Abstract
In textbooks of biochemistry, nucleoside diphosphate conversion to a triphosphate by nucleoside diphosphate ‘kinases’ (NDPKs, also named NME or NM23 proteins) merits a few lines of text. Yet this essential metabolic function, mediated by a multimeric phosphotransferase protein, has effects that lie beyond a simple housekeeping role. NDPKs attracted more attention when NM23-H1 was identified as the first metastasis suppressor gene. In this review, we examine these NDPK enzymes from a developmental perspective because of the tractable phenotypes found in simple animal models that point to common themes. The data suggest that NDPK enzymes control the availability of surface receptors to regulate cell-sensing cues during cell migration. NDPKs regulate different forms of membrane enclosure that engulf dying cells during development. We suggest that NDPK enzymes have been essential for the regulated uptake of objects such as bacteria or micronutrients, and this evolutionarily conserved endocytic function contributes to their activity towards the regulation of metastasis.
Keywords: Nucleoside diphosphate kinases, NME, Metastasis inhibitor, Endocytosis, Dynamin, Engulfment
Introduction
Nucleoside diphosphate kinases (NDPKs) were identified more than 50 years ago as ubiquitous housekeeping enzymes that catalyze the phosphorylation of nucleoside diphosphates to nucleoside triphosphates at the expense of ATP [1]. In the human genome, ten paralogous genes encode NDPKs, which belong to two distinct groups based on sequence homology and NDPK activity [2, 3]. Group I genes encode proteins (NME1–4; non-metastatic) with NDPK activity and show high similarity to one another, whereas group II members (NME5–9 and RP2, retinitis pigmentosa 2) are more divergent and display low or no NDPK activity. NDPKs gained initial attention when Patricia Steeg identified a group I NDPK, NME1 (also known as NM23-H1), as the first metastasis suppressor protein [4] (NM23 stands for non-metastatic clone 23); the cDNA of Nm23 was found to be downregulated in a highly metastatic derivative of a murine melanoma cell line [4]. Mathieu Boissan and colleagues [5] demonstrated the anti-metastatic effect of NME1 in mouse models: nme1 knockout mice where crossed with a mouse strain prone to hepatocellular carcinoma and the double transgenic mice had a higher incidence of lung metastases. Next, human and murine Nm23 were shown in cancer cell xenografts to inhibit metastasis without influencing primary tumor growth [6]. The level of NM23-H1 expression was inversely correlated with the metastatic potential for human solid tumors of epithelial origin such as breast, liver, colorectal, ovarian and lung carcinoma and for melanoma [7]. However, in other tumor types, such as ovarian cancers, neuroblastoma and hematological malignancies, upregulated NM23 levels have been detected from patient samples that correlate with a poor prognosis [8–10]. Thereafter, understanding the biological function of the Nm23 gene family was brought into focus through diverse results, suggesting that the regulation of many physiological processes was associated with this gene family (reviewed in [11]), such as cell migration [12], growth and differentiation [13, 14], signal transduction, transcriptional regulation [15, 16], and apoptosis [17]. Correspondingly, multiple protein interaction partners were reported [12]. These multiple functions assigned to the NM23 family of proteins have thus far been difficult to reconcile because the exact cellular function of NM23 has remained unclear. However, beyond nucleoside diphosphate kinase activity several other molecular activities have been linked to NDPKs, such as histidine-dependent protein kinase (histidine phosphotransferase) [18, 19] and nuclease activity [20, 21], as well as lipid bilayer binding [22–26].
NDPKs are evolutionary highly conserved from yeast to human [3, 27], suggesting critical cellular and developmental functions for members of this protein family. Although, studies in mammalian cell cultures have identified numerous functions and interaction partners of NDPKs, many of these findings have not been directly verified in vivo. Studies on model organisms helped in understanding the biological functions of NDPKs, and could confirm functions derived from in vitro and cell line studies.
In this review, we focus on this issue and give an overview of NDPKs’ developmental functions in different model systems such as the slime mold Dictyostelium discoideum, the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster, the zebrafish Danio rerio, the frog Xenopus laevis, and the mouse Mus musculus, where NDPKs have now been extensively studied.
Developmental functions of AWD/NM23 in the fruit fly Drosophila melanogaster
Early studies: the isolation of K-pn and awdb3 alleles
Amongst genetic model systems, the earliest and most extensive analysis of the homolog of human NM23, abnormal wing discs (AWD), was carried out in Drosophila melanogaster where the first mutant allele of awd, killer-of-prune (K-pn), was isolated in the 1950s by the famous fly geneticist Alfred Sturtevant [28]. He performed genetic crosses to demonstrate X-linkage of the eye color influencing gene, prune. Loss-of-function mutation of prune leads to brownish-purple eye color in otherwise normal adult flies instead of the wild-type bright red eye. Sturtevant’s seminal observation was that the K-pn mutation, which was itself viable, when crossed into viable prune loss-of-function mutants, resulted in lethality even in the presence of a single K-pn mutant allele. He reasoned that K-pn might, therefore, carry a dominant, gain-of-function mutation, which exerts synthetic lethality in the prune mutant background. However, the molecular lesion underpinning the K-pn allele and the cause of synthetic lethality phenotype were to remain unknown for almost three decades.
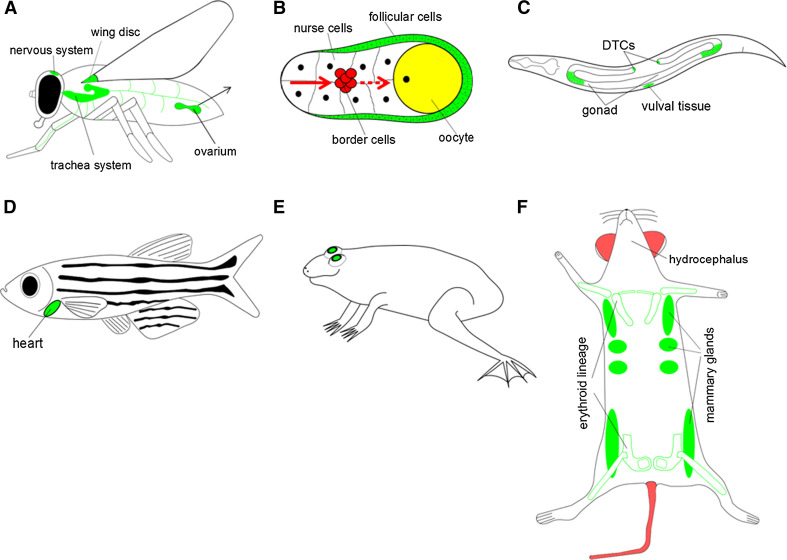
In the 1980s, the laboratory of Allen Shearn conducted a genetic screen for late larval/pupal lethality to identify genes regulating imaginal disc development [29]. Imaginal discs are epithelial infoldings in Drosophila larvae that develop into adult structures (such as legs, antennae, wings, etc.). In this developmental screen, alleles of 12 different disc-defective genes were found [29, 30]. One of them was named awd [30], as initially only the paired wing discs appeared to be defective in awd b3 mutant larvae. Further analysis revealed that other imaginal discs of awd b3 mutants, including leg and eye-antenna discs, also do not differentiate properly, and show increased cell death (Fig. 1a). Subsequently it turned out that the awd b3 allele is a genetic null mutation of the gene awd as a consequence of P-element insertion into the first exon. In addition, because awd b3 mutants also had defective ovaries, Dearolf and colleagues [30] performed the following elegant organ transplantation experiments. The Drosophila ovaries are composed of cells of both somatic and germline origin. If premature awd b3 mutant ovaries containing both somatic cells and germ cells were transplanted into wild-type females, the grafted flies remained sterile. However, if isolated germ cell precursors (pole cells) of awd b3 mutant embryos were introduced into wild-type embryos, this gave rise to fertile flies with normally developed ovaries, which produce gametes. This series of experiments indicated that the ovarian sterile phenotype of awd mutants is caused by defects in somatic (follicular) cells of the ovaries. The fact that awd activity is required in follicular cells became important in later studies (Fig. 1a, b).
Fig. 1.
NDPK-related developmental functions in different model systems. Note that many signaling events, where the functions of NDPKs were observed actually operate during embryogenesis or later developmental stages, but the locations of relevant cells (or their descendants), tissues or organs are shown here schematically in the appropriate adult organism. Green labeling indicates the site of NDPK’s function. a The Drosophila NDPK homolog AWD is involved in the development of imaginal discs, also in the wing disc. The absence of awd function leads to defects in neurotransmission, ectopic migration of tracheal cells and female sterility. Thus, AWD’s function is linked to the nervous system, the developing trachea and the ovarium (detailed in b). b The fly egg chamber is composed of the germ cell complex (nurse cells and the oocyte), which is surrounded by follicular (epithelial) cells. Throughout Drosophila oogenesis, awd is expressed in follicular epithelial cells to ensure epithelial integrity. A group of follicular cells delaminating from the epithelium called border cells (marked by red) migrate towards the oocyte to form the micropyle. In the border cells awd expression is downregulated. c In C. elegans, NDK-1 functions during vulval and gonadal development, as well as participates in the migration of distal tip cells (DTCs) and engulfment of apoptotic germ cell corpses. d Knockdown of NME2 in zebrafish embryos leads to decreased cardiac contractility and impaired vessel formation. e Frog NM23-X4 is needed for retinal development. f Nme1 −/− mutant female mice display nursing disability because of growth retardation of the mammary glands and defects in the final step of mammary duct maturation of the nipple. Nme2 −/− deficient mice have no obvious defects, but show an altered immune response, reduced Ca2+ reuptake in the kidney and impaired pathological angiogenesis. Nme1 −/−/Nme2 −/− double knockout mice die perinatally due to failure of erythroid lineage development. Nme7 knockout mice present defects, like hydrocephalus, which can be linked to impaired motility of cilia causing abnormal ependymal flow
AWD is the homolog of NM23-H1/H2 and displays NDPK activity
awd alleles were isolated and the gene awd was cloned cotemporaneously by Patricia Steeg and her colleagues, discovering Nm23 as the first non-metastatic (i.e., metastasis inhibitor) gene from experiments performed on a highly metastatic murine melanoma cell line [4]. Results obtained from Drosophila converged with human studies, and it became clear that AWD is the Drosophila homolog of NM23, given that AWD shows 78 % amino acid identity with the two closely related human proteins NM23-H1/H2 [31]. Next, several parallel studies set out to determine the enzymatic ‘activities’ of NM23/AWD family proteins. First, Allen Shearn, Patricia Steeg and their colleagues found that an antibody originally generated against bovine nucleoside diphosphate kinases [32] also recognized the fly AWD protein. Correspondingly, awd loss-of-function mutant larvae displayed dramatically decreased NDPK levels detected by the bovine NDPK-specific antibody, as compared to wild-type flies [33]. Second, from D. discoideum an NDPK-like enzyme was isolated, which showed 60 % sequence identity with AWD [34], and later was used as a source to crystalize hexameric cellular form of NDPK. All these data suggested that AWD, similar to its human and social amoeba counterparts, possess NDPK activity. It was also demonstrated that AWD provides the main source of NDPK activity (98 %) in Drosophila embryos [33].
