Abstract
Mitochondria are dynamic organelles whose shape is regulated by the opposing processes of fission and fusion, operating in conjunction with organelle distribution along the cytoskeleton. The importance of fission and fusion homeostasis has been highlighted by a number of disease states linked to mutations in proteins involved in regulating mitochondrial morphology, in addition to changes in mitochondrial dynamics in Alzheimer’s, Huntington’s and Parkinson’s diseases. While a number of mitochondrial morphology proteins have been identified, how they co-ordinate to assemble the fission apparatus is not clear. In addition, while the master mediator of mitochondrial fission, dynamin-related protein 1, is conserved throughout evolution, the adaptor proteins involved in its mitochondrial recruitment are not. This review focuses on our current understanding of mitochondrial fission and the proteins that regulate this process in cell homeostasis, with a particular focus on the recent mechanistic insights based on protein structures.
Keywords: Mitochondria, Fission, Dynamin-related protein 1, Adaptors, Lipids, Outer mitochondrial membrane, Endoplasmic reticulum
Introduction
Mitochondria are integral to cellular function and are responsible for energy production in eukaryotes, synthesis of metabolites, phospholipids and heme, and maintenance of calcium homeostasis [1–3]. While these are well-established functions, mitochondria have recently been shown to be anti-viral signalling platforms and contribute to apoptotic activation and cell death [4–6]. Given the diverse role of mitochondria in cellular physiology, it is not surprising that alterations in mitochondrial function are associated with disease states including Alzheimer’s, Huntington’s Parkinson’s diseases [7–12]. These and other diseases (e.g. Charcot–Marie Tooth 2A disease and optic atrophy) can be attributed to defects in known morphology proteins [13]. The opposing processes of fission and fusion maintain mitochondrial morphology and it is this equilibrium that ensures maintenance of mtDNA and metabolic mixing, bioenergetic functionality and organelle number [14, 15].
An imbalance in fission and fusion events can lead to a marked shift in morphology and viability of the organelle. Together these processes maintain the steady-state mitochondrial morphology needed for normal cell function, yet individually fission and fusion perform distinct roles. Mitochondrial fusion maintains a homogenous organellar population and ensures complete complementation of mtDNA [14]. In mammals, fusion is controlled by large GTPases on both the inner (OPA1) and the outer (MFN1/MFN2) mitochondrial membranes [16, 17]. The signals that trigger activation and co-ordination of a double membrane fusion event are currently unknown. As mitochondria cannot be made de novo, mitochondrial fission facilitates inheritance of mitochondria in dividing cells; but left unregulated, fission can lead to a heterogeneous population of organelles with disproportional mtDNA distribution, increased capacity to generate reactive oxygen species and altered viability of the organelle [18, 19]. Mammalian cells that are unable to undergo fission harbour mtDNA in irregularly clustered nucleoids, contributing to a non-uniform cristae structure that encloses the pro-apoptotic protein cytochrome c—resulting in a delay in programmed cell death [20]. Fission is one of the many important steps in mitophagy (a mitochondrial-specific pathway of organelle turnover) since it is stimulated by activation of pro-fission proteins [21, 22].
Dynamin-related protein 1 (Drp1)
Dynamins are a large superfamily of GTPases that have a number of fundamental roles in cellular homeostasis, including membrane fission events, anti-viral signalling, chloroplast biogenesis and plant cell plate formation [23, 24]. The master fission mediator conserved throughout evolution is dynamin-related protein (DRP) family member, Drp1 (or Dnm1 in yeast) [25, 26]. Drp1-mediated fission in different organisms is supported by the specialised architecture of adaptor proteins, including Fis1, Mff, MiD49 and MiD51 in mammals and Mdv1/Caf4 in yeast, as well as cytoskeletal and endoplasmic reticulum (ER) contacts [27, 28]. The importance of Drp1 and fission in mammalian physiology has been recently highlighted. A patient harbouring a heterozygote dominant-negative Drp1 allele presented with broad metabolic defects (both mitochondrial and peroxisomal), abnormal brain development, optic atrophy and died 37 days after birth [29]. In addition, Drp1 knockout in mice results in hypoplasia of the forebrain and is ultimately embryonic lethal [30, 31]. However, mouse embryonic fibroblasts isolated from Drp1 knockout mice are viable in culture, emphasising the hierarchical effect of loss of fission in cell lines, tissue or whole organism. In the case of the Drp1 knockout mouse, global loss leads to specific nervous system impairment. This is augmented in highly polarized cells such as neurons as they have an absolute reliance on oxidative phosphorylation with limited capacity for glycolysis [32, 33].
Post-translational regulation of Drp1
Drp1 can be modified by variety of proteins and cellular signals, and each of these can affect activity in different ways. These modifications include phosphorylation, S-nitrosylation, ubiquitination and sumoylation [34–36]. Cdk1/cyclin phosphorylates Drp1 at S585 and is thought to be an important step during mitosis as it can lead to increased fragmentation to ensure efficient segregation of mitochondria into daughter cells [37]. Phosphorylation of Drp1 at S367 by cAMP-dependant protein kinase A (PKA) inhibits the protein’s GTPase activity and impairs fission [38, 39]. Dephosphorylation of Drp1 is equally important in regulation of the protein—calcineurin removes the phosphate at S367 and in turn causes an increase in fragmentation following an increase in intracellular calcium levels [39]. Recently mitochondrial fission was shown to be required for RAS-induced cellular transformation. In this case, Erk2 phosphorylates Drp1 at S616 to activate fission. Furthermore, expression of a Drp1 S616A mutant that prevents phosphorylation, and blocks Ras-induced tumour growth [40, 41]. The mechanism by which mitochondrial fission is required for cell transformation and tumorigenesis is not clear but may involve aspects of mitochondrial reprogramming.
Drp1 domain structure
The domain structure of Drp1 consists of an amino terminal GTPase domain, a middle domain and the GTPase effector domain (GED), decorated with further structural elements that direct dynamin function (Fig. 1) [42–44]. Drp1 is predominantly cytosolic; however, a considerable quantity is found at mitochondria, specifically in foci that represent future or previous sites of fission [45]. Upon recruitment to mitochondrial constrictions, Drp1 polymerises into spirals and through GTP-dependent conformational changes, constricts the organelle leading to membrane scission [46]. Drp1 is also involved in peroxisomal fission, recruited to the similarly dynamic organelle by adaptor proteins Mff and Fis1 [47, 48].
