Abstract
The highly conserved Cdc6 protein is required for initiation of eukaryotic DNA replication and, in yeast and Xenopus, for the coupling of DNA replication to mitosis. Herein, we show that human Cdc6 is rapidly destroyed by a p53-independent, proteasome-, and ubiquitin-dependent pathway during early stages of programmed cell death induced by the DNA-damaging drug adozelesin, or by a separate caspase-dependent pathway in cells undergoing apoptosis through an extrinsic pathway induced by tumor necrosis factor-α and cycloheximide. The proteasome-dependent pathway induced by adozelesin is conserved in the budding yeast Saccharomyces cerevisiae. The destruction of Cdc6 may be a primordial programmed death response that uncouples DNA replication from the cell division cycle, which is reinforced in metazoans by the evolution of caspases and p53.
INTRODUCTION
Initiation of DNA replication in eukaryotic cells is regulated by a highly conserved “origin licensing” mechanism that requires the sequential assembly of proteins into prereplicative complexes (pre-RCs) at origins of replication, the sites on chromosomes where DNA synthesis begins (reviewed in Dutta and Bell, 1997; Diffley, 2001). Cdc6, a protein that has been shown to be rate limiting for initiation of DNA replication in a number of eukaryotic organisms, is essential for the establishment of pre-RCs. In addition to Cdc6, pre-RC formation requires the six-subunit origin recognition complex (ORC) and other proteins, including minichromosome maintenance (MCM) proteins 2–7. Pre-RCs are assembled in late mitosis or early G1 and are maintained by Cdc6 and other pre-RC proteins until initiation occurs in S phase mediated by cyclin-dependent kinase (CDK) activity. Activation of initiation of DNA replication and the establishment of DNA replication forks at origins of replication coincide with the conversion of pre-RCs to inactive post-RCs, and activated mitotic cyclin-dependent kinases block the formation of new pre-RCs until the end of mitosis, when this activity declines and pre-RCs can once again be established. This stringent regulatory mechanism coordinates initiation of DNA replication with cell growth and mitosis and protects the integrity of the genome by ensuring that DNA is replicated just once per cell cycle.
Initiation of DNA replication is also regulated by checkpoints that inhibit this process in response to DNA damage or stalled replication forks. In the budding yeast Saccharomyces cerevisiae, the inhibitory effects on initiation of DNA replication of methylmethane sulfonate (Shirahige et al., 1998) or hydroxyurea (Weinberger et al., 1999) are reduced in a strain with a mutation in the licensing protein ORC2 (orc2-1), indicating that Orc2p is required for the intra-S-phase checkpoint in this organism. Initiation proteins have also been implicated in checkpoints that restrain progression to, or exit from, mitosis. For example, some budding yeast strains harboring temperature-sensitive mutations in initiation proteins undergo mitosis with unreplicated DNA when shifted to the nonpermissive temperature during G1 (Toyn et al., 1995; Tavormina et al., 1997). S. cerevisiae Cdc6 and its homolog Cdc18 in fission yeast (Schizosaccharomyces pombe) similarly inhibit mitosis in G1 cells (Bueno and Russell, 1992; Kelly et al., 1993a,b; Piatti et al., 1995; Detweiler and Li, 1997; Tavormina et al., 1997). In addition, the depletion of Cdc6 (Hekmat-Nejad et al., 2000) or addition of the origin licensing inhibitor geminin (Michael et al., 2000) to Xenopus egg extracts causes nuclei formed in these extracts to undergo a catastrophic mitosis.
Adozelesin is an experimental DNA-alkylating antitumor agent that profoundly inhibits initiation of simian virus 40 (SV40) viral DNA replication in SV40-infected monkey cells, and this effect occurs in trans, suggesting it corresponds to a checkpoint (Cobuzzi et al., 1996). Adozelesin also inhibits initiation of cellular DNA replication in S. cerevisiae, and this effect is reduced in the same orc2-1 strain that harbors a defective intra-S-phase checkpoint response to methylmethane sulfonate and hydroxyurea (HU) (Weinberger et al., 1999). As expected for a strain with a defective intra-S-phase checkpoint, the orc2-1 strain is hypersensitive to adozelesin. In addition, a mutation in Orc1p that causes a more severe initiation defect compared with the orc2-1 mutation partially suppresses sensitivity to adozelesin specifically in G1 cells, and both wild-type and orc2-1 cells are completely resistant to adozelesin's lethal effects when they are in G2/M or are driven out of the cell cycle into the G0 state (Weinberger et al., 1999), where pre-RCs are absent (Diffley et al., 1994; Trabold and Burhans, unpublished observations). These findings suggest the existence of proliferation-dependent lethal effects of adozelesin in S. cerevisiae that are related to drug-induced changes in the function of ORC and/or other proteins in pre-RCs.
Adozelesin (Bhuyan et al., 1992a,b; our unpublished observations) and many other DNA-damaging drugs (Tannock and Hill, 1998) induce similar cell cycle-specific and proliferation-dependent lethal effects in mammalian cells as part of a programmed cell death response to irreparable DNA damage. The nature of the relationship between sensitivity to DNA-damaging agents and proliferative state in mammals remains unclear. However, similar to yeast, pre-RCs may be absent from cells in G0 (Williams et al., 1998; Stoeber et al., 2001), and entry into the proliferative state coincides with the licensing of origins via the expression of Cdc6 and other proteins through the E2F family of transcription factors, downstream of the synthesis of cyclin D1 and the subsequent phosphorylation of members of the retinoblastoma protein family (Hateboer et al. 1998; Yan et al. 1998). These facts and the high degree of conservation of origin licensing mechanisms in yeast and mammals suggest that the proliferation-dependent lethality of adozelesin in both these organisms might be related to effects of DNA damage that alter the function of origin licensing proteins.
In this study, we assessed the effects of adozelesin on mammalian chromosomal DNA replication and on mammalian and budding yeast proteins required for initiation and cell proliferation. Adozelesin inhibited initiation of mammalian chromosomal DNA replication, and this inhibition was accompanied by the specific proteasome-dependent degradation of the licensing protein Cdc6, as well as cyclin D1, Cdc25A, and other proteins required for cell cycle checkpoints and/or the establishment and maintenance of the proliferative state. Although the destruction of Cdc25A occurs as part of a DNA damage checkpoint that blocks DNA synthesis (Mailand et al., 2000; Falck et al., 2001), the destruction of Cdc6 did not seem to play a role in this checkpoint. Instead, Cdc6 destruction occurred at an early stage of programmed cell death upstream of, or parallel to, the function of caspases. Cdc6 was also destroyed in cells undergoing apoptosis induced by tumor necrosis factor (TNF)-α and cycloheximide, although this destruction was mediated by caspases instead of by the proteasome. Thus, two separate pathways target Cdc6 for destruction in mammalian cells in response to different agents that induce programmed cell death. The comparable cytotoxic effects of adozelesin in S. cerevisiae that we previously reported were also found to be accompanied by the specific proteasome-dependent destruction of S. cerevisiae Cdc6, as well as of Cdc6 protein from Arabidopsis thaliana, a model plant, when this protein is expressed in S. cerevisiae. This suggests that the proteasome-dependent destruction of Cdc6 induced by adozelesin is conserved throughout eukaryotes. Programmed cell death in proliferating mammalian cells is frequently associated with the uncoupling of DNA replication from mitosis due to the premature activation of mitotic cyclin-dependent kinases (reviewed in Guo and Hay, 1999). We propose that the destruction of Cdc6 by adozelesin contributes to this uncoupling by abrogating checkpoints that require Cdc6 and other origin licensing proteins.
MATERIALS AND METHODS
Cell Culture, and Treatment with Adozelesin, TNF-α, UV irradiation, and Inhibitors
All cell lines were maintained in DMEM containing 10% fetal calf serum, except K562 erythroleukemia cells, which were maintained in RPMI containing 10% fetal calf serum. Primary cultures of human pulmonary fibroblasts were prepared from residual lung tissues derived from surgical pneumectomy specimens and provided by the Tissue Procurement Service at Roswell Park Cancer Institute. Use of lung tissue was approved by the Institute Review Board, and prior informed consent of the participating patients was obtained. TNF-α (gift from Genetics Institute, Cambridge, MA) was used at 100 ng/ml final concentration, and adozelesin (gift from Pharmacia-Upjohn, Kalamazoo, MI) was used at 4 nM final concentration. Protein synthesis was inhibited by 10 μg/ml cycloheximide (Sigma-Aldrich, St. Louis, MO), proteasome activity by 10 μM MG132 (Calbiochem, San Diego, CA), caspase activity by 10 μM caspase inhibitor I (Calbiochem), protein kinase activity by 1 μM UCN-01 (National Cancer Institute, Bethesda, MD), lysosome activity by 100 μM chloroquine (Sigma-Aldrich), and calpain activity by 50 μM acetyl-Leu-Leu-Met (Calbiochem). All compounds were dissolved in dimethyl sulfoxide (Sigma-Aldrich) for stock solutions except for adozelesin, which was dissolved in dimethyl acetamide (Sigma-Aldrich). For UV radiation experiments, MDA cultures from which medium was briefly removed were irradiated with a UV Stratalinker 1800 (Stratagene, La Jolla, CA), followed by replacement of the medium and continued culture for indicated times before harvesting of proteins for Western blotting as described below. Effects of UCN-01 were analyzed by adding it to culture medium 1 h before UV exposure or addition of adozelesin.
