameters were set as recommended in the official protocol (https://yezhengstat.github.io/CUTTag_tutorial/). Low-quality mapped reads were filtered out using SAMtools (v1.3.1), followed by tagging duplicated reads utilizing Picard tools MarkDuplicates (v1.90). Peak calling of transcription factor and histone modifications were carried out using MACS2 (v2.1.0) and SICER2, respectively. Peaks (blacklist region excluded) with -log(q-value) >10 from MACS2 and score >200 from SICER2 were kept for downstream analysis. Bigwig files were generated by DeepTools (v3.4.3) module bamCoverage, computeMatrix, plotHeatmap, and plotProfile modules were adopted to visualize them. Annotation of peaks relied upon the annotatePeak function of R package ChIPseeker (v1.26.0) on transcript level, and tssRegion was set as c (-3000,3000). R package clusterProfiler (v3.18.0) provides effective ways to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Differentially binding regions were identified by the Differential Peaks module, and motif analysis was conducted by the Motifs Genome module, both are affiliated in Homer.
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism and Microsoft Excel software. For samples at least three biological repeats, values in all figures represent means ± SD (standard deviation). The P values were calculated by two-tailed unpaired Student's t-test. Differences with a P value <0.05 were considered statistically significant. No samples were excluded from any analysis.
Results
HBO1−/− hESCs undergo spontaneous neural fate specification
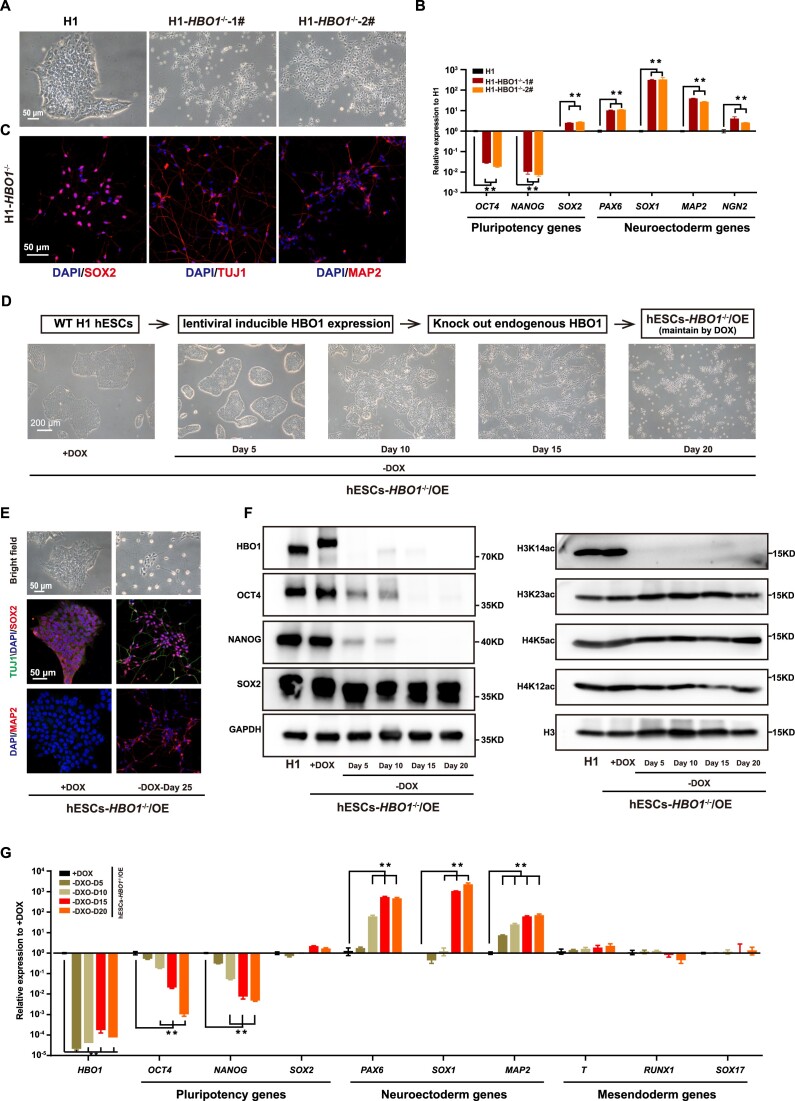
To examine the role of HBO1 in human ESCs, we generated HBO1 gene knock-out in human ESCs (Supplementary Figure S1A–C). Strikingly, HBO1−/− hESCs displayed a complete differentiation phenotype resembling neural fate cells, even under typical culture conditions that support pluripotency and suppress differentiation in human pluripotent stem cells (PSCs) (Figure 1A). Indeed, neural fate related genes were significantly up-regulated in HBO1−/− hESCs based on RT-qPCR or immunostaining assay (Figure 1B, C). Since HBO1−/− hESCs could not be further maintained and expanded, to extensively analyze the role of HBO1, we transfected a exogenous lentiviral based DOX inducible HBO1 expression in hESCs and performed endogenous HBO1 knock-out (hESCs-HBO1−/−/OE) (Supplementary Figure S1D–H). HBO1 expression and the undifferentiated state of hESCs-HBO1−/−/OE were maintained by DOX treatment (Figure 1D). Upon DOX withdrawal, hESCs-HBO1−/−/OE underwent spontaneous differentiation towards a neural fate morphology (Figure 1D). Upon longer culture at the same hESC culture condition, we could detect substantial MAP2+/TUJ1+ neurons in hESCs-HBO1−/−/OE (Figure 1E). As a control, we failed to detect MAP2+/TUJ1+ neurons in WT hESCs under the same culture condition (Figure 1E). Western-blot data confirms that HBO1 as well as pluripotency associated factors OCT4 and NANOG gradually disappeared upon DOX withdrawal, while SOX2, the known neuroectoderm factor were sustained expression consistent to the neural fate differentiation of these cells (Figure 1F). Furthermore, Since that HBO1 acetylates histones H3 and H4 (40), we examined H3 or H4 histone acetylation and showed that H3K14ac while not other histone acetylation was impaired HBO1 deficient cells (Figure 1F). Based on expression analysis, neural fate genes while not mesendoderm genes were up-regulated in hESCs-HBO1−/−/OE (Figure 1G). Together, these data show that hESCs-HBO1−/− failed to maintain pluripotency under typical culture conditions that support hESC pluripotency and underwent spontaneous neural fate specification.
Figure 1.
HBO1 −/− hESCs exit pluripotency and spontaneously differentiate into neuroectoderm fate. (A) Representative images of wild-type (WT) and targeted deletion of HBO1 in H1 hESCs. The scale bar represents 50 μm. (B) Expression of pluripotency and neuroectoderm genes in HBO1−/− hESCs, detected by quantitative reverse transcription PCR (RT-qPCR). WT H1 hESCs serve as a control. (C) Immunostaining analysis of neuroectoderm markers SOX2, TUJ1 and MAP2 expression in HBO1−/− hESCs. The scale bar represents 50 μm. (D) Top: Schematic of the strategy of endogenous HBO1 knockout in H1 hESCs with HBO1 overexpression, referred to as hESCs-HBO1−/−/OE. Bottom: The morphology of hESCs-HBO1−/−/OE with or without DOX in mTeSR1 medium. The scale bar represents 200 μm. (E) Representative bright field and immunostaining analysis of neuroectoderm markers SOX2 and MAP2 expression in hESCs-HBO1−/−/OE with or without DOX after 25 days (-DOX-Day 25). The scale bar represents 50 μm. (F) Western blot of pluripotency associated genes (OCT4, SOX2 and NANOG), HBO1 and indicated histone modifications in the indicated cell lines. GAPDH and H3 were used as loading control. (G) RT-qPCR analysis of HBO1, pluripotency, neuroectoderm, and mesendoderm associated genes expression in the indicated cell lines. hESCs-HBO1−/−/OE with DOX serve as control. In B and G, the data represent mean ± SD from three biological repeats with three technical replicates. The significance level was determined using unpaired two-tailed Student's t-tests. **P< 0.01.
