Abstract
We analyzed the signal that directs the outer membrane protein with the C-terminal transmembrane segment (TMS) to mammalian mitochondria by using yeast Tom5 as a model and green fluorescent protein as a reporter. Deletions or mutations were systematically introduced into the TMS or the flanking regions and their intracellular localization in COS-7 cells was examined using confocal microscopy and cell fractionation. 1) Three basic amino acid residues within the C-terminal five-residue segment (C-segment) contained the information required for mitochondrial-targeting. Reduction of the net positive charge in this segment decreased mitochondrial specificity, and the mutants were distributed throughout the intracellular membranes. 2) Elongation of the TMS interfered with the function of the C-segment and the mutants were delivered to the intracellular membranes. 3) Separation of the TMS and C-segment by linker insertion severely impaired mitochondrial targeting function, leading to mislocalization to the cytoplasm. 4) Mutations or small deletions in the region of the TMS flanking the C-segment also impaired the mitochondrial targeting. Therefore, the moderate length of the TMS, the positive charges in the C-segment, and the distance between or context of the TMS and C-segment are critical for the targeting signal. The structural characteristics of the signal thus defined were also confirmed with mammalian C-tail–anchored protein OMP25.
INTRODUCTION
Most eucaryotic membrane proteins are inserted into translocation-competent membranes and then transported to organelles that do not contain a translocation apparatus. All integral membrane proteins of the mitochondrial outer membrane identified to date are encoded by nuclear DNA, synthesized by cytoplasmic ribosomes as mature size precursors, and posttranslationally integrated into the membrane (Shore et al., 1995; Mihara, 2000). Unlike the matrix-targeted preproteins with cleavable presequences, the signals that target the outer membrane proteins are contained within the mature protein sequence. The import receptors of the preprotein translocase of the mitochondrial outer membrane (TOM complex; Pfanner and Geissler, 2001), Tom70 (Hurt et al., 1985; McBride et al., 1992), and Tom20 (Schneider et al., 1991; Kanaji et al., 2000) are anchored to the membrane through the N-terminal transmembrane segment (TMS) in the Nin-Cout orientation (“N-anchored protein”). Tom22, which functions as the preprotein receptor and organizer of the TOM complex, is anchored to the outer membrane in the Nout-Cin orientation through a TMS in the middle portion of the molecule (Keil and Pfanner, 1993; Kiebler et al., 1993; Rodriguez-Cousino et al., 1998; Egan et al., 1999). Tom40 (Vestweber et al., 1989; Keil et al., 1993; Hill et al., 1998; Rapaport and Neupert, 1999; Ahting et al., 2001) and porin (Mihara and Sato, 1985; Stanley et al., 1995; Mannella et al., 1996) are β-barrel proteins spanning the outer membrane by 12–14 antiparallel β-strands and function as the transport channels of preproteins or small molecules, respectively. The N-terminal TMS, with moderate hydrophobicity and a net positive charge within five residues of the C-terminal flanking region, functions as the mitochondrial targeting signal of Tom20 (Kanaji et al., 2000). During translation, the signal recognition particle (SRP; Walter and Johnson, 1994) recognizes the TMS of Tom20; basic amino acid residues in the C-terminal flanking region, however, interfere with the function of the SRP, thus preventing its SRP-dependent endoplasmic reticulum (ER) targeting (Kanaji et al., 2000). There are similar structural features in rat and Neurospora crassa Tom70 and several other outer membrane proteins (Mihara, 2000).
Another class of membrane proteins, the “C-anchored” proteins, are composed of three domains: an N-terminal hydrophilic functional domain exposed to the cytosol; a TMS; and the following short hydrophilic segment (C-segment), probably extruding to the intermembrane space. Cytochrome b5 (Mitoma and Ito, 1992; Borgese et al., 2001), microsomal aldehyde dehydrogenase (Masaki et al., 1994), syntaxin 1 (Masaki et al., 1998), and vesicle-associated membrane proteins VAMP-1A and VAMP-2 (Kim et al., 1999) are destined for the ER, whereas mitochondrial outer membrane cytochrome b5 (OMb; Kuroda and Ito, 1998), a splice isoform of VAMP-1A, VAMP-1B (Isenmann and Wattenberg, 1998), and OMP25 (Nemoto and Camilli, 1999) are destined for the mitochondria. Figure 1 depicts several of these proteins. The length of the TMS is critical and hydrophobic segments that are 20 residues or longer function as the ER-targeting signal (Kim et al., 1999). The importance of the N- or C-terminal flanking regions has also been demonstrated (Mitoma et al., 1992; Masaki et al., 1994; Kim et al., 1999; Borgese et al., 2001). Isenmann and Wattenberg (1998) demonstrated that mitochondrial localization of the mitochondrial isoform VAMP-1B is determined by the C-terminal positive charge and the length of the TMS (17 residues). Kuroda and Ito (1998) demonstrated that basic amino acid residues at the C-terminal tail are critical for OMb; when either one of two basic amino acid residues in the C-terminal tail was replaced by alanine, the mutant OMb then became targeted to the ER. Despite these findings, the precise character of the mitochondrial targeting signals, as well as the component(s) that recognize the signal, remain to be analyzed. As a first step toward understanding the targeting and insertion mechanisms of the C-anchored proteins, we used yeast Tom5 as a model and characterized the mitochondrial targeting signal of mammalian cells in detail. Tom5 is tightly associated with the TOM core complex and represents the connecting link between the import receptors and Tom40 (Dietmeier et al., 1997), although the mammalian homolog of Tom5 is not known. It is composed of the cytosolic N-terminal half of 27 residues carrying a net negative charge, the TMS at the following region consisting of 18 amino acid residues (residues 28–45), and the C-terminal hydrophilic region of five amino acid residues, which contains three positive charges (referred to as the C-segment). We constructed a series of green fluorescent protein (GFP)-Tom5 fusion proteins carrying deletions, insertions, or point mutations within the TMS or its flanking regions and examined their intracellular localization in COS-7 cells by using confocal microscopy and defined the mitochondrial targeting signals. These defined structural features of the signal were confirmed with authentic mammalian C-tail anchor protein OMP25. We also characterized the targeting and membrane anchorage of the GFP-Tom5 fusions and GFP-OMP25 by using cell fractionation, alkaline carbonate extraction, and blue native (BN)-PAGE, and compared them with those expressed in yeast cells.
Figure 1.
Amino acid sequences of the C-terminal portion of several C-anchored proteins and their intracellular localization. The TMSs are bolded, and basic and acidic amino acid residues are shown in red and blue letters, respectively. Hydrophobicity, hydropathic index/residue. Mt, mitochondria; PM, plasma membranes.
