Abstract
Natural killer (NK) cells are innate immune effectors that provide first line of defence against viruses. Human NK cells are heterogeneous in nature, and their functions rely on a dynamic balance between germ-line-encoded activating and inhibitory receptors. HIV-1 infection results in altered NK cell receptor repertoire and impaired effector functions including the ability to lyse virus-infected cells and secretion of antiviral cytokine IFN-γ. Over the last decade, additional NK cell subset-specific molecules have been identified, leading to emergence of a more complex cellular diversity than previously thought. Herein, we discuss NK cell subset redistribution, altered receptor repertoire and influence of interaction of polymorphic leucocyte antigen (HLA) and killer cell immunoglobulin-like receptors (KIR) on HIV-1 disease progression.
Keywords: Innate immunity, CD56 dim and bright, Granzyme B, Antibody-dependent cellular cytotoxicity, Highly active antiretroviral therapy
Introduction
Metazoans have evolved intrinsic defence systems to confront invading microbes and ensure their elimination from the body. The first line of natural protection is provided by the innate immune system and later by pathogen-specific adaptive immunity. The innate immune responses against viral infections are largely provided by NK cells, representing typically 5–15 % of human peripheral blood lymphocytes [1–3]. Phenotypically, human NK cells are characterised as CD3− CD56+ lymphocytes expressing CD16, the FcγRIIIA receptor. According to the newly adapted nomenclature of innate lymphoid cells (ILC), NK cells are placed into group 1 of ILC family, based on T-box transcription factor, EOMES and/or T-bet expression and interferon-gamma (IFN-γ) production [4]. Before activation, NK cells need interaction between inhibitory receptors and MHC class-1 molecules; otherwise, they remain hyporesponsive. This crucial step in NK cell development is called education and is accompanied by changes in the NK cell repertoire. However, the educational stage of an individual NK cell can be assessed only by functional means since markers have not been identified [5]. Unlike cytotoxic CD8+ T lymphocytes (CTLs), NK cells selectively recognise and kill target cells lacking MHC class-1 expression. NK cell’s direct or CD16-mediated antibody-dependent recognition of target cells triggers release of immune-regulatory cytokines, such as IFN-γ, tumour necrosis factor-alpha (TNF-α) and granulocyte–macrophage colony-stimulating factor (GM-CSF), and cytolytic factors including perforins (Pfr) and granzymes (Gr), which cause apoptosis via caspase-8/Bid or by death receptor pathways using Fas-FasL and TRAIL [6].
In addition to their presence in the peripheral blood, NK cells also exhibit diverse tissue distribution, referred to as tissue-resident NK (trNK) cells [7]. For example, the uterine, mucosal and liver-resident NK cells phenotypically differ from their blood counterpart [8]. Peripheral blood NK cell diversity is highly complex; recent studies have described more than a thousand phenotypes [9] sharing NK cell receptors (NKRs), across the leucocyte lineages [10]. However, their significance in human pathophysiology is elusive. Major NKRs consist of natural cytotoxicity receptors (NCR-NKp30, NKp44, NKp46), CD16, the killer-immunoglobulin (Ig)-like receptors; inhibitory (iKIR) and activating (aKIR), and C-type lectin-like receptors (NKG2D, CD94/NKG2-A, -C). Binding of activating receptors including CD16 to their putative ligands triggers activating signal either via cytoplasmic immune-receptor tyrosine-based activating motifs (ITAM), ITAM-bearing adaptor molecules, such as DAP10 and DAP12, or phosphorylation of intracellular domains lacking any ITAMs as in the case of DNAM-1 [11]. In contrast, inhibitory receptors transduce their signals via immune-receptor tyrosine-based inhibitory motifs (ITIM) [12]. The balance of activating receptors including CD16 and inhibitory receptor signalling determines whether the cell will be activated and can thus lyse the target cell.
HIV-1 infection and host immune responses
HIV-1 virus infection is responsible for causing acquired immunodeficiency syndrome (AIDS), manifested by massive CD4+ T cell depletion associated with AIDS-defining illness. This includes multiple infections by viruses and bacteria including tuberculosis, and lymphomas. However, individuals with greater than 350 cells/μl are at lower risk of developing AIDS-defining diseases. A perturbed immune system is the hallmark of HIV-1/AIDS, in particular CD4+ T cells which are preferentially targeted by HIV-1. Virus replication within CD4+ T cells causes suppressed proliferation, IL-2 production and cell death, resulting in the absence of help provided to CD8+ T cells, B cells and others cell types, including NK cells. During early infection, HIV-specific cytotoxic CD8+ T lymphocytes (CTLs) are able to suppress virus replication but fail to contain for prolong period of time [13] with the exception of long-term non-progressors (LTNPs) and Elite controllers (ECs). It is believed that the prolonged infection may result in viral mutation enabling them to escape from CTL-mediated lysis of infected cells.
The humoral immune response is mediated by B cell-produced antibodies against HIV-1 antigens [14]. Non-neutralizing antibodies can control or eradicate a viral infection by multiple ways such as antibody-dependent cellular cytotoxicity (ADCC) and phagocytosis of infected cells. ADCC involves the activation of FcγRIIIA of NK cells by binding the Fc portions of antibodies bound to antigens expressed on the surface of target cells [15]. However, HIV-1-specific antibodies develop at an early stage of the infection but are unable to neutralise the virus due to acquisition of rapid sequence variation by virus [16].
DC (dendritic cell) plays a pivotal role in bridging the innate and adaptive immune responses against HIV-1, and the cross-talk between NK cell and DC is found to be essential for optimal NK cell activity. Priming of DC leads to NK cell activation and killing of immature DC (iDC) [17]. Activation is largely triggered by DC-secreted cytokines including IL-12 and IL-15. Thus, a perturbed NK–DC interaction results in poor anti-HIV activity [18]. In fact, HIV-1-infected individuals show a decrease in plasmacytoid DC number and IFN-α production that potentially interferes with the NK–DC interaction. In addition, HIV-1 patient’s derived activated autologous NK cells are inefficient at lysing iDC [19]. Conversely, HIV-1-infected DC interactions with healthy control NK cells led to impaired receptor repertoire [20]. These studies suggest that NK–DC interactions play a key role in regulating each other’s function and antiviral defence.
Implementation of highly active antiretroviral therapy (HAART) has significantly reduced HIV-1-associated mortality and morbidity across the globe, mainly by suppressing virus replication and CD4+ T cell recovery. However, the poor immune reconstitution in the gut-associated lymphoid tissues (GALT) and persistent immune activation remain major therapeutic challenges despite effective HAART regimen. Although the majority of individuals respond to therapy, a significant population constituting around 20 % are unable to achieve optimal immune recovery referred as immunological non-responders. Thus, these patients are at higher risk of developing AIDS-like illness compared with responders. Despite improved prognosis in the era of HAART, non-AIDS related co-morbidities, such as cardiovascular diseases, remain one of the major therapeutic challenges.
Mechanisms of NK cell control of HIV-1 infection
Over the years, seminal contributions have been made to our understanding of NK cells mediating immune protection against human viruses including, human cytomegalovirus (HCMV) [21], hanta virus [22], hepatitis-B virus (HBV) [23], hepatitis-C virus (HCV) [24] and HIV-1 [3]. Early evidence of antiviral NK cell activity was reported in murine cytomegalovirus (MCMV) infection [25], where virus-induced type-1 interferons (IFNs) produced by DCs resulted in NK cell-dependent protection [26]. As mentioned above, NK cell effector functions are severely impaired, and viremia has been suggested as one of the key factors of cellular defects [27]. It is well appreciated that NK cells from HIV-1 patients respond poorly towards target cells and produce low levels of IFN-γ and Gr. In addition to lysis, NK cell production of C–C chemokine CCL3 (MIP-1α), CCL4 (MIP-1β) and CCL5 (RANTES) has been shown to block the entry of HIV-1 R5 strains [28, 29]. Therefore, less production of these molecules by functionally impaired NK cells may result in accelerated virus replication. In addition to controlling viral replication, NK cells can also kill non-infected CD4+T cells, which start expressing NKp44L (a ligand of the activating NK receptor NKp44). CD4+T cells expressing NKp44L became susceptible to NK lysis mediated by NKp44+NK cells thereby playing a role in the CD4+T cell depletion that happens throughout the HIV-1 disease progression [30].
