Abstract
Glioblastoma is a particularly resilient cancer, and while therapies may be able to reach the brain by crossing the blood–brain barrier, they then have to deal with a highly invasive tumor that is very resistant to DNA damage. It seems clear that in order to kill aggressive glioma cells more efficiently and with fewer side effects on normal tissue, there must be a shift from classical cytotoxic chemotherapy to more targeted therapies. Since the epidermal growth factor receptor (EGFR) is altered in almost 50 % of glioblastomas, it currently represents one of the most promising therapeutic targets. In fact, it has been associated with several distinct steps in tumorigenesis, from tumor initiation to tumor growth and survival, and also with the regulation of cell migration and angiogenesis. However, inhibitors of the EGFR kinase have produced poor results with this type of cancer in clinical trials, with no clear explanation for the tumor resistance observed. Here we will review what we know about the expression and function of EGFR in cancer and in particular in gliomas. We will also evaluate which are the possible molecular and cellular escape mechanisms. As a result, we hope that this review will help improve the design of future EGFR-targeted therapies for glioblastomas.
Keywords: Glioblastoma, Mouse modeling, EGFR signaling pathway, EGFR stability, Therapy resistance
Introduction
Glial tumors are primary tumors that resemble astrocytes and/or oligodendrocytes. Grade IV astrocytomas (known as glioblastomas—GBM) are the most common glial tumors and they have a terrible prognosis, with a median survival rate of only 12–15 months. Among the main features of this type of cancer are a high mitotic index, diffuse infiltration, a tendency for necrosis, significant angiogenesis, resistance to apoptosis, and widespread genomic aberrations [1]. Standard treatment currently consists of surgery followed by radiotherapy and cytotoxic chemotherapy with the alkylating agent temozolomide [2], although treatment is generally palliative for most patients. Another important feature of GBM is the high degree of intra and intertumoral heterogeneity. For decades these tumors have been classified as primary or secondary GBM: the former displaying no evidence of a pre-existing, less malignant lesion, and comprising more than 90 % of the cases; with the latter represent a progression from a lower-grade glioma and they usually affect younger patients [3]. Nevertheless, no histological differences between these two entities have been described.
The expression of multiple genes and/or proteins is altered in glioblastomas, affecting the activation of oncogenes and/or the silencing of tumor-suppressor genes. Based on copy number and expression analyses, and on DNA sequencing studies, three signaling pathways have commonly been seen to be disrupted in GBM: (i) the receptor tyrosine kinases (RTK)/Ras/phosphoinositide 3-kinase (PI3 K) pathway, which includes the alterations in epidermal growth factor receptor (EGFR) (amplification and/or mutation in 40 % of cases), phosphatase and tensin homologue (PTEN: a PI3K inhibitor that is inactivated in 36 % of cases) and neurofibromatosis 1 (NF1: a Ras inhibitor inactivated in 23 % of cases); (ii) the p53 pathway, where the TP53 gene is mutated in 35 % of cases; (iii) the Rb pathway, where the cell-cycle inhibitors CDKN2A (p16INK4A) and CDKN2B are inactivated alternatively in about 50 % of cases [4, 5]. A relatively high frequency of mutations in the isocitrate dehydrogenase 1 and 2 (IDH1/2) has also been found in GBM, although these mutations appear to be preferentially associated with secondary GBM (80 % of the cases), in which they fulfill a significant pathogenic role [3]. By contrast, primary GBMs are characterized by a high proportion of mutations and/or the overexpression of EGFR gene. The EGFR is a 170-kDa glycosylated receptor with tyrosine kinase activity and alterations in EGFR function have not only been associated with GBM tumor initiation and growth but also with cell invasion, angiogenesis, and resistance to chemo- and radiotherapy [6, 7]. However, while EGFR kinase inhibitors have proven to be useful in treating other types of tumors, they offer poor outcomes in GBM patients. Moreover, contrary to what could be expected, there is some controversy about the correlation between the EGFR amplification and overexpression, and the clinical response to EGFR kinase inhibitors in GBM patients [8–11]. These results underline the special nature of the EGFR oncogenic network in these neoplasms.
Here we will try to draw up a comprehensive picture of EGFR signaling (in particular those aspects linked to cell proliferation and survival), based on classical models and more recent findings, in order to explain its oncogenic action in aggressive gliomas. We hope that this review will shed some light on the development of targeted therapies for EGFR-dependent GBMs and that as a result, better synergistic approaches and/or possible predictive markers might arise.
EGFR expression in gliomas
The frequent amplification of the EGFR gene in GBM was initially reported in 1985 [12] and it has been confirmed in many subsequent studies. It has been estimated that the EGFR gene is amplified in 30–40 % of GBMs and nearly 50 % of them overexpress the receptor [1, 4, 5]. Although not well understood, high levels of EGFR mRNA are also found in less malignant astrocytomas and oligodendrogliomas, with no gene amplification [13]. These observations underlie the relevant function of EGFR in glial cells and suggest that other oncogenic events may lead to increased transcription of this gene. Indeed, EGFR amplification has only been reported in 3 % of anaplastic (grade III) astrocytomas [14] and it is infrequent in secondary GBMs (only 8 %), whereas 60 % of primary GBMs show EGFR overexpression and 40 % of them contain EGFR amplifications. Moreover, EGFR amplification is rare in GBM patients younger than 35 years of age and the median age of patients with such alterations is 62 years [3]. From a histopathological point of view, EGFR gene amplification is relatively common in small cell GBMs (69 %) but rare in gliosarcomas (0 %) and giant cell GBMs (6 %) [15].
Several studies have correlated EGFR status and changes in other common GBM pathways. In general, there is a tendency for mutually exclusive changes in EGFR alterations and mutations in the tumor suppressor p53 [16–18]. Indeed, these modifications are considered as hallmarks of primary and secondary GBM, respectively [3]. On the other hand, the RB1 pathway seems to be important in both primary and secondary GBM, although homozygous deletions of the INK4A-ARF locus are more frequent in primary than in secondary tumors, this locus encoding two gene products (p16INK4A and p19ARF) involved in cell-cycle arrest and apoptosis [3]. Moreover, there is a frequent association of INK4A/ARF loss of function and EGFR activation in GBM [19], raising the possibility that critical functional interactions between these mutations are necessary for cellular transformation, as recently corroborated in several in vivo mouse models (see below).
From the neuropathological point of view, identification of EGFR amplification or the presence of EGFR mutations represents strong evidence that the tumor is a GBM, or at least that it should be treated like one, even in the absence of necrosis and microvascular proliferation in the biopsy [20]. However, while the diagnostic value of EGFR analysis is not questioned in GBM, there are some discrepancies regarding the prognostic value of EGFR amplification/overexpression, especially when patients of all ages are analyzed together. In fact, EGFR amplification has been associated with a worse prognosis in younger patients but with a better prognosis among older patients [18, 21–23]. Moreover, the EGFR amplification only predicted worse prognosis among younger patients in tumors with no p53 mutations, suggesting that the oncogenic potential of this receptor could be overcome by alterations in the tumor-suppressor pathway [20]. EGFR amplification is also evident in 26 % of long-term GBM survivors (patients surviving longer than 3 years [24]), suggesting that this oncogenic pathway is not much more aggressive than other GBM-related alterations.
EGFR and GBM molecular profiling
In recent years, high-throughput profiling techniques have made it possible to identify molecular subclasses of otherwise apparently uniform tumors. As a proof of principle, specific expression profiles were found to distinguish robustly primary from secondary GBM [25, 26], demonstrating that they are two different entities. More recently, The Cancer Genome Atlas (TCGA) used unsupervised clustering of global transcriptional data to define four GBM subclasses: proneural, neural, classical, and mesenchymal [27]. An earlier transcriptional study analyzing a set of grade III and IV gliomas established three molecular variants of malignant glioma, with at least two of them (also named proneural and mesenchymal) being very similar to those of the TCGA analysis [28, 29]. Interestingly, the proneural subgroup has a better prognosis and this gene-expression signature is enriched in anaplastic astrocytomas as well as in tumors with oligodendroglioma histology [30]. In accordance with these observations, proneural tumors are more common in younger patients. In fact, the beneficial effect of a younger age at diagnosis can be entirely attributed to the higher proportion of proneural GBM tumors detected [31].
The different GBM groups defined are correlated with defined genomic abnormalities. More specifically, proneural tumors have been strongly associated with alterations in platelet-derived growth factor receptor alpha (PDGFRA) and IDH1,2, while mesenchymal tumors are associated with mutations in NF1 [27]. Regarding EGFR, the TCGA analysis indicates that gene amplification, and in particular the presence of the vIII isoform, is enriched in the classic GBMs, although it may also occur in the other subtypes [27]. However, it has also been suggested that chromosome 7 amplifications (where the EGFR gene is located) are more frequent in the mesenchymal subtype [28]. Apart from the genomic studies, further support for the three basic subdivisions of malignant glioma have come from proteomic analyses, whereby NF1 expression, the upregulation of PDGF signaling, or of EGF signaling are highly reminiscent of the genomic abnormalities enriched in mesenchymal, proneural, and classical GBMs, respectively [32]. Regarding the classification of GBM cells when grown in vitro as primary cultures, most groups define only two subtypes, characterized by the differential expression of markers like CD133 and Olig2 in one group, and CD44 in the other. They are also referred to as proneural and mesenchymal GBM cells based on their expression profiles [33–36], although EGFR amplification has been described in both cases [34]. However, elsewhere the existence of three different subgroups of GBM cells was proposed, with EGFR being amplified and expressed in neurospheres expressing the signature of the classical subtype and MET (encoding the hepatocyte growth factor receptor, HGFR) being present in the mesenchymal and proneural subtypes [37]. Likewise, it was suggested that the gene expression profiles of glioma subtypes overexpressing EGFR are distinct from the rest [38], indicating that EGFR alterations drive a specific program of tumor development and that EGFR-addicted GBM might behave differently to other aggressive gliomas.
EGFR structure and mutations in GBM
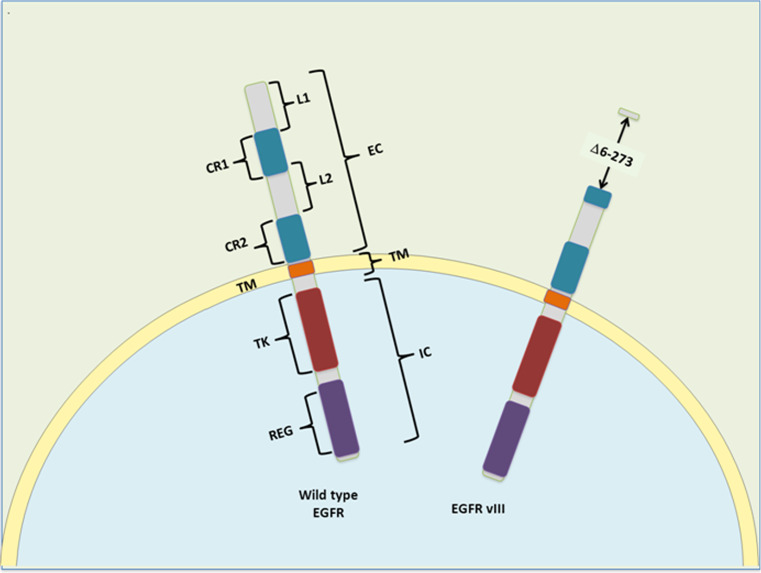
Epidermal growth factor (EGF) was identified by the embryologist Stanley Cohen in the early 1960s and its receptor (EGFR), which is also known as HER-1 or c-erbB-1, was identified a decade later [39]. EGFR is one of four transmembrane growth factor receptor proteins (c-erbB) that share similarities in structure and function, the other members being HER2 (c-erbB-2), HER3 (c-erbB-3), and HER4 (c-erbB-4). The EGFR is the receptor for members of the EGF family of extracellular ligands, including EGF, transforming growth factor-α (TGF-α), amphiregulin (AR), betacellulin, epiregulin, and the Heparin-binding EGF-like growth factor (HB-EGF [40]. EGFR is a 1,186-amino acid single polypeptide chain with three main regions: an extracellular (EC) receptor domain, a transmembrane region (TM), and an intracellular domain (IC) with tyrosine kinase (TK) activity (Fig. 1). The EC amino-terminal end can be divided into four domains, with the L1 and L2 domains responsible for ligand binding. Cysteine-rich (CR) domains 1 and 2 contain N-linked glycosylation sites and they form disulfide bonds that determine the tertiary conformation of the external portion of the molecule. EGFR can form homo- and hetero-dimers with other members of the c-erbB family, which result in differences in ligand affinity and downstream signaling [40]. Indeed, a large loop that protrudes from the back of CR2 makes contact with the respective domain of the other receptor [41]. The kinase activity of EGFR is stimulated by ligand engagement in a manner that depends on intermolecular interactions [42]. In contrast to other kinases, the trans-phosphorylation of the activation loop is not a critical event for EGFR activation [43]. In fact, recent structural studies revealed that the EGFR tyrosine-kinase domain has two different conformations. In the inactive one, it can inhibit its own activity but after EGF induces dimerization, the increase in the local concentration of the kinase domain provokes an allosteric change that drives the activation of EGFR [44, 45].
Fig. 1.
Structural motifs and regulatory domains of EGFR and its commonly mutated form vIII. Wild-type EGFR comprises and extracellular (EC), a transmembrane (TM), and an intracellular (IC) region. The EC comprises four domains: L1 and L2 form the ligand-binding pocket upon folding and CR1 (cystein-rich 1) domain includes the dimerization arm. In the IC region, there is a tyrosine-kinase (TK) domain and the regulatory (REG) region, which includes the autophosphorylation sites and the internalization domain. In the EGFRvIII, amino acids 6–273 are lost and the ligand-binding pocket cannot be formed
There are several mechanisms that could justify the activation of the EGFR signaling pathway in GBM. Overexpression on its own could provoke a local accumulation of the kinase domain that would provoke its activation. Moreover high expression of EGFR ligands has been reported in high-grade gliomas [46–48] and there are reports of TGFα amplification, mainly in recurrent gliomas [49]. However, it is also well known that many GBMs with EGFR amplification also carry mutations in EGFR [41, 50]. The most common EGFR mutations found in GBMs are in-frame deletion of regions in the extracellular domain like EGFRvIII (present in 30–40 % of GBMs with EGFR amplification). Oncogenic missense point mutations in the extracellular domain of the receptor were also recently reported [51], presumably promoting receptor dimerization. Another common mutation is a truncation of the intracellular region at amino acid 958, EGFRvV, which is present in 15 % of GBMs with EGFR amplification. This mutant receptor is internalization-deficient and therefore, it has enhanced ligand-dependent kinase activity [41, 50]. Mutations of the intracellular portion of EGFR are more common in other neoplasms and in fact, the tyrosine mutations in the kinase domain found in lung cancer that respond to specific EGFR inhibition [52] have not been detected in GBMs [53]. Notably, multiple mutations can sometimes be seen in the same amplified EGFR gene, a finding unique to GBM [54]. Indeed, a recent genomic study has revealed the presence of recurrent in-frame fusions involving EGFR (in 7.6 % of GBMs), with the most recurrent partners being septin 14 (SEPT14) and phosphoserin phosphatase (PSPH). Interestingly, the EGFR-SEPT14 fusions produce mitogen-independent growth and they constitutively activate signal transducer and activator of transcription 3 (STAT3) signaling, as well as imposing sensitivity to EGFR kinase inhibitors [55].
