Abstract
Initiation of DNA replication in eukaryotic cells is regulated through the ordered assembly of replication complexes at origins of replication. Association of Cdc45 with the origins is a crucial step in assembly of the replication machinery, hence can be considered a target for the regulation of origin activation. To examine the process required for SpCdc45 loading, we isolated fission yeast SpSld3, a counterpart of budding yeast Sld3 that interacts with Cdc45. SpSld3 associates with the replication origin during G1–S phases and this association depends on Dbf4-dependent (DDK) kinase activity. In the corresponding period, SpSld3 interacts with minichromosome maintenance (MCM) proteins and then with SpCdc45. A temperature-sensitive sld3-10 mutation suppressed by the multicopy of the sna41+ encoding SpCdc45 impairs loading of SpCdc45 onto chromatin. In addition, this mutation leads to dissociation of preloaded Cdc45 from chromatin in the hydroxyurea-arrested S phase, and DNA replication upon removal of hydroxyurea is retarded. Thus, we conclude that SpSld3 is required for stable association of Cdc45 with chromatin both in initiation and elongation of DNA replication. The DDK-dependent origin association suggests that SpSld3 is involved in temporal regulation of origin firing.
INTRODUCTION
In eukaryotic DNA replication, replication complexes assemble at origins of replication through a cell cycle-regulated process. In budding and fission yeast, origins of replication are recognized and bound by the origin recognition complex (ORC) throughout the cell cycle. As cells exit mitosis, Cdc6/Cdc18 and Cdt1 associate with origins and then recruit Mcm2–7 proteins to form a prereplicative complex (pre-RC) (Baker and Bell, 1998; Leatherwood, 1998; Lygerou and Nurse, 2000).
Cdc45/Sna41 is a factor with genetic interactions with minichromosome maintenance (MCM) proteins in budding and fission yeast (Hardy, 1997; Zou et al., 1997; Miyake and Yamashita, 1998). Cdc45 associates with chromatin in an MCM-dependent manner (Zou and Stillman, 1998; Mimura et al., 2000) and the resulting complex is termed preinitiation complex. The tight association of replication protein A with chromatin, which is a prerequisite for loading of Pol α occurs only in the presence of Cdc45 on chromatin (Mimura et al., 2000). In addition, Cdc45 associates with the Pol α complex (Mimura and Takisawa, 1998; Kukimoto et al., 1999; Uchiyama et al., 2001) and Pol ε (Zou and Stillman, 2000). Thus, Cdc45 is considered to have a critical function related to assembly of replication forks at replication origins (Takisawa et al., 2000).
The timing of origin association of Cdc45 corresponds to that of origin firing, yet the timing of MCM association does not vary among origins in budding yeast (Tanaka and Nasmyth, 1998; Aparicio et al., 1999). Cdc45 loading onto late-firing origins is inhibited, when the S-phase checkpoint is activated by depletion of the nucleotide pool. In Xenopus egg extracts, Cdc45 loading is abolished when the ATM-mediated checkpoint system is activated by DNA damage (Costanzo et al., 2000). Therefore, Cdc45 loading can be considered a target for the regulation of origin activation.
Chromatin association of Cdc45 requires actions of S-phase cyclin-dependent kinase (S-CDK) and Cdc7/Dbf4 kinase (Dbf4-dependent kinase, DDK) (Mimura and Takisawa, 1998; Arata et al., 2000; Jares and Blow, 2000; Walter, 2000; Zou and Stillman, 1998, 2000). Because several MCM subunits are phosphorylated by DDK in vitro and in vivo (Jares et al., 2000), the MCM complex seems to be the major physiological target of DDK kinase. However, the consequence of the phosphorylation of MCM subunits is not well defined. On the other hand, how CDK prevents the rereplication has been reported (Nguyen et al., 2001; Vas et al., 2001), yet little is known about what component of the replication machinery must be phosphorylated by CDK before initiation of replication.
DPB11 is isolated as a high-copy suppressor for mutations in two essential subunits of DNA polymerase II (ε) (Araki et al., 1995). Dpb11 interacts with Pol ε during the S phase and the origin association of these proteins is a prerequisite for Pol α-primase loading (Masumoto et al., 2000). Furthermore, an elevated dosage of DPB11 suppresses mutation in the carboxyl-terminal domain of Pol ε, which is essential for S-phase checkpoint (Navas et al., 1995; Dua et al., 1999; Kesti et al., 1999). Thus, the Dpb11–Pol ε complex may be required for DNA replication and for S-phase checkpoint. During screening for synthetic lethal mutants with the dpb11-1 (sld mutants), sld4-1 was found to be an allele of CDC45 (Kamimura et al., 1998). A novel gene, SLD3, isolated as one of the sld mutants, encodes a protein that interacts with Cdc45 in a two-hybrid system (Kamimura et al., 2001), hence Sld3 may be a factor functioning with Cdc45 and Dpb11.
To elucidate the mechanisms of origin activation and its regulation, the process of Cdc45 loading should be clarified. In this study, we focused on the roles of SpSld3, a fission yeast counterpart of Sld3. Based on our results, we suggest that SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in both initiation and elongation of DNA replication. The DDK-dependent origin association of SpSld3 suggests that SpSld3 may be involved in temporal regulation of origin activation.
MATERIALS AND METHODS
Strains and Media
The Schizosaccharomyces pombe haploid strains used were 972 h− and 975 h+ (Gutz and Doe, 1975), h− sna41-1 (Miyake and Yamashita, 1998; Masukata, unpublished data), h− cdc25-22, h− cdc10-129, h− cdc17-K42, h− cdc20-M10 (Nurse et al., 1976; Fantes, 1981), and h− nda3-KM311 (Hiraoka et al., 1984). H− cds1::ura4 leu1 ura4-D18 was a generous gift from Dr. A. Carr (University of Sussex, Sussex, United Kingdom) and h+ hsk1-89 leu1 from Dr. H. Masai (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). For gene disruption, a diploid strain was obtained by mating between haploids JY741 h− ade6-M216 leu1-32 ura4-D18, and JY746 h+ ade6-M210 leu1-32 ura4-D18. Complete YPD medium (1% yeast extract, 2% BactoPeptone, and 2% glucose) and the minimal medium (Alfa et al., 1993) were used.
Isolation of Fission yeast sld3+ Gene and Construction of Plasmids
A hypothetical gene encoding Sld3 homolog was found on a cosmid SPAC24.H6 in the fission yeast database. A 2.1-kb fragment containing the entire open reading frame (ORF) of the predicted sld3+ gene was polymerase chain reaction (PCR) amplified and was cloned into the pBluescriptII SK(+), to make pSLD3-PCR. pSLD3-Xb carrying the 6.2-kb XbaI genomic fragment containing the sld3+ gene was isolated by colony hybridization from a pUC119-XbaI library containing 6.0–6.5-kb XbaI fragments of 972 genomic DNA, by using the ORF fragment of pSLD3-PCR, as a probe. The nucleotide sequences of pSLD3-PCR were confirmed and the fragment containing the entire ORF of the sld3+ was inserted into pREP81-NotI, a derivative of thiamine-inducible expression vector pREP81 (Maundrell, 1993) carrying a NotI site, resulting in pSLD3. A 48-base pair intron from position +43 to +90 predicted in the ORF was confirmed by reverse transcription-PCR, by using the total cellular RNA as the template (Uchida and Masukata, unpublished data). pSNA41 is a derivative of pARS2004 (Okuno et al., 1999) carrying a 3.1-kb Sau3AI genomic fragment that contains the sna41+ gene (Masukata, unpublished data).
