ABSTRACT
Acinetobacter baumannii is highly resistant to antimicrobial agents, and XDR strains have become widespread. A. baumannii has developed resistance to colistin, which is considered the last resort against XDR Gram-negative bacteria, mainly caused by lipooligosaccharide (LOS) phosphoethanolamine (pEtN) and/or galactosamine (GalN) modifications induced by mutations that activate the two-component system (TCS) pmrAB. Although PmrAB of A. baumannii has been recognized as a drug resistance factor, its function as TCS, including its regulatory genes and response factors, has not been fully elucidated. In this study, to clarify the function of PmrAB as TCS, we elucidated the regulatory genes (regulon) of PmrAB via transcriptome analysis using pmrAB-activated mutant strains. We discovered that PmrAB responds to low pH, Fe2+, Zn2+, and Al3+. A. baumannii selectively recognizes Fe2+ rather than Fe3+, and a novel region ExxxE, in addition to the ExxE motif sequence, is involved in the environmental response. Furthermore, PmrAB participates in the phosphoethanolamine modification of LOS on the bacterial surface in response to metal ions such as Al3+, contributing to the attenuation of Al3+ toxicity and development of resistance to colistin and polymyxin B in A. baumannii. This study demonstrates that PmrAB in A. baumannii not only regulates genes that play an important role in drug resistance but is also involved in responses to environmental stimuli such as metal ions and pH, and this stimulation induces LOS modification. This study reveals the importance of PmrAB in the environmental adaptation and antibacterial resistance emergence mechanisms of A. baumannii.
IMPORTANCE
Antimicrobial resistance (AMR) is a pressing global issue in human health. Acinetobacter baumannii is notably high on the World Health Organization’s list of bacteria for which new antimicrobial agents are urgently needed. Colistin is one of the last-resort drugs used against extensively drug-resistant (XDR) Gram-negative bacteria. However, A. baumannii has become increasingly resistant to colistin, primarily by modifying its lipooligosaccharide (LOS) via activating mutations in the two-component system (TCS) PmrAB. This study comprehensively elucidates the detailed mechanism of drug resistance of PmrAB in A. baumannii as well as its biological functions. Understanding the molecular biology of these molecules, which serve as drug resistance factors and are involved in environmental recognition mechanisms in bacteria, is crucial for developing fundamental solutions to the AMR problem.
KEYWORDS: Acinetobacter baumannii, PmrAB, lipopolysaccharide, colistin, LPS modification
INTRODUCTION
Although antimicrobial agents have been used to treat Acinetobacter baumannii (a common Gram-negative bacterium) infections occurring in medical facilities, particularly in intensive care units (1–6), the prevalence of XDR A. baumannii strains has been increasing, limiting effective treatment options (7). Although broad-spectrum carbapenems are widely used to treat such infections (8), the prevalence of carbapenem-resistant A. baumannii (9) required a shift in therapeutic strategies (10).
Colistin, a cationic polypeptide antimicrobial agent, is critical in treating XDR A. baumannii infections, including those resistant to carbapenems (11, 12). Colistin binds to lipopolysaccharide (LPS) on the outer membrane of Gram-negative bacteria, destabilizing the cell membrane and leading to bactericidal activity (13). As the LPS in A. baumannii lacks O antigen and is instead characterized as LOS, the emergence of colistin-resistant A. baumannii strains has been reported, complicating treatment against these strains (11–15). The primary mechanism of colistin resistance in A. baumannii involves mutations in the two-component pmrAB gene system (16), which increase LOS modifications (phosphoethanolamine [pEtN] via pmrC and galactosamine [GalN] via naxD) and decrease colistin binding (17–20). However, the LOS modification that is crucial for colistin resistance is unclear.
PmrAB regulates the expression of specific genes (regulons) by activating (phosphorylating) the response regulator PmrA in response to environmental factors (21). A PmrAB regulon in Salmonella has been identified (22), with PmrB in Salmonella responding to extracellular environmental factors such as Fe3+, Al3+, and low pH (23–27), suggesting that PmrB responds to metal ions and pH changes. A. baumannii responds to low pH but does not respond to metal ions such as Fe3+ (28, 29). The environmental factors that PmrAB of A. baumannii responds to, excluding pH, are poorly understood. Moreover, the PmrAB regulon in A. baumannii has not been fully identified. Thus, PmrAB establishes drug resistance and regulates the specific regulons in response to unknown environmental factors but its biological mechanism remains undetermined. This study aimed to elucidate the molecular biology of the PmrAB regulon and its activators in A. baumannii to understand the role of PmrAB in A. baumannii response to environmental factors.
RESULTS
Elucidation of the PmrAB regulon in A. baumannii
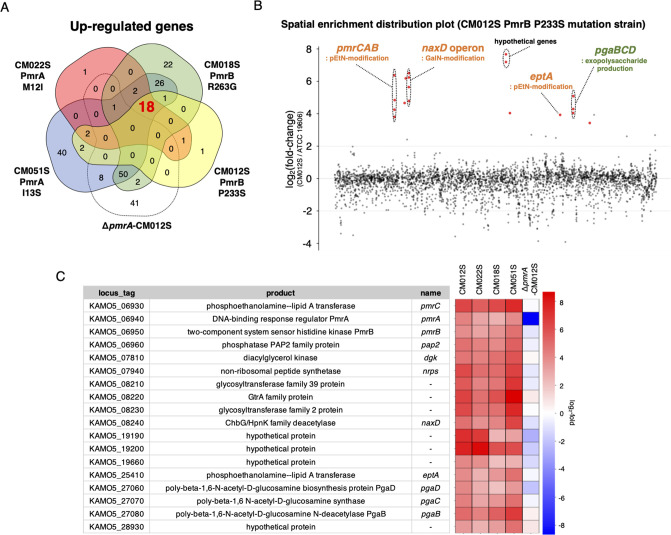
Previous research on the A. baumannii PmrAB system has predominantly utilized pmrAB mutant clinical isolates to deduce its regulon (30–32). However, this approach is limited by the lack of whole-genome sequences and the varying genetic backgrounds of different strains. A library of pmrAB-activating mutants with identical genetic backgrounds of A. baumannii has been successfully established using specific drug selection on the standard strain ATCC 19606 and the drug-resistant clinical isolate ATCC BAA-1605 (33). This library was utilized to select representative mutants for both pmrA and pmrB that were previously reported in clinical strains (PmrA: M12I, PmrB: P233S) (34–36) and those not reported (PmrA: I13S, PmrB: R263G). In addition to these four strains (Fig. S1), an artificial disruption of the pmrA gene was created in a pmrB-activating mutant strain (ΔpmrA-CM012S), thus forming a set of five representative strains in total. For ATCC BAA-1605, one representative strain was chosen from each pmrA- and B-activating mutant (PmrA: L20F and PmrB: P233S). Whole-genome analysis confirmed that these strains harbored no mutations other than those in pmrA and B. RNA-seq analysis of these strains identified 18 genes upregulated in the pmrAB mutant strain derived from ATCC 19606 strain, including those involved in autoregulation of the pmrCAB operon, and in addition to other LOS modification genes (eptA: pEtN modification gene and naxD), in the biosynthesis of polysaccharide poly-β-1,6-N-acetyl-D-glucosamine (pgaB, C, D; Fig. 1). All genes upregulated in the ATCC 19606 strain were found to be upregulated in the ATCC BAA-1605 pmrAB mutant strains as well, except for eptA that was not on the chromosome, thus resulting in 17 upregulated genes. Additional genes, including csuAB and three unknown genes, were upregulated in ATCC BAA-1605 (Fig. S2).
Fig 1.
Elucidation of the PmrAB regulon in A. baumannii by RNA-seq. (A) Genes with log2 (fold change) ≥ 2 and P value < 0.05 compared with the WT strain were considered significantly upregulated, and Venn diagrams were generated to illustrate this. The 18 genes common to the pmrA and B mutants but not to ΔpmrA-CM012S were identified as regulons. (B) A spatial enrichment distribution plot of CM012S (pmrB-activating mutant strain) is presented. The vertical axis indicates log2 (fold change), and the horizontal axis represents gene loci in the genome sequence. Red dots indicate regulon genes (18 genes). (C) List of the upregulated 18 regulon genes in the ATCC 19606 pmrAB-activating mutant strain is provided, which shows the locus tag of the ATCC 19606 strain (GenBank accession number: AP025740.1), its product, gene name, and expression levels of each gene in each strain, depicted on a heatmap.
