Abstract
Eukaryotic translation initiation factor 3 (eIF3) is a multisubunit complex that plays a central role in translation initiation. We show that fission yeast Sum1, which is structurally related to known eIF3 subunits in other species, is essential for translation initiation, whereas its overexpression results in reduced global translation. Sum1 is associated with the 40S ribosome and interacts stably with Int6, an eIF3 component, in vivo, suggesting that Sum1 is a component of the eIF3 complex. Sum1 is cytoplasmic under normal growth conditions. Surprisingly, Sum1 is rapidly relocalized to cytoplasmic foci after osmotic and thermal stress. Int6 and p116, another putative eIF3 subunit, behave similarly, suggesting that eIF3 is a dynamic complex. These cytoplasmic foci, which additionally comprise eIF4E and RNA components, may function as translation centers during environmental stress. After heat shock, Sum1 additionally colocalizes stably with the 26S proteasome at the nuclear periphery. The relationship between Sum1 and the 26S proteasome was further investigated, and we find cytoplasmic Sum1 localization to be dependent on the 26S proteasome. Furthermore, Sum1 interacts with the Mts2 and Mts4 components of the 26S proteasome. These data indicate a functional link between components of the structurally related eIF3 translation initiation and 26S proteasome complexes.
INTRODUCTION
The initiation of translation is a multistep process involving the binding of Met-tRNA and mRNA to ribosomes and is promoted by a large number of translation initiation factors (eIFs; reviewed by Merrick and Hershey, 1996). Translation initiation factor eIF3 is the largest of the eukaryotic initiation factors and is involved in a number of different aspects of the initiation step. eIF3 forms a stable complex with the 40S ribosomal subunit, which helps prevent premature association with the 60S subunit (Goumans et al., 1980). eIF3 stabilizes the binding of the ternary complex eIF2-GTP-Met-tRNA to the ribosomal 40S subunit (Benne and Hershey, 1978) and functions to bring the 40S subunit to the mRNA through interactions with the eIF4G subunit of the cap binding complex eIF4F (Etchison and Smith, 1990; Lamphear et al., 1995). Additionally, eIF3 has been shown to interact with eIF4B (Méthot et al., 1996), eIF5 (Asano et al., 1999), and eIF1 (Fletcher et al., 1999), suggesting that eIF3 plays a central role in initiation of translation by interacting with different initiation factors (Asano et al., 2000, Phan et al., 2001).
The mammalian eIF3 complex has been purified (Brown-Luedi et al., 1982) and consists of at least 10 subunits, with molecular masses ranging from 35 to 170 kDa (Asano et al., 1997a, 1997b; Méthot et al., 1997). The eIF3 complex from the yeast Saccharomyces cerevisiae has also been purified and can replace mammalian eIF3 in an in vitro assay for initiation, indicating a strong conservation of function in eukaryotes (Naranda et al., 1994). Biochemical interactions have been detected between the conserved p170 (eIF3a), p116 (eIF3b), p110 (eIF3c), p44 (eIF3g), and p36 (eIF3i) subunits in both mammalian and yeast systems (for eIF3 nomenclature, see Browning et al., 2001), indicating that these factors are likely to comprise a conserved core eIF3 complex (Asano et al., 1997a, 1998; Verlhac et al., 1997; Block et al., 1998; Phan et al., 1998). The eIF3 complex from S. cerevisiae contains additional proteins, p135 (TIF31) and p62 (GCD10; Garcia-Barrio et al., 1995; Vornlocher et al., 1999), for which corresponding homologues are not found in mammalian or plant eIF3 (Hershey and Merrick, 2000; Burks et al., 2001). An additional five subunits, p66 (eIF3d), p48 (eIF3e), p47 (eIF3f), p40 (eIF3h), and p28 (eIF3k) have been identified in the mammalian eIF3 complex, which are absent from S. cerevisiae. However, most of these subunits do appear to be highly conserved in Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, and the fission yeast Schizosaccharomyces pombe, suggesting that the eIF3 complex may consist of a highly conserved core domain, together with associated factors that appear to be evolutionarily divergent (Asano et al., 1997c; Phan et al., 1998).
We report here the characterization of the fission yeast sum1+ gene. sum1+ was originally isolated as a high copy suppressor of a class of S-M cell cycle checkpoint mutants in S. pombe and can also inhibit the normal cell cycle response to osmotic stress (Humphrey and Enoch, 1998). Sum1, a WD-repeat protein, shares a predicted 53% amino acid sequence identity to S. cerevisiae eIF3-p39 and a predicted 49% amino acid sequence identity to the human TRIP-1 protein (Humphrey and Enoch, 1998). The S. cerevisiae homologue, eIF3-p39, encoded by the TIF34 gene, is essential for cell viability and for maintaining and stabilizing the eIF3 complex (Naranda et al., 1997). It is also required for both cell cycle progression and mating (Verlhac et al., 1997). Surprisingly, the mammalian homologue, TRIP-1, has been independently identified both as a modulator of TGF-β response (Chen et al., 1995; Choy and Derynck, 1998) and as the p36 subunit of eIF3 (Asano et al., 1997a). We present evidence that Sum1 functions as an essential component of the eIF3 complex in fission yeast. We have examined the response of Sum1 to stress, and find, surprisingly, that it is rapidly relocalized to multifactor complexes in response to both osmotic and thermal stress. Finally, we demonstrate that Sum1 interacts with components of the 26S proteasome complex in vivo. These findings suggest that the eIF3 complex may integrate signals from multiple sources to modulate cell growth.
MATERIALS AND METHODS
Yeast Strains and General Techniques
S. pombe strains used in this study are listed in Table 1. Growth media and general methods for studying fission yeast have been described previously (Moreno et al., 1991). Cells were transformed by electroporation using a Bio-Rad gene pulser (Cambridge, MA). Cells were grown at 30°C in yeast extract medium (YE5S) or in Edinburgh minimum medium (EMM) containing appropriate amino acid supplements, unless otherwise stated. To repress/induce the expression of the nmt1 promoter, cells were grown in the presence/absence of thiamine at 20 μM, as indicated in the text. For viability studies, cells were grown to logarithmic phase in YE5S medium at 30°C, subjected to thermal stress (42°C) for times indicated and plated on YE5S plates. Numbers of colonies were determined after 3 d of incubation at 30°C.
Table 1.
Yeast strains used in this study
| Strain no. | Genotype | Source |
|---|---|---|
| TH9 | 972 h− | Lab stock |
| TH32 | leu1-32 h−pREP3X sum1(LEU2) | Lab stock |
| TH33 | leu1-32 h−pREP81X (LEU2) | Lab stock |
| TH62 | DIPLOID sum1+/sum1∷ura4+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | Lab stock |
| TH477 | sum1∷ura4+ ura4-D18 leu1-32pIRT2-6xHis-Myc-sum1(LEU2) | This study |
| TH496 | sum1∷ura4+ leu1+∷GFP-sum1 ura4-D18 ade− h− | This study |
| TH498 | sum1∷ura4+ leu1:HA-sum1 ura4-D18 ade−h+ | This study |
| TH504 | int6∷int6(HA3) leu1-32 KanMX | Crane et al. (2000) |
| TH505 | int6∷int6-HA3 KanMX sum1∷ura4+ leu1∷GFP-sum1 ade− | This study |
| TH528 | mts4-1 leu1-32 ura4-D18 h− | Wilkinson et al. (1997) |
| TH529 | mts2-1 leu1-32 ura4-D18 h− | Gordon et al. (1993) |
| TH548 | mts4-1 leu1-32 ura4-D18 pREP3X h− | This study |
| TH549 | mts4-1 leu1-32 ura4-D18 pREP3X-sum1 h− | This study |
| TH553 | mts2-1 leu1-32 ura4-D18 pREP3X h− | This study |
| TH554 | mts2-1 leu1-32 ura4-D18 pREP3X-sum1 h− | This study |
| TH580 | int6∷int6(HA3)leu1-32KanMXpIRT2-6xHis-Myc-sum1(LEU2) | This study |
| TH604 | sum1∷ura4+ leu1+∷3xHA-sum1 ura4-D18 ade− | This study |
| TH605 | sum1∷ura4+ leu1:6xHis-Myc-sum1 ura4-D18 ade− | This study |
| TH831 | GFP-Int6 leu1-32 ura4-d18 h− | Bandyopadhyay et al. (2000) |
| TH842 | cut8-563 leu1 h− | Tatebe and Yanagida (2000) |
| TH866 | ade6-D1 leu1-32 ura4-D18 his3-D1 h−pREP3X-FLAG-p116 | Crane et al. (2000) |
| TH917 | cut8-563 leu1 sum1∷ura4+ leu1+∷GFP-sum1 ura4-D18 ade− | This study |
| TH1117 | mts4-1 leu1-32 ura4D18 h-pIRT2GFP-sum1(LEU2) | This study |
| TH1118 | mts2-1 leu1-32 ura4D18 h- pIRT2GFP-sum1(LEU2) | This study |
| TH1119 | pad1-1 leu1-32 ura4D18 h-pIRT2GFP-sum1(LEU2) | This study |
Generation of HA/3xHA/GFP/6xHis-Myc–tagged sum1
Genomic DNA encoding sum1+ was obtained by PCR amplification from S. pombe total DNA using the 5′ oligonucleotide GTAATGGAGGACAGTG and the 3′ oligonucleotide CTGGGGCTGTTCTCC to generate a 2.25-kb product. This fragment was then subcloned into the pCR2.1 vector (Invitrogen, Carlsbad, CA). A BamHI to EcoRV DNA fragment was cloned into the BamHI and SmaI sites of the expression vector pIRT2, to form plasmid pIRT2-sum1. The 5′ oligonucleotide TATCTTTATGATGTTCCTGATTATGCT and the 3′ oligonucleotide AGCATAATCAGGAACATCATAAAGATA were hybridized to generate a blunt end sequence encoding the HA epitope tag. A 97-base pair DNA fragment encoding the epitope 3xHA was amplified by PCR from plasmid pREP41X-3xHA, using the 5′ oligonucleotide TACCCGTACGATGTTCCTG and the 3′ oligonucleotide CATATGAGCGTAATCTGGAACG. GFP was amplified by PCR from plasmid pGEM-GFP (F64L-S65T) using the 5′ oligonucleotide AGTAAAGGAGAAGAACTTTTC and the 3′ oligonucleotide TTTGTATAGTTCATCCATGCC to generate a 714-base pair product. 6xHis-Myc was amplified by PCR from pREP42MH, using the 5′ oligonucleotide GGTAGCAGCCACCATCATC and the 3′ oligonucleotide CATATGTAAGTCCTCCTCGCTG to generate a 112-base pair product. Resulting fragments were cloned into the StuI site located in-frame at the 5′ end of sum1+ to form plasmids pIRT2-HA-sum1, pIRT2-3xHA-sum1, pIRT2-GFP-sum1, and pIRT2-6xHis-Myc-sum1. To test the ability of these strains to functionally complement the sum1::ura4+ disruption, strain TH62 was transformed with these plasmids. Leu+ transformants were selected and sporulated using standard conditions. Leu+ spores were germinated on EMM+A+U plates and replica-plated to EMM+A plates. Only haploid cells transformed with plasmids that can complement the sum1::ura4+ deletion and that can grow in the absence of uracil were selected. To integrate the tagged Sum1 at the leu1 locus, BamHI and SacI DNA fragments of plasmids pIRT2-HA-sum1, pIRT2-3xHA-sum1, pIRT2-GFP-sum1, and pIRT2-6xHis-Myc-sum1 were subcloned into the BamHI and SacI sites of the pJK148 vector (Keeney and Boeke, 1994) to form plasmids pJK148-HA-sum1, pJK148-3xHA-sum1, pJK148-GFP-sum1, and pJK148-6xHis-Myc-sum1. These integrating vectors were linearized with NruI and transformed into TH62. Leu+ stable integrants were selected on EMM+A plates and sporulated using standard conditions. Leu+ spores were germinated and haploids were selected on EMM+A plates. Stable integration of the tagged sum1 gene at the leu1 locus was confirmed by Southern blot analysis. Normal growth and morphology was observed for integrant strains. Multiple independent integrant strains containing 3xHA-Sum1 were all found to be temperature-sensitive at 35.5°C, and this strain (TH604) is subsequently referred to in the text as sum1-ts.
