Abstract
The schizophrenia susceptibility gene, Rgs4, is one of the most intensively studied regulators of G-protein signaling members, well known to be fundamental in regulating neurotransmission. However, little is known about its role in the developing nervous system. We have isolated zebrafish rgs4 and shown that it is transcribed in the developing nervous system. Rgs4 knockdown did not affect neuron number and patterning but resulted in locomotion defects and aberrant development of axons. This was confirmed using a selective Rgs4 inhibitor, CCG-4986. Rgs4 knockdown also attenuated the level of phosphorylated-Akt1, and injection of constitutively-activated AKT1 rescued the motility defects and axonal phenotypes in the spinal cord but not in the hindbrain and trigeminal neurons. Our in vivo analysis reveals a novel role for Rgs4 in regulating axonogenesis during embryogenesis, which is mediated by another schizophrenia-associated gene, Akt1, in a region-specific manner.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1178-z) contains supplementary material, which is available to authorized users.
Keywords: Rgs4, Axonogenesis, Akt, Zebrafish
Introduction
G-protein-mediated signaling is initiated when G-protein-coupled receptors (GPCRs) bind their ligands, triggering GDP bound to the Gα subunits to be replaced with GTP. The signaling is deactivated when the GTPase hydrolyzes the GTP. Hydrolysis of GTP can be regulated by members of the regulator of G-protein signaling (RGS) family which accelerate the intrinsic GTPase activity and consequently negatively regulate G-protein signaling and act as G protein GTPase-activating proteins (GAPs). Emerging evidence also reveals that RGS proteins can bind to proteins other than Gα (for example, GPCRs) to regulate G-protein mediated signaling [1, 2]. To date, more than 20 RGS members have been identified, recognized by a conserved RGS domain that is responsible for hastening the intrinsic GTPase activity. These proteins can be further classified into several subfamilies according to sequence homology and the presence of domains additional to the RGS domain [3].
RGS4 belongs to the R4/B subfamily which includes RGS1-5, 8, 13, 16, 18, and 21. Like most of the R4/B members, RGS4 regulates G proteins of the Gαi/o and Gαq families [2]. Mammalian RGS4 is highly expressed in adult brain, most abundantly in the cerebral cortex [4–6] and has been intensively studied for its role in modulating both pre- and postsynaptic neurotransmission, such as calcium homeostasis neurotransmitter release and opioid, cholinergic, serotonergic, and acetylcholine signaling [7, 8]. More recently, RGS4 has been revealed as a potential etiological factor for schizophrenia. Several independent genetic studies have demonstrated an association between schizophrenia and the RGS4 gene, and a relative decrease in the levels of RGS4 mRNA and protein have been found in samples from schizophrenia patients. A current hypothesis therefore states that aberrant RGS4 expression may lead to an abnormal neurotransmission that is responsible for schizophrenia. However, the physiological role of RGS4 in the progression of this disease is currently not clear [9]. Indeed, although previous studies have demonstrated abundant expression of Rgs4 in the developing nervous system, very few studies of the prenatal role of RGS4 have been reported. The Xenopus orthologue of Rgs4 is expressed in the neural fold and developing central nervous system [10], and mouse Rgs4 is specifically expressed in central and peripheral neuronal precursors [6]. Ectopic expression of the neuronal differentiation regulators, PHOX2b and MASH1, induces Rgs4 expression in chicken spinal cord [6], suggesting that RGS4 is involved in neurogenesis. The Rgs4 knockout mouse exhibited normal formation of neurons, but it had behavioral defects, such as subtle positive symptoms, although whether axonogenesis was affected has not been investigated [11]. Therefore, the role of RGS4 in the developing nervous system remains unclear.
In order to dissect the role of Rgs4 during neurodevelopment, we isolated its zebrafish homologue and showed that the gene is transcribed in the developing nervous system. Functional disruption of Rgs4 by morpholino knockdown and an inhibitory chemical compound resulted in aberrant axon formation and motility defects. We also found phosphorylated Akt1 was downregulated in Rgs4 morphants. Strikingly, the axonal phenotypes seen when RGS4 was inhibited could be rescued by phosphorylated AKT1 in the spinal cord but not in the hindbrain and trigeminal neurons. Our results therefore reveal a novel role of Rgs4 in the regulation of axonogenesis during embryogenesis, which can be mediated by Akt signaling.
Materials and methods
Ethics statement
All experiments were performed in strict accordance to standard guidelines for zebrafish work and approved by the Institutional Animal Care and Use Committee of Chang Gung University (IACUC approval number: CGU04-57 and CGU08-86).
Sequence comparisons and phylogeny
Amino acid sequences were aligned and displayed using Vector NTI (Invitrogen). The GenBank accession numbers of the compared proteins are as follows: zebrafish Rgs4 (BC154780.1); Xenopus laevis rgs4 (NM_001087536); chicken Rgs4 (NM_204385); mouse RGS4 (NM_009062.3); rat RGS4 (NM_017214); and human RGS4 (NM_005613.5).
Fish maintenance
Tü (wild-type) zebrafish embryos were purchased from the Zebrafish International Resource Center (ZIRC, OR, USA). The Tg(huC:eGFP) transgenic line was a kind gift from Chang-Jen Huang [12] and the Tg(islet1:GFP) line was obtained from Hitoshi Okamoto [13]. Zebrafish were raised, maintained, and paired under standard conditions. The embryos were staged according to the number of somites, hours post-fertilization, and days post-fertilization [14].
Construct generation
The open reading frame of zebrafish rgs4 was PCR amplified with high fidelity Pfu polymerase (Fermentas) and primers (5′-AAGGATCCGGATCCATGTGTAAAGGGCTTGCTGCTC-3′ and 5′-TAATCGATATCGATAGGCATAACTAGGCAAACACTGAC-3′) which were designed according to the zebrafish genome database (http://www.ensembl.org, version Zv6) (GenBank Accession Number BC154780.1). rgs4 MO1 and MO2 binding sequences were inserted upstream of an enhanced green fluorescent protein reporter in the pCS2 vector to create a 5′rgs4-EGFP construct to evaluate the specificity and efficiency of morpholinos. The open reading frame of mouse Rgs4 was amplified with primers (5′-ATCGATATGTGCAAAGGACTTGCAGGTCTGC-3′ and 5′-ATCGATTTAGGCACACTGGGAGACCAGGGAAG-3′) which were designed according to the NCBI database (GenBank Accession Number NM_009062.3).
RNA and morpholino injection
Capped RNA encoding the full coding sequence of zebrafish rgs4, mouse Rgs4, and constitutively activated AKT1 was prepared as described previously [15]. The human constitutive-active AKT1 (ca-AKT1) was composed of a src myristoylation signal sequence fused to the N-terminus of AKT1 (kindly provided by Chung-Der Hsaio [16]). Since Akt1 plays crucial roles in multiple cellular processes during development, and over-expression of ca-AKT into zebrafish blastomeres could result in multiple developmental defects ([16] and unpublished observation), the amount for ca-AKT1 injection was titrated and optimized for a concentration that did not cause significant behavior or axonal defects but was able to rescue the phenotype in rgs4 morphants (25 pg). Antisense morpholino oligonucleotides were purchased from Gene Tools (OR, USA). Two morpholinos against rgs4 were used: MO1 (AGCAGCAAGCCCTTTACACATGTTG) that overlaps the ATG start codon (+4 to −21), and MO2 (TCCAAAGCAAGAGAAAAGAAGTCTG) that corresponds to −22 to −47 5′ to the translation start site. A control morpholino designed to a random sequence of nucleotides not found in the zebrafish genome (5′-CCTCTTACCTCAGTTACAATTTATA-3′; Gene Tools) or a morpholino with 5 bases mismatch to MO2 (5′-TCgAAAcCAAGAcAAAAcAAcTCTG-3′; mismatched bases are indicated by small letters) was injected in an equal amount of MO2 as a control experiment. All injections were performed at the one- to two-cell stage and cRNAs or morpholinos were introduced into blastomeres.
Behavioral tests and video recording
All tests were performed in three independent experiments. Embryonic and larval behaviors were observed using a Leica Z16 APO stereomicroscope. Video images were captured using an Olympus C-7070 CCD camera and analyzed with VirtualDub and ImageJ (National Institutes of Health). For in-chorion coiling contraction, total contractions of ten embryos in 3 min for each test were counted and displayed as the number of times per second (Hz). Touch and escape responses were analyzed at 48 hpf and embryos were gently stimulated with a thin tungsten probe near the region of trigeminal neurons. A total of 20 tactile stimuli with 5-s intervals were applied in batches of ten embryos per experiment, and embryos with or without response were shown in percentages.
Confocal microscopy and time-lapse imaging
Embryos were mounted in 1 % low gelling temperature agarose (Sigma), and for time-lapse imaging, images were taken at 30-min intervals at 28.5 °C using a Zeiss LSM 510 confocal microscope.