The C-terminal MYST domain is essential for the function of HBO1
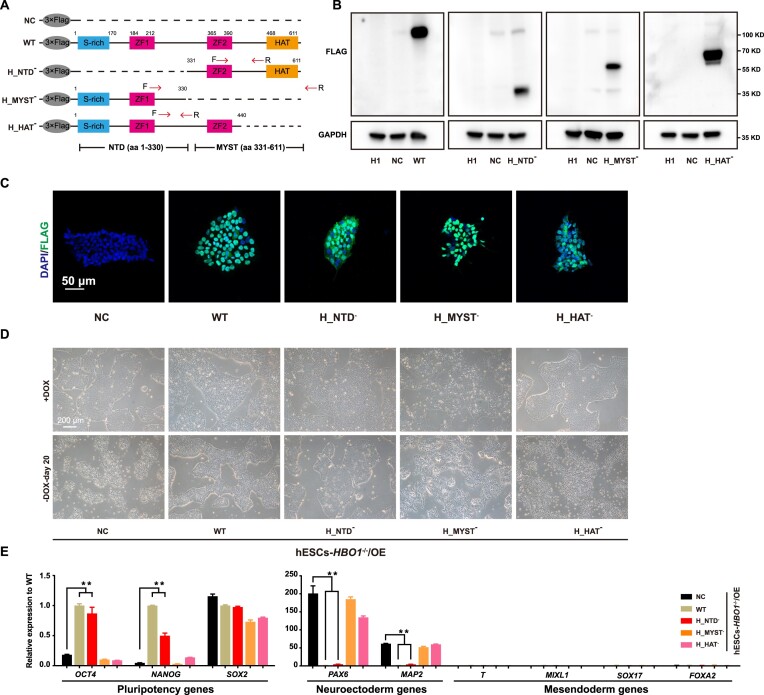
HBO1 is a core catalytic subunit of MYST acetyltransferase complexes and contains a C-terminal MYST acetyltransferase domain and a N-terminal domain (NTD) (41) (Figure 2A). The N-terminal domain contains a short zinc fingure (ZF) motif that is considered to bind co-factors to regulate HBO1 functions (41). To analyze the role of these domains in HBO1 functions, we generated lenti-viral based expression vector of different mutant forms of HBO1 deleted by either NTD or MYST domain (Figure 2A). The expression of different mutant forms of HBO1 could be detected western-blot as well as immunostaining (Figure 2B, C). Based on the hESCs-HBO1−/−/OE system, NTD deleted HBO1 (H_NTD−) largely rescued phenotype of hESCs-HBO1−/–(Figure 2D). In contrast, MTST domain deleted (H_MYST−) or HAT domain deleted HBO1 (H_HAT−) showed impaired function to rescue HBO1 deficiency in hESCs (Figure 2D). Analysis on the gene expression confirmed that H_NTD− largely maintained expression of pluripotency associated genes and suppressed up-regulation of neuroectoderm genes in HBO1 deficient hESCs, while H_MYST− or H_HAT− lost this function (Figure 2E). Together, these data demonstrate that the C terminal MYST acetyltransferase domain is essential to HBO1 functions in hESCs.
Figure 2.
The MYST domain is essential for HBO1 function. (A) Schematic of Wild-type (WT) and truncated human HBO1 constructs tagged by 3 × Flag. S-rich, serine-rich domain; ZF, zinc finger; HAT, histone acetyltransferase. (B) Immunoblotting analysis of expression of each mutant by anti-FLAG. (C) Immunofluorescence analysis of expression of each mutant. The scale bar represents 50 μm. (D) Representative images of expression of WT or each truncated mutants of HBO1 in hESCs-HBO1−/−/OE with or without DOX. The scale bar represents 200 μm. (E) RT-qPCR analysis of pluripotency, neuroectoderm, and mesendoderm associated genes expression in the indicated cell lines after 20 days of withdrawal DOX. Overexpression 3 × FLAG blank plasmid in hESCs-HBO1−/−/OE (NC) serve as control. The data represent mean ± SD from three biological repeats with three technical replicates. The significance level was determined using unpaired two-tailed Student's t-tests. **P< 0.01.
HBO1 binds transcriptionally active or poised chromatin of pluripotency and embryonic patterning genes in hESCs
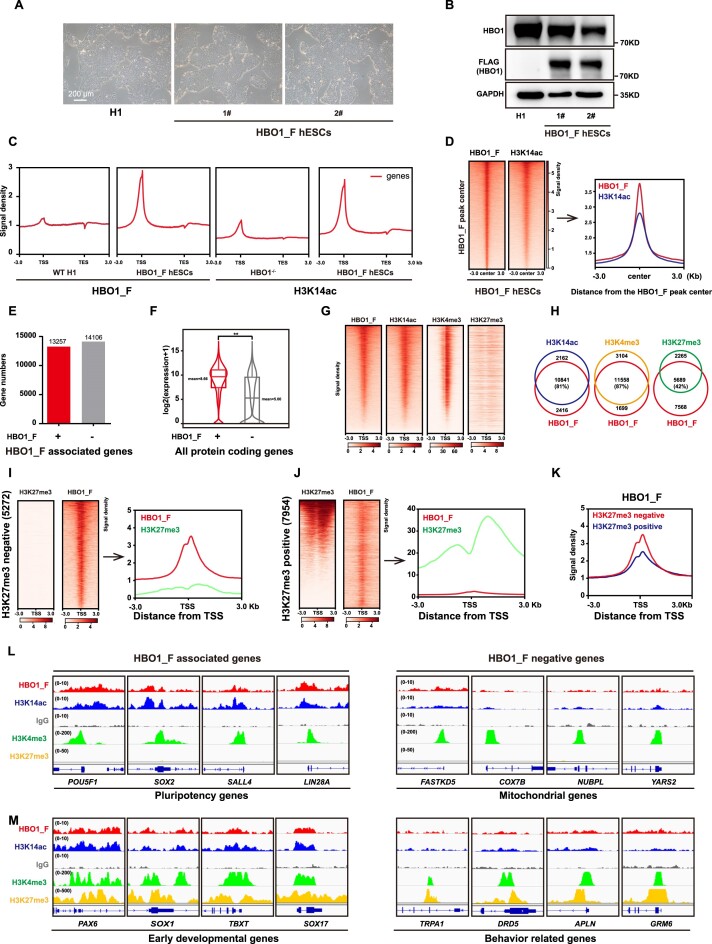
We then sought to examine the molecular role of HBO1 in hESCs. We firstly generated a triple-FLAGs knock-in at the C terminal of endogenous HBO1 in hESCs (HBO1_F hESCs) (Supplementary Figure S2A–D). HBO1_F hESCs showed typical undifferentiated morphology and HBO1_F expression was validated by western blot (Figure 3A, B). We then performed CUT&Tag-seq assay by anti-FLAG and also anti-H3K14ac in HBO1_F hESCs (Figure 3C). HBO1_F binding peaks greatly co-localized with H3K14ac perks on chromatin (Figure 3D), consistent to previous reports and the data shown in Figure 1F that HBO1 catalyzed H3K14ac (42,43). HBO1 and H3K14ac co-bound genes were highly enriched in functions related stem cell regulations (Supplementary Figure S2E). Roughly 50% of HBO1 and H3K14ac peaks localized on promoter region, and a portion of previously identified enhancers (44) was also associated with HBO1 and/or H3K14ac (Supplementary Figure S2F, G). Generally, HBO1 highly enriched genes showed more elevated expression levels compared with HBO1 less-enriched genes (Figure 3E, F). We and others have shown that the pluripotency and homeostasis genes in hESCs have active chromatin associated with H3K4me3 while the developmental genes are repressed with H3K27me3 (26,27). HBO1 highly enriched genes showed a high level of H3K4me3 but less H3K27me3 enrichment (Figure 3G, H, Supplementary Figure S2H). Conversely, H3K27me3 less-enriched genes exhibited higher HBO1 enrichment compared with H3K27me3 high-enriched genes (Figure 3I–K). Together, these data indicate that HBO1 bound transcriptionally active or poised genes rather than inactive genes in human ESCs. For example, genes related to self-renewal, stem cell functions or other critical homeostasis functions, such as POU5F1, SOX2, LIN28A, etc. were highly enriched in HBO1 binding (Figure 3L, Supplementary Figure S2I). However, some housekeeping genes that known as stem cell genes were less bound by HBO1, such as mitochondrial genes (Figure 3L, Supplementary Figure S2K). In addition, HBO1 also preferentially bound early essential embryonic patterning genes in hESCs such as PAX6, SOX1, SOX17, T, etc. (Figure 3M, Supplementary Figure S2J), rather than other H3K27me3 associated genes such as behavior related genes (Figure 3M, Supplementary Figure S2L). Together, these data suggest that HBO1 preferentially binds transcriptionally active genes that are related to pluripotency in hESCs.
