Abstract
Although cholesterol is essential for membrane fluidity and deformability, the level of its lateral heterogeneity at the plasma membrane of living cells is poorly understood due to lack of appropriate probe. We here report on the usefulness of the D4 fragment of Clostridium perfringens toxin fused to mCherry (theta*), as specific, non-toxic, sensitive and quantitative cholesterol-labeling tool, using erythrocyte flat membrane. By confocal microscopy, theta* labels cholesterol-enriched submicrometric domains in coverslip-spread but also gel-suspended (non-stretched) fresh erythrocytes, suggesting in vivo relevance. Cholesterol domains on spread erythrocytes are stable in time and space, restricted by membrane:spectrin anchorage via 4.1R complexes, and depend on temperature and sphingomyelin, indicating combined regulation by extrinsic membrane:cytoskeleton interaction and by intrinsic lipid packing. Cholesterol domains partially co-localize with BODIPY-sphingomyelin-enriched domains. In conclusion, we show that theta* is a useful vital probe to study cholesterol organization and demonstrate that cholesterol forms submicrometric domains in living cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1951-x) contains supplementary material, which is available to authorized users.
Keywords: His-mCherry-theta-D4, 125I-toxin, Lateral membrane heterogeneity, Vital confocal imaging, Membrane tension, Temperature, C2C12 myoblasts, BODIPY-sphingomyelin
Introduction
Cholesterol is a major, essential constituent of the mammalian plasma membrane [1]. Its lateral distribution is known to be asymmetric, as exemplified by transient nanometric rafts and caveolin-stabilized invaginations [2–4]. In contrast, cholesterol transbilayer distribution remains unclear; several studies, however, reached the same conclusion that a significant part (50–75 %) is distributed in the inner leaflet [5, 6]. Membrane cholesterol plays key regulatory roles including (1) membrane fluidity via lipid ordering; (2) membrane deformability by modulation of plasma membrane protein interactions at the interface with cortical cytoskeleton [7]; (3) formation and stabilization of nanometric lipid assemblies, rafts and caveolae [2, 4], as important signaling platforms [8–10]; and (4) phase coexistence in artificial membranes [11–13]. Cholesterol is, thus, a key component of membrane biology and the concept of its clustering into membrane domains is attractive to explain its different functions. However, cholesterol clustering in large submicrometric domains has not been demonstrated so far. Interestingly, ergosterol-enriched micrometric protein domains have been reported at the yeast plasma membrane [14, 15].
Addressing the genuine lateral distribution of cholesterol at the plasma membrane requires vital imaging with probes combining specificity, non-toxicity, high sensitivity for tracer use, and easy quantitation. These conditions were not simultaneously met by the three approaches used so far: (1) labeling with low-molecular weight fluorescent “reporters”; (2) binding of labeled cholesterol-specific polypeptides derived of pore-forming toxins; and (3) metabolic isotopic labeling followed by high-resolution imaging mass spectroscopy. The first approach is based on small fluorescent reporters, among which the most popular is filipin, a polyene antibiotic that forms specific yet membrane-disrupting complexes with cholesterol, thus limiting its use to fixed cells [16]; it is also quite photosensitive. As alternative low molecular weight reporters, fluorescent sterols such as dehydroergosterol are difficult to incorporate in the plasma membrane (our unpublished data) and not sensitive enough because of low quantum yield and rapid photobleaching [17]. The second approach is based on cholesterol-specific polypeptides derived of pore-forming toxins. Among these stands perfringolysin O, also known as theta toxin, secreted by the gram-positive bacterium, Clostridium perfringens. Theta toxin is composed of four domains (D1–D4), where D4 is the cholesterol-recognition domain and D1 the pore-forming domain, responsible for toxicity [18–22]. In the D4 domain, two conserved amino acid residues (Thr490 and Leu491) specifically interact with membrane cholesterol hydroxyl group [21]. Binding to cholesterol clusters induces configuration changes in the D1 domain, which leads to toxin oligomerization [23] and causes cell lysis [19]. Genetic engineering of pore-forming toxins paved the way to new non-toxic approaches. Using a 125I-labeled mutant of perfringolysin O, not cytolytic at low temperature, Radhakrishnan and colleagues recently provided quantitative evidence for the compartmentation of fibroblast membrane cholesterol into three distinct pools [24]. An alternative elegant approach is based on truncated perfringolysin O, limited to its C-terminal domain D4 (theta-D4), preserving cholesterol-binding specificity but abolishing cytotoxicity [19]. However, theta-D4 preserves a high threshold value for binding to cholesterol (~30 mol %; [25, 26]). Fusion of theta-D4 with the fluorescent protein Dronpa (best suited for super-resolution microscopy) allowed to evidence cholesterol-enriched domains on fixed HeLa cells [27] but this promising tool was not adapted for vital imaging because Dronpa is a reversibly switchable photoactivatable fluorescent protein. The third approach, high-resolution imaging mass spectroscopy, exploits metabolically labeled endogenous lipids and is, thus, neither invasive nor toxic, but requires cell fixation. Using this method, Kraft and colleagues evidenced 15N-sphingolipid-enriched stable submicrometric domains at the fibroblast plasma membrane, but such domains could not be evidenced after metabolic labeling with 18O-cholesterol [28, 29].
Altogether, despite their potential, these various probes/approaches are limited by the need of fixation, which is far from trivial for membrane lipids, certainly not achieved by formaldehyde, and may lead to artifactual redistribution [30]. Moreover, it is often difficult to interpret enriched “patches” as genuine lipid-enriched membrane domains vs structural membrane protrusions such as microvilli that are not resolved in the z axis. This concern is especially relevant for cholesterol, known to preferentially associate with membrane ruffles [27, 31].
We recently reported on the usefulness of a non-toxic domain of the lysenin toxin fused to a monomeric red fluorescent protein (mCherry) as proof-of-concept of toxin-based vital probe for confocal imaging and confirmed the existence of sphingomyelin (SM) submicrometric domains at the human red blood cell (RBC) plasma membrane [32], previously suggested based on the insertion of BODIPY-SM analogs [33–35]. In the present study, we extended this approach to cholesterol, using detoxified theta-D4 fragment [27] fused to mCherry (hereafter referred to as theta*) to address for the first time the lateral plasma membrane organization of endogenous cholesterol on unfixed RBCs.
RBCs represent an ideal model to study lateral membrane cholesterol organization. First, they are the simplest, best characterized [36, 37] and most robust eukaryotic cell system, due to membrane stabilization by a strong tangential spectrin network connected by two types of non-redundant anchorage complexes, based on 4.1R and ankyrin, respectively [38]. Second, mature RBCs display a featureless smooth surface, obviating concerns of surface projections as confounding source of enriched signal. Third, mature RBCs show neither metabolic nor vesicular membrane turn-over. Of note, RBC cholesterol content is exceptionally high and exclusively located at the plasma membrane (~45 mol % [39]).
We first verified the binding specificity and innocuity of theta*. Binding isotherm of the radio-iodinated probe indicated that trace labeling yielded a visible signal by confocal microscopy. Confocal vital imaging revealed abundant submicrometric domains of endogenous cholesterol, enriched (up to ~20-fold) over the rest of the plasma membrane. These abundant domains were not only seen on RBCs partially spread onto coverslips as usual system, but also on RBCs suspended as biconcave objects in a 3D-gel, ruling out spreading artifacts. We, thus, suggest that cholesterol submicrometric domains may represent genuine features of circulating RBCs. Our study also paves the way to (1) the regulation of cholesterol domains by showing dependence on cortical cytoskeleton anchorage and endogenous SM; and (2) the spatial relation between cholesterol- and SM-enriched domains. We finally extended the demonstration of heterogeneous cholesterol distribution at the plasma membrane of living nucleated mammalian cells, C2C12 myoblasts.
Materials and methods
Expression and purification of His-mCherry-theta-D4
The expression plasmid pET28/His-mCherry-theta-D4 encodes for a N-terminal 6xHis-tag followed by the monomeric red fluorescent protein mCherry and the C-terminal, non-toxic domain D4 of the cholesterol-specific theta toxin (theta-D4); this protein construct is named theta*. The plasmid was generated by swapping Dronpa for mCherry from pET28/His-Dronpa-theta-D4 [27], as previously described for a lysenin fragment [32]. Expression, purification, biochemical characterization and storage of theta* were performed as described [32].
Red blood cell isolation and pharmacological treatments
Red blood cells (RBCs) were isolated from healthy volunteers. This study was approved by the Medical Ethics Institutional Committee; each donor gave written informed consent. Blood was collected on K+/EDTA-coated tubes, diluted and washed as previously described [32]. To deplete cholesterol and sphingomyelin (SM), washed RBCs were pre-incubated in suspension in medium (DMEM; Invitrogen; washed RBCs:medium ratio of ~1:12, v/v) supplemented with 1 mg/ml defatted-bovine serum albumin (DF-BSA) and containing either 0–1.5 mM methyl-β-cyclodextrin (mβCD; Sigma-Aldrich) at 37 °C for 30 min or 0–10 mU/ml Bacillus cereus sphingomyelinase (SMase; Sigma-Aldrich) at 20 °C for 10 min. Efficiency of cholesterol depletion was measured using Amplex Red Cholesterol kit (Invitrogen) with omission of cholesterol esterase [35]. Innocuity of theta* and mβCD was verified by lack of hemolysis, as previously described [32, 33]. To phosphorylate 4.1R complexes by protein kinase C (PKC), washed RBCs were pre-incubated in suspension with DMEM containing 1 mg/ml DF-BSA, supplemented by 5 µM of the PKC activator, phorbol 12-myristate 13-acetate (PMA), and 20 nM of the phosphatase inhibitor, calyculin A (CalA; both from Sigma), at 37 °C for 20 min.
After treatment, RBCs were further pelleted at 130g × 2 min, then labeled with theta* (see below) in the continued presence of pharmacological agents as indicated in the figure caption. Alternatively, RBCs were first labeled with theta* in suspension at 20 °C for 20 min, washed and incubated with mβCD at 37 °C, either in suspension before spreading onto coverslips, or on-stage after spreading onto coverslips.
Theta* binding to multilamellar liposomes and RBCs
Before each experiment, theta* was diluted in DMEM containing 1 mg/ml DF-BSA and cleared of aggregates by centrifugation at 20,000g × 10 min at 4 °C. Multilamellar vesicles (MLVs) were generated from SM (chicken egg; Avanti polar lipids), l-α-phosphatidylcholine (PC; Sigma-Aldrich) and cholesterol (Sigma-Aldrich) essentially as described [32]. MLVs were assembled from either SM and PC (1:1 molar ratio) or SM, PC and cholesterol (3:4:5.5 molar ratio), then incubated with theta* (1:2000, toxin:total lipid molar ratio) at 37 °C for 30 min and isolated by two centrifugations (60,000g × 30 min each). Alternatively, washed RBCs were incubated with theta* (~1:25, toxin:RBC total lipids estimated from [40]) at the indicated temperature for 20 min and washed by centrifugation at 130g × 2 min. Pellets containing MLVs or RBCs and supernatants were analyzed by Western blotting as described [32].
Theta* radio-iodination and binding to RBCs
Theta* radio-iodination was performed as described [32], to a specific radioactivity of ~20 Bq/ng protein. Binding of 125I-theta* to RBCs was performed in suspension (washed RBCs:medium ratio of ~1:4, v/v) as previously described [32]. When indicated, RBCs were pre-treated with mβCD or SMase as above, prior to labeling with 5 × 103Bq 125I-theta* mixed with 1 µM cold theta*, in the continuous presence of pharmacological agents.