Quantification of axons
Affected embryos which showed at least 80 % truncated axons were included in the percentage of defective embryos and at least 30 embryos per batch were counted to calculate embryos with or without phenotype. Control and Rgs4-deficient groups were always processed in parallel from embryos delivered from the same parents to avoid differences in overall development from influencing results. To count spinal cord axons, the axon tracks in 10 somitic segments from the level between the 9th to the 18th somite were counted in each embryo. For primary motor axons, axon tracts that failed to reach the choice point were considered as a positive phenotype. Counts for Rohon–Beard axons collectively comprise single axons, fascicles of axons, and axonal branches. The acetylated α-tubulin-labeled reticulospinal axons in the developing hindbrain were too condensed and therefore only the commissures between the two medial longitudinal fasciculus in rhombomere segments 1–5 were counted. All tests were performed in three independent experiments. Mean and standard deviation were calculated from three independent experiments and represented as mean ± SD.
Embryo dissociation and fluorescence activated cell sorting
Embryos were dissociated using Nathan Lawson’s protocol (available at http://lawsonlab.umassmed.edu) with slight modification. In general, Tg(huC:eGFP) embryos (mixed stages ranging from 16 to 22 hpf) were dechorionated and the yolks were removed by gently pipetting in 100 μl calcium-free Ringer’s solution, following digestion in a solution containing 0.1 % collagenase P and 0.25 % trypsin for 25 min at 28 °C. Cells were dissociated by pipetting and resuspended in suspension medium (1 % calf serum, 0.8 mM CaCl2, 50 U/ml penicillin, and 0.05 mg/ml streptomycin in DMEM) and then separated into GFP-positive and GFP-negative fractions using a flow cytometer (BD FACSAria; BD Biosciences).
Western blot analysis
Embryos were homogenized in SDS lysis buffer. 60 μg of protein was loaded on 12 % SDS polyacrylamide gel and transferred to a PVDF membrane and detected with antibody for phospho-Akt1 or total Akt1 (1:1,000; Cell Signaling). After washes, membranes were incubated with goat anti-Mouse HRP-conjugated secondary Ab (Cell Signaling) and developed with ECL (Millipore). Band intensities were quantified using Multi Gauge analysis software. The results are representative of three separate experiments performed in duplicate.
Statistical analysis
For spontaneous contraction, touch response, and axon counts, statistical analysis was performed by unpaired Student’s t test using Microsoft Excel 2007. p values less than 0.05 were described as statistically significant. All graphs show mean and standard deviation and represented as mean ± SD in the text.
Chemical treatment
CCG-4986 was applied as a selective RGS4 inhibitor [17, 18, 19]. Embryos were treated with CCG-4986 (15 μM) at 14 hpf, 1 h before the onset of first axon extension in the spinal cord [20]. Vehicle control was 1 % dimethylsulfoxide (DMSO).
Histological analysis
For whole mount in situ hybridization, the full length coding sequence of zebrafish rgs4 cDNA was cloned into pCS2+ vector and linearized with EcoRI. Riboprobes for rgs4, neurogenin1 and pax2a were generated by in vitro transcription with T7 RNA polymerase and labeled with digoxigenin-11-UTP (Roche) and then purified using Illustra Microspin G-25 Columns (GE Healthcare) according to the manufacturer’s instructions. Zebrafish embryos were fixed with 4 % paraformaldehyde and permeabilized by 100 % methanol for at least 10 h at −20 °C. Subsequently, embryos were rehydrated and digested with proteinase K as recommended [21]. Embryos were then hybridized with riboprobe for a minimum of 15 h at 68 °C. Unbound and excessive probe was removed by three washes in 50 % formamide in 2× SSC 0.1 % Tween-20, followed by three washes in 0.2× SSC 0.1 % Tween-20. After three rinses with PBST (PBS plus 0.1 % Tween-20), embryos were blocked in 5 % sheep serum for 1 h at room temperature and then incubated with sheep-anti-digoxigenin-alkaline phosphatase (1:2,000; Roche) overnight at 4 °C. Unbound antibody was removed by washing six times in PBST for 24 h with gentle agitation. Color was developed in the dark at room temperature using NBT/BCIP substrate (Roche). The reaction was stopped by washing in TE (10 mM Tris–HCl pH 7.8, 10 mM EDTA).
For immunohistochemistry, the embryos were blocked in 5 % goat serum and antibodies of HuC/HuD (1:500 dilution; Invitrogen), Islet1/2 (1:250 dilution; Developmental Studies Hybridoma Bank), znp-1 (1:100 dilution; Zebrafish International Resource Center), 3A10 (1;200 dilution; Developmental Studies Hybridoma Bank) or acetylated α-tubulin (1:1,000 dilution; Sigma) were applied. Fluorochrome conjugated antibodies Alexa Fluor 488 (or 594) goat anti-mouse IgG (Invitrogen) were used to detect the primary antibodies. Embryos were mounted with Vectashield mounting medium with DAPI (Vector Laboratories). Confocal Microscopy was performed using a Zeiss LSM 510 microscope, and a z-stack of fluorescence images was acquired. The black-and-white fluorescent signals were inverted to negative film for a clearer presentation.
Quantitative real time PCR
For quantitative real-time PCR (qPCR), embryos were homogenized in TRIzol reagent (Invitrogen) and total RNA was extracted using a standard method. cDNA was synthesized from total RNA with random hexamer priming using RevertAid First Strand cDNA Synthesis Kit (Fermentas). qPCR was performed on an ABI StepOne™ Real-Time PCR System (Applied Biosystems) with SYBR green fluorescent label (Fermentas). Primers for neurogenein1 (F: 5′-CGCACACGGATGATGAAGACTCGCG-3′; R: 5′-CGGTTCTTCTTCACGACGTGCACAGTGG-3′), pax2a (F: 5′-ACCGGGTGAAGCTTGTGAAC-3′; R: 5′-CGTCATGCYCGTCCAGAAGTT-3′) were used. Gene expression levels were normalized to gapdh and assessed using the comparative CT (40 cycles) according to the manufacturer’s instructions (Applied Biosystems).
Birefringence
Embryos were examined using a Leica DM 2500 upright microscope. Muscle birefringence was analyzed by placing embryos between two polarizing filters adjusted until only the light refracting through the striated muscle was visible. In order to observe the shape and boundaries of the somite simultaneously, low level of transmitted light with Nomarski interference contrast optics was also used.
Results
Characterization of zebrafish rgs4
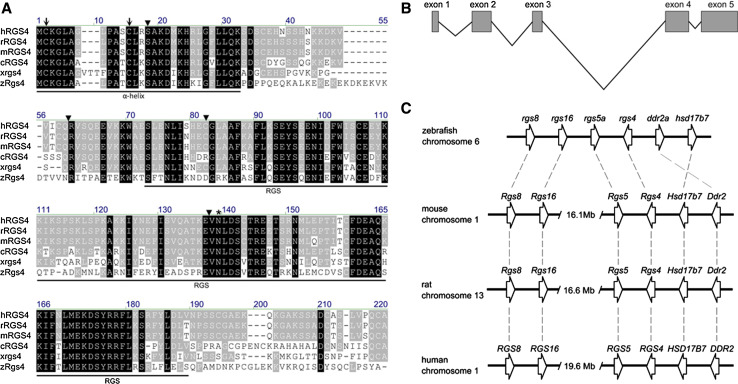
A 645-bp sequence encoding a 215 residue peptide of the open reading frame of rgs4 was identified on the zebrafish genome database (http://www.ensembl.org) and cloned by PCR (Fig. 1a). Alignment with the genomic sequence showed the open reading frame was contained within five exons (Fig. 1b). The amino acid sequence of Rgs4 shows structural features of the B/R4 subfamily that contains a relatively short coding sequence in comparison to other RGS members [2], an α-helix domain at the amino-terminus (that has been shown to facilitate direct binding to membrane phospholipids [22, 23]), and a highly conserved RGS domain (Fig. 1a). Compared to other orthologues of the B/R4 subfamily, zebrafish Rgs4 showed the highest degree of similarity to chicken RGS4 with 51 % identical and 67 % conserved amino acids and slightly weaker homology with Xenopus rgs4 (49 % identical and 66 % conserved amino acids) and mouse RGS4 (47 % identical and 69 % conserved amino acids). Five RGS4 splice variants were identified in human [24], and the zebrafish Rgs4 described here is most similar to isoforms 1 and 2 which have different first exons but encode the same protein [24] (47 % identical and 69 % conserved amino acids) (Fig. 1a, b). Within the RGS domain, the identity increases to 58 % for chicken, 54 % for Xenopus, and 52 % for mammalian homologues. Comparison of the exon–intron structure of the vertebrate Rgs4 homologues revealed that all intron positions have been strictly conserved, which provides strong evidence for evolution from a common ancestor gene specific to this subgroup (Fig. 1a). In addition, zebrafish Rgs4 also contains conserved cysteines at positions 2 and 12 which have been shown to be responsible for palmitoylation [22, 23]. Syntenic comparison with mammalian homologues showed the gene loci flanking the zebrafish rgs4 gene are highly conserved with those gene sets in human chromosome 1, mouse chromosome 1, and rat chromosome 13 (Fig. 1c). This result further substantiates the orthology of the zebrafish rgs4 homologue to mammalian Rgs4.
Fig. 1.
