Abstract
The spindle checkpoint prevents anaphase from occurring until all chromosomes have attached properly to the mitotic spindle. The checkpoint components Mad1 and Mad2 associate with unattached kinetochores and are probably involved in triggering the checkpoint. We now demonstrate that in Xenopus egg extracts Mad1 and Mad2 form a stable complex, whereas a fraction of Mad2 molecules is not bound to Mad1. The checkpoint establishment and maintenance are lost upon titrating out free Mad2 with an excess of Mad1 or a truncated Mad1 (amino acids 326–718, Mad1C) that contains the Mad2-binding region. Mad1N (amino acids 1–445) that binds kinetochores, but not Mad2, reduces Mad1 and Mad2 at kinetochores and abolishes checkpoint maintenance. Furthermore, the association between Mad2 and Cdc20, the activator for the anaphase-promoting complex, is enhanced under checkpoint-active condition compared with that at metaphase. Immunodepletion analysis shows that the Mad1-free Mad2 protein is unable to bind Cdc20, consistent with the model that kinetochore localization of Mad2 facilitates the formation of Mad2–Cdc20 complex. This study demonstrates that the ratio between Mad1 and Mad2 is critical for maintaining a pool of Mad1-free Mad2 that is necessary for the spindle checkpoint. We propose that Mad2 may become activated and dissociated from Mad1 at kinetochores and is replenished by the pool of Mad1-free Mad2.
INTRODUCTION
Segregation of newly duplicated sister chromatids into daughter cells during anaphase is a critical event in each cell division cycle. Any mishap in this process gives rise to aneuploidy that is common in human cancers and some forms of genetic disorders. In eukaryotic cells, anaphase initiates only after all chromosomes have established stable attachment to spindle microtubules emanating from opposite spindle poles. Defects in spindle assembly or chromosome attachment prevent the onset of anaphase by activating the spindle checkpoint. The checkpoint signal is generated at kinetochores that are not occupied by microtubules (Rieder et al., 1995) or lack tension that normally comes from bipolar attachment of microtubules (Li and Nicklas, 1995).
Some of the spindle checkpoint components are evolutionarily conserved from yeast to human. They include Mad1, Mad2, Mad3/BubR1 (Bub1-related), Bub1, Bub3, and Mps1 (reviewed in Shah and Cleveland, 2000). Genetic analysis shows that Mad1, Mad2, Mad3, Bub1, and Bub3 lie in the same checkpoint pathway that controls anaphase onset by monitoring spindle integrity. On the other hand, Bub2 lies in a distinct pathway that controls mitotic exit by monitoring the position of the mitotic spindle (Gardner and Burke, 2000; reviewed in Hoyt, 2000). Yeast Mad1 is a nuclear protein that becomes hyperphosphorylated during normal mitosis and when spindle assembly is disrupted (Hardwick and Murray, 1995). The upstream kinase for Mad1 is likely to be Mps1 (Hardwick et al., 1996; Weiss and Winey, 1996), and its overexpression results in hyperphosphorylation of Mad1 and a mitotic arrest (Hardwick et al., 1996). Besides Mps1, phosphorylation of Mad1 requires Mad2, Bub1, and Bub3 (Hardwick and Murray, 1995; Hardwick et al., 1996; Weiss and Winey, 1996). Mad2 forms a complex with Mad1 and this interaction is essential for Mad1 phosphorylation (Chen et al., 1999).
Some of the checkpoint components are localized to kinetochores in metazoans and are probably involved in generating the checkpoint signal. These components include Mad1 (Chen et al., 1998; Jin et al., 1998), Mad2 (Chen et al., 1996; Li and Benezra, 1996), Bub1 (Taylor and McKeon, 1997), BubR1 (Chan et al., 1999), Bub3 (Taylor et al., 1998), and Mps1 (Abrieu et al., 2001). These proteins locate to kinetochores at the end of prophase. Mad1 and Mad2 dissociate from kinetochores that have attached to spindle microtubules (Chen et al., 1996, 1998; Waters et al., 1998), whereas Bub1 and Bub3 remain at kinetochores until early anaphase (Jablonski et al., 1998; Basu et al., 1999; Sharp-Baker and Chen, 2001). Xenopus Mad1 forms a complex with Mad2 and functions, at least in part, to recruit Mad2 to unattached kinetochores (Chen et al., 1998). Bub1 is also required for Mad1 and Mad2 to bind kinetochores (Sharp-Baker and Chen, 2001).
The downstream target of the spindle checkpoint is the anaphase-promoting complex (APC), the ubiquitin protein ligase involved in ubiquitination and degradation of the anaphase inhibitor Pds1 and cyclin B (reviewed in Page and Hieter, 1999). Degradation of Pds1 and cyclin B triggers anaphase onset and exit from mitosis, respectively. The specificity of APC to different substrates is conferred by its associated specificity factor/activator (Visintin et al., 1997). APC bound with Cdc20 targets Pds1, whereas the APC–Cdh1 complex recognizes cyclin B (Visintin et al., 1997). When the spindle checkpoint is activated, Mad2 probably binds and inhibits Cdc20 (Fang et al., 1998; Hwang et al., 1998; Kim et al., 1998), thus preventing Pds1 degradation and sister chromatid segregation. BubR1 has also been shown recently to have a similar inhibitory effect on APCCdc20 in vitro (Tang et al., 2001). A hypothesis suggests that unattached kinetochores convert checkpoint proteins, such as Mad2, to their active form (Chen et al., 1998; Gorbsky et al., 1999). On activation, these molecules may then leave kinetochores to inhibit the APCCdc20. The active molecule probably loses its activity with time and needs to be replenished through unattached kinetochores. This hypothesis has been supported by fluorescence recovery after photobleaching (FRAP) analysis that demonstrates a turnover rate of 24–28 s for Mad2 at kinetochores (Howell et al., 2000). The unattached kinetochore may facilitate assembly of active Mad2 along with other spindle checkpoint components. Interestingly, it has been shown recently that a complex containing Mad2, Bub3, and BubR1 is a more potent inhibitor for APCCdc20 in vitro than Mad2 alone (Sudakin et al., 2001).
Previous immunodepletion analysis suggests that a fraction of Mad2 molecules in Xenopus egg extract might not be associated with Mad1 (Chen et al., 1998). In this study, we explore the possibility that the Mad1-free Mad2 may play a role in the spindle checkpoint.
