Abstract
Dystrophin is widely thought to mechanically link the cortical cytoskeleton with the muscle sarcolemma. Although the dystrophin homolog utrophin can functionally compensate for dystrophin in mice, recent studies question whether utrophin can bind laterally along actin filaments and anchor filaments to the sarcolemma. Herein, we have expressed full-length recombinant utrophin and show that the purified protein is fully soluble with a native molecular weight and molecular dimensions indicative of monomers. We demonstrate that like dystrophin, utrophin can form an extensive lateral association with actin filaments and protect actin filaments from depolymerization in vitro. However, utrophin binds laterally along actin filaments through contribution of acidic spectrin-like repeats rather than the cluster of basic repeats used by dystrophin. We also show that the defective linkage between costameric actin filaments and the sarcolemma in dystrophin-deficient mdx muscle is rescued by overexpression of utrophin. Our results demonstrate that utrophin and dystrophin are functionally interchangeable actin binding proteins, but that the molecular epitopes important for filament binding differ between the two proteins. More generally, our results raise the possibility that spectrin-like repeats may enable some members of the plakin family of cytolinkers to laterally bind and stabilize actin filaments.
INTRODUCTION
Costameres are assemblies of cytoskeletal and integral membrane proteins that physically connect the sarcolemmal membrane to the force-generating sarcomeric apparatus at the Z-line in striated muscle (Craig and Pardo, 1983; Pardo et al., 1983). The dystrophin–glycoprotein complex is one element of costameres (Ervasti et al., 1990; Ervasti and Campbell, 1991; Porter et al., 1992; Ervasti and Campbell, 1993; Williams and Bloch, 1999) that is thought to mechanically stabilize the sarcolemmal membrane from shear stresses imposed during eccentric muscle contraction (Petrof et al., 1993; Straub et al., 1997). Biochemical studies have demonstrated that dystrophin contains two distinct and spatially separated actin binding sites located at the amino terminus and within the middle third of the large rod domain (Rybakova et al., 1996; Amann et al., 1998). The two actin binding sites of dystrophin form an extended lateral contact with actin filaments and protect them from depolymerization in vitro (Rybakova et al., 1996; Rybakova and Ervasti, 1997). More recently, we demonstrated that dystrophin is necessary for a mechanically strong physical link between the sarcolemma and actin filaments of costameres (Rybakova et al., 2000).
Utrophin is a widely expressed autosomal gene product with high sequence similarity to dystrophin (Tinsley et al., 1992). Utrophin is distributed throughout the sarcolemma in fetal and regenerating muscle, but is down-regulated in normal adult muscle and restricted to the myotendinous and neuromuscular junctions (Blake et al., 1996). Because utrophin and dystrophin bind the same complement of proteins (Matsumura et al., 1992; Kramarcy et al., 1994; Winder et al., 1995), it was hypothesized that utrophin may be capable of compensating for dystrophin deficiency. Indeed, transgenic overexpression of utrophin in dystrophin-deficient mdx mice resulted in full recovery for all known parameters of the dystrophic phenotype (Tinsley et al., 1998). Based on these promising results, utrophin up-regulation or gene therapy is under intense investigation as a potential therapy for Duchenne muscular dystrophy. Recent results, however, have raised concern about whether utrophin can effect the same mechanically strong link with costameric actin as provided by dystrophin (Rybakova et al., 2000). Although endogenous utrophin expression is up-regulated in striated muscle of mdx mice (Matsumura et al., 1992; Porter et al., 1998) and partially attenuates the phenotype associated with dystrophin deficiency (Deconinck et al., 1997a; Grady et al., 1997), this level of utrophin expression was not sufficient to retain costameric actin filaments on mechanically isolated sarcolemma (Rybakova et al., 2000). In addition, sequence comparisons (Winder, 1997; Amann et al., 1999) and biochemical analysis of recombinant protein fragments (Winder et al., 1995; Amann et al., 1998, 1999; Renley et al., 1998; Moores and Kendrick-Jones, 2000) have indicated that dystrophin and utrophin may bind F-actin through distinct mechanisms. Most notably, utrophin lacks the actin binding region composed of basic spectrin-like repeats that is present in the middle rod domain of dystrophin (Amann et al., 1999). These data suggested that utrophin may bind actin filaments solely through its amino-terminal calponin homology domain, and with an order of magnitude lower affinity (Winder et al., 1995; Moores and Kendrick-Jones, 2000) compared with dystrophin (Rybakova et al., 1996).
Herein, we have expressed full-length recombinant utrophin and show that the protein exhibits a native molecular weight and molecular dimensions indicative of a monomer. Contrary to expectations, full-length utrophin bound laterally along actin filaments with high affinity and protected filaments from depolymerization in vitro. Compared with dystrophin, utrophin made less extensive lateral contact with actin filaments and through distinct molecular epitopes. We also demonstrate that costameric actin is rescued on mechanically isolated sarcolemma from transgenic mdx mice that overexpress utrophin (Tinsley et al., 1998). Our results indicate that utrophin can perform all the actin binding functions documented for dystrophin. These data strongly support the continued exploration of therapies aiming to treat Duchenne muscular dystrophy through interventions that increase utrophin expression.
MATERIALS AND METHODS
Recombinant Utrophin
The 11-kb BamHI/XbaI fragment from a vector encoding full-length mouse utrophin with an amino-terminal FLAG epitope (Guo et al., 1996) was ligated into the BamHI/XbaI site of pFASTBAC1 donor plasmid. The recombinant plasmid was transformed into DH10BAC cells for site-specific transposition into bMON14272 bacmid DNA. High-titer viral stocks were used to infect 5 × 177-cm2 Sf21 cell monolayers, which were harvested 72 h postinfection and resuspended in 10 ml of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, and a cocktail of protease inhibitors (Rybakova et al., 1996). The lysate was circulated over a 2-ml anti-FLAG M2 agarose column (Sigma-Aldrich, St. Louis, MO), which was washed extensively with buffer A (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Triton X-100) and bound utrophin eluted with buffer A containing 100 μg/ml FLAG peptide (Sigma-Aldrich). For protein used in rotary shadowing, the M2 column was washed and eluted as described above except that Triton X-100 was omitted. Purified utrophin was concentrated in a Centricon 100 (Amicon, Beverly, MA) and assayed for protein with the Bio-Rad DC protein assay kit by using bovine serum albumin as standard. The typical yield of pure utrophin was 700 μg from 5 × 177 cm2 of cell monolayer. Recombinant utrophin was analyzed on Coomassie blue-stained SDS-polyacrylamide gels and by Western blot analysis with the utrophin-specific monoclonal antibodies MANCHO3 (Man et al., 1991) and DRP-2 (Novocastra, New Castle, UK), and with anti-FLAG clone M2 (Sigma-Aldrich).