ADCC plays a critical role in NK cell-mediated killing of HIV-infected cells. HIV-specific antibodies can trigger NK cell’s activation by binding to both the HIV-1 antigens and FcγRIIIA leading to NK cell lysis of target cells. ADCC potentially offers an effective, adaptive immune response to HIV-1 infection, and broader ADCC responses may play a role in long-term control of HIV-1 disease progression [31]. ADCC has also been suggested as a key factor in HIV-1 protection conferred by EC owing to higher levels of ADCC antibodies than viraemic subjects [32]. Strong evidence in support of antibody-dependent NK cell-mediated protection against HIV-1 has recently been observed in Thai RV144 HIV-1 vaccine trials [33] where a majority of the treated individuals exhibited profound antiviral NK cell effector activity. Although ADCC is found to be an effective way of eliminating HIV-1-infected cells, still this is not always the case; very recently, a longitudinal study has shown diminished HIV-specific effector antibody responses, including ADCC in infected individuals receiving HAART [34]. This study suggests that despite successful treatment, therapeutic approaches may require improved antibody-mediated HIV-1 control.
NK cell evasion by HIV-1
Similar to other human intracellular pathogens, HIV-1 has likewise evolved mechanisms to circumvent NK cell activity, including escape from ADCC owing to mutation in HIV Env glycoprotein (gp120 and gp41), epitope masking, trimerization of gp120/gp41 spikes and shedding of HIV-1 Env proteins [15, 35]. Although both CD8+ T cells and NK cells have the ability to efficiently lyse HIV-1-infected cells, they experience great difficulty in cases where virus selectively downregulates surface HLA-A and HLA-B, while protecting the expressions of HLA-C and HLA-E molecules [36, 37]. Therefore, only those NK cells, which lack the inhibitory receptors specific for HLA-C and HLA-E, and HIV-1-infected cells with downregulated expressions of HLA-A and HLA-B, became susceptible to NK cell-mediated cytotoxicity [38]. Another mechanism involves downregulation of NKG2D ligands MHC class-related chain-A (MICA), -B (MICB) and HCMV UL-16 binding proteins (ULBPs) potentially by the HIV-1 protein Nef [39], thereby, preventing the binding of NKG2D receptors and their downstream signalling, a key step required for efficient NK cell killing.
In addition, HIV accessory protein Vpu has been suggested to influence the HIV-specific ADCC [40]. For example, Vpu downmodulates NK cell co-activating ligand NTB-A on the surface of infected CD4+ T cells, thereby protecting the infected cell’s lysis by NK cells [41]. Further, Vpu has been shown to downregulate and degrade tetherin, a cellular host restriction factor that enables the release of free virus aggregates [42, 43]. As a result, the recognition of these virus aggregates by HIV-specific antibodies and subsequent NK cell-mediated viral clearance is hampered [44]. In contrast to Vpu, HIV-accessory protein Vpr upregulates the expression of NK cell-activating ligands ULBP-1, -2 and -3 in HIV-1-infected primary CD4+T lymphocyte, thereby enhancing their susceptibility to NK cell-mediated killing [45]. Another evasion mechanism involving Vpu and Nef proteins is the downregulation of PVR (CD155, Necl-5), the ligand for NK cell-activating receptor DNAM-1 (CD226), thereby, preventing NK cell-mediated lysis of HIV-1-infected CD4+ T cells [46]. These studies explain how HIV-1 uses sophisticated strategies to circumvent antiviral immune response and ensures their survival in host body.
NK cell subsets in HIV-1 infection
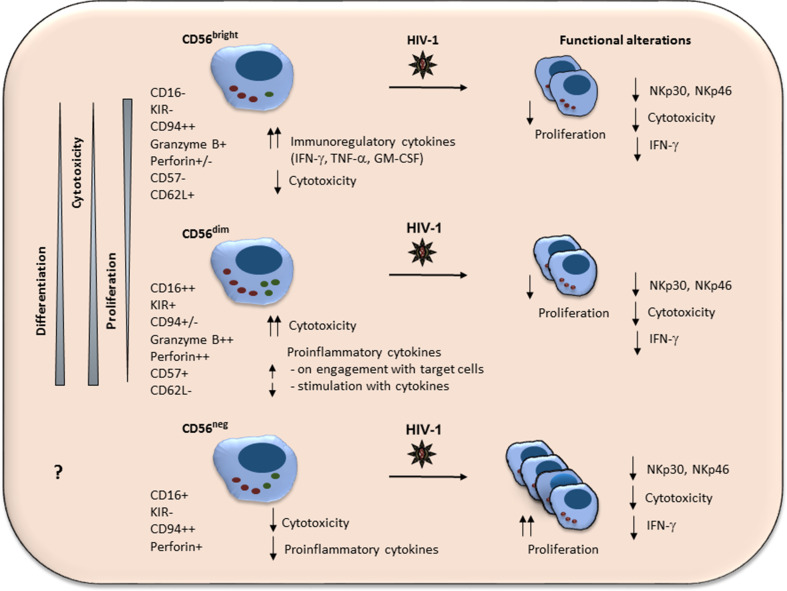
Based on surface CD56 density, NK cells are categorised into three distinct subsets: the cytokine producing CD56bright CD16− subset, the cytotoxic CD56dim CD16+ subset, and a minor CD56neg CD16+ NK cell subset with poor antiviral activity (Fig. 1). The figure shows an elaborated view of each subset in relation to HIV-1 pathogenesis.
Fig. 1.
Diverse phenotypic and functional attributes of NK cell subsets in HIV-1 infection. Schematic representation of major human peripheral blood NK cell subsets, CD56bright, CD56dim and CD56neg, expressing different levels of antigens. Also, depicted are the expression levels and pattern of NK cell subset-specific markers, representing their differentiation and functional attributes. For example, the highly differentiated CD56dim population expresses higher levels of CD16, KIR, NKG2A, CD57 and Gr than less-differentiated CD56bright subset. In addition to expression of differentiation markers, the CD56dim subset is less proliferative but more cytolytic than CD56bright NK cells. During HIV-1 infection, there is redistribution of these subsets and the display of low levels of NCR associated with diminished cytotoxicity and IFN-γ production across the subsets. The unusual CD56neg subset expresses substantial levels of CD16 and Gr, but is functionally impaired in HIV-1 infection and more proliferative than CD56dim and CD56bright subsets. However, the differentiation pathway of CD56neg NK subset is not fully understood
HIV-1 and CD56bright NK subsets
A subset of CD56bright NK cells represents approximately 5–10 % of the total peripheral blood NK cells. However, they record relatively higher frequency in secondary lymphoid organs (SLO) such as lymph nodes. CD56bright NK cells are less cytotoxic but potent producers of immuno-regulatory cytokines. In addition to their low levels of CD16 and KIR, CD56bright NK cells express the inhibitory receptor CD94/NKG2A, the lymph-node-homing receptor CCR7 and L-selectin CD62L [47–49]. From the developmental point of view, CD56bright NK cells are considered precursors to highly differentiated CD56dim NK counterparts [50], and are suggested to pass through several intermediate stages comprising CD16+CD56bright [51], CCR7− CD56bright [49], CD94-expressing CD94bright CD56dim [52] and CD62L+ CD56dim cells [53], before acquiring the fully mature CD56dim NK phenotype (Fig. 1). Moreover, the environmental signalling either homeostatic and/or pathogen-induced pro-inflammatory, and receptor engagement might be the governing force in driving the NK cell differentiation and associated phenotypic diversity. Apart from secreting immuno-regulatory cytokines, CD56bright NK cells proliferate more in response to cytokine stimuli than their CD56dim NK counterparts, a feature of less-differentiated NK cells. The role of CD56bright NK cell in HIV-1 pathogenesis is less studied compared to CD56dim and CD56neg subsets. Clinical data support a sharp decline in the overall CD56+ NK population accompanied by expanding CD56neg NK cells in early stage of infection, a phenomenon believed to be a compensatory mechanism to overcome the loss of CD56+ cells [54].
