Abstract
It is widely assumed that a taxonomic core community emerges among microbial communities from similar habitats because similar environments select for the same taxa bearing the same traits. Yet, a core community itself is no indicator of selection because it may also arise from dispersal and neutral drift, i.e. by chance. Here, we hypothesize that a core community produced by either selection or chance processes should be distinguishable. While dispersal and drift should produce core communities with similar relative taxon abundances, especially when the proportional core community, i.e. the sum of the relative abundances of the core taxa, is large, selection may produce variable relative abundances. We analyzed the core community of 16S rRNA gene sequences of 193 microbial communities occurring in tiny water droplets enclosed in heavy oil from the Pitch Lake, Trinidad and Tobago. These communities revealed highly variable relative abundances along with a large proportional core community (68.0 ± 19.9%). A dispersal-drift null model predicted a negative relationship of proportional core community and compositional variability along a range of dispersal probabilities and was largely inconsistent with the observed data, suggesting a major role of selection for shaping the water droplet communities in the Pitch Lake.
Keywords: core microbiome, microbial community assembly, microbial islands, neutral processes, species composition, subsurface
Analysis of microbial communities in tiny water droplets enclosed in oil allows to infer underlying community assembly processes of the taxonomic core community.
Research of the past decade has shown that microbial life thrives in microliter-sized water droplets that are enclosed in heavy oil in the Pitch Lake, the world's largest natural asphalt lake in Trinidad and Tobago. These water droplets are densely populated with metabolically active, anaerobic microorganisms and originate from the oil reservoir in the deep subsurface (Meckenstock et al. 2014, Pannekens et al. 2020, 2021, Voskuhl et al. 2021). The microbial communities in the water droplets have exceptional ecological features, which make them interesting for addressing fundamental questions in ecology. Importantly, they share a similar origin and they have been exposed to similar and rather constant environmental conditions for a presumably long time. These features suggest that unidirectional selection pressure by the environment has played an important role in shaping these communities, while the enclosure in oil suggests that microbial dispersal has played a minor role. Microbial community assembly in the water droplet communities is thus likely to be less complex than in other systems, which can provide an important advancement in ecology if clear links between community-wide patterns and the underlying assembly processes can be detected.
In this study, we therefore made use of the water droplets to tackle the core community, a widely used concept in microbial ecology. The core community is a community-wide pattern, which is defined as the group of taxa that occurs in all or almost all microbial communities within a given data set (Hamady and Knight 2009, Shade and Handelsman 2012, Neu et al. 2021, Sharon et al. 2022, Custer et al. 2023). It is commonly considered to be formed by those taxa that are true residents of a certain habitat, i.e. the core community exists because similar environmental conditions at the sampling localities select for the same taxa bearing the same traits. In contrast, the variable part of a community may contain taxa that are functionally redundant or adapted to the very specific conditions of only one or a few of the sampling localities, as well as transient taxa that have ended up in a community by chance. The perception of core taxa as habitat natives is at the basis of a plethora of studies that identify microbial core taxa with the ultimate aim to better understand their ecological role in an environment. Taxonomic core communities have therefore been identified for a variety of habitats including soils (Gschwend et al. 2021), compost (Wang et al. 2020), soda lakes (Zorz et al. 2019), marine water bodies (Caporaso et al. 2012), wastewater (Palanisamy et al. 2021) and host-associated habitats such as plants (Lundberg et al. 2012, Zhou et al. 2019) and animals (Ainsworth et al. 2015, Henderson et al. 2015, Astudillo-Garcia et al. 2017, Hernandez-Agreda et al. 2017), including humans (Turnbaugh et al. 2009, Zaura et al. 2009, Caporaso et al. 2011, Tong et al. 2019).
Yet, a number of more recent studies have emphasized that a taxonomic core community per se is no indicator of selection because it can also emerge by chance. On the theoretical side, it has been shown that a core community arises in an agent-based computational neutral model for host-associated microbial communities (Zeng and Rodrigo 2018). Moreover, Sloan et al. (2006) developed an analytical neutral model specifically for microbial communities, which predicts that in neutral communities all taxa form an S-shaped abundance-occupancy relationship. By fitting this model to data, two recent studies demonstrated that the core taxa fell into the confidence interval of Sloan's neutral model, and pointed at the possibility of a neutral core community (Shade and Stopnisek 2019, Sieber et al. 2019). On the empirical side, two studies have provided evidence for a non-adaptive core community by showing that the core taxa were not adapted to the local environment but had been repeatedly introduced from the outside. The first of these studies demonstrated that the healthy human lung appears to consist largely of core and non-core taxa that are introduced by dispersal from the oral cavity rather than being true lung residents (Venkataraman et al. 2015). The second study demonstrated that caterpillars lack resident gut microorganisms, but contain microbial core and non-core gut taxa which are most likely transient because these taxa are typically associated with leaves and are thus food-derived (Hammer et al. 2017). Thus, both theoretical and empirical studies make clear that in certain systems, the idea of identifying core taxa in order to study their ecological meaning might be misleading.
Here, we hypothesize that core communities resulting from either selection or from dispersal and neutral drift may be distinguishable based on specific patterns. Dispersal and neutral drift should produce core and non-core taxa with rather similar relative abundances across samples, especially when dispersal rates are high and the resulting proportional core community is large (Mouquet and Loreau 2002, Cadotte 2006). We here define the proportional core community as the sum of the relative abundances of the core taxa, whereas the classical core community is often defined as the number of core taxa (Shade and Handelsman 2012). Selection, on the other side, may also produce core and non-core taxa with similar relative abundances, but this should only occur in the special case where selection forces a community to converge to a stable taxonomic composition (Coyte et al. 2015, Faust et al. 2015, Goldford et al. 2018). In many cases, selection can force microorganisms to fluctuate transiently or even continuously, either through periodic oscillations or chaotic dynamics, as has been shown for many microbial systems ranging from wastewater to human stool (Becks et al. 2005, Graham et al. 2007, Beninca et al. 2008, Ofiteru et al. 2010, Yurtsev et al. 2016, Faust et al. 2018, Liu et al. 2019). Thus, selection may often produce core taxa with variable relative abundances, even when the proportional core community is large.
In this study we investigated whether the core community contains such imprints of selection or dispersal and neutral drift with the help of 193 water microbial communities form the Pitch Lake. We analyzed the composition of 16S rRNA gene sequences and investigated the roles of dispersal, neutral drift and selection on the proportional core community and the compositional variability with the help of a computational null model.
Material and methods
Sampling, DNA-extraction, and sequencing
The procedures for oil and water droplet sampling, DNA-extraction and 16S rRNA gene library preparation have been described previously (Pannekens et al. 2020, Voskuhl et al. 2021). In short, oil was sampled from different locations on the Pitch Lake surface in Trinidad and Tobago in February 2017 and March 2018 and 2019 using sterile equipment, and shipped to Germany under nitrogen atmosphere. Because the Pitch Lake is dynamic, the exact locations where freshly seeping oil could be sampled differed from year to year (see Fig. S1). Single water droplets were isolated from oil by gentle pipetting and subjected to DNA-extraction using a 2-step lysis protocol (Pannekens et al. 2020). Further sample processing occurred in two technical replicates. To detect a broad range of bacteria and archaea we amplified the V3-V4 region of the 16S rRNA gene using the primers 341f and 805r (Takahashi et al. 2014) and prepared technical replicates for sequencing with Illumina MiSeq according to the Illumina 16S metagenomics sequencing library preparation guide (Part 15 044 223 Rev. B) (Pannekens et al. 2020). To control for potential contaminations during handling, negative controls were processed throughout the entire work flow, from DNA extraction over amplicon PCR and all purification steps until sequencing, for each batch of samples that was processed. The controls did not provide indication of important cross contamination or contamination by foreign sequences.
