Abstract
Parkinson’s disease (PD) is characterized by progressive midbrain dopaminergic neuron degeneration and the formation of intracellular protein aggregates, referred to as Lewy bodies. F-box only protein 7 (FBXO7) gene mutations are closely associated with progression of the autosomal recessive form of familial PD. FBXO7 encodes a component of Skp1, cullin, F-box ubiquitin ligase complexes; however, its cellular targets, including substrates and regulators, are not yet clarified. To identify potential substrates of FBXO7, we performed a yeast two-hybrid screen of a human fetal brain library and identified neurotrophin receptor-interacting MAGE protein (NRAGE) as a novel FBXO7-binding partner. We found that FBXO7 interacts with NRAGE and mediates Lys-63-linked poly-ubiquitination of NRAGE in mammalian cells. FBXO7 overexpression accelerates formation of NRAGE–TAK1–TAB1 complexes, whereas FBXO7 knockdown correspondingly decreases complex formation. In addition, BMP4 stimulation enhances NRAGE ubiquitination through FBXO7 and facilitates endogenous NRAGE–TAK1–TAB1 complex formation. Furthermore, FBXO7 positively regulates formation of the BMP receptor–NRAGE–TAK1–TAB1 complex, and up-regulates NF-κB activity. Taken together, our results suggest that FBXO7 affects BMP4-mediated signaling through proteasome-independent ubiquitination of NRAGE and augments formation of downstream signaling components.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-014-1665-5) contains supplementary material, which is available to authorized users.
Keywords: BMP, FBXO7, NF-κB, NRAGE, Parkinson disease, TAB1, TAK1, Ubiquitination
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease after Alzheimer’s disease. PD is neuropathologically defined by dopaminergic cell loss in the substantia nigra pars compacta and formation of α-synuclein-containing protein aggregates, termed Lewy bodies, in surviving neurons [1, 2]. Although defective ubiquitin–proteasome system, mitochondrial dysfunction, oxidative stress, and autophagy impairment have been suggested to play roles in PD pathogenesis, mutations in at least seven genes have also been individually linked to familial forms of PD. These genes include autosomal dominant mutations in α-synuclein, ubiquitin hydroxylase-L1, and leucine-rich repeat kinase 2, as well as autosomal recessive mutations in parkin, PTEN-induced putative kinase 1, DJ-1, and ATP13A2 [3]. Recently, mutations in the F-box only protein 7 (FBXO7) gene were reported to be the cause of an autosomal recessive neurodegenerative disease presenting with severe juvenile levodopa-responsive Parkinsonism and additional pyramidal signs [4–6].
Human FBXO7 was originally identified through a screen for Skp1-interacting proteins [7, 8]. FBXO7 is a member of the F-box protein (FBP) family, which contains 40–50 amino acid F-box domains that bind to Skp1 [9]. FBP is also a substrate-recognition component of Skp1–cullin1–F–box protein (SCF) ubiquitin E3 ligase, the largest group of multi-subunit E3 ligases within the RING finger domain family [10]. In SCF, FBP is fundamental for flexible substrate recognition and acts as an interchangeable docking site for the ligase [11]. Currently, the ability of FBXO7 to function as a component of SCFFBXO7 is known for three substrates: HURP, cIAP1, and TRAF2 [12–14]. Lys-48-linked ubiquitination of HURP by SCFFBXO7 is preceded by HURP phosphorylation by cyclin B/Cdk1, followed by HURP undergoing proteasome-dependent degradation. It has also been reported that SCFFBXO7 presumably negatively regulates NF-κB activity by promoting Lys-63-linked ubiquitination of cIAP1 and TRAF2 [14]. According to a recent quantitative analysis of the cullin1 proteome, SCFFBXO7 is the fifth most abundant SCF ligase in cultured cells [15]. However, it is little known about its ubiquitination substrates, function, and role in diseases.
MAGE is a gene family that encodes melanoma-associated antigens [16]. MAGE proteins have been classified into two subfamilies: MAGE-I and MAGE-II [16]. MAGE-I genes show cancer- and testis-specific expression pattern as well as antigenic properties that render them ideal candidates for cancer immunotherapies. MAGE-II family members are ubiquitously expressed in somatic tissue. Neurotrophin receptor-interacting MAGE protein (NRAGE; also known as MAGED1, Dlxin-1) was originally characterized in a screen examining proteins that interacted with the intracellular domain of the p75 neurotrophin receptor [17]. NRAGE consists of more than 25 members and shares a conserved MAGE homology domain (MHD). NRAGE is a MAGE-II gene and plays an important role in neuronal progenitor cell (NPC) apoptosis during development [18, 19]. NRAGE is highly expressed during early corticogenesis, specifically in neural progenitors of the ventricular zone and in differentiating neuroblasts of the cortex, concomitant with the spatial and temporal occurrence of bone morphogenetic protein (BMP)-mediated apoptosis [20]. This BMP-mediated signaling is an alternative signaling cascade that utilizes NRAGE to regulate mitotic arrest and apoptosis of NPCs [20, 21].
NPC differentiation or proliferation is regulated in part by BMPs, which are members of the transforming growth factor β (TGF-β) superfamily [22]. BMPs regulate key events during neurogenesis by governing the paradoxical switch between morphogenesis and apoptosis within the developing cortex. BMP-mediated signaling goes through two representative pathways: the “canonical” and “alternative” pathways [22]. The “canonical” pathway involves the Smad1, Smad5, and Smad4 proteins that activate downstream signaling. In the “alternative” pathway, BMP receptor (BMPR) activation and NRAGE facilitate TAK1–TAB1–XIAP complex and drives p38 activation [20]. In addition, NF-κB activation is up-regulated by the BMP-initiated alternative pathway, and the TAK1–TAB1–XIAP complex is responsible for phosphorylation of IKK-α/β. As such, NRAGE plays a critical role in the alternative BMP pathway through binding BMPR and facilitating TAK1–TAB1–XIAP complex formation, and subsequently transmitting downstream pro-apoptotic signals in NPCs or during NR-κB activation in HEK293 cells [23].
