Abstract
The agricultural fungicide cymoxanil (CMX) is commonly used in the treatment of plant pathogens, such as Phytophthora infestans. Although the use of CMX is widespread throughout the agricultural industry and internationally, the exact mechanism of action behind this fungicide remains unclear. Therefore, we sought to elucidate the biocidal mechanism underlying CMX. This was accomplished by first performing a large-scale chemical-genomic screen comprising the 4000 haploid non-essential gene deletion array of the yeast Saccharomyces cerevisiae. We found that gene families related to de novo purine biosynthesis and ribonucleoside synthesis were enriched in the presence of CMX. These results were confirmed through additional spot-test and colony counting assays. We next examined whether CMX affects RNA biosynthesis. Using qRT-PCR and expression assays, we found that CMX appears to target RNA biosynthesis possibly through the yeast dihydrofolate reductase (DHFR) enzyme Dfr1. To determine whether DHFR is a target of CMX, we performed an in-silico molecular docking assay between CMX and yeast, human, and P. infestans DHFR. The results suggest that CMX directly interacts with the active site of all tested forms of DHFR using conserved residues. Using an in vitro DHFR activity assay we observed that CMX inhibits DHFR activity in a dose-dependent relationship.
Subject terms: Chemical biology, Chemical genetics, Enzyme mechanisms, Mechanism of action
Introduction
Plant pathogens are an ever-increasing concern for crop security and yield. To this point, crop security and destruction have been recently cited as major geopolitical and humanitarian concerns by the Food and Agriculture Organization (FAO) of the United Nations1. To combat destructive plant pathogens, the agricultural industry commonly employs biocidal compounds including pesticides (herbicides, insecticides, etc.) and antimicrobials (fungicides, antibiotics, antiprotozoals, etc.)2. While use of these biocides directly increases crop yield and security3, the bioactivity for many of these compounds remain unknown. One such compound is the fungicide cymoxanil (CMX; 1-(2-cyano-2-methoxyiminoacetyl)-3-ethylurea). This synthetic acetamide is currently used as foliar-applied fungicide for treatment of the causal agent of potato blight Phytophthora infestans, to which it displays specificity4,5. CMX is additionally used as a treatment for tomato blight and grape mildew6,7.
Little work has been invested into investigating the mechanism behind CMX, with many studies rather focused on assessing the impact of the fungicide on P. infestans infected crops. In a study performed by Ziogas and Davidse, CMX was found to broadly target DNA and RNA biosynthesis8. The authors did note that this was likely a secondary effect - an observation later supported by Andrieu and colleagues8,9. In terms of human toxicity, CMX is thought to be rapidly absorbed in the blood and plasma with maximum concentrations reached within four hours of dosage10. A 2021, CMX is reported to meet the current health and environmental safety standards of Canada and Europe nor is considered to be carcinogenic - a conclusion supported by a recent study pertaining to Iranian fruit crop consumption11–13. In south Asia, grapes or raisins contaminated with CMX were found to pose no risk for human consumption14,15. Despite its prevalent international use, details underlying the mechanism of action of CMX remain ambiguous.
A common approach to study the mechanism of action for a compound is the use of chemical-genomic screening. This type of screening is based on the notion that the presence of parallel compensatory pathways can compensate for the genetic inactivation of a single pathway, thereby resulting in no obvious phenotypic consequence16,17. Inactivation of the second functionally overlapping pathway using a single-gene deletion mutation18 or a chemical inactivation19 can cause an unexpected phenotypic effect that can be measured. A common approach in such screens is to challenge a library of non-essential gene deletion mutants in Escherichia coli20 or Saccharomyces cerevisiae21,22 to a sub-inhibitory concentration of the target bioactive compound. Specifically, single-gene deletion libraries are challenged by the compound of interest with the resulting biological responses providing indirect evidence of genetic targets (Fig. 1a)23–25. This approach has been used to elucidate the molecular activity of natural bioactive compounds26,27, pharmaceuticals28, nanoparticles29 and herbal extracts30.
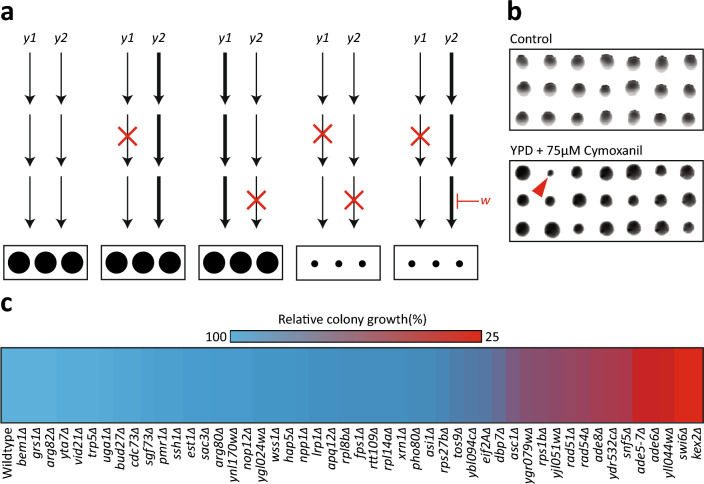
Figure 1.
Chemical-genomic screening reveals 49 gene candidates involved in the response to CMX. (a) A schematic depicting the theory underlying chemical genomic screening. From left to right: two parallel genetic pathways (y1 and y2) produce a growth phenotype. When either pathway is mutated, the parallel pathway compensates. When both pathways contain mutations, no compensation can occur resulting in a decrease in growth phenotype. Coupling a single mutation with an inhibitory compound (w) mimics the effect of two mutations, resulting in a decreased growth phenotype. (b) Representative images from large-scale chemical genomic screening. The red arrow indicates a sensitive colony. (c) The sensitivities of each gene candidate to CMX derived from follow up colony size sensitivity analysis.
