Abstract
Background:
Until recently most testing algorithms in the United States (US) utilized Western blot (WB) as the supplemental test. CDC has proposed an algorithm for HIV diagnosis which includes an initial screen with a Combo Antigen/Antibody 4th generation-immunoassay (IA), followed by an HIV-1/2 discriminatory IA of initially reactive-IA specimens. Discordant results in the proposed algorithm are resolved by nucleic acid-amplification testing (NAAT).
Objectives:
Evaluate the results obtained with the CDC proposed laboratory-based algorithm using specimens from men who have sex with men (MSM) obtained in five metropolitan statistical areas (MSAs).
Study design:
Specimens from 992 MSM from five MSAs participating in the CDC’s National HIV Behavioral Surveillance System in 2011 were tested at local facilities and CDC. The five MSAs utilized algorithms of various screening assays and specimen types, and WB as the supplemental test. At the CDC, serum/plasma specimens were screened with 4th generation-IA and the Multispot HIV-1/HIV-2 discriminatory assay was used as the supplemental test. NAAT was used to resolve discordant results and to further identify acute HIV infections from all screened-non-reactive missed by the proposed algorithm. Performance of the proposed algorithm was compared to site-specific WB-based algorithms.
Results:
The proposed algorithm detected 254 infections. The WB-based algorithms detected 19 fewer infections; 4 by oral fluid (OF) rapid testing and 15 by WB supplemental testing (12 OF and 3 blood). One acute infection was identified by NAAT from all screened-non-reactive specimens.
Conclusions:
The proposed algorithm identified more infections than the WB-based algorithms in a high-risk MSM population. OF testing was associated with most of the discordant results between algorithms. HIV testing with the proposed algorithm can increase diagnosis of infected individuals, including early infections.
Keywords: HIV-1, Diagnostics, Algorithms, Laboratory
1. Background
Since 1989, the recommended HIV testing algorithm has consisted of screening with an HIV antibody immunoassay (IA) followed by Western blot (WB) or immunofluorescence assay (IFA) to confirm repeatedly reactive specimens [1]. The supplemental/confirmatory assays use viral lysate antigens and are designed for IgG detection only (1st generation IA). The latest FDA-approved laboratory-based screening assays can detect HIV p24 Ag and IgM and IgG antibodies against HIV-1/2 (4th generation IA) and have better sensitivity than other generation IAs, including WB, during early HIV-1 infection [2–8]. In early stages of infection, from the acute period to the appearance of IgG, WB fails to confirm infection detected by nucleic acid amplification testing (NAAT) or p24 or containing only IgM antibodies. In early HIV-1 infections, reports indicate that NAAT is positive 26 days before the WB becomes positive and 4th generation IAs detect infection approximately 19 days before the WB becomes positive [2,6].
It is important to detect acute HIV infections as soon as possible when viral loads are known to be high and likelihood of transmission is increased [9–14]. In high-risk populations where the number of acute infections may be high, early detection would facilitate earlier initiation of care and potentially reduce HIV transmission. An alternative algorithm that involves screening with a sensitive 4th generation-IA, followed by HIV-1/HIV-2 antibody differentiation IA, has been proposed by the the Centers for Disease Control & Prevention [15–17]. Specimens reactive on a 4th generation IA are tested with a differentiation assay which has been shown to be more sensitive than WB during early HIV infection and allows discrimination of HIV-1 from HIV-2 infections [6,18,19]. Specimens testing negative or indeterminate (i.e., discordant results) in the differentiation assay are resolved by NAAT.
The proposed algorithm has detected more HIV-1 infections in different populations [6,20]. In addition, the sensitivity and specificity of the algorithm in established infections has been shown to be higher than 99% [6]. Initial screening with 3rd generation IA in the context of the proposed algorithm also showed high sensitivity and specificity in persons with established infections, blood donors, high-risk populations, and has allowed correct classification of specimens [21,22]. The current study was initiated to obtain additional performance data for the CDC proposed algorithm in high risk individuals.
