Abstract
The mammalian neocortex is a sheet of cells covering the cerebrum that provides the structural basis for the perception of sensory inputs, motor output responses, cognitive function, and mental capacity of primates. Recent discoveries promote the concept that increased cortical surface size and thickness in phylogenetically advanced species is a result of an increased generation of neurons, a process that underlies higher cognitive and intellectual performance in higher primates and humans. Here, we review some of the advances in the field, focusing on the diversity of neocortical progenitors in different species and the cellular mechanisms of neurogenesis. We discuss recent views on intrinsic and extrinsic molecular determinants, including the role of epigenetic chromatin modifiers and microRNA, in the control of neuronal output in developing cortex and in the establishment of normal cortical architecture.
Keywords: Cortex, Growth, Surface/radial expansion, Progenitor diversity, Molecular determinants, Epigenetic regulation, MicroRNA
Introduction
The neocortex (NCX), a brain structure specific to mammals, is a layered sheet of cells covering the cerebrum that is composed of multiple subtypes of neuronal and glial cells. In the NCX, neurons with specific morphological, connective, and transmission characteristics and functions are distributed radially into six laminae and tangentially in distinct functional domains, whose compositions increase progressively in phylogenetically recent species. During development, the lateral and radial growth of the cortex determines the size of the cortical surface and its thickness, respectively. The expansion of the size of the cerebral hemispheres, and the appearance of a highly complex multi-laminated and folded NCX, were critical events in mammalian evolution. Although the basic principles for NCX development in mammals with lissencephalic NCX (in general, showing a small cortical surface and no folding) or gyrencephalic NCX (an expanded cortical surface with folding) are similar, the modulation of the developmental processes in different species causes important variations in neocortical architecture, surface, size, and thickness. It is therefore still not possible to reconcile the available wide range of experimental data in one single model. Here, we introduce recently discovered key features of cortical neurogenesis including key cellular and molecular mechanisms involved in cortical tangenitial and radial expansion.
Progenitor diversity
The majority of the NCX neurons are glutamatergic projection neurons generated by neuroepithelial (NE) stem cells and progenitors, located in the germinative ventricular (VZ) and subventricular zones (SVZ) of the dorsal telencephalon. Based on their apicobasal polarized morphology during the interphase of the mitotic cycle, distinct types of neuronal progenitors have been identified: (1) bipolar progenitors, the NE cells, and the ventricular radial glial cells (vRGCs); (2) nonpolar progenitors, the intermediate progenitor cells (IPs, also known as basal progenitors, BP); and (3) monopolar progenitors, the short neural precursors (SNPs) and the outer radial glial cells (oRG) [1–4] (Fig. 1; Table 1).
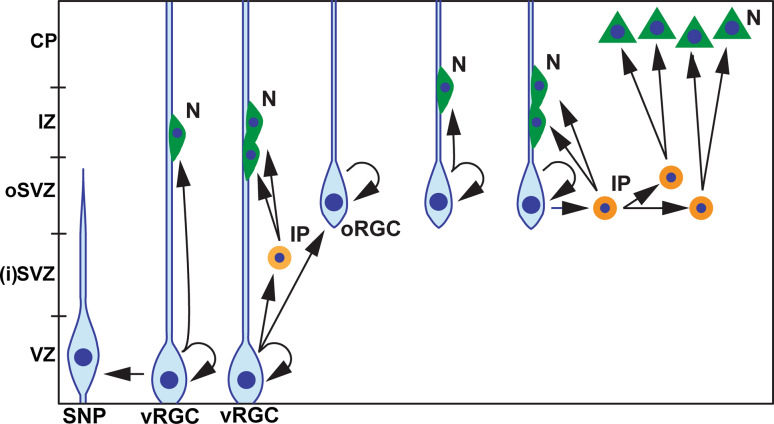
Fig. 1.
Main progenitor subtypes in developing neocortex. The scheme illustrates the main cortical progenitor subtypes. The ventricular (apical) Pax6(+) radial glial cell (vRGC) self renews in the ventricular zone (VZ) and generates a neuron (dark green cell, N) either directly or through produced Tbr2(+) intermediate progenitor (IP) that after a symmetrical division in the subventricular zone (SVZ), amplifies the neuronal output. Mostly in the rodent cortex, vRGCs generate neurogenic short Pax6(+) radial glial progenitors (SNP), attached to the apical ventricular surface. Predominantly in gyrencephalic species, vRGCs produce abundant set of Pax6(+) outer radial glial cells (oRGC), located more densly in the outer subventricular zone (oSVZ) where they divide either symmetrically to expand their pool or asymmetrically to self-renew and generate IPs-like cells, robustly amplifying the generated from oSVZ neurons. The IP cells (light orange) in the rodent cortex derive from vRGCs, and are located in SVZ (equivalent to inner, iSVZ, in human cortex) while the IP-like cells (dark orange) in ferret/human cortex originate predominantly from oRG located in oSVZ (based on literature data: [22, 38, 39, 43, 94, 196])
Table 1.
Progenitor subtypes in mouse cortex
| Type | Stage | Location | Apicobasal morphology | Marker | Position at mitosis | Progeny | References |
|---|---|---|---|---|---|---|---|
| NE | E9.5–E13.5 | VZ | Bipolar | Prominin 1 (CD133), Nestin | Apical | NEs, RGCs, IPs, Ns | [2, 23] |
| vRGCs | E10.5–P0 | VZ | Bipolar | Pax6, Sox2, Glast, Blbp, S100β, Vimentin, RC2, Nestin | Apical | vRGCs, IPs, Ns, oRGCs | [2] |
| bpRGCs | SVZ/VZ | Bipolar | RC2, Glast, hGFAP, Sox2, Pax6 | Sub apical | bpRGCs, oRGCs | [24, 25] | |
| IPs | E10.5–P0 | SVZ | Nonpolar | Tbr2 | Basal | IPs, Ns | [4, 23, 26, 27] |
| oRGC | E12.5–P0 | oSVZ/iSVZ | Monopolar | Pax6, Sox2, Blbp, Nestin, Glast | Basal | oRGCs, IPs, Ns | [35–39] |
| SNPs | E12.5–E16.5 | VZ | Monopolar | Tubulin α1 promoter | Apical | SNPs, Ns | [20, 22] |
NE neuroepithelial cells, vRGCs venrticular radial glial cells, oRG outer radial glial cells, IPs intermediate progenitors, Ns neuron, SNPs short neural precursors, bpRGCs bipolar radial glial cells, LL lower layers, UL upper layers, VZ ventricular zone, SVZ subventricular zone, oSVZ outer SVZ, iSVZ inner SVZ
Bipolar progenitors
The NE cells are polarized along their apicobasal axis showing apical and basal attachments to the ventricular surface and the pial (basal) surface, respectively. They specifically express the intermediate filament protein Nestin and show apical localization of the membrane protein prominin-1 (CD133) [5, 6]. Their tight and adherents junctions are associated with complexes such as the partition-defective Par protein complex [7], and they express intergrin alpha as basal lamina component [2].
After the onset of neurogenesis [in the mouse around embryonic day (E) 10.5], the NE cells in VZ progressively transform into bipolar radial glial cells (bpRGCs) that extend an apical process to the ventricular surface, and a long basal process across the enire thickness of the developing NCX, reaching the basement membrane at the pial surface. As the apical plasma membrane of the RGCs corresponds to the ventricular surface where they undertake mitosis, these progenitors are named ventricular radial glial cells (vRGCs) (or apical, aRGCs). While losing few early neuroepithelial features, vRGCs gain some astroglial characteristics, namely expression of the astrocyte-specific glutamate transporter Glast/Scl1A3, the calcium binding protein S100β, Vimentin, glial fibrillary acidic protein Gfap, brain-lipid-binding protein (Blbp), and transcription factor (TF) Pax6 [8, 9]. Extending their long, bipolar fibers between the VZ and the pial surface, vRGCs were considered to belong to the astroglial cell lineage. They were thought to provide only guidance for newly born neurons during their migration towards their final locations in the cortical plate (CP), and to generate astrocytes and oligodendrocytes at the end of neurogenesis [10]. Current evidence has shown, however, that vRGCs are pluripotent progenitors, generating both cortical neurons and astroglia [11–17]. During the mitotic cell cycle, NEs and vRGCs undergo interkinetic nuclear migration (INM), a mitosis phase-dependent migration of the cell nucleus through which the nuclei end up at the apical or basal surface of the VZ during the M- or S-phase, respectively [18]. The apicobasal polarity and self-renewal of vRGCs is maintained by the Rho-GTPase cdc42, which activates the Par-complex to maintain adherent junction coupling and VZ progenitor fate. Upon disruption of cdc42 gene function, the vRGC in VZ gradually convert into IPs [19].
Similar to the vRGCs, a small subset of the progenitors with radial morphology, the short neural progenitors, SNPs, also divide at the apical surface; however, they retract their basal processes during the M-phase of the mitotic cycle [20, 22], thus extending only one apical process to the ventricular surface. Collectively, the NE cells, vRG progenitors, and SNPs are referred to as apical progenitors [23]. The recently described bipolar radial glia cells (bpRGCs) in cortical VZ, and even more prominently in SVZ, of the gyrencephalic ferret/human cortex, do not contact the apical or pial surfaces, and divide at subapical positions, generating progeny intensively proliferating in species with higher gyrifiaction [24, 25].
Nonpolar progenitors
The IPs are non-epithelial unpolarized cells assumed to be principal progenitors for the generation of cortical neurons. Being the progeny of vRGCs already produced during the early stages of neurogenesis [23], IPs migrate away from the VZ to form a secondary proliferative zone, the SVZ, that becomes visible after E12.5 in the mouse cortex [23, 26, 27]. Progenitors, dividing at the basal surface of the VZ (and in later stages of neurogenesis in the SVZ including oRG, as described below), are collectively referred to as basal progenitors [23]. The specification of IP fate depends on the sequential expression of a transcription factor cascade (Pax6–Ngn2–Tbr2) in vRGCs and in the IPs derived from them (reviewed in [30]). Differential expression of the TFs Pax6 (Paired box 6) and Tbr2 (T-brain gene-2)/EOMES identifies the IPs as Tbr2(+)/Pax6(−) cells, in contrast to vRGCs that are Pax6(+)/Tbr2(−) cells [1, 28, 29]. In addition, expression of molecular markers such as Svet1 (intron region of the putative Netrin gene receptor Unc5d), vGlut2, NeuroD1/NeuroD2 and TFs Cux1/Cux2 [31, 32, 34] and Insm1 (insulinoma-associated 1) distinguish the IPs from vRGCs [33].
