Abstract
Higher eukaryotic organisms have a variety of specific and nonspecific defense mechanisms against viral invaders. In animal cells, viral replication may be limited through the decrease in translation. Some viruses, however, have evolved mechanisms that counteract the response of the host. We report that infection by HIV-1 triggers acute decrease in translation. The human protein kinase GCN2 (eIF2AK4) is activated by phosphorylation upon HIV-1 infection in the hours following infection. Thus, infection by HIV-1 constitutes a stress that leads to the activation of GCN2 with a resulting decrease in protein synthesis. We have shown that GCN2 interacts with HIV-1 integrase (IN). Transfection of IN in amino acid-starved cells, where GCN2 is activated, increases the protein synthesis level. These results point to an as yet unknown role of GCN2 as an early mediator in the cellular response to HIV-1 infection, and suggest that the virus is able to overcome the involvement of GCN2 in the cellular response by eliciting methods to maintain protein synthesis.
Keywords: HIV-1, Integrase, GCN2, Translation
Introduction
Following cell infection by retroviruses, genomic viral RNA is copied by reverse transcriptase (RT) in a double-stranded proviral DNA that is inserted into the host-cell nuclear genome by the retroviral encoded enzyme integrase (IN) [1]. In addition to RT and IN, cellular and viral proteins are associated with proviral DNA in the preintegration complex (PIC). IN is necessary and sufficient in vitro for the first two steps of DNA integration: 3′-end processing, where two nucleotides are removed from each 3′-end of the two viral DNA LTRs; and strand transfer, where each 3′-processed viral DNA end is integrated in the host-cell DNA.
Retroviral replication is a complex process with a network of interactions involving viral and cellular proteins, where protein kinases in particular play an important regulatory role. Specific protein phosphorylation may lead to activation, inactivation or other functional modifications, which are important determinants of virus-host cell inter-regulation [2]. In mammals, four different protein kinases responding to an environmental stress have been described: heme-regulated inhibitor (HRI), double-stranded RNA-dependent protein kinase (PKR), general control non-derepressible-2 (GCN2 or eIF2AK4) and PKR-like endoplasmic reticulum kinase (PERK). These enzymes regulate protein synthesis in response to environmental stress. Each of these kinases is activated upon a specific cellular stress. PKR is activated in the presence of dsRNA upon viral infection, HRI is activated by heme starvation and PERK is sensitive to the presence of mis-folded proteins in the endoplasmic reticulum.
GCN2 was originally characterized in the yeast Saccharomyces cerevisiae as being required for the control of amino acid levels in GCN4 mRNA translation. GCN2 is activated by autophosphorylation in amino acid-starved cells. In these cells, uncharged tRNA accumulate and bind to a region of GCN2 homologous to histidyl-tRNA synthetase (HisRS), located at the C-terminus of the kinase domain [3]. This leads to conformational changes of the kinase to trigger autophosphorylation and increase eIF2 kinase activity. Activated GCN2 phosphorylates the alpha subunit of the translation initiator factor 2 (eIF2α) leading to a decrease in protein translation in response to amino acid starvation. A GCN2 ortholog, also activated under conditions of amino acid starvation, was identified in drosophila and mammals [4–6]. Moreover, the above-mentioned mechanism for transcriptional activation of gene expression is conserved throughout eukaryotic evolution [7, 8].
These four kinases specifically phosphorylate eIF2α on Ser-51. Then eIF2α binds to and inhibits the guanine nucleotide exchange factor eIF2B leading to a reduction in general translation.
The decrease in cellular protein translation limiting viral replication is observed in response to viral infection [11]. Nevertheless, viruses adopt various strategies to overcome this handicap and maintain viral protein translation by limiting eIF2α phosphorylation. Activated PKR exerts a strong antiviral activity on HIV-1 expression in cell culture [9]. However, cellular and viral mechanisms prevent PKR from being activated upon HIV-1 infection, except when viral replication is high [9]. PKR and GCN2 are involved in inhibition of Vesicular Stomatitis Virus (VSV) replication independently of eIF2α phosphorylation [10]. The mammalian GCN2 is activated upon infection by the Sindbis Virus (SV) [11]. The fact that mammalian GCN2 is also activated in response to specific stress signals such as serum deprivation [4], glucose limitation or UV irradiation [12] suggests that GCN2 might be involved in other stress-induced pathways, via the phosphorylation of eIF2α or not.
In this study, we show that in HIV-1 infected cells, RNA silencing of GCN2 leads to an increase in HIV-1 replication, suggesting that GCN2 has a restriction role in HIV-1 infection. Our results show that infection by HIV-1 constitutes a stress that leads to the activation of GCN2 with the resulting decrease in protein synthesis. We previously showed the interaction of GCN2 with HIV-1 integrase (IN) [13]. Addition of IN to GCN2-activated cells restores translation of proteins. We propose that IN interacts with GCN2 by acting as a pseudosubstrate of kinase, thus maintaining a normal cellular and viral protein translation level upon HIV-1 infection.
Viral infection of human cells activates the cellular stress response. In this work we investigated whether the activity of kinases other than PKR involved in environmental stress response was affected by HIV-1 infection. Interrogation of the resource allowing the querying of cellular responses to HIV infection (http://www.peachi.labtelenti.org/, [14]) showed that GCN2 transcription was up-regulated 10–14 h post HIV-1 infection, which corresponds to the integration step of the HIV-1 cycle. Transcription of HRI and PERK was not affected in comparison with the RNA synthesis in non-infected cells. PKR was down-regulated during this period. We decided to study the importance of GCN2 in viral replication.
Materials and methods
Cell culture
Cells and virus production: HIV-1Laï was obtained as described in [15]. HeLa P4 cells [16] used for infection experiments were maintained in DMEM medium (Invitrogen) supplemented with 10 % inactivated fetal calf serum (FCS), 1 mg/ml geneticin (G418, Gibco-BRL). They encode a Tat-inducible β-galactosidase whose expression driven by the HIV-1 LTR is linked to the expression of the viral Tat protein. HeLa P4 infection by HIV-1Laï is then measured by monitoring β-galactosidase activity (4-MUG) [15]. HEK-293T cells were used for overexpression of DDK-GCN2 in immunoprecipitation experiments.
siRNA experiments
GCN2 siRNA or control siRNA (Santa Cruz Biotechnology) were transfected in HeLa P4 cells using the siRNA transfection reagent from SantaCruz Biotechnology. The siRNA used are composed of three different siRNA against GCN2. Cells were plated using 400 μl of DMEM medium/10 % FCS in 48-multiwell plates at 50,000 cells per well. The optimal siRNA amount for GCN2 extinction was determined after extraction of proteins in RIPA buffer (25 mM Tris–HCl (pH 7.6), 150 mM NaCl, 1 % NP-40, 1 % sodium deoxycholate, 0.1 % SDS) and Western blot using anti-GCN2 antibody from Cell Signaling Technology™ according to the manufacturer’s instructions. 2.5 pmol of GCN2 siRNA and control siRNA were used for the silencing experiments. When transfection of 500,000 cells was performed, 20 pmol of siRNA were used.
HIV-1Laï infectivity after siRNA transfection was assayed on HeLa P4 cells. Cells were plated at 50,000 cells per well. When cells reached 60–80 % confluence, transfection of siRNA was performed. HIV-1Laï (M.O.I. = 1) was added to HeLa P4 cells 24 h post-transfection of siRNA for 24 h. After 24 h of infection, the supernatant was discarded and the wells were washed three times with 400 μl of NaCl 0.9 %. Each well was refilled with 400 μl of a reaction buffer containing 50 mM Tris–HCl pH 8.5, 100 mM β-mercaptoethanol, 0.05 % Triton X-100 and 5 mM 4-methylumbelliferyl-B-D-galactoside (4-MUG) (Sigma). After 24 h, the reaction was measured in a fluorescence microplate reader (Cytofluor II, Applied Biosystems) at 360/460 nm Excitation/Emission.
