Abstract
Ohno [Ohno, S. (1970) in Evolution by Gene Duplication, Springer, New York] proposed that gene duplication with subsequent divergence of paralogs could be a major force in the evolution of new gene functions. In practice the functional differences between closely related homologues produced by duplications can be subtle and difficult to separate experimentally. Here we show that DNA microarrays can distinguish the functions of two closely related homologues from the yeast Saccharomyces cerevisiae, Yap1p and Yap2p. Although Yap1p and Yap2p are both bZIP transcription factors involved in multiple stress responses and are 88% identical in their DNA binding domains, our work shows that these proteins activate nonoverlapping sets of genes. Yap1p controls a set of genes involved in detoxifying the effects of reactive oxygen species, whereas Yap2p controls a set of genes over represented for the function of stabilizing proteins. In addition we show that the binding sites in the promoters of the Yap1p-dependent genes differ from the sites in the promoters of Yap2p-dependent genes and we validate experimentally that these differences are important for regulation by Yap1p. We conclude that while Yap1p and Yap2p may have some overlapping functions they are clearly not redundant and, more generally, that DNA microarray analysis will be an important tool for distinguishing the functions of the large numbers of highly conserved genes found in all eukaryotic genomes.
INTRODUCTION
DNA microarrays can reveal functional similarities between genes with little or no sequence homology. This is because the whole-genome mRNA expression patterns that result from the mutation of genes with similar functions are often very similar and can be thought of as “molecular phenotypes” (Hughes et al., 2000b). As a case study to determine whether these molecular phenotypes are sensitive enough to discriminate between the functions of closely related transcription factors, we chose to study Yap1p and Yap2p. Although previous experiments with DNA microarrays demonstrated that a number of genes involved in stress response show Yap1p-dependent expression (Gasch et al., 2000), little is known about the differences between genes regulated by Yap1p versus Yap2p. Yap1p and Yap2p are 88% identical in their DNA binding regions and have both been shown to bind the same consensus site (TTAGTAA; Fernandes et al., 1997). Furthermore, overexpression of either protein induces resistance to multiple cellular stresses (Schnell et al., 1992; Bossier et al., 1993; Wu et al., 1993; Hirata et al., 1994b; Stephen et al., 1995). Whether Yap1p and Yap2p exert these similar phenotypic effects by controlling the same or different sets of genes has remained unclear. One study did identify three genes whose expression are dependent on Yap1p but not Yap2p (Stephen et al., 1995). However, no targets for Yap2p have yet been identified. If and how Yap1p and Yap2p show specificity toward different regulons are also unresolved questions because both proteins bind to and activate transcription from the same consensus sequence (Hirata et al., 1994b; Fernandes et al., 1997). To begin to answer these questions we used whole-genome microarrays to measure the expression of all the genes in the genome in wild-type, yap1Δ , yap2Δ , and yap1Δ yap2Δ cells grown in minimal medium. Because Yap1p and Yap2p are implicated in the response to cellular stresses we also measured expression in cells treated with the oxidizing agent hydrogen peroxide (H2O2) and the metal cadmium (Cd2+). In this report we focus on the response to H2O2, but the full dataset is available at http://arep.med.harvard.edu/ExpressDB.
MATERIALS AND METHODS
Yeast Manipulations
Strain BY4740 (MATa, leu2Δ0, lys2Δ0, ura3Δ0) was used as the control strain in this study and yap1Δ, yap2Δ, and yap1Δ yap2Δ derivatives were constructed as described (Brachmann et al., 1998). For RNA extractions all strains were grown to mid log phase in minimal media and induced for 1 h with either 0.6 mM H2O2, 1 μM CdCl2, or mock treated, and mRNA was extracted, labeled and hybridized to oligonucleotide arrays as described (Wodicka et al., 1997). All experiments were repeated at least twice (sometimes three times) and the average expression level of the independent experiments was used for the analysis.
For plating assays, all strains were grown to OD600 of 0.4, dilutions were made and 5 μL of each dilution was spotted onto the appropriate medium
β-Galactosidase assays were performed as described (Dudley et al., 1999).