Figure 3.
HBO1 and H3K14ac co-localize on pluripotency and embryonic lineage genes in hESCs. (A) Representative images of WT and H1 hESCs with a triple-FLAGs knock-in at the C terminal of endogenous HBO1, referred to as HBO1_F hESCs. The scale bar represents 200 μm. (B) Western blot analysis of HBO1 and FLAG expression in WT H1 and HBO1_F hESCs. GAPDH was used as a loading control. (C) Signal densities at the gene body of FLAG (HBO1_F) in WT H1 and HBO1_F hESCs (left), and H3K14ac in HBO1−/− hESCs and HBO1_F hESCs (right) from CUT&Tag-seq data. (D) Heatmap and signal densities analysis of FLAG (HBO1_F) and H3K14ac at FLAG (HBO1_F) peak center around ± 3 kb in HBO1_F hESCs. (E) The bar diagram analysis of the number of FLAG (HBO1_F) associated (+) and negative (–) genes in HBO1_F hESCs. (F) The Box-plot analysis of FLAG (HBO1_F) associated (+) and negative (–) protein coding genes transcription levels in HBO1_F hESCs. (G) The heatmap analysis of the binding of HBO1_F, H3K14ac, H3K4me3 and H3K27me3 around chromatin TSS (transcriptional start site). The heatmap was ordered by HBO1_F peak from highest to lowest. H3K4me3 (23) and H3K27me3 (24) ChIP-seq data were published. (H) Overlap between HBO1_F targeted and H3K14ac, H3K4me3, and H3K27me3 respectively targeted genes in hESCs. (I) Heatmap and signal densities analysis of FLAG (HBO1_F) and H3K27me3 in H3K27me3 negative but HBO1_F, H3K14ac and H3K4me3 all positive genes. (J) Heatmap and signal densities analysis of FLAG (HBO1_F) and H3K27me3 in H3K27me3 positive gengs. (K) Signal densities analysis of FLAG (HBO1_F) in H3K27me3 negative or positive gengs. (L) Genomic views of the FLAG (HBO1_F), H3K14ac associated (left) or negative (right) with H3K4me3 positive but H3K27me3 negative of genes in the indicated cell lines. (M) Genomic views of the FLAG (HBO1_F), H3K14ac associated (left) or negative (right) with bivalent genes in the indicated cell lines.
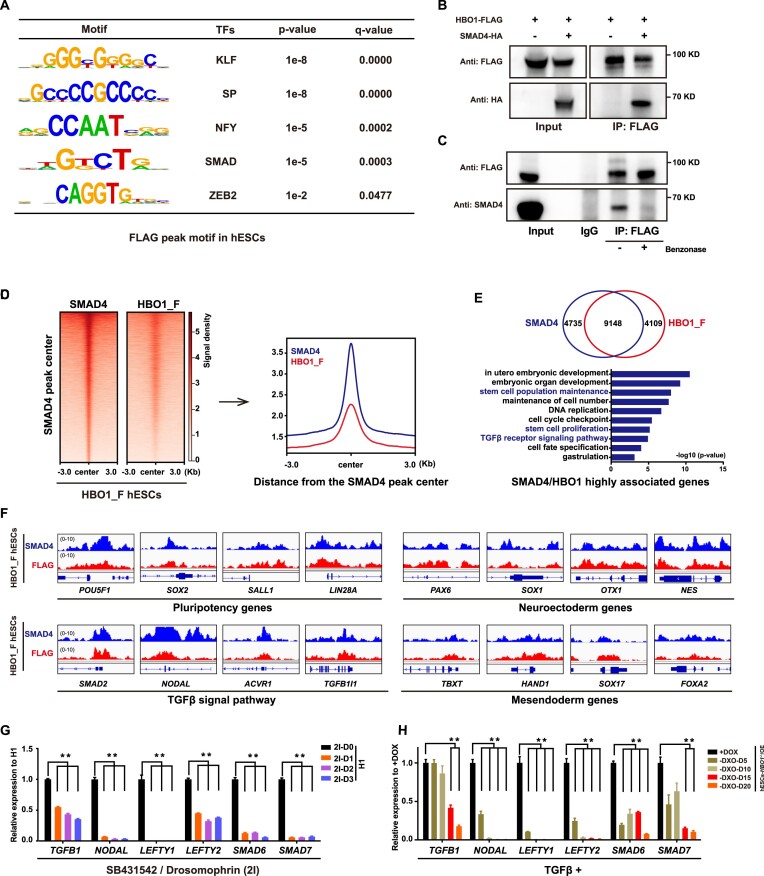
HBO1 and SMAD4 assoaciate and co-occupy on pluripotency related genes in hESCs
Since dual-SMAD (SB431542/Dorsomorphin) inhibition in hESCs leads to pluripotency exit and neural fate differentiation (10), resembling the phenotype of spontaneous neural differentiation in HBO1 deficient hESCs. To further examine whether HBO1 regulates TGF-β functions in hESCs, we searched enriched transcription factor (TF) motifs in HBO1 peak regions and revealed that the SMAD motif was among the top enriched motifs (Figure 4A). Indeed, we further showed that HBO1 and SMAD4, the co-SMAD for both TGF-β branches were associated based on co-immunoprecipitation assay (Figure 4B). Furthermore, using HBO1_F hESCs, we showed that the endogenous SMAD4 and HBO1 were interacted and this interaction could be impaired by the nuclease treatment (Figure 4C), indicating that their interaction depends on chromatin binding. We then performed CUT&Tag-seq assay by anti-SMAD4 in HBO1_F hESCs. SMAD4 and HBO1 showed substantial overlap on chromatin occupancy in hESCs (Figure 4D). The top genes bound by both factors were highly enriched in genes related to pluripotency, early embryonic patterning, self-renewal etc. (Figure 4E, F). These data indicate that HBO1 might be a downstream effector recruited by SMAD4 to mediate TGF-β actions at the chromatin level. It's known that TGF-β family member genes are also direct targets of TGF-β signaling to form a feedback auto-regulatory mechanism (45,46). These genes were suppressed by the dual-SMAD inhibition in human ESCs (Figure 4G). We showed that these genes were also suppressed in HBO1 deficient hESCs albeit in the presence of TGF-β signaling (Figure 4H), indicating that HBO1 is important for the TGF-β signaling target gene expression in human ESCs.
Figure 4.