One such report on impact of HIV-1 on the CD56bright NK cell subset has recently been reported from our laboratory, describing an altered phenotype with respect to the homing receptor CCR7 [49]. The chronically infected therapy naïve individuals show a significant loss of CCR7+ CD56bright NK cells associated with relevant increased CCR7− subpopulation. Interestingly, the loss of the CCR7+ subpopulation positively correlates with HIV-1 viremia, and their frequency is moderately restored with HAART. These observations argued most likely downstream effects of chronic immune activation are caused by rapid HIV-1 replication. The role of CD56bright NK cells in antiviral host defence is also reported in HIV/HCV co-infection, where CD27-expressing CD56bright NK cells are suggested to resolve acute HCV infection in HIV-1 sero-positive individuals [55] potentially via IFN-γ-mediated HCV suppression. Further investigations are required to understand the contribution of various CD56bright intermediate stages mentioned above, in human pathophysiology including acute and chronic HIV-1 infections.
HIV-1 and CD56dim NK subsets
CD56dim NK cells constitute the major proportion (approximately 90 %) of the peripheral blood NK cells. Accumulating data support the linear differentiation of CD56dim NK cells from CD56bright precursors following a sequential and gradual process involving loss of surface CD94/NKG2A and gains in CD16, KIR and CD57 expressions [56] before achieving terminally matured status (Fig. 1). Functionally, CD56dim NK cells effectively lyse the virus-infected cell as they produce large quantity of executioner proteins, Gr and pfr. However, the notion that CD56bright NK cells are mainly cytokine producers and CD56dim NK cells act as killers may not hold true since both subsets produce IFN-γ. In fact, CD56dim subset has been shown to produce higher quantity of IFN-γ compared to CD56bright NK subset on engagement with target cells rather than with IL-2, IL-12, IL-15 and IL-18 stimulation [53]. As mentioned before, the overall loss of CD56+ NK cells including CD56dim is suggested to be a consequence of ongoing virus replication, and this loss can be compensated by rapidly dividing aberrant CD56neg NK cells [57].
Our own laboratory has observed preferential loss of less-differentiated CD56dim NK cells, specifically those that produce low levels of IFN-γ in response to IL-12 and IL-18 [58]. Of note, the loss was restricted mostly to less-differentiated, CD57− and CD57dim subpopulations compared with CD57bright CD56dim NK subsets [59]. Furthermore, the increased levels of terminal differentiation marker CD57, along with KIR and Gr expressions on CD56dim NK, reflect the features exhibited by mature NK cells. However, the mechanisms of preferential loss of less-differentiated CD57− CD56dim NK cells during chronic HIV-1 infection are unknown. During the differentiation process, NK cells acquire CD57 and KIRs expressions to gain a mature phenotype [60]. Among them, CD56dim CD57+ NK cells are found to exhibit better ADCC than CD56dim CD57− NK cells [59], suggesting again the superior functional attributes of mature NK cells. While educated KIR3DL1+ NK cells show higher activation against autologous targets coated with HIV-1 antigens and antiviral antibodies compared with KIR3DL1− NK cells [61]. These higher capacities of CD57+ and KIR3DL1+ NK cells to exhibit elevated anti-HIV antibody-dependent activation suggest that education and differentiation contribute independently to ADCC-mediated NK cell activation [62]. The functional impairment of CD56dim NK cells has been demonstrated in NK cell–DC co-culture experiments. Studies show poor NK cell lysis, when healthy control CD56dim NK cells were co-cultured with HIV-1 viremic and aviremic-derived DCs, potentially due to defective NK–DC interaction [63], involving impaired maturation and cytokine production by iDC [64].
HIV-1 and CD56neg NK subsets
The loss of CD56+ NK cells during HIV-1 infection is associated with expansion of a poorly defined aberrant CD56− CD16+ (CD56neg) NK cell population [65], expressing low levels of natural cytotoxicity receptor, NKp30 and NKp46 [57] without changing NKG2D [66], reduced IFN-γ production, and impaired cytotoxicity [54] (Fig. 1). However, the data on inhibitory receptors are conflicting: one study reported increases expressions of KIR2DL2, 3 and LILRB1 [57], while another found lower KIR2DL1 expression on CD56neg NK cells [66]. Further, compared with CD56dim NK cells, CD56neg NK cells respond poorly to ADCC on stimulation [65]. However, both HIV-1 and HCV patient’s derived CD56neg NK cells have been shown to produce significant amount of antiviral chemokine MIP-1β [66, 67]. In contrast to CD56dim and CD56bright NK cells, the lack of major NK cell lineage-specific markers on CD56neg NK cells hinders our understanding of their origin and developmental pathways. Further, sharing of characteristic phenotypes of both less-differentiated (low CD57 expression and high proliferative capacity) and highly differentiated CD56dim NK cells (low CD94/NKG2A and elevated KIR expressions) on CD56neg NK cells make them phenotypically more complex to define their origins [57, 65, 68]. Nonetheless, based on phenotypic and functional characteristics, one may argue that CD56neg cells might have originated from early CD56dim stage. However, further studies are required to specifically address this issue.
Under physiological conditions, CD56neg NK cells represent only a minor subset. However, their population increases during chronic viral infections including HIV-1 [54, 57, 65, 68, 69] and HCV [67, 70]; therefore, their relevant increases may act as a potential pathophysiological indicator. Impaired HIV-1-associated CD56neg NK cell functional defects were also seen in ex vivo co-culture experiments with DC [63], reflecting their inability to communicate effectively with DC. These alterations may not be considered a consequence of HIV-1 infection since the occurrence of productive infection of NK cell is a rare event, except in one study [71]. Importantly, CD56neg NK cell subsets show substantial heterogeneity. For example, activated CD7+ CD56neg NK cells, but not CD7− CD56neg NK cells, expressing CD95, KIRs and NKG2A/C, selectively expand during HIV-1 infection [72]. In addition to these, two additional subpopulations reported to emerge during infection are CD122− CCR7+ and CD122+ CCR7− subsets. Of note, the latter subset expands significantly and shows positivity for the terminal differentiation marker CD57, reflecting their closeness with mature CD56dim NK cells [69].
As mentioned before, the expansion of CD56neg NK cells during HIV-1 infection has been suggested to be a mechanism to compensate for the loss of CD56+ NK cells in order to maintain overall NK cell homeostasis in infected individual [54]. Moreover, the rapid CD56neg NK cell expansion has also been argued as a consequence of high viremia, since both parameters strongly correlate with each other but not with virally suppressed LTNPs [57]. In this context, HAART-mediated suppression of HIV-1 viremia causes considerable reduction of CD56neg NK cells, supporting the notion that viremia indeed remains a major inducing factor for aberrant cellular phenotype. Similarly, antiretroviral and pegylated IFN plus ribavirin treatment of HIV/HCV co-infection has led to reduced numbers of aberrant CD56neg NK cells [70].
Apart from the reduced CD56 expression during infection, Siglec-7, an inhibitory C-lectin-type molecule has been suggested as one of the early surface markers downregulated with HIV-1 infection [73]. Interestingly, Siglec-7 expression remains suppressed throughout the course of infection, and their regulation substantially relies on viremia, as LTNPs and ECs display normal cell distribution. All these studies, therefore, describe accumulation of an aberrant and “anergic” CD56neg NK cell population in response to CD56+ NK cell loss, which could be responsible for overall NK cell redistribution and associated dysfunctions. Considering the immune correlates of protection against HIV-1, our laboratory has recently described a new polyfunctional CD8+ NK cell phenotype in a large cohort of HIV-1 positive individuals, which inversely correlates with the diseases progression [74]. Therefore, a higher frequency of these cells may reflect an improved antiviral NK cell activity, and this may potentially serve as a non-genetic determinant of HIV-1 disease progression.
Changes in the NK cell receptor repertoire in HIV-1 immuno-pathogenesis
Given that NK cell effector function depends on the balance between activating and inhibitory receptors, it is largely unknown how the relative NKR alterations in HIV-1 infection affect NK cell antiviral activity. Discussed below are the major NKRs and their potential role in HIV-1 immuno-pathogenesis.
