Abstract
Links between cancer and stem cells have been proposed for many years. As the cancer stem cell (CSC) theory became widely studied, new methods were developed to culture and expand cancer cells with conserved determinants of “stemness”. These cells show increased ability to grow in suspension as spheres in serum-free medium supplemented with growth factors and chemicals. The physiological relevance of this phenomenon in established cancer cell lines remains unclear. Cell lines have traditionally been used to explore tumor biology and serve as preclinical models for the screening of potential therapeutic agents. Here, we grew cell-forming spheres (CFS) from 25 established colorectal cancer cell lines. The molecular and cellular characteristics of CFS were compared to the bulk of tumor cells. CFS could be isolated from 72 % of the cell lines. Both CFS and their parental CRC cell lines were highly tumorigenic. Compared to their parental cells, they showed similar expression of putative CSC markers. The ability of CRC cells to grow as CFS was greatly enhanced by prior treatment with 5-fluorouracil. At the molecular level, CFS and parental CRC cells showed identical gene mutations and very similar genomic profiles, although microarray analysis revealed changes in CFS gene expression that were independent of DNA copy-number. We identified a CFS gene expression signature common to CFS from all CRC cell lines, which was predictive of disease relapse in CRC patients. In conclusion, CFS models derived from CRC cell lines possess interesting phenotypic features that may have clinical relevance for drug resistance and disease relapse.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1160-9) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, Colon cancer cell lines, Spheres, Microarray
Introduction
The cancer stem cell (CSC) theory has generated much interest in both the research and clinical communities, notably for colorectal cancer (CRC) [1–3]. According to this hierarchical model, CSCs are defined by their ability to self-renew indefinitely, while also being able to differentiate and generate both tumorigenic and non-tumorigenic daughter cells that constitute the bulk of the tumor [4]. CSCs are thought to have a low rate of division and proliferation that helps them resist various chemotherapies and radiation. Both these forms of treatment preferentially affect highly proliferative cells, thus potentially making CSCs a major reason for the failure of anticancer treatment [5]. In tumors including CRC, the presence and survival of CSCs has been suggested as a key mechanism underlying chemoresistance and disease relapse [6, 7]. This original interpretation of the CSC theory has recently been challenged, however. Some authors have highlighted the plasticity of the CSC phenotype and suggested that it could be induced through dedifferentiation processes influenced by the tumor cell environment [8].
Cancer cell lines have been widely used to explore tumor biology and as preclinical models for the screening of potential therapeutic agents. They are a valuable resource that can be used repeatedly and have also been well characterized with respect to mutational and gene expression profiles [9]. Similar frequencies of gene mutation have been reported in primary tumors and in cancer cell lines derived from the same primary site. Cancer cell lines are not contaminated with stromal tissue, which can sometimes affect the interpretation of data obtained from primary tumors [10]. Furthermore, cancer cell lines often faithfully represent the tumor from which they were isolated [11, 12]. It remains to be determined whether cancer cell lines are relevant biological tools to study the role of CSCs in tumorigenesis. A number of authors have hypothesized the existence of cancer stem-like cells in these cellular models [13, 14]; however, their phenotype is still poorly characterized. Moreover, it is not known whether cancer stem-like cells from cell lines have any clinical relevance [15, 16].
In order to study cells from cancer cell lines that could display a stem-like phenotype, the first requirement is to have a system in which they can be propagated. The ability to grow in suspension as spheres in serum-free medium supplemented with specific growth factors and chemicals has been described for the expansion of neuronal stem cells [17]. Sphere culture has also been proposed as a valuable method for isolating cancer cells with conserved stemness determinants that are able to propagate in defined media [18–21]. In the present study, we have used this method to grow cell-forming spheres (CFS) from a panel of 25 CRC cell lines. These cell lines were selected to reflect the heterogeneity of CRC in terms of showing microsatellite stability (MSS) or instability (MSI). CRC is a complex tumor entity that includes distinctive molecular phenotypes associated with different clinical features, including response to chemotherapy [22–24]. We investigated the cellular and molecular phenotypes of CFS derived from this panel of CRC cell lines, with particular reference to treatment resistance and CSC features.
Materials and methods
Tissue collection and preparation of xenografts
Human colon tissue fragments were obtained in accordance with the ethical standards of the institutional committee on human experimentation from 15 patients undergoing a colon resection for CRC at the Saint-Antoine hospital in Paris. A biobank collection of 30 tumors stored at −80 °C was used to obtain fresh tumor tissue after engraftment in 5-week-old female nude mice (nu/nu). Tumor implantation procedures were performed as previously described [38]. Twelve tumor xenografts were grown for between 1 and 4 months after engraftment. Cancer tissues were intensively washed four times in PBS solution containing antibiotics and then incubated overnight in DMEM/F12 (PAA) containing penicillin (500 U/ml), streptomycin (500 μg/ml), amphotericin B (0.25 μg/ml), and ceftazidime (50 μg/ml). Enzymatic digestion was performed using collagenase (1.5 mg/ml; Sigma) and hyaluronidase (20 μg/ml; Sigma) in PBS for 1 h. These digests were used for FACS analysis.
Cell culture
Colon cancer cell lines were cultured in DMEM media supplemented with 10 % FCS (20 % for Caco-2 cell line), 100 U/ml penicillin G, and 100 μg/ml streptomycin. For the culture of CFS, cell lines were grown in serum-free DMEM/F12 media supplemented with 100 U/ml penicillin G, 100 μg/ml streptomycin, 6 g/l glucose, 1 mg/ml NaHCO3, 5 mM HEPES, 2 mM l-glutamine, 4 μg/ml Heparin, 4 mg/ml BSA, 60 mmol/l putrescine, 20 nmol/l progesterone, 30 nmol/l sodium selenite, 25 μg/ml, insulin, 100 μg/ml apo-transferrin, and human recombinant EGF and FGF-2 (Sigma), both at a final concentration of 20 μg/ml (sphere-medium).
CFS formation assay
The CFS capacity of colon cancer cell lines tested in this study was derived from monolayer culture or floating culture (for Colo320 colon cancer cell line). It was assessed by plating 2 × 105 cells in a T25 flask (8,000 cells/cm2). In CFSPositive cell lines, CFS were observed 3–7 days after plating. To obtain pure CSC-like cells, the culture period in sphere-medium was extended to 10 passages. To evaluate the CFS capacity of sorted CD166+CD44+EpCAMhigh or CD166−CD44−EpCAMlow HCT116 cell subpopulations (see Fig. 4c, below), 1,000 cells/well were plated in 96-well culture dishes in 200 μl of sphere-medium. The number of CFS in each well was evaluated after 5 days.
Fig. 4.