National HIV Behavioral Surveillance (NHBS) is a behavioral surveillance system used to monitor HIV-related risk, testing, and prevention behaviors and HIV prevalence among populations at high risk for acquiring HIV in different cycles: men who have sex with men (MSM), injection-drug users, and heterosexuals at increased risk for HIV infection [23]. In the 2008 and 2011 cycles, NHBS enrolled only MSM in metropolitan statistical areas (MSA) using a venue-based, time-space sampling approach [24]. HIV testing conducted in 2008 and 2011 showed stable overall prevalence of 19% and 18% respectively [25,26]. In 2011, to improve HIV testing for future rounds of NHBS, a pilot was conducted in five MSAs (hereafter referred to as sites) with known high HIV prevalence that agree to participate in the study to evaluate the performance of the CDC proposed algorithm and to determine the feasibility of using blood-based testing in this type of survey.
2. Objective
This study used serum/plasma specimens collected from MSM during the 2011 NHBS cycle to evaluate the performance of the proposed laboratory-based algorithm and compared results to site-specific results that used WB as supplemental assay.
3. Study design
3.1. Specimen collection
In the 2011 cycle, MSM that agreed to participate were enrolled to collect specimens at five sites with historically high, but varying prevalence [24–26]. These sites were located in five states in different geographic areas. Table 1 describes the specimen types collected. At four sites, OF or whole blood was collected according to the package insert instructions for rapid testing with OraQuick Advance (OraSure Technologies, Inc., Bethlehem, PA). At these sites, whole blood, dried blood spot (DBS), or OF specimens were collected from individuals who screened preliminary positive and/or were self-reported HIV-positive per the site specific NHBS protocols and sent to a local laboratory for confirmation. At the remaining site, whole blood was collected in EDTA vacutainers and sent to a local laboratory for processing and testing. Informed consent was obtained for all HIV testing. If individuals consented for storage of samples for additional testing in the pilot study, blood was collected in EDTA vacutainers and shipped to CDC at ambient temperature for processing within 48 hours (n = 304) or sent to a local laboratory for processing (n = 688). Remnant frozen specimens from local laboratories were also sent to CDC for testing. The sample size achieved at each site varied due to the timing of the implementation of the pilot, availability of a phlebotomist at the venue, and NHBS participants consent to enroll in the study.
Table 1.
HIV testing algorithms.
| Sample type for rapid test | Initial rapid test screening | Sample type sent to the laboratory | Screening tests | Supplemental tests | |
|---|---|---|---|---|---|
| Site 1 | Oral fluid | OraQuick Advancea | Whole blood | GS HIV-1/2 Plus Ob | Bio-Rad HIV-1 WB |
| Site 2 | Oral fluid | OraQuick Advancea | Oral fluid | Not performed | OraSure HIV-1 WB |
| Site 3 | Whole blood | OraQuick Advancea | Dried blood spot | GS HIV-1/2 Plus Oc | Bio-Rad HIV-1 WB |
| Site 4 | Oral fluid | OraQuick Advancea | Oral fluid | GS HIV-1/2 Plus Oc | OraSure HIV-1 WB |
| Site 5 | Not performed | Whole blood | GS HIV-1/2 Plus Ob | Bio-Rad HIV-1 WB | |
| CDC | Whole blood or | GS HIV-1/2 Combo Ag/Abb | Multispot | ||
| frozen plasma | HIV-1/HIV-2b | ||||
| Aptima NAATd |
OraSure Technologies.
BioRad Laboratories.
Validated protocol in Bio-Rad assay.
Gen-Probe.
HIV testing was offered to all NHBS participants and enrollment was anonymous so each individual was assigned a unique study identifier. Rapid tests results were returned to the participants at the testing site. Participants were able to obtain confirmatory results using their unique study identifier and a designated contact. IRB approval that included consent for additional testing and storage was obtained by each of the five sites that participated in the pilot study. Information collected during the NHBS interviews about self-reported HIV status and current antiretroviral (ARV) therapy use were considered during interpretation of site-specific results. Self-reported HIV-positive (SRP) individuals were considered positive.