DNA Synthesis Measurements and Two-Dimensional Analysis of Replicating DNA
DNA synthesis was determined by labeling nascent DNA with 2 μCi/ml [3H]thymidine (Amersham Biosciences, Piscataway, NJ) for 20 min and measuring incorporation into perchloric acid or glacial acetic acid/alcohol-precipitable DNA by scintillation counting. Exponentially proliferating populations of K562 cells were treated with 40 nM adozelesin for indicated times and replication intermediates harvested on the nuclear matrix (Dijkwel et al., 1991) were separated from nonreplicating DNA on neutral-neutral two-dimensional gels as described previously (Brewer and Fangman, 1987). Gels were blotted onto Hybond N+ membranes and probed with radiolabeled sequences specific for a 5.6-kb EcoRI fragment of DNA in the human rDNA locus. Signals were detected by PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Plasmids and Cell Transfection
The pCMV-Cdc6-wt plasmid and plasmids expressing the destruction box mutant (CDC6 dl58-61) and KEN box mutant (CDC6 3A) were a generous gift of Kristian Helin (European Institute of Oncology, Milan, Italy) and are described in Petersen et al. (2000). Cells were transfected with 4 μg of plasmid DNA in 6 μl of FuGENE 6 (Roche Applied Science, Indianapolis, IN) per 10-cm2 cell culture monolayer following the manufacturer's recommendation.
Immunoprecipitation, Western Blotting, and Cell Fractionation
For immunoprecipitations, cell monolayers were lysed in radioimmunoprecipitation buffer (50-μl/cm2 monolayers) containing 10 μg/ml each of leupeptin and aprotinin (Sigma-Aldrich). Lysates were incubated with antibodies to MYC (Upstate Biotechnology, Lake Placid, NY) for 2 h at 4°C. Immunocomplexes were recovered by incubation with protein G-conjugated Sepharose (Amersham Biosciences), washed three times with radioimmunoprecipitation buffer, and boiled in SDS sample buffer. For whole cell lysates analysis, cell monolayers were dissolved directly in the culture by adding boiling SDS sample buffer. To separately isolate chromatin-bound and extractable Cdc6, MDA cells were scraped, resuspended in 100 μl of cold lysis buffer (0.5% Triton X-100m CSK containing proteases inhibitors), and immediately centrifuged. Pellets were resuspended with 100 μl of cold lysis buffer and treated with DNase I (1000 U/ml) for 30 min at room temperature. Supernatants and pellets resuspended with 100 μl of cold lysis buffer were diluted 1:1 with SDS sample buffer and denatured by boiling as described above. Immunoprecipitates or aliquots of lysates were separated on 7.5–12% SDS-polyacrylamide gels. Proteins were transferred to Protean membranes (Schleicher & Schuell, Keene, NH). The membranes were probed with antibodies to Cdc6, Cdc25A, cyclin D1, and p53, leukemia inhibitory factor receptor α (LIFRα) (Santa Cruz Biotechnology, Santa Cruz, CA), p21 (Calbiochem), hemagglutinin (HA) (Sigma-Aldrich), and RPA32 and RPA70 (generously provided by Dr. T. Melendy, SUNY, Buffalo, NY), Mcm2 and appropriate secondary antibodies (ICN Biomedicals, Aurora, OH) in phosphate-buffered saline containing 0.1% Tween 20, 5% milk, or 3% albumin. Results were visualized by enhanced chemiluminescence reaction according to the manufacturer (Amersham Biosciences). Gel loading was controlled by parallel probing with antibodies against Mcm2 or by Ponceau staining of protein on blots.
Detection of Apoptosis
YO-PRO and propidium iodide reagents were used as directed by the manufacturer (Molecular Probes, Eugene, OR). For DNA laddering experiments, ∼2.5 × 105 cells were scraped and resuspended in 0.5 ml of 10 mM Tris, 1 mM EDTA, 0.6% SDS, pH 7.4. The resulting lysates were treated with RNaseA and proteinase K, and genomic DNA was isolated by phenol/chloroform extraction. DNA was analyzed on 1.6% agarose gels containing ethidium bromide.
S. cerevisiae Experiments
Wild-type cells expressing scCdc6 (YLD 15 containing GAL1,10-CDC6, a gift from J. Diffley, Cancer Research UK, United Kingdom) were grown at 30°C in YP medium containing raffinose, and to induce Cdc6, cells were transferred to medium containing galactose. Adozelesin was used at a final concentration of 4 μM, and MG132 at a final concentration of 250 μM. Total S. cerevisiae protein extracts were prepared by resuspending 1 × 108 cells in 50 μl of buffer containing 2% SDS, 80 mM Tris pH 6.8, 10% glycerol, 1.5% dithiothreitol, and 0.1 mg/ml bromphenol blue. Cdc6 and Mcm2 were detected with antibodies generously provided by J. Diffley. The AtCDC6a cDNA (accession no. AJ271606) was cloned in-frame into the BamHI site of pMHTgal vector to produce a C-terminal Myc-tagged (9E10 epitope) AtCDC6 protein. This construction was digested with StuI and used to transform S. cerevisiae W303-1a. Extracts of cells expressing AtCDC6 were made as described in Drury et al. (2000) and AtCDC6 detected by immunoblotting with an anti-Myc antibody. As a loading control, immunoblots were probed in parallel with antibodies against Orc2p (gift from J. Diffley).
RESULTS
Adozelesin Inhibits Initiation of Cellular DNA Replication in Mammals
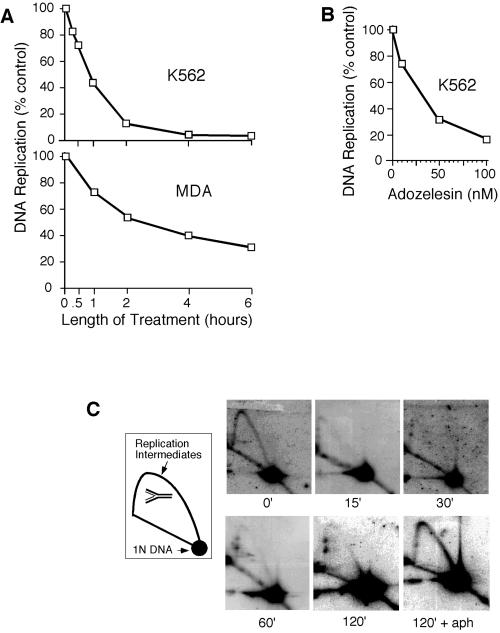
We first asked whether adozelesin inhibits initiation of cellular DNA replication in mammalian cells, similar to its effects on replication of SV40 DNA and S. cerevisiae chromosomes (Cobuzzi et al., 1996; Weinberger et al., 1999). Pulse-labeling with tritiated thymidine showed that adozelesin inhibited DNA synthesis in cultured human cells in a time- and dose-dependent manner (Figure 1, A and B). Inhibition of DNA synthesis by adozelesin in K562 cells was accompanied by a rapid decline in numbers of replication intermediates (RIs) in the repeated rDNA locus of these cells detected by neutral-neutral two-dimensional gel electrophoresis (Figure 1C). The more rapid decline in numbers of RIs induced by adozelesin in K562 cells compared with inhibitory effects on DNA synthesis (Figure 1A) suggests the absence of significant inhibitory effects on elongation of nascent chains at early times of drug exposure, similar to the effects of adozelesin on SV40 DNA replication (Cobuzzi et al., 1996). The number of RIs did not diminish in cells that were also treated with the DNA polymerase inhibitor aphidicolin to block the maturation of replication forks (Figure 1C, 120′+aph). Therefore, the decline in rDNA RIs was caused by the maturation of replication forks in the absence of new initiation events, and not by their collapse or destruction by nucleases.
Figure 1.
Adozelesin inhibits initiation of mammalian chromosomal DNA replication. (A) K562 and MDA cells were treated with 40 nM adozelesin for indicated times and DNA synthesis was assessed by measuring tritiated thymidine incorporation into acid-precipitable DNA. (B) K562 cells were treated with indicated concentrations of adozelesin for 2 h and DNA synthesis was assessed as in A. (C) Replicating and nonreplicating DNA isolated from K562 cells treated with adozelesin or adozelesin + aphidicolin (aph) for the indicated times were separated from one another by neutral-neutral two-dimensional gel electrophoresis, and blots of gels were probed with a fragment of DNA from the tandemly repeated human rDNA locus. Schematic shows migration of single branched replication intermediates and nonreplicating DNA molecules (1N DNA).