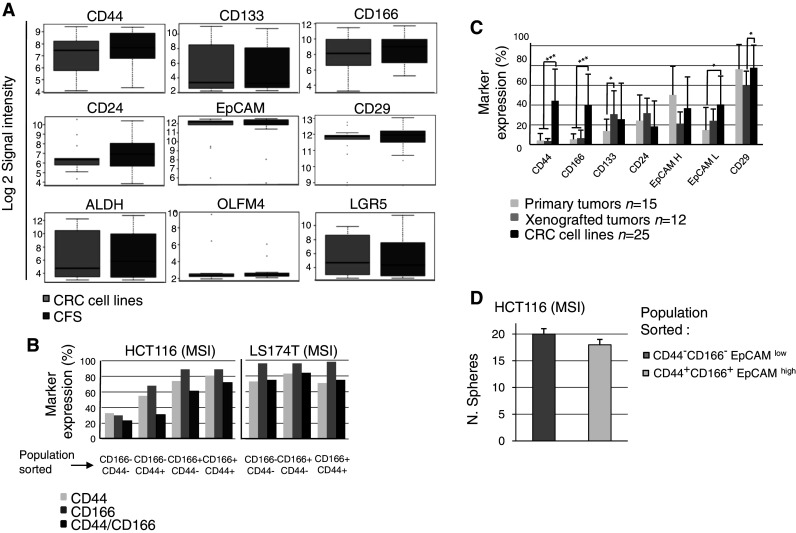
a Expression of putative colorectal CSC markers (CD44, CD133, CD166, CD24, CD29, EPCAM, ALDH, OLFM4, LGR5) were investigated using gene expression arrays in 13 CFS/parental cell lines (8 MSI: LIM2405, HCT8, HCT116, HCT15, TC-7 CO115, RKO, LS411; 5 MSS: V9P, HT29, SW620, Colo320, FET). b Expression of CD166 and CD44 markers was investigated by flow cytometry in different subpopulations of sorted cells from the HCT116 and LS174T parental cells. CD166−/CD44−, CD166+/CD44−, CD166+/CD44+ and CD166−/CD44+ (HCT116 only) cells were sorted by FACS and re-inoculated in medium. After a few days of growth, cells from sorted subpopulations were analyzed again for marker expression. The CD expression pattern was compared to the cell sorted profile. c Expression of six markers in CRC cell lines (n = 25), primary CRCs (n = 15) and/or tumor xenografts established directly from the primary CRC (n = 12). d CFS ability of HCT116 after sorting of two subpopulations of CD166+CD44+EpCAMhigh and CD166−CD44−EpCAMlow cells
Proliferation and chemosensitivity assay
Rates of proliferation and sensitivity to 5-FU were assessed using the cell proliferation reagent WST-1 (Roche). Briefly, 104 cells of each cell line or from CFS cultures were plated per well in 24-well plates in 2 ml of media with or without 5-FU. After 5 days, WST-1 reagent was added at a 1:10 final dilution and incubated for 4 h at 37 °C. The relative survival fraction of cells was compared between treated and untreated cells.
CFS assay following 5-FU treatment
Inoculation of 2 × 106 cells into a T75 flask was made with different concentrations of 5-FU to obtain 10 % cell survival after 5 days of incubation. Cells were then washed detached and 105 cells were inoculated into a T25 flask with sphere-medium (4,000 cells/cm2). After 3 days observation, photographs were taken to determine the proportion of CFS amongst the 5-FU resistant cells. For experimental controls, untreated cells were plated at the same concentration in sphere-medium.
Chemical screening
The multi-step strategy used to screen the Institut Curie/CNRS chemical library is shown in Fig. 3a (see below). This bank contains 8,560 compounds in a 96-well format at 10 mg/ml in DMSO (i.e. mean concentration 10 mM). Screening was performed at 10 and 1 μM final concentrations in 96-well plates and in a final volume of 200 μl of medium. CRC cell lines were incubated with the chemical bank at 700 cells/well for 5 days in standard culture conditions. The Wst-1 assay was used to indirectly estimate cell survival according to the indicated procedure (Roche). Confirmation of the 15 compounds (validation step) was obtained by starting with drug powders in order to reach the correct initial concentration and then performing a cell survival test in 24-well plates with 2 ml of medium. CRC cell lines and CFS were incubated with the chemical bank at 700 and 1,500 cells/well, respectively, for 5 days in standard culture conditions.
Fig. 3.
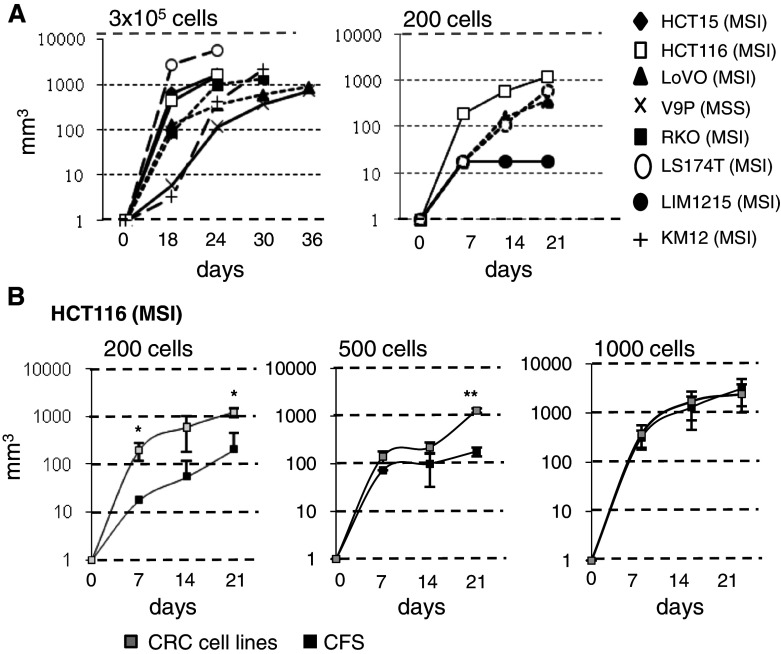
a Analysis of tumor growth following injection of a high number (n = 3 × 105) of cells from CRC cell lines (5 CFSpositive and 3 CFSnegative, left top panel) or low number (n = 200) of cells (2 CFSpositive and 2 CFSnegative, right top panel). b Comparison of tumor growth following injection of CRC cell line HCT116 and the corresponding CFS at 200 cells (*P = 0.034 at 7 days and P = 0.012 at 21 days), 500 cells (**P = 0.006 at 21 days) and 1,000 cells
Subcutaneous transplantation of colon cancer cell lines
Colon cancer cell lines and CFS were suspended in 200 μl PBS-Matrigel (1:1) mixture. They were injected subcutaneously in the flank of 5-week-old nude mice (nu/nu; 1 injection/flank). Experiments were performed in triplicates (3 mice/each sample). Tumor formation was evaluated using a caliper starting on the third week after injection and then weekly for 4 weeks. Animals were sacrificed when the tumor size was between 15 and 20 mm in diameter, or 7 weeks after the injection.
Microsatellite analysis
Non-coding microsatellite repeat markers were used to detect instability in five microsatellites (NR27, NR21, NR24, Bat25, and Bat26 comprising the pentaplex PCR system) in CRC cell lines and their CFS counterparts, as previously described [39]. The TGFBR2, BAX, MSH3, and MSH6 genes containing coding repeats were amplified as previously described [40]. Four other genes containing coding mononucleotide repeat were also amplified with specific primers (sequences available on request). Amplified PCR products were run on an Applied Biosystems PRISM 3100 Genetic Analyzer automated capillary electrophoresis DNA sequencer. Allelic sizes were estimated using gene mapper software (Applied Biosystems).