Thus, to investigate the mechanism of action of CMX, we employed a sensitivity screen using roughly 4000 haploid non-essential gene deletion strains of the yeast S. cerevisiae against CMX. Using a combination of yeast chemical genomics, molecular genetics, in-silico docking simulations, and protein activity assays, our observations suggest that CMX may disrupt RNA biosynthesis through targeting the dihydrofolate reductase (DHFR) enzyme. DHFR is a folate reductase protein that functions to convert dihydrofolate (DHF) to tetrahydrofolate (THF). The resulting THF acts as a carbon donor to key metabolites including purines. Thus, by preventing the reducing activity of DHFR, CMX indirectly inhibits purine biosynthesis through preventing the conversion of DHF to THF. The results presented in this study therefore contribute to the overall physiological and biochemical understanding of CMX.
Results
Chemical genomic profiling of CMX
To investigate the mechanism of action of CMX, we first determined a suitable sub-inhibitory concentration of the fungicide on the growth of S. cerevisiae. We subjected a random collection of 384 single-gene deletions to several concentrations of CMX (0, 12.5, 25, 37.5, 75, 150 µM). The results indicate that for the investigated yeast non-essential gene deletions, 75 µM of CMX results in an approximately 5% of the colonies showing a noticeable reduction in their relative growth. This frequency of sensitive mutants can represent a meaningful balance for investigating the mode of activity of a compound19,31. We next subjected the entire yeast single-gene deletion set to YPD media with or without 75 µM CMX (Fig. 1b – representative image). After incubating the yeasts at 30 °C for 48 hours, we found that 49 yeast colonies displayed a relative growth defect of 25% or greater. These candidates were then transferred onto a manageable single plate and subjected to follow up studies that included the repeat of the sensitivity screen. Figure 1c represents a heat map corresponding to the average colony size for the 49 strains obtained from the large-scale screen.
To confirm the sensitivity of the obtained strains we next subjected these strains to spot-test analysis. Spot-test analysis offers a refined screening approach to study strain sensitivity which is complementary to the colony size approach32. A serial dilution of each candidate yeast (in addition to a WT) was spotted on plates with or without 75 µM CMX (Fig. 2a). To quantify colony sensitivity, each spot was measured with a 4x4 region of interest for grey value intensity (Fig. 2b). This analysis resulted in 13 candidates displaying sensitivity to CMX relative to the WT control.
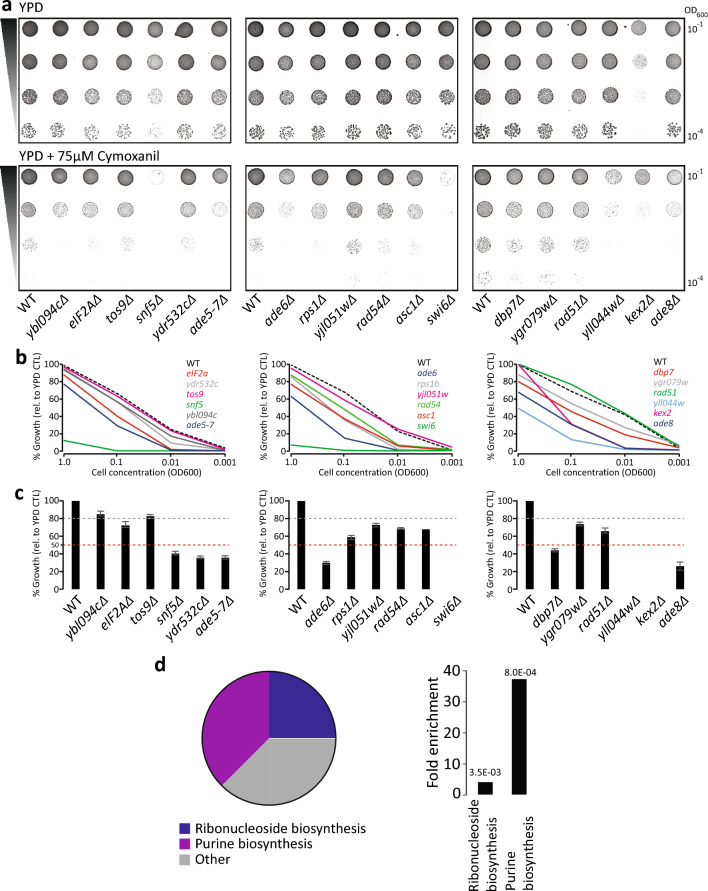
Figure 2.
Small-scale verification of 18 gene candidates to CMX. (a) Spot-test analysis of the 18 candidates in the presence and absence of 75 µM CMX. (b) Quantification of spot-tests in (a). (c) Colony-count analysis of the 18 candidates. Grey bar represents a 20% reduction in colony growth. The red bar represents a 50% reduction in colony growth. (d) GO-analysis of the 8 most-sensitive gene candidates reveals two statistically significant gene families: de novo purine biosynthesis and ribonucleoside biosynthesis.
In parallel, we performed a colony-counting assay. We diluted the candidate yeasts from overnight culture to 1.0E–03 OD and transferred them onto plates in the presence or absence of 75 µM CMX. We observed that many of the yeasts displayed a growth sensitivity relative to the WT control and appear to either be partially or extremely sensitive (Fig. 2c). We observed that 8 yeast deletion strains showed at least 50% reduction in growth (Fig. 2c). Table 1 shows the list of the gene deletion strains that were sensitive in all investigated assays. In the GO term enrichment analysis of the affected genes, de novo purine nucleotide biosynthesis and ribonucleoside synthesis appeared to be significantly enriched (Fig. 2d).
Table 1.