NDPKs are thought to exert their nucleoside diphosphate kinase (phosphotransferase) activity through a high energy N-phosphate linkage on histidine118 (histidine119 residue in AWD). The Shearn laboratory was the first to demonstrate in vivo that histidine119 is key to one of AWD’s functions, as awd loss-of-function mutants can only be complemented by a wild-type awd cDNA-containing construct, but not by a transgene encoding the H119A allele devoid of the N–P link [35]. Interestingly, larval lethality of awd mutants could be completely rescued by the more ancient human Nm23-H2, but not its closely related Nm23-H1 isoform that is thought to have arisen by subsequent gene duplication. In the 1990s, it was eventually realized that K-pn in fact is an allele of awd (and was hence renamed awd K-pn), which carries a proline to serine substitution in position 97 (which corresponds to P96 in NM23-H1/2) that ensures structural integrity of the multimeric forms of NDPK [36, 37]. It is thought that the K-pn (P97S) mutation lies in a mutant loop that imparts an increased flexibility but reduced stability to NDPK protein’s hexameric quaternary structure. This might enable the protein to interact with additional partners and explain the complexity of the phenotype-mutation axis that differentially perturbs proliferation and differentiation (see Xenopus below). This is also consistent with the idea that awd K-pn might be a neomorphic allele. Although the Prune/NM23-H1 interaction was confirmed in human systems [38, 39], the molecular mechanism of how a single awd K-pn allele is able to confer lethality to prune mutants remains an unresolved issue.
AWD regulates neurotransmission through its endocytic function
Ramaswami’s laboratory was interested in identifying genes that regulate neurotransmission at synaptic junctions using forward genetic approaches [40]. In synaptic vesicle recycling and cellular trafficking, endocytosis plays a key role. Pioneering work in Ramaswami’s laboratory revealed that AWD/NM23 is an essential component of endocytosis: AWD was identified as a factor required for dynamin-dependent synaptic vesicle recycling [40]. Dynamin is a GTPase functioning in a later stage of endocytosis in eukaryotic cells where it pinches off the nascent vesicle by constricting its neck. Drosophila mutants carrying a thermosensitive allele of dynamin/shibire are paralyzed at the restrictive temperature due to defective uptake of neurotransmitters at the neuromuscular junctions. Genetic screens were conducted for mutations, which enhance the phenotype of shibire (shi) mutants. The thermosensitive shi mutant flies were further mutagenized at the permissive (lower) temperature, and second-site mutations causing paralysis at lower temperatures were looked for [40]. Remarkably, three alleles of awd were isolated in this screen. They confirmed the genetic interaction, using existing awd alleles: these mutations also enhanced the shi phenotype, but the histidine119 (H119A) mutant could not. Together, these data suggest the existence of a highly specific interaction between dynamin/shibire and awd.
Very recently, the group of Mathieu Boissan [41] has explored the specific interaction between dynamins and nucleoside diphosphate kinases at the molecular level during membrane remodeling events. Dynamin superfamily proteins are molecular motors, which use GTP as energy source. NDPKs locally fuel GTP towards dynamins, thus these motor proteins are able to work with high thermodynamic efficiency [41].
AWD’s function in epithelial tube morphogenesis during trachea development
Using tracheogenesis as a model of tube generation, the laboratory of Tien Hsu was initially interested in characterizing the function of the Drosophila von Hippel-Lindau (VHL) tumor suppressor homolog during development of the fly respiratory system [42]. Germline inactivation of the VHL gene is associated with an increased risk of renal cell carcinoma [43]. As renal cancer has a tubular origin, and the Drosophila trachea is a tractable epithelial tubule system, the fact that dVHL mutants exhibit a tracheal defect is highly interesting. Development of the fly trachea is both linked to matching oxygen demand with supply, and is used to model tubular morphogenesis (also called branching morphogenesis; reviewed in [44, 45]). In brief, during early stages of embryogenesis tracheal placodes (also termed sacs) arise through invagination of specialized clumps of ectodermal cells expressing certain transcription factors. Subsequently, branches are formed from these invaginating placodes. After elongation of branches, some of them fuse to generate an interconnected network of the tracheal tubes. Importantly, immediately post-placode formation, cell division ceases and branching morphogenesis happens as a series of coordinated cell migration events. FGF (fibroblast growth factor)/FGFR (fibroblast growth factor receptor) signaling is a key chemotactic system that drives coordinated tracheal migration: tracheal cells express (transcription-factor-induced) breathless/FGFR, while the target tissues ahead of the advancing tube provide the ligand, branchless/FGF. The combination drives cytoskeletal remodeling of the advancing tracheal tips as they penetrate into an increasingly hypoxic tissue.
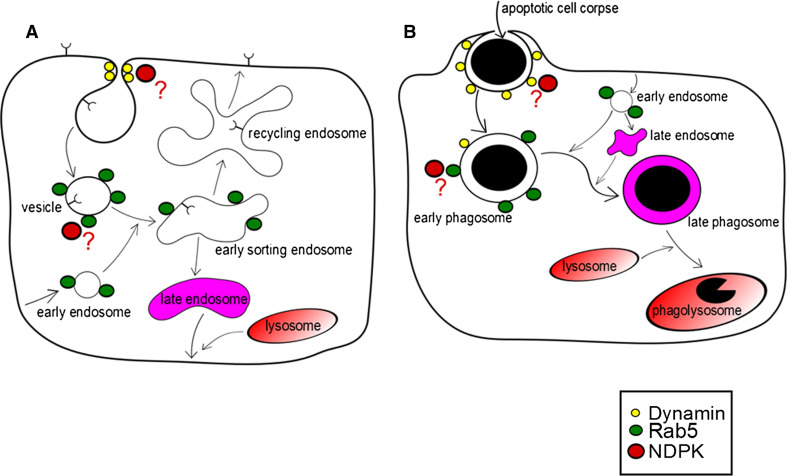
The Hsu laboratory reported AWD as an interaction partner of Drosophila VHL in a yeast two-hybrid screen; later this interaction was further investigated and VHL, which encodes an E3 ubiquitin ligase, was found to stabilize the AWD protein through its non-canonical function [46, 47]. Detailed analysis of awd loss-of-function mutants showed ectopic tracheal migration accompanied by overaccumulation of Breathless/FGFR on tracheal cell surfaces [48]. As the awd mutant phenotype was exacerbated in the shi/dynamin mutant background [48], it was concluded that AWD regulates tracheal cell motility by modulating FGFR levels through a dynamin-mediated pathway (Fig. 1a). Thus, this study reaffirmed the endocytic function of AWD and suggested that internalization of membrane-bound FGFR is the mechanism, whereby AWD and dynamin are able to downregulate FGFR levels on tracheal cell surfaces resulting in suppression of directed cell migration (Fig. 2a). A plausible explanation was proposed for NDPK’s potential involvement in metastasis inhibition: NDPK/AWD activity facilitates the downregulation of activated growth factor receptors via receptor internalization, whereas the absence of NDPK function causes increased levels of cell surface receptors and results in oversensitised cells to chemotactic signals and elevated metastatic potential.
Fig. 2.
NDPKs in endocytosis and vesicular trafficking. Dynamin is necessary for membrane remodeling events. NDPKs (Drosophila AWD and C. elegans NDK-1) show a genetic interaction with dynamin and are proposed to supply GTP for dynamin. In both cases (a, b), the release of endocytic vesicles/phagosomes from plasma membrane is related to dynamin’s membrane fission activity, and then Rab5 GTPase promotes the fusion of these vesicles/early phagosomes with early endosomes. a During Drosophila development levels of cell surface receptors levels on migrating cell types (for example, FGFR on tracheal cells) are regulated through receptor internalization. Dynamin’s membrane fission activity is required for the release of receptor-containing endocytic vesicles from plasma membranes. Rab5 then promotes the fusion of these vesicles with early endosomes. AWD is proposed to act in these processes: it interacts genetically with dynamin and its activity is required for Rab5 function in early endosome formation. Early endosomes communicate with both lysosomes and recycling endosomes: their cargo will be either degraded in lysosomes or recycled back to the plasma membrane. b In C. elegans, DYN-1/dynamin is involved in both engulfment and clearance of apoptotic cells. NDK-1/NDPK shows a genetic interaction with DYN-1. During apoptotic corps removal, the dying cell (such as a germ cell in the worm), also called apoptotic corps is first engulfed by a neighboring cell (gonadal sheath cell). DYN-1 organizes vesicle transport and promotes fusion of early endosomes to ensure the extension of pseudopods. Recruitment of the early endosomal marker RAB-5 also relies on DYN-1. DYN-1 is also essential for the degradation of apoptotic corpses, through its role in phagosome maturation: DYN-1 is necessary for recruitment and fusion of endosomes and lysosomes to phagosomes. The exact relationship of NDK-1/NDPK, DYN-1/dynamin and the GTPase RAB-5 is not yet known
AWD regulates border cell migration, and is essential for epithelial integrity during Drosophila oogenesis
The function of AWD was further investigated in another migrating cell type of epithelial follicular origin, a cluster of co-migrating invader cells found within ovaries, called border cells. Border cell migration during fly oogenesis is considered as an important model of vectorial epithelial cell migration [49], and, in addition, as a model of epithelial-to-mesenchymal transition (EMT) [50]. The Drosophila egg chamber is composed of the germ cell complex (nurse cells and the oocyte), which is surrounded by follicular (epithelial) cells (Fig. 1b). These outer lining cells ensure the integrity of the egg chamber and also provide positional information to the developing oocyte. AWD is expressed in these follicular epithelial cells throughout oogenesis (Fig. 1b), but not in the border cells despite their derivation from the same follicular cells [51]. This suggests cell-type-specific regulation of AWD levels. Border cells are a group of 6–10 anterior follicular cells, which delaminate together and move away from the epithelium during oogenesis, invading through the adjacent nurse cells within the ovary until they reach the anterior of the oocyte. Border cells, on arrival at their final position, have an essential function in forming the opening of the micropyle, a structure necessary for future sperm entry. Border cell migration is regulated by the Drosophila PDGF (platelet derived growth factor)/VEGF (vascular endothelial growth factor) signaling (the PDGF homolog ligand, Pvf comes from the oocyte and is received by the Pvr receptor on the border cells) [52, 53]. The JAK (Janus kinase)/STAT (signal transducer and activator of transcription) pathway is also involved [54], and other factors beyond the scope of this review such as integrins and EGF are also important for border cell movement. Nallamothu and colleagues showed that AWD, expressed in follicular cells, functions as a negative regulator of cell migration because it downregulates the cell surface receptors Pvr and Domeless, receptors of the PDGF and JAK/STAT pathways, respectively, most probably through receptor internalization in cooperation with Shi/dynamin, similar to the regulation of FGFR levels in tracheal cells discussed above. Thus, to drive border cell migration, AWD expression is downregulated in border cells (Fig. 1b). Conversely, ectopic expression of AWD causes stalled migration of border cells [51].