HBO1 co-localizes with SMAD4 on chromatin and important for TGFβ signal function in hESCs. (A) Enriched motifs analysis of the HBO1_F peaks in HBO1_F hESCs. (B) Western blots analysis of FLAG (HBO1) and HA (SMAD4) co-immunoprecipitated from total protein extracts in 293T that were transfected with protein overexpression plasmids for FLAG-tagged HBO1 and/or HA-tagged SMAD4. (C) Western blots analysis of FLAG (HBO1) and SMAD4 co-immunoprecipitated from total protein extracts in HBO1_F hESCs. The Benzonase super nuclease was employed to break down DNA and RNA nucleic acids. (D) Heatmap and signal densities of SMAD4and FLAG (HBO1_F) at SMAD4 peak center around ±3 kb in HBO1_F hESCs. (E) Top: Overlap between HBO1_F and SMAD4 targeted genes in HBO1_F hESCs. Bottom: GO terms of biological processes analysis of SMAD4 and FLAG (HBO1_F) highly enriched genes. (F) Genomic views analysis of the SMAD4 and FLAG (HBO1_F) binding of pluripotency, neuroectoderm, TGFβ signal pathway, and mesendoderm associated genes in HBO1_F hESCs. (G) RT-qPCR analysis of TGFβ signal pathway associated genes expression in WT hESCs treated with 2I (SB431542/Dorsomorphin inhibitors). WT H1 hESCs serve as a control. (H) RT-qPCR analysis of TGFβ signal pathway associated genes expression in the indicated cell lines. hESCs-HBO1−/−/OE (+DOX) serve as a control. In G and H, the data represent mean ± SD from three biological repeats with three technical replicates. The significance level was determined using unpaired two-tailed Student's t-tests. **P< 0.01.
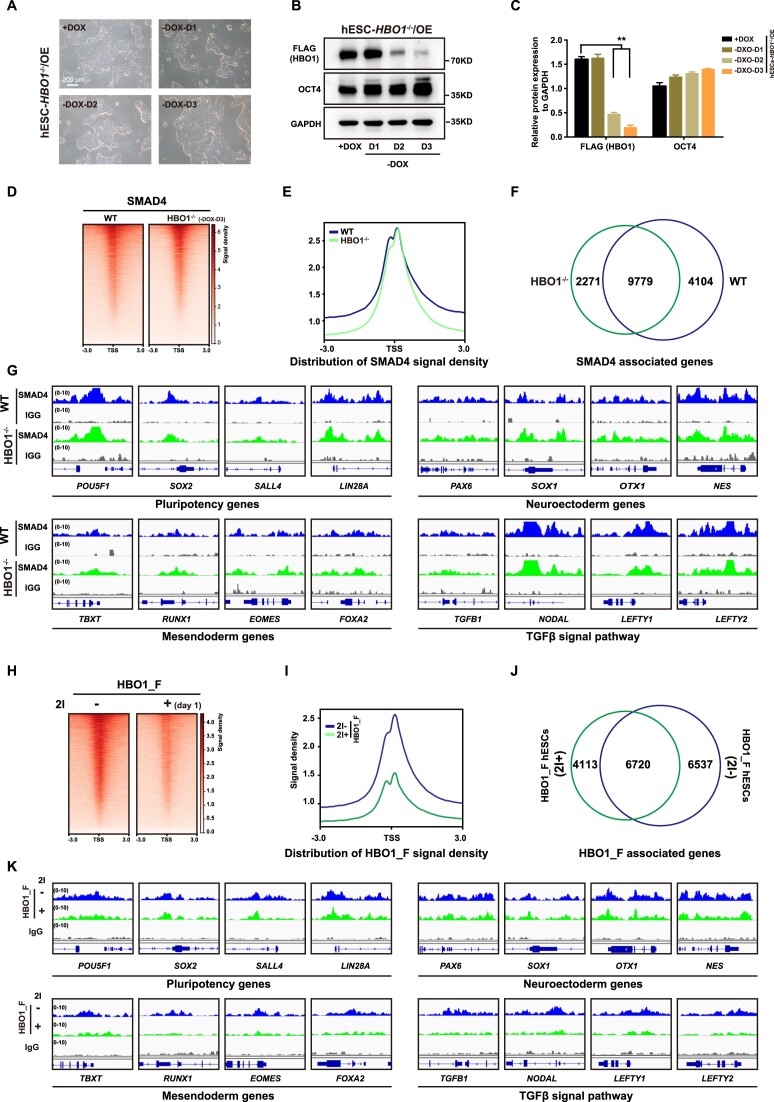
SMAD inhibition alleviates HBO1 chromatin occupancy in hESCs
We next examined interdependency of SMAD4 and HBO1 in chromatin occupancy. We firstly analyzed SMAD4 chromatin bindings in HBO1 deficient hESCs. Upon DOX withdrawal, hESCs-HBO1−/−/OE cells lost most of HBO1 protein at day 3 of DOX withdrawal (HBO1−/− hESCs) but still maintained undifferentiated phenotype with a high level of OCT4 protein (Figure 5A–C, Supplementary Figure S3A–C). SMAD4 chromatin occupancy showed no significant impairment in HBO1−/− hESCs compared with WT hESCs (Figure 5D, E). Over 90% of SMAD4 associated genes were overlapped in between WT and HBO1−/− hESCs (Figure 5F). Examples for SMAD4 associated genes in WT and HBO1−/− hESCs were shown in Figure 5G. On the other hand, we analyzed HBO1 chromatin bindings in hESCs with dual-SMAD inhibition (2i) (Figure 5H). Strikingly, HBO1 chromatin enrichment was significantly reduced in 2i-treated hESCs (Figure 5I). About 50% of HBO1 associated genes in WT hESCs significantly reduced HBO1 enrichment down below the peaking-calling bar in 2i-treated hESCs (Figure 5J). Examples for these HBO1 associated genes in WT or HBO1−/− hESCs were shown in Figure 5K. These genes include pluripotency associated genes as well as other linage genes (Figure 5K). Together, these data suggest that SMAD4 is important for HBO1 chromatin occupancy in hESCs.
Figure 5.
SMAD4 is important for HBO1 chromatin occupancy. (A) The morphology of hESCs-HBO1−/−/OE with or without DOX of days 1, 2 or 3 in mTeSR1 medium. The scale bar represents 200 μm. (B) Expression of OCT4 and HBO1 by western blot in the indicated cell lines. GAPDH was used as a loading control. (C) Histogram statistical analysis of the FLAG (HBO1) and OCT4 protein expression relative to GAPDH. The data represent mean ± SD from three biological repeats. The significance level was determined using unpaired two-tailed Student's t-tests. **P< 0.01. (D and E) Heatmap and signal densities of SMAD4 chromatin binding in WT and hESCs-HBO1−/−/OE upon DOX withdrawal for three days (HBO1−/−). (F) Overlap between SMAD4 targeted genes in WT and HBO1−/- hESCs. (G) Examples of genomic view of SMAD4 associated genes in WT and HBO1−/- hESCs. (H and I) Heatmap and signal densities of FLAG (HBO1_F) chromatin binding in WT hESCs treated with (+) or without (–) dual-SMAD inhibitors (SB431542/Dorsomorphin, 2I). (J) Overlap between FLAG (HBO1_F) associated genes in WT or 2I treated hESCs. (K) Examples of genomic view of HBO1_F associated genes in in WT or 2I treated hESCs.