Candidate functional descriptions derived from chemical-genomic screening in the presence of CMX.
| Gene name | Gene function |
|---|---|
| ADE5-7 | Catalyzes a step in de novo purine biosynthesis pathway |
| ADE6 | Catalyzes a step in de novo purine biosynthesis pathway |
| ADE58 | Catalyzes a step in de novo purine biosynthesis pathway |
| SWI6 | Transcription regulation; forms complex with Swi4 and Mbp1 |
| SNF5 | Transcription regulation; forms complex with Snf2 and Snf6 |
| DBP7 | RNA processing |
| KEX2 | Calcium-dependent serine protease |
| YLL044w | Dubious ORF |
Due to the nature of chemical-genomic screening, the enriched families likely display an indirect effect of CMX. As purine biosynthesis is heavily enriched, the primary target of CMX is likely a protein involved in this process. DFR1 is an essential gene in yeast that encodes for an evolutionary conserved DHFR, a key enzyme in purine biosynthesis. Being an essential gene, it is absent from the non-essential gene knockout library used here. Interestingly it is co-expressed with four of the target genes identified in our screen, namely ADE5,7, ADE6, ADE8 and DBP733. Also, it genetically interacts with ADE6 and is predicted to functionally interact with YLL044W33. Consequently, we further studied nucleotide biosynthesis and Dfr1 as a potential target for CMX.
CMX attenuates RNA biosynthesis
To study the effect of CMX on RNA synthesis, total RNA was extracted from yeast cells treated with different concentrations of CMX. Indicated in Fig. 3a, we observed a dose-response correlation between CMX concentration and the reduction in rRNA content obtained from total RNA extraction. As the concentration of CMX increases, we observed a corresponding decrease in rRNA band intensity (Fig. 3a). This is in accord with an activity for CMX to target RNA biosynthesis. Compared to the control (no CMX), rRNA content was reduced by approximately 80% at 37.5 µM CMX (Fig. 3b).
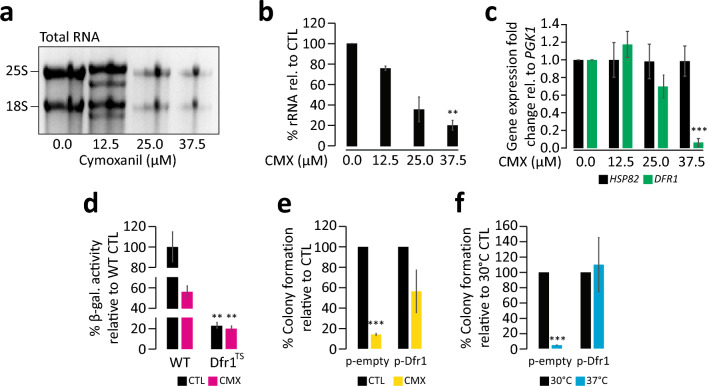
Figure 3.
The yeast DHFR enzyme Dfr1 is a target of CMX. (a) total RNA was extracted from yeasts subjected to several concentrations of CMX, showing a dose-dependent reduction in rRNA band intensity obtained from a total RNA extraction, quantified in (b). (c) qRT-PCR analysis of DRF1 and HSC82 from RNA extracted from yeasts grown in the presence of increasing concentrations of CMX. (d) β -gal activity of WT and a temperature-sensitive knockdown of Dfr1 in the presence and absence of CMX. (e) Colony-counting analysis of yeasts expressing DFR1 on a plasmid in the presence or absence of CMX. An empty vector not containing Drf1 was used as a control under the same conditions. (f) Using the same strains in (e), both were subject to 30 °C and 37 °C as a control. **Denotes significance values less than 0.01. ***Denotes significance values less than 0.001. The original gel seen in “A” can be found in Supplemental Fig. 1.
We then assessed the expression of DFR1 and HSC82 (used as a control) in the presence of CMX (Fig. 3c). The DFR1 gene encodes the yeast DHFR protein and HSC82 encodes a heat shock chaperone often utilized as a control34–37. This was achieved using quantitative reverse transcriptase-PCR (qRT-PCR) on yeasts subjected to varying concentrations of CMX (Fig. 3c). PGK1 was used as a housekeeping gene where all values were related to it. We found that as the concentration of CMX increases, DFR1 expression decreases. At first, we see a possible increase in DFR1 expression at a very mild concentration 12.5 µM CMX. However, at the sub-inhibitory concentration of 37.5 µM DFR1 mRNA content was reduced by approximately 90%. No difference in the expression for HSP82 mRNA was observed.
We then used a reporter system to further study the effect of CMX on the yeasts (Fig. 3d). If CMX affects ribonucleotide synthesis, it is likely that the pool of nucleotides will be compromised in cells treated with CMX thereby resulting in a lower rate of gene expression. Indicated by a lower activity of β-galactosidase, CMX was observed to reduce the level of gene expression measured by a β-gal reporter system (Fig. 3d). A similar effect was observed using a temperature sensitive DFR1 knockdown strain. Shifting the cells to an inhibitory temperature of 37 °C resulted in reduced β-gal activity. Under this condition, the addition of CMX had no additional effect in β-gal expression suggesting that the effect of CMX seems to be DFR1-dependent.
We also examined the effect of overexpression of DFR1 on cell sensitivity to CMX (Fig. 3e) Using a colony count assay, at the concentration of 50 µM CMX control cells had significantly reduced growth compared to the cells grown without CMX. When cells were transformed with a plasmid that carries a DFR1 gene, cell sensitivity was partially compensated. This was not true for the cells transformed with a control plasmid. As an additional control, we also examined the ability of the DFR1 expressing plasmid to compensate for the temperature sensitive mutant for DFR1 (Fig. 3f). At the inhibitory temperature of 37°C, colony formation was severely hindered in temperature sensitive mutants. However, introduction of the yeast DFR1 using a plasmid returned the cell growth to the WT levels.
