Abstract
Infectious salmon anemia virus (ISAV; Isavirus salaris) causes an economically important disease of Atlantic salmon (Salmo salar L.). ISA outbreaks have resulted in significant losses of farmed salmon globally, often with a sudden onset. However, 2 phenotypically distinct variants of ISAV exist, each with divergent disease outcomes, associated regulations, and control measures. ISAV-HPRΔ, also known as ISAV-HPR deleted, is responsible for ISA outbreaks; ISAV-HPR0, is avirulent and is not known to cause fish mortality. Current detection methodology requires genetic sequencing of ISAV-positive samples to differentiate phenotypes, which may slow responses to disease management. To increase the speed of phenotypic determinations of ISAV, we developed a new, rapid multiplex RT-qPCR method capable of 1) detecting if a sample contains any form of ISAV, 2) discriminating whether positive samples contain HPRΔ or HPR0, and 3) validating RNA extractions with an internal control, all in a single reaction. Following assay development and optimization, we validated this new multiplex on 31 ISAV strains collected from North America and Europe (28 ISAV-HPRΔ, 3 ISAV-HPR0). Finally, we completed an inter-laboratory comparison of this multiplex qPCR with commercial ISAV testing and found that both methods provided equivalent results for ISAV detection.
Keywords: infectious salmon anemia, Isavirus salaris, multiplex PCR, salmon isavirus
Infectious salmon anemia (ISA) is a serious disease of Atlantic salmon, Salmo salar L. Outbreaks can occur unpredictably and have resulted in significant economic loss in farmed salmon globally. 34 First discovered in Norway in 1984, 40 ISA has now been diagnosed in all major salmon farming countries including Canada,4,26,31 Scotland, 37 the United States, 5 Chile,19,22 and the Faroe Islands. 3 The disease is characterized by lethargy, acute anemia, hemorrhagic liver necrosis, ascites, and renal tubular necrosis.16,36 Horizontal transmission (fish-to-fish) is thought to be the main form of transmission, with the gill serving as the primary portal of entry.1,33,42 The course of disease can be prolonged within a population and lead to high cumulative mortality. 34
The causative agent of ISA is infectious salmon anemia virus (ISAV; syn. salmon isavrius; Orthomyxoviridae, Isavirus salaris), which shares several properties with well-studied influenza viruses. 41 Specifically, ISAV is pleomorphic, consisting of particles enveloped in 90–130-nm spheres with 13–15-nm long spike proteins; disruption of the virion releases filamentous nucleocapsid structures. The 8 genomic segments of the ISAV single-stranded, negative-polarity RNA genome have been completely sequenced,11,12,28,29 revealing 10 predicted proteins and 2 distinct genotypes: genotype 1 (European) and genotype 2 (North American) based on polymorphisms in the hemagglutinin gene.8,14,18,27,32 Two phenotypically distinct variants of ISAV have been identified within both European and North American genotypes. One is the well-characterized highly virulent ISAV–high polymorphic region (HPR)Δ variant associated with ISA, commonly referred to as ISAV-HPR deleted. The second is the avirulent variant known as ISAV-HPR0, which is not associated with ISA or indeed any other notable disease. 8 Both HPRΔ and HPR0 hemagglutinin-esterase (HE) proteins have functional receptor-binding and receptor-destroying activities, with a specific 5N-4O-acetylated sialic acid on the host cell surface identified as the viral receptor. 17 The sole consistent difference in these highly divergent phenotypes is in the length of the stalk of the HE protein coded by the HPR within segment 6 of the ISAV genome. 35
Whether the avirulent HPR0 phenotype can lead to virulent HPRΔ remains debated. Some authors regard the risk of a deletion occurring in HPR0 leading to HPRΔ as low, but non-negligible.25,33 The catalyst and mode of evolution for this event to occur have yet to be fully studied. 2 It is clear, however, that both ISAV phenotypes are commonly found in regions where Atlantic salmon farming occurs, with the HPR0 phenotype being more geographically widespread. 27
ISA, and/or even the detection of any variant of ISAV, is notifiable to the World Organisation for Animal Health (WOAH) and is required to be reported by a country’s competent authority. 43 As of October 2022, ~54 countries have mandated some level of ISAV testing or freedom (USDA Export/Import staff veterinarian, pers. comm., 2022 Oct). In salmon-producing countries, regulatory measures for the control of ISA are variable and can include passive or active surveillance, mandatory fish health reporting, regulations for transportation of live fish, fallowing of production sites between fish generations, and biosecurity best management practices to help minimize ISA outbreaks. HPRΔ detection and subsequent confirmation can initiate control strategies that can involve regulatory zonation, movement restrictions, and depopulation of infected cages and sites. 43
The WOAH Manual of Diagnostic Tests for Aquatic Animals 43 outlines the typical steps used to confirm the presence of ISAV in a population. Surveillance strategies aim to detect HPRΔ target fish with typical clinical signs, such as anemia, and test only internal organs (kidney and heart); screening for HPR0 requires the random collection of gill samples. According to EU legislation on animal health 2017/429, and specifically the Commission Implementing Regulation (EU) 2018/1882, infection with HPRΔ is a category C disease. Hence, samples are to be tested with RT-qPCR 39 and sequenced for the segment 6 HPR to discriminate between HPRΔ and HPR0, based on fragment length. However, sequencing all ISAV-positive samples can be a laborious process, which may not happen until days or weeks after the initial ISAV screening, and may delay regulatory and disease mitigation responses. Being able to rapidly differentiate phenotypic variants decreases confirmation time, facilitates robust surveillance monitoring, and avoids delays in regulatory response decisions, which, in turn, establishes production efficiency.