Alignment of Rgs4 homologues and synteny comparison. a Amino acid alignment of human, rat, mouse, chicken, Xenopus laevis, and zebrafish Rgs4 sequences. Residues identical in all proteins are marked in black boxes and similarity is shown in grey boxes. The α-helix domain and RGS domains are indicated. The intron positions are marked with arrowheads and the conserved cysteines that are putative targets for palmitoylation are shown with arrows. A conserved asparagine (N) within the RGS domain necessary for GAP activity for Gα is indicated with an asterisk. Initial letters: h human, r rat, m mouse, c chicken, x Xenopus laevis, z zebrafish. b Schematic representation shows the genomic organization of the rgs4 gene in zebrafish. c Synteny comparison between the human, mouse, rat, and zebrafish Rgs4/rgs4 loci reveals that the genomic organization of the genes is largely conserved. Genes are not drawn to scale.
rgs4 is expressed in the developing nervous system
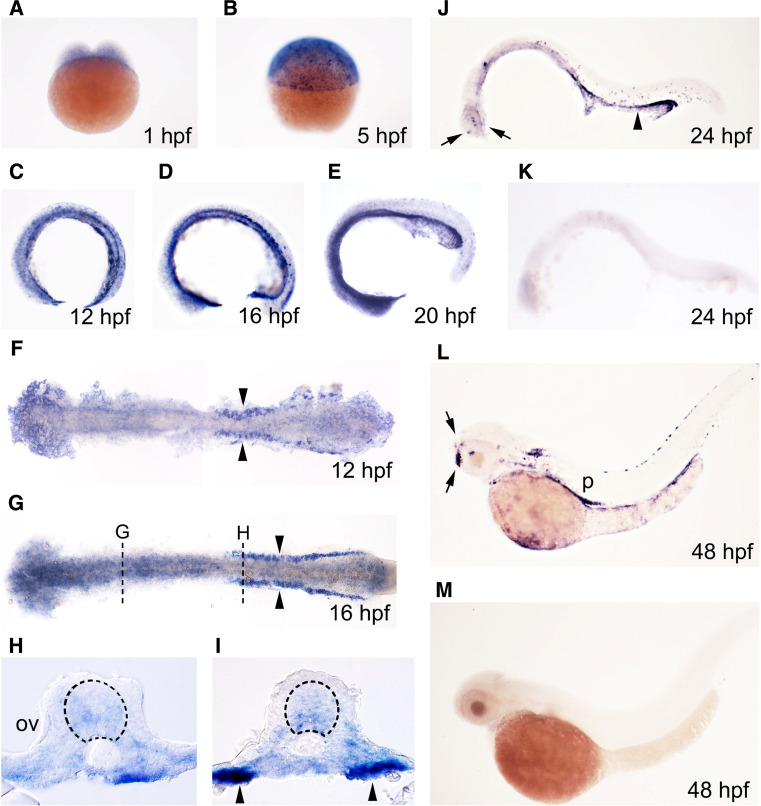
Expression of rgs4 was analyzed by whole mount in situ hybridization on zebrafish embryos age from 1-cell stage to 48 h post-fertilization (hpf). Ubiquitous weak expression through the whole embryo before and during gastrulation suggests that the gene is expressed as a maternal mRNA (Fig. 2a, b). Continued expression in the developing nervous system was detected in the brain and spinal cord from 12 hpf, but this general expression gradually disappeared after 24 hpf (Fig. 2c–j) and was replaced by clusters of neurons expressing much higher levels of rgs4 in the telencephalon and diencephalon (Fig. 2j, l). In addition to the developing nervous system, significant expression could be seen in the developing pronephric ducts from 12 hpf (Fig. 2c–l) and in the pancreas from 48 hpf (Fig. 2l). In comparison, expression of mouse Rgs4 was identified in motor neurons in the hindbrain, cranial ganglia, and dorsal root ganglia, but expression in developing kidneys and pancreas have not been described [6, 11], revealing evolutionary diversity of expression between Rgs4 homologues.
Fig. 2.
rgs4 expression in the developing zebrafish. rgs4 expression was detected during zebrafish embryogenesis analyzed by in situ hybridization. Stages of embryos shown in bottom right corner of each panel. a–d, i–m Lateral view, dorsal to the top. f, g Dorsal view, anterior to the left, yolk removed and embryos flat-mounted. rgs4 expression is expressed as a maternal mRNA (a, b) and then first appears in the developing nervous system at 12 hpf (c, f) and is retained in the brain until 24 hpf (a–j). Transverse sections as marked in (g) showing expression in the developing brain (h) and spinal cord (i) (dashed circle). Strong expression, restricted to compact clusters of neurons, can be detected from 24 hpf (arrows in j, l). k, m No signal was detected using sense riboprobe. Arrowheads in all panels indicate rgs4 expressing cells in the pronephros. OV otic vesicle, P pancreas
Rgs4 knockdown resulted in motility defects
In order to delineate the role of Rgs4 in zebrafish, the morpholino (MO) knockdown approach was used to interfere with its expression. Two 25-bp antisense morpholinos (MO1 and MO2) were synthesized to target different regions 5′ to the translation start site of the rgs4 mRNA to block protein production. Blast analysis revealed homology of less than 20 bp identity for MO1 or MO2 to other genomic sequences, none of which corresponded to 5′ UTR or exon–intron splicing site of predicted or characterized genes, suggesting that MO1 and MO2 will act specifically on rgs4. This specificity was confirmed by rescue experiments in which the MOs were coinjected with cRNA for rgs4, as described for each experiment below. To confirm the efficacy of the morpholino knockdown approach, each of the two rgs4 morpholinos was coinjected with the cRNA of a reporter construct that contained the rgs4 MO1 and MO2 binding sequences upstream of an enhanced green fluorescent protein reporter (5′rgs4-EGFP). Effective knockdown of the translation of this construct (evident by loss of EGFP protein) was observed upon coinjection with either of the two rgs4 morpholinos but unaffected by coinjection with control morpholino (Supplementary Fig. 1a).
Embryos injected with MO1 or MO2 were analyzed at 24 h post-fertilization (hpf), 3 days post-fertilization (dpf) and 5 dpf for morphological defects. Amounts of 8 ng of MO1 or 1.5 ng of MO2 injection exhibited identical phenotypes, and therefore only embryos injected with 1.5 ng of MO2 are shown. Although Rgs4 knockdown resulted in no significant morphological abnormalities (Supplementary Fig. 1b), locomotor defects were observed at all stages analyzed (see below).
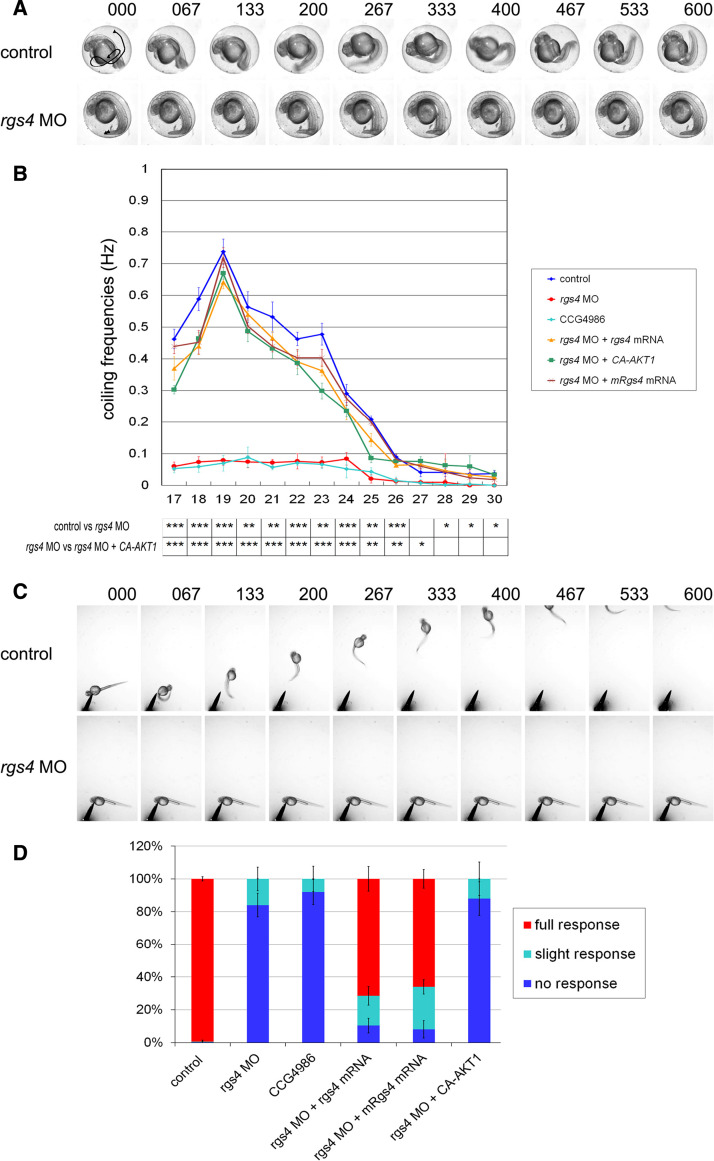
We first examined the earliest motor behavior, spontaneous coiling, in the rgs4 morpholino-injected embryos. Spontaneous coiling consists of alternating side-to-side metronome-like contractions of the tail, a behavior that does not require forebrain, midbrain, or hindbrain input [20]. rgs4 morpholino-injected embryos exhibited only a very slight coiling angle and a single one-sided coiling contraction in comparison to the alternating contractions observed in controls (Fig. 3a, b). Time-lapse analysis showed that the coiling behavior first appeared in control embryos around 17 hpf (0.46 ± 0.03 Hz), and the frequency peaked at 19 hpf (0.74 ± 0.04 Hz) and gradually decreased as reported previously (Fig. 3b) [25], whereas embryos injected with rgs4 morpholino displayed severely reduced coiling frequencies at all stages examined (Fig. 3b). In comparison to the control embryos, the coiling frequency in rgs4 morphants dropped to 0.06 ± 0.01 Hz at 17 hpf (p = 0.00046) and the maximum frequency was 0.08 ± 0.02 Hz at 19 hpf (p = 0.00029), after which the frequency decreased and the morphants stopped coiling almost completely after 30 hpf (Fig. 3b). Previous studies have shown that spinal cord activity was sufficient to generate spontaneous coiling and that this locomotion is independent of sensory stimulation [25]. Therefore, defective coiling motion and decreased coiling frequency in Rgs4 knockdown embryos suggests that Rgs4 is necessary for proper function of the spinal motor circuit.