Plasmid Constructions
To create the wild-type YKL086W reporter gene primers BC248 (5′-CGGAATTCTATGTAAAATAGAGACGAATGAAAA-3′) and BC249 (5′-GCCCTTATTGTGGCCACCATTGCGTC-3′) were used to amplify the YKL086W promoter region and this fragment was cloned into the EcoRI and BamHI sites of pSEYC102 (Gift of Fred Winston). The resulting plasmid was named pBC266. All mutant constructs were derived from pBC266 using sequential PCR mutagenesis (Ausubel et al., 1994). For mutation of the core base pairs in the extended site we used primers BC285 (5′-CGATTGCTTTTTCCCTGATccGcAAGCTACATCATTTATAC-3′) and BC286 (5′-GTATAAATGATGTAGCTTgCggATCAGGGAAAAAGCAATCG-3′) and for mutation of the flanking residues in the extended sited we used primers BC283 (5′-CGATTGCTTTTTCCCTGgTTAGTAAcaTACATCATTTATAC-3′) and BC284 (5′-GTATAAATGATGTAtgTTACTAAcCAGGGAAAAAGCAATCG-3′). For mutation of the core base pairs within the core site we used primers BC292 (5′-CCCAGAAGTCGCCATTATTTcTAGctATTACAGTAGCCCTGTT-GGG-3′) and BC293 (5′-CCCAACAGGGCTACTGTAATagCTAgA-AATAATGGCGACTTCTGGG-3′).
Data Analysis
Genes with low expression and low variance were filtered from the dataset as described (Cohen et al., 2000). The dataset was then divided into clusters of coexpressed genes using the computer program QTClust (Heyer et al., 1999) using a correlation threshold of 0.7. A detailed description of all the clusters produced from this analysis can be found at http://genetics.med.harvard.edu/∼cohen/yaps/Yaps.html. A Yap binding site weight matrix (Stormo et al., 1982) was constructed using sites from four promoters known to be regulated by Yap1p (Kuge and Jones, 1994; Wu and Moye-Rowley, 1994; Wemmie et al., 1994; Grant et al., 1996). This weight matrix was used as an input to the computer program ScanACE (Hughes et al., 2000a) to determine the distribution of Yap sites among all of the expression clusters. Only sites that scored at least as well as the average site in the matrix were counted as Yap sites. The significance of clusters in which a high proportion of the promoters within the cluster contained at least one Yap site was assessed using the hypergeometric probability distribution, without correction for multiple hypotheses, as follows:
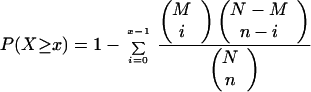 |
where x is the number of promoters in a particular cluster with at least one Yap site, n is the number of promoters in the genome with at least one Yap site, M is the number of promoters in a particular cluster, and N is the number of promoters in the genome. Only clusters where p < 0.05 were considered significant. The statistical significance of clusters in which promoters that had Yap sites tended to contain multiple sites was assessed using the chi-square test by comparing the distribution of genes in the genome that had (1,2 … n) sites to the distribution of genes in a particular cluster that had (1,2 … n) sites. Again clusters were considered significant when p < 0.05.
The significance of groups of genes that were enriched for particular MIPS functional annotations was tested as described (Tavazoie et al., 1999).