Recombinant Utrophin N-Terminal Actin Binding Domains
To generate an expression vector encoding mouse utrophin amino acids 1–261 fused with an amino-terminal FLAG epitope (FLAG-UTR261), the oligonucleotide primers 5′-TATTCCGGATTATTCA-TACC-3′ and 5′-GCACCTCTCGAGTTCAATCTAT-3′ were used to polymerase chain reaction-amplify a cDNA encoding FLAG-UTR261 from the pFASTBAC1 donor plasmid containing full-length, FLAG-utrophin. The polymerase chain reaction product was subcloned into pCR-Blunt (Invitrogen, Carlsbad, CA), cut out with BamHI and XhoI, and inserted between the BamHI and XhoI sites of pET23 (Novagen, Madison, WI). Both FLAG-UTR261 and an untagged construct (UTR261+) were expressed in Escherichia coli strain BL21 (DE3) and purified by serial ion exchange and gel filtration chromatography essentially as described previously (Moores and Kendrick-Jones, 2000).
Hydrodynamic Analysis
Measurement of the sedimentation coefficient and Stokes radius and calculation of the native molecular weight and frictional coefficient of recombinant utrophin were performed as described previously (Rybakova and Ervasti, 1997; Amann et al., 1999).
Electron Microscopy
Recombinant utrophin was diluted 10-fold into 0.15 M ammonium bicarbonate/acetate, pH 7.4, and adjusted to 55% glycerol. Low-angle rotary shadowing and electron microscopy were performed as described previously (Yurchenco and Cheng, 1993).
Actin Binding Analysis
Recombinant utrophin binding to muscle and nonmuscle F-actin (Cytoskeleton, Denver, CO) was measured as described previously (Rybakova et al., 1996). Briefly, purified recombinant utrophin or utrophin fragments were incubated with 6 μM F-actin in actin binding buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM ATP, 2 mM MgCl2, 0.2 mM dithiothreitol, and 0.1% Triton X-100) also containing 100 mM NaCl and centrifuged at 100,000 × g for 20 min. The resulting supernatants and F-actin pellets were resolved on Coomassie blue-stained SDS-polyacrylamide gels and the amount of bound and free protein measured by densitometry. The effect of utrophin on F-actin depolymerization was assessed by high-speed cosedimentation after dilution into low ionic strength buffer as described previously (Rybakova et al., 1996; Rybakova and Ervasti, 1997). Briefly, various concentrations of utrophin were preincubated for 20 min with F-actin in actin binding buffer containing 30 mM NaCl and then diluted with actin binding buffer to final actin and NaCl concentrations of 2 μM and 4.5 mM, respectively. At various times postdilution, samples were centrifuged and analyzed for the fraction of actin remaining in the pellet as described above.
Analysis of Mechanically Peeled Sarcolemma and Myofibers
Sarcolemma and peeled myofibers were isolated from the extensor digitorum longus muscles of age-matched mdx, and Fiona transgenic mdx mice (Tinsley et al., 1998) and visualized by confocal microscopy as described previously (Rybakova et al., 2000). F-actin was detected with Alexa568-phalloidin (Molecular Probes) and utrophin was stained with rabbit 56 antiserum (Rybakova et al., 2000) or monoclonal antibody (mAb) DRP1 (Novocastra).
Quantitation of Utrophin Protein Expression in Muscle
Trunk and limb muscles of C57BL/10ScSn control (Jackson Laboratories, Bar Harbor, ME), mdx, and Fiona transgenic mdx mice were snap frozen in liquid nitrogen, and stored at −80°C. Frozen muscle (0.5 g) was pulverized in a mortar and pestle, cooled with liquid nitrogen, and solubilized in 2 ml of 1% SDS, 5 mM EGTA, and a cocktail of protease inhibitors. The samples were incubated for 2 min at 100°C and centrifuged at 12,000 × g. The protein concentration of the supernatants was measured with the Bio-Rad DC protein assay kit by using bovine serum albumin as standard. Nitrocellulose transfers containing various amounts of protein were incubated with a 1:200 dilution of mAb MANCHO3 (Man et al., 1991) and immunoreactivity was detected with 125I-goat anti-mouse IgG and autoradiography. The intensities of immune signal were analyzed densitometrically. A standard curve of purified recombinant utrophin was included on all gels/transfers.
RESULTS
Expression, Purification, and Characterization of Recombinant Utrophin
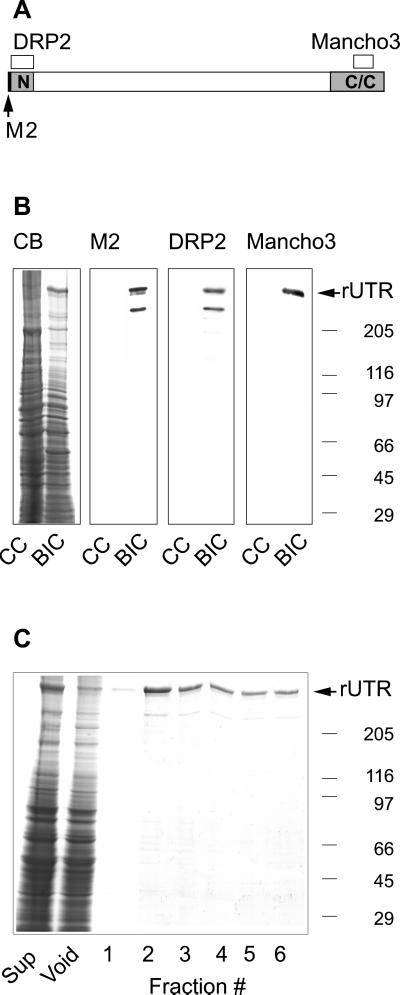
The actin binding properties of utrophin have only been extrapolated from studies of small recombinant fragments (Winder et al., 1995; Amann et al., 1999; Moores and Kendrick-Jones, 2000; Zuellig et al., 2000). Therefore, we generated a baculovirus construct encoding full-length mouse utrophin with an amino-terminal FLAG epitope (Figure 1A). Coomassie blue-stained gels of infected cell lysates (Figure 1B) revealed a novel protein with an average molecular weight of 379,000. The protein was confirmed as intact FLAG-tagged utrophin on Western blots stained with antibodies to either the amino or carboxyl termini of utrophin and by M2 antibody to the FLAG epitope (Figure 1, A and B). Both the anti-FLAG and utrophin amino-terminal, but not the carboxy-terminal antibodies also reacted with a second band of ∼300,000 molecular weight (Figure 1B), which was probably a proteolytic fragment. Recombinant utrophin was purified using anti-Flag M2 agarose chromatography (Figure 1C). Densitometry indicated that full-length utrophin comprised ∼95% of the recovered protein. The remaining ∼5% was due to the ∼300,000 molecular weight proteolytic fragment (Figure 1C). From the measured Stokes' radius (9.1 nm) and sedimentation coefficient (10.4 S), we calculated a native molecular weight of 404,000 for recombinant utrophin, which was within ∼3% of its predicted molecular weight of 393,000. The calculated frictional coefficient (1.86) suggested that purified recombinant utrophin assumed an asymmetric rod shape as predicted previously (Tinsley et al., 1992).