Monopolar progenitors
In mammals with enlarged and folded NCX, particularly in primates and especially in the human brain, the SVZ expands and becomes clearly subdivided into an inner (iSVZ) and outer (oSVZ) zone, delineated by fiber tracts [42–44]. The oRGC are a novel RGC subtype, identified initially in the outer subventricular zone (oSVZ) of gyrencephalic species [24, 35–40]. Although scarce in number, oRGC (also named basal RGCs, bRGCs) have been detected in smooth brain species in the SVZ just above the band of Tbr2(+) IPs [35, 36, 41]. However, a substantial difference is that, in mouse, oRGC divide asymmetrically to self-renew and directly generate neurons, whereas in human cortex, oRGC in oSVZ generate transamplifying cells (IP-like cells) that in turn produce neurons [36]. The oRGC extend only one basal process that is used by the generated neurons in oSVZ to migrate by somal translocation into the CP. As a progeny of the vRGCs [36], oRG show characteristics similar to those of vRGCs, including the expression of vRGC markers such as Pax6, Sox2, Nestin, Glast, and Blbp. Despite that, oRGC do not express the vRGC markers prominin1 (CD133), Par3 or aPKC (atypical protein kinase C) and they do not undergo INM during mitosis [37–39]. Based on cellular and marker expression characteristics, it is assumed that, in primate brain, progenitors located in iSVZ are close to the rodent IPs, while the enlarged set of oSVZ progenitors (oRGC, and their trans-amplifying IP-like cells), particularly in the human cortex, contributes to increased thickness of upper layer (UL) neuronal identities [45] (Fig. 3). Since only a few oRGC exist in the SVZ of the rodent cortex, the pronounced presence of oRGC in iSVZ/oSVZ is considered to be a cellular mechanism underlying the cortical expansion and folding in phylogenetically advanced mammals [3, 38] (Figs. 1, 3).
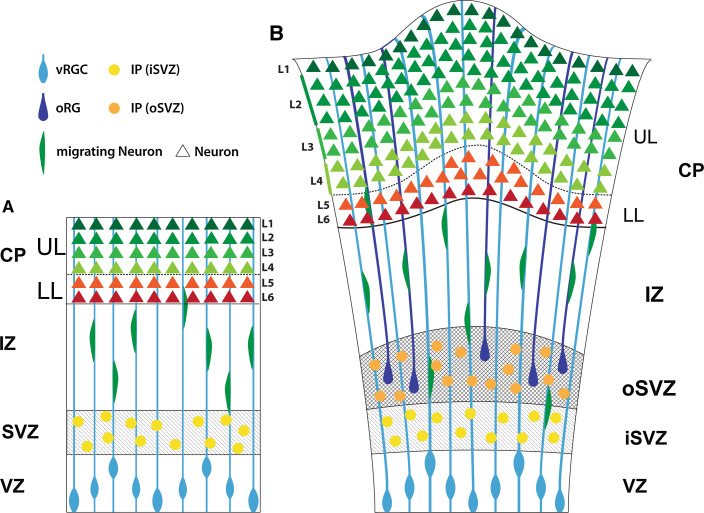
Fig. 3.
Features of cortical neurogenesis in rodent and human brain. A In lissencephalic mouse brain, the ventricular radial glial cells (vRGCs) divide at the apical surface in the ventricular zone (VZ) to self-renew and generate either neurons or more often, intermediate (IPs) progenitors (hell orange) that divide in the subventricular zone (SVZ, rodents) or inner subventricular zone (iSVZ, ferret/human) to generate pairs of neurons. The neurons use the RG radial fibers as a scaffold to migrate into cortical plate (CP). In rodents, only few outer radial glial cells (oRG) are found at the basal surface of the SVZ, which undergo direct neurogenesis and generate neurons [36]. B In gyrencephalic folded human brain, the SVZ is augmanted, and subdivided into inner (iSVZ) and outer subventricular zones (oSVZ). The oSVZ contains highly abundant outer radial glial cells (oRGC) extending only one basal fiber towards the pia. The oRG cells self-renew and generate neurogenic Tbr2(+) IPs-like cells (dark-orange) which continue dividing for a limited number of cycles, before generating increased number of neurons. These neurons use the basal process of oRGC as additional scaffold, helping their lateral dispersion during cortical folding. Different from IPs in the mouse SVZ, the IPs in iSVZ and oSVZ undergo multiple mitotic cycle, increasing the output from the original vRGCs. The composition of oSVZ in different gyrencephalic species (ferret, macaque, human) is highly heterogeneous and requires more conclusive views on their molecular features, lineage relationships and functions. At the end of neurogenesis, the vRGCs generate cells from the glial cell lineage, astrocytes and oligodendrocytes (not shown in the figure; for a review, see [197])
Modes of progenitor division
During cortical development, progenitors undergo progressive cell divisions, leading from the expansion of the progenitor pool to the generation of neurons. It is the mode of cell division, as well as the role of the spindle rotation in the selection of the cleavage plane, which finally determines the size of the mammalian neocortex [11, 16]. Before initiation and during the early stages of neurogenesis, the NE/RGCs divide mostly in a symmetric manner to yield daughter cells with identical apical and basal components, thus supplying the progenitor pool and inducing the tangential growth of the cortex (Fig. 2a) [46, 47]. Although evidence from a fixed- and time-lapsed imaging study has convincingly demonstrated that basal processes of the dividing vRGCs in mouse, chick, and zebrafish embryonic cortex may split during the symmetric proliferative divisions (reviewed by Fietz and Huttner [1]), there are data indicating that, on symmetric divisions of RGCs, the basal process is inherited by only one of the RGC deughters, while the second one re-grows its basal process. However, whether, during the asymmetric RGCs division, the basal process is strictly inherited either by the RGC daughter (and required for its self-renewal) or by the generated neuron (using it as a structure for migration), is still an unresolved issue.
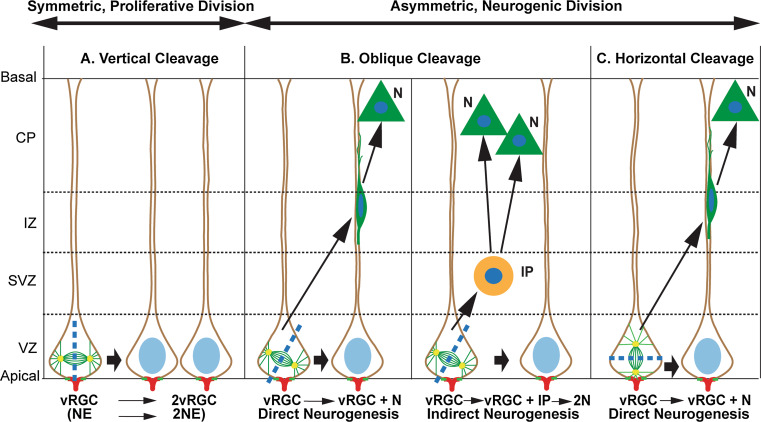
Fig. 2.
Divison modes of cortical progenitors in rodent cortex. A Before beginning and during the early stages of neurogenesis, the neuroepithelial cells (NE) and ventricular radial glial cells (vRGCs), divide symmetrically through vertical (apicobasal) cleavage to self renew, enlarging the stem cells/progenitor pools. Consequently, the two daughter cells receive equal amount of apical membrane components. The basal process of symmetrically dividing RGC can either split or can be inherited by the older RGC while the new RGC daughter re-grows a basal process (reviewed in [1]). B vRGCs/NE cells have a small apical domain (red) and tight junctions (green) that anchor them to each other as well as to the ventricular surface. During neurogenesis, vRGCs divides asymmetrically, preferably through oblique/vertical cleavage, which results in unequal distribution of apical cell fate determinants. Consequently, vRGCs self-renew and generate either neuron (direct mode of neurogenesis) or another type of progenitor, the intermediate progenitor (IP) which produces two neurons after a symmetrical division in SVZ (rodent cortex) or more neurons after multiple divisions in the iSVZ (ferret/human cortex, not presented in the schema). Thus, IPs act as amplifiers allowing through indirect mode of neurogenesis to increase the number of neurons generated from an RG unit in VZ at a particular developmental stage. Because of still controversial view on mode of inheritance of the basal processes especially in asymmetric division (reviewed in [1]), this issue is not included in the figure. C Rarely, vRGCs in VZ (rodents) can divide asymmetrically through horizontal (planar) cleavage generating RG progenitor and neuron. This is the preferable division mode for oRGC in oSVZ/iSVZ (ferret/human). Recent evidence suggests that the daughter cell that maintains apical contact inherits the basal fiber of the RGC through which it receives Notch and Integrin signalling and becomes a vRGC. The daughter cell that lacks a basal process express Notch ligands and activates Notch signalling in neighboring vRGCs, and starts expression of IP specific markers (based on literature data: [2, 3, 51, 196])
The cortical thickness, on the other hand, depends on two different modes of progenitor proliferation. During early neurogenesis, although infrequently, the vRGCs undergo asymmetric divisions through which the apical and basal cell elements are unevenly distributed between the two daughter cells. This leads either to the production of one neuron or to a neurogenic basal progenitor (IPs/oRGC), in addition to the self-renewal of vRGCs [48, 50, 51] (Fig. 2c). Before entering terminal neurogenic divisions in the SVZ, these progenitors undergo 1–3 (IPs in rodents) or multiple (oRGC) subsequent proliferative divisions with horizontal or random cleavage planes to amplify their pools, thereby expanding their neurogenic output and thus leading to expansion of cortical size (surface and thickness) (Fig. 2b) [23, 26, 27, 52]. Later studies have strongly suggested that the acquisition of distinct cell fates after the RGC division critically depends on the inheritance of the tiny apical domain containing putative cell fate determinants [49] (Fig. 2b).
Mechanisms of cortical surface expansion
Mechanisms that may control specifically either surface area size or radial cortical thickness and folding are still not well understood. According to the classical radial unit hypothesis [46], proliferating vRG progenitors in the VZ generate neurons, which use the RGC processes as a scaffold to migrate into the cortex where they reach distinct functional areas (Fig. 3). Thus, neurons with the same ontogeny form radial columns with specific functions. It has been proposed that the size of cerebral surface critically depends on the number of radial units in the VZ, whereas the cortical thickness is determined by the neuronal output from the VZ progenitors of each column. Although being interconnected, certain cellular and molecular mechanisms show a tendency to preferably influence one or the other process.
Transition from NE to RGC fate and onset of neurogenesis
During development, the timing of the onset of the transition from NEs to vRGCs decides whether the progenitors will cease self-renewal and begin generation of neurons. The delayed initiation of the asymmetric mode of NE/RGCs division results in amplification of the progenitor pool [44]. Such a prolonged phase depends on cell-cycle lengthening [53, 54] and correlates with excessive expansion of cortical size and surface, as seen in phylogenetically advanced mammals [10, 55–57]. Maintenance of the stem-like properties of RGCs is critically dependent on Notch signaling. The transformation of NE cells into vRGCs is delayed in mice deficient for both receptors, Notch1 and Notch3 [58], while expression of activated Notch1 in NE cells promotes acquisition of generation of premature vRGC identity [59]. Similarly, loss of Notch activation as seen in Mib1 (Mind bomb 1) conditional KO mutant mice leads to depletion of the RGC number, and premature differentiation into IPs and neurons [60]. Interestingly, the cellular source for the Notch signaling turned out to be the SVZ IPs as well as migrating neurons that express Mib, a RING-type E3 ubiquitin ligase which induces endocytosis of the Delta ligand in RGCs [60].