MATERIALS AND METHODS
Preparation of Xenopus Egg Extracts, Spindle Checkpoint Assay, Immunoblot, and Immunodepletion
CSF-arrested extracts were obtained from the cytoplasm of unfertilized Xenopus eggs that are arrested at metaphase of the second meiotic division by the cytostatic factor (CSF). CSF-arrested extract and demembranated sperm nuclei were prepared as described previously (Murray, 1991). The spindle checkpoint was activated in the CSF-arrested extract after incubation with 10 μg/ml nocodazole and sperm nuclei at 9,000–15,000 nuclei/μl of extract (Minshull et al., 1994). Once the checkpoint is activated, addition of calcium is unable to induce the mitotic exit as determined by condensed chromosomes and a sustained Cdc2-associated histone H1 kinase activity. Spindle checkpoint assays, immunoblot, and immunodepletion were performed as described (Chen et al., 1996, 1998). For interphase extract shown in Figure 9A, the CSF-arrested extract was driven into interphase by incubation with 0.5 mM calcium chloride for 30 min at 22°C, followed by addition of 100 ng/μl cycloheximide to prevent synthesis of cyclin B and mitotic entry. The checkpoint extract was prepared in CSF-arrested extract incubated with 15,000 sperm nuclei/μl of extract at 22°C for 10 min and another 20-min incubation with 10 ng/μl nocodazole. All three types of extract shown in Figure 9A contained the same number of nuclei and were incubated at 22°C for the same amount of time.
Figure 9.
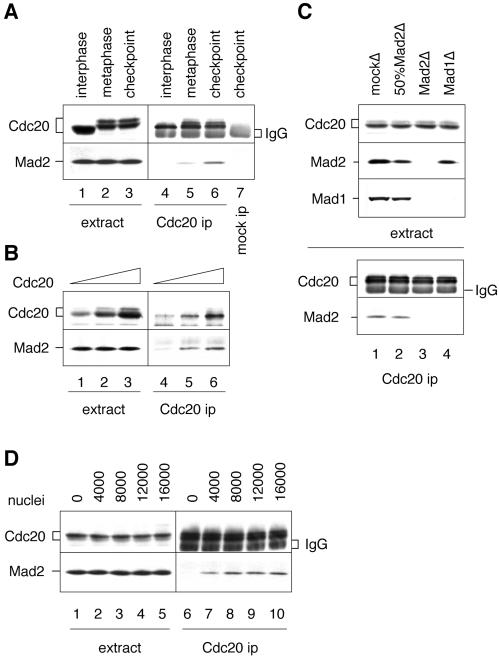
Unattached chromosomes enhance Mad2–Cdc20 interaction. (A) Spindle checkpoint enhances Mad2–Cdc20 interaction. Interphase (lanes 1 and 4), CSF-arrested (metaphase, lanes 2 and 5), or spindle checkpoint-active (lanes 3, 6, and 7) extracts containing the same number of nuclei were subjected to immunoprecipitation with anti-Cdc20 antibody (lanes 4–6) or with a control antibody (lane 7). Total protein in the extract (lanes 1–3) or the immunoprecipitates (lanes 4–7) was then immunoblotted for Cdc20 (top) or Mad2 (bottom). The band below Cdc20 in lanes 4–7 is the IgG heavy chain. (B) Cdc20 is the limiting factor in the complex of Mad2–Cdc20. Extracts containing increasing amounts of Cdc20 translation as indicated on top were incubated with sperm nuclei and nocodazole to activate the spindle checkpoint, followed by immunoprecipitation with anti-Cdc20 antibody. Total protein in the extract (lanes 1–3) or the immunoprecipitates (lanes 4–6) was then immunoblotted for Cdc20 (top) or Mad2 (bottom). Cdc20 in lane 1 is at the endogenous level. (C) Preventing kinetochore binding of Mad2 abolishes Mad2–Cdc20 interaction. CSF-arrested extract was immunodepleted with control antibody (lane 1), with anti-Mad2 antibody (lane 3), or with anti-Mad1 antibody (lane 4). Mad2 depletion removed both Mad2 and Mad1, whereas Mad1 depletion left behind >50% of Mad2. Lane 2 is a mix of mock- and Mad2-depleted extract (1:1), which contained 50% of Mad1 and Mad2. After incubation with sperm nuclei and nocodazole, the extracts were subjected to anti-Cdc20 immunoprecipitation, followed by immunoblot for Cdc20 or Mad2 as indicated (bottom two panels). Proteins in the extracts were also immunoblotted for Cdc20, Mad2, or Mad1 as indicated on the left (top three panels). (D) Level of Mad2–Cdc20 interaction is proportional to the number of unattached chromosomes. Extracts were incubated with nocodazole and sperm nuclei at density indicated on top (per microliter of extract), followed by Cdc20 immunoprecipitation and immunoblot analysis. The amount of Mad2 associated with Cdc20 levels off at the nuclear density of 8,000–16,000.
Gel Filtration Analysis
The CSF-arrested or spindle checkpoint-active extracts were diluted with an equal volume of 2× lysis buffer (20 mM KPO4, pH 7.5, 2 mM EDTA, 10 mM EGTA, 100 mM β-glycerophosphate, 2 mM MgCl2) plus 20% glycerol, 0.2% Triton X-100, and 0.2 μM microcystin-LR (Calbiochem, San Diego, CA). The diluted samples were spun at 50,000 rpm for 1 h at 4°C in a TLA100.3 rotor (Beckman Coulter, Fullerton, CA). The supernatant (100 μl) was passed through a 0.2-μm filter (ultra-free MC centrifugal filter units; Millipore, Bedford, MA) before being loaded onto a Superose 6 HR 10/30 column (Amersham Biosciences, Piscataway, NJ) that has been equilibrated with 2 column volumes of 1× lysis buffer containing 10% glycerol. The column was subsequently eluted with 1.5 column volume of lysis buffer plus 10% glycerol and 1-ml fractions were collected. It took ∼2 h from dilution of the extracts to the end of column elution. The eluates were concentrated by trichloroacetic acid precipitation and one-fifth of each fraction was used for immunoblot analysis.
In Vitro Transcription and Translation in Egg Extracts
Xenopus MAD1, MAD2, or CDC20 was inserted into a modified pGEM transcription vector that contains 5′- and 3′-untranslated regions of Xenopus cyclin B1 and the Kozak consensus sequence (ACCATGG) to facilitate translation in egg extracts. The plasmids used in this article encode Mad1-HA2 (tagged with two copies of hemagglutinin [HA] epitope, pRC309), Mad1-HA (pRC304), Mad1 (without any tag, pRC341), Mad2 (without any tag, pRC285), Mad1N (amino acids 1–445, pRC291), Mad1C (amino acids 326–718, pRC288), and full-length Cdc20 (pRC474). The plasmids were first linearized by cleavage at a unique restriction site 3′ to the polyadenylation signal. The linearized plasmids were used to produce transcripts by using the mMESSAGE mMACHINE T7 transcription kit (Ambion, Austin, TX). We typically reconstitute the transcripts from a 20-μl transcription reaction with 5 μl of water, yielding transcripts of ∼4 mg/ml. To remove secondary structures, the transcripts were heated to 65°C for 3 min and left on ice immediately before use. Translation reaction was performed as described (Sharp-Baker and Chen, 2001) in CSF-arrested extracts that were intact or immunodepleted of endogenous Mad1 by anti-Mad1 antibodies, or both Mad1 and Mad2 by anti-Mad2 antibodies. Mad1 and Mad2 produced in the standard reactions were usually two- to fourfold over the endogenous level, and Cdc20 at 40-fold endogenous level. For results shown in Figures 2, 3, 6, and 7, mock translation was added to samples that did not receive Mad1 or Mad2 translation, so that all samples contained equal amount of the translation reactions.