Figure 1.
Expression and purification of recombinant utrophin. (A) Schematic of utrophin with the locations of epitopes for anti-FLAG M2, DRP2, and MANCHO3 monoclonal antibodies. (B) Coomassie blue-stained SDS-polyacrylamide gel (CB) and identical nitrocellulose transfers containing uninfected Sf21 cells (CC), and Sf21 cells infected with recombinant baculovirus encoding full-length utrophin (BIC). The arrow identifies the high molecular weight novel protein apparent in BIC stained with Coomassie blue and reactive with utrophin and anti-FLAG antibodies. (C) Coomassie blue-stained gel loaded with equivalent volumes of infected insect cell proteins solubilized with 1% Triton X-100 (Sup), the anti-FLAG M2 agarose void, and fractions eluted with FLAG peptide. The molecular weight standards (× 10−3) are indicated on the right.
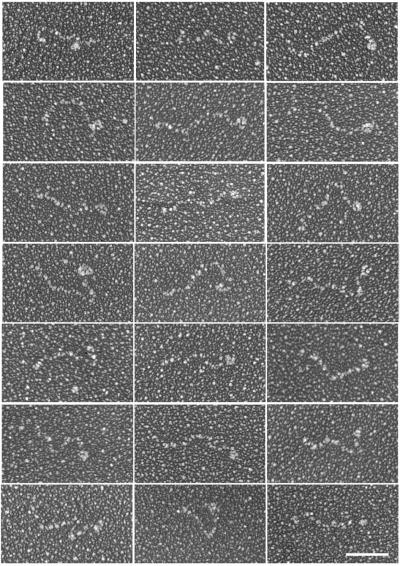
Although dystrophin and utrophin are widely envisioned as highly flexible rod-shaped molecules, the experimental evidence in support of such a model is minimal (Pons et al., 1990). Therefore, we examined recombinant utrophin by electron microscopy after rotary shadowing (Figure 2). The distribution of molecules was sparse due to the marginal solubility of utrophin in the buffer necessary for rotary shadowing. However, the observed molecules predominantly appeared as highly elongated structures with an average contour length of 118 ± 22 nm (± SD, n = 59). Based on available molecular dimensions (Yan et al., 1993; Keep et al., 1999), the amino-terminal actin binding domain and 22 spectrin-like repeats of utrophin would be expected to span a length of 115 nm. The globular structure near one end of most molecules may represent the cysteine-rich and C-terminal domains of utrophin because these domains were absent from the proteolytic fragment in purified utrophin (Figure 1) and the cysteine-rich domain of dystrophin adopts a globular structure (Huang et al., 2000). Finally, the molecules displayed random bends or kinks throughout their length, suggesting that utrophin is very flexible.
Figure 2.
Rotary shadowed utrophin. Shown are images selected to display the array of molecular configurations observed. The average contour length was 118 ± 22 nm (n = 59) and most molecules exhibited a globular structure at one end. Bar, 50 nm.
Actin Binding Properties of Recombinant Utrophin
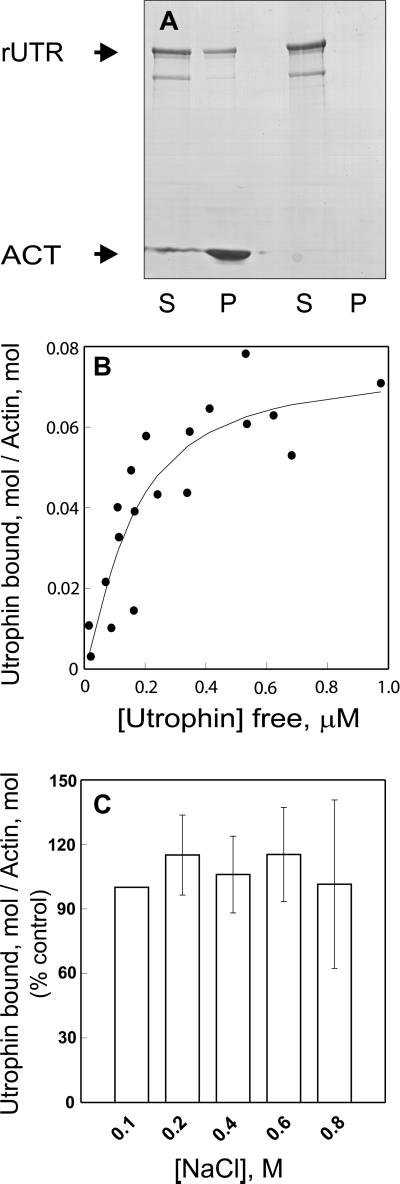
High-speed cosedimentation analysis demonstrated that recombinant utrophin bound saturably to skeletal muscle F-actin with a Kd of 0.15 ± 0.06 μM and a Bmax of 1 mol of utrophin/14 mol of actin, whereas virtually no utrophin sedimented in the absence of F-actin (Figure 3, A and B). Utrophin bound nonmuscle actin with a similar Kd of 0.24 ± 0.03 μM, indicating no marked preference for different actin isoforms. A recombinant protein encoding utrophin amino acids 1–261 fused with an amino-terminal FLAG epitope (FLAG-UTR261) and an untagged construct (UTR261+) both bound F-actin with 1:1 stoichiometry and Kd values of 16.5 ± 5.1 and 7.1 ± 4.1 μM, respectively. Thus, the FLAG epitope cannot account for the dramatic differences in actin binding properties between full-length utrophin and the isolated amino-terminal actin binding domain. Most interesting, the surprisingly high affinity and low stoichiometry of full-length utrophin binding to F-actin indicate that the rod domain also participates in utrophin binding to actin.