During mitosis of vRGCs, the apical polarity protein 3 (Par3) becomes dispersed in the proliferating cells. The mode of Par3 distribution (even vs. polarized) in the daughters determines the symmetric or asymmetric inheritance of the polarized components. Consequently, the cell with the greater amount of Par3 parallels high Notch signaling activity in the daughter cell that renews as vRGC [7]. In contrast, cells with inherited less Par3 express Notch signaling activity and acquire cell fate of either neuron or IP.
Environmental signals regulate progenitor proliferation
Cortical growth and patterning depends on signaling molecules emanated from inductive centers of the cortical primordium, formed after closure of the neural tube. Such determinants belong to morphogen families like Wnts, bone morphogenic proteins (Bmp), transforming growth factors (Tgfs), retinoic acid (RA), sonic hedgehog (Shh), and fibroblast growth factors (Fgfs) [61] (Table 2). For instance, in vRGCs, β-catenin is a constituent of the adherent junctional complexes and participates in Wnt signal transduction. Due to enforced re-entering of the progenitors into mitosis, transgenic expression of a stabilized β-catenin in NE/RGCs results in higly increased progenitor number and folding of the VZ, enlarged and folded cortical surface area, and increased thickness [62]. Similar expansion of cortical surface was observed following a decrease in cell death through elimination of Caspase 9 [63] or EphrinA7 (reviewed in [64, 65]).
Table 2.
Molecular determinants of cortical surface area (CS), thickness (CT) and folding (CF)
| Genes/Mouse | Affected progenitors | Cortical phenotype | References |
|---|---|---|---|
| Signaling factors | |||
| Lrp6 (KO) | IPs | CT, CS | [204] |
| β-Catenin (cKO/cOE) | IPs | CT, CS, CF | [62, 205] |
| GSK-3 (cKO) | vRGCs, IPS | CT, CS, CF | [206] |
| Mib1 (cKO) | vRGCs, IPS | CT, CS | [60] |
| Fgfr1/2/3 (cKO) | IPs | CT, CS | [207] |
| Fgf10 (KO) | vRGCs, IPS | CT, CS | [66] |
| Smoothened (KO) | IPs | CT, CS | [208] |
| Shh (cKO) | IPs | CT | [209] |
| TFs affecting earlier vs. later born neuronal fate | |||
| Foxg1 (KO, cKO) | IPs | CT, CS | [106, 115] |
| Ngn1/2 (KO) | IPs | CT | [110] |
| Pax6 (cKO, cOE) | IPS, oRGs | CT, CS | [86, 210–213] |
| Tlx1 (KO) | IPs | CT | [112] |
| Id4 (KO) | IPs | CT, CS | [113, 214] |
| Sall 1 (putative TF) (cKO) | IPs | CT, CS | [114] |
| TFs affecting the expansion of IP pool | |||
| Tbr2 (cKO) | IPs | CT, CS | [90, 91] |
| Insm1 (KO) | IPs | CT | [33] |
| Arx (cKO) | IPs | CT, CS | [92] |
| Cux2 (KO) | IPs | CT | [120] |
| AP2y/Tcfap2 (KO) | IPs | CT | [95] |
| Trnp1 (KD) | IPS, oRG, bpRGCs | CF | [24, 25] |
| NFIB (KO) | oRG | CT | [126] |
| TFs involved in progenitor specification and layer identity | |||
| Fezf2 (KO) | CT | [99, 215] | |
| Otx1 | CT | [118, 119] | |
| Cux1/2 (KO) | CT | [31, 32, 120] | |
| POU3F2/POU3F3 (KO) | CT, CS | [121–123] | |
| TFs acting as postmitotic determinants | |||
| Bcl11b (Ctip2) | CT | [117] | |
| Sox5 (KO) | CT | [128, 129] | |
| Tbr1 (KO) | CT | [216, 217] | |
| Satb2 (KO) | CT | [164, 165] | |
| Chromatin and epigenetic factors | |||
| Brg1 (cKO) | vRGCs | CT, CS | [158, 219] |
| BAF170 (cKO, cOE) | IPs | CT, CS | [159, 160] |
| Ezh2 (cKO) | IPs, oRG | CT | [151] |
| Querkopf (cKO) | CT, CS | [141] | |
| AF9 (KO) | vRGCs, IPs | CT | [218] |
| Dnmt1 (cKO) | CT, CS | [146] | |
CT cortical thickness (referring to affected upper layers, lower layers or both), CS cortical surface, CF cortical folding, KO knockout, cKO conditional knockout, cOE conditional overexpression, KD knockdown, IPs Intermediate progenitors, oRG outer radial glia, vRGCs ventricular radial glia cells, bpRGCs bipolar radial glia cells
The timely transition from NE cells into vRGCs is also dependent on Fgfs and Notch signaling [66, 67] (Table 2). The powerful morphogen Fgf8, secreted from the anterior forebrain inductive center, controls area location across the anterio-posterior cortical axis [68, 69]. Fgf8 specifies anterior cortical fate via transcriptional regulation of TFs Er81, Foxg1, Nkx2.1, Pea, and SP8, but inhibits caudal fate through repression of TFs COUP-TF1, Emx2, and Wnt8b [70–72]. Nonetheless, Fgf8 also plays a role in the maintainance of normal proliferation and survival of cortical progenitors [73]. Fgf10 exerts a similar function mostly within the frontal cortex [66, 67]. Targeted deletion of Fgf10 delays the transformation of NE cells into vRGCs, leading to profound increase of neuronal output, laminar thickness, and the expansion of cortical size [66]. On the other hand, Fgf15 and Fgf2 support the differentiation of NE cells to vRGCs, promoting neurogenesis [66, 67, 74]. Moreover, an increase in the Fgf signaling via Fgf2 before the beginning of neurogenesis (and without affecting the IP numbers) leads to gyrus formation in the smooth mouse cortex with largely preserved layering and morphology [75]. In contrast, loss of function of all Fgf receptors during the earliest stages of corticogenesis leads to the severe loss of vRGCs and premature termination of neurogenesis, involving the Notch signaling pathway [76].
Spindle orientation and neurogenesis
Recent data suggest that spindle orientation exerts control on NE/RGC proliferation versus differentiation, thus influencing the progenitor pool and cortical size [51, 77–80]. While randomly orientated at the onset of mitosis, the spindle rotates during the metaphase to acquire planar orientation, thus becoming truly parallel to the surface of the NE/RGC until the anaphase of the cell cycle. After a vertical cleavage, the generation of identical daughter cells (self-renewal), and the enlargement of the progenitor pool are permitted, which is of crucial importance for maintenance of normal cortical size and surface.
Mutations of MCPH1, CDK5RAP2, and NDE1 genes in human affect spindle orientation and progenitor proliferation [81, 82], causing microcephaly [83]. An important regulator of symmetric proliferative progenitor division during early neurogenesis is the protein phosphatase PP4c, which phosphorylates the microtubule binding protein Ndel1, assuring the planar orientation of mitotic spindle [88]. Consequently, deletion of PP4c specifically during early neurogenesis leads to random spindle orientation and premature asymmetric neurogenic division, diminishing the progenitor pool and the final neuronal output. Maintenance of a precise alignment of the mitotic spindle critically depends on Aspm (abnormal spindle-like microcephaly-associated protein) that is localized at the poles of the mitotic spindle [84, 85]. Similarly, the microtubule-associated protein gene Spag5 (or Astrin), a direct target of TF Pax6, is also localized in the spindle poles [86]. The elimination of Spag5 function promotes non-vertical (differentiative oblique) instead of vertical symmetric (proliferative) divisions, depleting the progenitor pool for normal neurogenesis, and contributing to the reduced radial thickness of the cortex in Pax6-deficiency [86]. Spindle randomization in vRGCs has also been shown for the LGN protein complex, which, acting together with the dynein complex binding partner lissencephaly1 (Lis1), gathers pulling force in order to control the plane of cell division [77, 79].
Other facts imply, however, that spindle orientation may influence predominantly cell fate decisions in the neurogenic progenitor population [87]. Thus, conditional knockout and overexpression of Incuteable (Incs) in mice revealed a profound effect on the fraction of RGCs undergoing oblique divisions: mutation of mInsc almost abolishes oblique mitotic spindle while mInsc overexpression promotes oblique divisions and specifically increases the number of Tbr2(+) IP cell lineage [87]. This results in premature transition of RGCs to IPs, and severely reduced cortical thickness, due to apoptotic death of the progenitors in VZ that contains prematurely differentiated neurons [88]. Overall, these new data support the view that a change in the spindle orientation in a critical phase during early neurogenesis is directly related to the transition between symmetric and asymmetric progenitor division and has a potential to control cortical surface growth.
Mechanisms of radial cortical expansion
Regulation of IP production
As a framework for understanding the developmental expansion of cortical surface and thickness, the modified radial unit hypothesis provides the basis to understand how a molecularly specified protomap of cortical arealization in VZ progenitors is transferred to the CP [46, 55, 70, 71]. Since IPs are already present at the beginning of neurogenesis, it has been predicted that alteration of IP numbers would mostly change radial thickness and not surface area [46]. The relative abundance of IPs and/or oRGC compared to vRGCs number is assumed to determine both cortical size and folding in gyrencephalic species [4, 57, 89].
The genesis of IPs from vRGCs and their proliferative capacity is dependent on both intrinsic and extrinsic factors, including diffusible morphogenes (Table 2) [21, 205–209]. Most of the studied intrinsic determinants of IP fate have turned out to be transcription factors. The specific expression of TF Tbr2 in IPs is necessary and sufficient for IP specification: conditional ablation of Tbr2 leads to loss of IPs and decrease of cortical thickness affecting all cortical layers [90, 91]. Furthermore, TF Insm1 controls the conversion of RG progenitors into IPs, and, accordingly, Insm1 deletion profoundly diminishes the IP number leading to marked reduction of cortical radial thickness [33]. Expressed in a subset of vRGCs, and showing an enhanced level at the start of IP production, the TF AP2gamma/Tcfap2c has a crucial role in the upregulation of important IP molecular determinants (e.g., Tbr2 and NeuroD1), placing this factor as an upstream regulator of IP progenitor fate, acting in a region-specific manner (in occipital cortex) [95]. Cortical progenitors in the VZ (RGCs) and SVZ (IPs) express either Cyclin D1 or CyclinD2, two G1-phase active cyclins, respectively [93, 94]. Although not required for IPs fate specification, Cyclin D2 plays a crucial role in maintaining normal size of the IP pool and cortical size and thickness [93].
Role of oRG progenitors
The selective influence of IPs on radial cortical growth and gyrification in primates has been challenged by Nonaka-Kinoshita et al. [94]. They have provided proof that a massive expansion of IPs is not sufficient to induce folding in transgenic mice overexpressing cyclin D4 to promote IP proliferation, but instead leads only to surface expansion. However, in the gyrencephalic cortex of ferret, which shows abundant presence of oRG, the same manipulation causes both cortical surface area expansion and folding, indicating a specific contribution of oRGC over IPs in the radial complex organization in phylogenetically distinct species [94]. Moreover, the expanded set of oRG in iSVZ provided additional scaffold for the oRGC-neuronal progeny, also allowing lateral dispersion when reaching the CP. Overall, these findings support the view that the two types of abventricular progenitors, IPs (in SVZ) and oRGC (in iSVZ/oSVZ), have specific functions in the expansion of surface versus radial thickness/folding, respectively, across the mammalian phylogeny [3, 41, 43] (Fig. 3).