RBC labeling with theta* alone or in combination with BODIPY-SM, confocal vital imaging and fluorescence recovery after photobleaching (FRAP)
Except when otherwise stated, freshly isolated RBCs were incubated in suspension with 0.5–0.75 µM theta* at 20 °C for 20 min, pelleted and resuspended in DMEM containing 5 mg/ml BSA then again in DMEM alone, and immobilized onto 2-cm2 poly-l-lysine-coated coverslips. Coverslips were coated with 0.1 mg/ml poly-l-lysine (70–150 kDa, Sigma-Aldrich):DMEM (1:1, v/v) at 37 °C for 30 min, drained and let to dry at 37 °C for at least 1 h. Labeled and washed RBCs (~107 cells in 25 µl) were plated onto coated coverslips at 20 °C for exactly 4 min, replaced by fresh medium, and attached RBCs were further allowed to spread for another 4 min. This resulted into variable levels of stretching, monitored by the projected individual RBC areas (see Fig. 5). After two rapid washings, coverslips were placed upside–down into Lab-Tek chambers for immediate examination of RBCs in the living state. For on-stage experiments, coverslips bearing theta*-labeled RBCs were placed into Lab-Tek chambers with right-side up for easy access of reagents.
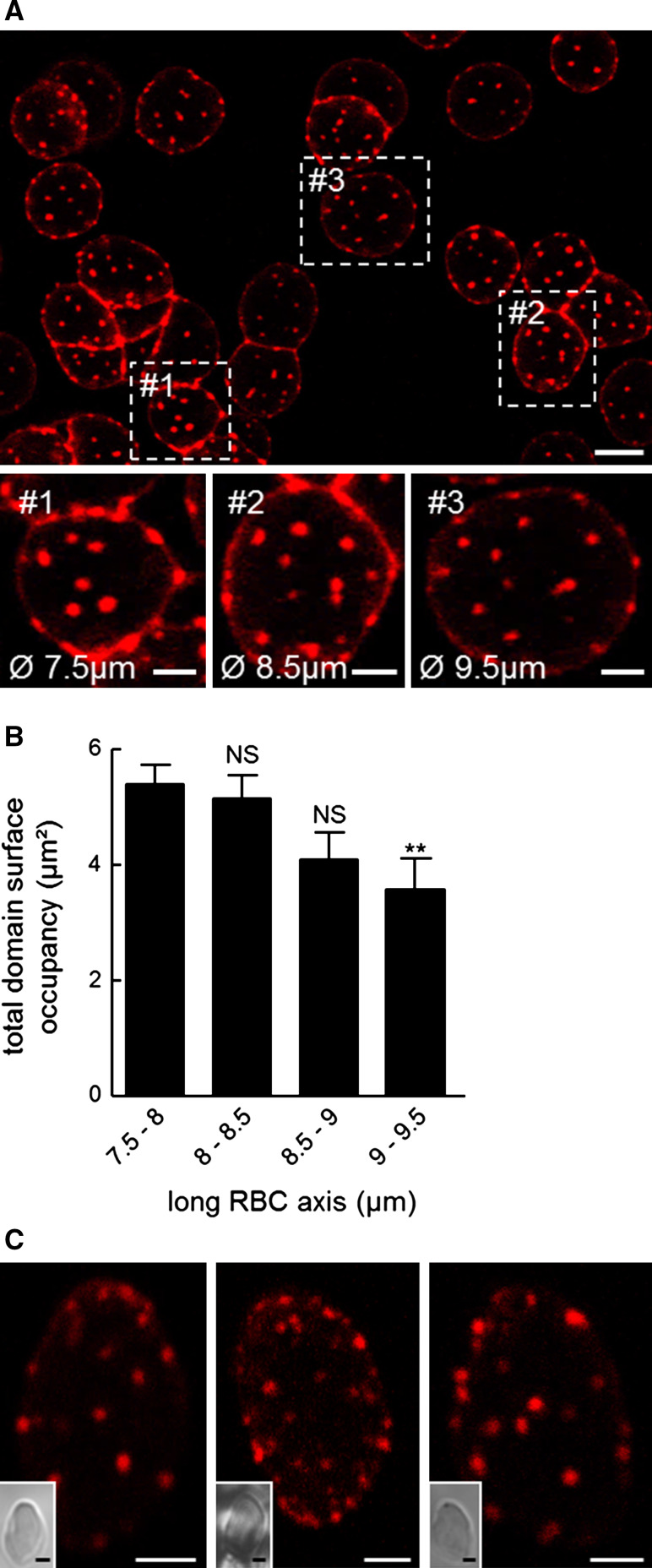
Fig. 5.
Cholesterol submicrometric domains largely resist membrane stretching and are similarly abundant in gel-suspended (3D), non-stretched RBCs. A Confocal imaging of domains on differentially spread RBCs. RBCs were labeled with theta*, washed, attached-spread and visualized at 20 °C. RBC attachment on poly-l-lysine leads to variable stages of RBC spreading (boxed areas #1 to #3 are enlarged at insets). Theta*-enriched domains resist membrane stretching but decrease in size. Scale bars 5 µm at general view and 2 µm at insets. B Morphometry of domain surface occupancy. Surface of theta*-labeled domains was determined per hemi-cell surface and values were classified according to RBC long diameter. Mean ± SEM of total domain area recorded on 8–26 cells from two independent experiments. Kolmogorov–Smirnov test was performed using the 7.5–8 µm class as reference; NS not significant, **p < 0.01. C Confocal imaging of domains on RBCs trapped in a 3D-gel. RBCs were labeled with theta*, washed and trapped as suspension in hydrogel, then directly observed by confocal microscopy at 37 °C. Insets show preservation of RBC ovoid shape. Images representative of six independent experiments. Scale bars 2 µm
Double-labeling with theta* and BODIPY-SM (Invitrogen) was performed as follows: RBCs were labeled in suspension with theta* at 20 °C for 15 min, washed, attached onto poly-l-lysine-coated coverslips for 4 min and labeled with 0.75 µM BODIPY-SM at 20 °C for 15 min in the absence of theta*.
For 3D examination, theta* labeled RBCs were washed by centrifugation and the pellet was resuspended in a cold hydrogel (CyGel Sustain Biostatus) at 4 °C (liquid at this temperature). This solution was dropped onto a glass slide, placed at 37 °C to allow the CyGel to reset, covered by a coverslip and directly observed at 37 °C [32]. For confocal imaging of theta*-labeled RBCs after fixation, cells were first incubated with 0.5 % glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, at 20 °C for 20 min, washed in PBS, and mounted onto a glass slide in aqueous mounting medium (Dako). This glutaraldehyde concentration did not produce any autofluorescence signal on unlabeled RBCs at the settings used to analyze theta*-labeled RBCs (data not shown).
All preparations were examined with a Zeiss LSM510 confocal microscope using a plan-Apochromat 63× NA 1.4 oil immersion objective in a thermostated chamber (XL/LSM incubator, Zeiss; Tempcontrol 37-2, PeCon) [33–35] or with a Zeiss wide-field fluorescence microscope (Observer.Z1) using a plan-Apochromat 100×/1.4 oil Ph3 objective. FRAP was performed at 37 °C as described [32–35], by photobleaching of ~2.5-µm2 regions of interest (ROI) to ~40–50 % residual fluorescence intensity; this required 6 iterations at 100 % laser power.
C2C12 myoblast culture and pharmacological treatments
Myoblasts were propagated in DMEM medium (Lonza) supplemented with 10 % fetal calf serum (FCS), two antibiotics and an antimycotic (all from Life Technologies). For experiments, cells were seeded on IBIDI chambers (µ-Dish 35 mm; IBIDI) at ~7000 cells/cm2. After 2 days, cells were concomitantly labeled with 0.6–0.8 µM theta* and 0.3 µM BODIPY-SM in DMEM supplemented with DF-BSA at 10 or 20 °C for 30 min. To block endocytosis, cells were ATP-depleted by incubation with 2.5 mM sodium azide and 12.5 mM deoxyglucose during labeling with theta* and BODIPY-SM. To inhibit F-actin assembly, cells were pre-incubated with 50 nM latrunculin B (LatB) at 37 °C for 30 min before labeling. When appropriate, cells were pre-incubated with 5 mM mβCD to deplete cholesterol. After labeling, cells were washed and examined with a Zeiss LSM510 confocal microscope at 10 or 20 °C (in the continuous presence of sodium azide and deoxyglucose, when appropriate).
Scanning electron microscopy
RBCs were labeled or not with 0.5 µM theta*, spread onto poly-l-lysine-coated coverslips, extensively washed with DMEM, rinsed twice in 0.14 M cacodylate buffer, pH 7.4, then fixed at 20 °C by carefully increasing glutaraldehyde (Fluka-Sigma) concentration in 0.1 M cacodylate buffer from 0.1 to 0.5 to 1 % (v/v), for 30 min each, followed by 2 % glutaraldehyde at 4 °C overnight. Samples were extensively washed in 0.1 M cacodylate buffer and post-fixed with 1 % (w/v) osmium tetroxide at 4 °C for 2 h, dehydrated in graded ethanol series and critical point dried (Leica EM CPD030, Vienna, Austria). A 10-nm gold film was sputter-coated (Leica EM MED020, Vienna, Austria), and specimens were observed in a CM12 electron microscope (Philips, Eindhoven, Netherlands) at 80 kV with the use of the secondary electron detector as described [35].
Morphometry
Measurements of number and size of cholesterol-enriched domains and of total RBC projected area (referred to hemi-RBC) were performed on high-resolution confocal images, using the AxioVision 4.8.2 software (Zeiss). All acquired images were first thresholded strictly in parallel, to remove in a standardized manner areas that would not be analyzed because of too low intensity. Therefore, we isolated only a fixed window of pixel intensities (between 50 and 255 in an 8-bit image). Then, all small dots at the limit of microscopic detection (surface <4 pixels) were automatically removed by the scrap function. A binary image was prepared for analysis in which all small holes were filled. This resulted in the final image for analysis. On visual inspection of these binary images, we could see that these areas corresponded to the domains in the original acquired image. The area and diameters (smallest and largest) of each domain as well as the longest diameter of the RBC were determined. When indicated, 3D-deconvolution was performed on reconstructed x–z vertical sections acquired with the wide-field fluorescence microscope, using the 3D-deconvolution module in AxioVision with the “Inverse Filter” algorithm. When depth pseudo-coloration was added, domain colors indicate their position in 3D: from red (top, free surface) to blue (bottom, coverslip-attached side).
Distribution of cholesterol at the lateral plasma membrane of C2C12 myoblasts was quantified based on signal heterogeneity, measured by the standard deviation of pixel intensity along the plasma membrane. Briefly, specific fields of lateral plasma membrane from high-resolution confocal images were manually encircled and the mean intensity as well as its associated standard deviation was determined for red (theta*) and green (BODIPY-SM) signals using the AxioVision 4.8.2 software (Zeiss). Standard deviations were then normalized as percentage of the corresponding mean intensity to yield variation coefficients that were finally expressed as ratios between theta* and BODIPY-SM.
Statistical analyses
Values are presented as mean ± SEM. The significance of differences was tested by Kolmogorov–Smirnov test (NS, not significant; **, p < 0.01 and ***, p < 0.001).