RNA and DNA extraction
Total RNA from CFS and CRC cell lines was extracted using Trizol (Invitrogen) and DNA was extracted using a standard phenol–chloroform procedure. Both RNA and DNA were assessed for integrity and quantity following stringent quality control criteria (CIT program protocols http://cit.ligue-cancer.net).
Flow cytometry
Flow cytometry was performed on adherent cell lines or CFS cultures after dissociation with accutase (PAA). Cells were washed once in PBS supplemented with 1 % BSA (Sigma) and resuspended in PBS/1 % BSA at a concentration of 106 cells/100 μl. Cells were stained with IgG-PE/PECy5/FITC/APC (BD Biosciences) to detect non-specific binding of antibodies and autofluorescence. Primary antibodies used were: CD44-FITC 1:75 (clone G44-26; BD Biosciences), CD133-PE 1:100 (clone AC133; Miltenyi Biotec), EpCAM-APC 1:200 (clone HEA-125; Miltenyi Biotec), CD24-FITC 1:100 (clone ML5; BD Biosciences) and CD29-PECy5 1:100 (clone MAR4; BD Biosciences). Cells were incubated for 30 min at 4 °C in the dark and then washed in buffer (PBS, 1 % BSA, 1 mM EDTA). Expression of cell surface markers was detected with a FACScan flow cytometer (BD Biosciences). Cell line suspensions were sorted according to their CD166 and/or CD44 and/or EpCAM expression with a FACS Coulter (BD Biosciences). Separated subpopulations were reanalyzed for purity.
Genomic and gene expression arrays and analysis
Data preparation
Gene expression analysis using arrays was carried out on the IGBMC microarray platform (Strasbourg, France). Total RNA was amplified, labeled, and hybridized to Affymetrix Human Genome U133 plus2 GeneChips following the manufacturer’s protocol (Affymetrix, Santa Clara, CA, USA). The chips were scanned with the Affymetrix GeneChip Scanner 3000 and raw intensities were quantified from subsequent images using GCOS 1.4 software (Affymetrix). Data were normalized using the Robust Multi-array Average method and implemented in the R package affy [41].
Genomic arrays were performed on the Integragen Platform (Evry, France). DNAs from 13 CFS/adherent cell lines (i.e. LIM2405, HCT8, HCT116, HCT15, TC-7, CO115, RKO, LS411, V9P, HT29, SW620, Colo320, FET) were hybridized on IlluminaSNP HumanCNV610 chips according to instructions provided by the array manufacturer (Illumina, San Diego, CA, USA). Data were normalized and processed as described in supplemental methods [42]. Data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress).
All analyses were performed using R software (http://www.R-project.org)
Unsupervised analysis of gene expression data
To evaluate the distance between CRC cell lines and CFS, PCA and consensus hierarchical clustering analysis of the 13 pairs (26 samples) were performed on probe-sets present [log2 intensity > log2(3.5)] in at least 5 % of the samples and having a robust coefficient of variation significantly different from the median variance of all probe sets (rCV > 0.05 and P value variance test <0.01).
CFS signature
Genes differentially expressed between CRC cell lines and CFS were assessed using Limma paired moderated t test [43]. Genes having a P value <0.001 define the CFS signature (n = 359 probe sets).
CFS functional analysis
All KEGG pathways and gene sets functionally related to stemness from KEGG, Biocarta, GeneOntology, Molecular Signatures database and Stanford Microarray database were tested for enrichment of up- and down-regulated genes in CFS. Enrichment of the top 100–500 up/down deregulated probe sets was evaluated by computing a hypergeometric test. The median P value across up/down top probe set lists was used to select pathways and gene sets of interest (P value <0.05).
Analysis of deregulated regions
To define up/down regulation regions separately for each CFS/CRC cell line pair, the genome was segmented into overlapping windows of 5 Mb. In each window, the enrichment of up- or down-regulated genes (log2(FC) > 0.5) for the given pair was assessed by a Fisher test between up/down genes in the windows and up/down regulated genes in the rest of the genome. To compute the frequency across pairs, each region was assigned −1(down-regulated)/0 (not modified)/1 (up-regulated) depending on the significance (P value <0.05) of the enrichment.
Survival analysis
Two publicly available Affymetrix U133P2 datasets with Recurrence Free Survival annotations were used: [25] dataset GSE17536 and GSE17537 comprising 148 samples of Stage II/III CRC), and [26] GSE14333 comprising 99 samples of Stage B and C CRC not contained in the previous dataset. These were normalized by RMA and by clinical center. To evaluate the survival impact of the CFS signature in those datasets, a subset of genes from the original signature (see Supplementary Table S1) was selected based upon their high fold-change [|log2(CFS/cell line)| > 0.8; n = 55]. To define a prognostic CFS signature, the 55-gene signature was reduced to probe sets significantly associated with prognosis in the first dataset using univariate Cox models (log rank P value <0.05; n = 8). For both CFS signatures, an average expression score per sample was then defined. Normalized intensity values of selected genes were each centered to zero by subtracting the median expression of each gene. Genes that were down-regulated in CFS were multiplied by (−1), thus both up- and down-regulated genes can be used in the score. All genes from the given signature were then averaged. A higher score corresponded to a higher deregulation of CSF genes. The score was then divided into high and low score groups by taking tails of the score distribution in the considered dataset, i.e. 30 % of the highest and 30 % of the lowest scores.
Survival curves were calculated according to the Kaplan–Meier method with an end-point at 5 years. Differences between curves were assessed using the log-rank test. Univariate and multivariate associations for outcome were performed using the Cox regression model.
Results
The ability of CRC cell lines to grow as CFS in serum-free medium enriched with growth factors and chemical supplements is highly variable
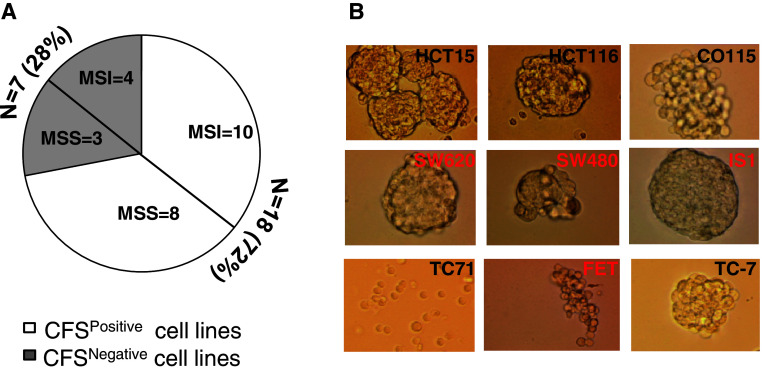
We tested the ability of established MSI and MSS CRC cell lines (n = 25) to grow as CFS in sphere-medium. CFS were obtained from 10 MSI and 8 MSS cell lines (total 18/25, 72 %) (Fig. 1a). In the remaining 7 CRC cell lines (4 MSI, 3 MSS), a small number of cells remained afloat but died after 1–4 passages (1–6 weeks) in sphere-medium (data not shown). Morphologically, the CFS were quite heterogeneous and ranged from densely packed spheres with almost indiscernible individual cells to more loosely packed spheres or individual floating cells (Fig. 1b).
