Abstract
Viruses are master remodelers of the host cell environment in support of infection and virus production. For example, viruses typically regulate cell gene expression through modulating canonical cell promoter activity. Here, we show that Epstein Barr virus (EBV) replication causes ‘de novo’ transcription initiation at 29674 new transcription start sites throughout the cell genome. De novo transcription initiation is facilitated in part by the unique properties of the viral pre-initiation complex (vPIC) that binds a TATT[T/A]AA, TATA box-like sequence and activates transcription with minimal support by additional transcription factors. Other de novo promoters are driven by the viral transcription factors, Zta and Rta and are influenced by directional proximity to existing canonical cell promoters, a configuration that fosters transcription through existing promoters and transcriptional interference. These studies reveal a new way that viruses interact with the host transcriptome to inhibit host gene expression and they shed light on primal features driving eukaryotic promoter function.
Graphical Abstract
Graphical Abstract.
Introduction
By definition, viruses utilize the molecular resources of a living host to replicate. The use of host resources, however, does not simply entail the passive theft of cell materials and molecular processing machinery. Rather, through millions of years of evolution, viruses have developed the ability to exert impressive alterations in the host cell environment to enhance virus replication and viral fitness. Remodeling of the host cell environment extends not only to the manipulation of host cell regulatory pathways to, for example, subvert innate immune responses or to promote an S-phase environment in support of DNA virus replication, but also extends to remodeling of cellular substructures and overall cell architecture. The Epstein Barr virus (EBV), a human gammaherpesvirus linked to lymphoma, stomach cancer, nasopharyngeal carcinoma and multiple sclerosis (1–3), induces a DNA damage response, triggering entry into a pseudo-S-phase environment that supports viral DNA replication (4–6). Like other herpesviruses, EBV also encodes a nuclease, BGLF5, that causes host shut off through the degradation of host mRNAs that are destined for translation (7). This is thought to be a mechanism for decreasing the burden of cell mRNAs on the cell translation machinery, thereby enhancing dedicated production of viral proteins. Morphologically, EBV replication causes profound changes to the nuclear architecture including the formation of large biomolecular condensates called viral replication compartments that ultimately encompass the majority of the nuclear space (8). In addition, EBV replication causes extensive reorganization of cellular chromatin that facilitates eventual chromosome compaction and repositioning to the nuclear lamina (6,8).
Viruses can alter cell signaling through multiple mechanisms including the manipulation of existing cell transcription factor activity. For example, the small DNA tumor viruses SV40, adenovirus, and the human papilloma virus each encode a viral protein that binds the retinoblastoma protein (RB1) to disrupt its interaction with the cell cycle transcription factor E2F and drive cells into S-phase (9). Classically, the manipulation of cellular transcription factor activity results in the reprogramming of transcriptional networks that alter the expression of existing cell genes. Here, we show that through the action of three viral transcription factors/complexes, EBV replication spawns the formation of tens of thousands of new transcription initiation sites throughout the cell genome. Further, we show that unique chromatin characteristics of active cell promoters facilitates a bias for some classes of de novo transcription to initiate near, but not within, existing active cell promoters. For this group of de novo promoters, directionally biased transcription leads to elongation through canonical promoters and transcriptional interference. This represents a new concept for how a virus can interact with the cell transcriptome and a new way in which a virus can suppress cell gene expression in support of viral protein production. Further, we hypothesize that these de novo promoters are rudimentary and are present throughout the cell genome independent of cell evolutionary pressures that typically drive the design of complex cell promoter sequences/structures. The rudimentary properties of tens of thousands of these promoters may facilitating future investigations into the fundamental nature of chromatin context and other basic features of eukaryotic promoter function.
Materials and methods
Cell culture
EBV+ Burkitt's Lymphoma lines, Mutu I (obtained from the laboratory of Samuel H Speck) and Akata (obtained from Kenzo Takada), and EBV- Burkitt's Lymphoma line, DG75 (ATCC, cat no. CRL-2625) were cultured in RPMI (Fisher Scientific, cat no. SH30027) supplemented with 10% FBS (ThermoFisher Scientific, cat no. 10437) and 1% Pen/Strep (Fisher Scientific, cat no. 15140-122) in an incubator set to 37°C with 5% CO2.
Nucleofection
Three million Mutu or DG75 cells were transfected using an Amaxa Nucleofector II machine (Lonza). CMVp-GFP (0.3 μg) and SV40p-Zta (or SV40p-Cntl) or pLVX-Rta (or pLVX-Cntl) (2.7 μg) were added to each transfection reaction, along with 100 μl of Amaxa Cell Line Nucleofector Kit R (Lonza, cat no. VCA-1001). Samples were loaded into a cuvette and electroporated using the setting, ‘G016’, on the device. Cells were then immediately transferred to plates with growth media and placed in the incubator.
BCR crosslinking
Goat α-Human IgM (Sigma-Aldrich, cat no. 10759) or Affinipure goat α-Human IgG (Jackson ImmunoResearch, cat no. 109-005-003) were added to cell culture media of Mutu and Akata cells, respectively, to make a final concentration of 10 μg/ml. Cells were incubated for 24 h prior to analysis.
Fluorescence activated cell sorting
Mutu, Akata, or DG75 cells were collected and centrifuged at 1000g for 5 min. Cells were resuspended in 500 μl of PBS with 1 mM EDTA and passed through a 35 μM mesh filter (Genesee Scientific, cat no. 28-154). Approximately 500 000 GFP+ cells were isolated from each sample using a BD FACSAria III (BDBioSciences) machine. Cells were pelleted via centrifugation at 1000g for 5 min, then flash frozen.
α-gp350 pulldown of reactivating cells
24 h after either BCR-crosslinking or Zta nucleofection, cells in culture media were washed in PBS, then resuspended in 1 ml solution of PBS with 1% BSA (Cell Signaling Technologies, cat no. 9998) and a 1:100 dilution of α-gp350 antibody (ThermoFisher, cat no. MAB10219). Samples were rotated at 4C for 2 h, then washed twice with PBS. To prepare goat α-mouse Dynabeads (Invitrogen, cat no. 11-033), 50 μl per sample of Dynabead slurry was transferred to a microfuge tube on a magnetic rack. After 2 min, the supernatant was removed and beads were washed with PBS, and resuspended in PBS with 1% BSA. The Dynabead solution was added to the cells and samples were placed on a rocker for 1 h at 25°C. To purify reactivating cells from induced populations, the samples were placed on a magnetic rack for 2 min. The supernatant was discarded, and cell-Dynabead complexes were washed twice with PBS. After the final wash, the gp350+ cells were pelleted via centrifugation at 1000g. Latent cells from uninduced populations were subjected to a negative gp350 selection to remove spontaneously reactivating cells. Latent samples were placed on a magnetic rack for 2 min then the supernatant was transferred to another tube and beads were discarded. This step was repeated to remove all traces of the magnetic beads. Next, the cells were centrifuged at 1000g for 5 min and resuspended in PBS. Finally, gp350− cells were pelleted by centrifugation for 5 min at 1000g.
SDS-PAGE
Cells were lysed in RIPA buffer supplemented with cOmplete Protease Inhibitor Cocktail (Roche, cat no. 11836170001). Briefly, cell pellets were resuspended in in lysis buffer and left on ice for 10 min. Lysates were then spun at 15 000g in 4C for 10 min, then the soluble fraction was transferred to another tube. Protein concentrations were measured using the Rapid Gold BCA Protein Assay kit (Pierce, cat no. A53225). Equal amounts of protein were loaded on 4–20% Mini-PROTEAN polyacrylamide gels (BioRad, cat no. 4568093) and electrophoresed at 150 V for 45 min. Protein was then transferred to nitrocellulose membranes (BioRad, cat no. 1704159) using a Trans-Blot Turbo Transfer System (BioRad, cat no. 1704150). Membranes were blocked in TBS with 5% BSA and 0.1% Tween (BioRad, cat no. 1706531). α-GAPDH (Santa Cruz Biotechnology, cat no. sc-47724, 1:1000), α-Zta (Santa Cruz Biotechnology, cat no. sc-53904, 1:500), α-IRF4 (Cell Signaling Technologies, cat no. 62834, 1:1000), and α-VCA (ThermoFisher Scientific, cat no. PA1-73003, 1:1000) antibodies were diluted in TBST with 5% BSA. Membranes were incubated with the antibody solution on a rocker at 4°C overnight. The next day, membranes were washed 3 times in TBST, then incubated with a 1:10 000 dilution of either Donkey Anti-Rabbit 800CW (Licor, cat no. 926-68070), Donkey Anti-Goat 800CW (Licor, cat no. 926-32214), Goat Anti-Mouse 680RD (Licor, cat no. 926-32213), or Goat Anti-Mouse 800CW (Licor, cat no. 926-32210) at room temperature for 1 hour. Membranes were washed 3 times for 10 min each then exposed on an Odyssey CLx Imager (Licor).
RNA extraction and cDNA synthesis
Cells were resuspended in 1ml of TRIzol Reagent (Invitrogen, cat no. 15596026) and processed according to the manufacturer's protocol. RNA pellets were reconstituted in 42 μl of ddH2O. 5 μl of 10× DNase buffer and 3 μl of recombinant DNase I (New England Biotechnology, cat no. M0303L) were added and tubes were incubated at 37°C for 15 min. DNase I was then removed from samples using the Monarch RNA Cleanup Kit (New England Biotechnology, cat no. T2030L). 1μg of RNA was reverse transcribed using LunaScript (New England Biotechnology, cat no. M3010), according to the manufacturer's protocol.
PCR
cDNA was diluted 1:10 in ddH2O. 2 μl of diluted cDNA was added to each reaction. PCR was performed using GoTaq Green Mastermix (Promega, cat no. M7122) in 20 μl reaction volumes (10 μl GoTaq Green, 2 μl diluted cDNA, 0.5 μl of 10 μM primer combo, and 7.5 μl ddH2O). Primer sequences are listed in Supplemental Table S1. An initial 95°C melting step for 2 min was followed by 25–40 cycles of PCR, each consisting of the following steps: 95°C for 30 s, 60°C for 30 s, then 72°C for 60 s, with a final extension at 72°C for 5 min at the end. PCR products were then run on a 1% agarose gel with ethidium bromide and imaged using a gel documentation system.