Detection of GCN2 and phosphorylated GCN2
HeLa P4 cells were plated at 10,000 cells/well in 96-well plates in DMEM/10 % FCS/gentamicin. 24 h later, HIV-1Laï was added to the cells (M.O.I = 0.3). Extraction of cellular proteins was performed in RIPA buffer 2, 4, 8, 24, and 48 h post-infection. Irrespective of whether GCN2 was phosphorylated or not, GCN2 was detected by western blot using a GCN2 antibody from Cell Signaling Technology™ according to the manufacturer’s instructions. Phospho-GCN2 was detected by Western blot using a Phospho-GCN2 (Thr898) antibody from Cell Signaling Technology™ according to the manufacturer’s instructions. Actin was revealed using a Sigma anti-actin antibody. Normalisation of samples was done by measure of the ratio Phospho-GCN2/actin and GCN2/actin.
Detection of neo-synthesized proteins
HeLa P4 cells were grown in DMEM medium/10 % FCS in 25 cm2 flasks (500,000 cells) for 24 h. Methionine depletion was performed by incubation of cells for 1 h in DMEM without methionine (Sigma). Azido-homoalanine (L-AHA, Invitrogen) was added to the cells at 50 μM final concentration. Upon addition of L-AHA, cells were either infected with HIV-1Lai particles or transfected with IN (purified as described in [13]) or BSA using the peptide delivery carrier Chariot (Active Motif™) according to the manufacturer’s instructions. Analysis by SDS-PAGE was performed as follows. Labeling of L-AHA incorporated in proteins was done using TAMRA-alkyne according to the manufacturer’s instructions (Click-iT Protein Analysis Detection Kits, Invitrogen) followed by UV-detection (neo-synthesized proteins) and Coomassie Blue staining (input) of the gel. When using flow cytometry, labeling was performed with Alexa Fluor® 647 alkyne (Invitrogen) and cells were analyzed on Fortessa flow cytometer (Becton–Dickinson) (Flow cytometry platform, SFR TransBiomed, Bordeaux Segalen University, France). Normalisation of samples was made by the quantification of total proteins using Coomassie blue staining. Quantification of protein was performed by Image J software.
Two-hybrid experiment
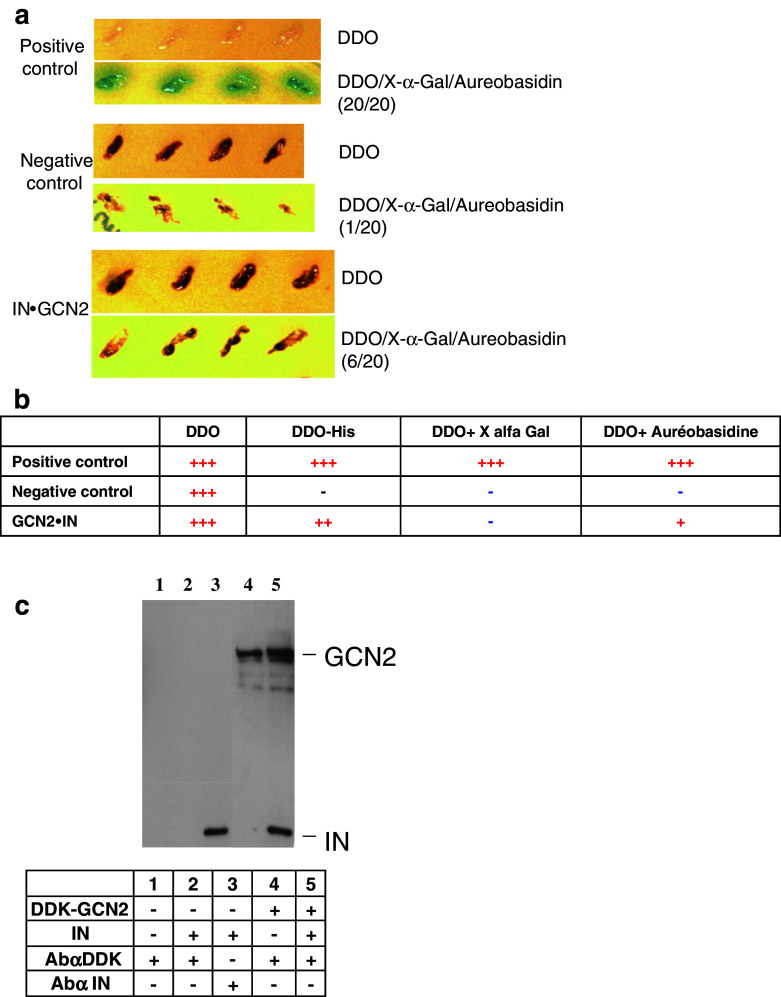
A two-hybrid experiment was performed using the Matchmaker™ Gold Yeast two-hybrid system from Clontech. Three reporter genes were used to detect interaction: HIS3 (yeasts should grow on -His minimal medium), AUR1-C (interaction leads to the synthesis of inositol phosphoryl ceramide synthase and confers strong resistance to the toxic drug Aureobasidin A), and MEL1 (yeast colonies that express Mel1 turn blue in the presence of the chromogenic substrate X-α-Gal). pGBKT7-53 (encodes the Gal4 DNA-BD fused with murine p53) and pGADT7-T (encodes the Gal4 AD fused with SV40 large antigen) are used as positive control since p53 and large T antigen are known to interact in a yeast two-hybrid assay. Negative control is performed using pGBKT7-Lam (encodes the Gal4 BD fused with lamin) and pGADT7-T. No colonies should grow on SD/-Leu/-Trp (DDO) + Aureobasidin.
Plasmids constructions and the two-hybrid experiment were performed as described in [13]. pGBT9.IN (Leu-Trp+) was used as prey and pGAD.GCN2 (Leu + Trp−) as bait. Selection of diploids was performed on SD/-Leu/-Trp (DDO). Selection of interaction was done on SD/-Leu/-Trp/X-α-Gal/AbA (DDO/X/A) for activation of AUT1-C and MEL1 reporters. Selection on SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA (QDO/X/A) was performed for activation of HIS3.
Immunoprecipitation assay
Overexpression of DDK-tagged human GCN2 was performed by transfection of HEK/293T cells (50 000 cells/well, 48 multiwell plates) with a human cDNA clone (1 μg) from OriGene (Rockville, MD, USA) using lipofectamine 2000 (Invitrogen) as the transfection reagent. Cells lysates were prepared 48 h post-transfection in RIPA buffer. Recombinant IN (12 pmol) was added for 6 h. Immunoprecipitation was performed by addition overnight of an anti-DDK monoclonal antibody (OriGene) followed by addition of beads coated with polyclonal sheep anti-mouse antibodies (Dynabeads, Invitrogen) according to the manufacturer’s instructions. Samples were separated by SDS-PAGE and transferred to PVDF membranes before immunodetection of GCN2 and IN. Detection of DDK-GCN2 was performed with anti-DDK antibody according to the manufacturer’s instructions at a 1/1000e dilution. Detection of IN was performed using anti-HIV-1 integrase antibody from Bioproducts MD (dilution 1/2000).
Results
GCN2 is phosphorylated in HIV-1 infected cells
The increase in GCN2 transcription first raises the question whether GCN2 synthesis is increased upon HIV-1 infection. Secondly, to be active, GCN2 needs to be autophosphorylated. Experiments were performed to measure the quantity and the phosphorylation level of GCN2 during viral infection in comparison with uninfected cells.