Preparation of Cell Extracts for Analysis of Total Protein
Cells were lysed with glass beads (425–600 μm; Sigma, St. Louis, MO) in a Bio-Savant Fast Prep 120 machine and protein extracts prepared in 2× sample buffer (Laemmli, 1970).
For [35S]methionine and [35S]cysteine incorporation experiments, 100 μCi of Promix l-[35S] in vitro cell labeling mix (Anachem, Luton, Bedfordshire, United Kingdom) was added to 6 × 107 cells, and mixtures were incubated for 20 min at the appropriate temperature. Labeled cells were then harvested by centrifugation, and the pellet was drained well, frozen in liquid nitrogen, and kept at −80°C. Lysates were prepared in 2× sample buffer and analyzed by SDS-PAGE. Gels were stained with Coomassie Blue, dried, and autoradiographed using a PhosphorImager (Bio-Rad).
Nickel-Agarose Affinity Purification and Immunoprecipitation
For nickel-agarose affinity purification, strain TH504 was transformed with pIRT2-6xHis-Myc-sum1 to generate strain TH580, and cells were grown at 30°C in EMM medium to logarithmic phase. Pelleted cells were lysed with glass beads into lysis buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 3% Triton X-100, 10% glycerol, 1 mM PMSF) containing 1× complete protease inhibitors (Roche Laboratories, Nutley, NJ). Strain TH605 and TH9 were grown at 30°C in YE5S medium to logarithmic phase. Pelleted cells were lysed with glass beads into lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol) containing 1× complete protease inhibitors.
Lysates were clarified by centrifugation, and the protein concentration was determined (Bradford, 1976). Proteins were partially purified by affinity chromatography on Ni+-NTA beads (QIAGEN, West Sussex, UK) according to the manufacturer's instructions. Affinity-purified proteins were resuspended in sample buffer and were resolved by SDS-PAGE and immunoblotting.
For immunoprecipitation, cell extracts were prepared as described previously (Moreno et al., 1991) in the following buffer: 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, 10% glycerol containing 1× complete protease inhibitors. Anti-Mts4 polyclonal antiserum, 10 μl, was first incubated with 50 μl of protein A-Sepharose (Sigma). Two milligrams of protein extract was incubated with the serum for 1 h at 4°C. After several washes in the above buffer, the proteins bound to the Mts4 antibodies were released by boiling in sample buffer, and samples were analyzed by SDS-PAGE and immunoblotting.
SDS-PAGE and Immunoblotting
Samples were fractionated by SDS-PAGE and transferred electrophoretically to a nitrocellulose membrane (Millipore, Bedford, MA). Membranes were subsequently immunoblotted with mouse monoclonal Myc, HA, and GFP antibodies at 1/1000 dilution (BabCo, Richmond, CA), or rabbit polyclonal Mts4 antibody at 1/1000 dilution. Detection was performed using peroxidase-conjugated anti-mouse or anti-rabbit IgGs (Amersham, Amersham, UK) and chemiluminescence visualization (ECL; Amersham) according to the manufacturer's instructions.
Polysome Profile Analysis
Polysomes were obtained using a protocol modified from Tzamarias et al. (1989). Cycloheximide, 50 μg/ml, was added to 50 ml of cultures at OD595 of 0.5. Cells were rapidly chilled, washed once in breaking buffer (10 mM Tris-HCl, 100 mM NaCl, 30 mM MgCl2, 50 μg/ml cycloheximide, and 200 μg/ml heparin) and harvested by centrifugation. Cells were lysed with glass beads in 200 μl of breaking buffer. Cell lysates were clarified by centrifugation at 13,000 × g for 2 min in a microfuge. Supernatants were fractionated on 7–47% sucrose gradients (made in 50 mM Tris-acetate, pH 7, 50 mM NH4Cl, and 12 mM MgCl2) for 105 min at 40,000 rpm using a SW40-Ti rotor in a Beckman L70 centrifuge (Fullerton, CA). Polysome profiles were obtained by monitoring the absorbance at 254 nm along the gradient using an LKB 2238 Uvicord SII, and the output was recorded using a Picolog analog-to-digital converter and data logging software (Pico Technology, Cambridge, United Kingdom). Proteins in the polysome profile fractions were concentrated by trichloroacetic acid (TCA) precipitation, and samples were analyzed by SDS-PAGE and immunoblotting.
Cell Staining, Confocal Microscopy, and Immunofluorescence
GFP-Sum1 and GFP-Int6 cells were subjected to stress, as indicated in the text, and subsequently harvested, resuspended in a small volume of YE5S, and observed using a Bio-Rad MRC 600 confocal laser scanning system attached to a Nikon Diaphot inverted microscope (Tokyo, Japan). For DAPI staining, strain TH496 was grown to logarithmic phase, and after exposure to stress, cells were washed in PBS, resuspended in a small volume medium, and stained with DAPI, as described in Moreno et al. (1991). Nuclear staining was observed by confocal microscopy. For DNA/RNA staining, strain TH496 was grown to logarithmic phase, washed, and centrifuged. Cell pellets were resuspended in 1 ml of medium containing 10 μg/ml dihydroethidium (Molecular Probes, Eugene, OR). Cells were then incubated at 30°C for 45–60 min in the dark and washed two or three times in 1 ml of medium. After the final wash, cells were resuspended in a small volume of medium and observed using confocal microscopy.
For immunofluorescence microscopy, cells were grown to logarithmic phase in selective medium and were fixed in 100% methanol for 10 min. The cells were then prepared as described in Hagan and Hyams (1988). Monoclonal mouse anti-FLAG (clone M2; Sigma) and monoclonal mouse anti-eIF4E (a generous gift from J. McCarthy) were used at 1:1000. Polyclonal rabbit anti-Mts4 was used at 1/100 dilution. Detection was performed using Texas Red–conjugated anti-mouse or Cy-3 conjugated anti-rabbit secondary antibodies at a 1/50 dilution. Stained cells were observed by confocal microscopy as described above.
RESULTS
Sum1 Is a Translation Initiation Factor
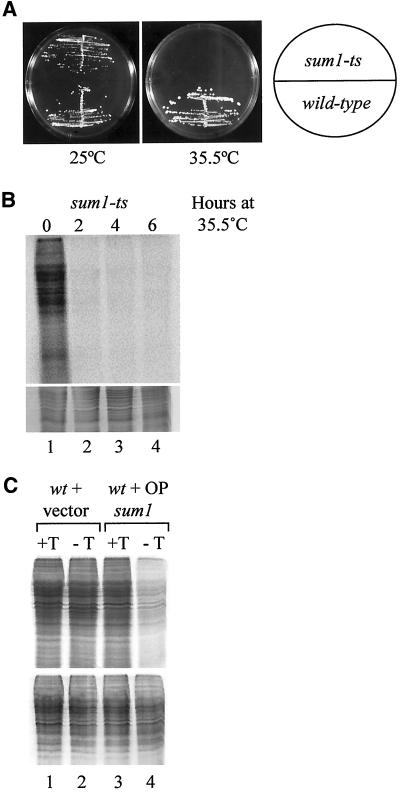
To characterize the role of Sum1 in translation, a temperature-sensitive allele (sum1-ts) was generated (see MATERIALS AND METHODS). The sum1-ts allele supports growth at the permissive temperature of 25°C, but cells fail to form colonies at the restrictive temperature of 35.5°C (Figure 1A). To determine whether Sum1 is required for general translation, sum1-ts cells were exposed to a pulse of [35S]methionine at different times after a shift to the restrictive temperature. The levels of de novo protein synthesis were determined by detection of incorporated [35S]methionine and cysteine, after SDS-PAGE analysis of total protein samples. General protein synthesis was found to be strongly reduced in a sum1-ts strain after 2 h at 35.5°C (Figure 1B, compare lanes 1 and 2). This decrease in protein synthesis was not observed in a wild-type strain where levels of labeled amino acid incorporation remain unchanged after shifting to the restrictive temperature (our unpublished results). These data strongly suggest that Sum1 is essential for general translation of proteins in fission yeast.
Figure 1.