AWD is expressed and functions in the follicular cells of the egg chamber during oogenesis (Fig. 1b). This expression pattern of AWD is consistent with the work of the Shearn laboratory (discussed earlier), as the ovarian sterile phenotype of awd mutants originates from defects of the follicular epithelial cells. The Hsu laboratory demonstrated that AWD is required to maintain epithelial integrity [55]. Lack of AWD and excess AWD activity both disturb epithelial integrity. In the absence of AWD activity, accumulation and spreading of adherens junction components, such as Drosophila E-cadherin, beta-catenin and alpha-spectrin, were observed, which resulted in disintegrated epithelial structure (piling up of epithelial cells). Overexpression of awd also perturbs epithelial characteristics, but in a different manner. Epithelial cells overexpressing awd lose adherens junction components on their surface and undergo a morphological change: they start to adopt a spindle-like shape instead of the epithelial polygonal morphology. This morphological change is a hallmark of EMT, indicating that overexpression of AWD has a proto-oncogenic effect in follicular epithelial cells. Together, these data are also consistent with the idea that an optimal level of AWD is needed to appropriately regulate the turnover of adherens junction components through its endocytic controlling function towards multiple receptors. Consistently, a subtly regulated turnover of dynamin is also necessary to maintain epithelial integrity [56, 57].
AWD is required for Notch signaling in follicular and wing disc cells
Next, the Hsu laboratory linked AWD’s function to the Notch pathway [58]. Endocytosis and endosomal trafficking are required for proper processing of Notch signaling and intracellular trafficking in both Drosophila follicle cells and imaginal discs. In homozygous awd mutant follicular cells and wing disc cells, which were generated by genetic mosaic techniques, Notch signaling was found to be defective. The authors observed that loss of awd function blocks Notch signaling by altering the receptor processing after the S2 cleavage step that normally liberates an intracellular Notch signal to the nucleus. Defective AWD causes Notch accumulation in early endosomes, but acting beyond dynamin on this occasion. Thus, another (a later) endocytic step at which AWD is necessary has also been identified: AWD functions in early endosome maturation by influencing Rab5 function. This adds further complexity as follows: AWD, a GTP-GDP balancing enzyme is linked to GTPase function as both dynamin and Rab5 proteins are GTPases. However, it must be noted that they are two very different GTPases with different biochemical properties. Rab5 is a small GTPase with high affinity for GTP and a low basal GTP hydrolysis rate whereas dynamin is a large atypical GTPase with very low affinity for GTP and a high basal GTP hydrolysis rate.
The worm NDK-1: the C. elegans group I NDPK promotes activation of the Ras/MAPK cascade and participates in DTC migration and apoptotic engulfment
NDK-1/NM23H1/H2 is required for full activation of Ras signaling in the nematode C. elegans
In the genome of C. elegans, three NDPK paralogs were identified by BLAST searches [27]. The open reading frame (ORF) F25H2.5 shows high sequence similarity to group I NDPKs, therefore it was named ndk-1 (C. elegans ortholog of nucleoside diphosphate kinase-1; [59]) whereas the other two paralogs (ORFs F55A3.6 and Y48G8AL.15) belong to the more divergent group II. Functions of the single group I NDPK homolog NDK-1 in Ras signaling have recently been characterized by our laboratory [59]. ok314 is a deletional allele of ndk-1, which removes the entire ORF. 50 % of ndk-1(ok314) homozygous mutants die as embryos, but the remainder develop into sterile adults, which display a protruding vulva phenotype (Fig. 1c).
First, the vulval phenotype of viable ndk-1(–) mutants was examined. Four signaling pathways including the RTK (receptor tyrosine kinase)/Ras/MAPK (mitogen activated protein kinase), Notch, Wingless/Wnt and synMuv (for synthetic Multivulva) molecular systems are involved in the development of the hermaphrodite reproductive organ, the vulva, which is an intensively studied example of animal organogenesis [60]. The EGFR/Ras/MAPK pathway plays an essential role during vulval development, as an EGF ligand emanating from a specific cell of the somatic gonad, called the anchor cell (AC), provides an inductive signal for a subset of epidermal blast cells, the six vulval precursor cells (VPCs, also called P(3–8).p). At the early L3 larval stage, the three VPCs closest to the AC, P(5–7).p, receive the inductive signal, which activates the EGFR/Ras/MAPK pathway in these cells (reviewed in [60, 61]). P6.p, the VPC closest to the AC, adopts the primary vulval fate due to LET-23/EGFR activation at high levels. In the neighboring VPCs, P5.p and P7.p, LET-23/EGFR activation occurs at lower levels, and these VPCs adopt the secondary vulval fate as a result of lateral signaling mediated by the LIN-12/Notch pathway. Each induced VPC divides three times, generating 22 daughter cells at the L4 larval stage. These cells undergo cell migration events, and fuse into 7 toroids to form the adult vulval structure. Mutations causing reduced EGFR/Ras/MAPK signaling lead to a Vulvaless phenotype, whereas increased Ras signaling causes a Multivulva phenotype (multiple ventral protrusions, reviewed in [61]). Protruding vulva (Pvl) mutants display a single protrusion, which is caused by eversion of the vulval tissue [62].
Using specific markers which label individual vulval cell types, it was demonstrated that the number of vulva cells in ndk-1(ok314) mutants is decreased, as compared with the 22 cells observed in the wild type. Misspecification of both primary and secondary vulval cell fates occurred in ndk-1(ok314) mutants [59]. In addition, one of the markers examined, egl-17/FGF, is a known MAPK target [63], showing reduced expression in P6.p granddaughter cells in the ndk-1 mutant background, suggesting that Ras/MAPK signaling is dampened in the absence of NDK-1. Next, the epistatic relationships of ndk-1 with Ras pathway components were established to further understand the mechanisms by which NDK-1 influences Ras/MAPK signaling. Epistasis analysis revealed that ndk-1 affects vulval patterning downstream of lin-45/Raf but upstream of mek-2/MEK and mpk-1/MAPK [59]. Concordant with these results, a dramatic decrease was detected in activated phospho-MAPK levels in somatic tissues of ndk-1 knockouts, whereas total MAPK levels were unchanged, as compared with wild-type animals. These data suggest that NDK-1 is necessary for full activation of MAPK signaling in somatic tissues of the worm. Because the scaffold proteins KSR-1/KSR-2 (kinase suppressor of Ras) are indispensable for MAPK activation [64], and because several studies on human cell lines have linked NM23’s function to KSR scaffolds [65, 66], ndk-1(–);ksr-1(–) and ndk-1(–)ksr-2(–) double knockouts were generated to test potential genetic interactions between ndk-1 and ksr genes. In these double mutants, ndk-1 single-mutant phenotypes were enhanced. For example, embryonic lethality of homozygous ndk-1(–) mutants (50 %) became fully penetrant in ndk-1(–) ksr-2(–) double mutant background. NDK-1 was also shown to directly interact with worm KSR-2 and murine KSR-1 by in vitro pulldown assays.
Thus, a previously proposed link between an NDPK protein and KSR scaffolds [65, 66] was confirmed in vivo [59]. However, as NDK-1 is necessary for proper MAPK activation, one can conclude that it promotes the activation of Ras/MAPK signaling. In human studies, however, NDPKs were shown to attenuate Ras/ERK signaling through phosphorylation of KSR scaffolds [65, 66]. Moreover, cancer invasion induced by NM23-H1 silencing was reverted by a pharmacological inhibitor of the Ras/ERK pathway, further showing an inverse relationship between NDPK activity and Ras/ERK activation in human [67]. Structural differences in worm and mammalian KSRs might lead to altered regulation of these scaffold proteins [68, 69]. Furthermore, the mechanisms of Raf activation, which is proposed to occur through Raf dimerization [70], might not be evolutionary conserved as the nematode possess only a single B-Raf homolog whereas in mammalian systems, 3 Raf isoforms are present [69].
NDK-1 regulates distal tip cell migration and apoptotic engulfment
Defects in NDK-1 functions also cause abnormal gonadal development in C. elegans [59]. The viable homozygous ndk-1(–) mutant worms are sterile. The development of their germ cells is arrested at the mitotic phase. In addition, gonad arms in these mutants display an abnormal shape [71]. Wild-type C. elegans hermaphrodites possess two U-shaped gonad arms whose morphogenesis is directed by specialized leader cells called distal tip cells (DTCs), which are located at the distal edges of either arm [72]. During larval development, DTCs migrate in response to three-dimensional attractive and repulsive cues to properly form mirror image U-shaped gonad arms. DTC migration provides a tractable system to monitor genes influencing guided cell migration [72]. In a transgenic strain expressing a translational fusion NDK-1 reporter, NDK-1 accumulation was detected in the DTCs [71]. Consistent with this NDK-1 expression profile, an incomplete DTC migration was observed in ndk-1 loss-of-function mutants; DTCs stopped their migration prematurely in the third (or dorsal) phase of migration, resulting in a “J-shaped” gonad arm [71] (Fig. 1a). In C. elegans, integrins also act in the third phase of DTC migration [72, 73]. Therefore, an epistasis analysis was performed with genes involved in integrin-mediated signaling (members of the CED-10/Rac signaling pathway such as vab-3/Pax6, ina-1/alpha-integrin, unc-73/Trio, mig-2/RhoG, ced-5/Dock180, ced-12/Elmo, ced-10/Rac) and with abl-1/ABL and abi-1/ABI which have been shown to act in parallel to the CED-10 pathway [74]. Epistasis analysis revealed that NDK-1 functions downstream of CED-10/Rac and in parallel to ABI-1/ABI in DTC migration [71].
In multicellular organisms, unnecessary or dangerous cells are often eliminated by type I programmed cell death, apoptosis. Dying cells are removed by neighboring cells or specialized cell types, which engulf the apoptotic cells and destroy their debris to avoid inflammation. Pioneering studies in C. elegans identified and extensively characterized genes of the killing step of programmed cell death and also genes participating in cell corpse engulfment [75].