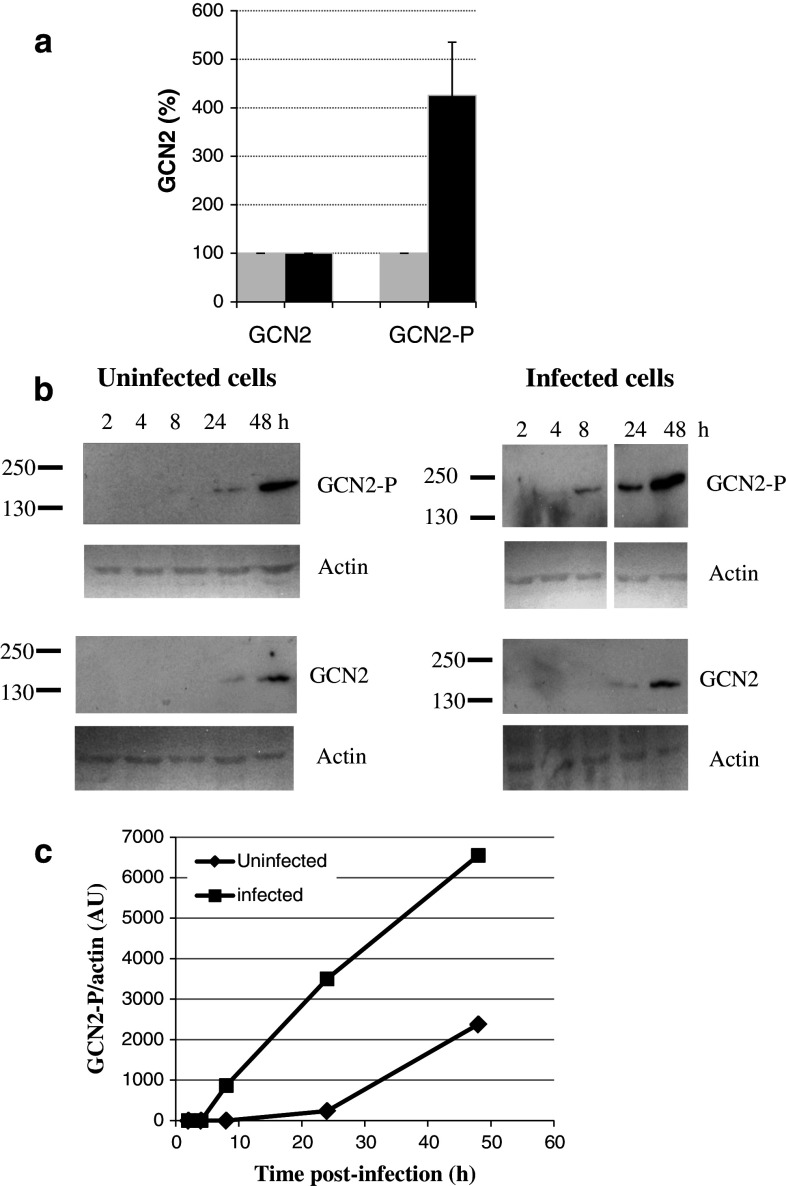
HeLa P4 cells were grown in the absence or presence of HIV-1. Cells were harvested, lysed, and proteins were separated by SDS-PAGE analysis and transferred to nitrocellulose membranes. Immunodetection of GCN2 was performed with anti-GCN2 antibodies independently of the enzyme phosphorylation status (GCN2), while phosphorylated-GCN2 was detected using an antibody that specifically recognizes the phosphorylated T898 of GCN2 (GCN2-P).
When cells were harvested 24 h post-infection, phosphorylation of GCN2 was strongly increased in infected cells when compared with non-infected cells (Fig. 1a), though the level of endogenous GCN2 was the same in infected and non-infected cells.
Fig. 1.
Effect of HIV-1 infection on GCN2 phosphorylation. a Cellular proteins were extracted 24 h after infection (filled black square) or without infection (filled grey square) with HIV-1. Immunodetection of endogenous GCN2 independently of phosphorylation and of phosphorylated GCN2 using antibodies against phosphorylated T898 was performed by Western blot. b Cellular proteins were extracted at several times after infection or without infection and analyzed by Western blot. Phosphorylated GCN2, GCN2, and actin were detected using antibodies against phosphorylated GCN2 (T898), GCN2 and actin respectively. A typical experiment is shown in b. c Ratio phospho-GCN2/actin is shown for infected (filled black square) and uninfected (filled black diamond) cells. Quantification of phosphorylated GCN2 and actin was performed by Image J software. Results are derived from blots performed together and with the same time of exposure. A typical experiment is shown in c
Next, HeLa P4 cells were grown from 0 to 48 h in the absence or the presence of HIV-1 (Fig. 1b). Detection of endogenous GCN2 was performed by western blot. Increasing amounts of GCN2 were detected upon the time. Similar amounts of GCN2 were detected in infected and uninfected cells at 24 and 48 h post-infection. Detection of GCN2-P was performed. Phosphorylation was detected as early as 8 h post-infection, while it was not detected at that time in non-infected cells. Thereafter, GCN2 phosphorylation increased over time in both infected and non-infected cells.
Detection of actin was performed as a loading control. The ratio GCN2/actin was plotted versus the time post infection in infected and non infected cells (Fig. 1c). GCN2 phosphorylation increased over time with GCN2-P level higher in infected cells than in non-infected ones. We conclude that GCN2 is phosphorylated in infected cells, and is potentially active in HIV-1 infected cells. Infection by HIV-1 may constitute a stress leading to early activation of GCN2.