RESULTS
Characterization of Molecular Phenotypes
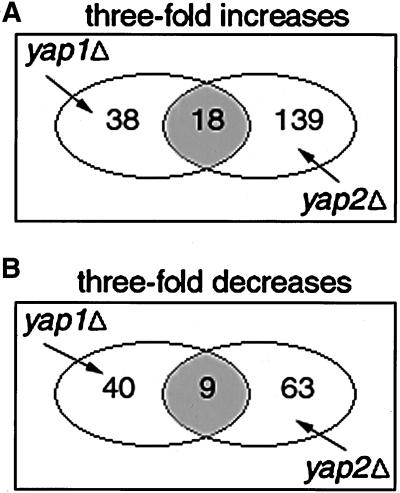
In this study we define the molecular phenotype of a gene to be the constellation of changes in gene expression that take place upon deletion of the gene. Because Yap1p and Yap2p have both been implicated in stress response, we determined their molecular phenotypes in H2O2. We arbitrarily chose to include only genes whose expression changed by more than threefold in our molecular phenotypes. For example, the Yap1p molecular phenotype is composed of genes that do not vary in wild-type cells grown in H2O2, but whose expression changes at least threefold in yap1Δ cells grown in H2O2. Although Yap1p and Yap2p have partially overlapping molecular phenotypes (Figure 1, A and B), it is clear that there are a significant number of genes whose expression changes only in yap1Δ mutants and other changes that occur only in yap2Δ mutants. Aside from the changes shown in Figure 1, there were also 82 genes whose expression changed in yap1Δ yap2Δ cells but not in either of the single mutants. This result suggests that there may be some functional redundancy between Yap1p and Yap2p. However the fact that these two proteins have different molecular phenotypes implies that they also have separable functions.
Figure 1.
Venn diagrams representing the molecular phenotypes of the yap1Δ and yap2Δ mutants. The number of genes that showed either a threefold increase (A) or threefold decrease (B) in the mutant strains are shown in the diagrams. The overlap between the yap1Δ and yap2Δ ovals in the diagrams represents the genes that change in either mutant. The lists of genes that occur in each category are available on our web site at http://genetics.med.harvard.edu/∼cohen/yaps/Yaps.html.
Identification of Yap1p- and Yap2p-dependent Genes
Because our preceding results suggested that Yap1p and Yap2p have separable functions and because these proteins are themselves transcription factors, we hypothesized that there would be separate groups of genes whose transcription were directly regulated by either Yap1p or Yap2p.
We observed 250 genes whose expression changed by more than threefold in wild-type cells upon addition of H2O2 to the growth medium. Fifty-three percent of these changes depended on the presence of Yap1p, Yap2p, or Yap1p and Yap2p, underscoring the importance of these proteins in the response to oxidative stress. However from these data alone it was not possible to discern how many of these changes were primary targets of Yap1p or Yap2p regulation. We reasoned that genes showing Yap-dependent regulation that also contained Yap binding sites in their promoters would be more likely to be direct targets of Yap1p or Yap2p.
To find sets of genes whose expression depended on either Yap1p or Yap2p, we first partitioned our dataset into 29 groups of coexpressed genes using the clustering algorithm QTClust (Heyer et al., 1999). For a full description of all clusters see http://genetics.med.harvard.edu/∼cohen/yaps/Yaps.html. Next we determined the distribution of Yap binding sites among the different expression clusters. To do this we constructed a multiple alignment of Yap1p binding sites from promoters known to be regulated by Yap1p. We then ran the computer program ScanACE (Hughes et al., 2000a), which uses a multiple alignment to search for additional matching sequences, to identify all of the occurrences of Yap1p binding sites in the genome. Finally, we used two different statistical tests to search for clusters in which Yap sites were statistically over represented in the promoters of the genes within those clusters. First we looked for clusters in which a high proportion of the promoters contained at least one Yap binding site (Table 1). We also looked for clusters in which promoters tended to have multiple Yap sites (Table 2). Using these criteria six clusters were deemed to contain more Yap sites in the promoters of their genes then expected by chance.
Table 1.
Percentage of genes whose promoters have at least one Yap site is shown for those clusters in which the percentage is significantly higher than the percentage of genes in the genome that have at least one Yap site
| Percentage of genes with sites | P value | |
|---|---|---|
| Cluster 79 | 50 | 0.02 |
| Cluster 30 | 37 | 0.03 |
| Genome | 20 | 1.0 |
See MATERIALS AND METHODS.
Table 2.
The average number of Yap sites per promoter for promoters with Yap sites, is shown for clusters in which the average is significantly higher than that found in the genome at large
| No. of sites/genes with sites | P value | |
|---|---|---|
| Cluster 33 | 1.9 | 8.0 × 10−4 |
| Cluster 51 | 1.9 | 2.0 × 10−4 |
| Cluster 31 | 1.8 | 2.0 × 10−3 |
| Cluster 39 | 1.6 | 7.0 × 10−3 |
| Genome | 1.2 | 1.0 |
See MATERIALS AND METHODS.