3.2. HIV testing
Each NHBS site performed a different diagnostic algorithm (Table 1). Briefly, preliminary positive or SRP participants were confirmed using a WB-based algorithm with either whole blood (2 sites), DBS (1 site), or OF (2 sites). Two laboratories used a validated protocol for screening with DBS and OF with the GS HIV-1/2 Plus O (Bio-Rad Laboratory, Redmond, WA) that showed comparable results to the original protocols. All other tests were performed as indicated in the manufacturer’s package insert.
To evaluate the proposed laboratory algorithm at CDC, 992 serum/plasma specimens were initially tested with GS HIV-1/2 Combo Ag/Ab IA (GS Combo; Bio-Rad Laboratories, Redmond, WA) as indicated in the package insert. IA-repeatedly reactive specimens were tested with Multispot HIV-1/HIV-2 rapid test (Multispot; Bio-Rad Laboratories, Redmond, WA) as supplemental test (Table 1). Specimens that were repeatedly reactive on the 4th generation IA GS Combo and non-reactive on the supplemental test were subjected to NAAT with APTIMA HIV-1 RNA Qualitative assay (Gen-Probe, Inc., San Diego, CA). In order to identify acute HIV-1 infections potentially missed by the proposed algorithm, all available specimens at CDC which screened-non-reactive were subjected to NAAT. All tests were performed according to the manufacturer’s package insert. Plasma specimens from individuals that were identified at the sites as negative using OF on the Oraquick Advance Rapid HIV-1/2 antibody test (OraQuick, OraSure Technologies, Inc., Bethlehem, PA) were tested on OraQuick at CDC.
3.3. Algorithm performance
After testing at CDC was completed, the final interpretation of HIV results from the testing performed at the local laboratories and CDC were compared overall and by site. Differences in the number of positive results obtained with the proposed laboratory-based algorithm and the site-specific WB-based algorithms were analyzed using the McNemar’s test with one degree of freedom and continuity correction (95% confidence interval) when the sample size was >50 pairs. Participant self-reported HIV status and antiretroviral (ARV) use were considered when interpreting the results.
4. Results
4.1. Test results from CDC laboratory testing
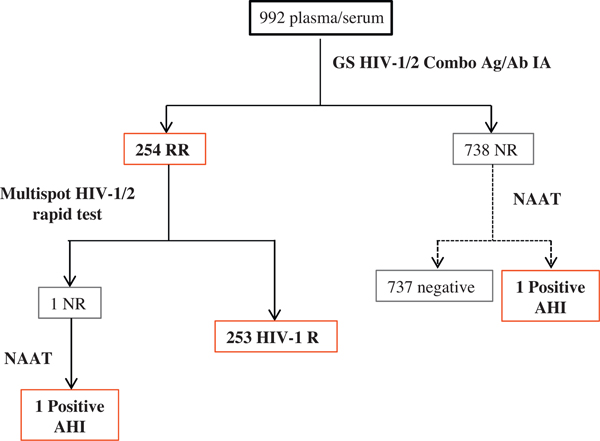
A total of 992 serum/plasma specimens were tested at CDC (169 from site 1, 98 from site 2, 32 from site 3, 332 from site 4 and 361 from site 5). Fig. 1 shows the flow of testing and results for the alternative algorithm (solid line) and additional testing by NAAT (dotted line). Multispot was performed on 254 4th generation IA-repeatedly reactive specimens: 250 were reactive on the HIV-1 peptide and HIV-1 recombinant protein, 3 were reactive only on the HIV-1 recombinant protein (all SRP individuals currently taking ARV drugs), and one was non-reactive. Five specimens from 2 sites also showed reactivity against the HIV-2 peptide initially, but were not confirmed when performing the dilution protocol described in the Multispot package insert, therefore no HIV-2 infections were detected in this sample set. NAAT resolved the IA-repeatedly reactive/Multispot-non-reactive discordant specimen as HIV acute infection. Among 738 screened-non-reactive plasma specimens, NAAT detected one acute infection. The prevalence of acute infection in this population was 0.2% (2/992) and the alternative algorithm failed to detect one specimen that was only reactive by NAAT.
Fig. 1.