Fig. 1.
a Results of the CFS assay in 25 CRC cell lines (MSS = 11; MSI = 14). b Morphological features of CFS from 9 CRC cell lines (MSI black, MSS red) grown in sphere-medium. The morphology of CFS derived from T71 is peculiar and these grow in suspension as single cells
CFS display identical clonal genomic alterations to their parental CRC cells
Extensive analysis using SNP microarrays revealed that genomic alterations due to chromosomal instability (CIN) were similar between parental CRC cell lines and their related CFS. This was observed both with MSI and MSS CRC cell lines displaying low and high levels of CIN, respectively. SNP analysis was performed on 13 CFS/adherent cell lines (LIM2405, HCT8, HCT116, HCT15, TC-7 CO115, RKO, LS411, V9P, HT29, SW620, Colo320) (Fig. 2a; and data not shown). The status of DNA microsatellites that constitute accurate markers of the ‘history’ of each tumor cell in CRC cell lines displaying MSI was also analyzed. Mutation analysis of DNA microsatellite sequences in 10 CFSPositive MSI cell lines (HCT116, CO115, HCT15, HCT8, ISHI, LIM2405, LS411, RKO, TC-7, TC71) revealed identical patterns of alteration in non-coding (BAT26, BAT25, NR21, NR25, NR27) and coding (TGFBR2, RAD50, MSH6, MSH3, MBD4, BAX, ATR, BLM) repeats between parental cells and their corresponding CFS (Fig. 2b). Thus, CRC cell lines and their corresponding CFS progeny are clonally identical, indicating the CFS phenotype arises from the selection of pre-existing clones in the parental cell lines that are able to grow under specific conditions of serum deprivation.
Fig. 2.
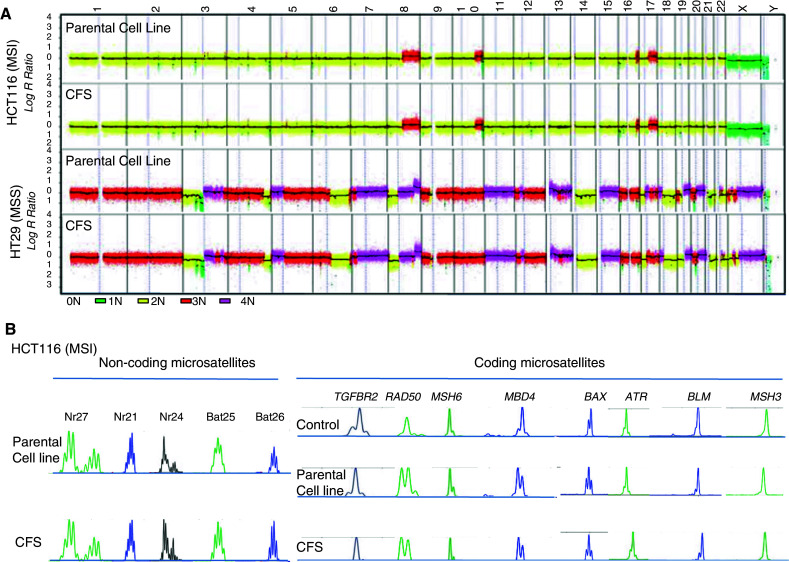
a Extensive microarray analysis revealed that chromosomal aberrations due to CIN were similar between 13 parental CRC cell lines (LIM2405, HCT8, HCT116, HCT15, TC-7, CO115, RKO, LS411, V9P, HT29, SW620, Colo320, FET) and their corresponding CFS. Examples of chromosomal instability in HCT116 (MSI cell line), HT29 (MSS cell line) and their corresponding CFS. b Microsatellite instability in HCT116 parental cells and their corresponding CFS. Mutation analysis of DNA microsatellites showed identical patterns of alteration in both non-coding (BAT26, BAT25, NR21, NR25, NR27) and coding (TGFBR2, RAD50, MSH6, MSH3, MBD4, BAX, ATR, BLM) repeats in the parental CRC cell lines and their corresponding CFS. This was observed in HCT116 and in 10 CFSPositive MSI CRC cell lines (HCT116, CO115, HCT15, HCT8, ISHI, LIM2405, LS411, RKO, TC-7, TC71, not shown)
Acquisition of the CFS phenotype by CRC cells is associated with specific changes in gene expression
The gene expression profiles of 13 CFSPositive CRC cell lines (8 MSI: LIM2405, HCT8, HCT116, HCT15, TC-7 CO115, RKO, LS411; 5 MSS: V9P, HT29, SW620, Colo320, FET) and their corresponding CFS populations were compared using microarrays. Principal component analysis of the profiles revealed the CFS grouped together with their parental CRC cell lines (Supplementary Fig. S1A). A total of 264 genes displayed significant down- or up-regulation in CFS (P value <0.001, paired moderated t test) and are listed in Supplementary Table S1. Specific signaling and metabolic pathways were associated with the CFSPositive gene expression signature (Table 1). As expected, a number of genes from this signature reflected the different culture conditions used for adherent and CFS cells and were linked mainly to cell metabolism and growth factor pathways (Table 1). The CFS signature also included genes related to stemness and to cellular mechanisms associated with treatment resistance, such as transmembrane transporters, apoptosis and DNA damage (Table 1). Specific chromosomal regions were enriched with genes from the CFS signature (Supplementary Fig. S1B). These regions displayed a similar genomic status (DNA copy-number) in CFS and the parental CRC cell line, suggesting that gene deregulation occurred via epigenetic processes that were independent of DNA copy number. Regions that were frequently down or up-regulated in CFS (>30 %) are shown in the lower panel of Figure S1B.
Table 1.
