Abstract
Activation of the epidermal growth factor receptor (EGFR) triggers multiple signaling pathways and rapid endocytosis of the epidermal growth factor (EGF)–receptor complexes. To directly visualize the compartmentalization of molecules involved in the major signaling cascade, activation of Ras GTPase, we constructed fusions of Grb2, Shc, H-Ras, and K-Ras with enhanced cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP), and used live-cell fluorescence imaging microscopy combined with the fluorescence resonance energy transfer (FRET) technique. Stimulation of cells by EGF resulted in the accumulation of large pools of Grb2-CFP and YFP-Shc in endosomes, where these two adaptor proteins formed a complex with EGFR. H-Ras and K-Ras fusion proteins were found at the plasma membrane, particularly in ruffles and lamellipodia, and also in endosomes independently of GTP/GDP loading and EGF stimulation. The relative amount of endosomal H-Ras was higher than that of K-Ras, whereas K-Ras predominated at the plasma membrane. On application of EGF, Grb2, and Ras converge in the same endosomes through the fusion of endosomes containing either Grb2 or Ras or through the joint internalization of two proteins from the plasma membrane. To examine the localization of the GTP-bound form of Ras, we used a FRET assay that exploits the specific interaction of GTP-bound CFP-Ras with the YFP-fused Ras binding domain of c-Raf. FRET microscopy revealed that GTP-bound Ras is located at the plasma membrane, mainly in ruffles and at the cell edges, as well as in endosomes containing EGFR. These data point to the potential for endosomes to serve as sites of generation for persistent signaling through Ras.
INTRODUCTION
The interaction of the epidermal growth factor receptor (EGFR) with epidermal growth factor (EGF) or other ligands at the cell surface triggers EGFR dimerization, activates receptor tyrosine kinase, and causes phosphorylation of the tyrosine residues in the carboxyl terminus of the receptor (Yarden and Sliwkowski, 2001). These phosphotyrosines serve as binding sites for proteins that contain Src homology (SH) 2 or phosphotyrosine-binding domains and that either possess enzymatic activity or link the receptor to enzymes. The activation of Ras GTPase is the major signaling event initiated by the EGFR (Joneson and Bar-Sagi, 1997). The initial step in this pathway is the recruitment of growth factor binding protein 2 (Grb2) to the receptor (Lowenstein et al., 1992). Grb2 is a modular protein consisting of one SH2 domain flanked by two SH3 domains. Grb2 is constitutively associated with Ras guanine-nucleotide exchange factor mSOS1/2 via its SH3 domains, whereas the Grb2 SH2 domain mediates binding to EGFR phosphotyrosines Tyr1068 and Tyr1086 (Li et al., 1993; Rozakis-Adcock et al., 1993; Batzer et al., 1994; Okutani et al., 1994). Grb2 can also bind to the phosphorylated tyrosines of another adaptor protein, Shc, that itself directly interacts with the EGFR mainly via receptor Tyr1148 and Tyr1173 (Okutani et al., 1994). Recruitment of the Grb2–SOS complex to the EGFR positions SOS adjacent to membrane-associated Ras, thus facilitating release of GDP from Ras and replacement by GTP. GTP-bound Ras binds to and alters the activity of Raf kinase, phosphotidylinositol-3 (PI3) kinase, Ral GTPase, and other “effector” proteins (Joneson and Bar-Sagi, 1997).
There are three ubiquitously expressed forms of Ras: H-Ras, K-Ras, and N-Ras (Reuther and Der, 2000). Despite almost identical amino acid sequences, these forms have different functions in vivo (Umanoff et al., 1995; Johnson et al., 1997; Yan et al., 1998). One attractive explanation for specificity of signaling in the Ras family is the difference in cellular localization of the Ras isoforms. All Ras isoforms undergo farnesylation and carboxylmethylation, which allows stable association of Ras with the membrane of endoplasmic reticulum (Choy et al., 1999). In addition, H-Ras and N-Ras are palmitoylated, probably in the Golgi apparatus, which provides a signal for targeting to the plasma membrane. In contrast, K-Ras4B (hereafter referred as to K-Ras) lacks a palmitoylation motif and instead relies on a polybasic motif in its carboxyl terminus for targeting to the plasma membrane without a Golgi-trafficking step (Choy et al., 1999; Apolloni et al., 2000). Furthermore, sequence differences in the carboxyl terminus may be responsible for distinct compartmentalization of Ras proteins within the plasma membrane subdomains. H-Ras and N-Ras are mostly confined to lipid raft/caveolae areas, whereas K-Ras tends to reside outside of rafts (Prior and Hancock, 2001; Prior et al., 2001). The activation of H-Ras seems to be accompanied by its exit from rafts (Prior and Hancock, 2001).
Immediately after activation of the EGFR at the cell surface, the ligand–receptor complexes are rapidly internalized through clathrin-dependent and -independent endocytic pathways (reviewed in Carpenter, 2000; Waterman and Yarden, 2001). After internalization EGF–receptor complexes accumulate in early endosomes from where they are either recycled back to the cell surface or targeted to the late endosomal and lysosomal compartments for degradation. Accelerated internalization and degradation of activated EGFR result in the down-regulation of EGFR, which leads to attenuation of growth factor signaling.
In many types of cells rapid endocytosis of activated surface EGFR in combination with a slower receptor degradation results in the prolonged concentration of receptors in endosomes. Several lines of evidence suggest that signals can be generated by endosomal receptors and that these receptors may be important for persistent signaling processes (Baass et al., 1995; Carpenter, 2000). Thus, despite the fact that the acidic pH of endosomes might be expected to cause ligand-receptor dissociation and deactivation of the receptor, it has been demonstrated that EGF–receptor complexes remain intact, dimerized, and phosphorylated during early stages of endocytosis (Cohen and Fava, 1985; Lai et al., 1989; Nesterov et al., 1990; Sorkin and Carpenter, 1991). Furthermore, the association of EGFR with effector proteins was demonstrated in endosomal fractions from rat liver (Di Gugliemo et al., 1994) and mammary cells (Burke et al., 2001). Colocalization of EGFR with effector molecules in endosomes has been shown by immunohistochemistry (Oksvold et al., 2000, 2001; Wang et al., 2001). However, biochemical analyses of protein–protein interactions require cell disruption and are performed in the presence of detergents, which can lead to dissociation of preexisting complexes or formation of new complexes, and therefore incorrect estimation of the extent of association. On the other hand, microscopic detection of protein colocalization in chemically fixed cells suffers from poor spatial resolution as well as inherent problems of artifactual redistribution of proteins.
In our work, we have attempted to visualize the movement of fluorescently tagged EGFR and effector proteins during endocytosis in living cells (Sorkin et al., 2000; Sorkin, 2001). These proteins were made fluorescent by fusing them to spectral variants of green fluorescent protein (GFP), specifically yellow (YFP) and cyan fluorescent (CFP) proteins. GFP-tagged EGFR interactors have been used recently by other groups (Brock and Jovin, 2001; Matsuda et al., 2001; Wang et al., 2001). Moreover, we applied fluorescence resonance energy transfer (FRET) microscopy to directly visualize protein–protein interactions during EGFR endocytosis. FRET between CFP and YFP can occur if the distance between CFP and YFP is <5 nm, which typically requires direct interaction of tagged proteins. Various FRET-based techniques have been developed recently to monitor protein–protein interactions (reviewed in Bastiaens and Pepperkok, 2000). The modified version of FRET microscopy that we used (Sorkin et al., 2000) is designed to obtain high-resolution FRET images of cells, thus allowing visualization of the compartmentalization of interacting molecules.
FRET imaging microscopy was used herein to simultaneously visualize receptor complexes with adaptor proteins that are involved in Ras activation and Ras itself during EGFR endocytosis. Fusion proteins of Ras with YFP and CFP were used to directly compare the localization of H-Ras and K-Ras with that of EGFR and Grb2, and to detect activated GTP-bound H-Ras in living cells. The data of imaging microscopy and FRET analysis show that the endosomal compartment is the major cellular location of EGFR signaling complexes and Ras proteins.
MATERIALS AND METHODS
Reagents
EGF was obtained from Collaborative Research (Bedford, MA) and Sigma-Aldrich (St. Louis, MO). EGF conjugated to rhodamine (EGF-Rh) was from Molecular Probes (Eugene, OR). Pfu polymerase was from Stratagene (La Jolla, CA). Glutathione-agarose beads were from Amersham Biosciences (Piscataway, NJ). Monoclonal antibodies that recognize all Ras isoforms, RAS10, were purchased from Upstate Biotechnology (Lake Placid, NY). A polyclonal antibody to GFP was from CLONTECH (Palo Alto, CA).