An important difference between the mouse smooth and gyrencephalic ferret cortex is the timely specific distribution of oRGC in SVZ. Although few oRGC (approximately 3–10 % of all progenitors) have been identified in the mouse cortex during early neurogenesis (at E12.5), in the ferret, a large set of oRG (approximately 50 %) and Tbr2(+) IPs are located in iSVZ [36, 98]. This results in a substantial increase of the progenitor pool during early neurogenesis, extensive radial growth with increased UL neuronal numbers, and cortical folding in the ferret brain [98]. It has therefore been proposed that cortical radial growth reflects specific expansion of the germinative zones, containing distinct primary sources for neurogenesis: the VZ in the smooth cortex, and iSVZ or oSVZ in species with low-folded (ferret) or highly folded cortex (non-human primates and humans), respectively [40, 42, 98]. Along the same lines, despite the scarce presence of oRGC in the lyssencephalic mouse brain [35, 36], the Götz laboratory have recently induced augmented radial cortical thickness and gyrification by manipulating the expression of the Trnp1 (TMF1-regulated nuclear protein 1) gene, which encodes for a DNA-binding protein [24, 25], controlling the cell proliferation [96]. In vivo manipulation of the Trnp1 expression level (normally high in vRGCs and low in IPs/oRGC) disclosed specific effects in cortical architecture. Overexpression of Trnp1 promotes the NE/vRGC renewal and, accordingly, the tangential/lateral expansion of the cortical surface. Lowering the Trnp1 level causes radial expansion due to increased numbers of both IPs and oRGC in SVZ and folding of the lyssencephalic cortex. These interesting findings support the view that establishment of the capacity for cortical folding during mammalian evolution may involve superimposed layers of cellular decisions including modulation of the pool size of not only oRGC but also IPs, as predicted in the IPs hypothesis [97].
Reminiscent to IPs that are a source of Notch signaling for vRGCs [60], the oRGC express Hes1, the main effector of Notch [38]. Pharmacological inhibition of Notch signaling in cultured human cortical tissue induces premature neuronal differentiation, implying that Notch signaling prevents oRG differentiation, thereby regulating the neuronal output [38]. Similarly, integrin signaling, via contact between the basal fiber of oRGC and cortical basal lamina, maintains the oRGC identity [37].
Selective expansion of distinct cortical layers affect cortical thickness
Neurons with distinct layer identities are produced following the intrinsically encoded “inside-first–outside-last” developmental program, according to which early born neuronal subtypes populate the lower (infragranular) cortical layers (LL, L6, L5) while later born neurons progressively reach the upper (supragranular) layers (UL, L4–L2) (reviewed in [99]). Early work proposed that expansion of the thickness of the ULs compared to LLs in the human brain contributes to the increase of the NCX surface and formation of gyri [100]. In the macaque cortex, the period of neurogenesis includes 28 cell cycles compared to only 11 cycles in the rodent brain [101, 102]. This causes a substantial enlargement of the progenitor pool and increased production of UL neurons, mostly from iSVZ/oSVZ progenitors coordinated with an enlarged cortical surface in gyrencephalic species [103].
A distortion of the balance between the available progenitor pools (apical vs. basal progenitor subtypes) leads to specific alterations in cortical thickness and surface area [55, 56, 104, 105]. A recent report suggests that, even before the start of neurogenesis, expression of TF Cux2 delineates two sets of Cux2(−) and Cux2(+) RGCs that produce LL and UL neuronal identities, respectively [107]. Even when forced to differentiate prematurely, the Cux2(+) RGCs still produce neurons with only UL identities [107, 108]. Nevertheless, this issue is still controversial since transgenic lineage analyses have indicated that Cux2 + RGCs could generate both UL and LL neurons [109].
A wealth of experiments have demonstrated that TFs play important intrinsic roles in many aspects of corticogenesis, including specification of neuronal layer identity. One molecular mechanism that determines earlier versus later born neuronal fate includes TF Foxg1, which prevents the generation of Cajal–Retzius neurons after their normal birthday (at E9.5), allowing subsequent generation of normally sized LL and UL [106]. Expression of proneuronal bHLH TFs Ngn1 and Ngn2 in the progenitors seems to determine predominantly LL neuronal fate, while TFs Pax6 and Tlx regulate the generation of UL neuronal fates by maintaining RGCs proliferation during early or late neurogenesis, respectively [110–112] (Table 2). Interestingly, at different stages of corticogenesis, TF Id4 exerts specific functions, at early stages for early stem cell proliferation and at later stages for differentiation of UL neuronal fate, thereby controlling both lateral expansion and thickness of NCX [112, 113]. Likewise, expression of TF Sall1 in early RGCs promotes proliferative over neurogenic division, whereas, later on, Sall1 supports both the exit of IPs from mitosis and the differentiation of UL neuronal identities [114]. Other studies indicate that TFs FoxG1 [106, 115], Tbr2 [90, 91], and Insnm1 [33] promote cortical growth/thickness mostly by enabling the expansion of the IPs pool (Table 2). Acting as a direct regulator of Cdkn1, an inhibitor of cell cycle progression, the TF Arx (Aristaless-related homeobox) controls the proliferation of IPs. Following targeted mutation, the ArxKO brains show strong reduction of radial thickness, due mostly to UL misbuilding [92].
Cortical thickness is also influenced by TFs that specify sets of neuronal layer identities [99, 110]. Both the zinc-finger TF Fezf2 (formely Fezl or Zfp312) [116, 117] and the orthodenticle homeobox TF Otx1 [118], are expressed primarily in early vRGCs, and subsequently in subsets of their L6/L5 or L5 neuronal progenies. Despite this correlation, Otx1 expression does not affect the establishment of LL neuronal fate [119], while Fezf2 is required for the generation and specification of cortocospinal motor neurons (CSMNs) [116]. In the adult brain of Fezf2 null mutant mice, CSMNs are absent; however, the total cortical thickness is identical with control brains due to the expanded generation of L6 neurons and L6 thickness, suggesting a fate switch (L5 into L6) of the early cortical progenitors upon abolishment of Fezf2 function [116, 125].
Expression of TFs Cux1/Cux2 and Svet1-RNA label progenitors in SVZ and UL neurons, whilst POU3F2/POU3F3 (formerly Brn1 and Brn2) and Tle1 (Groutcho1) are expressed in subsets of late RG progenitors and L3–L2 neurons [31, 32, 34, 99]. Notably, targeted deletion of Cux2 leads to increased progenitor proliferation in SVZ and increased UL cortical density and cortical thickness [120]. In contrast, double disruption of POU3F2/POU3F3 in mice causes a dramatic reduction in cortical thickness due to the specific loss of UL neurons and defective neuronal migration [121–123]. The Nuclear Factor One B (Nfib) is expressed in both vRGCs and LL neurons [124]. Upon targeted mutation of the Nfib gene, oRG progenitors and late basal progenitors are absent, the ULs become thinner, and later born UL neurons show delayed migration towards the CP, thus contributing to the reduction of cortical thickness [126].
Concomitant with their role in regulating layer-specific patterns of molecular identity, a number of TFs are required for correct neuronal migration from the germinative zones into the CP (reviewed in [127]). Such a role has been proposed even for TFs acting only as post-mitotic determinants in neuronal sets in distinct layers, e.g., Bcl11b (formerly Ctip2) expressed in L5, Sox5 expressed in ULs [128, 129], and TF Tbr1, which co-regulates multiple aspects of both early and later born neurons [127]. Similarly, targeted mutation of the adhesion molecule Mdga1 is necessary for L2/L3 neurons to migrate and “build” the normal thickness of the ULs in the cortex [130].
Emerging roles of epigenetic control in cortical development
The generation of the astonishing variety of neuronal subtypes in the developing cortex is coordinated by distinct transcriptional programs, in which epigenetic and chromatin regulations are believed to play decisive roles [131–133]. There is increasing evidence that a number of genetic neurodevelopmental disorders, which lead to intellectual disability, are caused by mutations in the genes encoding epigenetic factors and chromatin remodeling proteins [133, 134]. Here, only some key epigenetic mechanisms related to the determination of cortical size, thickness, and neural fate determination are considered, focusing mainly on the role of chromatin status modification and the recently discovered functions of microRNAs.
Chromatin modifiers
Epigenetic machinery controls heritable changes in gene expression programs in NSCs without altering the genomic sequence, thereby mediating a switch in the state of chromatin structure [135]. Epigenetic modifications and chromatin regulation display dual roles: they exert direct effects on gene transcription, and serve as platforms for the recruitment of TFs, allowing for persistent changes in chromatin states [131–133].
The basic structural unit of the eukaryotic chromatin is the nucleosome that consists of 145–147 base pairs DNA in 1.8 helical turns around an octamere of four evolutionary highly conserved histone proteins [136, 137]. The accessibility of TFs to their regulatory elements in downstream target genes is achieved by two main mechanisms: (1) covalent histone modification, and DNA methylation; (2) non-covalent, energy-dependent chromatin modifications involving ATP-dependent chromatin remodeling complexes, such as SWI/SNF (SWItch/Sucrose NonFermentable) [131–133]. Histone acetylation and deacetylation is mediated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. Histone acetylation results in relaxing chromatin condensation and activation of gene transcription while decrease of histone acetylation causes heterochromatin condensed state and suppression of transcription [138, 139].
Proliferation and self-renewal of NSCs depends on interaction of TF TLX-HDAC, that lead to transcriptional repression of its target genes, p21 (cyclin-dependent kinase inhibitor) and PTEN (a tumor suppressor gene) [140]. The first in vivo data that histone acetylation controls cortical development came from phenotypic analyses of the Querkopf knock-out mice, in which the gene encoding for K (lysine) acetyltransferase 6B (Kat6b) was deleted [141]. Querkopf-deficient mutants display severe defects in the central nervous system, specifically displaying a small-sized and thin cortex, and predominant dysgenesis of Otx1(+) L5 neurons [141]. Several recent studies have shown that mouse embryonic stem cell (mESCs) can recapitulate the sequential appearance of cortical layering in vitro; however, with a deficiency in generation of UL neuronal fate [142, 143]. Transient inhibition of HDAC by valproic acid (VPA) in mESC-derived neural progenitors not only induces neuronal differentiation but also selectively enriches the Cux1(+) UL vs Ctip2(+) LL neuronal fate, suggesting that histone acetylation promotes a temporal progression of neurogenesis leading to a fate switch from LL to UL producing progenitors [144].
Covalent chemical modification of DNA (more specifically methylation of the fifth position cytosine, 5mC) by DNA methyltransferases (Dnmt1, Dnmt2, Dnmt3) plays important roles in the regulation of gene expression, chromatin structure, gene imprinting, X-chromosome inactivation, and genomic stability [145]. Conditional deletion of Dnmt1 in both dividing neural precursor cells and postmitotic neurons in mouse brain resulted in severe thinning of CP [146]. Dnmt3b is robustly expressed during early neurogenesis (E10.5–13) whereas Dnmt3a is expressed in neural stem cells in postnatal brains [147]. However, their individual roles in cortical neurogenesis are still not clear. Additionally, simultaneous elimination of the proteins TET2 and TET3 involved in DNA demethylation causes impairment of progenitor differentiation and cell accumulation [148].