Results
Theta* specifically binds to cholesterol in artificial and natural membranes
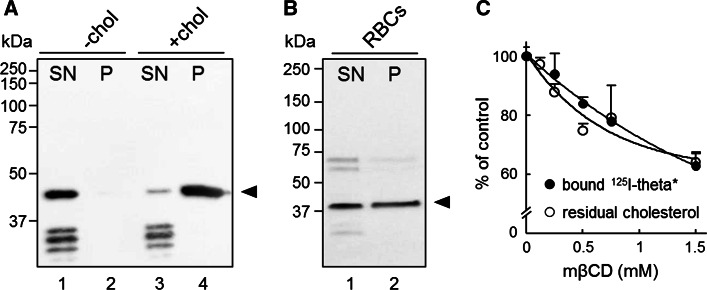
We received the plasmid encoding for theta-D4 fragment fused to the fluorescent protein Dronpa. Dronpa is a reversibly switchable photoactivatable fluorescent probe, thus suitable for super-resolution microscopy, which, however, requires cell fixation. Since lipids are poorly fixable, we preferred vital confocal imaging and thus exchanged the protein Dronpa for the highly photostable (thus best suited for confocal microscopy) red fluorescent monomeric protein mCherry. By swapping Dronpa for mCherry gene in the pET28 expression vector, we generated a fluorescent probe consisting of N-terminal His tag-bearing monomeric mCherry fused with the non-toxic C-terminal domain D4 of theta toxin (His-mCherry-theta-D4; in short theta*), as the minimal fluorescent polypeptide reporter able to bind cholesterol [19]. Western blot analysis of recombinant protein purified from the nickel column revealed a major band at ~41 kDa (arrowheads at Fig. 1A, B), corresponding to the expected position of theta*, as well as other minor bands of lower molecular weights (<37 kDa, lane 1 at Fig. 1A). These bands, representing ~half of total purified recombinant protein, were attributed to spontaneous proteolysis of theta*, as recently reported using a similar approach to generate a fluorescent lysenin toxin fragment specific to sphingomyelin (SM) [32]. Upon incubation with theta* of liposomes prepared without cholesterol and washing by centrifugation, all bands remained in the supernatant (SN, lane 1 at Fig. 1A). In contrast, using cholesterol-bearing liposomes or red blood cells (RBCs), only the ~41 kDa-band corresponding to the intact theta* was recovered in the pellet (P, lanes 4 and 2 at Fig. 1A, B, respectively), while all minor bands remained in the supernatant (SN, lane 3 at Fig. 1A). We concluded that intact theta* specifically interacted with cholesterol in model and natural membranes.
Fig. 1.
His-mCherry-theta (theta*) specifically binds to cholesterol. A, B Specific binding of intact theta* to liposomes containing cholesterol and to red blood cells. Theta* was incubated with multilamellar vesicles (A MLVs, 1:2000 molar ratio by reference to total lipids) containing cholesterol [+chol; lanes 3 and 4] or not (−chol; lanes 1 and 2), or with freshly isolated red blood cells (B RBCs, ~1:25 molar ratio) in suspension. After centrifugation, pellets (P) containing MLVs (B) or RBCs (B) and supernatants (SN) were analyzed by Western blotting for the His-tag on theta*. SN fractions were diluted 5 and 10 times vs P fractions, in panel A and B respectively. Arrowheads (right) correspond to the expected position of intact theta* (~41 kDa). Representative blots from three independent experiments. C Correspondence between theta* binding and cholesterol level in RBCs. Freshly isolated RBCs were either kept untreated or cholesterol-depleted by methyl-β-cyclodextrin (mβCD) prior to labeling in suspension with 125I-theta* (5 × 103Bq) mixed with 1 µM cold theta* (in the continued presence of mβCD if appropriate). After washing by iterative centrifugation/resuspension, 125I-theta* bound to RBCs was measured and expressed as percentage of control cells (filled circles; mean ± SEM of 3–9 samples pooled from three independent experiments). Decrease of theta* binding closely parallels residual cholesterol level (open circles; mean ± SEM from four independent experiments)
We next verified the cholesterol-binding specificity in RBCs by measurement of 125I-labeled theta* adsorption upon graded cholesterol extraction by empty methyl-β-cyclodextrin (mβCD; ~40 % depletion at 1.5 mM mβCD). This treatment was not toxic, based on hemolysis assay (data not shown). Figure 1C showed that decrease of 125I-theta* binding to RBCs paralleled residual total cholesterol membrane content in the range of 0–1.5 mM mβCD. At infinite mβCD concentrations, residual cholesterol content in the entire plasma membrane would reach a ~60 % plateau value (one-phase decay fitting), whereas theta* binding apparently continued to decrease. Such contrast might be explained by the differential cholesterol content addressed in both curves, i.e., total (including outer and inner leaflets; Fig. 1C, open symbols) vs outer leaflet cholesterol (only accessible to theta*; Fig. 1C, closed symbols). Based on a ~50–75 % distribution of cholesterol in the inner leaflet of human RBCs [5, 6], total residual membrane cholesterol curve would probably not reach a zero value upon mβCD extraction. In contrast, when cholesterol at the outer leaflet falls below the threshold required for theta* binding (~30 mol % [25, 26]), the fitting curve could decrease until a zero value. These data established that (1) endogenous cholesterol accessible at the outer leaflet of RBC membrane could be selectively and quantitatively probed by theta*; and (2) the high residual cholesterol level at high mβCD concentrations, which could be attributed to the mβCD-inaccessible inner leaflet cholesterol, could not be probed by theta*.
To test for the possibility of using tracer concentration of theta* for vital confocal imaging, we determined theta* binding parameters to RBCs at increasing concentrations (isotopic dilution) causing no detectable toxicity by hemolysis assay (Fig. S1A). Scatchard plot analysis indicated a curvilinear plot, suggesting two classes of theta* binding sites with high- and low-affinity (Fig. S1B, inset), as previously shown using another theta derivative [41]. According to these two classes of binding sites, we chose the 0.5–1 µM concentration range (corresponding to 15–30 pmol/106RBCs, see Fig. S1B) to define the appropriate theta* concentration for vital confocal imaging experiments (see Fig. S3).
Theta* reveals clustering of cholesterol in numerous submicrometric domains on partially spread living RBCs
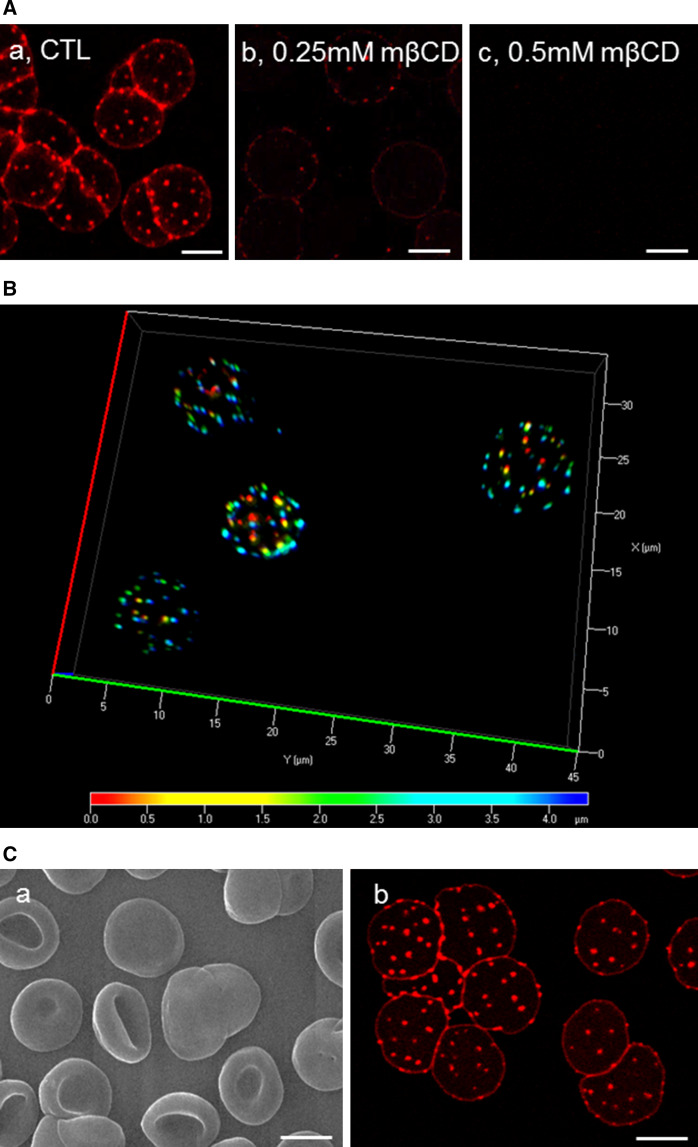
When RBCs were labeled in suspension with theta* at 20 °C, then partially spread onto poly-l-lysine-coated coverslips, numerous well-defined, mostly round submicrometric fluorescent domains were observed (Figs. 2Aa and S2). These were similar in shape and size, but outnumbered submicrometric domains enriched in SM we recently described [32–35]. Theta*-labeled domains did not reflect induction of protrusions from the RBC membrane, as evidenced by the preservation of a smooth featureless surface of theta*-labeled RBCs using scanning electron microscopy (SEM; Fig. 2Ca). We also verified that theta*-labeled domains resisted glutaraldehyde fixation used for SEM (Fig. 2Cb). Domains were visualized on both poly-l-lysine-attached and -free sides (Figs. 2B and S2), ruling out induction by poly-l-lysine coating.
Fig. 2.
Theta* reveals numerous cholesterol submicrometric membrane domains on RBCs partially spread onto poly-l-lysine coverslips. A Submicrometric domains are numerous and cholesterol-dependent on living spread RBCs. Freshly isolated RBCs were either kept untreated (CTL; a) or cholesterol-depleted by 0.25 mM (b ~15 % cholesterol depletion, see Fig. 1C) or 0.5 mM (c ~25 % cholesterol depletion) mβCD at 37 °C, labeled in suspension with theta* in the continuous presence of mβCD, washed, attached-spread onto poly-l-lysine-coated coverslips and directly visualized by confocal microscopy at 20 °C. B Cholesterol submicrometric domains are present on both sides of living spread RBCs. Fresh RBCs were labeled and spread as at A and observed with wide-field fluorescence microscope. 3D-deconvolution and depth pseudo-coloration were then applied to visualize domain position in 3D (red corresponds to poly-l-lysine-free side, blue to poly-l-lysine-attached side). For rotating view, see Fig. S2. C Scanning electron and confocal microscopy of theta*-labeled, glutaraldehyde-fixed RBCs. RBCs were labeled and spread as above, then fixed with 0.5 % glutaraldehyde and processed either for scanning electron microscopy (a) or confocal microscopy (b). Notice that theta* labeling does not ultrastructurally alter the smooth RBC plasma membrane (a) and that glutaraldehyde fixation preserves theta* labeling of cholesterol submicrometric domains (b). All scale bars 5 µm
To further address whether theta* was a passive probe or would instead trigger flat membrane domain formation, we looked at the effect of a fourfold theta* concentration range on the fluorescence intensity and abundance of labeled domains (Fig. S3) using identical settings for image acquisition. Line intensity profiles indicated that (1) domain enrichment reached up to ~20-fold as compared with the RBC membrane outside domains at all concentrations used; (2) average fluorescence intensity first increased proportionally with concentration of theta* added; and (3) saturation was reached before saturation of the CCD camera (Fig. S3), indicating reciprocal steric hindrance. The abundance of submicrometric domains slightly increased from 0.5 to 0.75 µM and then remained stable (~12 domains/hemi-RBC at 0.5 µM vs ~17 at 0.75 and 1 µM). This increase might be explained by the chosen concentration range, i.e., within the first binding site at 0.5 µM (corresponding to 15 pmol/106RBCs, see Fig. S1B) then bestride the first and second binding sites at 1 µM (30 pmol/106RBCs). We, thus, chose the 0.5–0.75 µM concentration range, for all subsequent experiments.