HIV test results from the NHBS pilot study at CDC. Solid line represents the flow of the CDC proposed algorithm for laboratory HIV diagnostics, the assays were performed as indicated in the package insert. Dotted line represents the flow of testing to identify acute infections that would be missed by the CDC proposed algorithm. IA: immunoassay; NAAT: nucleic acid amplification test; R: reactive; RR: repeatedly reactive; NR: non-reactive; AHI: acute, HIV infection.
The overall positivity in the specimens tested at CDC from the 5 sites in this high-risk population was 25.7% (255/992), including two acute HIV-1 infections. However, the number of specimens collected and the number of HIV infections detected varied by site. Positivity for each site was 6.5% (11/169), 12.2% (12/98), 18.8% (6/32), 23.2% (77/332), and 41.3% (149/361) for sites 1, 2, 3, 4, and 5, respectively.
4.2. Comparison of the CDC proposed laboratory algorithm and site-specific WB-based algorithms
At site 1 eight participants were preliminary positive by OF-OraQuick and confirmed by blood-WB, whereas 11 participants were classified as HIV-positive with the proposed algorithm (Table 2). Three HIV-1 infections were missed during the OF screening. However, the plasma specimens from these three individuals who were unaware of their HIV status were reactive using plasma on OraQuick at CDC (data not shown). No statistically significant differences were observed between algorithms (p = 0.2482).
Table 2.
Comparison of HIV testing algorithms.
| Site | No. of samples tested | CDC proposed algorithm No. of positive | Site-specific algorithm No. of positive | Site-specific algorithm % False negatived | Differences | |
|---|---|---|---|---|---|---|
| 1 | 169 | 11 (6.5%) | 8 (4.7%) | 27.3 | 3 OF-OraQuick negative | 3 Unaware of HIV status |
| 2 | 98 | 11a (11.2%) | 10 (10.2%) | 8.3 | 1 OF-OraQuick negative | 1 Unaware of HIV status |
| 3 | 32 | 6 (18.8%) | 6 (18.8%) | 0 | Not observed | |
| 4 | 332 | 77 (23.2%) | 65 (19.6%) | 15.6 | 12 OF-OraSure WB | 10 SRPc (8 IND, 2 NEG) 2 unaware of HIV status (IND) |
| 5 | 361 | 149 (41.3%) | 146b (40.6%) | 2 | 3 Blood-Bio-Rad WB | 3 Unaware of HIV status (2 IND, 1 NEG) |
| Total | 992 | 254 | 235 | 19 | 9 Unaware of HIV status, 10 SRP |
OF: oral fluid; SRP: self-reported HIV-positive; IND: WB-indeterminate; NEG: WB-negative.
Nuosmbers do not include the acute infection identified as NAAT-positive only.
146 positive in a total of 360 participants, one individual identified as HIV-negative by the CDC proposed algorithm had no results from the site
All participants currently receiving antiretroviral therapy
The percent false negative was calculated based on HIV positivity by the CDC proposed algorithm and NAT results.
At site 2 rapid testing was only done for participants unaware of their HIV status, but OF was collected from all self-reported HIV-positive and OF-OraQuick preliminary-positive participants for HIV WB testing at a local laboratory (Table 1). A total of 10 infections were identified, seven were self-reported positive with positive OF-WB and three were preliminary positive and confirmed by OF-WB. One plasma specimen from an individual unaware of their status was non-reactive with OF screening, but was classified as HIV-positive by the proposed algorithm (Table 2). Plasma from this individual was also reactive by OraQuick at CDC (data not shown). No statistically significant differences were observed between algorithms (p = 1). One acute infection was identified at CDC by NAAT screening of IA non-reactive specimens and was missed by both algorithms.
At site 3 all participants were screened by blood-OraQuick regardless of their self-reported status and DBS were collected from all SRP and blood-OraQuick preliminary-positive participants for HIV supplemental testing at a local laboratory (Table 1). Six participants were confirmed positive by DBS-WB at the site and were also detected by the proposed algorithm (Table 2). No differences were observed between the final interpretation from the site and CDC (no statistical analysis was performed due to the limited sample size).