The polycomb group (PcG) proteins function within multiprotein complexes (named Polycomb repressive complexes, PRC1 and PRC2), which modify histones and generally repress transcriptional target activity [149]. Recent studies have disclosed that PRC2 controls the balance between progenitor self-renewal versus differentiation in the developing cortex. An enhancer of the Zeste homolog (Ezh2) is a catalytic component of PRC2, which catalyzes the addition of methyl groups in histone H3 at Lys27 (H3K27) in target gene promoters, causing epigenetic silencing [150]. More specifically, conditional abolishment of Ezh2 function before the onset of neurogenesis in the developing mouse cortex causes removal of the H3K27me3 repressive mark, and a shift towards differentiation affecting both the direct and indirect mode of neurogenesis [151]. Specifically, upon Ezh2 deletion, enhanced generation of IPs/early born LL neurons and depletion of IPs/later born UL neurons was found. This is most probably due to acceleration of the timing of developmental program that triggers glia differentiation inappropriately early [151].
In contrast to the PcG protein complex, the trithorax (TrxG) chromatin-remodeling complex mostly activates gene expression [152], regulating different developmental processes including stem cell proliferation [153]. Activation of gene expression via the methylation of lysine 4 on histone H3 is the main mechanism involved [154]. Recently, TF ZNF335/NIF-1 was identified as a gene disrupted in severe microcephaly in humans, exerting an important role in progenitor self-renewal and differentiation [155]. As a part of TrxG complex, ZNF335/NIF-1 interacts with the H3K4 chromatin methyltransferase complex, activating brain-specific genes that promote neuronal differentiation through control of REST repressor element 1 (RE1)-silencing transcription factor [156], acting through recruitment of HDACs [157]. Accordingly, brain-specific knockout leads to severely reduced forebrain structures [155].
The vertebrate mSWI/SNF chromatin remodeling complex, also known as the BAF (Brg1/Brm associated factor) complex, contains at least 15 different subunits, including two interchangeable core ATPase subunits (Brg1 or Brm), invariant core subunits (BAF47, BAF155, BAF170), and a variety of lineage-restricted subunits [133]. By using the energy of ATP to disrupt nucleosome-DNA contact, chromatin remodeling moves, removes, or exchanges nucleosomes along the DNA, and makes DNA–chromatin interactions accessible to transcriptional regulators allowing binding to their targets. Biologically distinct BAF assemblies are active in distinct neural cell types, e.g., the npBAF complex in neural progenitors and the nBAF complex in differentiated neurons [158]. Furthermore, the progressive transition from embryonic stem cells (ESCs) to neural progenitors is accompanied by the exchange of one of the two BAF155 subunits of the (esBAF) complex for BAF170 in the npBAF complex [133]. In the developing mammalian cortex, the BAF170 subunit is only transiently expressed in non-neurogenic (Pax6+/prominin−) vRGCs during the period of early neurogenesis (E10.5–E14.5) when neurons with LL identities are generated [159]. Analyses of the functional consequences of the in vivo ablation or overexpression of BAF170 in transgenic mice have revealed that BAF170 is an important player in the control of cortical surface size and thickness [159]. During early (E12.5–E13.5) neurogenesis, the loss of BAF170 function leads to a transient decrease in early born neuronal progenies (L6, L5 neuronal subsets) in conjunction with a surplus of generated Tbr2(+) IPs as well as premature (heterochronic) activation of UL fate specification genes (e.g., Cux1, Tle1) [159]. At later stages, however, the enlarged pool of IPs produces a modest surplus of LL neurons and large excess of UL neuronal fates. Together, the available data indicate that a dynamic expression of the BAF170 subunit in the BAF chromatin remodeling complex may control distinct aspects of cortical development, including temporal specification of a subpopulation of vRGCs as non-neurogenic, and maintaining an accurate timing for the initiation of the indirect (via IPs) mode of neurogenesis. Mechanistically, it was found that, during the early cortical neurogenesis (E10.5–E14.5) in the Brm-based BAF complex, TF Pax6 binds to BAF170 subunits, which recruit the REST-corepressor complex to the Pax6 downstream target gene promoters that are involved in the specification of IPs (Tbr2) and upper layer neuronal fates (Cux1, Tle1/Groucho1) [159] (Fig. 4). In addition, the loss of BAF170 in the BAF complex results in its substitution by the BAF155 subunit inducing euchromatin state, thus facilitating the accessibility of TF Pax6 to corresponding targets [160]. Consequently, upon elimination of the BAF170 form of the complex, the ultimate biological outcome of the BAF subunit exchange is the premature promotion of the indirect (via augmented IP pool) mode of neurogenesis. Interestingly, in a recent report, Ninkovic et al. [161] described that, during neurogenesis in adult olfactory bulb, instead of interacting with the Brm subunit, TF Pax6 recruits specifically Brg1 (the alternative ATPase subunit in the BAF chromatin remodeling complex), which promotes the Pax6-mediated program for adult neurogenesis. Thus, specific interactions of Pax6 and BAF chromtin modifiers in distinct cellular contexts of developing and adult brain facilitate transcriptional programs of neurogenesis.
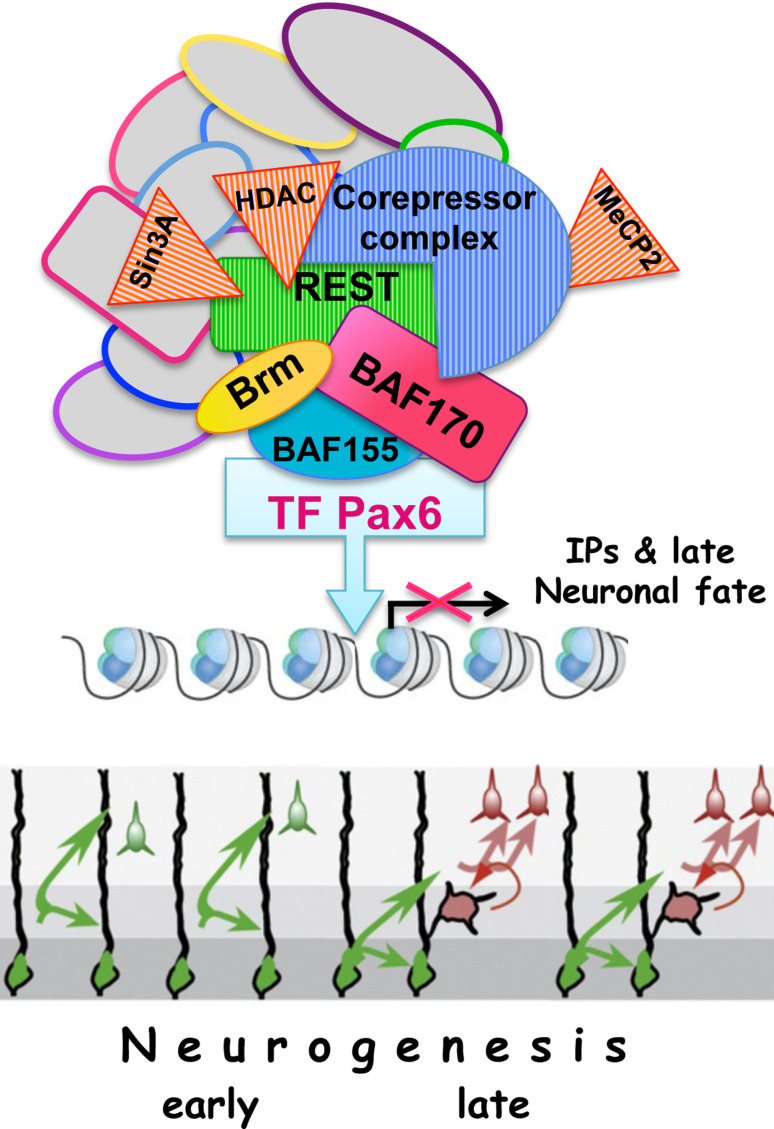
Fig. 4.
Hypothetical model proposing how epigenetic mechanism based on BAF chromatin remodeling complex controls the modes (direct vs. indirect) of cortical neurogenesis defining the cortical size, thickness and neuron layer subtype composition in the adult brain (according to [159, 160]). In the mouse cortex, coupling between TF Pax6 and the BAF complex subunits (BAF170, BAF155, Brm) and the REST–corepressor complex controls expression of genes involved in specification of IPs and late neurogenesis. During early stages of cortical neurogenesis, the restricted expression of BAF170 in non-neurogenic vRGCs recruits the REST-corepressor complex to promoters of TF Pax6 target genes (Tbr2, Cux1, Tle1 and other still unknown targets) that are involved in specification of both Tbr2(+) IPs, and genes specifying late (UL) neuronal fate. In BAF170 deficiency, BAF170 is replaced by additional BAF155 subunit, leading to loss of the REST-corepressor complex and induction of euchromatin state that relieves the repression of these Pax6 target genes, inducing a premature (heterochronic) induction of late (mostly indirect) neurogenesis. Bisulfite sequencing analysis of CpG methylation on Cux1 and Tle1 promoters using genomic DNA isolated from BAF170cKO and control cortices revealed a substantial decrease of CpG methylation in BAF170cKO and increase of these marks upon BAF170 overexpression in vivo. In opposite, BAF170 loss of function resulted in an increase in active H3K9Ac chromatin marks on Cux1 and Tle1 promoters (and reduction of repressive modifications H3K27Me3), BAF170cOE had opposite effects. BAF170cKO: cortex-specific conditional knock-out of BAF170; BAF170cOE: cortex specific conditional overexpression of BAF170, The differently colored circles indicate subunits of the BAFcomplex
Recent studies have elucidated some interesting molecular mechanisms that specify the projection trajectory of the two major classes of cortical pyramidal neurons: corticocortical (callosal) projection neurons located mostly in ULs and subcortical projection neurons having LL identity [99, 162]. Notably, chromatin remodeling involving the BAF complex also controls the projection subtype identity. TFs Ctip2/Bcl11b, a newly identified BAF chromatin remodeling subunit [133, 163] that labels all L5 SCMN, and Satb2 (AT-rich sequence-binding protein 2 (Satb2), that binds to DNA, are essential for the specification of subcortical (L5) and callosal (L3–L2) projection neurons, respectively [117, 164, 165]. In the absence of Satb2, active chromatin markers (H3K4me2) and histone H3 acetylation in the Ctip2 locus were markedly increased, permitting the ectopic activation of Ctip2 in Satb2-deficient callosal neurons [164]. Mechanistically, TF Satb2 promotes callosal projection fate by antagonizing the expression of Ctip2 in an epigenetic-like manner [164, 165], mediated by the proto-oncogene Ski [166] involving nucleosome chromatin remodeling [165]. Overall, the available, yet scarce, data suggest that interplay between TFs and epigenetic chromatin modifications is an important regulatory mechanism of different aspects of fate specification in neural stem and progenitor cells during cortical neurogenesis, profoundly affecting cortical size/thickness.