Plasmid Construction
Human K-Ras2 (K-Ras4B) cDNA was obtained from Guthrie cDNA Resource Center (Guthrie Research Institute, Sayre, PA). Human H-Ras cDNA and H-Ras mutant Ser17Asn (H-RasN17) were provided by Dr. G.L. Johnson (University of Colorado Health Sciences Center, Denver, CO), whereas Gly12Val mutant of H-Ras (H-RasV12) was provided by Dr. M. Dell'Acqua (University of Colorado Health Sciences Center). To generate YFP/CFP-fusion proteins, full-length H-Ras, K-Ras, H-RasV12, H-RasN17, and K-Ras were amplified by polymerase chain reaction and ligated into pECFP-C1 or pEYFP-C1 (CLONTECH) by using EcoRI and BamHI (H-Ras, V12, and N17), or BglII and HindIII restriction sites (K-Ras). cDNA encoding human c-Raf-1 was provided by Dr. F. Tebar (University of Barcelona, Barcelona, Spain). To generate Raf YFP/CFP-fusion proteins, full-length c-Raf-1, and a fragment corresponding to amino acid residues 1–153 of c-Raf-1 (Ras binding domain, RBD) were amplified by polymerase chain reaction, and ligated into pEYFP-N1 by using XhoI and BamHI sites. Grb2-YFP, Grb2-CFP, and YFP-Shc were described previously (Sorkin et al., 2000; Sorkin, 2001). To replace arginine 86 by alanine in Grb2-CFP, corresponding point mutation was made using QuickChange site-directed mutagenesis kit according to the manufacturer's protocol (Strategene). Similarly, tyrosines 239, 240, and 317 were replaced by phenylalanines in YFP-Shc to generate YFP-Shc-F. All constructs were verified by dideoxynucleotide sequencing.
Cell Culture and Transfections
Human epidermal carcinoma A-431 cells were grown in DMEM containing 10% calf serum, antibiotics, and glutamine. Mouse NIH 3T3 cells (clone WT8) expressing 4 × 105 human EGFRs/cell (Sorkin et al., 1996) were grown in DMEM containing 10% newborn calf serum, and supplemented with antibiotics, glutamine, and geneticin (G418). Porcine aortic endothelial (PAE) cells (clone B11) expressing 2 × 105 EGFRs/cell (Carter and Sorkin, 1998) were grown in F12 medium containing 10% fetal bovine serum, and supplemented with antibiotics, glutamine, and G418.
The transfections of CFP or YFP fusion protein constructs were carried out using Effectene (QIAGEN, Hilden, Germany) in six-well plates. For microscopy, cells were replated 24 h after transfection onto 25-mm glass coverslips or glass microscope chambers (Bioptechs, Butler, PA). Cells were starved in serum-free and phenol red-free medium containing 0.1% bovine serum albumin (BSA) for 6–16 h before experiments. Expression of YFP- and CFP-fused proteins was confirmed by Western blotting as described previously (Huang et al., 2001).
Living Cell Fluorescence Imaging
For experiments at room temperature, glass coverslips with cells were mounted in a microscopy chamber (Molecular Probes). For experiments at 37°C, a microscopy chamber was mounted onto the temperature-controlled adapter on the microscope stage. The fluorescence imaging workstation consisted of a Nikon inverted microscope equipped with a 100× oil immersion objective lens, cooled charge-coupled device SensiCam QE-16 MHz (Cooke, Germany), z-step motor, dual filter wheels, and a Xenon 175-W light source, all controlled by SlideBook 3.0 software (Intelligent Imaging Innovation, Denver, CO).
With the exception of time-lapse experiments, imaging was carried out at room temperature to avoid image disalignment owing to movement of organelles during multichannel data acquisition. The detection of YFP, CFP, and rhodamine fluorescence was performed using YFP, CFP, and Cy3 filter channels, respectively, and an 86004BS dichroic mirror (Chroma, Brattleboro, VT). Time-lapse images (150-ms integration time) were acquired at 30-s intervals. Photobleaching diminished signal by ∼20% at the end of the experiment. QuickTime movies were created from the original SlideBook time-lapse images using Director 8.
In most of experiments the excitation intensity was attenuated down to 20% of the maximum power of the light source. Images were acquired using 2 × 2 binning mode
FRET Measurements
The method of sensitized FRET measurement has been described previously (Sorkin et al., 2000; for detailed description, see Gordon et al., 1998). Briefly, images were acquired sequentially through YFP, CFP, and FRET filter channels. Filter sets used were YFP (excitation, 500/20 nm; emission, 535/30 nm), CFP (excitation, 436/10 nm; emission, 470/30 nm), and FRET (excitation, 436/10 nm; emission, 535/30 nm). An 86004BS dichroic mirror (Chroma) was used. Binning 2 × 2 modes and 125–250-ms integration times were used. The background images were subtracted from the raw images before carrying out FRET calculations. Corrected FRET was calculated on a pixel-by-pixel basis for the entire image by using equation 1: FRETC = FRET − (0.50 × CFP) − (0.02 × YFP), where FRET, CFP, and YFP correspond to background-subtracted images of cells coexpressing CFP and YFP acquired through the FRET, CFP, and YFP channels, respectively. The 0.5 and 0.02 are the fractions of bleed-through of CFP and YFP fluorescence, respectively, through the FRET filter channel. Corrected FRET (FRETC) images are presented in pseudocolor mode. FRETC intensity is displayed stretched between the low and high renormalization values, according to a temperature-based lookup table with blue (cold) indicating low values and red (hot) indicating high values. To eliminate the distracting data from regions outside of cells, the YFP channel is used as a saturation channel, and the FRETC images are displayed as YFP intensity-modulated images. In these images, data with YFP values greater than the high threshold of the saturation channel are displayed at full saturation, whereas data values below the low threshold are displayed with no saturation (i.e., black).
An alternative approach was also used to calculate FRETC and the apparent efficiency of FRET (Ea) for selected regions of images, for instance, individual ruffles or endosomes. Mean FRETC values were calculated from mean fluorescence intensities for each selected subregion according to equation 1, and Ea values were calculated according to equation 2: Ea = FRETC/YFP, where FRETC and YFP are the mean intensities of FRETC and YFP fluorescence in the selected subregion.
All calculations were performed using the Channel Math and FRET modules of the SlideBook software.
Immunofluorescence Staining
Cells were grown on glass coverslips coated with entactin-collagen IV-laminin cell attachment matrix (Upstate Biotechnology). Cells were treated with 100 ng/ml EGF-Rh for 15 min, washed with Ca2+, Mg2+-free phosphate buffered saline (CMF-PBS), fixed with freshly prepared 3% paraformaldehyde (Electron Microscopy Sciences) for 30 min at 4°C, and permeabilized using a 3-min incubation in CMF-PBS containing 0.1% Triton X-100 and 0.5% BSA at room temperature. The cells were then washed three times with CMF-PBS, blocked in CMF-PBS containing 3% BSA for 30 min, and incubated with RAS10 antibodies in the presence of 0.1% Triton X-100 and 0.5% BSA. Incubation with the secondary antibody (donkey anti-rabbit IgG labeled with Cy5) was in the same solution. Both primary and secondary antibody solutions were precleared by centrifugation at 100,000 × g for 10 min. The coverslips were mounted in Fluoromount-G (Fisher Scientific, Pittsburgh, PA) containing 1 mg/ml p-phenylenediamine. To obtain high-resolution three-dimensional images of cells, a Z-stack of 200-nm optical sections was obtained and deconvolved using a modification of the constrained iteration method. Final arrangement of all images was performed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Expression of GST-Fusion Proteins and GST Pull-Down Experiments
Glutathione S-transferase–fused Ras binding fragment of Raf-1 (GST-RBD) in pGEX-4T3 was provided by Dr. N. Agell (University of Barcelona). GST-RBD was made in Escherichia coli BL21 pLysS. Cells were harvested and washed once in 50 mM Tris-HCl, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride pH 8.0, and frozen at −80°C for 20 min. Cell pellets were resuspended and sonicated in 50 mM Tris-HCl, 4 mM EDTA, 2 mM EGTA, 1 mM dithiothreitol, 25% wt/vol sucrose, pH 8.0, supplemented with 1% Triton X-100 and protease inhibitors. The lysates were centrifuged at 100,000 × g for 20 min, and the supernatants were bound to glutathione-Sepharose 4B beads (Amersham Biosciences) by incubating for 3 h at 4°C at a ratio of 320 μl of 50% slurry per 50 ml of starting culture. The beads were washed and stored at 4°C.