Chromatin immunoprecipitation
To obtain 100 million reactivated cells, 500 million Akata cells were reactivated via BCR crosslinking, then reactivated cells were purified after 24 h using anti-gp350 immunoprecipitation. Both 100 million reactivated cells and 100 million latent cells were crosslinked using a truChIP Chromatin Shearing Kit (Covaris, cat no. 520154) following the manufacturer's protocol, with formaldehyde crosslinking for 10 min. Chromatin was sheared to approximately 300bp fragments using an M220 Focused-ultrasonicator (Covaris, cat no. 500295) with the following settings: 10% duty factor, 75 W PIP, 200 cycles per burst, 7°C setpoint temperature, with a treatment time of 14 min. After shearing, 1 ml chromatin was diluted 1:1 in 2× Covaris Dilution Buffer, then divided into separate tubes for IP with H3K4Me3 antibody (1:50; Cell Signaling Technologies, cat no. 9751S). Chromatin was incubated with the antibody at 4°C on a rotator overnight. 25 μl each of Protein A (ThermoFisher Scientific, cat no. 10002D) and Protein G Dynabeads (ThermoFisher Scientific, cat no. 10004D) were combined, washed, resuspended in 25 μl of PBS per reaction, then added to each chromatin sample. Samples were incubated at room temperature for 1 h on a rocker. To isolate Protein A/G-antibody-chromatin complexes, tubes were then placed on a magnetic rack for two min. Supernatant was discarded and beads were washed in Mixed Micelle buffer (150 mM NaCl, 20 mM Tris–Cl, 5 mM EDTA, 5% sucrose, 1% Triton X-100, 0.2% SDS) twice, then buffer 500 (0.1% deoxycholic acid, 1 mM EDTA, 50 mM HEPES, 500 mM NaCl, 1% Triton X-100) twice, then LiCl detergent solution (0.5% deoxycholic acid, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 10 mM Tris–Cl) once, TE (10 mM Tris, 1 mM EDTA) once, then resuspended in reverse crosslinking buffer (1% SDS, 0.1 M NaHCO3) and incubated at 65°C overnight. The next day, 5 μl of proteinase K (ThermoFisher Scientific, cat no. 25530049) was added to each sample and samples were incubated at 37C for 2 h. Then, 40 μl of 5 M LiCl and 1.5 μl GlycoBlue (ThermoFisher Scientific, cat no. AM9515) was added. DNA was then extracted by adding 300 μl of 25:24:1 phenol–chloroform–isoamyl alcohol (ThermoFisher Scientific, cat no. AC327111000), vortexed, and centrifuged for 5 min at 15 000g. The aqueous phase was transferred to a separate tube and 1 ml of 100% ethanol was added. Samples were incubated at −20°C overnight before DNA was precipitated via centrifugation at 15 000g for 30 min. DNA pellets were washed in 1 ml of 70% ethanol then pellets were air dried and resuspended in 300 μl ddH2O. DNA samples were sent to the Beijing Genomics Institute (BGI) for sequencing.
RNA sequencing
RNA integrity was confirmed using a bioanalyzer with an RNA integrity number (RIN) of >8 required for sequencing.
Mutu and Akata BCR-induction: Cells were nucleofected with a BMRF1-driven GFP reporter (pCEP4-BMRF1p-GFP) containing the hygromycin resistance gene. Stable cells were obtained after growing cells in 250 μg/ml hygromycin for 7 days. Cells were induced with 10 μg/ml of anti-IgM (Mutu) or anti-IgG (Akata) for 24 h, followed by FACS isolation of GFP+ cells. Uninduced GFP– control cells from untreated samples were isolated in parallel (N = 3 for each group). Stranded, paired end, polyA-selected libraries were constructed and sequenced by Novogene (Mutu) or BGI (Akata).
Mutu Zta induction: Mutu cells were co-transfected with CMVp-GFP (0.3μg) and either SV40p-Cntl or SV40p-Zta vectors (2.7 μg) using a Nucleofecter device. After 24 h, GFP+ cells were isolated from each sample via FACS (N = 3 for each group). Stranded, paired end, polyA-selected libraries were constructed and sequenced by Novogene.
Mutu Zta induction timecourse: Mutu cells were co-transfected with CMVp-GFP (0.3 μg) and either SV40p-Cntl or SV40p-Zta vectors (2.7 μg). GFP + cells were isolated via FACS at 6, 12 and 24-h timepoints (N = 4 for each group at each timepoint). Unstranded, polyA-selected libraries, and sequencing was performed by BGI.
DG75 ectopic Zta or Rta expression: For Zta, DG75 cells were co-transfected with CMVp-GFP (0.3μg) and either SV40p-Ctl or SV40p-Zta vectors (2.7 ug). For Rta, DG75 cells were co-transfected with CMVp-GFP (0.3 μg) and either pLVX-Cntl or pLVX-Rta vectors (2.7 μg). After 24 h, GFP + cells were isolated via FACS. Stranded, ribodepleted libraries and sequencing was performed by BGI. De novo transcript expression was determined by measuring the coverage at each de novo TSS, from positions +1 to +300 (or +1 to first base of an annotated TSS, if it was closer than 300 bases from the de novo TSS). Expression values were compared between Zta and Cntl or Rta and Cntl using DESeq2 (10). De novo transcripts that were enriched in Zta or Rta- transfected DG75 cells were considered statistically significant if they had a P-adjusted value <0.05.
Sequencing reads were aligned to the combined human (GRCh38) and Akata EBV genomes using STAR (11) v2.6.1 to capture positional coverage information. BAM alignment files were converted to bigWig format using the bamCoverage function of deepTools (12). Transcript quantification was performed by pseudo-aligning sequencing reads to the combined human (Ensembl v.90) and EBV transcriptomes using kallisto (13) (v0.46.0). Sleuth (14) (v0.30.0) was used to calculate differential expression.
CAGE sequencing
RNA from triplicate samples of Akata (Cntl and BCR) and Mutu (Cntl and Zta) cells were sent to KKDNAFORM, where CAGE libraries were prepared and sequenced. Reads were aligned to the combined human and Akata EBV genomes using STAR (v2.6.1), with positional coverage of only the first base of each read output in bigWig format.
Identification of de novo transcripts
The first base of CAGE-Sequencing reads was used to mark TSSs for all samples. A TSS was considered ‘de novo’ if it was (i) supported by at least one CAGE-Sequencing read in all three replicates of reactivated Mutu or Akata cells, (ii) had a sum of at least seven total CAGE-Sequencing reads across replicates, (iii) was undetected in any of the three replicates of latent Mutu or Akata cells and (iv) was at a position at least 25 bp away from the nearest annotated TSS.
ATAC sequencing
24 h after induction, Akata (Cntl and BCR) and Mutu (Cntl and Zta) cells were processed as described in Buenrostro et al. (15) by the Tulane ATAC sequencing core. Seventy-five base paired end sequencing was performed on an Illumina NextSeq 2000 sequencing machine.
Sequencing reads were aligned to the combined Human and EBV genomes using bowtie2 (16) (v2.5.1) using the parameter ‘-X 2000’. The ‘callpeak’ command of MACS2 (17) (v2.2.7.1) was used to call peaks from the resulting coverage files, using the following parameters: ‘–broad, -g hs, –call-summits, -t sample.bam’. Peaks from uninduced and induced conditions were merged using the bedtools (18) (v2.29.2) ‘merge’ command. The resulting BAM alignment files and peaks were then processed using TOBIAS bindetect (19) (v0.13.2) to account for ATAC insertion bias and to normalize coverage. For nucleosomal positioning scores and binding footprints, aligned reads were processed via Nucleoatac (20) (v0.3.4), using the broad peaks output from MACS2 and the bam files output from bowtie2. Transcription factor binding scores were obtained by running the ‘process atac’ command of DAStk, using the broad peaks output from MACS2 and the HOCOMOCO transcription factor binding model (v11) (21). Differential MD-scores were calculated using the ‘differential_md_score’ command, with an accepted P-value of 0.00001.
Whole genome bisulfite sequencing
Quadruplicate Akata cntl and BCR samples were purified using either positive (BCR) or negative (cntl) anti-gp350 selection. DNA was isolated using a Monarch Genomic DNA Purification Kit (New England Biolabs, cat no. T3010L). Bisulfite treatment and library preparation was done by BGI. One hundred base paired-end sequencing was performed on a DNBSeq machine. Sequencing reads were aligned to the combined GRCh38 and Akata genomes using bismark (22) (v0.24.0) with bowtie2.
ChIP sequencing
Immunoprecipitated DNA fragments were sent to BGI for library preparation and sequencing. One hundred base paired end sequencing was done on a DNBSeq machine. Sequencing reads were aligned to the combined GRCh38 and Akata genomes using bowtie2 (v2.5.1). Duplicates were marked and removed using the MarkDuplicates command of Picard (https://broadinstitute.github.io/picard/; v2.20.3), with the parameter, ‘REMOVE_DUPLICATES = true’. Alignment coverage bigWig files were generated using the bamCoverage command of deepTools (23) (v3.3.2), with the following parameters: ‘–binSize 1, –blackListFileName hg38-blacklist.v2.bed’. The hg38 blacklist regions file was obtained from Amemiya et al. (24).
CRISPR/Cas9 de novo promoter deletions
Recombinant Cas9 and modified sgRNAs targeting either side of the denovo promoter were synthesized by Synthego (sequences in Supplemental Table S1). 2 μl of each sgRNA was combined with 1 μl of Cas9 and 100 μl of Nucleofector Solution R, and incubated at room temperature for 10 min prior to transfection. Mutu cells were then electroporated using protocol ‘G016’. Cells were then allowed to recover for 72 h. Clones of transfected cells were obtained via limiting dilution plating in 96-well plates.