In this study, we evaluated new substrates of FBXO7 using a yeast two-hybrid assay and identified a novel binding partner, NRAGE. Based on the known role of NRAGE to regulate the NPC differentiation and FBXO7 in PD pathogenesis, we examined whether FBXO7 regulates NRAGE-mediated BMP signaling.
Experimental procedures
Materials
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), LipofectAMINE PLUS reagent, Lipo-2000 reagent, secondary goat anti-IgG horseradish peroxidase conjugated anti-rabbit, and anti-mouse IgGs were purchased from Life Technologies (Grand Island, NY, USA). Protein A-Sepharose was obtained from GE Healthcare (Piscataway, NJ, USA). Enhanced chemiluminescence (ECL) reagent was purchased from Perkin-Elmer Life and Analytical Sciences (Waltham, MA, USA). Anti-Flag and all other chemicals were commercially available, analytical grade products purchased from Sigma-Aldrich (St. Louis, MO, USA). Anti-FBXO7, anti-NRAGE, anti-TAK1, anti-HA, anti-Myc, anti-Histon-H1, anti-tubulin, anti-HSP90, and rabbit immunoglobulins (IgGs) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-ubiquitin, anti-Lys-48-specific, and anti-Lys-63-specific ubiquitin antisera were purchased from Merck Millipore (Philadelphia, PA, USA).
DNA constructs and RNA interference
Mammalian expression vectors for HA- or Flag-tagged human wild-type FBXO7 (isoform-1) as well as deletion mutants fused with Flag, such as FBXO7 lacking the C-terminal F-box (denoted as FBXO7-ΔF) or PRR domain (FBXO7-ΔC), were kindly provided by Kuiken (The Netherlands Cancer Institute, Amsterdam, Netherlands). The plasmid encoding HA-tagged human FBXO7 isoform-2 that lacks the N-terminal Ubl domain (FBXO7-ΔU) was kindly provided by You (Kaohsiung Medical University, Kaohsiung, Taiwan). To increase transfection efficiency, wild-type FBXO7 isoform-1 and its F-box deletion mutant were PCR amplified using the forward primer, 5′-ATA ACG CGT ATG AGG CTG GGG GTG-3′, and the reverse primer, 5′-GCC ATA GCG GCC GCT CAC ATG AAT GAC AG-3′. PCR products were sub-cloned into the modified pRK5-Flag vector (Stratagene) containing MluI and NotI sites. FBXO7 isoform-2 was PCR amplified using the forward primer, 5′-ATA ACG CGT ATG GCC CGG CCT CCC-3′, and the same reverse primer used for the FBXO7 isoform-1. PCR products were sub-cloned into pRK5-Flag and pRK5-HA vectors. The FBXO7 mutant lacking the N-terminal Ubl and FP domains (nucleotide 1119–1569 of FBXO7 isoform-1 sequence; FBXO7-ΔN) was PCR amplified using the forward primer, 5′-GCC ACG CGT GTC CTC CCA TTG GAA-3′, and the same reverse primer used for FBXO7 isoform-1. PCR products were sub-cloned into the pRK5-Flag vector. The plasmids encoding NRAGE in pEGFP-N3 vector was a gift from Verdi (Maine Medical Center Research Institute, Scarborough, ME, USA). The plasmid encoding Myc-tagged wild-type NRAGE was generated by PCR amplification of NRAGE using the forward primer, 5′-GCC ATA ACG CGT ATG GCT CAG AAA ATG GAC-3′, and the reverse primer, 5′-ATA GCG GCC GCT CAC TCA ACC CAG AA-3′. PCR products were then sub-cloned into the pRK5-Myc vector containing MluI and NotI sites. The plasmids encoding Flag-tagged TAK1 and HA-tagged TAB1 were kindly provided by Takausu (Keio University, Tokyo, Japan). FBXO7- and NRAGE-specific siRNAs were purchased from Bioneer (Seoul, Korea). The FBXO7 siRNA duplex sequences were 5′-ACA UCU GGU AGG UUC AGU G(dTdT)-3′. The NRAGE siRNA duplex sequences were 5′-AUC UCA GAC UUG UUA GCA G(dTdT)-3′. A control siRNA (NT-siRNA) with no known mammalian homology (siCONTROL non-targeting siRNA #1, Bioneer) was used as a negative control. siRNA duplexes were transfected into cells using Lipofectamine 2000, according to the manufacturer’s protocol.
Yeast two-hybrid assay
Screening was performed using a 13-week human fetal brain Matchmaker cDNA library sub-cloned into the activation domain (AD) in pACT2 vector (Promega) and a yeast L40 strain carrying the pHybLex/Zeo-plasmid encoding full-length wild-type FBXO7 isoform-2 as bait. All yeast two-hybrid screening protocols were performed as described previously [24].
Cell culture and DNA transfection
P19 embryonic carcinoma (P19) cells were provided by Lee (College of Medicine, The Catholic University of Korea, Seoul, Korea). Human embryonic kidney 293 (HEK293) and P19 cells were maintained in DMEM containing 10 % FBS and 100 U/ml penicillin–streptomycin (Life Technologies). Cells were transfected with various expression vectors using LipofectAMINE PLUS reagent, Lipo-2000 reagent, and TransIT-2020 reagent (Mirus Bio LLC.), according to the manufacturer’s protocols. In all experiments, cells were serum-starved for approximately 4 h prior to the addition of 10 ng/ml BMP4 (R&D systems, Minneapolis, MN, USA).
Immunoprecipitation and immunoblot assay
Cells were rinsed twice with ice-cold phosphate-buffered saline (PBS), scraped with 1 % Nonidet P-40 lysis buffer (50 mM Tris, pH 7.5, containing 1.0 % Nonidet P-40, 150 mM NaCl, 10 % glycerol, 1 mM sodium orthovanadate, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mM EGTA, 1 mM sodium fluoride and 0.2 mM phenylmethylsulfonyl fluoride), and briefly sonicated. Lysates were collected by centrifugation at 13,000×g for 20 min at 4 °C. All the immunoprecipitation and immunoblot assay protocols were performed as described previously [24].