The EGFRvIII isoform
The EGFRvIII deletion represents 60–70 % of EGFR mutations in GBM, involving exons 2–7 of the extracellular domain (Fig. 1), with the multiple Alu repeats in introns 1 and 7 potentially mediating susceptibility to such specific gene rearrangement [54]. This mutated gene encodes for a receptor that lacks amino acids 6–273 with a novel tertiary conformation of the extracellular domain. It has been proposed that the oncogenic action of EGFRvIII is due to the constitutive activation of its kinase activity, resembling the behavior of the viral EGFR homologue, v-erbB, which exists primarily in dimers [56]. In fact, such altered EGFRvIII kinetics could produce a distinct set of downstream signals to those associated with wild-type EGFR [57] and indeed, there are reports of selective and/or constitutive activation of the signaling pathways involving: PI3K [58, 59], Ras [60], c-jun N-terminal kinase (JNK [61]), Src family kinases (SFK [62]), urokinase-type plasminogen activator receptor (uPAR [63]), and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB [64]). Moreover, EGFRvIII confers resistance to drug and ionizing radiation (IR) on GBM cells [65, 66].
The study of several mouse glioma models has indicated that the vIII variant is more tumorigenic that the wild-type receptor (see below). In fact, a multivariate analysis revealed that EGFRvIII overexpression was an independent and significant prognostic factor for poor overall survival [23]. However, in patients, this mutation occurs almost exclusively together with EGFR amplifications [54], suggesting a crosstalk between the mutant and the wild-type receptors in human GBM cells. This cooperation could be explained in a cell-autonomous manner, as the vIII isoform might be a substrate for EGFRwt and this phosphorylation would trigger the nuclear translocation of EGFRvIII and STAT3 activation [67]. Moreover, it has been suggested that EGFRvIII can directly bind to and activate the wild-type receptor [68], although autocrine and/or paracrine crosstalk has also been proposed. Indeed, it has been shown that EGFRvIII induces the expression of EGFR ligands (e.g., HB-EGF), whereas EGFRwt can activate the mutant isoform by facilitating EGFRvIII dimerization [69], suggesting a feed-forward loop that regulates the oncogenic action of these receptors. More recently, it was reported that cells expressing EGFRvIII release cytokines that activate neighboring EGFRwt-expressing cells [like interleukin 6 (IL6) and leukemia inhibitory factor (LIF)], favoring the formation of heterogeneous gliomas in mice [70]. These non-cell autonomous effects could explain the coexistence of the mutation with gene amplification in the same tumor, yet not necessarily in the same cells. Intriguingly, the oncogenic capacity of EGFRvIII may be propagated to non-expressing cells by cell-to-cell transfer of microvesicles (also called exosomes [71]). Exosomes are membrane-enclosed vesicles (30–100 nm in diameter) that are derived from endosomes during the formation of multivesicular bodies [72]. In GBM it has been shown that exosomes can mediate the horizontal transfer of EGFR (both protein and mRNA), altering the proliferation of receptor cells [73]. In addition, EGFR ligands have been seen to accumulate in exosomes isolated from cancer patients, which could reflect the dialogue between the different tumor cells, as well as between the tumor cells and their niche [74]. This exosome-mediated communication could add more complexity to the role of the EGFR in gliomas.
EGFR and GBM tumor initiation: mouse models of GBM related to EGFR
The high frequency of alterations to EGFR in primary GBMs suggests that this receptor not only participates in tumor growth but also in tumor initiation. In fact, numerous studies carried out on animal models have indicated that the enhanced EGFRwt expression in neural stem cells (NSCs) or more committed neuronal or glial precursor cells, and more particularly that of truncated EGFRvIII, can cooperate with other genetic alterations to induce primary brain cancer initiation and progression.
In accordance with the strong correlation between the alterations to the EGFR gene and the loss of the INK4A-ARF locus [19], most of the glioma models based on the overexpression of wild-type or vIII EGFR have been developed in cells that are deficient for this tumor suppressor. The first model described made use of avian retroviral vectors to transfer EGFRvIII into Inka-Arf null mice expressing tv-a, a gene encoding the TVA retroviral receptor, under the control of brain cell type-specific promoters [75]. Interestingly, the astrocytic lesions observed were much more frequent when the nestin promoter (progenitor-specific) was used in preference to the astrocyte-specific glial fibrillary acidic protein (GFAP) promoter. By contrast, EGFRvIII appeared to be incapable of generating gliomas on a p53-deficient background, unless CDK4 was also overproduced [76], possibly explaining the mutual exclusivity of the mutations in EGFR and P53 found in GBM. When a different approach was used, using retroviral vectors to overexpress EGFR in vitro in well-defined astrocyte or NSCs cultures from Ink4a-Arf-deficient mice, and then reintroducing these cells into the brains of SCID mice, both compartments were seen to be equally permissive for the generation of high-grade gliomas [77]. Using a similar approach, in vitro EGFRvIII expression in PTEN-deficient NSCs synergistically induced chromosomal instability and the formation of astrocytic tumors [78]. These data suggest that the deregulation of specific genetic pathways, rather than the cell-of-origin, dictates the emergence and phenotype of high-grade gliomas. However, low-grade oligodendrogliomas appeared in 20 % of transgenic mice that express the v-erbB oncogene under the control of S100b promoter, lesions that were more aggressive and that displayed higher penetrance in the context of p53 or Ink4-Arf heterogeneity [79]. S100b is expressed by oligodendroglia and astrocytes during early brain development, although it is also present in NSCs, and thus it is difficult to conclude which cells originate the tumors observed [80]. However, the differences with previous models suggest that oligodendrocytes are more readily transformed by v-erbB, at least during early neural development.
EGFRvIII overexpression can also cooperate with oncogenic Ras mutations, as witnessed with the Ras astrocytoma-prone model (RasB8 mice: GFAP-V12Ha-ras transgenic mice [81]). In transgenic mice that express both EGFRvIII and mutated Ras in the same cells, there was a higher penetrance of the tumors than in single Ras transgenic mice. However, the phenotype was also different in these mice, as overexpression of EGFRvIII led to the appearance of oligodendroglial and mixed oligoastrocytomas [82]. Interestingly, GFAP-EGFRvIII astrocytes forced to express mutated Ras in vitro generated oligodendroglioma-like tumors when inoculated back into immunodeficient brains. By contrast, injection of adenovirus-expressing EGFRvIII into adult RasB8 brains induced low-grade and high-grade astrocytomas with a high penetrance [83]. These results, together with those from Ink4-Arf −/- mice confirm that the expression of EGFRvIII is not sufficient to initiate gliomagenesis, although it cooperates with other genetic alterations to induce glioma formation. They also suggest that the same EGFR mutation can generate tumors with an astrocytic or oligodendrocytic phenotype, depending on the cell type and the developmental stage in which the alteration takes place. However, it is still unclear if EGFRvIII activates different signaling pathways in the germline and somatic context that could explain the changes in the phenotypic outcomes.
An important corollary from these mouse models is that EGFRwt cannot substitute for EGFRvIII to drive infiltrative glioma formation [76, 77], perhaps suggesting that sustained EGFR signaling is necessary for glial transformation. The mitogenic effect of EGFRvIII has been proposed to be due to a weak but constitutive kinase activity, amplified by the failure to attenuate signaling by down-regulating the receptor [84]. However, overexpression of self-activating EGFRwt may not be sufficient to induce cell transformation due to its constant lysosomal targeting. Indeed, using a conditional transgenic model in which somatic vIII or EGFRwt expression is induced in adult animals by stereotactic injection of an adenovirus expressing Cre recombinase, EGFRvIII expression promotes the formation of aggressive gliomas in conjunction with the loss of the cdkn2a and/or PTEN locus [59]. By contrast, overexpression of EGFRwt to levels similar to those observed in human GBMS is very inefficient at inducing tumor formation under the same conditions [59]. Similar studies were later carried out using bicistronic lentiviral vectors designed to express TGFα and Cre recombinase so that EGFR expression could be induced in Ink4-Arf -/- and/or PTEN-deficient mice. In this case, somatic, ligand-mediated activation of EGFR was necessary for gliomagenesis [85, 86], further evidence that persistent EGFR signaling is a necessary oncogenic event. These results are consistent with clinical observations that EGFR ligands are commonly overexpressed in receptor-amplified GBM. Interestingly, in the presence of PTEN, the tumors formed resembled the classical GBM subtype, while in the absence of PTEN expression they were more similar to the mesenchymal subtype in molecular terms [86]. It remains to be confirmed if a similar combination of mutations occurs in human tumors classified in this subgroups.
EGFR-dependent downstream signaling
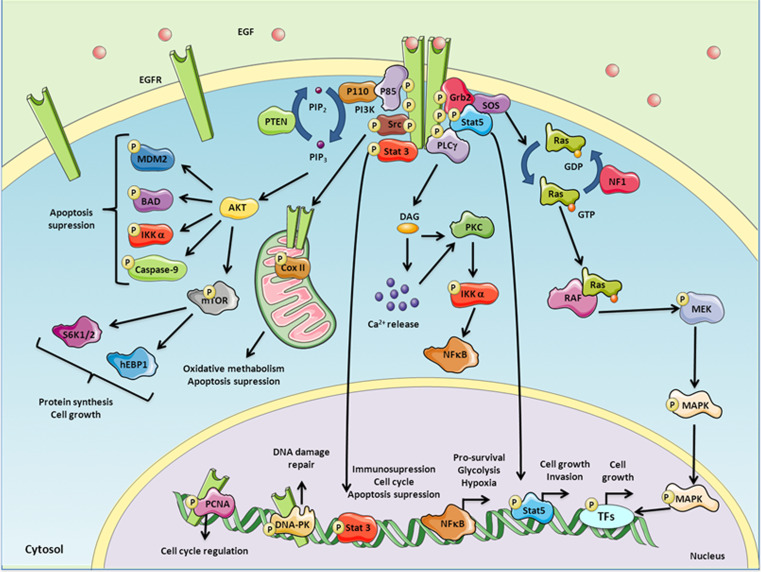
EGFR signaling is activated by a three-step mechanism. First, the binding of a specific ligand to the receptor induces dimerization of the ligand-binding domains. Second, this dimerization results in the auto-phosphorylation of five specific tyrosine residues at the carboxy-terminus of the intracellular domain of EGFR (Y992, Y1045, Y1068, Y1148, and Y1173), of which Y1173 is the major auto-phosphorylation site [87, 88]. Third, the activated EGFR recruits several signaling molecules that associate with the phosphorylated tyrosines through their Src homology domain 2 (SH2) or phospho-tyrosine binding (PTB) domains, many of which become phosphorylated by the receptor. These associations link a series of important signaling cascades to this activated EGFR tyrosine kinase (Fig. 2).
Fig. 2.
EGFR kinase-dependent signaling. The interaction between EGF and EGFR triggers the phosphorylation of several residues in the intracellular domain of the receptor and the recruitment of several adapter molecules that in turn activates a variety of intracellular pathways which involves MAPK/ERK, PI3K, STAT-3, PLCγ-PKC-NFκB and Src, among other downstream signal transducers. These signaling events result in changes in protein synthesis, cell growth, apoptosis and immune suppression, and cellular metabolism. EGFR can be also translocated into the nucleus, where it has an effect on cell cycle regulation and DNA damage repair
MAPK/ERK cascade
EGFR has been classically associated with cell proliferation through the recruitment of growth factor receptor bound protein 2 (Grb2) and activation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK). Grb2 recruits the guanine nucleotide exchange factor Son of Sevenless (SOS) to the receptor and it promotes binding of GTP to Ras, which then binds to and activates RAF. Activated RAF in turn binds to and phosphorylates MEK, which then phosphorylates ERK1/2. Upon activation, ERK kinases can translocate to the nucleus and activate several transcription factors (TFs) that enhance the transcription of genes involved in cellular proliferation [89], including Elk-1 (ETS domain-containing protein), peroxisome-proliferator-activated receptor γ (PPARγ), STAT1 and STAT3, C-myc and activating protein-1 (AP-1).
The expression of ERK and activated phospho-ERK has been correlated with proliferation and shorter survival time in gliomas [90]. However, while constitutively activated, mutated forms of Ras are found in nearly 50 % of all human tumors, few Ras mutations have been found in gliomas. By contrast, the GTPase-activating NF1 that inhibits Ras is inactivated in 23 % of cases [5]. Nevertheless, even in wild-type NF1 tumors, high levels of active Ras-GTP are found [91], suggesting that ERK-dependent mitogenic signaling in GBM is likely to be mostly mediated by the inappropriate activation of EGFR and/or other membrane molecules: RTKs, integrins, vascular endothelial growth factor (VEGF).
PI3K signaling
EGFR can modulate the balance between senescence and apoptosis by recruiting the p85 subunit of PI3K, and subsequently activating the p110 subunit. PI3K phosphorylation of phosphatidylinositol-4, 5-bisphosphate (PIP2) yields the second messenger phosphatidylinositol [3–5] -triphosphate (PIP3). This PIP3 serves as a membrane-docking site for the serine/threonine protein kinase AKT, although PIP3 is dephosphorylated to yield PIP2 by the tumor suppressor protein PTEN, which attenuates AKT signaling. Phosphorylated AKT appears to be able to prevent programmed cell death through targeted inhibition (phosphorylation) of Bad (a pro-apoptotic member of the Bcl-2 family) and caspase-9, and through the activation of murine double minute 2 homolog (MDM2) and inhibitor of nuclear factor kappa-B kinase subunit alpha (IKKα [92]). In turn, activated IKKα phosphorylates inhibitor of κB (IκB), targeting it for ubiquitination and proteosomal degradation, and provoking the activation and nuclear translocation of NF-κB. NF-κB plays and important role in inflammation and cancer, and it can induce pro-survival genes like Bcl-XL or caspase inhibitors [93]. Activated AKT also promotes cell growth through the activation of mammalian target of rapamycin (mTOR), a master integrator of growth factor signals, and of the sensing of nutrients and ATP [94]. The downstream effectors of mTOR are the eukaryotic initiation factor 4E (eIF4E) and ribosomal protein S6 kinase (S6K1/2) that induces translation of mRNA by phosphorylation of S6 [95].