Processing of raw sequences
Forward reads were processed by the R package DADA2 version 1.22.0 (Callahan et al. 2016). Processing included removal of primers and trimming of reads using standard settings with length truncation at 240 bases, followed by sequencing error correction, denoising, chimera removal, and annotation using the SILVA 138.1 database version. Resulting sequences were amplicon sequence variants (ASV) with 100% sequence identity and formed the basis of further analysis. Based on visual inspection of the ASV-based rarefaction curves, technical replicates with more than 10 000 reads were further processed and merged by calculating mean relative ASV abundances. 13 samples were removed from the dataset because they contained more than 10% ASVs with presumably aerobic metabolism, indicating a possible contamination from the outside (Fig. S2). Classification of ASVs according to their oxygen tolerance was based on a self-designed database (Data S1). The final data set contained 193 water droplet communities, with 64 communities from 2017, 86 from 2018, and 43 from 2019. For some further analyses, ASVs obtained by DADA2 were clustered with 97% identity using the cluster command algorithm of mothur software (Schloss et al. 2009), delivering operational taxonomic units (OTUs).
qPCR and total community size
The determination of absolute number of 16S rRNA gene abundances in the water droplet communities by quantitative PCR (qPCR) has been described by Voskuhl et al. (2021), who applied the protocol of Takai and Horikoshi (2000) with minor modifications. In short, forward primer Uni340F (CCT ACG GGR BGC ASC AG), reverse primer Uni806R (GGA CTA CNN GGG TAT CTA AT) and the FAM-TAMRA-labelled Probe Uni516F (YCA GCM GCC GCG GTA AHA CVN RS) were used to amplify the V3-V4 region of the 16S rRNA genes of bacteria and archaea. qPCR runs were performed in duplicates on a C1000 Touch Thermal Cycler (CFX96™ Real-Time System, Bio-Rad Laboratories GmbH, Feldkirchen, Germany). Absolute 16S rRNA gene quantification in genes/µl was performed by comparing cycle quantification values (cq-values) to dsDNA-standards from Bacillus alkalidiazotrophicus strain MS 6 that were included in every qPCR run. The volume of each water droplet was estimated by converting its mass using a specific gravity of 1.0 g ml−1. Total size of each water droplet community was calculated as the product of droplet volume and concentration of 16S rRNA genes. Total community sizes were not corrected for differences in the numbers of 16S rRNA genes per cell between different microbial taxa.
Radiographic imaging of oil
To investigate the probability of collision and fusion of entire water droplets moving upwards through the oil we first measured size and distribution of the water droplets in the oil. Oil from different sampling sites on the Pitch Lake was filled into 15 cm long plastic cylinders with a diameter of 1.5 cm. Cylinders were sealed at both ends with hot glue, placed on plastic stands and stored at 4°C until analysis. The oil-filled columns were scanned with X-ray tomography (X-tek XCT 225, Nikon Metrology, Japan) at an energy of 90 kV, a beam current of 155 µA, and exposure time of 1 s and 2 frames per projection. Raw radiographic images were converted into 8-bit grey scale images with 12.5 µm spatial resolution using the software X-tec CT Pro. Converted images were then filtered with a median filter with a radius of 3 voxels, image contrast was improved using the CLAHE routine of Fiji software version 1.52i (Schindelin et al. 2012). Segmentation into water and oil phases were carried out with the hysteresis segmentation method (Vogel and Kretzschmar 1996), whereby lower and upper threshold levels were selected for each image individually. An additional median filter with a radius of 3 voxels was applied to the segmented images to remove small objects. Sizes of water inclusions were determined using the 3D MorphoLibJ plugin into ImageJ (Legland et al. 2016).
Determination of water content and density of oil
Water was separated from oil by transferring between 28 and 48 g of heavy oil sampled at the Pitch Lake in the years 2017 and 2018 into several 50 ml-Falcon-tubes and centrifuging them repeatedly for 1 h at 3214 x g and 4°C until no more water was extractable. The water had a lower density than the oil so that its volume was determined by pipetting it from the surface. Water content was determined in triplicate for each sampling year and eventually expressed as mean volume proportion (v/v). The density of the oil was determined by measuring the volume of 40 and 45 g oil in glass beakers.
Computational fluid-dynamics model for the movement of water droplets through oil
We used Ansys Fluent version 19.0 to simulate the movement of water droplets through oil in a computational fluid-dynamics model and to calculate the probability of water droplet collision and fusion. Since the water droplets are very small compared to the oil area, they were modelled as discrete particle phase. Droplet fusion was enabled using the collision and breakup functions embedded in the discrete particle model (DPM) (Taylor 1963, O'Rourke 1981). Droplets were introduced into the oil area using the injection function embedded in the software. It is expected that in the absence of external forces the droplets move upward with the terminal velocity of a rising bubble due to buoyancy, u:
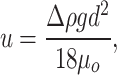 |
where Δρ is the difference between oil density, ρo, and water density, ρw, g is the gravitational acceleration, d is the initial diameter of the droplet, and µo is the viscosity of the oil. Two different cases were studied, namely (i) with uniform initial droplet sizes, and (ii), with distributed initial droplet sizes, whereby an exponential size distribution was assumed based on the radiographic measurements of droplet sizes (Pannekens et al. 2021). Simulations were carried out for a total water content of 0.1% (v/v), which was experimentally determined. The oil area was modelled as a 2D-geometry with dimensions of 0.2 m x 2.0 m and contained a uniform square mesh with 0.01 m mesh size for optimized calculations. The 2D area served as a cross-sectional view of a real 3D system. A small-scale 3D system was also tested for one scenario, where no significant difference was observed compared to the 2D system. The system was initialized with droplets distributed uniformly, and the upper boundary at 2.0 m height was set to constant pressure. Two transport scenarios were studied. In the conductive scenario, the oil phase was stationary, and droplet transport was controlled by gravity forces only. In the convective scenario, the oil phase entered the area at constant velocity through the lower boundary. The effect of turbulence was studied using the k-ε model (Launder and Spalding 1974), which allows considering buoyancy forces. Model parameters are summarized in Table S1.
Computer simulations of dispersal and neutral drift
Microbial dispersal may not only proceed through collisions of entire droplets but also through movements of individual cells between droplets, which may happen e.g. as long as droplets in the reservoir are connected. We investigated the possible influence of individual dispersal and neutral drift on microbial community composition with computer simulations using R 4.1.2 (Supplementary Code S1). We only considered sampling sites for which both qPCR and 16S rRNA gene data were available for a minimum of nine communities, resulting in 103 communities belonging to nine different sampling sites. The initial composition of each community was given by the measured ASV composition. Absolute community sizes as calculated from qPCR data remained constant throughout the simulations. Four outliers with excessively large community size were not investigated in the simulations.
During the simulations, individuals in a local community were sequentially removed and replaced by other individuals that were randomly picked from within the local community and that were allowed to double. These random death and birth events caused random fluctuations of the relative abundances in each community over time, i.e. they caused neutral drift (Hubbell 2001). An immigration event due to dispersal was simulated by replacing a randomly removed individual by an individual that was randomly chosen from the metacommunity, which was calculated by integrating relative taxon abundances across samples. Because microbial communities differed between sampling sites, we implemented distinct metacommunities for each site. We also investigated dispersal from one large metacommunity based on all samples. The dispersal probability defined for a simulation run determined the percentage of randomly removed individuals that were replaced by individuals from the metacommunity (Hubbell 2001). Dispersal was neutral when dispersal probabilities were equal for all taxa, or adaptive when certain taxa had higher dispersal probability than others.