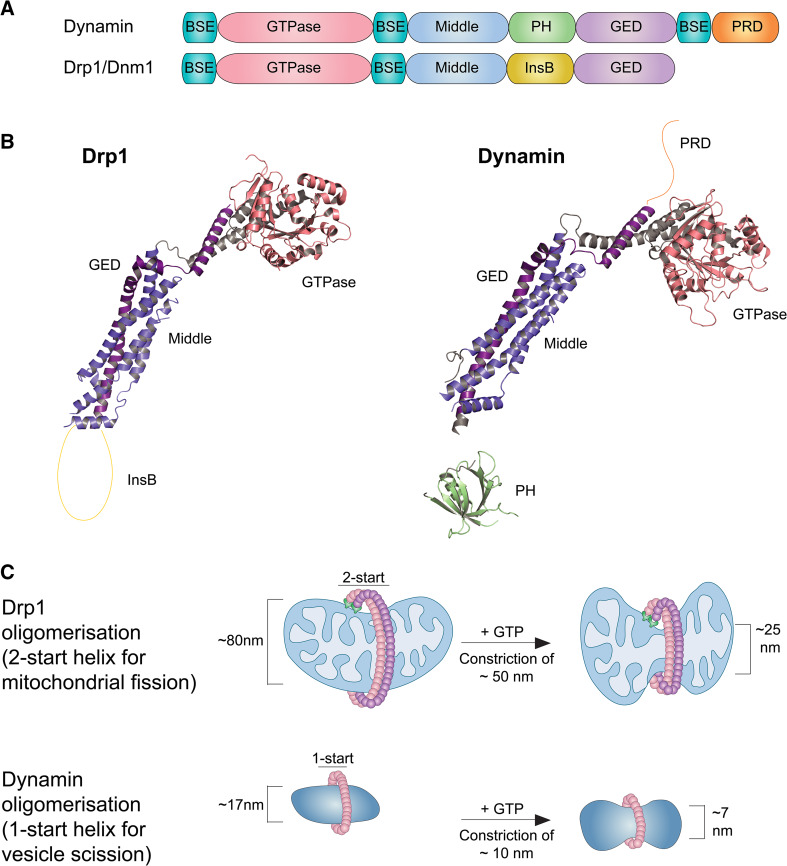
Fig. 1.
Drp1/Dnm1 and dynamin assembly and contractile mechanisms. a Structural motifs present in Drp1/Dnm1 and dynamin. BSE bundle signalling element, GTPase GTPase domain, Middle middle domain, PH pleckstrin homology domain, InsB insert B, GED GTPase effector domain, PRD proline rich domain. b Crystal structures of Drp1 and dynamin (PDB accession numbers: Drp1: 4BEJ; dynamin: 3ZVR). c Models of 2- and 1-start helices formed by Drp1 and dynamin, respectively. While dynamin interacts with the lipid bilayer of membranes directly (via a PH domain), Drp1 requires the presence of membrane adapter proteins at the mitochondrial surface
Drp1 and Dnm1 share the same structural features and consist of a GTPase domain, a bundle-signalling element (BSE), a stalk, and an Insert B (InsB) domain (Fig. 1). Unlike dynamins, Drp1 and Dnm1 lack a lipid-interacting pleckstrin homology (PH) domain or a compensating transmembrane region such as those found in the other morphology-mediating GTPases OPA1 and MFN1/2 [49]. Instead they harbour a variable InsB region that was predicted to interact with mitochondrial adapter proteins [50, 51]. Additionally, while the solved structure of the PH domain reveals a compact, β-strand-rich fold, loosely attached to the dynamin stalk, InsB is predicted to be largely unstructured [42, 43, 50] (Fig. 1). Recently, it was demonstrated that InsB in Drp1 is not absolutely required for fission activity but can modulate Drp1 oligomerisation and activity through post-translational modifications [50].
Cryo-electron microscopy of Dnm1-decorated liposomes has shed light on its oligomerisation and constriction mechanisms [52]. Both proteins assemble on liposomes in the absence of nucleotide in a helical fashion; however, while dynamin forms a one-start helix, the more loosely packed Dnm1 forms a two-start helix, resulting in a larger helical pitch as well as a larger structure diameter (Fig. 1) (129 nm for Dnm1, 50 nm for dynamin) [52]. Additionally, while the three-dimensional reconstruction of the dynamin oligomer identified an interaction with the underlying lipid bilayer, a 3–4 nm gap was observed between Dnm1 and the lipid tubule, indicating that the InsB region that substitutes in the position of the PH domain of dynamin-1 does not directly interact with the membrane [52]. While the typical diameter of the neck of a clathrin-coated vesicle of 40 nm can be encircled by around 14 dynamin dimers, the larger mitochondrial constriction sites of around 110 nm require a more substantial constriction and is calculated to require 48 Dnm1 dimers or 24 Drp1 tetramers per turn of the helix [44, 52–54]. Analysis of the conformational changes produced by GTP hydrolysis revealed notably larger decrease in the helical diameter of Dnm1 than what has been described for dynamin [55, 56]. Dnm1 produces a 50-nm outer membrane diameter decrease, as compared to 10 nm observed for dynamin, a constriction associated with increased axial spacing resulting from a sliding mechanism between adjacent strands. The proximity between two single opposing lipid surfaces required for spontaneous membrane fission has been calculated to be 1–2 nm [57, 58]. As the average luminal diameter of Dnm1-lipid tubules falls outside this range, it is arguable whether GTP hydrolysis of Dnm1 alone is sufficient to cause membrane scission, giving rise to the possibility that membrane receptors for Dnm1/Drp1 have a function in promoting further oligomer constriction in addition to their role in recruitment [52].
The crystal structure of Drp1 was recently solved and reveals important insight into oligomerisation interfaces and mechanisms [44]. Similar to what has been previously shown for dynamin, the stalk domains of Drp1 mediate dimerisation and higher-order oligomerisation via three interfaces [42–44]. The crystal structure identified a novel fourth assembly interface on the stalk of Drp1, resulting in a parallel stacking of the dimer in the crystal lattice. This Drp1-specific interface was proposed to assemble two neighbouring Drp1 filaments, stabilising a more rigid oligomerisation system [44] (Fig. 1).