Immunocytochemistry
After transfection, cell were washed twice with PBS (pH 7.4) and immediately fixed for 15 min at room temperature using 3.7 % formaldehyde diluted in PBS. After fixation, cells were permeabilized for 20 min using 0.2 % Triton X-100. Cells were blocked for 30 min at room temperature in 1 % bovine serum albumin, and incubated overnight at 4 °C with the corresponding antibodies. Depending on the primary antibodies, cells were incubated for 2 h with Rhodamine-conjugated, TRITC-conjugated, or FITC-conjugated secondary antibodies, respectively. Samples were stained with 4,6-diamidino-2-phenylindole (DAPI), and mounted using the Slow Fade Light Antifade kit with DAPI (molecular probes). Immunostained cells were visualized using a LSM-700 confocal microscope (Carl Zeiss). Data were obtained at wavelength of 405 nm for DAPI, 488 nm for GFP, and 546 nm for rhodamine.
Preparation of cytosolic and nuclear fractions
Cells were washed with ice-cold PBS and re-suspended in hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl) supplemented with protease inhibitors, including dithiothreitol, aprotinin, and leupeptin. Cells were incubated for 30 min on ice, lysed with a disposable syringe, followed by centrifugation at 1,000×g for 15 min at 4 °C. The supernatants were used as the cytosolic fractions. The nuclear pellet fractions were washed with hypotonic buffer, and lysed using 1.0 % NP-40 lysis buffer. Supernatants from each fraction were collected after centrifugation 15,000×g for 15 min at 4 °C.
Luciferase reporter assays
HEK293 cells were seeded at a density of 3 × 105 cells per well and cultured overnight in 6-well plates. NF-κB-dependent firefly luciferase reporter and effector plasmids were co-transfected along with the Renilla luciferase plasmid into cells. Forty-eight hours after transfection, cells were harvested in passive lysis buffer (Promega), and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega). Relative luciferase activity was calculated by dividing firefly luciferase activity by Renilla luciferase activity, and all experiments were performed in triplicate.
Apoptosis and cell survival assay
Live cells were collected 48 h after BMP4 addition or without supplementation. The numbers of viable and nonviable cells were determined by trypan blue exclusion. An equal volume of 0.4 % trypan blue solution (Sigma-Aldrich) was added to the media and incubated for 1 min. The numbers of total and stained cells were counted within 5 min using a hemocytometer. The relative cell death ratio was calculated by dividing the number of nonviable cells by total cells. The data shown are three independent experiments performed in triplicate.
Statistical analysis
Group means were compared using unpaired two-tailed t tests. P values less than 0.05 were considered significant.
Results
FBXO7 interacts with NRAGE
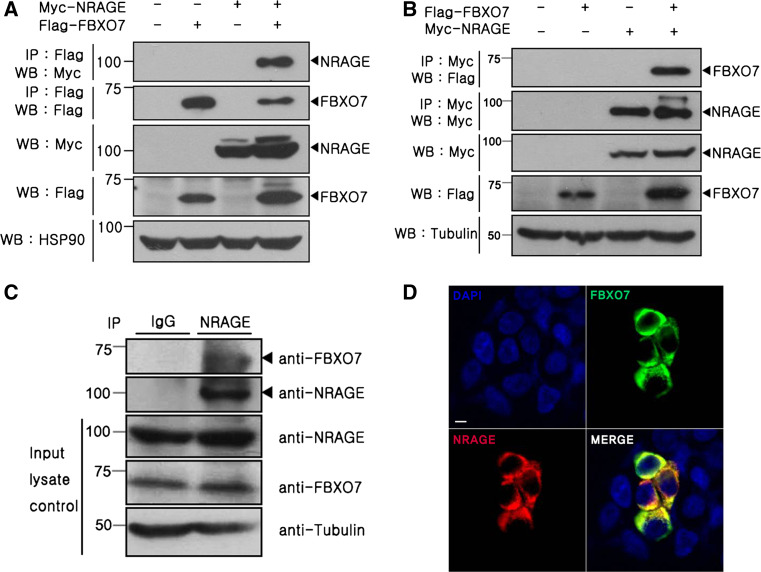
It has been reported that two protein isoforms are expressed from the FBXO7 gene in normal human cells. The FBXO7 isoform-1 is longer than isoform-2 because of the presence of an N-terminal Ubl domain [25]. To identify potential FBXO7-interacting proteins, a yeast two-hybrid assay was performed using a human fetal brain cDNA library and FBXO7 isoform-2 as bait. After screening 5 × 106 independent library clones, a previously unreported binding target, NRAGE, and a number of previously identified FBXO7-binding partners (i.e., proteasome inhibitor subunit 1 (denoted as PI31 or PSMF1); [26] and Skp1; [9]) were obtained (data not shown). To gain further insight and to determine how FBXO7 and NRAGE are linked, we first determined whether FBXO7 binds NRAGE in mammalian cells. After HEK293 cells were transfected with Flag-tagged FBXO7 and Myc-tagged NRAGE, immunoprecipitation assays were performed. As shown in Fig. 1a, transiently transfected FBXO7 selectively binds NRAGE in HEK293 cells. When the co-immunoprecipitation assays were performed in a reverse order, we observed the same result (Fig. 1b). These data suggest that ectopically expressed NRAGE binds FBXO7 in HEK293 cells.
Fig. 1.