Analysis of radiolabel incorporation in different sum1 alleles. (A) Growth of wild-type (TH9) and sum1-ts (TH604) strains on YE5S at the permissive and restrictive temperatures. (B) [35S]methionine incorporation in sum1-ts mutant at the permissive and restrictive temperatures. Cells were shifted from the permissive temperature (25°C) to the restrictive temperature (35.5°C) for 0, 2, 4, and 6 h (lanes 1–4, respectively). Cells were labeled for 20 min before harvesting. Incorporation was determined by SDS-PAGE analysis (top panel). Coomassie stain of above gel showing total protein levels is presented (bottom panel). (C) [35S]methionine incorporation is reduced in cells overexpressing sum1. Wild-type cells transformed with vector alone (TH33) or pREP3X-sum1 (TH32) were grown in EMM in the presence (+T) or absence (−T) of thiamine for 24 h to derepress the nmt1 promoter. Cells were labeled for 20 min before harvesting. Incorporation was determined by SDS-PAGE analysis (top panel). Coomassie stain of above gel showing total protein levels is presented (bottom panel).
To determine whether sum1 overexpression effects are associated with a role in translation, the effect of sum1 overexpression on protein synthesis was examined. Wild-type cells were transformed with a control plasmid or a plasmid in which the sum1 cDNA is placed under the control of the nmt1 promoter. The nmt1 promoter is repressed in the presence of thiamine, and high expression levels of sum1 cDNA were obtained by derepressing the nmt1 promoter in the absence of thiamine for 24 h. Strains TH32 (wild-type + OP sum1) and TH33 (wild-type + vector; see Table 1) were grown in the presence or absence of thiamine for 24 h. Levels of de novo protein synthesis were determined by SDS-PAGE as described above. As shown in Figure 1C (compare lanes 3 and 4), radiolabel incorporation into newly synthesized proteins was reduced after overexpression of sum1 in the absence of thiamine (−T). The same strains transformed with vector alone showed efficient radiolabel incorporation in the presence or absence of thiamine (Figure 1C, lanes 1 and 2). These results indicate that sum1 overexpression leads to a reduction in translation.
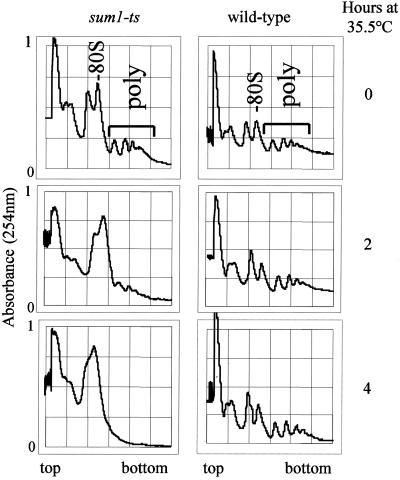
We next wanted to determine whether Sum1 plays a role in translation initiation. To test this possibility, polysome profiles were compared in wild-type and sum1-ts strains at the permissive and restrictive temperatures. At 25°C, an increase in the monosome (80S) to polysome ratio was observed in the sum1-ts strain compared with wild-type cells (Figure 2, top panels). This finding suggests that even at the permissive temperature (25°C), the efficiency of translation initiation is mildly reduced in sum1-ts compared with wild type. After a shift of sum1-ts to 35.5°C for 2 h, a significant increase in the monosome peak was observed, together with a reduction in polysome levels (Figure 2, middle left panel). After incubation of sum1-ts at 35.5°C for 4 h, the monosome peak had accumulated further, and the polysome peaks were completely absent (Figure 2, bottom left panel). In contrast, no significant difference in the wild-type polysome profile was observed under these conditions (Figure 2, right panels). These results indicate an essential role for Sum1 in translation initiation in fission yeast.
Figure 2.
Sum1 is required for translation initiation. Polysome profile analysis of wild-type (TH9) and sum1-ts (TH604) cells. Exponentially growing cultures of TH9 and TH604 in YE5S were incubated at the permissive and restrictive temperatures for the indicated times. Samples were collected, and polysome profile analysis was performed after velocity sedimentation of whole cell extracts on sucrose gradients (7–47%). Fractions were scanned at 254 nm, and absorbance profiles are shown with sedimentation from left (top) to right (bottom). The positions of 80S ribosomes and polysomes are indicated.
Sum1 Is a Component of the eIF3 Complex
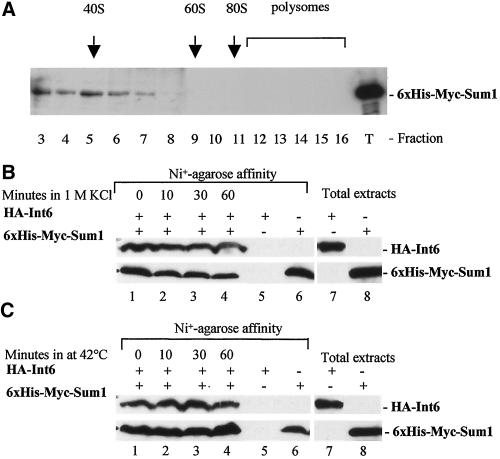
Because components of the eIF3 complex are associated with ribosomal subunits (Benne and Hershey, 1978), the possibility that Sum1 is associated with ribosomal subunits was examined. Extracts from a strain encoding a 6xHis-Myc–tagged Sum1 (TH605) were subjected to sucrose gradient centrifugation. Fractions were collected, precipitated with 10% TCA, and analyzed by SDS-PAGE. From Western blot analysis, the 6xHis-Myc-Sum1 protein (size, 43 kDa) was detected in low-density fractions but was found to be enriched in fraction 5, corresponding to the 40S ribosome fraction (Figure 3A). These results are consistent with the sedimentation patterns observed for the mammalian p36 homolog (Asano et al., 1997a) and fission yeast Int6 eIF3 subunit (Bandyopadhyay et al., 2000,; Crane et al., 2000; Akiyoshi et al., 2001).
Figure 3.
Sum1 is associated with 40S ribosomes and Int6. (A) Sucrose density gradient separation of whole cell extract of S. pombe strain TH605 (6xHis-Myc-Sum1). Whole cell extract was resolved by velocity sedimentation across a 7–47% sucrose density gradient (see MATERIALS AND METHODS). Proteins were concentrated by TCA precipitation and 6xHis-Myc-Sum1 protein detected by Western blotting, using anti-myc antibodies. The positions at which the 40S, 60S, and 80S ribosomes and polysomes migrated are indicated, as determined by monitoring the absorbance profiles at 254 nm across the sucrose gradients during fraction collection. Fraction numbers are presented. Total extract from strain TH605 was loaded as control (T). (B) Cells expressing HA-Int6 and 6xHis-Myc-Sum1 (TH580) were grown to midlog phase at 30°C followed by addition of 1 M KCl for the times indicated. 6xHis-Myc-Sum1 was isolated by affinity purification, as described in MATERIALS AND METHODS. Total extracts (as indicated) or affinity-purified proteins were subjected to Western blotting. HA-Int6 was detected using anti-HA antibody (top panels) or 6xHisMyc-Sum1 detected using anti-Myc antibody (bottom panels) from strain TH580 expressing 6xHis-Myc-Sum1 and HA-Int6 (lanes 1–4), from strain TH504 expressing HA-Int6 only (lane 5), or from strain TH605 expressing 6xHis-Myc-Sum1 only (lane 6). (C) Strain TH580 was grown to midlog phase in EMM at 30°C followed by a shift to 42°C for the times indicated, and affinity purification experiments were performed as outlined above.
To further test the possibility that Sum1 is a component of eIF3, we examined whether Sum1 and Int6 interact in vivo. To this end, a strain (TH580) was constructed that expresses a 6xHis-Myc–tagged Sum1 and HA-tagged Int6. In this strain, 6xHis-Myc-Sum1 is expressed from a plasmid under its own promoter, which can complement a sum1 deletion strain. 6xHis-Myc-Sum1 thus appears to be fully functional. The construct HA-Int6 has been previously described (Crane et al., 2000). 6xHis-Myc–tagged Sum1 was affinity-purified using nickel beads, and copurified proteins were analyzed by Western blots. As shown in Figure 3B, a protein of ∼62 kDa, corresponding to HA-Int6 protein, copurified with 6xHis-Myc-Sum1 (43 kDa) under normal conditions (lane 1). Interactions were confirmed by reciprocal immunoprecipitations using strain TH505, which encodes both GFP-Sum1 and HA-Int6 (Crane et al., 2000). The association of Sum1 with the 40S ribosome, the physical interaction between Sum1 and Int6, together with its sequence similarity to TIF34 strongly suggests that Sum1 is a component of the eIF3 complex in fission yeast.