Results
Cell transcriptome changes during EBV reactivation
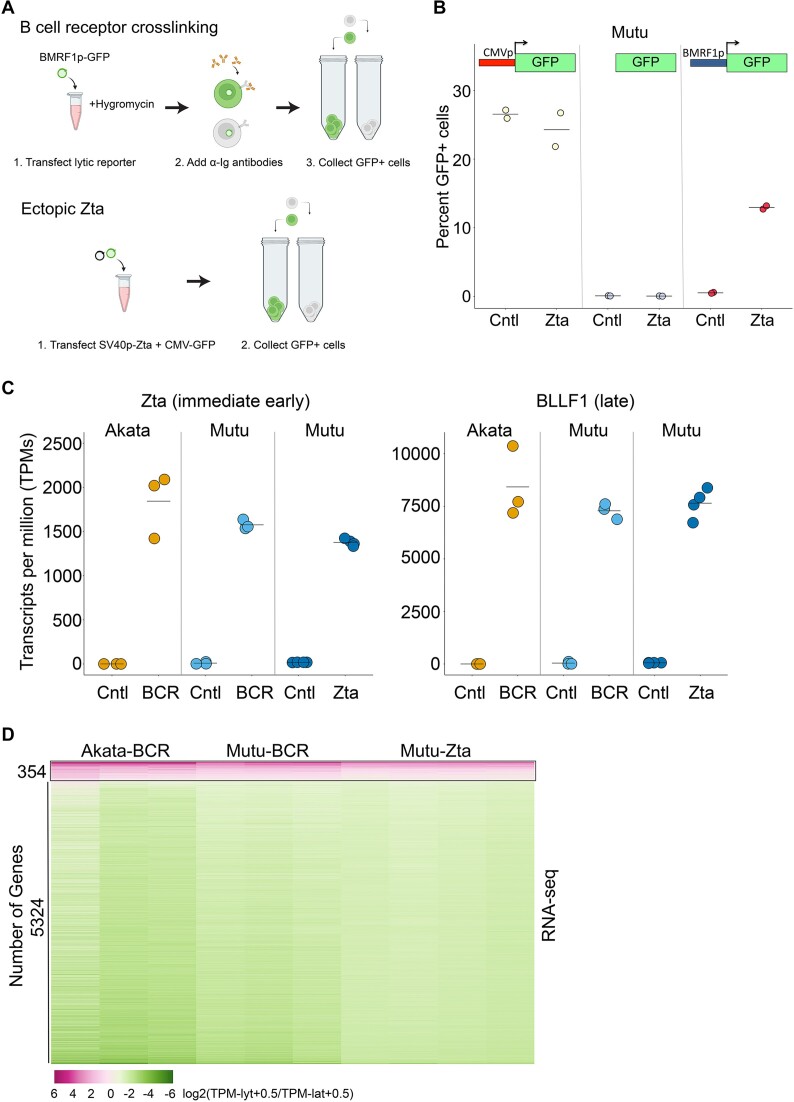
The objective of this study was to carry out a detailed analysis of host cell transcriptome remodeling by EBV lytic replication. In most EBV reactivation models, however, reactivation occurs in only a subpopulation of cells. To minimize obscuring of virus induced cell gene expression changes by background non-reactivating cells, we purified lytic cells using fluorescence activated cell sorting (FACS) (Figure 1A). Reactivation was analyzed in two EBV positive Burkitt's lymphoma cell lines, Akata and Mutu, treated with anti-immunoglobulin (anti-Ig) antibodies to induce B-cell receptor (BCR) crosslinking and viral lytic replication. To facilitate isolation of reactivating cell subpopulations, cells were first stably transfected with an autonomously replicating reporter plasmid with a green fluorescent protein (GFP) marker controlled by the EBV early BMRF1 promoter (Figure 1A). This reporter was found to be robust and specific when tested through transient co-transfection with either a control or an expression vector containing the EBV immediate early transcription factor, Zta, whose ectopic expression can trigger the full EBV lytic cascade (Figure 1B). Because BCR signaling can regulate gene expression independently of changes caused by EBV lytic replication, we also analyzed changes in cell gene expression following ectopic expression of Zta (co-transfected with a CMV-GFP reporter) (Figure 1A).
Figure 1.
Regulation of the cell transcriptome by EBV lytic replication. (A) The EBV positive Burkitt's lymphoma cell lines, Akata and Mutu, were transfected with the autonomously replicating pCEP4 plasmid containing the EBV early BMRF1 promoter upstream from a green fluorescent protein (GFP) reporter and stable transfectants were selected using hygromycin (upper panel). Stably transfected BMRF1p-GFP Akata or Mutu cells were left untreated or treated with anti-IgG or anti-IgM for 24 h. GFP+ cells were collected for the treated cells and GFP- cells were collected for the untreated cells. To induce reactivation directly through ectopic expression of the EBV transactivator, Zta, Mutu cells were co-transfected with a CMV-GFP reporter plasmid plus a control or an SV40p-Zta expression vector (lower panel) and GFP+ cells were collected for analyses. This panel was created with BioRender.com. (B) Mutu cells were co-transfected with a control or an SV40p-Zta expression vector plus either a CMV-GFP (left panel), a promoter-less GFP (middle panel), or a BMRF1p-GFP reporter plasmid (right panel) for 24 h and the percent GFP cells were counted. (C) Transcripts per million (TPM) values for Zta or the late gene, BLLF1, from RNA-seq analysis of 24 h Akata-BCR, Mutu-BCR or Mutu-Zta models. (D) Heatmap of log2(TPMlyt + 0.5/TPMlat + 0.5) values for common statistically significant changes across all three induction models.
Altogether, we induced reactivation in Akata and Mutu cells through BCR activation and we induced reactivation in Mutu cells through ectopic expression of Zta. Lytic reactivating cells were isolated by FACS 24 h after induction and RNA preparations were subjected to RNA-sequencing of polyadenylated RNAs. In all three cases, high levels of viral gene expression were observed in reactivating cell populations (for example, see results for EBV encoded Zta and the late BLLF1 transcripts (Figure 1C)). It is noteworthy, however, that expression of Zta in ectopically transfected Mutu cells was slightly lower than that detected in BCR activated Akata and Mutu cells indicating that transfection with the Zta expression vector did not result in super-physiological Zta RNA levels (Figure 1C).
Assessing common cell gene expression changes in all three models, we identified 5324 genes that were downregulated and 354 genes that were upregulated in cells undergoing lytic replication (adj P-val < 0.05) (Figure 1D). The finding that the majority of genes display reduced expression is consistent with the known role of host shut off activity that broadly degrades cell mRNAs (7,25,26). The finding that some genes are induced (Figure 1D) despite the backdrop of host shut off is in-line with previous studies (27) and suggests that some transcripts can either overcome or avoid host shut off. The detection of genes with induced expression is not purely the results of sequencing partial transcript fragments undergoing host shut off since we detected increased levels of full-length transcripts using Oxford Nanopore sequencing (unpublished). Further, Western blot analysis of the induced gene, interferon response factor 4 (IRF4) showed induction at the protein level (Supplemental Figure S1). Together, this data indicates that for genes showing statistically significant changes across all three reactivation models, most are likely downregulated through host shut off while some genes are induced during reactivation and may play some role in viral replication.
De novo transcription initiation at cellular genes during EBV reactivation
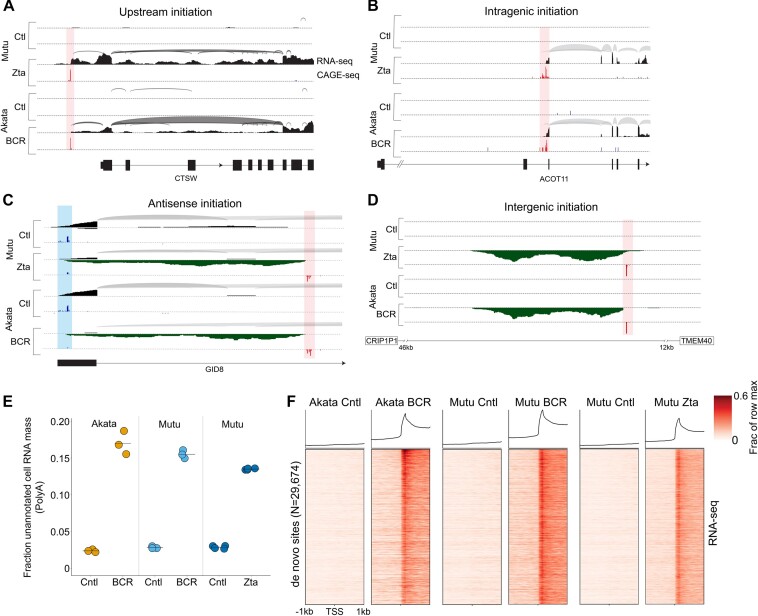
Despite the finding that some genes are induced during reactivation to produce full length proteins, a survey of induced gene coverage revealed striking anomalies in the majority of cases. Specifically, visualization of genomic coverage plots revealed apparent transcription initiation occurring outside of known transcription start sites (TSSs), either upstream of annotated promoters (Figure 2A) or within gene bodies (Figure 2B). To confirm these findings, we performed Cap Analysis of Gene Expression-seq (CAGE-seq) (28) to precisely determine transcription start sites in FACS separated Akata-BCR and Mutu-Zta models. This analysis confirmed that approximately 63% of induced genes have noncanonical transcription initiation that contributes to expression during reactivation (e.g. Figure 2A and B). Novel transcription initiation was not limited to induced genes but also extended to transcripts that showed either little change or statistically significant decreases in transcription (e.g. see Supplemental Figure S2A and B).
Figure 2.
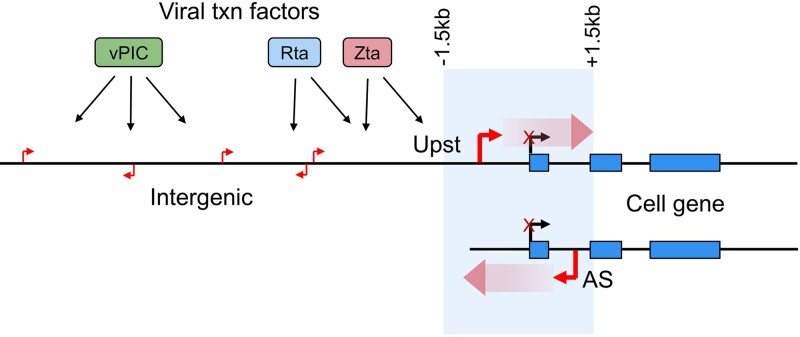
EBV reactivation causes transcription initiation at novel, previously unannotated transcriptional start sites throughout the cell genome. (A–D) Plotting of splice junction reads (arches – thicker arches represent higher number of splice junction reads) and single base level coverage (height represents number of reads identified at each position) from RNA-seq data and CAGE-seq coverage (height represents number of reads starting at each position) in the Mutu-Zta and Akata-BCR models. Blue coverage/shading represents canonical transcription initiation and red coverage/shading represents ‘de novo’ transcription initiation. Exon structures of associated genes are indicated at the bottom of each plot. (E) Relative levels of RNA-seq read coverage mapping outside of annotated cell genes (i.e. de novo, dark matter transcription) in latency and reactivation across all three induction models. Y-values represent fraction of RNA-seq reads that map outside of or antisense to the start and end positions of annotated cell gene units relative to all reads mapping to the cell genome. (F) De novo start sites identified in either Mutu-Zta or Akata-BCR models were collated and used to plot heatmaps of RNA-seq read coverage spanning -1kb to plus 1kb from each de novo promoter start site (vertical axis). Coverages were normalized by row across all 6 sample groups.