MATERIALS AND METHODS
Plasmid Construction
All of the constructs were inserted into a mammalian expression vector pRc/cytomegalovirus (CMV; ver. 1, Invitrogen, Carlsbad, CA). pRc/CMV was digested with HindIII and XbaI, treated with a Klenow fragment, and self-ligated using T4-DNA ligase as described previously (Kanaji et al., 2000). cDNA coding for enhanced green fluorescent protein (EGFP) was isolated from pEGFP (Kanaji et al., 2000) by polymerase chain reaction (PCR) by using the following primers: 5′-AGCTCTAGACCACCATGGTGAGCAAGG-GCGAGGAG-3′ (XbaI site and initiation codon of EGFP are underlined and the Kozac sequence is bolded) and 5′-ACTGGGGCCCCCGCGG CTTGTACAGCTCGTCCATGCC-3′ (ApaI site is underlined and SacII site is bolded). The obtained DNA fragment was digested with XbaI and ApaI and subcloned in the XbaI and ApaI sites of pRc/CMV to generate pRc/CMV-EGFP. To construct pRc/CMV EGFP-Tom5, the cDNA of yeast Tom5 was amplified by PCR from the yeast genome by using the following primers: 5′-ATATATCCGCGGATGTTTGGTCTACCTC AACAG-3′ (SacII site is underlined and the initiation codon of Tom5 is bolded) and 5′-ATAGGGCCCTTATTTCCATTGCTTTTTCAC-3′ (ApaI is underlined and the stop codon is bolded). To construct pRc/CMV EGFP-b5, the cDNA of rat cytochrome b5 coding 95–134 residues were amplified by PCR from rat liver cDNA by using the following primers: 5′-CAAGCCGCGGCCTTCGGAAACCCT TATCACT-3′ (SacII site is underlined) and 5′-ATAGGGCCCTTAATCTTCT GCCATGTAGAG-3′ (ApaI site is underlined and the stop codon is bolded). To construct pRc/CMV EGFP-OMP25, the cDNA of rat OMP25 coding 170–206 residues was amplified by PCR from rat liver cDNA by using the following primers: 5′-AAGCCGCGGCATCGAGGCGACGGAGAGGCC-3′ (SacII site is underlined) and 5′-ATAGGGCCCTCAGAGCTGCTTTCGGTATC-3′ (ApaI site is underlined and the stop codon of OMP25 is bolded). The PCR products thus prepared were cut with SacII and ApaI, and ligated into the pRc/CMV-EGFP vector. To construct pMID2-EGFP-Tom5 and pMID2-EGFP-OMP25, the PCR fragments described above were ligated to the 3′ terminus of EGFP cDNA and the fusion constructs were inserted into the yeast expression vector pMID2.
Most mutant constructs were generated using two-step PCR. The first PCR step was performed using the appropriate oligonucleotides carrying mutated codons and the SP6 promoter as primers and pRc/CMV EGFP-Tom5 or pRc/CMV EGFP-OMP25 as the template. The second PCR step was performed using the first PCR product and the T7 promoter sequence as the primers and pRc/CMV EGFP-Tom5 or pRc/CMV EGFP-OMP25 as the template. The second PCR product was digested with XbaI and ApaI and subcloned into the XbaI and ApaI sites of pRc/CMV.
Cell Culture and Transfection
COS-7 cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal calf serum (Biosciences, Lenexa, KS) in an atmosphere of 5% CO2 at 37°C. DNA transfection was performed according to the manufacturer's instructions using FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN) or LipofectAMINE reagent (Invitrogen). Plasmid DNA (1 μg) was transfected to COS-7 cells on a coverslip in a 3.5-cm dish (Falcon Plastics, Oxnard, CA) and the cells were incubated for ∼18 h.
Fluorescence Microscopy
When mitochondria were to be stained, 100 nM MitoTracker (Molecular Probes, Eugene, OR) was added to the medium and incubated for 20 min before fixation. The cells on the coverslips were fixed with acetone/methanol (1:1) at room temperature for 5 min. To stain the ER, the fixed cells on coverslips were incubated with 1% bovine serum albumin in phosphate-buffered saline then incubated with rabbit anti-calnexin antibodies (Stress Gen, Victoria, Canada) and Texas-Red–conjugated goat anti-rabbit IgG (BioSource International, Camarillo, CA) in phosphate-buffered saline containing 1% bovine serum albumin for 1 h. Fluorescent images were taken and analyzed using a confocal laser microscope (Radiance 2000; Bio-Rad, Hercules, CA).
Subcellular Fractionation
Fractionation of COS-7 cells was performed according to the methods described by Kuroda et al. (1998). Briefly, cells in a 10-cm dish were washed with phosphate-buffered saline. Collected cells were precipitated by centrifugation at 600 × g for 5 min and washed with HES (10 mM HEPES-KOH buffer pH 7.5 containing 1 mM EDTA and 10% sucrose). The cells were suspended into 1 ml of HES containing 20 μg/ml α2-macroglobulin and homogenized in a tight-fitted Potter-type homogenizer for 10 strokes. The homogenate was centrifuged at 600 × g for 5 min and the supernatant was centrifuged at 6000 × g for 10 min to obtain mitochondria (P1). The supernatant fraction was then centrifuged at 100,000 × g for 30 min to separate the microsomal (P2) and supernatant (S) fractions. Subcellular fractionation of yeast cells was performed as follows. Yeast cells (SEY6210) were transformed with the pMID2-based vectors by using lithium acetate method and grown in SD-medium containing tryptophan at 27°C to 1.0 OD600. They were then treated with Zymolyase (Seikagaku America, Rockville, MD) to obtain spheroplasts and fractionated according to the method of Daum et al. (1982). The subcellular fractions were subjected to SDS-PAGE and immunoblotting. The immunoblots were visualized by ECL (Amersham Biosciences, Piscataway, NJ) and the images were analyzed by LAS-1000 (Fuji Film, Tokyo, Japan).
Blue Native-PAGE
Blue native-PAGE was performed essentially as described previously (Schägger and von Jagow, 1991). Mitochondria were isolated from COS-7 cells expressing the GFP-Tom5 or the OMP-25-GFP fusion constructs by the method described above. The mitochondria were solubilized in 50 μl of 10 mM HEPES-KOH buffer pH 7.4 containing 2% digitonin, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 10% glycerol, and insoluble material was removed by centrifugation for 15 min at 100,000 × g. The supernatant was mixed with 5 μl of sample buffer (5% Coomassie brilliant blue G-250, 100 mM bis-Tris pH 7.0, 500 mM 6-aminocaproic acid), and electrophoresed through 5–16% polyacrylamide gradient gels. The gel slots were excised and subjected to immunoblotting by using antibodies against EGFP (Miyazaki et al., 2001) or rTom40 (Suzuki et al., 2000).