GFP-Sum1 Protein Relocalizes under Stress Conditions
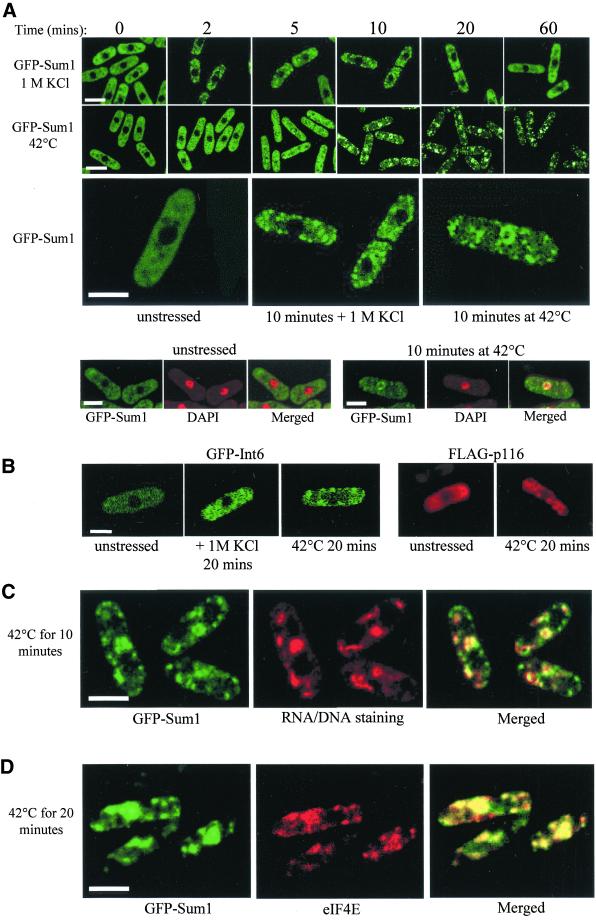
Because Sum1 overexpression leads to significant cell cycle delay after osmotic stress (Humphrey and Enoch, 1998), we examined the regulation of Sum1 under conditions of stress. No changes in Sum1 protein levels were observed under conditions of osmotic, oxidative, or thermal stress, and changes in Sum1 protein mobility were not observed by SDS-PAGE (our unpublished results). The cellular localization of Sum1 protein was additionally examined under stress conditions. Sum1 was tagged at the N-terminus with the green fluorescent protein (GFP-Sum1) and integrated into a sum1::ura4 strain (TH496). The GFP-Sum1 fusion protein expression is under the control of the sum1+ promoter and can functionally complement the sum1 deletion strain. Additionally, GFP-Sum1 can suppress the HU sensitivity of cdc2–3w when overexpressed (our unpublished results). GFP-Sum1 thus appears to be fully functional. Under nonstressed conditions, GFP-Sum1 protein was present in the cytoplasm of the cell and was excluded from the nucleus (Figure 4A, time 0; see also GFP-Sum1 with nuclear [DAPI] staining, bottom panels). This observation is consistent with a role for Sum1 in translation initiation. The smaller unstained areas in the cytoplasm of the cell from which Sum1 is excluded correspond to vacuoles (our unpublished results). Expressing GFP protein alone in fission yeast shows a uniform localization pattern of the protein across the cell (Chen et al., 1999). Because overexpression of sum1 cDNA can inhibit the normal cell cycle response to osmotic stress, GFP-Sum1 localization was examined under conditions of osmotic stress. After exposure of cells to 1 M KCl, GFP-Sum1 protein was observed to rapidly relocalize to cytoplasmic foci (Figure 4A, top panels; see also magnified picture). Similar results were observed after exposure to 1.2 M sorbitol (our unpublished results). The diffuse cytoplasmic localization pattern was resumed after extended periods of exposure to these agents, indicating that cells adapt to osmotic stress. The localization of GFP-Sum1 was further examined under additional forms of stress. GFP-Sum1 was found to rapidly relocalize to cytoplasmic foci after exposure to thermal stress (42°C; Figure 4A, middle panels; see also magnified picture). GFP-Sum1 was additionally present within a nuclear “ring”-shaped structure after 10 min at 42°C, and this localization pattern is sustained at this temperature (Figure 4A, middle panels). Colocalization of GFP-Sum1 with the nuclear stain DAPI revealed a striking yellow ring structure, consistent with GFP-Sum1 localizing to the inner side of the nuclear envelope after heat shock (Figure 4A, bottom panels). Formation of such stress-dependent foci is reversible, because if cells were incubated at 30°C for 1 h after heat shock, GFP-Sum1 became localized within the cytoplasm again (our unpublished results). We further tested whether GFP-Sum1 was relocalized after exposure to hydrogen peroxide, HU, or ionizing radiation; however, GFP-Sum1 relocalization was not observed under these conditions (our unpublished results).
Figure 4.
Analysis of GFP-Sum1 localization under conditions of stress. Strain TH496 was constructed in which a fully complementing GFP-Sum1 fusion protein was integrated into the sum1::ura4 strain. Strain TH496 was grown to midlog phase in YE5S, and GFP-Sum1 was observed by confocal microscopy. (A) Fluorescence images of strain TH496 were obtained, after exposure to 1 M KCl (top panel) and at 42°C (middle panel) for the times indicated. Scale bar, 10 μm. Further magnified fluorescence images of TH496 exposed to the environmental stress indicated are shown. Scale bar, 5 μm. Colocalization of GFP-Sum1 (green) and DAPI staining (red) was examined in strain TH496 grown at 30°C (unstressed) or at 42°C for 10 min, as indicated. Scale bar, 5 μm. (B) Fluorescence images of GFP-Int6 were obtained (left panels) after growth under normal conditions, osmotic stress (1 M KCl for 20 min), or thermal stress (42°C for 20 min). Immunofluorescence images ofFLAG-p116 were obtained (right panels) under growth conditions as described for GFP-Int6. Scale bar, 5 μm. (C) Colocalization of GFP-Sum1 with RNA/DNA stain under thermal stress. Fluorescence images of strain TH496 were obtained during midlog phase growth in YE5S at 30°C, after a shift to 42°C for 10 min, and staining with dihydroethidium (see MATERIALS AND METHODS). Scale bar, 5 μm. (D) Colocalization of GFP-Sum1 with eIF4E under thermal stress. Strain TH496 (GFP-Sum1) was grown to midlog phase in YE5S at 30°C, followed by a shift to 42°C for 20 min. Methanol fixed GFP-Sum1 cells were stained with an anti-eIF4E antibody and the Texas Red–conjugated anti-mouse antibody. Texas Red (eIF4E) and GFP (Sum1) fluorescence was visualized using confocal microscopy. Scale bar, 5 μm.
To determine whether GFP-Sum1 foci were formed as a result of eIF3 relocalization under conditions of osmotic or thermal stress, we tested whether Sum1 was associated with Int6 under these stress conditions. Cells were exposed to osmotic or thermal stress for increasing periods of time, and the ability of 6xHis-Myc-Sum1 to interact specifically with HA-Int6 was determined by affinity purification and Western blot analysis. From these experiments, HA-Int6 was found to be stably associated with 6xHis-Myc-Sum1 under both osmotic stress (1 M KCl; Figure 3B, lanes 1–4) and thermal stress (42°C; Figure 3C, lanes 1–4). These data are consistent with Sum1 relocalizing as part of the eIF3 complex in response to environmental stress. To test this hypothesis further, GFP-Int6 localization was examined under stress conditions. GFP-Int6 was observed to rapidly relocalize to cytoplasmic foci after exposure to osmotic or thermal stress (Figure 4B, GFP-Int6). However, in contrast to Sum1, GFP-Int6 was not tightly associated with a nuclear ring-like structure at 42°C (compare Figure 4A and 4B). The localization of the fission yeast ortholog of p116 (eIF3b) was also examined. p116 was expressed from a plasmid as a FLAG epitope–tagged form (Crane et al., 2000). Immunofluorescence studies using anti-FLAG antibodies, revealed p116 to be cytoplasmic under normal conditions (Figure 4B, FLAG-p116, unstressed) and to rapidly relocalize to cytoplasmic foci after thermal stress (Figure 4B, FLAG-p116, 42°C). FLAG-p116 was not tightly associated with a nuclear ring-like structure at 42°C. These data are consistent with Sum1 relocalizing to cytoplasmic foci as part of a dynamic eIF3 complex during environmental stress.
The finding that components of the eIF3 complex are relocalized to cytoplasmic foci under conditions of stress raised the possibility that additional components of the translational machinery may also be localized to these foci after environmental stress in fission yeast. To test this possibility, colocalization of GFP-Sum1 with rRNA was examined. rRNA is strongly stained by dihydroethidium, an RNA/DNA stain. This cytoplasmic staining pattern is distinct from that observed from the weaker mitochondrial DNA/RNA staining (our unpublished results). After exposure to thermal stress for 10 min, GFP-Sum1 colocalizes with areas of DNA/RNA staining in the cytoplasm in addition to the nuclear periphery (Figure 4C). These data suggest that GFP-Sum1 is associated with rRNA at large cytoplasmic foci after thermal stress. A striking yellow ring structure is also observed within the nucleus (Figure 4C, right panel). The translation initiation subunit eIF4E has been identified as a cap-binding protein in fission yeast (Ptushkina et al., 1996). To determine whether eIF4E was also observed at specific foci after thermal stress, immunofluorescence was performed using anti-eIF4E antibodies. After exposure of TH496 to thermal stress for 20 min, eIF4E was observed to localize to both cytoplasmic and nuclear foci (Figure 4D, middle panel). A merged picture of GFP-Sum1 and eIF4E staining revealed a striking colocalization pattern to both cytoplasmic foci and additionally within the nucleus after thermal stress (Figure 4D, right panel). These data indicate that the GFP-Sum1, together with rRNA and eIF4E colocalize to specific foci under conditions of thermal stress. These data together suggest that the eIF3 complex is rapidly associated with multiple large multifactor complexes after exposure to environmental stress.
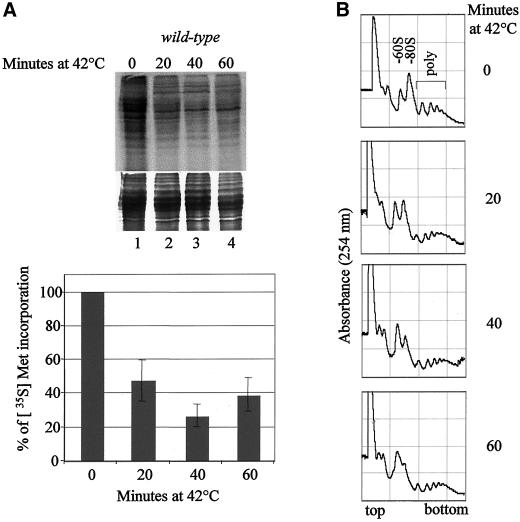
Because Sum1 is essential for translation initiation, we next wanted to determine whether translation was still observed under conditions in which Sum1 is localized to both cytoplasmic and nuclear foci. Wild-type cells were shifted to 42°C for increasing lengths of time, and levels of de novo protein synthesis were determined as described above. After exposure to thermal stress, radiolabel incorporation into newly synthesized proteins was reduced to <50% of that observed in unstressed conditions within 20 min (Figure 5A, compare lanes 1–4). Importantly, a significant degree of radiolabel incorporation was still observed, indicating that protein synthesis still occurs under these conditions. Polysome profiles were also examined from wild-type cells under conditions of thermal stress (Figure 5B). The reduced levels of RNA associated with both monosomes and polysomes, together with an increased 60S ribosomal peak, are consistent with the observation that bulk polyA+ RNA is localized to the nucleus under these conditions (Tani et al., 1996; our unpublished results). These data indicate that protein synthesis still occurs, albeit at reduced levels under conditions in which GFP-Sum1 and additional subunits of translation factors are observed to localize to specific cytoplasmic and nuclear foci. Importantly, no loss of viability is observed under these conditions in either wild-type or in strain TH496, in which GFP-Sum1 performs an essential function (Figure 6C). These data strongly suggest that the cytoplasmic foci described above are likely to function as translation centers under conditions of thermal stress.