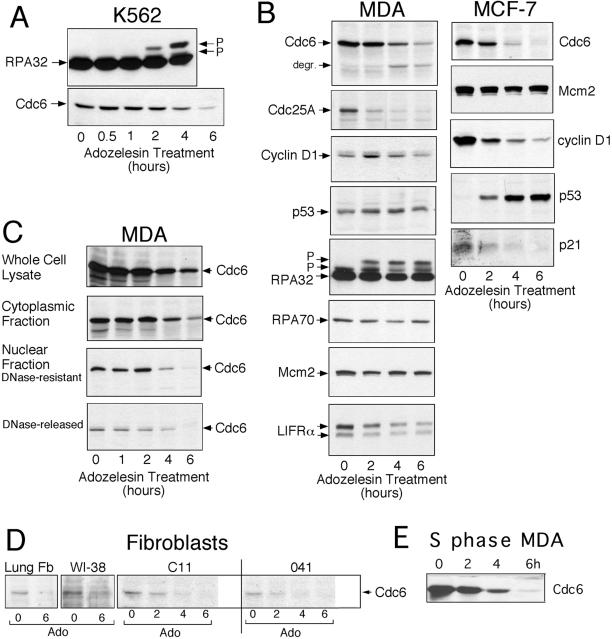
Levels of Cdc6 and Other Proteins Related to Cell Proliferation Are Reduced by Adozelesin Treatment
To determine whether adozelesin alters the biochemical properties or relative amounts of proteins required for initiation of DNA replication and cell proliferation, we analyzed proteins extracted from drug-treated K562 cells by immunoblotting. Levels of Cdc6, but not the 32-kDa subunit of the single-strand DNA binding replication protein RPA (RPA32), declined in response to adozelesin treatment (Figure 2A). The loss of Cdc6 paralleled the appearance of slower mobility forms of RPA32 that were identical to those previously shown to be produced by hyperphosphorylation of this protein in cells exposed to adozelesin or UV radiation (Carty et al., 1994; Liu et al., 2000).
Figure 2.
Levels of Cdc6 and other proteins are reduced by adozelesin treatment. (A–D) Cells were treated for the indicated times with 40 nM adozelesin and cellular proteins were extracted, separated on SDS-PAGE gels, and immunoblotted to detect the indicated proteins. (C) Nuclei were separated from cytoplasm of drug-treated cells to obtain a “cytoplasmic fraction” and a “nuclear fraction” and isolated nuclei were treated with DNase I to obtain the “DNase I-resistant” and “DNase I-released” fractions. (D) Cultures of normal lung fibroblasts (Lung Fb) and established fibroblast cell lines were treated for 16 h with 5 μg/ml aphidicolin to accumulate large numbers of cells in S phase. Then 40 nM adozelesin (Ado) was added for the additional times indicated before proteins were extracted and analyzed by immunoblotting. (E) MDA cells were treated with aphidicolin for 16 h to synchronize them in S phase and then treated with 40 nM adozelesin and aphidicolin for an additional 2–6 h.
The reduction in the level of Cdc6 induced by adozelesin was not limited to K562 cells. A comparable or even more rapid decline in the amount of Cdc6 was induced by adozelesin in MDA and MCF-7 human breast cancer cells (Figure 2B). In many experiments, the loss of the 62-kDa Cdc6 protein was accompanied by the appearance of a faster migrating form at ∼48 kDa that seemed to be a proteolytic degradation product (Figure 2B, degr). Levels of Mcm2, a protein that interacts with Cdc6 in pre-RCs, remained stable. However, other cell cycle regulatory proteins also declined in abundance in MDA and MCF-7 cells exposed to adozelesin, although with significantly different kinetics compared with Cdc6. These included the phosphatase Cdc25A, which regulates initiation of DNA replication upstream of the cyclin-dependent kinase Cdk2 (Vigo et al., 1999), and cyclin D1, which activates CDK 4 and 6 phosphorylation of retinoblastoma protein (reviewed in Lipinski and Jacks, 1999) and subsequent E2F-regulated expression of Cdc6 and other licensing proteins as cells reenter the cell cycle. Both Cdc25A and cyclin D1 were recently shown to be degraded by the proteasome in response to DNA damage as part of p53-independent late G1- or S-phase checkpoints (Agami and Bernards, 2000; Mailand et al., 2000; Falck et al., 2001). Although MCF-7 cells produce wild-type p53 and this protein increased in levels upon adozelesin treatment, MDA cells produce a mutant p53 protein (Thor et al., 1992) at constitutively elevated levels that were not altered upon exposure to adozelesin (Figure 2B). Thus, similar to the DNA damage-induced destruction of Cdc25A and cyclin D1, the adozelesin-induced decline in Cdc6 levels did not seem to require p53. In parallel with the increase in p53 induced by adozelesin in MCF-7 cells, low levels of the cyclin-dependent kinase inhibitor p21 observed in these cells were reduced even further by adozelesin treatment (Figure 2B). A similar reduction in levels of p21 has been reported to occur during apoptosis (Levkau et al., 1998).
The loss of proteins induced by adozelesin was not confined to the nuclear compartment. For instance, adozelesin treatment of MDA cells also induced loss of the processed 190-kDa form of LIFRα, which resides in the plasma membrane (Figure 2B). In certain cell types such as embryonic stem cells (Stewart, 1994) and breast cancer cells (Kellokumpu-Lehtinen et al., 1996), LIFRα plays a role in stimulating proliferation upstream of Cdc25A, cyclin D1, and Cdc6. Levels of two other DNA replication proteins analyzed in parallel, the 32- and 70-kDa subunits of RPA (RPA32 and RPA70), remained stable in MDA and/or MCF-7 cells (Figure 2B).
Cdc6 can be found in two fractions in mammalian cells, one that is readily extracted from chromatin, and a second that remains tightly bound to chromatin and resists digestion of DNA with DNase I (Fujita et al., 1999). To determine whether adozelesin induces a decline in levels of one or both of these populations of Cdc6, chromatin-bound proteins in lysates of adozelesin-treated MDA cells were separated from nonchromatin-bound proteins and separately analyzed on immunoblots (Figure 2C). The loss of Cdc6 that was apparent in whole cell lysates occurred in both the extractable population of Cdc6 and the fraction bound to chromatin, including that which resisted DNase I digestion. The chromatin fraction presumably includes Cdc6 found in licensing complexes at origins of replication.
Cdc6 levels were also diminished by adozelesin treatment in primary lung fibroblasts and in the normal diploid fibroblast cell line WI-38 (Figure 2D). Basal levels of Cdc6 were much lower in these cells compared with K562, MDA, and MCF-7 cancer cells. Therefore, to facilitate detection of Cdc6, these cells were first treated with the DNA polymerase inhibitor aphidicolin to accumulate large numbers of cells at the G1/S boundary, where Cdc6 expression is expected to be high (Mendez and Stillman, 2000; Petersen et al., 2000). Because adozelesin caused a reduction in the level of Cdc6 in these cells while they remained blocked in S phase (Figure 2D), the loss of Cdc6 was not related to an adozelesin-induced block of cell cycle progression in early G1, which is the only portion of the cell cycle where Cdc6 levels are normally very low (Mendez and Stillman, 2000; Petersen et al., 2000). Similarly, adozelesin caused a decrease in levels of Cdc6 in MDA cells that were blocked in S phase before and during adozelesin treatment (Figure 2E). Redistribution by adozelesin of large numbers of cells to early G1 in log phase cultures was ruled out by fluorescence-activated cell sorting (FACS) analysis of MDA (our unpublished data) and MCF-7 cells (Figure 5C), which showed that after 6 h of adozelesin treatment, only 10–12% of these cells had proceeded through mitosis. Cdc6 levels also declined in the Li-Fraumeni fibroblast cell lines c11 and MDA041 blocked in S phase (Figure 2D). Because MDA041 cells are functionally p53-null (Yin et al., 1992), the decline in levels of Cdc6 induced by adozelesin cannot depend on p53.
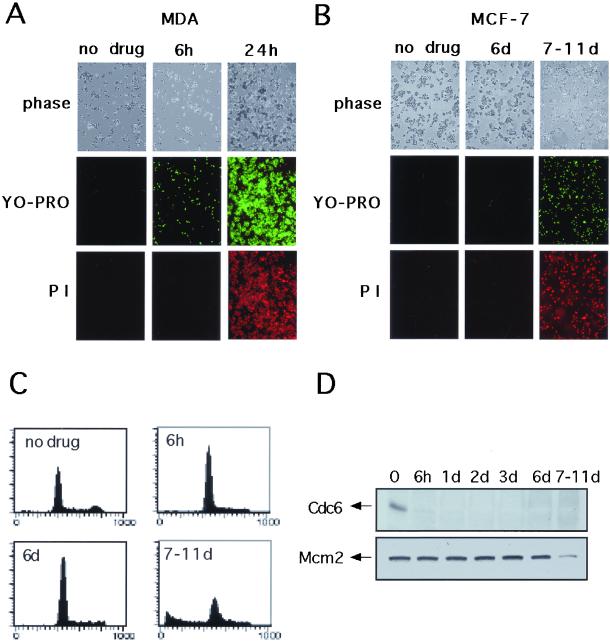
Figure 5.