Our 4 aims were to: 1) design a reverse-transcription quantitative real-time PCR (RT-qPCR) assay with the ability to rapidly differentiate ISAV HPR0 from HPRΔ without the need for follow-up sequencing, 2) create a new RT-qPCR multiplex that combines this new phenotyping assay with those for generic ISAV identification and an internal positive control, 3) evaluate the analytical specificity and sensitivity of the ISAV testing components of this new multiplex, and 4) complete an inter-laboratory comparison using the new multiplex to evaluate performance compared to traditional ISAV screening methods.
Materials and methods
Our research proposal was assessed by the National Cold Water Marine Aquaculture Center (NCWMAC) Animal Care and Use Committee (IACUC). All experimental design and protocols were reviewed and approved prior to any research being conducted. The NCWMAC is registered as a research facility in accordance with the U.S. Department of Agriculture Animal Welfare Act USDA certificate 11-G-0001. The S. salar gill clips that we used were not collected from live animals at NCWMAC; however, the care and use of salmon were covered by IACUC-approved holding and spawning standard operating procedures.
Identification of HPR0-specific sequences in the ISAV HPR and assay development
First, we gathered and aligned over 600 ISAV segment 6 HPR sequences publicly available in GenBank 10 (National Center for Biotechnology Information) for both the HPRΔ and HPR0 genotypes (Suppl. Table 1) in Geneious Prime v.2023.0.2. 21 We included variants from most geographic regions where ISAV is endemic to evaluate genotypic diversity during assay development. Unsurprisingly, we found considerable variation in the HPR of virulent strains. In contrast, we identified a 51-bp region in the HPR0 HPR that was nearly monomorphic for all HPR0 strains and consistently deleted in one form or another in HPRΔ, as reported by previous researchers.9,17
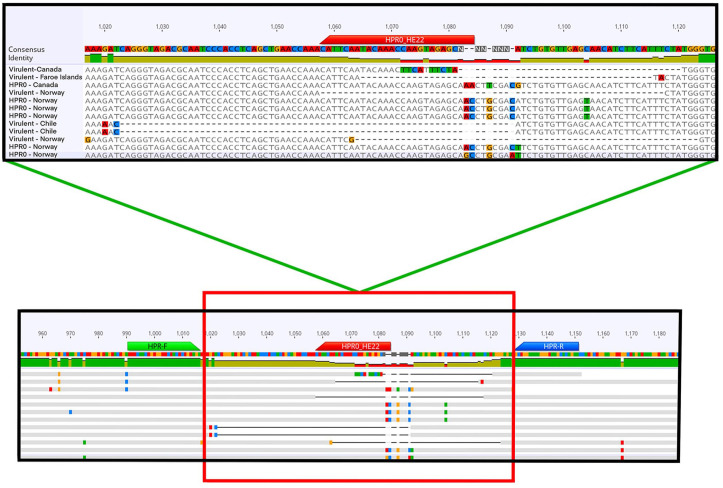
Upon further investigation of this 51-bp region in HPR0, we found a 13-bp portion that was not present in any of the HPRΔ sequences in our alignment (Fig. 1). Both upstream and downstream of this 13-bp region, we found relatively conserved sequences that were suitable for binding primers for PCR. We predicted that these outer primers would create PCR products of 161-bp for HPR0 and 110–130-bp for HPRΔ. We then targeted this region for use in developing a hydrolysis probe (TaqMan) assay specifically for HPR0 to incorporate it into existing ISAV RT-qPCR workflows already used to detect both HPR0 and HPRΔ. Thus, our goal was to create a multiplex RT-qPCR that could do the following in a single reaction: 1) test for the presence of any strain of ISAV, 2) test for the presence of HPR0, and 3) validate the RNA extraction process.
Figure 1.
Graphical representation of the location of the infectious salmon anemia virus (ISAV) segment 6 high polymorphism region (HPR) we selected to use for our HPR0-specific assay. Here, we show the binding locations of the HPR-F and HPR-R primers and the HPR0-HE22 probe of our assay. We designed the probe to bind to a section of sequence that is unique to ISAV-HPR0; the outer primers (HPR-F and HPR-R) will amplify both HPR0 and virulent strains (HPRΔ).
In our initial RT-qPCR multiplex, we included the following: an ISAV segment 8 assay to test for any ISAV commonly used in regulatory testing, 39 our newly developed HPR0-specific assay, and a S. salar elongation factor Fα assay 30 to validate RNA extractions. However, during assay optimization, we found that the efficiency of the ISAV segment 8 assay that we originally selected 39 was severely impacted by multiplexing and instead we switched to the 404F_ISA8, 583R_ISA8, and 491_ISA8 ISAV segment 8 assay. 7 To increase assay reproducibility and reduce sample handling time, we chose to use a one-step master mix that combined the RT and PCR steps. Our final optimized RT-qPCR multiplexes were completed in 10-μL reactions comprised of the following: 2.5 μL of master mix (UltraPlex 1-Step ToughMix, Quantabio; 4×), 1.5 μL of primer and probe mixture (Table 1), 4 μL of RNAse-free water, and 2 μL of RNA extract. We completed thermal cycling using the following protocol: 10 min at 50°C for RT, 3 min at 95°C to inactivate RT, followed by 45 cycles of 95°C denaturation for 10 s, and 60°C annealing and extension for 90 s.
Table 1.