Construction of a PmrAB reporter assay system for A. baumannii
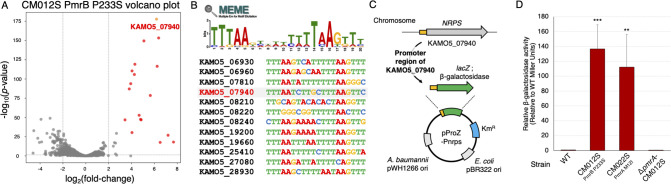
To elucidate the response of PmrAB in A. baumannii, we developed a reporter assay system using β-galactosidase activity. We identified KAMO5_07940, a gene locus encoding a non-ribosomal peptide synthase (NRPS), whose expression is significantly upregulated by PmrAB [P value = 2.20 × 10−178, log2 (fold change) = 6.18 in CM012S; Fig. 2A]. Analysis of the 200-bp upstream regions of the 18 upregulated genes using MEME Suite (37) revealed a DNA consensus sequence (-TTTAAN6TTTAA-) upstream of all genes (except those forming operons; Fig. 2B). This motif is also present upstream of the NRPS gene. Based on these findings, we constructed a reporter plasmid that utilizes the upstream region of the NRPS gene to precisely evaluate PmrAB activity (Fig. 2C). When the reporter plasmid was introduced into PmrAB-active mutant strains (CM022S PmrA M12I and CM012S PmrB P233S), the β-galactosidase activity drastically increased. This effect was not observed for the PmrA knockout strain (ΔpmrA-CM012S; Fig. 2D). This result confirmed the establishment of the reporter assay system for evaluating PmrAB activation in A. baumannii.
Fig 2.
Construction of the PmrAB reporter assay system for A. baumannii. RNA-seq analysis was conducted on the pmrB-activating mutant strain CM012S (P233S mutant), established from ATCC 19606 (WT strain). (A) A volcano plot comparing the CM012S strain with the WT strain is presented. The red dots (representing 17 genes) signify the PmrAB regulon. The gene KAMO5_07940, which is significantly upregulated [P value = 2.20 × 10−178, log2 (fold change) = 6.18], is highlighted with a yellow dot. (B) The 200-bp upstream region of the PmrAB regulon genes was extracted, and a consensus sequence predicted to bind PmrA was identified using the MEME Suite. (C) A schematic diagram of the reporter plasmid pProZ-Pnrps. Based on shuttle vectors for A. baumannii and E. coli, the lacZ gene encoding β-galactosidase was introduced downstream of the promoter region upstream of the KAMO5_07940 gene. (D) The pProZ-Pnrps plasmid was introduced into CM012S (pmrB mutation strain), CM022S (pmrA mutation strain), and pmrA disruption in pmrB-activating strain (pmrA-CM012S), and relative β-galactosidase activity was measured. Experiments were conducted independently in triplicate, and the results are presented as mean values with standard deviations represented by error bars. Statistical significance is shown as a reference of the WT strain mRNA levels using Dunnett’s test; ***P < 0.001, **P < 0.01.
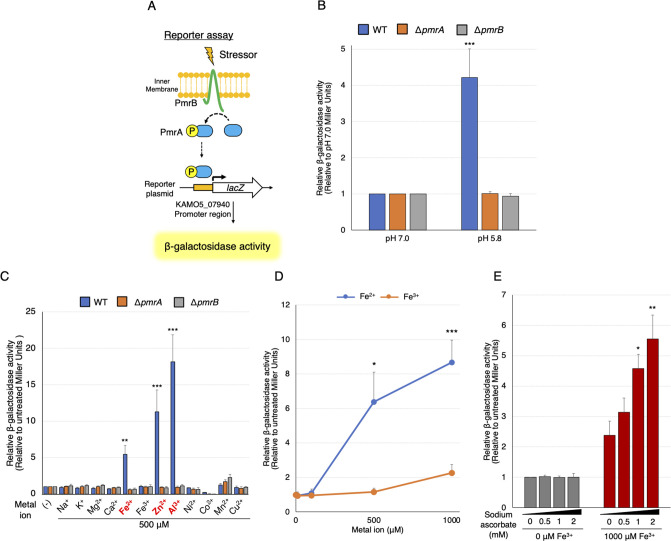
A. baumannii PmrAB responds to Fe2+, Zn2+, and Al3+
The wild-type (WT) ATCC 19606 strain and the ΔpmrA and ΔpmrB strains were transformed with the reporter plasmid. These strains were exposed to low pH (pH 5.8) and to 500 µM each of the various indicated metal ions, following which the β-galactosidase activity was measured (Fig. 3A). We detected high β-galactosidase activity at low pH (Fig. 3B). Furthermore, we discovered that PmrAB of A. baumannii responds to metal ions such as Fe2+, Zn2+, and Al3+ (Fig. 3C). The mRNA expression level of autoregulated pmrCAB increased in response to Fe2+, Zn2+, and Al3+ (Fig. S3A). A similar trend was observed for pH induction, although the difference was not statistically significant (Fig. S3B). Another strain of A. baumannii, ATCC 17978, similarly responded to pH 5.8, Fe2+, Zn2+, and Al3+ (Fig. S3C and S3D).
Fig 3.
Identification of PmrAB response factors in A. baumannii. (A) A schematic diagram of the PmrAB reporter assay in A. baumannii. Upon activation of PmrB, PmrA is phosphorylated and binds to the upstream region of the KAMO5_07940 gene, which induces lacZ expression and produces β-galactosidase. The β-galactosidase activity was evaluated after incubation at pH 7.0 or 5.8 (B) or with 500 µM NaCl, KCl, MgCl2, CaCl2, FeSO4, FeCl3, ZnSO4, AlCl3, NiSO4, Co(NO3)2, MnCl2, or CuSO4 (C) for 60 min. The values are expressed as relative values, with those at pH 7.0 or without metal ions set at 1.0. (D) Relative β-galactosidase activity is shown after 60 min of incubation with Fe2+ (FeSO4) or Fe3+ (FeCl3) at concentrations of 0, 100, 500, or 1,000 µM for the ATCC 19606 strain harboring the reporter plasmid. (E) β-galactosidase activity was evaluated after 60 min of incubation of the ATCC 19606 strain transfected with the reporter plasmid with or without 1,000 µM of FeCl3, and with 0, 0.5, 1, or 2 mM of ascorbic acid used as a reducing agent. The values are shown as relative values, with the value without FeCl3 addition set at 1.0. All experiments were performed in triplicate. Mean values and error bars represent standard deviations. Statistical significance is shown using Tukey’s test as a reference of pH 7.0 or untreated values (B, C), unpaired t-test compares Fe2+ and Fe3+ (D), and Dunnett’s test as a reference of the untreated value (E); ***P < 0.001, **P < 0.01, *P < 0.05.
Notably, PmrAB of A. baumannii did not respond to Fe3+, suggesting selective responsiveness to Fe2+. Next, we examined whether PmrAB discriminates between Fe2+ and Fe3+. The activity of β-galactosidase increased in a concentration-dependent manner in response to Fe2+ treatment but was comparatively low for Fe3+ (Fig. 3D). Furthermore, when we employed sodium ascorbate as a reducing agent to reduce Fe3+ to Fe2+ (38), β-galactosidase activity increased in a concentration-dependent manner (Fig. 3E). These results suggest that PmrAB of A. baumannii TCS selectively recognizes Fe2+ rather than Fe3+.
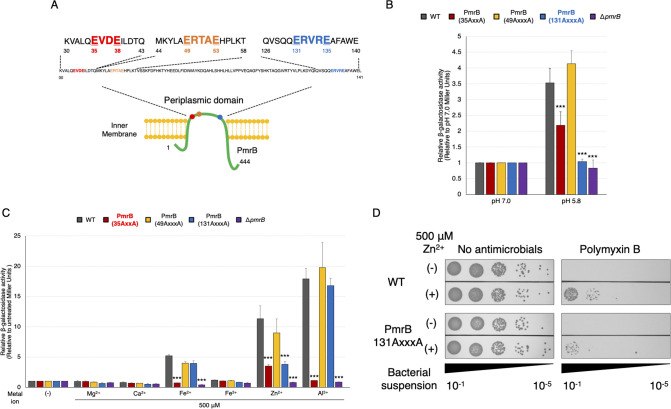
PmrB 35-ExxE-38 and 131-ExxxE-135 motifs in the periplasmic region are essential for response to low pH and metal ions in A. baumannii
In other species of PmrAB, the ExxE motif sequence in the periplasmic region of PmrB is essential for response to metal ions and low pH (25, 27). In A. baumannii PmrB, one ExxE (35-EVDE-38) motif was identified in the periplasmic region as predicted by TMHMM (39). Additionally, two ExxxE motifs (49-ERTAE-53 and 131-ERVRE-135), each with an extra-amino acid between the glutamate residues, were found in the periplasmic region (Fig. 4A). To determine which motifs are critical for the environmental response of A. baumannii PmrB, we generated mutants in which the glutamate in the ExxE/ExxxE motifs was replaced by alanine and conducted a reporter assay. These mutant strains were exposed to low pH, Fe2+, Zn2+, or Al3+, following which the activity of β-galactosidase was compared with that of the wild-type strain (PmrB WT). The 35-ExxE-38 motif was important for stimulation by low pH, Fe2+, Zn2+, or Al3+, as expected (Fig. 4B and C). Notably, the strain with alanine substituted for glutamate at 131-ExxxE-135 (strain 131-AxxxA-135) exhibited a considerably reduced response to low pH (Fig. 4B). Conversely, in the strain 131-AxxxA-135, responsiveness to Fe2+ and Al3+ did not change; however, responsiveness to Zn2+ was reduced to approximately 60% of that of the parent strain (Fig. 4C). Furthermore, the results showed that strain 131-AxxxA-135 stimulated by Zn2+ showed reduced resistance to polymyxin B, which was less responsive compared to the WT strain (Fig. 4D; Fig. S4). Hence, we found a functional motif 131-ExxxE-135 of PmrB. Moreover, the 49-ExxxE-53 motif had little effect on these environmental responses. These findings suggest that the 35-ExxE-38 and 131-ExxxE-135 motifs are the major amino acid sequences involved in the response to metal ions and low pH in PmrB.