Natural cytotoxicity receptors (NCRs)
NCRs are germ-line-encoded major NK cell-activating receptors comprising NKp30, NKp44, and NKp46. Binding of these receptors to their putative ligands mostly expressed on stressed, tumour and virus-infected cells triggers lysis of these cells [75, 76]. Some of the important human viral ligands recognised directly by NCRs are summarised (Table 1). The vital role of NCRs in antiviral host defence has been reported both in animals and humans. For instance, NKp46 is found to be essential to clear influenza virus in mice [77]. NCRs exhibit a certain degree of redundancy as they can recognise the same ligand of a different viral origin. For example, influenza, Sendai, and New Castle disease virus-derived haemagglutinin (HA) and HN (HA-neuraminidase) have affinity towards both NKp44 and NKp46 receptors [78–80]. Contrary to this, despite being an activating receptor, NKp30’s engagement with HCMV-pp65, Pox and vaccinia-derived HA inhibits NK cell activity [81, 82]—a viral evasion mechanism to avoid lysis of infected cells. That means NK cell binding of viral ligands is more or less dependent on individual engaging NCR and not necessarily all NCRs deliver activation signals.
Table 1.
NK cell-activating receptors and their identified viral ligands
| Receptors | Virus and their ligands | Activation | Refs. |
|---|---|---|---|
| NKp46 (NCR1/CD335) | Influenza, New Castle disease, Pox and Sendai virus HA and HN | Yes | [77, 79, 81] |
| NKp44 (NCR2/CD336) | Influenza, New Castle disease and Sendai virus HA and HN | Yes | [77, 78, 80] |
| NKp30 (NCR3/CD337) | HCMV-pp65, Pox and Vaccinia virus HA | No | [81, 82] |
| NKG2D | HCMVMIC -A,-B and ULBPs | Yes | [111] |
HA haemagglutinin, HN HA neuraminidase, MIC -A and -B MHC class I chain-related protein (MIC)-A and -B, ULBPs UL16-binding proteins
Antiviral activity by NCR has also been shown in chronic hepatitis C virus (HCV) infection, where NKp46hi NK cell subset suppresses virus replication in vitro [83]. The anti-HCV characteristics of these cells were due to high levels of IFN-γ secretion and increased cytolytic activity. Accumulating data support abnormal NCR repertoire associated with impaired cytotoxicity and immunoregulatory functions in HIV-1 infection [73, 84]. Notably, HIV-1 viremia greatly impacts NCR-triggered NK cell effector function as observed in HIV-1 viremic individuals, who display a marked reduction in NKp30 and NKp46 receptor associated with poor cytotoxicity and IFN-γ production compared with aviremic individuals [27, 57, 85]. This is supported by another study where LTNP and EC, with suppressed viremia, successfully maintain NCR and cellular functionality in contrast to antiretroviral-treated aviremic progressors [86]. These studies suggest that active viral replication profoundly impacts NCR regulation and their downstream effector function. In contrast, two recent studies have demonstrated downregulation of NKp46 receptors on NK cells resulting in impaired cytolytic activity and diminished innate immunity during chronic viral infection [87, 88].
Killer cell immunoglobulin (Ig)-like receptors (KIRs)
KIRs are differentially expressed on NK cell subsets [89], and every single human NK cell expresses at least one to several MHC-1-specific NK cell receptors (on average 2–9 different KIRs and/or KLRs per cell) resulting in a diverse NK cell repertoire [90–92]. Both KIR and HLA polymorphisms play a central role in defining NK cell-mediated outcome of HIV-1 disease. One of the early reports of KIR:HLA interaction on virus control demonstrates resolution of HCV infection in individuals positive for KIR2DL3 and homozygous for HLA-C1 [24], while individuals, expressing KIR3DS1 and positive for HLA-Bw4-80I, are protected against developing HCV-induced hepato-cellular carcinoma [93]. Furthermore, the KIR and HLA combination also plays a role in determining the HIV-1 transmission between sexual partners. In this regard, inhibitory KIR/HLA incompatibility appears to confer protection against HIV-1 transmission [94]. Moreover, the viral peptide selectivity of KIRs is found to be much broader compared with adaptive T cell receptors [95] since KIRs recognise peptide motifs, rather than individual peptides. This means that KIR binding essentially depends on the sequence and structure of epitope presented by HLA class-1 molecules. In addition, KIR-associated amino-acid polymorphism also contributes to NK cell control of HIV-1. For example, HIV-1-selected sequence polymorphism in KIR2DL2 confers strong binding with infected CD4+ T cell, resulting in NK cell inhibition [96].
Recent population genetic studies have revealed the influence of HLA–KIR association in HIV-1 disease and clinical outcome (Table 2). For example, individuals positive for HLA-Bw4-80I show delayed progression towards AIDS than those without this allele, mostly due to their interaction with both activated KIR3DS1 and inhibitory KIR3DL1 receptors on NK cells [97, 98]. Notably, NK cells positive for KIR3DL1 confer better cellular function in individuals with HLA-Bw4 than HLA-Bw6 homozygous alleles [99, 100], while the protective effect conferred by KIR3DS1-expressing NK cells [101–103] is due to their efficient capability to suppress HIV-1 replication [104]. Further, HLA-C-educated NK cells positive for inhibitory KIR2DL1-3, appear to modulate NK cell functionality during primary HIV-1 [105]. In addition to HLA–KIR interaction, KIR copy number greatly impacts the outcome of the disease. For example, a recent study has shown strong association of activating receptor KIR2DL4 copy number with better CD4+ T cells recovery in SIV-infected Mamu-A*01-negative rhesus macaques [106], suggesting KIR copy number is a strong protective determinant of the disease. In the same line, a genome-wide screening of structural variants has demonstrated association of high KIR3DS1 copy number with lower viral set point in the presence of a putative ligand, while NK cells from individual with high copy number of KIR3DL1 in the presence of KIR3DS1 and ligand show efficient inhibition of HIV-1 replication [107].
Table2.
Influence of KIR–HLA interaction on chronic viral diseases
| KIR | HLA | Function | Virus | Consequence | Refs. |
|---|---|---|---|---|---|
| KIR3DS1 | HIA-Bw4-80I | Activating | HIV-1 | Delay progression to AIDS | [97] |
| KIR3DL1 | HUVBw4-80I | Inhibitory | HIV-1 | Delay progression to AIDS | [98] |
| KIR3DS1 | HLA-Bw4 | Activating | HIV-1 | Protection from infection | [101] |
| KIR2DL3 | HIA-C1 | Inhibitory | HCV | Infection resolution | [24] |
| KIR3DS1 | HLH-Bw4-BDI | Activating | HCV | Protection from HCC | [93] |
KIR killer immunoglobulin-like receptor, HLA human leucocyte antigen, HCC hepatocellular carcinoma
As discussed above, while the amino acid sequences do influence the HLA–KIR interaction, recent studies have revealed how a minimal sequence variability within HLA-C allele-restricted HIV-1 Gag24 epitopes can significantly affect NK cell functions. Recognition of HLA-C03:04- and HLA-Cw*0102-restricted peptide by inhibitory KIR2DL1 exhibits reduced NK cell functions [108, 109]. Interestingly, NK cells that are educated through KIR3DL1 mediate potent anti-HIV ADCC against autologous HLA-Bw4+ target cells. However, in HIV-1 infection, no functional differences were observed between KIR3DL1+ and KIR3DL1− NK cells in HLA-Bw4+-infected individuals, suggesting abrogation of functionally competent educated NK cells may have resulted in poor ADCC, and thus contributed to HIV-1 disease progression [61]. Taken together, the above studies suggest that NK cells’ ability to control HIV-1 disease is significantly dependent on the HLA allele and to a certain extent on KIR polymorphism of the host.