Primer and probes, targets, sequences, fluorophores, and concentration for the new infectious salmon anemia virus (ISAV) multiplex RT-qPCR assay.
| Oligonucleotide name | Target | Sequence (5′–3′) | Fluorophore (quencher) | Concentration in each 10-μL reaction, nM | Reference |
|---|---|---|---|---|---|
| 404F_ISA8 | ISAV segment 8, HPR0 & HPRΔ | TGGGCAATGGTGTATGGTATGA | – | 700 | 7 |
| 583R_ISA8 | GAAGTCGATGAACTGCAGCGA | – | 700 | ||
| 491_ISA8 | CAGGATGCAGATGTATGC | FAM (MGB) | 350 | ||
| HPR-F | ISAV segment 6 HPR, HPR0 specific | AAACTTCAGAGGAACATCACAGATGT | – | 400 | Our study |
| HPR-R | AACAGAGCAATCCCAAAACCTGC | – | 400 | ||
| HPR0-HE22 | TTTGCTCTACTTGGTTTGTATTGAATG | Yakima yellow (VIC) (BHQ) | 200 | ||
| Salsa_ELF-F | Salmo salar elongation factor Fα mRNA | CCCCTCCAGGACGTTTACAAA | – | 200 | 30 |
| Salsa_ELF-R | CACACGGCCCACAGGTACA | – | 200 | ||
| Salsa_ELF-P | ATCGGTGGTATTGGAAC | Cy5 (MGB) | 100 |
Dash (–) indicates oligonucleotides that are unlabeled and have no fluorophore.
ISAV-HPR0 assay specificity testing
We evaluated the specificity of our new HPR0 assay on a set of 38 samples including HPRΔ and HPR0 strains collected from North America and Europe, as well as other viruses of farmed Atlantic salmon, and known ISAV-negative fish. Of these isolates, we subsampled 18 ISAV samples at the Technical University of Denmark (DTU; Kgs. Lyngby, Denmark), which were kindly provided by Dr. Torfinn Moldal at the Norwegian Veterinary Institute (NVI), and applied them to FTA (Flinders Technology Associates) cards using the manufacturer’s protocol, along with the other non-target viruses and negative controls. We then mailed the FTA cards for primary evaluation at the University of Maine Cooperative Extension Diagnostic and Research Laboratory (UMCEDRL; Orono, ME, USA). We extracted the viral isolates from the FTA cards using TRIzol (Invitrogen), followed by an organic extraction, 38 and then evaluated the specificity of the newly developed multiplex qPCR assay on these samples. Following the primary evaluation of the method, we ran the same 18 ISAV subsamples (plus 2 negative controls) at DTU for an inter-laboratory compa-rison.
Multiplex assay sensitivity testing and inter-laboratory testing
To begin, we evaluated the analytical sensitivity of the new HPR0 assay in the previously described multiplex. We obtained a synthetic nucleic acid artificial positive control (APC) gBlock (Integrated DNA Technologies), which contained the segment 6 HPR of HPR0 and segment 8 of HPRΔ. Next, we completed serial dilutions of this artificial nucleic acid to determine the limit of detection (LOD) and calculate the reaction efficiency of the new HPR0 assay.
We also directly compared the sensitivity of the new HPR0 assay and multiplex to an assay frequently used in the aquaculture industry to screen fish for ISAV, 39 which is the same test we determined earlier to be unacceptable for multiplexing. During the 2021 salmon spawning season at the U.S. Department of Agriculture–Agricultural Research Service (USDA-ARS) National Cold Water Marine Aquaculture Center (NCWMAC; Franklin, ME, USA) we collected gill clips from adult Atlantic salmon and selected 182 for additional testing. We then divided each gill clip into 3 subsamples. We sent one subset to Kennebec River Biosciences (KRB; Richmond, ME, USA) for commercial ISAV segment 8 RT-qPCR screening. We retained one of the remaining sample subsets at the NCWMAC and provided the final subset to the UMCEDRL. We stored all collected gill clips at −80°C post-collection until used in laboratory procedures.
For the sample extractions at both the UMCEDRL and NCWMAC, we used a ~30-mg piece of gill clip and homogenized it (TissueLyser II; Qiagen). KRB homogenized their subsamples (Bead Mill 24; FisherBrand) in 1 mL of Leibovitz L15 medium with 2% fetal bovine serum and 350 µg/mL of gentamicin. At the UMCEDRL, we extracted RNA (RNeasy mini kit; Qiagen) using the manufacturer’s recommended protocol. At the NCWMAC, we completed sample extractions on the homogenates with Tri Reagent (MilliporeSigma) using the manufacturer’s protocol. KRB extracted their subsamples (MagMAX viral RNA isolation kit, KingFisher Flex; Thermo Fisher) and completed RT-qPCR (VetMAX-Plus Multiplex One-Step kit; Thermo Fisher) and an industry standard ISAV segment 8 assay 39 on either a QuantStudio 3 (Thermo Fisher) or a 7500 fast real-time PCR system (Applied Biosystems). Once extracted, we ran the separate RNA extracts at NCWMAC and UMCEDRL using the multiplex assay as described herein in at least duplicate on a QuantStudio 5 (Thermo Fisher) at UMCEDRL and a CFX 96/384 (Bio-Rad) at NCWMAC. We considered samples producing no Cq in 2 consecutive replicates for the same sample to be negative for an assay. In contrast, we considered samples producing Cq values in 2 consecutive runs to be positive for an assay. We reran any sample producing a positive result for only one replicate an additional 2 times. We considered samples inconclusive if they failed to produce a positive result in at least one of the 2 additional runs.
Results
Multiplex assay specificity
Of the 38 samples we used to test the specificity of the new HPR0 assay, only the 31 containing any strain of ISAV tested positive with the generic ISAV segment 8 assay of our new multiplex (Table 2). In contrast, we found that the HPR0 assay only amplified 3 isolates that we had previously identified as HPR0 via sequencing of the segment 6 HPR. We also found that both of the ISAV testing components worked together in multiplex and that samples containing HPR0 produced Cq values for both the generic ISAV assay and HPR0 assay, in contrast to HPRΔ, which only produced Cq values for the generic ISAV assay. In addition, we successfully replicated the ISAV assay specificity testing results at DTU on the same 18 ISAV sample subset (plus 2 negative controls) that was sent to the UMCEDRL for initial evaluation (Suppl. Table 2). Finally, we determined that the elongation factor Fα assay was successful in amplifying endogenous S. salar nucleic acid from tissue extracts, but had difficulty amplifying material from some ISAV cell culture–infected supernatants that we also tested.