Specific signaling and metabolic pathways significantly associated with the CFS gene signature
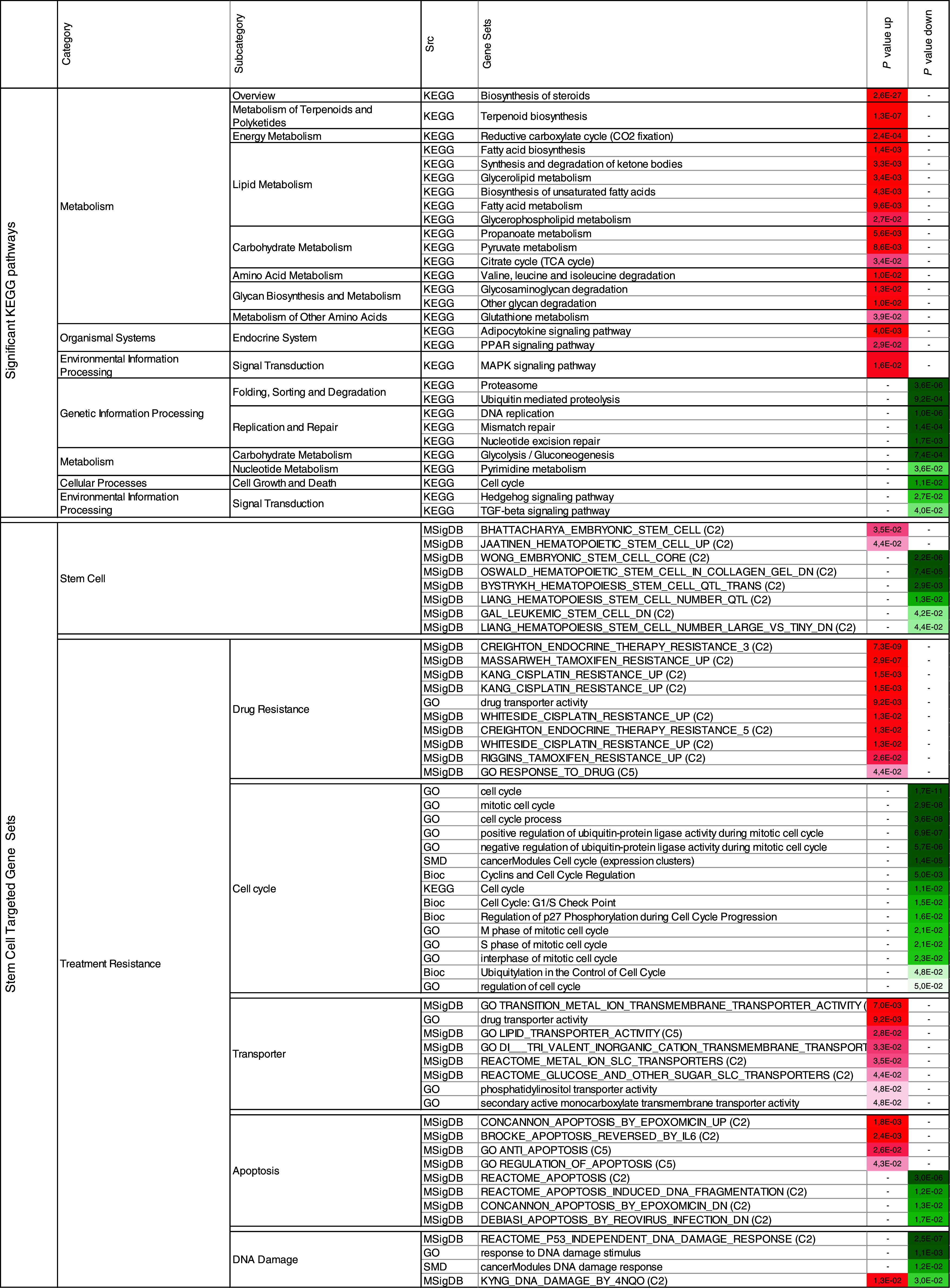
P value up (down): hypergeometric test P value using top up (down) regulated genes in CFS versus cell line
Bioc biocarta, GO geneontology, MSigDB molecular signatures database, SMD stanford microarray database, KEGG kyoto encyclopedia of genes and genomes, C2 MSigDB curated gene sets, C5 MSigDB GO gene sets
Both CFS and their parental CRC cell lines are highly tumorigenic and they show similar expression of CSC markers
Serial engraftments of 8 CRC cell lines (5 CFSPositive: HCT15, HCT116, LoVo, V9P, RKO; 3 CFSNegative: LS174T, LIM1215, KM12) were performed in nude mice. All 8 cell lines tested were found to be highly tumorigenic, even when only 200 CRC cells were injected (Fig. 3a). CFS cells derived from HCT116 and LoVo cell lines were also highly tumorigenic. Low numbers of injected CFS cells (200 or 500 cells) derived from both these MSI CRC cell lines were less tumorigenic compared to the parental cell lines (Fig. 3b; and data not shown). However, no difference was apparent when 1000 cells were injected. The expression of putative colorectal CSC markers (CD44, CD133, CD166, CD24, CD29, EPCAM, ALDH, OLFM4, LGR5) evaluated using arrays was not significantly different between 13 pairs of CFS cells and their corresponding parental cell lines (8 MSI: LIM2405, HCT8, HCT116, HCT15, TC-7 CO115, RKO, LS411; 5 MSS: V9P, HT29, SW620, Colo320, FET) (Fig. 4a). The expression of 6 putative colorectal CSC markers (CD44, CD166, CD133, CD24, EpCAM, CD29) was also compared using flow cytometry in the 25 parental CRC cell lines (Supplementary Table S2). Overall, the expression of these markers was highly variable and none was expressed exclusively in CFSpositive cell lines. Moreover, their expression was highly variable over time, as shown for CD44 and CD166 expression in cell sub-populations sorted by FACS from LS174T and HCT116 parental cells (Fig. 4b). Finally, we quantified the expression of putative colorectal CSC markers (CD44, CD133, CD166, CD24, CD29, EPCAM) in 15 primary CRCs and 12 CRC tumor xenografts established from these primary CRCs and grown in nude mice. Most markers were expressed at significantly higher levels in CRC cell lines compared to primary CRCs and/or tumor xenografts (Fig. 4c). Overall, these results demonstrate that the CFS phenotype derived from CRC cell lines is only weakly related to the putative CSC phenotype from primary tumors. In line with this, a CFS assay showed similar results using sorted populations of CD166+CD44+EpCAMhigh or CD166−CD44−EpCAMlow HCT116 cells (Fig. 4d).
The ability of CRC cells to grow as CFS is strongly increased by prior treatment with 5-Fluorouracil
The ability of LoVo and HCT116 cell lines to grow as CFS in sphere-medium increased following treatment with the chemotherapeutic agent 5-Fluorouracil (5-FU) for 5 days at IC10 % (5 μM for HCT116 and 7.5 μM for LoVo cells) (Fig. 5a, b; and data not shown). In contrast, 5-FU-resistant clones from the LIM1215 and LS174T CFSNegative cell lines remained unable to grow in sphere-medium after 5-FU treatment at IC10 (Fig. 5a). CRC cell lines were also compared to their CFS counterparts for resistance to 5-FU. In 6 CRC cell lines tested (FET, HCT116, LS411, V9P, TC71, LIM2405), the CFS displayed greater resistance to 5-FU than their parental cells (Fig. 5c). Both CFSPositive and CFSNegative CRC cell lines displayed marked differences in resistance to this drug (data not shown). LIM2405 showed a strong predilection to grow as CFS in sphere-medium (data not shown), yet the adherent cells and CFS showed similar resistance to 5-FU (Fig. 5c, bottom and right panels).
Fig. 5.