C-type lectin-like receptors
Most characterised human C-type lectin-like receptors mainly include activating NKG2D and CD94/NKD2C, and inhibitory CD94/NKG2A and NKR-P1A (CD161). Human CD94/NKG2A and CD94/NKG2C receptors are heterodimers that recognise non-classical, loaded HLA-E molecules and deliver inhibitory and activating signals to NK cells, respectively. NKR-P1A (CD161) binds to a non-MHC-coded ligand and lectin-like transcript-1 (LLT-1) [110]. Among all the C-type lectin-like family members, NKG2D is the most studied receptor, since many of its ligands including MICA, MICB, and ULBP 1–4 of HCMV have been identified [76, 111]. Like other intracellular human pathogens, HIV-1 employs well-developed evasion strategies to circumvent the immune system. In this context, HIV-1 has been shown to downregulate NKG2D ligands on virus-infected cells potentially via protein Nef to evade NK cell-mediated killing [39]. Similarly, soluble NKG2D ligands present in the plasma of HIV-1-infected individuals has been demonstrated to suppress NKG2D-mediated NK cell impairment [46]. Nonetheless, modulation of NKG2D ligands by viral proteins Vif, Nef, Vpu significantly contribute to poor recognition of infected cells, and thus in poor target cell lysis. Moreover, it is also believed that certain NKG2D ligands are induced by HIV-1-infected CD4+ cells and that NK cells effectively lyse them. In this regard, HIV-1-infected CD4+ T cells have been shown to express elevated NKG2D ligand via Vpr-APOBEC3G interaction-induced DNA-damage response and are effectively lysed by NK cells [112].
Emerging concept of memory-like NK cells
Generation and persistence of memory T and B cell lymphocytes have been extensively studied in various animal and human infection models [113], but in recent years, several studies have come up with ‘memory-like’ properties of NK cells in pathogenic and non-pathogenic models. In this context, one of the early observations described hapten-specific liver NK cell recall response to ear swelling in T- and B-cell-deficient mice [114]. In another murine infection model, MCMV m157-specific clonal expansion and recall response were observed in Ly49H+ NK cells [59, 115]. Later, several groups have reported memory-like NK cell behaviour in other viral infections, including chronic hepatitis viruses [116], Hanta [22], and Chikungunya [117] in humans, and vaccinia [118], HSV [119] and influenza [120] in mice. However, due to lack of concrete evidence, this phenomenon is yet to be fully proven in humans. In addition to in vivo detection of memory-like NK cells, NK cells pre-activated with IL-12/IL-15 and IL-18 have been shown to induce memory-like phenotype in re-stimulation experiments as determined by high IFN-γ production [121]. Similar observations were shown in an adoptive transfer model of cytokine pre-activated NK cells into mice following re-stimulation experiments [122]. Currently, we do not have reports of memory-like like NK cells in HIV-1 infection, an area that needs thorough investigation. NK cell memory is gaining immense interest pertaining to their generation in acute infection and long-term persistence in pathogenic conditions. Whether innate lymphocytes such as gamma-delta T (γδ), natural killer T (NKT) and mucosal-associated invariant T (MAIT) cells possess this property is an interesting area that needs to be investigated.
Concluding remarks
The increasing body of evidence strongly supports the role for NK cell-mediated immune protection against HIV-1. Identification of new KIR alleles through population-based studies has significantly improved our understanding of HIV-1 disease outcome. However, a detailed mechanistic aspect of HLA–KIR associations could prove beneficial in designing NK cell-based immunotherapy. With the emergence of memory-like NK cells, our focus should be directed towards potential implications in immunotherapy against viral infection. Perhaps non-human primate (NHP) models of SIV/SHIV infection can provide better insight into our understanding in designing NK cell-targeted vaccines. The use of sophisticated tools such as mass cytometry-based investigation on NK cell may provide in-depth cellular and molecular insights on roles in human pathophysiology. Certainly, more experimental models and investigations are required to understand the mechanisms that translate the NK cell function which protects from HIV-1 infection and suppression of disease progression.
Acknowledgments
The authors would like to thank Dr. Scott Hale, Emory University School of Medicine, USA for critically reading the manuscript. This study was supported by the University of Malaya Research Grant (RG 501-13HTM) of the Health and Translational Medicine Cluster awarded to AWA, and DMO received funding from Bundesministerium fur Bildung und Forschung, and DZIF TTU 04.802, Germany.
Contributor Information
A. Wahid Ansari, Phone: +60 322 463 383, Email: wahid.ansari@gmail.com.
Dirk Meyer-Olson, Phone: +49 528 16210 1022, Email: dirk.meyer-olson@fachklinik-bad-pyrmont.de.
References
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 3.Jost S, Altfeld M. Control of human viral infections by natural killer cells. Annu Rev Immunol. 2013;31:163–194. doi: 10.1146/annurev-immunol-032712-100001. [DOI] [PubMed] [Google Scholar]
- 4.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E. Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13(2):145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 5.Hoglund P, Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat Rev Immunol. 2010;10(10):724–734. doi: 10.1038/nri2835. [DOI] [PubMed] [Google Scholar]
- 6.Zamai L, Ahmad M, Bennett IM, Azzoni L, Alnemri ES, Perussia B. Natural killer (NK) cell-mediated cytotoxicity: differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J Exp Med. 1998;188(12):2375–2380. doi: 10.1084/jem.188.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sojka DK, Tian Z, Yokoyama WM. Tissue-resident natural killer cells and their potential diversity. Semin Immunol. 2014;26(2):127–131. doi: 10.1016/j.smim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi FD, Ljunggren HG, La Cava A, Van Kaer L. Organ-specific features of natural killer cells. Nat Rev Immunol. 2011;11(10):658–671. doi: 10.1038/nri3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, Davis MM, Norman PJ, Guethlein LA, Desai M, Parham P, Blish CA (2013) Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 5(208):208ra145. doi:10.1126/scitranslmed.3006702 [DOI] [PMC free article] [PubMed]
- 10.Strauss-Albee DM, Horowitz A, Parham P, Blish CA. Coordinated regulation of NK receptor expression in the maturing human immune system. J Immunol. 2014 doi: 10.4049/jimmunol.1401821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Andrade LF, Smyth MJ, Martinet L. DNAM-1 control of natural killer cells functions through nectin and nectin-like proteins. Immunol Cell Biol. 2014;92(3):237–244. doi: 10.1038/icb.2013.95. [DOI] [PubMed] [Google Scholar]
- 12.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283(5403):857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 14.Mouquet H. Antibody B cell responses in HIV-1 infection. Trends Immunol. 2014;35(11):549–561. doi: 10.1016/j.it.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Huber M, Trkola A. Humoral immunity to HIV-1: neutralization and beyond. J Intern Med. 2007;262(1):5–25. doi: 10.1111/j.1365-2796.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- 16.Moir S, Fauci AS. Insights into B cells and HIV-specific B-cell responses in HIV-infected individuals. Immunol Rev. 2013;254(1):207–224. doi: 10.1111/imr.12067. [DOI] [PubMed] [Google Scholar]
- 17.Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, Mingari MC, Moretta A. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006;214:219–228. doi: 10.1111/j.1600-065X.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- 18.Altfeld M, Fadda L, Frleta D, Bhardwaj N. DCs and NK cells: critical effectors in the immune response to HIV-1. Nat Rev Immunol. 2011;11(3):176–186. doi: 10.1038/nri2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tasca S, Tambussi G, Nozza S, Capiluppi B, Zocchi MR, Soldini L, Veglia F, Poli G, Lazzarin A, Fortis C. Escape of monocyte-derived dendritic cells of HIV-1 infected individuals from natural killer cell-mediated lysis. Aids. 2003;17(16):2291–2298. doi: 10.1097/00002030-200311070-00003. [DOI] [PubMed] [Google Scholar]