Drp1, cardiolipin and reactive oxygen species
The mechanistic interplay between Drp1 and mitochondrial phospholipids has been a topic of intense research recently. This originates from the fact that the activity of yeast Drp1 was shown to be stimulated in the presence of liposomes that mimic the mitochondrial outer membrane (MOM) [46]. Further research into identifying individual phospholipids important for this stimulation reveals cardiolipin (CL) as the most potent stimulator of Drp1 GTPase activity [59, 60]. CL is a mitochondrial-specific phospholipid that contributes 12–17 % of the total mitochondrial phospholipid content, present largely in the inner membrane (14–23 %) and a smaller proportion in the outer membrane (3–10 %) [61]. Conical-shaped CL has a non-bilayer forming propensity and may facilitate the formation of an intermediate state during membrane remodelling events including fission and fusion [62–65]. A direct interaction between Drp1 and CL-containing membranes was observed in vitro using a Trp-Dansyl FRET assay [60] whereas lipid dot-blot analysis found that direct Drp1–CL interactions were mediated by four lysine residues in the InsB region of Drp1 that are conserved among vertebrates [59] Disruption of an essential cardiolipin synthase gene (cls-1) in Arabidopsis thaliana leads to a block in fission resulting in unbiased fusion [64]. The block in fission is attributed to the loss of the Drp3 (the plant Drp1 ortholog) complex at mitochondria which is stabilised by cardiolipin thus highlighting the functional consequence of the interaction in vivo [64]. Thus a growing body of research lays emphasis on the Drp1–CL interactions as an important step for Drp1-dependent mitochondrial fission.
One of the factors that can affect the mechanism of Drp1-induced mitochondrial fragmentation from within mitochondria is reactive oxygen species (ROS). ROS are damaging by-products of the electron transport chain (ETC), housed with the mitochondrial inner membrane (MIM). ROS are initially formed by the premature release of electrons from the ETC and the reduction of molecular oxygen to form the superoxide radical ·O2 − which can damage mitochondrial components including mtDNA, respiratory chain complexes and lipid membranes [66, 67]. Increases in mitochondrial fragmentation and ROS have been observed in many disease states including diabetes, ischaemia reperfusion injury (IRI), AD and PD [68–72]. In IRI, the production of ROS in the reperfusion stage results in fragmentation of the mitochondrial network [73]. Interestingly, inhibiting mitochondrial fission or promoting fusion in a HL-1 model can attenuate the onset on IRI and delay cell death, suggesting other complex factors are at play [73]. Although mitochondria house proteins are capable of detoxifying superoxide (such as manganese superoxide dismutase), excessive levels of ROS can overwhelm such systems. In order to reduce the levels of ROS, exogenous antioxidants have been employed to minimise the lipid peroxidation induced by ROS [74, 75]. Studies in both cell and animal models for PD, AD, cardiac disease, metabolic syndrome and type I diabetes suggest that treatment with MitoQ, a lipophilic ubiquinone molecule that selectively accumulates within mitochondria in vivo, can protect against oxidative damage and the downstream effects that follow [69, 76–79]. Although often seen occurring concomitantly in disease models such as IRI, the relationship between ROS and mitochondrial fission is not well understood. Superoxide is produced at seven different sites associated with proteins across the MIM, yet only two of these sites release superoxide into the IMS [80]. Although these superoxide molecules will directly damage lipids of either the inner leaflet of the MOM or the MIM, exactly how lipid peroxidation triggers Drp1 recruitment and subsequent fission remains unknown.
Adaptor proteins for Drp1/Dnm1 recruitment and fission
Fis1 and Mdv1/Caf4
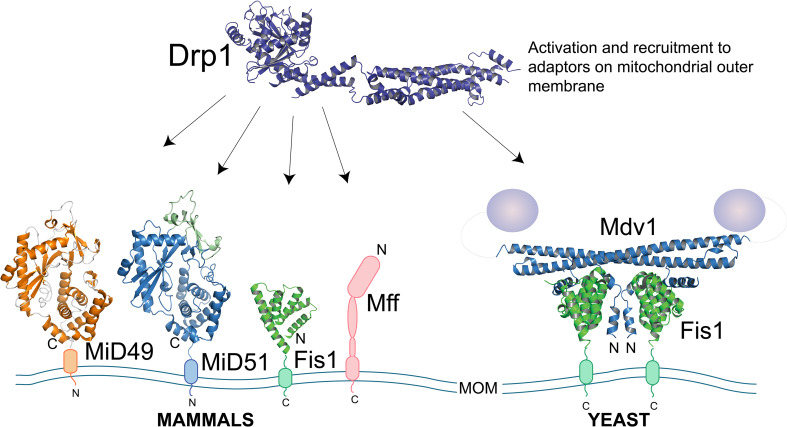
Yeast screens identified Fis1 as the first integral membrane protein to play a role in mitochondrial fission [81, 82]. Fis1 is C-terminally tethered to the mitochondrial outer membrane and contains an α-helix, involved in oligomerisation, followed by two tetratrico-peptide repeat (TPR) motifs [83]. This well-defined protein–protein interaction motif is commonly associated in multiprotein complexes and comprises helix–turn–helix repeats arranged in an antiparallel fashion [84]. Fis1 is also expressed on peroxisomes and has been shown, along with Drp1, to mediate fission of peroxisomal membranes [85–87]. In yeast, Fis1 forms a central component of the mitochondrial dynamin adaptor complex, where it stabilises the peripheral membrane proteins Mdv1 and Caf4 to the mitochondrial outer membrane, which in turn recruit Dnm1 to mitochondria [81, 88–90]. Mdv1 and Caf4 are orthologous proteins with WD40 repeat β-propeller motifs that form foci with Dnm1. Crystal structures revealed that the αA and αB helices of both Mdv1 and Caf4 pack into a concave hydrophobic groove on Fis1 (Fig. 2) [82, 91]. Furthermore, the structure of the Fis1–Mdv1 complex, including the Mdv1 coiled-coil region, reveals formation of a compact dimer stabilised by a scaffolding contact between Fis1 and the Mdv1 coiled-coil, which would position the β-propeller toward the cytosol. It was recently shown that yeast Fis1 is dispensable for recruitment of Dnm1 for fission when Mdv1 is tethered to the membrane, which indicates that the role of Fis1 in Dnm1 recruitment may simply present a protein fold for docking Dnm1 adaptors onto the mitochondrial surface [92]. Interestingly, the Mdv1–Dnm1 interaction is mediated through a number of critical residues on the WD40 repeat and InsB presenting the first described function of InsB in a dynamin-related protein–adaptor interaction [51].
Fig. 2.