Elevated phosho AKT levels have been observed in up to 85 % of GBM cell lines and samples from patients [96]. Activation of the PI3K pathway is significantly associated with increasing tumor grade, dampened apoptosis, and with an adverse clinical outcome of human gliomas [97]. In fact, AKT activation is correlated with EGFR amplification [98]. Alterations to this pathway are frequent in GBM, and they include activating mutations and amplifications of p110α [4, 99, 100], and p110δ [101], as well as gain of function mutations in the p85α regulatory subunit [4, 102]. Moreover, as we have already mentioned, the PTEN gene is lost, mutated, or epigenetically silenced in 40–50 % of gliomas, resulting in enhanced PI3 K activity and downstream signaling [4, 5, 92]. Interestingly, like EGFR amplifications, PTEN mutations are found almost exclusively in primary GBMs where there is a frequent association between the EGFR amplification and loss of 10q (where the PTEN suppressor gene is located [103]). However, a significant correlation between the presence of EGFR amplification and PTEN mutations has yet to be described [104]. Nevertheless, the deregulation of PI3K-PTEN could influence the effectiveness of certain molecular therapies targeting the EGFR (see below).
STAT3 activation
STAT3 is a latent transcription factor that is activated by tyrosine phosphorylation, which leads to its dimerization, nuclear translocation, and DNA binding. STAT3 is constitutively active in a wide variety of primary hematological and epithelial tumors, and in astrocytomas [105]. STAT3 tyrosine phosphorylation is induced by stimulated EGFR although it can also be induced by stimulation of other upstream receptors and/or non-receptor kinases including PDGFR, Src, and JAK2 (Janus kinase 2 [106]). Interestingly, it was recently indicated that STAT3 is further activated in GBM by enhancer of Zeste homolog 2 (EZH2)-mediated methylation. This alternative mechanism is induced by AKT phosphorylation of EZH2, indicative of RTK-independent means to activate STAT3 in GBM when the PI3K pathway is mutated [107].
Like ERK, there are no reports of STAT3 gain-of-function mutations in GBM but rather, STAT-3 activation is thought to be the consequence of either the deregulation of upstream kinases or the loss of endogenous inhibitors [108, 109]. STAT3 has been associated with cell-cycle progression, apoptosis, and immunosuppression in GBM [108, 110], and many studies indicate the anti-neoplastic potential of STAT3 inhibitors in GBMs [111]. Regarding EGFR-mediated activation, it has been reported that STAT3 constitutive activation coexists with EGFR expression in almost 30 % of high-grade gliomas and that targeting STAT3/JAK2 sensitizes these tumors to anti-EGFR agents [109]. However, elsewhere it was indicated that STAT-3 phosphorylation is only correlated with the presence of EGFRvIII, suggesting that it is specifically activated in the presence of such mutations. These findings could be related to the increased gliomagenic potential of this mutant receptor form (see below).
PLCγ-PKC signaling
Phospho-lipase Cγ (PLC-γ) is recruited to and phosphorylated by EGFR. Activated PLCγ in turn interacts with the plasma membrane where it cleaves PIP2 to inositol triphosphate (IP3) and diacylglycerol (DAG). Together with DAG, IP3-mediated induction of calcium (Ca2+) release can activate protein kinase C (PKC), which in turn can phosphorylate a plethora of substrates regulating proliferation, apoptosis, cell survival, and cell migration [112]. In GBM, the survival of patients with tumors expressing PKC or PLCγ was significantly shorter [90]. A novel study indicated that PLCγ signaling in response to GBM EGFR activation induces IKKβ and it promotes NF-κB nuclear translocation [113]. As mentioned above, NF-κB activity has been linked to the suppression of apoptotic signals, and in fact, this transcription factor cooperates with EGFR in breast [114] and lung cancer [115], and aberrant constitutive activation of NF-κB has been observed in glioblastomas [116]. Moreover, it was recently demonstrated that NFκ I1A, the gene that codes for the NF-κB inhibitor (IκBα), is often deleted in these tumors. Indeed, deletion of IκBα has a similar effect to that of EGFR amplification in the pathogenesis of GBM and it is associated with comparatively short survival [117]. These results suggest that activation of NF-κB is another fundamental pathway for glioma progression and that it can be achieved either by genetic deletion of its inhibitor or by EGFR amplification.
Apart from its role in suppressing apoptosis, nuclear NF-κB also cooperates with other transcription factors like hypoxia-inducible factor 1α (HIF1α). This interaction can lead to the overexpression of pyruvate kinase M2 (PKM2) [113], which catalyzes the last step in glycolysis, and is responsible for net ATP production and the accumulation of lactate within the glycolytic sequence [118]. Tumor cells have elevated rates of glucose uptake and higher lactate production, even in the presence of oxygen. This phenomenon, known as aerobic glycolysis or the Warburg effect, supports tumor cell growth. Accordingly, PKM2 expression is increased in cancer cells in which it facilitates lactate production [119, 120], and stronger PKM2 expression has been identified in GBM than in lower-grade gliomas and normal tissue [121]. In fact, it was postulated that PKM2 upregulation is the key step in EGFR-promoted glycolysis, demonstrating a good correlation between PKM2 expression and EGFR and IKKβ activity in GBM samples [113].
PKM2 is also located in the nucleus where it has been shown to function as a co-factor for HIF-1α, facilitating the transcription of hypoxia responsive genes and further promoting glucose uptake and lactate production [122]. Therefore, through the activation of NF-κB and the overexpression of PKM2, PLCγ-PKC signaling links EGFR activation to the regulation of glycolysis and the hypoxic response, contributing to the survival of GBM cells in their harsh tumorigenic niche.
EGFR–Src interaction
Although the major tyrosine sites in the EGFR C-terminal domain appear to be auto-phosphorylated, some tyrosine residues are phosphorylated by other intracellular tyrosine kinases. For example, EGFR Y845 is phosphorylated by Src, also known as tyrosine-protein kinase CSK [123, 124]. Co-overexpression of EGFR and Src frequently occurs in human tumors, and this has been linked to enhanced tumor growth. Src is capable of potentiating receptor-mediated tumorigenesis, causing synergistic increases in EGF-induced DNA synthesis, soft agar colony growth, and tumor formation in nude mice [125–127]. One of the proposed binding targets for EGFR phospho-Y845 is the transcription factor STAT5b, which is overexpressed in GBM compared to normal tissue or lower-grade gliomas [128] and where it seems to be preferentially activated by EGFRvIII [129]. Recent work indicates that SFKs are frequently coactivated with EGFR in GBM cell lines and patients [130], and it has been shown that dasatinib (a SFK inhibitor) enhances the efficacy of EGFR-targeted therapies in these tumors [62]. Moreover, it has been proposed that Src-dependent EGFR activation is induced by IR in glioma cells [131]. All the above provide a rationale for the combined use of anti-EGFR and anti-SFK therapies
Another pathway that appears to be regulated through Y845 following EGF stimulation is that mediating EGFR trafficking to mitochondria and interaction with the cytochrome-c oxidase subunit II (CoxII) [132]. The catalytic activity of EGFR and Src, as well as endocytosis and a mitochondrial localization signal, are required for these events. Rapamycin, apoptosis inducers, and EGFR inhibition can further enhance EGFR mitochondrial transport [133, 134]. Once in the mitochondria, CoxII can be phosphorylated by both EGFR and c-Src, reducing Cox activity and cellular ATP, regulating cell survival [132, 135]. Although the relevance of the EGFR-CoxII interaction in GBM remains to be determined, the Src-induced localization of EGFRvIII is stimulated in conditions of low glucose, and this mutant EGFR reduced glucose dependency by stimulating mitochondrial oxidative metabolism [136]. Interestingly, the amount of mitochondrial EGFR seems to be fine-tuned by the balance between autophagy and apoptosis, and inhibition of the former or induction of the latter provokes an accumulation of EGFR in this organelle as a pro-survival mechanism [134]. Taken together, these studies suggest that tumor cells reprogram the intracellular trafficking of EGFR/EGFRvIII by increasing its mitochondrial accumulation as a mechanism to escape from therapy- and stress-induced apoptosis and growth suppression.
Nuclear EGFR signaling
Most of the EGFR-activated signaling pathways end up in the nuclear translocation of second messengers and in the modulation of the activity of several TFs. However, EGFR itself has often been detected in the nuclei of cancer cells, primary tumor specimens, and other highly proliferative tissues [137]. Increased nuclear EGFR localization correlates with poor clinical outcome in several types of cancer [138], although this analysis has not been performed in gliomas. However, both EGFRwt and EGFRvIII have been detected in the nucleus of normal glial cells and primary GBM specimens, where they cooperate with STAT3 [139, 140].
A novel nuclear localization sequence (NLS) at amino acids 645–657 has recently been characterized EGFR, adjacent to the transmembrane domain [141], which allows nuclear translocation of members of this receptor family via binding to importin β [142]. Furthermore, there is cumulative evidence indicating that EGFR internalization serves to transport the receptor from the cell surface to the nucleus [143]. Once there, EGFR still functions as a tyrosine kinase, phosphorylating and stabilizing PCNA, and thus enhancing the proliferative potential of cancer cells [144]. This could explain the strong correlation between the nuclear localization of EGFR and the highly proliferative status of tissues [137]. However, it remains to be determined if there are other substrates for EGFR and/or EGFRvIII in the nucleus.
Nuclear EGFR and DNA damage regulation
EGFR overexpression has been implicated in radioresistance in a variety of human cancers, including GBM. Moreover, it has been correlated with a poor radiographic response of some patients with this tumor to radiotherapy [7, 15]. EGFR itself can be activated by radiation in a ligand-independent way, promoting cancer cell survival and proliferation. Furthermore, EGFR and EGFRvIII have been linked by several authors to the repair of double-strand breaks (DSB), the most lethal DNA lesions induced by ionizing radiation [7, 145]. In fact, the use of EGFR inhibitors in GBM cell lines and intracranial xenografts caused tumor regression when combined with radiotherapy [146, 147]. It appears that both PI3K and ERK pathways mediate the signals downstream of the receptor that activate DNA-dependent serine/threonine protein kinase (DNA-PK), a kinase required for non-homologous end joining (NHEJ) of DSBs [148, 149]. Moreover, the disruption of PI3K/AKT signaling by small-molecule inhibitors blocks DSB repair in GBM, whereas PTEN loss promotes it, resulting in radioresistance [150]. Furthermore, the radioresistance conferred by EGFRvIII seems to be a consequence of hyperactivated PI3K/AKT signaling [148]. However, more recent discoveries indicate that nuclear EGFR can influence DNA repair directly through a physical interaction with DNA-PK. The EGFR antibody cetuximab decreased nuclear DNA-PK protein and kinase activity by reducing its physical interaction with the receptor [151]. Moreover, cetuximab blocks nuclear shuttling of EGFR and prevents DNA-PK phosphorylation and DSB repair [152, 153]. Together, these data suggest that DNA-PK inhibitors and/or EGFR inhibitors may represent an effective strategy for radiosensitizing GBM tumors.
Targeting EGFR in GBMs
Several anti-EGFR-based therapeutic strategies have been assessed in pre-clinical and clinical trials as monotherapy, or in combination with radiotherapy and conventional chemotherapy. Some of the most promising results and ongoing clinical trials in GBM patients are resumed here (Table 1), including treatments with antibodies against EGFR (like cetuximab and nimotuzumab) or the vIII isoform (mAb806). The aim of these approaches is to provoke fewer side effects than traditional chemotherapy, although it is still not clear if their capacity to cross the blood–brain barrier will be sufficient to improve the results obtained with small molecules, and the cost-effectiveness ratio is still very high. Vaccination against EGFR vIII (rindopepimut) is also an attractive alternative, although due to the intratumoral heterogeneity of GBMs, it is still not known if this immunotherapy will induce a long-term reduction of the tumors. The most advanced EGFR-based therapies currently used clinically are the small-molecule tyrosine kinase inhibitors (TKIs). The best-studied TKIs are quinazoline-derived synthetic molecules with a low molecular weight, which can block the magnesium-ATP-binding pocket of the intracellular TK domain. This union prevents the activation of the kinase domain by ligand-induced auto-phosphorylation and the downstream activation of survival signaling pathways. The first EGFR-specific TKIs used in the clinic to treat newly diagnosed and recurrent gliomas were gefitinib, erlotinib, and lapatinib. However, despite the promising pre-clinical results obtained in vitro and in vivo with these first-generation inhibitors, they have not lived up to expectation [154]. Hence, second-generation TKIs have been designed that can bind irreversibly to the ATP binding site of various HER receptors, such as afatinib and dacomitinib. Most of these TKIs have proven some efficacy in other tumors and they are currently being tested clinically on recurrent GBM patients (Table 1). However, while we wait for the results of these next-generation TKIs, we need to reconsider the possible explanations for the lack of therapeutic response observed until now with EGFR-directed strategies in GBM patients.
Table 1.
Main EGFR-targeted agents being tested in preclinical and clinical trials for GBM patients
| Agent | Brand name | Company | Target | Class | Selected references/trial identifier | |
|---|---|---|---|---|---|---|
| Monoclonal antibodies |
Cetuximab (C225) |
Erbitux | ImClone Systems Inc. | EGFR/HER1 | Mouse-human chimeric antibody | [155, 156] |
|
Nimotuzumab (h-R3) |
TheraCIM | YM Biosciences | EGFR/HER1 | Human antibody | [157] | |
| Panitumumab | Vectibix | Amgen | EGFR/HER1 | Human antibody | NCT01017653 | |
| 125I-MAb 425 | Fox Chase Cancer Center | EGFR | Radiolabeled murine antibody | [158] | ||
| mAb 806 | ABT-806 | Abbott | EGFR vIII | Human antibody | [159] | |
| Small-molecule tyrosine kinase inhibitors |
Gefitinib (ZD1839) |
Iressa | Astra Zeneca Pharmaceuticals | EGFR/HER1 | Aniliquinazoline-based reversible inhibitor | [8] |
|
Erlotinib (OSI-774) |
Tacerva | Genentech Inc. | EGFR/HER1 | Aniliquinazoline-based reversible inhibitor | [160, 161] | |
|
Lapatinib (GW572016) |
Tykerb | GlaxoSmithKline | EGFR/HER1, HER2 | Thiazolylquinazoline-based reversible inhibitor | [162] | |
| Afatinib | Gilotrif | Boehringer Ingelheim | EGFR/HER1, HER2, HER4 | Anilinoquinazoline-based irreversible inhibitor | NCT00977431 | |
|
Dacomitinib (PF-00299804) |
Pfizer | EGFR/HER1, HER2, HER4 | Anilinoquinazoline-based irreversible inhibitor | NCT01520870 | ||
| Vandetanib (ZD6474) | Zactima | AstraZeneca Pharmaceuticals | EGFR/HER1, VEGFR | Aniliquinazoline-based inhibitor | [163] | |
|
Pelitinib (EKB-569) |
Wyeth Pharmaceuticals | EGFR/HER1 | Cyanoquinoline-based irreversible inhibitor | |||
| Vaccines |
Rindopepimut (CDX-110) |
Celldex Therapeutics | EGFRvIII | Peptide vaccination | ACT VI |
Conclusions from TKI studies in GBM: synergistic approaches and predictive markers
There are several possible explanations for the failure of EGFR-targeted therapy in GBM patients. Deficient tumor drug penetration and reduced systemic TKI availability due to antiepileptic drugs used in most GBM clinical trials could explain the lack of success of these compounds [6, 154, 164]. However, beyond the drug-delivery limitations, some molecular features of GBMs could be responsible for the limited benefits provided by these therapeutic agents.