Figure 2.
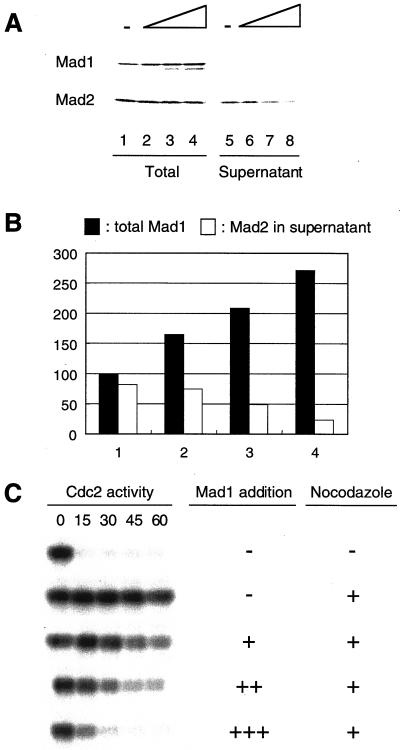
Excess Mad1 titrates out Mad1-free Mad2 and abolishes spindle checkpoint. (A) Immunoblot analysis of Mad1 and Mad2 in extracts containing increasing amount of Mad1 translation. The samples were immunoprecipitated with anti-Mad1 antibodies. Total proteins before immunoprecipitation (lanes 1–4) or supernatants left after immunoprecipitation (lanes 5–8) were analyzed by immunoblot with anti-Mad1 or anti-Mad2 antibodies as indicated. The level of Mad2 left in the supernatant decreases with increasing amount of Mad1 added. Mock translation was added, so that all samples contained the same amount of translation reaction. A graph of the result is shown in B. The relative level of total Mad1 (lanes 1–4 in A) and Mad2 in the supernatant (lanes 5–8 in A) is shown. The amount of Mad1 and Mad2 in lane 1 of A is designated as 100. (C) Autoradiograms of histone H1 kinase assay. The same samples shown in lanes 1–4 of A were incubated with sperm nuclei and nocodazole for 30 min. Calcium chloride was then added to trigger mitotic exit. Samples were taken every 15 min for histone H1 kinase assay as indicated on top. In the presence of excess Mad1, the extract failed to activate the checkpoint.
Figure 3.
Addition of Mad2 reverses the effect of excess Mad1 and restores the checkpoint. Extracts were incubated with nuclei and nocodazole in the presence of Mad1 translation alone, Mad2 translation alone, or both. Samples were processed for immunoblot analysis (A) and for H1 kinase measurement (B) as described in Figure 2. Restoration of the checkpoint by the addition of Mad2 indicates that the ratio between Mad1 and Mad2 levels is critical.
Figure 6.
Mad1N is recruited onto kinetochores that have bound with endogenous Mad1 and Mad2. Sperm nuclei and nocodazole were incubated in mitotic extract for 30 min followed by the addition of mock, full-length Mad1, Mad1N, or Mad1C translated in extract depleted for both Mad1 and Mad2. Samples were incubated for another 30 min and then processed for immunofluorescence staining as described for Figure 5.
Figure 7.
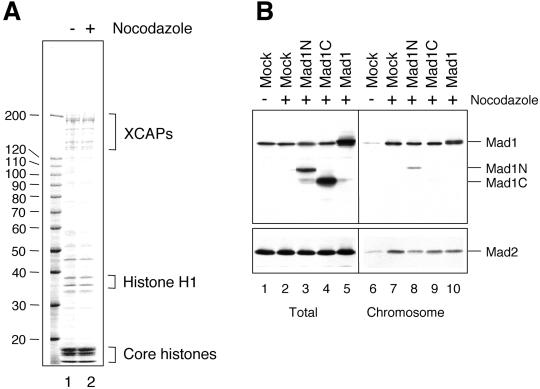
Mad2 levels in the chromosomal fractions are reduced by additional Mad1N, Mad1C, and Mad1. (A) Coomassie staining of proteins associated with chromosomes that were isolated from extracts treated with (lane 2) or without (lane 1) nocodazole. The migration of molecular weight standards is indicated on the left. The predominant proteins are labeled on the right. (B) Immunoblot of total proteins or chromosomal fractions with anti-Mad1 or anti-Mad2 antibodies. As described for Figure 6, mitotic chromosomes were assembled in extracts with (lanes 2–5 and 7–10) or without (lanes 1 and 6) nocodazole, followed by the addition of mock, Mad1N, Mad1C, or Mad1 translation as indicated on top. Chromosomes were isolated and analyzed by immunoblot for Mad1 (top) or Mad2 (bottom). The addition of Mad1N reduces chromosomal Mad2 to 43% of that on mock-challenged chromosomes. Those challenged with Mad1C or Mad1 contained ∼70% of Mad2.
Immunofluorescence
To assemble mitotic chromosomes in egg extracts, 20 μl of CSF-arrested extract was incubated with sperm nuclei (∼1000/μl of extract) at 23°C for 10 min followed by addition of nocodazole to 10 μg/ml and incubation for another 20 min at 23°C to disrupt microtubules. The samples were fixed by diluting 10-fold with XB (10 mM HEPES, pH 7.8, 50 mM sucrose, 100 mM KCl, 10 mM MgCl2, and 1 mM CaCl2) containing 0.1% Triton X-100 and 1% formaldehyde, and incubated for 10 min at room temperature. Samples were then layered over 5 ml of a 30% glycerol cushion made in XB plus 0.1% Triton X-100, and spun in a HB-4 rotor at 10,000 rpm for 10 min at 4°C to collect chromosomes onto a coverslip. Immunofluorescence staining was performed as described previously (Chen et al., 1998). Images were collected using a charge-coupled device camera (MicroMAX-5 MHz; Princeton Instruments, Princeton, NJ) attached to a fluorescence microscope (E800; Nikon, Melville, NY). Images were collected and processed with the MetaMorph Imaging System (version 4.0; Universal Imaging, Downingtown, PA) and converted to Photoshop format (Adobe Systems, Mountain View, CA).