Fig 4.
Altered responsiveness of A. baumannii PmrB motif sequence mutants to low pH and metal ions. (A) A schematic diagram of the amino acid sequence in the PmrB periplasmic region of A. baumannii is presented. Mutant strains with alanine substitutions for glutamate in the ExxE and ExxxE motifs were generated and transfected with a reporter plasmid. These were then incubated at pH 7.0 or 5.8 (B), and β-galactosidase activity was measured after incubation with 500 µM NaCl, KCl, MgCl2, CaCl2, FeSO4, FeCl3, ZnSO4, AlCl3, NiSO4, Co(NO3)2, MnCl2, or CuSO4 for 60 min (C). The values are presented as relative values, with those at pH 7.0 or without metal ions set at 1.0. All experiments were performed in triplicate. Mean values and error bars represent standard deviations. Statistical significance is shown as a reference of the ATCC 19606 (WT) strain value using Dunnett’s test; ***P < 0.001. (D) The WT and PmrB (131AxxxA) strains in the logarithmic growth phase were exposed to 500 µM ZnSO4 for 2 h. Subsequently, the bacterial suspension was adjusted to an OD600 of 0.2. After incubating the bacterial suspension with 7 µg/mL polymyxin B or without antimicrobial agents for 2 h, 10 µL of the 10-fold serial dilution of the bacterial suspension was plated onto LB agar plates, which were then incubated at 37°C for 24 h. Experiments were conducted in triplicate, and images of representative agar plates are shown.
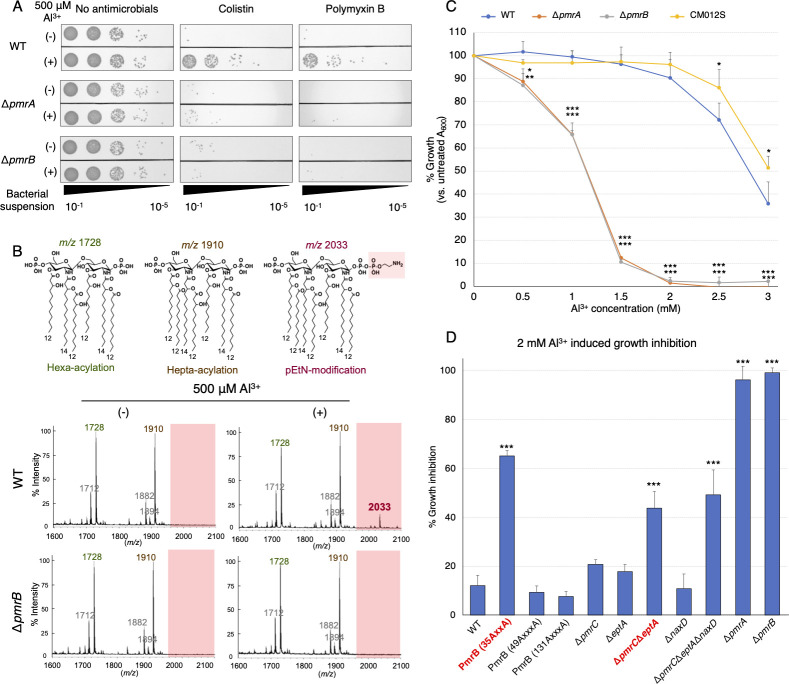
Mechanism of PmrAB-dependent lipid A modification and Al3+ toxicity avoidance in A. baumannii
Among the environmental factors, the most significant response, as detected by β-galactosidase activity for PmrAB, was elicited by Al3+; therefore, we evaluated whether PmrAB induces phenotypic changes in response to Al3+, including tolerance to colistin and polymyxin B, and lipid A modification. Stimulation by Al3+ induced resistance to colistin and polymyxin B in WT but not in the ΔpmrA and ΔpmrB strains (Fig. 5A). Next, we investigated the Al3+-induced LOS modification using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS). Al3+-stimulated lipid A exhibited a peak at m/z 2033, indicating phosphoethanolamine (pEtN) modification (Fig. 5B; Fig. S5). The lipid A modification peak was absent in the ΔpmrB strain. Furthermore, the minimum inhibitory concentrations (MICs) of colistin were assessed both in the presence and absence of Al3+ (Table 1). The results showed that the addition of 500 µM Al3+ led to an increase in MICs for both the WT and the ΔnaxD strain with a disrupted GalN modification gene. In contrast, the ΔeptA strain, associated with the pEtN modification, exhibited only a minor increase in colistin resistance. Notably, the ΔpmrC, ΔpmrCΔeptA, and ΔpmrCΔeptAΔnaxD strains did not show any increase in colistin resistance. These results indicated that PmrAB is activated by Al3+ stimulation, which results in phenotypic changes.
Fig 5.
A. baumannii PmrAB-dependent phenotypic changes induced by Al3+. (A) The ATCC 19606, ΔpmrA, and ΔpmrB strains in the logarithmic growth phase were exposed to 500 µM AlCl3 for 2 h. Subsequently, the bacterial suspension was adjusted to an OD600 of 0.2. After incubating the bacterial suspension with 10 µg/mL colistin or 7 µg/mL polymyxin B, or without any antimicrobial agent (control) for 2 h, 10 µL of a 10-fold serial dilution (ranging from 10−1 to 10−5) of the bacterial suspension was plated onto LB agar plates, which were then incubated at 37°C for 24 h. Experiments were conducted in triplicate, and images of representative agar plates are shown. (B)The ATCC 19606 (WT; middle row) and ΔpmrB strain (bottom row) in the logarithmic growth phase were exposed to 500 µM AlCl3 for 2 h. Lipid A was extracted from the bacteria, and MALDI-TOF/MS was used to analyze its structure. The predicted structure of lipid A and its mass-to-charge ratio peaks are shown at the top. (C) The WT, ΔpmrA, ΔpmrB, and CM012S (PmrB P233S-activated mutant strain) strains were prepared at an OD600 of 0.001 and were incubated with the various indicated concentrations of AlCl3 for 24 h. Absorbance A600 was monitored, and growth ability was expressed as a relative value, with the value without AlCl3 addition set at 100%. (D) PmrB motif mutant strains (ExxE/ExxxE alanine substitution strain) and various LOS modification gene disruption strains (ΔpmrCΔeptA, ΔnaxD, ΔpmrCΔeptAΔnaxD) were grown in LB broth containing 2 mM AlCl3 for 24 h. A600 of the bacteria was measured, and the relative growth inhibition was calculated by subtracting the value from 100%. All experiments were performed in triplicate. Mean values and error bars represent standard deviations. Statistical significance is shown as a reference of the WT strain value using Dunnett’s test; ***P < 0.001, **P < 0.01, *P < 0.05.
TABLE 1.