MicroRNA as regulators of corticogenesis
MicroRNA (miRNAs) are endogenous single-stranded noncoding RNA molecules of 22–24 nucleotides in length that are involved in posttranscriptional regulation of protein expression during various developmental processes including cortical neurogenesis [167–172]. In the cell nucleus, cleavage of the primary-miRNA by RNase III enzyme Drosha and DGCR8/Pasha (a binding partner) generates a 70-nucleotide-long precursor (pre-miRNA) that is exported into the cytoplasm, and is further processed into the mature double-stranded miRNA by RNAse III enzyme Dicer. Upon unfolding, one strand is included in a silencing complex and guided to seed sequences usually located in the 3′-untranslated regions of target mRNAs causing transcript degradation or translational repression [173, 174].
Because miRNAs target multiple mRNAs, and the encoding genes are often included in clusters or families, generation of genomic miRNA loss- and gain-of-function transgenic models are challenging procedures. At present, our knowledge about the role of specific miRNAs in the regulation of cortical size and thickness in transgenic mice in vivo is still fragmentary. Global loss of miRNA regulation through conditional knockout of Dicer at different stages of corticogenesis driven by the Cre-strains, Emx1-Cre [175, 176], Nestin-Cre [176], Foxg1-Cre [177, 178], CamKII-Cre [179], and Nex-Cre [180] have revealed specific phenotypes as a result of time-dependent regulation of cell-fate determination of the progenitors (reviewed in [172, 181]). Deletion of Dicer function at the onset of neurogenesis (via Emx1-Cre line) leads to the most severe cortical phenotype at postnatal stages, affecting the cortical thickness and lateral cortical expansion and showing overproduction of early born LL neuronal fates (as a result of precocious LL differentiation), that leads to a severe reduction of later born UL neurons [176]. However, slightly later elimination of Dicer activity (via the Nestin-Cre line) results in a milder reduction of cortical thickness showing affected generation and migration of the later born neurons [176]. In addition, ablation of Dicer in neuronal progenitors [175, 176] triggers severe apoptosis and differentiation defects, while loss of Dicer activity in postmitotic neurons [179, 180] leads to mild apoptosis, suggesting that the role of miRNAs for cell survival is stage- and cell type-specific.
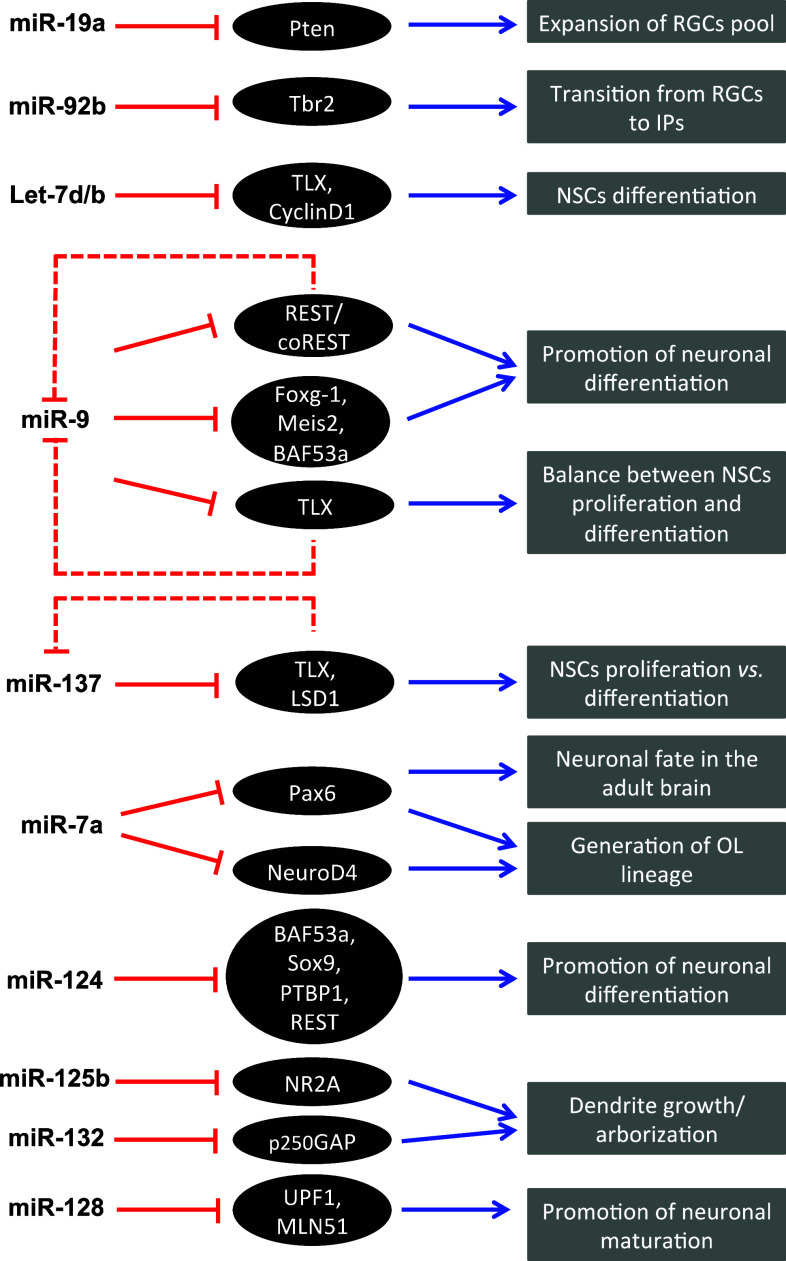
As discussed, growth of cortex with normal size and thickness critically depends on positive and negative factors for NSCs, RGCs/IPs self-renewal, and subsequent differentiation. Conditional deletion of miR-17-19 family (consisting of miR-17, miR-18, mir-19, and miR-92 subfamilies with distinct seed sequences) at the onset of neurogenesis in mouse cortex leads to pronounced reduction of cortical thickness [182]. Interestingly, miR-19a positively influences the expansion of RGC pool and regulates the transition RGC–IPs by inhibiting the tumor suppressor gene Pten, thus controlling the RGCs expansion [182]. As recently shown by Nowakowski et al. [178], the miR-92b, another member of the cluster, plays a restrictive role for generation of Tbr2(+) IPs from RGCs in the developing mouse cortex (Fig. 5) (reviewed in [182]).
Fig. 5.
miRNAs involved cortical neurogenesis. MiRNAs target multiple mRNAs in order to implement their regulatory action, also known as “fine-tuning”, in a variety of processes, as indicated. Some miRNAs establish feedback circuits. An example of this is miR-9 and Tlx, which form a negative feedback loop to induce precise spatiotemporal orchestration of NSC proliferation/differentiation in mouse telencephalon. At the same time, miRNA let-7d regulates the Tlx–miR-9 cascade to control neural cell fate and neurogenesis. MiR-19a and miR-92b, which belong to the same cluster (miR-17-92), control the expansion of RGCs pool and the transition to IPs by negatively regulating Pten and Tbr2, respectively. In embryonic neurogenesis, miR-7a inhibits the neurogenic factors Pax6 and NeuroD1 to promote the generation of oligodendroglial (OL) lineage. In the forebrain niche for adult neurogenesis, miR-7a-mediated regulation of Pax6 expression promotes the dopaminergic neural fate of the interneurons of olfactory bulb. The expression of miR-124 is neural-specific and increases upon differentiation of NSCs into neurons. Both miR-124 and miR-9* repress REST, which restricts the expression of neuronal genes in non-neuronal cells as well as the progenitor-specific BAF complex subunit BAF53a. This triple negative genetic circuit is specifically involved in the regulation of mitotic exit of cortical progenitors and the beginning of neuronal differentiation (based on data from [177–204])
Recent application of techniques for focal overexpression or inhibition of specific miRNAs indicates important roles in neuronal differentiation (reviewed in [172, 181]). By targeting both the nuclear receptor TLX that promotes NSC-fate and the Cyclin D1, the miRNA Let-7b regulates neural stem cell differentiation [183]. Several studies indicate that, in vertebrates, miR-124 promotes neuronal differentiation and cell cycle exit of NSC cells in the SVZ of adult brain, a niche for postnatal neurogenesis. Forced expression of miR-124 in SVZ promotes neuronal differentiation while knockdown of miR-124 maintains the NSC fate (Fig. 5) [184, 185]. Although the majority of studies indicate that miRNAs are in general more important for induction of neuronal differentiation, a recent work has discovered a role of miR-7a in forebrain neuronal subtype specification, promoting dopaminergic neural fate of the interneurons of olfactory bulb, due to inhibition of TF Pax6 expression (Fig. 5) [186].
It should be noted that the function of only a few of these miRNAs as possible regulators of cortical size and thickness has so far been studied in transgenic mouse models. Targeted deletion of miR-9-1 together with miR-9-3, two precursors of miR-9/miR-9* (*miRNA synthesized from the opposite strand) show a marked reduction of the germinative VZ and diminished thickness of all cortical layers. In fact, miR-9 suppresses NSC proliferation and promotes neuronal differentiation by modulation of the expression of several important factors, Foxg-1, Meis2, Nr2e1, and TLX (Fig. 5) [187, 188].
Interestingly, when involved in cell fate control, many miRNAs forms feedback regulatory loops with their target’s genes. The mechanism of miR-9-dependent promotion of neuronal differentiation includes a negative feedback regulation of the REST complex by silencing REST/CoREST genes [189–191]. Such regulatory interactions have been reported for miR-9 and TLX in determination of neural stem fate [183, 200], and for miR-137 with TLX and histone deacetylase LSD1 in controlling the proliferation versus differentiation during corticogenesis [192]. Similarly, in NSCs, miRNA let-7d binds the 3′UTR of TLX and acts upstream of the TLX/miRNA-9 cascade, controlling neural cell fate and neurogenesis (Fig. 5) [183]. Recent data also suggest that a double-negative feedback loop of miR-9 and Hes1, a Notch signalling effector that exerts short-period expression oscillations, is essential for maintenance of progenitor proliferation and cells exit from mitosis [193].
Among the targets of miR-9/9* and miR-124 are REST, which restricts the expression of neuronal genes in non-neuronal cells [189], and BAF53a, a component of the npBAF complex [158]. Notably, before NSCs start to exit from mitosis, miR-9* and miR-124 are activated and bind to the BAF53a 3′ UTR to fully repress BAF53a activity which is replaced by BAF53b subunit in the neural-specific BAF complex (nBAF) (Fig. 5) [177, 194]. Together, these data suggest that regulation of mitotic exit of cortical progenitors and the beginning of neuronal differentiation is under a triple-negative genetic circuit involving miR-9*, miR-124, and REST as the main players [185]. Even more intriguingly, the synergistic effect of the expression of miR9/miR9* and miR-124 was found to convert human fibroblasts to neurons [195]. More detailed analyses in transgenic models in future are expected to shed light on the specific roles of individual miRNAs in determining cortical size and thickness.