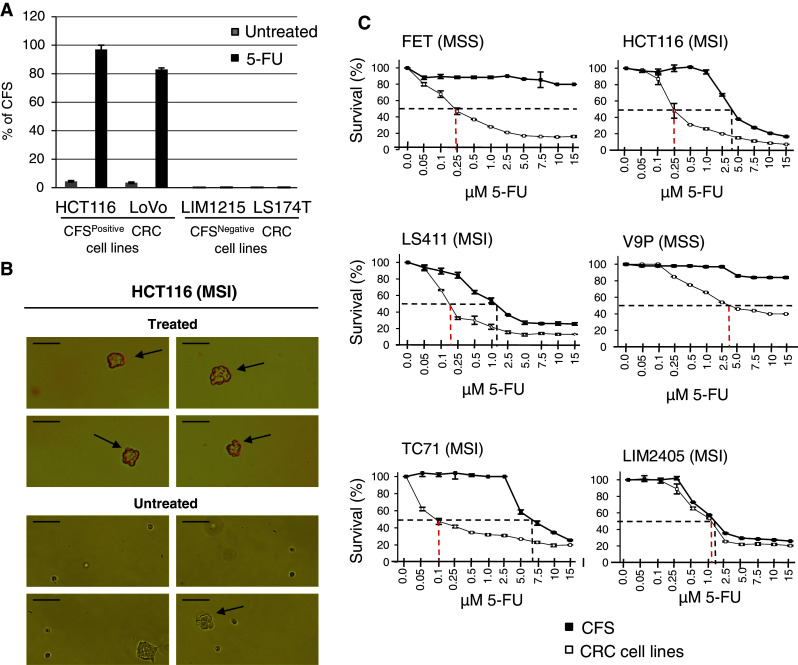
a Percentage of CFS in sphere-medium from LoVo, HCT116, LIM1215 and LS174T cells before and after treatment with 5-Fluorouracil for 5 days at IC10 % (7.5, 7.5, 5 and 20 μM, respectively). b Representative image of spheres (arrows) from HCT116 cells treated with 5-fluorouracil for 5 days at IC10 % (top) and untreated (bottom) after 3 days in sphere-medium (size bar 100 μm). c Cell survival curves for 6 CRC cell lines and their corresponding CFS following 5 days of exposure to increasing concentrations of 5-Fluorouracil. Dotted lines indicate the IC50 value in each case
CFS established from CRC cell lines share a gene signature that predicts disease relapse in CRC patients
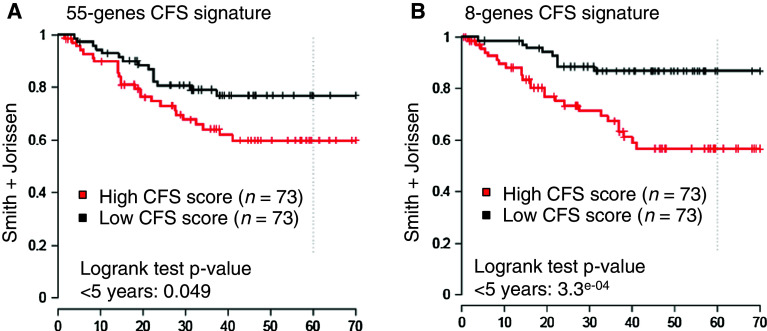
Fifty-five genes that were differentially expressed between CFS and their corresponding CRC cell lines displayed a high level of up- or down-regulation (log2-fold change for CFS/parental cell line >0.8; Supplementary Table S1; Fig. 6). This 55-gene CFS expression signature predicted disease relapse in a previously described, retrospective series of stage II and III CRC patients [25]. Tumors were classified into two groups according to the expression level of the 55 genes (T-CFSHigh or T-CFSLow; for further details, see “Materials and methods” and Fig. 6a), where T-CFSHigh and T-CFSLow correspond to tumors displaying high or low levels of expression of the CFS signature, respectively. Patients with T-CFSHigh tumors showed significantly shorter disease-free survival (DFS) compared to T-CFSLow patients (Supplementary Figure S3A, bottom and left panel). To confirm these clinical findings, the same analysis was performed on another, independent cohort of stage II and III CRC patients [26]. A trend for similar clinical impact of the 55-gene CFS expression signature was observed (Supplementary Figure S2A, bottom and right panel). Following normalization of the data, the two patient series were combined to achieve greater statistical power. In the overall series and in univariate analysis, the survival of stage II and III CRC patients was associated with the expression level of the 55-gene CFS signature (Fig. 7, left panel; Supplementary Figure S2A, top and left panel).
Fig. 6.
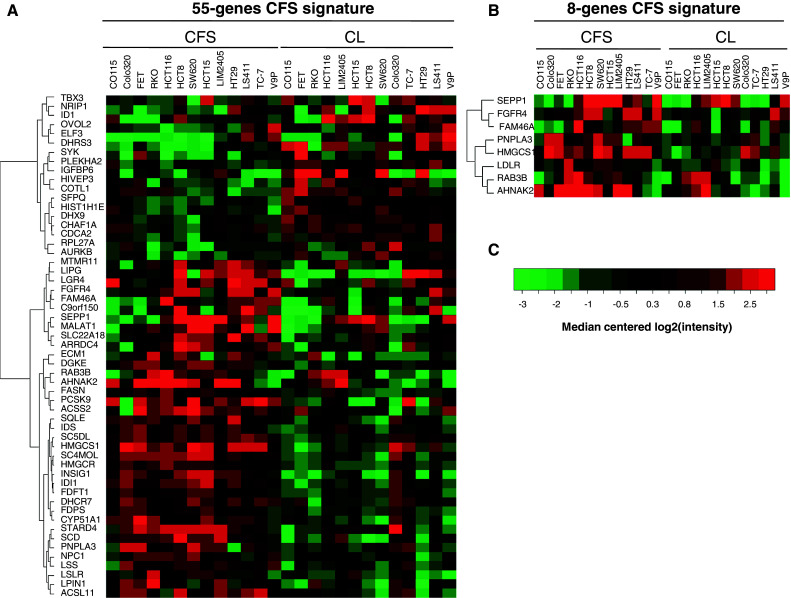
Heatmaps of the 55-gene (a) and 8-gene signatures (b). Visualization of the expression level for each gene in each sample relative to the median gene intensity (c) across all samples. Green corresponds to down-regulation and red to up-regulation. CL parental CRC cell line
Fig. 7.
Kaplan–Meier analysis of disease-free survival for CRC patients classified according to expression levels for the 55-gene (left panel) or 8-gene (right panel) CFS signature
Since the 55-gene signature showed a trend for association with patient outcome in the second dataset, we sought to identify a novel prognostic signature by selecting genes that were individually associated with outcome in the first dataset. This allowed us to define an 8-gene CFS prognostic signature (Fig. 6b; Supplementary Table S1) that showed stronger associations with disease relapse in both the individual and combined patient cohorts (Fig. 7, right panel; Supplementary Figure S2B, top and right panel). To ensure this finding was related to the CFS 55-gene selection and not to the methodology, the same approach was repeated 1,000 times using signatures from 55 genes selected at random. This analysis did not increase the false positive rate in the second dataset (data not shown). In the overall series and in multivariate Cox analysis that included TNM stage, the survival of patients with stage II or III CRC was confirmed to be associated with the expression level of 8 genes from the CFS signature (Table 2).
Table 2.
Association of the 8-gene CFS high/low score and TNM stage and prognosis in the combined dataset (Smith + Jorissen)
| Chi2 test P value | CFSHigh (n = 73) | CFSLow (n = 73) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TNM Stage | 2 (n = 82) | 0.097 | 35 (49 %) | 46 (63 %) | |||||||
| 3 (n = 65) | 38 (51 %) | 27 (37 %) | |||||||||
| Cox univariate analysis | Cox multivariate analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Valuea | n samples | n events | H.R. | 95 % C.I. | P value | H.R. | 95 % C.I. | P value | Model P value | |
| Jorissen + Smith 8-genes signature | TNM stage | 3 | 145 | 34 | 2.1 | 1–4.2 | 0.037 | 1.8 | 0.88–3.5 | 0.11 | 4.10E−04 |
| SC score | CFSLow | 145 | 34 | 0.27 | 0.13–0.58 | 8.0E−04 | 0.29 | 0.14–0.64 | 1.8E−03 | ||
H.R. Cox hazard ratio, 95 % C.I. 95 percent confidence interval of H.R.