Table 2.
Samples used for specificity testing of our new infectious salmon anemia virus (ISAV) multiplex RT-qPCR assay.
| Sample ID or GenBank accession | Isolates | Geographic origin | Strain | Generic ISAV assay result | HPR0 assay result |
|---|---|---|---|---|---|
| MK216303, MK216305, MK216306, MK216307, MK216308, MK216310, MK216321, MK216313, MN901913/MN901918, MN901916/MN901921, MN901917/MN901922, MT413436/MT413443, MT413437/MT413444, MT613037/MT613041, MT990389/MT997778, OM366041, AF378180, DQ785248, FM203261 | 19 | Norway | ISAV-HPRΔ | + | – |
| AF388581 | 1 | Scotland | ISAV-HPRΔ | + | – |
| EF105375, AF294870, MH397905 | 3 | Canada | ISAV-HPRΔ | + | – |
| MT990384/MT997773 | 1 | Norway | ISAV-HPR0 | + | + |
| 2018-50-202_1*+ 2020-02-10_4* | 1 | Norway | Mix of ISAV-HPRΔ & ISAV-HPR0 | + | + |
| KX424587, KU587561 | 2 | North America | ISAV-HPRΔ | + | – |
| MH397893, AY601904, MH397910 | 3 | Europe | ISAV-HPRΔ | + | – |
| KX823932 | 1 | Europe | ISAV-HPR0 | + | + |
| AM889221/AF342728 | 1 | Denmark | IPNV | – | – |
| Trial 19-13082* | 1 | Ireland | PMCV | – | – |
| AY546597 | 1 | Denmark | VHSV | – | – |
| Trial 15-10833-27* | 1 | Denmark | PRV1 | – | – |
| IHNV BLK94 (sample 1 from IHN panel)* | 1 | United States | IHNV | – | – |
| Negative Salmo salar gills and cells* | 2 | Denmark | Negative controls | – | – |
IHNV = infectious hematopoietic necrosis virus; IPNV = infectious pancreatic necrosis virus; negative controls = samples previously tested with no detected ISAV; PMCV = piscine myocarditis virus; PRV1 = piscine orthoreovirus 1; VHSV = viral hemorrhagic septicemia virus. GenBank isolate number or sample ID, the number of isolates, geographic origin, strain, generic ISAV results (404F_ISA8/583R_ISA8/491_ISA8) 7 primers and probe and HPR0 results for our newly developed assay. Isolates with positive and negative qualitative results are denoted by + and –, respectively.
Samples provided from the National Institute of Aquatic Resources at the Technical University of Denmark (DTU) without sequences uploaded to GenBank.
Multiplex assay sensitivity
Using the APC, we determined that, in multiplex, the amplification efficiencies of the generic ISAV assay was 95.0% (slope −3.45) and of the new HPR0 assay was 101% (slope −3.30; Figs. 2, 3). We calculated these values by analyzing 10 replicates for 7 different 10-fold dilutions from ~2.3 × 106 copies of the APC down to 2.3 copies per reaction. We used the generally accepted LOD as the lowest quantity of analyte that produced a positive result for 95% of replicates. 24 We determined that the LOD for the generic ISAV assay in multiplex was between 23 and 10 copies of APC. Through a series of LOD experiments, we found that 10 of 10 (100%) replicates with 23 copies were successful, compared to the 27 of 30 (90%) replicates with 10 copies. The performance of the HPR0 assay was similar, and we also determined the LOD to be between 23 and 10 copies of APC. However, this assay appeared to be less sensitive because only 24 of 30 replicates of 10 copy APC (80%) were positive, whereas 10 of 10 (100%) replicates were successful at 23 copies.
Figure 2.
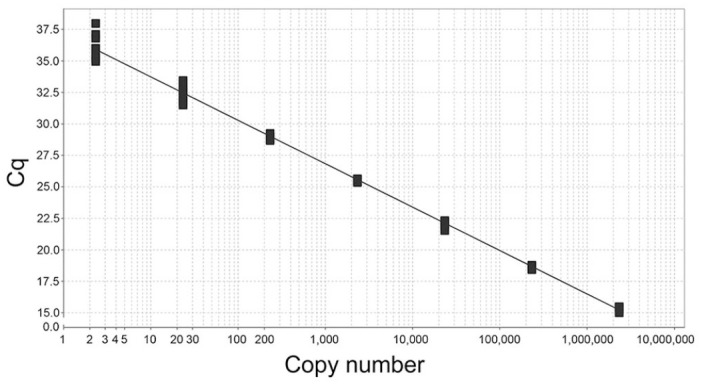
Standard curve of the generic infectious salmon anemia virus assay 7 in multiplex. The y-axis contains the Cq values of the corresponding number of copies of artificial positive control included in that particular assay. Each set of copy numbers tested contained 10 replicates.
Figure 3.
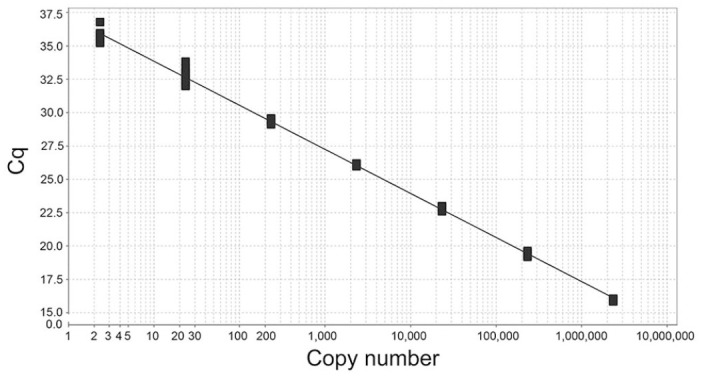
Standard curve of our new infectious salmon anemia virus (ISAV)-HPR0 assay in multiplex. The y-axis contains the Cq values of the corresponding number of copies of artificial positive control included in that particular assay. Each set of copy numbers tested contained 10 replicates.