Fig. 3.
Rgs4 knockdown resulted in locomotor phenotypes. a Spontaneous coiling behavior is defective in rgs4 morphants. Individual frames from a time-lapse video showing two consecutive coils by a control embryo at 24 hpf (top panels). Time of each frame is shown at the top. Tail movement is shown by a solid line in the first panel by tracking analyses. Both the frequency and angle of tail coiling is reduced in rgs4 morpholino-injected embryos (lower panels). b Kinetic analysis of the spontaneous coiling between 17 and 30 hpf. The frequency of coiling was reduced at all stages in rgs4 morphants (red) and CCG-4686 treated embryos (cyan) compared with the controls (blue). The defect in coiling frequency in rgs4 morpholino injected embryos could be rescued by concomitant injection of either zebrafish rgs4 (orange), mouse Rgs4 (brown), or ca-AKT1 (green) cRNA. Each data point represents the average frequency of contractions and standard errors for all freely moving embryos examined. Note that all p values were significant before 26 hpf. *p < 0.05, **p < 0.01, ***p < 0.001. c Video capture of aberrant touch and swim responses in Rgs4 knockdown embryos at 48 hpf. Tactile stimuli were applied at a region close to trigeminal ganglia. Control embryos responded to stimuli with a rapid escape response contraction followed by swimming. In contrast, Rgs4 morphants did not respond and were not able to escape. Times indicated on the top are milliseconds. d Proportion of embryos swimming after a tactile stimulus. Aberrant responses were observed in rgs4 morphants and CCG-4686 treated embryos whereas injection of zebrafish rgs4 or mouse Rgs4 cRNA in rgs4 morphants could restore the touch and swim responses. Note that injection of ca-AKT failed to rescue the touch and swim phenotype in Rgs4 deficient embryos. Measurements in (b) and (d) were performed on 20 embryos per batch, and 3 batches were used per time point. rgs4 MO, rgs4 morpholino; rgs4 zebrafish rgs4; mRgs4, mouse Rgs4
We next analyzed touch response and swimming behavior in the Rgs4 knockdown embryos. Wild-type zebrafish embryos become sensitive to touch and respond with vigorous coiling (after 21 hpf) and later swimming (from 28 hpf); both responses require intact neural circuits in hindbrain and spinal cord, and this is also one of the first indications that afferent connections have formed [25, 26]. Later, from 48 to 60 hpf, zebrafish embryos hatch from their chorions and react to tactile stimuli with a fast escape response. Embryos at 48 hpf were dechorionated manually and tested for their response to stimuli applied close to the region of the trigeminal ganglia; 84 ± 7.19 % of rgs4 morphants showed no respond to stimuli and 17 ± 7.19 % showed a slight response to stimulus but were not able to escape (in comparison to 99.8 ± 1.23 % response to stimuli for control embryos; Fig. 3c, d). Since touch response requires the activation of both hindbrain and spinal neuron, these results suggest that the spinal neural networks are defective in Rgs4 knockdown embryos and that the hindbrain networks might also be impaired.
Early neuronal differentiation appears normal in Rgs4 knockdown embryos
The locomotor phenotypes seen in Rgs4 knockdown embryos suggested a possible defect in neuronal differentiation. Therefore, the generation of neurons was analyzed in rgs4 morphants. Markers for neuronal precursors (neurogenin1), pan-post-mitotic neurons (HuC/D), interneurons (pax2a), primary motor neurons (Islet1/2), and cell apoptosis (active caspase-3) were investigated. No significant alteration in the number or localization of different types of neurons was observed in rgs4 morphants by 24 hpf (Supplementary Fig. 2), indicating that knockdown of Rgs4 does not affect neural patterning nor neuronal differentiation and survival in the hindbrain and spinal cord. This result is consistent with the previous observation in RGS4 knockout mice which also showed normal neuronal differentiation [11]. In conjunction with the results from our knockdown approach, we suggest that endogenous RGS4/Rgs4 is not required for normal neuronal differentiation under physiological conditions.
Loss of Rgs4 is sufficient to cause an axonogenic phenotypes in spinal cord
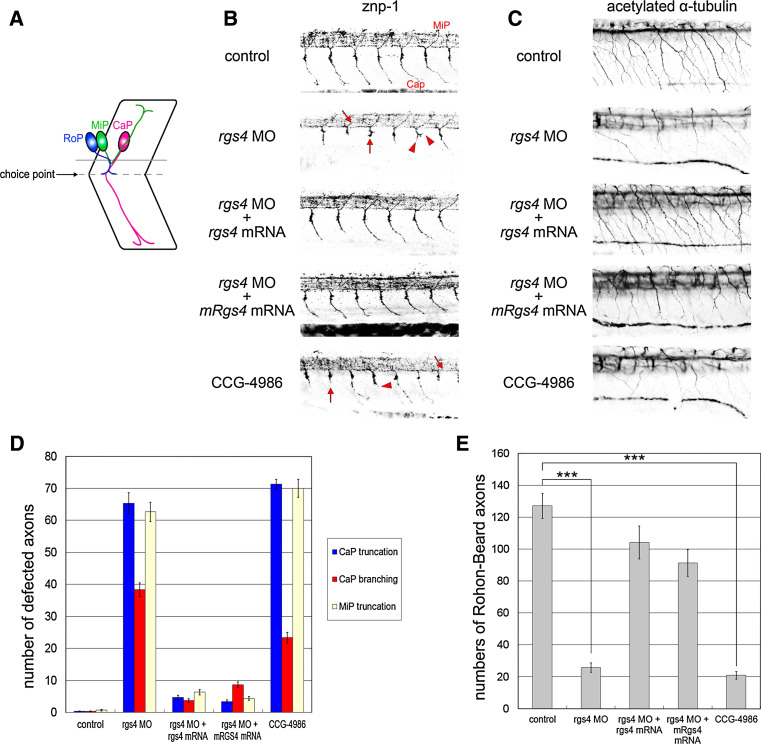
Since the rgs4 morphants showed abnormal motility, despite apparently normal neuronal differentiation, we questioned whether axon formation was affected. The zebrafish has recently emerged as an excellent vertebrate model to study axon outgrowth, guidance, and branching due to the highly stereotypical trajectory of axons, which can be conveniently examined in the transparent embryos [27, 28]. In wild-type zebrafish embryos, three primary motor neurons, CaP (caudal primary), MiP (middle primary), and RoP (rostral primary), are located in the middle of each spinal cord hemisegment that project axons in a common path from the spinal cord to the medial surface of the myotomes, where they encounter a somitic choice point. At this choice point, the axons of the three primary motor neurons make cell type-specific decisions and continue on divergent pathways (Fig. 4a). Motor axons were detected using the znp-1 monoclonal antibody and, as shown in a control embryo at 24 hpf, CaP axon projections navigate to the choice point and project into ventral myotome (Fig. 4b). In contrast, 90.8 % (n = 153) of Rgs4 knockdown embryos displayed truncated motor axons (with an average of 65.33 ± 3.27 % truncated axons in each embryo counted; see “Materials and methods” for details) that did not fully project into the ventral myotome and stalled at or before the choice point (Fig. 4b, d). In addition to truncation, aberrant CaP axon branching was observed (Fig. 4b, d) in 85.0 % of embryos (n = 153, with an average of 38.33 ± 2.15 % axon with branches; Fig. 4d). Dorsally projecting motor nerves (MiP) also exhibited truncation defects as the result of Rgs4 knockdown (62.67 ± 2.99 %; Fig. 4b, d; Supplementary Fig. 6). These defects suggest that motoneuron projections are affected by reduction of Rgs4 protein. To determine if Rgs4 knockdown also affected other spinal cord axons, we examined the Rohon–Beard sensory axons detected by acetylated α-tubulin antibody. In control embryos, Rohon–Beard neurons were located in the dorsal spinal cord and sent axons ventrally into the peripheral muscle and skin [29] (Fig. 4c). In contrast, Rohon–Beard axon fibers were both fewer and tended to be truncated in rgs4 morpholino injected embryos (Fig. 4c, e; 25.7 ± 2.90 compare to 127.2 ± 7.83 in the controls, p = 1.22 × 10−40). The defects of motor and sensory axon caused by Rgs4 knockdown could be rescued by concomitant injection of zebrafish rgs4 or mouse Rgs4 cRNA, indicating that the morpholino-induced defects were the result of specific inhibition of Rgs4 function (Fig. 4b–e), and that Rgs4/RGS4 has a similar role in different organisms. In contrast, widespread over-expression of rgs4 cRNA alone did not cause an axonal defect (data not shown). Taken together, these results reveal that Rgs4 knockdown is sufficient to cause axonogenic phenotypes in both motor and sensory axons in spinal cord.