EGFR vIII mutations and the AKT pathway
It has been proposed that EGFR vIII-positive cells are resistant to gefitinib because they need larger amounts of the drug and a longer exposition to it in order to dampen EGFR downstream signaling, especially the PI3K/AKT signaling pathway [165]. Moreover, tumors bearing EGFRvIII respond worse to cetuximab [154, 166]. By contrast, in a wild-type PTEN context, EGFRvIII expression was associated with GBM responsiveness to EGFR kinase inhibitors [167]. However, these findings could not be confirmed in subsequent trials, including a randomized phase II trial [160]. Nevertheless, high basal AKT activation (due to activation by other RTKs or by PTEN deletion) seems to be one of the causes that could explain the therapeutic failure of EGFR inhibitors in several types of cancer. For example, it was reported that restoring PTEN to a PTEN-deficient cell line augments the response to EGFR TKIs, inducing higher levels of apoptosis [168]. In GBMs, it was proposed that patients carrying wild-type PTEN tumors or low levels of phosphorylated PKB/AKT would have a better outcome in response to anti-EGFR treatments [10, 167]. Moreover, it was also shown that tyrosine phosphorylation of PTEN by SFKs and the fibroblast growth factor receptor (FGFR) can modulate its activity. In particular, PTEN phosphorylation at Y240 by FGFR is linked to EGFR-TKI resistance and reduced survival in GBM patients [169]. These results suggest that checking for the presence of the vIII isoform and the PTEN status (both at the genetic and at the protein level) might be important surrogate markers for EGFR-directed clinical trials. Moreover, they indicate that targeting the PI3K-AKT pathway could enhance the beneficial effects of EGFR TKIs.
Different EGFR conformations in lung cancer and GBM
The EGFR mutations detected in GBM and non-small cell lung cancer (NSCLC) have oncogenic transforming potential and they promote strong basal phosphorylation of the receptor in vitro. However, the different locations of these lung and brain tumor mutations have been associated with the diverse response of these cancers to EGFR inhibitors. Crystallography studies have indicated that when coupled to gefitinib and erlotinib, the receptor adopts an active conformation, also known as “type I” conformation, which is related to the mutations in the intracellular kinase domain that are frequent in NSCLC, which sensitizes the tumor cells to EGFR-targeted therapies [52, 53, 170]. By contrast, when complexed with lapatinib, EGFR is in an inactive configuration, also called the “type II” conformation, which is typical of the ectodomain mutations found in GBM samples, including missense and in frame deletions such as the EGFR vIII variant. It is therefore not surprising that glioma cells carrying extracellular EGFR mutants were poorly inhibited by erlotinib, whereas type II inhibitors induced cell death in the same cells [170]. Furthermore, it was confirmed that EGFRvIII releases erlotinib more rapidly than wild-type or lung cancer mutants, and that kinase-site occupancy was directly correlated with cell-cycle arrest [171]. The tropism of TKIs for a particular receptor conformational state could explain why some agents are more effective in lung cancer than in GBM patients, supporting the use of type II EGFR inhibitors for GBM, although they should have better brain penetrance than lapatinib.
Redundancy of RTK signals
Treatment of GBM cells in vitro with gefitinib provoked dephosphorylation of the EGFR but also of most of the aforementioned pathway regulators (Fig. 2). However, analysis of the in vivo effects on established GBM xenografts or tissue from treated patients demonstrated that gefitinib efficiently dephosphorylates its target without exerting a significant effect on other components of this pathway [172]. These data suggest that compensatory mechanisms exist in vivo, probably mediated by the activation of common signaling molecules by other RTKs [173]. One RTK that is ubiquitously expressed in cancer cells is the insulin-like growth factor receptor (IGF-1R), a receptor that engages in functional crosstalk with the EGFR as IGF-1R-deficient cells are resistant to transformation by EGFR [174]. IGF-1R has been linked to GBM resistance to gefitinib due to the increased signaling of PI3K/AKT and the ribosomal protein S6 kinase. In addition, the pharmacological inhibition of IGF-R1 results in the sensitization of tumor cells to EGFR-TKIs [175].
Another RTK expressed in GBMs is PDGFR and interestingly the results from two different groups indicate that there is genetic heterogeneity in aggressive gliomas, with EGFR and PDGFR being amplified and activated simultaneously in adjacent intermingled cells [176, 177]. These results could suggest that combinations of different inhibitors could be more efficient than EGFR-TKIs alone. However, some recent evidence suggests that this is not necessarily the case, as the addition of sunitinib (capable of inhibiting several RTKs, including PDGFR and VEGFR) to gefitinib only improved the anti-GBM efficacy in vitro but not in xenograft models [178]. However, it was recently reported that inhibition of EGFR signaling de-represses the transcription of PDGFRβ and that combined inhibition of both receptors potently suppresses tumor growth in vivo [179]. Therefore, there is still space for a synergistic approach targeting both signaling pathways.
Crosstalk EGFR–MET
The MET RTK is amplified in 5 % of GBM although it is overexpressed in 30 % of these tumors, representing a poor prognostic factor [180]. Moreover, MET is activated in GBM cells with increased levels of EGFR/EGFRvIII [173, 181]. In fact, there is evidence of autocrine and paracrine crosstalk between both signaling pathways [182]. In line with these results, resistance to EGFR inhibition can be overcome by using small molecule inhibitors of MET [173, 181] or neutralizing antibodies to hepatocyte growth factor (HGF), the MET ligand [182, 183]. Treatment of EGFR amplified mouse GBM cells with EGFR TKIs induces a cytostatic response that is characterized, among other changes, by enhanced MET expression. Moreover, pharmacological inhibition of MET overcomes the resistance to EGFR inhibition in these cells by inducing a cytotoxic response [86]. Together, these results underline the importance of evaluating the MET status in EGFR-directed approaches and support the need for synergistic therapies.
Hypoxia and cell metabolism
As mentioned above, there is an important relationship between EGFR and tumor cell metabolism, which could be particularly relevant in the context of the GBM niche. A histopathological hallmark of GBM (particularly in primary tumors) is the presence of large fields of necrosis, which are related to a worse outcome [184]. Interestingly, EGFR inhibitors have a protective effect against cell death induced by acute hypoxia, opposite to their pro-apoptotic effects observed under normoxia [185]. It has been proposed that under low oxygen conditions, EGFR inhibition reduces glucose intake, delays ATP exhaustion and maintains the integrity of the mitochondrial membrane potential, probably through the dephosphorylation of ribosomal protein S6. The authors hypothesized that EGFR inhibition may simulate nutrient deprivation, preparing cells for low oxygen and starvation conditions [185] and accordingly, EGFR-TKIs could be counteractive in a highly necrotic context. However, despite the relevance of this hypothesis, a correlation between hypoxic and/or necrotic markers and the lack of response to EGFR inhibitors is still missing.
Kinase-independent functions of EGFR
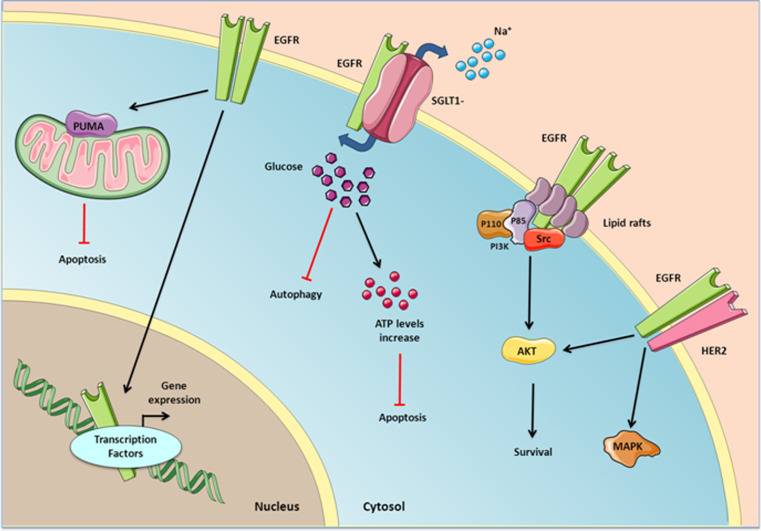
Studies on EGFR have mainly focused on conventional signal transduction pathways, yet it has long been known that many functions of EGFR require other mechanism besides those early transient responses. In fact, there is compelling evidence that EGFR can mediate cellular processes independent of its kinase activity in several types of cancer [138]. For example, the expression of a mutant EGFR receptor (D813A) with no kinase activity¡ can induce MAPK activation and DNA synthesis [186]. Moreover, another kinase-dead EGFR mutant (K721M) can activate survival signals through the interaction with other proteins like HER2 [187, 188]. Moreover, knocking down EGFR but not inhibiting its tyrosine kinase activity has been proposed to sensitize prostate and liver cancer cells to the apoptosis inducer adriamycin [189, 190]. These kinase-independent functions of EGFR could be an additional explanation for the failure of the TKI strategies, given that alternative downstream signal transducers could be regulated in a manner independent of phosphorylation. Here we summarize some of the non-catalytic actions of EGFR in GBM and other cancers (Fig. 3).
Fig. 3.
Kinase-independent EGFR signaling. Glucose uptake and mitochondrial-mediated apoptosis inhibition are the main processes resulting from kinase-independent EGFR signaling. EGFR can also associate with HER2 or Src and activate downstream survival signals independently of its kinase activity. Nuclear EGFR can serve as a cofactor to activate the transcription of several genes
EGFR and glucose uptake
The EGFR was found to prevent autophagic cell death by maintaining a cell’s intracellular glucose levels through the stabilization of the sodium/glucose cotransporter 1 (SGLT1 [191]). Interestingly, the EGFR-SGLT1 interaction does not respond to EGF stimulation or EGFR tyrosine kinase inhibition [192]. SGLTs can mediate glucose uptake into tumor cells even against a strong chemical gradient [193] and this seems to protect these cells from inducers of apoptosis. Interestingly, such apoptosis could be inhibited by increased extracellular glucose, perhaps reflecting that intracellular glucose deficiency mediates the sensitization to apoptosis induced by EGFR downregulation [189]. It was also demonstrated that EGFR and SGLT1 co-localize in prostate cancer tissue, and that inhibition of SGLT1 sensitized prostate cancer cells to EGFR inhibitors (gefitinib and erlotinib [192] providing an alternative synergistic approach to cure EGFR-addicted cancers.
SGLTs are overexpressed in several tumor entities and in some cases this overexpression has been correlated with EGFR expression [194, 195]. Furthermore, an irradiation-stimulated and EGFR-mediated increase in SGLT1 that provoked glucose uptake has been proposed to be required for the survival of genotoxically stressed tumor cells, a modification that could counteract the ATP crisis due to chromatin remodeling [194]. Importantly, SGLT1 inhibition could radio-sensitize tumor cells [194, 196]. Indeed, while there are no reports of SGLT1 expression in gliomas (with or without EGFR amplification), this is one of the cancers with the highest glucose consumption and thus one would expect these cells to express significant amounts of glucose transporters. In fact, metabolic reprogramming has been detected in more aggressive GBM, with cells expressing GLUT3, the high affinity neuronal glucose transporter, which enables them to survive in nutrient-restrictive environments [197]. It will be interesting to test whether EGFR modulates GLUT3 stability in glioma cells. However, it is in any case tempting to propose a synergistic effect of increased glucose uptake (mediated by the stabilization of SGLT1 and/or other glucose transporters) with the PKM2-mediated glycolytic upregulation in EGFR amplified and/or mutated high-grade gliomas (see above), a circumstance that would reinforce the receptor addiction in those GBMs.
Lipid raft activation of EGFR
TKIs targeting EGFR have also failed to be effective in breast cancer, even if the cells still depend on EGFR expression for growth. Interestingly, the receptor was localized to plasma membrane lipid rafts in the TKI-resistant cell lines, specialized membrane microdomains enriched in cholesterol, sphingolipids, and proteins. Moreover, interfering with cholesterol biosynthesis or lowering cholesterol levels produced a synergistic effect with gefitinib [198]. It was postulated that lipid rafts provide a platform that facilitates the interaction of EGFR, c-Src, and PI3K, leading to AKT activation and pro-survival signals, independent of EGFR kinase activity [199]. Furthermore, in colorectal cancer there is evidence that HIF1,2-directed transcriptional activation of caveolin 1 (CAV1), an essential structural constituent of caveolae (specialized lipid raft microdomains), increases EGFR dimerization and signaling [200]. However, there are no reports of lipid raft-related activation of EGFR signaling in GBMs. Moreover, it has been proposed that caveolae-enriched cellular fractions sequester EGFR and block signaling through this receptor [201]. Nevertheless, EGF-induced phosphorylation of the wild-type receptor results in EGFR dissociation from the caveolae, whereas EGFRvIII is predominantly cytoplasmic and does not associate with CAV1 unless cells are exposed to TKIs [202]. Therefore, while CAV1 is overexpressed in GBM cells and it seems to act as a tumor suppressor for EGFR-dependent cells [203], it is still possible that lipid rafts (other than caveolae) activate EGFR and that cholesterol modulation of membrane EGFR localization could have different outcomes on survival depending on the tumor cell type or the presence of different EGFR isoforms.
Inhibition of mitochondrial apoptosis
Another kinase-independent role for EGFR in GBM survival is related to the mitochondrial control of apoptosis. Both EGFR and EGFRvIII associate with the p53-upregulated modulator of apoptosis (PUMA), a pro-apoptotic member of the Bcl-2 family of proteins primarily located on the mitochondria [204]. PUMA strongly induces apoptosis in colorectal cancer, malignant gliomas, and in adult stem cells [205]. The EGFR-PUMA interaction is independent of EGF stimulation or kinase activity, and induces the sequestration of PUMA in the cytoplasm where it cannot initiate apoptosis. These observations are consistent with the co-expression of PUMA with EGFR/EGFRvIII in cell lines and patients’ samples, and with the strong resistance to apoptosis-inducting agents of GBMs [204].