Significant p value in italics (<0.05)
aValue: modality of the annotation associated to H.R.
CFS can be used as models to identify new drugs that are more efficient at killing CFS than 5-fluorouracil
Because of the current clinical interest in CFS, the Institut Curie-CNRS chemical library (8,560 compounds) was screened using the HCT116 (MSI) and FET (MSS) CRC cell lines (Supplementary Fig. S3A). Fifteen new compounds were identified with the ability to kill both parental CRC cells and CFS from HCT116 and/or FET at low concentration (IC50 % < 1 μM; Supplementary Fig. S3B). Of note, 8/15 (53 %) of these new compounds belong to the Nitrofurans’ family previously reported to display anticancer and antioxidant properties (Supplementary Table S3, entries 1–8) [29, 30]. Since the library contains 240 of this class of compound (2.8 % of the total), Nitrofurans were significantly over-represented amongst the 15 drugs with high effectiveness against CFS (P = 2.4 × 10−9, Fisher’s exact test). In contrast to results obtained with 5-FU (see Fig. 5b), the ability of HCT116 and FET cell lines to grow in serum-free medium did not increase following treatment with three of the new compounds for 5 days at IC10 % (Supplementary Fig. S3C).
Discussion
Not all CRC cell lines contain CFS. This result may reflect the true heterogeneity of CRC but may also be due to events that occurred in vitro during or after the establishment of CRC cell lines. All CRC cell lines tested here were highly tumorigenic in mouse xenograft assays. We did not observe increased tumorigenicity of CFS-positive compared to CFS-negative CRC cell lines, nor of CFS compared to their parental cell line counterpart. Surprisingly, at lower numbers of injected cells, CFS were less tumorigenic compared to their parental cell line. We have no obvious explanation for this result. In any case, CRC cell lines are not ideal models for evaluating tumorigenicity because of their high level of heterogeneity. Compared to primary colorectal tumors, aberrant expression of putative CSC markers was observed in the CRC cell lines investigated here, but was not different to that of CFS, as already reported for HCT116 [16]. Moreover, the current transcriptome analyses revealed only weak correlations between CFS and the expression of stem cell gene markers. These results highlight the fact that CSC markers are not specific for CFS. The CFS phenotype is therefore quite different to that of putative CSCs from primary tumors. It would be interesting to perform similar experiments relating to the expression of CSC markers, tumorigenicity and gene signatures using primary tumors and their sphere counterparts.
Although clearly different to putative CSCs from primary tumors, an interesting question raised by this study is whether the CFS models established from cancer cell lines have clinical relevance. The differential expression of genes in CFS relative to their parental cells is partly a result of the presence of growth factors in one medium and not in the other. This could involve growth factor pathways as well as CSC-like genes. In support of this, it was recently shown that CFS cultures contain only a fraction of CSCs and therefore cannot be regarded as pure CSC models. With this in mind, our data highlight that CFS have retained interesting phenotypical characteristics, including increased resistance to 5-FU in standard culture conditions. Moreover, the 55-gene CFS signature identified here was common to all CRC cell lines and was predictive for disease relapse in CRC patients. Although validated in two independent CRC series, these findings require confirmation in additional studies using larger cohorts of patients. In summary, CFS models derived from CRC cell lines have interesting phenotypical features and may have clinical relevance for drug resistance and disease relapse, but are unlikely to serve as models for putative CSCs in primary CRC.
The genomic and mutational analyses of CRC cell lines and CFS performed here, including the study of microsatellite DNA repeats in MSI cell lines, help to explain the origin of CFS. The status of DNA microsatellites constitutes an accurate marker of the ‘history’ of each tumor cell. These genetic markers, including the non-coding microsatellite repeats (BAT26, BAT25, NR21, NR25, and NR27), were identical between CFS and their parental CRC cells. The CFS phenotype therefore corresponds to a cellular state achieved only by some clones under specific culture conditions or exposure to drugs. Considering the transcriptome analyses, our results also indicate that only a fraction of cells from CRC cell lines have the capacity to rapidly adjust the expression of specific genes and hence to persist as CFS under challenging growth conditions. These data agree with other recent studies demonstrating widespread plasticity and dedifferentiation processes that affect some tumor cells under specific environmental conditions, particularly CRC cells [4, 31–35].
The expression signature of normal intestinal stem cells, also shared by cells with a stem-like cell phenotype within primary CRC, was recently found to be predictive of disease relapse in CRC patients [36]. However, another group reported the CSC gene signature in CRC may reflect the differentiation status of malignant tissue [37], rather than reflecting the number of CSCs as suggested in the former study. The results presented here relate only to the properties of CFS cultures derived from CRC cell lines, not from primary tumors. We speculate that the gene expression pattern observed in our CFS model corresponds to that of poorly differentiated ‘progenitor cells’, or dedifferentiated CRC cells, due to the altered cell culture conditions. This pattern could be expressed in an important fraction of cancer cells within the primary tumor. The risk of developing a recurrence of CRC might be associated with the expression of specific genes in these tumor cells and which are required for tumor regeneration following cancer therapy. Interestingly, 5 of the genes included in the limited 8-gene CFS signature are predicted to have indirect associations with signaling pathways. LDLR and HMGCS1 participate in SREBP control of the lipid pathway by stimulating lipid synthesis, while FGFR4 is a growth factor reported to be associated with poor prognosis and aggressive disease in many different cancer types including colon cancer [27]. Although still poorly described, AHNAK2 and FAM46A are both upregulated in cisplatin-resistant gastric cancer cell lines [28].
The clinical and molecular heterogeneity of CRC is a major limitation of this type of study. Principal component analyses of the transcriptomic profiles showed that considerable heterogeneity remains between CFS displaying MSI or MSS. The response to chemotherapy is thought to be different between patients with MSI and MSS colon tumors, even when considering tumors with the same stage of disease [37]. The CFS gene signature identified here was shared by both MSI and MSS CRC. Nevertheless, we could not evaluate the clinical relevance of MSI-specific or MSS-specific CFS signatures for patients with these tumor subgroups because MSI status is not contained in the publicly available Affymetrix U133P2 datasets with Recurrence Free Survival annotations used for this study. This is a subject for future investigation.
In conclusion, the current findings support the existence of CFS subpopulations within CRC cell lines and provide a framework to explain the origin of these tumor cells. They also suggest that CRC cells acquire the CFS phenotype through mechanisms that are only weakly related to CSC, but which could nevertheless be important for the development of chemoresistance by CRC cells in vivo in primary tumors. The screening of 8,560 potential anticancer agents using an assay involving CFS populations derived from CRC cell lines may therefore be a useful approach to identify novel drugs for clinical application. The 15 new drugs identified in this study, including several new compounds belonging to the Nitrofurans’ family, are currently being tested for toxicity and efficacy in pre-clinical studies using animal models.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the ‘Carte d’Identité des Tumeurs’ (CIT) program (http://cit.ligue-cancer.net) from the Ligue Nationale Contre le Cancer and by grants from the ‘Institut National du Cancer’ (INCa) (to A.D.). The A.D. group has the label “La Ligue Contre le Cancer”. A.C. is a recipient of an INCa fellowship (Institut National du Cancer). A.L. is a recipient of a MESR fellowship (Ministère de l’Enseignement Supérieur et de le Recherche.