For pull-down experiments, cells expressing wild-type CFP-H-Ras, YFP-H-RasV12, and YFP-H-RasN17 were lysed in buffer A (150 mM KCl, 2 mM MgCl2, 10% glycerol, 20 mM HEPES, 1 mM orthovanadate, 1% Triton X-100, 50 mM NaCl, 2 mM dithiothreitol, and protease and phosphatase inhibitors) for 10 min at 4°C, and the lysates were cleared at 12,000 × g for 10 min. Beads loaded with GST-RBD were incubated with cell lysates for 2 h at 4°C. After extensive washes of beads with buffer A, the beads were heated in sample buffer for 5 min at 95°C. Bound proteins were resolved by 12% SDS-PAGE, transferred to nitrocellulose membranes, probed by Western blotting with RAS10 antibodies, and visualized using chemiluminescence detection (Pierce Chemical, Rockford, IL).
RESULTS
EGF-induced Recruitment of Grb2 and Shc to Endosomes
To study trafficking and interactions of molecules involved in EGF-induced Ras activation, YFP and CFP fusion proteins of EGFR, Grb2, Shc, and Ras were generated. The dynamic localization of these proteins was monitored in three types of cells: A-431 epidermal carcinoma cells expressing high level of endogenous EGFRs, PAE cells stably expressing either human EGFRs or EGFR-CFP fusion proteins, and mouse NIH 3T3 cells stably expressing human EGFRs. Despite differences in EGFR levels, cell morphology and levels of heterologous expression of fusion proteins, similar results were obtained in all three cell lines.
The interaction of EGFR with Grb2 and Shc is the first step in Ras activation. To examine cellular localization of Grb2/Shc complexes with EGFR, we inspected trafficking of Grb2-CFP and YFP-Shc coexpressed in cells bearing endogenous EGFRs. A-431 cells were stimulated with EGF-Rh to allow simultaneous visualization of the movement of EGFRs and YFP/CFP-fused proteins in living cells during endocytosis. Before EGF stimulation Grb2-CFP was seen distributed in the cytosol and nucleus, whereas YFP-Shc was restricted to cytosol (Figure 1A). Nuclear localization of endogenous Grb2 was observed in PAE (Sorkin et al., 2000), A-431, and NIH 3T3 cells (our unpublished data). To test for possible interaction between Grb2-CFP and YFP-Shc, the efficiency of energy transfer between CFP and YFP was measured by calculating the sensitized FRET signal (FRETC) on a pixel-by-pixel basis as described in MATERIALS AND METHODS. Despite overlapping localization of these fusion proteins in the cytosol, only a very low FRETC signal was detected, indicative of minimal interaction. After stimulation of cells with EGF-Rh for 20 min at 37°C, both Grb2-CFP and YFP-Shc were seen colocalized with EGF-Rh in vesicular compartments, presumably, endosomes (Figure 1A). Colocalization of all three proteins in membrane ruffles and peripheral lamellipodia was more clearly seen at early time points (our unpublished data). The strongest FRETC signals were detected in endosomes. The recruitment of YFP-Shc and Grb2-CFP to ruffles and endocytic organelles was readily detected when cells were stimulated with low concentrations of EGF (1–2 ng/ml), although the extent of adaptor translocation to membranes was proportional to the amount of activated EGFR (our unpublished data). Essentially identical results were obtained in A-431, NIH 3T3, and PAE cells.
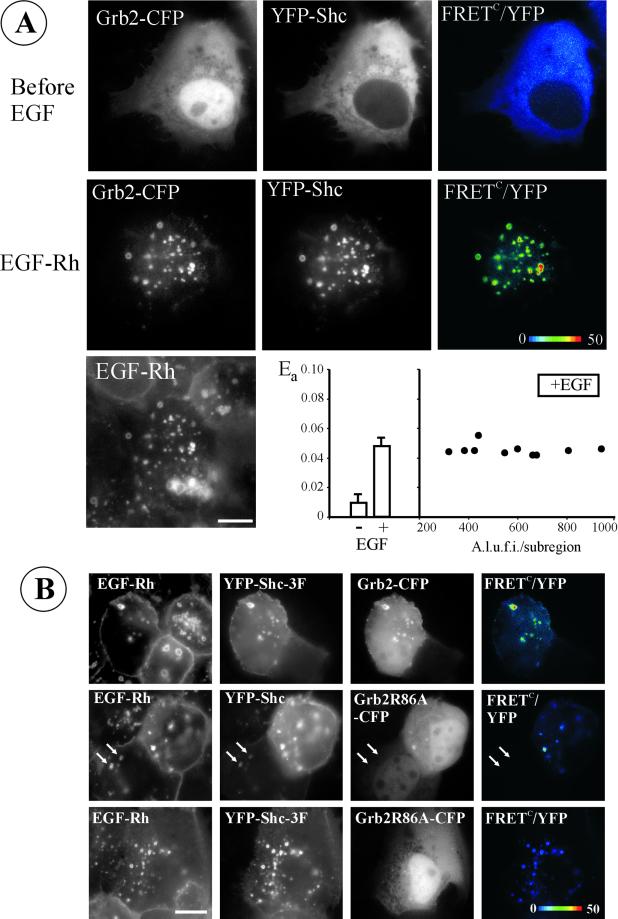
Figure 1.
Interaction of Grb2-CFP and YFP-Shc in endosomes induced by EGF. (A) Grb2-CFP and Shc-YFP were transiently expressed in A-431 cells. YFP, CFP, and FRET images were acquired before and after incubation of cells with 200 ng/ml EGF-Rh for 20 min at 37°C. EGF-Rh was detected using Cy3 filter channel. FRETC was calculated as described in MATERIALS AND METHODS, and is presented as pseudocolor intensity-modulated image (FRETC/YFP). Bar, 10 μm. On the graph, the apparent efficiencies of energy transfer between Grb2-CFP and YFP-Shc, Ea, calculated for multiple subregions of the images are shown. Left, averaged values of Ea are presented. Right, Ea values obtained for individual endosomes were plotted against mean intensities of YFP in each endosome. A.l.u.f.i., arbitrary linear units of fluorescence intensity. (B) Wild-type YFP-Shc and mutant YFP-Shc-3F (Y239F, Y240F, and Y317F mutations) were coexpressed with Grb2-CFP or Grb2R86A-CFP mutant in A-431 cells. The incubation with EGF-Rh and FRET imaging was performed as described in A. Arrows point to an example of the endosomes that contain EGF-Rh and YFP-Shc but lack Grb2R86A-CFP in the cell that expresses low levels of YFP-Shc. Bar, 10 μm.
Positive values of FRETC are indicative of energy transfer, but for comparison of FRET efficiencies, the sensitized FRETC signal must be related to the amount of acceptor (YFP) present. Therefore, the apparent efficiency of energy transfer (Ea) was calculated by dividing the mean intensity of FRETC by that of YFP in the selected regions of interest. Figure 1A shows that the mean value of Ea was significantly higher in EGF-treated cells than in control cells. The Ea values calculated for individual endosomes were similar to one another and independent of the amount of YFP-tagged protein in individual endosomes, thus validating this method of FRET measurement (Figure 1A).
The very high degree of colocalization of Grb2-CFP and YFP-Shc in endosomes combined with the high efficiency of FRET suggests that Grb2 and Shc fusion proteins are recruited to endosomes as a complex, either with each other or through their binding to the same EGFR molecule. In these experiments Grb2-CFP was present in excess of YFP-Shc, so that the acceptor was the limiting component of the energy transfer reaction. When the ratios of YFP/CFP fluorescence were calculated for individual endosomes, these ratios were strikingly similar within each cell examined. For example, in the cell presented in Figure 1A, this ratio was 0.170 ± 0.001, which corresponds approximately to the 1:2 M ratio for Shc-YFP/Grb2-CFP in endosomes. The excess of Grb2-CFP over Shc-YFP in endosomes suggests that at least one of two molecules of Grb2-CFP was bound directly to the EGFR, whereas another was bound to YFP-Shc.
To examine whether Grb2 can move to endosomes independently of binding to Shc, mutant YFP-Shc-3F was generated in which all three Grb2 binding sites (tyrosines 239, 240, and 317) were replaced by phenylalanines. When this mutant was coexpressed with Grb2-CFP, both proteins were efficiently recruited to endosomes, where they were present in proximity as evidenced by the positive FRETC signal (Figure 1B). Thus, FRET can be observed between Grb2-CFP and YFP-Shc that are not directly coupled to each other but are both bound to the EGFR. Measurements of Ea values for individual endosomes in several cells with various levels of Grb2-CFP and YFP-Shc-3F expression yielded mean Ea of 0.188 ± 0.110, suggesting that the efficiency of energy transfer between these two proteins is lower than that of wild-type Shc (0.48, Figure 1A). Grb2 mutant with the substitution of a key arginine in the phosphotyrosine binding pocket of the SH2 domain (Grb2R86A-CFP) did not bind to triple mutant Shc and displayed no endosomal localization when coexpressed with mutant YFP-Shc (Figure 1B). Interestingly, in the presence of very high levels of wild-type YFP-Shc, small amounts of Grb2R86A-CFP could be seen in endosomes containing YFP-Shc (Figure 1B). This observation implies that there is a residual direct or indirect interaction of Grb2 with Shc in the absence of a functional phosphotyrosine binding pocket in the Grb2 SH2 domain.