Colistin MIC of various mutant strains in the presence of Al3+
| Colistin (μg/mL) | WT | ΔpmrA | ΔpmrB | ΔpmrC | ΔeptA | ΔnaxD | ΔpmrC ΔeptA | ΔpmrC ΔeptA ΔnaxD |
|---|---|---|---|---|---|---|---|---|
| 0 µM Al3+ | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 500 µM Al3+ | 8 | 2 | 2 | 2 | 4 | 8 | 2 | 2 |
Treatment with excess Al3+ causes cytotoxicity in bacteria (40). Wild-type or PmrB-active strains (WT and CM012S) of A. baumannii also showed growth inhibition in an Al3+-dose-dependent manner; however, this inhibition effect was more pronounced for the PmrAB-knockout strains (Fig. 5C). Modification of LPS is crucial for avoiding metal toxicity via PmrAB activation in Salmonella spp. and Escherichia coli (27, 41, 42). We investigated the capacity of PmrAB to reduce the cytotoxicity caused by excessive Al3+ by employing an altered metal ion-responsive strain with alanine substitutions in the ExxE/ExxxE motif and a strain deficient in the pEtN and/or GalN modification genes, both of which are regulated by PmrAB. The PmrB 35-AxxA-38 strain, which is not responsive to metal ions, exhibited decreased growth under excess Al3+. Furthermore, disruption of the pEtN modification genes (ΔpmrCΔeptA) also led to significant growth inhibition (Fig. 5D). Conversely, the extent of growth inhibition in the strain with only the disruption of ΔpmrC, ΔeptA, and the GalN modification gene (ΔnaxD) was similar to that observed for the WT strain. These results suggested that PmrB senses excess Al3+ and modifies pEtN in LOS to reduce its toxicity.
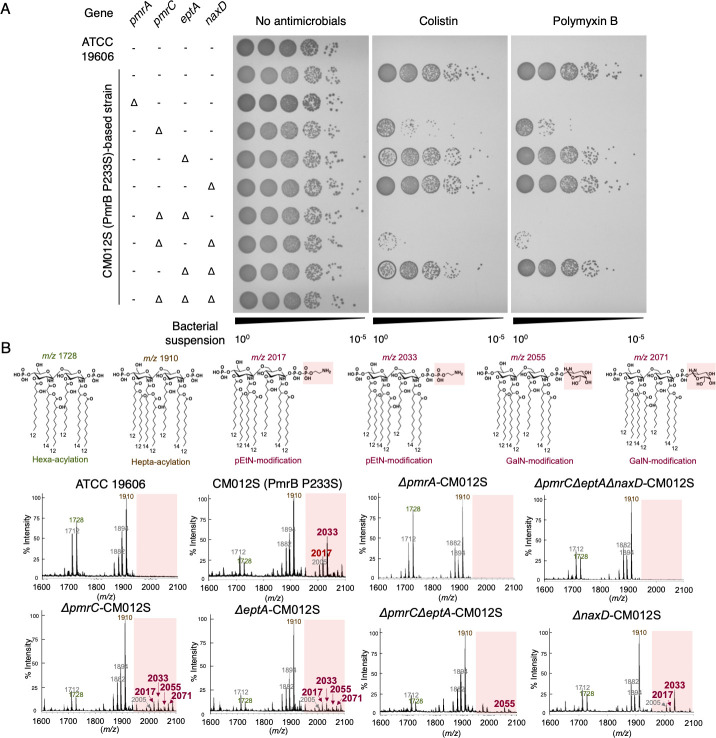
Phosphoethanolamine modification of lipid A is more important than galactosamine modification for colistin and polymyxin B resistance in A. baumannii
As described in the previous section, Al3+ stimulation of WT strain resulted in the appearance of a pEtN-modified peak at m/z 2033. In Salmonella spp., changes in surface charge due to LPS modification are crucial for avoiding metal toxicity and for polymyxin B resistance following PmrAB activation (24). Therefore, we investigated which among the two—pEtN or GalN—modifications of LOS is more crucial for colistin and polymyxin B resistance in PmrAB-active mutants of A. baumannii. Finally, we generated strains with various disrupted LOS modification-related genes from the colistin-resistant pmrB-activating mutant strain CM012S (Fig. S6A). We evaluated their survival on Luria-Bertani (LB) agar containing colistin and polymyxin B, and analyzed their LOS modifications using MALDI-TOF/MS. The results revealed that colistin and polymyxin B resistance was entirely lost in strains lacking pEtN modification-related genes (pmrC and eptA). In contrast, resistance was not diminished in strains lacking the GalN modification-related gene (naxD; Fig. 6A; Fig. S6B). The peak of pEtN modification of lipid A (m/z 2033), observed for strain CM012S, was absent for strains ΔpmrA-CM012S, ΔpmrCΔeptA-CM012S, and ΔpmrCΔeptAΔnaxD-CM012S. However, this peak was present for strain ΔnaxD-CM012S (Fig. 6B; Fig. S5), suggesting that lipid A modification of pEtN, and not that of GalN, plays a significant role in colistin/polymyxin B resistance.
Fig 6.
Effect of phosphoethanolamine and galactosamine modifications on colistin and polymyxin B resistance in A. baumannii. (A) Resistance to colistin and polymyxin B was evaluated for ATCC 19606 and CM012S (PmrB P233S-activating mutant strain) strains following the disruption of various LOS modification genes (pmrC, eptA, naxD), and in CM012S strain following the disruption of pmrA. The strains were cultured to reach a steady state, washed with PBS, and prepared in PBS to achieve an OD600 of 0.2. Subsequently, a 10-fold serial dilution (from 10−1 to 10−5) of the bacterial suspension was prepared. This diluted suspension was then plated onto LB agar medium containing 10 µg/mL colistin and 10 µg/mL polymyxin B, which was incubated for 24 h at 37°C. Experiments were performed independently in triplicate, and representative agar plates are shown. (B) Lipid A was extracted from each strain, and MALDI-TOF/MS was used to analyze its structure. The predicted structures of lipid A and their corresponding peak mass-to-charge ratios are displayed at the top.
There were two genes associated with pEtN modification: pmrC and eptA, each with the solely disrupted strains ΔpmrC-CM012S and ΔeptA-CM012S showing a pEtN modification peak (Fig. 6B). This suggests that both genes are involved in pEtN modification of LOS.
DISCUSSION
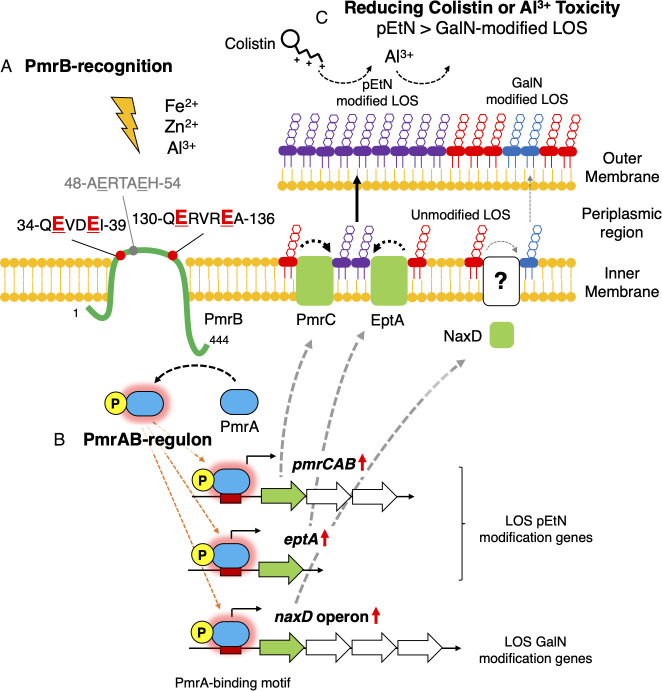
In this study, we aimed to elucidate the PmrAB regulon in A. baumannii. The analyses identified 18 upregulated genes (Fig. 1) that encode LOS modification and polysaccharide poly-β-1,6-N-acetyl-D-glucosamine-related enzymes. Strains with disrupted LOS modification genes were created and analyzed in this study, but there is a possibility that the extracellular polysaccharide synthase genes also change the bacterial surface layer (43). These findings align with those of previous studies that used colistin-resistant clinical isolates from diverse genetic backgrounds and suggest that the regulons identified here are broadly represented across A. baumannii strains (31, 32). Furthermore, KAMO5_07940, which encodes an NRPS, was upregulated in PmrAB-activated mutants. Although the crystal structure of this protein has been determined, its function remains unknown (44). NRPSs typically comprise an operon and synthesize a single compound via multiple NRPSs (45). However, the locus of KAMO5_07940 does not comprise an operon, which suggests that it may function independently. Future studies should elucidate the biological significance of this NRPS, which is upregulated by PmrAB. Additionally, a DNA consensus sequence was found upstream of these upregulated genes, excluding those forming an operon. This sequence correlates with the DNA-binding sequence predicted based on the PmrA protein structure (46), and PmrA is thought to bind to this motif sequence. Conversely, 12 genes were observed to be downregulated upon PmrAB activation (Fig. S7). Notably, this downregulation was not evident for the ATCC BAA-1605 strain (Fig. S8), indicating that the genetic background of individual strains influences the expression of these genes. The downregulated genes in the ATCC 19606 strain are predominantly involved in regulating intracellular iron concentrations. In contrast, the ATCC BAA-1605 strain exhibited decreased expression for a range of pili-related genes, suggesting that genes that are downregulated vary from strain to strain. Therefore, it cannot be discounted that genes downregulated by PmrAB activation may also play roles in modulating colistin resistance and virulence in A. baumannii (47, 48).