Figure 5.
Effect of thermal stress on translation in fission yeast. (A) [35S]methionine incorporation in wild-type strain (TH9) at 42°C for 0, 20, 40, and 60 min (lanes 1–4, respectively). Cells were labeled for 20 min before harvesting. Incorporation was determined by SDS-PAGE analysis (top panel). Coomassie stain of above gel showing total protein levels is presented (bottom panel). Graph represents the quantification of above gel using PhosphorImager. (B) Polysome profile analysis of wild-type (TH9). Exponentially growing cultures of TH9 in YE5S were incubated at 42°C for the indicated times. Samples were collected and polysome profile analysis performed after velocity sedimentation of whole cell extracts on sucrose gradients (7–47%). Fractions were scanned at 254 nm and absorbance profiles are shown with sedimentation from left (top) to right (bottom). The positions of 80S ribosomes and polysomes are indicated.
Figure 6.
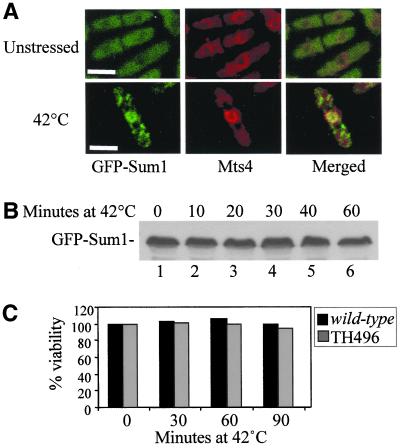
GFP-Sum1 colocalizes stably with Mts4 under thermal stress. (A) Colocalization of GFP-Sum1 and Mts4 under thermal stress. Strain TH496 (GFP-Sum1) was grown to midlog phase in YE5S at 30°C, followed by a shift to 42°C for 20 min. Methanol fixed GFP-Sum1 cells were stained with an anti-Mts4 antibody, and the Cy-5-conjugated anti-rabbit antibody. Cy-5 (Mts4) and GFP (Sum1) fluorescence was visualized using confocal microscopy. Scale bar, 5 μm. (B) Western blot analysis of GFP-Sum1. Strain TH496 (GFP-Sum1) was grown to midlog phase in YE5S at 30°C, followed by a shift to 42°C for the times indicated. Total extracts were fractionated by SDS-PAGE, and GFP-Sum1 was detected using anti-GFP antibody. (C) Viability assay of wild-type and GFP-Sum1 cells. Strains TH9 (wild-type) and TH496 (containing GFP-Sum1) were subjected to heat shock at 42°C for the times indicated, and cells were plated on complete medium to determine the number of viable cells.
Sum1 Interacts with Components of the 26S Proteasome
The 26S proteasome complex is a large multisubunit complex involved in degrading both cytoplasmic and nuclear proteins that have been targeted for destruction by the ubiquitin pathway. The 26S proteasome complex is localized predominantly at the nuclear periphery as a ring shaped structure, both in interphase and throughout mitosis in fission yeast (Wilkinson et al., 1998, 1999). Because GFP-Sum1 was observed to localize as a ring structure to the nuclear periphery under thermal stress (Figure 4, A and C), we further tested the possibility that GFP-Sum1 may colocalize with the 26S proteasome at the nuclear periphery after thermal stress. To this end, colocalization of GFP-Sum1 with Mts4 was examined. Mts4, the fission yeast homolog of RPN1, is a non-ATPase subunit of the 19S regulatory particle of the 26S proteasome (Wilkinson et al., 1997). As shown in Figure 6A, Mts4 is localized at the nuclear periphery at both 25 and 42°C. At 25°C, GFP-Sum1 is cytoplasmic, although some overlap with Mts4 is observed at the nuclear periphery under these conditions. After exposure to thermal stress for 10 min, GFP-Sum1 colocalization with Mts4 staining at the nuclear periphery is observed, as depicted by a strong yellow ring structure (Figure 6A, bottom right panel). These data indicate that GFP-Sum1 colocalizes with the 26S proteasome at 42°C. This finding raised the possibility that GFP-Sum1 was a substrate for the 26S proteasome under these conditions. However, GFP-Sum1 protein remained stable, and no loss of viability of this strain was observed under these conditions (Figure 6, B and C). Therefore, the possibility that Sum1 may interact with components of the proteasome was examined further.
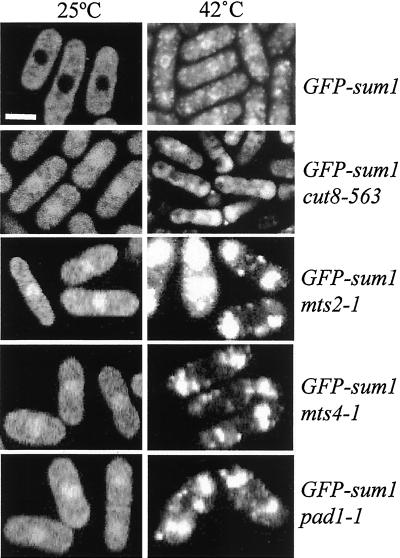
The localization of the 26S proteasome to the nuclear periphery is dependent on Cut8 in fission yeast: Loss of Cut8 results in proteasome subunits becoming enriched within the cytoplasm (Tatebe and Yanagida, 2000). To determine whether the localization of GFP-Sum1 was dependent on the localization of the 26S proteasome, GFP-Sum1 localization was examined in a cut8-563 mutant at both the permissive temperature (25°C) and at the restrictive temperature (36°C). Surprisingly, at the permissive temperature, GFP-Sum1 in a cut8-563 background (TH917) was found to be largely localized within the nucleus compared with the exclusively cytoplasmic localization pattern observed a wild-type background (TH496; Figure 7, left panel, GFP-sum1 cut8-563). This result clearly indicates that Cut8 is required to maintain the cytoplasmic localization of GFP-Sum1. Analysis of GFP-Sum1 (TH496) incubated at the restrictive temperature followed by heat shock at 42°C for 20 min resulted in GFP-Sum1 accumulation at the nuclear periphery (Figure 7, right panel, GFP-sum1). In contrast, a largely diffuse cytoplasmic staining pattern was observed for GFP-Sum1 in a cut8-563 background (TH917) under these conditions (Figure 7, right panel, GFP-sum1 cut8-563). No increase in GFP-Sum1 protein levels was observed in a cut8-563background (our unpublished results). These findings clearly indicate that the localization of GFP-Sum1 is Cut8 dependent, strongly suggesting a link between the localization of Sum1 and that of the 26S proteasome. The relationship between the 26S proteasome and Sum1 localization was tested further by examining GFP-Sum1 localization in temperature-sensitive mutants, which disrupts 26S proteasome function at the restrictive temperature. Localization of GFP-Sum1 was examined in mts4-1, which contains a temperature-sensitive lesion in the essential mts4+ gene. Localization of GFP-Sum1 at 25°C (permissive temperature) in a mts4-1 background (TH1117) was found to be mainly nuclear at 25°C and thus resembled the staining observed in a cut8-563 background (Figure 7, left panel, GFP-sum1 mts4-1). Incubation of this strain at the restrictive temperature followed by heat shock at 42°C for 10 min resulted in cytoplasmic GFP-Sum1 staining, with no staining at the nuclear periphery observed (Figure 7, right panel, GFP-sum1 mts4-1). Mts2 was first isolated as a temperature-sensitive mutant, conferring resistance to the microtubule destabilizing drug methyl 2-benzimidazolecarbamate, and subsequently identified as the S. pombe homologue of the human S4 subunit of the 26S proteasome (Gordon et al., 1993). Nuclear localization of GFP-Sum1 was similarly observed at 25°C in an mts2-1 background (TH1118), and incubation of this strain at the restrictive temperature followed by heat shock at 42°C for 10 min again resulted in cytoplasmic GFP-Sum1 staining pattern, with no accumulation at the nuclear periphery (Figure 7, left and right panel, GFP-sum1 mts2-1). Pad1 is a component of the 26S proteasome, which when overexpressed confers multidrug resistance in fission yeast (Spataro et al., 1997; Penney et al., 1998). Localization of GFP-Sum1 in a pad1-1 background (TH1119) was additionally found to be largely nuclear at 25°C, and incubation of this strain at the restrictive temperature, followed by heat shock at 42°C for 10 min similarly resulted in a cytoplasmic GFP-Sum1 staining pattern only (Figure 7, left and right panel, GFP-sum1 pad1-1). These data indicate that mutation of components of the 26S proteasome result in significant levels of nuclear Sum1 localization, even at their permissive temperature. Therefore, the 26S proteasome is required for the cytoplasmic localization of GFP-Sum1. Importantly, no changes in GFP-Sum1 localization were observed in mutants defective in the nuclear export protein Crm1 (our unpublished results), indicating that control of Sum1 localization by Cut8 is specific and occurs through a Crm1-independent mechanism. No such changes in GFP-Int6 or FLAG-p116 localization were observed in cut8 mutant cells either at the permissive or nonpermissive temperature (our unpublished results).
Figure 7.
Cytoplasmic localization of GFP-Sum1 requires an intact 26S proteasome. Cells from strains TH496 (GFP-Sum1), TH917 (GFP-Sum1, cut8-563), TH1117 (GFP-Sum1, mts4-1), TH1118 (GFP-Sum1, mts2-1), and TH1119 (GFP-Sum1, pad1-1) were visualized after growth to mid log phase at 25°C (left panels) or shifted to 35°C for 1 h followed by a further shift to 42°C for 20 min (right panels). Scale bar, 5 μm.