Widespread de novo transcription initiation across the host cell genome
Upon further inspection of RNA-seq and CAGE-seq coverage across the cell genome, we found that de novo transcription initiation was not limited to regions upstream or within gene bodies but extended to antisense orientations and intergenic regions (e.g. Figure 2C, D, Supplemental Figure S3) and occurred broadly throughout all cell chromosomes. To gauge the overall extent of de novo cell transcription, we calculated the total read coverage mapping outside of the start and end coordinates of all annotated cell gene bodies relative to the total number of reads mapping to the cell genome. This analysis showed that up to approximately 17% of all polyadenylated cell transcripts were derived from outside of annotated transcript boundaries during reactivation (Figure 2E) illustrating the substantial contribution that de novo transcription makes to the cell transcriptome.
Conservation of de novo transcription across cell models
To facilitate further investigation into de novo transcription, we sought to globally identify de novo transcription start sites. Using CAGE-seq data from the Akata-BCR and Mutu-Zta reactivation models, we defined de novo promoters as genomic positions where CAGE reads were detected in all three replicates from reactivating cells and no reads were detected in any replicate from latent cells. Further, we required that genomic positions satisfying these criteria be at least 25bp from an annotated/known start site (using the comprehensive Ensembl v.90 transcript annotation file (29)). Combining de novo promoters identified in both systems, we found a total of 29 674 de novo start sites.
Using the coordinates of newly defined de novo TSSs, we plotted RNA-seq coverage centered around the start sites using RNA-seq data from each reactivation model (Figure 2F). Although there are differences across models, there was an overall high degree of conservation (Figure 2F). Together, this data indicates that EBV reactivation causes widespread novel transcription initiation throughout the cell genome and that induction of de novo transcription is a programmed feature of EBV reactivation.
De novo transcription initiation is driven by viral transcription factors
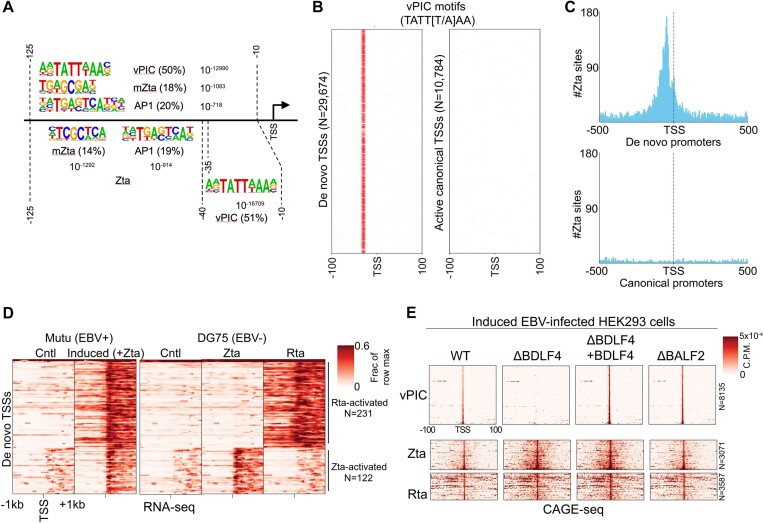
To investigate the mechanisms underlying de novo transcription, we sought to identify distinguishing sequence elements upstream from de novo promoter TSSs. Using the motif analysis software, HOMER (Hypergeometric Optimization of Motif EnRichment) (30) with default genomic DNA (hg38) background selection and GC content compensation, we first performed motif enrichment analysis of sequences between −125 and −10 bp relative to de novo promoter TSSs. The most significantly enriched motif identified by this analysis was the sequence, TATT[T/A]AA (Figure 3A). This is striking because this matches the distinctive ‘TATT’ binding motif through which gammaherpesvirus TATA-box-like binding complexes, referred to as vPIC (viral PreInitiation Complex), bind to activate viral late gene expression (31–34). Among the 6 viral factors that make up the EBV vPIC, the core BcRF1 factor interacts with the TATT motif to direct downstream transcription initiation (31–34). Consistent with its role in the positioning of transcription initiation (35), enrichment of the TATT[T/A]AA motif was even more significant when sequences spanning −40 and −10 bp from the TSSs were analyzed (Figure 3A). Further, plotting the distribution of TATT[T/A]AA motifs around de novo TSSs showed a constricted positioning around 34–30 bp upstream from the start sites, consistent with vPIC playing a determinative role in dictating the site of transcription initiation (Figure 3B). In contrast to findings at de novo promoters, there is an almost complete absence of the vPIC motifs at canonical/annotated cell promoters (Figure 3B) indicating that these motifs are likely uninvolved in canonical promoter function.
Figure 3.
De novo transcription is driven by viral transcription factors, vPIC (BcRF1), Zta, and Rta. (A) Motif analyses of regions upstream from de novo promoter start sites. The top three enriched motifs are shown for assessment of sequences between −125 and −10, the top two motifs are shown for assessment of sequences between −125 and −35 and the top motif is shown for sequences between −10 and −40. (B) Plotting of TATT[T/A]AA motifs across −100 to +100 bp from the de novo promoter start sites (left panel) and canonical/annotated start sites (right panel). Different start sites are positioned along the vertical axis. Positions with the TATT[T/A]AA motif are marked by a red line. (C) Plotting number of Zta binding sites from ChIP-seq (39) data with respect to de novo promoter start sites (left panel) and canonical (annotated) start sites (right panel). Counts are for number of Zta binding sites, using 5 bp bins. (D) Coverage heatmaps spanning −1 kb to +1 kb from start sites of de novo promoters found to be activated in DG75 cells transfected with either Zta or Rta (RNA-seq experiments). Corresponding heatmaps for respective de novo promoters in Mutu-Zta reactivation model shown for reference (left panel). The order of de novo regions on the y-axis is identical for each heatmap. Heatmap color intensity represents the mean signal of all replicates in each group. Positional coverage was normalized by row across both Mutu groups, and separately, across all DG75 cell sample groups. (E) CAGE-Seq plots of de novo TSSs from reactivated 293 cells infected with ΔBDLF4(vPIC factor), ΔBALF2 (DNA replication factor), or wildtype EBV (33).
In addition to the vPIC motif, the second and third most enriched motifs between −125 and −10 bp of de novo promoter TSSs were TCGCTCA and TGAGTCA (Figure 3A). The TCGCTCA motif is interesting because it is a high affinity Zta binding site and critically, Zta binding is dependent on cytosine methylation at the embedded CpG dinucleotide (36). The binary nature of Zta binding to the methylated vs unmethylated form of this motif plays a central role in regulating EBV reactivation (36). The TGAGTCA motif is an AP1 site to which Zta has also been shown to bind with high affinity (37). In keeping with the typical upstream localization of transcription factor binding at promoters, slightly higher statistically significant enrichment is observed for these Zta binding sequences when analyzing the region between positions −125 and −35 bp (Figure 3A). Consistent with Zta binding to cell DNA during reactivation, DAStK (38) analysis of differential chromatin accessibility at transcription factor motifs throughout the genome using Assay for Transposase-Accessible Chromatin-sequencing-seq (ATAC-seq) (15) data in the Mutu-Zta reactivation model showed enhanced occupancy at both the TCGCTCA and TGAGTCA motifs under reactivation conditions (Supplemental Figure S4). Lastly, using Zta ChIP-seq data from the Hammerschmidt lab (39), we found enriched Zta binding between −125 and −35 bp from de novo promoter TSSs (Figure 3C), providing further support of a role for Zta in activating a subset of de novo promoters.
To further address the role of Zta in activating de novo promoters and to begin to address a possible role for the other immediate early viral transcription factor, Rta, we transfected the EBV negative B-cell line, DG75 with either a Zta or an Rta expression vector and performed RNA-seq analysis of polyadenylated RNAs. We then identified de novo promoters that showed increased downstream coverage in response to either Zta or Rta expression in DG75 cells and plotted coverage centered around the respective TSSs. As shown in Figure 3D, a subset of de novo promoters was found to be responsive to either Zta or Rta in DG75 cells.
Because our inference of a role for vPIC in activating a subset of de novo promoters was based on motif enrichment, we next turned to provide experimental support for this contention. For this analysis, we utilized CAGE-seq data from Eric Johannsen's lab in which 293 cells infected with wild type EBV or an EBV mutant in which the essential vPIC component, BDLF4 was knocked out were induced for EBV reactivation (33). Strikingly, all de novo promoters with TATT motifs between −40 and −25 of the initiation site (excluding those that also have a Zta binding site or an Rta motif) showed a complete loss of activity in the BDLF4 knockout compared to wild type or the knockout with ectopically expressed BDLF4 (Figure 3E). In contrast, de novo promoters with Zta binding sites or Rta motifs showed no impact of BDLF4 knockout. These results show that de novo promoters with TATT motifs are activated by vPIC during reactivation. Notably, the general finding of de novo promoter activation during reactivation in 293 cells also extends our initial finding of de novo promoter activation to an epithelial system.
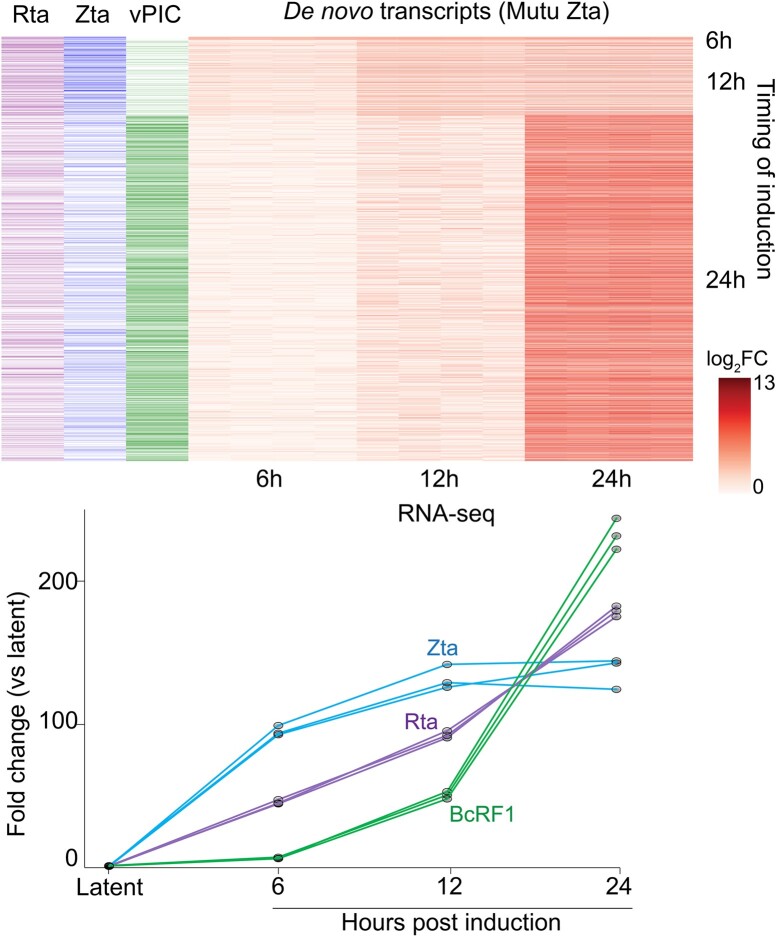
Lastly, we performed a time course experiment in the Mutu-Zta model to assess temporal relationships between de novo promoter expression and expression of Zta, Rta and the core vPIC DNA binding factor, BcRF1, which is expressed with later kinetics. As shown in Figure 4, whereas de novo promoters with delayed induction are enriched for the TATT[T/A]AA between −40 and −25 bp of the TSSs, de novo transcripts with earlier kinetics are enriched for Zta binding sites and slightly enriched for consensus Rta binding sequences (as defined by Heilmann et al. (40)) (between −200 and +1). Together this data supports a role for all three viral transcription factors in driving de novo transcription during reactivation.