RESULTS
C-Terminal Segment Is Required for Mitochondrial Targeting of Tom5 in Mammalian Cells
To define the mitochondrial targeting signal of the C-tail–anchored proteins, we chose yeast Tom5 as a model and constructed the GFP-Tom5 fusions carrying systematic deletions or mutations in the TMS or flanking regions. GFP-cytochrome b5 was also constructed as a control for a typical C-anchored ER membrane protein. These constructs were expressed in COS-7 cells under the control of the CMV promoter and intracellular localization was examined using a confocal microscope with MitoTracker staining or immunostaining with anti-calnexin IgG (ER marker) as the reference.
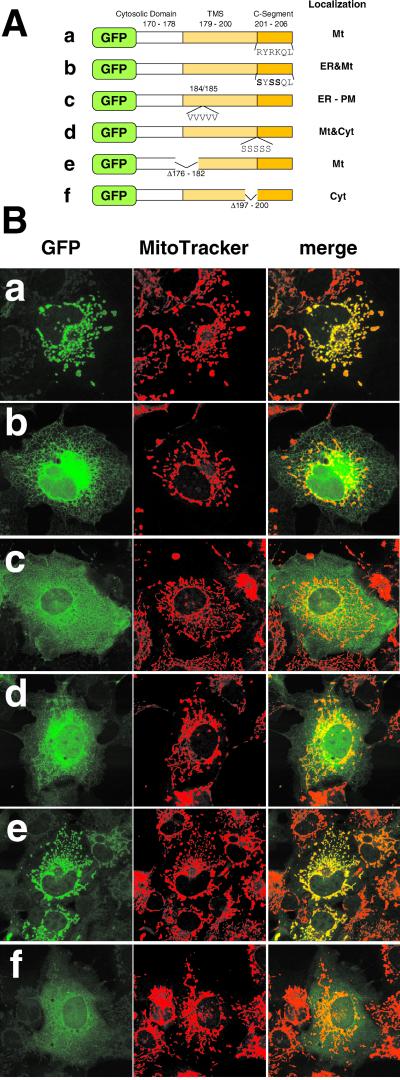
First, to probe the region responsible for mitochondrial targeting, the GFP-Tom5 fusions in which either the TMS or the C-segment of Tom5 had been deleted (ΔTM and ΔC, respectively; Figure 2A) were expressed in COS-7 cells. Wild-type GFP-Tom5 colocalized with MitoTracker as filamentous structures, whereas ΔTM localized diffusely in the cytosol. ΔC exhibited localization similar to ΔTM in the cytosol (Figure 2B). ΔTM (40–45) in which a segment of six residues in the TMS was deleted also localized to the cytosol (our unpublished data). Thus, the C-segment and a TMS are required for the mitochondrial targeting of Tom5. The mitochondria of the ΔTM- or ΔC-transfected cells exhibited well-dispersed filamentous structures, suggesting that there was little, if any, effect of overexpression of the constructs upon mitochondrial morphology and distribution.
Figure 2.
C-segment of Tom5 contains the mitochondrial targeting information. (A) Schematic structure of GFP-Tom5 and GFP-cytb5 fusion proteins and their intracellular localization. (B) Fluorescence microscopy of COS-7 cells expressing GFP fusion proteins. The cells expressing GFP-Tom5 and GFP-cytb5 constructs were incubated with MitoTracker or coimmunostained with IgG against calnexin (ER marker). Fluorescent images of GFP (green), MitoTracker (red), and calnexin (red) were taken using a confocal microscope. Other conditions are described in MATERIALS AND METHODS.
When GFP-cytochrome b5 fusion in which the C-terminal 40 residue-segment, including the TMS, was fused to the C-terminal of GFP (Figure 2A, cyt b5) was expressed in COS-7 cells, it colocalized with the ER marker calnexin (Figure 2B). Because the C-terminal 10-residue segment of cytochrome b5 functions as the ER-targeting signal (Mitoma and Ito, 1992), we examined intracellular localization of fusion protein b5Tom5C in which the C-segment of cytochrome b5 was replaced by that of Tom5 (Figure 2A). When expressed in COS-7 cells, it exhibited a typical mitochondrial fluorescent pattern (Figure 5B), indicating that the TMS of cytochrome b5 (18 residues) functioned correctly as the mitochondrial membrane anchor. On the other hand, Tom5b5C, in which the C-segment of Tom5 was replaced by the C-terminal eight residues of cytochrome b5, localized to the ER (Figure 2B). The GFP-Tom5 constructs carrying the C-segment, but lacking the TMS (ΔTM), localized in the cytosol (see above). Therefore, the TMS and the C-segment were both required for mitochondrial targeting. Furthermore, the C-segment of Tom5 functioned as a mitochondrial targeting signal when placed downstream of an appropriate TMS. When the C-terminal segment 23–50 of Tom5 (Figure 1) was fused to the N-terminal of GFP (Tom5TM-GFP; Figure 2A), the segment did not function as a mitochondrial targeting signal, but instead functioned as an ER-targeting signal and directed the construct to the ER (Figure 2). This indicates that hydrophobicity of the TMS was high enough to be recognized by SRP (Ng et al., 1996) when it was placed at the N terminus of a reporter protein; this segment functioned as the mitochondrial targeting signal only when it was placed at the C terminus of the reporter protein. These results also indicated that the structural requirement of the mitochondrial targeting signals for the N-anchored proteins and C-anchored proteins is clearly distinct.
Figure 5.
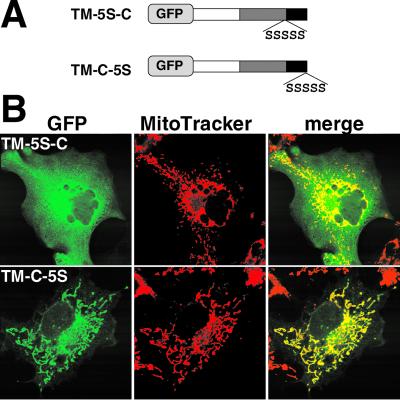
Separation of the TMS and C-segment by linker-insertion interferes with the mitochondrial targeting function. (A) Schematic representation and localization of GFP-Tom5 constructs in which serine residues were inserted in the TMS and C-segment (TM-5S-C) or added to the C-terminal (TM-C-5S). (B) Fluorescent images of GFP-Tom5 mutants and MitoTracker.