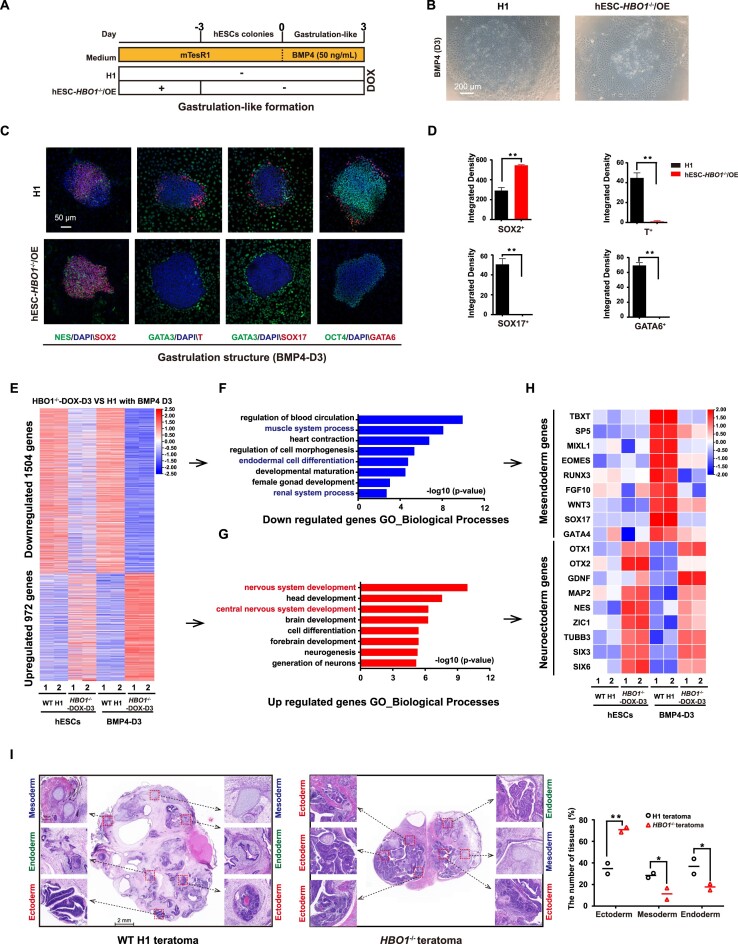
HBO1 specifies human mesendoderm
TGF-β branch and BMP branch of the TGF-β family play different roles to regulate human ESCs. TGF-β branch signaling supports self-renewal while the BMP branch triggers differentiation (14,47,48). BMP4 was also reported to induce hESCs to form a 2D gastruloid model containing typical three early germ layer lineages (39). To further examine whether HBO1 is essential for BMP action in hESC differentiation, we performed BMP4 induced gastruloid formation in the presence or absence of HBO1 (Figure 6A). We then triggered gastruloid formation on hESCs-HBO1−/−/OE cells by BMP4 at day 3 of DOX withdrawal based on the previously published protocol (39) (Figure 6B). As expected, we could clearly detect various early embryonic lineage fates in 2D gastuloids derived from wild-type (WT) hESCs, represented by immunostaining of lineage markers including SOX2/NES for neuroectoderm, GATA3 for trophectoderm, T for mesoderm and SOX17/GATA6 for endoderm (Figure 6C). In contrast, we failed to detect T+ or GATA6+/SOX17+ mesendoderm in HBO1 deficient hESCs, while the SOX2+ neuroectoderm lineage cells were enhanced (Figure 6D). Consistently, the mesendoderm genes were greatly suppressed in gastruloids formed in HBO1 deficient hESCs, according to RNA-seq data (Figure 6E–H). Lastly, we performed teratoma formation on WT and HBO1 deficient hESCs. We could easily detect tissue structures for all three germ layers in teratomas derived from WT hESCs (Figure 6I). In contrast, the mesendoderm tissue structure is very rare in teratomas derived from HBO1 deficient hESCs, while the neuroectoderm represented the majority of teratoma tissue (Figure 6I). In addition, we also performed directed differentiation of neural or mesoderm fate in hESCs-HBO1−/−/OE based on previously published protocol (37,38). Based on neural differentiation protocol through dual-SMAD inhibition (2i), we showed that HBO1 deficiency promoted neural differentiation while HBO1 over-expression (OE) suppressed neural differentiation in hESCs (Supplementary Figure S4A–F). Similarly, based on our previously published protocol to trigger hematopoietic progenitor cells (HPCs) differentiation through mesoderm, HBO1 deficient hESCs showed severe defect in generation of T+ mesoderm cells while HBO1 OE enhanced T+ mesoderm cell generation (Supplementary Figure S4G–I). In hESCs-HBO1−/−/OE derived T+ mesoderm cells, withdrawal of DOX for inactivation of HBO1 expression did not show severe cell death (Supplementary Figure S4J, K), indicating that impairment mesoderm specification in HBO1 deficient hESCs was not due to the cell death of these cells. Taken all the data together, we demonstrate HBO1 plays a critical role to specify mesoderm cell fate triggered by BMP4 signaling pathway.
Figure 6.
HBO1 deficiency impairs mesendoderm specification. (A) Scheme for hESCs based 2D gastruloid model differentiation. (B) Representative images of BMP4 (50 ng/ml) induction of 2D gastruloid model differentiation for 3 days in WT H1 and hESCs-HBO1−/−/OE (–DOX-day 3). The scale bar represents 200 μm. (C) Immunostaining analysis of SOX2/NES (neuroectoderm), GATA3 (trophectoderm), T (mesoderm), and SOX17/GATA6 (endoderm) expression in WT H1 and hESCs-HBO1−/−/OE (–DOX-day 3). The scale bar represents 50 μm. (D) The integrated density of SOX2+, T+, SOX17+ or GATA6+ cells were analyzed. The significance was determined by unpaired two-tailed Student's t-tests. **P < 0.01. The data represent the mean ± SD from three independent replicates. (E) Heatmap analysis of WT H1 hESCs, hESCs-HBO1−/−/OE (–DOX-day 3) and after BMP4 induction of 2D gastruloid model differentiation for 3 days. These significantly up- or down-regulated genes were from HBO1−/−/OE (BMP4-D3) compared with WT H1 (BMP4-D3). The data were from RNA-seq. (F) GO terms of biological processes enriched for down-regulated genes, as described in E. (G) GO terms of biological processes enriched for up-regulated genes, as described in E. (H) Heatmap analysis of selected neuroectoderm and mesendoderm genes from RNA-seq data in WT H1 hESCs, hESCs-HBO1−/−/OE (–DOX-day 3) and after BMP4 induction of 2D gastruloid model differentiation for 3 days. These significantly differential genes were from HBO1−/−/OE (BMP4-D3) compared with WT H1 (BMP4-D3). (I) H&E staining on sections of teratomas formed in WT H1 and hESCs-HBO1−/−/OE without DOX. Scale bar: 2 mm. the data represent mean ± SD from two biological repeats. The significance level was determined using unpaired two-tailed Student's t-tests. **P< 0.01. *P< 0.05.