- 20.Scott-Algara D, Arnold V, Didier C, Kattan T, Pirozzi G, Barre-Sinoussi F, Pancino G. The CD85j+ NK cell subset potently controls HIV-1 replication in autologous dendritic cells. PLoS One. 2008;3(4):e1975. doi: 10.1371/journal.pone.0001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuijpers TW, Baars PA, Dantin C, van den Burg M, van Lier RA, Roosnek E. Human NK cells can control CMV infection in the absence of T cells. Blood. 2008;112(3):914–915. doi: 10.1182/blood-2008-05-157354. [DOI] [PubMed] [Google Scholar]
- 22.Bjorkstrom NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, Michaelsson J, Malmberg KJ, Klingstrom J, Ahlm C, Ljunggren HG. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011;208(1):13–21. doi: 10.1084/jem.20100762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med. 2013;19(7):859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O’Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 25.Bukowski JF, Woda BA, Habu S, Okumura K, Welsh RM. Natural killer cell depletion enhances virus synthesis and virus-induced hepatitis in vivo. J Immunol. 1983;131(3):1531–1538. [PubMed] [Google Scholar]
- 26.Orange JS, Biron CA. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol. 1996;156(12):4746–4756. [PubMed] [Google Scholar]
- 27.Mavilio D, Benjamin J, Daucher M, Lombardo G, Kottilil S, Planta MA, Marcenaro E, Bottino C, Moretta L, Moretta A, Fauci AS. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci USA. 2003;100(25):15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliva A, Kinter AL, Vaccarezza M, Rubbert A, Catanzaro A, Moir S, Monaco J, Ehler L, Mizell S, Jackson R, Li Y, Romano JW, Fauci AS. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J Clin Invest. 1998;102(1):223–231. doi: 10.1172/JCI2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kottilil S, Shin K, Planta M, McLaughlin M, Hallahan CW, Ghany M, Chun TW, Sneller MC, Fauci AS. Expression of chemokine and inhibitory receptors on natural killer cells: effect of immune activation and HIV viremia. J Infect Dis. 2004;189(7):1193–1198. doi: 10.1086/382090. [DOI] [PubMed] [Google Scholar]
- 30.Vieillard V, Strominger JL, Debre P. NK cytotoxicity against CD4+ T cells during HIV-1 infection: a gp41 peptide induces the expression of an NKp44 ligand. Proc Natl Acad Sci USA. 2005;102(31):10981–10986. doi: 10.1073/pnas.0504315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wren LH, Chung AW, Isitman G, Kelleher AD, Parsons MS, Amin J, Cooper DA, Investigators Asc, Stratov I, Navis M, Kent SJ (2013) Specific antibody-dependent cellular cytotoxicity responses associated with slow progression of HIV infection. Immunology 138(2):116–123. doi:10.1111/imm.12016 [DOI] [PMC free article] [PubMed]
- 32.Lambotte O, Ferrari G, Moog C, Yates NL, Liao HX, Parks RJ, Hicks CB, Owzar K, Tomaras GD, Montefiori DC, Haynes BF, Delfraissy JF. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. Aids. 2009;23(8):897–906. doi: 10.1097/QAD.0b013e328329f97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madhavi V, Ana-Sosa-Batiz FE, Jegaskanda S, Center RJ, Winnall WR, Parsons MS, Ananworanich J, Cooper DA, Kelleher AD, Hsu D, Pett S, Stratov I, Kramski M, Kent SJ. Antibody-dependent effector functions against HIV decline in subjects receiving antiretroviral therapy. J Infect Dis. 2014 doi: 10.1093/infdis/jiu486. [DOI] [PubMed] [Google Scholar]
- 35.Chung AW, Isitman G, Navis M, Kramski M, Center RJ, Kent SJ, Stratov I. Immune escape from HIV-specific antibody-dependent cellular cytotoxicity (ADCC) pressure. Proc Natl Acad Sci USA. 2011;108(18):7505–7510. doi: 10.1073/pnas.1016048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10(6):661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 37.Ward JP, Bonaparte MI, Barker E. HLA-C and HLA-E reduce antibody-dependent natural killer cell-mediated cytotoxicity of HIV-infected primary T cell blasts. Aids. 2004;18(13):1769–1779. doi: 10.1097/00002030-200409030-00005. [DOI] [PubMed] [Google Scholar]
- 38.Bonaparte MI, Barker E. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood. 2004;104(7):2087–2094. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 39.Cerboni C, Neri F, Casartelli N, Zingoni A, Cosman D, Rossi P, Santoni A, Doria M. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J Gen Virol. 2007;88(Pt 1):242–250. doi: 10.1099/vir.0.82125-0. [DOI] [PubMed] [Google Scholar]
- 40.Alvarez RA, Hamlin RE, Monroe A, Moldt B, Hotta MT, Rodriguez Caprio G, Fierer DS, Simon V, Chen BK. HIV-1 Vpu antagonism of tetherin inhibits antibody-dependent cellular cytotoxic responses by natural killer cells. J Virol. 2014;88(11):6031–6046. doi: 10.1128/JVI.00449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, Barker E. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe. 2010;8(5):397–409. doi: 10.1016/j.chom.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6(5):409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giese S, Marsh M. Tetherin can restrict cell-free and cell-cell transmission of HIV from primary macrophages to T cells. PLoS Pathog. 2014;10(7):e1004189. doi: 10.1371/journal.ppat.1004189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramski M, Stratov I, Kent SJ. The role of HIV-specific antibody-dependent cellular cytotoxicity in HIV prevention and the influence of the HIV-1 Vpu protein. Aids. 2015;29(2):137–144. doi: 10.1097/QAD.0000000000000523. [DOI] [PubMed] [Google Scholar]
- 45.Richard J, Sindhu S, Pham TN, Belzile JP, Cohen EA. HIV-1 Vpr up-regulates expression of ligands for the activating NKG2D receptor and promotes NK cell-mediated killing. Blood. 2010;115(7):1354–1363. doi: 10.1182/blood-2009-08-237370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matusali G, Tchidjou HK, Pontrelli G, Bernardi S, D’Ettorre G, Vullo V, Buonomini AR, Andreoni M, Santoni A, Cerboni C, Doria M. Soluble ligands for the NKG2D receptor are released during HIV-1 infection and impair NKG2D expression and cytotoxicity of NK cells. FASEB J. 2013;27(6):2440–2450. doi: 10.1096/fj.12-223057. [DOI] [PubMed] [Google Scholar]
- 47.Beziat V, Descours B, Parizot C, Debre P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS One. 2010;5(8):e11966. doi: 10.1371/journal.pone.0011966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, Hughes TL, Marburger TB, Sung J, Baiocchi RA, Guimond M, Caligiuri MA. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22(3):295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Hong HS, Ahmad F, Eberhard JM, Bhatnagar N, Bollmann BA, Keudel P, Ballmaier M, Zielinska-Skowronek M, Schmidt RE, Meyer-Olson D. Loss of CCR7 expression on CD56(bright) NK cells is associated with a CD56(dim)CD16(+) NK cell-like phenotype and correlates with HIV viral load. PLoS One. 2012;7(9):e44820. doi: 10.1371/journal.pone.0044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luetke-Eversloh M, Killig M, Romagnani C. Signatures of human NK cell development and terminal differentiation. Front Immunol. 2013;4:499. doi: 10.3389/fimmu.2013.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beziat V, Duffy D, Quoc SN, Le Garff-Tavernier M, Decocq J, Combadiere B, Debre P, Vieillard V. CD56brightCD16+ NK cells: a functional intermediate stage of NK cell differentiation. J Immunol. 2011;186(12):6753–6761. doi: 10.4049/jimmunol.1100330. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Mao HC, Wei M, Hughes T, Zhang J, Park IK, Liu S, McClory S, Marcucci G, Trotta R, Caligiuri MA. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood. 2010;115(2):274–281. doi: 10.1182/blood-2009-04-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juelke K, Killig M, Luetke-Eversloh M, Parente E, Gruen J, Morandi B, Ferlazzo G, Thiel A, Schmitt-Knosalla I, Romagnani C. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood. 2010;116(8):1299–1307. doi: 10.1182/blood-2009-11-253286. [DOI] [PubMed] [Google Scholar]
- 54.Alter G, Teigen N, Davis BT, Addo MM, Suscovich TJ, Waring MT, Streeck H, Johnston MN, Staller KD, Zaman MT, Yu XG, Lichterfeld M, Basgoz N, Rosenberg ES, Altfeld M. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood. 2005;106(10):3366–3369. doi: 10.1182/blood-2005-03-1100. [DOI] [PubMed] [Google Scholar]