At site 4 initial screening was performed by OF-OraQuick and confirmation in the laboratory was done on OF (validated protocol with 3rd generation IA + OraSure HIV-1 WB) (Table 1). Sixty-five infections were confirmed by the site-specific WB-based algorithm, but 77 infections were identified by the proposed algorithm. Of 12 infections with inconsistent results between the testing algorithms, the OF-HIV-1 WBs were either WB-negative (n = 2) or WB-indeterminate (n = 10). Of the WB-indeterminate specimens, two were among participants who were unaware of their HIV status and eight were among SRP individuals who reported currently taking ARVs. The proposed algorithm detected statistically significant (p = 0.0015) more HIV-positive infections than the WB-based algorithm.
At site 5 plasma specimens were tested in the laboratory by a 3rd generation IA and HIV-1 WB (Table 1). One hundred forty-six specimens were identified as HIV WB-positive, two were WB-indeterminate and one was WB-negative, whereas 149 HIV infections were identified by the proposed algorithm (one HIV-negative specimen was not available for comparison). The three discordant results between site specific testing and CDC testing were among individuals unaware of their HIV status. The WB-negative specimen was from one HIV acute infection identified by the proposed algorithm as 4th generation IA-repeatedly reactive/Multispot-non-reactive and NAAT-positive. The two WB-indeterminate specimens identified by site specific testing were Multispot-reactive at CDC. While three additional infections were identified by the proposed algorithm the difference was not statistically significant (p = 0.2482).
Among all sites, the CDC proposed algorithm identified a total of 19 more specimens as HIV-positive that were either negative or indeterminate by the site-specific algorithms that relied on WB (p < 0.0001) (Table 2). Nine were among individuals who were unaware of their HIV status.
5. Discussion
NHBS is a behavioral surveillance system used to monitor prevalence and trends in HIV-related risk behaviors, HIV testing, and use of HIV prevention services among populations at high risk of acquiring HIV. Individual high-risk groups, MSM, injection-drug users, and heterosexuals at increased risk for HIV infection, are monitored in cycles [23–26]. The focus of this study was to evaluate the performance of the CDC proposed laboratory algorithm [15,16] for diagnostic yield in a subset of MSM from five NHBS sites with different HIV prevalence [26]. Factors such as return of results, or result turn-around time, were not assessed even though it has been shown that the proposed algorithm can reduce turn-around time compared to laboratory algorithms that use WB [27]. By applying the proposed algorithm, the number of individuals identified as HIV positive (including one acute HIV infection not detected by the WB-based site-specific algorithm) was significantly higher than the total at the five sites. In addition to one acute infection, the WB-based algorithms confirmed 19 fewer HIV infections with on-site screening by OF testing (n = 4) or by laboratory confirmation using WB with OF (n = 12) and blood (n = 3). For site 4, where the most differences were observed between a site algorithm and the proposed algorithm, testing was performed regardless of self-reported HIV status. Our findings from this site show that 10 SRP individuals currently taking ARV had negative or indeterminate results by the OF-WB in the laboratory. Furthermore, four individuals who were unaware of their HIV status and negative by screening with the OF-OraQuick rapid test were found to be HIV-positive using plasma on OraQuick and in the proposed algorithm. These results are consistent with previous studies that demonstrated limitations of testing with OF, during early infections and while taking ARV [28–32].
The use of CLIA waived rapid tests outside the laboratory accelerates turn around-time of results to individuals, allowing for an immediate referral to care, but does not currently maximize detection of individuals likely to be highly infectious. Conversely, the CDC proposed laboratory algorithm improves detection of infection during a highly infectious period, but the implementation of a laboratory-based algorithm in field settings presents several challenges, including requiring trained personnel for blood draws, sample handling and processing, and easy access to a laboratory for supplemental testing. So, in field settings such as the NHBS survey, the selection of sample type, HIV tests, and testing algorithms need careful consideration to balance the competing factors of maximized return of results and detection of individuals during a highly infectious period.