Conflict of interest
No potential conflicts of interests were disclosed.
Abbreviations
- 5-FU
5-Fluorouracil
- CFS
Cell-forming-spheres
- CIN
Chromosomal instability
- CRC
Colorectal cancer
- CSC
Cancer stem cells
- DFS
Disease-free survival
- MMR
Mismatch-repair
- MSI
Microsatellite instability
- MSS
Microsatellite stability
Contributor Information
Ada Collura, Phone: 33-1-49286672, FAX: 33-1-49286681, Email: ada.collura@inserm.fr.
Alex Duval, Phone: +33-1-49286680, FAX: +33-1-49286681, Email: alex.duval@inserm.fr.
References
- 1.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, Stassi G. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1(4):389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 3.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 6.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, Canty AJ, Danska JS, Bohlander SK, Buske C, Minden MD, Golub TR, Jurisica I, Ebert BL, Dick JE. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14(1):43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Medema JP, Vermeulen L. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature. 2011;474(7351):318–326. doi: 10.1038/nature10212. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Bodmer WF. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci USA. 2006;103(4):976–981. doi: 10.1073/pnas.0510146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volchenboum SL, Li C, Li S, Attiyeh EF, Reynolds CP, Maris JM, Look AT, George RE. Comparison of primary neuroblastoma tumors and derivative early-passage cell lines using genome-wide single nucleotide polymorphism array analysis. Cancer Res. 2009;69(10):4143–4149. doi: 10.1158/0008-5472.CAN-08-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, Tomlinson IP, Carter NP. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64(14):4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- 12.Willson JK, Bittner GN, Oberley TD, Meisner LF, Weese JL. Cell culture of human colon adenomas and carcinomas. Cancer Res. 1987;47(10):2704–2713. [PubMed] [Google Scholar]
- 13.Kondo T. Stem cell-like cancer cells in cancer cell lines. Cancer Biomark. 2007;3(4–5):245–250. doi: 10.3233/cbm-2007-34-508. [DOI] [PubMed] [Google Scholar]
- 14.Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci USA. 2010;107(8):3722–3727. doi: 10.1073/pnas.0915135107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittfeld C, Dietrich A, Peickert S, Hering S, Baumann M, Grade M, Ried T, Kunz-Schughart LA. CD133 expression is not selective for tumor-initiating or radioresistant cell populations in the CRC cell lines HCT-116. Radiother Oncol. 2009;92(3):353–361. doi: 10.1016/j.radonc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 16.Kai K, Nagano O, Sugihara E, Arima Y, Sampetrean O, Ishimoto T, Nakanishi M, Ueno NT, Iwase H, Saya H. Maintenance of HCT116 colon cancer cell line conforms to a stochastic model but not a cancer stem cell model. Cancer Sci. 2009;100(12):2275–2282. doi: 10.1111/j.1349-7006.2009.01318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 18.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 19.Fan X, Ouyang N, Teng H, Yao H. Isolation and characterization of spheroid cells from the HT29 colon cancer cell line. Int J Colorectal Dis. 2011;26(10):1279–1285. doi: 10.1007/s00384-011-1248-y. [DOI] [PubMed] [Google Scholar]
- 20.Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105(36):13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, Clevers H, Batlle E. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17(10):1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 22.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 23.Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62(9):2447–2454. [PubMed] [Google Scholar]
- 24.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138(6):2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, Eschrich S, Kis C, Levy S, Washington MK, Heslin MJ, Coffey RJ, Yeatman TJ, Shyr Y, Beauchamp RD. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhoffer M, Orntoft TF, Andersen CL, Gruidl M, Kamath VP, Eschrich S, Yeatman TJ, Sieber OM. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15(24):7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, Dragani TA. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23(29):7307–7311. doi: 10.1200/JCO.2005.17.350. [DOI] [PubMed] [Google Scholar]
- 28.Kang HC, Kim IJ, Park JH, Shin Y, Ku JL, Jung MS, Yoo BC, Kim HK, Park JG. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin Cancer Res. 2004;10(1 Pt 1):272–284. doi: 10.1158/1078-0432.CCR-1025-3. [DOI] [PubMed] [Google Scholar]
- 29.Hayakawa I, Shioya R, Agatsuma T, Furukawa H, Sugano Y. Thienopyridine and benzofuran derivatives as potent anti-tumor agents possessing different structure-activity relationships. Bioorg Med Chem Lett. 2004;14(13):3411–3414. doi: 10.1016/j.bmcl.2004.04.079. [DOI] [PubMed] [Google Scholar]
- 30.Baraldi PG, Romagnoli R, Giovanna Pavani M, del Carmen Nunez M, Bingham JP, Hartley JA. Benzoyl and cinnamoyl nitrogen mustard derivatives of benzoheterocyclic analogues of the tallimustine: synthesis and antitumour activity. Bioorg Med Chem. 2002;10(5):1611–1618. doi: 10.1016/S0968-0896(01)00425-4. [DOI] [PubMed] [Google Scholar]
- 31.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 32.Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7(9):1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, Ikeda Y, Mak TW, Suda T. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431(7011):997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 34.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 35.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Munoz P, Clevers H, Sancho E, Mangues R, Batlle E. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8(5):511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Sinicrope FA, Foster NR, Thibodeau SN, Marsoni S, Monges G, Labianca R, Kim GP, Yothers G, Allegra C, Moore MJ, Gallinger S, Sargent DJ. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103(11):863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poupon MF, Arvelo F, Goguel AF, Bourgeois Y, Jacrot M, Hanania N, Arriagada R, Le Chevalier T. Response of small-cell lung cancer xenografts to chemotherapy: multidrug resistance and direct clinical correlates. J Natl Cancer Inst. 1993;85(24):2023–2029. doi: 10.1093/jnci/85.24.2023. [DOI] [PubMed] [Google Scholar]
- 39.Buhard O, Suraweera N, Lectard A, Duval A, Hamelin R. Quasimonomorphic mononucleotide repeats for high-level microsatellite instability analysis. Dis Markers. 2004;20(4–5):251–257. doi: 10.1155/2004/159347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gayet J, Zhou XP, Duval A, Rolland S, Hoang JM, Cottu P, Hamelin R. Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines. Oncogene. 2001;20(36):5025–5032. doi: 10.1038/sj.onc.1204611. [DOI] [PubMed] [Google Scholar]
- 41.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letouze E, Sow A, Petel F, Rosati R, Figueiredo BC, Burnichon N, Gimenez-Roqueplo AP, Lalli E, de Reynies A. Identity by descent mapping of founder mutations in cancer using high-resolution tumor SNP data. PLoS ONE. 2012;7(5):e35897. doi: 10.1371/journal.pone.0035897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. doi:10.2202/1544-6115.1027 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.