We also determined the limit of accurate quantification (LOQ) for both assays to be ~233 copies (CVs of 10.3% for the generic ISAV assay and 9.0% for the HPR0 assay). We defined this as the lowest copy number tested during the creation of the standard curve where the CV was <25%. 24 We did not evaluate the LOD or LOQ of the elongation factor Fα assay internal control because the efficiency of this reaction was purposely inhibited to prevent it from consuming more reaction components than are needed for a simple qualitative result.
During the creation of the multiplex standard curves, we discovered that the Cq values for both the generic ISAV assay and the HPR0 assay were very similar in experiments using the same dilutions of the APC. The mean Cq values for the 10 replicates at each dilution of APC had a difference of <1 Cq between assays (Suppl. Table 3). If considering all Cq means from all APC replicate groups, the generic ISAV assay was ~0.5 Cq lower than the HPR0 assay, suggesting comparable sensitivity between these 2 assays.
Inter-laboratory comparison
Of the 182 gill clip subsamples that were screened at the 3 separate laboratories, we did not detect any HPRΔ in these fish, either by our HPR0 assay, or confirmatory ISAV segment 6 HPR sequencing conducted by KRB. We achieved agreement of 111 of 182 (61.0%) results between all laboratories when we categorized samples into ISAV positive, negative, or inconclusive (Suppl. Table 4). We found the overall agreement among laboratories to be lower than a pairwise comparison of categorical results: 137 of 182 (73.1%) for UMCEDRL-KRB, 125 of 182 (68.7%) for UMCEDRL-NCWMAC, and 128 of 182 (70.3%) for NCWMAC-KRB. When further categorizing ISAV-positive samples into “high-infected” (Cq < 30) and “low-infected” (Cq ≥ 30) groups, based on the generic ISAV assay results, we found that approximately the same number of samples were classified as “low-infected” at each of the 3 laboratories (176 of 546; 32.2%) as were classified as “high-infected” (182 of 546; 33.3%). However, the results varied at each laboratory, with 62 of 182 (34.1%) at UMCEDRL, 90 of 182 (49.5%) at KRB, and 24 of 182 (13.2%) at NCWMAC (Suppl. Table 5) classified as “low-infected” in contrast to the “high-infected” category with 73 of 182 (40.1%) at UMCEDRL, 36 of 182 (19.8%) at KRB, and 73 of 182 (40.1%) at NCWMAC. When comparing only the samples where at least one laboratory classified a sample as “high-infected,” we recorded a 76 of 86 (88.4%) agreement where all 3 laboratories determined that particular sample to be infected with ISAV. When considering just the ISAV HPR0 assay results between UMCEDRL and NCWMAC, we found an agreement for 138 of 182 (75.8%) samples.
ISAV HPR0 and HPRΔ mixtures
In addition to the specificity testing, sensitivity testing, and inter-laboratory comparison in our study, we evaluated hundreds of other ISAV-positive samples collected from farmed salmon and found 3 that appeared to have a mixed infection of HPR0 and HPRΔ (Suppl. Fig. 1). After encountering these samples, we created an artificial sample mixture with a higher ratio of HPRΔ to HPR0 (Suppl. Fig. 2) and a second mixture with a higher ratio of HPR0 to HPRΔ (Suppl. Fig. 3) to evaluate the impact of these mixtures on the RT-qPCR plots. We found that mixtures of HPR0 and HPRΔ in the same samples can lead to reduced reaction sensitivity or efficiency in our ISAV HPR0 assay.
Discussion
We developed a potentially important new multiplex RT-qPCR assay to differentiate virulent and avirulent strains of ISAV without the need for confirmatory sequencing. Although we found that the HPR0 assay did not amplify any HPRΔ strains that we tested, we believe that the method should be evaluated more widely with additional HPRΔ and HPR0 strains to ensure its reproducibility and specificity. If this continued evaluation proves successful, the incorporation of our new assay into routine ISAV screening procedures could shorten the response time to outbreaks because follow-up sequencing of every ISAV-positive sample would no longer be necessary. However, we believe that more work is needed before this method is ready to replace those assays currently in place for regular ISAV surveillance and regulatory testing, in the form of additional inter-laboratory comparisons or even split-sample proficiency testing and statistical analyses.6,7,13 Our study was intended to be descriptive in nature with the purpose of bringing the new HPR0 assay and paired multiplex to the attention of the scientific community for the purpose of widespread evaluation.
When directly comparing the new HPR0 assay to an established assay used for ISAV research and surveillance,7,15 we found that the analytical sensitivity was strikingly similar. Given that both the generic ISAV- and HPR0-specific assays are required to work in tandem for the quick differentiation of virulent and avirulent ISAV isolates, it is important that they both have similar analytical sensitivities, which we have successfully demonstrated with our dataset. Although a different generic assay 39 than the one we incorporated into the multiplex remains one of the most popular assays used for regulatory ISAV screening, we found it to be unsuitable for pairing with the HPR0 assay because it performed poorly in a multiplex reaction. Given that we are recommending that agencies and researchers switch to a completely different assay than the one that they are normally using for their ISAV surveillance and testing, we suggest using our newly described multiplex assay on the same tissues, eggs, and water as the traditional method to determine whether both methods produce equivalent results in each laboratory. However, our new method is advantageous because it does not require the sometimes lengthy follow-up sequencing to determine if ISAV-positive samples contain HPR0 or HPRΔ. This allows for a quicker response time and adds to production strategy efficiencies for the salmon farming industry.