It has been demonstrated that HIV acute infections often constitute a significant proportion of the new diagnoses in high-risk populations [33–35]. In this study, two acute HIV infections were identified. One acute infection was detected by the proposed laboratory algorithm and one was identified by NAAT testing of all screened IA-non-reactive specimens. Since this was not a random, cross-sectional sampling of all MSM in the five sites, it is not possible to estimate the true number of acute infections that might have been present at the time of the survey. However, identifying early HIV infections is beneficial not only for the individual, but also from a public health perspective. Individuals accessing care and treatment sooner would substantially reduce adverse health outcomes, increase life expectancy, and reduce risk of forward transmission. Data indicate that transmission is greater during acute HIV infection, therefore early diagnosis and rapid initiation of treatment are key for successful prevention strategies and emphasize the need for early and accurate diagnosis [14,36]. This study demonstrated that the proposed laboratory-based algorithm performs well in high-risk populations and the incorporation of NAAT improved the detection of acute infections. These data add to several previous studies [6,20–22] to further support the use of the CDC proposed algorithm. The results also substantiate previous findings regarding reduced sensitivity of OF testing during acute infection and in the presence ARVs [28–33] and highlight the importance of selecting the right sample type and diagnostic tests when choosing testing strategies in populations with high rates of acute infection and potential confounding effects of ARVs.
Acknowledgements
The authors would like to express their gratitude to the NHBS working group: Colin Flynn, Danielle German, Mark Thrun, Alia Al-Tayyib, Ralph Wilmoth, Trista Bingham, Ekow Kwa Sey, Marlene LaLota, Lisa Metsch, David Forrest, Manya Magnus, Irene Kuo, Tiffany West, Hugo Santacruz, Pierre Chambers, Irene Kuo, Anthony Rawls, Megan Condrey, Dano Beck, Cody Barnett, Gregory Phillips II, Luz Montanez, Richard Teran, Toby Leroux, Ashley Rowe, Charles Chen, Ann Vannavong, Brittani Robinson, and the CDC Behavioral Surveillance Team for the collaboration during the NHBS pilot study and Bernard Branson for his critical review of the manuscript.
Funding
This work was supported by the Centers for Disease Control and Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD and TB Prevention, Division of HIV/AIDS Prevention Funding Opportunity Announcement Number PS11-001: National HIV Behavioral Surveillance.
Footnotes
Competing interests
No financial disclosures were reported by the authors of this paper.
Ethical approval
The Centers for Disease Control and Prevention determined that this study is not human subjects research.
Disclaimer
The findings and conclusions in this report are ours and do not necessarily represent the views of the Centers for Disease Control and Prevention. Use of brand names is for identification purposes and does not imply endorsements by the US Department of Health and Human Services.
References
- [1].Centers for Disease Control and Prevention. Interpretation and use of the Western blot assay for serodiagnosis of human immunodeficiency virus type 1, infections. Morb Mortal Wkly Rep 1989;38(S7):1–7. [PubMed] [Google Scholar]
- [2].Owen SM, Yang C, Spira T, Ou CY, Pau CP, Parekh BS, et al. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. J Clin Microbiol 2008;46:1588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Barbe F, Kelin M, Badonnel Y. Early detection of antibodies to human immunodeficiency virus 1 by a third-generation enzyme immunoassay. A comparative study with the results of second-generation immunoassays and Western blot. Ann Biol Clin 1994;52:341–5. [PubMed] [Google Scholar]
- [4].Constantine NT, van der Groen G, Belsey E, Tamashiro H. Sensitivity of HIV antibody assays as determined by seroconversion panels. AIDS 1994;8:1715–20. [DOI] [PubMed] [Google Scholar]