Disruption of sld3
One-step gene replacement method (Rothstein, 1991) was used for disruption of the sld3+ gene. A 1.7-kb SpeI-XhoI segment of the sld3+ in pSLD3-Xb was replaced with the 1.8-kb HindIII fragment carrying the ura4+ gene. The 3.7-kb NdeI fragment carrying the sld3::ura4+ gene was used for transformation of JY741/JY746 diploid cells. Ura+ transformants were selected and replacement of one of the sld3+ genes with sld3::ura4+ was confirmed by Southern hybridization. Spores generated from the sld3+/sld3::ura4+ heterodisruptants were dissected.
Isolation of Temperature-sensitive sld3 Mutants
The PCR-based gene targeting method (Bahler et al., 1998) was used to construct temperature-sensitive sld3 mutants. The ura4+ gene was inserted at the NdeI site 1.2 kb downstream of the termination codon of the sld3+ gene in the 6.2-kb XbaI fragment, and the resulting pSLD3-mut was used as a template for PCR amplification. A 7.2-kb fragment was amplified using LA Taq DNA polymerase (TAKARA) with primers 5′-GCAACAATCAGAATGTCGTC-3′ and 5′-GTAGCCAAAAGCCTTCCATG-3′. HM21 h− ura4-D18 was transformed with the PCR fragments. Among 3000 Ura+ transformants grown at 23°C, three temperature-sensitive mutants that did not grow at 36°C were selected. Mutation sites were roughly mapped by an integration rescue method. Integration plasmids pSLD3-N and pSLD3-C were made by insertion of a 1.9-kb XbaI-BamHI and a 1.7-kb BamHI fragments encoding N- and C-terminal halves, respectively, of the sld3+ gene into pYC11, a derivative of pBluescript SK(+) carrying Saccharomyces cerevisiae LEU2 gene at the Nae I site (Takahashi et al., 1994). When leu1-32 derivatives of the sld3-10, -41, and -52 were transformed with either plasmid, the temperature-sensitive growth of all three mutants was rescued by the integration of pSLD3-N. The corresponding regions of the mutants were amplified by PCR and the nucleotide sequences were determined, using the ABI310 sequencing system.
Epitope Tagging of sld3+ and sna41+
To construct the sld3 gene with the carboxyl terminus tagged with FLAG-epitope, a 1-kb fragment containing the C-terminal region of sld3+ was PCR amplified using primers 5′-AACTGCAGGGGCTTTGCATACGAAGGAC-3′ and 5′-CGGGATCCTCACTTGTCATCGTCGTCCTTGTAGTCCATATGAGGACTGGCTGATTTTTTTTTA-ACAGGTG-3′, creating a FLAG-epitope sequence and an NdeI site upstream of the termination codon. Then, a 140-base pair NdeI fragment containing five FLAG-epitope sequences was inserted at the NdeI site. pPA-URA was prepared by insertion of the BamHI-EcoRI fragment containing the polyadenylation signal of the nmt1 gene (Maundrell, 1993) and the ura4+ gene into pUC119. pSLD3-FLAG5 was prepared by insertion of the sld3+ gene tagged with five FLAG-epitopes into pPA-URA and used for transformation of HM21 h− ura4-D18. A Ura+ transformant, RY122 h− sld3-flag-ura4+ ura4-D18, was obtained and integration of the plasmid at the sld3+ locus was confirmed by Southern hybridization.
A strain expressing SpCdc45-Myc was constructed, as described below. A 1.0-kb NotI fragment encoding nine tandem Myc-epitopes was inserted at the NotI site created before the stop codon of sna41+ gene of pSNA41. The resulting 3.5-kb SphI-XbaI fragment containing a 1.8-kb region upstream of the termination codon, Myc-epitopes, and a 650-base pair region downstream of the termination codon were inserted into pUC119 together with a 1.8-kb HindIII fragment carrying ura4+ gene, resulting in pSNA41-Myc. pSNA41-Myc was used for transformation of HM21 h− ura4-D18. A Ura+ transformant, RY46 h− sna41-myc9-ura4+ ura4-D18 was obtained and integration was confirmed by the Southern hybridization.
RY46 was backcrossed with 975 h+ to construct RY59 h+ sna41-myc9. RY80 h+ orp1-flag5 sna41-myc9 was obtained by crossing between RY59 and TTY15 (Ogawa et al., 1999). RY173 h− sld3-10 orp1-flag5 sna41-myc9 was obtained by crossing between RY80 and h− sld3-10. RY122 was crossed with 975 h+, resulting in RY124 h− sld3-flag5 and RY125 h+ sld3-flag5. RY124 or RY125 was mated with cdc mutants to construct RY163 h− cdc25-22 sna41-myc9 sld3-flag5, RY139 h− cdc10–129 sld3-flag5, RY144 h− cdc17-K42 sld3-flag5 and RY148 h− cdc20-M10 sld3-flag5. RY137 h− nda3-KM311 sld3-flag5 and RY199 h− hsk1–89 sld3-flag5 were made by crossing RY125 with h− nda3-KM311 leu1 and RY124 with h+ hsk1-89 leu1, respectively.
Preparation of Chromatin-enriched Fractions
Chromatin-enriched fractions were prepared from haploid cells, as described previously (Ogawa et al., 1999) but with the following modifications. After the incubation in ice-cold stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3) for 5 min, cells were washed and resuspended to 5 × 108 cells/ml in PEMS buffer [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) pH 6.9, 1 mM EGTA, 1 mM MgSO4, 1 M sorbitol] containing 2% β-mercaptoethanol. Zymolyase 100T (Seikagaku, Tokyo, Japan) and Lysing enzyme (Sigma-Aldrich, St. Louis, MO) were added to a final concentration of 0.2 and 5 mg/ml, respectively. The suspension was incubated at 30°C for 60 min. Spheroplasts were washed twice with ice-cold PEMS and lysed in HB buffer [25 mM 3-(N-morpholino)propanesulfonic acid pH 7.2, 15 mM MgCl2, 15 mM EGTA, 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 0.5 mM Na3VO4, 1 mM dithiothreitol, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, 20 μg/ml aprotinin] containing 1% Triton X-100 at a concentration of 108 cells/ml.
Immunoprecipitation and Immunoblotting
The recombinant proteins encoding amino acid position 192–440 of SpCdc45 or N-terminal 1–211 amino acids of SpMcm7 were expressed in Escherichia coli and purified as described previously (Ogawa et al., 1999). Polyclonal rabbit antisera were raised against these proteins (Hokudo, Sapporo, Japan). Anti-SpCdc45 antiserum was further affinity-purified on an N-hydroxysuccinimide–activated column.