Although the inter-laboratory rate of result agreement (reproducibility) on the same samples was only 61.0%, we believe that this number was not unacceptably low, considering that roughly half of our HPR0-positive samples were infected with low quantities (high Cq values) of ISAV-HPR0. In addition, the pairwise comparisons of results between laboratories also produced similar values, suggesting that at extremely low concentrations of ISAV in a sample, that stochastic effects likely had a greater impact on the agreement of results between laboratories than the different methods employed. For instance, other researchers created models for evaluating the agreement of ISAV test results between laboratories based on real testing data they collected when evaluating the same generic ISAV assay we used in our multiplex.6,7 The median predicted proportion of inter-laboratory agreement (Cohen kappa) the authors generated for pairwise comparisons of non-homogenized samples and a situation similar to the sample pool we collected (~65% ISAV prevalence, with 50% of those being “low-infected” ISAV) was ~0.60–0.70. In addition, some variance in results between laboratories was expected given that all 3 laboratories used different RNA extraction methods and RT-qPCR instrumentation, and many discrepancies occurred at low viral loads where increased stochasticity is expected. However, the much higher percentage of agreement among all 3 laboratories (88.4%) of “high-infected” positive samples suggests that both our new multiplex and the traditional method 39 would have a similar level of success in screening suspect fish to monitor ISAV outbreaks, which matches a previous study that found inter-laboratory agreement greater at higher ISAV viral titers. 23
We selected the particular fluorophores for the multiplex assay probes with portability between instruments in mind; KRB and UMCEDRL have QuantStudio 5 instruments, in contrast to the CFX 96/384 used at NCWMAC. Yakima yellow, which we used for the HPR0-HE22 when paired with BHQ3, is a cost-effective equivalent of VIC that will work on many different qPCR instruments and has a narrower spectral profile than the commonly used HEX fluorophore. If Yakima yellow is not available for purchase, VIC with an MGB quencher might be used, but we did not evaluate this experimentally. However, both the 491_ISA8 and Salsa_ELF-P probes must be ordered with an MGB quencher because the melting temperatures of these probes would preclude them from working well at the 60°C annealing and extension step we used for our multiplex assay.
We included the elongation factor Fα internal control as a qualitative assay to validate the success of RNA extractions. We purposely selected a positive control that would perform poorly at the reaction temperatures we selected for the multiplex so that it would not consume too many reagents. In most cases, we would expect the amount of S. salar mRNA for this gene to greatly outnumber the quantity of ISAV in each reaction. We determined the optimal amount of primer and probe for the internal control based on screening S. salar gill tissues for ISAV. For samples taken from water or cell culture isolates, the amount of primer and probe of the internal control will likely need to be increased significantly for it to work reliably. Although we did not verify it experimentally, this set of primers and probe has also been used previously on brown trout (S. trutta) 20 and rainbow trout (Oncorhynchus mykiss), 44 and may function as an internal positive control on other salmonids.
Although we have demonstrated that our new multiplex containing the HPR0 assay can be used to quickly discern between virulent and avirulent viral types, we believe that this assay may have difficulty in situations in which mixtures of HPR0 and HPRΔ are present in the same sample. This is because the outer primers for the HPR0 assay will amplify both HPR0 and HPRΔ, whereas the probe is HPR0-specific. These 2 competing PCR reactions can reduce the efficiency of the HPR0 assay and may cause reduced sensitivity, reaction efficiency, or both, which can be visualized on the RT-qPCR plots. However, very little research has been completed on the topic of mixed infections, and we believe that the inclusion of this assay into regular testing workflows may shed light on this possibility. In the preliminary testing we completed on artificially created mixtures of HPRΔ and HPR0, we determined that, in situations in which the ratio of HPRΔ to HPR0 was ≥6:1, HPR0 was only barely detectable by our assay, similar to what we had seen for a sample we believed to have a mixed infection. However, when the situation was reversed (1:6 HPRΔ to HPR0) we found the amplification curve for the HPR0 assay was still flattened and the Cq shifted, when compared directly to that of the generic ISAV assay, which is also similar to what happened at a 1:1 mixture, but to a lesser extent. Our recommendation for dealing with sample mixtures, while partially subjective, is to sequence the HPR of any samples in which the amplification curves or Cq values of the HPR0 assay and generic ISAV assay are dissimilar (>2–3 Cq values apart), for secondary confirmation. We believe this step will be necessary until more information on the prevalence of HPR0 and HPRΔ sample mixtures in “real-world” samples is reported.
Supplemental Material
Supplemental material, sj-pdf-1-vdi-10.1177_10406387231223290 for Rapid differentiation of infectious salmon anemia virus avirulent (HPR0) from virulent (HPRΔ) variants using multiplex RT-qPCR by Thomas F. Rounsville, Mark P. Polinski, Alyssa G. Marini, Sarah M. Turner, Niccolò Vendramin, Argelia Cuenca, Michael R. Pietrak, Brian C. Peterson and Deborah A. Bouchard in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank Janet Warg from the USDA-APHIS National Veterinary Services Laboratories for providing ISAV isolates for the specificity testing. We thank Nellie Gagné of Fisheries and Oceans Canada for providing ISAV isolates for the specificity testing. We thank Torfinn Modal, National Veterinary Institute in Norway, for providing ISAV samples through the EURL for fish and crustacean diseases for the specificity testing. We also thank John Nugent, Brian Silva, Brendan Ward (UMCEDRL), and Demitri Lifgren (NCWMAC) for technical assistance with the inter-laboratory comparison.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Our research was supported by the U.S. Department of Agriculture–Agricultural Research Service NACA Agreement 58-8030-0-004 with the University of Maine’s Aquaculture Research Institute. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
Supplemental material: Supplemental material for this article is available online.