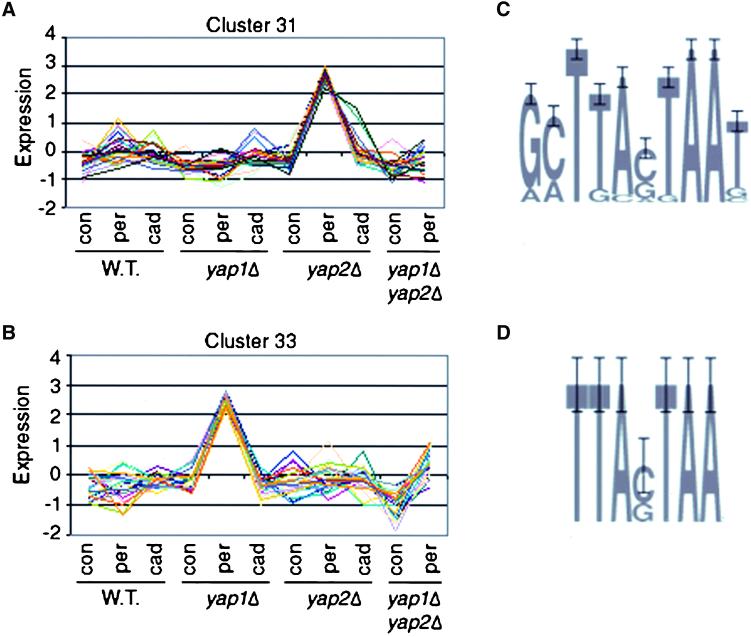
Among this set clusters 31 (25 genes) and 33 (24 genes) are of particular interest because they clearly separate genes controlled by Yap1p and Yap2p (Figure 2, A and B). The genes in cluster 31 show a Yap1p-dependent increase in expression in H2O2. The small increase in normalized expression in the wild-type cells in H2O2 actually corresponds to an average change of fourfold H2O2. What is striking, however, about the expression of the genes in this cluster is that the Yap1p-dependent increase in expression in H2O2 is greatly magnified in the absence of Yap2p. This expression is Yap1p dependent because it is absent in the yap1Δ yap2Δ mutant. Cluster 33 shows an almost opposite expression pattern from cluster 31. The genes in this cluster do not show a significant increase in expression in H2O2 in wild-type cells (1.8-fold on average), a result that might be expected because of the absence of H2O2 hypersensitivity in yap2Δ mutants (Hirata et al., 1994a). However, they do show a very large increase in expression in the absence of Yap1p. This expression is Yap2p dependent because it is absent in the yap1Δ yap2Δ mutant. Therefore, there is clearly a set of genes whose expression is dependent on Yap1p but not Yap2p and a set of genes whose expression is dependent on Yap2p but not Yap1p.
Figure 2.
The normalized average expression profiles for (A) cluster 31 and (B) cluster 33. Expression in each condition is graphed as the Z-score, the number of SDs the average expression level in any one condition is above or below the average expression over all of the conditions. Abbreviations: con, control, minimal media with no additions; per, peroxide, minimal media with 1 h induction in 0.6 mM H2O2; cad, cadmium, minimal media with 1 h induction in 1 μM CdCl2. Sequence logos (Schneider and Stephens, 1990) for the Yap sites found in the promoters of genes in cluster 31 (C) and cluster 33 (D). The height of each letter in a stack represents the degree of conservation of that base at a particular position in a multiple alignment of Yap sites from the promoters of coregulated genes. The number of different letters in a stack at a particular position in the site represents the different bases that were present at that position in the multiple alignment.