Localization H-Ras and K-Ras
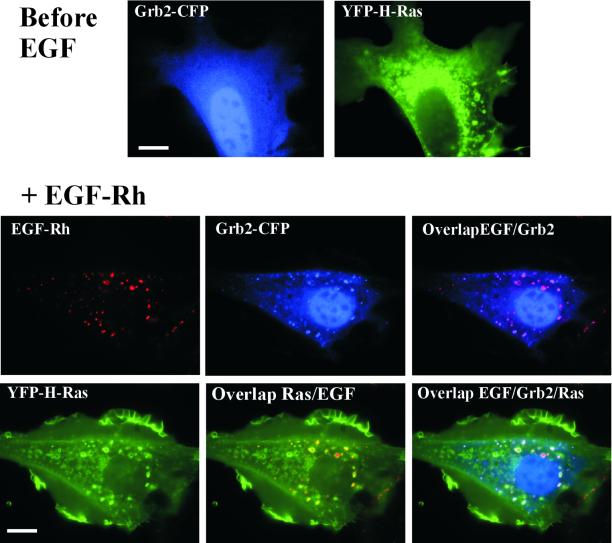
The data presented in Figure 1 suggest that the large pools of adaptor proteins involved in Ras activation move together with EGFR during endocytosis. Therefore, we next compared the localization of EGFR and Ras. To visualize Ras in living cells, we generated CFP- and YFP-fusion proteins of H-Ras and K-Ras (Figure 2). When CFP-H-Ras and YFP-K-Ras were transiently expressed in PAE/EGFR cells, they were found to be completely colocalized throughout the plasma membrane, in ruffles, lamellipodia, and intracellular vesicular structures (Figure 3A). Quantitative differences were, however, observed. More YFP-K-Ras than CFP-H-Ras was present at the plasma membrane, whereas relatively more CFP-H-Ras than YFP-K-Ras was associated with intracellular membranes. In cells stimulated with EGF-Rh, H-Ras and to a lesser extent K-Ras were seen in vesicular compartments that also contained internalized EGF-Rh, suggesting that these structures are indeed endosomes (Figure 3A). EGF-Rh visible in vesicles was not removed by acidic buffer (pH 4.5) treatment, which is known to dissociate EGF–receptor complexes at the cell surface, thus indicating that the punctate EGF-Rh fluorescence does represent internalized ligand (our unpublished data). Partial colocalization of YFP-H-Ras and a transferrin-Texas Red conjugate and Rab5a in cells treated or not treated with EGF suggests that at least some of these compartments are early or recycling endosomes (our unpublished data).
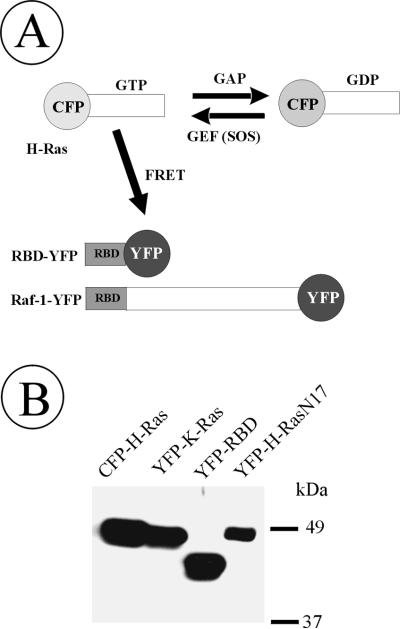
Figure 2.
Schematic representation and immunodetection of Ras- and Raf-YFP/CFP fusion proteins. (A) Depicted are Ras and Raf proteins fused to YFP/CFP at the amino and carboxyl terminus, respectively. CFP-Ras can bind GTP and convert it to GDP. GTPase activity of Ras is facilitated by GAP, whereas SOS, a guanidine nucleotide exchange factor (GEF), elevates association of CFP-Ras with GTP. Ras binding domain of c-Raf-1 (RBD) is tagged with YFP. RBD-YFP will specifically bind to GTP-loaded CFP-Ras, which would allow energy transfer from CFP-Ras to RBD-YFP. (B) Cells transiently expressing CFP-H-Ras, YFP-K-Ras, RBD-YFP, and YFP-H-RasN17 were lysed, and CFP/YFP-fusion proteins present in lysates were detected by Western blotting with GFP antibodies. All other fusions of Ras and Raf generated in this work migrated on SDS-PAGE according to predicted molecular mass.
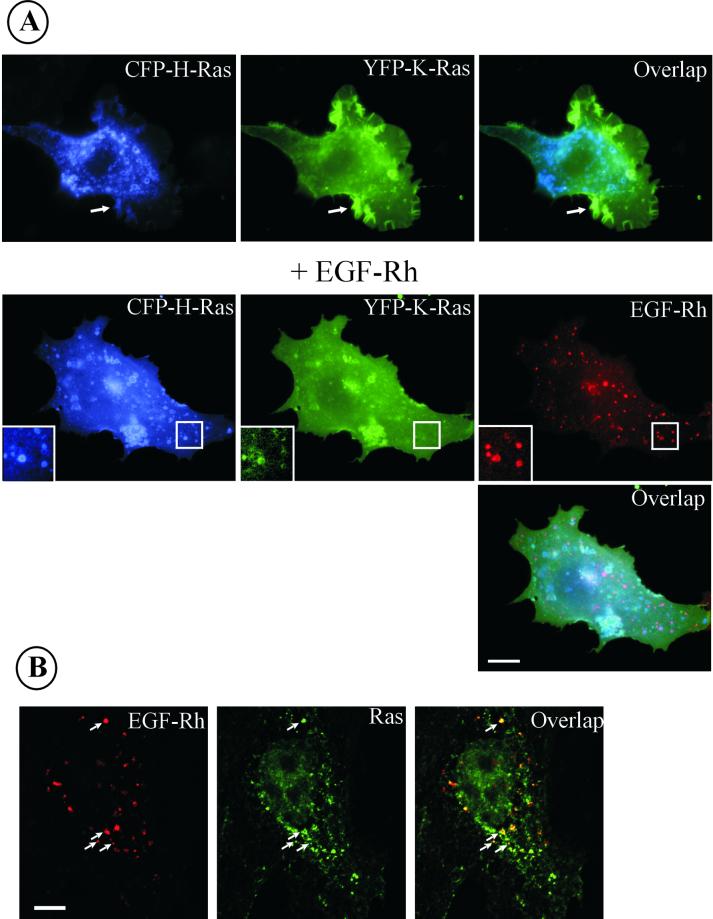
Figure 3.
Localization of expressed CFP-H-Ras, YFP-K-Ras, and endogenous Ras proteins. (A) CFP-H-Ras and YFP-K-Ras were transiently expressed in PAE/EGFR cells, and images were acquired through YFP, CFP, and Cy3 channels before and after the cells were stimulated with EGF-Rh for 15 min at 37°C. Insets, high-contrast images of the cell region indicated by white rectangle. Arrows point to an example of plasma membrane ruffles and lamellipodia. Bar, 10 μm. (B) A-431 cells were incubated with 200 ng/ml EGF-Rh for 15 min at 37°C. The cells were fixed with paraformaldehyde, permeabilized, and immunostained with RAS10 antibody followed by Cy5-conjugated secondary antibody. Z-stack of 200-nm optical sections was acquired through Cy3 (red) and Cy5 (green) filter channels. Images were deconvoluted. An optical section through the middle of the cell, where the most intense signal of EGF-Rh was observed, is shown. Arrows point to examples of colocalization of EGF-Rh and Ras in endosomes. Bar, 10 μm.
Previous studies by other groups have established that GFP-fused Ras proteins behave similarly to endogenous Ras (Choy et al., 1999). In agreement with these results, Figure 3B shows the immunofluorescence detection of endogenous Ras in endosomes containing EGF-Rh in paraformaldehyde-fixed and permeabilized A-431 cells. However, a significant background level of nonspecific fluorescence did not permit clear distinction between endogenous Ras present in the vesicular (Golgi/endosomes) compartments vs. in the plasma membrane. Moreover, vesicles containing YFP/CFP-Ras in live cells often collapse or fuse into larger, irregular-shaped compartments during cell fixation. We, therefore, carried out analyses of trafficking and interactions of YFP/CFP-fused Ras proteins in living cells. Furthermore, because H-Ras seemed to be present in endosomes at higher concentration than that of K-Ras, but no qualitative difference in H-Ras and K-Ras distribution was observed, subsequent experiments used the H-Ras fusion protein.