Proteasome-dependent destruction of Cdc6 occurs during early apoptosis. MDA (A) or MCF-7 (B) cells were treated or not treated with adozelesin for 6 h and adozelesin-containing medium was replaced with fresh drug-free medium for an additional 24 h. Apoptosis was assessed at the indicated times after addition of drug by phase contrast microscopy, YO-PRO staining, and staining with propidium iodide. (C) FACS analysis of MCF-7 cells after adozelesin treatment. (D) Western blot analysis of Cdc6 and Mcm2 in MCF-7 cells after adozelesin treatment.
Adozelesin Induces Proteasome- and Ubiquitin-dependent Destruction of Cdc6
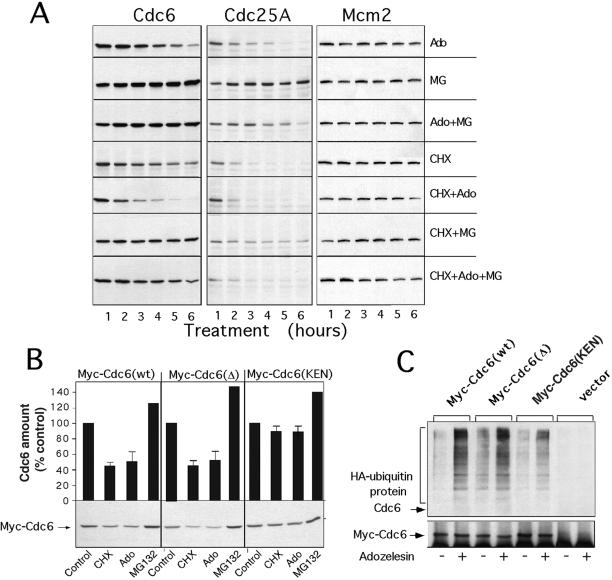
Three recent studies report that DNA damage induces the proteasome-dependent destruction of Cdc25A (Mailand et al., 2000; Falck et al., 2001) and cyclin D1 (Agami and Bernards, 2000) as part of late G1- or S-phase checkpoints that transiently block DNA replication. To determine whether the decline in Cdc6 levels induced by adozelesin in parallel with the decline in levels of these other proteins was also caused by a proteasome-dependent mechanism, we compared the kinetics of this decline in the presence or absence of the proteasome inhibitor MG132. The decline in Cdc6 levels observed in adozelesin-treated MDA cells (Figure 3A, Ado) did not occur in cells exposed to both adozelesin and MG132 (Figure 3A, Ado+MG), suggesting that adozelesin engages the proteasome. Moreover, treatment of cells with MG132 alone (Figure 3A, MG) caused a twofold increase in levels of Cdc6, supporting recent reports that the normal turnover of Cdc6 also requires the proteasome (Mendez and Stillman, 2000; Petersen et al., 2000). When the rate of Cdc6 degradation in cells treated with the protein synthesis inhibitor cycloheximide was determined, the results indicated that in the absence of drug treatment, Cdc6 has a half-life of 2–3 h (Figure 3A, CHX). This half-life was reduced to 1–2 h by adozelesin treatment (Figure 3A, CHX+Ado). In the presence of MG132, the normal (Figure 3A, CHX+MG) as well as adozelesin-mediated (Figure 3A, CHX+Ado+MG) degradation of Cdc6 was significantly reduced. The small, but detectable decline in levels of Cdc6 in cells treated with cycloheximide, MG132, and adozelesin (Figure 3A, CHX+Ado+MG) may reflect the contribution of proteasome-independent effects of adozelesin on levels of Cdc6 (see below). These proteasome-independent effects were not mediated by lysosomal proteases or calpain, because treatment of cells with chloroquine, a lysotropic agent, or N-acetyl-leucyl-leucyl-methional, an inhibitor of calpain, did not change the rate of Cdc6 degradation in adozelesin-treated cells (our unpublished data). Similar results were obtained in a parallel analysis of the effects of adozelesin, cycloheximide, and MG132 on levels of Cdc25A (Figure 3A, Cdc25A). In contrast, levels of Mcm2 remained stable throughout these treatments (Figure 3A, Mcm2).
Figure 3.
Adozelesin induces ubiquitination and proteasome-dependent degradation of Cdc6. (A) MDA cells were treated with 40 nM adozelesin (Ado) for the indicated times in presence or absence of 10 μM MG132 (MG), 10 μg/ml cycloheximide (CHX), or combinations as indicated. Cell extracts were analyzed for Cdc6, Cdc25A, or MCM2 levels by immunoblotting. (B) MCF-7 cells were transfected with vectors expressing Myc-Cdc6 wild-type [Myc-Cdc6 (wt)] or mutated destruction box [Myc-Cdc6(Δ)] or KEN motif [Myc-Cdc6(KEN)] proteins. Cells were treated 6 h with adozelesin (Ado), cycloheximide (CHX), or MG beginning 24 h after transfection and analyzed for the level of transfected Cdc6 by immunoblotting with anti-Myc antibodies. Signals from immunoblots in three independent experiments were quantitated by densitometry and the averages and SEs presented above each lane. (C) MCF-7 cells were cotransfected with expression vectors for HA-tagged ubiquitin and the Myc-tagged wild-type and mutant Cdc6 described above. Lysates were made from cells after adozelesin treatment for 6 h beginning 24 h after transfection, and Myc-tagged Cdc6 proteins were immunoprecipitated with Myc antibodies. Immunoprecipitated proteins were analyzed for HA-ubiquitin and then for Myc-tagged Cdc6 by immunoblotting with anti-HA (top) and anti-Myc (bottom) antibodies. hIg is the IgG light chain that reacts with the secondary antibody.
The normal turnover of mammalian Cdc6 involves ubiquitination and requires specific destruction box and KEN-box motifs (Petersen et al., 2000). Therefore, we next asked whether these motifs and/or ubiquitination play a role in the proteasome-dependent degradation of Cdc6 induced by adozelesin. Cells were transfected with plasmids expressing wild-type or mutant myc-tagged Cdc6 and levels of myc-tagged proteins in cell lysates were determined by immunoblotting with antibodies against the myc epitope. Levels of wild-type myc-Cdc6 were reduced by adozelesin treatment or when protein synthesis was inhibited with cycloheximide [Figure 3B, Myc-Cdc6 (wt)]. The less dramatic reduction in levels of myc-Cdc6 compared with endogenous Cdc6 (Figures 2 and 3A) induced by adozelesin may reflect saturation of the proteasome by the high levels of ectopic myc-Cdc6 in these cells. The level of myc-Cdc6 containing a mutation in a destruction box motif that previously had been shown to partially block Cdc6 turnover in early G1 (Petersen et al., 2000) was reduced by adozelesin to a similar extent compared with wild-type Cdc6 [Figure 3B, Myc-Cdc6(Δ)], suggesting that this destruction box was not required for adozelesin-induced degradation. Moreover, in our experiments, the decrease in the level of myc-Cdc6(Δ) that occurred in cycloheximide-treated cells was similar to that of wild-type Cdc6, suggesting that the mutation also did not interfere with normal degradation. The biochemical nature of the difference between our results and a previous study (Petersen et al., 2000) is not clear, but may be attributable to the use of different cell types. In contrast, the level of myc-Cdc6 protein containing a mutation in the KEN box motif did not decline significantly in cells treated with either cycloheximide or adozelesin [Figure 3B, Myc-Cdc6(KEN)]. Therefore, the KEN box motif plays a role in the normal turnover of Cdc6 as reported previously (Petersen et al., 2000), and also plays a role in the destruction induced by adozelesin.
To determine whether the proteasome-dependent destruction of Cdc6 was related to increased ubiquitination that might be altered by the KEN-box mutation, we next examined the levels of ubiquitin-conjugated wild-type and mutant Cdc6 proteins as a function of adozelesin treatment. Cells were transfected with the expression vectors for myc-tagged wild-type or mutant Cdc6 molecules described above and with a vector expressing HA-tagged ubiquitin. Transfected cells were treated with adozelesin for 6 h, and myc-tagged Cdc6 proteins were recovered by immunoprecipitation with antibodies against Myc. The immunoprecipitates were then analyzed for HA-ubiquitinated proteins by immunoblotting (Figure 3C).