Theta* reveals two pools of cholesterol with differential sensitivity to cholesterol depletion
To further confirm specificity of theta* binding to cholesterol by confocal microscopy, labeling and imaging were performed after cholesterol depletion. Surprisingly, even a modest (~15 %) cholesterol depletion induced by 0.25 mM mβCD strongly decreased number and size of theta*-labeled submicrometric domains (Fig. 2Ab). Domains fully disappeared at 0.5 mM mβCD (~25 % cholesterol depletion) (Fig. 2Ac). The contrast between the high sensitivity to mβCD of labeling cholesterol domains observed by confocal imaging and the minor decrease of 125I-theta* binding (proportional to cholesterol depletion up to 1.5 mM mβCD, see Fig. 1C) under identical conditions pointed to the occurrence of a second more “diffuse” cholesterol pool outside submicrometric domains, as revealed by confocal microscopy upon modification of acquisition settings (Fig. S4).
To further address the differential effect of cholesterol depletion on total 125I-theta* binding to RBCs and labeling of enriched submicrometric domains, we next looked at the effect of mβCD in RBCs pre-labeled by theta*. In these conditions also, whereas 125I-theta* binding was partially preserved (data not shown), cholesterol submicrometric domains fully disappeared supporting the existence of two cholesterol pools (Fig. S5A). The kinetics of domain disappearance on-stage could be further monitored by time-lapse imaging of RBCs pre-labeled with theta*, upon mβCD addition: domains underwent shrinking; then most of them disappeared within 15 min (arrowheads at Fig. S5B) and the few survivors became smaller (arrows at Fig. S5B).
Cholesterol submicrometric domains are promoted by endogenous sphingomyelin
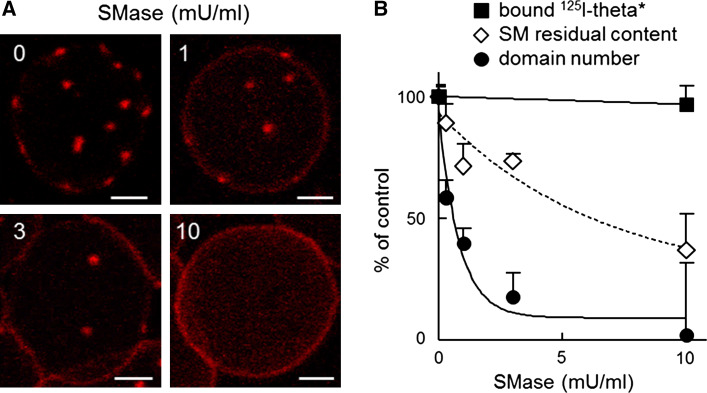
Lipid cohesiveness at the origin of rafts dwells on preferential interactions between cholesterol and sphingolipids. Since we previously found that spontaneous assembly of endogenous SM into submicrometric domains at the RBC plasma membrane depends on cholesterol [32, 34, 35], we conversely examined whether cholesterol submicrometric domains were also depending on SM. To this aim, we looked at the effect of SM conversion into ceramide by sphingomyelinase (SMase, up to 60 % SM depletion without detectable hemolysis; open diamonds at Fig. 3B [32]) on the abundance of theta*-labeled submicrometric domains. As shown at Fig. 3A, B (filled circles), cholesterol submicrometric domains strongly decreased upon moderate SM depletion (~25 % depletion by 3 mU/ml SMase) and became undetectable at higher SM loss (~60 % depletion by 10 mU/ml SMase). In remarkable contrast, total binding of theta* was almost unaffected by SMase, as shown by radio-labeling (filled squares at Fig. 3B) and by Western blotting for the His-tag on theta* (data not shown). This indicated that loss of cholesterol domains upon SMase was not due to impaired theta* binding but reflected a crucial role of SM for cholesterol domain stabilization and further suggested that, at least upon SM depletion, theta* by itself is unable to trigger domain formation.
Fig. 3.
Cholesterol submicrometric domains result from sphingomyelin-dependent clustering. A Cholesterol domains vanish upon sphingomyelin depletion. RBCs were either kept untreated or sphingomyelin (SM)-depleted by the indicated sphingomyelinase (SMase) concentrations, then incubated with theta* in the continuous presence of SMase as appropriate, attached-spread and observed by confocal microscopy, all at 20 °C. Representative of two independent experiments. Scale bars 2 µm. B Sphingomyelin depletion abrogates theta*-labeled cholesterol domains without affecting 125I-theta* binding. RBCs were depleted in SM by the indicated SMase concentrations as at A, then either evaluated for SM content (open diamonds), labeled by non-radioactive theta* as at panel A for quantification of domains by confocal imaging (filled circles), or labeled by 125I-theta* as at Fig. 1C for determination of 125I-theta* binding to RBCs (filled squares). All data are mean ± SEM and expressed as percentage of untreated controls
Altogether, results presented thus far indicated that (1) theta* labels numerous submicrometric domains enriched up to ~20-fold on the entire membrane of living RBCs without causing membrane alteration; (2) theta* labeling preserves accessibility to mβCD, which allows to follow domain erosion and disappearance on-stage; (3) submicrometric domains are much more sensitive to low concentrations of mβCD than total theta*-accessible pool, indicating a second pool; (4) submicrometric domains critically depend on endogenous SM; and (5) theta* by itself might detect pre-existing cholesterol-enriched domains rather than triggering domain formation.
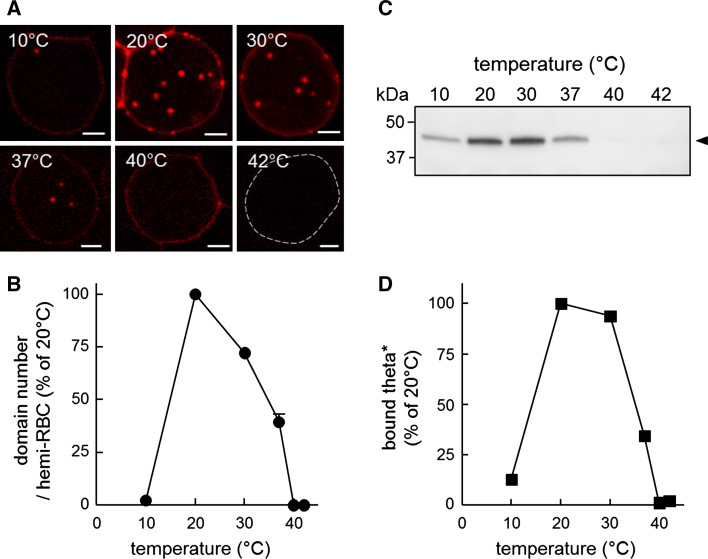
Cholesterol submicrometric domains are temperature-dependent
We then evaluated whether cholesterol domains showed a temperature-dependent behavior as domains enriched in polar lipids [32–35]. Figure 4A, B shows that abundance of submicrometric domains labeled by theta* increased from 10 to 20 °C, peaked at 20 °C, strongly decreased at 37 °C and abruptly vanished around 40 °C. Moreover, increasing temperature from 20 to 37 °C also decreased theta* binding to RBCs (Fig. 4C, D) and average domain size (Fig. S6). Due to the threshold value for theta* binding to cholesterol (~30 mol % [25, 26]), the above data suggested cholesterol submicrometric domain erosion together with a more random distribution upon increase of temperature. The hypothesis of domain erosion was further confirmed by time-lapse imaging of partially spread RBCs under incubation at supra-physiological temperature (Fig. S7). Altogether, these results could reflect a role of lipid phase behavior for cholesterol domain stabilization.
Fig. 4.
Cholesterol submicrometric domain abundance and theta* binding to cholesterol depend on temperature. A Confocal imaging. RBCs were labeled with theta*, washed, attached onto poly-l-lysine-coated coverslips and examined at the indicated temperatures. Dotted line at 42 °C indicates RBC periphery. Domains are more visible at 20–30 °C. All scale bars 2 µm. B Morphometry of domain abundance. Number of theta*-labeled domains was determined per hemi-cell surface at the indicated temperatures and is expressed as percentage of 20 °C. Values are mean ± SEM of domains recorded on 125–634 cells (except at 10 °C, 9 cells) from two independent experiments. C Binding of theta* to RBCs. RBCs were labeled in suspension with theta* at the indicated temperatures. After centrifugation, pellets containing RBCs were analyzed by Western blotting for the His-tag on theta*. Arrowhead (right) corresponds to the expected position of intact theta*. D Western blot quantification of theta* binding using ImageJ software, expressed as percentage of 20 °C. For temperature dependence of domain size, see Fig. S6 and for “on-stage” effect of temperature, see Fig. S7
Cholesterol submicrometric domains resist extensive RBC spreading on coverslips and appear equally abundant on gel-suspended (3D) RBCs
We recently reported that endogenous SM domains labeled by a fluorescent lysenin-derived polypeptide vanished by simply increasing membrane tension [32]. We, thus, next addressed the effect of tension on theta*-labeled cholesterol domains. This question was first examined on spread living RBCs by exploiting differential spreading on poly-l-lysine-coverslips. As shown at Fig. 5A, inset #3, cholesterol domains on highly stretched RBCs remained equally abundant albeit smaller (Fig. 5B), thus appeared much more resistant to increased lateral tension than SM domains. Since spreading onto poly-l-lysine coverslips induced an artificially flat highly stretched conformation, up to almost a flat rigid system far from the biconcave shape and plasticity of circulating RBCs, we looked for cholesterol submicrometric domains on theta*-labeled RBCs after gentle suspension in a 3D-hydrogel examined at physiological temperature. We found that theta*-labeled domains appeared similarly abundant on 3D-RBCs (Fig. 5C). This indicated that cholesterol submicrometric domains were not artificially induced by spreading on poly-l-lysine-coverslips but might be relevant in vivo for circulating human erythrocytes, as previously shown for endogenous SM domains [32].
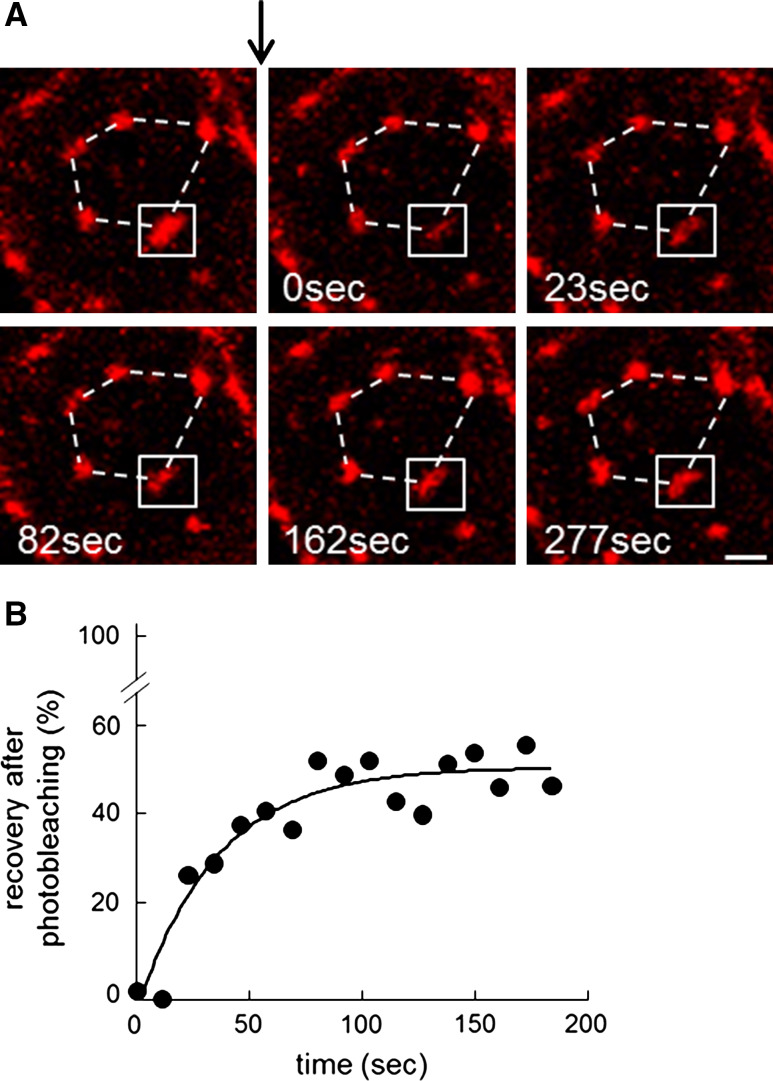
Cholesterol submicrometric domains on spread RBCs are immobile assemblies of exchangeable cholesterol
Having verified the relevance of cholesterol submicrometric domains on coverslips as in 3D support, we returned to poly-l-lysine coverslip analysis for its reproducibility and stability. To explore the biogenesis and maintenance of submicrometric cholesterol assemblies, we applied fluorescence recovery after photobleaching (FRAP) on theta*-labeled domains. Even though the high mCherry photostability was not a priori ideal for this approach, six flashes achieved adequate photobleaching to analyze recovery. As shown at Fig. 6A, unbleached domains retained a fixed position and a comparable size over an interval of ~5 min, compatible with immobilization by the spectrin cytoskeleton, at least in partially spread RBCs. By FRAP, fluorescence was recovered at its exact original position within the bleached boxed zone and reached ~50 % at infinite time with a t1/2 of ~25 s (Fig. 6B). In a cell devoid of any vesicular trafficking and kept in a theta*-free medium, such results indicated that cholesterol submicrometric domains were immobile assemblies composed of dynamic individual cholesterol molecules, or small clusters such as rafts not resolved by vital confocal imaging, able to laterally diffuse along the plane of the membrane. They further supported the existence of two cholesterol pools: the cholesterol domain pool in equilibrium with a second more “diffuse pool”.