Using the MIPS database (Mewes et al., 1999), we asked whether the Yap1p- and Yap2p-dependent genes were enriched for particular functions. Cluster 31 (Yap1p dependent) was enriched for genes in the category “detoxification” (p = 2.4 × 10−4). This cluster contained genes such as glutathione s-transferase and superoxide dismutase, which are clearly involved in the response to reactive oxygen species. By contrast cluster 33 (Yap2p dependent) was enriched for genes in the category “protein folding and stability” (p = 2.3 × 10−2) and contained genes such as chaperones and ubiquitin-conjugating enzymes. However, 54% of the genes in cluster 33 were of unknown function, suggesting that the Yap2p-dependent regulon may also have other functions besides affecting protein turnover. In response to oxidative stress a cell must deal directly with reactive oxygen species as well as stabilize its correctly folded proteins and degrade its misfolded proteins. Directly comparing cluster 31 to cluster 33 shows that the functions of the genes enriched in each cluster are significantly different (z = 2.8, p < 0.01) and suggests that detoxification is controlled by the Yap1p regulon, whereas protein turnover is affected by the Yap2p regulon. These results may also help explain the relationship between the Yap1p- and Yap2p-dependent genes. For example, in the absence of Yap2p, the Yap1p response to H2O2 is magnified (Figure 2A). This may be because in the absence of the Yap2p-dependent protein turnover response there is a greater need for the detoxification response. Thus, although the detoxification genes are dependent only on Yap1p they can sense the absence of Yap2p, perhaps because Yap1p is activated by the presence of unfolded proteins. Alternatively, Yap2p may directly repress the promoters of Yap1p-dependent genes.
Clusters 31 and 33 were both identified because the genes within those clusters were over represented for Yap binding sites. We also used the program AlignACE (Hughes et al., 2000a) to search the promoters of genes within Clusters 31 and 33 for other DNA sequences that might be over represented within these sets. Aside from the Yap sites themselves no other motifs showed significant over representation within these promoters, suggesting that it is the Yap sites that are responsible for the expression patterns of the clusters. How can the same binding site control the expression of genes with such different expression patterns? To address this question we reconstructed binding site matrices (Stormo et al., 1982) for the Yap sites from clusters 31 and 33 (Figure 2, C and D). Although sites from both clusters 31 and 33 tended to conform to the known Yap site (TTAGTAA), many sites from cluster 31 contained additional conserved bases flanking this “core” sequence. Although the core sequence is distributed widely across the genome and is present in almost all of our expression clusters, the “extended” yap site is found almost exclusively in the promoters of genes within the Yap1p-dependent cluster 31. Three genes (TRX2, YCF1, and GLR1), which were previously identified as being Yap1p dependent, contain sites in their promoters with strong matches to the extended site we identified. One other Yap1p-dependent gene (GSH1) does not contain an extended site in its promoter. However, the regulation of this gene has already been shown to be different from that of other Yap1p-dependent genes (Stephen et al., 1995). Taken together these results raised the possibility that the flanking bases found in the extended site are important for Yap1p-dependent expression.
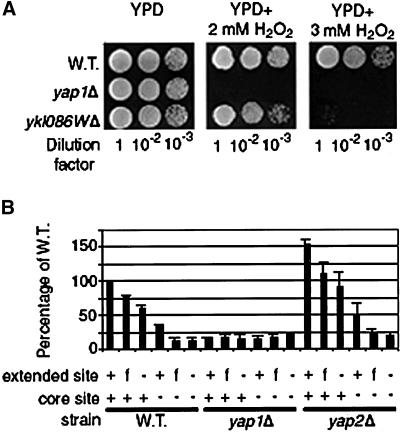
We chose to test this hypothesis on YKL086W, a novel member of the Yap1p regulon from cluster 31. Although the function of YKL086W is currently unknown, when we deleted it from the genome the resulting cells were hypersensitive to H2O2 (Figure 3A), demonstrating that this gene is involved in the response to reactive oxygen species. This also suggests that other genes in cluster 31 with unknown functions that contain Yap binding sites are involved in the response to oxidative stress. We fused the promoter region of YKL086W to the LacZ gene from Escherichia coli. This reporter gene mimicked the Yap1p-dependent expression pattern observed on the microarray for YKL086W. The promoter region of this gene contains one extended Yap site, GTTAGTAACA (flanking bases shown in bold), and one core Yap site, TTAGTAA. Although the analysis was complicated by the presence of the core site, it was clear from various mutant derivatives that mutating the flanking bases in the extended site caused a reproducible reduction in activity from the reporter (Figure 3B). In constructs where the core site had been mutated so as to be inactive, mutating the flanking bases in the extended site was equivalent to mutating bases within the core sequence of the extended site. These results suggest that the flanking bases in the extended site are required for the function of that site and for specification of Yap1p-dependent transcription.