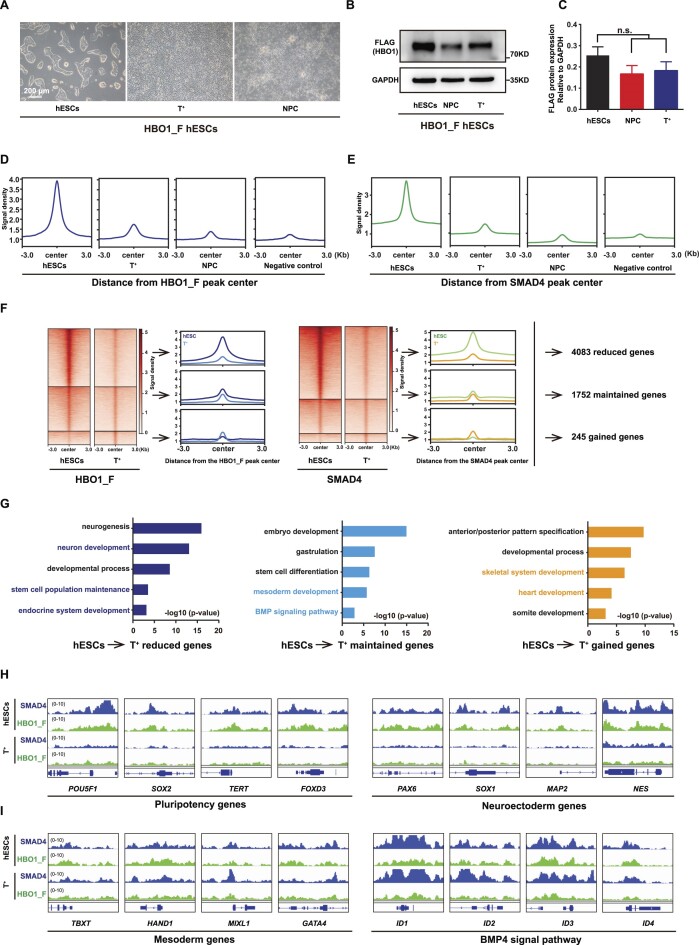
HBO1 and SMAD4 specify the active chromatin for mesoderm genes upon pluripotency exit
To further investigate the role of HBO1 in mesendoderm specification, we examined its chromatin occupancy switch in mesoderm or neural progenitor cells (NPCs) derived from hESCs. Firstly, we triggered HBO1_F hESCs into T+ mesoderm lineages based on our previously reported hematopoietic differentiation protocol through which hESCs could be converted into nearly 100% T+ mesoderm lineage cells at Day 2 of differentiation (38). hESCs derived NPCs were generated by the previously published typical dual-SMAD signaling inhibition strategy to suppress both branches of TGF-β signaling (37) (Figure 7A, Supplementary Figure S5A, B). Substantial levels of HBO1 protein could be detected in both hESCs derived T+ mesoderm cells and NPCs, albeit a slight down-regulation compared with undifferentiation hESCs (Figure 7B, C). We then examined genome wide occupancy by HBO1 and SMAD4 in mesoderm cells and NPCs. Chromatin binding intensities of HBO1 or SMAD4 showed significant reductions in both hESCs derived mesoderm cells and NPCs compared with undifferentiated hESCs (Figure 7D, E). However, a substantial number of confident HBO1 or SMAD4 chromatin binding peaks could be called out in mesoderm cells based the stringent peak-calling bar (Supplementary Figure S5C, D). In contrast, we failed to detect binding peaks of HBO1 and SMAD4 in NPCs based on the same algorithm (Supplementary Figure S5C, D). Since hESCs derived NPCs require dual-SMAD inhibition, it was not surprise that no confident SMAD4 chromatin binding peaks could be detected (Supplementary Figure S5E). Lastly, we examined chromatin occupancy of HBO1 and SMAD4 in mesoderm cells triggered by BMP4 signaling. Compared with their chromatin occupancy in undifferentiated hESCs, HBO1 or SMAD4 binding peaks were divided into three groups i.e. reduced, maintained or gained (Figure 7F). The genes that showed reduced HBO1/SMAD4 in mesoderm cells were enriched in neural developmental genes as well as pluripotency genes (Figure 7G, H). In contrast, the mesoderm lineage associated genes maintained or gained HBO1/SMAD4 bindings in mesoderm cells (Figure 7G, I). Together, these data indicate that HBO1/SMAD4 co-bind the mesoderm lineage associated genes upon pluripotency exit.
Figure 7.
HBO1 and SMAD4 specify the active chromatin in mesoderm cells. (A) Representative images of HBO1_F hESCs and mesoderm (T+) and ectoderm (neural progenitor cells, NPC) cells differentiation from HBO1_F hESCs. Scale bars: 200 μm. (B) Western blot analysis of FLAG (HBO1) in the indicated cells. GAPDH was used as a loading control. (C) Histogram statistical analysis of the FLAG protein expression relative to GAPDH. The data represent mean ± SD from three biological repeats. The significance level was determined using unpaired two-tailed Student's t-tests. n.s., no significance. (D) Signal densities analysis of FLAG (HBO1_F) bingding in the indicated cells at FLAG (HBO1_F) peak center around ± 3 kb. Anti-FLAG in WT H1 hESCs was used as a negative control. (E) Signal densities analysis of SMAD4 in the indicated cells at SMAD4 peak center around ±3 kb. Anti-FLAG in WT H1 hESCs was used as a negative control. (F) Left: Heatmap and signal densities analysis of reduced, maintained or gained FLAG (HBO1_F) peaks in T+ cells compared with hESCs. Middle: Heatmap and signal densities analysis of reduced, maintained, or gained SMAD4 peaks in T+ cells compared with HBO1_F hESCs. Right: The gene numbers for together reduced, maintained, or gained of FLAG (HBO1_F) and SMAD4 peaks in T+ cells compared with HBO1_F hESCs. (G) GO terms of biological processes analysis of reduced, maintained, or gained genes, as described in F. (H) Genomic views analysis of the SMAD4 and FLAG (HBO1_F) reduced binding of genes in T+ cells compared with HBO1_F hESCs. (I) Genomic views of the SMAD4 and FLAG (HBO1_F) maintained or gained binding of genes in T+ cells compared with HBO1_F hESCs.
Discussion
CRFs have been known to be important in lineage fate decision through establishing an lineage specific signature for epigenome (49,50). Early embryonic lineage decision was mainly instructed by the external signaling morphogens (51,52). How the morphogen signaling is intrinsically interpreted to establish the lineage specific epigenetic signature remains not fully elucidated. TGF-β family signaling represents the most important morphogen in early embryonic patterning and lineage decision (53,54). Here in this study, we reveal that HBO1, a known histone acetyltransferase (HAT) determines the action of SMAD in both human pluripotency and mesendoderm lineage specification. TGF-β fails to activate target genes and support pluripotency in hESCs in the absence of HBO1. HBO1−/− hESCs exit pluripotency but spontaneously give rise to neuroectoderm fate rather than the random fate choice. Moreover, HBO1 deficient hESCs failed to specify mesendoderm that were triggered mainly by BMP4 signaling. Our results highlight that CRFs are instructed by the external morphogen signaling to specify the poised lineage specific chromatin state. Indeed, HBO1 and SMAD4 co-occupy the chromatin of mesoderm lineage associated gene in hESCs derived mesoderm cells while reduce chromatin occupancy in NPCs with suppressed TGF-β signaling, indicating that HBO1 depends on SMAD4 to bind the target chromatin for the normal TGF-β signaling action.
HBO1 belongs to MYST family HAT that mainly catalyzes H3K14ac and is highly conserved from yeast to human (55). Compared with another HAT, CBP/P300 that were extensively investigated, the role of HBO1 has not been fully uncovered. Our findings reveal that HBO1 serves as a critical cell intrinsic CRF to for the function of TGF-β family signaling morphogens in early embryonic lineage patterning. Our data are consistent with previous studies that mouse embryos lacking Hbo1 were lethal at the early embryonic stage due to a widely disorganized tissue (56). Together, these data indicate that HBO1 plays an important role to regulate critical lineage decision in early embryonic development.
Besides, HBO1 was also shown to link to cancer genesis (41). HBO1 was found to be over-expressed in various cancers with gene amplification (57–59). The somatic mutation frequency in human HBO1 in cancer cells is also very high (41). HBO1 was shown to promote cell proliferation in bladder and breast cancers and also related to drug resistance in pancreatic cancer (60–62). Up-regulation of HBO1 was shown to connect to the poor prognosis in gastric cancer (58). However, on the other hand, HBO1 was also reported to be suppressed in acute myeloid leukemia (63), indicating that the role of HBO1 in cancer is largely context-dependent. The HBO1 function to embryonic lineage fate decision described here is largely dependent on TGF-β signaling. Interestingly, TGF-β signaling is also well known to play important role in cancer genesis (4,64). TGF-β induces an epithelial-mesenchymal transition (EMT) in cancer cells and promotes their metastasis (65,66). It will be interesting to elucidate whether HBO1 is also a cell intrinsic determinant for TGF-β signaling in cancer genesis.
Supplementary Material
Acknowledgements
We thank the lab members in GIBH for their kind help.
Author contributions: G.P. and C.Z. initiated and designed the project, and wrote the manuscript. C.Z. performed most experiments and analyzed result data. Y.S. and C.Z. performed the gene knockout of the HBO1 in hESCs. C.Z. and Q.X. Performed inducible over-expression or gene knockout cell lines in human ESCs. Y.Z., J.W. and J.Z. performed the FACS and RT-qPCR. T.Z., Q.C. and A.L. performed Teratoma Formation analysis. H.L. and C.Z. performed the RNA sequencing and CUT&Tag-seq analysis, and analyzed theese data. All authors read and approved the final manuscript.