FBXO7 interacts with NRAGE. a, b Where indicated, HEK293 cells were transfected for 24 h with plasmids encoding Flag-tagged FBXO7 and/or Myc-tagged NRAGE. Immunoprecipitation (IP) of cell lysates was performed with the Flag antibody, followed by Western blotting (WB) with the Myc antibody. The co-IP experiment was also performed in the reverse order, as indicated. Proper expression of transiently transfected proteins in cell lysates was examined using specific antibodies. To determine equal protein loading, cell lysates were analyzed by immunoblotting with anti-HSP90 or anti-tubulin IgG. A slightly bigger band detected for NRAGE is speculated to be possibly post-translationally modified form of NRAGE. c Where indicated, immunoprecipitation of HEK293 cell lysates was performed with the anti-NRAGE antibody, followed by immunoblotting with anti-FBXO7 antibodies. As a control, cell lysates were immunoprecipitated with preimmune IgG, as indicated. FBXO7 and NRAGE expression in cell extracts was determined by immunoblotting with each antibody. d HEK293 cells were fixed, permeabilized, and labeled using anti-Flag and anti-Myc antibodies. Cells were stained with TRITC-conjugated and FITC-conjugated secondary antibodies and DAPI. Immunostained preparations were examined using a confocal microscope (LSM-700 confocal microscope by Carl Zeiss). Scale bar 5 μm
To exclude the possibility that this binding occurred as an artifact of DNA transfection, we tested whether endogenous FBXO7 interacts with endogenous NRAGE in HEK293 cells. Immunoblot analyses of anti-NRAGE IgG co-immunoprecipitates using anti-FBXO7 antiserum demonstrated a specific interaction between the endogenous proteins in HEK293 cells (Fig. 1c). As a control, we observed no obvious bands in the immunocomplex samples prepared with preimmune IgG. Immunocytochemical analyses of HEK293 cells confirmed co-localization of these two proteins, which were mainly present in the cytoplasmic region (Fig. 1d). Overall, these data suggest that FBXO7 and NRAGE specifically interact in mammalian cells.
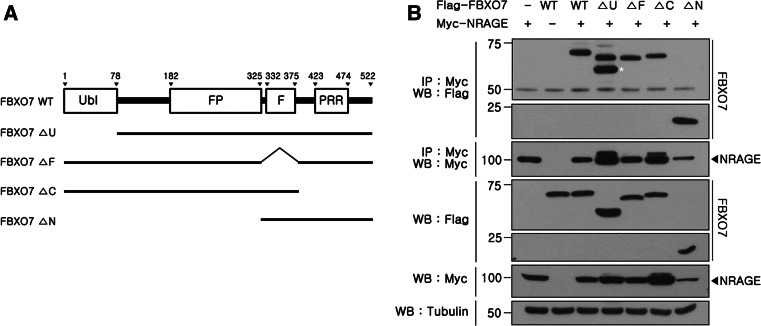
To determine which domain(s) within FBXO7 protein is responsible for the interaction with NRAGE, we generated several FBXO7 deletion constructs fused to Flag (Fig. 2a), and performed co-immunoprecipitation/immunoblot assays. As shown in Fig. 2b, immunoblot analyses of anti-NRAGE immunocomplexes with anti-Flag IgG revealed that NRAGE bound well to full-length FBXO7 as well as all FBXO7 peptides, including FBXO7-ΔU, FBXO7-ΔF, FBXO7-ΔC, and FBXO7-ΔN (Fig. 2b). This result suggests that there does not appear to be a single critical region of FBXO7 required for the interaction with NRAGE. Additionally, more than two domains could be independently responsible for NRAGE binding.
Fig. 2.
Mapping the binding domain of FBXO7 to NRAGE. a Diagram of Flag-tagged wild-type FBXO7 and its deletion mutants. FBXO7 consists of an N-terminal ubiquitin-like domain (UBL), PI31-interacting domain (FP), Skp1-binding F-box domain (FBD), and C-terminal proline-rich region (PRR). b HEK293 cells were transfected with Myc-tagged NRAGE alone or in combination with Flag-tagged wild-type FBXO7 or its deletion mutants, as indicated. The upper band of NRAGE in IP: Myc/WB: Flag experiment would be a non-specific band. While the calculated molecular weight of FBXO7-ΔC mutant is expected to be between that of FBXO7-ΔN and FBXO7-ΔU mutant, but its actual size was rather different, possibly due to the specific post-translational modification of the fragment. Total lysates and anti-Myc immunocomplexes were analyzed by Western blot using the anti-Flag antibody. To determine equal loading, cell lysates were analyzed by immunoblotting with the anti-tubulin antibody
Additionally, to evaluate the ability of various FBXO7 deletion mutants to assemble the functional SCF complex, we performed co-immunoprecipitation experiments of Flag-tagged wild-type FBXO7 or its deletion mutants, such as FBXO7-ΔU, FBXO7-ΔF, FBXO7-ΔC, or FBXO7-ΔN with endogenous Skp1 and cullin1. Western blot analysis of anti-Flag immunocompelxes with anti-Skp1 and anti-cullin1 antibodies revealed that FBXO7-ΔU, FBXO7-ΔC, and FBXO7-ΔN as well as full-length FBXO7 bind to those two proteins and formed SCF complex well, except FBXO7-ΔF (Supplementary figure S1). It is well known that the N-terminal F-box domain of F-Box protein links it to other components of SCF complex through Skp1 binding, which in turn associates with cullin1. Therefore, these results suggest that those three FBXO7 deletion mutants except FBXO7-ΔF mutant can properly form SCFFBXO7 in vivo.