CMX is predicted to interact with the DHFR catalytic domain
To further study the relationship between DHFR protein and CMX we used an in silico molecular dynamics assay. The results from the docking assay suggest that CMX is predicted to interact with the catalytic domain of the yeast DHFR (∆G = -6.82 kcal/mol, Fig. 4a). Three residues are predicted to mediate this interaction: GLY25, LEU27, and SER63 (Fig. 4b). We then assessed whether CMX is also predicted to interact with the human DHFR protein. While the human and yeast DHFR proteins share a 50% sequence similarity, the catalytic domain is highly conserved between the two as they share a 70% identity within this region. Using the same parameters, we found that CMX is also predicted to interact with the catalytic domain of human DHFR (∆G = − 7.17 kcal/mol, Fig. 4c). In addition, the predicted interaction appears to involve several residues including: LEU22, TRP24, LEU27, GLU30, and GLY31 (Fig. 4d). We then assessed whether CMX may interact with the DHFR protein of P. infestans. Congruent with the previous predictions, CMX is predicted to interact with the catalytic domain of P. infestans DHFR (∆G = − 7.51 kcal/mol, Fig. 4e). The results suggest that this predicted interaction utilizes LEU28 and ASP31 (Fig. 4f).
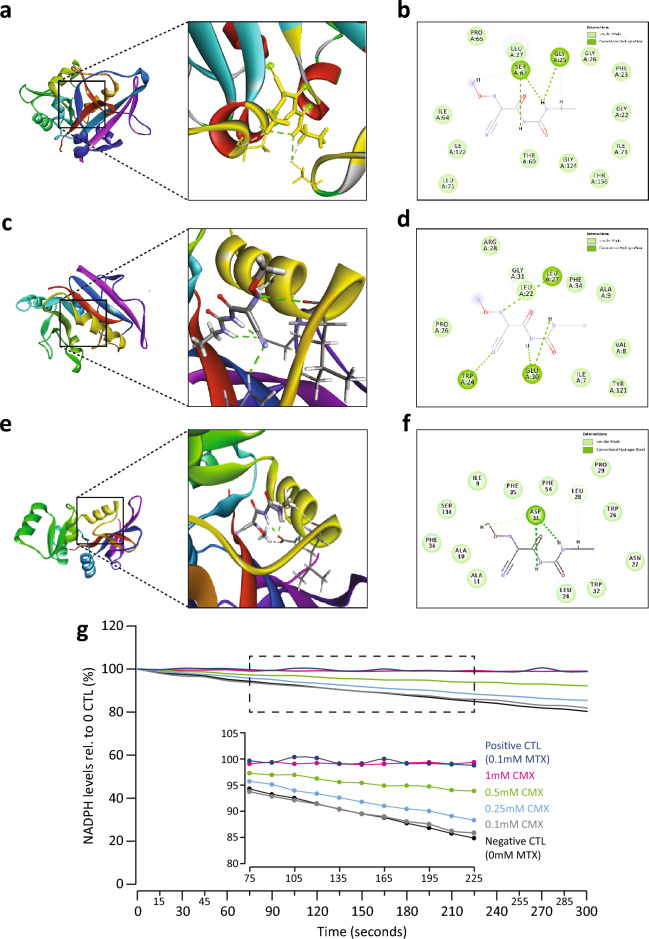
Figure 4.
CMX inhibits the activity of mammalian DHFR. (a) Molecular docking simulations predict an interaction between CMX and the catalytic domain of yeast Dfr1 (∆G = − 6.82 kcal/mol). (b) The residues implicated in the predicted interaction from panel (a). (c) CMX is also predicted to interact with the catalytic domain of mammalian DHFR (∆G = − 7.17 kcal/mol). (d) the residues implicated in the predicted interaction from panel (c). (e) CMX is predicted to interact with the catalytic domain of the P. infestans DHFR protein ((∆G = − 7.51 kcal/mol). (f) The residues implicated in the predicted interaction from panel (e). (g) A DHFR assay between increasing concentrations of CMX and mammalian DHFR. The presence and absence of MTX were used as experimental controls.
Next to investigate the ability of CMX to inhibit DHFR activity, we performed a standard DHFR assay between human DHFR and CMX. Methotrexate (MTX), a strong inhibitor of DHFR was used as a control. In a dose response manner, we found that DHFR activity decreases as the concentration of CMX increases (Fig. 4g). To this point, 0.5–1 mM of CMX inhibits the activity of DHFR to levels comparable to that for MTX (0.1 mM) control.
Discussion
The data presented in this study suggests that CMX interacts with the active site of the DHFR enzyme, revealing a new possible mechanism for the compound CMX. This is supported by several lines of evidence including chemical-genomic screening data, gene expression and RNA analyses, as well as DHFR modeling and activity assays. Specifically, chemical-genomic screening resulted in two related gene families that were found to be enriched in response to CMX treatment: namely de novo purine biosynthesis and ribonucleoside synthesis (Figs. 1, 2d and e). This is congruent with a previous study that suggested CMX may broadly affect DNA/RNA synthesis8. In our screening analysis, the calcium-dependent serine protease KEX2 single-gene deletion strain displayed sensitivity to CMX (Fig. 2a–c). However, it should be noted that this gene deletion mutant has been found to be sensitive to a variety of conditions presumably due to the lack of α-pheromone processing38,39. Therefore, the observed sensitivity of the KEX2 deletion strain to CMX may not be specific and should be interpreted with caution.
Considering the data thus far, we inferred that one of the possible targets of CMX may be the yeast DHFR enzyme Dfr1. We tested this hypothesis by first assessing the total RNA content of the cell in response to CMX and observed a dose-dependent reduction of rRNA in response to CMX. We also investigated the expression of the DFR1 gene in response to CMX. We found that CMX affects the DFR1 mRNA content in a dose-dependent manner (Fig. 3a–c). The β-gal activity assay demonstrated that loss of Dfr1 due to a shift to a restrictive temperature significantly reduces the activity of β-gal that are reminiscent of CMX treatment of the same strain (Fig. 3d). Addition of CMX to the cells in Dfr1 restrictive temperature had no additional effect on β-gal activity further connecting the activities of Dfr1 and CMX. In-silico molecular dynamics analysis also suggested an interaction between CMX with DHFR (Fig. 4a–f). Solidifying the prediction data, using an in vitro DHFR activity assay we found that CMX interfered with the activity of DHFR in a dose-dependent relationship (Fig. 4g).