Figure 3.
(A) Plating assays for the viability of wild-type, yap1Δ , and ykl086wΔ cells on rich medium with and without H2O2. All strains were grown to OD600 of 0.4, dilutions were made, and 5 μL of each dilution was spotted onto the appropriate medium. (B) β-Galactosidase assaysYKL086W reporter genes. All constructs were induced for 1 h in 0.6 mM H2O2. All constructs yielded < 20 U of activity in the absence of H2O2. Mutations in the reporter constructs are designated by either a (+) where the site has not been mutated, (−) where base pairs in the core sequence of the site have been mutated, or (f)¤ where base pairs flanking the core sequence of the extended site have been mutated. All values are normalized to the wild-type construct in the wild-type background, which had an expression level of 342 ± 46 U.
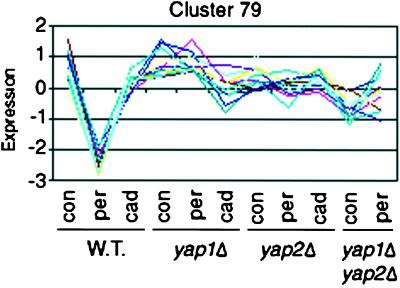
Another interesting cluster is number 79 (12 genes) in which the genes show a strong decrease in transcription in H2O2 that is dependent on both Yap1p and Yap2p (Figure 4). The promoters of these genes are over represented for Yap binding sites (Table 1), suggesting that this repression of transcription is direct. The genes in this cluster are enriched in the MIPS functional category “DNA synthesis and replication.” Although this enrichment is not statistically significant (p = 0.14), it makes sense that a cell would slow replication during oxidative stress while damage done to DNA is repaired. These results provide the first evidence that Yap1p and Yap2p may repress as well as activate transcription.
Figure 4.
Normalized average expression profile for the genes in cluster 79. Expression in each condition is graphed as the Z-score, the number of SDs the average expression level in any one condition is above or below the average expression over all of the conditions. Abbreviations: con, control, minimal media with no additions; per, peroxide, minimal media with 1 h induction in 0.6 mM H2O2; cad, cadmium, minimal media with 1 h induction in 1 μM CdCl2.
DISCUSSION
Our results demonstrate that although Yap1p and Yap2p are closely related homologues, with similar phenotypes, these proteins are clearly nonredundant. Yap1p and Yap2p activate distinct regulons involved in different aspects of the oxidative stress response, with Yap1p-dependent genes being involved directly in detoxifying the effects of reactive oxygen species, whereas Yap2p-dependent genes help stabilize and fold proteins in an oxidative environment. The molecular functions of Yap1p and Yap2p have diverged such that these homologues now activate different regulons, yet both proteins are involved in the response to cellular stresses. Selective pressures may have driven this divergence by increasing the scope and flexibility of the response to cellular stresses. Although we have focused here on oxidative stress, the divergence of Yap1p and Yap2p may allow different physiological responses to different cellular stresses. There may be conditions in which Yap1p and Yap2p are differentially expressed or activated. However, at the level of mRNA expression there is no significant difference between the YAP1 and YAP2 transcripts in 217 whole-genome expression data sets tested. Our working hypothesis is therefore that Yap1p and Yap2p both respond to similar cellular stresses, and they are both maintained in the genome because they activate different regulons.