- 55.Eisenhardt M, Glassner A, Wolter F, Kramer B, Kokordelis P, Nischalke HD, Boesecke C, Rockstroh JK, Spengler U, Nattermann J. CD27(+)CD56Bright natural killer cells may be involved in spontaneous clearance of acute hepatitis C in HIV-positive patients. Aids. 2014 doi: 10.1097/QAD.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 56.Moretta L. Dissecting CD56dim human NK cells. Blood. 2010;116(19):3689–3691. doi: 10.1182/blood-2010-09-303057. [DOI] [PubMed] [Google Scholar]
- 57.Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, O’Shea MA, Kinter A, Kovacs C, Moretta A, Fauci AS. Characterization of CD56-/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci USA. 2005;102(8):2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong HS, Eberhard JM, Keudel P, Bollmann BA, Ballmaier M, Bhatnagar N, Zielinska-Skowronek M, Schmidt RE, Meyer-Olson D. HIV infection is associated with a preferential decline in less-differentiated CD56dim CD16+ NK cells. J Virol. 2010;84(2):1183–1188. doi: 10.1128/JVI.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, Lanier LL. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 2010;116(19):3865–3874. doi: 10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Bjorklund AT, Flodstrom-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, Malmberg KJ. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116(19):3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 61.Parsons MS, Wren L, Isitman G, Navis M, Stratov I, Bernard NF, Kent SJ. HIV infection abrogates the functional advantage of natural killer cells educated through KIR3DL1/HLA-Bw4 interactions to mediate anti-HIV antibody-dependent cellular cytotoxicity. J Virol. 2012;86(8):4488–4495. doi: 10.1128/JVI.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parsons MS, Loh L, Gooneratne S, Center RJ, Kent SJ. Role of education and differentiation in determining the potential of natural killer cells to respond to antibody-dependent stimulation. Aids. 2014;28(18):2781–2786. doi: 10.1097/QAD.0000000000000489. [DOI] [PubMed] [Google Scholar]
- 63.Mavilio D, Lombardo G, Kinter A, Fogli M, La Sala A, Ortolano S, Farschi A, Follmann D, Gregg R, Kovacs C, Marcenaro E, Pende D, Moretta A, Fauci AS. Characterization of the defective interaction between a subset of natural killer cells and dendritic cells in HIV-1 infection. J Exp Med. 2006;203(10):2339–2350. doi: 10.1084/jem.20060894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donaghy H, Wilkinson J, Cunningham AL. HIV interactions with dendritic cells: has our focus been too narrow? J Leukoc Biol. 2006;80(5):1001–1012. doi: 10.1189/jlb.0306158. [DOI] [PubMed] [Google Scholar]
- 65.Hu PF, Hultin LE, Hultin P, Hausner MA, Hirji K, Jewett A, Bonavida B, Detels R, Giorgi JV. Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+CD56+ cells and expansion of a population of CD16dimCD56- cells with low lytic activity. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10(3):331–340. [PubMed] [Google Scholar]
- 66.Eller MA, Eller LA, Ouma BJ, Thelian D, Gonzalez VD, Guwatudde D, McCutchan FE, Marovich MA, Michael NL, de Souza MS, Wabwire-Mangen F, Robb ML, Currier JR, Sandberg JK. Elevated natural killer cell activity despite altered functional and phenotypic profile in Ugandans with HIV-1 clade A or clade D infection. J Acquir Immune Defic Syndr. 2009;51(4):380–389. doi: 10.1097/QAI.0b013e3181aa256e. [DOI] [PubMed] [Google Scholar]
- 67.Gonzalez VD, Falconer K, Bjorkstrom NK, Blom KG, Weiland O, Ljunggren HG, Alaeus A, Sandberg JK. Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: correlation with outcome of pegylated IFN-alpha and ribavirin treatment. J Immunol. 2009;183(10):6612–6618. doi: 10.4049/jimmunol.0901437. [DOI] [PubMed] [Google Scholar]
- 68.Bjorkstrom NK, Ljunggren HG, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol. 2010;31(11):401–406. doi: 10.1016/j.it.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Hong HS, Eberhard JM, Keudel P, Bollmann BA, Ahmad F, Ballmaier M, Bhatnagar N, Zielinska-Skowronek M, Schmidt RE, Meyer-Olson D. Phenotypically and functionally distinct subsets contribute to the expansion of CD56−/CD16+ natural killer cells in HIV infection. Aids. 2010;24(12):1823–1834. doi: 10.1097/QAD.0b013e32833b556f. [DOI] [PubMed] [Google Scholar]
- 70.Gonzalez VD, Falconer K, Michaelsson J, Moll M, Reichard O, Alaeus A, Sandberg JK. Expansion of CD56− NK cells in chronic HCV/HIV-1 co-infection: reversion by antiviral treatment with pegylated IFNalpha and ribavirin. Clin Immunol. 2008;128(1):46–56. doi: 10.1016/j.clim.2008.03.521. [DOI] [PubMed] [Google Scholar]
- 71.Valentin A, Rosati M, Patenaude DJ, Hatzakis A, Kostrikis LG, Lazanas M, Wyvill KM, Yarchoan R, Pavlakis GN. Persistent HIV-1 infection of natural killer cells in patients receiving highly active antiretroviral therapy. Proc Natl Acad Sci USA. 2002;99(10):7015–7020. doi: 10.1073/pnas.102672999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Milush JM, Lopez-Verges S, York VA, Deeks SG, Martin JN, Hecht FM, Lanier LL, Nixon DF. CD56negCD16(+) NK cells are activated mature NK cells with impaired effector function during HIV-1 infection. Retrovirology. 2013;10:158. doi: 10.1186/1742-4690-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brunetta E, Hudspeth KL, Mavilio D. Pathologic natural killer cell subset redistribution in HIV-1 infection: new insights in pathophysiology and clinical outcomes. J Leukoc Biol. 2010;88(6):1119–1130. doi: 10.1189/jlb.0410225. [DOI] [PubMed] [Google Scholar]
- 74.Ahmad F, Hong HS, Jackel M, Jablonka A, Lu IN, Bhatnagar N, Eberhard JM, Bollmann BA, Ballmaier M, Zielinska-Skowronek M, Schmidt RE, Meyer-Olson D. High frequencies of polyfunctional CD8+ NK cells in chronic HIV-1 infection are associated with slower disease progression. J Virol. 2014;88(21):12397–12408. doi: 10.1128/JVI.01420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285(5428):727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 76.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419(6908):734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 77.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, Zakay-Rones Z, Porgador A, Mandelboim O. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7(5):517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 78.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31(9):2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 79.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409(6823):1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 80.Jarahian M, Watzl C, Fournier P, Arnold A, Djandji D, Zahedi S, Cerwenka A, Paschen A, Schirrmacher V, Momburg F. Activation of natural killer cells by newcastle disease virus hemagglutinin-neuraminidase. J Virol. 2009;83(16):8108–8121. doi: 10.1128/JVI.00211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jarahian M, Fiedler M, Cohnen A, Djandji D, Hammerling GJ, Gati C, Cerwenka A, Turner PC, Moyer RW, Watzl C, Hengel H, Momburg F. Modulation of NKp30- and NKp46-mediated natural killer cell responses by poxviral hemagglutinin. PLoS Pathog. 2011;7(8):e1002195. doi: 10.1371/journal.ppat.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, Gazit R, Gonen-Gross T, Hanna J, Nahari E, Porgador A, Honigman A, Plachter B, Mevorach D, Wolf DG, Mandelboim O. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6(5):515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 83.Kramer B, Korner C, Kebschull M, Glassner A, Eisenhardt M, Nischalke HD, Alexander M, Sauerbruch T, Spengler U, Nattermann J. Natural killer p46High expression defines a natural killer cell subset that is potentially involved in control of hepatitis C virus replication and modulation of liver fibrosis. Hepatology. 2012;56(4):1201–1213. doi: 10.1002/hep.25804. [DOI] [PubMed] [Google Scholar]
- 84.Fauci AS, Mavilio D, Kottilil S. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat Rev Immunol. 2005;5(11):835–843. doi: 10.1038/nri1711. [DOI] [PubMed] [Google Scholar]