When assessing the results from the molecular docking assay, LEU22 is predicted to be involved in the interaction between CMX and human DHFR (Fig. 4c and d) - the same residue which mediates the interaction between human DHFR and MTX40. Previously, clinical isolates of human DHFR substituted LEU22 with arginine thereby significantly reducing the binding affinity of MTX40,41, indicating that LEU22 is important for the association of MTX to DHFR. In the future, a similar approach can be applied to further investigate the potential interaction of CMX to the human DHFR. CMX was also predicted to interact with LEU27 in both tested forms of DHFR (Fig. 4a–d). Previous studies have noted the interaction between MTX and LEU27 or LEU28 in bacterial DHFR (Lacticaseibacillus casei and E. coli respectively42). Specifically, the p-aminobenzoyl group of MTX was found to interact with these residues42, indicating that LEU27 might be an important residue for DHFR inhibitors. MTX is also reported to interact with the conserved TRP24 and GLU30 residues in human DHFR43. Our predicted results for the interaction between human DHFR and CMX also utilize these residues, further supporting our prediction data (Fig. 4c and d). When assessing SER63 being predicted to be an interaction residue, previous work by Cody and colleagues noted that SER64 of the fungus Pneumocystis carinii DHFR (pcDHFR) forms a contact site with the pcDHFR inhibitor PY101443.
We next found that CMX is predicted to interact with the P. infestans DHFR protein, specifically through LEU28 and ASP31 (Fig. 4e and f). Our results indicate that CMX is predicted to associate to the catalytic domain of human DHFR and can inhibit the enzymes activity in a dose-dependent manner. Inhibiting DHFR is a common anti-cancer strategy, as preventing folate metabolism results in anti-tumor proliferation. Before its widespread adoption after safety and regulatory review, CMX was postulated to be potentially carcinogenic or mutagenic to humans. Considering that CMX likely inhibits human DHFR, it is also likely that it may inhibit cellular proliferation. While further research would be required to determine physiologically relevant concentrations that would presumptively be detrimental to humans, the current agricultural application of CMX may need to undergo review considering the results presented here.
In light of these observations, we propose a new model pertaining to the biocidal activity of CMX (Fig. 5). In this model, CMX interacts with the catalytic domain of the yeast DHFR enzyme Dfr1. Interaction with the catalytic domain might be mediated by several residues including LEU27. This putative interaction possibly inhibits the activity of Dfr1, thereby preventing the conversion of DHF to THF - the latter being an important cofactor in purine biosynthesis and cell metabolism44. Consequentially, RNA biosynthesis is inhibited as purines are no longer synthesized. This culminates in several off-target effects including potentially affecting RNA biosynthesis. The data presented in this study therefore offer possible targets for future studies to dissect the proposed interaction between CMX and DHFR. Taken together, our results suggest that CMX disrupts RNA biosynthesis through a probable association with the active site of yeast and human DHFR.
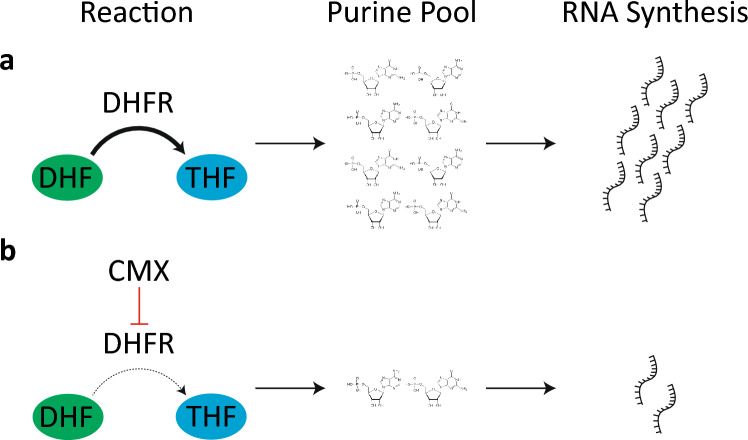
Figure 5.
A model outlining how CMX interferes with the activity of DHFR, culminating in downstream effects and decreases in RNA synthesis. (a) Under normal conditions, DHFR converts dihydrofolate (DHF) to tetrahydrofolate (THF). This conversion is critical for production of purines, which are later used in RNA synthesis. (b) CMX inhibits the activity of DHFR, thereby reducing the conversion of DHF to THF resulting in decrease in the number of purines and therefore decreasing RNA synthesis.
Methods
Strains and reagents
Deletion strains in BY4741 (MATa orf∆: kanMX4 his3∆1 leu2∆0 met15∆0 ura3∆0) were obtained from the yeast single-gene deletion collection, originally purchased from ThermoFisher Canada45. Single-gene deletion strains were verified using PCR analysis. All reagents were purchased from ThermoFisher Canada unless otherwise stated. Primers for qRT-PCR were obtained from Integrated DNA Technologies (IDT) Canada.
Chemical-genomic screen
To perform the chemical-genomic screen, we first subjected a random collection of 384 single-gene deletions (1 plate of the single-gene deletion set) to varying concentrations of CMX. Once a suitable sub-inhibitory concentration was determined, we then performed the large-scale screening using the entire deletion set in the presence of 75 µM CMX in duplicate. Any yeasts that displayed sensitivity were subsequently transferred onto a new single plate. We then repeated the screening to verify the gene-deletion candidates from the large-scale screening in biological triplicate. The plates were photographed, and the pictures were uploaded into Growth Detector (GD)32. Colony size was measured in GD by assessing the pixels corresponding to the colonies. These pixels were measured and normalized over the average growth of the colonies on that plate and related to that of the control plate as described in Ref.32. Reductions in colony growth are reported as a heat map across the 49 strains. The verified candidates were then subjected to drug sensitivity and colony-counting analysis.