Conclusions and future perspectives
The expansion of cortical surface and thickness in primate brain have been linked to the adoption of complex cognition. In past years, fundamental knowledge has accumulated on the progenitor diversity and specific proliferation behavior during cortical neurogenesis in phylognetically different species. Although showing striking conservation, the mechanisms controlling the cortical size appear to act in species-specific manners which await further clarification. Future studies should help to elucidate the lineage relationships between distinct progenitor subtypes with respect to their cell polarity, asymmetry, spindle orientation, mode of division, and acquired fates of the daughter cells during specific stages of cortical neurogenesis in different species. Available data suggest important roles of transcription factors and morphogenes in different aspects of these processes; however, the underlaying molecular mechanisms are still largely unknown. As multiple cortical developmental processes seems to be co-regulated by the same transcriptional program, future efforts should be aimed at the identification and regulation of specific pathways by particular TF. Epigenetic mechanisms mediated by various TFs could be of key importance for the establishment of transcriptional programs covering the enormous diversity of cortical progenitor and neuronal subtypes identities. An interesting prospective for future exprimentations will be to uncover gene regulatory pathways established by epigenetic machinery and miRNAs during cortical neurogenesis. Deep elucidation of the factors that controls these processes will be essential for understanding the pathogenesis of neurodevelopmental diseases with a great significance for the human mental health.
Acknowledgments
We apologize to colleagues whose work we may not have been able to include in this review due to space constraints. We thank J. Staiger for his support, T. Schneider and A. Dudek for proofreading and discussion. This work was supported by the Research Program, Faculty of Medicine, Georg-August-University of Göttingen and TU432/1-1 DFG grant (T.C.T), the Max Planck Gesellschaft (A.S. and A.P.); the Cluster of Excellence and DFG Research Center Nanoscale Microscopy and Molecular Physiology of the Brain (A.S. and M.A.T.). The authors declare no competing financial interests.
References
- 1.Fietz SA, Huttner WB. Cortical progenitor expansion, self-renewal and neurogenesis-a polarized perspective. Curr Opin Neurobiol. 2011;21:23–35. doi: 10.1016/j.conb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 3.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 5.Corbeil D, Joester A, Fargeas CA, Jaszai J, Garwood J, Hellwig A, Werner HB, Huttner WB. Expression of distinct splice variants of the stem cell marker prominin-1 (CD133) in glial cells. Glia. 2009;57:860–874. doi: 10.1002/glia.20812. [DOI] [PubMed] [Google Scholar]
- 6.Weigmann A, Corbeil D, Hellwig A, Huttner WB. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci USA. 1997;94:12425–12430. doi: 10.1073/pnas.94.23.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotz M, Steindler D. To be glial or not-how glial are the precursors of neurons in development and adulthood? Glia. 2003;43:1–3. doi: 10.1002/glia.10251. [DOI] [PubMed] [Google Scholar]
- 9.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 10.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 12.Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- 13.Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Gotz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 14.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 15.Tamamaki N, Nakamura K, Okamoto K, Kaneko T. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci Res. 2001;41:51–60. doi: 10.1016/s0168-0102(01)00259-0. [DOI] [PubMed] [Google Scholar]
- 16.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 17.Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, Brakebusch C, Gotz M. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- 20.Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 22.Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilz GA, Shitamukai A, Reillo I, Pacary E, Schwausch J, Stahl R, Ninkovic J, Snippert HJ, Clevers H, Godinho L, et al. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat Commun. 2013;4:2125. doi: 10.1038/ncomms3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stahl R, Walcher T, De Juan Romero C, Pilz GA, Cappello S, Irmler M, Sanz-Aquela JM, Beckers J, Blum R, Borrell V, et al. Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell. 2013;153:535–549. doi: 10.1016/j.cell.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 27.Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 28.Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- 32.Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
- 33.Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, Paabo S, Huttner WB. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron. 2008;60:40–55. doi: 10.1016/j.neuron.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- 35.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fietz SA, Kelava I, Vogt J, Wilsch-Brauninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 38.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 39.Reillo I, de Juan Romero C, Garcia-Cabezas MA, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Cerdeno V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, Walker AI, Noctor SC. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS ONE. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelava I, Reillo I, Murayama AY, Kalinka AT, Stenzel D, Tomancak P, Matsuzaki F, Lebrand C, Sasaki E, Schwamborn JC, et al. Abundant occurrence of basal radial glia in the subventricular zone of embryonic neocortex of a lissencephalic primate, the common marmoset Callithrix jacchus . Cereb Cortex. 2012;22:469–481. doi: 10.1093/cercor/bhr301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borrell V, Reillo I. Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev Neurobiol. 2012;72:955–971. doi: 10.1002/dneu.22013. [DOI] [PubMed] [Google Scholar]
- 44.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 45.Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, Kennedy H, Dehay C. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47:353–364. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 47.Rakic P. Developmental and evolutionary adaptations of cortical radial glia. Cereb Cortex. 2003;13:541–549. doi: 10.1093/cercor/13.6.541. [DOI] [PubMed] [Google Scholar]
- 48.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 49.Kosodo Y, Roper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamonica BE, Lui JH, Hansen DV, Kriegstein AR. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat Commun. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lancaster MA, Knoblich JA. Spindle orientation in mammalian cerebral cortical development. Curr Opin Neurobiol. 2012;22:737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calegari F, Haubensak W, Haffner C, Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25:6533–6538. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calegari F, Huttner WB. An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J Cell Sci. 2003;116:4947–4955. doi: 10.1242/jcs.00825. [DOI] [PubMed] [Google Scholar]
- 55.Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- 56.Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 57.Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 58.Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 2005;19:1028–1033. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 60.Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 61.Tiberi L, Vanderhaeghen P, van den Ameele J. Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr Opin Cell Biol. 2012;24:269–276. doi: 10.1016/j.ceb.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 63.Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 64.Haydar TF, Kuan CY, Flavell RA, Rakic P. The role of cell death in regulating the size and shape of the mammalian forebrain. Cereb Cortex. 1999;9:621–626. doi: 10.1093/cercor/9.6.621. [DOI] [PubMed] [Google Scholar]
- 65.Depaepe V, Suarez-Gonzalez N, Dufour A, Passante L, Gorski JA, Jones KR, Ledent C, Vanderhaeghen P. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature. 2005;435:1244–1250. doi: 10.1038/nature03651. [DOI] [PubMed] [Google Scholar]
- 66.Sahara S, O’Leary DD. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. doi: 10.1016/j.neuron.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoon K, Nery S, Rutlin ML, Radtke F, Fishell G, Gaiano N. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci. 2004;24:9497–9506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 69.Garel S, Huffman KJ, Rubenstein JL. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- 70.Mallamaci A, Stoykova A. Gene networks controlling early cerebral cortex arealization. Eur J Neurosci. 2006;23:847–856. doi: 10.1111/j.1460-9568.2006.04634.x. [DOI] [PubMed] [Google Scholar]
- 71.O’Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 72.Sahara S, Kawakami Y, Izpisua Belmonte JC, O’Leary DD. Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior-posterior cortical area patterning. Neural Dev. 2007;2:10. doi: 10.1186/1749-8104-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- 74.Borello U, Cobos I, Long JE, McWhirter JR, Murre C, Rubenstein JL. FGF15 promotes neurogenesis and opposes FGF8 function during neocortical development. Neural Dev. 2008;3:17. doi: 10.1186/1749-8104-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rash BG, Tomasi S, Lim HD, Suh CY, Vaccarino FM. Cortical gyrification induced by fibroblast growth factor 2 in the mouse brain. J Neurosci. 2013;33:10802–10814. doi: 10.1523/JNEUROSCI.3621-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rash BG, Lim HD, Breunig JJ, Vaccarino FM. FGF signaling expands embryonic cortical surface area by regulating Notch-dependent neurogenesis. J Neurosci. 2011;31:15604–15617. doi: 10.1523/JNEUROSCI.4439-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 78.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 79.Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 80.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 81.Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- 82.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 83.Manzini MC, Tambunan DE, Hill RS, Yu TW, Maynard TM, Heinzen EL, Shianna KV, Stevens CR, Partlow JN, Barry BJ, et al. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker–Warburg syndrome. Am J Hum Genet. 2012;91:541–547. doi: 10.1016/j.ajhg.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr Opin Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 85.Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asami M, Pilz GA, Ninkovic J, Godinho L, Schroeder T, Huttner WB, Gotz M. The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development. 2011;138:5067–5078. doi: 10.1242/dev.074591. [DOI] [PubMed] [Google Scholar]
- 87.Postiglione MP, Juschke C, Xie Y, Haas GA, Charalambous C, Knoblich JA. Mouse inscuteable induces apical-Basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron. 2011;72:269–284. doi: 10.1016/j.neuron.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xie Y, Juschke C, Esk C, Hirotsune S, Knoblich JA. The phosphatase PP4c controls spindle orientation to maintain proliferative symmetric divisions in the developing neocortex. Neuron. 2013;79:254–265. doi: 10.1016/j.neuron.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Molnar Z, Metin C, Stoykova A, Tarabykin V, Price DJ, Francis F, Meyer G, Dehay C, Kennedy H. Comparative aspects of cerebral cortical development. Eur J Neurosci. 2006;23:921–934. doi: 10.1111/j.1460-9568.2006.04611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnar Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Colasante G, Simonet JC, Calogero R, Crispi S, Sessa A, Cho G, Golden JA, Broccoli V (2013) ARX regulates cortical intermediate progenitor cell expansion and upper layer neuron formation through repression of Cdkn1c. Cereb Cortex. doi:10.1093/cercor/bht222 [DOI] [PMC free article] [PubMed]
- 93.Glickstein SB, Monaghan JA, Koeller HB, Jones TK, Ross ME. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J Neurosci. 2009;29:9614–9624. doi: 10.1523/JNEUROSCI.2284-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nonaka-Kinoshita M, Reillo I, Artegiani B, Martinez-Martinez MA, Nelson M, Borrell V, Calegari F. Regulation of cerebral cortex size and folding by expansion of basal progenitors. EMBO J. 2013;32:1817–1828. doi: 10.1038/emboj.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pinto L, Drechsel D, Schmid MT, Ninkovic J, Irmler M, Brill MS, Restani L, Gianfranceschi L, Cerri C, Weber SN, et al. AP2gamma regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat Neurosci. 2009;12:1229–1237. doi: 10.1038/nn.2399. [DOI] [PubMed] [Google Scholar]
- 96.Volpe M, Shpungin S, Barbi C, Abrham G, Malovani H, Wides R, Nir U. trnp: A conserved mammalian gene encoding a nuclear protein that accelerates cell-cycle progression. DNA Cell Biol. 2006;25:331–339. doi: 10.1089/dna.2006.25.331. [DOI] [PubMed] [Google Scholar]
- 97.Hevner RF, Haydar TF. The (not necessarily) convoluted role of basal radial glia in cortical neurogenesis. Cereb Cortex. 2012;22:465–468. doi: 10.1093/cercor/bhr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poluch S, Juliano SL (2013) Fine-tuning of neurogenesis is essential for the evolutionary expansion of the cerebral cortex. Cereb Cortex. doi:10.1093/cercor/bht232 [DOI] [PMC free article] [PubMed]