For immunoprecipitation, exponentially growing cells (5 × 108) were suspended in 300 μl of HB buffer. The cells were disrupted using glass beads and extracts were centrifuged at 4°C at 14,000 rpm for 20 min. Extracts were preincubated with magnetic beads (Dynal Biotech, Oslo, Norway) in the presence of 150 U of DNase I (Takara, Kyoto, Japan) at 4°C for 1 h then the precleared lysate was incubated with magnetic beads associated with mouse anti-FLAG monoclonal antibody M2 (Sigma-Aldrich), anti-SpCdc45 polyclonal antibody, or rabbit anti-SpMcm6 polyclonal antisera (Ogawa et al., 1999) for 2 h. Immunoblotting was carried out as described previously (Ogawa et al., 1999). In addition to the anti-FLAG antibody, anti-SpMcm6, anti-SpMcm7, anti-SpOrc4, and anti-SpCdc45 polyclonal serum, rabbit anti-Myc polyclonal antibody (MBL, Nagoya, Japan) was used for immunoblotting. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies (1000-fold dilution; Amersham Biosciences, Piscataway, NJ) were the second antibodies used and the binding was visualized using the Amersham ECL system. Calf intestinal phosphatase (CIP) treatment of immunoprecipitates was done as described previously (Lindsay et al., 1998). Phosphatase inhibitors used were 60 mM β-glycerophosphate and 15 mM p-nitrophenylphosphate.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation assay was done as described previously (Ogawa et al., 1999). Briefly, cells were fixed with formaldehyde and DNA fragments associated with SpSld3-FLAG and SpMcm6 were immunoprecipitated with anti-FLAG antibody and anti-SpMcm6 polyclonal antisera, respectively. The ars2004 and non-ARS regions were PCR amplified from the total cellular DNA (Total) and the immunoprecipitated DNA by using the ars2004 and two non-ARS primer sets and subjected to agarose gel electrophoresis. The ars2004 primers amplify a 239-base pair fragment in the origin. The non-ARS (1) and non-ARS (2) primers amplify 289- and 195-base pair fragments ∼30 and 18 kb distant from the ars2004, respectively.
Flow Cytometric Analysis
Flow cytometric analyses were made after propidium iodide staining of cells, by using BD Biosciences FACScan as described previously (Ogawa et al., 1999).
RESULTS
Fission Yeast sld3+ Is Essential for Cell Growth
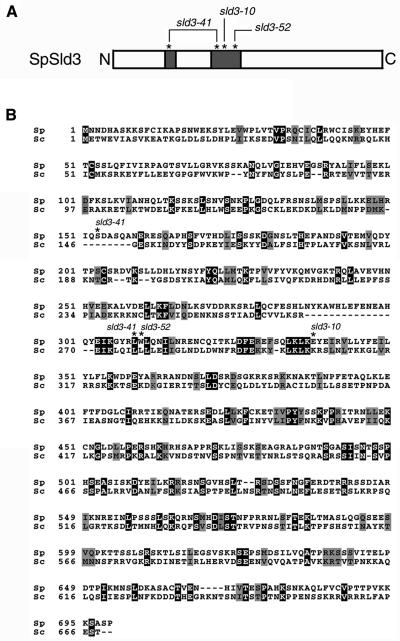
We identified a hypothetical protein (GenBank CAA90850.2) in a fission yeast genome database. This protein shares 14% identity and 24% similarity with budding yeast Sld3 (Figure 1B). The gene encoding the protein was named as sld3+, as a potential counterpart of budding yeast SLD3. The sld3+ encodes a 699-amino acid polypeptide with a calculated molecular mass of ∼79 kDa. This protein does not contain characteristic protein motifs, except for putative coiled-coil motifs in regions 142–165 and 253–341 of amino acid positions (Figure 1A).
Figure 1.
Structure of S. pombe SpSld3. (A) Schematic representation of SpSld3 is shown. The shaded boxes indicate the coiled-coil regions predicted by COILS program. Asterisks indicate the positions of amino acids changed by the sld3-10, -41, and -52 mutations. (B) Predicted amino acid sequences are aligned with those of S. cerevisiae Sld3, by using the ClustalW method. Identical amino acids are shaded and similar amino acids are hatched by boxshade. Asterisks indicate the positions of amino acids altered by the sld3-10, -41, and -52 mutations as described in the text.
We replaced one of the sld3+ copies in a ura− diploid with the sld3::ura4+. Sporulation and tetrad dissection of the sld3+/sld3::ura4 heterodisruptant yielded two viable and two lethal spores and all viable spores were Ura−. To confirm that the lethality was due to disruption of the sld3+, the sld3+/sld3::ura4+ diploids transformed with pSLD3 plasmid carrying the sld3+ gene were sporulated. Tetrad dissection yielded three and four viable spores, including Ura+ spores. Thus, the sld3+ is indeed essential for cell growth.
Temperature-sensitive sld3 Mutants Are Defective in DNA Replication
To investigate the essential role of Sld3 in cell growth, we isolated temperature-sensitive mutants of sld3. Mutations were introduced by PCR into a fragment carrying the sld3+ and the ura4+ genes and the PCR products were used for transformation of a ura− haploid strain. Among 3000 Ura+ transformants grown at 23°C, three mutants that did not grow at 36°C were isolated. Growth of the mutants at the restrictive temperature was restored by transformation with pSLD3, which suggested that the sld3 gene has the mutations.
Determination of the nucleotide sequence of the mutant sld3 genes revealed that the glutamic acid residue at position 338 is replaced with glycine in the sld3-10, tryptophan at position 310 with arginine in the sld3-52, and the serine at position 153 and the leucine at position 309 with leucine and proline, respectively, in the sld3-41. It should be noted that all of these changes reside in predicted coiled-coil regions, although these amino acids are not conserved between the budding yeast Sld3 and SpSld3, except for one of two sites in the sld3-41 (Figure 1A).
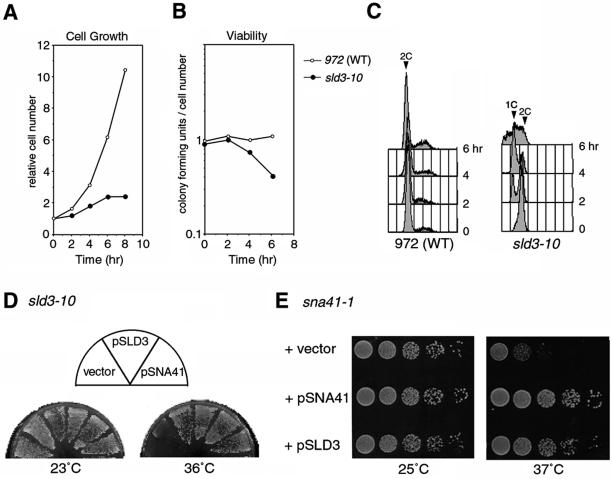
We first examined growth of the sld3-10 at the restrictive temperature. The increase of the cell number was retarded during 2–4 h at 36°C and arrested after 6 h (Figure 2A). Cell viability decreased to ∼40% at 6 h (Figure 2B). To determine effects of sld3-10 mutation on DNA replication, we analyzed DNA contents at the restrictive temperature, by using flow cytometry. sld3-10 cells, initially containing 2C DNA as the wild type, produced cells with 1C DNA content after 2 h at 36°C and the population of these cells increased at 4 h (Figure 2C). In addition, cells with <1C DNA content appeared at 6 h. The other two mutants, sld3-41 and sld3-52, also accumulated cells with 1C DNA content at the restrictive temperature (our unpublished data). Therefore, chromosome DNA is not duplicated in these mutants. SpSld3 is apparently required for DNA replication or for G1–S progression of cell cycle.