Fig. 6.
Cholesterol submicrometric domains are immobile assemblies of exchangeable cholesterol. Analysis by fluorescence recovery after photobleaching (FRAP) at 37 °C of ~2.5 µm2 fields centered on a theta*-labeled submicrometric domain (boxed in A), at the indicated time intervals after photobleaching. A Representative vital imaging. Topological relation between domains over time is indicated by polygonal broken lines. Arrow indicates time of bleaching of the boxed square. Notice immobility in space of all domains (fixed positions as in constellation, dotted lines). Scale bar 1 µm. B Quantification of fluorescence recovery. Fluorescence recovery is expressed as percentage of signal before photobleaching, after correction of residual fluorescence immediately after bleaching. Curve derived by monoexponential fitting indicates a mobile fraction of ~50 % at infinite time corresponding to t1/2 ~25 s (mean of 8 bleached domains pooled from four experiments)
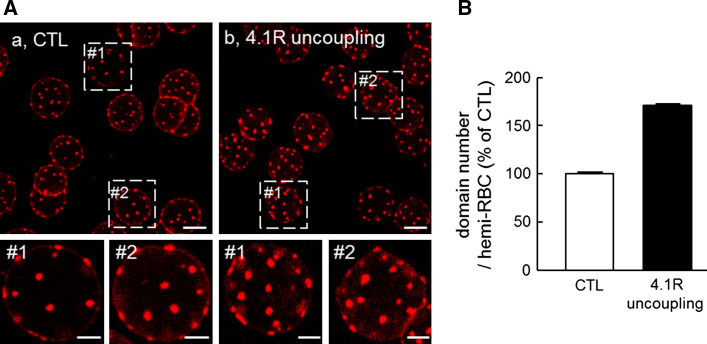
Cholesterol submicrometric domains are restricted by strong membrane:cytoskeleton anchorage via 4.1R-based complexes
Since theta*-labeled cholesterol domains assumed fixed positions in time and space, we next looked at their relation with the underlying spectrin network, in particular at membrane:spectrin anchorage. Among the two types of such anchors, 4.1R complexes can be acutely uncoupled by phosphorylation of one of their key component, α-adducin, upon protein kinase C (PKC) activation by phorbol 12-myristate 13-acetate (PMA) reinforced by calyculin A (CalA) as phosphatase inhibitor [33, 42]. We found that acute uncoupling of membrane:cytoskeleton anchorage increased the abundance of theta*-labeled domains by ~70 % (Fig. 7), suggesting that, in untreated RBCs, 4.1R complexes act as lipid fences that normally restrict the number of submicrometric cholesterol domains.
Fig. 7.
Cholesterol submicrometric domains are restricted by spectrin cytoskeleton via 4.1R-based complexes. A Confocal imaging. RBCs were kept untreated (a, CTL) or treated with PMA as PKC activator combined with the phosphatase inhibitor CalA, to uncouple membrane:cytoskeleton at 4.1R complexes (b, 4.1R uncoupling). Cells were further labeled in suspension with theta*, attached-spread and directly analyzed at 20 °C in continuous presence of PMA + CalA if appropriate. Boxed areas are enlarged in insets. Scale bars 5 μm at general views and 2 µm at insets. B Morphometry. Abundance of theta*-labeled domains per hemi-RBC in PMA + CalA-treated RBCs (filled bar) is expressed as percentage of untreated cells (open bar). Data are mean ± SEM of 495 RBCs (CTL) and 622 RBCs (4.1R uncoupling) and pooled from two independent experiments
Cholesterol- and BODIPY-SM-enriched submicrometric domains partially co-localize
The mutual dependence of cholesterol and SM submicrometric domains for endogenous SM and cholesterol, respectively (Fig. 3; [32]) raised the issue of their spatial relation. We, thus, performed double labeling between endogenous cholesterol, decorated by theta* toxin, and SM, by insertion in the RBC plasma membrane of BODIPY-SM. As shown in Fig. 8, almost all domains enriched in BODIPY-SM coincided with theta*-labeled domains (see yellow arrowheads in Fig. 8A, filled column at Fig. 8B and peaks #2, 3 and 4 at line intensity profile in Fig. 8C), while more than half of cholesterol-enriched submicrometric domains did not coincided with BODIPY-SM enriched domains (see red arrowheads in Fig. 8A, white column at Fig. 8B and peak #5 at Fig. 8C). Such results indicated that the cholesterol submicrometric domains pool should include at least two different types of domains with a differential SM enrichment.
Fig. 8.
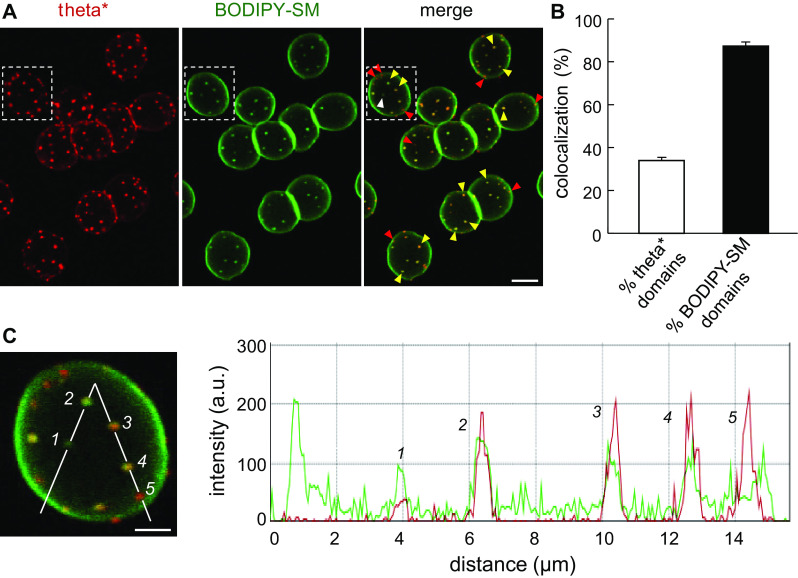
Topological relation between cholesterol- and BODIPY-SM-enriched domains. A Representative vital imaging. Erythrocytes were labeled in suspension with theta* (red), washed, attached onto poly-l-lysine-coverslips and then further labeled with BODIPY-SM (green), all at 20 °C. Notice partial co-localization of theta* domains with BODIPY-SM (yellow arrowheads). White and red arrowheads indicate domains decorated only by BODIPY-SM or theta*, respectively. Representative of six independent experiments. Scale bar 5 µm. B Quantification. Number of co-localization expressed as percentage of either total theta* domains (open bar) or total BODIPY-SM domains (filled bar). Mean ± SEM of 361 RBCs from six independent experiments. C Intensity profile. Right panel shows line intensity profile of red (theta*) and green (BODIPY-SM) signals along path indicated at left by white broken line on the RBC boxed at panel A. Numbers refer to the indicated domains. We consider domains #2, 3 and 4 as perfect co-localization, domain #1 as specifically enriched in BODIPY-SM and domain #5 as specifically enriched in cholesterol. Scale bar 2 µm
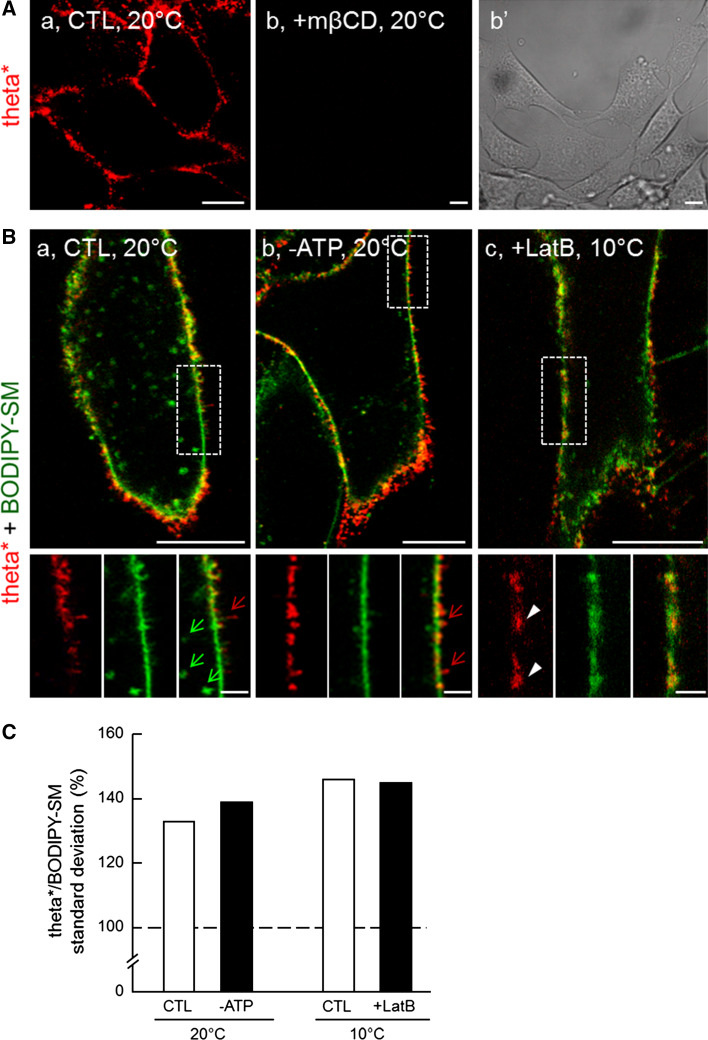
Theta* also reveals heterogeneous cholesterol distribution at the lateral plasma membrane of living C2C12 myoblasts
To extend our analyses to mammalian nucleated cell membranes, we further examined cholesterol distribution at the plasma membrane of living C2C12 myoblasts. Because the large size of the toxin* was not favorable to the analysis of the ventral cell surface, we limited our examination to the lateral plasma membrane (see control at 20 °C in Fig. 9Aa). We first confirmed theta* binding specificity to cholesterol in these cells, based on disappearance of labeling upon ~35 % cholesterol depletion by 5 mM mβCD (Fig. 9Ab, b′). In the absence of mβCD, a discontinuous labeling with theta* was observed (Fig. 9Aa and red at Fig. 9Ba), contrasting with a more uniform signal for BODIPY-SM (green at Fig. 9Ba), raising the question as to whether theta* dots could merely reflect cholesterol concentration at endocytic structures, membrane extensions such as filopodia, microvilli and ruffles, or focal adhesion plaques. To first examine the role of endocytic structures, cells were ATP-depleted by incubation in medium containing sodium azide and deoxyglucose, a treatment that effectively arrested endocytosis ([35]; b; compare with green arrows at a), and surface-labeled with theta* together with BODIPY-SM. Theta* heterogeneous labeling was preserved upon ATP depletion (compare red signal at insets a vs b in Fig. 9B and quantification at Fig. 9C), ruling out that theta* patches were associated with endocytic structures. Second, since large plasma membrane patches of dehydroergosterol have been reported to merely reflect increased lipid content in proportion to increased membrane surface in actin-driven foldings and ruffles rather than lipid enrichment in flat micrometric domains [43], we looked for relation between theta*-labeled domains and the actin cytocortex. To this aim, microfilaments were depolymerized by G-actin sequestration using the membrane-permeant latrunculin B (LatB) and then surface-labeled with theta* and BODIPY-SM. Whereas stress fibers and focal adhesion plaques completely vanished upon 50 nM LatB (data not shown) and filopodia were strongly decreased at the lateral face (c; compare with red arrows at a and b), theta* patches remained unaffected by this treatment (Fig. 9Bc and quantification at Fig. 9C). These experiments thus reasonably ruled out the possibility that theta* submicrometric patches could simply represent endocytic vesicles or actin-dependent surface extensions, and further indicated that they at least partially reflected intrinsic plasma membrane domains.