Fig. 4.
Rgs4 knockdown caused axon trajectory defects in the spinal cord. a Schematic representation of axon trajectories of primary motor neurons at 24 hpf. Axons from three types of spinal neurons share a common path from the spinal cord to the choice point (dashed line). At the choice point, CaP axons extend into the ventral myotome, MiP axons sprout dorsally, and RoP axons pause. b Spinal cord axon tracts immunostained with znp-1 at 24 hpf. Truncated (red arrows) and branched motor axons (red arrowheads) can be detected in rgs4 morpholino-injected and CCG-4986-treated embryos. c The axons of Rohon–Beard sensory cells stained for acetylated α-tubulin at 27 hpf were also defective following Rgs4 knockdown and CCG-4986 treatment. d Quantitative data showing significant increased motor axonal defects caused by Rgs4 deficiency. e Rohon–Beard axon counts confirm significantly reduced axon numbers presented as average number of axons in somitic segments 9–18. The defects of motor and sensory axon caused by morpholino injection were rescued by concomitant injection of zebrafish rgs4 or mouse Rgs4 cRNA (b–e). CaP caudal primary motor neuron; MiP middle primary motor neuron; RoP rostral primary motor neuron. a–c are lateral views (rostral to the left; dorsal to the top). rgs4 MO, rgs4 morpholino. z-stacks of fluorescence images were acquired by confocal microscopy. The black-and-white fluorescent signals were inverted to negative film for a clearer presentation. Data in (d, e) are mean ± SD. ***p < 0.01. rgs4, zebrafish rgs4; mRgs4, mouse Rgs4
Since the phenotype of rgs4 morphants showed a defect of movement, we also examined the development and patterning of somitic trunk muscles. Birefringence of polarized light through the parallel fibrillar arrays of the trunk muscle [30–32] revealed normal patterns in rgs4 morphants compared to control embryos (Supplementary Fig. 3). In addition, no defect was detected in the somite/muscle morphology or patterning between controls and rgs4 morpholino-injected embryos analyzed by differential interference contrast optics with transmitted light (Supplementary Fig. 3). These data suggest that knockdown of Rgs4 does not affect myotome differentiation and organization, but specifically regulates axon formation.
Knockdown of Rsg4 affects hindbrain axon formation and pathfinding
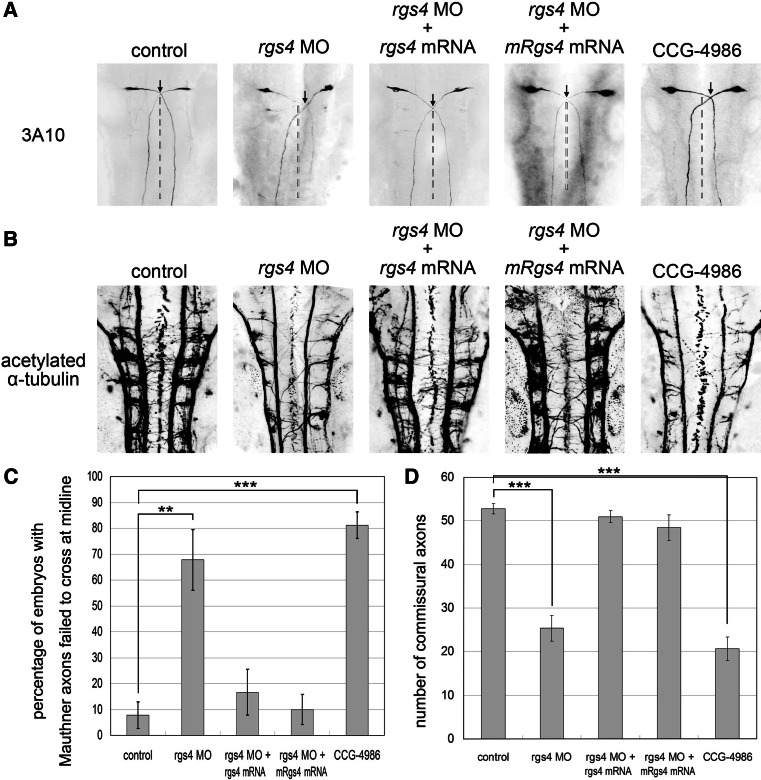
The axonal defects observed in the spinal cord of rgs4 morphants suggested this could be the cause for the aberrant spontaneous coiling and swimming behavior. We next studied whether the hindbrain neural circuits were also affected. We first analyzed the bilaterally paired Mauthner cells located in rhombomere 4, since these are the most prominent members of the reticulospinal population, which have been shown to control the touch-escape response in teleost fish and amphibians [33]. Immunohistochemistry with the 3A10, anti-neurofilament, antibody-stained Mauthner neurons revealing that the axonal trajectory was affected. Wild-type Mauthner cells project axons symmetrically to the contralateral spinal cord and form the medial longitudinal fasciculus starting at 17 hpf [28]. In embryos injected with rgs4 morpholino, axons of the paired Mauthner neurons were able to project to the contralateral spinal cord, but were deflected sharply or far more laterally, so failing to cross each other precisely at the ventral midline in comparison to the control (Fig. 5a, b; p = 0.005431), indicating a defect in Mauthner axon pathfinding. Axons of other hindbrain neurons were examined with the pan-axonal maker acetylated α-tubulin antibody revealing that the ladder-like arrays of commissures were much less dense and less well organized in the hindbrain of Rgs4 knockdown embryos (Fig. 5c, d; 52.7 ± 2.90 compared to 25.4 ± 2.97 in the controls, p = 1.68 × 10−38). These knockdown phenotypes could be largely rescued by concomitant injection of zebrafish rgs4 or mouse Rgs4 cRNA (Fig. 5a–d). Over-expression of rgs4 cRNA alone did not cause axonal defect in the hindbrain (data not shown). In conclusion, our results show that Rgs4 is essential for correct formation of the hindbrain commissural trajectories.
Fig. 5.
Knockdown of Rgs4 resulted in aberrant axon growth in the hindbrain. a Mauthner neuron cell bodies and axons were stained with 3A10 antibody in 24-hpf embryos. The crossing points of the bilateral projected Mauthner fibers are shown by arrows and midlines of neural tube are indicated with dashed lines. The Mauthner axons failed to project symmetrically to the contralateral side and did not cross each other at the midline in Rgs4 knockdown or CCG-4986 treated embryos. Hindbrain midlines are judged by the location of the central canal. b Quantification of embryos with aberrant Mauthner axon trajectory, which shows significantly increased numbers of defective embryos in Rgs4 knockdown or CCG-4986 treated embryos. c Pan-axonal marker, acetylated α-tubulin-labeling, shows significantly reduced number of axons in rgs4 morpholino-injected or CCG-4986-treated embryos in comparison to the control confirmed by axon counts. d The axonal phenotypes detected by 3A10 and acetylated α-tubulin in Rgs4 morphants was rescued by co-injection with zebrafish rgs4 or mouse Rgs4 cRNA (a–d). Dorsal view, anterior to the top. rgs4 MO, rgs4 morpholino. **p < 0.01; ***p < 0.001
Rgs4 knockdown resulted in defective trigeminal axons
The essential components of the touch response include the trigeminal sensory neurons of the head, Mauthner neurons in the hindbrain, and motor neurons in the spinal cord. In addition to the axonal defects in the hindbrain and spinal cord, we found that the growth of trigeminal axons was also severely impaired in Rgs4-deficient embryos (Supplementary Fig. 7), and that this phenotype could be rescued by injection with zebrafish rgs4 or mouse Rgs4 cRNA. The axons of the trigeminal neurons and hindbrain were both defective, so our results demonstrated that loss of Rgs4 produced various axonal phenotypes in the hindbrain and trigeminal neurons, and deviant touch and escape responses.