Positive Charges in C-Segment Determine Mitochondria Localization of GFP-Tom5 Fusions
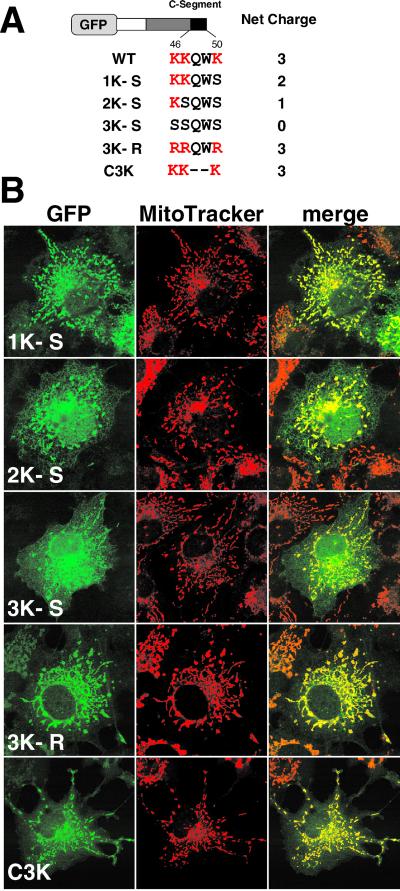
Tom5 has three positively charged residues, Lys-46, -47, and -50, in the C-segment. To assess the importance of these basic residues, they were replaced by serine residues by using site-directed mutagenesis (Figure 3). 1K-S exhibited a mitochondrial pattern with a slight background of the reticular ER pattern. On reduction of the positive charges in the C-segment, the efficiency of mitochondrial localization decreased and, conversely, the extent of ER-targeting increased. These three positive charges functioned normally irrespective of their position in the C-segment (our unpublished data). The construct in which the three lysine residues were all replaced by arginine (3K-R) localized exclusively to mitochondria, indicating that it is the positive charges that determine mitochondrial localization. When Gln-48 and Trp-49 were deleted, the construct (C3K) localized to the mitochondria with a trace amount mislocalized to the cytoplasm (Figure 3), indicating that the TMS and the following three basic amino acid residues functioned as the minimal mitochondrial targeting signal. In this relation, when Trp-49, but not Gln-48, was deleted or replaced by the other amino acid residues, the mitochondrial targeting efficiency was slightly decreased for unknown reasons (our unpublished data).
Figure 3.
Basic amino acid residues in the C-segment are critical for mitochondrial targeting of the GFP-Tom5 fusions. (A) Schematic representation of GFP-Tom5 constructs in which lysine residues were replaced by alanine (3K-S, 2K-S, and 1K-S) or arginine (3K-R) residues, or the construct in which Gln-48 and Trp-49 in the C-segment were deleted (C3K). Amino acid sequences of the C-segment are shown. (B) Localization of GFP-Tom5 constructs. The cells were incubated with MitoTracker and fluorescent images of GFP and MitoTracker were taken using a confocal microscope.
Length or Hydrophobicity of TMS Is Critical for Mitochondrial Targeting
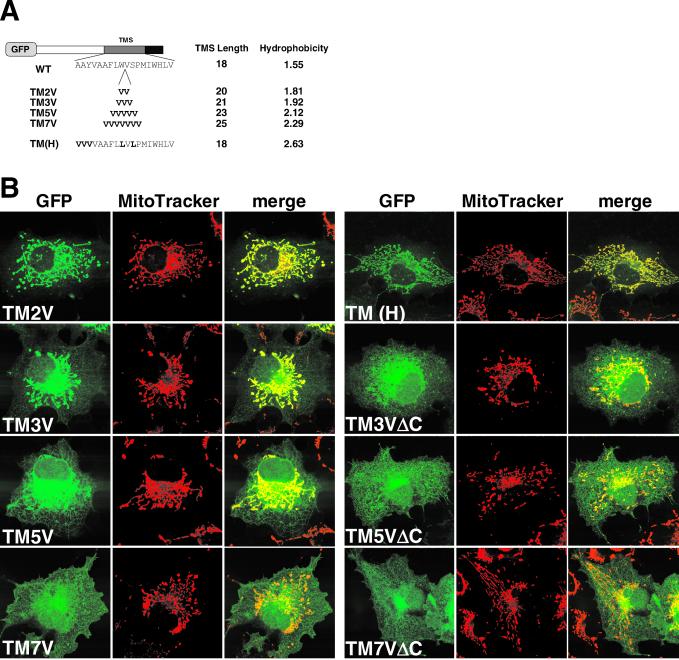
Mitochondrial targeting of N-anchored membrane proteins requires that the TMS has a moderate length or hydrophobicity (Kanaji et al., 2000). A shortened TMS with VAMP-1 is critical for the mitochondrial targeting of a spliced variant, VAMP-1B (Isenmann et al., 1998; Figure 1). To examine this, we created GFP-Tom5 constructs whose TMS was elongated by valine residues (Figure 4A). TM2V, in which the TMS was elongated by two residues with valine (TMS, 20 residues), colocalized with MitoTracker (Figure 4B). When the TMS was elongated by three valine residues (TMS, 21 residues) or more, the mitochondrial targeting efficiency gradually decreased and, conversely, the ER-targeting efficiency increased. Thus, elongation of the TMS impaired mitochondrial specificity. The mitochondrial targeting was not completely abolished, however, even after seven valine residues were introduced (TM7V). We then examined the effect of TMS length on subcellular localization of the C-segment–deleted constructs. ΔC (Figure 2) and TM1VΔC (our unpublished data) localized to the cytosol; however, upon elongation of the TMS by valine insertion, the constructs localized to both the ER and mitochondria, suggesting that the TMS with increased length functioned as a nonspecific membrane-anchor sequence. To examine which feature of the TMS, length or hydrophobicity, is the major determinant, we constructed TM(H) in which five residues in the TMS were replaced by valine and leucine residues without changing the length (Figure 4A). It was precisely transported to the mitochondria (Figure 4B). These results indicate that basic amino acid residues in the C-segment, in cooperation with the TMS of an appropriate length (18–20 residues), function as the specific mitochondrial targeting signal. The TMS, when elongated, functioned dominantly over the mitochondrial targeting activity of the C-segment and the constructs were distributed nonspecifically throughout the intracellular membranes, including the mitochondria and ER. Localization of the TMS-elongated constructs to organelle membranes other than the ER and mitochondria, however, remains to be confirmed.