In this study, we found that the response of PmrB to iron ions is species dependent. Although Salmonella PmrB selectively recognizes Fe3+ (27), PmrB of A. baumannii is activated by Fe3+ under low pH conditions but not by Fe3+ alone (28). This suggests that because Fe3+ efficiently reduces to Fe2+ under acidic solutions, it is the reduced Fe2+ that activates PmrB. Unlike Salmonella PmrB, which does not recognize Zn2+, E. coli PmrB does, and A. baumannii PmrB recognizes Zn2+ as well (27, 41). We identified regions in PmrB motif sequences essential for responsiveness to metal ions and pH. The results revealed the importance of the A. baumannii PmrB-specific ExxxE sequence, in addition to the previously considered ExxE motif, for eliciting responses to pH and Zn2+. In future studies, it remains to be elucidated in detail whether these differences in metal selectivity are due to differences in amino acid sequences in and around the motif sequence or in the number of amino acid residues in the periplasmic region.
We found that PmrAB protects against excess Al3+ by changing the LOS in response to Al3+, which is widely distributed in the environment and is mainly found in soil and rivers after acid rain (49). PmrAB senses excess Al3+ in the environment and upregulates the expression of the regulon’s pEtN modification genes. This modification of LOS counteracts the negative charge on the bacterial surface and suppresses the effects of cations like Al3+ and colistin. Metal ion-induced PmrAB activation further induces LPS modification in Salmonella spp. (27). Although both pEtN and 4-amino-4-deoxy-L-arabinose (L-Ara4N) glycosylation modifications of LPS have been reported for Salmonella spp. and E. coli, L-Ara4N modification is more crucial for polymyxin B resistance than pEtN modification (24, 50). A. baumannii does not possess the L-Ara4N modification genes (such as aminoarabinose transferase arnT gene); however, it possesses a similar glycosylation (GalN) modification gene—the naxD. NaxD is a deacetylase and is unlikely to perform GalN modification alone. Activated PmrAB also enhanced the expression of three genes (KAMO5_08210, KAMO5_08220, and KAMO5_08230) downstream of the naxD gene (KAMO5_08240), named the naxD operon, suggesting that these genes are responsible for GalN modification in a coordinated manner. The mRNA levels of the naxD operon increased during Al3+ stimulation (Fig. S9A). We thus concluded that, for colistin and polymyxin B resistance and the avoidance of Al3+ toxicity in A. baumannii, pEtN modification is more critical than GalN modification by the naxD operon. These differences could be attributed to structural differences in the LPS of Salmonella spp. and E. coli and that of A. baumannii, such as the presence or absence of the O antigen (51–53), or differences in the number of acyl groups (12, 54, 55). A recent study on lipid A structure analysis of polymyxin B-resistant A. baumannii clinical strains reported the presence of pEtN modification but not that of GalN modification (56), which aligns with our results on PmrAB mutant strains (Fig. 6; Fig. S7). Although a small GalN modification peak (m/z 2071 and 2055) was observed for the pEtN modification gene disruption strains (ΔpmrC-CM012S, ΔeptA-CM012S, ΔpmrCΔeptA-CM012S; Fig. 6B), the involvement of the naxD gene in LOS modification cannot be completely excluded. Because a GalN modification peak was observed in a strain in which the pEtN modification genes were disrupted, pEtN and GalN modification may competitively compete for the phosphate group, which is the modification site of lipid A. Therefore, the naxD operon, regulated by PmrAB, may have roles in LOS modification and other functions.
The eptA gene is activated following insertion into its promoter region by the insertion sequence ISAba1, a process associated with the acquisition of resistance to colistin and polymyxin B (57, 58). The expression of eptA may be regulated by PmrAB, but this was not experimentally demonstrated. Increased eptA expression in PmrAB-activating mutants and Al3+-stimulated WT strain (Fig. 1; Fig. S9B) and the presence of a putative PmrA-binding sequence in the eptA promoter region was confirmed (Fig. 2B). Moreover, a pEtN modification peak of lipid A was observed when only ΔpmrC or ΔeptA was disrupted, but this modification peak disappeared in the strain with double disruption of ΔpmrCΔeptA (Fig. 6B). This suggests that both pmrC and eptA are genes involved in the pEtN modification of lipid A. However, based on differences in the growth of pmrC and eptA mutants in colistin and polymyxin B agar, pmrC may contribute more to resistance to those antimicrobials (Fig. 6A). The reason may be due to differences in the expression levels of both genes, whereby pmrC is more highly expressed in PmrAB-active mutants than eptA (Fig. 1C). In the A. baumannii reference strain ATCC 19606, both the pmrC and eptA genes are present on the chromosome, but ATCC BAA-1605 does not have an eptA gene. Conversely, some clinical isolates possess multiple eptA genes (57, 58). The detailed relationship between pmrC and eptA and the impact of eptA copy number on colistin resistance are topics for future research.
In this study, we comprehensively analyzed the PmrAB regulon and its responses in A. baumannii (Fig. 7). We revealed that PmrAB, previously considered to be solely involved in the drug resistance mechanism, also exhibits biological functions in response to various environmental factors, such as metal ions, in addition to colistin resistance. The biological functions of PmrAB, as revealed in this study, are essential for understanding drug resistance, environmental adaptation, and infection in bacteria, which contributes to a more comprehensive understanding of A. baumannii.
Fig 7.
Schematic diagram of phosphoethanolamine modification of lipopolysaccharide by PmrAB of A. baumannii in response to metal ions. (A) PmrAB of A. baumannii is a two-component system that governs responses to Fe2+, Zn2+, and Al3+. The 35-ExxE and 131-ExxxE motif sequences located in the periplasmic region of PmrB, a sensor kinase, are critical to these responses. (B) Among the genes activated by PmrAB are those responsible for LOS modification, which include pmrC and eptA genes for pEtN modification and naxD operon for GalN modification. (C) The pEtN modification of LOS is crucial for reducing Al3+ toxicity and for resistance to colistin. In A. baumannii, PmrAB is activated in response to metal ions, which leads to an upregulation of pEtN modification genes. This upregulation contributes to colistin and Al3+ toxicity avoidance by facilitating the modification of LOS with pEtN.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The type strain of A. baumannii ATCC 19606 and drug-resistant clinical isolate ATCC BAA-1605 were acquired from ATCC (Manassas, VA, USA). The bacteria were incubated in LB broth (BD Biosciences, San Diego, CA, USA) at 37°C for 18 h with shaking at 135 rpm. Colistin-resistant strains of A. baumannii were established in a previous study (33).
Plasmid construction
The pSBKT3 (kanamycin resistance gene; counter-selection gene: sacB) and pSBKT5v2 (kanamycin resistance gene; counter-selection gene: tdk) plasmids were used for homologous recombination. The reporter assay also used the pProZ-Pnrps plasmid (kanamycin resistance gene; reporter marker: lacZ). Each target gene of A. baumannii was subcloned into these plasmids using In-Fusion cloning (Takara Bio, Shiga, Japan). The primers used for this process are listed in Table S1 and S2. These plasmids were then transformed either into A. baumannii by electroporation or into S17-1 λpir by heat shock.
Generation of A. baumannii mutant strains by two-step homologous recombination
Genetic mutants were created using a two-step homologous recombination method. S17-1 λpir harboring plasmids pSBKT3 or pSBKT5v2 and A. baumannii were conjugated on LB agar for 3–6 h. Subsequently, the bacteria were harvested and selected for A. baumannii on M9 agar (Sigma-Aldrich, St. Louis, MO, USA) containing 0.3% citric acid as the carbon source and 50 µg/mL kanamycin (both from Fujifilm Wako Pure Chemical, Osaka, Japan). The colonies formed were collected by blue-white selection on X-gal agar (Shimadzu Diagnostics, Tokyo, Japan) containing 50 µg/mL kanamycin. For pSBKT3, the counter-selection was conducted by growing the recombinant A. baumannii on LB agar containing 20% sucrose (Fujifilm Wako Pure Chemical) and incubating for 18–20 h at 37°C. Strains with pSBKT5v2 were counter-selected after induction by isopropyl β-D-1-thiogalactopyranoside (IPTG) for tdk. For strains that completed the first recombination using pSBKT5v2, A. baumannii was inoculated into LB broth containing 50 µg/mL kanamycin and was cultured at 37°C for 18–20 h with shaking at 135 rpm. This bacterial culture was further cultivated in LB broth without antibiotics until the OD600 reached 0.5–0.8, at which point IPTG (Fujifilm Wako Pure Chemical) was added to a final concentration of 1 mM and was cultured for 3 h. Counter-selection was then performed using LB agar coated with 100 µL of 200 µg/mL azidothymidine (Tokyo Chemical Industry, Tokyo, Japan).