Contributor Information
Cong Zhang, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Yongli Shan, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Huaisong Lin, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Yanqi Zhang, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Qi Xing, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Jinmin Zhu, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Tiancheng Zhou, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Aiping Lin, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Qianyu Chen, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Junwei Wang, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Guangjin Pan, Key Laboratory of Immune Response and Immunotherapy, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, 510530,China; Guangzhou Medical University, Guangzhou 511436, China; University of Chinese Academy of Sciences, Beijing 100049, China; Centre for Regenerative Medicine and Health, Hong Kong Institute of Science and Innovation, Chinese Academy of Sciences, Hong Kong; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong-Hong Kong Joint Laboratory for Stem Cell and Regenerative Medicine, Center for Cell Lineage and Cell Therapy, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China; GIBH-HKU Guangdong-Hong Kong Stem Cell and Regenerative Medicine Research Centre, GIBH-CUHK Joint Research Laboratory on Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530, China.
Data availability
The RNA-Seq and CUT&Tag-seq generated data have been deposited in the Genome Sequence Archive (67) in National Genomics Data Center (68) for Human database under the accession codes HRA005583 and HRA006159 that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human. All data supporting the findings of this study are available within the article and its supplemental information files or from the corresponding authors upon reasonable request.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Key Research and Development Program of China, Stem Cell and Translational Research [2022YFA1105001]; Research Funds from Health@InnoHK Program launched by Innovation Technology Commission of the Hong Kong SAR, P. R. China; the National Natural Science Foundation of China [32270624, 31971374]; Science and Technology Planning Project of Guangdong Province, China [2023B1212060050, 2023B1212120009]; the Youth Innovation Promotion Association of the Chinese Academy of Sciences [2022360 to Y.S.]; China Postdoctoral Science Foundation Funded Project [2023M733516]; Fountain-Valley Life Sciences Fund of University of Chinese Academy of Sciences Education Foundation [ZXXM202201]; Guangzhou Key Research and Development Program [202206010041]; Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine [2020B1212060052]; Guangdong Province Special Program for Outstanding Talents [2019JC05Y463 to G.P.]. Funding for open access charge: National Key Research and Development Program of China, Stem Cell and Translational Research [2022YFA1105001].
Conflict of interest statement. None declared.
References
- 1. Arnold S.J., Robertson E.J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Bio. 2009; 10:91–103. [DOI] [PubMed] [Google Scholar]
- 2. Mole M.A., Weberling A., Zernicka-Goetz M. Comparative analysis of human and mouse development: from zygote to pre-gastrulation. Gastrulation. 2020; 136:113. [DOI] [PubMed] [Google Scholar]
- 3. Richardson L., Wilcockson S.G., Guglielmi L., Hill C.S. Context-dependent tgfβ family signalling in cell fate regulation. Nat. Rev. Mol. Cell Biol. 2023; 24:876–894. [DOI] [PubMed] [Google Scholar]
- 4. David C.J., Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018; 19:419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012; 13:616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Boxtel A.L., Economou A.D., Heliot C., Hill C.S. Long-range signaling activation and local inhibition separate the mesoderm and endoderm lineages. Dev. Cell. 2018; 44:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greenfeld H., Lin J., Mullins M.C. The BMP signaling gradient is interpreted through concentration thresholds in dorsal-ventral axial patterning. PLoS Biol. 2021; 19:e3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang P.B., Li J.A., Tan Z.J., Wang C.Y., Liu T., Chen L., Yong J., Jiang W., Sun X.M., Du L.Y. et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008; 111:1933–1941. [DOI] [PubMed] [Google Scholar]
- 9. Nostro M.C., Cheng X., Keller G.M., Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008; 2:60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009; 27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003; 21:183–186. [DOI] [PubMed] [Google Scholar]
- 12. Ying Q.L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003; 115:281–292. [DOI] [PubMed] [Google Scholar]
- 13. Xu R.H., Sampsell-Barron T.L., Gu F., Root S., Peck R.M., Pan G.J., Yu J.Y., Antosiewicz-Bourget J., Tian S.L., Stewart R. et al. NANOG is a direct target of TGF beta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008; 3:196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vallier L., Alexander M., Pedersen R.A. Activin/nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005; 118:4495–4509. [DOI] [PubMed] [Google Scholar]
- 15. Massagué J., Seoane J., Wotton D Smad transcription factors. Genes Dev. 2005; 19:2783–2810. [DOI] [PubMed] [Google Scholar]
- 16. Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007; 447:407–412. [DOI] [PubMed] [Google Scholar]
- 17. Dalton S. Signaling networks in human pluripotent stem cells. Curr. Opin. Cell Biol. 2013; 25:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Janknecht R., Wells N.J., Hunter T. TGF-beta-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Gene Dev. 1998; 12:2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xi Q.R., Wang Z.X., Zaromytidou A.I., Zhang X.H.F., Chow-Tsang L.F., Liu J.X., Kim H., Barlas A., Manova-Todorova K., Kaartinen V. et al. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011; 147:1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kahata K., Hayashi M., Asaka M., Hellman U., Kitagawa H., Yanagisawa J., Kato S., Imamura T., Miyazono K. Regulation of transforming growth factor-beta and bone morphogenetic protein signalling by transcriptional coactivator GCN5. Genes Cells. 2004; 9:143–151. [DOI] [PubMed] [Google Scholar]
- 21. Bai R.Y., Koester C., Ouyang T., Hahn S.A., Hammerschmidt M., Peschel C., Duyster J. SMIF, a Smad4-interacting protein that functions as a co-activator in TGF beta signalling. Nat. Cell Biol. 2002; 4:181–190. [DOI] [PubMed] [Google Scholar]
- 22. Atlasi Y., Stunnenberg H.G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017; 18:643–658. [DOI] [PubMed] [Google Scholar]
- 23. Zhang C., Lin H., Zhang Y., Xing Q., Zhang J., Zhang D., Liu Y., Chen Q., Zhou T., Wang J. et al. BRPF1 bridges H3K4me3 and H3K23ac in human embryonic stem cells and is essential to pluripotency. iScience. 2023; 26:105939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shan Y.L., Zhang Y.Q., Zhao Y., Wang T.Y., Zhang J.Y., Yao J., Ma N., Liang Z.C., Huang W.H., Huang K. et al. JMJD3 and UTX determine fidelity and lineage specification of human neural progenitor cells. Nat. Commun. 2020; 11:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017; 171:34–57. [DOI] [PubMed] [Google Scholar]
- 26. Pan G.J., Tian S.L., Nie J., Yang C.H., Ruotti V., Wei H.R., Jonsdottir G.A., Stewart R., Thomson J.A. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007; 1:299–312. [DOI] [PubMed] [Google Scholar]
- 27. Bernstein B.E., Mikkelsen T.S., Xie X.H., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 28. Shan Y.L., Liang Z.C., Xing Q., Zhang T., Wang B., Tian S.L., Huang W.H., Zhang Y.Q., Yao J., Zhu Y.L. et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nat. Commun. 2017; 8:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collinson A., Collier A.J., Morgan N.P., Sienerth A.R., Chandra T., Andrews S., Rugg-Gunn P.J. Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in Human embryonic stem cells. Cell Rep. 2016; 17:2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tian T.V., Di Stefano B., Stik G., Vila-Casadesus M., Sardina J.L., Vidal E., Dasti A., Segura-Morales C., De Andres-Aguayo L., Gomez A. et al. Whsc1 links pluripotency exit with mesendoderm specification. Nat. Cell Biol. 2019; 21:824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y.L., Xie S.G., Zhou Y., Xie Y.Y., Liu P., Sun M.M., Xiao H.S., Jin Y., Sun X.J., Chen Z. et al. H3K36 Histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Rep. 2014; 8:1989–2002. [DOI] [PubMed] [Google Scholar]