Figure 3.
Utrophin binding to F-actin. (A) Coomassie blue-stained SDS-polyacrylamide gel of the supernatant (S) and pellet (P) recovered after 0.6 μM recombinant utrophin (rUTR) was centrifuged at 100,000 × g in the presence or absence of 6 μM skeletal muscle F-actin (ACT). (B) Increasing concentrations of recombinant utrophin were incubated with 6 μM F-actin with subsequent centrifugation. The amount of free and actin-bound utrophin was determined densitometrically from Coomassie blue-stained gels of supernatant and pellet fractions. Symbols represent data from two independent experiments performed with different utrophin preparations. Nonlinear regression analysis yielded a Kd value of 0.2 μM and a Bmax value of 1 utrophin:14 actin monomers. (C) Utrophin (0.2 μM) was incubated with 6 μM F-actin over a range of NaCl concentrations (0.1–0.8 M) and subjected to centrifugation at 100,000 × g. The binding data (n = 3) were normalized against the amount of actin pelleted and expressed as the percentage of utrophin cosedimented with F-actin in 0.1 M NaCl.
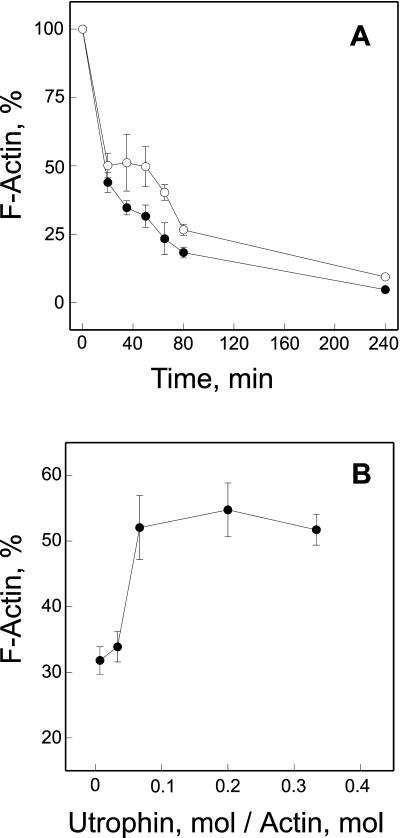
Because only two spectrin repeats in utrophin are basic, it seemed likely that the utrophin rod domain participates in actin binding through a nonelectrostatic mechanism. We found that utrophin binding to F-actin was insensitive to NaCl concentrations up to 0.8 M (Figure 3C). In contrast, dystrophin binding to F-actin was significantly inhibited by 0.5 M NaCl (Rybakova et al., 1996). Utrophin would also be expected to significantly slow the depolymerization of actin filaments as shown previously for dystrophin (Rybakova et al., 1996). As predicted, we found that F-actin depolymerization was significantly slowed in the presence of utrophin (Figure 4A). However, the protective effect of utrophin on actin depolymerization was more transient, lasting only 80 min compared with dystrophin, which persisted for at least 4 h (Rybakova et al., 1996). The protective effect of utrophin on F-actin depolymerization saturated at a utrophin: actin molar ratio of 1 utrophin:14 actin monomers (Figure 4B), which is highly consistent with the stoichiometry measured at equilibrium. In total, our results suggest that utrophin binds with high affinity along side an actin filament and can stabilize F-actin in vitro in a manner analogous to dystrophin. However, the decreased stoichiometry of utrophin binding to F-actin and its insensitivity to increased ionic strength further suggests that utrophin binds laterally along actin filaments through molecular contacts that are distinct from those used by dystrophin.
Figure 4.
Effect of utrophin on F-actin depolymerization. (A) Time course of actin depolymerization in the absence (●) or presence of utrophin (○). F-actin was preincubated alone or in a 10:1 M ratio with utrophin and filament depolymerization was induced by dilution to final actin and NaCl concentrations of 2 μM and 4.5 mM, respectively. At various times postdilution, samples were centrifuged at 100,000 × g for 20 min and the fraction of F-actin remaining was determined densitometrically from Coomassie blue-stained gels loaded with equal volumes of supernatants and pellets. Time points include centrifugation time. (B) F-actin was preincubated with increasing amounts of utrophin to give the indicated utrophin/actin monomer ratio and depolymerization was initiated by dilution as described above. Thirty minutes after initiation of depolymerization, samples were centrifuged at 100,000 × g and the percentage of F-actin remaining was determined as in A. The data represent the average (± SEM) from five (A) and three (B) independent experiments.
Utrophin Overexpression Rescues Costameric Actin Defect of mdx Muscle
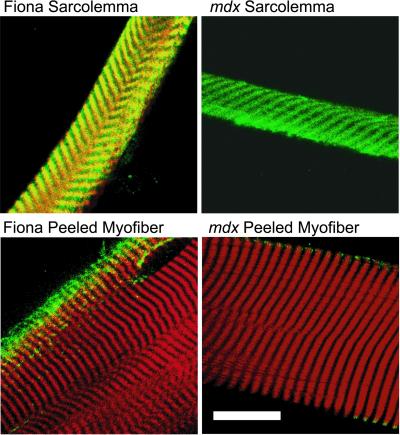
We recently demonstrated that a population of actin filaments colocalized with dystrophin in a costameric pattern on sarcolemma peeled from single myofibers of normal mouse muscle (Rybakova et al., 2000). In contrast, costameric actin was uniformly absent from sarcolemma of dystrophin-deficient mdx muscle even though utrophin was markedly up-regulated and retained in a costameric pattern (Rybakova et al., 2000). We have now peeled sarcolemma from myofibers of a transgenic mdx mouse line (Fiona) that overexpresses full-length utrophin to levels that correct all other phenotypic parameters associated with dystrophin deficiency (Tinsley et al., 1998). Strikingly, 18 of 19 sarcolemma from two different Fiona mice displayed bright phalloidin staining in a well organized costameric pattern that closely overlapped with utrophin (Figure 5). The one Fiona sarcolemma without phalloidin staining also failed to exhibit any utrophin immunoreactivity. In contrast to the uniform retention of costameric actin on sarcolemma from the Fiona line, phalloidin staining was absent in nine of nine sarcolemma from two age-matched mdx mice, although costameric utrophin was present on all specimens (Figure 5). Thus, when overexpressed to sufficiently high levels, utrophin can substitute for dystrophin in retaining costameric actin on mechanically peeled sarcolemma.