Fig. 9.
Theta* labels submicrometric domains at the lateral plasma membrane of C2C12 myoblasts. A Theta* labels clusters sensitive to cholesterol depletion. C2C12 myoblasts in IBIDI chambers were pre-incubated or not (a) with 5 mM mβCD at 37 °C (b, b′), then labeled with theta* and visualized by confocal imaging at 20 °C. b′ shows presence of myoblasts in b. B Cholesterol heterogeneity does not primarily reflect endocytotic structures nor ruffles. C2C12 were pre-incubated (c) or not (a, b) with latrunculin B (LatB; to inhibit actin polymerization), then labeled with theta* and BODIPY-SM in presence (b) or not (a, c) of sodium azide and deoxyglucose (-ATP; to prevent endocytosis by energy depletion) and visualized at 20 °C (a, b) or 10 °C (to prevent endocytosis by low temperature, c). Green and red arrows indicate deep endocytotic structures and membrane ruffles respectively; white arrowheads point to cholesterol-enriched lateral domains. Scale bars 10 µm at general views and 2 µm in insets. C Quantification. Mean intensity and associated standard deviations (SDs) were quantified for both theta* and BODIPY-SM signals. SDs were then normalized as percentage of the corresponding mean intensity to yield variation coefficients that were further presented as ratios between theta* and BODIPY-SM. Results are mean of two to three independent experiments pooled from 29 to 50 area per condition. Dotted line indicates theoretical level of equal variation coefficient between theta* and BODIPY-SM
Discussion
Overview
This paper reports on the use of His-mCherry-theta-D4 (theta*) as a vital probe for cholesterol organization at the plasma membrane of human red blood cells (RBCs) as paradigmatic cell model. First, we show that theta* is a specific, non-toxic, sensitive and quantitative probe of cholesterol accessible at the outer plasma membrane leaflet of living cells. Second, we report for the first time that cholesterol organizes into stable submicrometric domains at the living RBC membrane. The abundance of cholesterol domains in gel-suspended RBCs at physiological temperature supports their existence in circulating RBCs in vivo. Third, combination of either moderate cholesterol extraction by methyl-β-cyclodextrin (mβCD) or sphingomyelin (SM) depletion by sphingomyelinase (SMase) with vital fluorescence imaging vs quantitative 125I-theta* binding identifies a second less organized cholesterol pool. Fourth, we provide (1) mechanistic insights on cholesterol submicrometric domain composition and dependence, based on membrane:cytoskeleton anchorage, temperature and SM as well as (2) spatial relation with domains labeled by BODIPY-SM.
Theta* validation as membrane cholesterol probe
In comparison with generally available cholesterol probes, filipin, dehydroergosterol and full-length pore-forming toxins, lipid-binding toxin fragments coupled to a photostable monomeric fluorescent protein, such as theta*, combine several advantages. First, theta* specifically targets endogenous cholesterol in the outer leaflet, consistent with previous studies [44], without deep plasma membrane insertion nor induction of cytolysis upon pore formation. Indeed, whereas full-length perfringolysin O readily forms SDS-resistant oligomers upon binding to membrane cholesterol [45, 46] due to its D1 domain [19, 23, 45, 46], deletion of this domain in theta* prevents oligomerization and cytolysis (no detectable hemolysis). Second, theta* is suitable at low tracer concentration for confocal vital imaging, i.e., without use of fixatives that casts doubt on the genuine lipid organization in intact cells and allows labeling of cells in 3D-hydrogels. Third, theta* is a quantitative probe based on both fluorescence recording and radio-iodination. The main drawback of fluorescent toxin fragments, such as theta*, is their size, much greater than the targeted lipid. For example, theta* molecular weight is about ~100-fold higher than cholesterol and its projected diameter is ~33-fold larger than cholesterol. However, even if size discrepancy predicts steric hindrance, it does not preclude specificity. Indeed, we recently demonstrated that non-saturating concentration of the large mCherry-linked lysenin toxin fragment (similar in size to theta*) labeled the same submicrometric domains as upon insertion at trace level of the small fluorescent analog BODIPY-SM at the RBC plasma membrane [32], whatever the order of labeling. It, thus, validated a posteriori the latter tracer as a bona fide qualitative probe and confirmed that the large mCherry-toxin fragment does not trigger, but rather reveals pre-existing submicrometric domains. This was also beautifully illustrated by the validity of EGF-ferritin to mimic the fate of the 75-fold smaller EGF [47].
We could confirm theta* specificity for cholesterol, based on (1) selective binding of the intact theta* to cholesterol-containing liposomes; (2) abrogation of theta*-labeled submicrometric domains in RBCs as well as in C2C12 myoblasts by confocal imaging upon partial cholesterol depletion by mβCD; and (3) proportionate loss of 125I-theta* binding and membrane cholesterol upon graded RBC cholesterol extraction up to 1.5 mM mβCD. At low theta* concentrations (0.75 µM), sufficient for visualization of cholesterol domains by confocal microscopy, ~1.6 × 105 theta* molecules are bound per RBC, i.e., ~0.3 % of outer plasma membrane leaflet cholesterol content. This tracer level of occupancy suggests minimal perturbation if any.
We estimate the maximal binding of 125I-theta* at ~4.5 × 106/human RBC, in agreement with binding on sheep RBCs of a nicked derivative of perfringolysin O produced by proteolysis with subtilisin Carlsberg (Cθ) (1.05 × 106 sites/RBC [41]). Both estimates diverge by one order of magnitude from the cholesterol content measured at the RBC outer leaflet (~5 × 107 per RBC [40]). This low labeling efficiency of theta* can be explained by several reasons. First, whereas theta* oligomerization can reasonably be excluded (see above), cholesterol molecules might not all be accessible to the toxin* because of the larger surface projection of theta* as compared to cholesterol (~41 kDa, ~10 nm2 [48] vs ~0.3 nm2). Second, labeling efficiency of theta* to cholesterol is dictated by the cholesterol concentration, i.e. the threshold value (~30 mol % cholesterol required for theta* binding [25, 26]). Third, cholesterol embedding in domains enriched with lipids bearing large polar heads or tightly packed in the lo-phase [22] could make it inaccessible to the toxin*. Whereas this theta*-inaccessible cholesterol pool remains to be clarified, its existence is in agreement with a recent study showing that only one cholesterol pool among three is accessible to perfringolysin O in fibroblast plasma membrane [24].
Submicrometric domains as a first pool of cholesterol at the outer plasma membrane leaflet
Submicrometric cholesterol domains revealed here by confocal microscopy on unfixed, partially spread, featureless RBCs are rather abundant (~17 per hemi-RBC at 20 °C at 0.75 µM), large (average diameter of ~600–700 nm) and round. It is reasonable to suggest that round shape of domains is due to minimization of free energy at domain boundary and that their size is controlled by strong membrane:cytoskeleton coupling at regularly spaced anchorage points, thus by membrane tension.
Submicrometric cholesterol domains showed an up to ~20-fold enrichment in cholesterol over the rest of the plasma membrane. This high enrichment, which could be surprising at first, can be justified as follows. Assuming (1) a RBC hemi-surface of ~50 µm2, (2) a total surface occupancy by domains of ~5 µm2 (as determined at Fig. 5B), (3) constitution of domains by 100 % cholesterol, and thus (4) a ~20-fold lower enrichment of the “diffuse pool” (i.e., 5 % cholesterol), it follows that the average plasma membrane cholesterol concentration accessible to theta* (i.e., outer plasma membrane cholesterol) should be ~15 mol % ([5 µm2 × 100] + [45 µm2 × 5]/50 µm2). Since cholesterol at the outer plasma membrane leaflet of human RBCs [5, 6] is estimated at 25–50 %, one can further estimate the cholesterol concentration in the entire RBC plasma membrane (including inner and outer leaflets) at ~40 mol %, in agreement with cholesterol measurement (~45 mol %; [39]).
We also found cholesterol submicrometric domains at the lateral plasma membrane of C2C12 myoblasts. Surprisingly, very few papers have described cholesterol-enriched domains. Among these, submicrometric domains were previously inferred by another group on unfixed ghosts by high-resolution atomic force microscopy upon cholesterol extraction by mβCD (100–300 nm in diameter [49]). Alternatively, cholesterol submicrometric domains have been directly visualized on HeLa cells by super-resolution microscopy (PALM) using Dronpa-theta-D4 (~240 nm in diameter) at 20 °C [27], but not in fibroblasts by high-resolution ion mass spectrometry based on 18O-cholesterol at 37 °C [28]. Our results provide some reasons as to why cholesterol domains may have been absent or dismissed by other investigators. First, visualization of submicrometric cholesterol domains may depend on a critical threshold concentration (see above and [25, 26]), and cholesterol abundance varies considerably between cell plasma membranes (~50 mol % in HeLa cells [50] vs ~45 mol % in RBCs [39] vs ~40 mol % in myoblasts [51] vs ~30 mol % in fibroblasts [52]). Second, domain abundance also strongly varies with temperature and declines at low and high temperatures, a possible cause of non-reproducibility. Third, fixation must be considered as a serious limitation, since even high formaldehyde concentrations do not actually arrest membrane protein long-range movement [30]. Fourth, since we found that cholesterol domain number was increased upon uncoupling membrane from the underlying spectrin cytoskeleton at 4.1R complexes, we speculate that unequal membrane:cytoskeleton anchorage (particularly strong in RBCs and myoblasts due to spectrin and dystrophin, respectively), could also potentially explain differences in supramolecular cholesterol plasma membrane distribution among cell types.