Figure 2.
Temperature-sensitive growth of the sld3-10. Exponentially growing wild-type (972) and the sld3-10 cells at 23°C were shifted to 36°C and cells collected at indicated time points were analyzed. The cell number (A) and the cell viability (B) are shown. (C) Flow cytometry profiles are shown. (D) Suppression of sld3-10 by elevated gene dosage of sna41+ is shown. The sld3-10 cells transformed with a vector, pSLD3, or pSNA41 plasmid were streaked out on EMM2 plates and incubated at 23 or 36°C. (E) Suppression of sna41-1 by sld3+ is shown. The logarithmically growing sna41-1 cells carrying a vector, pSNA41, or pSLD3 plasmid were spotted after serial dilutions and incubated at 25 or 37°C.
With 4,6-diamidino-2-phenylindole staining, we observed that ∼10% of sld3-10 cells contained unequally divided nucleus or undivided nucleus cleaved by the septum after 6 h at 36°C, yet such cells were not observed in the wild type (our unpublished data). These cells would correspond to cells with <1C DNA content in flow cytometry analysis and these are presumably formed as a result of nuclear division without completion of DNA replication. These results suggest that the sld3-10 cells have a defect in generation of S-M checkpoint signal.
Budding yeast SLD3 has a genetic interaction with SLD4/CDC45 (Kamimura et al., 2001). To investigate such interaction between the sld3+ and the sna41+ gene encoding SpCdc45, sld3 mutants were transformed with a high-copy plasmid carrying sna41+. The growth of sld3-10, -41, and -52 at the restrictive temperature was restored by the sna41+ plasmid (Figure 2D; our unpublished data). In a reciprocal experiment, growth of the sna41-1, a temperature-sensitive mutant of the sna41, was restored by transformation with pSLD3 (Figure 2E). Therefore, strong genetic interactions exist between sld3+ and sna41+. Taken together with the sequence similarity, sld3+ seems to be a counterpart of budding yeast SLD3.
Cell Cycle-dependent Interactions of SpSld3 with SpCdc45 and SpMcm6
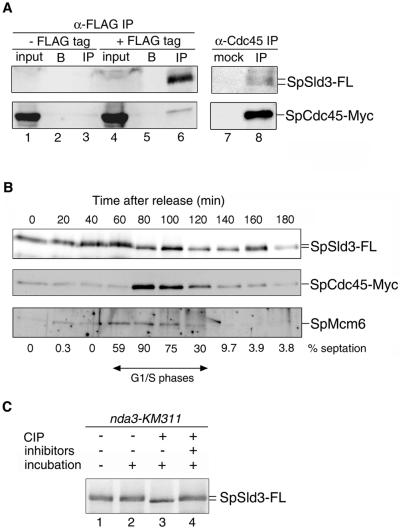
Based on genetic interactions between the sld3+ and sna41+, we examined physical interactions between SpSld3 and SpCdc45. Cells expressing SpCdc45-Myc or both SpCdc45-Myc and SpSld3-FLAG from chromosome loci were arrested in the early S phase by adding hydroxyurea (HU), which inhibits deoxyribonucleotide synthesis. SpSld3 was immunoprecipitated in the presence of DNase I and this would reduce nonspecific coprecipitation mediated by DNA. SpCdc45 was coprecipitated from the cells expressing SpSld3-FLAG (Figure 3A, lane 6), but not from the untagged strain (lane 3) or without anti-FLAG antibody (lane 5). In a reciprocal experiment using an anti-SpCdc45 antibody, SpSld3-FLAG coprecipitated with SpCdc45 (Figure 3A, lane 8). These observations mean that SpSld3 associates with SpCdc45 in the early S phase.
Figure 3.
Coimmunoprecipitation of SpCdc45 and SpMcm6 with SpSld3 in G1–S phases. (A) Cells expressing both SpSld3-FLAG5 and SpCdc45-Myc9 (lanes 4–6 and 7 and 8) or only SpCdc45-Myc9 (lanes 1–3) were cultured in the presence of 10 mM HU for 3 h at 30°C to arrest in the early S phase. Whole cell extracts (lanes 1 and 4) were prepared and subjected to immunoprecipitation with anti-FLAG antibody (lanes 3 and 6), mock treatment without antibody (lanes 2 and 5), with normal rabbit serum (lane 7), or anti-SpCdc45 antibody (lane 8). Ten times more immunoprecipitates compared with the input were used for Western blotting with anti-FLAG and anti-Myc antibodies. We did not detect SpSld3-FLAG without immunoprecipitation, probably because the amount was below detection limits. (B) cdc25-22 cells expressing SpSld3-FLAG5 and SpCdc45-Myc9 were arrested for 4 h at 36°C and then released at 25°C. At the indicated time points, extracts were immunoprecipitated with anti-FLAG antibody. SpSld3-FLAG, SpMcm6, and SpCdc45 in the immunoprecipitates were analyzed by immunoblotting. Cell cycle progression was monitored by the percentage of cells containing a septum (septation index), as shown below the panels. In fission yeast, cytokinesis occurs about one-quarter of cell cycle later than nuclear division (Alfa et al., 1993). Thus, the septation index becomes maximum approximately at G1–S transition in wild-type cells, under the conditions we used. (C) SpSld3 was immunoprecipitated using an anti-FLAG antibody from the nda3-KM311 sld3-flag5 cells cultured for 4 h at 20°C (lanes 1–4). Immunoprecipitates were treated with CIP in the presence (lanes 4) or the absence (lanes 3) of phosphatase inhibitors.
Next, we examined the association of SpSld3 with SpCdc45 during the cell cycle. Temperature-sensitive cdc25-22 cells expressing SpSld3-FLAG and SpCdc45-Myc were arrested at the G2/M boundary and released into the cell cycle at the permissive temperature. Analyses of the SpSld3-immunoprecipitates (IPs) by Western blotting showed that SpCdc45 coprecipitated 80–120 min after release (Figure 3B). The timing of the coprecipitation was shortly after increase in the septation index, which suggests that SpSld3 associates with SpCdc45 mainly in the S phase. On the other hand, SpMcm6 was detected in SpSld3-IPs at 60–100 min, such being earlier than SpCdc45. SpMcm7 also coprecipitated with SpSld3 during the same period as SpMcm6 (our unpublished data). SpSld3 probably associates with MCM proteins before the association with SpCdc45.