ORCID iDs: Thomas F. Rounsville  https://orcid.org/0000-0002-5279-5074
https://orcid.org/0000-0002-5279-5074
Mark P. Polinski  https://orcid.org/0000-0001-7237-3808
https://orcid.org/0000-0001-7237-3808
Sarah M. Turner  https://orcid.org/0000-0001-5743-9725
https://orcid.org/0000-0001-5743-9725
Niccolò Vendramin  https://orcid.org/0000-0002-9217-7887
https://orcid.org/0000-0002-9217-7887
Michael R. Pietrak  https://orcid.org/0000-0002-9301-342X
https://orcid.org/0000-0002-9301-342X
Contributor Information
Thomas F. Rounsville, Jr, Pest Management Unit, University of Maine Cooperative Extension Diagnostic and Research Laboratory, Orono, ME, USA.
Mark P. Polinski, National Cold Water Marine Aquaculture Center, U.S. Department of Agriculture–Agricultural Research Service, Franklin, ME, USA
Alyssa G. Marini, Pest Management Unit, University of Maine Cooperative Extension Diagnostic and Research Laboratory, Orono, ME, USA University of Maine School of Biology and Ecology, Orono, ME, USA.
Sarah M. Turner, Aquatic Animal Health Laboratory, University of Maine Cooperative Extension Diagnostic and Research Laboratory, Orono, ME, USA
Niccolò Vendramin, Unit for Fish and Shellfish Diseases, National Institute of Aquatic Resources, Technical University of Denmark, Kgs. Lyngby, Denmark.
Argelia Cuenca, Unit for Fish and Shellfish Diseases, National Institute of Aquatic Resources, Technical University of Denmark, Kgs. Lyngby, Denmark.
Michael R. Pietrak, National Cold Water Marine Aquaculture Center, U.S. Department of Agriculture–Agricultural Research Service, Franklin, ME, USA
Brian C. Peterson, National Cold Water Marine Aquaculture Center, U.S. Department of Agriculture–Agricultural Research Service, Franklin, ME, USA
Deborah A. Bouchard, Aquatic Animal Health Laboratory, University of Maine Cooperative Extension Diagnostic and Research Laboratory, Orono, ME, USA
References
- 1. Aamelfot M, et al. Infectious salmon anaemia virus (ISAV) mucosal infection in Atlantic salmon. Vet Res 2015;46:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aamelfot M, et al. Localised infection of Atlantic salmon epithelial cells by HPR0 infectious salmon anaemia virus. PLoS One 2016;11:e0151723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anonymous. ISA hits the Faroes. Fish Farm Int 2000; 27:47. [Google Scholar]
- 4. Bouchard D, et al. Isolation of infectious salmon anemia virus (ISAV) from Atlantic salmon in New Brunswick, Canada. Dis Aquat Organ 1999;35:131–137. [DOI] [PubMed] [Google Scholar]
- 5. Bouchard DA, et al. First report of Infectious Salmon Anaemia (ISA) in the United States. Bull Eur Ass Fish Pathol 2001;21:86–88. [Google Scholar]
- 6. Caraguel C, et al. Traditional descriptive analysis and novel visual representation of diagnostic repeatability and reproducibility: application to an infectious salmon anaemia virus RT-PCR assay. Prev Vet Med 2009;92:9–19. [DOI] [PubMed] [Google Scholar]
- 7. Caraguel C, et al. A modelling approach to predict the variation of repeatability and reproducibility of a RT-PCR assay for infectious salmon anaemia virus across infection prevalence and infection stages. Prev Vet Med 2012;103:63–73. [DOI] [PubMed] [Google Scholar]
- 8. Christiansen DH, et al. A low-pathogenic variant of infectious salmon anemia virus (ISAV-HPR0) is highly prevalent and causes a non-clinical transient infection in farmed Atlantic salmon (Salmo salar L.) in the Faroe Islands. J Gen Virol 2011;92:909–918. [DOI] [PubMed] [Google Scholar]
- 9. Christiansen DH, et al. First field evidence of the evolution from a non-virulent HPR0 to a virulent HPR-deleted infectious salmon anaemia virus. J Gen Virol 2017;98:595–606. [DOI] [PubMed] [Google Scholar]
- 10. Clark K, et al. GenBank. Nucleic Acids Res 2016;44:D67–D72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clouthier SC, et al. Genomic organization of infectious salmon anaemia virus. J Gen Virol 2002;83:421–428. [DOI] [PubMed] [Google Scholar]
- 12. Cottet L, et al. Infectious salmon anemia virus—genetics and pathogenesis. Virus Res 2011;155:10–19. [DOI] [PubMed] [Google Scholar]
- 13. Delphino MKVC, et al. Bayesian analysis of diagnostic sensitivity and specificity for detecting infectious salmon anaemia virus (ISAV) using IFAT and real-time RT-PCR testing from laboratories in Atlantic Canada. Aquaculture 2023;563:739006. [Google Scholar]
- 14. Devold M, et al. Sequence analysis of the fusion protein gene from infectious salmon anemia virus isolates: evidence of recombination and reassortment. J Gen Virol 2006;87:2031–2040. [DOI] [PubMed] [Google Scholar]
- 15. Ditlecadet D, et al. First report of successful isolation of a HPR0-like variant of the infectious salmon anaemia virus (ISAV) using cell culture. J Fish Dis 2022;45:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evensen O, et al. A morphological study of the gross and light microscopic lesions of infectious anaemia in Atlantic salmon (Salmo salar). Res Vet Sci 1991;51:215–222. [DOI] [PubMed] [Google Scholar]
- 17. Fourrier M, et al. Deletions in the highly polymorphic region (HPR) of infectious salmon anaemia virus HPR0 haemagglutinin-esterase enhance viral fusion and influence the interaction with the fusion protein. J Gen Virol 2014;95:1015–1024. [DOI] [PubMed] [Google Scholar]