- 32. Aloia L., Di Stefano B., Sessa A., Morey L., Santanach A., Gutierrez A., Cozzuto L., Benitah S.A., Graf T., Broccoli V. et al. Zrf1 is required to establish and maintain neural progenitor identity. Gene Dev. 2014; 28:182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Verdin E., Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Bio. 2015; 16:258–264. [DOI] [PubMed] [Google Scholar]
- 34. Sheikh B.N., Akhtar A. The many lives of KATs - detectors, integrators and modulators of the cellular environment. Nat. Rev. Genet. 2019; 20:7–23. [DOI] [PubMed] [Google Scholar]
- 35. Xiao Y.H., Li W.J., Yang H., Pan L.L., Zhang L.W., Lu L., Chen J.W., Wei W., Ye J., Li J.W. et al. HBO1 is a versatile histone acyltransferase critical for promoter histone acylations. Nucleic Acids Res. 2021; 49:8037–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Voss A.K., Thomas T. MYST family histone acetyltransferases take center stage in stem cells and development. Bioessays. 2009; 31:1050–1061. [DOI] [PubMed] [Google Scholar]
- 37. Su Z.H., Zhang Y.Q., Liao B.J., Zhong X.F., Chen X., Wang H.T., Guo Y.P., Shan Y.L., Wang L.H., Pan G.J. Antagonism between the transcription factors NANOG and OTX2 specifies rostral or caudal cell fate during neural patterning transition. J. Biol. Chem. 2018; 293:4445–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang T., Huang K., Zhu Y.L., Wang T.Y., Shan Y.L., Long B., Li Y.H., Chen Q.Y., Wang P.T., Zhao S.Y. et al. Vitamin C-dependent lysine demethylase 6 (KDM6)-mediated demethylation promotes a chromatin state that supports the endothelial-to-hematopoietic transition. J. Biol. Chem. 2019; 294:13657–13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014; 11:847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lalonde M.E., Avvakumov N., Glass K.C., Joncas F.H., Saksouk N., Holliday M., Paquet E., Yan K., Tong Q., Klein B.J. et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Gene. Dev. 2013; 27:2009–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lan R.F., Wang Q.Q. Deciphering structure, function and mechanism of lysine acetyltransferase HBO1 in protein acetylation, transcription regulation, DNA replication and its oncogenic properties in cancer. Cell. Mol. Life Sci. 2020; 77:637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tao Y., Zhong C., Zhu J.J., Xu S.T., Ding J.P. Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2. Nucleic Acids Res. 2017; 45:5707–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mishima Y., Miyagi S., Saraya A., Negishi M., Endoh M., Endo T.A., Toyoda T., Shinga J., Katsumoto T., Chiba T. et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011; 118:2443–2453. [DOI] [PubMed] [Google Scholar]
- 44. Gao T., Qian J. EnhancerAtlas 2.0: an updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020; 48:D58–D64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miyazawa K., Miyazono K. Regulation of TGF-beta Family signaling by inhibitory smads. Csh Perspect. Biol. 2017; 9:a022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan X., Xiong X., Chen Y.G. Feedback regulation of TGF-β signaling. Acta Biochim. Biophy. Sin. 2018; 50:37–50. [DOI] [PubMed] [Google Scholar]
- 47. Oshimori N., Fuchs E. The harmonies played by TGF-β in stem cell biology. Cell Stem Cell. 2012; 11:751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bernardo A.S., Faial T., Gardner L., Niakan K.K., Ortmann D., Senner C.E., Callery E.M., Trotter M.W., Hemberger M., Smith J.C. et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011; 9:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morey L., Santanach A., Di Croce L. Pluripotency and epigenetic factors in mouse embryonic stem cell fate regulation. Mol. Cell. Biol. 2015; 35:2716–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kraushaar D.C., Zhao K.J. The epigenomics of embryonic stem cell differentiation. Int. J. Biol. Sci. 2013; 9:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith J.C., Hagemann A., Saka Y., Williams P.H. Understanding how morphogens work. Philos. Trans. Roy. Soc. Lond. Ser. B, Biol. Sci. 2008; 363:1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zorn A.M., Wells J.M. Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 2009; 25:221–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schier A.F. Nodal morphogens. Cold Spring Harb. Perspect. Biol. 2009; 1:a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Economou A.D., Hill C.S. Temporal dynamics in the formation and interpretation of nodal and BMP morphogen gradients. Curr. Top. Dev. Biol. 2020; 137:363–389. [DOI] [PubMed] [Google Scholar]
- 55. Thomas T., Voss A.K. The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle. 2007; 6:696–704. [DOI] [PubMed] [Google Scholar]
- 56. Kueh A.J., Dixon M.P., Voss A.K., Thomas T. HBO1 Is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 2011; 31:845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. MacPherson L., Anokye J., Yeung M.M., Lam E.Y.N., Chan Y.C., Weng C.F., Yeh P., Knezevic K., Butler M.S., Hoegl A. et al. HBO1 is required for the maintenance of leukaemia stem cells. Nature. 2020; 577:266–270. [DOI] [PubMed] [Google Scholar]
- 58. Wang Y., Chen S.F., Tian W., Zhang Q., Jiang C.Y., Qian L., Liu Y. High-expression HBO1 predicts poor prognosis in gastric cancer. Am. J. Clin. Pathol. 2019; 152:517–526. [DOI] [PubMed] [Google Scholar]
- 59. Zhong W.H., Liu H.P., Deng L., Chen G.H., Liu Y.B. HBO1 overexpression is important for hepatocellular carcinoma cell growth. Cell DeathDis. 2021; 12:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen Z.H., Zhou L.J., Wang L.W., Kazobinka G., Zhang X.P., Han X.M., Li B., Hou T. HBO1 promotes cell proliferation in bladder cancer via activation of wnt/-catenin signaling. Mol. Carcinogen. 2018; 57:12–21. [DOI] [PubMed] [Google Scholar]
- 61. Hu X., Stern H.M., Ge L., O’Brien C., Haydu L., Honchell C.D., Haverty P.M., Peters B.A., Wu T.D., Amler L.C. et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol. Cancer Res. 2009; 7:511–522. [DOI] [PubMed] [Google Scholar]
- 62. Song B., Liu X.S., Rice S.J., Kuang S., Elzey B.D., Konieczny S.F., Ratliff T.L., Hazbun T., Chiorean E.G., Liu X. Plk1 phosphorylation of orc2 and hbo1 contributes to gemcitabine resistance in pancreatic cancer. Mol. Cancer Ther. 2013; 12:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sauer T., Arteaga M.F., Isken F., Rohde C., Hebestreit K., Mikesch J.H., Stelljes M., Cui C., Zhou F., Göllner S. et al. MYST2 acetyltransferase expression and histone H4 lysine acetylation are suppressed in AML. Exp. Hematol. 2015; 43:794–802. [DOI] [PubMed] [Google Scholar]
- 64. Massagué J. TGFbeta in cancer. Cell. 2008; 134:215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moustakas A., Heldin C.H. Mechanisms of tgfβ-induced epithelial-mesenchymal transition. J. Clin. Med. 2016; 5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang J., Antin P., Berx G., Blanpain C., Brabletz T., Bronner M., Campbell K., Cano A., Casanova J., Christofori G. et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020; 21:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Dong L., Zhang Z., Yu C., Sun Y. et al. The genome sequence archive family: toward explosive data growth and diverse data types. GenomicsProteomics Bioinformatics. 2021; 19:578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. CNCB-NGDC Members and Partners Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2023. Nucleic Acids Res. 2023; 51:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq and CUT&Tag-seq generated data have been deposited in the Genome Sequence Archive (67) in National Genomics Data Center (68) for Human database under the accession codes HRA005583 and HRA006159 that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human. All data supporting the findings of this study are available within the article and its supplemental information files or from the corresponding authors upon reasonable request.