For drug sensitivity analysis, yeasts were grown from independent colonies for 17 hours at 30 °C in liquid YPD. Spot test analysis of serial dilutions of cell suspensions were spotted onto solid media in the presence or absence of CMX. For growth sensitivity to CMX, 75 µM was used in media as described previously46. Sensitivity to the compound was assessed by comparing the number and size of the colonies formed on each plate after 48 hours in comparison with wild type. To measure colony size, the corresponding plates were photographed and uploaded into Fiji/ImageJ software. The pictures were converted to grey-scale and colony size was measured using a circular region of interest (outlining the circumference of each yeast spot) using pixel white/black intensity. The resulting values were then normalized to the WT of each plate.
To perform the colony counting assay, yeasts from independent colonies were grown for 17 hours at 30 °C in liquid YPD. 100 µL of diluted (10−3) cell cultures were subsequently transferred onto YPD plates in the absence and presence of CMX. The colonies were counted two days after incubation at 30 °C. Each experiment was repeated at least three times. A t-test analysis (p ≤ 0.05) was used to determine statistically significant differences. The remaining candidates were then subjected to Gene Ontology (GO) analysis to determine enrichment and significance.
Quantitative realtime-PCR (qRT-PCR)
The content of mRNAs was evaluated using qRT-PCR analysis. Deletion mutants in BY4741 were grown in YPD overnight with or without CMX treatment. Total RNA was extracted with Qiagen® RNeasy Mini Kit as described previously47. Complementary DNA (cDNA) was synthesized using iScript Select cDNA Synthesis Kit (Bio-Rad®) according to the manufacturer’s instructions. cDNA was then used as a template for quantitative PCR. qRT-PCR was carried out using Bio-Rad® iQ SYBR Green Supermix and the CFX connect real-time system (Bio-Rad®), according to the manufacturer’s instructions. PGK1 was used as a constitutive reference gene (internal control). The procedure and data analysis were performed according to MIQE guidelines48. In brief, total RNA extraction was performed in biological triplicate, with the quality and quantity of RNA assessed via spectrophotometry and gel electrophoresis. Approximately 1 µg of RNA was used to synthesize cDNA. CT values were normalized to the PGK1 internal control through subtraction with the subsequent values averaged and normalized to the control for HSP82 or DFR1 respectively. These values were then transformed to ΔΔCT scores. The primers used were designed to 60 nucleotides prior to the sequence in the flanking region of the indicated gene. PGK1 Forward: CAGACCATTCTTGGCCATCT; PGK1 Reverse: CGAAGATGGAGTCACCGATT; HSC82 Forward: CTTGTTTTCTTTTTCTTGAAACGCTAC; HSC82 Reverse: GTCAATCGTAAGTGTACACTAAACTTT DFR1 Forward: AGTTAACATTATGCTTTGCATGATAAT; DFR1 Reverse: GAGGCTTATCAGTTCTATCACTATTTA. PGK1 primer sequences were designed previously described49. Significance was determined with a t-test analysis (p ≤ 0.05). Error bars are derived using a standard deviation (3 replicates).
Quantitative β-galactosidase assay
The effect of CMX on translation in different yeast strains were examined using LacZ reporter systems. To evaluate the activity of LacZ expression cassettes, quantitative β-galactosidase assay was performed using ONPG (O-nitrophenyl-α-d-galactopyranoside) and the p416GAL1-LacZ plasmid as previously described50,51. LacZ expression was induced by incubating yeasts in YP-Galactose for 6 hours at 37 °C. Each experiment was repeated at least three times. T-test analysis (p ≤ 0.05) was used to determine statistically significant differences.
Molecular docking
The crystal structure and homologues of the DHFR proteins with appropriate domains were obtained from the Protein Data Bank database using the protein-specific local alignment search method utilizing Position-Specific Iterated BLAST. The homology model of the DHFR domains was created using homologs with the highest sequence similarity using the Swiss Model, a fully automated platform for modeling protein structure homology. Using 3DRefine and Phyre2, protein refinement and homology sequencing studies are conducted to develop credible protein models for detecting the likelihood of binding site predictions. The 3D structure of CMX is generated and docked to the protein structure for docking predictions using PyMol and the High Ambiguity Driven Protein-protein Docking tool. NAMD version 2.11b was used for molecular dynamics simulations, which utilizes an Amber force field including substrate peptide and additional enzyme cofactors. To exclude any overlaps in the simulation data, the interference ratio contains a minimum of 10,000 steps with a time stage of 2 fs and a temperature of 298 K using the Generalized Born Implicit Solvent. Path data were examined using PyMol and extra plug-ins to determine further hydrogen bonds, RMSD, RMSF, salt bridges, and other interacting bond forms during the DHFR-CMX interaction.
DHFR activity assay
DHFR activity in the presence of CMX was assessed using the Dihydrofolate Reductase assay kit from Sigma-Aldrich Canada according to the manufacturer’s instructions. Briefly, 1.5E–03 units of DHFR were subjected to increasing concentrations of CMX. The DHFR enzyme was incubated with the CMX for a total of 5 minutes per concentration. DHFR in the presence and absence of 0.1 mM MTX were used as controls. The assay was repeated in technical triplicate with T-test analysis (p ≤ 0.05) used to determine significance.
Supplementary Information
Acknowledgements
Thank you to Dr. Michael Downey for supplying strains for phenotype confirmation in Fig. 2, and to Dr. Dominic Hoepfner for critical discussions relating to this project. TDDK is supported by a MITACs Accelerate Post-Doctoral Fellowship. This work is dedicated to the loving memory of Abbas Golshani.