In the absence of adozelesin treatment, immunoprecipitates of cells transfected with plasmids expressing either wild-type Cdc6 [Figure 3C, Myc-Cdc6(wt)] or Cdc6 that harbored a destruction box mutation [Figure 3C, Myc-Cdc6-(Δ)] showed modest levels of a HA-ubiquitin–containing protein that migrated slower than intact Myc-Cdc6. Adozelesin treatment dramatically increased the amount of this protein in both cases. Slower migrating HA-ubiquitin–containing protein was also observed in immunoprecipitates of adozelesin-treated cells transfected with the plasmid expressing myc-Cdc6 containing the KEN-box mutation [Figure 3C, Myc-Cdc6(KEN)]. However, in several repetitions of this experiment, the amount of this protein was reproducibly reduced compared with immunoprecipitates of cells expressing wild-type myc-tagged Cdc6. Reprobing of immunoblots with an antibody against myc to detect the unmodified myc-tagged Cdc6 (Figure 3C, bottom) indicated that the changes in amount of HA-ubiquitin–containing protein were not related to differences in the efficiency with which cells were transfected or myc-tagged Cdc6 was immunoprecipitated. Because slower migrating HA-ubiquitin–containing proteins were only observed in immunoprecipitates from cells expressing myc-tagged Cdc6 molecules, and not from empty vector-transfected cells (Figure 3C, vector), these proteins correspond to polyubiquitinated Cdc6- or Cdc6-interacting proteins that were specifically precipitated by the anti-myc antibody. These results strongly suggest that adozelesin stimulates the ubiquitination of Cdc6, and this stimulation is partially prevented by the KEN-box mutation. Because this mutation also partially stabilizes Cdc6 in adozelesin-treated cells (Figure 3C), the degradation of Cdc6 induced by adozelesin seems to be largely ubiquitin dependent.
Destruction of Cdc6 Occurs in Association with Programmed Cell Death
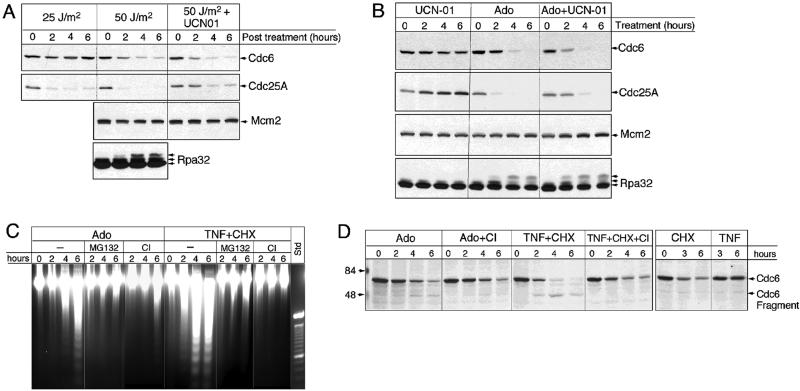
The inhibitory effect of adozelesin on initiation of DNA replication and the similarity of the proteasome-dependent and p53-independent degradation of Cdc6, Cdc25A, and cyclin D1 induced by adozelesin and other DNA-damaging agents suggests the possibility that the proteolytic destruction of all three proteins is part of a DNA damage checkpoint response that inhibits initiation and occurs independently of the type of DNA damage lesion. To test this hypothesis, we analyzed the effects of UV radiation on levels of Cdc6 and Cdc25A in MDA cells (Figure 4A). Exposure to 25 J/m2 UV radiation did not appreciably affect Cdc6 levels, but caused a rapid loss of Cdc25A that was comparable with the loss recently reported in UV-irradiated U2OS cells (Mailand et al., 2000). However, when MDA cells were subjected to 50 J/m2 UV radiation, both Cdc25A and Cdc6 levels rapidly declined. This decline occurred in parallel with the apparent hyperphosphorylation of the 32-kDa subunit of RPA, similar to that induced by adozelesin in these cells (Figure 2B). Under the same conditions, the amount of Mcm2 protein in MDA cells was not altered (Figure 4A).
Figure 4.
Cdc6 destruction is induced by UV radiation and TNF-α and coincides with a caspase-dependent apoptotic response. (A) MDA cells were irradiated (25 or 50 J/m2 UV radiation) and returned to normal culture conditions in the presence or absence of 1 μM of the kinase inhibitor UCN-01 for 0–6 h. Proteins extracted at indicated times after irradiation were analyzed by immunoblotting. (B) MDA cells were treated with 1 μM UCN01, 40 nM adozelesin (Ado), or both (Ado+UCN01) for indicated times and extracted proteins analyzed by immunoblotting. (C) DNA was isolated from MDA cells treated with various combinations of Ado, 100 ng/ml TNF-α (TNF), and 10 μg/ml cycloheximide (TNF-CHX) with or without 10 μM MG132 (MG) or 10 μM caspase inhibitor I (CI) for indicated times and then size-fractionated on ethidium bromide-containing agarose gels. The standard (std) is a 100-base pair ladder. (D) Cells were treated with the indicated compounds for 0–6 h and protein extracts analyzed by immunoblotting.
The failure of the lower dose of 25 J/m2 UV radiation to induce the destruction of Cdc6 at the same time that it induced the destruction of Cdc25A suggests these two proteins are either targeted by different DNA damage response pathways or are fundamentally different in their sensitivity to a common pathway. To test these possibilities further, we asked whether the destruction of Cdc6 induced by UV radiation could be blocked by treating cells with the kinase inhibitor UCN-01, which previously had been shown to abrogate the destruction of Cdc25A and the resulting DNA damage checkpoint induced by UV radiation in U2OS cells (Mailand et al., 2000). Although UCN-01 partially prevented the UV-induced destruction of Cdc25A in MDA cells, the level of Cdc6 in cells irradiated with 50 J/m2 UV was reduced even further in cells also treated with UCN-01 (Figure 4A). Therefore, the destruction of Cdc6 induced by UV radiation is accomplished at least in part by a pathway that is different from the one that targets Cdc25A. To determine whether adozelesin treatment also engages these different pathways, we asked whether UCN-01 had a similar effect on levels of these proteins in adozelesin-treated cells. Indeed, similar to its effects in UV-irradiated cells, UCN-01 partially stabilized the level of Cdc25A, but enhanced the reduction in the level of Cdc6 (Figure 4B).
These results are not consistent with the hypothesis that the destruction of Cdc6 occurs as part of a checkpoint that inhibits initiation of DNA replication and requires the destruction of Cdc25A. The destruction of Cdc6 induced by high, but not low doses of UV radiation suggests it might occur instead as part of a programmed death response to irreparable DNA damage. This was also suggested by the coincidental hyperphosphorylation of RPA and loss of p21 (Figure 2B), both of which have been shown to occur during apoptosis (Levkau et al., 1998; Treuner et al., 1999; Chai et al., 2000). Therefore, we asked whether Cdc6 destruction by adozelesin occurs in association with an apoptotic response that results in DNA laddering and includes a caspase inhibitor-sensitive component. Within 4–6 h of adozelesin treatment, DNA laddering was apparent in MDA cells (Figure 4C, Ado). Moreover, adozelesin-induced DNA laddering was significantly reduced by treating MDA cells with the proteasome inhibitor MG132 or caspase inhibitor I. Therefore, adozelesin induces a proteasome- and caspase-dependent apoptotic response in MDA cells that occurs coincidently with the destruction of Cdc6. However, parallel analysis of Cdc6 levels in these cells when they were also treated with caspase inhibitor I indicated that, unlike inhibition of the proteasome (Figure 3), inhibition of caspases did not prevent a significant loss of Cdc6 (Figure 4D, Ado+CI). It did, however, reduce the apparent Cdc6 cleavage product of ∼48 kDa produced by adozelesin treatment (Figure 4D, Cdc6 fragment). Therefore, although Cdc6 is primarily degraded by the proteasome in response to adozelesin treatment, adozelesin induces a minor, but detectable cleavage of Cdc6 by caspases as well.
To determine whether other triggers of apoptosis would induce a similar proteasome- and/or caspase-dependent destruction of Cdc6, MDA cells were treated with a combination of TNF-α and cycloheximide. This treatment induced a strong apoptotic response that resulted in extensive DNA laddering within 4–6 h (Figure 4C, TNF+CHX), which could be suppressed by either proteasome or caspase inhibitors (Figure 4C). Immunoblot analysis of Cdc6 indicated that the combined treatment with TNF-α and cycloheximide reduced the amount of Cdc6 below levels induced by TNF-α (Figure 4D, TNF) or cycloheximide (Figure 4D, CHX) alone. The prominent appearance of the caspase inhibitor-sensitive 48-kDa Cdc6 breakdown product suggested a more extensive involvement of caspase-dependent pathways in this decline. In fact, comparison of Cdc6 levels in cells treated with TNF-α/cycloheximide and caspase inhibitor (Figure 4D, TNF+CHX+CI) with levels in cells treated with cycloheximide alone (Figure 4D, CHX) indicated that caspase cleavage could completely account for the degradation of Cdc6 induced by TNF-α/cycloheximide, in contrast to the predominantly proteasome-dependent destruction of Cdc6 induced by adozelesin.