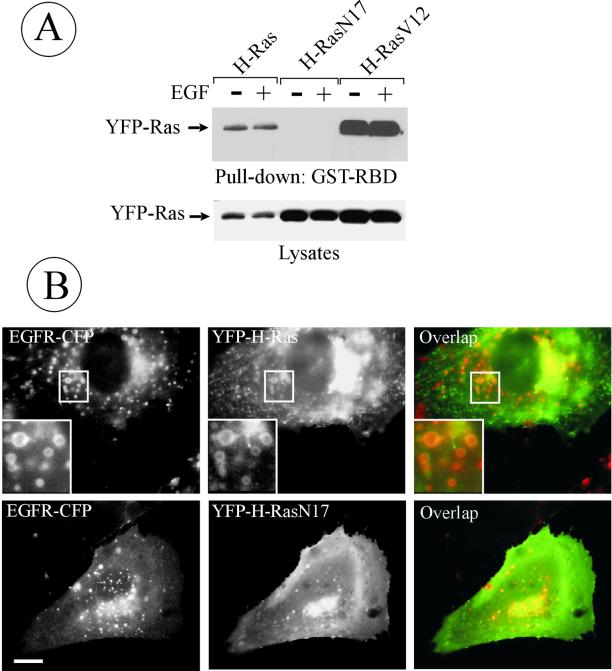
To test whether the localization of H-Ras in endosomes depends on Ras activity, G12V and S17N H-Ras mutants were studied. Constitutively active H-RasV12 is capable of binding to GTP but cannot hydrolyze GTP to GDP. The H-RasN17 mutant has a 100-fold higher affinity for GDP than for GTP and, when overexpressed in cells, can inhibit Ras effects in a dominant-negative manner. These YFP-fused mutants were transiently expressed in PAE/EGFR cells, and the amount of GTP-bound Ras in cell lysates was determined by affinity precipitation with the GST-fused Ras-binding fragment of Raf-1 (GST-RBD) that has several times higher binding affinity to GTP·Ras than to GDP·Ras (Wennstrom and Downward, 1999). As seen in Figure 4A, wild-type and H-RasV12 but not H-RasN17 were pulled down by GST-RBD, thus validating the use of these proteins in imaging experiments.
Figure 4.
Colocalization of dominant-negative H-RasN17 with endosomal EGFRs. (A) Cells were transiently transfected with YFP-H-Ras, YFP-H-RasN17, or YFP-H-RasV12; lysed; and the GTP-bound Ras proteins were affinity precipitated (pulled down) from the lysates by using GST-RBD beads. Isolated proteins and aliquots of lysates were separated by electrophoresis, transferred to nitrocellulose membrane, and YFP-fused proteins were detected by blotting with GFP antibodies. (B) PAE/EGFR-CFP cells were transiently transfected with wild-type YFP-H-Ras or YFP-H-RasN17. Living cells were imaged through CFP and YFP filter channels. Insets, high-magnification images of the regions indicated by white rectangles. Bar, 5 μm.
To compare the localization of Ras mutants with that of internalized EGFR in living cells, YFP-tagged mutants and wild-type H-Ras were transiently expressed in the PAE/EGFR-CFP cells. H-RasN17 was seen colocalized with EGFR-CFP in endosomes to the same extent as wild-type H-Ras (Figure 4B) and H-RasV12 (our unpublished data), suggesting that the localization of H-Ras does not depend on its GTP/GDP-binding status. Interestingly, in contrast to wild-type H-Ras and H-RasV12, overexpressed H-RasN17 did not produce enlarged endosomes. No FRET signal was detected between EGFR-CFP and YFP-H-Ras, suggesting that these proteins either do not form a stable complex or are positioned unfavorably for energy transfer.
Grb2 and H-Ras Are Found in the Same Endosomes
The experiments presented in Figures 3 and 4 demonstrated that YFP/CFP-H-Ras was located in endosomes; a pool of endosomes in cells exposed to EGF also contained EGFR. Because Grb2 moves to endosomes together with activated EGFR, it is possible that Grb2, presumably in association with SOS, and H-Ras converge in the same endocytic compartments, which may increase Ras activity. To directly compare the localization of Grb2 and H-Ras in living cells, NIH 3T3 cells were transiently cotransfected with Grb2-CFP and YFP-H-Ras. In cells not exposed to EGF, the two proteins displayed distinct distributions (Figure 5). When cells were stimulated with EGF-Rh, Grb2-CFP was found colocalized to a large extent with EGF-Rh in endosomes, whereas Grb2-CFP with YFP-H-Ras were only partially colocalized. No FRET signal was detected between Grb-CFP and YFP-H-Ras. Because YFP-H-Ras is present in endosomes in cells not treated with EGF, it is possible that endosomes containing internalized EGFRs and Grb2-CFP fuse with endosomes containing YFP-H-Ras. Another possibility for encounter in endosomes could be through the internalization of the EGFR–Grb2 complexes and H-Ras via the same plasma membrane-derived vesicle.
Figure 5.
Colocalization of Grb2-CFP and YFP-H-Ras in NIH 3T3 cells. NIH 3T3/EGFR cells were transiently transfected with Grb2-CFP and YFP-H-Ras. Live-cell YFP and CFP images were acquired before and after incubation of cells with 200 ng/ml EGF-Rh for 20 min at 37°C. EGF-Rh was detected using Cy3 filter channel. Bar, 10 μm.
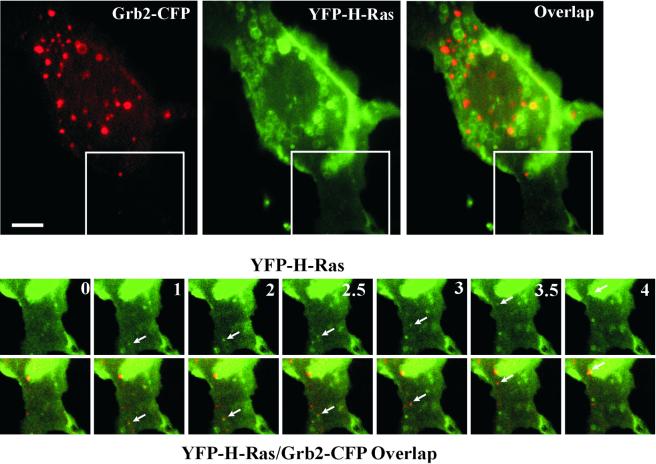
To visualize the dynamics of H-Ras and Grb2 movement in cells treated with EGF, PAE/EGFR cells were transiently transfected with Grb2-CFP and YFP-H-Ras, stimulated with EGF for 5 min at 37°C, and then time-lapse imaging was performed (Figure 6). At time “0” a large amount of intracellular YFP-H-Ras was seen; some endosomes also contained Grb2-CFP (Figure 6, top). Numerous profiles of tethered endosomes were seen and the fusion of endosomes containing Grb2-CFP and YFP-H-Ras were frequently observed in the perinuclear area. At the cell periphery the formation and movement of small endocytic vesicles that contain both YFP-H-Ras and Grb2-CFP was observed (Figure 6, bottom; see Quick-Time Movie in supplementary materials). In the example shown in the figure, the vesicle first appears in the peripheral region of the cell, moves toward the cell center, and eventually coalesces with the larger vesicle. Time-lapse imaging thus provides evidence for both joint internalization of Ras and Grb2, as well as the fusion of Grb2-containing endocytic vesicles with preexisting H-Ras–containing endosomes.
Figure 6.
Visualization of the formation and movement of endocytic vesicles containing YFP-H-Ras and Grb2-CFP. PAE/EGFR cells transfected with Grb2-CFP and YFP-H-Ras were grown in microscope chambers. Serum-starved cells were incubated with 100 ng/ml EGF for 5 min at 37°C, and 20 images were acquired with 30-s intervals at 37°C (time is indicated in minutes). Large images are taken at time 0. Selected time-lapse YFP and YFP/CFP overlapped images of the region of the cell indicated by the white rectangle are presented at the bottom. Arrows point to the vesicle containing both Grb2-CFP and YFP-H-Ras that appears at time 1 min and moves toward the perinuclear area of the cell. Quick time movie is available in the supplemental materials. Bar, 5 μm.