- [5].Louie B, Pandori M, Wong E, Klausner JD, Liska S. Use of an acute seroconversion panel to evaluate a third-generation enzyme-linked immunoassay for detection of human immunodeficiency virus-specific antibodies relative to multiple other assays. J Clin Microbiol 2006;44:1856–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol 2011;52S:S17–22. [DOI] [PubMed] [Google Scholar]
- [7].Chavez P, Wesolowski L, Patel P, Delaney K, Owen SM. Evaluation of the performance of the Abbott ARCHITECT HIV Ag/Ab combo assay. J Clin Virol 2011;52S:S51–5. [DOI] [PubMed] [Google Scholar]
- [8].Bentsen C, McLaughlin L, Mitchell E, Ferrera C, Liska S, Myers R, et al. Performance evaluation of the Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA, a 4th generation HIV assay for the simultaneous detection of HIV p24 antigen and antibodies to HIV-1 (group M and O) and HIV-2 in human serum or plasma. J Clin Virol 2011;52S:S57–61. [DOI] [PubMed] [Google Scholar]
- [9].Pilcher CD, Joaki G, Hoffman IF, Martinson FEA, Mapanje C, Stewart PW, et al. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS 2007;21:1723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cohen MS, Gay CL, Bush MP, Hecht FM. The detection of acute HIV infection. J Infect Dis 2010;202(S2). S270–S77. [DOI] [PubMed] [Google Scholar]
- [11].Eshleman SH, Khaki L, Laeyendecker O, Piwowar-Manning E, Johnson-Lewis L, Husnik M, et al. Detection of individuals with acute HIV-1 infection using the ARCHITECT HIV Ag/Ab combo assay. J Acquir Immune Defic Syndr 2009;52:121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pandori MW, Hackett J, Louie B, Vallari A, Dowling T, Liska S, et al. Assessment of the ability of a fourth-generation immunoassay for human immunodeficiency virus (HIV) antibody and p24 antigen to detect both acute and recent HIV infections in a high-risk setting. J Clin Microb 2009;47:2639–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Patel P, Mackellar D, Simmons P, Uniyal A, Gallagher K, Bennet B, et al. Detecting acute human immunodeficiency virus infection using 3 different screening immunoassays and nucleic acid amplification testing for human immunodeficiency virus RNA, 2006–2008. Arch Intern Med 2010;170:66–74. [DOI] [PubMed] [Google Scholar]
- [14].Owen SM. Testing for acute HIV infection: implications for treatment as prevention. Curr Opin HIV AIDS 2012;7:125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Association of Public Health Laboratories and the Centers for Disease Control & Prevention. HIV Testing Algorithms — A Status Report. Available at: http://www.aphl.org/aphlprograms/infectious/hiv/Documents/StatusReportFINAL.pdf
- [16].Pandori MW, Branson BM. 2010 HIV diagnostics conference. Expert Rev Anti Infect Ther 2010;8:631–3. [DOI] [PubMed] [Google Scholar]
- [17].Pilcher CD, Christopoulos KA, Golden M. Public health rationale for rapid nucleic acid or p24 antigen tests for HIV. J Infect Dis 2010;201(Suppl. 1): S7–15. [DOI] [PubMed] [Google Scholar]
- [18].O’Connell RJ, Peel SA. MultispotTM HIV-1/HIV-2 rapid test: advantages over the other rapid HIV tests. Expert Rev Mol Diagn 2007;7:499–505. [DOI] [PubMed] [Google Scholar]
- [19].Nasrullah M, Ethridge SF, Delaney KP, Wesolowski LG, Granade TC, Schwendemann J, et al. Comparison of alternative interpretive criteria for the HIV-1 Western blot and results of the multispot HIV-1/HIV-2 rapid test for classifying HIV-1 and HIV-2 infections. J Clin Virol 2011;52S:S23–7. [DOI] [PubMed] [Google Scholar]
- [20].Nasrullah M, Wesolowski LG, Meyer III WA, Owen M, Masciotra S, Vorwald C, et al. Performance of a fourth-generation HIV screening assay and an alternative HIV diagnostic testing algorithm. AIDS 2012;27:731–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wesolowski LG, Delaney KP, Hart C, Dawson C, Owen SM, Candal D, et al. Performance of an alternative laboratory-based algorithm for diagnosis of HIV infection utilizing a third generation immunoassay, a rapid HIV-1/HIV-2 differentiation test and a DNA or RNA-based nucleic acid amplification test in persons with established HIV-1 infection and blood donors. J Clin Virol 2011;52S: S45–9. [DOI] [PubMed] [Google Scholar]