Rgs4 regulated axonogenesis requires GAP activity for Gα subunits
We next studied whether Rgs4-regulated axonogenesis requires its Gα binding and GTPase activity. Methyl-N-[(4-chlorophenyl)sulfonyl]-4-nitrobenzenesulfinimidoate (CCG-4986) has been shown to be a selective RGS4 inhibitor that disrupts RGS4-Gα interaction and inhibits GTPase acceleration [17–19]. Therefore, 15 μM of CCG-4986 was applied to zebrafish embryos at one- to two-cell stage or at the 10-somite stage (just before the onset of axonogenesis), and embryos were analyzed at the same stages as for morpholino injections (Figs. 4, 5). CCG-4986-treated embryos showed motility defects very similar to rgs4 morphants (Fig. 3b for spontaneous coiling, and Fig. 3d for touch response). Axonal defects, such as truncation and branching in the spinal cord (Fig. 4b–e), irregular pathfinding in the hindbrain, and fewer axonal tracts in the hindbrain and trigeminal nerves, were also very similar to those observed in rgs4 morphants (Fig. 5; Supplementary Fig. 7). These results demonstrated that Rgs4 regulated axonogenesis requires GAP (GTPase-accelerating proteins) activity for Gα subunits, suggesting that the RGS domain was essential for its function. They also support the conclusion that the same phenotype seen following rgs4 morpholino injection was due to the specific knockdown of Rgs4.
Rgs4 regulated axonogenesis requires Akt activation
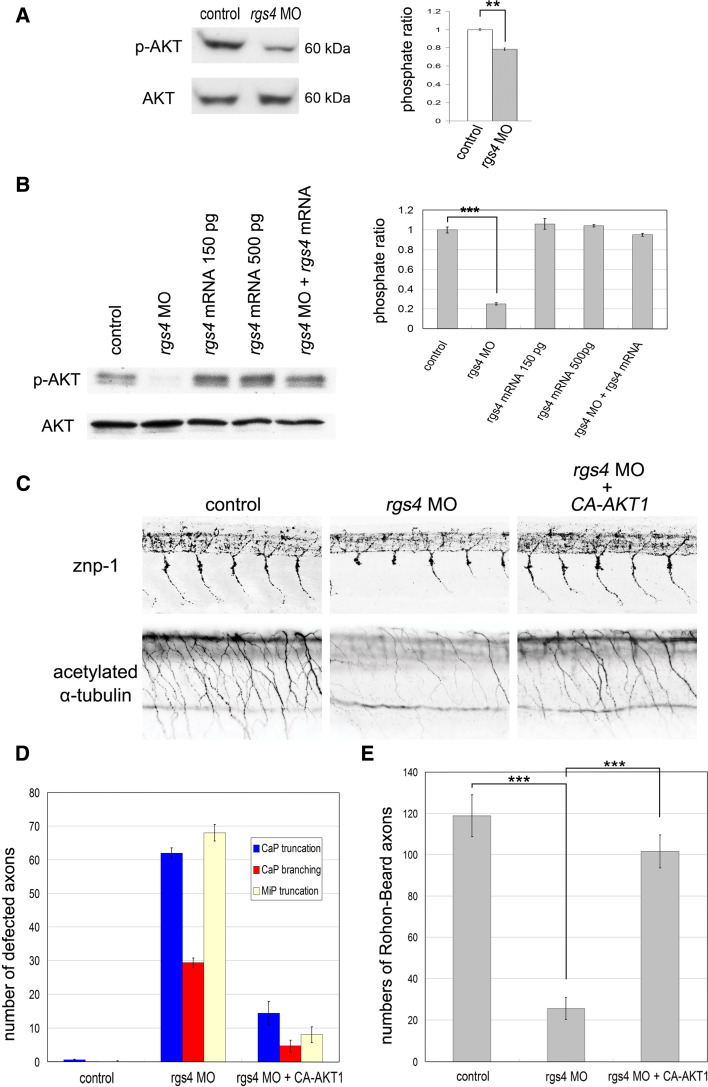
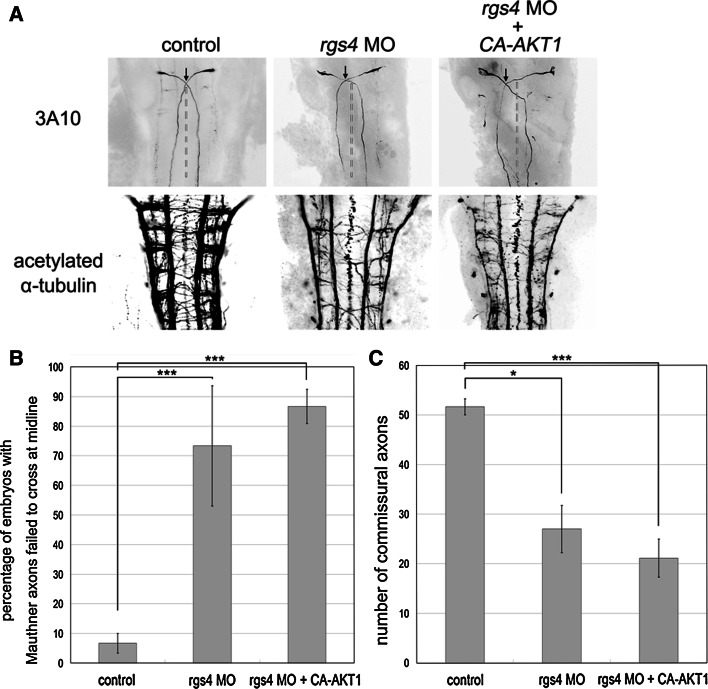
Our next question was how Rgs4 regulates axonogenesis at a molecular level. PI3K-Akt is one of the major signaling pathways downstream of receptor tyrosine kinases (RTKs) that have been intensively analyzed and shown to be fundamental in axonogenesis [34]. This pathway is also regulated by G-protein-coupled receptor (GPCR) signaling [35]. More recently, Leone et al. [36] showed that activation of Akt pathway by serotonin receptors can be inhibited by RGS4 in neuroblastoma cells. We therefore investigated whether Rgs4-regulated axonogenesis was mediated by PI3K-Akt pathways. Western blot analysis of whole embryo extracts at 14 hpf showed that knockdown of Rgs4 decreased the levels of phosphorylated Akt (Fig. 6a; p = 0.003396), suggesting that Rgs4 positively regulates the Akt signaling pathway. To show that this effect occurs in neurons, HuC-positive neurons were sorted from Tg(huC:eGFP) embryos using flow cytometry, followed by western blot analysis, and the results showed that downregulation of phosphorylated Akt occurred in the neurons of the rgs4 morphants (Fig. 6b). Injection of rgs4 cRNA into the rgs4 morphants restored the level of phosphorylated Akt, whereas injection with rgs4 cRNA alone did not induce the level of phosphorylated Akt (Fig. 6b). This suggested that an additional cofactor was required to further elevate the level of phosphorylated Akt. To further determine if this regulation could modulate motor behavior and axonogenesis, constitutively active AKT1 (ca-AKT1) was injected into rgs4 morphants. This resulted in restoration of spontaneous coiling (Fig. 3b; all p values showed statistically significant in comparison to rgs4 morpholino injection alone); however, this could not rescue the touch response (Fig. 3d). This suggests that the motor activity of the spinal cord might have been rescued but the hindbrain and trigeminal circuits were still impaired. Indeed, analysis of axonal markers showed that injection of ca-AKT1 cRNA in rgs4 morphants or CCG-4986-treated embryos rescued the axonal defects in the spinal cord (Fig. 6c–e), but was not able to restore the axonal phenotype observed in the hindbrain and trigeminal nerves (Fig. 7; Supplementary Figs. 5, 7). In conclusion, these results demonstrate that Rgs4-regulated axonogenesis could be positively mediated by the Akt signaling pathway in the spinal motor and sensory neurons.
Fig. 6.
Rgs4-regulated axonogenesis is mediated by Akt1 in the spinal cord. a Left western blot analysis with antibody for phospho-Akt1 (p-AKT) and total Akt1 (AKT) shows reduced p-AKT in rgs4 morphants analyzed at 14 hpf. Right densitometric analysis shows that knockdown of Rgs4 significantly reduced p-AKT/AKT levels. p = 0.003396. b Western blot analysis with antibodies for phospho-Akt1 (p-AKT) and total Akt1 (AKT) indicated the downregulation of p-AKT in HuC-positive neurons, which were sorted from the Tg(huC:GFP) embryos injected with the rgs4 morpholino by flow cytometry, when compared with the controls. The level of p-AKT was restored by co-injection with rgs4 cRNA, whereas injection with rgs4 cRNA alone did not significantly affect p-AKT expression. c Immunohistochemistry with znp-1 and acetylated α-tubulin shows the axonal defects observed in the spinal cord of rgs4 morphants could be rescued by injection of constitutively-activated AKT1 cRNA, and this was confirmed by quantitative analysis (d, e). The crossing points of Mauthner axons are shown by arrows, and dashed lines indicate hindbrain midlines. ca-AKT1, constitutively-activated AKT1; rgs4 MO, rgs4 morpholino. **p < 0.01; ***p < 0.001
Fig. 7.