Whole-genome sequencing
Genomic DNA was extracted from bacteria incubated for 18 h at 37°C using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) and Genomic-tip (Qiagen). Sequencing libraries were prepared by Azenta (Chelmsford, MA, USA) or Genome-Lead (Kagawa, Japan). The library was prepared using the NEBNext Ultra II FS DNA Library Preparation Kit for Illumina (New England BioLabs, Ipswich, MA, USA) or Illumina DNA Prep Tagmentation (Illumina, San Diego, CA, USA), following the manufacturer’s instructions. Sequencing was conducted on an Illumina HiSeq X Ten or NovaSeq 6000 sequencer (Illumina) using the 2 × 150 bp sequencing format.
To obtain the complete reference genome sequence of ATCC BAA-1605, long-read sequencing analysis was conducted using Oxford Nanopore sequencing technology (Oxford Nanopore Technologies, Oxford, UK). After preparing barcoded libraries with the Rapid Barcoding Kit (Oxford Nanopore Technologies), sequence analysis was performed on the MinION Mk1B (Oxford Nanopore Technologies), with base calls generated using Guppy v6.4.6 (Oxford Nanopore Technologies) (59). Whole-genome sequences were assembled using Unicycler v0.5.0 (60). Open reading frames and RNA regions were annotated using Prokka v1.14.5 (61) with GCA_025995075.1 as the primary database.
For mutation analysis, fastq data were quality checked and trimmed using TrimGalore v0.6.10. The sequences were then annotated using the ATCC 19606 genome sequence (GenBank accession number: AP025740.1) and the ATCC BAA-1605 genome sequence (GenBank accession number: AP029033.1-AP029034.1) as references. Mutation analysis was performed using breseq v0.36.0 (62).
RNA sequencing
Total RNA from A. baumannii showing logarithmic growth was extracted using NucleoSpin RNA (Macherey-Nagel) according to the manufacturer’s instructions (three biological replicates). The library for sequencing was prepared by the Genome-Lead using the MGIEasy Adapters-96 kit (MGI, Shenzhen, China), and 2 × 150 bp sequences were obtained using the DNBSEQ T7 sequencer (MGI).
Fastq files were quality assessed, and adapters were removed using TrimGalore v0.6.10. These trimmed reads were then mapped to the reference genomes of A. baumannii ATCC 19606 (GenBank accession number: AP025740.1) and ATCC BAA-1605 (GenBank accession number: AP029033.1-AP029034.1) using HISAT2 v2.2.1 (63). The mapping data were sorted with SAMtools v1.17 (64), and aligned reads in the open reading frames of the genome were counted using the Subread package featureCounts v2.0.1 (65). Differences in gene expression were analyzed using the DESeq2 v1.34.0 package (Bioconductor) (66). Genes with an adjusted P value < 0.05 and |log2 (fold change)| ≥ 2 were considered differentially expressed genes.
Venn diagrams, heatmaps, volcano plots, and genome distribution plots were generated using Python modules venn, venn2, Biopython, and seaborn, and by calculating and drawing custom Venn diagrams (https://bioinformatics.psb.ugent.be/webtools/Venn/). Homologs of ATCC 19606 and ATCC BAA-1605 were defined in BLASTp with an e-value ≤ 1e-10 and % identity ≥ 90%.
Real-time and endpoint PCR
Total RNA from the indicated A. baumannii strains was extracted using TRI-Reagent (Molecular Research Center, Cincinnati, OH, USA). The total RNA was treated with DNase I (Nippon Gene, Tokyo, Japan) to remove genomic DNA; reverse transcription from RNA to cDNA was performed using the PrimeScript RT Reagent Kit (Takara Bio); and real-time PCR was performed using SYBR Green ExTaq II (Takara Bio) with primers listed in Table S3. The amplification and detection of PCR products were carried out on a Thermal Cycler Dice Real Time System III (Takara Bio). Endpoint PCR was performed using GoTaq Master Mix (Table S4; Promega, Madison, WI, USA) in constant cycles.
Reporter assay
Strains harboring the reporter plasmid were incubated in LB broth containing 50 µg/mL kanamycin for 18 h at 37°C with shaking at 135 rpm. The culture was then diluted 100-fold in LB broth containing 50 µg/mL kanamycin and was incubated at 37°C with shaking at 135 rpm to reach an OD600 of 0.1–0.5. Subsequently, the strains were exposed to specific stresses [500 µM NaCl, KCl, MgCl2, CaCl2, FeSO4, FeCl3, ZnSO4, AlCl3, NiSO4, Co(NO3)2, MnCl2, or CuSO4 (Fujifilm Wako Pure Chemical), or pH 5.8] for 60 min at 37°C. The bacteria were then lysed using toluene (Fujifilm Wako Pure Chemical; 20-µL toluene for 700 µL of suspension), following which the toluene was evaporated by incubation at 37°C for 60 min. The lysate was mixed with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol; all from Fujifilm Wako Pure Chemical). The lysate was then mixed with 20 µL of o-nitrophenyl-β-D-galactopyranoside solution (4 mg/mL o-nitrophenyl-β-galactoside [Fujifilm Wako Pure Chemical] in 100-mM sodium phosphate buffer, pH 7.0), following which 100 µL of the solution was added to a 96-well plate for 15–60 min at 28°C. The reaction was stopped by adding 50 µL of 1 M Na2CO3 (Fujifilm Wako Pure Chemical). Absorbance A420 and A600 were measured using Varioskan (Thermo Fisher Scientific, Waltham, MA, USA), and β-galactosidase activity was calculated in Miller Units and then expressed as a relative value. The reaction equation used is as follows: Miller Units = 1,000 × [A420(x min) − A420(0 min)] / [reaction time (x min) × reaction vol (mL) × A600].
Short-term polymyxin bactericidal assay
The resistance of bacteria to short-term exposure to polymyxins was evaluated by exposing the bacteria to an antimicrobial agent for 2 h and subsequently detecting the bacteria that survived this exposure. Induction with Al3+ was performed by shaking at 135 rpm for 2 h, with or without the addition of 500 µM AlCl3. The bacteria were washed twice with phosphate-buffered saline (PBS) and were adjusted to an OD600 of 0.2. This solution was then incubated with 10 µg/mL colistin and 7 µg/mL polymyxin B for 2 h. After sterilization, a 10-fold dilution series of the bacterial solution was prepared, and 10 µL of each dilution was plated onto LB agar plates without any antimicrobial agents. The plates were incubated for 18–20 h at 37°C. The number of colonies on the plates was counted to evaluate the effectiveness of the sterilization process.
Long-term polymyxin bactericidal assay
The bacterial solution was incubated for 18 h in LB broth at 37°C with shaking at 135 rpm, washed with PBS, and prepared in PBS to achieve an OD600 of 0.2. A 10-fold dilution series of this solution was prepared, and 10 µL of each dilution was plated onto LB agar containing 10 µg/mL colistin and 10 µg/mL polymyxin B. These plates were incubated for 18–20 h at 37°C. The effectiveness of the bactericidal treatment was assessed by counting the number of colonies.
Metal-induced bacterial growth inhibition assay
The bacteria were initially incubated in LB broth for 18 h at 37°C with shaking at 135 rpm. Subsequently, these were prepared in LB broth containing varying concentrations of AlCl3 (0, 0.5, 1, 1.5, 2, 2.5, or 3 mM) to achieve an OD600 of 0.001. This bacterial suspension was incubated in 96-well plates for 24 h at 37°C. The absorbance at A600 was measured using Varioskan, and the relative values of A600 in the presence and absence of metal ions were calculated. This process was conducted to evaluate the bacteria’s growth and growth inhibition capabilities.
Analysis of lipid A by MALDI-TOF/MS
Lipid A of LOS was extracted using the ammonium hydroxide/isobutyric acid method (67), with some modifications. Briefly, lyophilized bacteria from 3 or 10 mL of culture medium that had been incubated for 18 h at 37°C with shaking were resuspended in 400 µL of isobutyric acid/1 M ammonium hydroxide (5:3, vol/vol; Fujifilm Wako Pure Chemical) and were heated for 2 h at 100°C. After cooling, the samples were centrifuged at 2,000 × g for 15 min at 4°C. An equal volume of ultrapure water was added to the supernatant, which was then lyophilized. The residue was washed twice with 1 mL of methanol (Fujifilm Wako Pure Chemical) and was centrifuged at 2,000 × g for 15 min at 4°C. The insoluble lipid A was solubilized in 1 mL of chloroform/methanol/water (3:1.5:0.25, vol/vol/vol; Fujifilm Wako Pure Chemical) and was centrifuged at 10,000 × g for 3 min at 4°C, following which the supernatant was collected. The collected supernatant was mixed with 1 mL of water and was centrifuged at 400 × g for 30 s at 4°C, following which the lipid A fraction (lower layer) was collected. The lipid A fraction from which the solvent was removed by flushing with N2 gas was dissolved in 100 µL of chloroform/methanol (2:1, vol/vol) and was mixed with an equal volume of a matrix (10 mg/mL 2,5-dihydroxybenzoic acid in water/methanol [7:3, vol/vol] containing 0.1% [wt/vol] trifluoroacetic acid) on a target plate. MALDI-TOF/MS analysis was performed using an UltrafleXtreme-DHS2 TOF/TOF (Bruker Daltonics, Billerica, MA, USA) instrument in negative-reflective mode. The chemical modifications of lipid A were evaluated based on the obtained MALDI-TOF mass spectra.