- [22].Delaney KP, Heffelfinger JD, Wesolowski LG, Owen SM, Meyer III WA, Kennedy S, et al. Performance of an alternative laboratory-based algorithm for HIV diagnosis in a high-risk population. J Clin Virol 2011;52S:S5–10. [DOI] [PubMed] [Google Scholar]
- [23].Gallaher KM, Sullivan PS, Lansky A, Onorato IM. Behavioral surveillance among people at risk for HIV infection in the US: the National HIV Behavioral Surveillance System. Public Health Rep 2007;122(Suppl. 1):32–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].MacKellar DA, Gallaguer KM, Finlayson T, Sanchez T, Lansky A, Sullivan PS. Surveillance of HIV risk and prevention behaviors of men who have sex with men – a national application of venue-based, time-space sampling. Public Health Rep 2007;122(Suppl. 1):39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wejnert C, Le B, Zhu J, Finlayson T, Oster A, Smith A, et al. HIV prevalence and awareness of infection in 2008 and 2011 among MSM: 20 US cities. In: Conference on Retroviruses and Opportunistic Infections; 2013. Link available online was deleted by the conference: http://www.retroconference.org/2013b/Abstracts/45701.htm [Google Scholar]
- [26].Prevalence and awareness of HIV infection among men who have sex with men – 21 cities, United States, 2008. Morb Mortal Wkly Rep 2010;59:1201–7. [PubMed] [Google Scholar]
- [27].Neumann D, Bennett B, Gillis L. Performance of the new HIV-1/2 diagnostic algorithm in Florida’s Public Health Testing Population: a review of the first five months of utilization. In: 2012 HIV Diagnostics Conference, 2012. Atlanta, GA. Available at: http://www.cvent.com/events/2012-hiv-diagnostics-conference/custom-128-7213a1d01a6341dbbf81115a21797240.aspx [DOI] [PubMed] [Google Scholar]
- [28].Pavie J, Rachline A, Loze B, Niedbalski L, Delaugerre C, Laforgerie E, et al. Sensitivity of five rapid HIV tests on oral fluid or finger-stick whole blood: a real-time comparison in a healthcare setting. PLoS ONE 2010;5:e11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hamers RL, de Beer IH, Kaura H, van Vugt M, Caparos L, Rinke de Wit TF. Diagnostic accuracy of two oral fluid-based tests for HIV surveillance in Namibia. J Acquir Immune Defic Syndr 2008;1:116–8. [DOI] [PubMed] [Google Scholar]
- [30].Sherman G, Lilian R, Coovadia A. Oral fluid tests for screening of human immunodeficiency virus-exposed infants. Pediatr Infect Dis J 2010;2:169–72. [DOI] [PubMed] [Google Scholar]
- [31].Luo W, Masciotra S, Delaney K, Charurat M, Croxton T, Constantine N, et al. Comparison of HIV oral fluid and plasma testing during early infection in a longitudinal Nigerian cohort. J Clin Virol 2013;58(Suppl. 1):e113–8. [DOI] [PubMed] [Google Scholar]
- [32].O’Connell RJ, Merritt TM, Malia JA, VanCott TC, Dolan MJ, Zahwa H, et al. Performance of the OraQuick rapid antibody test for diagnosis of human immunodeficiency virus type 1 infection in patients with various levels of exposure to highly active antiretroviral therapy. J Clin Microbiol 2003;41: 2153–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stekler JD, Swenson PD, Coombs RW, Dragavon J, Thomas KK, Brennan CA, et al. HIV testing in a high-incidence population: is antibody testing alone good enough? Clin Infect Dis 2009;49:444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Patel P, Klausner JD, Bacon OM, Liska S, Taylor M, Gonzalez A, et al. Detection of acute HIV infections in high-risk patients in California. J Acquir Immune Defic Syndr 2006;42:75–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Acute HIV infection: New York City, 2008. Morb Mortal Wkly Rep 2009;58:1296–9. [PubMed] [Google Scholar]
- [36].Smith K, Powers KA, Kashuba AD, Cohen MS. HIV-1 treatment as prevention: the good, the bad, and the challenges. Curr Opin HIV AIDS 2011;6:315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]