Titration of Mad1-free Mad2
In vitro-translated Mad1-HA or Mad1 was added to CSF-arrested extracts before spindle checkpoint activation. For experiments shown in Figure 2, the control sample contained 12 μl of CSF-arrested extract (60% of the total volume) mixed with 8 μl of mock translation (40%) that was made in extract depleted for both Mad1 and Mad2. For samples containing an increasing amount of Mad1-HA, 12 μl of CSF-arrested extract was mixed with 2, 4, or 8 μl of Mad1-HA translation and additional mock translation to bring the final concentration of the translation to 40% for all samples. For Figure 3, various translations made in Mad1- and Mad2-depleted extracts were added to 25% of the total volume. The extract and translation mix was incubated at 23°C for 15 min before checkpoint activation with sperm nuclei and nocodazole. Samples were taken for histone H1 kinase assays and for immunoblots. To detect the level of Mad1-free Mad2, 10 μl of the sample was incubated at 4°C for 1 h with Affi-prep protein A beads (Bio-Rad, Hercules, CA) coated with anti-Mad1 antibodies. At the end of incubation, the beads were pelleted by centrifugation to precipitate Mad1 and its associated Mad2, and the supernatants were taken for immunoblot analysis. The levels of the protein were quantitated with NIH Image (version 1.61).
Immunoblot of Chromosomal Proteins
Sperm nuclei were incubated at 23°C for 10 min with 40 μl of CSF-arrested extracts containing various translation reactions, followed by incubation with or without 10 μg/ml nocodazole for another 20 min. At the end of incubation, samples were diluted with 360 μl of ice-cold XB containing 0.1% Triton X-100 and leupeptin, pepstatin, and chymostatin (LPC), and mixed gently by inverting the tubes few times. They were then layered over 1 ml of XB containing 30% sucrose, 0.1% Triton X-100, and LPC, and spun in HB-6 rotor at 10,000 rpm for 15 min. After centrifugation, the supernatant was removed and the chromosome pellets were washed with 0.5 ml of XB containing 30% sucrose, 0.1% Triton X-100, and LPC, and spun again in HB-6 rotor at 10,000 rpm for 5 min. After removing the supernatant, the pellets were solubilized in SDS-PAGE sample buffer and analyzed by immunoblot. The intensity of the signals on the blot was measured by NIH Image (version 1.61).
Immunoprecipitation of Cdc20
The Xenopus CDC20 homology, also named Fizzy (Lorca et al., 1998), that lacks the N-terminal 69 amino acids was produced in bacteria as a glutathione S-tranferase (GST) fusion (plasmid provided by Dr. T. Lorca, CNRS, Montpellier, France). The expression of GST-Cdc20 fusion protein was induced in bacteria by the addition of 0.1 mM isopropyl β-d-thiogalactoside for 4 h, and the GST-Cdc20 fusion protein in the inclusion bodies was prepared as described for Xenopus Mad1 protein (Chen et al., 1998). The purified protein was used to produce antibodies in rabbits (Convance Research, Denver, PA) and to purify the antibodies. For immunoprecipitation of Cdc20, affinity-purified anti-Cdc20 antibodies were coupled to Affi-prep protein A beads and the beads were washed twice with lysis buffer containing 0.1% Triton-100, 200 nM microcystin-LR, and 10 μg/ml each of LPC. The beads were then mixed at 4°C for 1 h with 30 μl of extracts that had been incubated at 22°C for 30 min with nuclei at 15,000 nuclei/μl or at nuclear densities indicated in Figure 9D. After mixing, the beads were washed twice with the same buffer as described above, twice with the buffer plus 0.5 M NaCl, and once with buffer without additional NaCl. It took ∼30 min to complete the washing steps. The immunoprecipitates were then solubilized in SDS-sample buffer and subjected to immunoblot analysis.
RESULTS
Excess Mad1 Abolishes Checkpoint
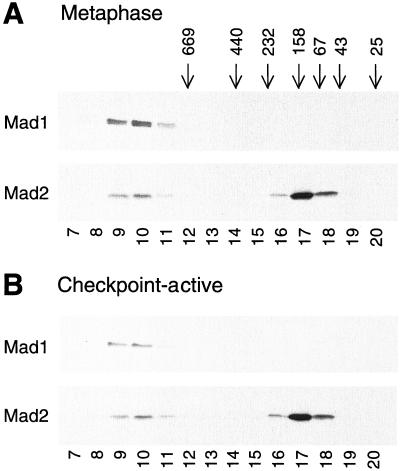
The spindle checkpoint proteins Mad1 and Mad2 coimmunoprecipitate from yeast (Chen et al., 1999) and Xenopus egg extracts (Chen et al., 1998). In egg extracts, Mad1 seems to be the limiting factor in the complex formation, because all of the Mad1 molecules were removed along with Mad2 when anti-Mad2 antibodies were used in the immunodepletion (Chen et al., 1998). However, only 20–40% of Mad2 was removed when Mad1 was immunodepleted (Chen et al., 1998), indicating that only a fraction of Mad2 was in a complex with Mad1. Gel filtration analysis of metaphase egg extracts showed that Mad1 eluted in one peak that corresponded to a size of >669 kDa (Figure 1A). On the other hand, Mad2 was fractionated into two peaks, the first of which overlapped with the peak of Mad1 (Figure 1A). The majority of Mad2 was eluted in the second peak at ∼158 kDa (Figure 1A), consistent with the notion that the majority of Mad2 is not complexed with Mad1. When the spindle checkpoint was activated in the extract with a high density of nuclei and nocodazole, the elution profile of Mad1 and Mad2 was similar to that seen in metaphase extract (Figure 1B).
Figure 1.
Gel filtration analysis of Mad1 and Mad2. Metaphase egg extract (A) or spindle checkpoint-active extract (B) was fractionated on Superose 6 column and fractions were immunoblotted for Mad1 (top) or Mad2 (bottom). The fraction number is indicated on the bottom. The fractionation of size standards is indicated on top.
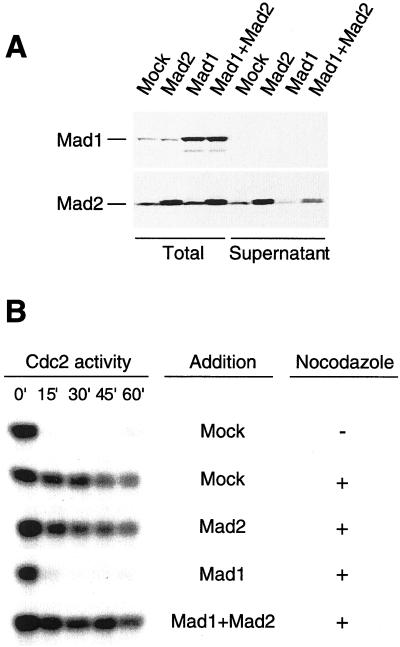
In extract depleted for Mad1, the residual Mad2 was unable to bind kinetochores and the checkpoint was impaired (Chen et al., 1998). To test whether the pool of Mad1-free Mad2 plays any role in the checkpoint, an excess of Mad1 was added to the extract to sequester Mad2 in the complex with Mad1. We overproduced epitope-tagged Mad1 directly in the egg extract from corresponding RNA synthesized in vitro. Purified recombinant proteins were not used for the following reasons. First, we were not able to generate soluble Mad1 protein either in bacterial or baculoviral expression systems, even when the protein was coexpressed with Mad2. Second, the protein produced directly in egg extract is more likely to be in its native conformations. Indeed, Mad1 and Mad2 produced in this manner restored the spindle checkpoint in extracts depleted for endogenous Mad1 and Mad2 (our unpublished data). To reduce the level of Mad1-free Mad2, Mad1 was first translated in extract depleted for Mad1 and Mad2 with anti-Mad2 antibody. The translation reaction was then added to a separate aliquot of egg extract to increase the ratio of Mad1 to Mad2. Immunoblot analysis showed that the addition of Mad1 reduced the level of free Mad2 left in the supernatant after immunodepletion of Mad1 (Figure 2A). When Mad1 was present at 2.7-fold over the endogenous level, the free Mad2 was reduced to 23% of normal concentration (Figure 2B). The extract containing excess Mad1 failed to support the spindle checkpoint in a dose-dependent manner, as evidenced by the decline of Cdc2-associated H1 kinase activity after adding calcium to inactivate the CSF activity (Figure 2C). Furthermore, the addition of translated Mad2 along with Mad1 restored the fraction of Mad1-free Mad2 as determined by the immunoblot (Figure 3A). The checkpoint function was also restored in this extract (Figure 3B), indicating that excess Mad1 interferes with the function of Mad2, rather than other molecules.