FBXO7 mediates NRAGE poly-ubiquitination
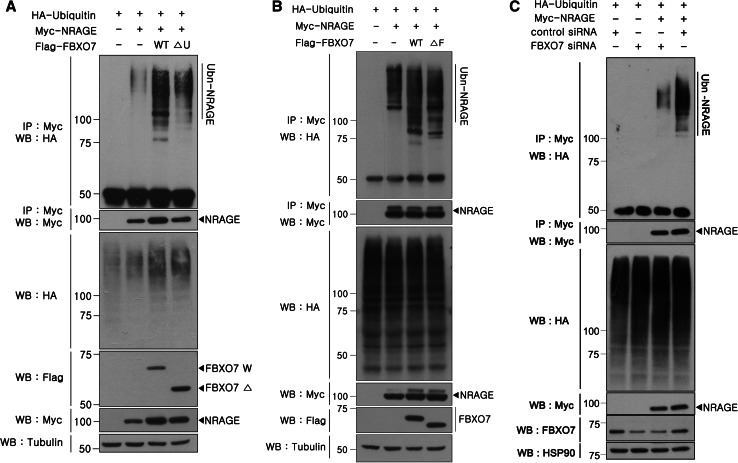
Because FBXO7 is a substrate-recognition component of the SCF ubiquitin E3 ligase complex, we addressed whether SCFFBXO7 promotes the conjugation of single- or poly-ubiquitin chains to NRAGE. After HEK293 cells were transiently transfected with Myc-tagged NRAGE, HA-tagged ubiquitin, or Flag-tagged wild-type FBXO7 alone or in combination, NRAGE was immunoprecipitated and samples were analyzed by Western blotting with the HA antibody. As shown in Fig. 3a, full-length FBXO7 does not promote mono-ubiquitination of NRAGE, but dramatically increased the extent of NRAGE poly-ubiquitination. To verify that FBXO7 mediated NRAGE poly-ubiquitination, the effect of two other FBXO7 vectors, FBXO7 lacking the N-terminal Ubl-domain (FBXO7-ΔU) or the N-terminal F-box domain (ΔF) was tested. Compared to wild-type FBXO7, NRAGE poly-ubiquitination was markedly diminished in the presence of the FBXO7-ΔU mutant, and less diminished in the presence of the FBXO7-ΔF mutant (Fig. 3a, b). Similarly, FBXO7 knockdown in HEK293 cells using FBXO7-specific siRNAs significantly reduced the amount of NRAGE poly-ubiquitination. However, this effect was not observed in cells treated with non-specific control siRNAs (Fig. 3c). These results suggest that the SCFFBXO7 complex acts as an ubiquitin E3 ligase for NRAGE and promotes its poly-ubiquitination.
Fig. 3.
FBXO7 mediates poly-ubiquitination of NRAGE. a, b HEK293 cells were transfected for 24 h with HA-tagged ubiquitin plus Myc-tagged NRAGE alone or in combination with Flag-tagged FBXO7 isoform-1 (WT), isoform-2 (FBXO7-ΔU), or the mutant deleting F-box (FBXO7-ΔF). Cell lysates were immunoprecipitated with the anti-Myc antibody, followed by immunoblotting with the anti-HA antibody. Protein expression in cell lysates was verified by immunoblotting with specific antibodies, as indicated. To determine equal loading, cell lysates were analyzed by immunoblotting with the anti-tubulin antibody. c HEK293 cells were transfected for 48 h with FBXO7-specific siRNAs (100 mM) or control siRNAs, alone or in the presence of HA-ubiquitin and Myc-NRAGE, as indicated. Co-IP experiments were performed as described. Hsp90 was used as a loading control. The intense band at 50 kDa denotes the heavy chains of IgG
FBXO7 mediates Lys-63-linked poly-ubiquitination of NRAGE without affecting NRAGE level
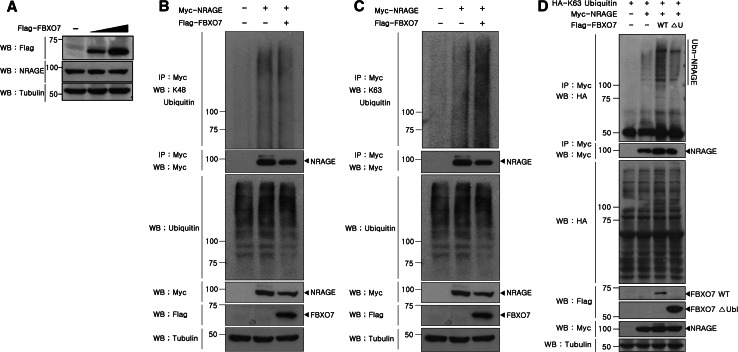
There are two different modes of reaction for covalent poly-ubiquitin conjugation to target proteins, named for which of the seven lysine amino acids are used to link the chain together, via Lys-48-linked or Lys-63-linked chains [27]. The first Lys-48-linked chains predominantly signal target proteins for proteasomal degradation, which consequently destroys and recycles the substrate proteins. The second Lys-63-linked chains do not result in protein degradation via the proteasome, but instead regulate the biochemical and functional properties of the targets and are closely linked to a number of physiological processes, such as endocytic trafficking, inflammation, translation, and DNA repair [28]. Therefore, we examined whether FBXO7-mediated poly-ubiquitination of NRAGE is via Lys-48- or Lys-63-linked chains. Because Lys-48-linked ubiquitination results in proteasomal degradation of substrates, we initially assessed whether FBXO7 causes NRAGE degradation. As shown in Fig. 1a, when compared with cells transfected with NRAGE alone, ectopically expressed FBXO7 did not remarkably affect the level of overexpressed NRAGE. We next examined the effect of FBXO7 overexpression on endogenous NRAGE levels. When HEK293 cells were transfected with increasing amounts of Flag-tagged FBXO7, endogenously expressed NRAGE levels were unaffected (Fig. 4a), suggesting that the FBXO7-mediated increase in NRAGE poly-ubiquitination may occur through Lys-63-linked ubiquitin chains.
Fig. 4.
FBXO7 mediates Lys-63-linked poly-ubiquitination of NRAGE without affecting NRAGE levels. a HEK293 cells were transfected with increasing amounts of Flag-FBXO7. Endogenous NRAGE expression was confirmed by immunoblotting with the anti-NRAGE antibody. To determine equal loading, cell lysates were analyzed by immunoblotting with the anti-tubulin antibody. b, c HEK293 cells were transfected for 24 h with Myc-NRAGE alone or in combination with Flag-FBXO7. Cell lysates were immunoprecipitated with the anti-Myc antibody and immunoblotted with anti-K63 or anti-K48-ubiquitin antibodies. Ubiquitin, FBXO7, or NRAGE expression in cell extracts was determined by immunoblotting with anti-K63, anti-K48-ubiquitin, anti-Myc, or anti-Flag antibodies. d HEK293 cells were transfected for 24 h with plasmids encoding HA-tagged K63-ubiquitin plus Myc-tagged NRAGE alone or together with Flag-tagged FBXO7. Co-IP experiments were performed as described
To confirm this speculation, HEK293 cells were co-transfected with plasmids encoding Myc-tagged NRAGE alone or together with Flag-FBXO7. Cell lysates were immunoprecipitated with the anti-Myc antibody, followed by immunoblot analyses with either anti-Lys-48- or anti-Lys-63-linked ubiquitin-specific antibodies. As expected, NRAGE ubiquitination significantly increased with the anti-Lys-63-linked ubiquitin-specific antibody, but not with anti-Lys-48-ubiquitin antiserum (Fig. 4b, c). We further investigated the pattern of Lys-63-linked NRAGE ubiquitination using a plasmid encoding K63-ubiquitin, in which the other six lysine residues are mutated into conjugation-defective arginines and thus linkage can proceed only through Lys-63. After cells were transfected for 24 h with plasmids encoding HA-tagged K63-ubiquitin and Myc-tagged NRAGE alone or in combination with Flag-tagged FBXO7, immunoprecipitation assay was performed using the anti-Myc antibody. Immunoblot analyses of the samples with the anti-HA antibody showed that FBXO7 enhances NRAGE poly-ubiquitination in Lys-63-linked ubiquitin chains (Fig. 4d). Taken together, these results indicate that NRAGE is a novel substrate of the SCFFBXO7 ubiquitin E3 ligase, and poly-ubiquitination by SCFFBXO7 is processed through Lys-63-linked ubiquitin chains.