Figure 4.
Time course heatmap of fold change in expression of de novo transcripts (quantified based on coverage from +1 to +300 from de novo promoter TSS). De novo transcripts are arranged along the vertical axis and clustered according to timing of first detection (6, 12 and 24 h – right side of heatmap). To left of heatmap are marks indicating the presence of Rta consensus motifs (GNCCN[8–10]GGNG) within −200 to +1 of the start sites, Zta binding sites (ChIP-seq) within −200 and +40 of start sites, and TATT[T/A]AA motifs within −40 to −25 of the start sites. Fold change in the DNA binding factor of vPIC (BcRF1), Zta, and Rta are plotted based on quantification from RNA-seq data.
Chromatin context in de novo promoter activity
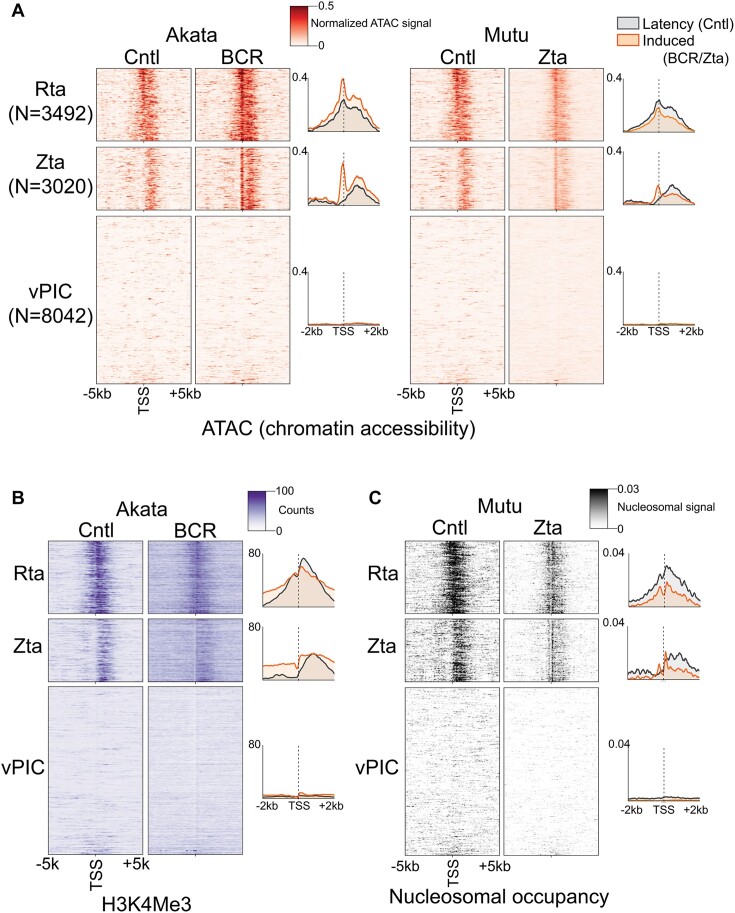
Scanning the human genome for vPIC and Zta binding motifs, we identified a total of 2.5 million TATT[T/A]AA and 0.5 million TGAGTCA or TGAGCGA motifs. The numbers of these motifs vastly outweigh the number of de novo promoters identified (29 674) indicating that de novo promoter formation is likely chromatin context dependent. We therefore sought to determine chromatin features that influence the establishment of de novo promoters. We first assessed surrounding chromatin accessibility during latency and reactivation using our ATAC-seq data from the Akata-BCR and Mutu-Zta models. For these experiments, we separately analyzed de novo promoters with Zta binding sites, Rta motifs, or vPIC motifs (Figure 5A), as well as those with no detectable motifs (referred to as ‘unclassified’, Supplemental Figure S5A). While no discernable pattern of chromatin accessibility was detected near de novo promoters with vPIC sites during latency (Figure 5A), a striking pattern of chromatin accessibility was observed downstream from de novo promoters with Zta binding sites, Rta motifs, and unclassified de novo promoters (Figure 5A and Supplemental Figure S5A). In reactivating cells, there appears to be a repositioning of open chromatin to just upstream from the de novo TSSs, consistent with the typical open chromatin characteristic of active promoters. Together, these analyses suggest that downstream open chromatin features play a role in spawning upstream de novo transcription and that open chromatin is newly positioned at de novo promoters that are created during reactivation. Because open chromatin is a hallmark of transcriptionally active promoters, this raised the possibility that a subset of de novo promoters tend to form upstream from existing expressed promoters.
Figure 5.
Zta and Rta (and ‘unclassified’, see Supplemental Figure S5) subclasses of de novo promoters are positioned upstream from chromatin features characteristic of active promoters. (A) Chromatin accessibility (alignment coverage from ATAC-seq data) is plotted for positions from −5 kb to +5 kb from the respective de novo promoter start sites in both latent (‘Cntl’) and reactivation conditions in the Akata-BCR and Mutu-Zta reactivation models. Heatmap color intensities represent processed coverage values (via TOBIAS ATACorrect (19); positional coverages were corrected for sequence bias and normalized using the ratio of reads in called peaks to total reads). (B) H3K4me3 histone mark coverage (from ChIP-seq data) is plotted for positions from −5 kb to +5 kb from the respective de novo promoter start sites in latent (‘Cntl’) and reactivation conditions in the Akata-BCR reactivation model. (C) Positioned nucleosomes (signal obtained using NucleoATAC analysis of ATAC-seq data) is plotted for positions from −5 kb to +5 kb from the respective de novo promoter start sites in both latent (‘Cntl’) and reactivation conditions in the Mutu-Zta reactivation model. The y-axis for each panel is sorted in ascending order according to the sum of all de novo CAGE-Seq counts across both Mutu and Akata cells. The order is identical in panels A to C.
We next performed a ChIP-seq experiment for the active H3K4me3 histone mark in the Akata-BCR model during latency and reactivation. To scale up the number of cells required for ChIP-seq experiments, we used bead-based selection of reactivating cells expressing the viral gp350 protein on the cell surface (41). Similar to the accessible chromatin findings, a striking positioning of H3K4me3 signal is observed downstream from de novo promoters with Zta binding sites, Rta motifs and those that are unclassified (Figure 5B and Supplemental Figure S5B), further supporting the propensity for some classes of de novo promoters to form upstream from active promoters.
To further assess proximity to active TSSs, we determined the relationship between de novo promoter TSSs and positioned nucleosomes since positioned nucleosomes are typically present within the first 150 bp downstream from TSSs (42). Information on positioned nucleosomes across a genome can be derived from ATAC-seq data based on nucleosome length fragment size enrichment. Using ATAC-seq data from the Mutu-Zta model, we found enrichment of positioned nucleosomes within 2 kb downstream from Zta, Rta and unclassified de novo promoter TSSs (Figure 5C and Supplemental Figure S5C). Under reactivating conditions, there is apparent redistribution of positioned nucleosomes to within 150bp downstream of Zta, Rta and unclassified de novo TSSs, consistent with repositioning of transcription initiation (Figure 5C and Supplemental Figure S5C). There is also apparent de novo nucleosome phasing upstream from de novo TSSs under reactivation conditions, consistent with bidirectional transcription at de novo promoters. Together, these data suggest that Zta, Rta and unclassified de novo promoters tend to form upstream and proximal to active cell promoters.
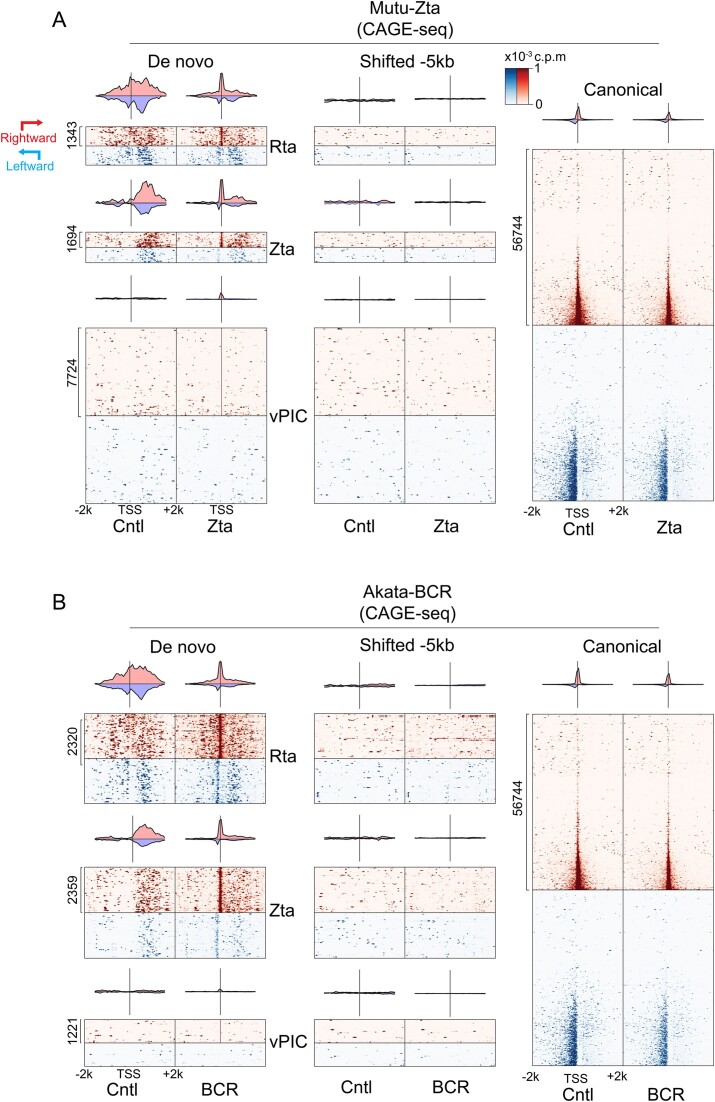
To directly assess the bias for upstream proximity to existing active promoters for Zta, Rta and unclassified de novo promoter formation, we plotted CAGE-seq signals surrounding de novo promoter TSSs in latency and reactivation conditions in the Akata-BCR and Mutu-Zta models. Because not all de novo promoters are active in both the Akata-BCR and Mutu-Zta models, we limited each analysis to de novo promoters with activity in the respective model. As a control, we plotted CAGE-seq signals at annotated (canonical) promoters which showed the expected signals in the forward as well as upstream anti-sense direction, consistent with the general bidirectional nature of transcription (Figure 6A, B, right panels). De novo promoters with Zta binding sites, Rta motifs (Figure 6A, B) and those that are unclassified (Supplemental Figure S6) showed enrichment for active transcription initiation downstream from the de novo promoter start sites under latency conditions, with apparent repositioning of the start site to the de novo TSS under reactivation conditions. Strikingly, the enrichment for active downstream transcription initiation was not limited to transcription in the same orientation as de novo transcription (Figure 6). Instead, enriched downstream transcription occurred in both the sense and antisense directions (Figure 6A, B, ‘Cntl’ panels). As a control, we also plotted CAGE-seq data centered at positions 5 kb upstream from each de novo promoter which showed no discernable features, indicating that the pattern found at de novo promoters is a specific and unique characteristic (Figure 6A, B, middle panels).