A second pool of cholesterol at the outer RBC plasma membrane
Using full-length perfringolysin O toxin binding, Radhakrishnan and colleagues recently discriminated three cholesterol membrane pools in fibroblasts [24]. Four lines of evidence indicate that at least two cholesterol pools must also coexist at the RBC surface. First, our data show that, in RBCs, submicrometric cholesterol domains occupy ~8 % of the cell surface and line intensity profiles indicate a uniform up to ~20-fold enrichment over signal outside domains. Incidentally, a diffuse pool of cholesterol was visible by confocal microscopy upon adapted laser acquisition settings. Moreover, despite abrogation of all cholesterol submicrometric domains after ~25 % depletion by 0.5 mM mβCD, 125I-theta* binding to RBCs was only decreased by ~15 %. Third, 125I-theta* binding was fully preserved upon suppression of cholesterol submicrometric domains by SMase, indicating cholesterol dynamic redistribution into this second “diffuse pool”. Fourth, by FRAP, photobleached domains could rapidly recover, suggesting replenishment of the bleached domains with mobile unbleached theta*:cholesterol complexes from outside of domains. Altogether, these data suggest the existence of at least two distinct cholesterol pools at the RBC plasma membrane: the cholesterol-enriched submicrometric domains surrounded by a more “diffuse pool”. Our preliminary data on cholesterol distribution in RBC membranes after detergent extraction and floatation in sucrose density gradients confirm the presence of two cholesterol-enriched pools and suggest that submicrometric cholesterol domains do not simply represent large clusters of non-resolved nanometric lipid rafts (data not shown).
Biogenesis of cholesterol submicrometric domains: restriction by spectrin vs promotion by sphingomyelin
Whereas increase by stretching on coated coverslips up to an almost two-dimensional object does not affect the abundance of cholesterol submicrometric domains for a given temperature, their size is reduced on maximally stretched RBCs. This suggests a crucial role of membrane tension for domain biogenesis and/or maintenance, as observed on giant lipid vesicles [53]. Cholesterol domains are also remarkably stable in time and space, as shown by fluorescence recovery after photobleaching (FRAP). Moreover, increase of cholesterol submicrometric domains upon acute uncoupling of membrane:spectrin anchorage at 4.1R complexes indicates restriction by protein:based fences [54, 55]. These three lines of evidence suggest that cholesterol submicrometric domains are restricted by membrane:spectrin anchorage, in perfect agreement with the picket-fence network which allows confined diffusion of lipids and proteins in mesoscale domains [54, 56] while preventing macroscopic domain formation [57, 58].
However, the strong dependence on the temperature of cholesterol submicrometric domains can hardly be exclusively explained by changes in the cytoskeleton and thus indicates a phase behavior, i.e., lipid-based cohesion. Indeed, domain abundance decreased between 20 and 37 °C, resulting in a modification of cholesterol distribution, random and/or in smaller domains undistinguishable at confocal microscopy resolution. Since there is a threshold value for binding of theta* to cholesterol (see above; [25, 26]) and because we still observed theta* binding to RBCs at 37 °C, both reorganizations could occur at this temperature. However, above 37 °C, cholesterol distribution would randomize. Moreover, all cholesterol domains disappeared upon SM depletion, which could not be explained by a decrease of theta* binding to cholesterol (unchanged as shown by 125I-theta* binding experiment). This observation not only indicates that theta* detects pre-existing domains rather than triggering membrane domain formation, but further points to a modification of cholesterol lateral organization under SM depletion in smaller domains undistinguishable at confocal microscopy resolution. It further suggests that local SM membrane composition represents a major stabilization factor for cholesterol submicrometric domains. Accordingly, although cholesterol domains were more abundant (~17 vs ~5/hemi-RBC for cholesterol- and SM-enriched domains, respectively), and less dependent to membrane tension than SM domains, both types of domains shared several features: (1) size and shape; (2) temperature dependence with a peak abundance at 20 °C; and (3) stability in time and space (see also [32]). All these indirect evidences of cholesterol and SM interactions are supported by the almost perfect coincidence of BODIPY-SM domains with theta*-labeled domains. However, a significant fraction of theta*-labeled domains did not show detectable BODIPY-SM enrichment. Since all theta*-labeled domains disappeared upon SMase treatment, one can postulate that all cholesterol-enriched submicrometric domains contain SM, yet at variable SM:cholesterol ratios. Altogether, these data indicate that segregation of cholesterol in submicrometric domains at the surface of living RBCs is prevented by strong anchorage to cytoskeleton but favored by the local lipid (cholesterol vs SM) composition. The recent findings of Honigmann and collaborators on model membranes [58] seem to support this view.
Biological relevance of distinct cholesterol pools
Visualization of apparently identical submicrometric cholesterol domains in RBCs gently suspended in a 3D-hydrogel at physiological temperature, as in attached-spread RBCs, suggests relevance for circulating biconcave RBCs. Three roles, yet not mutually exclusive, can be envisaged in vivo. First, by analogy to caveolae in endothelial cells [59], submicrometric cholesterol domains in RBCs may provide readily mobilizable membrane modules, allowing RBC deformability necessary to squeeze into the narrow splenic pores for ~10,000 times during their 120-day lifetime. Second, boundaries of submicrometric domains may represent high-tension fragility sites, i.e, reflect propensity to fragmentation and hemolysis during splenic filtration of senescent or diseased RBCs. To test these two first hypotheses will require biophysical studies [60, 61] or comparative vesiculation experiments [62–64]. Third, lipid clustering into stable submicrometric domains may help to organize proteins into functional domains, by specifying local intrinsic membrane thickness or fluidity that favors protein clustering, thus signaling or molecular exchanges. For example, SM clustering at the trans-Golgi network organizes transmembrane proteins into functional enzymatic domains [65]. Likewise, phosphatidylinositol-4,5-bisphosphate (PIP2) micrometric domains at the inner plasma membrane leaflet induce clustering of the SNAP receptor, syntaxin-1A [66]. Specifically at the RBC membrane, one could envisage that lipid domains impact on the recruitment and activity of channels and transporters involved in calcium regulation, a key regulator of RBC deformability and splenic clearance. Alternatively, lipid domains could help clustering CD47, a molecular switch that controls phagocytosis of senescent RBCs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (AVI 108863 kb)
Acknowledgments
We thank Drs. A. Miyawaki, M. Abe and T. Kobayashi at Riken Brain Science Institute (Saitama, Japan) as well as H. Mizuno (KU Leuven, Belgium) for generously supplying the Dronpa-theta-D4 plasmid, Dr. A. De Matteis (Naples, Italy) for mCherry-Rab5 plasmid and P. Gailly for C2C12 myoblasts; P. Henriet, H. Emonard and J. Lorent for help in producing toxin* and liposomes and T. Lac and N. Chevalier for technical support (UCL, Belgium). This work was supported by grants from UCL (FSR), the F.R.S.-FNRS, Salus Sanguinis foundation, ARC, IUAP and the Région Wallonne. DT and MVdC are Research associates at Fonds de la Recherche Scientifique (F.R.S.-FNRS).
Abbreviations
- CalA
Calyculin A
- DF-BSA
Defatted-bovine serum albumin
- FRAP
Fluorescence recovery after photobleaching
- LatB
Latrunculin B
- mβCD
Methyl-β-cyclodextrin
- MLVs
Multilamellar vesicles
- PC
Phosphatidylcholine
- PKC
Protein kinase C
- PMA
Phorbol 12-myristate 13-acetate
- RBC
Red blood cell
- SEM
Scanning electron microscopy
- SM
Sphingomyelin
- SMase
Sphingomyelinase
- theta*
His-mCherry-theta-D4
References
- 1.Lange Y, Swaisgood MH, Ramos BV, Steck TL. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J Biol Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- 2.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 3.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Parton RG, Way M, Zorzi N, Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroeder F, Nemecz G, Wood WG, Joiner C, Morrot G, Ayraut-Jarrier M, Devaux PF. Transmembrane distribution of sterol in the human erythrocyte. Biochim Biophys Acta. 1991;1066:183–192. doi: 10.1016/0005-2736(91)90185-B. [DOI] [PubMed] [Google Scholar]
- 6.Lange Y, Slayton JM. Interaction of cholesterol and lysophosphatidylcholine in determining red cell shape. J Lipid Res. 1982;23:1121–1127. [PubMed] [Google Scholar]
- 7.Sun M, Northup N, Marga F, Huber T, Byfield FJ, Levitan I, Forgacs G. The effect of cellular cholesterol on membrane-cytoskeleton adhesion. J Cell Sci. 2007;120:2223–2231. doi: 10.1242/jcs.001370. [DOI] [PubMed] [Google Scholar]
- 8.Laurenzana A, Fibbi G, Chilla A, Margheri G, Del Rosso T, Rovida E, Del Rosso M, Margheri F. Lipid rafts: integrated platforms for vascular organization offering therapeutic opportunities. Cell Mol Life Sci. 2015;72:1537–1557. doi: 10.1007/s00018-014-1814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumann P, Thiele W, Cremers N, Muppala S, Krachulec J, Diefenbacher M, Kassel O, Mudduluru G, Allgayer H, Frame M, Sleeman JP. CD24 interacts with and promotes the activity of c-src within lipid rafts in breast cancer cells, thereby increasing integrin-dependent adhesion. Cell Mol Life Sci. 2012;69:435–448. doi: 10.1007/s00018-011-0756-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chichili GR, Rodgers W. Cytoskeleton-membrane interactions in membrane raft structure. Cell Mol Life Sci. 2009;66:2319–2328. doi: 10.1007/s00018-009-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veiga MP, Arrondo JL, Goni FM, Alonso A, Marsh D. Interaction of cholesterol with sphingomyelin in mixed membranes containing phosphatidylcholine, studied by spin-label ESR and IR spectroscopies. A possible stabilization of gel-phase sphingolipid domains by cholesterol. Biochemistry. 2001;40:2614–2622. doi: 10.1021/bi0019803. [DOI] [PubMed] [Google Scholar]
- 12.Fidorra M, Heimburg T, Bagatolli LA. Direct visualization of the lateral structure of porcine brain cerebrosides/POPC mixtures in presence and absence of cholesterol. Biophys J. 2009;97:142–154. doi: 10.1016/j.bpj.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bali R, Savino L, Ramirez DA, Tsvetkova NM, Bagatolli L, Tablin F, Crowe JH, Leidy C. Macroscopic domain formation during cooling in the platelet plasma membrane: an issue of low cholesterol content. Biochim Biophys Acta. 2009;1788:1229–1237. doi: 10.1016/j.bbamem.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossmann G, Opekarova M, Malinsky J, Weig-Meckl I, Tanner W. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 2007;26:1–8. doi: 10.1038/sj.emboj.7601466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malinsky J, Opekarova M, Grossmann G, Tanner W. Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu Rev Plant Biol. 2013;64:501–529. doi: 10.1146/annurev-arplant-050312-120103. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton-Miller JM. Chemistry and biology of the polyene macrolide antibiotics. Bacteriol Rev. 1973;37:166–196. [PMC free article] [PubMed] [Google Scholar]