Detection of GTP-bound Form of Ras by FRET Microscopy
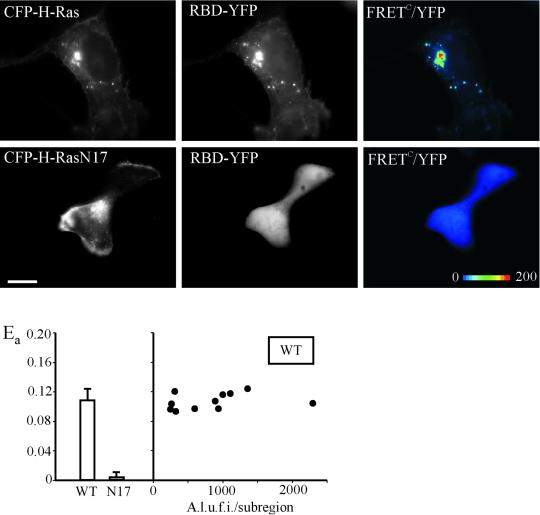
The data described above and previous observations in living cells suggest that Grb2, possibly associated with SOS, may be positioned adjacenct to Ras at the plasma membrane and in endosomes. These observations imply that Ras can be activated at the cell surface as well as in endosomes. To analyze the localization of active H-Ras in living cells, we established an assay based on monitoring the interaction of CFP-H-Ras with the YFP-fused Ras-binding domain of Raf-1 (RBD-YFP). As schematically shown in Figure 2A, RBD-YFP is capable of binding to GTP-bound CFP-Ras, resulting in colocalization of these two fusion proteins and, potentially, energy transfer between CFP and YFP. When RBD-YFP was expressed alone, it was diffusely distributed throughout the cytosol and nucleus. When CFP-H-Ras and RBD-YFP were cooverexpressed a pool of RBD-YFP was seen colocalized with CFP-H-Ras at the plasma membrane, ruffles, cell edges, Golgi area, and endosomes. A cell expressing large amounts of CFP-H-Ras (excess over RBD-YFP) is shown in Figure 7. In such cells RBD-YFP was mostly colocalized with CFP-H-Ras. FRETC images reveal strong energy transfer between CFP-Ras and RBD-YFP, suggesting that these two proteins form a complex at the membrane. In contrast, coexpression of CFP-H-RasN17 with RBD-YFP did not result in the membrane translocation of RBD-YFP to the sites of localization of CFP-H-Ras. No FRETC was observed. To compare apparent FRET efficiencies, Ea values were measured in the selected subregions of the cells. Figure 7 shows that the mean value of Ea was negligible in cells expressing the N17 mutant of H-Ras, whereas this value was >10% in cells expressing wild-type H-Ras. As in the case of the Grb2 and Shc pair, Ea values for H-Ras/RBD interaction calculated for individual fluorescent compartments varied very little and were independent of the amount of tagged proteins (Figure 7). When full-length Raf-1 fusion protein (Raf-YFP, Figure 2) was coexpressed with CFP-H-Ras, Raf-YFP was well colocalized with H-Ras; however no FRET was detected, despite colocalization of Raf-YFP and CFP-H-Ras, suggesting that YFP and CFP moieties in this interaction pair are not favorably positioned.
Figure 7.
Detection of GTP-bound Ras in living cells. CFP-H-Ras or CFP-H-RasN17 was coexpressed with RBD-YFP in A-431 cells. FRETC images were obtained as described in MATERIALS AND METHODS, and are presented as pseudocolor intensity-modulated images (FRETC/YFP). The apparent efficiencies of energy transfer between CFP and YFP, Ea, calculated for multiple subregions of the images are presented in the graph. Left, averaged values of Ea are compared for cells expressing wild-type CFP-H-Ras (wt) or CFP-H-RasN17 (N17). Right, Ea values were calculated for several subregions corresponding to the individual vesicular compartments in wild-type CFP-H-Ras–expressing cells and plotted against mean intensity of YFP in each compartment (subregion). A.l.u.f.i., arbitrary linear units of fluorescence intensity. Bar, 10 μm.
To confirm FRET between CFP-H-Ras and YFP-RBD, the method of donor (CFP) emission recovery due to acceptor (YFP) photobleaching was used (Bastiaens and Pepperkok, 2000). Cells were paraformaldehyde fixed, and the fluorescence intensity of CFP was measured before and after photobleaching of YFP by 3-min excitation through 535/10-nm filter. Photobleaching YFP typically increased the mean intensity of the total cellular CFP fluorescence by 5–10%, as expected for photobleaching suppression of energy transfer (our unpublished data). This method, however, could not be used in our imaging system to measure FRET with spatial resolution in living cells due to the movement of cellular organelles during photobleaching and the toxicity of continuous irradiation. In summary, the experiments described in Figure 7 and photobleaching experiments validate the use of FRET measurements between Ras and RBD fusion proteins to detect GTP-bound form of Ras.
By using RBD-YFP as a sensor, the effect of EGF stimulation on the localization of activated CFP-H-Ras was examined. A large pool of GTP-bound CFP-H-Ras was detected in cell edges, membrane ruffles, and endosomes in unstimulated cells expressing high levels of CFP-H-Ras (Figure 7), which makes it difficult to follow the movement of active CFP-H-Ras during EGF stimulation. Therefore, cells with a moderate/low expression of CFP-H-Ras were used in these experiments.
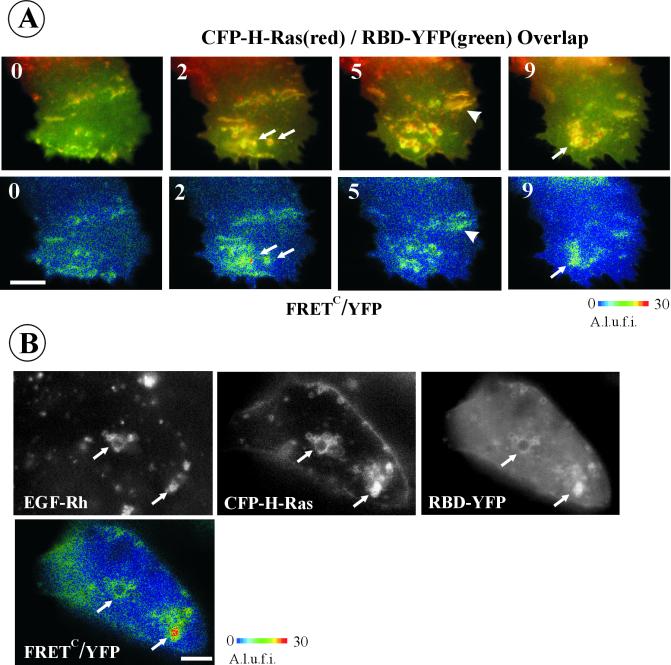
Stimulation of A-431 cells coexpressing CFP-H-Ras and RBD-YFP with EGF caused redistribution of activated CFP-H-Ras (Figure 8A). Repeated imaging of the same cell at various elapsed times after stimulation demonstrates that a pool of activated Ras together with decorating RBD-YFP moves to extensively ruffled regions and to endosome-like structures. Neither the total intensity of cellular FRETC signal nor Ea values were changed significantly after EGF stimulation. That the total amount of GTP·Ras was not altered significantly by EGF stimulation is also evident from the pull-down experiments presented in Figure 4A.
Figure 8.
Localization of GTP-bound H-Ras in EGF-stimulated cells. (A) CFP-H-Ras was coexpressed with RBD-YFP in A-431 cells. Cells were grown on the microscopy chamber. YFP, CFP, and FRET images were acquired from the same cell before and after 2-, 5-, and 9-min incubation of the cells with 200 ng/ml EGF at 37°C. FRETC images were calculated as described in MATERIALS AND METHODS, and is presented as pseudocolor intensity-modulated images (FRETC/YFP). Arrows point to membrane structures with the typical shape of endosomal compartments. Arrowheads point on plasma membrane ruffles. A.l.u.f.i., arbitrary linear units of fluorescence intensity. Bar, 5 μm. (B) A-431 cells expressing CFP-H-Ras and RBD-YFP were stimulated with 200 ng/ml EGF-Rh for 15 min at 37°C. YFP, CFP, and FRET images were acquired at room temperature, FRETC were calculated and presented as YFP intensity-modulated images (FRETC/YFP). EGF-Rh was detected through Cy3 filter channel. Arrows point to examples of colocalization of FRETC and EGF-Rh signals in endosomal compartments. A.l.u.f.i., arbitrary linear units of fluorescence intensity. Bar, 5 μm.