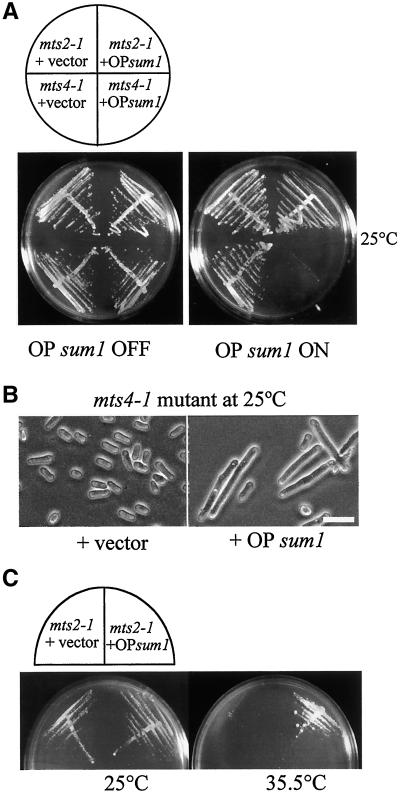
To further examine the relationship between Sum1 and the 26S proteasome, possible genetic interactions were sought between OP Sum1 and temperature-sensitive alleles of components of the proteasome. To identify any genetic interactions between mts4 and sum1, TH528, which contains a temperature-sensitive lesion in the essential mts4+ gene, was transformed with a control plasmid (pREP3X) or a plasmid from which the sum1 cDNA was highly expressed (pREP3X-sum1) to form TH548 and TH549, respectively. Overexpression of sum1 (OP sum1 ON) in mts4-1, at the permissive temperature, resulted in synthetic dosage lethality of mts4 cells (Figure 8A, right panel, lower right quadrant). Normal growth for these strains was observed at the permissive temperature when sum1 expression was repressed (OP sum1 OFF) in the presence of thiamine (Figure 8A, left panel, lower right quadrant). mts4-1 cells overexpressing sum1 were found to be highly elongated compared with wild-type cells and failed to grow even at the permissive temperature of 25°C, indicating a cell cycle arrest (Figure 8B, right panel). This phenotype is distinct from that of mts4-1 at the restrictive temperature, in which cells die with a metaphase arrest phenotype (Wilkinson et al., 1997). These data indicate a genetic interaction between overexpression of sum1 and the mts4 component of the proteasome complex. Mts2, interacts physically with Mts4, and genetic interactions have been identified between mts2− and mts4− (Wilkinson et al., 1997). Genetic interactions between Mts2 and Sum1 were therefore also examined. To this end, strain TH529, which contains a temperature-sensitive mutation in the essential mts2+ gene, was transformed with a control plasmid (pREP3X) or a plasmid from which the sum1 gene was highly expressed (pREP3X-sum1) to form TH553 and TH554, respectively. Expression of sum1 in the presence (OP sum1 OFF) or absence of thiamine (OP sum1 ON) had no effect on the growth of mts2-1 mutants at the permissive temperature (Figures 8A, top quadrants, and 8C, left panel). However, mts2-1 cells failed to form colonies at the restrictive temperature of 35.5°C (Figure 8C, right panel, left quadrant). In contrast, overexpression of sum1 was able to rescue the temperature sensitivity of the mts2-1 mutant cells and allowed them to form colonies at the restrictive temperature of 35.5°C (Figure 8C, right panel, right quadrant). These data clearly indicate strong genetic interactions between Sum1 and the Mts2 and Mts4 components of the 26S proteasome in fission yeast. These interactions are likely to be specific to Mts2 and Mts4 components of the 26S proteasome, because no genetic interactions were observed after overexpression of Sum1 in mts3–1 (our unpublished results).
Figure 8.
Genetic interactions between OP sum1, mts4-1, and mts2-1. (A) Overexpression of sum1 in mts2 and mts4 mutants. mts4-1 (TH528) and mts2-1 (TH529) were transformed with pREP3X (+ vector) or pREP3X-sum1 (+ OP sum1) to form TH548, TH549, TH553 and TH554, respectively. Cells were grown on plates at the permissive temperature of 25°C in the absence (OP sum1 ON) or presence of thiamine (OP sum1 OFF). (B) Overexpression of sum1 in mts4-1 mutant. TH528 was transformed with pREP3X (+ vector) or pREP3X-sum1 (+ OP sum1). Cells were observed on EMM plates at the permissive temperature of 25°C. Scale bar, 10 μm. (C) Overexpression of sum1 in mts2-1 mutant. mts2-1 (TH529) was transformed with pREP3X (+ vector) or pREP3X-sum1 (+ OP sum1). Cells were observed on EMM plates at the permissive (left panel) and restrictive (right panel) temperatures.
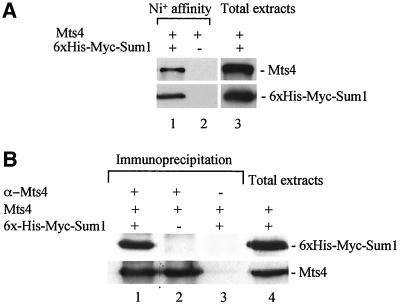
To determine whether Sum1 was physically associated with components of the proteasome in vivo, reciprocal coimmunoprecipitation experiments were performed between Sum1 and Mts4. To this end, a strain (TH605) was used in which an integrated construct encoding 6xHis-Myc–tagged Sum1 functionally replaced the endogenous sum1+ gene. 6xHis-Myc-Sum1 was affinity–purified using nickel beads, and copurified proteins were analyzed by western blots. As shown in Figure 9A, a band corresponding to Mts4 (97 kDa) was copurified with 6xHis-Myc-Sum1 under normal conditions (lane 1). To further test this interaction, reciprocal immunoprecipitations were also performed from strain TH605 using anti-Mts4 antibodies. As seen in Figure 9B, after immunoprecipitation with anti-Mts4 antibodies, a band corresponding to 6xHis-Myc-Sum1 was detected from the above strain, using anti-Myc antibodies (lane 1), but not in strains encoding HA-Sum1 (TH498; lane 2) or mock experiments (lane 3). These data demonstrate a specific physical interaction between Sum1 and Mts4 in vivo under normal growth conditions.
Figure 9.
Biochemical interactions between Sum1 and Mts4 proteins. (A) Copurification of Mts4 with tagged Sum1. Cells expressing 6xHis-Myc-Sum1 (TH605) or wild-type (TH9) were grown to midlog phase in YE5S at 30°C. 6xHis-Myc-Sum1 was isolated by affinity purification, as described in MATERIALS AND METHODS. Total extracts (as indicated) or affinity-purified proteins were subjected to Western blotting and Mts4 protein detected using anti-Mts4 antibody (top panels) or 6xHis-Myc-Sum1 detected using anti-Myc antibody (bottom panels) from either TH605, expressing 6xHis-Myc-Sum1 (lanes 1 and 3) or wild-type cells (TH9; lane 2). (B) Coimmunoprecipitation of 6xHis-Myc-Sum1 with Mts4. Strains were grown to midlog phase at 30°C. Mts4 was isolated by immunoprecipitation, as described in MATERIALS AND METHODS. Total extracts or immunoprecipitated proteins (as indicated) were subjected to Western blotting, Sum1–tagged protein was detected using anti-Myc antibody (top panels), and Mts4 protein was detected using anti-Mts4 antibodies from strain TH605, expressing 6xHis-Myc-Sum1 (lane 1); TH498, expressing HA-Sum1 (lane 2). Mock immunoprecipitation with no Mts4 antibody is shown (lane 3). The presence or absence of Mts4 or 6xHis-Myc-Sum1 in each strain is indicated.
DISCUSSION
Sum1 Is a Component of the eIF3 Translation Initiation Complex
We have identified Sum1 as an essential component of the eIF3 translation initiation complex. This conclusion is based on the following observations: First, Sum1 shares striking sequence similarity to eIF3-p39 (eIF3i), encoded by the TIF34 gene in S. cerevisiae, and TRIP-1 in humans (Chen et al., 1995; Naranda et al., 1997). Second, overexpression of sum1 resulted in reduced levels of global protein synthesis. Third, incubation of a temperature-sensitive allele of sum1 at the restrictive temperature resulted in complete loss of [35S]methionine incorporation, loss of polysomes, and an increased monosome fraction. Fourth, sucrose gradient analysis revealed Sum1 to be associated with 40S ribosomal fractions, as has previously been reported for other eIF3 subunits, including the mammalian p36 (Asano et al., 1997a) and Int6 in fission yeast (Bandyopadhyay et al., 2000; Crane et al., 2000; Akiyoshi et al., 2001). Finally, Myc-tagged Sum1 was found to stably associate in vivo with HA-tagged Int6, a known component of the fission yeast eIF3 complex. The identification of Sum1 as a component of the eIF3 complex indicates that this protein is functionally as well as structurally conserved among eukaryotes. Mammalian Int6/p48, has been demonstrated to interact with components of the eIF3 core (Asano et al., 1997b). The fission yeast Int6 has been reported to interact with the S. pombe ortholog of S. cerevisiae TIF35 (Akiyoshi et al., 2001). In addition, the fission yeast eIF3d/Moe1 and eIF3e/Int6 interact with the eIF3b-p116 subunit of the fission yeast eIF3 complex (Bandyopadhyay et al., 2001). Our data demonstrate a stable interaction in fission yeast between Sum1, a putative core subunit, and fission yeast Int6. From this, it would appear that the association between Int6 and components of the eIF3 core is maintained in fission yeast, suggesting that the general structure of the eIF3 complex is likely to be highly conserved among eukaryotes. Sum1 was originally identified as a high copy suppressor of S-M checkpoint mutants and inhibitor of the normal cell cycle response to osmotic stress (Humphrey and Enoch, 1998). The identification of Sum1 as a core component of the eIF3 complex suggests that the cell cycle response to stress can be translationally modulated in fission yeast.
Relocalization of Sum1 under Stress Conditions
We have examined the localization of Sum1 under conditions of stress. Surprisingly, we observed GFP-Sum1 to be rapidly relocalized to specific foci: After exposure to osmotic stress, GFP-Sum1 relocalizes transiently to multiple cytoplasmic foci. After exposure to thermal stress, GFP-Sum1 is rapidly relocalized and maintained at cytoplasmic foci and is additionally present at the inner nuclear periphery. Our data suggest that Sum1 is rapidly relocalized as part of a dynamic eIF3 complex to cytoplasmic foci in response to environmental stress. Because Sum1 was additionally found to colocalize with an RNA component and with eIF4E during thermal stress, these data strongly suggest that the translational machinery is spatially reorganized to specific foci under these conditions. Moreover, because translation was still observed under these conditions, albeit at reduced levels, these findings raise the possibility that the Sum1-associated foci perform a translational role under conditions of environmental stress. These stress-dependent foci observed in fission yeast resemble stress granules observed in plant and mammalian cells under conditions of environmental stress (Nover et al., 1983; Kedersha et al., 1999). Stress granules are ribonuclear aggregates at which untranslated mRNAs accumulate as a consequence of stress-induced translational arrest and have been proposed to be sites at which untranslated mRNAs are sorted and processed for either reinitiation, degradation, or packaging into stable nonpolysomal mRNP complexes (Nover et al., 1989; Kedersha et al., 1999, 2000). Our findings raise the possibility that the spatial reorganization of the translation machinery to cytoplasmic foci is a common response to environmental stress in all eukaryotes.