Transcriptional activity of EGFR
Once in the nucleus, EGFR can modulate the transcription of several genes and indeed, a kinase-dead EGFR mutant can stimulate DNA synthesis in a kinase-independent manner [206]. Nuclear EGFR has been defined as a transcriptional co-factor that contains a transactivation domain in its C-terminus and that is able to modulate cyclin D1 gene expression [137]. Since then, several other transcriptional targets of EGFR have been defined, mostly implicated in cell cycle progression and the nitric oxide pathway: nitric oxide synthase (iNOS), a protein involved in inflammation, tumor progression, and metastasis [207]; B-Myb, a protein controlling proliferation [208]; cyclooxygenase-2 (COX-2 [140]); aurora kinase A, a protein involved in chromosomal instability [209]; and c-Myc [210]. Given that EGFR lacks a DNA-binding domain, the mechanism of EGFR-mediated gene regulation involves direct interaction of EGFR with STAT3 to regulate iNOS and COX2 promoters, with STAT5 to regulate the Aurora Kinase A promoter, with E2F1 TFs to regulate the B-Myb promoter, and with Src and STAT3 to regulate the expression of c-Myc. The constitutive presence of EGFR in the tumor nuclei may be beneficial to the tumors that encounter EGFR-targeted antibodies and TKIs. In fact, cancer cells that have acquired resistance to cetuximab [211] or gefitinib [212] accumulate more nuclear EGFR. These observations provide a rationale for the combined use of inhibitors of this receptor with molecules that could block EGFR nuclear translocation or for the use of inhibitors that affect both processes. For example, AKT inhibitors may fulfill such dual effects given that AKT-mediated EGFR phosphorylation at Ser-229 has been shown to be required for EGFR nuclear entry [212]. Similarly, Dasatinib, a known Src inhibitor, has a synergistic effect with cetuximab by limiting EGFR translocation to the nucleus [211].
Targeting EGFR stability in GBM
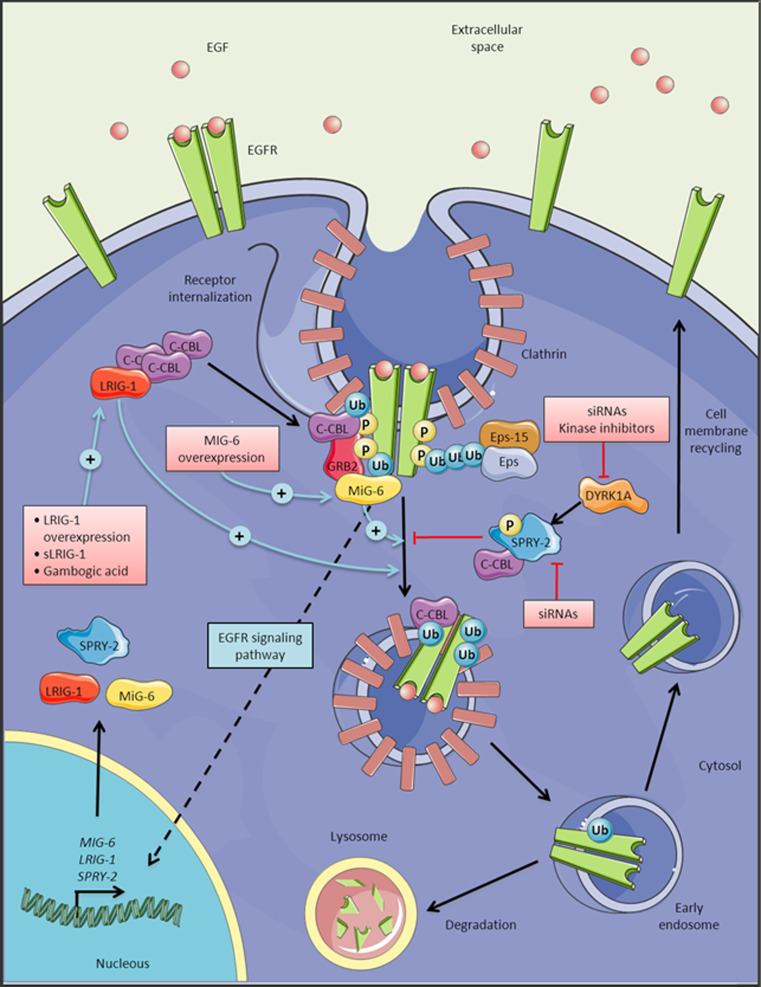
If kinase-independent functions of EGFR are responsible for tumor maintenance, we should look for alternative strategies that could downregulate receptor levels, alone or in combination with TKIs. Among the different strategies that could be used to regulate EGFR protein levels, the use of antisense RNAs against both wild-type and mutant receptors has been tested. This method impairs GBM cell growth in vitro and in vivo [213–215], although the absence of efficient and specific siRNA delivery tools hampers the possibility of its immediate clinical application. Here we review the mechanisms that control EGFR downregulation and its implications for the development of new GBM therapies (Fig. 4). EGFR signaling can be downregulated through a variety of cellular processes, such as receptor ubiquitination, dephosphorylation, restriction of ligand access, receptor trafficking to the lysosome and its subsequent degradation [216]. CBL is the primary E3 ubiquitin ligase that is recruited to the regulatory domain (REG) in the receptor’s tail after ligand stimulation (Fig. 1). This protein can bind directly to phospho-Y1045, or indirectly via Grb2, and it recruits E2 enzymes to its ring-finger domain to promote the ubiquitination and internalization of EGFR [216]. Receptor internalization seems to be a kinase-independent process and it is followed by efficient recycling to the plasma membrane [217]. In fact, the equilibrium between degradation and recycling determines the output of EGFR stimulation. Defective endocytotic downregulation of EGFR is associated with cancer. Indeed, dominant-negative forms of CBL are considered to be oncogenes in human myeloid neoplasms [218]. No such mutations have been found in GBM although the 19q13 allele containing the CBL sequence is frequently lost in these tumors [219]. Another way to manipulate the EGFR network is to maintain the level of activity just below the threshold required for CBL recruitment and receptor degradation. This is the case for several mutant forms detected in lung cancer [220] and also for EGFRvIII [84]. It is also noteworthy that EGF, but not TGFα (frequently overexpressed alongside EGFR [221] or amphiregulin [222]) triggers efficient EGFR degradation. Interestingly, co-expression of TGFα drives the tumorigenic potential of EGFR for tumor initiation [85]. These results suggest that GBM cells need to inhibit EGFR receptor degradation in order to enhance downstream signaling and promote cell growth and proliferation. In fact, this property has already been used to isolate GBM cells more capable of forming tumors by flow cytometry, which are those that express more EGFR at the plasma membrane [214].
Fig. 4.
Regulation of EGFR turnover. Clathrin-mediated endocytosis of EGFR internalization involves a variety of proteins, which results in the degradation of the receptor in the lysosomes or its recycling to the membrane. Activation of EGFR leads to the induction of LRIG1, MIG-6 and SPRY-2 expression, all of which are implicated in the regulation of EGFR turnover. Pink boxes indicate the possible strategies directed to target EGFR stability through the activation or inhibition of these proteins
LRIG1
Given the relevance of the EGFR signaling pathway, it is not surprising that its internalization involves a variety of positive and negative regulatory loops that ultimately influence the final response to EGFR activation. In fact, receptor activation drives the transcription of genes like leucine-rich repeats and immunoglobulin-like domains-1 (LRIG1) and mitogen-inducible gene 6 (MIG-6), which code for positive inducers of receptor degradation, and Sprouty 2 (SPRY2), an inhibitor of EGFR internalization [223]. The transmembrane glycoprotein LRIG1 has been proposed as a tumor suppressor protein as it increases the amount of CBL recruited to the EGFR, limiting its downstream signaling [224, 225], and it is also involved in EGFRvIII variant degradation in GBM cells in a CBL-independent manner [226]. Moreover, an increased EGFR/LRIG1 ratio has been detected in gliomas when compared to normal brain tissue, suggesting that LRIG1 downregulation is connected to tumor progression. The overexpression of LRIG1 in cultured glioma cells reduces EGFR at the cell surface, independent of its activation status, and it triggers cell growth inhibition and impaired invasion, enhancing apoptosis [227, 228]. It was recently demonstrated that soluble LIGR1 (sLIGR1) has an antitumoral effect in vitro and in vivo, promoting cell cycle arrest by downregulating ERK phosphorylation, with no effect on the levels and activation of the EGFR [228]. It was proposed that sLRIG1 may act through other RTKs or in an RTK-independent manner, which makes it a promising inhibitor and a more general antitumoral agent. A similar effect was recently observed for gambogic acid, which activates AMP-activated protein kinase (AMPK) and subsequently upregulates LRIG1, inhibiting EGFR signaling, increasing GBM cell apoptosis, and impairing tumor growth [229].
MIG-6
MIG-6 has been identified as a molecule that is induced following EGF stimulation and it is recruited to the activated receptor where it enhances the trafficking of the EGFR into late endosomes/lysosomes for degradation [230]. Endogenous EGFR is hyperactivated in Mig-6 knockout mice, resulting in hyperproliferation and impaired differentiation of epidermal keratinocytes [231]. High-resolution genomic profiling of GBMs allowed a highly recurrent (13 % of tumors) focal 1p36 deletion to be identified, the region in which MIG-6 lies. Moreover, it was demonstrated that MIG-6 expression is down-regulated in half of the GBMs tested and that there is a positive correlation between MIG-6 genomic alterations and the presence of EGFR amplification and/or the mutant EGFRvIII in GBM samples [230]. These results support the role of MIG-6 as a tumor suppressor in GBM, especially for EGFR-dependent tumors.
SPRY2-DYRK1A
The implication of the SPRY-2 protein in modulating EGFR stability is well known. SPRY-2 is an inducible regulator that is phosphorylated at a conserved tyrosine residue (Y55) after EGFR activation. This phosphotyrosine acts as a docking site for the SH2 domain of CBL and it competes with activated EGFR Y1045 phosphorylation. Hence, SPRY-2 removes CBL from activated EGFR and it blocks CBL-mediated EGFR ubiquitination, endocytosis, and degradation, which leads to sustained receptor signaling [232, 233]. Although SPRY2 is a tumor suppressor in different types of cancer, it also promotes tumor formation in colon cancer [234]. In GBM, several members of the SPRY family are included in a transcriptome module associated with EGFR amplification in GBMs [235], suggesting that they might act as oncogenes in at least a subset of glial tumors. Indeed, the role of the dual-specificity tyrosine phosphorylation-regulated kinase (DYRK1A) in regulating EGFR stability was recently described in neural progenitor and GBM cells [236, 237]. While the mechanisms through which DYRK1A might regulate EGFR stability have not been fully characterized, it has been proposed that DYRK1A may act upstream of SPRY-2, modulating EGFR targeting to the lysosomes. Accordingly, DYRK1A prevents EGFR degradation and favors the recycling of this receptor to the cell surface, which results in enhance EGFR signaling and tumor progression. Moreover, the expression of DYRK1A is correlated with that of EGFR in GBM suggesting that this kinase is necessary for the oncogenic action of EGFR [237]. The silencing of DYRK1A with siRNAs reduces the EGFR levels in vitro and in vivo, inhibiting self-renewal and proliferation, increasing apoptosis, and delaying tumor growth [237]. More interestingly, pharmacological inhibition of DYRK1A kinase activity also has a clear effect against tumors, indicating that it could be a good target to provoke EGFR degradation.
If we consider all the data presented, the notion that the EGFR tends to be stabilized in the membrane in GBMs by inhibiting receptor degradation is reinforced, either by downregulating positive modulators of internalization or by overexpressing negative effectors of this process. Alterations in the expression of these modulators could explain why there is no linear correlation between EGFR protein levels and the response to anti-EGFR therapy [8–11, 160]. In fact, several studies suggest that EGFR activity and erlotinib sensitivity may be more accurately predicted by the MIG-6/EGFR ratio in different tumors, and that resistance to TKIs is associated with an increase in MIG-6 expression and therefore, a decrease in EGFR activity [238, 239]. In fact, Mig-6 knock-out cells are unusually sensitive to gefitinib [231]. Therefore, measuring the membrane EGFR or analyzing the expression of modifiers of EGFR turnover might be relevant to predict the response to TKIs. On the other hand, it will be interesting to test if targeting EGFR could enhance the efficacy of current strategies that focus on inhibiting EGFR activity. As a proof of principle, green tea (-)-epigallocatechin-3-gallate (EGCG), a known DYRK1A inhibitor [240], has potent synergistic anti-tumor effects with erlotinib in head and neck [241] or lung [242] cancer.
Concluding remarks
Alterations in the EGFR gene occur in almost 50 % of GBMs, particularly in primary tumors. The studies of this receptor have focused mainly on the conventional signal transduction pathways that control cell proliferation and survival, such as MAPK and PI3K. However, data is accumulating that suggests this classical view cannot explain the true complexity of the cellular functions that are responsible for the tumorigenic activity of the EGFR in neural cells. There is compelling evidence linking EGFR activity with the regulation of cell metabolism and with the adaptative responses of GBM cells to their hypoxic microenvironment. Moreover, the localization of the receptor in different subcellular compartments (mainly the nucleus and the mitochondria) seems to be important in controlling the DNA damage and apoptotic responses, crucial steps in tumor initiation and survival. Crystallographic analyses have shed some light on the nature of GBM-associated EGFR mutations, indicating the prevalence of the inactive state of this receptor in tumor cells. Therefore, molecules that could bind to this conformation would be preferable to treat aggressive gliomas. All these studies are fundamental to orientate the therapeutic targeting of EGFR in GBMs, better defining the readouts of the activity of EGFR inhibitors and understanding more clearly why molecules working in other tumors fail to exert a beneficial effect in gliomas. While we await the results of clinical trials with second- and third-generation TKIs, we need to anticipate the possible compensatory mechanisms activated by other RTKs or downstream mutations that could identify bona fide predictive markers, as well as more effective synergistic approaches. Finally, we have to bear in mind that the response to EGFR activation may be independent of its kinase activity, and therefore targeting receptor stability could be more effective than the use of TKIs. Hopefully, this new complex and comprehensive picture of EGFR signaling in GBMs will allow us to reach satisfactory clinical results for at least a subset of patients with this terrible disease.