Author contributions
Conceptualization: D.J.B, A.G; Methodology: T.D.D.K, D.J.B, J.W, S.K.J, M.A.G; Formal analysis: T.D.D.K, D.J.B; Investigation: T.D.D.K, D.J.B, J.W, S.K.J, M.A.G, E.S, T.P, C.B.J, N.B.R, B.E, S.T; Writing—Original Draft: T.D.D.K, D.J.B, A.G; Writing—Review and Editing: T.D.D.K, M.H, H.M, K.B.S, B.S, E.F, A.G; Visualization: T.D.D.K, M.A.G, A.G; Supervision: A.G; Project administration: A.G; Funding acquisition: A.G.
Funding
The funding was supported by Natural Sciences and Engineering Research Council of Canada, 123456.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-62563-5.
References
- 1.Food and Agriculture Organization (FAO) Crop prospects and food situation—Quarterly global report no. 2. Tech. Rep. 2023 doi: 10.4060/cc6806en. [DOI] [Google Scholar]
- 2.Kankariya RA, Chaudhari AB, Gavit PM, Dandi ND. 2,4-Diacetylphloroglucinol: A Novel Biotech Bioactive Compound for Agriculture. Springer Singapore; 2019. pp. 419–452. [Google Scholar]
- 3.Rettinassababady C, Jeyalakshmi C. Bio-Fungicides: The Best Alternative for Sustainable Food Security and Ecosystem. Springer India; 2014. pp. 401–411. [Google Scholar]
- 4.Rekanovic E, et al. Toxicity of metalaxyl, azoxystrobin, dimethomorph, cymoxanil, zoxamide and mancozeb to Phytophthora infestans isolates from Serbia. J. Environ. Sci. Heal. B. 2012;47:403–409. doi: 10.1080/03601234.2012.657043. [DOI] [PubMed] [Google Scholar]
- 5.Zhu G-N, et al. Sensitivities of Phytophthora infestans to metalaxyl, cymoxanil, and dimethomorph. Agric. Sci. China. 2008;7:831–840. doi: 10.1016/S1671-2927(08)60120-0. [DOI] [Google Scholar]
- 6.Chacón MG, Caicedo E, Ordóñez ME. Sensitivity to metalaxyl, mefenoxam, and cymoxanil in Phytophthora andina isolates collected from tree tomato (Solanum betaceum) in ecuador. Trop. Plant Pathol. 2022;47:786–794. doi: 10.1007/s40858-022-00512-1. [DOI] [Google Scholar]
- 7.Balkan T, Kara K. Dissipation kinetics of some pesticides applied singly or in mixtures in/on grape leaf. Pest Manag. Sci. 2023;79:1234–1242. doi: 10.1002/ps.7299. [DOI] [PubMed] [Google Scholar]
- 8.Ziogas BN, Davidse LC. Studies on the mechanism of action of cymoxanil in Phytophthora infestans. Pestic. Biochem. Physiol. 1987;29:89–96. doi: 10.1016/0048-3575(87)90066-6. [DOI] [Google Scholar]
- 9.Andrieu N, Jaworska G, Genet J-L, Bompeix G. Biological mode of action of famoxadone on Plasmopara viticola and Phytophthora infestans. Crop. Prot. 2001;20:253–260. doi: 10.1016/S0261-2194(00)00156-3. [DOI] [Google Scholar]
- 10.PubChem Compound Summary for CID 5364079, Cymoxanil. https://pubchem.ncbi.nlm.nih.gov/compound/Cymoxanil (Accessed 3 August 2023).
- 11.Pest Management Regulatory Agency (PMRA). Re-evaluation Decision Cymoxanil and Its Associated End-use Products Final Decision. Tech. Rep. PPRS 1/52, Government of Canada (2021).
- 12.Anastassiadou M, et al. Evaluation of confirmatory data following the article 12 MRL review for cymoxanil. EFSA J. 2019 doi: 10.2903/j.efsa.2019.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoshnam F, et al. Investigation and probabilistic health risk assessment of pesticide residues in cucumber, tomato, and okra fruits from Khuzestan, Iran. Environ. Sci. Pollut. Res. 2022;29:25953–25964. doi: 10.1007/s11356-022-21249-9. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Ye Q, Wan K, Wang F. Residue behavior and risk assessment of cymoxanil in grape under field conditions and survey of market samples in Guangzhou. Environ. Sci. Pollut. Res. 2019;26:3465–3472. doi: 10.1007/s11356-018-3890-1. [DOI] [PubMed] [Google Scholar]
- 15.Shabeer TPA, Bhat RS, Rasool SN, Narayana Rao D. Residue dissipation and processing factor for dimethomorph, famoxadone and cymoxanil during raisin preparation. Food Chem. 2015;170:180–185. doi: 10.1016/j.foodchem.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Alamgir M, Erukova V, Jessulat M, Azizi A, Golshani A. Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chem. Biol. 2010 doi: 10.1186/1472-6769-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman JL, IV, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.1056072. [DOI] [PubMed] [Google Scholar]
- 18.Babu M, et al. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 2011;7:e1002377. doi: 10.1371/journal.pgen.1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galván I, et al. Antifungal and antioxidant activities of the phytomedicine pipsissewa, chimaphila umbellata. Phytochemistry. 2008;69:738–746. doi: 10.1016/j.phytochem.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Galván Márquez I, et al. Mode of action of nisin on Escherichia coli. Can. J. Microbiol. 2020;66:161–168. doi: 10.1139/cjm-2019-0315. [DOI] [PubMed] [Google Scholar]
- 21.Parsons AB, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 22.Hernández RB, Moteshareie H, Burnside D, McKay B, Golshani A. Manganese-induced cellular disturbance in the Baker’s yeast, Saccharomyces cerevisiae with putative implications in neuronal dysfunction. Sci. Rep. 2019;9:6563. doi: 10.1038/s41598-019-42907-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnside D, et al. Use of chemical genomics to investigate the mechanism of action for inhibitory bioactive natural compounds. In: Brahmachari G, et al., editors. Bioactive Natural Products: Chemistry and Biology. Wiley Blackwell; 2014. pp. 9–32. [Google Scholar]