Association of Sum1 with Components of the Proteasome
Recent studies indicate that components of the translation initiation complex, eIF3, the COP9 signalosome, and the proteasome complex have similar properties and structures (Asano et al., 1997c; Aravind and Ponting, 1998; Glickman et al., 1998; Hofmann and Bucher, 1998; Wei et al., 1998; Kim et al., 2001). Two structural motifs, the PCI (for Proteasome, COP9, Initiation factor) and MPN (for Mpr1 and Pad1 N terminal) appear to be present exclusively in these multisubunit complexes (Table 2). These findings suggest that these complexes are derived from a common ancestral origin and that components of these complexes may perform overlapping functions. Indeed, associations between subunits of the eIF3 and COP9 signalosome have been reported (Karniol et al., 1998; Yahalom et al., 2001). Although proteins structurally related to Sum1 have not been identified within the 26S proteasome or COP9 complexes, several lines of evidence presented here indicate an interaction between Sum1 and components of the proteasome complex: First, GFP-Sum1 stably colocalized with the Mts4 component of the 26S proteasome at the nuclear periphery following thermal stress. Second, we found GFP-Sum1 localization to be dependent on an intact 26S proteasome: Analysis of GFP-Sum1 in cut8-563, mts4-1, mts2-1, and pad1-1, at the permissive temperature, revealed a high degree of nuclear localization. Moreover, the rapid relocalization of GFP-Sum1 to the nuclear periphery during heat shock was also abolished in these mutants. Third, genetic interactions between Sum1 and mutations in components of the 26S proteasome were observed: OP sum1 exhibited synthetic-dosage lethality with a temperature-sensitive allele of mts4-1, resulting in cell cycle arrest at the permissive temperature. Further, OP sum1 could suppress a temperature-sensitive allele of mts2-1. Finally, copurification and coimmunoprecipitation experiments revealed physical interactions between Sum1 and Mts4 in vivo, under normal growth conditions. This finding is consistent with the overlap observed between GFP-Sum1 and Mts4 staining at the nuclear periphery under normal growth conditions. The finding that Sum1 localization is dependent on an intact 26S proteasome provides a functional insight into the interaction of these two structurally related complexes. However, the precise role of Sum1 within the nucleus under these conditions is unclear. In contrast to Sum1, Int6 and p116 did not localize to the nuclear periphery under stress, and Int6 and p116 localization was not dependent on the 26S proteasome (our unpublished results). Moreover, Int6 does not cosediment with components of the proteasome (Crane et al., 2000). These data suggest that Sum1 interacts with components of the proteasome independently of the eIF3 complex and therefore may perform a distinct function in this context. A role for p36-TIF34, the sum1+ homologue, has been proposed in both assembling and maintaining the eIF3 translation initiation complex in S. cerevisiae (Naranda et al., 1997; Verlhac et al., 1997). Given the structural similarity between components of the eIF3 and the 26S proteasome complexes, Sum1 could potentially function to maintain the stability of the 26S proteasome under stress conditions. Indeed the hypothesis that Sum1 functions to stabilize the 26S proteasome during heat shock is supported by the observation that GFP-Sum1 rapidly localizes to the 26S proteasome after heat shock and additionally from the finding that Sum1 overexpression suppressed the temperature-sensitivity of mts2-1. Alternatively, Sum1 may interact with components of the proteasome to increase turnover efficiency of misfolded proteins produced under heat shock conditions. Our observations also indicate, however, that Sum1 physically interacts with Mts4 under normal conditions. The cell cycle arrest phenotype resulting from sum1 overexpression in mts4-1 suggests the Sum1–Mts4 interaction may perform an essential function in cell cycle control. Alternatively, because Sum1 and Mts4 are required for protein synthesis and proteolytic functions, respectively, it is reasonable to consider the possibility that the Sum1–Mts4 interaction may function in coordinating these processes. Experiments are currently underway to further explore these exciting possibilities.
Table 2.
Mammalian and yeast subunits of the translation initiation complex eIF3
| Human | S. pombe | S. cerevisiae | Structurea |
|---|---|---|---|
| 075153b | SPBC530c | TIF31 | |
| p170 | SPBC17D11c | TIF32/RPG1 | |
| p116 | SPAC25G10c | PRT1/CDC63 | RRM |
| p110 | p110/SPAC4A8c | NIP1 | PCI |
| p66 | Moe1 | Absent | |
| AK000613b | CAB16250c | p62/GCD10 | |
| p48/Int6 | Int6 | Absent | PCI |
| p47 | SPBC4C3c | Absent | MPN |
| p44 | SPBC18H10c | TIF35 | RRM |
| p40 | c821c | Absent | MPN |
| p36/TRIP1 | Sum1 | TIF34 | WD40 |
| p35 | SPAC29B12c | HCR1 |
The comparison between human and S. cerevisiae eIF3 components shown above is based on that described previously in Asano et al. (1997c).
Motifs present in subunits: RRM, ṞNA ṟecognition m̱otif; WD40, WD repeat domains; PCI, P̱roteasome/C̱OP9/I̱nitiation factor; MPN, M̱pr1/P̱ad1 Ṉ-terminal. See text for references.
Accession numbers identified by BLAST searches using the S. cerevisiae sequences.
Cosmids encoding fission yeast homologues.
We have characterized the fission yeast Sum1 protein. We present evidence that it functions as a component of the eIF3 complex, that it rapidly relocalizes to specific foci under stress conditions as part of a multifactor complex, and that it interacts both genetically and biochemically with components of the 26S proteasome. Our findings indicate that Sum1, like its mammalian counterpart, TRIP-1, may perform functions in addition to its translational role as an eIF3 subunit. A diverse array of functions have been associated with other eIF3 subunits or orthologues: The mammalian eIF3-p40 and p48 subunits have been implicated in tumorigenesis (Marchetti et al., 1995; Desbois et al., 1996; Nupponen et al., 1999) and may interact with p110 and a nuclear protein, Rfp (Morris-Desbois et al., 1999). The eIF3/p48 has been recently shown to interact with the human protein HSPC021, which contains motifs found in subunits of the eIF3, 26S proteasome and of the COP9 signalosome (Morris-Desbois et al., 2001). Moreover, eIF3-p48 has recently been demonstrated to interact with P56, a protein induced by interferon and double-stranded RNA, indicating that eIF3 is a target for translational stress response (Guo et al., 2000). Multiple functions have now been ascribed to the fission yeast Int6 homolog, including roles in multidrug resistance, microtubule assembly, chromosome segregation, and spore formation (Bandyopadhyay et al., 2000; Crane et al., 2000; Yen and Chang, 2000; Akiyoshi et al., 2001). Cytoskeletal functions have also been ascribed to the mammalian eIF3-p170, eIF3-p44, and the fission yeast ortholog of p66 (Chen et al., 1999; Hou et al., 2000; Lin et al., 2001; Palecek et al., 2001; Pincheira et al., 2001). Further, the eIF3-p110 subunit is required for nuclear import in S. cerevisiae (Gu et al., 1992). Whether the additional functions associated with these eIF3 subunits are distinct from their role in translation remains to be determined. However, these findings raise the possibility that the eIF3 complex may play a central role in coordinating a number of diverse functions with cell growth. Genes encoding orthologues of most of the subunits of the mammalian eIF3 complex are present in the fission yeast genome (Table 2), suggesting that fission yeast will be a useful model system for studying the mammalian eIF3 complex. The identification of Sum1 as an essential component of the fission yeast eIF3 complex will provide an important molecular tool with which to further characterize the relationships between the functionally diverse subunits of the largest translation initiation factor.
ACKNOWLEDGMENTS
The authors thank S. Townsend at MRC Harwell and N. White, who is funded by the Wellcome Trust, at the Sir William Dunn School of Pathology, University of Oxford, for their expert help in confocal microscopy; J. McCarthy for the generous gift of the eIF4E antibody; and S. Kearsey and A. Pearce for helpful comments on the manuscript. This work was supported by the Medical Research Council.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–06–0301. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–06–0301.