Dominant Negative Mad1 Proteins
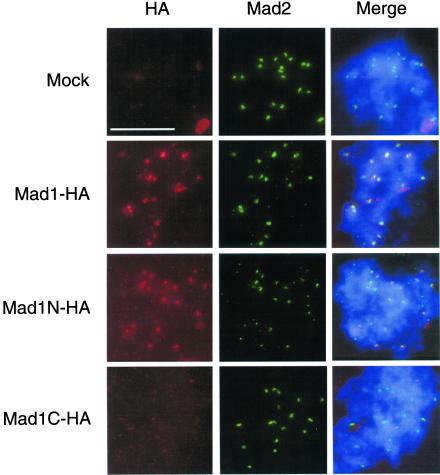
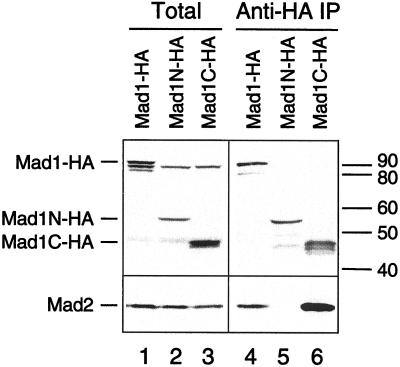
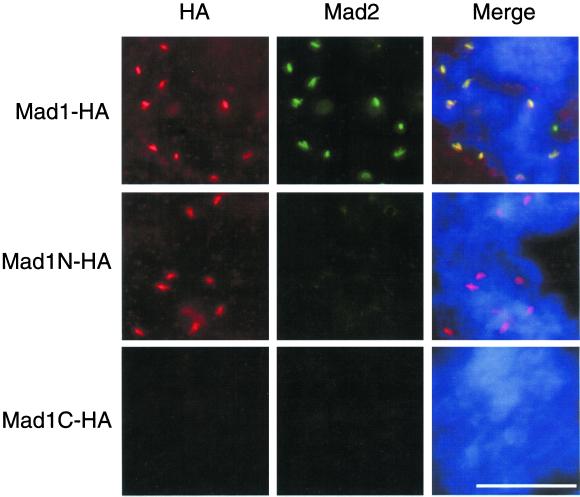
Do Mad1-free Mad2 molecules regulate the levels of Mad1 and Mad2 on kinetochores? Is the interaction between Mad1 and Mad2 required for Mad1 to bind kinetochores? To answer these questions we first tested whether Mad1 was able to localize to kinetochores in the absence of Mad2. Truncated Mad1 molecules were generated that contain or lack the binding region for Mad2, which is expected to reside within amino acids 467–586 based on analogy with the human Mad1 (Jin et al., 1998). HA-tagged full-length Mad1 or proteins corresponding to amino acids 1–445 (Mad1N) or 326–718 (Mad1C) were translated in egg extracts. Immunoprecipitation of these extracts with anti-HA antibodies showed that Mad2 coimmunoprecipitated with both full-length Mad1 and Mad1C (Figure 4), indicating that Mad1C contains the Mad2-binding region. To determine the kinetochore binding ability, HA-tagged full-length Mad1, Mad1N, or Mad1C were translated in extracts immunodepleted for the endogenous Mad1. After incubating the extracts with sperm nuclei and nocodazole, chromosomes were isolated and stained for anti-HA and anti-Mad2 antibodies. Both full-length Mad1 and Mad1N were found at kinetochores, whereas Mad1C was not (Figure 5), indicating that the kinetochore binding region resides within amino acids 1–445 of Mad1. As expected, Mad2 was absent at kinetochores when only Mad1N was present, due to a lack of interaction between Mad1N and Mad2 (Figure 5, middle). Consistent with the result of Mad1 depletion (Chen et al., 1998), Mad2 associated with Mad1C was not localized to kinetochores, because Mad1C was unable to bind kinetochores (Figure 5, bottom).
Figure 4.
Immunoblot analysis of truncated Mad1 proteins and their interaction with Mad2. Extracts containing translation of HA-tagged full-length Mad1 (lanes 1 and 4), amino acids 1–445 (Mad1N, lanes 2 and 5), or amino acids 326–718 (Mad1C, lanes 3 and 6) were immunoprecipitated with anti-HA antibodies. Total proteins in the extracts (lanes 1–3) and the proteins associated with the immunoprecipitates (lanes 4–6) were analyzed by immunoblot with anti-Mad1 (top) or anti-Mad2 (bottom) antibodies. Both full-length Mad1 and Mad1C coimmunoprecipitate with Mad2. No self-interaction of Mad1 is detected. The migration of various HA-tagged Mad1 proteins is indicated on the left, and molecular size standards are indicated on the right.
Figure 5.
Kinetochore binding domain resides within amino acids 1–445 of Mad1. Mitotic extracts were immunodepleted with anti-Mad1 antibodies to remove endogenous Mad1. The extracts were supplemented with HA-tagged full-length Mad1, Mad1N, or Mad1C that were translated in Mad1-depleted extracts. After incubating the extracts with sperm nuclei and nocodazole, chromosomes were isolated and stained with mouse anti-HA and rabbit anti-Mad2 antibodies. Fluorescein-conjugated anti-rabbit and Texas Red-conjugated anti-mouse IgG antibodies were used as secondary antibodies. The DNA was stained with the DNA-binding dye Hoechst 33258. The merges of all three fluorochromes are also shown (Merge). All pictures of the same fluorochrome were processed in the same way. Both full-length Mad1 and Mad1N associate with kinetochores. Similar results were obtained in the absence of Mad2 (our unpublished data). Bar, 10 μm.