Results
Collision probability of entire water droplets
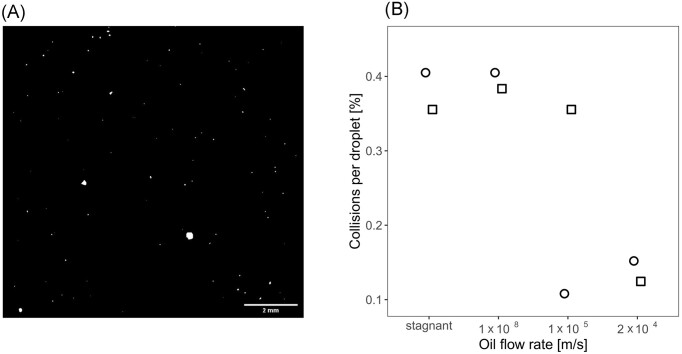
The water droplets in the Pitch Lake get transported by density-driven movement as well as advective movement of the oil from the deep oil reservoir to the surface of the Pitch Lake. Consequently, there is a chance that the taxonomic composition of the microbial communities in the water droplets is influenced by collision and fusion of entire droplets. Radiographic imaging of the oil in a squared column with an edge length of 11.25 mm and subsequent image segmentation revealed that water droplets were widely scattered and had volumes ranging from 0.001 nl to 2.33 µl diameter (Fig. 1A). We estimated the collision probability of these water droplets with the help of a computational fluid-dynamics model. Water droplets were modeled as discrete particles moving upward in a simple, two-dimensional area of 2 m length due to buoyancy and oil flow. No collision was observed under the assumption of a uniform droplet size and a water content of 0.1% as determined by measurement. Collisions increased when a range of droplet sizes was assumed. Larger droplets generally moved faster and merged with the smaller ones located at their upward trajectory, which was more pronounced for lower oil velocities where droplet upward movement was dominated by conduction (Movie S1). Overall, only 0.2% of the total volume of all droplets changed due to collisions, and collisions were particularly rare for larger droplets such as those that we have sampled in the Pitch Lake (Fig. S3). Collision probability was quantified by comparing the size distribution of droplets leaving the upper area boundary with the initial size distribution of the droplets when entering the lower area boundary. The maximum collision probability of 1.01 × 10−4 per droplet was observed in a laminar area with an oil flow velocity of 10−8 m/s. Collision probability increased linearly with oil area length increasing from 2 to 8 m (Fig. S4). Linear extrapolation of these results yielded a maximum collision probability of 6.16 × 10−3 per water droplet for the distance of 80 m between Pitch Lake bottom and surface (Kayukova et al. 2016) and 7.6% for the distance of 1500 m between oil reservoir and lake surface (Fig. 1B). Collision probability under laminar and turbulent oil flow was generally comparable, yet, a sudden drop of collision probability occurred when oil flow rates became similar to the terminal rising velocity of the droplets, which occurred at a slightly lower oil flow rate under laminar flow (around 1 × 10−5 m s−1) than turbulent flow (around 2 × 10−4 m s−1, Fig. 1B). The maximum collision probability estimated by this model was insufficient to cause significant changes in community composition, which we investigated by comparing the original data set with a modified data set, in which 7.6% of the water droplets had been merged randomly (Information S1). Based on the model, collisions can thus be considered rare and water droplets as physically isolated. The collision rate, i.e. the collision probability relative to the lifetime of the water droplet, along the entire reservoir-lake distance will be highest if oil rises fast through geological faults, but it will be very low if oil and water droplets are trapped in the sediment pores in the deep reservoir for long, possibly geological, time spans (Wilhelms et al. 2001).
Figure 1.
Distribution and collision probability of water droplets in Pitch Lake oil. (A) A cross-sectional segment of an oil column with dimensions of 11.25 x 11.25 mm. White objects represent water droplets, black background shows oil matrix. Water droplets may appear non-spherical because of the preparatory handling of the oil. (B) Probability of water droplet collisions along the vertical transect from the reservoir to the Pitch Lake surface, estimated by computer simulations. Collisions of water droplets were investigated in a fluid-dynamics model of water droplets rising through a 2 m long oil-area with different oil flow velocities and with laminar (circles) and turbulent (squares) oil flow. Collision probability was quantified by comparing the distribution of droplet sizes and their relative share in the total water volume at the beginning and end of the simulated 2 m-oil area and extrapolating the results to the presumed depth of the oil reservoir at 1500 m.
Microbial community composition and core community
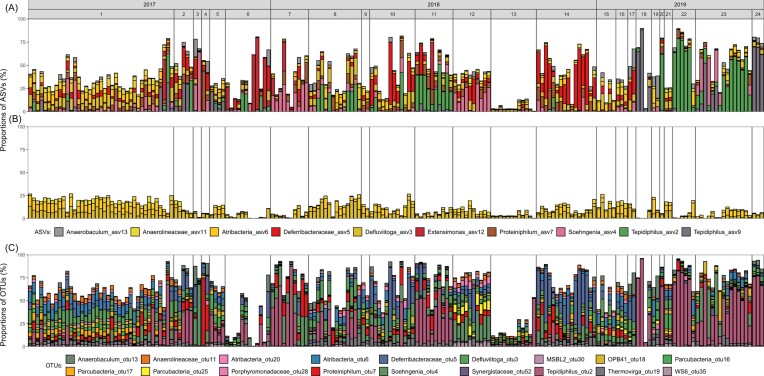
According to our hypothesis, communities primarily shaped by dispersal and neutral drift or by selection should be distinguishable based on two patterns, which are proportional core community and community variability. To determine these two patterns we analyzed the taxonomic composition of 193 water droplet communities that were sampled in three consecutive years (2017 to 2019) at different sites on the Pitch Lake. Communities consisted of prokaryotes typically found in anoxic and hydrocarbon-rich environments, as indicated by the taxonomic annotation of the 200 most abundant ASVs, which represented 89.0 ± 5.2% of the reads (median ± median absolute deviation, MAD) (Supplementary Data S1). Communities showed a high variability in composition, as illustrated by the relative abundances of the ten most abundant ASVs across all communities (Fig. 2A). Community composition was statistically related to sampling year and to a lesser extent also to sampling site, as tested by a two-sided W*d-Test, which is a Welch MANOVA robust to (Hamidi et al. 2019) (Supplementary Table S2). This relationship may indicate some degree of adaptation of the communities to the prevailing habitat conditions in each year, such as oil composition. Despite the high compositional variability, communities shared a large proportional core community. Three ASVs, annotated as Defluviitoga_ASV3, Atribacteria_ASV6, Anaerolinaceae_ASV11, where found in all 193 droplets and constituted 9.8 ± 9.2% of all reads within each community (median ± MAD; Fig. 2B). Because the calculation of the proportional core community is sensitive to several parameters, we considered different sequencing depths, sample sizes and levels of taxonomic resolution and occupancy, i.e. the minimum proportion of samples, in which core organisms need to occur (Neu et al. 2021; Supplementary Information S2). The proportional core community increased substantially when lowering sequence similarity from 100% to 97%, and occupancy from 100% to 95%, yielding an estimation of 68.0 ± 19.9% formed by 18 core taxa (Fig. 2C; see Supplementary Data S1 for taxonomic annotation). Further parameter changes caused only small increases of the proportional core community, showing that 68% is a robust estimation for the water droplet communities in the Pitch Lake (Supplementary Information S2, Supplementary Fig. S5). According to our hypothesis, a large proportional core community together with variable relative abundances indicates an important role of selection. This indication is supported by a BLAST alignment of the identified 18 core OTUs, which mostly revealed highest similarity to sequences in the NCBI-database previously detected in oil or anoxic habitats (Table 1). The large proportional core community means that communities consisted to a large part of the same taxa, which further implies that a major role of speciation in shaping the water droplet communities can be excluded, whereby we here consider speciation as a process that would lead to new 16S rRNA gene sequences with a sequence difference of > 3%.
Figure 2.
Taxonomic composition of microbial communities in water droplets enclosed in oil. Bars represent proportions of amplicon sequence variants (ASVs; A, B) or operational taxonomic units (OTUs; C) of individual water droplet communities, colors represent individual ASVs or OTUs. (A) Relative abundance of the ten most abundant ASVs across 193 droplet communities. (B) Core community with 100% sample occupancy and 100% sequence similarity (no clustering of ASVs into OTUs). (C) Core community with 95% sample occupancy and 97% sequence similarity (clustering of ASVs into OTUs). Numbers in top row show sampling years and sites.
Table 1.