REFERENCES
- Akiyoshi Y, Clayton J, Phan L, Yamamoto M, Hinnebusch AG, Watanabe Y, Asano K. Fission yeast homolog of murine Int-6 protein, encoded by mouse mammary tumor virus integration site, is associated with the conserved core subunits of eukaryotic translation initiation factor 3. J Biol Chem. 2001;276:10056–10062. doi: 10.1074/jbc.M010188200. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Kinzy TG, Merrick WC, Hershey JW. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997a;272:1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- Asano K, Merrick WC, Hershey JW. The translation initiation factor eIF3–p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997b;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- Asano K, Vornlocher HP, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JW. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem. 1997c;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebush AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Matsumoto T, Maitra U. Fission yeast Int6 is not essential for global translation initiation, but deletion of int6+causes hypersensitivity to caffeine and affects spore formation. Mol Biol Cell. 2000;11:4005–4018. doi: 10.1091/mbc.11.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Lakshmanan V, Matsumoto T, Chang EC, Maitra U. Moe1 and spInt6, the fission yeast homologues of mammalian translation initiation factor 3 subunits p66 (eIF3d) and p48 (eIF3e), respectively, are required for stable association of eIF3 subunits. J Biol Chem. 2002;277:2360–2367. doi: 10.1074/jbc.M107790200. [DOI] [PubMed] [Google Scholar]
- Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Block KL, Vornlocher HP, Hershey JW. Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. J Biol Chem. 1998;273:31901–31918. doi: 10.1074/jbc.273.48.31901. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown-Luedi ML, Meyer LJ, Milburn SC, Mo-Ping Yau P, Corbett S, Hershey JWB. Protein synthesis initiation factors from human HeLa cells and rabbit reticulocytes are similar: comparison of protein structure, activities, and immunochemical properties. Biochemistry. 1982;21:4202–4206. doi: 10.1021/bi00261a002. [DOI] [PubMed] [Google Scholar]
- Browning KS, Gallie DR, Hershey JWB, Hinnebusch AG, Maitra U, Merrick WC, Norbury C. Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci. 2001;26:284. doi: 10.1016/s0968-0004(01)01825-4. [DOI] [PubMed] [Google Scholar]
- Burks EA, Bezerra PP, Le H, Gallie DR, Browning KS. Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J Biol Chem. 2001;276:2122–2131. doi: 10.1074/jbc.M007236200. [DOI] [PubMed] [Google Scholar]
- Chen R, Miettinen PJ, Maruoka EM, Choy L, Derynck R. A WD-domain protein that is associated with and phosphorylated by the TGF-β type II receptor. Nature. 1995;377:548–552. doi: 10.1038/377548a0. [DOI] [PubMed] [Google Scholar]
- Chen C-R, Li Y-C, Chen J, Hou M-C, Papadaki P, Chang EC. Moe1, a conserved protein in Schizosaccharomyces pombe, interacts with a Ras effector, Scd1, to affect proper spindle formation. Proc Natl Acad Sci USA. 1999;96:517–522. doi: 10.1073/pnas.96.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy L, Derynck R. The type II transforming growth factor (TGF)-β receptor-interacting protein TRIP-1 acts as a modulator of the TGF-β response. J Biol Chem. 1998;273:31455–31462. doi: 10.1074/jbc.273.47.31455. [DOI] [PubMed] [Google Scholar]
- Crane R, Craig R, Murray R, Dunand-Sauthier I, Humphrey T, Norbury C. A fission yeast homologue of the mammalian oncoprotein and eIF3 subunit Int6 induces drug resistance when overexpressed. Mol Biol Cell. 2000;10:3993–4003. doi: 10.1091/mbc.11.11.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- Etchison D, Smith K. Variations in cap-binding complexes from uninfected and poliovirus-infected HeLa cells. J Biol Chem. 1990;265:7492–7500. [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CU, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999;18:2631–2637. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio MT, Naranda T, Vasquez de Aldana CR, Cuesta R, Hinnebusch AG, Hershey JWB, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Dillon P, Rosen C, Hastie ND. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature. 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- Goumans H, Thomas A, Verhoeven A, Voorma HO, Benne R. The role of eIF-4C in protein synthesis initiation complex formation. Biochim Biophys Acta. 1980;608:39–46. doi: 10.1016/0005-2787(80)90131-8. [DOI] [PubMed] [Google Scholar]
- Gu Z, Moerschell RP, Sherman F, Goldfarb DS. NIP1, a gene required for nuclear transport in yeast. Proc Natl Acad Sci USA. 1992;89:10355–10359. doi: 10.1073/pnas.89.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC. Translational Control of Gene Expression, (ed. Sonnenberg, N.) et al. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2000. Pathway, and mechanism of initiation of protein synthesis; pp. 33–88. [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Hou C-L, Tang C-j C, Roffler SR, Tang TK. Protein 4.1R binding to eIF3–p44 suggests an interaction between the cytoskeletal network and the translation apparatus. Blood. 2000;96:747–753. [PubMed] [Google Scholar]
- Humphrey T, Enoch T. Sum1, a highly conserved WD-repeat protein, suppresses S-M checkpoint mutants and inhibits the osmotic stress cell cycle response in fission yeast. Genetics. 1998;148:1731–1742. doi: 10.1093/genetics/148.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Yahalom A, Kwok S, Matsui M, Deng XW, Chamovitz DA. The Arabidopsis homologue of an eIF3 complex subunit associates with the COP9 complex. FEBS Lett. 1998;439:173–179. doi: 10.1016/s0014-5793(98)01367-2. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Hofmann K, von Arnim AG, Chamovitz DA. PCI complexes: pretty complex interactions in diverse signaling pathways. Trends Plant Sci. 2001;6:379–386. doi: 10.1016/s1360-1385(01)02015-5. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Lin L, Holbro T, Alonso G, Gerosa D, Burger MM. Molecular interaction between human tumor marker protein p150, the largest subunit of eIF3, and intermediate filament protein K7. J Cell Biochem. 2001;80:483–490. doi: 10.1002/1097-4644(20010315)80:4<483::aid-jcb1002>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R. Int6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Hershey JWB. In: Translational Control. Hershey JWB, Mathews MB, Sonenberg N, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. pp. 31–69. [Google Scholar]
- Méthot N, Soo Song M, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méthot N, Rom E, Olsen H, Sonenberg N. The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. J Biol Chem. 1997;272:1110–1116. doi: 10.1074/jbc.272.2.1110. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. In: Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods in Enzymology: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink GR, editors. New York: Academic Press; 1991. pp. 795–823. [DOI] [PubMed] [Google Scholar]
- Morris-Desbois C, Bochard C, Reynaud C, Jalinot P. Interaction between the Ret finger protein and the Int-6 gene product and co-localization into nuclear bodies. J Cell Sci. 1999;112:3331–3342. doi: 10.1242/jcs.112.19.3331. [DOI] [PubMed] [Google Scholar]
- Morris-Desbois C, Rety S, Ferro M, Garin J, Jalinot P. The human protein HSP021 interacts with Int-6 and is associated with eukaryotic translation initiation factor 3. J Biol Chem. 2001;276:45988–45995. doi: 10.1074/jbc.M104966200. [DOI] [PubMed] [Google Scholar]
- Naranda T, MacMillan SE, Hershey JWB. Purified yeast translational initiation factor eIF3 is an RNA binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- Naranda T, Kainuma M, MacMillan SE, Hershey JW. The 39-kilodalton subunit of eukaryotic translation initiation factor 3 is essential for the complex's integrity and for cell viability in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:145–153. doi: 10.1128/mcb.17.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;9:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nupponen NN, Porkka K, Kakkola L, Tanner M, Persson K, Borg A, Isola J, Visakorpi T. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol. 1999;154:1777–1783. doi: 10.1016/S0002-9440(10)65433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek J, Hasek J, Ruis H. Rpg1/Tif32p, a subunit of translation initiation factor 3, interacts with actin-associated protein Sla2p. Biochem Biophys Res Commun. 2001;282:1244–1250. doi: 10.1006/bbrc.2001.4721. [DOI] [PubMed] [Google Scholar]
- Penney M, Wilkinson C, Wallace M, Javerzat JP, Ferrell K, Seeger M, Dubiel W, McKay S, Allshire R, Gordon C. The Pad1+gene encodes a subunit of the 26S proteasome in fission yeast. J Biol Chem. 1998;37:23938–23945. doi: 10.1074/jbc.273.37.23938. [DOI] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Schoenfeld LW, Valasek L, Nielsen KH, Hinnebusch AG. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA (i)Met. EMBO J. 2001;20:2954–2965. doi: 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincheira R, Chen Q, Huang Z, Zhang JT. Two subcellular localizations of eIF3 p170 and its interaction with membrane-bound microfilaments: implications for alternative functions of p170. Eur J Cell Biol. 2001;80:410–418. doi: 10.1078/0171-9335-00176. [DOI] [PubMed] [Google Scholar]
- Ptushkina M, Fierro-Monti I, van den Heuvel J, Vasilescu S, Birkenhager R, Mita K, McCarthy JE. Schizosaccharomyces pombehas a novel eukaryotic initiation factor 4F complex containing a cap-binding protein with the human eIF4E C-terminal motif KSGST. J Biol Chem. 1996;271:32818–32824. doi: 10.1074/jbc.271.51.32818. [DOI] [PubMed] [Google Scholar]
- Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, Norbury C. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26S proteasome subunit. J Biol Chem. 1997;48:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- Tani T, Derby RJ, Hiraoka Y, Spector DL. Nucleolar accumulation of poly(A)+ RNA in heat-shocked yeast cells: implication of nucleolar involvement in mRNA transport. Mol Biol Cell. 1996;7:173–192. doi: 10.1091/mbc.7.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Yanagida M. Cut8, essential for anaphase, controls localization of 26S proteasome, facilitating destruction of cyclin and Cut2. Curr Biol. 2000;21:1329–1338. doi: 10.1016/s0960-9822(00)00773-9. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Roussou I, Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Verlhac MH, Chen RH, Hanachi P, Hershey JW, Derynck R. Identification of partners of TIF34, a component of the yeast eIF3 complex, required for cell proliferation and translation initiation. EMBO J. 1997;16:6812–6822. doi: 10.1093/emboj/16.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornlocher HP, Hanachi P, Ribeiro S, Hershey JW. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiaeTIF32 and TIF31 genes. J Biol Chem. 1999;274:16802–16812. doi: 10.1074/jbc.274.24.16802. [DOI] [PubMed] [Google Scholar]
- Wei N, Tsuge T, Dohmae N, Takio K, Matsui M, Deng XW. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr Biol. 1998;8:919–922. doi: 10.1016/s0960-9822(07)00372-7. [DOI] [PubMed] [Google Scholar]
- Wilkinson CRM, Wallace M, Seeger M, Dubiel W, Gordon C. Mts4, a non ATPase subunit of the 26S protease in fission yeast is essential for mitosis and interacts directly with the ATPase subunit Mts2. J Biol Chem. 1997;272:25768–25777. doi: 10.1074/jbc.272.41.25768. [DOI] [PubMed] [Google Scholar]
- Wilkinson C, Wallace M, Morphew M, Perry P, Allshire R, Javerzat J-P, Richard Mcintosh J, Gordon C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR, Penney M, McGurk G, Wallace M, Gordon C. The 26S proteasome of the fission yeast Schizosaccharomyces pombe. Philos Trans R Soc Lond B Biol Sci. 1999;354:1523–1532. doi: 10.1098/rstb.1999.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom A, Kim TH, Winter E, Karniol B, von Arnim AG, Chamovitz DA. Arabidopsis eIF3e (INT6) associates with both eIF3c and the COP9 signalosome subunit CSN7. J Biol Chem. 2001;276:334–340. doi: 10.1074/jbc.M006721200. [DOI] [PubMed] [Google Scholar]
- Yen HC, Chang EC. Yin6, a fission yeast Int6 homolog, complexes with Moe1 and plays a role in chromosome segregation. Proc Natl Acad Sci USA. 2000;26:14370–14375. doi: 10.1073/pnas.97.26.14370. [DOI] [PMC free article] [PubMed] [Google Scholar]