We have also provided evidence that the specification of Yap1p- versus Yap2p-dependent transcription occurs through an extended Yap site found only in the promoters of Yap1p-dependent genes. These extended sites are not found in Yap2p-dependent genes, and reporter gene assays confirmed that the additional base pairs in the extended sites are important for regulation by Yap1p. This mechanism for obtaining specificity is consistent with what is known about other families of transcription factors. For example, base pairs flanking the core binding site have also been shown to be important in specifying transcription between members in families of basic helix-loop-helix leucine zipper transcription factors (O'Hagan et al., 2000). How Yap2p-dependent transcription is specified is still unclear. Although the core Yap site is statistically over represented in the promoters of Yap2p-dependent genes, that site is also present in the promoters of many genes that do not show Yap2p-dependent transcription. The Yap sites in Yap2p-dependent promoters may work in combination with other transcription factor binding sites that fell below the threshold of detection of the search algorithms we used.
As demonstrated by the expression pattern of cluster 79, Yap1p and Yap2p may function as repressors as well as activators of transcription. One possible model to describe the Yap network is that during oxidative stress Yap1p and Yap2p homodimers activate distinct regulons, whereas Yap1p/Yap2p heterodimers collaborate to repress a separate regulon.
Our approach to separating the functions of the Yap1p and Yap2p transcription factors has several advantages. Focusing on expression clusters that are over represented for Yap binding sites helps distinguish direct versus indirect effects on transcription caused by transcription factors. This approach also does not require any a priori prediction of what a Yap-dependent cluster should look like and therefore allows the identification of clusters with unexpected expression patterns, such as cluster 79. Finally our approach is automatable and requires minimal curation, and is therefore easily scalable to larger genomes and larger protein families. This work should serve as a model to disentangle the functions of the huge number of paralogs being identified by the various genome sequencing projects.
ACKNOWLEDGMENTS
We thank Semyon Kruglyak for the computer code for the QTClust algorithm, Jason Hughes for the ScanACE and AlignACE programs, Fred Winston for strains and plasmids, and members of the Church lab for discussions and critical readings of the manuscript. B.A.C. was supported by a postdoctoral fellowship from the American Cancer Society (PF-98-159-01-MBC). This work was supported by the US Department of Energy (DE-FG02-87-ER60565), the Office of Naval Research and DARPA (N00014-97-1-0865), the Lipper Foundation, and Hoechst Marion Roussel.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0472. Article and publication date are at www.molbiolcell.org/cgi/10.1091/mbc.01–10–0472.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons, Inc; 1994. [Google Scholar]
- Bossier P, Fernandes L, Rocha D, Rodrigues-Pousada C. Overexpression of YAP2, coding for a new yAP protein, and YAP1 in Saccharomyces cerevisiae alleviates growth inhibition caused by 1,10-phenanthroline. J Biol Chem. 1993;268:23640–23645. [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- Dudley AM, Gansheroff LJ, Winston F. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4–912delta promoter in Saccharomyces cerevisiae. Genetics. 1999;151:1365–1378. doi: 10.1093/genetics/151.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Collinson LP, Roe JH, Dawes IW. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Yano K, Miyahara K, Miyakawa T. Saccharomyces cerevisiae YDR1, which encodes a member of the ATP-binding cassette (ABC) superfamily, is required for multidrug resistance. Curr Genet. 1994a;26:285–294. doi: 10.1007/BF00310491. [DOI] [PubMed] [Google Scholar]
- Hirata D, Yano K, Miyakawa T. Stress-induced transcriptional activation mediated by YAP1 and YAP2 genes that encode the Jun family of transcriptional activators in Saccharomyces cerevisiae. Mol Gen Genet. 1994b;242:250–256. doi: 10.1007/BF00280413. [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000a;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000b;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Heumann K, Kaps A, Mayer K, Pfeiffer F, Stocker S, Frishman D. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 1999;27:44–48. doi: 10.1093/nar/27.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, Meltzer P, DePinho RA. Gene-target recognition among members of the myc superfamily, and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell N, Krems B, Entian KD. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992;21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- Stephen DW, Rivers SL, Jamieson DJ. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- Stormo GD, Schneider TD, Gold L, Ehrenfeucht A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Szczypka MS, Thiele DJ, Moye-Rowley WS. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J Biol Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Wu A, Wemmie JA, Edgington NP, Goebl M, Guevara JL, Moye-Rowley WS. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]
- Wu AL, Moye-Rowley WS. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol. 1994;14:5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]