We then asked whether Mad1N and Mad1C had an effect on kinetochore-bound Mad1 or Mad2. Sperm nuclei and nocodazole were first incubated in mitotic extract to assemble kinetochores with endogenous Mad1 and Mad2. Mock, Mad1, Mad1N, or Mad1C translation was then added. Immunofluorescence staining of chromosomes using anti-HA and anti-Mad2 antibodies showed that both Mad1 and Mad1N were recruited onto kinetochores (Figure 6). The addition of Mad1N reduced Mad2 staining at kinetochores compared with mock addition, probably due to the replacement of endogenous Mad1 with Mad1N that was unable to bind Mad2. Interestingly, we consistently found that Mad2 staining decreased slightly upon the addition of Mad1C or Mad1 (Figure 6).
The effect of additional Mad1, Mad1N, and Mad1C on the level of Mad2 at kinetochores was also assessed by immunoblot of isolated chromosomes. Mitotic chromosomes assembled in crude egg extracts were isolated through a sucrose cushion. The predominant proteins in this chromosomal fraction were the known chromosomal proteins, including histones and XCAPs (Figure 7A), similar to that obtained from chromosomes assembled in a high-speed supernatant of the extract (Hirano and Mitchison, 1994). Immunoblot of the chromosomal fraction with anti-Mad1 and anti-Mad2 antibodies showed that there was a marked increase of these proteins associated with chromosomes isolated from nocodazole-treated extract compared with those from untreated extract (Figure 7B, compare lanes 6 and 7), consistent with the immunofluorescence study showing that Mad1 and Mad2 are localized to unattached kinetochores (Chen et al., 1998). The level of Mad2 on chromosomes that were first bound with endogenous Mad1/Mad2 and then challenged with Mad1N was ∼43% of that on chromosomes challenged with mock translation (Figure 7B, lane 8). Those challenged with Mad1C or Mad1 contained ∼70% of Mad2 (Figure 7B, lanes 9 and 10). These results demonstrate that titrating out Mad1-free Mad2 with Mad1C or full-length Mad1 decreases the level of Mad2 on kinetochores.
The reduction of Mad2 on kinetochores upon addition of either Mad1N or Mad1C during checkpoint maintenance suggests that these truncated proteins probably behave as dominant negatives for the spindle checkpoint. To test whether Mad1N or Mad1C interferes with the checkpoint establishment, we incubated nuclei and nocodazole in extracts containing Mad1N or Mad1C at approximately threefold and sevenfold, respectively, molar excess of the endogenous Mad1 (Figure 8B). To examine the effect on checkpoint maintenance, the checkpoint was first activated in extracts by incubation with nuclei and nocodazole, followed by addition of Mad1N or Mad1C and incubation for another 30 min. At the end of incubation, calcium chloride was added to all samples to induce the mitotic exit. Figure 8A shows that addition of Mad1N or Mad1C to egg extracts during the checkpoint establishment or maintenance resulted in a decline of Cdc2 activity upon addition of calcium to egg extracts, indicating that the checkpoint was impaired and that Mad1N and Mad1C are dominant negative for the checkpoints.
Figure 8.
Mad1N and Mad1C abolish checkpoint establishment and maintenance. (A) Autoradiograms of histone H1 kinase assay. For checkpoint establishment samples, the translation of mock, Mad1N, or Mad1C was added to CSF-arrested extract, followed by incubation with sperm nuclei and nocodazole for 20 min to activate the spindle checkpoint. For checkpoint maintenance, the checkpoint was first activated in CSF-arrested extract by incubation with nuclei and nocodazole for 20 min, followed by the addition of translation and another incubation for 30 min. Calcium chloride was then added to trigger mitotic exit. Samples were taken immediately before the addition of calcium chloride (time 0) and every 15 min thereafter for histone H1 kinase assay as indicated on top. (B) Immunoblot analysis of Mad1, Mad1N, Mad1C, and Mad2 in samples used in A.
Dependence of Mad2–Cdc20 Interaction on Unattached Chromosome
The target of Mad2 has been shown to be Cdc20. Binding of Mad2 to Cdc20 prevents Cdc20 from activating the APC. Deletion analysis has shown that Mad2 lacking C-terminal 10 amino acids is unable to bind Mad1 (our unpublished data; Sironi et al., 2001) and also fails to inhibit the APC (Fang et al., 1998; Sironi et al., 2001), raising the possibility that the same region of Mad2 may interact with both Mad1 and Cdc20. Excess Mad1 or Mad1C thus may abolish the spindle checkpoint simply by outcompeting Cdc20 in binding to Mad2. We thus examined the interaction between Mad2 and Cdc20 by coimmunoprecipitation. Mad2 was not detectable in the Cdc20 immunoprecipitate prepared from interphase extract, and was present in the immunoprecipitate from metaphase extract (Figure 9A). Cdc20 immunoprecipitated from spindle checkpoint-active extract contained a higher level of Mad2 compared with that from metaphase extract (Figure 9A, compare lanes 5 and 6). The difference in the degree of Mad2–Cdc20 association is not due to a change in the protein level, because both Cdc20 and Mad2 were present in similar levels under these conditions (Figure 9A, lanes 1–3). This result shows that the Mad2–Cdc20 interaction is enhanced when the checkpoint is activated.
The concentrations of Cdc20 and Mad2 in the egg extract are ∼10 and 200 nM, respectively (our unpublished data), indicating that Cdc20 is the limiting factor for its association with Mad2. Indeed, when various amounts of Cdc20 translation were incubated with checkpoint-active extract, the level of Mad2 in the Cdc20 immunoprecipitate increased with increasing concentration of Cdc20 (Figure 9B). Despite the vast excess of Mad2 over Cdc20 and a large fraction of Mad2 in the Mad1-free pool, Mad2 interacted only weakly with Cdc20 when the checkpoint was not active (Figure 9A, compare lanes 5 and 6). One possibility for the enhanced interaction induced by the checkpoint is that Mad2 is converted into an active form at unattached kinetochores and acquires a higher affinity for Cdc20. To determine whether binding of Mad2 to kinetochores facilitates its interaction with Cdc20, we examined the Mad2–Cdc20 interaction in extracts depleted for Mad1 or Mad2. In Mad1-depleted extract, Mad2 failed to localize to kinetochores (Chen et al., 1998) and the protein was not detectable in the Cdc20 immunoprecipitate even though the extract still contained >50% of Mad2 molecules after the depletion (Figure 9C, lanes 4). The lack of Mad2–Cdc20 interaction was not due to the reduced level of Mad2, because the interaction was observed in extract that contained 50% normal levels of Mad1 and Mad2 through partial depletion with anti-Mad2 antibody (Figure 9C, lane 2). This result supports the notion that recruitment of Mad2 to kinetochores enhances its ability to associate with Cdc20.