Apoptotic engulfment and DTC migration are analogous processes as both require precise reorganization of cytoskeleton and regulation of membrane trafficking/recruiting to extend cell surfaces [76, 77]. The CED-10 pathway and signaling through abi-1/abl-1 regulate both engulfment and migration processes [74, 78], whereas the parallel acting CED-1 signaling participates only in engulfment. The CED-10 pathway is linked to cytoskeleton remodeling, whereas the CED-1 pathway recruits membranes to extend the surface of the engulfing cell.
The above data prompted us to examine whether apoptosis is affected in ndk-1(–) mutants. Indeed, an increased number of apoptotic cell corpses were measured in the germline of ndk-1(RNAi) animals and in ndk-1(ok314) embryos, as compared to wild-type animals [71]. Furthermore, NDK-1 accumulation was detected in sheath cells around dying germ cell corpses (Fig. 1a). Gonadal sheath cells are specialized engulfing cells, which eliminate neighboring germ cell corpses [79]. Analysis of the number of apoptotic corpses in ced-10(–), ndk-1(–) single mutants and ndk-1(–); ced-10(–) double mutants led to the conclusion that NDK-1 functions downstream of CED-10/Rac, i.e., in the process of apoptotic engulfment as well. In addition, a genetic interaction was observed between ndk-1 and dyn-1/dynamin (e.g., single mutants show phenotypic similarities, moreover ndk-1; dyn-1 double mutants are not viable). dyn-1/dynamin is a downstream effector of the parallel acting CED-1 pathway, and its Drosophila homolog, shibire, shows a well-known genetic interaction with awd.
Together, these results suggest that NDK-1 plays a role in DTC migration and engulfment of apoptotic cells by influencing the CED-10 pathway. Moreover, NDK-1 interacts with the CED-1 pathway through DYN-1 in engulfment through DYN-1. Thus, this study is the first to link an NDPK to apoptotic engulfment [71] (Fig. 1c) and consistent with a large body of data confirming that NDPK and dynamin are ubiquitous functional partners across model systems.
NM23-X4 function in Xenopus retinal development
Cyclins control kinases that in turn regulate the cell cycle, but are themselves controlled by a family of inhibitor proteins, the cyclin-dependent kinase inhibitors (CDKIs) [80]. During frog retinal development, stem cells generate the neuronal precursor cells that exit the cell cycle immediately before their terminal and cell-fate determination step [81, 82]. Next, these cells differentiate to determine the final composition of mature retinal cells of different subtypes. Thus, the terminal cell cycle before cell specification of pluripotent precursors is when CDKI proteins become key players in determining a given cell’s fate irrespective of the nature of the final retinal cell types (i.e., irrespective of whether that cell will become neuronal or glial).
The emerging concept is that at a given time during development of the retina, once a set of factors (determinants) have accumulated to a threshold [83], this specifies which amongst many of the final retinal components are to be preferred. CDKI family members promiscuously enhance that role in phenotypic choice. For example, in mouse retina, deletion of Math5 [84], a driver of retinal ganglion cell fate, upregulates one CDKI protein, which then creates a glial alternative (Muller cell), i.e., non-neuronal cell fate. Such effects are driven by interaction with the N-termini of such CDKI family members, although their C-termini are equally important but play a different role, in which they interact with both the actin cytoskeleton and microtubular structures to drive differentiation. In summary, there exists promiscuity of action in these CDKI molecules towards a given cell fate depending on other factors (the cell context) [85].
To further understand the context relationship between cell cycle exit from G1, subsequent cell-fate determination and the contemporaneous regulation of CDKI components, Shin-Ichi Ohnuma and colleagues [86] tested the hypothesis that there may be other proteins that bind to the N-terminus of this regulator [14]. They used a bacterial 2-hybrid screen of the N-terminus of the CDKI p27XiC1. Since p27XiC1 is a low abundance protein that is normally rapidly degraded, they also inhibited the proteasome to raise CDKI levels finding that frog NM23-X4, X3, the human NM23-H1 and NM23-H4 all interacted with this CDKI (and with only some of the three human CDKI-equivalents, suggesting specificity of actions). Overexpression of NM23-X4 (less for other isoforms) all inhibited the pro-gliogenesis proclivity of p27XiC1, but inactive NM23 mutants that could not transfer p-histidine were unable to act as CDKI inhibitors. Interestingly, the K-pn-equivalent mutant, a mutant lying remote from the catalytic site but important for the quaternary stability of NDPK multimers, inhibited just as well as the wild type, but this mechanism of action may not be dependent on CDKI(s) per se because overexpression of NM23-X4, when the CDKI was pre-suppressed, retained the glial promoting effect [85, 87]. Interestingly, overexpression of NM23 in the absence of CDKI potentiates gliogenesis, suggesting that NM23 has also ability to modulate gliogenesis in a CDKI independent manner (Fig. 1e).
The Dictyostelium NDPK is implicated in macro- and micropinocytosis
To further explore the relationship between NDPK, cell migration, responses to nutrient supply, and substrates available to the developing organism, Fisher and colleagues applied the tractable social amoeba Dictyostelium, exploiting its unparalleled variety of cell types and cell morphology that are all amenable to genetic manipulation [88, 89]. This soil amoeba can exist either as single cells that each engulf bacteria/soluble nutrients or can form multicellular aggregates (primitive pre-cadherin epithelial coated slugs) that undergo directional, i.e., cue-driven vectorial mobility [88, 89]. When induced to starve by either mitochondrial depletion or by inducing a false starvation signal generated by stably expressing a constitutively overactivated form of the cellular fuel sensor, AMP-activated kinase (AMPK), the directional cue in response to light (but not the speed) of the motile slug form was abolished [89]. The consequent ‘blind’ random slug motion implied a sensor defect. However, light cue-dependent vectoring was restored by concomitant NDPK knockdown, suggesting that NDPK is a latent but positive regulator of an energy depleted state. Slugs use the front of their rounded transparent cell bodies to focus light, and this primitive ‘lens’ refracts light towards the back of the slug body as a means to navigate from dark soil to the light at the soil surface. At the soil surface, the aggregate slug can differentiate into a multicellular stalk and head structure that will make spores at its distal tip, to be carried in the wind to complete its life cycle [90]. Importantly, vectorial motion to light was not the only perturbed function during induced starvation. As with the retinal fate, during starvation cell differentiation was also profoundly altered with structural anomalies (abnormal head and stalk of the fruiting body), and once again, concomitant NDPK knockdown reversed the mal-differentiation observed in the starved state to an outwardly normal morphology [89]. Importantly, NDPK knockdown alone in the fully fed state could neither alter vectorial motion nor change gross cell morphology during differentiation. This interesting observation is consistent with the roles of NDPK in hypoxia responsiveness, nutrition and cell-fate specification [81]. Next, these authors studied nutrient uptake in single Dictyostelium cells either plated onto a flat-bed on top of bacterial lawns as a food source (macropinocytosis assay) or, by placing these single cells in a liquid environment, exposed to micronutrient broths of soluble factors such as amino acids (axenic culture) [88]. In axenic culture, NDPK knockdown alone now created a phenotype (despite substrate availability) whereby endocytosis and exocytosis of recycling vesicles budding from the plasma membrane were both unexpectedly accelerated, and yet these NDPKs knocked down single cells failed to retain sufficient imbibed nutrients and consequently displayed slower growth. Since NDPK/AWD is a very important regulator of GTP supply to enhance dynamin function (shi function, in flies) [48], the authors concluded that nutrient retention within the single cell interior was perturbed by lack of NDPK. Their measured reduction in the mass of nutrient accumulation implies a deficit in nutrient transfer from the plasma membrane to the machinery within the cell interior that utilizes imbibed nutrients. However, in the case of single cells exposed to two-dimensional bacterial lawns, the exact opposite was observed whereby NDPK knockdown increased the rate of phagocytic bacterial uptake (and hence enhanced growth rate). Thus single cells confined to a two-dimensional surface use a process that is negatively regulated by NDPK whereas single cells in suspension culture use NDPK to balance nutrient entry by micropinocytosis against cell growth. Once again, context is key for NDPK function.
NDPK knockout mice point to roles of different NME proteins in erythroid lineage development, CD4+ T cell activation, cardiac contractility and ciliar functions
The mouse genome encodes 8 NDPKs, which constitute 2 groups similar to human: NM23-M1-M4 isoforms belong to group I NDPKs, whereas group II is composed of M5–M8 isoforms [91]. Mouse and human orthologs of Nm23 family genes are strongly conserved: the structure of the orthologous genes, their expression pattern in different tissues and primary structure of the encoded proteins are all very similar. Human NM23-H1 and NM23-H2 show 94 and 98 % sequence identity with their mouse counterparts, respectively. Active homo- and heterohexamers are composed of NM23-M1 and NM23-M2 subunits, and the M1 and M2 isoforms are responsible for 80 % of the cell’s NDPK activity [92].
Constitutive knockout mice have been generated for Nm23-M1 (also known as Nme1; [93]), Nm23-M2 (also known as Nme2; [94]), a double knockout of Nme1 and Nme2 [95], and finally, Nm23-M5 (Nme5) and Nm23-M7 (also known as Nme7; [96]). Mice deficient for either NME1 or NME2 are able to develop until adulthood without gross developmental defects, however, Nme1 −/− mice obtained by Arnaud-Dabernat and co-workers [93] display a mild hypotrophy. Further studies on Nme1 −/− knockout mice have shown that mutant females cannot feed their pups due to growth retardation of the mammary glands and defects in the final step of mammary duct maturation of the nipple leading to duct obstruction [97] (Fig. 1f).
Experiments using Nme1 knockout mice in the laboratory of Marie-Lise Lacombe brought the strongest evidence for the metastasis suppressor effect of NME1. When Nme1 −/− mice were crossed with mice prone to develop liver tumors (ASV mice expressing the SV 40 large T antigen in the liver leading to hepatocellular carcinoma), the double transgenic mice showed a higher incidence of lung metastases compared to the single mutant and the single transgenic strains [5]. Interestingly, in Nme1 −/− mice, subjected to partial hepatectomy, liver regeneration was not affected, suggesting either that NME1 is not involved in this process or another NDPK paralog compensates for the absence of NME1 during liver regeneration [67].