Proteasome Destruction of Cdc6 Occurs in Early Apoptosis Independently of Caspase 3 Activity
The role of the proteasome in apoptosis may occur at an early precaspase stage, upstream of the mitochondrial events associated with a commitment to cell death (Dallaporta et al., 2000). To determine whether the proteasome-dependent destruction of Cdc6 occurs in early apoptosis, we used an assay that distinguishes earlier stage apoptotic cells from nonapoptotic and late-stage apoptotic or necrotic cells on the basis of permeability changes that result in the differential uptake of fluorescent dyes. Early apoptotic cells are detected by their permeability to the green fluorescent dye YO-PRO, but not propidium iodide, whereas late-stage apoptotic or necrotic cells stain positively with both YO-PRO and propidium iodide. Although untreated MDA cells were impermeable to either dye (Figure 4E, MDA, no drug), ∼50% of MDA cells treated with adozelesin for 6 h stained positively with YO-PRO, but not propidium iodide (Figure 5A, MDA, 6h). These cells were morphologically similar in appearance to the untreated controls (Figure 5A, phase). When adozelesin was withdrawn from the medium after 6 h and cells were cultured for an additional 18 h in the absence of drug, 100% of the cells stained positively with both dyes (Figure 5A, MDA, 24h) and large numbers had rounded up and/or detached from the substratum (Figure 5A, phase), indicating they were now in a late stage of apoptosis or completely inviable. Thus, in MDA cells, the proteasome-dependent destruction of Cdc6 during the first 6 h of drug treatment (Figures 2–4) occurs during an early stage of apoptosis.
We repeated this analysis in MCF-7 cells, which are relatively insensitive to genotoxic and nongenotoxic inducers of apoptosis due to mutational inactivation of the caspase-3 gene (Kurokawa et al., 1999). Caspase-3 seems to play an apical role among the effector caspases during caspase-dependent apoptosis (Hengartner, 2000), and a number of the downstream elements of caspase-dependent apoptotic responses, such as DNA laddering, do not occur in MCF-7 cells (Kurokawa et al., 1999). Despite the fact that Cdc6 was undetectable in MCF-7 cells 6 h after adozelesin treatment began (Figures 2B and 5D), these cells remained impermeable to both YO-PRO and propidium iodide and maintained a normal morphology for at least 6 days after adozelesin was withdrawn (Figure 5B). After a period of time that varied between 7 and 11 days after drug treatment in different experiments, MCF-7 cells detached from the substratum in large numbers and stained positively with both YO-PRO and propidium iodide (Figure 5B). FACS analysis indicated that, after an initial decline in the number of cells with a G2/M content of DNA during the 6 h of adozelesin treatment, cells remained arrested in G1 and/or S phase for up to 6 d. After 6 days, a population of cells with a sub-G1 content of DNA accumulated and the number of cells with a G1 content of DNA declined (Figure 5C). As expected for cells lacking caspase-3, adozelesin treatment did not cause DNA laddering in MCF-7 cells (our unpublished data). Parallel Western blot analysis of Cdc6 protein indicated that after the decline in Cdc6 during the initial 6-h drug treatment (Figures 2B and 5D), Cdc6 remained absent from adozelesin-treated MCF-7 cells for up to 11 days (Figure 5D, Cdc6). Levels of Mcm2 remained stable for 7–11 days after adozelesin treatment and then declined in parallel with the uptake of YO-PRO and PI and the degradation of DNA detected by FACS (Figure 5D, Mcm2). These results and the failure of a general caspase inhibitor to block the proteasome-dependent destruction of Cdc6 in MDA cells (Figure 4D) establish that the specific, proteasome-dependent destruction of Cdc6 induced by adozelesin occurs upstream of, or parallel to, the action of caspase-3 and other caspases during apoptosis. Furthermore, in MCF-7 cells, Cdc6 destruction precedes by several days an adozelesin-induced necrotic-like cell death accompanied by a more general destruction of proteins.
Proteasome-dependent Destruction of Cdc6 Induced by DNA-damaging Agents Is Conserved in Yeast
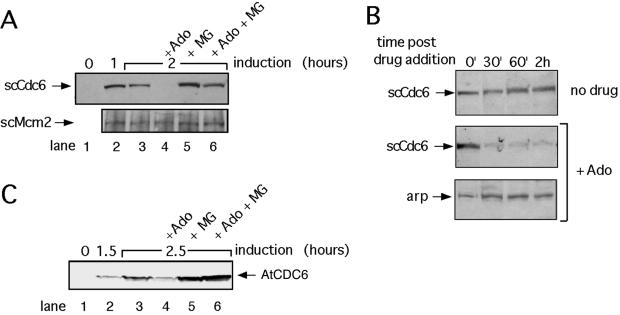
The cytotoxicity of adozelesin in S. cerevisiae cells is related at least in part to drug-induced alterations in the function of proteins that interact with Cdc6 in pre-RCs, because mutations in these proteins can result in resistance to adozelesin (Weinberger et al., 1999). To determine whether adozelesin cytotoxicity might be related to a proteasome-dependent destruction of scCdc6 in budding yeast cells, we asked whether adozelesin also induces the degradation of Cdc6 in this organism. To increase the level of detection of scCdc6, which is normally very low, these experiments used cells that were genetically altered to express scCdc6 at high levels from a galactose-inducible promoter (Drury et al., 2000). Although scCdc6 was not detectable by immunoblotting of proteins extracted from these cells before galactose induction (Figure 6A, scCdc6, lane 1), a significant amount of scCdc6 was detected in cells switched to galactose-containing medium for 1 or 2 h (Figure 6A, scCdc6, lanes 2 and 3). In contrast, levels of scCdc6, but not the pre-RC protein scMcm2, were greatly reduced in cells exposed to adozelesin for 1 h beginning an hour after galactose induction (Figure 6A, lane 4). Treatment of cells with the proteasome inhibitor MG132 for 1 h beginning an hour after induction of Cdc6 reproducibly increased the level of scCdc6 slightly, which presumably reflects inhibition of the normal proteasome-dependent turnover of scCdc6 (Figure 6A, scCdc6, lane 5). The combined treatment of cells with adozelesin and MG132 blocked the decline in scCdc6 levels induced by adozelesin alone (Figure 6A, scCdc6, lane 6), indicating that the proteasome was responsible for this decline.
Figure 6.
Adozelesin induces the proteasome-dependent destruction of Cdc6 in budding yeast. (A and C) S. cerevisiae cells harboring plasmids expressing the S. cerevisiae Cdc6 (scCdc6) or Arabidopsis CDC6 (AtCDC6) genes regulated by a galactose-inducible promoter were changed from raffinose- to galactose-containing medium to induce the ectopic expression of scCdc6 or AtCDC6. They were treated with 4 μM adozelesin and/or 250 μM MG132 beginning 1 h (scCdc6 experiments) or 1.5 h (AtCDC6 experiments) after galactose induction. Total protein lysates made at the indicated times after induction were analyzed by immunoblotting for S. cerevisiae Cdc6 (scCdc6) or arabidopsis Cdc6 (AtCDC6) proteins, or for scMcm2 protein as a control. (B) cdc4 cells were synchronized in early S phase by release from an α factor G1 block into HU and then cultured in the continued presence of HU at the nonpermissive temperature (37°C) for 2 h to destroy the activity of Cdc4p. Cycloheximide and adozelesin were then added for the times indicated before cells were lysed for Western blot analysis. Arp, actin-related protein.
To confirm that adozelesin induces the degradation of scCdc6, we analyzed the effects of adozelesin treatment on scCdc6 levels in the presence of cycloheximide to block protein synthesis (Figure 6B). This experiment used a strain with a temperature-sensitive mutation in CDC4 that causes a defect in the normal degradation of scCdc6 in S-phase cells shifted to the nonpermissive temperature (Drury et al., 1997). Cells remained trapped in early S phase with hydroxyurea for the duration of this experiment to rule out the possibility that Cdc6 destruction was caused by adozelesin-induced redistribution of cells to other points in the cell cycle where destruction of Cdc6 might normally occur. In the absence of adozelesin, scCdc6 remained stable for a period of at least 2 h after a shift to the nonpermissive temperature (Figure 6B, no drug). Addition of adozelesin and cycloheximide after galactose induction of scCdc6 resulted in a substantial decrease in levels of scCdc6 over this same time period (Figure 6B, scCdc6, +Ado), but not levels of an actin-related protein detected in the same extracts (Figure 6B, arp, +Ado). Thus, similar to its effect on mammalian Cdc6, adozelesin induces the proteasome-dependent degradation of scCdc6, and this degradation is specific. Furthermore, unlike the normal turnover of scCdc6, adozelesin-induced destruction does not require Cdc4p.
The similarity of the proteasome-dependent effects of adozelesin on Cdc6 in yeast and mammalian cells suggests they might be conserved in all eukaryotes. Cdc6 of A. thaliana (AtCDC6) shares a number of features with Cdc6 from mammals, including primary sequence, E2F-regulated expression, and degradation in a ubiquitin- and proteasome-dependent manner (Castellano et al., 2001). We have so far been unable to detect endogenous AtCDC6 in plant cells by immunoblotting. However, we hypothesized that if the mechanism underlying the proteasome-dependent destruction of Cdc6 induced by adozelesin is highly conserved, the same structural determinants in Cdc6 that regulate mammalian and S. cerevisiae Cdc6 destruction might also participate in the destruction of AtCDC6 expressed in S. cerevisiae cells. Therefore, we repeated the immunoblot analysis by using an S. cerevisiae strain that ectopically expresses a Myc-AtCDC6 protein under the control of a galactose-inducible promoter (Figure 6C, Arabidopsis). Levels of AtCDC6 were similarly reduced by adozelesin treatment in these experiments, and this decline could be reversed by treatment with MG132. Thus, AtCDC6 can also serve as a substrate for proteasome-dependent destruction induced by adozelesin in budding yeast. This suggests that the mechanism underlying the loss of Cdc6 induced by adozelesin may be conserved in plants as well.