Figure 5.
Utrophin overexpression in mdx mice rescues costameric actin on mechanically peeled sarcolemma. Shown are images of mechanically peeled sarcolemma, or myofibers stained with Alexa568-phalloidin (red) and rabbit 56 antiserum to utrophin (green), with areas of coincidence appearing yellow. Specimens were obtained from transgenic mdx mice overexpressing utrophin (Fiona), or from age-matched mdx mice. Bar, 20 μm.
Quantitation of Utrophin Expression in Control, mdx, and Fiona Mice
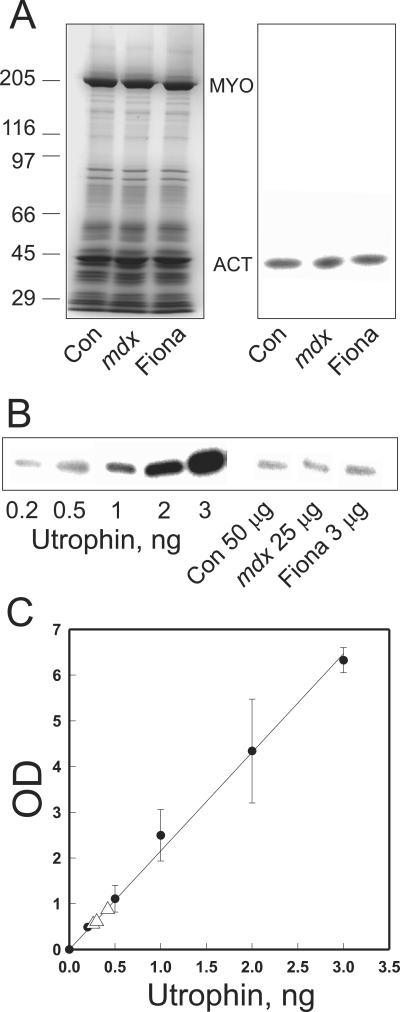
Based on our results, utrophin can perform all of the in vitro and in vivo actin binding functions previously documented for dystrophin. However, it remained unclear how much utrophin is required to correct the mdx phenotype relative to the amount of dystrophin normally expressed in wild-type muscle. Therefore, we measured utrophin abundance in skeletal muscle of control, mdx, and Fiona mice by quantitative Western blot analysis with recombinant utrophin as a standard (Figure 6). Both the 43-kDa actin-containing band and the 205-kDa myosin heavy chain band exhibited nearly identical densitometric intensities on Coomassie blue-stained gels when equal amounts of total protein were loaded for control, mdx, and Fiona muscle (Figure 6A). Furthermore, Western blots loaded with equal amounts of total muscle protein for each mouse line yielded nearly identical autoradiographic intensities when stained with a mAb specific for α-sarcomeric actin and detected with 125I-goat anti-mouse secondary (Figure 6A). For utrophin, however, we found it necessary to load different amounts of protein from the three mouse lines to ensure that immune signals from each were measured in the linear range (Figure 6B). Control muscle exhibited the lowest utrophin abundance (0.00058 ± 0.000053%), whereas the highest utrophin level was measured in Fiona muscle (0.014 ± 0.0015%). In good agreement with previous measurements of relative abundance (Matsumura et al., 1992; Porter et al., 1998), the utrophin content of mdx muscle (0.0013 ± 0.00014%) was approximately twofold greater than that of control muscle. Interestingly, the absolute utrophin abundance of mdx muscle was >60% of a previous estimate of dystrophin abundance in control muscle (Hoffman et al., 1987). Because the bulk of utrophin immunoreactivity in normal skeletal muscle cross sections is localized to nonmuscle cell types (Rivier et al., 1997; Peters et al., 1998), it could be argued that utrophin abundance in mdx muscle should be corrected by subtracting the amount of utrophin expressed in normal muscle (0.00058%). Even with this correction, however, our results suggest that the utrophin content of mdx muscle (0.00072%) approaches one-third of the measured dystrophin abundance (0.002%) in normal muscle (Hoffman et al., 1987).
Figure 6.
Utrophin protein abundance in control, mdx, and Fiona transgenic mdx mice. (A) Coomassie blue-stained gel (left) or nitrocellulose transfer (right) loaded with equal amounts of total protein from control, mdx, and Fiona muscle. The transfer was stained with a mAb specific for α-sarcomeric actin detected with 125I-anti-mouse IgG. Molecular weight standards (× 10−3) are shown on the left. (B) Autoradiogram from a nitrocellulose transfer containing the various amounts of purified utrophin, 50 μg of control, 25 μg of mdx, and 3 μg of Fiona total skeletal muscle extract stained with MANCHO3 and detected by 125I-anti-mouse IgG. (C) Standard curve of autoradiographic intensity vs. utrophin load from the autoradiogram in B. The utrophin signals obtained for muscle from the three lines of mice are indicated by open triangles. The utrophin content of control, mdx, and Fiona muscle was 0.00058 ± 0.000053, 0.0013 ± 0.00014, and 0.014 ± 0.0015%, respectively (n = 4).
DISCUSSION
Studies in knock-out and transgenic mice have provided compelling evidence that utrophin can compensate for many of the functions normally performed by dystrophin in skeletal muscle (Tinsley et al., 1996, 1998; Deconinck et al., 1997a, b; Grady et al., 1997). Herein, we have demonstrated that utrophin can retain costameric actin on mechanically peeled sarcolemma when overexpressed in mdx muscle to levels that also correct all other known parameters of the mdx phenotype. These results simultaneously validate the costameric actin defect of mdx sarcolemma (Rybakova et al., 2000) as a direct consequence of dystrophin deficiency and reinforce the hypothesis that utrophin and dystrophin are functionally interchangeable.