Nme2 −/− mice were generated by the laboratory of Ed Skolnik [94]. These knockout mice are phenotypically normal at birth and have a normal life span. The absence of NME2 influences the function of the immune system: although T and B cell development is normal in these mice, K+ channel KCa3.1 activity and cytokine production are defective in T helper (Th)1 and 2 cells. The Skolnik group showed in a previous study that activation of the K+ channel KCa3.1 is required for Ca2+ influx and the subsequent activation of B and T cells, therefore the proper function of NME2 is critical for activation of CD4+ T cells. KCa3.1 channel activation occurs by phosphorylation: a histidine residue located in the carboxyl terminus of KCa3.1 is directly phosphorylated by NME2 through its histidine kinase activity [98]. Interestingly, the KCa3.1 channel was recently linked to invasion in glioma, where high KCa3.1 expression is associated with poor patient prognosis [99]. Moreover, KCa3.1 channel activation is also implicated in proliferation of vascular smooth cells, linking NDPK dependent activation of the channel to atherosclerosis and vessel re-stenosis.
Nme1 −/−/Nme2 −/− double knockout mice were generated by Edith Postel and colleagues [95] by targeted deletion of both adjacent genes on chromosome 17q21.3. Nme1 −/−/Nme2 −/− newborn mice are three to five times smaller than wild-type mice, display hematological phenotypes including severe anemia, impaired maturation of erythrocytes, and die perinatally. These pleiotropic phenotypes show that NME1 and NME2 functions are necessary for erythroid lineage development (Fig. 1f), and it is suggested that the absence of both NM23 isoforms leads to insufficient triphosphonucleoside precursors for DNA and RNA synthesis in these double mutant mice. Tissue-specific targeted deletion is clearly needed to understand this issue further.
Mouse embryo fibroblasts (MEFs) from Nme1 −/−/Nme2 −/− mice and zebrafish have been used by Tom Wieland and colleagues to investigate the role of NME2 in heterotrimeric G protein signaling. The authors found that NME2 forms a complex with the G beta-gamma complex (Gβγ) [100] and directly passes its high energy phosphate onto Gβ. The high energetic phosphate on Gβ induces G protein activation without the need for receptor-ligand interactions on the cell surface [100, 101]. Thus, histidine phosphorylation of Gβγ dimers by NME2 can activate G proteins independently from GPCRs. In vitro experiments suggested that this phosphorelay can regulate Gs protein activity in cardiomyocytes, thereby stimulating cAMP synthesis and cardiac contractility through β-adrenergic receptor signaling [102, 103].
In mouse embryo fibroblasts (MEFs) derived from non-viable Nme1 −/−/Nme2 −/− knockout mice, the absence of NME1/2 in membranes was accompanied by a gradual loss of Gβ1 and Gβ2 as well as a reduction in basal cAMP levels. Re-expression of human NME2 in KO MEFs rescued the loss in G protein content and restored basal cAMP levels [104]. The Wieland laboratory showed that heart failure is associated with similar defects and in zebrafish, where morpholino-mediated knockdown of NME2, as well as Gβ1, both resulted in dramatically reduced cardiac contractility (Fig. 1d). They suggested that decreased α-subunit abundance alters adenylyl cyclase-regulating Gs and Gi protein subfamilies [104]. These defects were also rescued by injection of zebrafish Nme2 mRNA into morphants.
In a novel study, the Wieland laboratory identified the role of NME2/NDPK B in angiogenesis using Nme2 deficient mice and zebrafish nme2 morphants. They found that NDPK B depletion in endothelial cells disturbs VEGFR-2 cycling and impairs the formation of endothelial adherence junctions [105].
The expression of group II genes was described in axonemal structures of cilia and flagella [2, 106]. Constitutive Nme5 and Nme7 knockout mice were generated and described by Vogel and co-workers [96, 107, 108]. Nme7 knockout mice present defects like situs inversus (a congenital condition characterized by left–right transposition of thoracic and visceral organs and associated vasculature) and hydrocephalus [108] (Fig. 1f). Both situs inversus and hydrocephalus can be linked to impaired motility of cilia. Motile flagella and cilia normally create fluid flow over cell surfaces. Impaired ependymal flow contributes to the development of hydrocephalus, whereas the randomization of left–right body asymmetry is due to dysfunctional nodal cilia during early embryogenesis. All these data suggest that NME7 is involved in the biogenesis and function of motile cilia. Vogel and colleagues also reported that Nme5 −/− mice display a pathological phenotype highly suggestive of primary ciliary dyskinesia [107].
Discussion
NDPK-related molecular activities in animal models
Nucleoside diphosphate kinase enzymes display multiple molecular functions, such as nucleoside diphosphate kinase, histidine kinase (histidine phosphotransferase), and nuclease activities, as well as DNA and lipid bilayer binding. Developmental studies on mice and zebrafish reinforce the histidine kinase activity of NME2 [19]. Activation of CD4+ helper T cells is impaired in the immune system of Nme2 −/− mice due to the lack of NME2-mediated phosphorylation of the KCa3.1 channel [94]. Recently, in the kidney, in the process of urinary Ca2+ excretion a Ca2+-permeable channel, the transient receptor potential-vanilloid-5 (TRPV5) channel was found to be phosphorylated and thus activated by NME2 [109]. Experiments performed on fibroblasts of Nme1 −/−/Nme2 −/− knockout mice and zebrafish show that NME2 influences heterotrimeric G protein signaling through phosphorylation of Gβγ dimers, thereby stimulating cardiac contractility [104]. Data on the activation of heterotrimeric G proteins and different ion channels point to a role of NDPK as a histidine phosphotransferase (histidine kinase).
NME1 is ascribed to regulate the outcome of Ras/MAPK signaling through phosphorylation of a MAPK scaffold, KSR. Inactivation of KSR1 by NME1 occurs through phosphorylation of Ser392 of this scaffold protein [65]. Thus, KSR1 is also a target of NM23-H1’s histidine kinase activity, which was demonstrated in different cell lines [65, 66]. In C. elegans, a genetic interaction was observed between NDK-1/NDPK and the worm KSRs [59]. However, NDK-1 exerted an opposite effect on the outcome of Ras signaling compared to data obtained from cell lines, probably due to altered regulation of KSRs [69]. Discrepancy between results obtained from mammalian cell lines and from the worm might be resolved if we better understand the function, structure and regulation of KSR scaffolds.
In Xenopus, the cyclin-dependent kinase inhibitor p27Xic1 is a cell-fate determinant of retinal gliogenesis and neurogenesis. The frog NME4 homolog NM23-X4 functions as an inhibitor of p27Xic1-mediated gliogenesis in retinal development. NM23-X4 directly binds to p27Xic1, and this binding is dependent on amino acid residues (e.g., His148 and Ser150 of NM23-X4), which are critical for kinase activity [14].
Data from Dyctiostelium, Drosophila and C. elegans suggest that NDPKs participate in the processes of endocytosis and phagocytosis (reviewed in [51, 71, 88] and see below in this work). The endocytic role of AWD and NDK-1 is linked to the large GTPase dynamin. In human cells it was shown that in the NM23–dynamin interaction the nucleoside diphosphate kinase activity of NDPK is important to generate GTP from GDP and to ensure the energy supply of dynamin [41].
NDPKs as signal integrators of nutrient supply
Developmental studies on the slime mold Dictyostelium indicated a genetic interaction between AMPK and NDPK [89], knockdown of NDPK suppressed the phenotypic defects caused by AMPK hyperactivity. AMPK signaling acts as a main sensor of cellular energy status, and also influences processes related to cellular energy levels, including autophagy and pre-conditioning to hypoxia. Moreover, activation of AMPK with drugs such as metformin has been linked to lower incidences of certain cancers [110]. In recent experiments on human bronchial epithelial cells, AMPKalpha1 and NDPK-A (NM23-H1) were found to bind each other, and catalytically active NDPK-A was essential in the transmission of an inhibitory AMPK signal towards an energy consuming process, ion transport [111]. Here, AMPK retards the activity of the cystic fibrosis transmembrane conductance regulator, CFTR, an ion channel whose dysfunction causes cystic fibrosis [112]. This CFTR inhibitory signal requires functional NDPK-A. The authors also mutated a key residue close to the active site in NDPK at Ser120, a site that is mutated in the neonatal form of neuroblastoma (S120G) [113]. They found that the S120 mutation also abrogated the inhibitory AMPK signal, as observed for the catalytically dead H118F mutant [111].
It may be pertinent that the fidelity of NDPK and AMPK interaction requires an intact S120 on NDPK that is mutated in a developmental form of neuroblastoma. One unifying idea from Lascu and colleagues is that the S120G mutation affects the quaternary structure of the NDPK hexamer; e.g., it generates an unstable molten globule form of NDPK that cannot form the highly active hexamer [114, 115].
Another study in Drosophila performed by Onyenwoke and colleagues [116] also established a functional relationship between AMPK and NDPK. The researchers identified AWD/NDPK as a direct target of AMPK. The regulation of NDPK by AMPK is mediated through a phosphoserine switch on S120 residue of AWD. These authors propose that under unfavorable conditions such as starvation, AMPK inhibits ATP consumption of NDPK to conserve energy. Phosphorylation of S120 residue of NDPK blocks its functions, thus leading to inactivation of its anti-metastatic function in neuroblastoma.
Together, the relationship of NDPK and AMPK is not yet perfectly understood in different model systems, but studies in Dictyostelium and Drosophila confirm the existence of a genetic interaction between these factors. In the fruit fly, NDPK activity is directly regulated by AMPK through a phosphoserine switch but the exact mechanism remains to be determined.
Data from Drosophila, C. elegans and Dictyostelium converge on NDPKs’ role in endocytosis and phagocytosis
In Drosophila, awd function has been extensively studied and associated strongly with endocytosis. awd interacts genetically with shi/dynamin to promote endocytosis in various tissues. In neurons, for example, awd facilitates dynamin-mediated neurotransmitter uptake at neuromuscular junctions. AWD also functions in migrating cell types to ensure proper migration of tracheal cells and border cells by modulating appropriate receptor levels on cell surfaces through receptor internalization. AWD-mediated endocytosis also contributes to ensure epithelial integrity of follicular cells in the egg chamber by regulating the level of adherens junctions’ components. Recently, the endocytic function of AWD has been shown to be indispensible for Notch signaling. In addition, the authors suggested that AWD is required for Rab5 function during early endosome maturation.