FBXO7 promotes formation of the TAK1–TAB1–NRAGE complex
We further investigated how FBXO7-mediated poly-ubiquitination affects the functional role of NRAGE. NRAGE expression is required to mediate NPC apoptotic cell death and NF-κB activation in HEK293 cells through BMPR-dependent pathways [20, 23]. Activation of BMPRs leads to the formation of the TAK1–TAB1–XIAP signaling complex, and NRAGE is a crucial component regulating these signaling pathways. NRAGE facilitates TAK1–TAB1–XIAP–NRAGE complex formation, which drives apoptosis in NPCs or IKK-α/β phosphorylation in HEK293 cells [23, 29]. Given the importance of NRAGE in alternative BMP signaling, we examined the functional role of NRAGE ubiquitination mediated by SCFFBXO7 in BMP-induced downstream signaling, particularly on TAK1–TAB1–NRAGE complex formation.
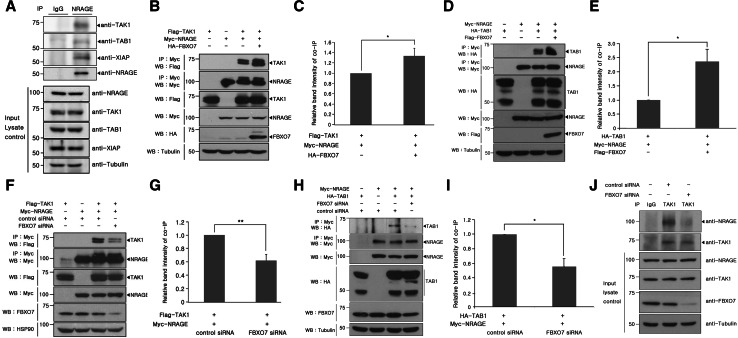
Firstly, we examined whether the formation of endogenous NRAGE–TAK1–TAB1–XIAP complex actually occurs. We performed co-immunoprecipitation experiments of using HEK293 cell lysates with anti-NRAGE antibody. The subsequent Western blot analysis of anti-NRAGE immunoprecipitates with the antibodies recognizing endogenous TAK1, TAB1, and XIAP protein, respectively, demonstrated that NRAGE are co-immunoprecipitated with TAK1, TAB1, and XIAP, altogether (Fig. 5a). To examine the role of NRAGE ubiquitination on this complex formation, HEK293 cells were then transiently transfected with Flag-tagged TAK1, HA-tagged TAB1, and Myc-tagged NRAGE in the absence or presence of HA- or Flag-tagged FBXO7. Immunoprecipitation of Myc-tagged NRAGE and subsequent immunoblotting with anti-Flag and anti-HA antibodies revealed that ectopic expression of TAK1 plus NRAGE or TAB1 plus NRAGE resulted in the association of TAK1-NRAGE and TAB1-NRAGE in the absence of FBXO7 (Figs. 5b–e). The addition of FBXO7 promoted the formation of these two complexes, respectively (Figs. 5b–e). Similarly, when HEK293 cells were transfected with either TAK1 plus NRAGE or TAB1 plus NRAGE and endogenous FBXO7 was selectively knocked down using FBXO7-specific siRNAs, formation of these two complexes significantly decreased (Fig. 5f–i). The non-specific control siRNAs had no effect on complex formation (Fig. 5f–i).
Fig. 5.
FBXO7 promotes formation of the TAK1–TAB1–NRAGE complex. a HEK293 cell lysates were immunoprecipitated with either anti-NRAGE antibody or rabbit preimmune IgG, followed by immunoblotting with the indicated antibodies. Tubulin was used as a loading control. b, d Where specified, HEK293 cells were transfected for 24 h with plasmids encoding Myc-tagged NRAGE, Flag-tagged TAK1 (b), or HA-tagged TAB1 (d) alone or in combination with either HA-FBXO7 (b) or Flag-FBXO7 (d). Cells were immunoprecipitated with the anti-Myc antibody, followed by immunoblotting with anti-Flag or HA antibodies. Proper expression of transfected proteins in cell lysates was examined using immunoblotting, as indicated. To determine equal loading, cell lysates were analyzed by immunoblotting with the anti-tubulin antibody. c, e Relative band intensity levels in the blots of (b) and (d) were quantified by three independent experiments. f, h Where specified, HEK293 cells were transfected for 48 h with plasmids for Myc-tagged NRAGE, Flag-tagged TAK1 (f), or HA-tagged TAB1 (h) alone or in combination with either FBXO7-siRNAs or control siRNAs. Co-IP experiments were performed as described in (b) and (d), respectively. The multiple bands in overexpressed TAB1 lanes may result from differential post-translational modifications and/or irreversible protein cleavage during the sample preparation. g, i Relative band intensity levels of the blots in e and g were quantified by three independent experiments. j HEK293 cells were transiently transfected for 48 h with FBXO7-siRNAs or control siRNAs. Immunoprecipitation of each cell lysates was performed with the anti-TAK1 antibody, followed by immunoblotting with the anti-NRAGE antibody. As a control, cell lysates were immunoprecipitated with rabbit preimmune IgG, as indicated. Endogenously expressed proteins in cell extracts were examined by immunoblotting with the appropriate antibodies. To examine equal loading, cell lysates were analyzed by immunoblotting with the anti-tubulin antibody
To exclude the effect of DNA transfection, we examined whether the association between endogenous NRAGE and TAK1 occurs in HEK293 cells, and if so, if complex formation is affected by FBXO7 knockdown. As shown in Fig. 4i, HEK293 cells were transfected for 24 h with FBXO7-specific siRNAs or control siRNAs and cell lysates were immunoprecipitated with the anti-TAK1 antibody. Immunoblot analyses with anti-NRAGE antibodies showed that endogenous TAK1 bound to NRAGE (Fig. 5j). The TAK1-NRAGE complex was diminished when FBXO7 was knocked down (Fig. 5j). Overall, these results suggest that FBXO7 positively regulates mutual interactions between TAK1–NRAGE and TAB1–NRAGE. These actions are expected to promote NRAGE-mediated downstream BMP signal transduction.