SpSld3 migrated as diffused bands during 0–60 min after release from the G2/M block, whereas it formed a sharp band with a higher mobility at 80 min and later time points. To examine the possibility that the slower mobility of SpSld3 was due to the phosphorylation, SpSld3 immunoprecipitated from the nda3-KM311 sld3-flag5 cells arrested in M phase (Hiraoka et al., 1984) was incubated with CIP. This phosphatase treatment yielded a sharp band with a higher mobility (Figure 3C, lane 3), whereas the slowly migrating bands remained in the presence of phosphatase inhibitors (Figure 3C, lane 4). Thus, SpSld3 is phosphorylated in M-phase–arrested cells. The higher mobility band observed during the S phase (Figure 3B) seems to be a hypophosphorylated form.
Localization of SpSld3 at Chromosomal Replication Origin during G1–S Phases
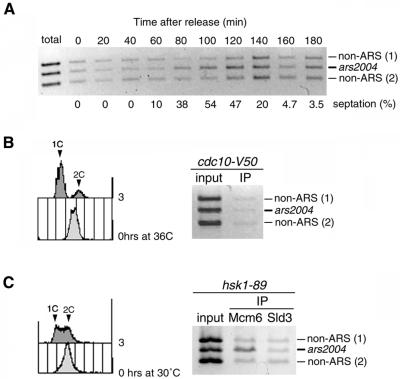
SpMcm6 localizes to the replication origin during G1–S phases (Ogawa et al., 1999). Because SpSld3 associates with SpMcm6, we asked whether SpSld3 is associated with the replication origin during a specific phase of the cell cycle. We did chromatin ChIP assays (Ogawa et al., 1999) by using cdc25-22 cells expressing SpSld3-FLAG, synchronized as described above. The cells were cross-linked with formaldehyde at indicated time points and DNA fragments associated with SpSld3-FLAG were recovered by immunoprecipitation. SpSld3 association was analyzed by PCR for the chromosomal replication origin ars2004 (Okuno et al., 1997), together with two non-ARS regions. The three fragments were amplified to a similar extent from total cellular DNA used as a template (Figure 4A). On the other hand, when immunoprecipitated DNA was used as a template, the ars2004 fragment was preferentially amplified during 80–120 min after release (Figure 4A) and this period corresponded to G1–S phases monitored by an increase in the septation index. Preferential amplification of the ars2004 fragment depended on the FLAG-tagged SpSld3 and on cross-linking treatment (our unpublished data). These observations mean that SpSld3 associates with the ars2004 origin during G1–S phases.
Figure 4.
SpSld3 associates with origin DNA in G1–S phases.(A) The cdc25-22 cells expressing SpSld3-FLAG5 and SpCdc45-Myc9 were synchronized, as described for Figure 3B. At every 20 min, cells were fixed with formaldehyde and DNA fragments associated with SpSld3-FLAG were immunoprecipitated with anti-FLAG antibody. The ars2004 and non-ARS regions were PCR amplified from the total cellular DNA (Total) and the immunoprecipitated DNA by using the ars2004 and two non-ARS primer sets and subjected to agarose gel electrophoresis. The percentages of cells containing a septum are shown below the panel. (B) cdc10-V50 sld3-flag5 cells grown at 23°C were incubated for 3 h at 36°C. Cells were subjected to chromatin-immunoprecipitation with anti-FLAG antibody as described above. The flow cytometry profiles at 0 and 3 h are shown (left). (C) hsk1-89 sld3-flag5 cells were shifted to 30°C and incubated for 3 h. Cells were subjected to chromatin-immunoprecipitation with anti-SpMcm6 antibody or anti-FLAG antibody. The flow cytometry profile shows that about half of cells contained unreplicated DNA (left).
To examine timing of the origin-association of SpSld3 more precisely, we did ChIP assays by using the cdc10 strain in which Cdc18 or Cdt1 is not expressed and MCM proteins are not loaded onto the chromatin at the restrictive temperature (Ogawa et al., 1999; Nishitani et al., 2000). cdc10-V50 cells expressing SpSld3-FLAG were incubated at 36°C for 3 h to arrest the cell cycle in the early G1 phase. The ars2004 fragment was not preferentially amplified from immunoprecipitates of SpSld3 (Figure 4B). To verify that the ChIP assay functioned for the cells incubated at the high temperature, cdc25-22 cells released from G2/M boundary were arrested in the early S phase in the presence of HU, and then incubated at 36°C for 1 h. The ars2004 fragment was selectively amplified from DNA immunoprecipitated with SpSld3 to an extent similar to the sample not incubated at 36°C (our unpublished data). These results suggest that the SpSld3 is not loaded onto the replication origin at the cdc10-arrest point in the G1 phase.
We then examined the origin association of SpSld3 in hsk1-89 mutant, a temperature-sensitive mutant of Hsk1 kinase (Takeda et al., 2001). It was reported that DDK activity is not required for chromatin loading of MCM but is required for Cdc45 loading in Xenopus egg extracts (Jares and Blow, 2000; Walter, 2000). Thus, in hsk1-89 cells, the cell cycle is expected to arrest before initiation, in late G1 phase. hsk1-89 mutant cells expressing SpSld3-FLAG were incubated at the restrictive temperature (30°C) for 2 h then subjected to ChIP assay. The ars2004 fragment was preferentially amplified from the SpMcm6-IPs (Figure 4C), thus confirming that DDK is not required for MCM loading in fission yeast. The specific amplification of ars2004 fragment was not observed using the SpSld3-IPs as a template (Figure 4C). Because DDK activity is reduced by the hsk1-89 mutation (Takeda et al., 2001), the association of SpSld3 with the replication origin depends on DDK activity. The SpSld3 associates with origins in the late G1 phase, after or on the DDK-execution point.
Chromatin Loading of SpCdc45 Is Impaired by the sld3-10 Mutation
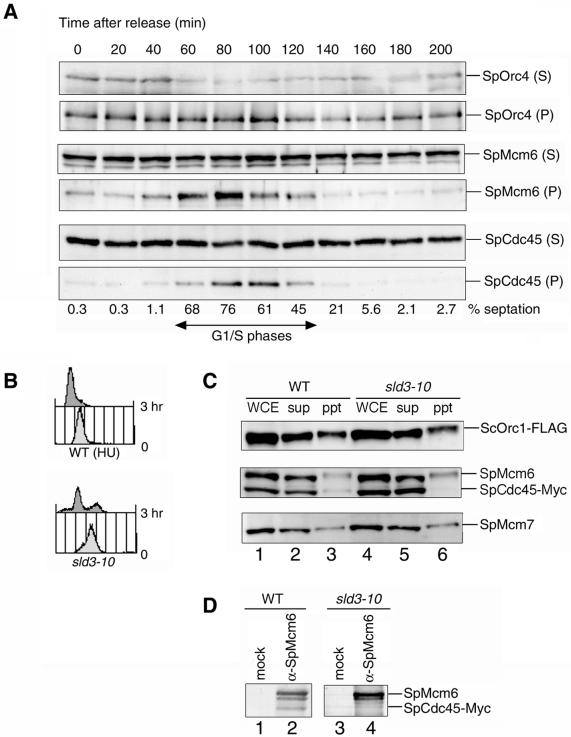
To determine the role of SpSld3 in initiation of DNA replication, we investigated whether the sld3-10 mutation would affect the loading of MCM and SpCdc45 onto chromatin. We first examined the association of MCM and SpCdc45 proteins with chromatin in cell cycle progression. We used cdc25-22 cells expressing SpCdc45-Myc and SpSld3-FLAG synchronized as described above. At the indicated time points, the chromatin-enriched fraction was separated from the soluble-protein fraction and analyzed by Western blotting. As shown in Figure 5A, SpOrc4, a subunit of SpORC, was present in the chromatin-enriched fraction throughout the cell cycle. SpMcm6 was associated with chromatin from 40 to 120 min after release, which corresponds to the G1–S phases. SpMcm7 was detected in the chromatin-enriched fraction during the same period as SpMcm6 (our unpublished data). On the other hand, SpCdc45 appeared in the chromatin-enriched fraction at 60 min, peaked at 80–100 min after release, and was then reduced (Figure 5A). Therefore, SpCdc45 associates with chromatin later than do MCM proteins. We did not detect SpSld3 in the chromatin-enriched fraction or in the soluble fraction, probably because the amounts of the protein in these fractions were below detection limits (our unpublished data).