Evidence for dynamin-NDPK interaction was also observed in C. elegans. The phenotypes of NDPK-defective ndk-1(–) and dynamin-defective dyn-1(–) single mutants are quite similar in the worm; the late-embryonic lethality with persistent cell corpses. Furthermore, ndk-1(ok314); dyn-1(ky51) double mutants are lethal indicating a genetic interaction between the two genes in C. elegans as well. NDK-1 has also been linked to apoptotic engulfment [71], which is mediated by two partially redundant pathways, the CED-10 and CED-1-mediated molecular systems. The large GTPase DYN-1/dynamin, which plays a pivotal role in various membrane-related cellular processes [117], is a downstream component of the CED-1-mediated pathway. Cells dying by apoptosis are eliminated in every eukaryotic system either by neighboring cells or by specialized cell types. Failure of this process in mammals results in inflammation [118], which favors tumor progression in certain circumstances [119]. Removal of apoptotic cells occurs in two steps: first, the dying cell is surrounded and engulfed by a so-called engulfing cell; second, the apoptotic cell (often called a corpse in the worm) is then degraded within the engulfing cell by the lysosomal system [120] (Fig. 2b). Detailed studies on dyn-1 revealed its function in both steps of apoptotic cell removal [121, 122]. During the engulfment phase, DYN-1 ensures the rapid expansion of phagocyte membrane along the surface of dying cells by organizing vesicle transport and promoting fusion of early endosomes to extending pseudopods [121, 122] (Fig. 2b). DYN-1 is also essential for the degradation of apoptotic cells through its fundamental role in phagosome maturation: DYN-1 is necessary for recruitment and fusion of endosomes and lysosomes to phagosomes [121, 123, 124] (Fig. 2b). Recruitment of two endosomal markers, RAB-5 and RAB-7, also relies on DYN-1 activity [122–124]. RAB-5 is important for promoting the maturation of phagosomes containing apoptotic cells [124, 125].
Studies in Dictyostelium reinforce the role of NDPKs in phagocytosis: pinocytosis, phagocytosis and exocytosis were first linked to NDPKs in this model [88].
Recently, it was shown that NDPK supplies GTP for dynamin’s functions [41]. Thus, we propose that data from Drosophila concerning receptor internalization and results related to C. elegans apoptotic engulfment imply a general role of NDPKs in endocytosis and vesicular trafficking (Fig. 2a, b). Dynamin acts as a main organizer of membrane remodeling events. The membrane fission activity of dynamin is essential for the release of endocytic vesicles from plasma membranes. Rab5 then promotes the fusion of these vesicles with early endosomes. Endosomes play a key role in endocytic pathways, as they communicate with both lysosomes and recycling endosomes. During receptor internalization, cell surface receptors are taken up into endocytic vesicles from where they are either targeted to the lysosome for degradation or recycled back to the plasma membrane through recycling endosomes, thereby regulating surface receptor levels (Fig. 2a). AWD is involved in this process in migrating cell types such as tracheal and border cells according to data obtained from Drosophila (Fig. 2a).
In C. elegans, DYN-1/dynamin is involved in both engulfment and clearance of apoptotic cells. The GTPase Rab-5 plays an essential role in phagosome maturation as it promotes fusion between early (sorting) endosomes and phagosomes (Fig. 2b). It is known that Rab5 function is dependent on dynamin activity during apoptotic cell corps removal. The exact relationship of NDK-1/NDPK, DYN-1/dynamin and the GTPase Rab5 should be further investigated during engulfment and phagosome maturation.
Commonality of developmental roles of NDPKs across model systems
As detailed above, NDPKs’ functions are related to nutrient supply, energy consumption and receptor internalization through endocytosis. These processes have great importance in tumor progression as well, as they are implicated in the development of tumor vasculature. Hypoxic conditions favor the development of tumor vasculature [119] driven by VEGF/FGF signaling pathways. The multifunctional tumor suppressor VHL is linked to hypoxia sensing, as it is able to degrade the α-subunit of hypoxia-inducible factor (HIF-α) under normal physiological conditions through its E3 ligase-related function [126]. Conversely, loss of VHL function results in increased HIF-α expression and VEGF production, which in turn leads to angiogenesis [127].
VHL binds to AWD/NM23, and this evolutionary conserved interaction has been recently characterized in detail in flies and cell lines [128]. VHL and NM23 are associated with early endosomes during endocytosis of activated FGFR1, and the loss of VHL disrupts the association of NM23 and endosomes [128]. This work proposed that VHL, this time through its non-canonical function, acts as an adaptor molecule that brings NM23 to the endosome.
Together, we propose that signals sensing hypoxia through VHL and signals sensing cellular energy status through AMPK converge on NDPKs. This unifying concept awaits confirmation but fits with proposed relationships between NDPK and hypoxia sensing, and links NDPKs to nutrition and cell proliferation [81].
Clarifying the link between phagocytosis, ion transport, AMPK signaling and NDPK function requires further studies, but combined data already obtained suggest that actual NDPK function depends on cellular context (single cells versus cell–cell aggregates). This is a recurring theme for this ancient and conserved protein family.
Acknowledgments
A.M. is supported by the Russell Trust and the Myrovlitis Trust and the work on NDPK was supported by previous grants from the Wellcome Trust. K.T.-V. and T.V. are supported by the OTKA grant K109349. K.T.-V. is a grantee of the János Bolyai Scholarship of the Hungarian Academy of Sciences.
References
- 1.Agarwal R, Parks R. Nucleoside diphosphokinases. In: Boyer PD, editor. The enzymes. New York: Academic Press; 1973. pp. 307–333. [Google Scholar]
- 2.Boissan M, Dabernat S, Peuchant E, et al. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem. 2009;329:51–62. doi: 10.1007/s11010-009-0120-7. [DOI] [PubMed] [Google Scholar]
- 3.Desvignes T, Pontarotti P, Fauvel C, Bobe J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol Biol. 2009;9:256. doi: 10.1186/1471-2148-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steeg PS, Bevilacqua G, Kopper L, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 5.Boissan M, Wendum D, Arnaud-Dabernat S, et al. Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:836–845. doi: 10.1093/jnci/dji143. [DOI] [PubMed] [Google Scholar]
- 6.Ouatas T, Salerno M, Palmieri D, Steeg PS. Basic and translational advances in cancer metastasis: Nm23. J Bioenerg Biomembr. 2003;35:73–79. doi: 10.1023/a:1023497924277. [DOI] [PubMed] [Google Scholar]
- 7.Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr. 2000;32:301–308. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- 8.Harłozińska A, Bar JK, Gerber J. nm23 expression in tissue sections and tumor effusion cells of ovarian neoplasms. Int J Cancer. 1996;69:415–419. doi: 10.1002/(SICI)1097-0215(19961021)69:5<415::AID-IJC11>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Niitsu N, Nakamine H, Okamoto M, et al. Expression of nm23-H1 is associated with poor prognosis in peripheral T-cell lymphoma. Br J Haematol. 2003;123:621–630. doi: 10.1046/j.1365-2141.2003.04668.x. [DOI] [PubMed] [Google Scholar]
- 10.Hailat N, Keim DR, Melhem RF, et al. High levels of p19/nm23 protein in neuroblastoma are associated with advanced stage disease and with N-myc gene amplification. J Clin Invest. 1991;88:341–345. doi: 10.1172/JCI115299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacombe ML, Boissan M. NME1 (NME/NM23 nucleoside diphosphate kinase 1) Atlas Genet Cytogenet Oncol Haematol. 2013;17:526–538. [Google Scholar]
- 12.Marino N, Marshall J-C, Steeg PS. Protein-protein interactions: a mechanism regulating the anti-metastatic properties of Nm23-H1. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:351–362. doi: 10.1007/s00210-011-0646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M-Y, Jeong W-J, Oh J-W, Choi K-Y. NM23H2 inhibits EGF- and Ras-induced proliferation of NIH3T3 cells by blocking the ERK pathway. Cancer Lett. 2009;275:221–226. doi: 10.1016/j.canlet.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki T, Bilitou A, Waters CT, et al. Xenopus NM23-X4 regulates retinal gliogenesis through interaction with p27Xic1. Neural Dev. 2009;4:1. doi: 10.1186/1749-8104-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 16.Thakur RK, Kumar P, Halder K, et al. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan Z, Beresford PJ, Oh DY, et al. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 18.Engel M, Véron M, Theisinger B, et al. A novel serine/threonine-specific protein phosphotransferase activity of Nm23/nucleoside-diphosphate kinase. Eur J Biochem. 1995;234:200–207. doi: 10.1111/j.1432-1033.1995.200_c.x. [DOI] [PubMed] [Google Scholar]
- 19.Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Sci Signal. 2009;2:pe13. doi: 10.1126/scisignal.261pe13. [DOI] [PubMed] [Google Scholar]
- 20.Ma D, Xing Z, Liu B, et al. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J Biol Chem. 2002;277:1560–1567. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, McCorkle JR, Novak M, et al. Metastasis suppressor function of NM23-H1 requires its 3′–5′ exonuclease activity. Int J Cancer. 2011;128:40–50. doi: 10.1002/ijc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokarska-Schlattner M, Boissan M, Munier A, et al. The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J Biol Chem. 2008;283:26198–26207. doi: 10.1074/jbc.M803132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baughman C, Morin-Leisk J, Lee T. Nucleoside diphosphate kinase B (NDKB) scaffolds endoplasmic reticulum membranes in vitro. Exp Cell Res. 2008;314:2702–2714. doi: 10.1016/j.yexcr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell KAP, Szabo G, de Otero SA. Direct binding of cytosolic NDP kinases to membrane lipids is regulated by nucleotides. Biochim Biophys Acta. 2009;1793:469–476. doi: 10.1016/j.bbamcr.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Epand RF, Schlattner U, Wallimann T, et al. Novel lipid transfer property of two mitochondrial proteins that bridge the inner and outer membranes. Biophys J. 2007;92:126–137. doi: 10.1529/biophysj.106.092353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlattner U, Tokarska-Schlattner M, Ramirez S, et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J Biol Chem. 2013;288:111–121. doi: 10.1074/jbc.M112.408633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilitou A, Watson J, Gartner A, Ohnuma S-I. The NM23 family in development. Mol Cell Biochem. 2009;329:17–33. doi: 10.1007/s11010-009-0121-6. [DOI] [PubMed] [Google Scholar]