DISCUSSION
The induction by adozelesin and TNF-α/cycloheximide of multiple independent pathways targeting the initiation protein Cdc6 for destruction may be important for understanding how adozelesin and other drugs exert their cytotoxic and antitumor effects. We previously showed that adozelesin profoundly inhibits initiation of SV40 DNA replication in trans, with little effect on elongation of nascent DNA strands at replication forks (Cobuzzi et al., 1996). Adozelesin also inhibits initiation of cellular DNA replication in budding yeast, and this inhibitory effect is reduced by a mutation in the origin licensing protein Orc2p (Weinberger et al., 1999). In this study, we show that adozelesin inhibits initiation of mammalian cellular DNA replication (Figure 1) in concert with the proteasome-dependent destruction of Cdc6, another origin licensing protein that interacts with ORC and is also required for initiation of DNA replication in all eukaryotes. Cdc6 destruction is also induced by UV radiation (Figure 4), and is accompanied by the destruction of other cell cycle regulatory proteins, including Cdc25A and cyclin D1 (Figures 2–4).
Both Cdc25A and cyclin D1 were recently shown to be destroyed in a proteasome-dependent manner in response to DNA damage as part of multiple checkpoints that inhibit DNA replication (Agami and Bernards, 2000; Mailand et al., 2000; Falck et al., 2001). This suggests that perhaps the proteasome-dependent destruction of Cdc6 also occurs as part of a checkpoint that inhibits DNA replication in response to adozelesin and UV radiation, and perhaps other DNA-damaging agents. However, both Cdc25A destruction and the initiation-inhibitory effect of adozelesin occur much faster than Cdc6 destruction. In addition, Cdc25A destruction induced by UV radiation occurs at lower doses that are insufficient to induce the destruction of Cdc6 observed at higher UV doses. Furthermore, the destruction of Cdc25A induced by either adozelesin or higher doses of UV radiation can be distinguished from the destruction of Cdc6 by the differential effects of the Chk1 kinase inhibitor UCN-01, which stabilizes Cdc25A (Figure 4; Mailand et al., 2000), but destabilizes Cdc6 (Figure 4) in cells subjected to these DNA-damaging agents. Therefore, although the inhibitory effect of adozelesin on initiation of cellular DNA replication in mammals probably involves the activation of a Cdc25A-dependent DNA damage checkpoint, similar to the effects of UV radiation (Mailand et al., 2000), it is unlikely that the destruction of Cdc6 is required for this checkpoint.
Instead, our data show that Cdc6 is destroyed in adozelesin-treated cells by a p53-independent, proteasome-dependent pathway in association with an irreversible arrest of DNA replication and programmed cell death. Cdc6 is also destroyed in cells undergoing a p53-independent programmed cell death through an extrinsic apoptotic pathway induced by TNF-α, although in this case, its destruction is mediated by an entirely different mechanism involving caspases (Figure 4). The proteasome-dependent destruction of Cdc6 induced by adozelesin occurs during an early stage of apoptosis (Figure 5A) upstream of, or parallel to, classical caspase-dependent apoptotic pathways, because a general caspase inhibitor does not block Cdc6 destruction by the proteasome (Figure 4D). Adozelesin also induces the specific destruction of Cdc6 in MCF-7 cells lacking an important effector of apoptosis, caspase 3 (Figure 2). In these cells, elimination of Cdc6 and arrest of DNA replication occur many days before they die coincidentally with a more generalized proteolysis (Figure 5). Thus, the proteasome-dependent destruction of Cdc6 is unlikely to be a simple consequence of the cellular disassembly that occurs during the latter stages of apoptosis or necrosis. This is consistent with the suggestion that the proteasome plays a role in apoptosis at an early stage, upstream of mitochondrial events (Dallaporta et al., 2000). Finally, S. cerevisiae Cdc6 and ectopically expressed Arabidopsis Cdc6 are also specifically targeted for destruction by the proteasome in budding yeast cells treated with adozelesin (Figure 6). Conservation of the proteasome-dependent destruction of Cdc6 in yeast and mammals and the evolution in mammals of two fundamentally different pathways that target Cdc6 for destruction, depending on the type of apoptotic trigger, suggest that the elimination of Cdc6 has an important function, most likely as part of a commitment to programmed cell death.
The complexity of the factors regulating Cdc6 stability has so far precluded the construction of stable mutants that could be used to ask whether Cdc6 destruction plays a participatory role in cell death pathways. However, a number of observations in yeast and Xenopus suggest how the destruction of Cdc6 or alterations to other licensing proteins might contribute to cell death. The experimental elimination of Cdc6 from budding (Piatti et al., 1995; Detweiler and Li, 1997) or fission (Kelly et al., 1993) yeast cells activates mitosis in cells with unreplicated chromosomes, producing a mitotic catastrophe. In S. cerevisiae, the mitotic catastrophe caused by eliminating Cdc6 can be blocked by the parallel elimination of mitotic cyclins (Piatti et al., 1995; Elsasser et al., 1996; Detweiler and Li, 1997; Desdouets et al., 1998), which indicates that it requires the induction of mitotic CDK activity. Furthermore, Cdc6 (Bueno and Russell, 1992; Calzada et al., 2000; Perkins et al., 2001) and the licensing protein Orc6 (Weinreich et al., 2001) interact with the mitotic form of the S. cerevisiae cyclin-dependent kinase Cdc28 and inhibit its activity. Inactivation of other proteins required for initiation of DNA replication also induces mitotic cyclin-dependent kinase activity and mitosis in the absence of DNA replication in S. cerevisiae (Toyn et al., 1995). Mitotic catastrophe was also recently observed in nuclei assembled in Xenopus extracts from which Cdc6 had been depleted (Hekmat-Nejad et al., 2000) or when pre-RC formation was blocked in these extracts by the addition of geminin, a protein that blocks pre-RC formation until the end of mitosis (Michael et al., 2000). Again, this catastrophic mitosis requires the activation of a mitotic cyclin-dependent kinase. Recently, Xenopus Cdc6 was shown to recruit cyclin-dependent kinase complexes required for initiation of DNA replication to origins of replication (Furstenthal et al., 2001). Recruitment of this complex by Cdc6 may serve to spatially constrain its regulation to sites where DNA replication is initiated (Furstenthal et al., 2001).
All of these findings suggest that coordination of DNA synthesis with mitosis requires the reciprocal regulation of licensing protein and cyclin-dependent kinase activity by Cdc6 and other proteins in complexes at origins of replication, and that loss of the integrity of these complexes interferes with this coordination, with lethal consequences. Although it is not yet known whether Cdc6 plays a role in regulating cyclin-dependent kinases in mammals, conservation of its structure and other aspects of its function from yeast to mammals suggest this may be the case. Moreover, a growing body of evidence suggests that disruption of the coordinate regulation of DNA synthesis and cyclin-dependent kinase activity is an important, although not obligatory, feature of programmed cell death that causes the unscheduled activation of cyclin-dependent kinases in cells with unreplicated or partially replicated chromosomes (reviewed in Guo and Hay, 1999). An intriguing possibility, therefore, is that the destruction of Cdc6 deregulates the cell cycle during programmed cell death by activating cyclin-dependent kinases at the same time that it disrupts functional origin licensing complexes in cells with unreplicated or incompletely replicated chromosomes. Subsequent inhibition of the reassembly of these complexes by activated mitotic cyclin-dependent kinases, which normally provides an irreversible block to reinitiation in the same cell cycle as part of the origin licensing mechanism, would ensure that these cells would never recover from the ensuing cell cycle arrest. The irreversible nature of this state may be important for the commitment to programmed cell death.
This model has important implications for understanding how adozelesin and other drugs might exert their cytotoxic and antitumor effects, particularly in cells with defective p53 responses to DNA damage. For instance, nonproliferating cells do not have fully assembled origin licensing complexes (Williams et al., 1998; Madine et al., 2000), and down-regulation of Cdc6 and Mcm proteins in particular may be important for establishing the quiescent state during differentiation and replicative senescence (Stoeber et al., 2001). Thus, cells that are not in the proliferative state should be refractile to a pathway that targets Cdc6 and perhaps other origin licensing proteins. Because the destruction of Cdc6 induced by DNA damage does not require p53, normal proliferating cells may be protected from the consequences of Cdc6 destruction by p53-dependent DNA damage checkpoints, which, despite the loss of Cdc6, continue to restrain cyclin-dependent kinases until this damage is repaired.
The complexity of mammalian genomes and the large extent to which they are altered in cancer cells (Folkman et al., 2000; Anderson et al., 2001) complicates efforts to delineate cell death pathways and their relationship to antitumor action. The conservation of a proteasome-dependent pathway that destroys Cdc6 in a simple, genetically tractable organism such as budding yeast should facilitate efforts to dissect its mechanisms and potential relationships to programmed cell death. Indeed, we predicted the existence of a DNA damage response that targets origin licensing in mammals in our study of adozelesin effects in S. cerevisiae (Weinberger et al., 1999). This highlights the utility of yeasts as model organisms for studying complex pathways related to cancer and its effective treatment.