Figure 5.
Association of SpCdc45 with chromatin is reduced in sld3-10 mutant. (A) Cell cycle-dependent association of SpCdc45 with chromatin is shown. The cdc25-22 cells expressing SpSld3-FLAG5 and SpCdc45-Myc9 were synchronized, as described for Figure 3B. Cells collected at the indicated times were subjected to chromatin fractionation. The supernatant (S) and chromatin-enriched fraction (P) were analyzed by Western blotting with anti-SpMcm6, anti-SpCdc45, and anti-SpOrc4 antibodies. The septation index is shown below the panels. (B) Wild-type and sld3-10 cells expressing SpCdc45-Myc9 and SpOrc1-FLAG5 were grown to log phase at 23°C. The sld3-10 cells were then incubated at 36°C for 3 h. The wild-type cells were incubated for 3 h at 36°C in the presence of 12 mM HU to arrest in the early S phase. Results of flow cytometry are shown. (C) Association of SpCdc45 with chromatin in sld3-10 was examined. Lysates were subjected to chromatin fractionation. Whole cell extracts (WCE), supernatant (sup), and chromatin-enriched fraction (ppt) were analyzed by immunoblotting with anti-FLAG (top), anti-SpMcm6 and anti-Myc (middle), and anti-Mcm7 (bottom) antibodies. (D) Association of SpMcm6 with SpCdc45 in sld3-10 at restrictive temperature was examined. The wild-type and sld3-10 cells were incubated at 36°C as in B. Cell lysates were immunoprecipitated with normal rabbit serum (mock IP) or anti-SpMcm6 antibody. Immunoprecipitates were analyzed by immunoblotting with anti-SpMcm6 and anti-Myc antibodies.
To examine the effect of the sld3-10 mutation on association of SpCdc45 with the chromatin, mutant cells expressing SpOrc1-FLAG and SpCdc45-Myc were incubated at 36°C for 3 h. As a control, the wild-type cells were arrested at the early S phase by incubating at 36°C for 3 h in the presence of HU. Flow cytometry revealed that the mutant and the HU-treated wild-type strains accumulated cells with a 1C DNA content (Figure 5B). The results of Western blotting showed that approximately one-third of SpOrc1 and one-tenth each of SpMcm6, SpMcm7, and SpCdc45 were in the chromatin-enriched fraction of the HU-arrested wild-type cells (Figure 5C). In contrast, SpCdc45 was not detected in the chromatin-enriched fraction of sld3-10 extracts, whereas SpOrc1, SpMcm6, and SpMcm7 were detected as the HU-arrested wild type (Figure 5C). Therefore, the sld3-10 mutant has a defect in loading or maintenance of SpCdc45 onto chromatin. Consistent with these results, SpCdc45 did not coprecipitate with SpMcm6 at the restrictive temperature of sld3-10 (Figure 5D). These results suggest that SpSld3 is required for loading of SpCdc45 onto the pre-RCs at replication origins.
Requirement of SpSld3 during S Phase
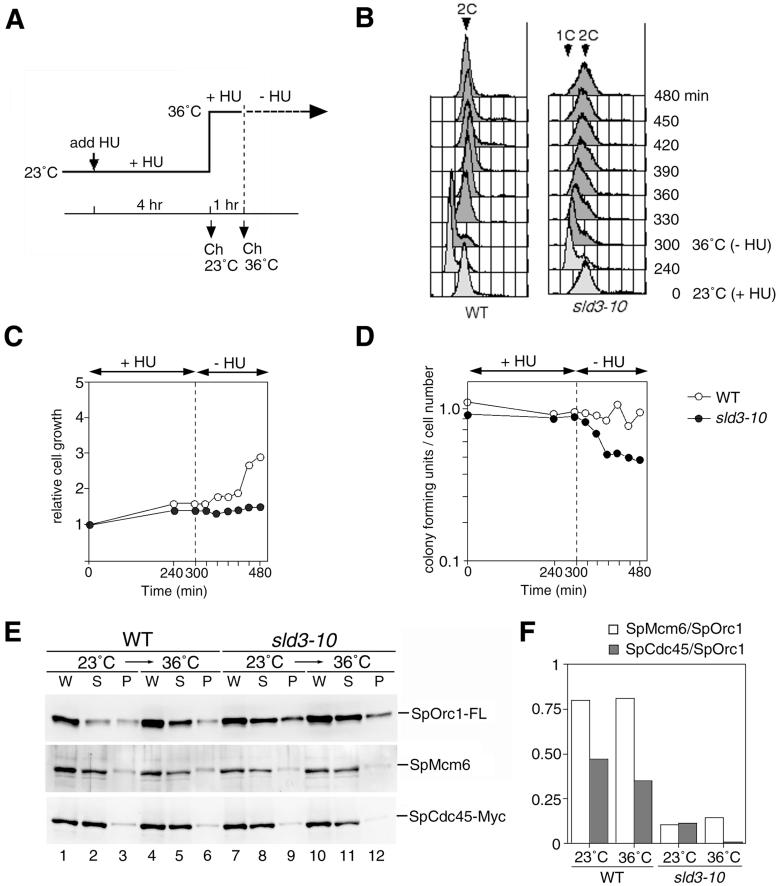
Because MCM and Cdc45 proteins are required for elongation of DNA replication (Labib et al., 2000; Tercero et al., 2000), we determined whether SpSld3 is required after entry into S phase. Wild-type and sld3-10 cells were arrested by HU at 23°C and then incubated at 36°C to inactivate the mutant protein. On removal of HU at 36°C, the DNA content of the wild-type cells increased from 1C to 2C within 30 min (Figure 6B). In contrast, sld3-10 cells increased the DNA content very slowly after release. Although the DNA content of sld3-10 cells increased to nearly 2C DNA after 3 h, the cell number did not increase (Figure 6C), probably because DNA synthesis was not completed, as suggested by the broad peak seen on flow-cytometry results. The severe retardation of DNA replication may result from a defect in elongation in addition to a defect in initiation of late-firing origins, which had not been activated in the HU-arrested cells. The gradual loss of viability after release (Figure 6D) might be due to irreversible defects in elongation of DNA replication such as aberrant DNA synthesis.