Mitochondrial division in mammals and yeast. In mammals, Drp1 is recruited to the mitochondrial outer membrane by adaptors MiD49, MiD51, Mff and possibly Fis1 (PDB accession numbers: Drp1: 4BEJ; MiD49: 4WOY, MiD51: 4NXV, Fis1: 1NZN). Division in yeast is controlled by Dnm1, which is recruited by the β-propeller domain of Mdv1, and stabilised in a dimer by the TPR domain of Fis1 at the outer mitochondrial membrane (PDB accession numbers: Fis1–Mdv1 complex, 3UUX)
Despite its evolutionary conservation, the role of Fis1 in mammalian mitochondrial fission remains controversial. Early work indicated that overexpression of human Fis1 results in mitochondrial fragmentation with loss inducing mitochondrial elongation [93–95]. More recently, however, deletion of Fis1 in mouse embryonic fibroblasts indicated only a minor role in mitochondrial fission with no obvious differences in mitochondrial morphology or fusion rates observed [47, 96]. Also, the lack of Fis1 did not affect Drp1’s association with mitochondria [16, 36, 47]. Recent co-immunoprecipitation studies have suggested that Fis1 forms part of a larger fission complex at the ER–mitochondrial interface, containing the Mitochondrion-Associated Membrane (MAM) ER proteins Bap31 and Calnexin [97, 98]. Fis1 has also been suggested to mediate stress-induced mitochondrial fission as part of an apoptotic response [99]. Further to this it has also been proposed that Fis1 may act in a fission sequence after Drp1 and other adaptors—perhaps in relation to a stress response rather than normal cell homeostasis [98]. Depending on the stimulus, Fis1 may contribute to division of mitochondria in the apoptotic or mitophagy pathway [98]. The lack of mammalian homologs of Mdv1 and Caf4 coupled to the increasing evidence that Fis1 is not the primary mammalian mitochondrial adaptor for fission suggests an evolutionary divergence in the fission process.
Mitochondrial fission factor, Mff
Mitochondrial fission factor (Mff) was the first mitochondrial adaptor shown to recruit Drp1 to mitochondria in a Fis1-independent manner [47, 85]. siRNA knockdown of Mff inhibited mitochondrial fission and delayed cytochrome c release during apoptosis [85]. Peroxisomal fission was also inhibited upon Mff depletion resulting in peroxisomal extensions [47, 85], indicating that Mff, like Drp1, is involved in both mitochondrial and peroxisomal fissions. Indeed, an interaction with the peroxisomal fission mediator Pex11p has been demonstrated by co-immunoprecipitation and was shown to require Drp1 [100, 101]. Mff localises to discrete foci on mitochondria and co-localises with Drp1 [47, 85]. It was shown that Mff-induced fission requires the presence of Drp1, and that Drp1 association with mitochondria is reduced in Mff-depleted cells [47]. Mff and Drp1 co-immunoprecipitate; however, the requirement for chemical crosslinking indicates a transient interaction [47]. It now appears that the Mff is a more potent mediator of mitochondrial fission than Fis1, as the presence of Mff and Drp1, in contrast to Fis1 and Drp1, was found to be sufficient to produce fission in a yeast system devoid of its own fission mediators (Dnm1, Fis1, Mdv1) [92]. In keeping with this, re-introduction of Mff in MEFs lacking endogenous Mff or Fis1, elicited a pronounced Drp1 recruitment to mitochondria [96].
Mff is C-terminally anchored to the mitochondrial outer membrane and contains two conserved short N-terminal amino acid repeats, suggested to be binding motifs (Fig. 2) [85]. It also contains a predicted coiled-coil domain, which is required for correct targeting to mitochondria. Investigation of the Mff–Drp1 interaction using BRET, co-immunoprecipitation and functional assays revealed a binding interface on Drp1 containing the highly conserved R376 of the stalk domain [50]. The interaction of Mff and Drp1 at this site is proposed to contribute to the spiral compaction of Drp1 oligomers on the mitochondrial outer membrane [50]. The involvement of Mff in fission in higher eukaryotes provides an intriguing starting point for the elucidation of complex assembly; however, specific binding mechanisms remain to be demonstrated with structural information.
Mitochondrial dynamics proteins MiD49 and MiD51
Mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51) are mitochondrial outer membrane proteins specifically found in chordates. MiD51 was identified as part of a random cellular localisation screen of uncharacterised human proteins, which following overexpression, resulted in changes in mitochondrial dynamics [102]. MiD49 was subsequently identified due to sequence homology [103]. MiD49 and MiD51 share 45 % sequence identity yet lack significant sequence homology to other proteins. In contrast to Fis1 and Mff, MiD49 and MiD51 appear to be exclusively mitochondrial, indicating a mechanism to ensure specificity and selectivity of Drp1-mediated fission at mitochondria [103]. MiD49 and MiD51 are N-terminally tethered to the mitochondrial outer membrane and consist of a transmembrane domain, a predicted disordered region of approximately 70 amino acids followed by a cytosolic domain (Fig. 2). Confocal microscopy analysis demonstrated the localisation of MiD51 seems to occur around mitochondrial constriction sites, much like Drp1 itself [103]. In cells overexpressing MiD49 or MiD51, the predominantly cytosolic Drp1 was enriched at mitochondria at sites containing MiD49/51-GFP [103]. Using yeast two-hybrid assays and co-immunoprecipitation/cross-linking, a direct interaction between Drp1 and MiD proteins was observed [92, 96, 103, 104]. Overexpression of MiD49/51 produces two distinct mitochondrial morphology states depending on MiD protein levels—either forming elongated tubules that project from a perinuclear collapsed network (high level expression), or a relatively normal morphology accompanied by MiD49/51 foci formation (low level expression). In the context of a role as fission mediators, elongation of mitochondrial tubules upon overexpression of MiD49/51 appears counterintuitive. However, given that the high levels of overexpressed protein far exceed physiological levels, it was proposed that the phenotype is a result of sequestration of Drp1, rendering the GTPase non-functional [103]. The observation was contradictorily interpreted as a role of MiD51 (independently identified as MIEF1) in the promotion of mitochondrial fusion rather than fission [105]. However, studies from other laboratories have confirmed that MiD49/51 mediate Drp1-driven mitochondrial fission [92, 96]. MiD49 and MiD51 were able to restore CCCP-induced mitochondrial fragmentation in Fis1/Mff-knockout cells and their knockdown was independently shown to cause elongation [96]. Furthermore, induction of MiD49/51 expression caused mitochondrial elongation through Drp1 inactivation [104], while MiD49 with Drp1 could drive fission events in a yeast line lacking endogenous fission machinery [92]. Interestingly, in vitro, addition of MiD49 was found to decrease the external diameters of Drp1-constricted liposomes from 31 to 15 nm, a distance amenable to membrane scission in the context of the mitochondrial double membrane [92]. This indicates that MiD49/51 may function downstream of Mff and that they are actively involved in facilitating Drp1-mediated mitochondrial scission.