Identification of 18 core OTUs based on sequence homology searches within NCBI's standard nucleotide (nr/nt) database using the nucleotide Basic Local Alignment Search Tool (BLASTn). The best match of the first three different studies appearing in the query hit list are shown.
| Name of core OTU | Phylum | Closest relative | Accession number | % identity | Source of sequence in ncbi database: | Reference |
|---|---|---|---|---|---|---|
| Tepidiphilus_otu1 | Proteobacteria | Uncultured Tepidiphilus sp. clone PAM1 | MK246951 | 99 | Enrichment culture from oil field produced water | Berdugo-Clavijo et al. 2019 |
| Uncultured Tepidiphilus sp. clone OTU4 | KP677516 | 99 | Thermophilic bioelectrical system inoculated with activated sludge | Zhang et al. 2015 | ||
| Uncultured bacterium clone MC3F-1 | GQ999964 | 99 | Thermophilic lignocellulose degrading consortium | Wongwilaiwalin et al. 2010 | ||
| Defluviitoga_otu2 | Thermotogota | Uncultured bacterium clone V4P-15 | JQ519713 | 100 | Water-flooded oil reservoir | Zhang et al. 2012 |
| Uncultured Thermotogae bacterium clone GBR520BAC95 | KX008288 | 100 | Methanogenic conversion of crude oil hydrocarbons in a bioreactor | |||
| Uncultured Thermotogaeceae bacterium clone YNB-12 | JF808036 | 98 | Indigenous oil microbial community producing methane | Kobayashi et al. 2012a | ||
| Soehngenia_otu3 | Firmicutes | Soehngenia sp. strain WJ1 | MK644260 | 100 | Sulfate-reducing strain from offshore high-temperature soured oilfield | |
| Uncultured Tissierella sp. clone 2a | MG802213 | 100 | Biological filter submerged in sewage sludge | |||
| Soehngenia saccharolytica strain BN-16 | MF188185 | 100 | Industrial sludge | |||
| Deferribacteraceae_otu4 | Deferribacterota | Uncultured Deferribacteraceae bacterium clone XJ6B | MH202726 | 100 | Production water from Xingjiang Kelamayi petroleum reservoir, China | |
| Uncultured Deferribacteres bacterium clone Z3ALLBAC43 | KX062831 | 100 | Mixed production waters of Shengli oil reservoir, China | |||
| Uncultured bacterium clone d5-27 | KT342257 | 100 | Oil field | |||
| JS1_otu5 | Caldatribacteriota | Uncultured candidate division JS1 bacterium clone D010011E02 | GU179739 | 100 | Oil well | |
| Uncultured candidate division JS1 bacterium clone NS2 | EU722240 | 100 | Production water from Alaskan mesothermic petroleum reservoir | Pham et al. 2009 | ||
| Uncultured bacterium clone 2b_83 738 | MG803490 | 99 | Biological filter submerged in sewage sludge | |||
| Proteiniphilum_otu6 | Bacteroidota | Uncultured bacterium clone V4P-27 | JQ519725 | 100 | Water-oil sample from oil production well | Zhang et al. 2012 |
| Bacterium enrichment culture clone ecb22 | HQ395216 | 100 | Oil reservoir fluid from biodegraded oil reservoir, Malaysia | |||
| Porphyromonadaceae bacterium enrichment culture clone B312134 | HQ133064 | 100 | Methanogenic hexadecane-degrading enrichment culture from crude oil-contaminated soil of Shengli oil field, China | Cheng et al. 2013 | ||
| Longilinea_otu7 | Chloroflexi | Uncultured bacterium | LR639805 | 100 | Wastewater treatment system | |
| Uncultured bacterium clone 2a | MG804725 | 100 | Sewage sludge, submerged biological filter | |||
| Uncultured bacterium clone | MG331445 | 100 | Anaerobic sludge | |||
| Acetomicrobium _otu8 | Synergistota | Uncultured Synergistaceae bacterium WD11 | LC378411 | 100 | Methanogenic communities derived from petroleum reservoirs | Kato et al. 2019 |
| Uncultured Acetomicrobium sp. Clone Z3C13 | KX063318 | 100 | C13 and C14 alkanes-degrading methanogenic enrichment culture | |||
| Uncultured Acetomicrobium sp. Clone XDZ3IFBAC52 | KU555244 | 100 | Thermophilic crude-oil degradation methanogenic consortium in bioreactor | |||
| Parcubacteria _otu9 | Patescibacteria | Uncultured bacterium clone MW-B43 | JQ088359 | 97 | Crude oil reservoir, Huabei oil field, China | Tang et al. 2012 |
| Uncultured bacterium clone bacOTU 229 | KR013524 | 88 | Anaerobic methanogenic reactor inoculated with sludge from mesophilic WWTP anaerobic reactor | |||
| Uncultured bacterium, isolate OTU 801 | LT624755 | 87 | Anaerobic digestions | |||
| Parcubacteria _otu10 | Patescibacteria | Uncultured bacterium clone MW-B43 | JQ088359 | 100 | Crude oil reservoir, Huabei Oilfield | Tang et al. 2012 |
| Uncultured bacterium clone B6a | HM245643 | 89 | Reduced pockmark sediment, Baltic Sea | Shubenkova et al. 2010 | ||
| Uncultured Microgenomates group bacterium clone OPd16 | AF047563 | 89 | Obsidian pool hot spring, Yellowstone National Park, USA | |||
| WCHB1-81_otu11 | Actinobacteriota | Uncultured bacterium | LR639718 | 96 | Wastewater treatment system | |
| Uncultured bacterium clone HL2 C08 | KC705364 | 96 | Hot lake hypersaline margin soil | Kilmer et al. 2014 | ||
| Uncultured bacterium clone HAW-RM37-2-b-650d-T | FN563186 | 96 | Mesophilic biogas digester | Krakat et al. 2011 | ||
| Thermovirga _otu12 | Synergistota | Uncultured Synergistaceae bacterium clone R2 S20 D119 F01 | MN414189 | 100 | Anaerobic granular sludge at high salinity | |
| Uncultured Synergistetes bacterium clone D021041F03 | GU180014 | 100 | Oil well, Alaska | |||
| Uncultured bacterium clone c49Bk26b | FJ941509 | 100 | Oil field production water, The Netherlands | van der Kraan et al. 2009 | ||
| JS1_otu13 | Caldatribacteriota | Uncultured bacterium glone GoM solidAsph Bac180 | LC424917 | 100 | Liquid and solid asphalt pieces from Chapopote asphalt volcano, Gulf of Mexico | |
| Uncultured candidate division JS1 bacterium clone GoM161 Bac81 | AM745161 | 100 | Marine sediments, Gulf of Mexico | Orcutt et al. 2010 | ||
| Uncultured bacterium clone SMI1-GC205-Bac2b | DQ521788 | 100 | Anaerobic methane-oxidizing community in hypersaline Gulf of Mexico sediments | Lloyd et al. 2006 | ||
| Parcubacteria _otu14 | Patescibacteria | Uncultured bacterium clone B6a | HM245643 | 87 | Reduced pockmark sediment, Baltic Sea | Shubenkova et al. 2010 |
| Uncultured Microgenomates group bacterium clone OPd16 | AF047563 | 87 | Obsidian pool hot spring, Yellowstone National Park, USA | |||
| Uncultured bacterium clone MW-B43 | JQ088359 | 86 | Crude oil reservoir, Huabei Oilfield | Tang et al. 2012 | ||
| Bacteroidales _otu15 | Bacteroidota | Porphyromonadaceae bacterium enrichment culture clone HB_96 | JN202698 | 100 | Methanogenic hexadecane-degrading consortium enriched with crude oil-contaminated soil | |
| Uncultured bacterium | LR639987 | 94 | Wastewater treatment system | |||
| Uncultured bacterium clone 46 783 | MF769183 | 94 | Biogas plant | |||
| MSBL2_otu16 | Cloacimonadota | Uncultured Spirochaetes bacterium clone NRB7 | HM041924 | 96 | Produced fluid from Niiboli oilfield, Japan | Kobayashi et al. 2012b |
| Uncultured bacterium clone PS3SGXI1245 | FJ469340 | 94 | Thermophilic microbial community, oil processing facility primary separator, USA | Duncan et al. 2009 | ||