Injection of ca-AKT1 in rgs4 morphants could not rescue axonal defects in the hindbrain. a Defects such as failure of Mauthner axons to cross each other at the midline and fewer acetylated α-tubulin stained commissural axons could be detected in Rgs4 knockdown embryos, and neither of these phenotypes could be restored by injection of constitutively-activated AKT1. b Quantification confirms number of embryos with aberrant Mauthner axon trajectory are significantly increased in either rgs4 morphants or embryos coinjected with rgs4 morpholino and constitutively-activated AKT1. c Quantification of axons indicates significant loss of hindbrain commissural axons in rgs4 morpholino-injected embryos, and injection of constitutively-activated AKT1 failed to restore the hindbrain phenotypes. ca-AKT1, constitutively-activated AKT1; rgs4 MO, rgs4 morpholino. *p < 0.05; ***p < 0.001
Discussion
Rgs4 is essential for axon formation
During development, neurons become assembled into functional networks by dentritic and axonal outgrowths (broadly termed neurite outgrowth) and connect synaptically. Many downstream molecules in the G-protein signaling pathway, such as small GTPases Rap1 and Rac, Src, STAT3, and MAP kinases, have been described that control neurite outgrowth [37]. Although RGS proteins are well known for regulating G-protein signaling, there is little direct evidence of a role for RGS proteins in neurite outgrowth. RGSZ1 and RGS6 directly interact with the microtubule-destabilizing protein, SCG10, and regulate the SCG10-induced microtubule organization [38, 39]. The Ras-binding domain of RGS14 facilitates the formation of the Ras/Raf/MEK/ERK complex to regulate H-Ras-dependent neuritogenesis [40], and RGS2 directly interacts with tubulin, enhancing microtubule polymerization and neurite outgrowth [41]. However, these studies were based on in vitro analyses. We have shown that knockdown of Rgs4 in zebrafish embryos is sufficient to cause aberrant axon formation, and this may be responsible for the locomotor defects we observed. To our knowledge, this is the first report demonstrating the role of a RGS protein specifically in axon formation. In addition, unlike the studies describe above showing that RGS proteins regulate neurite outgrowth through the domains other than RGS, we showed that Rgs4-regulated axonogenesis requires GAP activity of the RGS domain. Therefore, our results not only provide in vivo evidence of a critical role of RGSs in neuritogenesis but also reveal a novel function of the RGS domain that might apply to other RGS family members.
Rgs4-regulated axonogenesis is mediated by Akt signaling
We found that Rgs4 could regulate Akt signaling during axon formation. Akt is phosphorylated by PI3K signaling following activation of a tyrosine kinase or GPCR at the plasma membrane, and subsequently regulates a number of substrates that are involved in aspects of neurite outgrowth [42]. The ability of RGS proteins to accelerate GTPase activity suggests that RGSs may negatively modulate GPCR-induced Akt activation. Indeed, a previous study by Leone et al. [36] demonstrated that RGS4 is able to attenuate serotonin receptor-initiated AKT signaling in neuroblastoma cells. Intriguingly, our results demonstrated that phosphorylated Akt1 is essential for Rgs4-regulated G-protein signaling in axon formation. Therefore, Akt may regulate, or respond to, Rgs4-regulated G-protein signaling in axonogenesis by a mechanism yet to be identified. Although this seems to contradict the previous in vitro data [36], this can be explained by the fact that G-proteins can regulate Akt by two opposing mechanisms, either activating PI3K-dependent AKT signaling by Gβγ subunits, or by inhibiting AKT activation by Gαq [43]. Indeed, we show that Rgs4 requires the GAP activity of its Gα subunits to regulate axonogenesis, and that this requires Akt activation. This, therefore, favors the latter mechanism that this regulation may be mediated by a Gα subunit (Rgs4 inactivates GTP-bound Gα signaling and relieves the inhibition of Akt signaling). Unfortunately, this issue is currently difficult to address due to very limited knowledge about G-protein subunits with only a few homologues identified in zebrafish. Our results also suggest that Rgs4 may regulate axonogenesis via a GPCR-modulated mechanism that differs from its mechanism in neurotransmission.
The trigeminal nerves, hindbrain and spinal cord play key roles in the generation of motor activities that are evolutionary conserved across species [20]. We show that rgs4 morphants exhibit defects in both touch responses and swimming capability which suggests impaired neural circuits in the trigeminal nerves, hindbrain, and spinal cord. However, distinct aspects of axonal defects were observed in Rgs4 knockdown embryos. The axons of the spinal cord showed aberrant elongation and branching, whereas relatively few axonal tracts were observed in the hindbrain and trigeminal nerves, as well as mis-oriented pathfinding of the hindbrain commissures nerves. Akt has been shown to be essential for neurite elongation, branching, and caliber [42, 44–46]. We found that co-injection with constitutively-active AKT1 attenuated the effect of rgs4 morpholinos in the spinal cord, indicating that Akt activation is required for Rgs4-modulated axonogenesis. Strikingly, activation of Akt could not restore the axonal phenotypes in the hindbrain and trigeminal nerves of rgs4 morphants, suggesting a different pathway is involved. One candidate is MEK-Erk1/2 signaling, since this signaling can also be activated by GPCR and has been shown to be essential for neurite pathfinding [47, 48]. Indeed, we found the level of phosphorylated Erk1/2 was down-regulated in Rgs4 knockdown embryos (unpublished observation). In summary, our work shows that Rgs4 regulates axonogenesis and may regulate processes by different downstream mechanisms.
The pathophysiological role of Rgs4 in axonal defects and schizophrenia
Several genes, including RGS4, have been found to have a significant genetic association with schizophrenia [9]. Aberrant expression has been described, and defects in these genes may lead to abnormal neurodevelopment and dysregulation of dopamine and/or glutamine signaling, which provides a possible pathological mechanism for schizophrenia at a molecular level [49]. In the neurodevelopmental hypothesis, susceptibility genes, such as AKT1, ErbB4, neuregulin-1 (NRG1), dysbindin-1, and disrupted-in-schizophrenia-1 (DISC1), that regulate progenitor cell proliferation, neuronal migration, and synaptic connectivity, have been proposed [50]. Recent studies have revealed that these genes are also implicated in the process of neurite formation, providing a model in which abnormalities in neurite formation could have potential etiologic implications for schizophrenia [51]. Although direct evidence of abnormal neurite formation in patients is still missing, there is evidence that at least some neuropathological defects may be explained by defects in dendrites and/or axons [51, 52]. In the present study, we showed that in zebrafish embryos a schizophrenic susceptibility gene, rgs4, is essential for axon formation, and that this regulation is mediated by another schizophrenia candidate gene, Akt1. This result provides further evidence for the neuritogenetic model for schizophrenia and suggests a possible intracellular signaling cascade for the association of axonogenesis with schizophrenia worthy of further investigation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Jin-Chung Chen, Hung-Li Wang and Rong-Chi Huang for discussions, Chang-Jen Huang for Tg(huC:eGFP) line, Hitoshi Okamoto for Tg(islet1:GFP) line, Min-Chi Chen for statistical analysis, Pierre Drapeau and Hey-Jen Tsay for behavior analysis, Jim-Tong Horng for western blot, Chung-Der Hsaio for ca-AKT, and David Wilkinson for neurogenin1 and pax2a constructs for riboprobes. We are also grateful to Taiwan Zebrafish Core facility at ZeTH and Zebrafish Core in Academia Sinica for providing fish. This work was supported by grants from Chang Gung Memorial Hospital (CMRPD170513 and CMRPD1B0251) and the National Science Council of Taiwan (96-2745-B-182-003-URD).
Footnotes
S.-J. Lin, H.-Y. Shih and F.-Y. Hsieh contributed equally to this work.