The lack of MiD49/51 sequence homology to known protein structures or obvious structural motifs has made deduction of functional domains or a protein-binding site difficult. Recently, the MiD51 cytosolic domain was independently solved by two groups and found to harbour a nucleotidyltransferase fold (Fig. 2) [106, 107]. However, MiD51 lacks the key catalytic triad of residues that define a protein as a bone fide nucleotidyltransferase. The distal cytosolic loop of MiD51 contains a region of the protein responsible for the interaction with Drp1, termed the Drp1 recruitment region (DRR). Deletion of this loop, five residues in length, does not impair the overall fold of the protein, but does block the ability of MiD51 to recruit Drp1 to mitochondria [107]. Interestingly, although MiD51 can bind ADP and GDP in the fold, mutations that disrupt nucleotide binding in this region did not abrogate recruitment of Drp1, indicating adaptation of the nucleotidyltransferase fold for a novel purpose [107]. However, Chan and colleagues demonstrated that binding ADP promotes Drp1 assembly into spirals and enhances GTP hydrolysis—leading to the fact that ADP is a co-factor for MiD51-dependent Drp1 fission [108]. Structural analysis of MiD49, like MiD51, revealed it also harbours a nucleotidyltransferase fold; however, MiD49 does bind nucleotides like in homolog MiD51 (Fig. 2) [109]. Further to this, the crucial residues needed to bind nucleotides are not conserved, suggesting functional disparity between the two proteins. However, the Drp1 surface-binding loop is conserved between the two proteins [109]. Although the physiological consequences of MiD51-binding dinucleotide diphosphates and MiD49 not binding are unclear, it has provided a starting point for determining the mechanistic contribution of the MiD proteins in mitochondrial fission.
The role of fission in apoptosis and mitophagy
Tissue and organ maintenance requires elimination of dysfunctional cells though apoptosis. By housing and releasing pro-apoptotic molecules such as a cytochrome c, mitochondria are central to the regulation of the intrinsic apoptotic pathway [4]. Fragmentation of the mitochondrial network is often seen in apoptosis and precedes caspase activation, mitochondrial outer membrane permeabilization, cristae remodelling (such as Opa1 processing) and cytochrome c release [110]. Central to this is the interaction of proteins belonging to the Bcl-2 family (which regulate apoptotic progression) and morphology proteins—these interactions have been observed during the progression of programmed cell death [93, 111–113]. Under normal homeostasis, Mfn1 and Mfn2 interact with Bak and VDAC2 at MOM [111, 114]. The interaction differs in apoptotic cells where Bak becomes dissociated from Mfn2 and VDAC2, but remains in complex with Mfn1 [111]. At the MOM, Drp1 interacts with Bak and translocated Bax, where it becomes sumoylated [36]. This modification results in a stable association of Drp1 with the MOM. The role of modified Drp1 in apoptotic fission, however, is not well understood, as a mutant of Drp1 (that is unable to be sumoylated) can still be recruited to the MOM following apoptotic stimuli, suggesting that sumoylation is not a requirement/or specific to apoptotic fission [36]. Further to this, a role for Drp1 in Bax oligomersiation, subsequent pore formation and cytochrome c release has been suggested [115]. Independent of its role as a GTPase, Drp1 has been shown localised to areas rich in CL in artificial membranes [115]. These regions containing Drp1 oligomers and CL appear to be a hot spot for membrane tethering and hemifusion in vitro [115]. These events may allow Bax oligomerisation and pore formation crucial for cytochrome c release and subsequent caspase activation. If indeed Drp1 does play such a pivotal role in the progression of apoptosis, this may explain the delay in programmed cell death and reduction in cytochrome c release when Drp1 is inhibited or deleted [30, 31, 116, 117].
Like apoptosis, mitophagy is a selective process—removing damaged mitochondria from the cell to prevent death on a larger scale. Modulation of mitochondrial morphology is one of the many important steps in mitophagy as fragmented mitochondria are often (but not always) observed prior to mitophagy [22]. In keeping with this, many studies have linked the rate of mitophagy to the size of mitochondria. Mitochondria visualised within autophagosomes have been reported to be <1 μm in diameter [22, 118–120]. However, this is not seen exclusively, making it complex to delineate. Both overexpression of Opa1 and inhibition of fission in a pancreatic β-cell line result in a ~70 % reduction in mitophagy rates [22]. These conflicting results suggest that organelle length may not be a rate-limiting step in mitophagy. Fis1 has recently been implicated in the disposal of defective mitochondria [98, 121]. Although once thought to be the main MOM adaptor of Drp1, it now appears that Fis1 plays a key role in stress-induced fission, as it can be immunoprecipitated with Drp1 when fission is chemically induced [98]. Mutations or knockout of Fis1 in C. elegans or mammalian cells show increased amounts of LGG-1/LC3 aggregates, suggesting that Fis1 augments the removal of these aggregates in times of cellular stress [98].
The involvement of both division and fusion prior to mitophagy highlights the importance of mitochondrial homeostasis in cellular health. In contrast to fission, fusion requires an intact membrane potential (ΔΨm) making this a selective process and excluding mitochondria that are deemed ‘unfit’. Termed ‘kiss-and-run’ events, a brief fusion event is followed by division of the mitochondrion. These fusion/fission events often result in daughter mitochondria with unequal ΔΨm [22]. If the ΔΨm is significantly low in one of these mitochondria, PINK1 is unable to be imported into mitochondria and remains at the MOM for signalling; as such PINK1 is often referred to as a sensor of mitochondrial fitness [122]. In keeping with this role as a sensor, outer membrane accumulated PINK1 phosphorylates ubiquitin directly, which in turn activates the E3 ubiquitin ligase activity of Parkin [123–125]. Activation and recruitment of Parkin allows autophagosome adaptors to assemble, recruit and help form the mature autophagosome [126–128]. This process results in degradation of the daughter mitochondrion with a lower ΔΨm [22]. However, if the mitochondrion can recover its ΔΨm before the recruitment of Parkin, it can re-enter the fission/fusion cycle [22, 129]. This suggests that mitochondrial fission, which is often a cellular response to stress, is a mechanism utilised by the cell to segregate and eliminate damaged mitochondria from an otherwise healthy network.