Figure 4.
Length of the TMS is critical for mitochondrial targeting of GFP-Tom5. (A) Schematic representation of the TMS-elongated GFP-Tom5 constructs. The indicated number of valine residues was inserted between Trp-36 and Val-37 of Tom5 (TM2V, TM3V, TM5V, and TM7V). The mutant in which five residues were replaced by valine and leucine residues is also shown [TM(H)]. (B) Localization of the TMS-elongated GFP-Tom5 or ΔC mutants. COS-7 cells expressing the indicated constructs were incubated with MitoTracker and fluorescent images were taken as described for Figure 2.
Separation of TMS and Basic C-Segment by a Hydrophilic Linker Peptide Interferes with Mitochondrial Targeting Function
During studies with the elongated TMS constructs, we found that insertion of amino acid residues between the TMS and basic C-segment significantly affected mitochondrial targeting efficiency. As shown in Figure 5B, when five serine residues were inserted between Val-45 and Lys-46, the construct TM-5S-C lost the mitochondrial targeting ability and localized to the cytosol. In contrast, when the same sequence was fused to the C-terminal of GFP-Tom5, the construct TM-C-5S localized correctly to the mitochondria, indicating that the TMS and basic amino acid residues in the C-segment must be in a suitable context or distance for correct mitochondrial-targeting.
Single Deletions in Latter Half of TMS Strongly Influence Mitochondrial Targeting Function
To further examine the importance of the TMS of Tom5, we constructed a series of deletion mutants in the TMS. When one hydrophobic amino acid residue in the C-terminal side of Pro-39 was deleted (Δ45V, Δ44L, Δ42W, Δ41I, and Δ40 M; Figure 6A), the mutated constructs lost their mitochondrial targeting activity and diffused throughout the cytosol (Figure 6B). In contrast, when one or two amino acid residues at the N-terminal side of Pro-39 (Δ39P, Δ37V, and Δ31,37V) or hydrophilic amino acid at the C-terminal side (Δ43H) were deleted, the mutation only moderately affected the targeting function; the constructs partly mislocalized to the cytosol. Similar results were obtained with the L44-S mutant. These results suggest that the TMS could be functionally divided into two segments at Pro-39; hydrophobic residues in the C-terminal side of Pro-39 (40 M-45V) strongly contributed to the mitochondrial targeting efficiency, whereas those in the former half of the TMS affected function only moderately.
Figure 6.
Deletion of a single residue in the latter half of the TMS strongly affects mitochondrial targeting. (A) Schematic representation of the GFP-Tom5 constructs in which deletion or point mutations were introduced into the TMS. Amino acid sequences of the TMS are shown. (B) Localization of GFP-Tom5 constructs in COS-7 cells.
Intracellular Localization of GFP-Tom5 Fusions in COS-7 Cells as Assessed by Cell Fractionation
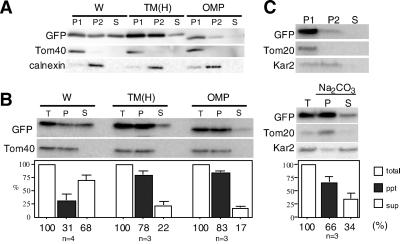
We performed cell fractionation for wild-type GFP-Tom5 (W) by using calnexin as the ER marker and TOM40 as the mitochondrial marker proteins. GFP-Tom5 was enriched in the mitochondria (P1); however, in contrast to the results obtained with confocal microscopy, a significant fraction localized in microsomal (P2) and cytosolic (S) fractions (Figure 7A). We then examined membrane anchorage of the construct by alkaline carbonate extraction; ∼70% of GFP-Tom5 was extractable with the treatment (Figure 7B). As a control, we examined subcellular localization of GFP-Tom5 expressed in yeast cells; it was correctly targeted to mitochondria and anchored efficiently to the membrane in an alkaline carbonate-resistant manner (Figure 7C). We therefore speculated that a fraction of GFP-Tom5 that had been targeted to COS-7 mitochondria was redistributed to microsomal membranes or cytosol during cell fractionation, probably because mammalian mitochondria required a TMS with higher hydrophobicity than yeast for membrane anchorage. To address this point, COS-7 cells expressing TM(H) (mean hydrophobicity 2.63; Figure 4A) were subjected to subcellular fractionation. As expected, the mutant exhibited increased recovery to the mitochondrial fraction (Figure 7A) and increased efficiency of membrane insertion (Figure 7B). We concluded that mammalian mitochondria required a TMS with higher hydrophobicity than for yeast mitochondria for the membrane anchorage of the C-tail anchor proteins.
Figure 7.
Subcellular fractionation of COS-7 cells expressing GFP-Tom5, TM(H), or GFP-OMP25 in which the C-terminal 37-residue segment of OMP25 was fused to the C-terminal of GFP, and comparison of the targeting specificity with yeast. (A) Fractionation of COS-7 cells expressing wild-type GFP-Tom5 (W), TM(H), or GFP-OMP25 (OMP). COS-7 cells were fractionated into the mitochondria (P1), microsomes (P2), and cytosolic (S) fractions. They were subjected to SDS-PAGE and subsequent immunoblotting by using antibodies against GFP, Tom40 (mitochondrial marker), or calnexin (ER marker). (B) Membrane anchorage of the expressed fusion constructs as examined by alkaline carbonate extraction. Total membrane fractions were prepared from COS-7 cells expressing GFP-Tom5, TM(H), or GFP-OMP25 and divided into two aliquots. One aliquot (T) was untreated, whereas the other was treated with 100 mM sodium carbonate pH 11.5 and ultracentrifuged to separate the membrane (P) and supernatant (S) fractions. All fractions were subjected to SDS-PAGE followed by immunoblot analysis by using antibodies against GFP and rTOM40 (mitochondrial membrane marker). (C) Subcellular localization of wild-type GFP-Tom5 expressed in yeast and the membrane anchorage as examined by alkaline carbonate extraction. Yeast cells expressing GFP-Tom5 were fractionated into the mitochondria (P1), microsome (P2), and cytosol (S) fractions, which were subjected to SDS-PAGE and subsequent immunoblot analysis by using antibodies against GFP, yeast Tom20 (mitochondria marker), or Kar2p (microsome marker). Other conditions are described in MATERIALS AND METHODS.