If unattached kinetochores facilitate assembly of a Mad2–Cdc20 complex, we expect that the level of Mad2 associated with Cdc20 will be dependent on the concentration of unattached kinetochores. We tested this hypothesis by adding various amounts of sperm nuclei into egg extracts in the presence of nocodazole, followed by Mad2-Cdc20 coimmunoprecipitation analysis. Figure 9D shows that there was indeed a dose dependency between Mad2–Cdc20 interaction and the nuclear density. The level of Mad2 in the Cdc20 immunoprecipitate leveled off at 8000–16000 nuclei/μl (Figure 9D, lanes 8–10), consistent with our finding that Cdc20 is limiting for Mad2–Cdc20 interaction.
DISCUSSION
Mad1-free Mad2 Molecules Are Important for Spindle Checkpoint
The initial observation that immunodepletion of Mad1 from egg extracts removes only a fraction of Mad2 has prompted us to examine the pool of Mad1-free Mad2 molecules. Gel filtration analysis reveals that Mad1 coelutes with a fraction of Mad2 with a size >669 kDa, and most of the Mad2 peaks at ∼158 kDa (Figure 1). This result demonstrates that there are at least two pools of Mad2 in the cytosol and that the inability of anti-Mad1 to remove all Mad2 is not due to disruption of the Mad1–Mad2 complex by the antibody. Instead, it reflects the existence of Mad1-free Mad2 molecules. The mitotic and checkpoint-active extracts give indistinguishable gel filtration profile of Mad1 and Mad2, indicating that both pools of Mad2 exist regardless of whether the checkpoint is active or not. In fact, these pools of Mad2 and the interaction between Mad1 and Mad2 are constitutive, because immunodepletion analysis with anti-Mad1 and anti-Mad2 antibodies in interphase extract yielded the same result as that in metaphase extracts (our unpublished data). In budding yeast, the association between Mad1 and Mad2 is also constant during the cell cycle, and the interaction is found to be important for Mad1 phosphorylation and the checkpoint (Chen et al., 1999). Interestingly, gel filtration analysis of the yeast cell lysate shows that Mad1 cofractionates with a pool of Mad2 in a complex >670 kDa and that the majority of Mad2 molecules elutes with a mass <67 kDa (Chen et al., 1999). It remains to be determined whether the large complex contains multiple copies of Mad1 or Mad2, or additional checkpoint molecules. The small complex may contain Mad2 multimer or other checkpoint proteins. Despite the difference in the size of the Mad1-free Mad2 complexes from yeast and Xenopus, the similarity in the gel filtration profile of Mad1 and Mad2 indicates that the action and the regulatory mechanisms of these molecules are likely to be evolutionarily conserved.
We now provide evidence that the Mad1-free Mad2 molecules are important for the checkpoint. Addition of excess Mad1 to extracts reduces the level of free Mad2 and abolishes the checkpoint function. This effect is reversed by the addition of more Mad2 molecules, indicating that the ratio between Mad1 and Mad2 is critical and that the checkpoint functions only when there is enough of the Mad1-free Mad2 molecules. Interestingly, the effect of excess Mad1 on the checkpoint is opposite from that of excess Mad2. It has been shown previously that the addition of excess Mad2 to egg extract or overexpression of the protein in fission yeast blocks the metaphase-to-anaphase transition without an apparent effect on the structure of the mitotic spindle or chromosome congression (He et al., 1997; Chen et al., 1998). In egg extract, this metaphase arrest is independent of chromosomes (Chen et al., 1998), indicating that excess Mad2 probably activates constitutively downstream checkpoint event. In contrast, an excess of Mad1 inhibits, rather than activates, the checkpoint. These findings suggest that even though Mad1 and Mad2 form a tight complex, they probably play a very different role in the spindle checkpoint.
An excess of Mad1C that contains the Mad2-binding region also titrates out the free Mad2 molecules and inhibits the establishment and maintenance of the checkpoint. Interestingly, addition of Mad1C or full-length Mad1 during checkpoint maintenance results in a reduction of Mad2 molecules at kinetochores, indicating that Mad1-free Mad2 molecules are important for maintaining the Mad2 level at kinetochores. It also shows that Mad1C is a dominant negative for the checkpoint (Figure 8A).
Unattached Chromosomes Enhance Assembly of Mad2–Cdc20 Complex
The downstream target of Mad2 is Cdc20 that activates and directs the APC toward Pds1. Deletion analysis of human Mad2 has shown that a C-terminally truncated mutant that is unable to bind Mad1 also fails to interact with Cdc20 (Sironi et al., 2001), indicating that the same region of Mad2 may recognize both Mad1 and Cdc20. We provide several lines of evidence that the loss of the spindle checkpoint in the presence of excess Mad1 or Mad1C is not simply due to a competition between Mad1 and Cdc20 in binding to Mad2. First, we demonstrate that Mad2–Cdc20 interaction is a regulated process. Coimmunoprecipitation between these two proteins is detectable during M phase, but not at interphase (Figure 9A). The interaction is further enhanced when the spindle checkpoint is activated (Figure 9A). This result is consistent with a recent study in a human cancer cell line (Zhang and Lees, 2001). Second, Mad2 in the egg extract is in ∼20-fold molar excess of Cdc20, indicating that there is also a vast excess of Mad1-free Mad2 over Cdc20. Even without competition from Mad1, Mad2 interacts only weakly with Cdc20 when the checkpoint is not active, suggesting that the checkpoint signal facilitates the interaction by modulating Mad2 and/or Cdc20. Third, we show that binding of Mad2 to unattached kinetochore correlates with the assembly of Mad2–Cdc20 complex. In Mad1-depleted extract, Mad2 fails to localize to kinetochores and to associate with Cdc20 (Figure 9C). This result argues against the idea that Mad1 prevents Mad2 from binding to Cdc20 until the checkpoint signal is generated. Mad1 is unlikely to mediate the interaction between Mad2 and Cdc20, because Mad1 is not detectable in the Cdc20 immunoprecipitate (our unpublished data). One possible mechanism is that Mad2 needs to be recruited to kinetochores to acquire the ability to bind Cdc20. This notion is supported by the finding that Mad2–Cdc20 interaction is greatly reduced in Bub1-depleted extract that also fails to recruit Mad2 to kinetochore (our unpublished data). In addition, the level of Mad2–Cdc20 interaction is proportionally increased at the nuclear density of 0–8000 nuclei/μl of extract in the presence of nocodazole (Figure 9D), indicating that unattached chromosomes facilitate the assembly of Mad2–Cdc20. However, we cannot exclude the possibility that unattached chromosomes may affect another target that indirectly modulates the interaction between Mad2 and Cdc20. Interestingly, a recent study shows that partially purified chromosomes inhibit the APC in vitro (Sudakin et al., 2001).