| Uncultured spirochete clone C9001C_B13_4_C033 | AB645307 | 87 | Subseafloor sediments off Shimokita Peninsula | |||
| Dojkabacteria_otu17 | Patescibacteria | Uncultured bacterium | LR638767 | 100 | Wastewater treatment system | |
| Uncultured bacterium clone HAW-RM37-2-B-877d-A17 | FN563221 | 100 | Mesophilic biogas digester, anaerobic digestions | |||
| Uncultured bacterium clone bacOTU_303 | KR013597 | 100 | Anaerobic reactor inoculated with sludge coming from a mesophilic WWTP anaerobic reactor | Goux et al. 2015 | ||
| JGI-0000079-D21_otu18 | Synergistota | Uncultured bacterium | LR637332 | 100 | Wastewater treatment system | |
| Uncultured bacterium clone 39 666 | MF769100 | 100 | Biogas plant | |||
| Uncultured Synergistetes bacterium clone XDZ3IFBAC62 | JU555254 | 100 | Thermophilic crude-oil degrading methanogenic consortium in a simulated reactor |
Dispersal and neutral drift in the water droplet communities
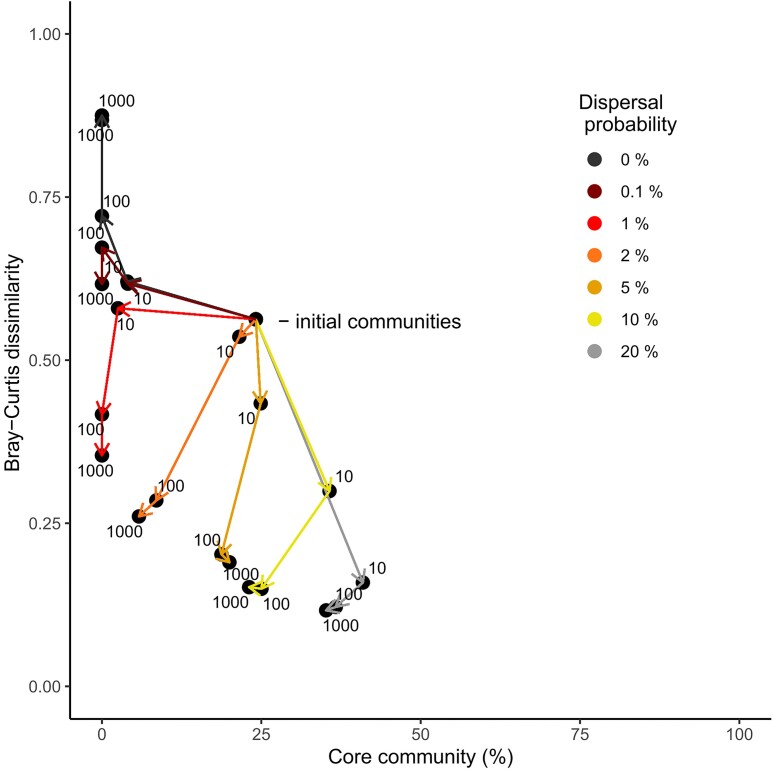
We used computer simulations to test our hypothesis that core communities driven by dispersal and neutral drift and by selection should be distinguishable based on the patterns proportional core community and community variability. We thereby used the computational model as null model, i.e. we investigated the effects of dispersal and neutral drift on said patterns, and compared the predicted community patterns with those observed in the water droplet communities (see e.g. Dornelas et al. 2006, Rosindell et al. 2012). As the effects of dispersal and drift depend on absolute community size (Hubbell 2001, Burns et al. 2016), we performed computer simulations using a subset of 103 droplet communities, for which qPCR data were available (Fig. S6). The initial compositions in the simulations were given by the real compositions observed in the Pitch Lake. These showed rather high community variability, measured as Bray-Curtis dissimilarity, and a high median proportional core community of 24% (Fig. 3), here calculated for an occupancy level of 95% and a sequence similarity of 100% (see Supplementary Information S2 and Fig. S5 for the sensitivity analysis of the proportional core community with respect to sequence similarity and other parameters). We ran simulations for a broad range of dispersal probabilities and for different number of generations. The divergence of Bray-Curtis dissimilarities and proportional core community from the original state was highly significant already after ten generations (Fig. 3; Tables S3, S4). Both patterns diverged further with increasing number of generations, the direction depending on the exact dispersal probability. Between 100 and 1000 generations, however, little additional changes were observed, indicating that both patterns had approached equilibrium values. At this stage compositional variability and proportional core community were negatively correlated along a gradient of dispersal probabilities. The correlation connected a state with high community variability and no core community at one end where there was no dispersal, with a state with low community variability and a large core community at the other end where there was much dispersal. Thus, a core community arising from dispersal and neutral drift had rather similar relative abundances as was stated in the hypothesis, especially when the proportional core community was large. The large proportional core community along with the high compositional variability observed in the Pitch Lake could not be maintained by any configuration of the neutral model, which indicates that selection has played a major role. The simulations results were qualitatively consistent when individuals dispersed from one common metacommunity rather than separate metacommunities at each sampling site, and when initial communities were given by random assemblies from the metacommunity (Figs S7, S8). The results were also qualitatively similar when dispersal probabilities were non-neutral but differed between taxa, which was implemented by giving a dispersal advantage of 10% to the ten most abundant taxa across all communities (Fig. S9).
Figure 3.
Development of average Bray-Curtis dissimilarity and proportional core community in computer simulations with stochastic drift and dispersal. Symbols represent the medians of Bray-Curtis-dissimilarities and relative core community sizes. Numbers indicate the length of the simulation run in terms of microbial generations, arrows indicate the direction of the development along time. Different colors indicate different dispersal probabilities. Note that Bray-Curtis dissimilarities were calculated separately for each sampling site because of separate metacommunities. Symbols show results of independent simulation runs.
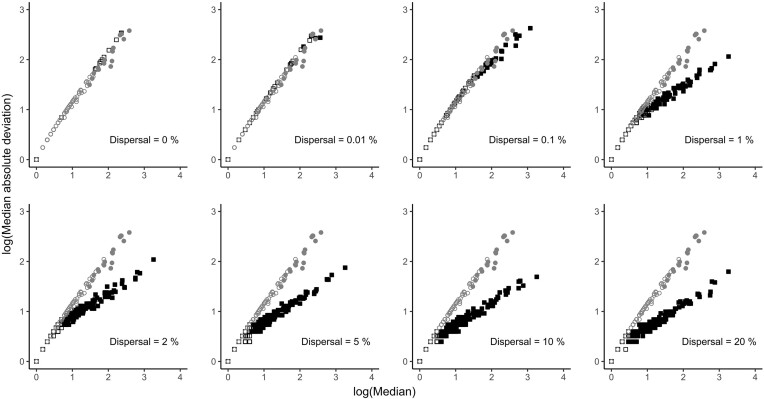
The divergence of the neutral model was also evident for each individual taxon. To facilitate inspection, Fig. 4 presents results from simulations based on one large metacommunity, but qualitatively consistent results are produced when assuming separate metacommunities for each sampling site (Supplementary Fig. S10). The taxa observed in the water droplet communities exhibited a steep relationship between median taxon relative abundance and the variability of relative abundance measured as MAD in a log-log plot. Observed core taxa were mostly, but not exclusively, those taxa with the highest median relative abundance (Fig. 4, grey circles). In the neutral model after 1000 generations with drift and without dispersal, all neutral taxa fell on the line formed by observed taxa, but there were zero neutral core taxa (Fig. 4, black squares). With increasing dispersal probability more and more neutral core taxa emerged falling on a separate line with an increasingly flat slope (Fig. 4). At a neutral dispersal probability of 1%, which produced a similarly sized core community as in the real water droplets (see Supplementary Fig. S7), neutral core taxa had clearly lower MADs than observed core taxa. Neutral dispersal together with neutral drift thus produced taxa with less variable relative abundances than was observed in the real communities, especially at moderate to high dispersal probabilities. This was particularly evident for abundant taxa such as the core taxa.
Figure 4.
Relationship between median relative taxon abundance and median absolute deviation (MAD) in observed and simulated communities. Grey symbols represent sequence data observed in the Pitch Lake, black symbols represent data resulting from simulations with neutral dispersal and drift. Filled symbols show taxa belonging to a core community with an occupancy level of 90%, open symbols show non-core taxa. Median and MAD are calculated across 103 water droplet communities, taxa represent amplicon sequence variants. Simulated data were produced assuming one large metacommunity for all communities.