To test whether vesicular structures containing activated CFP-H-Ras represent endosomes carrying EGFR, A-431 cells expressing CFP-H-Ras and BRD-YFP were stimulated with EGF-Rh. FRETC was measured after 15 min of continuous endocytosis at 37°C simultaneously with the detection of EGF-Rh as a marker of EGFR. Figure 8B demonstrates colocalization of EGF-Rh and the CFP-H-Ras/RBD-YFP interacting pair in endosomal compartments. The overlap of EGF-Rh and activated H-Ras was only partial, because some endosome-like structures had detectable levels of either rhodamine fluorescence or FRETC. Partial colocalization of EGF-Rh with active CFP-H-Ras was also observed in NIH 3T3 and PAE cells. Together, the microscopy and FRET analyses in living cells define the “motility-active” areas of the plasma membrane and the endosomal compartment as the major sites of residence for Ras signaling complexes.
DISCUSSION
Subcellular targeting and trafficking play a major role in coordination of the duration and the specificity of signaling processes. In this study, we examined trafficking of proteins involved in the EGF-dependent activation of the Ras signaling pathway. The initial step of this pathway is the recruitment of Grb2 to phosphorylated EGFR. Previous studies in living cells (Sorkin et al., 2000) and this work (Figures 1, 5, and 6) demonstrate that Grb2 interacts with the EGFR at the cell surface and in endosomes either directly or through binding to Shc. The complete colocalization of Grb2 and Shc, and the striking consistency of the molar ratio of these two proteins in individual endosomes suggest that Grb2/Shc/receptor complexes are of uniform nature in endosomal compartments (Figure 1). The relative contribution of the direct interaction of Grb2 with EGFR may depend on the stoichiometric ratio of Grb2 and Shc in the cell. The interaction of heterologously expressed Grb2 with the receptor is mostly direct. The direct interaction is sufficient to elicit most of the cell response to growth factors mediated by Grb2 (Lai and Pawson, 2000). When Grb2 and Shc are coexpressed these two proteins are either bound to each other or bound to the same EGFR molecule as suggested by the invariable FRET signal in endosomes. FRET between Grb2 and Shc associated with different monomers in the receptor dimer is less likely because no energy transfer was detected between dimerized carboxyl-terminus–tagged EGFRs (Sorkin et al., 2000).
The finding of large amounts of YFP/CFP-Ras in endosomes was surprising. When expressed individually or coexpressed, both H-Ras and K-Ras were frequently seen in the endosomal compartments, although the extent of accumulation of H-Ras in endosomes was relatively higher than that of K-Ras. It is possible that H-Ras is delivered to endosomes by means of vesicular transport from the Golgi apparatus. Indeed, perinuclear endosomes decorated with YFP-H-Ras could be found in every cell where YFP-H-Ras was also seen in the Golgi area regardless of the level of YFP-H-Ras expression. In addition, as revealed by time-lapse imaging, H-Ras can be internalized from the plasma membrane (Figure 6). The internalization of GFP-H-Ras in insulin-stimulated Rat-1 cells was recently demonstrated by evanescent wave microscopy (Rizzo et al., 2001). The ability of H-Ras to traffic through endosomes may play an important role in Rab5 activation, which leads to extensive endosome fusion and formation of large endosomes (Tall et al., 2001). Correct trafficking of Ras does not seem to require GTP binding because the distribution of the H-RasN17 in the cells was essentially similar to that of wild-type H-Ras. However, expression of N17 mutant did not result in endosome fusion (Figure 4), suggesting that GTP loading of Ras is required for Rab5 activation.
Previous studies of Ras localization defined the plasma membrane as the main cellular location of Ras (Willingham et al., 1980). More recent studies explored Ras microdomain compartmentalization within the plasma membrane (Prior et al., 2001). These previous results do not contradict our observations. Although a significant amount of transfected H-Ras and K-Ras was seen in the intracellular membranes, a larger pool of Ras fusion proteins was associated with the plasma membrane, particularly, in the areas with ruffling activity. It is conceivable that the accumulation of YFP/CFP-Ras in endosomes, Golgi, and endoplasmic reticulum observed in our experiments was due to overexpression of these proteins. However, previous studies and this work (Figure 3) showed that endogenous Ras is also detected in the membrane compartments in the Golgi area (Choy et al., 1999) and in endosomes of the insulin-treated cells (Rizzo et al., 2000). Thus, it can be proposed that trafficking of tagged Ras proteins reflects the behavior of endogenous Ras, although the intensity of trafficking through some pathways may be exaggerated.
The detection of GTP·Ras by FRET microscopy in living cells enables us to define the cellular locations where Ras effectors can be activated. This assay, however, relies on the expression of tagged Ras. The overexpression of H-Ras leads to activation of a substantial pool of H-Ras regardless of the conditions of serum starving or stimulation with EGF. In fact, stimulation of cells with EGF results in very little, if any, increase in the total cellular Ras-RBD FRETC signal. Significant activity of heterologously expressed Ras in serum-starved cells has also been reported by others (Lockyer et al., 1999). The existence of a large pool of GTP-bound but effector-inaccessible endogenous Ras has been demonstrated (Hamilton et al., 2001). Therefore, this technique does not allow monitoring of the activation of total cellular Ras by EGFR, but rather, it is designed to analyze the movement of activated Ras within the cell. A recent study by Mochizuki et al. (2001) made use of a chimeric protein consisting of H-Ras and the RBD fragment flanked with YFP and CFP to follow the activation of this chimera by FRET microscopy. The increase in the intramolecular Ras–RBD interaction in the chimera was detected upon EGF stimulation. However, it is not clear whether the fatty acid modifications and cellular localization of the chimeric protein are similar to that of wild-type Ras. For instance, although the latter chimera was reported to be localized at the plasma membrane, our results demonstrate that significant amounts of YFP/CFP–H-Ras and K-Ras are present in the intracellular membrane compartments.
A number of studies have used various technical approaches to explore the hypothesis of endosomal signaling by EGFRs (reviewed in Baass et al., 1995, Ceresa and Schmid, 2000; Di Fiore and De Camilli, 2001). These approaches mainly included the isolation of endosomes, biochemical separation of internalized from surface EGFRs, and immunohistochemical studies in fixed cells (Lai et al., 1989; Sorkin and Carpenter, 1991; Di Gugliemo et al., 1994; Oksvold et al., 2000; Burke et al., 2001; Oksvold et al., 2001; Wang et al., 2001). The requirement of EGFR endocytosis for activation of mitogen-activated protein kinase was revealed in cells in which the endocytosis was inhibited by mutant dynamin (Vietra et al., 1996). What insights do imaging microscopy of GFP-tagged proteins and FRET analysis in living cells provide? One type of information that FRET microscopy can offer is a quantitative description of the cellular distribution of interacting complexes and the extent of interaction. Our experiments indicate that the bulk of SH2/phosphotyrosine-binding domain adaptors can be transported together with EGFR to endosomes. In fact, as much as 60% of the total cellular pool of expressed Grb2 and Shc is bound to the endosomal EGFR in variety of cell types. In contrast, coimmunoprecipitation experiments yield only very low amounts of Grb2 and Shc associated with the EGFR, probably due to instability of complexes in detergent solutions (Di Gugliemo et al., 1994; Okutani et al., 1994; our unpublished data). Consequently, our results imply that Grb2–SOS complexes become unavailable to Ras located at the plasma membrane. Thus, massive endocytosis of Grb2 may lead to physical proximity of Grb2–SOS complex to endosomal Ras at the expense of the separation of this complex from plasma membrane Ras and its effectors. One Ras effector is thought to be the p85/p110 PI3 kinase, which is also detected in endosomes and can translocate to endosomes with growth factor receptors (Kapeller et al., 1993; Christoforidis et al., 1999). In this case, preferential localization of H-Ras in endosomes may explain its greater capacity to activate PI3 kinase compared with that of K-Ras.
The demonstration of a protein–protein interaction in living cells is a critical test of hypotheses originating from biochemical or other experiments requiring cell disruption. The ability to assign protein–protein interactions to specific compartments of the cell opens the way to more complete understanding of the regulation of signaling processes. For instance, in addition to regulation of output signals by Ras, Ras activity itself may be determined by its distribution. GTP/GDP status of Ras is regulated by exchange factors and GTPase-activating proteins (GAPs). Therefore, the increased GTP loading of Ras may be not only be due to proximity to Grb2-SOS but also due to the separation of Ras from GAP. Although in this study multicolor fluorescence imaging shows that EGFR–Grb2 complexes traverse cellular compartments containing Ras, additional experiments are needed to directly compare the compartmentalization of GAP, SOS, and different isoforms of Ras to explain how the specificity of input/output Ras signaling is spatially and temporally regulated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. W. Sather (University of Colorado Health Sciences Center) for critical reading of the manuscript, and Drs. Agell, Dell'Acqua, Johnson, and Tebar for Raf and Ras construct. This work was supported by grants from CA-089151 from National Cancer Institute, RPG-00-247-01-CSM from the American Cancer Society, and BC-995456 from the Department of Defense.