GCN2 silencing increases viral infectivity
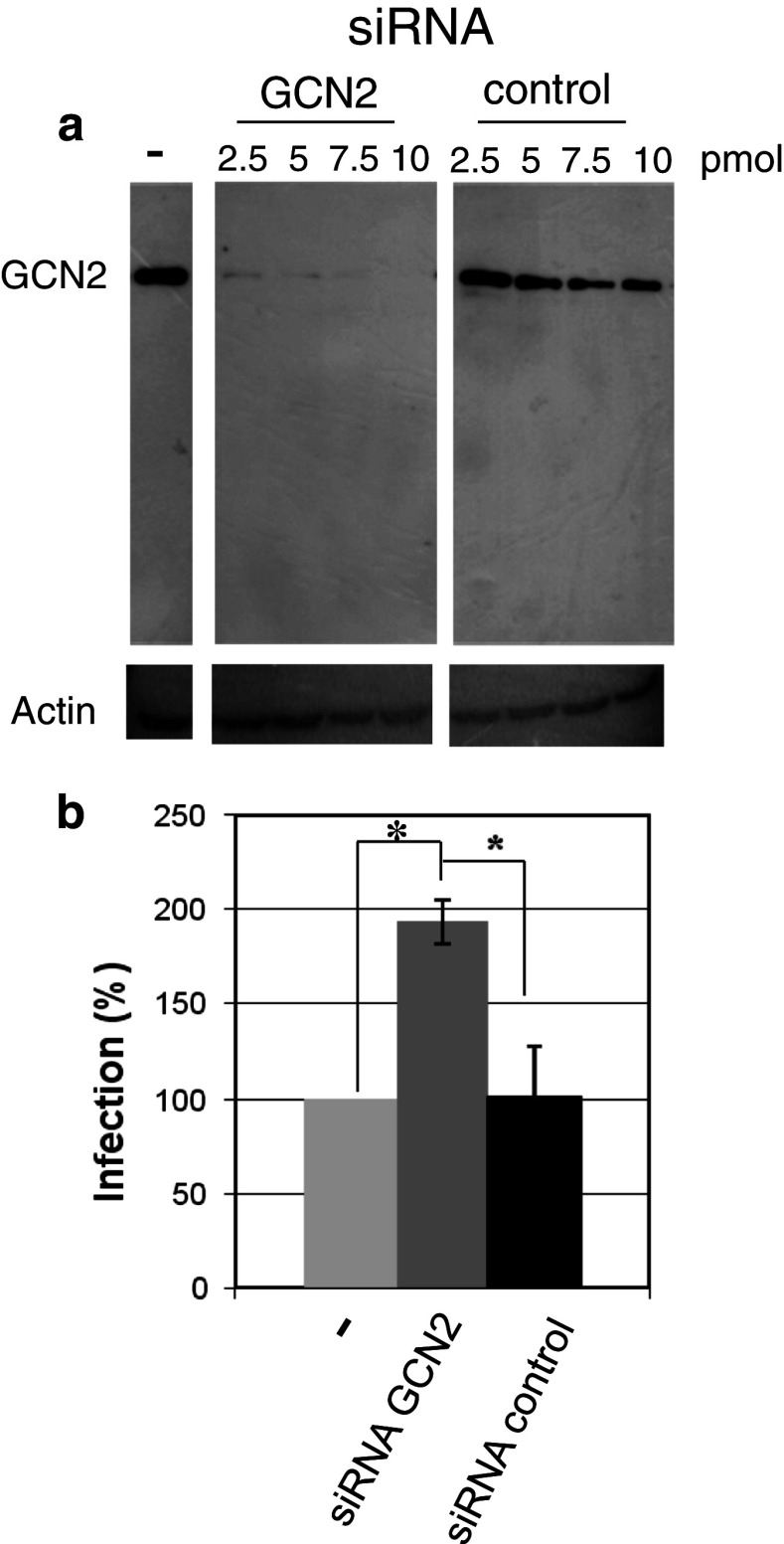
The increase in GCN2 transcription raises the question whether this kinase is involved in viral infectivity. We investigated the effect of GCN2 on HIV-1 infectivity in infected cells where GCN2 expression was decreased by RNA silencing. Within the Western blot limit of detection, GCN2 was strongly decreased 48 h after transfection of siRNA (Fig. 2a) in HeLa P4 cells. To measure the effect of GCN2 silencing on viral infectivity, HIV-1 was added to the cells 24 h post-transfection with the siRNA. Infectivity was measured 24 h post-infection (48 h post-transfection). Viral infectivity was increased in cells where GCN2 expression was silenced, in comparison with infectivity in non-transfected cells or cells transfected with a control siRNA (Fig. 2b). A significant 80 % increase in viral infectivity 24 h post-infection was observed. These results suggest a restriction role of GCN2 on viral infectivity.
Fig. 2.
Effect of GCN2 extinction by siRNA on HIV-1 infectivity. a Hela P4 cells were transfected with increasing quantities of GCN2 siRNA or control siRNA. GCN2 was detected by Western blot 48 h after transfection with anti-GCN2 antibodies. Control of input was performed with anti-actin antibodies. b HeLa P4 cells were transfected with 2.5 pmol of siRNA. Cells were infected with HIV-1 24 h post-transfection. Infectivity was measured 24 h post-infection (48 h post-transfection) after transfection of cells with GCN2 siRNA (dark grey bar), control siRNA (dark bar), or without transfection (light grey bar). P-values (using Student’s test) calculated from three experiments were 0.038 for GCN2 siRNA transfection versus untransfected cells, 0.015 for GCN2 siRNA transfection versus control siRNA, and 0.43 for control siRNA versus untransfected cells
Translation is decreased upon HIV-1 infection
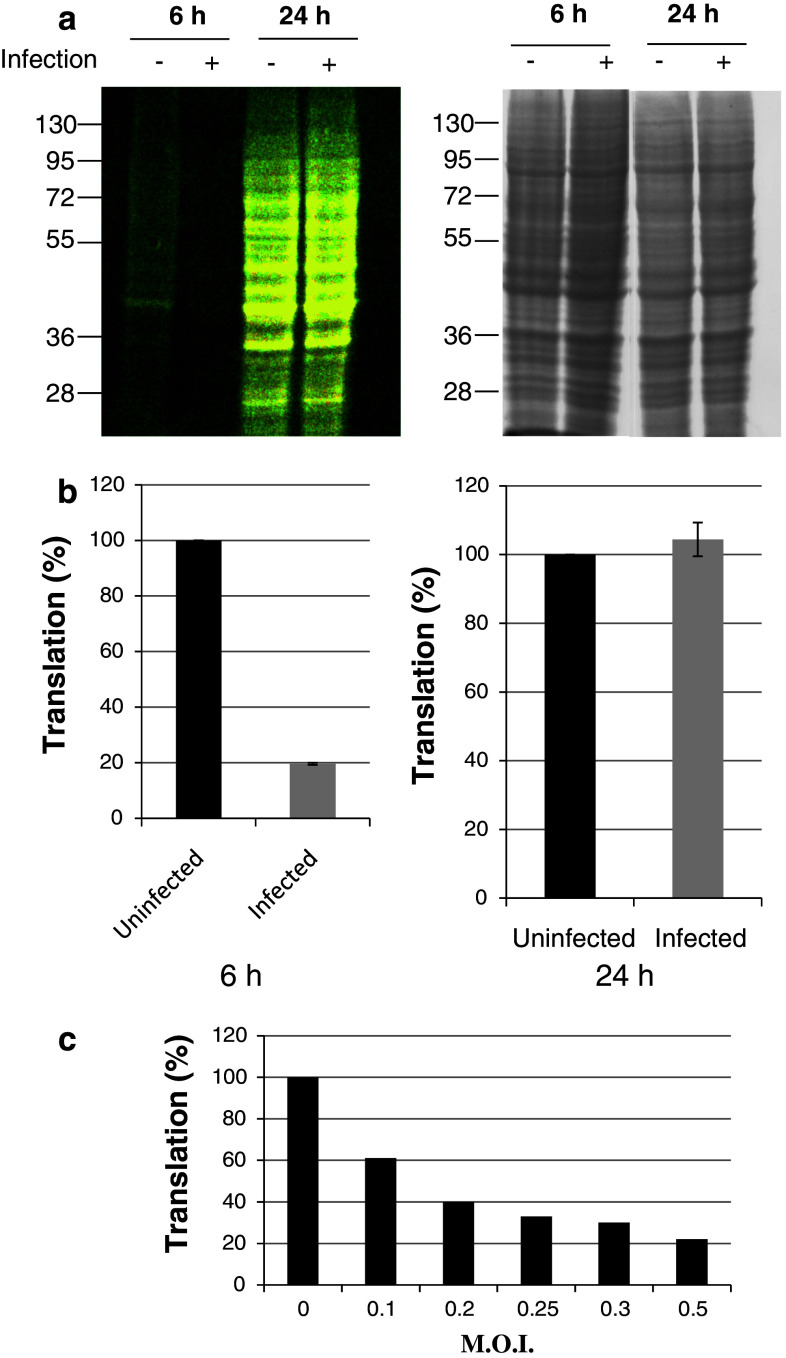
In non-infected cells, GCN2 activation is associated with a decrease in protein synthesis. As shown above, stimulation of GCN2 phosphorylation is observed upon infection of cells by HIV-1. To investigate whether activation of GCN2 following HIV-1 infection is associated with a decrease in translation, protein synthesis was monitored upon infection (Fig. 3). Cells were grown in a complete medium and then incubated in the absence of methionine for 1 h. Incorporation of the amino acid analog azido-homoalanine (L-AHA) into newly synthesized proteins was allowed for 6 and 24 h. After cell lysis, labeling of newly synthesized proteins was performed as described in the methods section with TAMRA alkyne monitored by SDS-PAGE analysis and visualization at 470 nm. When performed, cells were infected at the time of addition of L-AHA for 6 and 24 h. Six hours post-addition of L-AHA, fluorescence associated with protein translation was observed (Fig. 3a, lane 1). As expected, a decrease in translation was observed in the presence of HIV-1 (Fig. 3a, lane 2). Translation of proteins in the presence of HIV-1 24 h post-infection was restored to the level of synthesis observed without infection (Fig. 3a, lane 3 and 4). Equivalent levels of gel staining using Coomassie blue confirmed that similar amounts of protein were present in each lane (Fig 3 right panel).