Discussion
Our study shows that the large core community observed in the water droplet communities is an indicator of selection, because dispersal and neutral drift produced a similar large core community only when dispersal probability was high, leading to much more homogenous relative abundances than observed in the Pitch Lake. This implies that the proportional core community in general can help to infer underlying community assembly process.
Inference of community assembly processes in a transient state
In this study, we made inferences about the underlying community assembly processes based on the comparison of observed and simulated water droplet communities that have experienced drift and dispersal for ten microbial generations. Thus, we took into account the possibility that the observed water droplet communities did not represent final equilibrium outcomes but transient states. For the inferences to be valid, it is necessary to have an approximate idea about the age of the water droplets in terms of number of microbial generations. Pannekens et al. (2021) used anaerobic microcosms filled with equal amounts of Pitch Lake oil and artificial medium mimicking the chemical composition of the droplet water to measure growth and respiration rates over three years. In these experiments, microorganisms from the water droplets were able to enter the water phase from the oil because the water droplets, which are lighter than the heavy oil, rose to the surface of the oil phase where they came in contact with the overlaying water. The microbial communities in these microcosms showed rather slow exponential growth rates between 0.002–0.0039 day−1 as calculated from the cell densities, i.e. they had generation times between 38 and 82 days. Assuming this to be representative for generation times within in the Pitch Lake, a period of 10 microbial generations as investigated in the neutral model would correspond to 1.0–2.2 years. The water droplets are probably much older than that. They are known to be made of formation water (Meckenstock et al. 2014), and empirical evidence suggests that formation water has been populated by microorganisms already before or during genesis of the oil reservoir (Wilhelms et al. 2001, Head et al. 2003). It is conceivable that the inclusion of distinct water droplets in the interstitial pores of the deep subsurface is a very slow process, and that the water droplets get trapped there for geological time periods before they detach and rise to the surface through the geological fault producing the oil seep of the Pitch Lake. Thus, although direct proof is missing, there are strong reasons to believe the water droplet communities are (much) older than 10 microbial generations, meaning that the inferences from the model appear robust.
Dispersal, drift, and selection in the water droplet communities
Because the statistical patterns in the water droplet communities and the null model were largely inconsistent, we consider it unlikely that dispersal and neutral drift have significantly shaped the assembly of these communities. There are two alternative explanations. One possibility is that microbial community assembly was primarily driven by selection. The fact that the core community represented a large share of the individuals in each community but only a small share of the taxa indicates that only few taxa became established successfully in these habitats, either because of strong environmental filtering, i.e. strong selection pressure imposed by abiotic conditions, or a combination of environmental filtering and biotic interactions (Kraft et al. 2014, Aguilar-Trigueros et al. 2017, Cadotte and Tucker 2017). Strong environmental filtering is conceivable as the Pitch Lake represents an extreme environment, for which consistent dominance of only a few taxa is typical (Schulze-Makuch et al. 2011, Shu and Huang 2022). The fact that the relative abundances of the core taxa were variable may have resulted from variable selection due to small-scale environmental heterogeneity or due to biotic interactions. In a recent analysis of 43 water droplets from the Pitch Lake we found that 20.4% of the variability in community composition could be explained by local differences in the concentrations of chloride, sulfate, and potassium (Voskuhl et al. 2021). The so far unexplained 79.6% of the variability may have been caused by local differences of other, unmeasured environmental variables. The unexplained part may also have been caused by biotic interactions and may reflect multiple stable states or internally generated fluctuations of taxa. Such outcomes of e.g. competition, predation, or cross-feeding may occur even when the local environmental conditions are exactly the same and constant over time (Fussmann et al. 2000, Becks et al. 2005, Beninca et al. 2008, Sun et al. 2019). The only prerequisite for such internally generated variability would be minimal differences in community composition arising in the moment of water droplet formation (Huisman and Weissing 1999, 2001).
Another possibility is that microbial community assembly was driven by a combination of selection, dispersal and neutral drift. For instance, low dispersal rates and neutral drift may generate a noise level in the relative abundances sufficient to cause large community divergence due to biotic interactions. It might also be that low dispersal rates and neutral drift acted on top of the outcome of selection and have jumbled the relative taxon abundances, which would otherwise have been more similar due to selection-driven stable coexistence. Selection may also have created variability on top of a large core community caused by strong dispersal, though the possibilities for dispersal in the subsurface are in general very low (Louca 2022). Currently, we cannot make detailed inferences about microbial community assembly in the water droplets, but we can conclude that selection played a strong role because divergence of the neutral model was fast and widespread.
Dispersal probability of microbial cells in oil
The results of the null model suggest that the water droplet communities have not been significantly influenced by dispersal. Indeed, also from a biological point of view dispersal due to movement of individual cells through the oil phase is not very likely because the oil is toxic and has a very low water activity, which is at or below the border for microbial life (Schulze-Makuch et al. 2011). Dispersal might occur in the deep reservoir before water droplet detachment, if the reservoir is water-wet, i.e. if a thin water film covers the sediment grains and connects the water filling the pore spaces. In most reservoirs, however, oil is the continuous fluid phase filling the pore volume with discontinuous water filling only a smaller portion of the interstitial pores (Head et al. 2003). Moreover, Voskuhl et al. (2021, 2022) revealed strong variability in community composition as well as in ion composition in the water droplets, which by itself provides further evidence for considerable limitation of dispersal as well as diffusion. Together, this indicates that dispersal has had little influence on microbial community composition in the water droplets.
Incorporating the core community into studies of microbial community assembly
Adopting the concept of Vellend (2010, 2020) that all possible community assembly processes can be assigned to one of the four high-level processes selection, dispersal, neutral drift and speciation, we conclude that the water droplet communities were mainly influenced by selection. This conclusion results from an approach, which focuses on the relationship between community variability and proportional core community, thereby emphasizing the importance of the dominant taxa in the communities. In our study, the core taxa were only 18 OTUs out of a total of 37 343, but they represented on average 68% of all sequences in each community, a wide majority. Our conclusion appears robust given that the deviation of the null model became evident already at a young droplet age of only 10 microbial generations. Thus, even if dispersal of individuals happened in the reservoir while droplets were connected, a relatively short time of physical isolation appeared to be sufficient for selection-driven changes to become evident. It is interesting to note that a different conclusion may be obtained when fitting Sloan's neutral model to data, which focuses on different patterns, namely the relationship between occurrence and mean relative taxon abundance, and which puts equal weight to all taxa in the Pitch Lake water droplets, also the many rare ones (Sloan et al. 2006). The model explains 47.9% of the observed data (R²-value), and about half of the core taxa fall inside the 95% confidence interval and about half fall outside (Fig. S11). Although being no prove of neutrality (Sieber et al. 2019, Leroi et al. 2020, 2021), on may be tempted to interprete the degree of model fit as percentage measure of the relative importance of neutral processes, and those taxa within the confidence interval as neutral. The model of Sloan has been shown to be very insightful in comparative studies, where changes in the model fit matched differences in environmental conditions or experimental settings that could be correlated with dispersal ability (e.g. Burns et al. 2016, Sieber et al. 2019). Yet, as has been discussed elsewhere (McGill 2003, McGill et al. 2006) in our case a conclusion based on a single curve fitting would be rather weak model test and would have to be taken with care. Instead, our approach, which confronts the observations with various realizations of a null model, illustrates that the consideration of additional patterns such as the proportional core community together with community variability can provide new insights into the underlying mechanisms of microbial community assembly.
Supplementary Material
Acknowledgements
We thank F. T. Mbow for help during sample collection. Funding was provided by the European Research Council (ERC)[grand number 666952–EcOILogy] and the German Research Foundation (DFG)[grand number BR 5493/1-1].
Contributor Information
Verena S Brauer, Aquatic Microbiology, Environmental Microbiology and Biotechnology, Faculty of Chemistry, University of Duisburg-Essen, 45141 Essen, Germany; Centre for Water and Environmental Research (ZWU), University of Duisburg-Essen, 45141 Essen, Germany.