FBXO7 accelerates formation of the TAK1–NRAGE complex upon BMP4 treatment
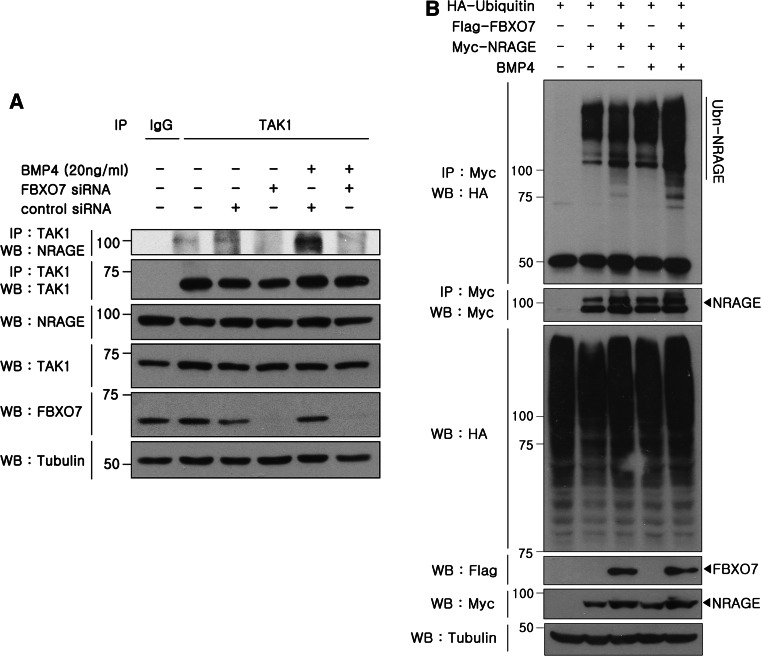
Based on the previous finding that NRAGE-mediated formation of the TAK1–TAB1–NRAGE complex is significantly enhanced in NPCs upon BMP treatment [29], we addressed whether BMP treatment and BMPR stimulation affect the binding affinity of endogenous NRAGE to TAK1 in the presence of FBXO7. HEK293 cells were mock-transfected or transfected with FBXO7-specific siRNAs or control siRNAs for 24 h and stimulated with 10 ng/ml BMP4 for another 24 h. As shown in Fig. 6a, immunoblot analyses of anti-TAK1 immunocomplexes with anti-NRAGE antiserum revealed that endogenous TAK1 and endogenous NRAGE interact in the absence of BMP4 stimulation. In addition, BMP4 stimulation caused a stronger interaction between TAK1 and NRAGE (Fig. 6a). Moreover, the BMP4-mediated increase of NRAGE and TAK1 binding is significantly decreased when FBXO7 is knocked down (Fig. 6a). These results demonstrate that BMP4 treatment and subsequent BMPR activation further facilitates the stimulatory effect of FBXO7 on formation of the TAK1–NRAGE complex in HEK293 cells.
Fig. 6.
FBXO7 accelerates formation of the TAK1–NRAGE complex and NRAGE poly-ubiquitination upon BMP4 treatment. a HEK293 cells were mock-transfected or transfected for 24 h with FBXO7-siRNAs or control siRNAs, and treated with 10 ng/ml BMP4 for 24 h. Cell lysates were immunoprecipitated with the anti-TAK1 antibody and immunoblotted with the anti-NRAGE antibody. Control immunoprecipitations were performed with rabbit IgG. To determine equal loading, cell lysates were analyzed by immunoblotting with anti-tubulin. b HEK293 cells were transfected for 24 h with HA-tagged ubiquitin and Myc-tagged NRAGE alone or in combination with Flag-tagged FBXO7, and treated with 10 ng/ml BMP4 for additional 24 h. Cell lysates were immunoprecipitated with the anti-Myc antibody and immunoblotted with the anti-HA antibody. Ubiquitin, NRAGE, and FBXO7 expression in cell extracts was determined by immunoblotting with the appropriate antibodies
We next assessed whether BMP4 stimulation affects the activity of FBXO7 ubiquitin E3 ligase to ubiquitinate NRAGE. After HEK293 cells were transfected with HA-ubiquitin plus Myc-tagged NRAGE alone or in combination with Flag-tagged FBXO7 and treated with 10 ng/ml BMP4, cell lysates were immunoprecipitated with the Myc antibody. Immunoblot analyses of anti-Myc-NRAGE immunoprecipitates with HA-antiserum showed that BMP4 stimulation further promotes NRAGE poly-ubiquitination by FBXO7 compared to cells treated with vehicle (Fig. 6b).