- 85.De Maria A, Fogli M, Costa P, Murdaca G, Puppo F, Mavilio D, Moretta A, Moretta L. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur J Immunol. 2003;33(9):2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 86.Marras F, Nicco E, Bozzano F, Di Biagio A, Dentone C, Pontali E, Boni S, Setti M, Orofino G, Mantia E, Bartolacci V, Bisio F, Riva A, Biassoni R, Moretta L, De Maria A. Natural killer cells in HIV controller patients express an activated effector phenotype and do not up-regulate NKp44 on IL-2 stimulation. Proc Natl Acad Sci USA. 2013;110(29):11970–11975. doi: 10.1073/pnas.1302090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parsons MS, Tang CC, Jegaskanda S, Center RJ, Brooks AG, Stratov I, Kent SJ. Anti-HIV antibody-dependent activation of NK cells impairs NKp46 expression. J Immunol. 2014;192(1):308–315. doi: 10.4049/jimmunol.1301247. [DOI] [PubMed] [Google Scholar]
- 88.Pembroke T, Christian A, Jones E, Hills RK, Wang EC, Gallimore AM, Godkin A. The paradox of NKp46+ natural killer cells: drivers of severe hepatitis C virus-induced pathology but in vivo resistance to interferon alpha treatment. Gut. 2014;63(3):515–524. doi: 10.1136/gutjnl-2013-304472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, Sykora KW, Schmidt RE. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol. 2001;31(10):3121–3127. doi: 10.1002/1521-4141(2001010)31:10<3121::aid-immu3121>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 90.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 91.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7(6):739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 92.Uhrberg M, Valiante NM, Young NT, Lanier LL, Phillips JH, Parham P. The repertoire of killer cell Ig-like receptor and CD94:NKG2A receptors in T cells: clones sharing identical alpha beta TCR rearrangement express highly diverse killer cell Ig-like receptor patterns. J Immunol. 2001;166(6):3923–3932. doi: 10.4049/jimmunol.166.6.3923. [DOI] [PubMed] [Google Scholar]
- 93.Lopez-Vazquez A, Rodrigo L, Martinez-Borra J, Perez R, Rodriguez M, Fdez-Morera JL, Fuentes D, Rodriguez-Rodero S, Gonzaez S, Lopez-Larrea C. Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2005;192(1):162–165. doi: 10.1086/430351. [DOI] [PubMed] [Google Scholar]
- 94.Jensen SS, Hartling HJ, Tingstedt JL, Larsen TK, Nielsen SD, Pedersen C, Fomsgaard A, Karlsson I. HIV-specific ADCC improves after antiretroviral therapy and correlates with normalization of the NK cell phenotype. J Acquir Immune Defic Syndr. 2014 doi: 10.1097/QAI.0000000000000429. [DOI] [PubMed] [Google Scholar]
- 95.Cassidy SA, Cheent KS, Khakoo SI. Effects of peptide on NK cell-mediated MHC I recognition. Front Immunol. 2014;5:133. doi: 10.3389/fimmu.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, Carrington M, Allen TM, Altfeld M. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476(7358):96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 98.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39(6):733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM, Bernard NF. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J Immunol. 2010;184(4):2057–2064. doi: 10.4049/jimmunol.0902621. [DOI] [PubMed] [Google Scholar]
- 100.Kamya P, Boulet S, Tsoukas CM, Routy JP, Thomas R, Cote P, Boulassel MR, Baril JG, Kovacs C, Migueles SA, Connors M, Suscovich TJ, Brander C, Tremblay CL, Bernard N, Canadian Cohort of HIVISP Receptor-ligand requirements for increased NK cell polyfunctional potential in slow progressors infected with HIV-1 coexpressing KIR3DL1*h/*y and HLA-B*57. J Virol. 2011;85(12):5949–5960. doi: 10.1128/JVI.02652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boulet S, Kleyman M, Kim JY, Kamya P, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, Bernard NF. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. Aids. 2008;22(12):1487–1491. doi: 10.1097/QAD.0b013e3282ffde7e. [DOI] [PubMed] [Google Scholar]
- 102.Long BR, Ndhlovu LC, Oksenberg JR, Lanier LL, Hecht FM, Nixon DF, Barbour JD. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol. 2008;82(10):4785–4792. doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ravet S, Scott-Algara D, Bonnet E, Tran HK, Tran T, Nguyen N, Truong LX, Theodorou I, Barre-Sinoussi F, Pancino G, Paul P. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109(10):4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 104.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204(12):3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Korner C, Granoff ME, Amero MA, Sirignano MN, Vaidya SA, Jost S, Allen TM, Rosenberg ES, Altfeld M. Increased frequency and function of KIR2DL1-3(+) NK cells in primary HIV-1 infection are determined by HLA-C group haplotypes. Eur J Immunol. 2014;44(10):2938–2948. doi: 10.1002/eji.201444751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hellmann I, Letvin NL, Schmitz JE. KIR2DL4 copy number variation is associated with CD4+ T-cell depletion and function of cytokine-producing NK cell subsets in SIV-infected Mamu-A*01-negative rhesus macaques. J Virol. 2013;87(9):5305–5310. doi: 10.1128/JVI.02949-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, Ge D, De Luca A, Martinez-Picado J, Wolinsky SM, Martinson JJ, Jamieson BD, Bream JH, Martin MP, Borrow P, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Carrington M, Goldstein DB, Alter G, Immunology NCfHAV Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9(11):e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van Teijlingen NH, Holzemer A, Korner C, Garcia-Beltran WF, Schafer JL, Fadda L, Suscovich TJ, Brander C, Carrington M, Evans DT, van Baarle D, Altfeld M. Sequence variations in HIV-1 p24 Gag-derived epitopes can alter binding of KIR2DL2 to HLA-C*03:04 and modulate primary natural killer cell function. Aids. 2014;28(10):1399–1408. doi: 10.1097/QAD.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fadda L, Korner C, Kumar S, van Teijlingen NH, Piechocka-Trocha A, Carrington M, Altfeld M. HLA-Cw*0102-restricted HIV-1 p24 epitope variants can modulate the binding of the inhibitory KIR2DL2 receptor and primary NK cell function. PLoS Pathog. 2012;8(7):e1002805. doi: 10.1371/journal.ppat.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol. 2005;175(12):7796–7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- 111.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3(10):781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 112.Norman JM, Mashiba M, McNamara LA, Onafuwa-Nuga A, Chiari-Fort E, Shen W, Collins KL. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat Immunol. 2011;12(10):975–983. doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33(4):451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11(12):1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457(7229):557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Beziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A, Hervier B, Theodorou I, Martinot M, Debre P, Bjorkstrom NK, Malmberg KJ, Marcellin P, Vieillard V. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur J Immunol. 2012;42(2):447–457. doi: 10.1002/eji.201141826. [DOI] [PubMed] [Google Scholar]
- 117.Petitdemange C, Becquart P, Wauquier N, Beziat V, Debre P, Leroy EM, Vieillard V. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog. 2011;7(9):e1002268. doi: 10.1371/journal.ppat.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gillard GO, Bivas-Benita M, Hovav AH, Grandpre LE, Panas MW, Seaman MS, Haynes BF, Letvin NL. Thy1+ NK [corrected] cells from vaccinia virus-primed mice confer protection against vaccinia virus challenge in the absence of adaptive lymphocytes. PLoS Pathog. 2011;7(8):e1002141. doi: 10.1371/journal.ppat.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abdul-Careem MF, Lee AJ, Pek EA, Gill N, Gillgrass AE, Chew MV, Reid S, Ashkar AA. Genital HSV-2 infection induces short-term NK cell memory. PLoS One. 2012;7(3):e32821. doi: 10.1371/journal.pone.0032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Helden MJ, Zaiss DM, Sijts AJ. CCR2 defines a distinct population of NK cells and mediates their migration during influenza virus infection in mice. PLoS One. 2012;7(12):e52027. doi: 10.1371/journal.pone.0052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, Cooper MA, Fehniger TA. Cytokine activation induces human memory-like NK cells. Blood. 2012;120(24):4751–4760. doi: 10.1182/blood-2012-04-419283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci USA. 2009;106(6):1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]