ACKNOWLEDGMENTS
We thank John Diffley and Lucille Drury for yeast strains and for advice and help in constructing a strain expressing AtCDC6, and Kristian Helin for plasmids. This research was supported by Public Health Service grants CA-84086, CA-81326 (to W.B.), and CA-08558 (to H.B.), and by shared resources funded by the Roswell Park Cancer Center support grant P30CA-16056. Research in the laboratory of C.G. is supported by grants BMC2000-1004 from MCyT, 07G/0033/2000, and an institutional grant from Fundacion Ramon Areces.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–02–0010. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–02–0010.
REFERENCES
- Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- Anderson GR, Stoler DL, Brenner BM. Cancer: the evolved consequence of a destabilized genome. Bioessays. 2001;23:1037–1046. doi: 10.1002/bies.1149. [DOI] [PubMed] [Google Scholar]
- Bhuyan BK, Smith KS, Adams EG, Petzold GL, McGovren JP. Lethality, DNA alkylation, and cell cycle effects of adozelesin (U-73975) on rodent and human cells. Cancer Res. 1992a;52:5687–5692. [PubMed] [Google Scholar]
- Bhuyan BK, Smith KS, Adams EG, Wallace TL, Von Hoff DD, Li LH. Adozelesin, a potent new alkylating agent: cell-killing kinetics and cell-cycle effects. Cancer Chemother Pharmacol. 1992b;30:348–354. doi: 10.1007/BF00689961. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Bueno A, Russell P. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992;11:2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada A, Sanchez M, Sanchez E, Bueno A. The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J Biol Chem. 2000;275:9734–9741. doi: 10.1074/jbc.275.13.9734. [DOI] [PubMed] [Google Scholar]
- Carty MP, Zernik-Kobak M, McGrath S, Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C. Expression, and stability of Arabidopsis CDC6 is associated with endoreplication. Plant Cell. 2001;13:2671–2686. doi: 10.1105/tpc.010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai F, Evdokiou A, Young GP, Zalewski PD. Involvement of p21(Waf1/Cip1) and its cleavage by DEVD-caspase during apoptosis of colorectal cancer cells induced by butyrate. Carcinogenesis. 2000;21:7–14. doi: 10.1093/carcin/21.1.7. [DOI] [PubMed] [Google Scholar]
- Cobuzzi RJ, Jr, Burhans WC, Beerman TA. Inhibition of initiation of simian virus 40 DNA replication in infected BSC-1 cells by the DNA alkylating drug adozelesin. J Biol Chem. 1996;271:19852–19859. doi: 10.1074/jbc.271.33.19852. [DOI] [PubMed] [Google Scholar]
- Dallaporta B, Pablo M, Maisse C, Daugas E, Loeffler M, Zamzami N, Kroemer G. Proteasome activation as a critical event of thymocyte apoptosis. Cell Death Differ. 2000;7:368–373. doi: 10.1038/sj.cdd.4400661. [DOI] [PubMed] [Google Scholar]
- Desdouets C, Santocanale C, Drury LS, Perkins G, Foiani M, Plevani P, Diffley JF. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase alpha. EMBO J. 1998;17:4139–4146. doi: 10.1093/emboj/17.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detweiler CS, Li JJ. Cdc6p establishes and maintains a state of replication competence during G1 phase. J Cell Sci. 1997;110:753–763. doi: 10.1242/jcs.110.6.753. [DOI] [PubMed] [Google Scholar]
- Diffley JFX. DNA replication: building the perfect switch. Curr Biol. 2001;11:R367–R370. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Vaughn JP, Hamlin JL. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991;11:3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Lou F, Wang B, Campbell JL, Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- Folkman J, Hahnfeldt P, Hlatky L. Cancer: looking outside the genome. Nat Rev Mol Cell Biol. 2000;1:76–79. doi: 10.1038/35036100. [DOI] [PubMed] [Google Scholar]
- Fujita M, Yamada C, Goto H, Yokoyama N, Kuzushima K, Inagaki M, Tsurumi T. Cell cycle regulation of human CDC6 protein. Intracellular localization, interaction with the human mcm complex, and CDC2 kinase-mediated hyperphosphorylation. J Biol Chem. 1999;274:25927–25932. doi: 10.1074/jbc.274.36.25927. [DOI] [PubMed] [Google Scholar]
- Furstenthal L, Kaiser BK, Swanson C, Jackson PK. Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J Cell Biol. 2001;152:1267–1278. doi: 10.1083/jcb.152.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Hay BA. Cell proliferation and apoptosis. Curr Opin Cell Biol. 1999;11:745–752. doi: 10.1016/s0955-0674(99)00046-0. [DOI] [PubMed] [Google Scholar]
- Hateboer G, Wobst A, Petersen BO, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Kellokumpu-Lehtinen P, Talpaz M, Harris D, Van Q, Kurzrock R, Estrov Z. Leukemia-inhibitory factor stimulates breast, kidney and prostate cancer cell proliferation by paracrine and autocrine pathways. Int J Cancer. 1996;66:515–519. doi: 10.1002/(SICI)1097-0215(19960516)66:4<515::AID-IJC15>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993a;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Nurse P, Forsburg SL. Coupling DNA replication to the cell cycle. Cold Spring Harb Symp Quant Biol. 1993b;58:637–644. doi: 10.1101/sqb.1993.058.01.071. [DOI] [PubMed] [Google Scholar]
- Kurokawa H, Nishio K, Fukumoto H, Tomonari A, Suzuki T, Saijo N. Alteration of caspase-3 (CPP32/Yama/apopain) in wild-type MCF-7, breast cancer cells. Oncol Rep. 1999;6:33–37. doi: 10.3892/or.6.1.33. [DOI] [PubMed] [Google Scholar]
- Levkau B, Koyama H, Raines EW, Clurman BE, Herren B, Orth K, Roberts JM, Ross R. Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: role of a caspase cascade. Mol Cell. 1998;1:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Jacks T. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, McHugh MM, Beerman TA, Melendy T. Adozelesin triggers DNA damage response pathways and arrests SV40 DNA replication through replication protein A inactivation. J Biol Chem. 2000;275:1391–1397. doi: 10.1074/jbc.275.2.1391. [DOI] [PubMed] [Google Scholar]
- Madine MA, Swietlik M, Pelizon C, Romanowski P, Mills AD, Laskey RA. The roles of the MCM, ORC, and Cdc6 proteins in determining the replication competence of chromatin in quiescent cells. J Struct Biol. 2000;129:198–210. doi: 10.1006/jsbi.2000.4218. [DOI] [PubMed] [Google Scholar]
- Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis [In Process Citation] Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Ott R, Fanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- Perkins G, Drury LS, Diffley JF. Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 2001;20:4836–4845. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM, Helin K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA replication origins during cell cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- Stewart CL. Leukemia inhibitory factor and the regulation of pre-implantation development of the mammalian embryo. Mol Reprod Dev. 1994;39:233–238. doi: 10.1002/mrd.1080390217. [DOI] [PubMed] [Google Scholar]
- Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH. DNA replication licensing and human cell proliferation. J Cell Sci. 2001;114:2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- Tannock IF, Hill RP. The Basic Science of Oncology. 3rd ed. New York: McGraw-Hill Book Company; 1998. [Google Scholar]
- Tavormina PA, Wang Y, Burke DJ. Differential requirements for DNA replication in the activation of mitotic checkpoints in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3315–3322. doi: 10.1128/mcb.17.6.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor AD, et al. Accumulation of p53 tumor suppressor gene protein: an independent marker of prognosis in breast cancers. J Natl Cancer Inst. 1992;84:845–855. doi: 10.1093/jnci/84.11.845. [DOI] [PubMed] [Google Scholar]
- Toyn JH, Johnson AL, Johnston LH. Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M-phase checkpoint. Mol Cell Biol. 1995;15:5312–5321. doi: 10.1128/mcb.15.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuner K, Okuyama A, Knippers R, Fackelmayer FO. Hyperphosphorylation of replication protein A middle subunit (RPA32) in apoptosis. Nucleic Acids Res. 1999;27:1499–1504. doi: 10.1093/nar/27.6.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Trabold PA, Lu M, Sharma K, Huberman JA, Burhans WC. Induction by adozelesin and hydroxyurea of origin recognition complex-dependent DNA damage and DNA replication checkpoints in Saccharomyces cerevisiae. J Biol Chem. 1999;274:35975–35984. doi: 10.1074/jbc.274.50.35975. [DOI] [PubMed] [Google Scholar]
- Williams GH, Romanowski P, Morris L, Madine M, Mills AD, Stoeber K, Marr J, Laskey RA, Coleman N. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci USA. 1998;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Chen HH, Stillman B. Inaugural article: binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc Natl Acad Sci USA. 2001;98:11211–11217. doi: 10.1073/pnas.201387198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, Williams RS. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]