Figure 6.
Zta and Rta (and ‘unclassified’, see Supplemental Figure S6) subclasses of de novo promoters are positioned upstream from active promoters. Transcription start site coverage (from CAGE-seq data) is plotted for positions from −2 kb to +2 kb from the respective de novo promoter start sites in both latent (‘Cntl’) and reactivation conditions in the Mutu-Zta (A) and Akata-BCR (B) reactivation models. Start site coverage in the same direction as each de novo promoter is plotted in red while start site coverage on the opposite strand is plotted on blue. As a control, start site coverage with respect to positions −5 kb from the de novo promoter start sites are shown in middle panels. In addition, start site coverage with respect to annotated canonical promoters is shown as a reference (right panels). Heatmaps only include de novo start sites detected in the cell line plotted (for example, only de novo TSSs detected in reactivated Mutu cells were plotted in Figure 6A, B). For each cell line, the y-axis is arranged in ascending order by the sum of all CAGE-Seq reads at the de novo TSS. The order is the same in Cntl and reactivation conditions for panels A and B.
In contrast to the findings with Zta, Rta or unclassified de novo promoters, those with vPIC motifs showed no bias for positioning near constitutively active promoters. Overall, these data suggest that while vPIC is capable of initiating transcription on its own and without the context of proximal active transcriptional hubs, de novo transcription initiated through Zta, Rta or undetermined factors have a reliance on existing transcriptional machinery and/or chromatin characteristics of active promoters. Further, de novo promoters with Zta, Rta or undetermined factors cause apparent proximal repositioning of transcription start sites, oriented with transcription proceeding in the direction of constitutively active promoters.
Distribution of de novo TSSs and Zta binding at expressed canonical promoters
The bias for Zta, Rta, and unclassified de novo promoters to form near active cell promoters may partly result from support provided by accessibility to phase separation environments that are enriched for RNA polymerase and other transcriptional resources. In addition, however, the striking directionality of promoter proximal de novo TSS positioning that orients de novo transcription to proceed through, rather than away from active promoters is also notable. While the basis for directionality is not clear, it may be related, in part, to the open chromatin environment of active promoters that favors Pol II processivity.
Based on these findings, we hypothesized that EBV exploits fundamental characteristics of active promoters to enhance the formation of nearby de novo transcription that proceeds through these promoters to cause transcriptional interference and inhibition of cell gene expression. Notably, the principle of transcriptional interference has been known for years with one of the first examples being the silencing of downstream retroviral LTRs by upstream LTR initiated transcription proceeding through the downstream LTR (43). Since these early studies, transcriptional interference was found to play an important role in turning off downstream promoters during promoter switching at cell genes (44–48). Although multiple mechanisms of transcriptional interference have been described, the most well accepted mechanism is promoter clearing caused by transcription through the respective downstream promoter (46–49).
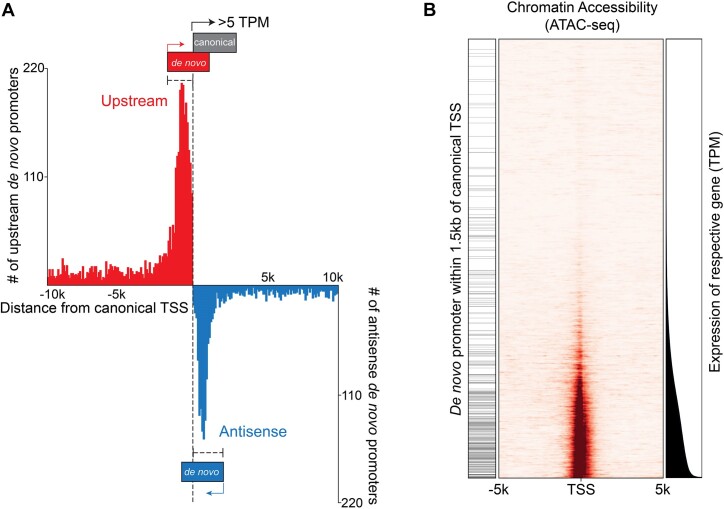
Plotting the positional distribution of upstream and antisense de novo promoters with respect to previously annotated start sites, we see that both upstream and antisense de novo promoters show enriched distribution within 1.5 kb of canonical promoter TSSs (Figure 7A). This positional relationship is likely not related to sequence characteristics of canonical promoters per se but rather transcriptional activity because the frequency of proximal (within 1.5 kb) upstream and antisense de novo TSSs correlates with expression of the cognate canonical genes (in Figure 7B, note the de novo promoter enrichment (left bars) at more active promoters at the bottom of the graph (canonical expression displayed to the right) which correspondingly show greater levels of open chromatin (middle part of figure).
Figure 7.
Enrichment of de novo promoters near expressed canonical promoters is dependent on canonical gene expression. (A) Histogram of upstream and antisense de novo promoters as a function of distance to canonical gene promoters. Counts are based on 200 bp bins. (B) Chromatin accessibility (middle panel) and expression levels (right panel) of annotated genes are plotted as a function of their respective expression levels (lowest to highest) in latent (‘Cntl’) Mutu cells. The left panel marks those annotated genes with a de novo promoter within 1.5 kb of the canonical gene TSS.
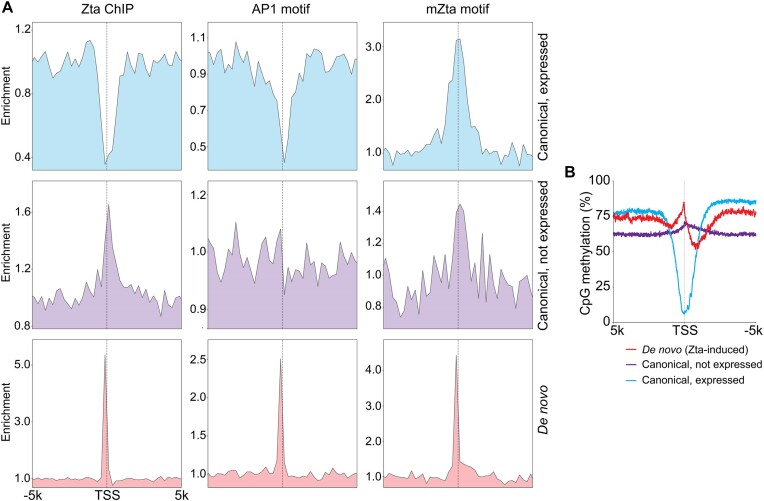
We next sought to examine why Zta induced transcription is prone to occur in proximity to rather than predominantly within active cell promoters. To investigate this issue, we first assessed Zta binding frequency at canonical promoters. As expected, there is tight enrichment of Zta binding just upstream from the TSSs of the Zta class of de novo promoters along with a corresponding enrichment of both AP1 and methyl-Zta motifs (Figure 8A). In contrast, there is a severe depletion of Zta binding at expressed canonical promoters but not at canonical promoters that are not active (Figure 8A). Strikingly, there is also a substantial depletion of AP1 sites at expressed canonical promoters, possibly resulting from negative selection pressures to prevent AP1 activation of the majority of genes that are uninvolved in AP1 signaling (Figure 8A). Therefore, the depleted frequency of AP1 sites within but not surrounding expressed canonical promoters may play a role in driving Zta to bind (and activate transcription) proximally rather than within active canonical promoters, causing de novo promoter activity rather than activation of existing canonical promoters.
Figure 8.
Basis for Zta binding outside of the body of active but not inactive canonical promoters. (A) Distribution of Zta binding (ChIP-seq) and AP1 and mZta motifs surrounding active (>3 TPM) and inactive (0 TPM) canonical promoters and the Zta class of de novo promoters. (B) Distribution of CpG methylation at active and inactive canonical promoters and de novo promoters in latent Mutu cells.
In contrast to the depleted AP1 site frequency at expressed canonical promoters, methyl-Zta binding motifs show enrichment, which may be partly explained by enrichment of CpG islands at promoters. Despite the enrichment of methyl-Zta binding sites at canonical promoters, active promoters typically display low levels of CpG methylation which would prevent binding of Zta to methyl-Zta binding sites.