Figure 6.
SpCdc45 is dissociated from the S-phase–arrested chromatin in sld3-10. (A) Wild-type and sld3-10 cells expressing SpOrc1-FLAG5 and SpCdc45-Myc9 were cultured for 4 h in the presence of 10 mM HU at the permissive temperature (23°C) and then incubated at 36°C for 1 h. Cells were washed and resuspended into the complete medium without HU. (A) Scheme of the experiment is shown. (B) DNA contents of cells after release from HU arrest were analyzed by flow cytometry. The cell numbers (C) and the viability obtained by colony-forming units/cell number (D) are shown. (E) Chromatin fraction was prepared from the HU-arrested cells before and after the incubation for 1 h at 36°C. Whole cell extracts (W), supernatant (S), and chromatin-enriched fraction (P) were analyzed by immunoblotting with anti-FLAG for SpOrc1-FL (top), anti-SpMcm6 (middle), and anti-Myc antibodies for SpCdc45-Myc (bottom). (F) Images in E were quantitated using the LAS1000 plus image analyzer (Fuji Film, Tokyo, Japan) and ratios of SpMcm6/SpOrc1 and SpCdc45/SpOrc1 are presented.
To investigate the role of SpSld3 during the S phase, we examined effects of sld3-10 mutation on stability of chromatin association of MCM and SpCdc45 proteins. In HU-arrested wild-type cells, the amounts of chromatin-bound SpOrc1, SpMcm6, and SpCdc45 remained unchanged after subsequent incubation at 36°C for 1 h (Figure 6E, lane 6). In contrast, in the sld3-10 cells, the amount of SpCdc45 associated with the chromatin was decreased by incubation at 36°C (Figure 6E, lane 12). The amount of SpMcm6 in the chromatin fraction was not significantly affected, as shown by quantification of immunostained signals (Figure 6F). These results show that the SpSld3 is required for maintenance of SpCdc45 on chromatin during the S phase.
DISCUSSION
We obtained evidence that SpSld3, a counterpart of budding yeast Sld3, is required for the initiation of DNA replication. SpSld3 that interacts with MCM and SpCdc45 proteins in G1–S phases is required for the chromatin loading of SpCdc45. Thus, SpSld3 is a factor essential for conversion of pre-RC to preinitiation complex. Moreover, SpSld3 is required for maintenance of SpCdc45 on chromatin in cells arrested in the early S phase. SpSld3 may function in elongation as well as in initiation of DNA replication in fission yeast.
SpSld3 Is a Novel Component of Preinitiation Complex
The sld3-10 and sna41-1 mutations are mutually suppressed with sna41+ and sld3+, respectively. sna41-1 mutation has been isolated as a suppressor for nda4-108, the mutation of SpMcm5 (Miyake and Yamashita, 1998). These genetic interactions suggest that SpSld3, SpCdc45, and MCM proteins function in the same or closely related steps in initiation of DNA replication.
ChIP analyses show that SpSld3 associates with the replication origin during G1–S phases (Figure 4B). In the corresponding period, SpSld3 associates with SpMcm6 (Figure 3B). Because SpMcm6 is localized to the origin during G1–S phases (Ogawa et al., 1999), SpSld3 may form a complex with MCM proteins at the replication origin. On the other hand, SpSld3 associates with SpCdc45 later than the association with SpMcm6 (Figure 3B, 80 min). This timing corresponds to that of the SpCdc45 loading onto chromatin, which has been observed later than SpMcm6 (Figure 5A). Because SpCdc45 is coprecipitated with SpMcm6 during G1–S phases (our unpublished data), SpCdc45 may associate with SpSld3 and MCM proteins at the origin, the result being formation of preinitiation complex.
The association of SpCdc45 with chromatin is abolished by the sld3-10 mutation, yet loading of MCM proteins is not affected (Figure 5C). Thus, chromatin loading of SpCdc45 depends on SpSld3. In light of all the above-mentioned points, we conclude that SpSld3 is required for SpCdc45 loading during initiation. Suppression of the sld3-10 mutation by elevated gene dosage of the sna41+ is consistent with this conclusion. Because all the mutations in three sld3 mutants reside in predicted coiled-coil regions, the coiled-coil regions of SpSld3 may be interfaces for the interaction with SpCdc45 or another factor, which may stabilize the association with SpCdc45.
Requirement for Origin Association of SpSld3
The origin-association of SpSld3 depends on DDK activity (Figure 4C). Although the consequence of the phosphorylation by DDK is currently unknown, the most preferable substrate for DDK is considered to be Mcm2. SpMcm2 in the MCM complex is phosphorylated by DDK in fission yeast (Brown and Kelly, 1998). The phosphorylation of SpMcm2 may possibly be required for interaction of the MCM complex with SpSld3.
The relative timing of the origin association of Sld3 correlates with timing of origin activation in budding yeast (Kamimura et al., 2001). Because DDK is required for activation of each origin (Bousset and Diffley, 1998; Donaldson et al., 1998), it is likely that loading of Sld3 onto each origin is regulated by DDK. Moreover, DDK activity is down-regulated through checkpoint kinase Rad53 (Dohrmann et al., 1999; Weinreich and Stillman, 1999), which inhibits firing of late origins in response to perturbation of DNA replication. Activation of the S-phase checkpoint inhibits association of Cdc45 with late-firing origins (Aparicio et al., 1999), an event possibly mediated by regulation of Sld3 loading by DDK. Together, loading of Sld3 may be a target for both temporal and checkpoint regulation of origin firing.
In budding yeast, loading of Sld3 onto replication origins depends on Cdc45 and vice versa. It has been suggested that a fragile complex of Sld3 and Cdc45, which is detected after cross-linking treatment, is loaded onto the origins (Kamimura et al., 2001). In contrast, the association of SpSld3 with SpMcm6 precedes that with SpCdc45. This raises the possibility that chromatin loading of SpSld3 does not depend on SpCdc45. At present, this possibility remains an open question because the sna41-1 mutation does not impair the loading of either SpSld3 or SpCdc45 (our unpublished data). Although the essential roles of these proteins should be conserved, precise regulation of loading might have diverged between the two yeasts.
Possible Role of SpSld3 in Elongation of DNA Synthesis
Cdc45 and MCM proteins play essential roles in elongation as well as initiation of DNA replication (Labib et al., 2000; Tercero et al., 2000). The sld3-10 mutation affects association of SpCdc45 in early S-phase–arrested cells, the result being slow progression of DNA replication after release (Figure 6). Therefore, SpSld3 plays an essential role after entry into S phase, presumably in elongation of DNA replication.
In the replication machinery of E. coli, tau protein is considered to mediate the association of DnaB helicase with DNA polymerase III holoenzyme (Kim et al., 1996a,b). In eukaryotic cells, several features of MCM proteins suggest that they are likely candidates for a replicative helicase (Labib and Diffley, 2001). Because Cdc45 associates with both Pol α and MCM proteins (Dalton and Hopwood, 1997; Mimura and Takisawa, 1998; Kukimoto et al., 1999; Uchiyama et al., 2001), Cdc45 is a candidate that coordinates DNA helicase with the Pol α-primase complex at replication forks (Baker and Bell, 1998). On the other hand, it has been observed in a two-hybrid system that Sld3 interacts with Dpb11, which forms a complex with Pol ε (Masumoto et al., 2000; Kamimura et al., 2001). Fission yeast sld3-10 mutation shows synthetic lethality with the cut5 mutation (our unpublished data), a possible counterpart of DPB11 (Saka et al., 1994), which suggests that the interaction between SLD3/sld3 and DPB11/cut5 is conserved. Therefore, Sld3 might be a factor that coordinates DNA helicase with Pol ε complex at the replication fork.