Determination of colistin MICs
The MICs for colistin were determined using the microdilution method in LB broth (BD Biosciences) as previously described (68). Briefly, the bacterial culture was adjusted to an OD600 of 0.001 using freshly prepared LB broth, with or without the addition of 500 µM Al3+ from overnight cultures. The MICs for colistin were monitored using a twofold dilution series starting at 256 µg/mL.
Statistical analysis
Statistical analysis was performed using R software (R Foundation for Statistical Computing, Vienna, Austria). Data are expressed as means ± standard deviation and were compared using unpaired t-test or the one-way analysis of variance and Dunnett’s test or Tukey’s test. Differences with P values of < 0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank Atsushi Yokotani (Kyoto Pharmaceutical University) and Tohru Miyoshi-Akiyama (National Center for Global Health and Medicine) for their helpful discussions regarding this study. We also thank Tomoka Nakamura, Tomoko Okada, Sayaka Nagamine, Yurika Hishikawa, Mitsuki Okuda (Kyoto Pharmaceutical University), and Yu Sakurai (National Center for Global Health and Medicine) for their technical assistance. We thank the Open Facility, Global Facility Center, Creative Research Institution, Hokkaido University for allowing us to conduct the lipid A analysis using UltrafleXtreme-DHS2 TOF/TOF (Bruker Daltonics). We thank Editage for English language editing.
This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Science Research Promotion Fund to G.K. (17K16230, 20K17475), the Kanehara Ichiro Memorial Foundation to G.K., the Morinomiyako Medical Research Foundation to G.K., the Kyoto Pharmaceutical University Fund for the Promotion of Scientific Research to G.K., and a grant from Agency for Medical Research and Development (AMED) to K.Y. (22fk0108611h0902). The funders had no role in the study design, data collection, interpretation, or the decision to submit the work for publication.
Methodology: N.Y. and G.K.; formal analysis: N.Y. and G.K.; investigation: N.Y., G.K., T.S., D.Y., M.M., R.Y., N.K., R.K., and N.T.; writing – original draft: N.Y. and G.K.; supervision: Y.M., M.F., S.Y., and K.Y.
The authors declare no competing financial interests.
Contributor Information
Go Kamoshida, Email: kamoshida@my-pharm.ac.jp.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health, Bethesda, Maryland, USA
DATA AVAILABILITY
The original data sets are available in publicly accessible repositories. The genome sequencing data of representative A. baumannii isolates from this study have been deposited in the DNA Data Bank of Japan under BioProject accession numbers PRJDB12922 and PRJDB16715. The RNA sequencing data of A. baumannii isolates were also registered (BioProject accession number PRJDB16717). Additionally, the GenBank file for the whole-genome sequence data of ATCC BAA-1605 (BioProject accession number PRJDB16714) has been registered with the DNA Data Bank of Japan.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jb.00435-23.
Figures S1 to S9.
Tables S1 to S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Mea HJ, Yong PVC, Wong EH. 2021. An overview of Acinetobacter baumannii pathogenesis: motility, adherence and biofilm formation. Microbiol Res 247:126722. doi: 10.1016/j.micres.2021.126722 [DOI] [PubMed] [Google Scholar]
- 2. Bassetti M, Vena A, Battaglini D, Pelosi P, Giacobbe DR. 2020. The role of new antimicrobials for Gram-negative infections in daily clinical practice. Curr Opin Infect Dis 33:495–500. doi: 10.1097/QCO.0000000000000686 [DOI] [PubMed] [Google Scholar]
- 3. Gonzalez-Villoria AM, Valverde-Garduno V. 2016. Antibiotic-resistant Acinetobacter baumannii increasing success remains a challenge as a nosocomial pathogen. J Pathog 2016:7318075. doi: 10.1155/2016/7318075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bassetti M, Righi E. 2015. New antibiotics and antimicrobial combination therapy for the treatment of gram-negative bacterial infections. Curr Opin Crit Care 21:402–411. doi: 10.1097/MCC.0000000000000235 [DOI] [PubMed] [Google Scholar]
- 5. Giamarellou H, Antoniadou A, Kanellakopoulou K. 2008. Acinetobacter baumannii: a universal threat to public health? Int J Antimicrob Agents 32:106–119. doi: 10.1016/j.ijantimicag.2008.02.013 [DOI] [PubMed] [Google Scholar]
- 6. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- 7. Doi Y, Husain S, Potoski BA, McCurry KR, Paterson DL. 2009. Extensively drug-resistant Acinetobacter baumannii. Emerg Infect Dis 15:980–982. doi: 10.3201/eid1506.081006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi: 10.1128/AAC.00296-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- 10. Nguyen M, Joshi SG. 2021. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review. J Appl Microbiol 131:2715–2738. doi: 10.1111/jam.15130 [DOI] [PubMed] [Google Scholar]
- 11. Almutairi MM. 2022. Synergistic activities of colistin combined with other antimicrobial agents against colistin-resistant Acinetobacter baumannii clinical isolates. PLoS One 17:e0270908. doi: 10.1371/journal.pone.0270908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Powers MJ, Trent MS. 2018. Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Mol Microbiol 107:47–56. doi: 10.1111/mmi.13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cafiso V, Stracquadanio S, Lo Verde F, Gabriele G, Mezzatesta ML, Caio C, Pigola G, Ferro A, Stefani S. 2018. Colistin resistant A. baumannii: genomic and transcriptomic traits acquired under colistin therapy. Front Microbiol 9:3195. doi: 10.3389/fmicb.2018.03195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qureshi ZA, Hittle LE, O’Hara JA, Rivera JI, Syed A, Shields RK, Pasculle AW, Ernst RK, Doi Y. 2015. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin Infect Dis 60:1295–1303. doi: 10.1093/cid/civ048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Z, Chen Y, Fang Y, Wang X, Chen Y, Qi Q, Huang F, Xiao X. 2015. Meta-analysis of colistin for the treatment of Acinetobacter baumannii infection. Sci Rep 5:17091. doi: 10.1038/srep17091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karakonstantis S. 2021. A systematic review of implications, mechanisms, and stability of in vivo emergent resistance to colistin and tigecycline in Acinetobacter baumannii. J Chemother 33:1–11. doi: 10.1080/1120009X.2020.1794393 [DOI] [PubMed] [Google Scholar]
- 17. Gogry FA, Siddiqui MT, Sultan I, Haq QMR. 2021. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front Med (Lausanne) 8:677720. doi: 10.3389/fmed.2021.677720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chin CY, Gregg KA, Napier BA, Ernst RK, Weiss DS. 2015. A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 59:7911–7914. doi: 10.1128/AAC.00515-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock REW. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751. doi: 10.1128/AAC.00256-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16:284–290. doi: 10.1016/j.tim.2008.03.007 [DOI] [PubMed] [Google Scholar]
- 23. Kato A, Higashino N, Utsumi R. 2017. Fe(3+)-dependent epistasis between the CpxR-activated loci and the PmrA-activated LPS modification loci in Salmonella enterica. J Gen Appl Microbiol 62:286–296. doi: 10.2323/jgam.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 24. Kato A, Chen HD, Latifi T, Groisman EA. 2012. Reciprocal control between a bacterium’s regulatory system and the modification status of its lipopolysaccharide. Mol Cell 47:897–908. doi: 10.1016/j.molcel.2012.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez JC, Groisman EA. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol 63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nishino K, Hsu F-F, Turk J, Cromie MJ, Wösten M, Groisman EA. 2006. Identification of the lipopolysaccharide modifications controlled by the Salmonella PmrA/PmrB system mediating resistance to Fe(III) and Al(III). Mol Microbiol 61:645–654. doi: 10.1111/j.1365-2958.2006.05273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wösten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113–125. doi: 10.1016/s0092-8674(00)00092-1 [DOI] [PubMed] [Google Scholar]