Fig. 3.
Effect of infection on protein translation. HeLa P4 cells were grown for 1 h in medium without methionine, then azido-homoalanine (L-AHA) was added upon infection with HIV-1Laï (M.O.I = 0.3). Translation was measured 6 and 24 h after infection (+) or without infection (−) as described in methods. Proteins were analyzed by SDS-PAGE. a Left newly synthesized proteins were visualized using Safe Illuminator blue light exposure; right gels were stained with Coomassie blue to verify appropriate protein loading (25 μg). b HeLa P4 cells were grown for 6 and 24 h in presence of L-AHA after infection or without infection. Same protocol as described above in (a) was used but newly synthesized proteins were labeled with Alexa fluor® 647 alkyne instead of TAMRA alkyne and quantification was done by flow cytometry using fluorescence mean. c HeLa P4 cells were infected with HIV-1Lai at several M.O.I. for 4 h. Neo-synthesized proteins level was measured as described in methods after analyze by SDS-PAGE
A similar experiment was performed followed by quantification of translation by flow cytometry instead of SDS-PAGE analysis. For this purpose, incorporation of L-AHA was allowed at the time of infection, and labeling of newly synthesized proteins was obtained using Alexa Fluor 647® alkyne instead of TAMRA alkyne (Fig. 3b). Translation was decreased upon infection by HIV-1 at 6 h post-infection, while at 24 h post-infection the translation level in infected cells was similar to protein synthesis in non-infected cells.
Detection of translation was followed after infection by HIV-1 at different MOI. Cells were infected at different M.O.I. for 4 h, and translation level was analyzed by SDS-PAGE. Quantification performed by densitometry of the gel (Fig. 3c) showed that the highest M.O.I. used (0.3) led to the lowest level of translation. Translation level increased when the M.O.I. decreased, and was correlated to the M.O.I. used.
GCN2 silencing increases the translation level in infected cells
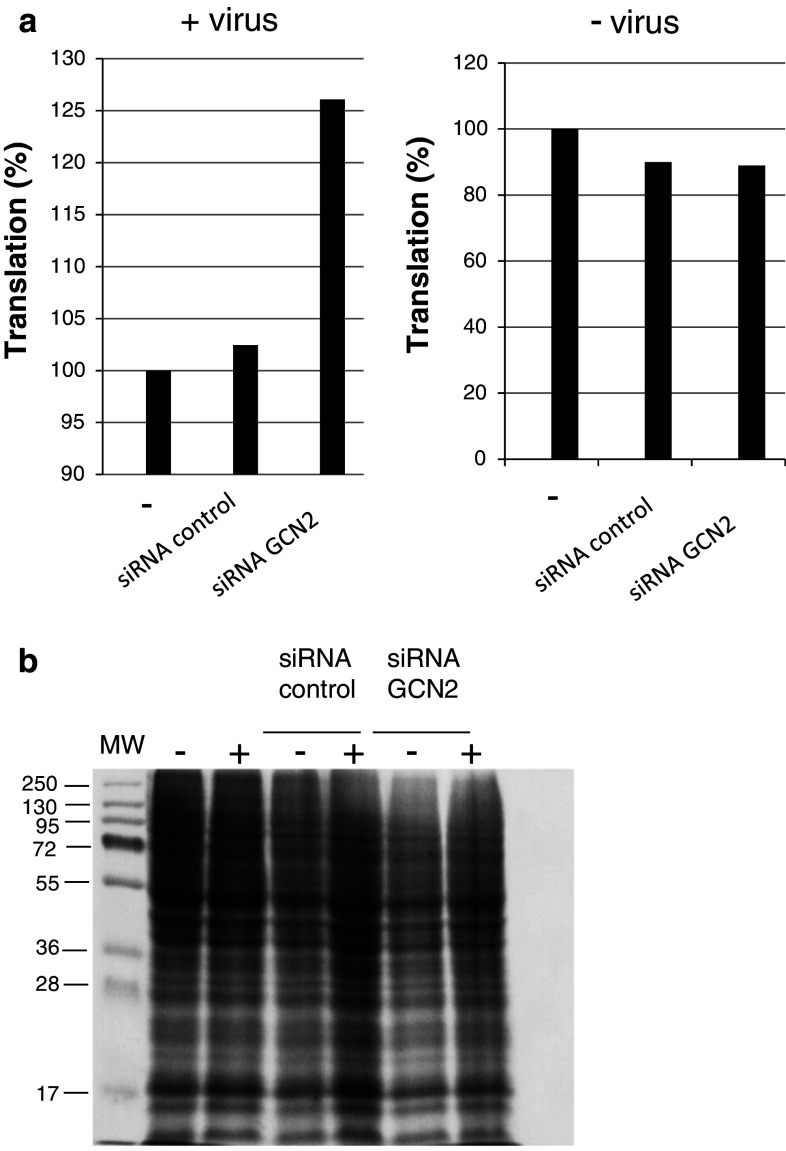
Our results show that GCN2 was phosphorylated upon HIV-1 infection. Second, translation was decreased upon HIV-1 infection. We wondered whether the decrease in translation upon infection was related to GCN2 activity. For this purpose, silencing of GCN2 in HeLa P4 cells was performed as described in materials and methods. 72 h post-transfection, cells were infected with HIV-1 for 4 h and translation was measured. Quantification of proteins performed after SDS-PAGE analysis is shown in Fig. 4.
Fig. 4.
Effect of GCN2 silencing on translation. HeLa P4 cells were transfected with 20 pmol of GCN2 siRNA or control siRNA as described in Methods. Cells were infected with HIV-1Laï (M.O.I. 0.3) 72 h post-transfection for 4 h, and neo-synthesized proteins were labeled as described in Methods. Appropriate protein loading (25 μg) was checked by Coomassie blue. staining of the gels. Translation in infected cells and non-infected cells is shown in (a) Verification of protein loading after Coomassie staining (b) is shown in infected (+) and non-infected (−) cells. MW, molecular weight standards
Protein synthesis in cells infected after silencing of GCN2 was increased by about 25 % in comparison with non-transfected cells (Fig. 4a left). No increase was observed when infection of cells transfected with a control siRNA was performed. In non-infected cells, translation was not affected by transfection of siRNA control or siRNA GCN2 (Fig. 4a right). Coomassie staining (Fig. 4b) confirmed that equivalent quantities of total proteins were loaded on the gel. These results establish a link between the presence of GCN2 and the decrease in translation in the first hour following HIV-1 infection. Moreover, these results are probably underestimated since GCN2 transcription is up-regulated upon HIV-1 infection.
GCN2 interacts with HIV-1 IN
Inhibition of viral expression and replication may be counteracted by mechanisms that involve viral products, cellular factors, or cellular proteins induced by the virus. For example, within the HIV-1 components, the viral trans-activator Tat and large amounts of the viral TAR RNA contribute to PKR inhibition [9].