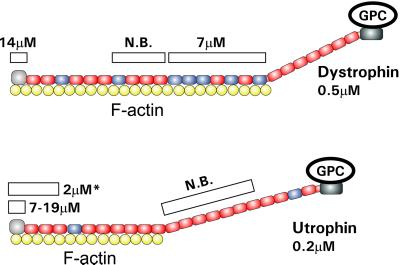
Given the differences between analogous domains in primary structure (Winder, 1997; Amann et al., 1999) and actin binding properties (Winder et al., 1995; Amann et al., 1998, 1999; Renley et al., 1998; Moores and Kendrick-Jones, 2000), it was expected that utrophin would bind actin filaments through a mechanism distinct from the lateral attachment used by dystrophin. Surprisingly, we have demonstrated that recombinant utrophin bound actin filaments with significantly higher affinity and through a more extensive lateral association than was anticipated. Our experiments indicate that full-length utrophin can occupy 14 actin monomers when saturating an actin filament compared with the one-to-one association consistently observed with the isolated utrophin amino-terminal actin binding domain (Moores and Kendrick-Jones, 2000). Our previous studies (Rybakova et al., 1996; Rybakova and Ervasti, 1997; Amann et al., 1999) indicated that 17 spectrin repeats allow dystrophin to associate with 24 monomers in an actin filament (Figure 7). Assuming that a similar repeat/actin monomer ratio holds for utrophin, our data predict that perhaps the first 10 spectrin-like repeats participate in the actin binding activity of utrophin (Figure 7). Interestingly, a recombinant protein corresponding to the amino-terminal actin binding domain and first 2.5 spectrin-like repeats of utrophin (Zuellig et al., 2000) bound F-actin with an affinity (2 μM) and stoichiometry (1:5) intermediate to that observed for the N-terminal actin binding domain alone (7–19 μM, 1:1) and full-length utrophin (0.2 μM, 1:14). Although we are not aware of any studies that have explicitly tested whether the C-terminal region of utrophin binds to F-actin, the homologous region of dystrophin failed to bind F-actin as assessed by high-speed cosedimentation (Corrado et al., 1994). Moreover, we previously detected no F-actin cosedimentation by the C-terminal dystrophin fragment or dystrophin associated proteins after calpain digestion of purified dystrophin–glycoprotein complex (Rybakova et al., 1996). Finally, dystrophin and utrophin would be expected to bind actin filaments with similar stoichiometries if they bound filaments through N- and C-terminal binding sites. However, our data indicate that utrophin interacts with fewer actin monomers in filaments compared with dystrophin. Based on current data, we propose that the actin binding region of utrophin spans from its amino terminus through repeat 10, whereas that of dystrophin extends out through repeat 17 (Figure 7). The participation of 10 spectrin-like repeats in utrophin binding to F-actin may explain why a utrophin construct lacking repeats 4–19 was less effective than full-length utrophin in ameliorating the phenotypes associated with dystrophin deficiency in mdx mice (Deconinck et al., 1997b; Tinsley et al., 1998).
Figure 7.
Side binding model for dystrophin and utrophin binding to actin filaments. Shown are schematic diagrams of dystrophin and utrophin complexed with F-actin. Red spectrin-like repeats are acidic, and blue repeats are basic. The white bars above dystrophin and utrophin represent the recombinant fragments for each protein that have been analyzed for actin binding activity with the measured affinities indicated. N.B., no binding observed. The fragment marked with an asterisk was described in Zuellig et al. (2000).
Because the utrophin rod domain lacks a cluster of basic, spectrin-like repeats (Amann et al., 1999), it is likely that utrophin repeats interact with actin filaments through a molecular mechanism that is distinct from the electrostatic interaction used by the dystrophin middle rod domain (Amann et al., 1998). Indeed, we observed that utrophin binding to F-actin was insensitive to NaCl concentrations (Figure 4) that significantly inhibited dystrophin binding to F-actin (Rybakova et al., 1996). A contribution by the spectrin-like repeats most proximal to the N-terminal actin binding activity of utrophin and dystrophin may explain why dystrophin constructs containing only five spectrin-like repeats can rescue the mdx phenotype (Wang et al., 2000), whereas constructs lacking the entire rod domain failed to provide functional correction (Hartigan-O'Connor and Chamberlain, 2000). Our results further imply that the actin side binding function now demonstrated for vertebrate dystrophin and utrophin may be more widely conserved across invertebrate dystrophin family members than could be predicted by sequence comparisons (Greener and Roberts, 2000; Neuman et al., 2001). More generally, we speculate that the combined presence of a CH-type actin binding domain and spectrin-like repeats may enable some members of the plakin family of cytolinkers to laterally bind and stabilize actin filaments (Leung et al., 2001). In support of this possibility, actin filaments associated with microtubule-bound microtubule actin crosslinking factor (MACF) were recently found to be more resistant to depolymerization induced by latrunculin B (Karakesisoglou et al., 2000).
We have also made use of purified recombinant utrophin as a standard to estimate the absolute utrophin protein content in control, mdx, and the Fiona line of transgenic mice. The utrophin content of Fiona muscle was ∼10-fold greater than that of mdx muscle and also approximately sevenfold greater than the dystrophin content of control muscle (Hoffman et al., 1987). A 50-fold overexpression of dystrophin was previously shown to correct the mdx phenotype without any toxic side effects (Cox et al., 1993). At present, it is not possible to determine the minimal level of utrophin expression necessary for retention of costameric actin on isolated sarcolemma, or for full correction of the dystrophic phenotype. However, utrophin expression in mdx muscle was greater than 60% of a similarly determined estimate of dystrophin content in striated muscle from normal mice (Hoffman et al., 1987). Even after correction for nonmuscle utrophin expression, our current measurements indicate that mdx muscle up-regulates utrophin expression to 36% of dystrophin levels in normal muscle, which may explain its milder phenotype compared with mice deficient in both dystrophin and utrophin (Deconinck et al., 1997a; Grady et al., 1997). On the other hand, transgenic expression of dystrophin to 20% of its normal levels was sufficient to prevent essentially all dystrophic symptoms in the mdx mouse (Phelps et al., 1995). Thus, the presence of even mild phenotype despite significant levels of utrophin in mdx muscle can be interpreted several ways. First, the increased utrophin expression of mdx muscle may be preferentially concentrated within regenerating fibers although weak sarcolemmal utrophin staining is sometimes apparent in large diameter mdx muscle fibers with peripheral nuclei (Peters et al., 1997). It is also possible that utrophin is less efficient in coupling the sarcolemma to costameres. Based on our depolymerization experiments, utrophin/F-actin complexes may be kinetically less stable compared with dystrophin/F-actin complexes (Rybakova et al., 1996), whereas other results (Lumeng et al., 1999; Imamura et al., 2000) suggest that the utrophin/β-dystroglycan interaction may be weaker. Alternatively, correction of the mdx phenotype by transgenic dystrophin expression to 20% of wild-type levels (Phelps et al., 1995) may instead have been due to functional additivity with concomitant up-regulation of utrophin. Likewise, rescue of the mdx phenotype by truncated dystrophins (Wang et al., 2000) may be due in part to additivity with the high utrophin levels endogenous to mdx muscle. Functional additivity further suggests that even low-level dystrophin expression induced by gene therapy when combined with pharmacological up-regulation of endogenous utrophin expression may yield an additive therapeutic benefit in patients with dystrophinopathies.