Lisa Voskuhl, Aquatic Microbiology, Environmental Microbiology and Biotechnology, Faculty of Chemistry, University of Duisburg-Essen, 45141 Essen, Germany.
Sadjad Mohammadian, Aquatic Microbiology, Environmental Microbiology and Biotechnology, Faculty of Chemistry, University of Duisburg-Essen, 45141 Essen, Germany.
Mark Pannekens, Aquatic Microbiology, Environmental Microbiology and Biotechnology, Faculty of Chemistry, University of Duisburg-Essen, 45141 Essen, Germany; IWW Water Center, 45476 Mülheim an der Ruhr, Germany.
Shirin Haque, Department of Physics, Faculty of Science and Technology, The University of the West Indies, St. Augustine, Trinidad and Tobago.
Rainer U Meckenstock, Aquatic Microbiology, Environmental Microbiology and Biotechnology, Faculty of Chemistry, University of Duisburg-Essen, 45141 Essen, Germany; Centre for Water and Environmental Research (ZWU), University of Duisburg-Essen, 45141 Essen, Germany.
Author contributions
Verena S. Brauer (Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing), Lisa Voskuhl (Investigation, Methodology, Writing – original draft, Writing – review & editing), Sadjad Mohammadian (Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing), Mark Pannekens (Investigation, Methodology, Writing – original draft), Shirin Haque (Resources, Writing – original draft, Writing – review & editing), and Rainer U. Meckenstock (Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Writing – original draft, Writing – review & editing)
Competing interests
The authors declare no competing interests.
Data and materials availability
Raw sequence data are deposited at the NCBI database in Bioprojects PRJNA1010525 and PRJNA642708.
References
- Aguilar-Trigueros CA, Rillig MC, Ballhausen MB. Environmental filtering is a relic. A response to Cadotte and Tucker. Trends Ecol Evol. 2017;32:882–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth TD, Krause L, Bridge T et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9:2261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astudillo-Garcia C, Bell JJ, Webster NS et al. Evaluating the core microbiota in complex communities: a systematic investigation. Environ Microbiol. 2017;19:1450–62. [DOI] [PubMed] [Google Scholar]
- Becks L, Hilker FM, Malchow H et al. Experimental demonstration of chaos in a microbial food web. Nature. 2005;435:1226–9. [DOI] [PubMed] [Google Scholar]
- Beninca E, Huisman J, Heerkloss R et al. Chaos in a long-term experiment with a plankton community. Nature. 2008;451:822–5. [DOI] [PubMed] [Google Scholar]
- Berdugo-Clavijo C, Sen A, Seyyedi M et al. High temperature utilization of PAM and HPAM by microbial communities enriched from oilfield produced water and activated sludge. AMB Expr. 2019;9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, Stephens WZ, Stagaman K et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016;10:655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadotte MW, Tucker CM. Should environmental filtering be abandoned?. Trends Ecol Evol. 2017;32:429–37. [DOI] [PubMed] [Google Scholar]
- Cadotte MW. Dispersal and species diversity: a meta-analysis. Am Nat. 2006;167:913–24. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D et al. The Western English Channel contains a persistent microbial seed bank. ISME J. 2012;6:1089–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Rui J, Li Q et al. Enrichment and dynamics of novel syntrophs in a methanogenic hexadecane-degrading culture from a Chinese oilfield. FEMS Microbiol Ecol. 2013;83:757–66. [DOI] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–6. [DOI] [PubMed] [Google Scholar]
- Custer GF, Gans M, van Diepen LTA et al. Comparative analysis of core microbiome assignments: implications for ecological synthesis. Msystems. 2023;8:e01066–01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas M, Connolly S, Hughes T. Coral reef diversity refutes the neutral theory of biodiversity. Nature. 2006;440:80–2. [DOI] [PubMed] [Google Scholar]
- Duncan KE, Gieg LM, Parisi VA et al. Biocorrosive thermophilic microbial communities in Alaskan North Slope oil facilities. Environ Sci Technol. 2009;43:7977–84. [DOI] [PubMed] [Google Scholar]
- Faust K, Bauchinger F, Laroche B et al. Signatures of ecological processes in microbial community time series. Microbiome. 2018;6:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Lahti L, Gonze D et al. Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr Opin Microbiol. 2015;25:56–66. [DOI] [PubMed] [Google Scholar]
- Fussmann GF, Ellner SP, Shertzer KW et al. Crossing the Hopf bifurcation in a live predator-prey system. Science. 2000;290:1358–60. [DOI] [PubMed] [Google Scholar]
- Goldford JE, Lu N, Bajic D et al. Emergent simplicity in microbial community assembly. Science. 2018;361:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goux X, Calusinska M, Lemaigre S et al. Microbial community dynamics in replicate anaerobic digesters exposed sequentially to increasing organic loading rate, acidosis, and process recovery. Biotechnol Biofuels. 2015;8:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DW, Knapp CW, Van Vleck ES et al. Experimental demonstration of chaotic instability in biological nitrification. ISME J. 2007;1:385–93. [DOI] [PubMed] [Google Scholar]
- Gschwend F, Hartmann M, Mayerhofer J et al. Site and land-use associations of soil bacteria and fungi define core and indicative taxa. FEMS Microbiol Ecol. 2021;97:fiab165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi B, Wallace K, Vasu C et al. W*d-test: robust distance-based multivariate analysis of variance. Microbiome. 2019;7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer TJ, Janzen DH, Hallwachs W et al. Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci USA. 2017;114:9641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head IM, Jones DM, Larter SR. Biological activity in the deep subsurface and the origin of heavy oil. Nature. 2003;426:344–52. [DOI] [PubMed] [Google Scholar]
- Henderson G, Cox F, Ganesh S et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Agreda A, Gates RD, Ainsworth TD. Defining the core microbiome in corals' microbial soup. Trends Microbiol. 2017;25:125–40. [DOI] [PubMed] [Google Scholar]
- Hubbell SP. The Neutral Theory of Biodiversity and Biogeography. Princeton, NJ: Princeton University Press, 2001. [Google Scholar]
- Huisman J, Weissing FJ. Biodiversity of plankton by species oscillation and chaos. Nature. 1999;402:407–10. [Google Scholar]
- Huisman J, Weissing FJ. Fundamental unpredictability in multispecies competition. Am Nat. 2001;157:488–94. [DOI] [PubMed] [Google Scholar]
- Kato S, Wada K, Kitagawa W et al. Conductive iron oxides promote methanogenic acetate degradation by microbial communities in a high-temperature petroleum reservoir. Microbes Environ. 2019;34:95–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayukova GP, Uspensky BV, Abdrafikova IM et al. Characteristic features of the hydrocarbon composition of Spiridonovskoe (Tatarstan) and Pitch Lake (Trinidad and Tobago) asphaltites. Pet Chem. 2016;56:572–9. [Google Scholar]
- Kilmer BR, Eberl TC, Cunderla B et al. Molecular and phenetic characterization of the bacterial assemblage of Hot Lake, WA, an environment with high concentrations of magnesium sulphate, and its relevance to Mars. Int J Astrobiol. 2014;13:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Endo K, Sakata S et al. Phylogenetic diversity of microbial communities associated with the crude-oil, large-insoluble-particle and formation-water components of the reservoir fluid from a non-flooded high-temperature petroleum reservoir. J Biosci Bioeng. 2012a;113:204–10. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kawaguchi H, Endo K et al. Analysis of methane production by microorganisms indigenous to a depleted oil reservoir for application in Microbial Enhanced Oil Recovery. J Biosci Bioeng. 2012b;113:84–7. [DOI] [PubMed] [Google Scholar]
- Kraft NJB, Adler PB, Godoy O et al. Community assembly, coexistence and the environmental filtering metaphor. Funct Ecol. 2014;29:592–9. [Google Scholar]
- Krakat N, Schmidt S, Scherer P. Potential impact of process parameters upon the bacterial diversity in the mesophilic anaerobic digestion of beet silage. Bioresour Technol. 2011;102:5692–701. [DOI] [PubMed] [Google Scholar]
- Launder BE, Spalding DB. The numerical computation of turbulent flows. Comput Meth Appl Mech Eng. 1974;3:269–89. [Google Scholar]
- Legland D, Arganda-Carreras I, Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–4. [DOI] [PubMed] [Google Scholar]
- Leroi AM, Lambert B, Rosindell J et al. Neutral syndrome. Nat Hum Behav. 2020;4:780–90. [DOI] [PubMed] [Google Scholar]
- Leroi AM, Lambert B, Rosindell J et al. Neutral theory is a tool that should be wielded with care. Nat Hum Behav. 2021;5:809. [DOI] [PubMed] [Google Scholar]
- Liu Z, Cichocki N, Hubschmann T et al. Neutral mechanisms and niche differentiation in steady-state insular microbial communities revealed by single cell analysis. Environ Microbiol. 2019;21:164–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Lapham L, Teske A. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl Environ Microbiol. 2006;72:7218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S. The rates of global bacterial and archaeal dispersal. ISME J. 2022;16:159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill B, Maurer BA, Weiser MD. Empirical evaluation of neutral theory. Ecology. 2006;87:1411–23. [DOI] [PubMed] [Google Scholar]
- McGill B. Strong and weak tests of macroecological theory. Oikos. 2003;102:679–85. [Google Scholar]
- Meckenstock RU, von Netzer F, Stumpp C et al. Water droplets in oil are microhabitats for microbial life. Science. 2014;345:673–6. [DOI] [PubMed] [Google Scholar]
- Mouquet N, Loreau M. Coexistence in metacommunities: the regional similarity hypothesis. Am Nat. 2002;159:420–6. [DOI] [PubMed] [Google Scholar]
- Neu AT, Allen EE, Roy K. Defining and quantifying the core microbiome: Challenges and prospects. Proc Natl Acad Sci USA. 2021;118:e2104429118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke PJ. Collective Drop Effects on Vaporizing Liquid Sprays. Thesis, NM (USA): Los Alamos National Lab, 1981. [Google Scholar]
- Ofiteru ID, Lunn M, Curtis TP et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA. 2010;107:15345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt BN, Joye SB, Kleindienst S et al. Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep-Sea Res II. 2010;2008–21.