Phosphorylation of SpSld3 in Cell Cycle Progression
Although CDK activity is required for chromatin loading of Cdc45, the target of phosphorylation required for the step is unknown. SpSld3 has potential CDK phosphorylation sites in the C terminus. Indeed, the phosphorylation status changes during the cell cycle are consistent with the idea that SpSld3 is phosphorylated by CDK. However, the phosphorylation of SpSld3 by S-CDK may not be required for loading of SpCdc45 for the following reasons. The majority of SpSld3 is hypophosphorylated when it associates with SpCdc45 in an unperturbed cell cycle (Figure 3B). In addition, alterations of three potential CDK phosphorylation sites of SpSld3 did not affect cell growth (our unpublished data). Appearance of the hypophosphorylated form of SpSld3 in G1–S phases raises the possibility that hypophosphorylation of SpSld3 might be a prerequisite for interaction with SpCdc45. In a cell-free replication system in Xenopus egg extracts, the activity of protein phosphatase 2A is required for initiation of DNA replication but not for the binding of ORC or MCM to chromatin (Lin et al., 1998). Whether protein phosphatase 2A or another phosphatase is involved in initiation of DNA replication in fission yeast is unknown, but it may be that phosphatase regulates SpSld3 for SpCdc45 loading or possible later steps.
ACKNOWLEDGMENTS
We thank H. Araki and Y. Kamimura for providing information before publication and for critical discussion, T. Nakagawa and T. Takahashi for critical reading of the manuscript, A. Carr and H. Masai for providing fission yeast strains, and T. Uchida for raising anti-SpCdc45 antibody. This study was supported in part by a grant-in-aid from the Ministry of Education, Science, Technology, Sports and Culture, Japan (to H.M.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–01–0006. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–01–0006.
REFERENCES
- Alfa C, Fantes P, Hyams J, Mcleod M, Warbrick E. Experiments with Fission Yeast, A Laboratory Course Manual. Cold Spring Harbor, NY.: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Aparicio OM, Stout AM, Bell SP. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA. 1999;96:9130–9135. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata Y, Fujita M, Ohtani K, Kijima S, Kato JY. Cdk2-dependent and -independent pathways in E2F-mediated S phase induction. J Biol Chem. 2000;275:6337–6345. doi: 10.1074/jbc.275.9.6337. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Baker TA, Bell SP. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GW, Kelly TJ. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Ying CY, Kim E, Avvedimento E, Gottesman M, Grieco D, Gautier J. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol Cell. 2000;6:649–659. doi: 10.1016/s1097-2765(00)00063-0. [DOI] [PubMed] [Google Scholar]
- Dalton S, Hopwood B. Characterization of Cdc47p-minichromosome maintenance complexes in Saccharomyces cerevisiae: identification of Cdc45p as a subunit. Mol Cell Biol. 1997;17:5867–5875. doi: 10.1128/mcb.17.10.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann PR, Oshiro G, Tecklenburg M, Sclafani RA. RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics. 1999;151:965–977. doi: 10.1093/genetics/151.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiaepol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- Fantes PA. Isolation of cell size mutants of a fission yeast by a new selective method: characterization of mutants and implications for division control mechanisms. J Bacteriol. 1981;146:746–754. doi: 10.1128/jb.146.2.746-754.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H, Doe FJ. On homo- and heterothallism in Schizosaccharomyces pombe. Mycologia. 1975;67:748–759. [PubMed] [Google Scholar]
- Hardy CF. Identification of Cdc45p, an essential factor required for DNA replication. Gene. 1997;187:239–246. doi: 10.1016/s0378-1119(96)00761-5. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Jares P, Blow JJ. Xenopuscdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jares P, Donaldson A, Blow JJ. The Cdc7/Dbf4 protein kinase: target of the S phase checkpoint? EMBO Rep. 2000;1:319–322. doi: 10.1093/embo-reports/kvd076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001;20:2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996a;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. tau couples the leading- and lagging-strand polymerases at the Escherichia coliDNA replication fork. J Biol Chem. 1996b;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- Kukimoto I, Igaki H, Kanda T. Human CDC45 protein binds to minichromosome maintenance 7 protein and the p70 subunit of DNA polymerase alpha. Eur J Biochem. 1999;265:936–943. doi: 10.1046/j.1432-1327.1999.00791.x. [DOI] [PubMed] [Google Scholar]
- Labib K, Diffley JF. Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr Opin Genet Dev. 2001;11:64–70. doi: 10.1016/s0959-437x(00)00158-1. [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- Lin XH, Walter J, Scheidtmann K, Ohst K, Newport J, Walter G. Protein phosphatase 2A is required for the initiation of chromosomal DNA replication. Proc Natl Acad Sci USA. 1998;95:14693–14698. doi: 10.1073/pnas.95.25.14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJ, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-Phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Nurse P. Cell cycle. License withheld–geminin blocks DNA replication. Science. 2000;290:2271–2273. doi: 10.1126/science.290.5500.2271. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Mimura S, Masuda T, Matsui T, Takisawa H. Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopusegg extracts. Genes Cells. 2000;5:439–452. doi: 10.1046/j.1365-2443.2000.00340.x. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. XenopusCdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Yamashita S. Identification of sna41 gene, which is the suppressor of nda4 mutation and is involved in DNA replication in Schizosaccharomyces pombe. Genes Cells. 1998;3:157–166. doi: 10.1046/j.1365-2443.1998.00177.x. [DOI] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Okazaki T, Masukata H. Identification of a predominant replication origin in fission yeast. Nucleic Acids Res. 1997;25:530–537. doi: 10.1093/nar/25.3.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Satoh H, Sekiguchi M, Masukata H. Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol Cell Biol. 1999;19:6699–6709. doi: 10.1128/mcb.19.10.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants miscause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Tatebayashi K, Ikeda H, Arai K, Masai H. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol Biol Cell. 2001;12:1257–1274. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takisawa H, Mimura S, Kubota Y. Eukaryotic DNA replication. from pre-replication complex to initiation complex. Curr Opin Cell Biol. 2000;12:690–696. doi: 10.1016/s0955-0674(00)00153-8. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JF. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama M, Griffiths D, Arai K, Masai H. Essential role of Sna41/Cdc45 in loading of DNA polymerase alpha onto minichromosome maintenance proteins in fission yeast. J Biol Chem. 2001;276:26189–26196. doi: 10.1074/jbc.M100007200. [DOI] [PubMed] [Google Scholar]
- Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JC. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopusegg extracts. J Biol Chem. 2000;275:39773–39778. doi: 10.1074/jbc.M008107200. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]