ER–mitochondrial interactions in fission
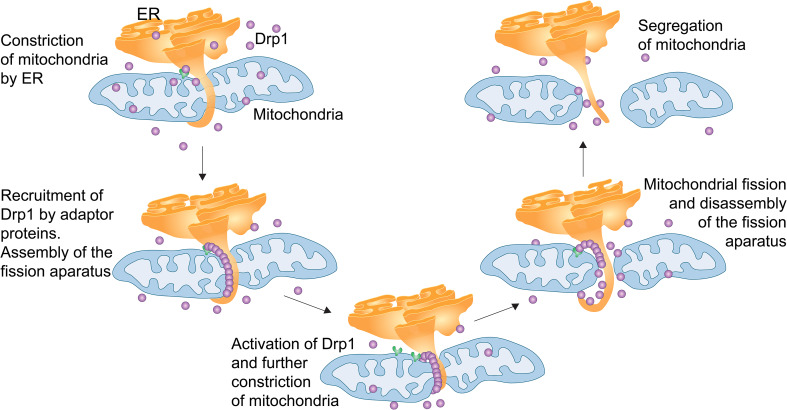
Mitochondria and the endoplasmic reticulum are engaged in a close spatial relationship that mediates Ca2+, apoptotic signalling and lipid metabolism between membranes [130, 131]. The close contacts these organelle networks make has been measured by electron tomography (ET) to be 9–30 nm wide and to usually occur in clusters, allowing bidirectional trafficking of factors across membranes as well as communication between organelles [132]. More recently, evidence has revealed links between ER–mitochondrial contacts with mitochondrial fission [133] (Fig. 3). Electron microscopy and tomography were used to visualise ER–mitochondrial contacts in yeast, and demonstrated that the ER forms rings around mitochondria, where tubule constriction sites with smaller diameters were bordered by a higher percentage of ER wrapping [133]. The study also showed that Drp1-mediated mitochondrial division in mammalian cells occurs at ER–mitochondrial contact sites and that this contact precedes interaction with Drp1 and Mff. This marking of fission sites by the ER suggests that ER proteins may be closely linked to fission or that the ER presents a physical constriction element, perhaps crudely reshaping the mitochondrial tubule before fission adaptors and effectors assemble and execute division [131].
Fig. 3.
ER–mitochondrial contacts in mitochondrial division. The ER forms extensions around mitochondria and physically constricts the organelle. Adaptor proteins such as MiD49/51 and Mff recruit cytosolic Drp1 to this scission site, where the organelle is constricted further. The fission apparatus is assembled and division of mitochondria occurs
Two recently characterised proteins have been identified to mediate the ER–mitochondrial connections related to fission. INF2 (inverted formin 2) was originally identified as a protein capable of bundling microtubules and interacting with actin, INF2 is an ER protein that drives actin polymerisation (in conjunction with myosin II) and stimulates Drp1 at fission sites [134–136]. The myosin link is intriguing as it accumulates on mitochondria in an actin/INF2-dependent manner. Further to this, inhibition of myosin 2 with blebbistatin, decreases the Drp1 association with mitochondria [135]. Taken together, these results suggest an important mechanistic role in which actin polymerisation, mediated by INF2 leads to myosin II recruitment and constriction of mitochondria at the fission site, thus enhancing Drp1 accumulation and subsequent fission. An ER-associated soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) receptor (SNARE) protein syntaxin 17 (Syn17) was also recently found to direct Drp1 to these contact sites (by binding specifically to Mff), thus promoting Drp1-driven division of mitochondria [137]. This suggests that the Syn17–Mff complex may be involved in the initial assembly of Drp1 at the constriction site. In order to regulate this direction of Drp1 to contact sites, Syn17 also interacts with Rab32, a mitochondrial-associated membrane-localised A-kinase anchor protein. Rab32 has been shown to determine targeting of PKA to the mitochondrial and ER membranes, phosphorylating Drp1 at S637, resulting in a block in the fission process [138]. This interaction between Syn17 and Rab32 appears to be a regulatory checkpoint as the Syn17/Rab32 interaction prevents post-translational modification of Drp1, while depletion of Syn17 increases phosphorylation [137]. Upon starvation conditions, Syn17 preferentially binds ATG14L over Rab32 and Drp1, causing mitochondrial elongation in the initial phase of autophagy [137]. This mechanism is particularly intriguing as it sheds light on how the ER tubule is directed to the future fission site on mitochondria—through the Syn17–Mff complex.
Concluding remarks
Regulation of mitochondrial fission is a complex process, made more convoluted by the fact that there are clear differences in the machineries between yeast and animals. There are many proposed mitochondrial Drp1 adaptors—how exactly these function together (and how they co-ordinate the scission process with fission mediators of the inner membrane) remains to be clarified. A possibility is that divergent regulators are required for different aspects of fission and/or tissue specific fission regulators. For example, while Fis1 and Mff have a dual mitochondrial and peroxisomal location, MiD49 and MiD51 appear to be specific mitochondrial fission factors [47, 87, 104]. Fis1 once thought to be integral to Drp1 recruitment in both yeast and mammalian mitochondria now appears to have a highly specialised role in fission [34, 98]. While Mff is largely ubiquitously expressed, MiD49 and MiD51 display differential tissue expression.
The future challenge to understand the precise mechanistic detail into the assembly of the mitochondrial fission apparatus revolves around cementing the roles of adaptor proteins along with the involvement of the cytoskeleton and ER in executing membrane scission with Drp1. The exact triggers for productive assembly of the fission apparatus are not yet clear, nor is the involvement of the inner membrane and correct positioning of mtDNA [20, 139]. A better understanding of lipid remodelling/composition at the site of scission is also required to help deduce how fission is executed.