- Palanisamy V, Gajendiran V, Mani K. Meta-analysis to identify the core microbiome in diverse wastewater. Int J Environ Sci Technol. 2021;19:5079–96. [Google Scholar]
- Pannekens M, Voskuhl L, Meier A et al. Densely populated water droplets in heavy-oil seeps. Appl Environ Microb. 2020;86:e00164–00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannekens M, Voskuhl L, Mohammadian S et al. Microbial degradation rates of natural bitumen. Environ Sci Technol. 2021;55:8700–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VD, Hnatow LL, Zhang S et al. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol. 2009;11:176–87. [DOI] [PubMed] [Google Scholar]
- Rosindell J, Hubbell SP, He F et al. The case for the ecological neutral theory. Trends Ecol Evol. 2012;27:203–8. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Makuch D, Haque S, de Sousa Antonio MR et al. Microbial life in a liquid asphalt desert. Astrobiology. 2011;11:241–58. [DOI] [PubMed] [Google Scholar]
- Shade A, Handelsman J. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol. 2012;14:4–12. [DOI] [PubMed] [Google Scholar]
- Shade A, Stopnisek N. Abundance-occupancy distributions to prioritize plant core microbiome membership. Curr Opin Microbiol. 2019;49:50–8. [DOI] [PubMed] [Google Scholar]
- Sharon I, Quijada NM, Pasolli E et al. The core Human microbiome: does it exist and how can we find it? A Critical Review of the Concept. Nutrients. 2022;14:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu WS, Huang LN. Microbial diversity in extreme environments. Nat Rev Micro. 2022;20:219235. [DOI] [PubMed] [Google Scholar]
- Shubenkova OV, Likhoshvai AV, Kanapatskii TA et al. Microbial community of reduced pockmark sediments (Gdansk Deep, Baltic Sea). Microbiol. 2010;79:799–808. [PubMed] [Google Scholar]
- Sieber M, Pita L, Weiland-Brauer N et al. Neutrality in the metaorganism. PLoS Biol. 2019;17:e3000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S et al. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–40. [DOI] [PubMed] [Google Scholar]
- Sun Z, Koffel T, Stump SM et al. Microbial cross-feeding promotes multiple stable states and species coexistence, but also susceptibility to cheaters. J Theor Biol. 2019;465:63–77. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Tomita J, Nishioka K et al. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS One. 2014;9:e105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Horikoshi K. Rapid detection and quanitification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microb. 2000;66:5066–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YQ, Li Y, Zhao JY et al. Microbial communities in long-term, water-flooded petroleum reservoirs with different in situ temperatures in the Huabei Oilfield, China, PLoS ONE. 2012;7:e33535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GI. The shape and acceleration of a drop in a high speed air stream. In: Batchelor GK (ed.), The Scientific Papers of G I Taylor, Vol. 3. Cambridge: Cambridge University Press, 1963, 457–64. [Google Scholar]
- Tong X, Leung MHY, Wilkins D et al. Neutral processes drive seasonal assembly of the skin mycobiome. Msystems. 2019;4:e00004–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kraan GM, Bruining J, Lomans BP et al. Microbial diversity of an oil-water processing site and its associated oil field: the possible role of microorganisms as information carriers from oil-associated environments. FEMS Microbiol Ecol. 2009;71:428–43. [DOI] [PubMed] [Google Scholar]
- Vellend M. Conceptual synthesis in community ecology. Q Rev Biol. 2010;85:183206. [DOI] [PubMed] [Google Scholar]
- Vellend M. The Theory of Ecological Communities. Princeton: Princeton University Press, 2020. [Google Scholar]
- Venkataraman A, Bassis CM, Beck JM et al. Application of a neutral community model to assess structuring of the human lung microbiome. mBio. 2015;6:e02284–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ, Kretzschmar A. Topological characterization of pore space in soil—sample preparation and digital image-processing. Geoderma. 1996;73:23–38. [Google Scholar]
- Voskuhl L, Akbari A, Muller H et al. Indigenous microbial communities in heavy oil show a threshold response to salinity. FEMS Microbiol Ecol. 2021;97:fiab157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuhl L, Brusilova D, Brauer VS et al. Inhibition of sulfate-reducing bacteria with formate. FEMS Microbiol Ecol. 2022;98:fiac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gong J, Li J et al. Insights into bacterial diversity in compost: core microbiome and prevalend of potential pathogenic bacteria. Sci Total Environ. 2020;718:137304. [DOI] [PubMed] [Google Scholar]
- Wilhelms A, Larter SR, Head I et al. Biodegradation of oil in uplifted basins prevented by deep-burial sterilization. Nature. 2001;411:1034–7. [DOI] [PubMed] [Google Scholar]
- Wongwilaiwalin S, Rattanachomsri U, Laothanachareon T et al. Analysis of a thermophilic lignocellulose degrading microbial consortium and multi-species lignocellulolytic enzyme system. Enzyme Microb Technol. 2010;47:283–90. [Google Scholar]
- Yurtsev EA, Conwill A, Gore J. Oscillatory dynamics in a bacterial cross-protection mutualism. Proc Natl Acad Sci USA. 2016;113:6236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaura E, Keijser BJ, Huse SM et al. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Rodrigo A. Neutral models of short-term microbiome dynamics with host subpopulation structure and migration limitation. Microbiome. 2018;6:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, She YH, Chai LJ et al. Microbial diversity in long-term water-flooded oil reservoirs with different in situ temperatures in China. Sci Rep. 2010;2:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shen D, Feng H et al. Cooperative role of electrical stimulation on microbial metabolism and selection of thermophilic communities for p-fluoronitrobenzene treatment. Bioresour Technol. 2015;189:23–9. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Liang B, Wang L-Y et al. Identify the core bacterial microbiome of hydrocarbon degradation and a shift of dominant methanogenesis pathways in oil an aqueous phases of petrolium reservoirs with different temperatures from China. Biogeosciences. 2019;16:4229–41. [Google Scholar]
- Zorz JK, Sharp C, Kleiner M et al. A shared core microbiome in soda lakes separated by large distances. Nat Commun. 2019;10:4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.