- 24.Jagadeesan SK, et al. DBP7 and YRF1-6 are involved in cell sensitivity to LiCl by regulating the translation of PGM2 mRNA. Int. J. Mol. Sci. 2023;24:1785. doi: 10.3390/ijms24021785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galván Márquez I, et al. Disruption of protein synthesis as antifungal mode of action by chitosan. Int. J. Food Microbiol. 2013;164:108–112. doi: 10.1016/j.ijfoodmicro.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 26.Darvishi E, Omidi M, Bushehri A, Golshani A, Smith M. Thymol antifungal mode of action involves telomerase inhibition. Med. Mycol. 2013;51:826–834. doi: 10.3109/13693786.2013.795664. [DOI] [PubMed] [Google Scholar]
- 27.Darvishi E, Omidi M, Bushehri A, Golshani A, Smith M. The antifungal eugenol perturbs dual aromatic and branched-chain amino acid permeases in the cytoplasmic membrane of yeast. PLoS One. 2013;8:e76028. doi: 10.1371/journal.pone.0076028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baetz K, et al. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4525–4530. doi: 10.1073/pnas.0307122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galván Márquez I, et al. Zinc oxide and silver nanoparticles toxicity in the baker’s yeast, Saccharomyces cerevisiae. PLoS One. 2018;13:e0193111. doi: 10.1371/journal.pone.0193111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mir-Rashed N, et al. Disruption of fungal cell wall by antifungal echinacea extracts. Med. Mycol. 2010;48:949–958. doi: 10.3109/13693781003767584. [DOI] [PubMed] [Google Scholar]
- 31.Alamgir M, Eroukova V, Jessulat M, et al. Chemical-genetic profile analysis in yeast suggests that a previously uncharacterized open reading frame, YBR261C, affects protein synthesis. BMC Genom. 2008;9:583. doi: 10.1186/1471-2164-9-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memarian N, et al. Colony size measurement of the yeast gene deletion strains for functional genomics. BMC Bioinform. 2007;8:117. doi: 10.1186/1471-2105-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warde-Farley D, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paysan-Lafosse T, et al. InterPro in 2022. Nucleic Acids Res. 2023;51:D418–D427. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barclay BJ, Zeng Y, Turner JH, Segel IH, Taylor RC. Mapping and sequencing of the dihydrofolate reductase gene (DFR1) of Saccharomyces cerevisiae. Gene. 1988;63:175–185. doi: 10.1016/0378-1119(88)90523-9. [DOI] [PubMed] [Google Scholar]
- 36.Carvalho FA, Vieira da Silva L, Collins JF, Pereira ME, Arraiano CM. Hsp90 and metal-binding J-protein family chaperones are not critically involved in cellular iron–sulfur protein assembly and iron regulation in yeast. FEBS Lett. 2023;597:1718–1732. doi: 10.1002/1873-3468.14612. [DOI] [PubMed] [Google Scholar]
- 37.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919-3930.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bader O, Krauke Y, Hube B. Processing of predicted substrates of fungal Kex2 Proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris. BMC Microbiol. 2008;8:116. doi: 10.1186/1471-2180-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine- arginine-cleaving endopeptidase required for processing of yeast prepro-α-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- 40.Cody V, Luft JR, Pangborn W. Understanding the role of Leu22 variants in methotrexate resistance: Comparison of wild-type and Leu22Arg variant mouse and human dihydrofolate reductase ternary crystal complexes with methotrexate and NADPH. Acta Crystallogr. D Biol. Crystallogr. 2005;61:147–155. doi: 10.1107/S0907444904030422. [DOI] [PubMed] [Google Scholar]
- 41.Bolins JT, Filman DJ, Matthews DA, Hamlin RC, Kraut J. Crystal structures of Escherichia coli and Lactobacillus dihydrofolate reductase refined at 1.7 Å resolution. J. Biol. Chem. 2004;257:13650–13662. doi: 10.1016/S0021-9258(18)33497-5. [DOI] [PubMed] [Google Scholar]
- 42.Cody V, Luft JR, Pangborn W, Gangjee A, Queener SF. Structure determination of tetrahydroquinazoline antifolates in complex with human and Pneumocystis carinii dihydrofolate reductase: Correlations between enzyme selectivity and stereochemistry. Acta Crystallogr. D Biol. Crystallogr. 2004;60:646–655. doi: 10.1107/S0907444904004650. [DOI] [PubMed] [Google Scholar]
- 43.Cody V, Pace J, Stewart E. Structural analysis of Pneumocystis carinii dihydrofolate reductase complexed with NADPH and 2,4-diamino-6-[2-(5-carboxypent-1-yn-1-yl)-5-methoxybenzyl]-5-methylpyrido[2,3-d]pyrimidine. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:418–423. doi: 10.1107/S1744309112011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Field M, Szebenyi D, Stover P. Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J. Biol. Chem. 2006;281:4215–4221. doi: 10.1074/jbc.M510624200. [DOI] [PubMed] [Google Scholar]
- 45.Giaever G, Nislow C. The yeast deletion collection: A decade of functional genomics. Genetics. 2014;197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hajikarimlou M, et al. Lithium chloride sensitivity in yeast and regulation of translation. Int. J. Mol. Sci. 2020;21:5730. doi: 10.3390/ijms21165730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takallou S, et al. The involvement of YNR069C in protein synthesis in the Baker’s yeast, Saccharomyces cerevisiae. Biology. 2024;13:138. doi: 10.3390/biology13030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustin S, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 49.Chambers A, et al. Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Mol. Cell. Biol. 1989;9:5516–5524. doi: 10.1128/mcb.9.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogan N, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stansfield I, Akhmaloka, Tuite M. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet. 1995;27:417–426. doi: 10.1007/bf00311210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.