Acknowledgments
The work in the authors’ laboratory is funded by the Spanish Ministerio de Economía y Competitividad (Instituto de Salud Carlos III: PI12/00775 and RD12/0036/0027). We thank Angel Ayuso-Sacido and Juan Sepúlveda-Sánchez for their critical review of the manuscript.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan Cì, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19:764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas (TCGA) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riddick G, Fine HA. Integration and analysis of genome-scale data from gliomas. Nat Rev Neurol. 2011;7:439–450. doi: 10.1038/nrneurol.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpel-Massler G, Schmidt U, Unterberg A, Halatsch ME. Therapeutic inhibition of the epidermal growth factor receptor in high-grade gliomas: where do we stand? Mol Cancer Res. 2009;7:1000–1012. doi: 10.1158/1541-7786.MCR-08-0479. [DOI] [PubMed] [Google Scholar]
- 7.Squatrito M, Holland EC. DNA damage response and growth factor signaling pathways in gliomagenesis and therapeutic resistance. Cancer Res. 2011;71:5945–5949. doi: 10.1158/0008-5472.CAN-11-1245. [DOI] [PubMed] [Google Scholar]
- 8.Rich JN, Reardon DA, Peery T, Dowell JM, Quinn JA, Penne KL, Wikstrand CJ, Van Duyn LB, Dancey JE, McLendon RE, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22:133–142. doi: 10.1200/JCO.2004.08.110. [DOI] [PubMed] [Google Scholar]
- 9.Yung WK, Vredenburgh JJ, Cloughesy TF, Nghiemphu P, Klencke B, Gilbert MR, Reardon DA, Prados MD. Safety and efficacy of erlotinib in first-relapse glioblastoma: a phase II open-label study. Neuro Oncol. 2010;12:1061–1070. doi: 10.1093/neuonc/noq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, Baumber R, Lamborn KR, Kapadia A, Malec M, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 11.Lassman AB, Rossi MR, Raizer JJ, Abrey LE, Lieberman FS, Grefe CN, Lamborn K, Pao W, Shih AH, Kuhn JG, et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01-03 and 00-01. Clin Cancer Res. 2005;11:7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- 12.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield MD, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 13.Reifenberger J, Reifenberger G, Ichimura K, Schmidt EE, Wechsler W, Collins VP. Epidermal growth factor receptor expression in oligodendroglial tumors. Am J Pathol. 1996;149:29–35. [PMC free article] [PubMed] [Google Scholar]
- 14.von Deimling A, Fimmers R, Schmidt MC, Bender B, Fassbender F, Nagel J, Jahnke R, Kaskel P, Duerr EM, Koopmann J, et al. Comprehensive allelotype and genetic analysis of 466 human nervous system tumors. J Neuropathol Exp Neurol. 2000;59:544–558. doi: 10.1093/jnen/59.6.544. [DOI] [PubMed] [Google Scholar]
- 15.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12:675–684. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe K, Tachibana O, Sata K, Yonekawa Y, Kleihues P, Ohgaki H. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996;6:217–223. doi: 10.1111/j.1750-3639.1996.tb00848.x. [DOI] [PubMed] [Google Scholar]
- 17.Bouvier-Labit C, Chinot O, Ochi C, Gambarelli D, Dufour H, Figarella-Branger D. Prognostic significance of Ki67, p53 and epidermal growth factor receptor immunostaining in human glioblastomas. Neuropathol Appl Neurobiol. 1998;24:381–388. doi: 10.1046/j.1365-2990.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- 18.Batchelor TT, Betensky RA, Esposito JM, Pham LD, Dorfman MV, Piscatelli N, Jhung S, Rhee D, Louis DN. Age-dependent prognostic effects of genetic alterations in glioblastoma. Clin Cancer Res. 2004;10:228–233. doi: 10.1158/1078-0432.ccr-0841-3. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi Y, Ueki K, Waha A, Wiestler OD, Louis DN, von Deimling A. Association of EGFR gene amplification and CDKN2 (p16/MTS1) gene deletion in glioblastoma multiforme. Brain Pathol. 1997;7:871–875. doi: 10.1111/j.1750-3639.1997.tb00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 21.Simmons ML, Lamborn KR, Takahashi M, Chen P, Israel MA, Berger MS, Godfrey T, Nigro J, Prados M, Chang S, et al. Analysis of complex relationships between age, p53, epidermal growth factor receptor, and survival in glioblastoma patients. Cancer Res. 2001;61:1122–1128. [PubMed] [Google Scholar]
- 22.Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 23.Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 24.Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, et al. Long-term survival with glioblastoma multiforme. Brain. 2007;130:2596–2606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 25.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tso CL, Freije WA, Day A, Chen Z, Merriman B, Perlina A, Lee Y, Dia EQ, Yoshimoto K, Mischel PS, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- 27.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59:1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 30.Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, Saltz JH, Brat DJ, Moreno CS. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Scheck AC, Cloughesy TF, Lai A, Dong J, Farooqi HK, Liau LM, Horvath S, Mischel PS, Nelson SF. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunther HS, Schmidt NO, Phillips HS, Kemming D, Kharbanda S, Soriano R, Modrusan Z, Meissner H, Westphal M, Lamszus K. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 34.Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, Junker M, Oefner PJ, Bogdahn U, Wischhusen J, et al. Transcriptional profiles of CD133+ and C. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 35.Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Bacco F, Casanova E, Medico E, Pellegatta S, Orzan F, Albano R, Luraghi P, Reato G, D’Ambrosio A, Porrati P, et al. The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Cancer Res. 2012;72:4537–4550. doi: 10.1158/0008-5472.CAN-11-3490. [DOI] [PubMed] [Google Scholar]
- 38.Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 39.Carpenter G, King L, Jr, Cohen S. Epidermal growth factor stimulates phosphorylation in membrane preparations in vitro. Nature. 1978;276:409–410. doi: 10.1038/276409a0. [DOI] [PubMed] [Google Scholar]
- 40.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 41.Zawrocki A, Biernat W. Epidermal growth factor receptor in glioblastoma. Folia Neuropathol. 2005;43:123–132. [PubMed] [Google Scholar]
- 42.Yarden Y, Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 43.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 45.Endres NF, Engel K, Das R, Kovacs E, Kuriyan J. Regulation of the catalytic activity of the EGF receptor. Curr Opin Struct Biol. 2011;21:777–784. doi: 10.1016/j.sbi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlegel U, Moots PL, Rosenblum MK, Thaler HT, Furneaux HM. Expression of transforming growth factor alpha in human gliomas. Oncogene. 1990;5:1839–1842. [PubMed] [Google Scholar]
- 47.Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- 48.Mishima K, Higashiyama S, Asai A, Yamaoka K, Nagashima Y, Taniguchi N, Kitanaka C, Kirino T, Kuchino Y. Heparin-binding epidermal growth factor-like growth factor stimulates mitogenic signaling and is highly expressed in human malignant gliomas. Acta Neuropathol. 1998;96:322–328. doi: 10.1007/s004010050901. [DOI] [PubMed] [Google Scholar]
- 49.Yung WK, Zhang X, Steck PA, Hung MC. Differential amplification of the TGF-alpha gene in human gliomas. Cancer Commun. 1990;2:201–205. doi: 10.3727/095535490820874416. [DOI] [PubMed] [Google Scholar]
- 50.Nicholas MK, Lukas RV, Jafri NF, Faoro L, Salgia R. Epidermal growth factor receptor-mediated signal transduction in the development and therapy of gliomas. Clin Cancer Res. 2006;12:7261–7270. doi: 10.1158/1078-0432.CCR-06-0874. [DOI] [PubMed] [Google Scholar]
- 51.Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minna JD, Gazdar AF, Sprang SR, Herz J. Cancer. A bull’s eye for targeted lung cancer therapy. Science. 2004;304:1458–1461. doi: 10.1126/science.1099578. [DOI] [PubMed] [Google Scholar]
- 53.Pines G, Kostler WJ, Yarden Y. Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS Lett. 2010;584:2699–2706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 55.Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Downward J, Yarden Y, Mayes E, Scrace G, Totty N, Stockwell P, Ullrich A, Schlessinger J, Waterfield MD. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 57.Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- 58.Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res. 2002;62:6764–6769. [PubMed] [Google Scholar]
- 59.Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci USA. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prigent SA, Nagane M, Lin H, Huvar I, Boss GR, Feramisco JR, Cavenee WK, Huang HS. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J Biol Chem. 1996;271:25639–25645. doi: 10.1074/jbc.271.41.25639. [DOI] [PubMed] [Google Scholar]
- 61.Antonyak MA, Moscatello DK, Wong AJ. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J Biol Chem. 1998;273:2817–2822. doi: 10.1074/jbc.273.5.2817. [DOI] [PubMed] [Google Scholar]
- 62.Lu KV, Zhu S, Cvrljevic A, Huang TT, Sarkaria S, Ahkavan D, Dang J, Dinca EB, Plaisier SB, Oderberg I, et al. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009;69:6889–6898. doi: 10.1158/0008-5472.CAN-09-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu J, Jo M, Cavenee WK, Furnari F, VandenBerg SR, Gonias SL. Crosstalk between the urokinase-type plasminogen activator receptor and EGF receptor variant III supports survival and growth of glioblastoma cells. Proc Natl Acad Sci USA. 2011;108:15984–15989. doi: 10.1073/pnas.1113416108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puliyappadamba VT, Chakraborty S, Chauncey SS, Li L, Hatanpaa KJ, Mickey B, Noorani S, Shu HK, Burma S, Boothman DA, et al. Opposing effect of EGFRWT on EGFRvIII-mediated NF-kappaB activation with RIP1 as a cell death switch. Cell Rep. 2013;4:764–775. doi: 10.1016/j.celrep.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lammering G, Hewit TH, Valerie K, Contessa JN, Amorino GP, Dent P, Schmidt-Ullrich RK. EGFRvIII-mediated radioresistance through a strong cytoprotective response. Oncogene. 2003;22:5545–5553. doi: 10.1038/sj.onc.1206788. [DOI] [PubMed] [Google Scholar]
- 66.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci USA. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA, Chen J, Lau J, Knobbe-Thomsen C, Weller M, et al. EGFR phosphorylates tumor-derived EGFRvIII Driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24:438–449. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luwor RB, Zhu HJ, Walker F, Vitali AA, Perera RM, Burgess AW, Scott AM, Johns TG. The tumor-specific de2-7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene. 2004;23:6095–6104. doi: 10.1038/sj.onc.1207870. [DOI] [PubMed] [Google Scholar]
- 69.Li,L., Chakraborty,S., Yang,C.R., Hatanpaa,K.J., Cipher,D.J., Puliyappadamba,V.T., Rehman,A., Jiwani,A.J., Mickey,B., Madden,C. et al. 2013. An EGFR wild type-EGFRvIII-HB-EGF feed-forward loop regulates the activation of EGFRvIII. Oncogene [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 70.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 72.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 73.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Higginbotham JN, Demory BM, Gephart JD, Franklin JL, Bogatcheva G, Kremers GJ, Piston DW, Ayers GD, McConnell RE, Tyska MJ, et al. Amphiregulin exosomes increase cancer cell invasion. Curr Biol. 2011;21:779–786. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 76.Holland EC, Hively WP, Gallo V, Varmus HE. Modeling mutations in the G1 arrest pathway in human gliomas: overexpression of CDK4 but not loss of INK4a-ARF induces hyperploidy in cultured mouse astrocytes. Genes Dev. 1998;12:3644–3649. doi: 10.1101/gad.12.23.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 78.Li L, Dutra A, Pak E, Labrie JE, III, Gerstein RM, Pandolfi PP, Recht LD, Ross AH. EGFRvIII expression and PTEN loss synergistically induce chromosomal instability and glial tumors. Neuro Oncol. 2009;11:9–21. doi: 10.1215/15228517-2008-081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weiss WA, Burns MJ, Hackett C, Aldape K, Hill JR, Kuriyama H, Kuriyama N, Milshteyn N, Roberts T, Wendland MF, et al. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 2003;63:1589–1595. [PubMed] [Google Scholar]
- 80.Rickmann M, Wolff JR. S100 immunoreactivity in a subpopulation of oligodendrocytes and Ranvier’s nodes of adult rat brain. Neurosci Lett. 1995;186:13–16. doi: 10.1016/0304-3940(95)11269-3. [DOI] [PubMed] [Google Scholar]
- 81.Ding H, Roncari L, Shannon P, Wu X, Lau N, Karaskova J, Gutmann DH, Squire JA, Nagy A, Guha A. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61:3826–3836. [PubMed] [Google Scholar]
- 82.Ding H, Shannon P, Lau N, Wu X, Roncari L, Baldwin RL, Takebayashi H, Nagy A, Gutmann DH, Guha A. Oligodendrogliomas result from the expression of an activated mutant epidermal growth factor receptor in a RAS transgenic mouse astrocytoma model. Cancer Res. 2003;63:1106–1113. [PubMed] [Google Scholar]
- 83.Wei Q, Clarke L, Scheidenhelm DK, Qian B, Tong A, Sabha N, Karim Z, Bock NA, Reti R, Swoboda R, et al. High-grade glioma formation results from postnatal pten loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res. 2006;66:7429–7437. doi: 10.1158/0008-5472.CAN-06-0712. [DOI] [PubMed] [Google Scholar]
- 84.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 85.Acquaviva J, Jun HJ, Lessard J, Ruiz R, Zhu H, Donovan M, Woolfenden S, Boskovitz A, Raval A, Bronson RT, et al. Chronic activation of wild-type epidermal growth factor receptor and loss of Cdkn2a cause mouse glioblastoma formation. Cancer Res. 2011;71:7198–7206. doi: 10.1158/0008-5472.CAN-11-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, Bronson RT, Pfannl R, White F, Housman DE, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–3050. doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Downward J, Parker P, Waterfield MD. Autophosphorylation sites on the epidermal growth factor receptor. Nature. 1984;311:483–485. doi: 10.1038/311483a0. [DOI] [PubMed] [Google Scholar]
- 88.Hsuan JJ. Oncogene regulation by growth factors. Anticancer Res. 1993;13:2521–2532. [PubMed] [Google Scholar]
- 89.Pearson G, Robinson F, Beers GT, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 90.Mawrin C, Diete S, Treuheit T, Kropf S, Vorwerk CK, Boltze C, Kirches E, Firsching R, Dietzmann K. Prognostic relevance of MAPK expression in glioblastoma multiforme. Int J Oncol. 2003;23:641–648. [PubMed] [Google Scholar]
- 91.Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15:2755–2765. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 92.Koul D. PTEN signaling pathways in glioblastoma. Cancer Biol Ther. 2008;7:1321–1325. doi: 10.4161/cbt.7.9.6954. [DOI] [PubMed] [Google Scholar]