- 18. Gagné N, LeBlanc F. Overview of infectious salmon anaemia virus (ISAV) in Atlantic Canada and first report of an ISAV North American-HPR0 subtype. J Fish Dis 2018;41:421–430. [DOI] [PubMed] [Google Scholar]
- 19. Godoy MG, et al. First detection, isolation and molecular characterization of infectious salmon anaemia virus associated with clinical disease in farmed Atlantic salmon (Salmo salar) in Chile. BMC Vet Res 2008;4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen BH, et al. Gill metal binding and stress gene transcription in brown trout (Salmo trutta) exposed to metal environments: the effect of pre-exposure in natural populations. Environ Toxicol Chem 2007;26:944–953. [DOI] [PubMed] [Google Scholar]
- 21. Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kibenge FSB, et al. Isolation and identification of infectious salmon anaemia virus (ISAV) from Coho salmon in Chile. Dis Aquat Organ 2001;45:9–18. [DOI] [PubMed] [Google Scholar]
- 23. Kibenge FSB, et al. Infectious salmon anaemia virus (ISAV) ringtest: validation of the ISAV diagnostic process using virus-spiked fish tissues and ISAV TaqMan® real-time RT-PCR. J Aquac Res Dev 2011;2:110. [Google Scholar]
- 24. Kralik P, Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol 2017;8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LeBlanc F, et al. In vivo virulence and genomic comparison of infectious salmon anaemia virus isolates from Atlantic Canada. J Fish Dis 2018;41:1373–1384. [DOI] [PubMed] [Google Scholar]
- 26. Lovely JE, et al. First identification of infectious salmon anaemia virus in North America with haemorrhagic kidney syndrome. Dis Aquat Organ 1999;35:145–148. [DOI] [PubMed] [Google Scholar]
- 27. Madhun AS, et al. Prevalence and genotypes of infectious salmon anaemia virus (ISAV) in returning wild Atlantic salmon (Salmo salar L.) in northern Norway. J Fish Dis 2019;42:1217–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mérour E, et al. Completion of the full-length genome sequence of the infectious salmon anemia virus, an aquatic orthomyxovirus-like, and characterization of mAbs. J Gen Virol 2011;92:528–533. [DOI] [PubMed] [Google Scholar]
- 29. Mjaaland S, et al. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J Virol 1997;71:7681–7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moore LJ, et al. Characterisation of salmon and trout CD8α and CD8β. Mol Immunol 2005;42:1225–1234. [DOI] [PubMed] [Google Scholar]
- 31. Mullins JE, et al. Infectious salmon anaemia in salt water Atlantic salmon (Salmo salar L.) in New Brunswick, Canada. Bull Eur Ass Fish Pathol 1998;18:110–114. [Google Scholar]
- 32. Nylund A, et al. Transmission of infectious salmon anaemia virus (ISAV) in farmed populations of Atlantic salmon (Salmo salar). Arch Virol 2007;152:151–179. [DOI] [PubMed] [Google Scholar]
- 33. Nylund A, et al. Wild and farmed salmon (Salmo salar) as reservoirs for infectious salmon anaemia virus, and the importance of horizontal- and vertical transmission. PLoS One 2019;14:e0215478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rimstad E, et al. Infectious salmon anaemia. In: Woo PTK, Bruno DW, eds. Fish Diseases and Disorders. Vol. 3: Viral, Bacterial and Fungal Infections. 2nd ed. CABI, 2011:143–165. [Google Scholar]
- 35. Rimstad E, Markussen T. Infectious salmon anaemia virus—molecular biology and pathogenesis of the infection. J Appl Microbiol 2020;129:85–97. [DOI] [PubMed] [Google Scholar]
- 36. Rimstad E, Mjaaland S. Infectious salmon anaemia virus. APMIS 2002;110:273–282. [DOI] [PubMed] [Google Scholar]
- 37. Rodger HD, et al. Infectious salmon anaemia (ISA) in the United Kingdom. Bull Eur Ass Fish Pathol 1998;18:115–116. [Google Scholar]
- 38. Skonieczna K, et al. RNA isolation from bloodstains collected on FTA cards—application in clinical and forensic genetics. Arch Med Sadowej Kryminol 2016;66:244–254. [DOI] [PubMed] [Google Scholar]
- 39. Snow M, et al. Development, application and validation of a Taqman real-time RT-PCR assay for the detection of infectious salmon anaemia virus (ISAV) in Atlantic salmon (Salmo salar). Dev Biol (Basel) 2006;126:133–145. [PubMed] [Google Scholar]
- 40. Thorud K, Djupvik HO. Infectious anaemia in Atlantic salmon (Salmo salar L.). Bull Eur Ass Fish Pathol 1988;8:109–111. [Google Scholar]
- 41. Toennessen R, et al. Comparative aspects of infectious salmon anemia virus, an orthomyxovirus of fish, to influenza viruses. Indian J Microbiol 2009;49:308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weli SC, et al. Infectious salmon anaemia virus infection of Atlantic salmon gill epithelial cells. Virol J 2013;10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. World Organisation for Animal Health (WOAH). Manual of Diagnostic Tests for Aquatic Animals. 8th ed. WOAH, 2021. [Google Scholar]
- 44. Zou J, et al. Identification and bioactivities of IFN-γ in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J Immunol 2005;175:2484–2494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-vdi-10.1177_10406387231223290 for Rapid differentiation of infectious salmon anemia virus avirulent (HPR0) from virulent (HPRΔ) variants using multiplex RT-qPCR by Thomas F. Rounsville, Mark P. Polinski, Alyssa G. Marini, Sarah M. Turner, Niccolò Vendramin, Argelia Cuenca, Michael R. Pietrak, Brian C. Peterson and Deborah A. Bouchard in Journal of Veterinary Diagnostic Investigation