In a previous study [13], we identified potential partners of HIV-1 IN by screening a yeast genomic library constructed in a two-hybrid vector using HIV-1 IN as bait. Among the candidates selected, interaction of yeast GCN2 with HIV-1 IN was observed by using the two-hybrid approach [13]. On the basis of the role of GCN2 in HIV-1 infection shown above, we wonder if the interaction IN/GCN2 might have a role in HIV-1 infection.
First, we checked the interaction between GCN2 and HIV-1 IN by a two-hybrid experiment using IN this time as a prey and GCN2 (residues 350–500) as bait. Diploids were selected on minimum medium SD/-Leu/-Trp (DDO). One hundred positive diploid clones were then selected and allowed to grow on His-deficient medium, in the presence of the antibiotic aureobasidin (A) or on medium supplemented with X-α-Gal (X). A typical experiment with 20 clones growing on DDO/X/A is presented in Fig. 5a. All positive clones and only one negative clone was able to grow on DDO/X/A. Diploids IN.GCN2 were able to grow on medium containing aureobasidin. Interaction of GCN2 and IN was not observed using MEL1 reporter gene, but was positive on His media (not shown). Results are summarized in Fig. 5b and confirm the interaction between IN and GCN2 protein kinase.
Fig. 5.
Interaction between IN and GCN2. Interaction between GCN2 and HIV-1 IN by two-hybrid experiment using IN as prey and GCN2 as bait was performed as described in Methods. A typical experiment is shown in a for selection on minimal media DDO, or on selective medium DDO/X/A containing aureobasidin and X-α-Gal. b Two-hybrid experiment results on reporter media minus His, plus X-α-Gal, and plus aureobasidin. Minus (−) means no interaction. Plus (+) sign means that an interaction was observed (+++ means strong interaction). c Interaction between IN and GCN2 by co-immunoprecipitation. 12 pmol of recombinant IN were added for 2 h to a cellular lysate of cells overexpressing DDK-tagged human GCN2. Immunoprecipitation of DDK-GCN2 and recovery of the immuno-complex were obtained as described in Methods. Immunodetection of GCN2 and IN was performed with anti-IN antibodies and anti-DDK antibodies. Components added during immunoprecipitation are summarized in the table inset. Lane 1, cell lysate plus anti-DDK antibodies. Lane 2, cell lysate plus IN plus anti-DDK antibodies. Lane 3, cell lysate plus IN plus anti-IN antibodies. Lane 4, cell lysate over-expressing DDK-GCN2 plus anti-DDK antibodies. Lane 5, cell lysate over-expressing DDK-GCN2 plus IN plus anti-DDK antibodies
The next issue we addressed was whether IN was able to interact with human protein kinase GCN2. For this purpose, human HEK-293T cells were transfected with a vector of expression of human GCN2 with a DDK-tag (DDK-GCN2). Lysis of cells was performed 2 days post-transfection and human cell extracts expressing DDK-GCN2 were then incubated for 2 h with purified IN. Immunoprecipitation was performed using an anti-DDK and proteins were analyzed by SDS-PAGE and Western blot (Fig. 5c). Co-immunoprecipitation of IN and GCN2 was observed (Fig. 5c, lane 5). IN was not detected when incubated with cellular lysate in the absence of over-expression of GCN2 after immunoprecipitation with anti-DDK antibodies (Fig. 5c, lane 2). No cellular proteins were detected after incubation with anti-tag antibodies (Fig. 5c, lane 1).
These results confirm the interaction between IN and human GCN2 kinase.
IN restores protein synthesis following amino acid starvation
We observed a decrease in protein translation upon HIV-1 infection. siRNA experiments showed a role of GCN2 in the decrease in translation. As interaction of IN with GCN2 was shown above, we investigated whether the presence of IN in human cells might have an effect on protein translation.
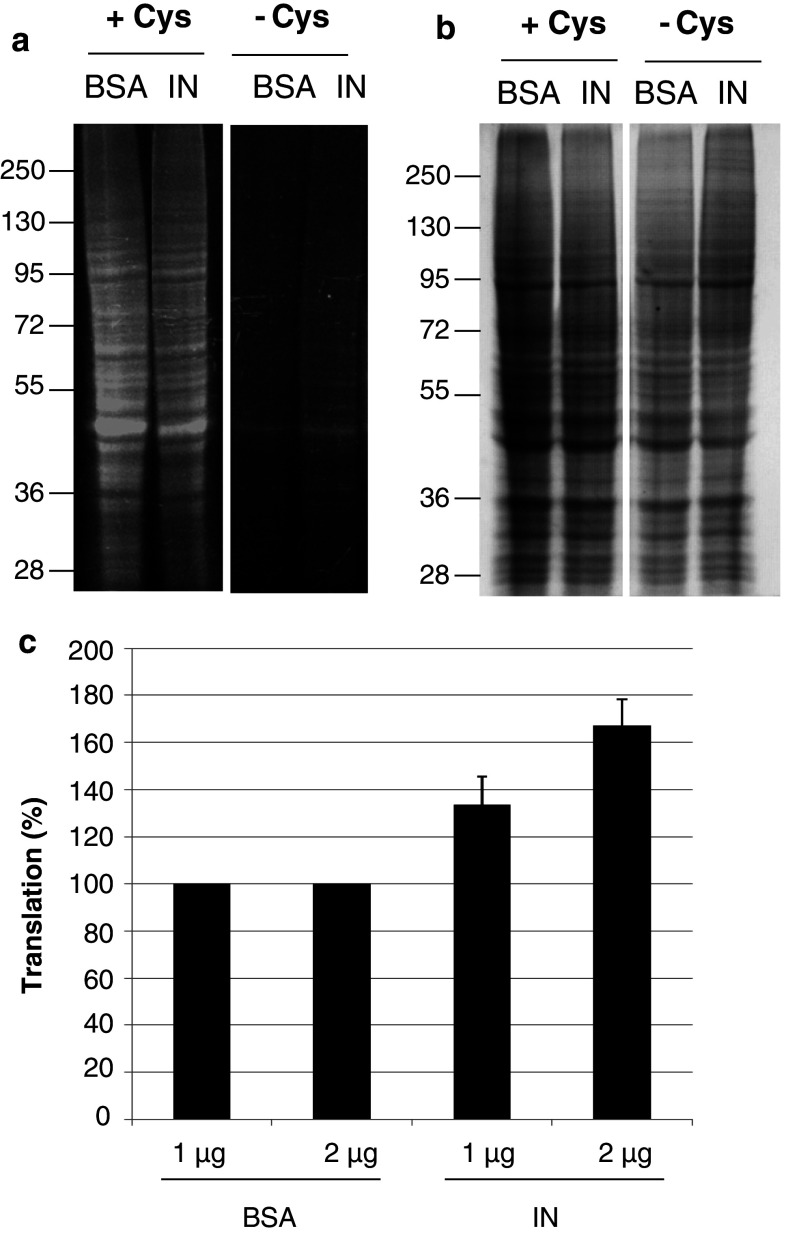
A GCN2 stress-specific decrease in translation by amino acid starvation was induced in HeLa P4 cells (Fig. 6). For this purpose, cells were grown for 1 h in media without cysteine and methionine to induce amino acid starvation and stress-specific GCN2 activation. After addition of L-AHA, cells were grown for 6 h in medium without cysteine and fluorescence was analyzed by SDS-PAGE. Transfection of recombinant IN, or of BSA as control, at the time of addition of L-AHA was performed using a peptide carrier. In parallel, the same experiment was conducted with cells grown in medium plus cysteine (Fig. 6, +cys).
Fig. 6.