Although TM(H) precisely localized to mitochondria and no ER localization signal was detectable under confocal microscopy (Figure 4B), a significant amount was still detected in the microsomal fraction (P2) after the cell fractionation (Figure 7A). We have not yet reconciled this discrepancy. It should be noted that GFP-Tom5 fusions and the GFP-OMP25 construct (see below) inserted into COS-7 mitochondria were sensitive to externally added proteinase K, indicating that they were anchored to the mitochondrial outer membrane, extruding the bulk portion into the cytosol (our unpublished data).
Structural Features of Signal Defined with Yeast Tom5 Are Conserved in Mammalian Mitochondrial C-Tail Anchor Proteins
We then examined whether the structural characteristics of the signal deduced with yeast Tom5 are also conserved in authentic mammalian C-tail anchor proteins. For this purpose, we chose OMP25. It is a C-tail anchor protein localizing in rat liver mitochondrial outer membrane and, as such, recovered to the particulate fraction after alkaline carbonate extraction (Nemoto and Camilli, 1999). We constructed GFP-OMP25, in which the C-terminal 37-residue segment, including the TMS, was fused to the C-terminal of GFP. On expression in COS-7 cells, it localized to mitochondria as observed under confocal microscopy (Figure 8B), confirming the previous report (Nemoto and Camilli, 1999). Cell fractionation revealed that it localized mostly in the mitochondrial fraction (Figure 7A). Removal of three positive charges in the C-segment, insertion of a linker peptide of five serine residues between the TMS and the C-segment, or deletion of a four-residue segment in the latter half, but not a seven-residue segment in the former half, of the TMS all induced mistargeting of the GFP-OMP25 constructs (Figure 8). Similar structural features were noted for VAMP-1B (Isenmann and Wattenberg, 1998). We thus concluded that the structural characteristics of the mammalian mitochondrial targeting signal deduced with yeast Tom5 were conserved in the mammalian outer membrane proteins.
Figure 8.
Structural characteristics of the targeting signal deduced with yeast Tom5 are all conserved in mammalian OMP25. (A) Schematic representation of GFP-OMP25 fusion constructs and the intracellular localization. (B) Confocal images of GFP-OMP25 fusion constructs expressed in COS-7 cells. The cells expressing GFP-OMP25 fusions were incubated with MitoTracker. Fluorescent images of GFP (green) and MitoTracker (red) were taken using a confocal microscope. Other conditions are described in Figure 2 and MATERIALS AND METHODS.
GFP-Tom5 Constructs Imported into Mammalian Mitochondria Are Not Assembled into TOM Complex but Are Dispersed in Membranes
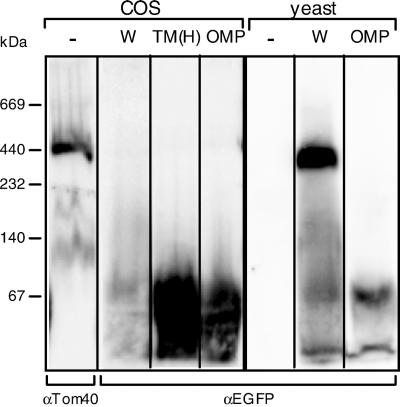
We then addressed the dispersal state in the membrane of the mitochondria-imported C-tail anchor proteins by using BN-PAGE that allows separation of the protein complex under native conditions. The mitochondria isolated from the GFP-Tom5 expressing yeast or COS-7 cells were subjected to BN-PAGE after solubilization with 2% digitonin. As shown in Figure 9, wild-type GFP-Tom5 (W) targeted to yeast mitochondria migrated as the TOM complex of ∼400 kDa (Model et al., 2001), indicating that GFP fused to the N terminus did not interfere with the assembly of the fusion construct into the TOM complex. Wild-type GFP-Tom5 or the TM(H) construct as well as GFP-OMP25 targeted to COS-7 mitochondria was not integrated into the TOM complex, but dispersed in the membrane, probably as dimeric forms (Figure 9). Therefore, in mammalian cells, GFP-Tom5 fusions were considered to behave as general C-tail anchor proteins that are not restricted to the TOM machinery.
Figure 9.
GFP-Tom5, TM(H), or GFP-OMP25 imported into mammalian mitochondria are dispersed in the membranes as dimeric forms. Mitochondria were prepared from COS-7 or yeast cells expressing wild-type GFP-Tom5 (W), TM(H), or GFP-OMP25 (OMP), and solubilized with 2% digitonin. The solubilized supernatants were subjected to BN-PAGE and subsequent immunoblot analysis by using antibodies against EGFP or rTOM40. Other conditions are described in MATERIALS AND METHODS. Marker proteins used were serum albumin, 67 kDa; lactate dehydrogenase, 140 kDa; catalase, 232 kDa; apoferritin, 440 kDa; and thyroglobulin, 669 kDa.
DISCUSSION
Mitochondrial targeting of C-anchored outer membrane proteins has not yet been thoroughly investigated. As the first step toward clarifying their targeting mechanisms, we characterized the mitochondrial targeting signal by using the GFP-Tom5 fusions as a model. Because fluorescent GFP fusions expressed in vivo possess a “tight fold” and the Tom5 peptide attached to the C-terminal of GFP can be as short as 50 residues, the C-terminal of Tom5 should always be exposed to the surface of the fluorescent active molecule. Thus, the fluorescence represents correctly folded proteins and, therefore, the present assay discounts nonspecific association of the unfolded proteins to various organelles. Our results demonstrated that the TMS with 18–20 hydrophobic residues and positive charges in the following C-segment are both important determinants for mitochondrial targeting. The importance of the C-segment as the mitochondrial targeting signal was most clearly shown by the experiment in Figure 2; the C-segment of Tom5 transplanted to the C-terminal of cytochrome b5, directed otherwise ER-targeted protein to the mitochondria, or vice versa. On reduction of the number of positive charges in the C-segment, the mutants gradually lost membrane specificity and were distributed not only to mitochondria but to the ER, indicating that at least three basic amino acids at the C-segment are required for the specific targeting of Tom5 to the mitochondria.
The membrane specificity was also lost when the TMS was elongated, even although the C-segment remained intact (Figure 4). In view of the report that hydrophobic forces drive spontaneous membrane insertion (Enoch et al., 1979; Rachubinski et al., 1980; Anderson et al., 1983), we expect that the elongated TMS functions as a dominant, nonspecific membrane insertion signal (Blobel, 1980) by its enhanced affinity with the lipid bilayer, and mutated proteins were distributed throughout the membranes, including mitochondria and ER, although whether these constructs also localized to the other membrane systems remains to be determined.