- 17.Gimpl G, Gehrig-Burger K. Probes for studying cholesterol binding and cell biology. Steroids. 2011;76:216–231. doi: 10.1016/j.steroids.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Waheed AA, Shimada Y, Heijnen HF, Nakamura M, Inomata M, Hayashi M, Iwashita S, Slot JW, Ohno-Iwashita Y. Selective binding of perfringolysin O derivative to cholesterol-rich membrane microdomains (rafts) Proc Natl Acad Sci USA. 2001;98:4926–4931. doi: 10.1073/pnas.091090798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimada Y, Maruya M, Iwashita S, Ohno-Iwashita Y. The C-terminal domain of perfringolysin O is an essential cholesterol-binding unit targeting to cholesterol-rich microdomains. Eur J Biochem. 2002;269:6195–6203. doi: 10.1046/j.1432-1033.2002.03338.x. [DOI] [PubMed] [Google Scholar]
- 20.Bavdek A, Gekara NO, Priselac D, Gutierrez Aguirre I, Darji A, Chakraborty T, Macek P, Lakey JH, Weiss S, Anderluh G. Sterol and pH interdependence in the binding, oligomerization, and pore formation of Listeriolysin O. Biochemistry. 2007;46:4425–4437. doi: 10.1021/bi602497g. [DOI] [PubMed] [Google Scholar]
- 21.Farrand AJ, LaChapelle S, Hotze EM, Johnson AE, Tweten RK. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc Natl Acad Sci USA. 2010;107:4341–4346. doi: 10.1073/pnas.0911581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson LD, Johnson AE, London E. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: insights into the origin of perfringolysin O-lipid raft interaction. J Biol Chem. 2008;283:4632–4642. doi: 10.1074/jbc.M709483200. [DOI] [PubMed] [Google Scholar]
- 23.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–692. doi: 10.1016/S0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 24.Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife. 2014;3:e02882. doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das A, Goldstein JL, Anderson DD, Brown MS, Radhakrishnan A. Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc Natl Acad Sci USA. 2013;110:10580–10585. doi: 10.1073/pnas.1309273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohno-Iwashita Y, Shimada Y, Waheed AA, Hayashi M, Inomata M, Nakamura M, Maruya M, Iwashita S. Perfringolysin O, a cholesterol-binding cytolysin, as a probe for lipid rafts. Anaerobe. 2004;10:125–134. doi: 10.1016/j.anaerobe.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Mizuno H, Abe M, Dedecker P, Makino A, Rocha S, Ohno-Iwashita Y, Hofkens J, Kobayashi T, Miyawaki A. Fluorescent probes for superresolution imaging of lipid domains on the plasma membrane. Chem Sci. 2011;2:1548–1553. doi: 10.1039/c1sc00169h. [DOI] [Google Scholar]
- 28.Frisz JF, Klitzing HA, Lou K, Hutcheon ID, Weber PK, Zimmerberg J, Kraft ML. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J Biol Chem. 2013;288:16855–16861. doi: 10.1074/jbc.M113.473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frisz JF, Lou K, Klitzing HA, Hanafin WP, Lizunov V, Wilson RL, Carpenter KJ, Kim R, Hutcheon ID, Zimmerberg J, Weber PK, Kraft ML. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc Natl Acad Sci USA. 2013;110:E613–E622. doi: 10.1073/pnas.1216585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziomek CA, Schulman S, Edidin M. Redistribution of membrane proteins in isolated mouse intestinal epithelial cells. J Cell Biol. 1980;86:849–857. doi: 10.1083/jcb.86.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimmer S, van Deurs B, Sandvig K. Membrane ruffling and macropinocytosis in A431 cells require cholesterol. J Cell Sci. 2002;115:2953–2962. doi: 10.1242/jcs.115.14.2953. [DOI] [PubMed] [Google Scholar]
- 32.Carquin M, Pollet H, Veiga-da-Cunha M, Cominelli A, Van Der Smissen P, N’Kuli F, Emonard H, Henriet P, Mizuno H, Courtoy PJ, Tyteca D. Endogenous sphingomyelin segregates into submicrometric domains in the living erythrocyte membrane. J Lipid Res. 2014;55:1331–1342. doi: 10.1194/jlr.M048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Auria L, Fenaux M, Aleksandrowicz P, Van Der Smissen P, Chantrain C, Vermylen C, Vikkula M, Courtoy PJ, Tyteca D. Micrometric segregation of fluorescent membrane lipids: relevance for endogenous lipids and biogenesis in erythrocytes. J Lipid Res. 2013;54:1066–1076. doi: 10.1194/jlr.M034314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Auria L, Van Der Smissen P, Bruyneel F, Courtoy PJ, Tyteca D. Segregation of fluorescent membrane lipids into distinct micrometric domains: evidence for phase compartmentation of natural lipids? PLoS One. 2011;6:e17021. doi: 10.1371/journal.pone.0017021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyteca D, D’Auria L, Van Der Smissen P, Medts T, Carpentier S, Monbaliu JC, de Diesbach P, Courtoy PJ. Three unrelated sphingomyelin analogs spontaneously cluster into plasma membrane micrometric domains. Biochim Biophys Acta. 2010;1798:909–927. doi: 10.1016/j.bbamem.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 36.Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem J. 1993;294(Pt 1):1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman SR, Kurdia A, Ammann L, Kakhniashvili D, Daescu O. The human red blood cell proteome and interactome. Exp Biol Med (Maywood) 2007;232:1391–1408. doi: 10.3181/0706-MR-156. [DOI] [PubMed] [Google Scholar]
- 38.Salomao M, Zhang X, Yang Y, Lee S, Hartwig JH, Chasis JA, Mohandas N, An X. Protein 4.1R-dependent multiprotein complex: new insights into the structural organization of the red blood cell membrane. Proc Natl Acad Sci USA. 2008;105:8026–8031. doi: 10.1073/pnas.0803225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper RA. Influence of increased membrane cholesterol on membrane fluidity and cell function in human red blood cells. J Supramol Struct. 1978;8:413–430. doi: 10.1002/jss.400080404. [DOI] [PubMed] [Google Scholar]
- 40.Cornwell DG, Heikkila RE, Bar RS, Biagi GL. Red blood cell lipids and the plasma membrane. JAOCS. 1967;45:297–304. doi: 10.1007/BF02667099. [DOI] [PubMed] [Google Scholar]
- 41.Ohno-Iwashita Y, Iwamoto M, Mitsui K, Ando S, Nagai Y. Protease-nicked theta-toxin of Clostridium perfringens, a new membrane probe with no cytolytic effect, reveals two classes of cholesterol as toxin-binding sites on sheep erythrocytes. Eur J Biochem. 1988;176:95–101. doi: 10.1111/j.1432-1033.1988.tb14255.x. [DOI] [PubMed] [Google Scholar]
- 42.Betz T, Lenz M, Joanny JF, Sykes C. ATP-dependent mechanics of red blood cells. Proc Natl Acad Sci USA. 2009;106:15320–15325. doi: 10.1073/pnas.0904614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wustner D. Plasma membrane sterol distribution resembles the surface topography of living cells. Mol Biol Cell. 2007;18:211–228. doi: 10.1091/mbc.E06-05-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maekawa M, Fairn GD. Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J Cell Sci. 2015;128:1422–1433. doi: 10.1242/jcs.164715. [DOI] [PubMed] [Google Scholar]
- 45.Iwamoto M, Ohno-Iwashita Y, Ando S. Effect of isolated C-terminal fragment of theta-toxin (perfringolysin O) on toxin assembly and membrane lysis. Eur J Biochem. 1990;194:25–31. doi: 10.1111/j.1432-1033.1990.tb19422.x. [DOI] [PubMed] [Google Scholar]
- 46.Sokolov A, Radhakrishnan A. Accessibility of cholesterol in endoplasmic reticulum membranes and activation of SREBP-2 switch abruptly at a common cholesterol threshold. J Biol Chem. 2010;285:29480–29490. doi: 10.1074/jbc.M110.148254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKanna JA, Haigler HT, Cohen S. Hormone receptor topology and dynamics: morphological analysis using ferritin-labeled epidermal growth factor. Proc Natl Acad Sci USA. 1979;76:5689–5693. doi: 10.1073/pnas.76.11.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Roe C, Courtoy PJ, Baudhuin P. A model of protein-colloidal gold interactions. J Histochem Cytochem. 1987;35:1191–1198. doi: 10.1177/35.11.3655323. [DOI] [PubMed] [Google Scholar]
- 49.Cai M, Zhao W, Shang X, Jiang J, Ji H, Tang Z, Wang H. Direct evidence of lipid rafts by in situ atomic force microscopy. Small. 2012;8:1243–1250. doi: 10.1002/smll.201102183. [DOI] [PubMed] [Google Scholar]
- 50.Bosmann HB, Hagopian A, Eylar EH. Cellular membranes: the isolation and characterization of the plasma and smooth membranes of HeLa cells. Arch Biochem Biophys. 1968;128:51–69. doi: 10.1016/0003-9861(68)90008-8. [DOI] [PubMed] [Google Scholar]
- 51.Perkins RG, Scott RE. Plasma membrane phospholipid, cholesterol and fatty acyl composition of differentiated and undifferentiated L6 myoblasts. Lipids. 1978;13:334–337. doi: 10.1007/BF02533724. [DOI] [PubMed] [Google Scholar]
- 52.Bezrukov L, Blank PS, Polozov IV, Zimmerberg J. An adhesion-based method for plasma membrane isolation: evaluating cholesterol extraction from cells and their membranes. Anal Biochem. 2009;394:171–176. doi: 10.1016/j.ab.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oglecka K, Rangamani P, Liedberg B, Kraut RS, Parikh AN. Oscillatory phase separation in giant lipid vesicles induced by transmembrane osmotic differentials. Elife. 2014;3:e03695. doi: 10.7554/eLife.03695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kusumi A, Suzuki KG, Kasai RS, Ritchie K, Fujiwara TK. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem Sci. 2011;36:604–615. doi: 10.1016/j.tibs.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 57.Ehrig J, Petrov EP, Schwille P. Near-critical fluctuations and cytoskeleton-assisted phase separation lead to subdiffusion in cell membranes. Biophys J. 2011;100:80–89. doi: 10.1016/j.bpj.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Honigmann A, Sadeghi S, Keller J, Hell SW, Eggeling C, Vink R. A lipid bound actin meshwork organizes liquid phase separation in model membranes. Elife. 2014;3:e01671. doi: 10.7554/eLife.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, Morone N, Parton RG, Raposo G, Sens P, Lamaze C, Nassoy P. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144:402–413. doi: 10.1016/j.cell.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Q, Park S, Ma H. Microfluidic micropipette aspiration for measuring the deformability of single cells. Lab Chip. 2012;12:2687–2695. doi: 10.1039/c2lc40205j. [DOI] [PubMed] [Google Scholar]
- 61.Bassereau P, Sorre B, Levy A. Bending lipid membranes: experiments after W. Helfrich’s model. Adv Colloid Interface Sci. 2014;208:47–57. doi: 10.1016/j.cis.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Willekens FL, Werre JM, Groenen-Dopp YA, Roerdinkholder-Stoelwinder B, de Pauw B, Bosman GJ. Erythrocyte vesiculation: a self-protective mechanism? Br J Haematol. 2008;141:549–556. doi: 10.1111/j.1365-2141.2008.07055.x. [DOI] [PubMed] [Google Scholar]
- 63.Salzer U, Zhu R, Luten M, Isobe H, Pastushenko V, Perkmann T, Hinterdorfer P, Bosman GJ. Vesicles generated during storage of red cells are rich in the lipid raft marker stomatin. Transfusion. 2008;48:451–462. doi: 10.1111/j.1537-2995.2007.01549.x. [DOI] [PubMed] [Google Scholar]
- 64.Bosman GJ, Lasonder E, Groenen-Dopp YA, Willekens FL, Werre JM. The proteome of erythrocyte-derived microparticles from plasma: new clues for erythrocyte aging and vesiculation. J Proteomics. 2012;76:203–210. doi: 10.1016/j.jprot.2012.05.031. [DOI] [PubMed] [Google Scholar]
- 65.van Galen J, Campelo F, Martinez-Alonso E, Scarpa M, Martinez-Menarguez JA, Malhotra V. Sphingomyelin homeostasis is required to form functional enzymatic domains at the trans-Golgi network. J Cell Biol. 2014;206:609–618. doi: 10.1083/jcb.201405009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, Dier M, Hell SW, Grubmuller H, Diederichsen U, Jahn R. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (AVI 108863 kb)