- 28.Sturtevant AH. A highly specific complementary lethal system in Drosophila melanogaster . Genetics. 1956;41:118–123. doi: 10.1093/genetics/41.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dearolf CR, Hersperger E, Shearn A. Developmental consequences of awdb3, a cell-autonomous lethal mutation of Drosophila induced by hybrid dysgenesis. Dev Biol. 1988;129:159–168. doi: 10.1016/0012-1606(88)90170-4. [DOI] [PubMed] [Google Scholar]
- 30.Dearolf CR, Tripoulas N, Biggs J, Shearn A. Molecular consequences of awdb3, a cell-autonomous lethal mutation of Drosophila induced by hybrid dysgenesis. Dev Biol. 1988;129:169–178. doi: 10.1016/0012-1606(88)90171-6. [DOI] [PubMed] [Google Scholar]
- 31.Rosengard AM, Krutzsch HC, Shearn A, et al. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 32.Nickerson JA, Wells WW. The microtubule-associated nucleoside diphosphate kinase. J Biol Chem. 1984;259:11297–11304. [PubMed] [Google Scholar]
- 33.Biggs J, Hersperger E, Steeg PS, et al. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell. 1990;63:933–940. doi: 10.1016/0092-8674(90)90496-2. [DOI] [PubMed] [Google Scholar]
- 34.Wallet V, Mutzel R, Troll H, et al. Dictyostelium nucleoside diphosphate kinase highly homologous to Nm23 and Awd proteins involved in mammalian tumor metastasis and Drosophila development. J Natl Cancer Inst. 1990;82:1199–1202. doi: 10.1093/jnci/82.14.1199. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Liu LZ, Deng XF, et al. The enzymatic activity of Drosophila AWD/NDP kinase is necessary but not sufficient for its biological function. Dev Biol. 1996;177:544–557. [PubMed] [Google Scholar]
- 36.Lascu I, Chaffotte A, Limbourg-Bouchon B, Véron M. A Pro/Ser substitution in nucleoside diphosphate kinase of Drosophila melanogaster (mutation killer of prune) affects stability but not catalytic efficiency of the enzyme. J Biol Chem. 1992;267:12775–12781. [PubMed] [Google Scholar]
- 37.Timmons L, Xu J, Hersperger G, et al. Point mutations in awdKpn which revert the prune/Killer of prune lethal interaction affect conserved residues that are involved in nucleoside diphosphate kinase substrate binding and catalysis. J Biol Chem. 1995;270:23021–23030. doi: 10.1074/jbc.270.39.23021. [DOI] [PubMed] [Google Scholar]
- 38.Reymond A, Volorio S, Merla G, et al. Evidence for interaction between human PRUNE and nm23-H1 NDPKinase. Oncogene. 1999;18:7244–7252. doi: 10.1038/sj.onc.1203140. [DOI] [PubMed] [Google Scholar]
- 39.D’Angelo A, Garzia L, André A, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5:137–149. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 40.Krishnan KS, Rikhy R, Rao S, et al. Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron. 2001;30:197–210. doi: 10.1016/s0896-6273(01)00273-2. [DOI] [PubMed] [Google Scholar]
- 41.Boissan M, Montagnac G, Shen Q, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344:1510–1515. doi: 10.1126/science.1253768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adryan B, Decker HJ, Papas TS, Hsu T. Tracheal development and the von Hippel-Lindau tumor suppressor homolog in Drosophila . Oncogene. 2000;19:2803–2811. doi: 10.1038/sj.onc.1203611. [DOI] [PubMed] [Google Scholar]
- 43.Kaelin WG. Treatment of kidney cancer: insights provided by the VHL tumor-suppressor protein. Cancer. 2009;115:2262–2272. doi: 10.1002/cncr.24232. [DOI] [PubMed] [Google Scholar]
- 44.Affolter M, Caussinus E. Tracheal branching morphogenesis in Drosophila: new insights into cell behaviour and organ architecture. Development. 2008;135:2055–2064. doi: 10.1242/dev.014498. [DOI] [PubMed] [Google Scholar]
- 45.Schottenfeld J, Song Y, Ghabrial AS. Tube continued: morphogenesis of the Drosophila tracheal system. Curr Opin Cell Biol. 2010;22:633–639. doi: 10.1016/j.ceb.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsouna A, Nallamothu G, Kose N, et al. Drosophila von Hippel-Lindau tumor suppressor gene function in epithelial tubule morphogenesis. Mol Cell Biol. 2010;30:3779–3794. doi: 10.1128/MCB.01578-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu T. Complex cellular functions of the von Hippel-Lindau tumor suppressor gene: insights from model organisms. Oncogene. 2012;31:2247–2257. doi: 10.1038/onc.2011.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dammai V, Adryan B, Lavenburg KR, Hsu T. Drosophila awd, the homolog of human nm23, regulates FGF receptor levels and functions synergistically with shi/dynamin during tracheal development. Genes Dev. 2003;17:2812–2824. doi: 10.1101/gad.1096903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol. 2012;13:631–645. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell GP, Thompson BJ. Colorectal cancer progression: lessons from Drosophila? Semin Cell Dev Biol. 2014 doi: 10.1016/j.semcdb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Nallamothu G, Woolworth JA, Dammai V, Hsu T. Awd, the homolog of metastasis suppressor gene Nm23, regulates Drosophila epithelial cell invasion. Mol Cell Biol. 2008;28:1964–1973. doi: 10.1128/MCB.01743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duchek P, Somogyi K, Jékely G, et al. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- 53.McDonald JA, Pinheiro EM, Montell DJ. PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development. 2003;130:3469–3478. doi: 10.1242/dev.00574. [DOI] [PubMed] [Google Scholar]
- 54.Silver DL, Geisbrecht ER, Montell DJ. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila . Development. 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]
- 55.Woolworth JA, Nallamothu G, Hsu T. The Drosophila metastasis suppressor gene Nm23 homolog, awd, regulates epithelial integrity during oogenesis. Mol Cell Biol. 2009;29:4679–4690. doi: 10.1128/MCB.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leibfried A, Fricke R, Morgan MJ, et al. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr Biol. 2008;18:1639–1648. doi: 10.1016/j.cub.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 57.Georgiou M, Marinari E, Burden J, Baum B. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr Biol. 2008;18:1631–1638. doi: 10.1016/j.cub.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 58.Ignesti M, Barraco M, Nallamothu G, et al. Notch signaling during development requires the function of awd, the Drosophila homolog of human metastasis suppressor gene Nm23. BMC Biol. 2014;12:12. doi: 10.1186/1741-7007-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masoudi N, Fancsalszky L, Pourkarimi E, et al. The NM23-H1/H2 homolog NDK-1 is required for full activation of Ras signaling in C. elegans . Development. 2013;140:3486–3495. doi: 10.1242/dev.094011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sternberg PW. Vulval development. WormBook. 2005 doi: 10.1895/wormbook.1.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sundaram MV. Canonical RTK-Ras-ERK signaling and related alternative pathways. WormBook. 2013 doi: 10.1895/wormbook.1.80.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenmann DM, Kim SK. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics. 2000;156:1097–1116. doi: 10.1093/genetics/156.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui M, Han M. Cis regulatory requirements for vulval cell-specific expression of the Caenorhabditis elegans fibroblast growth factor gene egl-17. Dev Biol. 2003;257:104–116. doi: 10.1016/s0012-1606(03)00033-2. [DOI] [PubMed] [Google Scholar]
- 64.Ohmachi M, Rocheleau CE, Church D, et al. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr Biol. 2002;12:427–433. doi: 10.1016/s0960-9822(02)00690-5. [DOI] [PubMed] [Google Scholar]
- 65.Hartsough MT, Morrison DK, Salerno M, et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 66.Tso PH, Wang Y, Yung LY, et al. RGS19 inhibits Ras signaling through Nm23H1/2-mediated phosphorylation of the kinase suppressor of Ras. Cell Signal. 2013;25:1064–1074. doi: 10.1016/j.cellsig.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 67.Boissan M, De Wever O, Lizarraga F, et al. Implication of metastasis suppressor NM23-H1 in maintaining adherens junctions and limiting the invasive potential of human cancer cells. Cancer Res. 2010;70:7710–7722. doi: 10.1158/0008-5472.CAN-10-1887. [DOI] [PubMed] [Google Scholar]
- 68.Yoder JH, Chong H, Guan K-L, Han M. Modulation of KSR activity in Caenorhabditis elegans by Zn ions, PAR-1 kinase and PP2A phosphatase. EMBO J. 2004;23:111–119. doi: 10.1038/sj.emboj.7600025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takács-Vellai K. The metastasis suppressor Nm23 as a modulator of Ras/ERK signaling. J Mol Signal. 2014;9:4. doi: 10.1186/1750-2187-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brennan DF, Dar AC, Hertz NT, et al. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 71.Fancsalszky L, Monostori E, Farkas Z, et al. NDK-1, the homolog of NM23-H1/H2 regulates cell migration and apoptotic engulfment in C. elegans . PLoS ONE. 2014;9:e92687. doi: 10.1371/journal.pone.0092687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lehmann R. Cell migration in invertebrates: clues from border and distal tip cells. Curr Opin Genet Dev. 2001;11:457–463. doi: 10.1016/s0959-437x(00)00217-3. [DOI] [PubMed] [Google Scholar]
- 73.Meighan CM, Schwarzbauer JE. Control of C. elegans hermaphrodite gonad size and shape by vab-3/Pax6-mediated regulation of integrin receptors. Genes Dev. 2007;21:1615–1620. doi: 10.1101/gad.1534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hurwitz ME, Vanderzalm PJ, Bloom L, et al. Abl kinase inhibits the engulfment of apoptotic [corrected] cells in Caenorhabditis elegans . PLoS Biol. 2009;7:e99. doi: 10.1371/journal.pbio.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reddien PW, Horvitz HR. The engulfment process of programmed cell death in Caenorhabditis elegans . Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- 76.Wu YC, Horvitz HR. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392:501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- 77.Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans . Nat Cell Biol. 2000;2:131–136. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- 78.Hsu T-Y, Wu Y-C. Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr Biol. 2010;20:477–486. doi: 10.1016/j.cub.2010.01.062. [DOI] [PubMed] [Google Scholar]