- 93.Bai D, Ueno L, Vogt PK. Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int J Cancer. 2009;125:2863–2870. doi: 10.1002/ijc.24748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 95.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 96.Wang H, Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest. 2004;84:941–951. doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- 97.Chakravarti A, Zhai G, Suzuki Y, Sarkesh S, Black PM, Muzikansky A, Loeffler JS. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22:1926–1933. doi: 10.1200/JCO.2004.07.193. [DOI] [PubMed] [Google Scholar]
- 98.Mizoguchi M, Betensky RA, Batchelor TT, Bernay DC, Louis DN, Nutt CL. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol. 2006;65:1181–1188. doi: 10.1097/01.jnen.0000248549.14962.b2. [DOI] [PubMed] [Google Scholar]
- 99.Gallia GL, Rand V, Siu IM, Eberhart CG, James CD, Marie SK, Oba-Shinjo SM, Carlotti CG, Caballero OL, Simpson AJ, et al. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol Cancer Res. 2006;4:709–714. doi: 10.1158/1541-7786.MCR-06-0172. [DOI] [PubMed] [Google Scholar]
- 100.Kita D, Yonekawa Y, Weller M, Ohgaki H. PIK3CA alterations in primary (de novo) and secondary glioblastomas. Acta Neuropathol. 2007;113:295–302. doi: 10.1007/s00401-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 101.Mizoguchi M, Nutt CL, Mohapatra G, Louis DN. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol. 2004;14:372–377. doi: 10.1111/j.1750-3639.2004.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK. Cancer-derived mutations in the regulatory subunit p85alpha of phosphoinositide 3-kinase function through the catalytic subunit p110alpha. Proc Natl Acad Sci USA. 2010;107:15547–15552. doi: 10.1073/pnas.1009652107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.von Deimling A, Louis DN, von Ammon K, Petersen I, Hoell T, Chung RY, Martuza RL, Schoenfeld DA, Yasargil MG, Wiestler OD, et al. Association of epidermal growth factor receptor gene amplification with loss of chromosome 10 in human glioblastoma multiforme. J Neurosurg. 1992;77:295–301. doi: 10.3171/jns.1992.77.2.0295. [DOI] [PubMed] [Google Scholar]
- 104.Liu W, James CD, Frederick L, Alderete BE, Jenkins RB. PTEN/MMAC1 mutations and EGFR amplification in glioblastomas. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- 105.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 106.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 107.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lo HW, Cao X, Zhu H, Ali-Osman F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res. 2008;14:6042–6054. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.See AP, Han JE, Phallen J, Binder Z, Gallia G, Pan F, Jinasena D, Jackson C, Belcaid Z, Jeong SJ, et al. The role of STAT3 activation in modulating the immune microenvironment of GBM. J Neurooncol. 2012;110:359–368. doi: 10.1007/s11060-012-0981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jackson C, Ruzevick J, Amin AG, Lim M. Potential role for STAT3 inhibitors in glioblastoma. Neurosurg Clin N Am. 2012;23:379–389. doi: 10.1016/j.nec.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 112.Mackay HJ, Twelves CJ. Targeting the protein kinase C family: are we there yet? Nat Rev Cancer. 2007;7:554–562. doi: 10.1038/nrc2168. [DOI] [PubMed] [Google Scholar]
- 113.Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, You MJ, Koh MY, Cote G, Aldape K, et al. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell. 2012;48:771–784. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Biswas DK, Iglehart JD. Linkage between EGFR family receptors and nuclear factor kappaB (NF-kappaB) signaling in breast cancer. J Cell Physiol. 2006;209:645–652. doi: 10.1002/jcp.20785. [DOI] [PubMed] [Google Scholar]
- 115.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, Moonsamy P, Dahlman K, Miller VA, Costa C, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nogueira L, Ruiz-Ontanon P, Vazquez-Barquero A, Moris F, Fernandez-Luna JL. The NFkappaB pathway: a therapeutic target in glioblastoma. Oncotarget. 2011;2:646–653. doi: 10.18632/oncotarget.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, Xian J, Cantley LC. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol. 2010;79:1118–1124. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23:560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. 2012;37:309–316. doi: 10.1016/j.tibs.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 121.Mukherjee J, Phillips JJ, Zheng S, Wiencke J, Ronen SM, Pieper RO. Pyruvate kinase M2 expression, but not pyruvate kinase activity, is up-regulated in a grade-specific manner in human glioma. PLoS One. 2013;8:e57610. doi: 10.1371/journal.pone.0057610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Luo W, Semenza GL. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2:551–556. doi: 10.18632/oncotarget.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 124.Sato K. Cellular functions regulated by phosphorylation of EGFR on Tyr845. Int J Mol Sci. 2013;14:10761–10790. doi: 10.3390/ijms140610761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maa MC, Leu TH, McCarley DJ, Schatzman RC, Parsons SJ. Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc Natl Acad Sci USA. 1995;92:6981–6985. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Song L, Morris M, Bagui T, Lee FY, Jove R, Haura EB. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006;66:5542–5548. doi: 10.1158/0008-5472.CAN-05-4620. [DOI] [PubMed] [Google Scholar]
- 127.Zhang Q, Thomas SM, Xi S, Smithgall TE, Siegfried JM, Kamens J, Gooding WE, Grandis JR. SRC family kinases mediate epidermal growth factor receptor ligand cleavage, proliferation, and invasion of head and neck cancer cells. Cancer Res. 2004;64:6166–6173. doi: 10.1158/0008-5472.CAN-04-0504. [DOI] [PubMed] [Google Scholar]
- 128.Liang QC, Xiong H, Zhao ZW, Jia D, Li WX, Qin HZ, Deng JP, Gao L, Zhang H, Gao GD. Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett. 2009;273:164–171. doi: 10.1016/j.canlet.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 129.Chumbalkar V, Latha K, Hwang Y, Maywald R, Hawley L, Sawaya R, Diao L, Baggerly K, Cavenee WK, Furnari FB, et al. Analysis of phosphotyrosine signaling in glioblastoma identifies STAT5 as a novel downstream target of DeltaEGFR. J Proteome Res. 2011;10:1343–1352. doi: 10.1021/pr101075e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, Burns M, Julian B, Peng XP, Hieronymus H, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol. 2009;27:77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park CM, Park MJ, Kwak HJ, Lee HC, Kim MS, Lee SH, Park IC, Rhee CH, Hong SI. Ionizing radiation enhances matrix metalloproteinase-2 secretion and invasion of glioma cells through Src/epidermal growth factor receptor-mediated p38/Akt and phosphatidylinositol 3-kinase/Akt signaling pathways. Cancer Res. 2006;66:8511–8519. doi: 10.1158/0008-5472.CAN-05-4340. [DOI] [PubMed] [Google Scholar]
- 132.Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol. 2004;24:7059–7071. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cao X, Zhu H, Ali-Osman F, Lo HW. EGFR and EGFRvIII undergo stress- and EGFR kinase inhibitor-induced mitochondrial translocalization: a potential mechanism of EGFR-driven antagonism of apoptosis. Mol Cancer. 2011;10:26. doi: 10.1186/1476-4598-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yue X, Song W, Zhang W, Chen L, Xi Z, Xin Z, Jiang X. Mitochondrially localized EGFR is subjected to autophagic regulation and implicated in cell survival. Autophagy. 2008;4:641–649. doi: 10.4161/auto.5971. [DOI] [PubMed] [Google Scholar]
- 135.Demory ML, Boerner JL, Davidson R, Faust W, Miyake T, Lee I, Huttemann M, Douglas R, Haddad G, Parsons SJ. Epidermal growth factor receptor translocation to the mitochondria: regulation and effect. J Biol Chem. 2009;284:36592–36604. doi: 10.1074/jbc.M109.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cvrljevic AN, Akhavan D, Wu M, Martinello P, Furnari FB, Johnston AJ, Guo D, Pike L, Cavenee WK, Scott AM, et al. Activation of Src induces mitochondrial localisation of de2-7EGFR (EGFRvIII) in glioma cells: implications for glucose metabolism. J Cell Sci. 2011;124:2938–2950. doi: 10.1242/jcs.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 138.Han W, Lo HW. Landscape of EGFR signaling network in human cancers: Biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318(2):124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, DePinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 142.Hubbard SR. The juxtamembrane region of EGFR takes center stage. Cell. 2009;137:1181–1183. doi: 10.1016/j.cell.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 143.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 144.Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 145.Chen DJ, Nirodi CS. The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–6560. doi: 10.1158/1078-0432.CCR-07-1610. [DOI] [PubMed] [Google Scholar]
- 146.Stea B, Falsey R, Kislin K, Patel J, Glanzberg H, Carey S, Ambrad AA, Meuillet EJ, Martinez JD. Time and dose-dependent radiosensitization of the glioblastoma multiforme U251 cells by the EGF receptor tyrosine kinase inhibitor ZD1839 (‘Iressa’) Cancer Lett. 2003;202:43–51. doi: 10.1016/j.canlet.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 147.Geoerger B, Gaspar N, Opolon P, Morizet J, Devanz P, Lecluse Y, Valent A, Lacroix L, Grill J, Vassal G. EGFR tyrosine kinase inhibition radiosensitizes and induces apoptosis in malignant glioma and childhood ependymoma xenografts. Int J Cancer. 2008;123:209–216. doi: 10.1002/ijc.23488. [DOI] [PubMed] [Google Scholar]
- 148.Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S, Hatanpaa KJ, Mickey B, Madden C, Maher E, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Golding SE, Morgan RN, Adams BR, Hawkins AJ, Povirk LF, Valerie K. Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol Ther. 2009;8:730–738. doi: 10.4161/cbt.8.8.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kao GD, Jiang Z, Fernandes AM, Gupta AK, Maity A. Inhibition of phosphatidylinositol-3-OH kinase/Akt signaling impairs DNA repair in glioblastoma cells following ionizing radiation. J Biol Chem. 2007;282:21206–21212. doi: 10.1074/jbc.M703042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bandyopadhyay D, Mandal M, Adam L, Mendelsohn J, Kumar R. Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J Biol Chem. 1998;273:1568–1573. doi: 10.1074/jbc.273.3.1568. [DOI] [PubMed] [Google Scholar]
- 152.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 153.Dittmann K, Mayer C, Wanner G, Kehlbach R, Rodemann HP. The radioprotector O-phospho-tyrosine stimulates DNA-repair via epidermal growth factor receptor- and DNA-dependent kinase phosphorylation. Radiother Oncol. 2007;84:328–334. doi: 10.1016/j.radonc.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 154.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Combs SE, Heeger S, Haselmann R, Edler L, Debus J, Schulz-Ertner D. Treatment of primary glioblastoma multiforme with cetuximab, radiotherapy and temozolomide (GERT)–phase I/II trial: study protocol. BMC Cancer. 2006;6:133. doi: 10.1186/1471-2407-6-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Neyns B, Sadones J, Joosens E, Bouttens F, Verbeke L, Baurain JF, D’Hondt L, Strauven T, Chaskis C, In’t VP, et al. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann Oncol. 2009;20:1596–1603. doi: 10.1093/annonc/mdp032. [DOI] [PubMed] [Google Scholar]
- 157.Bode U, Massimino M, Bach F, Zimmermann M, Khuhlaeva E, Westphal M, Fleischhack G. Nimotuzumab treatment of malignant gliomas. Expert Opin Biol Ther. 2012;12:1649–1659. doi: 10.1517/14712598.2012.733367. [DOI] [PubMed] [Google Scholar]
- 158.Li L, Quang TS, Gracely EJ, Kim JH, Emrich JG, Yaeger TE, Jenrette JM, Cohen SC, Black P, Brady LW. A Phase II study of anti-epidermal growth factor receptor radioimmunotherapy in the treatment of glioblastoma multiforme. J Neurosurg. 2010;113:192–198. doi: 10.3171/2010.2.JNS091211. [DOI] [PubMed] [Google Scholar]
- 159.Scott AM, Lee FT, Tebbutt N, Herbertson R, Gill SS, Liu Z, Skrinos E, Murone C, Saunder TH, Chappell B, et al. A phase I clinical trial with monoclonal antibody ch806 targeting transitional state and mutant epidermal growth factor receptors. Proc Natl Acad Sci USA. 2007;104:4071–4076. doi: 10.1073/pnas.0611693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF, Clement PM, Frenay M, Campone M, Baurain JF, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Raizer JJ, Abrey LE, Lassman AB, Chang SM, Lamborn KR, Kuhn JG, Yung WK, Gilbert MR, Aldape KA, Wen PY, et al. A phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy. Neuro Oncol. 2010;12:95–103. doi: 10.1093/neuonc/nop015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Thiessen B, Stewart C, Tsao M, Kamel-Reid S, Schaiquevich P, Mason W, Easaw J, Belanger K, Forsyth P, McIntosh L, et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol. 2010;65:353–361. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 163.Kreisl TN, McNeill KA, Sul J, Iwamoto FM, Shih J, Fine HA. A phase I/II trial of vandetanib for patients with recurrent malignant glioma. Neuro Oncol. 2012;14:1519–1526. doi: 10.1093/neuonc/nos265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hegi ME, Rajakannu P, Weller M. Epidermal growth factor receptor: a re-emerging target in glioblastoma. Curr Opin Neurol. 2012;25:774–779. doi: 10.1097/WCO.0b013e328359b0bc. [DOI] [PubMed] [Google Scholar]
- 165.Learn CA, Hartzell TL, Wikstrand CJ, Archer GE, Rich JN, Friedman AH, Friedman HS, Bigner DD, Sampson JH. Resistance to tyrosine kinase inhibition by mutant epidermal growth factor receptor variant III contributes to the neoplastic phenotype of glioblastoma multiforme. Clin Cancer Res. 2004;10:3216–3224. doi: 10.1158/1078-0432.ccr-03-0521. [DOI] [PubMed] [Google Scholar]
- 166.Pedersen MW, Pedersen N, Ottesen LH, Poulsen HS. Differential response to gefitinib of cells expressing normal EGFR and the mutant EGFRvIII. Br J Cancer. 2005;93:915–923. doi: 10.1038/sj.bjc.6602793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 168.Bianco R, Shin I, Ritter CA, Yakes FM, Basso A, Rosen N, Tsurutani J, Dennis PA, Mills GB, Arteaga CL. Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene. 2003;22:2812–2822. doi: 10.1038/sj.onc.1206388. [DOI] [PubMed] [Google Scholar]