Due mainly to its exceedingly low abundance in native tissues, the biochemical characterization of utrophin function has previously relied on analysis of recombinant protein fragments (Winder et al., 1995; Amann et al., 1999; Chung and Campanelli, 1999; James et al., 2000). Our actin binding studies of full-length recombinant utrophin suggest that the sum of the parts does not necessarily equal, or even accurately reflect the behavior of the whole. Future experiments will revisit models for the interaction between utrophin/dystrophin and β-dystroglycan, which currently rely almost exclusively on results with isolated protein fragments (Chung and Campanelli, 1999; Huang et al., 2000; James et al., 2000). Full-length recombinant utrophin will also be an invaluable probe to identify novel molecular partners. Finally, the utrophin prepared by these methods now make possible studies to characterize its mechanical properties at the level of single molecules (Rief et al., 1999).
ACKNOWLEDGMENTS
We are grateful to Drs. Ruslan Grishanin, Vadim Klenchin, and Paul Friesen for advice with the baculovirus expression system, and to Sarah Squire for technical assistance. We thank Drs. Kevin Campbell for rabbit 56 antiserum, Glenn Morris for MANCHO3 antibodies, and Steve Winder for the UTR261+ expression construct. This study was supported by National Institutes of Health grants AR42423 and AR01985 (to J.M.E.), the Muscular Dystrophy Association (to I.N.R.), and the Medical Research Council, United Kingdom (to K.E.D.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–09–0446. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–09–0446.
REFERENCES
- Amann KJ, Guo WXA, Ervasti JM. Utrophin lacks the rod domain actin binding activity of dystrophin. J Biol Chem. 1999;274:35375–35380. doi: 10.1074/jbc.274.50.35375. [DOI] [PubMed] [Google Scholar]
- Amann KJ, Renley BA, Ervasti JM. A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J Biol Chem. 1998;273:28419–28423. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- Blake DJ, Tinsley JM, Davies KE. Utrophin: a structural and functional comparison to dystrophin. Brain Pathol. 1996;6:37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x. [DOI] [PubMed] [Google Scholar]
- Chung W, Campanelli JT. WW and EF hand domains of dystrophin-family proteins mediate dystroglycan binding. Mol Cell Biol Res Commun. 1999;2:162–171. doi: 10.1006/mcbr.1999.0168. [DOI] [PubMed] [Google Scholar]
- Corrado K, Mills PL, Chamberlain JS. Deletion analysis of the dystrophin-actin binding domain. FEBS Lett. 1994;344:255–260. doi: 10.1016/0014-5793(94)00397-1. [DOI] [PubMed] [Google Scholar]
- Cox GA, Cole NM, Matsumura K, Phelps SF, Hauschka SD, Campbell KP, Faulkner JA, Chamberlain JS. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity. Nature. 1993;364:725–729. doi: 10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- Craig SW, Pardo JV. Gamma actin, spectrin, and intermediate filament proteins colocalize with vinculin at costameres, myofibril-to-sarcolemma attachment sites. Cell Motil. 1983;3:449–462. doi: 10.1002/cm.970030513. [DOI] [PubMed] [Google Scholar]
- Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, Watt DJ, Dickson JG, Tinsley JM, Davies KE. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997a;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- Deconinck N, Tinsley J, De Backer F, Fisher R, Kahn D, Phelps S, Davies K, Gillis JM. Expression of truncated utrophin leads to major functional improvements in dystrophin-deficient muscles of mice. Nat Med. 1997b;3:1216–1221. doi: 10.1038/nm1197-1216. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- Grady RM, Teng HB, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90:729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- Greener MJ, Roberts RG. Conservation of components of the dystrophin complex in Drosophila. FEBS Lett. 2000;482:13–18. doi: 10.1016/s0014-5793(00)02018-4. [DOI] [PubMed] [Google Scholar]
- Guo WXA, Nichol M, Merlie JP. Cloning and expression of full length mouse utrophin: the differential association of utrophin and dystrophin with AChR clusters. FEBS Lett. 1996;398:259–264. doi: 10.1016/s0014-5793(96)01216-1. [DOI] [PubMed] [Google Scholar]
- Hartigan-O'Connor D, Chamberlain JS. Developments in gene therapy for muscular dystrophy. Microsc Res Tech. 2000;48:223–238. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<223::AID-JEMT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with β-dystroglycan. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- Imamura M, Araishi K, Noguchi S, Ozawa E. A sarcoglycan-dystroglycan complex anchors Dp116 and utrophin in the peripheral nervous system. Hum Mol Genet. 2000;9:3091–3100. doi: 10.1093/hmg/9.20.3091. [DOI] [PubMed] [Google Scholar]
- James M, Nuttall A, Ilsley JL, Ottersbach K, Tinsley JM, Winder SJ. Adhesion-dependent tyrosine phosphorylation of β-dystroglycan regulates its interaction with utrophin. J Cell Sci. 2000;113:1717–1726. doi: 10.1242/jcs.113.10.1717. [DOI] [PubMed] [Google Scholar]
- Karakesisoglou I, Yang Y, Fuchs E. An epidermal plakin that integrates actin and microtubule networks at cellular junctions. J Cell Biol. 2000;149:195–208. doi: 10.1083/jcb.149.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keep NH, Winder SJ, Moores CA, Walke S, Norwood FLM, Kendrick-Jones J. Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer. Structure. 1999;7:1539–1546. doi: 10.1016/s0969-2126(00)88344-6. [DOI] [PubMed] [Google Scholar]
- Kramarcy NR, Vidal A, Froehner SC, Sealock R. Association of utrophin and multiple dystrophin short forms with the mammalian Mr 58,000 dystrophin-associated protein (syntrophin) J Biol Chem. 1994;269:2870–2876. [PubMed] [Google Scholar]
- Leung CA, Liem RKH, Parry DAD, Green KJ. The plakin family. J Cell Sci. 2001;114:3409–3410. doi: 10.1242/jcs.114.19.3409. [DOI] [PubMed] [Google Scholar]