The localization of spindle checkpoint proteins Mad1 and Mad2 to unattached kinetochores suggests that these molecules play a role in triggering and maintaining the checkpoint signal. However, it is not clear whether kinetochore binding of these proteins is essential for their checkpoint function. We now show that the checkpoint is abolished when Mad1 and Mad2 fail to bind kinetochores. By expressing truncated Mad1 molecules, we identify the kinetochore-binding region within amino acids 1–445 of Mad1. A truncated protein containing this region of Mad1 (Mad1N) is recruited onto the kinetochores that have been occupied with endogenous Mad1 and Mad2, resulting in a reduction of wild-type Mad1 on kinetochores. As expected, the level of Mad2 on kinetochores is also reduced, because Mad1N is unable to bind Mad2. Addition of Mad1N during the process of checkpoint establishment or maintenance abolishes the checkpoint, indicating that Mad1N is a dominant negative for the checkpoint. This effect is probably due to the ability of Mad1N to outcompete Mad1–Mad2 complex from binding to kinetochores (Figure 10C). Even though we cannot exclude the possibility that Mad1N may regulate other molecules, it is worth noting that Mad1N does not affect checkpoint proteins Bub1 and Bub3 whose kinetochore binding is independent of Mad1 (Sharp-Baker and Chen, 2001). Our result supports the model that Mad1 and Mad2 need to continuously bind kinetochores to maintain the checkpoint. It is possible that Mad2–Cdc20 complex dissociates with time and needs to be replenished through binding to kinetochores (Figure 10A). Consistent with the decrease of Mad2 at kinetochores, addition of excess Mad1, Mad1N, or Mad1C during checkpoint maintenance reduces Mad2–Cdc20 interaction (our unpublished data).
Figure 10.
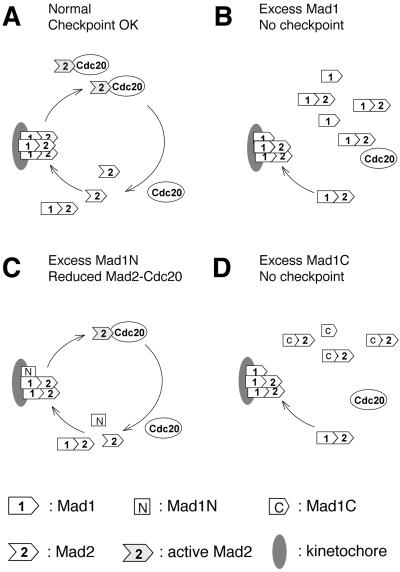
Models for how Mad1N and Mad1C may act as dominant negatives for the spindle checkpoint through different mechanisms. (A) Under normal condition, Mad1 targets its associated Mad2 to kinetochores. The pool of Mad1-free Mad2 protein ensures that Mad1 quickly binds new Mad2 when active Mad2 or Mad2–Cdc20 complex has dissociated, thus maintaining the checkpoint signal. (B and D) Excess Mad1 or Mad1C reduces the pool of Mad1-free Mad2 molecules, so that the kinetochores are not replenished with Mad2 once the previous Mad2 has left. (C) Mad1N replaces the full-length Mad1 on kinetochores. It fails to recruit Mad2, thus competing with wild-type Mad1 and Mad2 in binding kinetochores.
By expressing Mad1 alone in extracts depleted for the endogenous Mad1 and Mad2, we find that Mad1 binds kinetochores in the absence of Mad2 (our unpublished data). Mad1N that lacks the Mad2-binding region also localizes to kinetochores, suggesting that Mad2 is dispensable for kinetochore localization of Mad1. In combination with our previous observation that Mad2 fails to localize to kinetochores in extracts depleted for Mad1 (Chen et al., 1998), these results identify Mad1 as the protein that targets Mad1–Mad2 complex to kinetochores.
Possible Role of Mad1-free Mad2
The kinetochore is the site where spindle checkpoint signal is generated when the kinetochore lacks a stable microtubule attachment or tension exerted from microtubules. The checkpoint signal must be propagated from this locus into the cytosol to stop the anaphase. It has been hypothesized that checkpoint proteins may become activated upon binding to unattached kinetochores and then released into the cytosol to perform their function. This model predicts a dynamic interaction of these molecules with kinetochores and has been demonstrated first for Mad2 by using FRAP analysis (Howell et al., 2000). Binding of Mad2 to kinetochores may induce a conformational change in Mad2, allowing its interaction with Cdc20. Alternatively, unattached kinetochore may be the site where a stable complex containing Mad2 and Cdc20 is assembled.
Based on our findings, we hypothesize that the tight interaction between Mad1 and Mad2 is disrupted upon activation of Mad1 or Mad2 at kinetochores. The pool of Mad1-free Mad2 protein ensures that Mad1 remaining at kinetochores quickly binds new Mad2 after active Mad2 molecules or Mad2–Cdc20 complex have dissociated, thus maintaining the checkpoint signal (Figure 10A). This model predicts a reduced turnover rate of Mad2 at kinetochores upon titrating out Mad1-free Mad2 with Mad1C or Mad1 (Figure 10, B and D), which can be tested with FRAP analysis. The model also suggests a change in the size of the Mad1 and Mad2 protein complexes upon the checkpoint activation. However, our gel filtration analysis of Mad1 and Mad2 from metaphase and checkpoint-active extracts shows a similar elution profile. We think the state of active checkpoint complexes may not be captured by the gel filtration analysis due to their transient and unstable nature that may be necessary for the checkpoint signal to be silenced upon microtubule attachment to kinetochores. Alternatively, the active checkpoint complex may involve only a very small fraction of Mad1 and Mad2 and is masked by the bulk of the inactive molecules. In consistent with the latter possibility, we estimate that only <1% of Mad2 is coimmunoprecipitated with Cdc20 under checkpoint-active condition.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health and the David and Lucile Packard Foundation (to R.-H.C.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–01–0003. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–01–0003.
REFERENCES
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe J. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, Lopes C, Sunkel CE, Goldberg ML. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RD, Burke DJ. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ, Kallio M, Daum JR, Topper LM. Protein dynamics at the kinetochore: cell cycle regulation of the metaphase to anaphase transition. FASEB J. 1999;13(,suppl 2:S231–S234. doi: 10.1096/fasebj.13.9002.s231. [DOI] [PubMed] [Google Scholar]
- Hardwick K, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA. Exit from mitosis: spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Jin DY, Spencer F, Jeang KT. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Lorca T, Castro A, Martinez AM, Vigneron S, Morin N, Sigrist S, Lehner C, Doree M, Labbe JC. Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Sun H, Tonks NK, Murray AW. MAP-kinase dependent mitotic feedback arrest in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell Cycle Extracts. Methods Cell Biol. 1991;36:573–597. [PubMed] [Google Scholar]
- Page AM, Hieter P. The anaphase-promoting complex: new subunits and regulators. Annu Rev Biochem. 1999;68:583–609. doi: 10.1146/annurev.biochem.68.1.583. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Cleveland DW. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GKT, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of bub3 is required for kinetochore localization of bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]