Taken together, three basic amino acid residues positioned at the C terminus of the TMS with an appropriate length functioned as the mitochondrial targeting signal. This structural feature is also conserved in mitochondrial VAMP-1B (Figure 1). When both arginine residues or all three residues were changed to threonine, the mutants were transported to the membranes of the secretory organelles via the ER (Isenmann and Wattenberg, 1998). The present study demonstrated that length, rather than hydrophobicity, is the major determinant for TMS function [Figure 4, TM(H)]. In support of this, VAMP-1B has a TMS of 17 residues with a mean hydrophobicity of 3.20; nevertheless, it localized in the mitochondria. Therefore, it seems to be the length, rather than the hydrophobicity, that determines targeting to the mitochondrial outer membrane; the length might be required to adapt to the thickness of the lipid bilayer of the mitochondrial outer membrane.
Insertion of five serine residues between the TMS and C-segment severely interfered with mitochondrial targeting of Tom5, whereas their addition to the C-terminal end was ineffective. Thus, the distance between the hydrophobic TMS and the basic C-segment is a critical factor for mitochondrial targeting. This observation is consistent with the previous observation that the arginine locating just after the TMS in OMb is more critical for the mitochondrial targeting than another arginine located in the distal C-terminal side (Kuroda and Ito, 1998; Figure 1).
There was a difference between the former and the latter half of the TMS in the sensitivity to the introduced mutation. A single amino acid deletion within the latter half of the TMS (40 M-45V) interfered with the mitochondrial targeting function more strongly than did a single amino acid deletion within the former half of the TMS (Figure 6). These results again indicated that the TMS and the basic C-segment should be within a suitable distance or context. Taken together, some factors in the cytosol might recognize these structural features and direct them to the mitochondrial outer membrane.
The C-terminal domain of Tom5, consisting of the TMS and C-segment, when transplanted to the N-terminal of GFP, functioned as an ER-targeting signal, probably as the signal anchor (Kida et al., 2000). Therefore, these segments must be located at the C terminus to be recognized correctly as the mitochondrial targeting signal; the structural requirements of the mitochondrial targeting signals for N-anchored and C-anchored proteins are clearly distinct. Considering that the large ribosomal subunit houses the extended peptide of 39 residues (Blobel and Sabatini, 1970), the mitochondrial targeting signal of Tom5 thus characterized is almost completely protected within the large ribosomal subunit. Thus, the targeting reaction should proceed during posttranslational processing, which probably evades recognition by SRP, because recognition by SRP of the signal peptide occurs on the ribosome-nascent chain complex cotranslationally (Walter and Johnson, 1994).
The structural characteristics of the signal thus defined using yeast Tom5 were well conserved in mammalian C-tail anchor proteins Vamp1B (Isenmann and Wattenberg, 1998; discussed above) and OMP25. The membrane anchored GFP-Tom5 constructs and GFP-OMP25 were present in the dispersed state in the outer membranes and not integrated into the TOM complex. Therefore, GFP-Tom5 can be regarded as the model representing general C-tail anchor proteins that are not restricted to the TOM import machinery, but dispersed eventually into the lipid bilayers. These proteins seemed to be targeted through an identical pathway because they were imported into mitochondria that had been treated with trypsin to remove the outer membrane import receptors rTOM70, rTOM20, rTOM22, and OM37 (our unpublished data), although the involvement of the channel component rTOM40 remains to be analyzed.
The heterologous assay system with yeast Tom5 enabled us to distinguish between targeting and membrane integration steps in mammalian mitochondria. Wild-type GFP-Tom5 expressed in COS-7 cells was correctly targeted to mitochondria as observed under confocal microscopy, but was inefficiently integrated into the mitochondrial membrane, whereas the same construct expressed in yeast cells was efficiently integrated into the mitochondrial membrane. On increase of the hydrophobicity of the TMS, the fusion construct TM(H) was now firmly anchored to the mitochondrial membrane. These results suggest that the characteristics of the targeting signal of the C-tail anchor proteins are distinct between yeast and mammals. In fact, basic amino acid residues in the C-segment of GFP-Tom5 were not required for correct mitochondrial targeting and insertion of GFP-Tom5 in yeast (Horie, Sakaguchi, and Mihara, unpublished data). Characterization of the mitochondrial targeting signal of the C-tail anchor proteins in yeast is in progress.
How are these features of the signal recognized in the cytoplasm during posttranslational targeting? The nascent polypeptide associated complex (NAC), which has been characterized as the heterodimeric, ribosome-associated chaperone that prevents promiscuous interaction between SRP and the nascent polypeptides destined for cellular compartments other than the secretory pathway (Wiedmann et al., 1994), is involved in targeting of preproteins to the mitochondria in yeast (George et al., 1998; Fünfschilling and Rospert, 1999). Yeast Δegd2 mutants, lacking the NAC function, accumulate GFP-Tom22 and GFP-Bcl2 in the cytosol (Egan et al., 1999). The NAC seems to function as a general chaperone to maintain the organelle-targeting competence of the precursor in vivo. The present findings that the TMS and the basic C-segment must be within an appropriate context or distance for mitochondrial targeting function suggest that some factors in addition to the NAC that specifically recognize these features and stabilize the hydrophobic nascent protein in the cytosol, participate in the targeting.
ACKNOWLEDGMENTS
We thank M. Tokunaga (Kagoshima University) for providing yeast Kar2p antibodies. This work was supported by grants from the Ministry of Education, Science and Culture of Japan (to M. S. and K. M.), Human Frontier Science Program, and Core Research from Evolutional Science and Technology (to K.M.).
Abbreviations used:
- BN-PAGE
blue-native PAGE
- CMV
cytomegalovirus
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- NAC
nascent polypeptide associated complex
- PCR
PCR
- SDS-PAGE
SDS-PAGE
- SRP
signal recognition particle
- TMS
transmembrane segment
- TOM
translocater of outer membrane
- VAMP
vesicle-associated membrane protein
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–12–0570. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–12–0570.