To investigate CpG methylation status at active promoters in our model, we performed bisulfite sequencing in latent (and Zta reactivated) Mutu cells. Bisulfite sequencing appeared robust because high levels of CpG methylation were observed at viral lytic promoters but not the active viral latent Qp viral promoter in latently infected cells (Supplemental Figure S7). Further, an expected global loss of methylation across the EBV genome was observed in lytic reactivating cells due to the production of abundant unmethylated new viral genomes (Supplemental Figure S7). Assessing CpG methylation at Zta responsive cell de novo promoters, a peak in CpG methylation is seen just upstream from the start sites, consistent with a subset of these being activated through Zta binding to the methyl-Zta sites (Figure 8B). In contrast, there is a severe trough in CpG methylation at active (but not inactive) canonical promoters, supporting the idea that the unmethylated status of sites within active canonical promoters likely precludes Zta binding (Figure 8B). Together, these data indicate that Zta discriminates against inducing the expression of already active canonical promoters through the demethylated nature of expressed canonical promoters and a depleted AP1 motif frequency. This is consistent with the idea that while proximity to active promoters plays a role in driving this class of de novo promoters, a depleted frequency of AP1 sites and the unmethylated status of active promoters enforces sufficient distance to minimize Zta action through the canonical TSS.
Viral repositioning of start sites directs transcription through active promoters to facilitate transcriptional interference
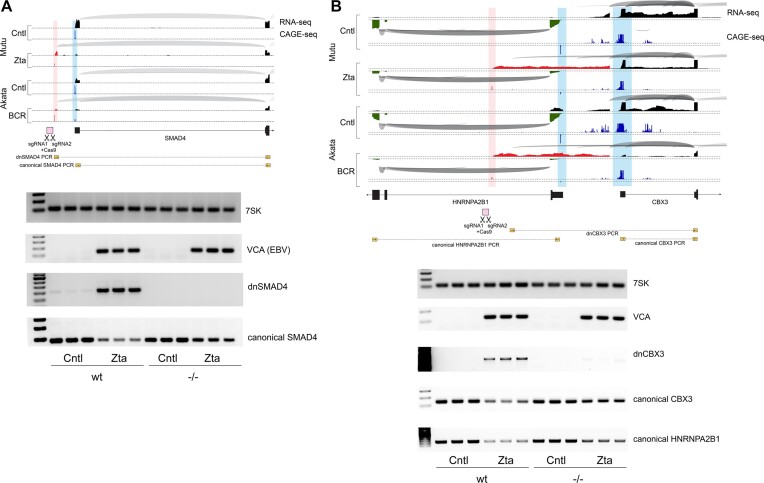
The formation of non-vPIC initiated de novo transcription appears to utilize active chromatin characteristics to promote nearby transcription that proceeds through active cell promoters. We therefore tested whether promoter proximal de novo transcription causes transcriptional interference. Because of the multifaceted and highly complex nature of interactions between the virus and the host transcriptome (including the impact of host shut off which concurrently reduces cell mRNA levels through ribonuclease degradation) (7), assessing enriched downregulation of transcriptional interference targets based on RNA-seq data is challenging. We therefore took a direct genetic approach to determine whether a loss of de novo transcription can alleviate suppression of canonical promoter activity. Specifically, we generated biallelic deletions of the upstream SMAD4 de novo promoter and a de novo promoter whose associated transcript spans the bidirectional HNRNPA2B1 and CBX3 promoters (Figure 9A and B). Wild type or de novo promoter knockout Mutu cells were co-transfected with a CMV-GFP and a control or a Zta expression vector and RNA was generated from FACS selected GFP positive cells. For both the SMAD4 and the HNRNPA2B1/CBX3 loci, deletion of the de novo promoter resulted in loss of de novo transcription during reactivation (Figure 9A and B). Assessing canonical SMAD4, HNRNPA2B1 and CBX3 transcripts, we found that reactivation caused substantial decreases in expression in wild type cells (Figure 9A and B). In cells with the de novo promoter deletions, the decreases in SMAD4, HNRNPA2B1, and CBX3 expression during reactivation was diminished, with the observed residual decreases likely caused by host shut off. While transcriptional interference is a well-established eukaryotic regulatory mechanism (50,51), these experiments show that similarly, de novo transcription can facilitate inhibition of canonical cell gene expression. Altogether, our studies show that EBV utilizes fundamental properties of active promoters to enhance de novo transcription near and through existing active cell promoters to cause downregulation of cell gene expression. This builds an additional layer to host shut off to presumably help free up translational resources for dedicated virus protein production.
Figure 9.
De novo transcription near canonical promoters cause transcriptional interference and suppression of cell gene expression. Upper panels show plots of splice junction reads (arches – thicker arches represent higher number of splice junction reads) and single base level coverage (height represents number of reads identified at each position) from RNA-seq data and CAGE-seq data (height represents number of reads identified at each position) in the Mutu-Zta and Akata-BCR models. Blue coverage/shading represents canonical transcription initiation and red coverage/shading represents ‘de novo’ transcription initiation. Exon structures of associated genes are indicated at the bottom of each plot. Lower panels show targeted deletions of de novo promoters proximal to SMAD4 (A) and HNRNPA2B1/CBX3 (B) canonical genes that were made in Mutu cells using CRISPR-Cas9 and two flanking sgRNAs (positions shown in upper panels). Knockout and wildtype cells were transfected with CMVp-GFP and SV40p-Zta or control vector and GFP + cells were isolated via FACS after 24 h. Gels are agarose gel electrophoresis of PCR fragments of the small RNA, 7SK, the viral late gene, VCA (BFRF3), the de novo transcript, and the canonical start site transcript for wild type cells and for de novo promoter knockout cells.
Discussion
Here, we show that EBV reactivation causes transcription initiation at new sites throughout the cell genome. For some of these promoters, the ability to initiate transcription at new start sites is likely due to unique properties of the viral pre-initiation complex, vPIC. Unlike the cellular analog, TATA binding protein (TBP), that requires dozens of accessory factors to engage Pol II as well as contributions from additional promoter bound transcription factors, vPIC can directly contact Pol II (52) and has been shown to elicit high level transcription through little more than the TATT[T/A]AA motif (32). Consistent with these unique properties of vPIC, we were unable to find any requirement for local chromatin characteristics nor a bias for positioning near existing active canonical promoters.
Contrasted with the lack of remarkable chromatin features near de novo promoters with vPIC sites, we found that the other classes of de novo promoters display a bias for proximity to and directionality towards chromatin features characteristic of active promoters. In these cases, EBV appears to leverage unique properties of active transcription to promote transcriptional interference and inhibit cell gene expression. At least for the Zta class of de novo promoters, there is a depleted frequency of Zta binding within active canonical promoters that is likely due to a depleted frequency of AP1 motifs and a lack of methylation at methyl-Zta binding sites (Figure 8). These properties bias Zta's influence proximal to, rather than within, active canonical TSSs, minimizing induction of canonical cell gene expression while favoring transcriptional interference. Whether similar mechanisms are guiding features of Rta and unclassified de novo promoters will require further investigation. Nevertheless, it is notable that the consensus Rta binding motif contains a CpG dinucleotide and further, we observe small spikes of CpG methylation just upstream from both Rta and unclassified de novo promoter TSSs (Supplemental Figure S8).
While promoter proximal de novo transcription may serve to help diminish cell gene expression during reactivation, we do not know whether vPIC driven transcription at intergenic and heterochromatin regions play functional role(s) during virus replication. Nevertheless, the widespread intergenic and antisense transcription that we observe during EBV reactivation has topological similarities to the RNA POL III mediated mouse B2 SINE (Short Interspersed Nuclear Element) element activation caused by gammaherpesvirus 68 (MHV68) lytic replication (53,54). In the case of MHV68 B2 SINE induction, the Glaunsinger group showed that some B2 transcripts form antisense hybrids with cell mRNAs to facilitate nuclear retention (54). By analogy, antisense de novo transcription during EBV replication may similarly alter the fate or function of cell mRNAs through hybrid formation.
Other consequences for genome-wide de novo transcription can also be envisioned. Herpesvirus lytic replication causes substantial reorganization of cell chromatin and its eventual repositioning to the nuclear lamina (8). In Figure 5, we show that de novo transcription causes alterations in local chromatin structure. It is conceivable that de novo transcription contributes to the observed chromatin reorganization during EBV lytic replication. Another consideration is the potential impact of de novo transcription on cell DNA replication. EBV causes reactivating cells to arrest at a pseudo-S-phase environment. This is thought to play a role in increasing nucleotide pools in support of viral DNA replication while the inhibition of cell DNA replication prevents competition for these resources from replication of cell DNA (55). De novo transcription interspersed throughout the cell genome could help enforce blockage at the G1/S boundary through causing DNA/RNA polymerase clashing.
As mentioned above, there are 2.5 million TATT[T/A]AA motifs and 0.5 million TGAGTCA or TGAGCGA scattered throughout the cell genome. Yet, we were only able to detect tens of thousands of de novo promoters during reactivation. This implies that de novo promoter formation relies on certain chromatin contexts. We hypothesize that EBV exploits the sheer frequency of 7 base sequence motifs through which transcription factors can potentially bind to achieve sufficient probability of encountering tens of thousands of chromatin contexts that are adequate to support de novo promoter formation. A unique aspect of these principles is that de novo promoters are formed based on the semi-stochastic occurrence of simple sequence motifs and existing chromatin environment rather than the complex and nuanced context of existing cell promoters that are controlled through myriads of cell signaling pathways. In essence, de novo promoters appear to be primal in nature and may therefore represent a unique opportunity to study some of the more fundamental principles of promoter formation and function. For example, while there are 223 017 thousand Zta binding sites (as defined by Buschle et al. (39)), there are only a handful that are associated with de novo promoter activity. Similarly, while there are 2.5 million TATT[T/A]AA motifs throughout the cell genome, only a small fraction of these lead to detectable de novo promoter activity. Therefore, EBV reactivation may provide a naturally occurring biological experiment through which chromatin context can be investigated to determine fundamental requirements of promoter formation.
Lastly, we recognize the possibility that some of the de novo promoters identified here may represent existing cell promoters that have not yet been identified. Our analyses utilized a fairly comprehensive Ensembl annotation the includes thousands of transcripts not included in commonly used RefSeq. In addition, our finding that almost no TATT motifs were found located within previously identified canonical TSSs (Figure 3B) makes it unlikely that this motif plays any role in normal cell transcription. Further, the methyl-Zta binding site and the Rta recognition sequences do not match known cell transcription factor binding sites. Altogether, while some of the ‘de novo’ promoters identified here might ultimately be found to occur naturally under some cell conditions, it is likely that the bulk of the de novo promoters identified here are specific to EBV reactivation.
Supplementary Material
Acknowledgements
This research was supported in part using high performance computing (HPC) resources and services provided by Technology Services at Tulane University, New Orleans, LA. Figure 1A was created with BioRender.com. We also acknowledge the use of computational resources and expertise from the Tulane Cancer Crusaders Next Generation Sequence Analysis Core at the Tulane Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Author contributions: N.A.U. and E.K.F. designed the study; N.A.U., E.K.F., C.R., T.O.G., E.I., M.C., M.L., J.B., N.V. and N.W. performed experiments; N.A.U., E.K.F., T.T.N., J.W., M.B. and T.O.G. performed bioinformatic analyses; D.T.N., T.M. and Y.D. provided consultations on experimental approaches and design; D.W. and M.M. performed FACS analyses; N.A.U. and E.K.F. wrote the manuscript.