Contributor Information
Michael T. Ryan, Email: Michael.Ryan@monash.edu
Laura D. Osellame, Email: laura.osellame@monash.edu
References
- 1.Duchen MR. Mitochondria and calcium: from cell signalling to cell death. J Physiol. 2000;529:57–68. doi: 10.1111/j.1469-7793.2000.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lodi R, Cooper JM, Bradley JL, Manners D, Styles P, Taylor DJ, Schapira AH. Deficit of in vivo mitochondrial ATP production in patients with Friedreich ataxia. Proc Natl Acad Sci USA. 1999;96(20):11492–11495. doi: 10.1073/pnas.96.20.11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 4.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281(5381):1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 5.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397(6718):441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 7.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21(9):3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 9.Mecocci P, MacGarvey U, Beal MF. Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann Neurol. 1994;36(5):747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 10.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5(8):731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 11.Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54(3):823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 12.Shirendeb U, Reddy AP, Manczak M, Calkins MJ, Mao P, Tagle D, Reddy H. Abnormal mitochondrial dynamics, mitochondrial loss and mutant Huntington oligomers in Huntington’s disease: implications for selective neuronal damage. Hum Mol Genet. 2011;20(7):1438–1455. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot–Marie–Tooth neuropathy type 2A. Nat Genet. 2004;36(5):449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Chan DC. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osellame LD, Blacker TS, Duchen MR. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab. 2012;26(6):711–723. doi: 10.1016/j.beem.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160(2):189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126(1):177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 18.Kotiadis VN, Duchen MR, Osellame LD. Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim Biophys Acta. 2014;1840(4):1254–1265. doi: 10.1016/j.bbagen.2013.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE. 2008;3(9):e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ban-Ishihara R, Ishihara T, Sasaki N, Mihara K, Ishihara N. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c . Proc Natl Acad Sci USA. 2013;110(29):11863–11868. doi: 10.1073/pnas.1301951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9(3):96–102. doi: 10.1016/S0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 24.Danino D, Hinshaw JE. Dynamin family of mechanoenzymes. Curr Opin Cell Biol. 2001;13(4):454–460. doi: 10.1016/S0955-0674(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 25.Pitts KR, Yoon Y, Krueger EW, McNiven MA. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol Biol Cell. 1999;10(12):4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smirnova E, Shurland D, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143(2):351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bui HT, Shaw JM. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol. 2013;23(19):R891–R899. doi: 10.1016/j.cub.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elgass K, Pakay J, Ryan MT, Palmer CS. Recent advances into the understanding of mitochondrial fission. Biochim Biophys Acta. 2013;1833(1):150–161. doi: 10.1016/j.bbamcr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356(17):1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 30.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11(8):958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 31.Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186(6):805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolanos JP, Almeida A, Moncada S. Glycolysis: a bioenergetic or a survival pathway? Trends Biochem Sci. 2010;35(3):145–149. doi: 10.1016/j.tibs.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolanos JP. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11(6):747–752. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- 34.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15(11):5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, Ehses S, Krause F, Chan DC, Alexander C, Bauer C, Youle R, Langer T, Martinou JC. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28(11):1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177(3):439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282(15):11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 38.Chang CR, Blackstone C. Drp1 phosphorylation and mitochondrial regulation. EMBO Rep. 2007;8(12):1088–1089. doi: 10.1038/sj.embor.7401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8(10):939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57(3):537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, Jabado O, Hoehn K, Kageyama Y, Sesaki H, Chipuk JE. mitochondrial division is requisite to ras-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57(3):521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477(7366):556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 43.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477(7366):561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013;32(9):1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12(8):2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325(5942):874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim Biophys Acta. 2006;1763(5–6):531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89(3):799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 50.Strack S, Cribbs JT. Allosteric modulation of Drp1 mechanoenzyme assembly and mitochondrial fission by the variable domain. J Biol Chem. 2012;287(14):10990–11001. doi: 10.1074/jbc.M112.342105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bui HT, Karren MA, Bhar D, Shaw JM. A novel motif in the yeast mitochondrial dynamin Dnm1 is essential for adaptor binding and membrane recruitment. J Cell Biol. 2012;199(4):613–622. doi: 10.1083/jcb.201207079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18(1):20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170(7):1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107(9):4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93(6):1021–1029. doi: 10.1016/S0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 56.Danino D, Moon KH, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J Struct Biol. 2004;147(3):259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135(7):1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozlovsky Y, Kozlov MM. Membrane fission: model for intermediate structures. Biophys J. 2003;85(1):85–96. doi: 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS ONE. 2014;9(7):e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, Ramachandran R. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell. 2014;25(12):1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822(1):1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 62.Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol. 2004;186(5):1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ortiz A, Killian JA, Verkleij AJ, Wilschut J. Membrane fusion and the lamellar-to-inverted-hexagonal phase transition in cardiolipin vesicle systems induced by divalent cations. Biophys J. 1999;77(4):2003–2014. doi: 10.1016/S0006-3495(99)77041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan R, Jones AD, Hu J. Cardiolipin-mediated mitochondrial dynamics and stress response in Arabidopsis. Plant Cell. 2014;26(1):391–409. doi: 10.1105/tpc.113.121095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unsay JD, Cosentino K, Subburaj Y, Garcia-Saez AJ. Cardiolipin effects on membrane structure and dynamics. Langmuir. 2013;29(51):15878–15887. doi: 10.1021/la402669z. [DOI] [PubMed] [Google Scholar]
- 66.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284(20):13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Solesio ME, Prime TA, Logan A, Murphy MP, Del Mar Arroyo-Jimenez M, Jordan J, Galindo MF. Galindo MF (2013) The mitochondria-targeted anti-oxidant MitoQ reduces aspects of mitochondrial fission in the 6-OHDA cell model of Parkinson’s disease. Biochim Biophys Acta. 2013;1:174–182. doi: 10.1016/j.bbadis.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 70.Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, Zhou J, Chen Q. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286(13):11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29(28):9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu T, Robotham JL, Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci USA. 2006;103(8):2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121(18):2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 74.Murphy MP. Targeting lipophilic cations to mitochondria. Biochim Biophys Acta. 2008;1777(7–8):1028–1031. doi: 10.1016/j.bbabio.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 75.Smith RA, Murphy MP. Animal and human studies with the mitochondria-targeted antioxidant MitoQ. Ann N Y Acad Sci. 2010;1201:96–103. doi: 10.1111/j.1749-6632.2010.05627.x. [DOI] [PubMed] [Google Scholar]