- 99.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 100.Richman DP, Stewart RM, Caviness VS., Jr Cerebral microgyria in a 27-week fetus: an architectonic and topographic analysis. J Neuropathol Exp Neurol. 1974;33:374–384. doi: 10.1097/00005072-197407000-00004. [DOI] [PubMed] [Google Scholar]
- 101.Takahashi T, Nowakowski RS, Caviness VS., Jr The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci USA. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Defelipe J. The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front Neuroanat. 2011;5:29. doi: 10.3389/fnana.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Caviness VS, Jr, Goto T, Tarui T, Takahashi T, Bhide PG, Nowakowski RS. Cell output, cell cycle duration and neuronal specification: a model of integrated mechanisms of the neocortical proliferative process. Cereb Cortex. 2003;13:592–598. doi: 10.1093/cercor/13.6.592. [DOI] [PubMed] [Google Scholar]
- 105.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 106.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 107.Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Muller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- 109.Guo C, Eckler MJ, McKenna WL, Mckinsey GL, Rubenstein JLR, Chen B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80:1167–1174. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guillemot F, Molnár Z, Tarabykin V, Stoykova A. Molecular mechanisms of cortical differentiation. Eur J Neurosci. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- 112.Land PW, Monaghan AP. Expression of the transcription factor, tailless, is required for formation of superficial cortical layers. Cereb Cortex. 2003;13:921–931. doi: 10.1093/cercor/13.9.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- 114.Harrison SJ, Nishinakamura R, Jones KR, Monaghan AP. Sall1 regulates cortical neurogenesis and laminar fate specification in mice: implications for neural abnormalities in Townes-Brocks syndrome. Dis Models Mech. 2012;5:351–365. doi: 10.1242/dmm.002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex. 2008;18:1865–1875. doi: 10.1093/cercor/bhm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 117.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 118.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Panto MR, Zappala A, Tuorto F, Cicirata F. Role of the Otx1 gene in cell differentiation of mammalian cortex. Eur J Neurosci. 2004;19:2893–2902. doi: 10.1111/j.0953-816X.2004.03326.x. [DOI] [PubMed] [Google Scholar]
- 120.Cubelos B, Sebastian-Serrano A, Kim S, Moreno-Ortiz C, Redondo JM, Walsh CA, Nieto M. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb Cortex. 2008;18:1758–1770. doi: 10.1093/cercor/bhm199. [DOI] [PubMed] [Google Scholar]
- 121.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 122.Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mason S, Pi McKenna per M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- 125.McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Betancourt J, Katzman S, Chen B (2014) Nuclear factor one B regulates neural stem cell differentiation and axonal projection of corticofugal neurons. J Comp Neurol 522(1):Spc1. doi:10.1002/cne.23490 [DOI] [PMC free article] [PubMed]
- 127.Kwan YK, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 130.Takeuchi A, O’Leary DD. Radial migration of superficial layer cortical neurons controlled by novel Ig cell adhesion molecule MDGA1. J Neurosci. 2006;26:4460–4464. doi: 10.1523/JNEUROSCI.4935-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.MuhChyi C, Juliandi B, Matsuda T, Nakashima K. Epigenetic regulation of neural stem cell fate during corticogenesis. Int J Dev Neurosci. 2013;31:424–433. doi: 10.1016/j.ijdevneu.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 132.Wen S, Li H, Liu J. Epigenetic background of neuronal fate determination. Prog Neurobiol. 2009;87:98–117. doi: 10.1016/j.pneurobio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 133.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Bokhoven H, Kramer JM. Disruption of the epigenetic code: an emerging mechanism in mental retardation. Neurobiol Dis. 2010;39:3–12. doi: 10.1016/j.nbd.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 135.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Van Holde KE. Chromatin: Springer series in molecular biology. New York: Springer; 1988. [Google Scholar]
- 137.Wolffe AP. Chromatin: structure and function. 3. San Diego: Academic; 1999. [Google Scholar]
- 138.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 139.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1079. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 140.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci USA. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thomas T, Voss AK, Chowdhury K, Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127:2537–2548. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- 142.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 143.Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, Passante L, Schiffmann SN, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 144.Juliandi B, Abematsu M, Sanosaka T, Tsujimura K, Smith A, Nakashima K. Induction of superficial cortical layer neurons from mouse embryonic stem cells by valproic acid. Neurosci Res. 2012;72:23–31. doi: 10.1016/j.neures.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 145.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 146.Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thal Relat Syst. 2005;3:227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 148.Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 150.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 151.Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 153.Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 154.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang YJ, Baltus AE, Mathew RS, Murphy EA, Evrony GD, Gonzalez DM, Wang EP, Marshall-Walker CA, Barry BJ, Murn J, et al. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell. 2012;151:1097–1112. doi: 10.1016/j.cell.2012.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 157.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 158.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 160.Tuoc TC, Narayanan R, Stoykova A. BAF chromatin remodeling complex: cortical size regulation and beyond. Cell Cycle. 2013;12:2953–2959. doi: 10.4161/cc.25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ninkovic J, Steiner-Mezzadri A, Jawerka M, Akinci U, Masserdotti G, Petricca S, Fischer J, von Holst A, Beckers J, Lie CD, Petrik D, Miller E, Tang J, Wu J, Lefebvre V, Demmers J, Eisch A, Metzger D, Crabtree G, Irmler M, Poot R, Götz M. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 165.Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 166.Baranek C, Dittrich M, Parthasarathy S, Bonnon CG, Britanova O, Lanshakov D, Boukhtouche F, Sommer JE, Colmenares C, Tarabykin V, et al. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc Natl Acad Sci USA. 2012;109:3546–3551. doi: 10.1073/pnas.1108718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 168.Kawahara H, Imai T, Okano H. MicroRNAs in neural stem cells and neurogenesis. Front Neurosci. 2012;6:30. doi: 10.3389/fnins.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 170.Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci. 2010;30:14931–14936. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lang MF, Shi Y. Dynamic roles of microRNAs in neurogenesis. Front Neurosci. 2012;6:71. doi: 10.3389/fnins.2012.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cremisi F. MicroRNAs and cell fate in cortical and retinal development. Front Cell Neurosci. 2013;7:141. doi: 10.3389/fncel.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hannon GJ, Rivas FV, Murchison EP, Steitz JA. The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:551–564. doi: 10.1101/sqb.2006.71.064. [DOI] [PubMed] [Google Scholar]
- 174.Pasquinelli AE. Birthing histone mRNAs by CSR-1 section. EMBO J. 2012;31:3790–3791. doi: 10.1038/emboj.2012.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nowakowski TJ, Mysiak KS, Pratt T, Price DJ. Functional dicer is necessary for appropriate specification of radial glia during early development of mouse telencephalon. PLoS ONE. 2011;6:e23013. doi: 10.1371/journal.pone.0023013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hong J, Zhang H, Kawase-Koga Y, Sun T. MicroRNA function is required for neurite outgrowth of mature neurons in the mouse postnatal cerebral cortex. Front Cell Neurosci. 2013;7:151. doi: 10.3389/fncel.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell Death Differ. 2012;19:1573–1581. doi: 10.1038/cdd.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, Garg V, Sun T. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3:1398–1406. doi: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhao C, Sun G, Ye P, Li S, Shi Y. MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci Rep. 2013;3:1329. doi: 10.1038/srep01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Akerblom M, Sachdeva R, Jakobsson J. Functional studies of microRNAs in neural stem cells: problems and perspectives. Front Neurosci. 2012;6:14. doi: 10.3389/fnins.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.de Chevigny A, Coré N, Follert P, Gaudin M, Barbry P, Béclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci. 2012;15:1120–1126. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- 187.Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal–Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Laneve P, Gioia U, Andriotto A, Moretti F, Bozzoni I, Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010;38:6895–6905. doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, Ma X, Shi Y. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bonev B, Stanley P, Papalopulu N. MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lamonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr Opin Neurobiol. 2012;22:747–753. doi: 10.1016/j.conb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol. 2005;17:639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 198.Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, Stallings RL, Henshall DC. miRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Aranha MM, Santos DM, Solá S, Steer CJ, Rodrigues CM. miR-34a regulates mouse neural stem cell differentiation. PLoS ONE. 2011;6:e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, Jin P, Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, Pfaff SL, Wilkinson MF. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci USA. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience. 2006;142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 205.Mutch CA, Schulte JD, Olson E, Chenn A. Beta-catenin signaling negatively regulates intermediate progenitor population numbers in the developing cortex. PLoS ONE. 2010;5:e12376. doi: 10.1371/journal.pone.0012376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kim WY, Wang X, Wu Y, Doble BW, Patel S, Woodgett JR, Snider WD. GSK-3 is a master regulator of neural progenitor homeostasis. Nat Neurosci. 2009;12:1390–1397. doi: 10.1038/nn.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kang W, Wong LC, Shi SH, Hebert JM. The transition from radial glial to intermediate progenitor cell is inhibited by FGF signaling during corticogenesis. J Neurosci. 2009;29:14571–14580. doi: 10.1523/JNEUROSCI.3844-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Komada M, Iguchi T, Takeda T, Ishibashi M, Sato M. Smoothened controls cyclin D2 expression and regulates the generation of intermediate progenitors in the developing cortex. Neurosci Lett. 2013;547:87–91. doi: 10.1016/j.neulet.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 209.Shikata Y, Okada T, Hashimoto M, Ellis T, Matsumaru D, Shiroishi T, Ogawa M, Wainwright B, Motoyama J. Ptch1-mediated dosage-dependent action of Shh signaling regulates neural progenitor development at late gestational stages. Dev Biol. 2011;349:147–159. doi: 10.1016/j.ydbio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 210.Georgala PA, Manuel M, Price DJ. The generation of superficial cortical layers is regulated by levels of the transcription factor Pax6. Cereb Cortex. 2011;21:81–94. doi: 10.1093/cercor/bhq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tuoc TC, Radyushkin K, Tonchev AB, Pinon MC, Ashery-Padan R, Molnar Z, Davidoff MS, Stoykova A. Selective cortical layering abnormalities and behavioral deficits in cortex-specific Pax6 knock-out mice. J Neurosci. 2009;29:8335–8349. doi: 10.1523/JNEUROSCI.5669-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Berger J, Berger S, Tuoc TC, D’Amelio M, Cecconi F, Gorski JA, Jones KR, Gruss P, Stoykova A. Conditional activation of Pax6 in the developing cortex of transgenic mice causes progenitor apoptosis. Development. 2007;134:1311–1322. doi: 10.1242/dev.02809. [DOI] [PubMed] [Google Scholar]
- 213.Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol. 2006;22:22. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bedford L, Walker R, Kondo T, van Cruchten I, King ER, Sablitzky F. Id4 is required for the correct timing of neural differentiation. Dev Biol. 2005;280:386–395. doi: 10.1016/j.ydbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 215.Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 217.Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Buttner N, Johnsen SA, Kugler S, Vogel T. Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc Natl Acad Sci USA. 2010;107:7042–7047. doi: 10.1073/pnas.0912041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Developmental Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]