Contributor Information
Nathan A Ungerleider, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Claire Roberts, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Tina M O’Grady, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Trang T Nguyen, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Melody Baddoo, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Jia Wang, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Eman Ishaq, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Monica Concha, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Meggie Lam, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Jordan Bass, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Truong D Nguyen, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Nick Van Otterloo, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Nadeeshika Wickramarachchige-Dona, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Dorota Wyczechowska, Stanley S. Scott Cancer Center, Louisiana State University Health Sciences Center, New Orleans, LA, USA.
Maria Morales, Tulane Cancer Center, New Orleans, LA, USA.
Tianfang Ma, Dana-Farber Cancer Institute, Boston, MA, USA.
Yan Dong, Department of Structural and Cellular Biology, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Erik K Flemington, Department of Pathology & Laboratory Medicine, Tulane University School of Medicine, Tulane Cancer Center, New Orleans, LA, USA.
Data availability
Bisulfite whole genome sequencing files were submitted to the Sequence Read Archive (accession: PRJNA1001943). All other raw sequencing files were uploaded to the NCBI Geo Archive with the following accession numbers: GSE240011 (CAGE-Seq), GSE240010 (ChIP-Seq), GSE240008 (RNA-Seq), GSE240007 (ATAC-Seq). Source code used in this study is available at https://doi.org/10.5281/zenodo.10709746. It can be found at: http://github.com/flemingtonlab/denovoTranscription.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health [R01CA262090, R01CA243793, R01 CA272142, P01CA214091 to E.K.F.]; Department of Defense [W81XWH-20-1-0517 to Y.D.]; N.A.U. is a Special Fellow of The Leukemia & Lymphoma Society. Funding for open access charge: NIH [R01CA262090].
Conflict of interest statement. None declared.
References
- 1. Farrell P.J. Epstein-Barr virus and cancer. Annu. Rev. Pathol. 2019; 14:29–53. [DOI] [PubMed] [Google Scholar]
- 2. Bjornevik K., Cortese M., Healy B.C., Kuhle J., Mina M.J., Leng Y., Elledge S.J., Niebuhr D.W., Scher A.I., Munger K.L. et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. 2022; 375:296–301. [DOI] [PubMed] [Google Scholar]
- 3. Soldan S.S., Lieberman P.M. Epstein-Barr virus and multiple sclerosis. Nat. Rev. Micro. 2023; 21:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hollingworth R., Grand R.J. Modulation of DNA damage and repair pathways by human tumour viruses. Viruses. 2015; 7:2542–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nikitin P.A., Luftig M.A. At a crossroads: human DNA tumor viruses and the host DNA damage response. Future Virol. 2011; 6:813–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chiu Y.F., Sugden A.U., Sugden B. Epstein-Barr viral productive amplification reprograms nuclear architecture, DNA replication, and histone deposition. Cell Host Microbe. 2013; 14:607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rowe M., Glaunsinger B., van Leeuwen D., Zuo J., Sweetman D., Ganem D., Middeldorp J., Wiertz E.J., Ressing M.E. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takagi S., Takada K., Sairenji T. Formation of intranuclear replication compartments of Epstein-Barr virus with redistribution of BZLF1 and BMRF1 gene products. Virology. 1991; 185:309–315. [DOI] [PubMed] [Google Scholar]
- 9. Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr. Opin. Genet. Dev. 1993; 3:63–70. [DOI] [PubMed] [Google Scholar]
- 10. Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016; 34:525–527. [DOI] [PubMed] [Google Scholar]
- 14. Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017; 14:687–690. [DOI] [PubMed] [Google Scholar]
- 15. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bentsen M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T. et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 2020; 11:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schep A.N., Buenrostro J.D., Denny S.K., Schwartz K., Sherlock G., Greenleaf W.J. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015; 25:1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kulakovskiy I.V., Vorontsov I.E., Yevshin I.S., Sharipov R.N., Fedorova A.D., Rumynskiy E.I., Medvedeva Y.A., Magana-Mora A., Bajic V.B., Papatsenko D.A. et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018; 46:D252–D259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amemiya H.M., Kundaje A., Boyle A.P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 2019; 9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Covarrubias S., Richner J.M., Clyde K., Lee Y.J., Glaunsinger B.A. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J. Virol. 2009; 83:9554–9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feederle R., Bannert H., Lips H., Muller-Lantzsch N., Delecluse H.J. The Epstein-Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 2009; 83:4952–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramasubramanyan S., Osborn K., Al-Mohammad R., Ijiel B., Zuo J., Balan N., Godfrey A., Patel H., Peters G., Rowe M. et al. Epstein–Barr virus transcription factor zta acts through distal regulatory elements to directly control cellular gene expression. Nucleic Acids Res. 2015; 43:3563–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takahashi H., Lassmann T., Murata M., Carninci P. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012; 7:542–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin F.J., Amode M.R., Aneja A., Austine-Orimoloye O., Azov A.G., Barnes I., Becker A., Bennett R., Berry A., Bhai J. et al. Ensembl 2023. Nucleic Acids Res. 2023; 51:D933–D941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wyrwicz L.S., Rychlewski L. Identification of Herpes TATT-binding protein. Antiviral Res. 2007; 75:167–172. [DOI] [PubMed] [Google Scholar]
- 32. Aubry V., Mure F., Mariame B., Deschamps T., Wyrwicz L.S., Manet E., Gruffat H. Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J. Virol. 2014; 88:12825–12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Djavadian R., Hayes M., Johannsen E. CAGE-seq analysis of Epstein-Barr virus lytic gene transcription: 3 kinetic classes from 2 mechanisms. PLoS Pathog. 2018; 14:e1007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nandakumar D., Glaunsinger B. An integrative approach identifies direct targets of the late viral transcription complex and an expanded promoter recognition motif in Kaposi's sarcoma-associated herpesvirus. PLoS Pathog. 2019; 15:e1007774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Serio T.R., Cahill N., Prout M.E., Miller G. A functionally distinct TATA box required for late progression through the Epstein-Barr virus life cycle. J. Virol. 1998; 72:8338–8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhende P.M., Seaman W.T., Delecluse H.J., Kenney S.C. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 2004; 36:1099–1104. [DOI] [PubMed] [Google Scholar]
- 37. Farrell P.J., Rowe D.T., Rooney C.M., Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989; 8:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tripodi I.J., Allen M.A., Dowell R.D. Detecting differential transcription factor activity from ATAC-Seq data. Molecules. 2018; 23:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Buschle A., Mrozek-Gorska P., Cernilogar F.M., Ettinger A., Pich D., Krebs S., Mocanu B., Blum H., Schotta G., Straub T. et al. Epstein-Barr virus inactivates the transcriptome and disrupts the chromatin architecture of its host cell in the first phase of lytic reactivation. Nucleic Acids Res. 2021; 49:3217–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heilmann A.M., Calderwood M.A., Portal D., Lu Y., Johannsen E. Genome-wide analysis of Epstein-Barr virus rta DNA binding. J. Virol. 2012; 86:5151–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ersing I., Nobre L., Wang L.W., Soday L., Ma Y., Paulo J.A., Narita Y., Ashbaugh C.W., Jiang C., Grayson N.E. et al. A temporal proteomic map of Epstein-Barr Virus lytic replication in B cells. Cell Rep. 2017; 19:1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang Z., Zhang B. On the role of transcription in positioning nucleosomes. PLoS Comput. Biol. 2021; 17:e1008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cullen B.R., Lomedico P.T., Ju G. Transcriptional interference in avian retroviruses–implications for the promoter insertion model of leukaemogenesis. Nature. 1984; 307:241–245. [DOI] [PubMed] [Google Scholar]
- 44. Shuman S. Transcriptional interference at tandem lncRNA and protein-coding genes: an emerging theme in regulation of cellular nutrient homeostasis. Nucleic Acids Res. 2020; 48:8243–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaikkonen M.U., Adelman K. Emerging roles of non-coding RNA transcription. Trends Biochem. Sci. 2018; 43:654–667. [DOI] [PubMed] [Google Scholar]
- 46. Palmer A.C., Ahlgren-Berg A., Egan J.B., Dodd I.B., Shearwin K.E. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol. Cell. 2009; 34:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pande A., Brosius J., Makalowska I., Makalowski W., Raabe C.A. Transcriptional interference by small transcripts in proximal promoter regions. Nucleic Acids Res. 2018; 46:1069–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pande A., Makalowski W., Brosius J., Raabe C.A. Enhancer occlusion transcripts regulate the activity of human enhancer domains via transcriptional interference: a computational perspective. Nucleic Acids Res. 2020; 48:3435–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hao N., Crooks M.T., Palmer A.C., Dodd I.B., Shearwin K.E. RNA polymerase pausing at a protein roadblock can enhance transcriptional interference by promoter occlusion. FEBS Lett. 2019; 593:903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Proudfoot N.J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986; 322:562–565. [DOI] [PubMed] [Google Scholar]
- 51. Martens J.A., Laprade L., Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004; 429:571–574. [DOI] [PubMed] [Google Scholar]
- 52. Castaneda A.F., Didychuk A.L., Louder R.K., McCollum C.O., Davis Z.H., Nogales E., Glaunsinger B.A. The gammaherpesviral TATA-box-binding protein directly interacts with the CTD of host RNA Pol II to direct late gene transcription. PLoS Pathog. 2020; 16:e1008843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karijolich J., Abernathy E., Glaunsinger B.A. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-kappaB pathway. PLoS Pathog. 2015; 11:e1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karijolich J., Zhao Y., Alla R., Glaunsinger B. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res. 2017; 45:6194–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kudoh A., Fujita M., Zhang L., Shirata N., Daikoku T., Sugaya Y., Isomura H., Nishiyama Y., Tsurumi T. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 2005; 280:8156–8163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bisulfite whole genome sequencing files were submitted to the Sequence Read Archive (accession: PRJNA1001943). All other raw sequencing files were uploaded to the NCBI Geo Archive with the following accession numbers: GSE240011 (CAGE-Seq), GSE240010 (ChIP-Seq), GSE240008 (RNA-Seq), GSE240007 (ATAC-Seq). Source code used in this study is available at https://doi.org/10.5281/zenodo.10709746. It can be found at: http://github.com/flemingtonlab/denovoTranscription.