References
- 1.Neitzel KL, Hepler JR. Cellular mechanisms that determine selective RGS protein regulation of G protein-coupled receptor signaling. Semin Cell Dev Biol. 2006;17(3):383–389. doi: 10.1016/j.semcdb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther. 2007;116(3):473–495. doi: 10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal. 2006;18(5):579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17(20):8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erdely HA, Lahti RA, Lopez MB, Myers CS, Roberts RC, Tamminga CA, Vogel MW. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci. 2004;19(11):3125–3128. doi: 10.1111/j.0953-816X.2004.03364.x. [DOI] [PubMed] [Google Scholar]
- 6.Grillet N, Dubreuil V, Dufour HD, Brunet JF. Dynamic expression of RGS4 in the developing nervous system and regulation by the neural type-specific transcription factor Phox2b. J Neurosci. 2003;23(33):10613–10621. doi: 10.1523/JNEUROSCI.23-33-10613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. The regulator of G protein signaling family. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 8.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 9.Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA. Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4) Biol Psychiatry. 2006;60(6):534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 10.Wu C, Zeng Q, Blumer KJ, Muslin AJ. RGS proteins inhibit Xwnt-8 signaling in Xenopus embryonic development. Development. 2000;127(13):2773–2784. doi: 10.1242/dev.127.13.2773. [DOI] [PubMed] [Google Scholar]
- 11.Grillet N, Pattyn A, Contet C, Kieffer BL, Goridis C, Brunet JF. Generation and characterization of Rgs4 mutant mice. Mol Cell Biol. 2005;25(10):4221–4228. doi: 10.1128/MCB.25.10.4221-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang KY, Chen GD, Cheng CH, Liao KY, Hung CC, Chang GD, Hwang PP, Lin SY, Tsai MC, Khoo KH, Lee MT, Huang CJ. Phosphorylation of the zebrafish M6Ab at serine 263 contributes to filopodium formation in PC12 cells and neurite outgrowth in zebrafish embryos. PLoS ONE. 2011;6(10):e26461. doi: 10.1371/journal.pone.0026461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20(1):206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 15.Chung PC, Lin WS, Scotting PJ, Hsieh FY, Wu HL, Cheng YC. Zebrafish Her8a is activated by Su(H)-dependent Notch signaling and is essential for the inhibition of neurogenesis. PLoS ONE. 2011;6(4):e19394. doi: 10.1371/journal.pone.0019394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh CM, Liu YC, Chang CJ, Lai SL, Hsiao CD, Lee SJ. Ptenb mediates gastrulation cell movements via Cdc42/AKT1 in zebrafish. PLoS ONE. 2011;6(4):e18702. doi: 10.1371/journal.pone.0018702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71(1):169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 18.Kimple AJ, Willard FS, Giguere PM, Johnston CA, Mocanu V, Siderovski DP. The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Galpha-interaction face. Biochim Biophys Acta. 2007;1774(9):1213–1220. doi: 10.1016/j.bbapap.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roman DL, Blazer LL, Monroy CA, Neubig RR. Allosteric inhibition of the regulator of G protein signaling-Galpha protein–protein interaction by CCG-4986. Mol Pharmacol. 2010;78(3):360–365. doi: 10.1124/mol.109.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drapeau P, Saint-Amant L, Buss RR, Chong M, McDearmid JR, Brustein E. Development of the locomotor network in zebrafish. Prog Neurobiol. 2002;68(2):85–111. doi: 10.1016/S0301-0082(02)00075-8. [DOI] [PubMed] [Google Scholar]
- 21.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein LS, Grillo AA, Loranger SS, Linder ME. RGS4 binds to membranes through an amphipathic alpha -helix. J Biol Chem. 2000;275(24):18520–18526. doi: 10.1074/jbc.M000618200. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasa SP, Bernstein LS, Blumer KJ, Linder ME. Plasma membrane localization is required for RGS4 function in Saccharomyces cerevisiae . Proc Natl Acad Sci USA. 1998;95(10):5584–5589. doi: 10.1073/pnas.95.10.5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding L, Mychaleckyj JC, Hegde AN. Full length cloning and expression analysis of splice variants of regulator of G-protein signaling RGS4 in human and murine brain. Gene. 2007;401(1–2):46–60. doi: 10.1016/j.gene.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Saint-Amant L, Drapeau P. Time course of the development of motor behaviors in the zebrafish embryo. J Neurobiol. 1998;37(4):622–632. doi: 10.1002/(SICI)1097-4695(199812)37:4<622::AID-NEU10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 26.Brustein E, Saint-Amant L, Buss RR, Chong M, McDearmid JR, Drapeau P. Steps during the development of the zebrafish locomotor network. J Physiol Paris. 2003;97(1):77–86. doi: 10.1016/j.jphysparis.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Hjorth J, Key B. Development of axon pathways in the zebrafish central nervous system. Int J Dev Biol. 2002;46(4):609–619. [PubMed] [Google Scholar]
- 28.Lewis KE, Eisen JS. From cells to circuits: development of the zebrafish spinal cord. Prog Neurobiol. 2003;69(6):419–449. doi: 10.1016/S0301-0082(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 29.Metcalfe WK, Myers PZ, Trevarrow B, Bass MB, Kimmel CB. Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development. 1990;110(2):491–504. doi: 10.1242/dev.110.2.491. [DOI] [PubMed] [Google Scholar]
- 30.Schapira G, Dreyfus JC, Joly M. Changes in the flow birefringence of myosin as a result of muscular atrophy. Nature. 1952;170(4325):494–495. doi: 10.1038/170494b0. [DOI] [PubMed] [Google Scholar]
- 31.Haskell RC, Carlson FD, Blank PS. Form birefringence of muscle. Biophys J. 1989;56(2):401–413. doi: 10.1016/S0006-3495(89)82686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- 33.Gahtan E, Baier H. Of lasers, mutants, and see-through brains: functional neuroanatomy in zebrafish. J Neurobiol. 2004;59(1):147–161. doi: 10.1002/neu.20000. [DOI] [PubMed] [Google Scholar]
- 34.Zhou FQ, Snider WD. Intracellular control of developmental and regenerative axon growth. Philos Trans R Soc Lond B. 2006;361(1473):1575–1592. doi: 10.1098/rstb.2006.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozengurt E. Mitogenic signaling pathways induced by G protein-coupled receptors. J Cell Physiol. 2007;213(3):589–602. doi: 10.1002/jcp.21246. [DOI] [PubMed] [Google Scholar]
- 36.Leone AM, Errico M, Lin SL, Cowen DS. Activation of extracellular signal-regulated kinase (ERK) and Akt by human serotonin 5-HT(1B) receptors in transfected BE(2)-C neuroblastoma cells is inhibited by RGS4. J Neurochem. 2000;75(3):934–938. doi: 10.1046/j.1471-4159.2000.0750934.x. [DOI] [PubMed] [Google Scholar]
- 37.He JC, Neves SR, Jordan JD, Iyengar R. Role of the Go/i signaling network in the regulation of neurite outgrowth. Can J Physiol Pharmacol. 2006;84(7):687–694. doi: 10.1139/y06-025. [DOI] [PubMed] [Google Scholar]
- 38.Nixon AB, Grenningloh G, Casey PJ. The interaction of RGSZ1 with SCG10 attenuates the ability of SCG10 to promote microtubule disassembly. J Biol Chem. 2002;277(20):18127–18133. doi: 10.1074/jbc.M201065200. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z, Chatterjee TK, Fisher RA. RGS6 interacts with SCG10 and promotes neuronal differentiation. Role of the G gamma subunit-like (GGL) domain of RGS6. J Biol Chem. 2002;277(40):37832–37839. doi: 10.1074/jbc.M205908200. [DOI] [PubMed] [Google Scholar]
- 40.Willard FS, Willard MD, Kimple AJ, Soundararajan M, Oestreich EA, Li X, Sowa NA, Kimple RJ, Doyle DA, Der CJ, Zylka MJ, Snider WD, Siderovski DP. Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector. PLoS ONE. 2009;4(3):e4884. doi: 10.1371/journal.pone.0004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heo K, Ha SH, Chae YC, Lee S, Oh YS, Kim YH, Kim SH, Kim JH, Mizoguchi A, Itoh TJ, Kwon HM, Ryu SH, Suh PG. RGS2 promotes formation of neurites by stimulating microtubule polymerization. Cell Signal. 2006;18(12):2182–2192. doi: 10.1016/j.cellsig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Read DE, Gorman AM. Involvement of Akt in neurite outgrowth. Cell Mol Life Sci. 2009;66(18):2975–2984. doi: 10.1007/s00018-009-0057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bommakanti RK, Vinayak S, Simonds WF. Dual regulation of Akt/protein kinase B by heterotrimeric G protein subunits. J Biol Chem. 2000;275(49):38870–38876. doi: 10.1074/jbc.M007403200. [DOI] [PubMed] [Google Scholar]
- 44.Zheng J, Shen WH, Lu TJ, Zhou Y, Chen Q, Wang Z, Xiang T, Zhu YC, Zhang C, Duan S, Xiong ZQ. Clathrin-dependent endocytosis is required for TrkB-dependent Akt-mediated neuronal protection and dendritic growth. J Biol Chem. 2008;283(19):13280–13288. doi: 10.1074/jbc.M709930200. [DOI] [PubMed] [Google Scholar]
- 45.Markus A, Zhong J, Snider WD. Raf and akt mediate distinct aspects of sensory axon growth. Neuron. 2002;35(1):65–76. doi: 10.1016/S0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 46.Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25(49):11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Yang F, Fu Y, Huang X, Wang W, Jiang X, Gritsenko MA, Zhao R, Monore ME, Pertz OC, Purvine SO, Orton DJ, Jacobs JM, Camp DG, 2nd, Smith RD, Klemke RL. Spatial phosphoprotein profiling reveals a compartmentalized ERK switch governing neurite growth and retraction. J Biol Chem. 2011;286(20):18190–18201. doi: 10.1074/jbc.M111.236133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soundararajan P, Fawcett JP, Rafuse VF. Guidance of postural motoneurons requires MAPK/ERK signaling downstream of fibroblast growth factor receptor 1. J Neurosci. 2010;30(19):6595–6606. doi: 10.1523/JNEUROSCI.4932-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lang UE, Puls I, Muller DJ, Strutz-Seebohm N, Gallinat J. Molecular mechanisms of schizophrenia. Cell Physiol Biochem. 2007;20(6):687–702. doi: 10.1159/000110430. [DOI] [PubMed] [Google Scholar]
- 50.Jaaro-Peled H, Hayashi-Takagi A, Seshadri S, Kamiya A, Brandon NJ, Sawa A. Neurodevelopmental mechanisms of schizophrenia: understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci. 2009;32(9):485–495. doi: 10.1016/j.tins.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bellon A. New genes associated with schizophrenia in neurite formation: a review of cell culture experiments. Mol Psychiatry. 2007;12(7):620–629. doi: 10.1038/sj.mp.4001985. [DOI] [PubMed] [Google Scholar]
- 52.Bellon A, Krebs MO, Jay TM. Factoring neurotrophins into a neurite-based pathophysiological model of schizophrenia. Prog Neurobiol. 2011;94(1):77–90. doi: 10.1016/j.pneurobio.2011.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.