Effect of IN on protein translation in amino acid starvation conditions. a Cells were grown in media without methionine, ±cysteine for 1 h. Transfection of 1 μg BSA or IN was performed using Chariot as transfection reagent and neo-synthesized proteins were labeled with TAMRA alkyne. Proteins after SDS-PAGE analysis were visualized under Safe Illuminator blue light exposure. +Cys and −Cys panels were issued from the same exposure of the same gel. b Same gel panels after Coomassie blue staining. c Cells were transfected with 1 and 2 μg of BSA or IN and grown for 6 h in medium plus L-AHA without methionine and with cysteine. Protein labeling was done using Alexa fluor® 647 alkyne and protein translation was monitored by flow cytometry
In cells grown in medium plus cysteine, the protein translation level was similar when BSA or IN were transfected in cells (Fig. 6a, lanes +Cys). This result indicates that IN is not responsible for the activation of GCN2 observed in Fig. 3 in the first hours following HIV-1 infection. Then, cells were grown in cysteine-free medium. As expected, cysteine starvation caused a decrease in protein synthesis (Fig. 6a,−Cys). Interestingly, translation in cells growing in conditions of cysteine starvation was slightly increased when IN was transfected (Fig. 6a,−Cys, IN). Equivalent gel staining confirmed that similar amounts of protein were present in each lane (Fig. 6b).
To measure this effect more precisely, the same experiment was performed and translation was quantified by flow cytometry instead of SDS-PAGE (Fig. 6c). When cells were grown in cysteine-free media, translation was dose-dependent with increasing quantities of IN (Fig. 6c), while transfection of BSA as a control showed no effect.
This suggests that IN can restore protein synthesis following a stress induced by amino acid starvation, which results in GCN2-specific activation.
Discussion
Protein synthesis was decreased in the first hours post-infection by HIV-1 and then the level of neo-synthesized proteins increased. GCN2 expression silencing partly restored the translation in infected cells, thus confirming the participation of GCN2 in the decrease in translation upon HIV-1 infection.
To our knowledge, activation of GCN2 upon infection by HIV-1 has not yet been reported. However, activation of eIF2α phosphorylation upon viral infection is well established in the case of PKR. This kinase is induced by interferon and is activated by double-stranded RNA (dsRNA) during viral infection, as described for alphaviruses [17]. Although PKR activation has not been measured in patients, PKR is transiently activated during HIV-1 infection of lymphatic cell lines [9, 18] depending upon several mechanisms, one of them involving the level of double-stranded TAR RNA element. Yet, while numerous studies have indicated that PKR plays a key role in IFN-mediated host-defense against some viruses [19, 20], the fact that PKR is not essential in the antiviral response suggests that alternative antiviral pathways related to protein phosphorylation exist.
Silencing of GCN2 in HIV-1 infected cells leads to an increase in HIV-1 infectivity. Our results suggest a restrictive role of GCN2 in HIV-1 infectivity. Consistent with this result, a recent work described an increased susceptibility to infection in mice carrying the mutation atchoum (atc) in GCN2, and demonstrates that GCN2 contributes to the translational arrest observed in response to viral infection in vivo [21].
GCN2 is phosphorylated in human cells infected by HIV-1. Therefore, in addition to uncharged tRNAs, GCN2 can be activated by viral factors. Similar results obtained with the Sindbis virus could throw light on the question concerning which factors are involved in such activation. Mammalian GCN2 (but not PKR) is activated in cells upon Sindbis virus (SV) infection [11]. In vitro activation was observed upon binding of two non-adjacent regions of the Sindbis virus (SV) genomic RNA to the histidyl-tRNA synthetase-related domain of the kinase.
Since it is widely accepted that GCN2 phosphorylation decreases the level of translation, we expected that the increase in phosphorylated GCN2 observed upon HIV-1 infection should lead to a decrease in protein synthesis. Decrease in translation upon HIV-1 infection can be rescued by GCN2 silencing, establishing a link between GCN2 and translation decrease in HIV-1 infected cells. The importance of GCN2 in the decrease in translation was probably underestimated in the silencing experiment since GCN2 transcription is itself up-regulated upon HIV-1 infection. Whether the translation of both cellular and viral proteins is decreased upon HIV-1 infection remains unknown. IRES-mediated translation is a great advantage for viruses harboring an IRES, as viral proteins can continue to be generated efficiently during cellular apoptosis or death. While cellular IRES-mediated translation is well documented, few studies have addressed the efficiency of viral IRES-mediated translation under cellular stresses. Licursi and collaborators [22] found that amino acid starvation promoted translation initiated by the IRES elements of encephalomyocarditis virus (EMCV) and foot-and-mouth disease virus (FMDV). In mouse cells, GCN2 inhibits replication of Sindbis virus by blocking early translation of the viral mRNA. However, subgenomic RNA (SV 26S RNA) is translated efficiently even in the presence of phosphorylated eIF2α. A stable RNA hairpin loop located downstream the AUG initiation codon can stall the ribosome on the correct translation initiation site of SV 26S mRNA. Thus, the requirement for the functional eIF2α needed for positioning the initiator Met-tRNA on the ribosome is bypassed [17]. A similar hypothesis can be proposed for HIV-1. Upon HIV-1 infection, activation of GCN2 might take place, leading to a decrease in protein synthesis in infected cells. At this early step, there is no need for HIV-1 to synthesize viral proteins, and reverse transcription catalyzed by RT carried by the infectious virus can synthesize DNA, which in turn is integrated in the host nuclear genome by IN carried in the virion. Nevertheless, after integration, transcription of viral genomic into mRNA must occur, leading to the synthesis of structural proteins and the assembly of new viral particles.
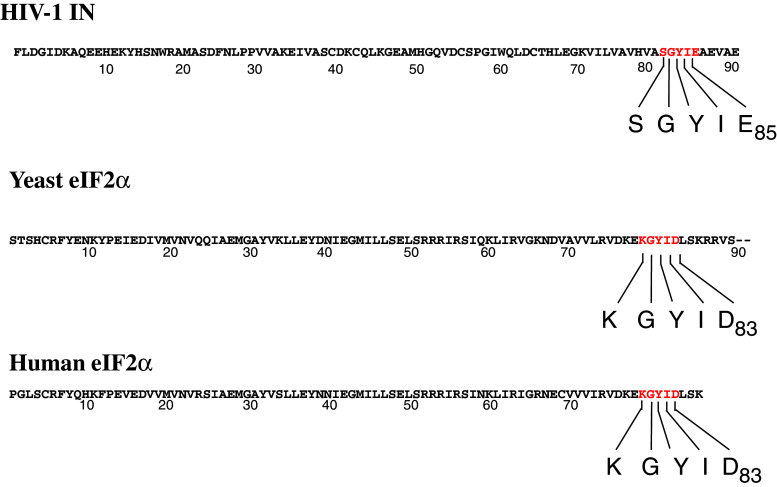
While translation is decreased shortly after infection, the level of protein synthesis 24 h post-infection was the same as in uninfected cells. In cells where translation was decreased after activation of GCN2 by amino acid starvation, addition of IN increased protein synthesis. As HIV-1 IN is able to interact with GCN2, IN could act as a pseudosubstrate interfering with the kinase phosphorylation of eIF2α. In yeast, GCN2 is known to interact with its substrate eIF2α through a KGYIE motif located in the N-terminal region of the elongation factor. This motif is also present in human eIF2α. Comparison of IN and eIF2α sequences showed the presence of a SGYIE motif in the HIV-1 integrase, which could be a site of interaction between IN and GCN2 (Fig. 7). The interaction between IN and GCN2 more than the phosphorylation of IN by the kinase seems to be the key step in the viral infection given that the unphosphorylated IN obtained in bacteria is fully active in the two reactions catalyzed by the enzyme.
Fig. 7.