- 28. Ko SY, Kim N, Park SY, Kim SY, Shin M, Lee JC. 2023. Acinetobacter baumannii under acidic conditions induces colistin resistance through PmrAB activation and lipid A modification. Antibiotics (Basel) 12:813. doi: 10.3390/antibiotics12050813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Silva PM, Kumar A. 2019. Signal transduction proteins in Acinetobacter baumannii: role in antibiotic resistance, virulence, and potential as drug targets. Front Microbiol 10:49. doi: 10.3389/fmicb.2019.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wright MS, Jacobs MR, Bonomo RA, Adams MD. 2017. Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. mBio 8:e02193-16. doi: 10.1128/mBio.02193-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park YK, Lee JY, Ko KS. 2015. Transcriptomic analysis of colistin-susceptible and colistin-resistant isolates identifies genes associated with colistin resistance in Acinetobacter baumannii. Clin Microbiol Infect 21:765. doi: 10.1016/j.cmi.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 33. Kamoshida G, Yamada N, Nakamura T, Yamaguchi D, Kai D, Yamashita M, Hayashi C, Kanda N, Sakaguchi M, Morimoto H, Sawada T, Okada T, Kaya Y, Takemoto N, Yahiro K. 2022. Preferential selection of low-frequency, lipopolysaccharide-modified, colistin-resistant mutants with a combination of antimicrobials in Acinetobacter baumannii. Microbiol Spectr 10:e0192822. doi: 10.1128/spectrum.01928-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Charretier Y, Diene SM, Baud D, Chatellier S, Santiago-Allexant E, van Belkum A, Guigon G, Schrenzel J. 2018. Colistin heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob Agents Chemother 62:e00788-18. doi: 10.1128/AAC.00788-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mustapha MM, Li B, Pacey MP, Mettus RT, McElheny CL, Marshall CW, Ernst RK, Cooper VS, Doi Y. 2018. Phylogenomics of colistin-susceptible and resistant XDR Acinetobacter baumannii. J Antimicrob Chemother 73:2952–2959. doi: 10.1093/jac/dky290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dafopoulou K, Xavier BB, Hotterbeekx A, Janssens L, Lammens C, Dé E, Goossens H, Tsakris A, Malhotra-Kumar S, Pournaras S. 2016. Colistin-resistant Acinetobacter baumannii clinical strains with deficient biofilm formation. Antimicrob Agents Chemother 60:1892–1895. doi: 10.1128/AAC.02518-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 38. Ainsaar K, Mumm K, Ilves H, Hõrak R. 2014. The ColRS signal transduction system responds to the excess of external zinc, iron, manganese, and cadmium. BMC Microbiol 14:162. doi: 10.1186/1471-2180-14-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 40. Jaiswal SK, Naamala J, Dakora FD. 2018. Nature and mechanisms of aluminium toxicity, tolerance and amelioration in symbiotic legumes and rhizobia. Biol Fertil Soils 54:309–318. doi: 10.1007/s00374-018-1262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee LJ, Barrett JA, Poole RK. 2005. Genome-wide transcriptional response of chemostat-cultured Escherichia coli to zinc. J Bacteriol 187:1124–1134. doi: 10.1128/JB.187.3.1124-1134.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chamnongpol S, Dodson W, Cromie MJ, Harris ZL, Groisman EA. 2002. Fe(III)-mediated cellular toxicity. Mol Microbiol 45:711–719. doi: 10.1046/j.1365-2958.2002.03041.x [DOI] [PubMed] [Google Scholar]
- 43. Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. 2012. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob Agents Chemother 56:59–69. doi: 10.1128/AAC.05191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Allen CL, Skiniotis G, Aldrich CC, Gulick AM. 2016. Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529:235–238. doi: 10.1038/nature16163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martínez-Núñez MA, López VELy. 2016. Nonribosomal peptides synthetases and their applications in industry. Sustain Chem Process 4. doi: 10.1186/s40508-016-0057-6 [DOI] [Google Scholar]
- 46. Palethorpe S, Milton ME, Pesci EC, Cavanagh J. 2022. Structure of the Acinetobacter baumannii PmrA receiver domain and insights into clinical mutants affecting DNA binding and promoting colistin resistance. J Biochem 170:787–800. doi: 10.1093/jb/mvab102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murdoch CC, Skaar EP. 2022. Nutritional immunity: the battle for nutrient metals at the host-pathogen interface. Nat Rev Microbiol 20:657–670. doi: 10.1038/s41579-022-00745-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alasfar RH, Isaifan RJ. 2021. Aluminum environmental pollution: the silent killer. Environ Sci Pollut Res Int 28:44587–44597. doi: 10.1007/s11356-021-14700-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roshini J, Patro LPP, Sundaresan S, Rathinavelan T. 2023. Structural diversity among Acinetobacter baumannii K-antigens and its implication in the in silico serotyping. Front Microbiol 14:1191542. doi: 10.3389/fmicb.2023.1191542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. DebRoy C, Fratamico PM, Yan X, Baranzoni G, Liu Y, Needleman DS, Tebbs R, O’Connell CD, Allred A, Swimley M, Mwangi M, Kapur V, Raygoza Garay JA, Roberts EL, Katani R. 2016. Comparison of O-antigen gene clusters of all O-serogroups of Escherichia coli and proposal for adopting a new nomenclature for O-typing. PLoS ONE 11:e0147434. doi: 10.1371/journal.pone.0147434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Reeves PR, Wang L. 2014. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol Rev 38:56–89. doi: 10.1111/1574-6976.12034 [DOI] [PubMed] [Google Scholar]
- 54. Steimle A, Autenrieth IB, Frick JS. 2016. Structure and function: lipid A modifications in commensals and pathogens. Int J Med Microbiol 306:290–301. doi: 10.1016/j.ijmm.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 55. Hoare A, Bittner M, Carter J, Alvarez S, Zaldívar M, Bravo D, Valvano MA, Contreras I. 2006. The outer core lipopolysaccharide of Salmonella enterica serovar Typhi is required for bacterial entry into epithelial cells. Infect Immun 74:1555–1564. doi: 10.1128/IAI.74.3.1555-1564.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim SH, Yun S, Park W. 2022. Constitutive phenotypic modification of lipid A in clinical Acinetobacter baumannii isolates. Microbiol Spectr 10:e0129522. doi: 10.1128/spectrum.01295-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Potron A, Vuillemenot J-B, Puja H, Triponney P, Bour M, Valot B, Amara M, Cavalié L, Bernard C, Parmeland L, Reibel F, Larrouy-Maumus G, Dortet L, Bonnin RA, Plésiat P. 2019. ISAba1-dependent overexpression of eptA in clinical strains of Acinetobacter baumannii resistant to colistin. J Antimicrob Chemother 74:2544–2550. doi: 10.1093/jac/dkz241 [DOI] [PubMed] [Google Scholar]
- 58. Trebosc V, Gartenmann S, Tötzl M, Lucchini V, Schellhorn B, Pieren M, Lociuro S, Gitzinger M, Tigges M, Bumann D, Kemmer C. 2019. Dissecting colistin resistance mechanisms in extensively drug-resistant Acinetobacter baumannii clinical isolates. mBio 10:e01083-19. doi: 10.1128/mBio.01083-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wick RR, Judd LM, Holt KE. 2019. Performance of neural network basecalling tools for Oxford nanopore sequencing. Genome Biol 20:129. doi: 10.1186/s13059-019-1727-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 62. Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 66. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. March C, Regueiro V, Llobet E, Moranta D, Morey P, Garmendia J, Bengoechea JA. 2010. Dissection of host cell signal transduction during Acinetobacter baumannii-triggered inflammatory response. PLoS One 5:e10033. doi: 10.1371/journal.pone.0010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kamoshida G, Akaji T, Takemoto N, Suzuki Y, Sato Y, Kai D, Hibino T, Yamaguchi D, Kikuchi-Ueda T, Nishida S, Unno Y, Tansho-Nagakawa S, Ubagai T, Miyoshi-Akiyama T, Oda M, Ono Y. 2020. Lipopolysaccharide-deficient Acinetobacter baumannii due to colistin resistance is killed by neutrophil-produced lysozyme. Front Microbiol 11:573. doi: 10.3389/fmicb.2020.00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S9.
Tables S1 to S4.
Data Availability Statement
The original data sets are available in publicly accessible repositories. The genome sequencing data of representative A. baumannii isolates from this study have been deposited in the DNA Data Bank of Japan under BioProject accession numbers PRJDB12922 and PRJDB16715. The RNA sequencing data of A. baumannii isolates were also registered (BioProject accession number PRJDB16717). Additionally, the GenBank file for the whole-genome sequence data of ATCC BAA-1605 (BioProject accession number PRJDB16714) has been registered with the DNA Data Bank of Japan.