Abbreviations used:
- BSA
bovine serum albumin
- CFP
cyan fluorescent protein
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EGF-Rh
epidermal growth factor conjugated to rhodamine
- FRET
fluorescence resonance energy transfer
- FRETC
corrected FRET
- GAP
GTPase-activating protein
- GFP
green fluorescent protein
- Grb2
growth factor binding protein 2
- PAE
porcine aortic endothelial
- PI3
phosphotidylinositol-3
- RBD
Ras binding domain
- YFP
yellow fluorescent protein
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–11–0552. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–11–0552.
REFERENCES
- Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baass PC, Di Guglielmo GM, Authier F, Posner BI, Bergeron JJM. Compartmentalized signal transduction by receptor tyrosine kinases. Trends Cell Biol. 1995;5:465–470. doi: 10.1016/s0962-8924(00)89116-3. [DOI] [PubMed] [Google Scholar]
- Bastiaens PI, Pepperkok R. Observing proteins in their natural habitat: the living cell. Trends Biochem Sci. 2000;25:631–637. doi: 10.1016/s0968-0004(00)01714-x. [DOI] [PubMed] [Google Scholar]
- Batzer AG, Rotin D, Urena JM, Skolnik EY, Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock R, Jovin TM. Heterogeneity of signal transduction at the subcellular level: microsphere-based focal EGF receptor activation and stimulation of Shc translocation. J Cell Sci. 2001;114:2437–2447. doi: 10.1242/jcs.114.13.2437. [DOI] [PubMed] [Google Scholar]
- Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G. The EGF receptor: a nexus for trafficking and signaling. Bioessays. 2000;22:697–707. doi: 10.1002/1521-1878(200008)22:8<697::AID-BIES3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Carter RE, Sorkin A. Endocytosis of functional epidermal growth factor receptor-green fluorescent protein chimera. J Biol Chem. 1998;273:35000–35007. doi: 10.1074/jbc.273.52.35000. [DOI] [PubMed] [Google Scholar]
- Ceresa BP, Schmid SL. Regulation of signal transduction by endocytosis. Curr Opin Cell Biol. 2000;12:204–210. doi: 10.1016/s0955-0674(99)00077-0. [DOI] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Cohen S, Fava RA. Internalization of functional epidermal growth factor:receptor/kinase complexes in A-431 cells. J Biol Chem. 1985;260:12351–12358. [PubMed] [Google Scholar]
- Di Fiore PP, De Camilli P. Endocytosis and signaling. an inseparable partnership. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- Di Gugliemo GM, Baass PC, Ou W-J, Posner B, Bergeron JJM. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. EMBO J. 1994;13:4269–4277. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton M, Liao J, Cathcart MK, Wolfman A. Constitutive association of c-N-Ras with c-Raf-1 and protein kinase C epsilon in latent signaling modules. J Biol Chem. 2001;276:29079–29090. doi: 10.1074/jbc.M102001200. [DOI] [PubMed] [Google Scholar]
- Huang F, Nesterov A, Carter RE, Sorkin A. Trafficking of yellow-fluorescent-protein-tagged mu1 subunit of clathrin adaptor AP-1 complex in living cells. Traffic. 2001;2:345–357. doi: 10.1034/j.1600-0854.2001.25020506.x. [DOI] [PubMed] [Google Scholar]
- Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joneson T, Bar-Sagi D. Ras effectors and their role in mitogenesis and oncogenesis. J Mol Med. 1997;75:587–593. doi: 10.1007/s001090050143. [DOI] [PubMed] [Google Scholar]
- Kapeller R, Chakrabarti R, Cantley L, Fay F, Corvera S. Internalization of activated platelet-derived growth factor receptor-phosphatidylinositol-3′ kinase complexes: potential interactions with the microtubule cytoskeleton. Mol Cell Biol. 1993;13:6052–6063. doi: 10.1128/mcb.13.10.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WH, Cameron PH, Doherty II J-J, Posner BI, Bergeron JJM. Ligand-mediated autophosphorylation activity of the epidermal growth factor receptor during internalization. J Cell Biol. 1989;109:2751–2750. doi: 10.1083/jcb.109.6.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KM, Pawson T. The ShcA phosphotyrosine docking protein sensitizes cardiovascular signaling in the mouse embryo. Genes Dev. 2000;14:1132–1145. [PMC free article] [PubMed] [Google Scholar]
- Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signaling [see comments] Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- Lockyer PJ, Wennstrom S, Kupzig S, Venkateswarlu K, Downward J, Cullen PJ. Identification of the ras GTPase-activating protein GAP1(m) as a phosphatidylinositol-3,4,5-trisphosphate-binding protein in vivo. Curr Biol. 1999;9:265–268. doi: 10.1016/s0960-9822(99)80116-x. [DOI] [PubMed] [Google Scholar]
- Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Paterson HF, Rodriguez R, Fensome AC, Ellis MV, Swann K, Katan M. Real time fluorescence imaging of PLC gamma translocation and its interaction with the epidermal growth factor receptor. J Cell Biol. 2001;153:599–612. doi: 10.1083/jcb.153.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Nesterov A, Reshetnikova G, Vinogradova N, Nikolsky N. Functional state of the epidermal growth factor-receptor complexes during their internalization in A-431 cells. Mol Cell Biol. 1990;10:5011–5014. doi: 10.1128/mcb.10.9.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksvold MP, Skarpen E, Lindeman B, Roos N, Huitfeldt HS. Immunocytochemical localization of Shc and activated EGF receptor in early endosomes after EGF stimulation of HeLa cells. J Histochem Cytochem. 2000;48:21–33. doi: 10.1177/002215540004800103. [DOI] [PubMed] [Google Scholar]
- Oksvold MP, Skarpen E, Wierod L, Paulsen RE, Huitfeldt HS. Re-localization of activated EGF receptor and its signal transducers to multivesicular compartments downstream of early endosomes in response to EGF. Eur J Cell Biol. 2001;80:285–294. doi: 10.1078/0171-9335-00160. [DOI] [PubMed] [Google Scholar]
- Okutani T, Okabayashi Y, Kido Y, Sugimoto Y, Sakaguchi K, Matuoka K, Takenawa T, Kasuga M. Grb2/Ash binds directly to tyrosines 1068 and 1086 and indirectly to tyrosine 1148 of activated human epidermal growth factor receptors in intact cells. J Biol Chem. 1994;269:31310–31314. [PubMed] [Google Scholar]
- Prior IA, Hancock JF. Compartmentalization of Ras proteins. J Cell Sci. 2001;114:1603–1608. doi: 10.1242/jcs.114.9.1603. [DOI] [PubMed] [Google Scholar]
- Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Kraft CA, Watkins SC, Levitan ES, Romero G. Agonist-dependent traffic of raft-associated Ras and Raf-1 is required for activation of the mitogen-activated protein kinase cascade. J Biol Chem. 2001;276:34928–34933. doi: 10.1074/jbc.M105918200. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Shome K, Watkins SC, Romero G. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem. 2000;275:23911–23918. doi: 10.1074/jbc.M001553200. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1 [see comments] Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- Sorkin A. Internalization of the epidermal growth factor receptor: role in signaling. Biochem Soc Trans. 2001;29:480–484. doi: 10.1042/bst0290480. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Carpenter G. Dimerization of internalized growth factor receptors. J Biol Chem. 1991;266:23453–23460. [PubMed] [Google Scholar]
- Sorkin A, Mazzotti M, Sorkina T, Scotto L, Beguinot L. Epidermal growth factor interaction with clathrin adaptors is mediated by the Tyr974-containing internalization motif. J Biol Chem. 1996;271:13377–13384. doi: 10.1074/jbc.271.23.13377. [DOI] [PubMed] [Google Scholar]
- Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc Natl Acad Sci USA. 1995;92:1709–1713. doi: 10.1073/pnas.92.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietra AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Liao HJ, Chattopadhyay A, Carpenter G. EGF-dependent translocation of green fluorescent protein-tagged PLC-gamma1 to the plasma membrane and endosomes. Exp Cell Res. 2001;267:28–36. doi: 10.1006/excr.2001.5241. [DOI] [PubMed] [Google Scholar]
- Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- Wennstrom S, Downward J. Role of phosphoinositide 3-kinase in activation of ras and mitogen-activated protein kinase by epidermal growth factor. Mol Cell Biol. 1999;19:4279–4288. doi: 10.1128/mcb.19.6.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham MC, Pastan I, Shih TY, Scolnick EM. Localization of the src gene product of the Harvey strain of MSV to plasma membrane of transformed cells by electron microscopic immunocytochemistry. Cell. 1980;19:1005–1014. doi: 10.1016/0092-8674(80)90091-4. [DOI] [PubMed] [Google Scholar]
- Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.