REFERENCES
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Nuepert W, Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Mostov KE, Blobel G. Mechanisms of integration of de novo-synthesized polypeptides into membranes: signal-recognition particle is required for integration into microsomal membranes of calcium ATPase and of lens MP26 but not of cytochrome b5. Proc Natl Acad Sci USA. 1983;80:7249–7253. doi: 10.1073/pnas.80.23.7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini DD. Controlled proteolysis of nascent polypeptide in rat liver cell fractions. I. Location of the polypeptides within ribosomes. J Cell Biol. 1970;45:130–145. doi: 10.1083/jcb.45.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Gazzoni I, Barberi M, Colombo S, Pedrazzini E. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol Biol Cell. 2001;12:2482–2496. doi: 10.1091/mbc.12.8.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmeier K, Hönlinger A, Bömer U, Dekker PJT, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Egan B, Beilharz T, George R, Isenmann S, Gratzer S, Wattenberg B, Lithgow T. Targeting of tail-anchored proteins to yeast mitochondria in vivo. FEBS Lett. 1999;451:243–248. doi: 10.1016/s0014-5793(99)00581-5. [DOI] [PubMed] [Google Scholar]
- Enoch HG, Fleming PJ, Strittmatter P. The binding of cytochrome b5 to phospholipid vesicles and biological membranes. Effect of orientation on intermembrane transfer and digestion by carboxypeptidase Y. J Biol Chem. 1979;254:6483–6488. [PubMed] [Google Scholar]
- Fünfschilling U, Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol Biol Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R, Beddoe T, Landl K, Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc Natl Acad Sci USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hurt EC, Müller U, Schatz G. The first twelve amino acids of a yeast mitochondrial outer membrane protein can direct a nuclear encoded cytochrome oxidase subunit to the mitochondrial inner membrane. EMBO J. 1985;4:3509–3518. doi: 10.1002/j.1460-2075.1985.tb04110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenmann S, Wattenberg BW. A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial-targeting signal. Mol Biol Cell. 1998;9:1649–1660. doi: 10.1091/mbc.9.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil P, Pfanner N. Insertion of MOM22 into the mitochondrial outer membrane strictly depends on surface receptors. FEBS Lett. 1993;321:197–200. doi: 10.1016/0014-5793(93)80107-6. [DOI] [PubMed] [Google Scholar]
- Keil P, Weinzierl A, Kiebler M, Dietmeier K, Söllner T, Pfanner N. Biogenesis of the mitochondrial receptor complex. Two receptors are required for binding of MOM38 to the outer membrane surface. J Biol Chem. 1993;268:19177–19180. [PubMed] [Google Scholar]
- Kida Y, Sakaguchi M, Fukuda M, Mikoshiba K, Mihara K. Membrane topogenesis of a type I signal-anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J Cell Biol. 2000;150:719–730. doi: 10.1083/jcb.150.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M, Keil P, Schneider H, van der, Klei IJ, Pfanner N, Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- Kim PK, Hollerbach C, Trimble WS, Leber B, Andrews DW. Identification of the endoplasmic reticulum signal in vesicle associated membrane proteins. J Biol Chem. 1999;274:36876–36882. doi: 10.1074/jbc.274.52.36876. [DOI] [PubMed] [Google Scholar]
- Kuroda K, Ito A. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J Biol Chem. 1998;273:31097–31102. doi: 10.1074/jbc.273.47.31097. [DOI] [PubMed] [Google Scholar]
- Mannella C, Nuewald A, Lawrence C. Detection of likely beta-strand region in sequences of mitochondrial preproteins using Gibbs sampler. J Bioenerg Biomembr. 1996;28:163–169. doi: 10.1007/BF02110647. [DOI] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Akagawa K, Tashiro Y. Important roles of the C-terminal portion of HPC-1/Syntaxin 1A in membrane anchoring and intracellular localization. J Biochem. 1998;124:311–318. doi: 10.1093/oxfordjournals.jbchem.a022113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. Microsomal aldehyde dehydrogenase is localized to the endoplasmic reticulum via its carboxy-terminal 35 amino acids. J Cell Biol. 1994;126:1407–1420. doi: 10.1083/jcb.126.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Millar DG, Li JM, Shore GC. A signal-anchor sequence selective for the mitochondrial outer membrane. J Cell Biol. 1992;119:1451–1457. doi: 10.1083/jcb.119.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara K. Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane. Bioessays. 2000;22:364–371. doi: 10.1002/(SICI)1521-1878(200004)22:4<364::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Mihara K, Sato R. Molecular cloning and sequencing of cDNA for yeast porin, an outer mitochondrial membrane protein. A search for targeting signal in the primary structure. EMBO J. 1985;4:769–774. doi: 10.1002/j.1460-2075.1985.tb03695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma J, Ito A. The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum. EMBO J. 1992;11:4197–4203. doi: 10.1002/j.1460-2075.1992.tb05513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki E, Sakaguchi M, Wakabayashi S, Shigekawa M, Mihara K. NHE6 protein possesses a signal peptide destined for endoplasmic reticulum membrane and localizes in secretory organelles of the cell. J Biol Chem. 2001;276:49221–49227. doi: 10.1074/jbc.M106267200. [DOI] [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DTWJ, Brown D, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- Rachubinski RA, Verma DP, Bergeron JJ. Synthesis of rat liver microsomal cytochrome b5 by free ribosomes. J Cell Biol. 1980;3:705–716. doi: 10.1083/jcb.84.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J Cell Biol. 1999;146:321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cousino N, Nargang FE, Baardman R, Neupert W, Lill R, Court DA. An import signal in the cytosolic domain of the Neurospora mitochondrial outer membrane protein TOM22. J Biol Chem. 1998;273:11527–11532. doi: 10.1074/jbc.273.19.11527. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schneider H, Söllner T, Dietmeier K, Eckerskorn C, Lottspeich F, Trülzsch B, Neupert W, Pfanner N. Targeting of the master receptor MOM19 to mitochondria. Science. 1991;254:1659–1662. doi: 10.1126/science.1661031. [DOI] [PubMed] [Google Scholar]
- Shore GC, McBride HM, Millar DG, Steenaart NAY, Nguyen M. Import and insertion of proteins into the mitochondrial outer membrane. Eur J Biochem. 1995;227:9–18. doi: 10.1111/j.1432-1033.1995.tb20354.x. [DOI] [PubMed] [Google Scholar]
- Stanley SJ, Davis A, D'Arcangelis D, Mannella CA. Peptide-specific antibodies as probes of the topography of the voltage gated channel of the mitochondrial outer membrane of Neurospora crassa. J Biol Chem. 1995;270:16694–16700. doi: 10.1074/jbc.270.28.16694. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Okazawa Y, Komiya T, Saeki K, Mekada E, Kitada S, Ito A, Mihara K. Characterization of rat TOM40, a central component of the preprotein translocase of the mitochondrial outer membrane. J Biol Chem. 2000;275:37930–37936. doi: 10.1074/jbc.M006558200. [DOI] [PubMed] [Google Scholar]
- Vestweber D, Brunner J, Baker A, Schatz G. A 42K outer-membrane protein is a component of the yeast mitochondrial protein import site. Nature. 1989;341:205–209. doi: 10.1038/341205a0. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]