N-terminal region sequence of HIV-1 IN, yeast eIF2α and human eIF2α. N-terminal region sequence of HIV-1 IN, yeast eIF2α and human eIF2α. Sequences SGYIE and KGYIE are in red
Retroviruses do not possess the necessary components for autonomous translation of viral proteins, so they use several methods to maintain translation using the cell protein synthesis machinery. In addition to their role in viral replication, some viral proteins are involved in such control mechanism(s). After being phosphorylated, the cellular factor eIF2α plays a crucial role in the initiation of protein synthesis. Several viral proteins act as pseudosubstrates for the eIF2α kinases. Thus, protein N of the respiratory syncytial virus binds to PKR kinase and sequesters it away from eIF2α [23]. Upon HIV-1 infection, the retroviral transactivation factor Tat inhibits PKR activity [24]. Expression in yeast of vaccinia protein K3L inhibits yeast GCN2 [25]. The K3L protein shares a KGYID motif with eIF2α, and it is thought to inhibit the eIF2α kinase by interaction with the catalytic domain of the enzyme. Sequence comparison of HIV-1 IN and eIF2α shows a 81SGYIE85 motif in the IN sequence. In IN, two residues (S81 and E85) are different from those described in the eIF2α and K3L sequence. The E83 residue of eIF2α was mutated and all the mutations that were assayed led to a decrease in eIF2α phosphorylation, except when the E residue was mutated in D, which interestingly is the residue found in the IN sequence. This observation strongly suggests that the SGYIE motif could be involved in the interaction between GCN2 and IN.
HIV-1 integrase has been shown recently to be phosphorylated by JNK kinase, an enzyme that is not expressed in resting CD4+ cells [26]. After activation of JNK, integrase is phosphorylated in a conserved serine that becomes a substrate of prolyl-isomerase (Pin1), leading to the stabilization of IN. However, the latter observation involves a very different mechanism to the one described here, which involves GCN2 kinase acting as a restriction factor, and HIV-1. Ligands of IN may become efficient therapeutic agents by preventing the interaction of the retroviral enzyme with GCN2, thus increasing the antiviral cellular response to HIV-1 infection. Thus, we hypothesize that increasing GCN2 activation might increase the efficiency of existing treatments. On the other hand, our findings indicate that the interaction between GCN2 and IN might become a new target in anti-HIV-1 therapy.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and the University Victor Segalen (Bordeaux 2). We thank Prof. R. Cooke (English Department. University Bordeaux 2) for his editorial assistance and Dr. S. Litvak for careful reading of the manuscript and fruitful discussions.
Conflict of interest
Authors declare that they have no conflict of interest.
Footnotes
Ophélie Cosnefroy and Anaïs Jaspart equally contributed to the work.
Sandrine Reigadas and Marie-Line Andréola equally contributed to the work.
Contributor Information
Sandrine Reigadas, Phone: +33-5-56795510, FAX: +33-5-56795673, Email: Sandrine.reigadas@chu-bordeaux.fr.
Marie-Line Andréola, Phone: +33-5-57571740, FAX: +33-5-57571766, Email: marie-line.andreola@reger.u-bordeaux2.fr.
References
- 1.Delelis O, Carayon K, Saib A, Deprez E, Mouscadet JF. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology. 2008;5:114. doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francis AC, Di Primio C, Allouch A, Cereseto A. Role of phosphorylation in the nuclear biology of HIV-1. Curr Med Chem. 2011;18:2904–2912. doi: 10.2174/092986711796150478. [DOI] [PubMed] [Google Scholar]
- 3.Wek RC, Jackson BM, Hinnebusch AG. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci USA. 1989;86:4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 5.Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics. 2000;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsen DS, Jordan B, Chen D, Wek RC, Cavener DR. Isolation of the gene encoding the Drosophila melanogaster homolog of the Saccharomyces cerevisiae GCN2 eIF-2alpha kinase. Genetics. 1998;149:1495–1509. doi: 10.1093/genetics/149.3.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clerzius G, Gelinas JF, Gatignol A. Multiple levels of PKR inhibition during HIV-1 replication. Rev Med Virol. 2011;21:42–53. doi: 10.1002/rmv.674. [DOI] [PubMed] [Google Scholar]
- 10.Krishnamoorthy J, Mounir Z, Raven JF, Koromilas AE. The eIF2alpha kinases inhibit vesicular stomatitis virus replication independently of eIF2alpha phosphorylation. Cell Cycle. 2008;7:2346–2351. doi: 10.4161/cc.6323. [DOI] [PubMed] [Google Scholar]
- 11.Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, et al. Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses. EMBO J. 2006;25:1730–1740. doi: 10.1038/sj.emboj.7601073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, et al. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–1286. doi: 10.1016/S0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- 13.de Soultrait VR, Caumont A, Durrens P, Calmels C, Parissi V, et al. HIV-1 integrase interacts with yeast microtubule-associated proteins. Biochim Biophys Acta. 2002;1575:40–48. doi: 10.1016/S0167-4781(02)00241-5. [DOI] [PubMed] [Google Scholar]
- 14.Mohammadi P, Desfarges S, Bartha I, Joos B, Zangger N et al (2013) 24 Hours in the life of HIV-1 in a T cell line. PLoS Pathog 9(1):e1003161. doi:10.1371/journal.ppat.1003161 [DOI] [PMC free article] [PubMed]
- 15.Ventura M, Tarrago-Litvak L, Dolle V, Nguyen CH, Legraverend M, et al. Effect of nucleoside analogs and non-nucleoside inhibitors of HIV-1 reverse transcriptase on cell-free virions. Arch Virol. 1999;144:513–523. doi: 10.1007/s007050050522. [DOI] [PubMed] [Google Scholar]
- 16.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, et al. HIV-1 reverse transcription. A termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 17.Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, et al. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clerzius G, Gelinas JF, Daher A, Bonnet M, Meurs EF, et al. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol. 2009;83:10119–10128. doi: 10.1128/JVI.02457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, et al. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/S1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 20.Stojdl DF, Abraham N, Knowles S, Marius R, Brasey A, et al. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J Virol. 2000;74:9580–9585. doi: 10.1128/JVI.74.20.9580-9585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Won S, Eidenschenk C, Arnold CN, Siggs OM, Sun L, et al. Increased susceptibility to DNA virus infection in mice with a GCN2 mutation. J Virol. 2011;86:1802–1808. doi: 10.1128/JVI.05660-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licursi M, Komatsu Y, Pongnopparat T, Hirasawa K. Promotion of viral internal ribosomal entry site-mediated translation under amino acid starvation. J Gen Virol. 2012;93:951–962. doi: 10.1099/vir.0.040386-0. [DOI] [PubMed] [Google Scholar]
- 23.Groskreutz DJ, Babor EC, Monick MM, Varga SM, Hunninghake GW. Respiratory syncytial virus limits alpha subunit of eukaryotic translation initiation factor 2 (eIF2alpha) phosphorylation to maintain translation and viral replication. J Biol Chem. 2010;285:24023–24031. doi: 10.1074/jbc.M109.077321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys. 2000;373:361–367. doi: 10.1006/abbi.1999.1583. [DOI] [PubMed] [Google Scholar]
- 25.Qian W, Zhu S, Sobolev AY, Wek RC. Expression of vaccinia virus K3L protein in yeast inhibits eukaryotic initiation factor-2 kinase GCN2 and the general amino acid control pathway. J Biol Chem. 1996;271:13202–13207. doi: 10.1074/jbc.271.44.27402. [DOI] [PubMed] [Google Scholar]
- 26.Manganaro L, Lusic M, Gutierrez MI, Cereseto A, Del Sal G, et al. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat Med. 2010;16:329–333. doi: 10.1038/nm.2102. [DOI] [PubMed] [Google Scholar]