- 169.Fenton TR, Nathanson D, de Ponte A, Kuga D, Iwanami A, Dang J, Yang H, Tanaka K, Oba-Shinjo SM, Uno M, et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc Natl Acad Sci USA. 2012;109:14164–14169. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, Kubek S, Oldrini B, Chheda MG, Yannuzzi N, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Barkovich KJ, Hariono S, Garske AL, Zhang J, Blair JA, Fan QW, Shokat KM, Nicolaides T, Weiss WA. Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer Discov. 2012;2:450–457. doi: 10.1158/2159-8290.CD-11-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hegi ME, Diserens AC, Bady P, Kamoshima Y, Kouwenhoven MC, Delorenzi M, Lambiv WL, Hamou MF, Matter MS, Koch A, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib—a phase II trial. Mol Cancer Ther. 2011;10:1102–1112. doi: 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- 173.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 174.Coppola D, Ferber A, Miura M, Sell C, D’Ambrosio C, Rubin R, Baserga R. A functional insulin-like growth factor I receptor is required for the mitogenic and transforming activities of the epidermal growth factor receptor. Mol Cell Biol. 1994;14:4588–4595. doi: 10.1128/mcb.14.7.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chakravarti A, Loeffler JS, Dyson NJ. Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res. 2002;62:200–207. [PubMed] [Google Scholar]
- 176.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 178.Joshi AD, Loilome W, Siu IM, Tyler B, Gallia GL, Riggins GJ. Evaluation of tyrosine kinase inhibitor combinations for glioblastoma therapy. PLoS One. 2012;7:e44372. doi: 10.1371/journal.pone.0044372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I, Villa GR, Tanaka K, Nael A, Yang H, et al. De-repression of PDGFRbeta transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3:534–547. doi: 10.1158/2159-8290.CD-12-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kong DS, Song SY, Kim DH, Joo KM, Yoo JS, Koh JS, Dong SM, Suh YL, Lee JI, Park K, et al. Prognostic significance of c-Met expression in glioblastomas. Cancer. 2009;115:140–148. doi: 10.1002/cncr.23972. [DOI] [PubMed] [Google Scholar]
- 181.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lal B, Goodwin CR, Sang Y, Foss CA, Cornet K, Muzamil S, Pomper MG, Kim J, Laterra J. EGFRvIII and c-Met pathway inhibitors synergize against PTEN-null/EGFRvIII + glioblastoma xenografts. Mol Cancer Ther. 2009;8:1751–1760. doi: 10.1158/1535-7163.MCT-09-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Barker FG, Davis RL, Chang SM, Prados MD. Necrosis as a prognostic factor in glioblastoma multiforme. Cancer. 1996;77:1161–1166. doi: 10.1002/(sici)1097-0142(19960315)77:6<1161::aid-cncr24>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 185.Steinbach JP, Klumpp A, Wolburg H, Weller M. Inhibition of epidermal growth factor receptor signaling protects human malignant glioma cells from hypoxia-induced cell death. Cancer Res. 2004;64:1575–1578. doi: 10.1158/0008-5472.can-03-3775. [DOI] [PubMed] [Google Scholar]
- 186.Coker KJ, Staros JV, Guyer CA. A kinase-negative epidermal growth factor receptor that retains the capacity to stimulate DNA synthesis. Proc Natl Acad Sci USA. 1994;91:6967–6971. doi: 10.1073/pnas.91.15.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Deb TB, Su L, Wong L, Bonvini E, Wells A, David M, Johnson GR. Epidermal growth factor (EGF) receptor kinase-independent signaling by EGF. J Biol Chem. 2001;276:15554–15560. doi: 10.1074/jbc.M100928200. [DOI] [PubMed] [Google Scholar]
- 188.Ewald JA, Wilkinson JC, Guyer CA, Staros JV. Ligand- and kinase activity-independent cell survival mediated by the epidermal growth factor receptor expressed in 32D cells. Exp Cell Res. 2003;282:121–131. doi: 10.1016/s0014-4827(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 189.Xu S, Weihua Z. Loss of EGFR induced autophagy sensitizes hormone refractory prostate cancer cells to adriamycin. Prostate. 2011;71(11):1216–1224. doi: 10.1002/pros.21337. [DOI] [PubMed] [Google Scholar]
- 190.Niu J, Li XN, Qian H, Han Z. siRNA mediated the type 1 insulin-like growth factor receptor and epidermal growth factor receptor silencing induces chemosensitization of liver cancer cells. J Cancer Res Clin Oncol. 2008;134:503–513. doi: 10.1007/s00432-007-0314-x. [DOI] [PubMed] [Google Scholar]
- 191.Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ren J, Bollu LR, Su F, Gao G, Xu L, Huang WC, Hung MC, Weihua Z. EGFR-SGLT1 interaction does not respond to EGFR modulators, but inhibition of SGLT1 sensitizes prostate cancer cells to EGFR tyrosine kinase inhibitors. Prostate. 2013;73:1453–1461. doi: 10.1002/pros.22692. [DOI] [PubMed] [Google Scholar]
- 193.Ganapathy V, Thangaraju M, Prasad PD. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol Ther. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 194.Dittmann K, Mayer C, Rodemann HP, Huber SM. EGFR cooperates with glucose transporter SGLT1 to enable chromatin remodeling in response to ionizing radiation. Radiother Oncol. 2013;107:247–251. doi: 10.1016/j.radonc.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 195.Hanabata Y, Nakajima Y, Morita K, Kayamori K, Omura K. Coexpression of SGLT1 and EGFR is associated with tumor differentiation in oral squamous cell carcinoma. Odontology. 2012;100:156–163. doi: 10.1007/s10266-011-0033-2. [DOI] [PubMed] [Google Scholar]
- 196.Huber SM, Misovic M, Mayer C, Rodemann HP, Dittmann K. EGFR-mediated stimulation of sodium/glucose cotransport promotes survival of irradiated human A549 lung adenocarcinoma cells. Radiother Oncol. 2012;103:373–379. doi: 10.1016/j.radonc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 197.Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, Weil RJ, Nakano I, Sarkaria JN, Stringer BW, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Irwin ME, Mueller KL, Bohin N, Ge Y, Boerner JL. Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol. 2011;226:2316–2328. doi: 10.1002/jcp.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Irwin ME, Bohin N, Boerner JL. Src family kinases mediate epidermal growth factor receptor signaling from lipid rafts in breast cancer cells. Cancer Biol Ther. 2011;12:718–726. doi: 10.4161/cbt.12.8.16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang Y, Roche O, Xu C, Moriyama EH, Heir P, Chung J, Roos FC, Chen Y, Finak G, Milosevic M, et al. Hypoxia promotes ligand-independent EGF receptor signaling via hypoxia-inducible factor-mediated upregulation of caveolin-1. Proc Natl Acad Sci USA. 2012;109:4892–4897. doi: 10.1073/pnas.1112129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Quann K, Gonzales DM, Mercier I, Wang C, Sotgia F, Pestell RG, Lisanti MP, Jasmin JF. Caveolin-1 is a negative regulator of tumor growth in glioblastoma and modulates chemosensitivity to temozolomide. Cell Cycle. 2013;12:1510–1520. doi: 10.4161/cc.24497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Abulrob A, Giuseppin S, Andrade MF, McDermid A, Moreno M, Stanimirovic D. Interactions of EGFR and caveolin-1 in human glioblastoma cells: evidence that tyrosine phosphorylation regulates EGFR association with caveolae. Oncogene. 2004;23:6967–6979. doi: 10.1038/sj.onc.1207911. [DOI] [PubMed] [Google Scholar]
- 203.Parat MO, Riggins GJ. Caveolin-1, caveolae, and glioblastoma. Neuro Oncol. 2012;14:679–688. doi: 10.1093/neuonc/nos079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhu H, Cao X, Ali-Osman F, Keir S, Lo HW. EGFR and EGFRvIII interact with PUMA to inhibit mitochondrial translocalization of PUMA and PUMA-mediated apoptosis independent of EGFR kinase activity. Cancer Lett. 2010;294:101–110. doi: 10.1016/j.canlet.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–S83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Eldredge ER, Korf GM, Christensen TA, Connolly DC, Getz MJ, Maihle NJ. Activation of c-fos gene expression by a kinase-deficient epidermal growth factor receptor. Mol Cell Biol. 1994;14:7527–7534. doi: 10.1128/mcb.14.11.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 208.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 209.Hung LY, Tseng JT, Lee YC, Xia W, Wang YN, Wu ML, Chuang YH, Lai CH, Chang WC. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jaganathan S, Yue P, Paladino DC, Bogdanovic J, Huo Q, Turkson J. A functional nuclear epidermal growth factor receptor, SRC and Stat3 heteromeric complex in pancreatic cancer cells. PLoS One. 2011;6:e19605. doi: 10.1371/journal.pone.0019605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 211.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, Chiu PC, Huang WP, Wang YN, Chen CH, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–20568. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kang CS, Zhang ZY, Jia ZF, Wang GX, Qiu MZ, Zhou HX, Yu SZ, Chang J, Jiang H, Pu PY. Suppression of EGFR expression by antisense or small interference RNA inhibits U251 glioma cell growth in vitro and in vivo. Cancer Gene Ther. 2006;13:530–538. doi: 10.1038/sj.cgt.7700932. [DOI] [PubMed] [Google Scholar]
- 214.Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi SL, Falini A, De Palma M, Bulfone A, Poliani PL, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 215.Verreault M, Weppler SA, Stegeman A, Warburton C, Strutt D, Masin D, Bally MB. Combined RNAi-mediated suppression of Rictor and EGFR resulted in complete tumor regression in an orthotopic glioblastoma tumor model. PLoS One. 2013;8:e59597. doi: 10.1371/journal.pone.0059597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 217.Wang Q, Villeneuve G, Wang Z. Control of epidermal growth factor receptor endocytosis by receptor dimerization, rather than receptor kinase activation. EMBO Rep. 2005;6:942–948. doi: 10.1038/sj.embor.7400491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 2010;70:4789–4794. doi: 10.1158/0008-5472.CAN-10-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mizoguchi M, Nutt CL, Louis DN. Mutation analysis of CBL-C and SPRED3 on 19q in human glioblastoma. Neurogenetics. 2004;5:81–82. doi: 10.1007/s10048-003-0164-x. [DOI] [PubMed] [Google Scholar]
- 220.Shtiegman K, Kochupurakkal BS, Zwang Y, Pines G, Starr A, Vexler A, Citri A, Katz M, Lavi S, Ben Basat Y, et al. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26:6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- 221.Ebner R, Derynck R. Epidermal growth factor and transforming growth factor-alpha: differential intracellular routing and processing of ligand-receptor complexes. Cell Regul. 1991;2:599–612. doi: 10.1091/mbc.2.8.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Stern KA, Place TL, Lill NL. EGF and amphiregulin differentially regulate Cbl recruitment to endosomes and EGF receptor fate. Biochem J. 2008;410:585–594. doi: 10.1042/BJ20071505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 224.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, III, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 226.Stutz MA, Shattuck DL, Laederich MB, Carraway KL, III, Sweeney C. LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Oncogene. 2008;27:5741–5752. doi: 10.1038/onc.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ye F, Gao Q, Xu T, Zeng L, Ou Y, Mao F, Wang H, He Y, Wang B, Yang Z, et al. Upregulation of LRIG1 suppresses malignant glioma cell growth by attenuating EGFR activity. J Neurooncol. 2009;94:183–194. doi: 10.1007/s11060-009-9836-1. [DOI] [PubMed] [Google Scholar]
- 228.Johansson M, Oudin A, Tiemann K, Bernard A, Golebiewska A, Keunen O, Fack F, Stieber D, Wang B, Hedman H, et al. The soluble form of the tumor suppressor Lrig1 potently inhibits in vivo glioma growth irrespective of EGF receptor status. Neuro Oncol. 2013;15:1200–1211. doi: 10.1093/neuonc/not054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.He XY, Liu XJ, Chen X, Bian LG, Zhao WG, Shen JK, Sun QF. Gambogic acid induces EGFR degradation and Akt/mTORC1 inhibition through AMPK-dependent-LRIG1 upregulation in cultured U87 glioma cells. Biochem Biophys Res Commun. 2013;435:397–402. doi: 10.1016/j.bbrc.2013.04.099. [DOI] [PubMed] [Google Scholar]
- 230.Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, Huang J, Dhakal S, Ivanova E, Xiao Y, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc Natl Acad Sci USA. 2010;107:6912–6917. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fassler R, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 232.Egan JE, Hall AB, Yatsula BA, Bar-Sagi D. The bimodal regulation of epidermal growth factor signaling by human Sprouty proteins. Proc Natl Acad Sci USA. 2002;99:6041–6046. doi: 10.1073/pnas.052090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Rubin C, Litvak V, Medvedovsky H, Zwang Y, Lev S, Yarden Y. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr Biol. 2003;13:297–307. doi: 10.1016/s0960-9822(03)00053-8. [DOI] [PubMed] [Google Scholar]
- 234.Barbachano A, Ordonez-Moran P, Garcia JM, Sanchez A, Pereira F, Larriba MJ, Martinez N, Hernandez J, Landolfi S, Bonilla F, et al. SPROUTY-2 and E-cadherin regulate reciprocally and dictate colon cancer cell tumourigenicity. Oncogene. 2010;29:4800–4813. doi: 10.1038/onc.2010.225. [DOI] [PubMed] [Google Scholar]
- 235.Ivliev AE, ‘t Hoen PA, Sergeeva MG. Coexpression network analysis identifies transcriptional modules related to proastrocytic differentiation and Sprouty signaling in glioma. Cancer Res. 2010;70:10060–10070. doi: 10.1158/0008-5472.CAN-10-2465. [DOI] [PubMed] [Google Scholar]
- 236.Ferron SR, Pozo N, Laguna A, Aranda S, Porlan E, Moreno M, Fillat C, de la Luna S, Sanchez P, Arbones ML, et al. Regulated segregation of kinase Dyrk1A during asymmetric neural stem cell division is critical for EGFR-mediated biased signaling. Cell Stem Cell. 2010;7:367–379. doi: 10.1016/j.stem.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 237.Pozo N, Zahonero C, Fernandez P, Linares JM, Ayuso A, Hagiwara M, Perez A, Ricoy JR, Hernandez-Lain A, Sepulveda JM, et al. Inhibition of DYRK1A destabilizes EGFR and reduces EGFR-dependent glioblastoma growth. J Clin Invest. 2013;123:2475–2487. doi: 10.1172/JCI63623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Naruo Y, Nagashima T, Ushikoshi-Nakayama R, Saeki Y, Nakakuki T, Naka T, Tanaka H, Tsai SF, Okada-Hatakeyama M. Epidermal growth factor receptor mutation in combination with expression of MIG6 alters gefitinib sensitivity. BMC Syst Biol. 2011;5:29. doi: 10.1186/1752-0509-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chang X, Izumchenko E, Solis LM, Kim MS, Chatterjee A, Ling S, Monitto CL, Harari PM, Hidalgo M, Goodman SN, et al. The relative expression of Mig6 and EGFR is associated with resistance to EGFR kinase inhibitors. PLoS One. 2013;8:e68966. doi: 10.1371/journal.pone.0068966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhang X, Zhang H, Tighiouart M, Lee JE, Shin HJ, Khuri FR, Yang CS, Chen Z, Shin DM. Synergistic inhibition of head and neck tumor growth by green tea (-)-epigallocatechin-3-gallate and EGFR tyrosine kinase inhibitor. Int J Cancer. 2008;123:1005–1014. doi: 10.1002/ijc.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Milligan SA, Burke P, Coleman DT, Bigelow RL, Steffan JJ, Carroll JL, Williams BJ, Cardelli JA. The green tea polyphenol EGCG potentiates the antiproliferative activity of c-Met and epidermal growth factor receptor inhibitors in non-small cell lung cancer cells. Clin Cancer Res. 2009;15:4885–4894. doi: 10.1158/1078-0432.CCR-09-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]