- Lumeng CN, Phelps SF, Rafael JA, Cox GA, Hutchinson TL, Begy CR, Adkins E, Wiltshire R, Chamberlain JS. Characterization of dystrophin and utrophin diversity in the mouse. Hum Mol Genet. 1999;8:593–599. doi: 10.1093/hmg/8.4.593. [DOI] [PubMed] [Google Scholar]
- Man N, Ellis JM, Love DR, Davies KE, Gatter KC, Dickson G, Morris GE. Localization of the DMDL gene-encoded dystrophin-related protein using a panel of nineteen monoclonal antibodies: presence at neuromuscular junctions, in the sarcolemma of dystrophic skeletal muscle, in vascular and other smooth muscles, and in proliferating cell lines. J Cell Biol. 1991;115:1695–1700. doi: 10.1083/jcb.115.6.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- Moores CA, Kendrick-Jones J. Biochemical characterization of the actin-binding properties of utrophin. Cell Motil Cytoskeleton. 2000;46:116–128. doi: 10.1002/1097-0169(200006)46:2<116::AID-CM4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Neuman S, Kaban A, Volk T, Yaffe D, Nudel U. The dystrophin/utrophin homologues in Drosphila and in sea urchin. Gene. 2001;263:17–29. doi: 10.1016/s0378-1119(00)00584-9. [DOI] [PubMed] [Google Scholar]
- Pardo JV, D'Angelo Siliciano J, Craig SW. A vinculin-containing cortical lattice in skeletal muscle: transverse lattice elements (“costameres”) mark sites of attachment between myofibrils and sarcolemma. Proc Natl Acad Sci USA. 1983;80:1008–1012. doi: 10.1073/pnas.80.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MF, Adams ME, Froehner SC. Differential association of syntrophin pairs with the dystrophin complex. J Cell Biol. 1997;138:81–93. doi: 10.1083/jcb.138.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MF, Sadoulet-Puccio HM, Grady RM, Kramarcy NR, Kunkel LM, Sanes JR, Sealock R, Froehner SC. Differential membrane localization and intermolecular associations of α-dystrobrevin isoforms in skeletal muscle. J Cell Biol. 1998;142:1269–1278. doi: 10.1083/jcb.142.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps SF, Hauser MA, Cole NM, Rafael JA, Hinkle RT, Faulkner JA, Chamberlain JS. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum Mol Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- Pons F, Augier N, Heilig R, Leger J, Mornet D, Leger JJ. Isolated dystrophin molecules as seen by electron microscopy. Proc Natl Acad Sci USA. 1990;87:7851–7855. doi: 10.1073/pnas.87.20.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter GA, Dmytrenko GM, Winkelmann JC, Bloch RJ. Dystrophin colocalizes with β-spectrin in distinct subsarcolemmal domains in mammalian skeletal muscle. J Cell Biol. 1992;117:997–1005. doi: 10.1083/jcb.117.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JD, Rafael JA, Ragusa RJ, Brueckner JK, Trickett JI, Davies KE. The sparing of extraocular muscle in dystrophinopathy is lost in mice lacking utrophin and dystrophin. J Cell Sci. 1998;111:1801–1811. doi: 10.1242/jcs.111.13.1801. [DOI] [PubMed] [Google Scholar]
- Renley BA, Rybakova IN, Amann KJ, Ervasti JM. Dystrophin binding to nonmuscle actin. Cell Motil Cytoskeleton. 1998;41:264–270. doi: 10.1002/(SICI)1097-0169(1998)41:3<264::AID-CM7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Rief M, Pascual J, Saraste M, Gaub HE. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- Rivier F, Robert A, Hugon G, Mornet D. Different utrophin and dystrophin properties related to their vascular smooth muscle distributions. FEBS Lett. 1997;408:94–98. [Google Scholar]
- Rybakova IN, Amann KJ, Ervasti JM. A new model for the interaction of dystrophin with F-actin. J Cell Biol. 1996;135:661–672. doi: 10.1083/jcb.135.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova IN, Ervasti JM. Dystrophin-glycoprotein complex is monomeric and stabilizes actin filaments in vitro through a lateral association. J Biol Chem. 1997;272:28771–28778. doi: 10.1074/jbc.272.45.28771. [DOI] [PubMed] [Google Scholar]
- Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub V, Rafael JA, Chamberlain JS, Campbell KP. Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J Cell Biol. 1997;139:375–385. doi: 10.1083/jcb.139.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley JM, Blake DJ, Roche A, Byth BC, Knight AE, Kendrick-Jones J, Suthers GK, Love DR, Edwards YH, Davies KE. Primary structure of dystrophin-related protein. Nature. 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- Wang B, Li J, Xiao X. Adeno-associated virus vector carrying human mindystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc Natl Acad Sci USA. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MW, Bloch RJ. Extensive but coordinated reorganization of the membrane skeleton in myofibers of dystrophic (mdx) mice. J Cell Biol. 1999;144:1259–1270. doi: 10.1083/jcb.144.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder SJ. The membrane-cytoskeleton interface: the role of dystrophin and utrophin. J Muscle Res Cell Motility. 1997;18:617–629. doi: 10.1023/a:1018627705273. [DOI] [PubMed] [Google Scholar]
- Winder SJ, Hemmings L, Maciver SK, Bolton SJ, Tinsley JM, Davies KE, Critchley DR, Kendrick-Jones J. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J Cell Sci. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- Yan Y, Winograd E, Viel A, Cronin T, Harrison SC, Branton D. Crystal structure of the repetitive segments of spectrin. Science. 1993;262:2027–2030. doi: 10.1126/science.8266097. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, Cheng Y-S. Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J Biol Chem. 1993;268:17286–17299. [PubMed] [Google Scholar]
- Zuellig RA, Bornhauser BC, Knuesel I, Heller F, Fritschy J-M, Schaub MC. Identification and characterization of transcript and protein of a new short N-terminal utrophin isoform. J Cell Biochem. 2000;77:418–431. doi: 10.1002/(sici)1097-4644(20000601)77:3<418::aid-jcb7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]