- 79.Gartner A, Boag PR, Blackwell TK. Germline survival and apoptosis. WormBook. 2008 doi: 10.1895/wormbook.1.145.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leitch AE, Haslett C, Rossi AG. Cyclin-dependent kinase inhibitor drugs as potential novel anti-inflammatory and pro-resolution agents. Br J Pharmacol. 2009;158:1004–1016. doi: 10.1111/j.1476-5381.2009.00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Love NK, Keshavan N, Lewis R, et al. A nutrient-sensitive restriction point is active during retinal progenitor cell differentiation. Development. 2014;141:697–706. doi: 10.1242/dev.103978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pacal M, Bremner R. Induction of the ganglion cell differentiation program in human retinal progenitors before cell cycle exit. Dev Dyn. 2014;243:712–729. doi: 10.1002/dvdy.24103. [DOI] [PubMed] [Google Scholar]
- 83.Bandura JL, Jiang H, Nickerson DW, Edgar BA. The molecular chaperone Hsp90 is required for cell cycle exit in Drosophila melanogaster . PLoS Genet. 2013;9:e1003835. doi: 10.1371/journal.pgen.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prasov L, Glaser T. Pushing the envelope of retinal ganglion cell genesis: context dependent function of Math5 (Atoh7) Dev Biol. 2012;368:214–230. doi: 10.1016/j.ydbio.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bilitou A, Ohnuma S. The role of cell cycle in retinal development: cyclin-dependent kinase inhibitors co-ordinate cell-cycle inhibition, cell-fate determination and differentiation in the developing retina. Dev Dyn. 2010;239:727–736. doi: 10.1002/dvdy.22223. [DOI] [PubMed] [Google Scholar]
- 86.Daniels M, Dhokia V, Richard-Parpaillon L, Ohnuma S-I. Identification of Xenopus cyclin-dependent kinase inhibitors, p16Xic2 and p17Xic3. Gene. 2004;342:41–47. doi: 10.1016/j.gene.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 87.Bilitou A, De Marco N, Bello AM, et al. Spatial and temporal expressions of prune reveal a role in Müller gliogenesis during Xenopus retinal development. Gene. 2012;509:93–103. doi: 10.1016/j.gene.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 88.Annesley SJ, Bago R, Bosnar MH, et al. Dictyostelium discoideum nucleoside diphosphate kinase C plays a negative regulatory role in phagocytosis, macropinocytosis and exocytosis. PLoS ONE. 2011;6:e26024. doi: 10.1371/journal.pone.0026024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Annesley SJ, Bago R, Mehta A, Fisher PR. A genetic interaction between NDPK and AMPK in Dictyostelium discoideum that affects motility, growth and development. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:341–349. doi: 10.1007/s00210-011-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Annesley SJ, Fisher PR. Dictyostelium discoideum—a model for many reasons. Mol Cell Biochem. 2009;329:73–91. doi: 10.1007/s11010-009-0111-8. [DOI] [PubMed] [Google Scholar]
- 91.Massé K, Dabernat S, Bourbon P-M, et al. Characterization of the nm23-M2, nm23-M3 and nm23-M4 mouse genes: comparison with their human orthologs. Gene. 2002;296:87–97. doi: 10.1016/s0378-1119(02)00836-3. [DOI] [PubMed] [Google Scholar]
- 92.Boissan M, Lacombe M-L. Learning about the functions of NME/NM23: lessons from knockout mice to silencing strategies. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:421–431. doi: 10.1007/s00210-011-0649-3. [DOI] [PubMed] [Google Scholar]
- 93.Arnaud-Dabernat S, Bourbon PM, Dierich A, et al. Knockout mice as model systems for studying nm23/NDP kinase gene functions. Application to the nm23-M1 gene. J Bioenerg Biomembr. 2003;35:19–30. doi: 10.1023/a:1023561821551. [DOI] [PubMed] [Google Scholar]
- 94.Di L, Srivastava S, Zhdanova O, et al. Nucleoside diphosphate kinase B knock-out mice have impaired activation of the K+ channel KCa3.1, resulting in defective T cell activation. J Biol Chem. 2010;285:38765–38771. doi: 10.1074/jbc.M110.168070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Postel EH, Wohlman I, Zou X, et al. Targeted deletion of Nm23/nucleoside diphosphate kinase A and B reveals their requirement for definitive erythropoiesis in the mouse embryo. Dev Dyn. 2009;238:775–787. doi: 10.1002/dvdy.21887. [DOI] [PubMed] [Google Scholar]
- 96.Vogel P, Read R, Hansen GM, et al. Situs inversus in Dpcd/Poll−/−, Nme7−/−, and Pkd1l1−/− mice. Vet Pathol. 2010;47:120–131. doi: 10.1177/0300985809353553. [DOI] [PubMed] [Google Scholar]
- 97.Deplagne C, Peuchant E, Moranvillier I, et al. The anti-metastatic nm23-1 gene is needed for the final step of mammary duct maturation of the mouse nipple. PLoS ONE. 2011;6:e18645. doi: 10.1371/journal.pone.0018645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Srivastava S, Li Z, Ko K, et al. Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells. Mol Cell. 2006;24:665–675. doi: 10.1016/j.molcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 99.Turner KL, Honasoge A, Robert SM, et al. A proinvasive role for the Ca(2+)-activated K(+) channel KCa3.1 in malignant glioma. Glia. 2014;62:971–981. doi: 10.1002/glia.22655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cuello F, Schulze RA, Heemeyer F, et al. Activation of heterotrimeric G proteins by a high energy phosphate transfer via nucleoside diphosphate kinase (NDPK) B and Gbeta subunits. Complex formation of NDPK B with Gbeta gamma dimers and phosphorylation of His-266 IN Gbeta. J Biol Chem. 2003;278:7220–7226. doi: 10.1074/jbc.M210304200. [DOI] [PubMed] [Google Scholar]
- 101.Hippe H-J, Lutz S, Cuello F, et al. Activation of heterotrimeric G proteins by a high energy phosphate transfer via nucleoside diphosphate kinase (NDPK) B and Gbeta subunits. Specific activation of Gsalpha by an NDPK B.Gbetagamma complex in H10 cells. J Biol Chem. 2003;278:7227–7233. doi: 10.1074/jbc.M210305200. [DOI] [PubMed] [Google Scholar]
- 102.Hippe H-J, Luedde M, Lutz S, et al. Regulation of cardiac cAMP synthesis and contractility by nucleoside diphosphate kinase B/G protein beta gamma dimer complexes. Circ Res. 2007;100:1191–1199. doi: 10.1161/01.RES.0000264058.28808.cc. [DOI] [PubMed] [Google Scholar]
- 103.Hippe H-J, Wolf NM, Abu-Taha HI, et al. Nucleoside diphosphate kinase B is required for the formation of heterotrimeric G protein containing caveolae. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:461–472. doi: 10.1007/s00210-011-0618-x. [DOI] [PubMed] [Google Scholar]
- 104.Hippe H-J, Wolf NM, Abu-Taha I, et al. The interaction of nucleoside diphosphate kinase B with Gbetagamma dimers controls heterotrimeric G protein function. Proc Natl Acad Sci USA. 2009;106:16269–16274. doi: 10.1073/pnas.0901679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feng Y, Gross S, Wolf NM, et al. Nucleoside diphosphate kinase B regulates angiogenesis through modulation of vascular endothelial growth factor receptor type 2 and endothelial adherens junction proteins. Arterioscler Thromb Vasc Biol. 2014;34:2292–2300. doi: 10.1161/ATVBAHA.114.304239. [DOI] [PubMed] [Google Scholar]
- 106.Munier A, Serres C, Kann M-L, et al. Nm23/NDP kinases in human male germ cells: role in spermiogenesis and sperm motility? Exp Cell Res. 2003;289:295–306. doi: 10.1016/s0014-4827(03)00268-4. [DOI] [PubMed] [Google Scholar]
- 107.Vogel P, Hansen G, Fontenot G, Read R. Tubulin tyrosine ligase-like 1 deficiency results in chronic rhinosinusitis and abnormal development of spermatid flagella in mice. Vet Pathol. 2010;47:703–712. doi: 10.1177/0300985810363485. [DOI] [PubMed] [Google Scholar]
- 108.Vogel P, Read RW, Hansen GM, et al. Congenital hydrocephalus in genetically engineered mice. Vet Pathol. 2012;49:166–181. doi: 10.1177/0300985811415708. [DOI] [PubMed] [Google Scholar]
- 109.Cai X, Srivastava S, Surindran S, et al. Regulation of the epithelial Ca2+ channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol Biol Cell. 2014;25:1244–1250. doi: 10.1091/mbc.E13-04-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.King JD, Lee J, Riemen CE, et al. Role of binding and nucleoside diphosphate kinase A in the regulation of the cystic fibrosis transmembrane conductance regulator by AMP-activated protein kinase. J Biol Chem. 2012;287:33389–33400. doi: 10.1074/jbc.M112.396036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kunzelmann K, Mehta A. CFTR: a hub for kinases and crosstalk of cAMP and Ca2+ . FEBS J. 2013;280:4417–4429. doi: 10.1111/febs.12457. [DOI] [PubMed] [Google Scholar]
- 113.Chang CL, Zhu XX, Thoraval DH, et al. Nm23-H1 mutation in neuroblastoma. Nature. 1994;370:335–336. doi: 10.1038/370335a0. [DOI] [PubMed] [Google Scholar]
- 114.Georgescauld F, Moynié L, Habersetzer J, et al. Intersubunit ionic interactions stabilize the nucleoside diphosphate kinase of Mycobacterium tuberculosis . PLoS ONE. 2013;8:e57867. doi: 10.1371/journal.pone.0057867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Giraud M-F, Georgescauld F, Lascu I, Dautant A. Crystal structures of S120G mutant and wild type of human nucleoside diphosphate kinase A in complex with ADP. J Bioenerg Biomembr. 2006;38:261–264. doi: 10.1007/s10863-006-9043-0. [DOI] [PubMed] [Google Scholar]
- 116.Onyenwoke RU, Forsberg LJ, Liu L, et al. AMPK directly inhibits NDPK through a phosphoserine switch to maintain cellular homeostasis. Mol Biol Cell. 2012;23:381–389. doi: 10.1091/mbc.E11-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Praefcke GJK, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 118.Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 120.Lu N, Zhou Z. Membrane trafficking and phagosome maturation during the clearance of apoptotic cells. Int Rev Cell Mol Biol. 2012;293:269–309. doi: 10.1016/B978-0-12-394304-0.00013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu X, Odera S, Chuang C-H, et al. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–757. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 122.He B, Yu X, Margolis M, et al. Live-cell imaging in Caenorhabditis elegans reveals the distinct roles of dynamin self-assembly and guanosine triphosphate hydrolysis in the removal of apoptotic cells. Mol Biol Cell. 2010;21:610–629. doi: 10.1091/mbc.E09-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kinchen JM, Doukoumetzidis K, Almendinger J, et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kitano M, Nakaya M, Nakamura T, et al. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:241–245. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 126.Kaelin WG. The von Hippel-Lindau tumor suppressor protein: roles in cancer and oxygen sensing. Cold Spring Harb Symp Quant Biol. 2005;70:159–166. doi: 10.1101/sqb.2005.70.001. [DOI] [PubMed] [Google Scholar]
- 127.Van Rooijen E, Voest EE, Logister I, et al. von Hippel-Lindau tumor suppressor mutants faithfully model pathological hypoxia-driven angiogenesis and vascular retinopathies in zebrafish. Dis Model Mech. 2010;3:343–353. doi: 10.1242/dmm.004036. [DOI] [PubMed] [Google Scholar]
- 128.Lin C-H, Dammai V, Adryan B, Hsu T. Interaction between Nm23 and the tumor suppressor VHL. Naunyn Schmiedebergs Arch Pharmacol. 2014 doi: 10.1007/s00210-014-1002-4. [DOI] [PubMed] [Google Scholar]