- 76.Adlam VJ, Harrison JC, Porteous CM, James AM, Smith RA, Murphy MP, Sammut IA. Targeting an antioxidant to mitochondria decreases cardiac ischemia-reperfusion injury. FASEB J. 2005;19(9):1088–1095. doi: 10.1096/fj.05-3718com. [DOI] [PubMed] [Google Scholar]
- 77.Chacko BK, Reily C, Srivastava A, Johnson MS, Ye Y, Ulasova E, Agarwal A, Zinn KR, Murphy MP, Kalyanaraman B, Darley-Usmar V. Prevention of diabetic nephropathy in Ins2(+/)(−)(AkitaJ) mice by the mitochondria-targeted therapy MitoQ. Biochem J. 2010;432(1):9–19. doi: 10.1042/BJ20100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graham D, Huynh NN, Hamilton CA, Beattie E, Smith RA, Cocheme HM, Murphy MP, Dominiczak AF. Mitochondria-targeted antioxidant MitoQ10 improves endothelial function and attenuates cardiac hypertrophy. Hypertension. 2009;54(2):322–328. doi: 10.1161/HYPERTENSIONAHA.109.130351. [DOI] [PubMed] [Google Scholar]
- 79.Manczak M, Mao P, Calkins MJ, Cornea A, Reddy AP, Murphy MP, Szeto HH, Park B, Reddy PH. Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer’s disease neurons. J Alzheimers Dis. 2010;20(Suppl 2):S609–S631. doi: 10.3233/JAD-2010-100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol. 2010;45(7–8):466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151(2):367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci USA. 2007;104(47):18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jofuku A, Ishihara N, Mihara K. Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission-stimulating protein. Biochem Biophys Res Commun. 2005;333(2):650–659. doi: 10.1016/j.bbrc.2005.05.154. [DOI] [PubMed] [Google Scholar]
- 84.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21(11):932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 85.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19(6):2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M. Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278(10):8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 87.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16(11):5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170(2):237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J Biol Chem. 2006;281(4):2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 90.Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151(2):353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y, Chan NC, Ngo HB, Gristick H, Chan DC. Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (Mdv1) coiled coil. J Biol Chem. 2012;287(13):9855–9861. doi: 10.1074/jbc.M111.329359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, Shaw JM. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci USA. 2013;110(15):E1342–E1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278(38):36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 94.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci. 2004;117(Pt 7):1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 95.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23(15):5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49 and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iwasawa R, Mahul-Mellier AL, Datler C, Pazarentzos E, Grimm S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011;30(3):556–568. doi: 10.1038/emboj.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25(1):145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang B, Nguyen M, Chang NC, Shore GC. Fis1, Bap31 and the kiss of death between mitochondria and endoplasmic reticulum. EMBO J. 2011;30(3):451–452. doi: 10.1038/emboj.2010.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Itoyama A, Michiyuki S, Honsho M, Yamamoto T, Moser A, Yoshida Y, Fujiki Y. Mff functions with Pex11pbeta and DLP1 in peroxisomal fission. Biol Open. 2013;2(10):998–1006. doi: 10.1242/bio.20135298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koch J, Brocard C. PEX11 proteins attract Mff and human Fis1 to coordinate peroxisomal fission. J Cell Sci. 2012;125(16):3813–3826. doi: 10.1242/jcs.102178. [DOI] [PubMed] [Google Scholar]
- 102.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1(3):287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12(6):565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, Ryan MT. MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem. 2013;288(38):27584–27593. doi: 10.1074/jbc.M113.479873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30(14):2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37(22):7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Richter V, Palmer CS, Osellame LD, Singh AP, Elgass K, Stroud DA, Sesaki H, Kvansakul M, Ryan MT. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J Cell Biol. 2014;204(4):477–486. doi: 10.1083/jcb.201311014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Loson OC, Liu R, Rome ME, Meng S, Kaiser JT, Shan SO, Chan DC. The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure. 2014;22(3):367–377. doi: 10.1016/j.str.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Loson OC, Meng S, Ngo H, Liu R, Kaiser JT, Chan DC. Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1. Protein Sci. 2015;24(3):386–394. doi: 10.1002/pro.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arnoult D, Grodet A, Lee YJ, Estaquier J, Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J Biol Chem. 2005;280(42):35742–35750. doi: 10.1074/jbc.M505970200. [DOI] [PubMed] [Google Scholar]
- 111.Brooks C, Wei Q, Cho SG, Dong Z. Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J Clin Invest. 2009;119(5):1275–1285. doi: 10.1172/JCI37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li H, Chen Y, Jones AF, Sanger RH, Collis LP, Flannery R, McNay EC, Yu T, Schwarzenbacher R, Bossy B, Bossy-Wetzel E, Bennett MV, Pypaert M, Hickman JA, Smith PJ, Hardwick JM, Jonas EA. Bcl-xL induces Drp1-dependent synapse formation in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2008;105(6):2169–2174. doi: 10.1073/pnas.0711647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shroff EH, Snyder CM, Budinger GR, Jain M, Chew TL, Khuon S, Perlman H, Chandel NS. BH3 peptides induce mitochondrial fission and cell death independent of BAX/BAK. PLoS ONE. 2009;4(5):e5646. doi: 10.1371/journal.pone.0005646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lazarou M, Stojanovski D, Frazier AE, Kotevski A, Dewson G, Craigen WJ, Kluck RM, Vaux DL, Ryan MT. Inhibition of Bak activation by VDAC2 is dependent on the Bak transmembrane anchor. J Biol Chem. 2010;285(47):36876–36883. doi: 10.1074/jbc.M110.159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, Martinou JC. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142(6):889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cereghetti GM, Costa V, Scorrano L. Inhibition of Drp1-dependent mitochondrial fragmentation and apoptosis by a polypeptide antagonist of calcineurin. Cell Death Differ. 2010;17(11):1785–1794. doi: 10.1038/cdd.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Estaquier J, Arnoult D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007;14(6):1086–1094. doi: 10.1038/sj.cdd.4402107. [DOI] [PubMed] [Google Scholar]
- 118.Chen H, Chomyn A, Chan C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280(28):26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 119.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci USA. 2004;101(45):15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205(2):143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460(1):127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 126.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 127.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117(Pt 13):2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 128.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29(11):1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191(5):933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 2010;29(16):2715–2723. doi: 10.1038/emboj.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13(10):607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174(7):915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell. 2011;22(23):4575–4587. doi: 10.1091/mbc.E11-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24(4):409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339(6118):464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arasaki K, Shimizu H, Mogari H, Nishida N, Hirota N, Furuno A, Kudo Y, Baba M, Baba N, Cheng J, Fujimoto T, Ishihara N, Ortiz-Sandoval C, Barlow LD, Raturi A, Dohmae N, Wakana Y, Inoue H, Tani K, Dacks JB, Simmen T, Tagaya M. A role for the ancient SNARE Syntaxin 17 in regulating mitochondrial division. Dev Cell. 2015;32:1–14. doi: 10.1016/j.devcel.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 138.Bui M, Gilady SY, Fitzsimmons RE, Benson MD, Lynes EM, Gesson K, Alto NM, Strack S, Scott JD, Simmen T. Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties. J Biol Chem. 2010;285(41):31590–31602. doi: 10.1074/jbc.M110.101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]