BMP4 does not affect the protein stability, subcellular localization, and binding affinity of FBXO7 and NRAGE
Next, we examined whether BMP4 affects the levels of endogenous FBXO7 and NRAGE, as well as their cellular localization and binding affinities. After HEK393 cells were stimulated with BMP4, immunoblot analyses of cell lysates with anti-FBXO7 and anti-NRAGE antibodies showed that BMP4 treatment did not significantly influence endogenous FBXO7 and NRAGE protein expression (Supplementary Fig. S2A). We addressed whether BMP4 regulates the intracellular localization of these two proteins. After HEK293 cells were treated with vehicle or BMP4 for the indicated times, cells were fractionated into cytoplasmic and nuclear fractions. Immunoblot analyses of each fraction revealed that FBXO7 and NRAGE are localized predominantly to the cytoplasmic compartment, and their subcellular localization is unaffected by BMP4 treatment (Supplementary Fig. S2B).
We then assessed whether the binding affinity between NRAGE and FBXO7 is regulated by BMP4 stimulation. After HEK293 were transfected for 24 h with Flag-FBXO7 and/or Myc-NRAGE, cells were treated with BMP4. Immunoblot analyses of anti-Myc immunocomplexes with anti-Flag antibodies showed that overexpressed NRAGE binds to FBXO7, and their interaction is not changed upon BMP4 treatment (Supplementary Fig. S2C). To further confirm this finding, co-immunoprecipitation assays were performed to assess the binding between endogenous NRAGE and overexpressed Flag-FBXO7. Similarly, BMP4 stimulation did not significantly influence the binding between endogenous NRAGE and exogenous FBXO7 (Supplementary Fig. S2D). Taken together, our findings suggest that BMP4 treatment and subsequent BMPR stimulation does not change the expression levels of NRAGE and FBXO7, their subcellular localization, or their binding affinity, but rather selectively and positively affects FBXO7-mediated protein complex formation between TAK1 and NRAGE.
FBXO7 enhances BMP4-induced and NRAGE-mediated NF-κB activity
In addition to regulating TAK1–TAB1 complex formation, NRAGE is required for IKK-α/β phosphorylation and subsequent transcriptional activation of the NF-κB p65 subunit via the XIAP–TAK1–TAB1 complex [23]. An inhibitory role of FBXO7 has also been reported based on modulating the regulatory ubiquitination events in NF-κB signaling [14]. Thus, we examined whether FBXO7-mediated poly-ubiquitination of NRAGE affects NF-κB activation through the BMP-initiated alternative signaling pathway.
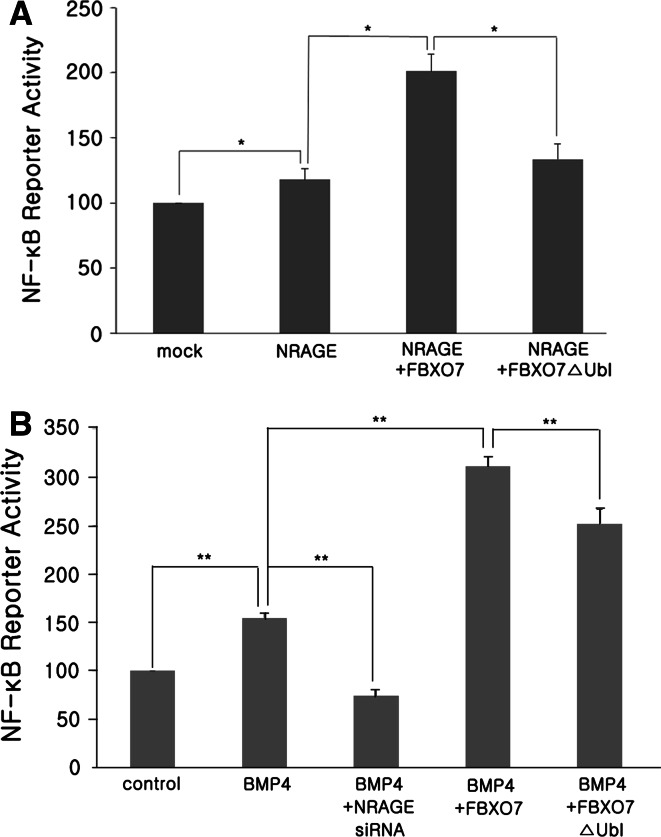
Using NF-κB luciferase reporter gene-based assays, we found that NRAGE overexpression stimulates NF-κB activity (Fig. 7a), as previously reported [23]. Furthermore, co-transfection of wild-type FBXO7 and NRAGE remarkably enhanced NF-κB activity, whereas the FBXO7-ΔU mutant did not (Fig. 7a). To explore the regulatory role of FBXO7 on NF-κB activity through BMP4- and NRAGE-mediated signaling pathways, HEK293 cells were mock-transfected or transfected with plasmids encoding either wild-type FBXO7 or FBXO7-ΔU, and stimulated with BMP4. NF-κB luciferase reporter activity showed that BMP4 stimulation causes activation of NF-κB activity (Fig. 7b), accompanied by nuclear translocation of the NF-κB p65 subunit (data not shown). However, NRAGE knockdown using NRAGE-specific siRNAs prior to BMP4 treatment abrogated this effect (Fig. 7b). This positive regulatory effect of FBXO7 on BMP signaling was verified by assaying cells transfected with plasmids encoding wild-type FBXO7 or FBXO7-ΔU followed by BMP4 stimulation. Consistent with the results shown in Fig. 7a, overexpression of wild-type FBXO7 dramatically enhanced BMP4-mediated NF-κB activity (Fig. 7b). However, the FBXO7 mutant lacking the Ubl domain failed to show this effect (Fig. 7b). These results suggest that FBXO7 acts as a positive regulator of the BMP4-mediated alternative signaling pathway and this effect occurs through FBXO7-mediated NRAGE ubiquitination. FBXO7 then stimulates NF-κB activity by promoting formation of the TAK1–TAB1–NRAGE complex.
Fig. 7.
FBXO7 enhances BMP4-induced and NRAGE-mediated NF-κB activity. a HEK293 cells were transfected for 48 h with the firefly luciferase reporter plasmid and pRL plasmid expressing Renilla luciferase alone or in combination with plasmids encoding Myc-tagged NRAGE and Flag-tagged wild-type FBXO7 or FBXO7-ΔU, as indicated. Cells were lysed and NF-κB activity was analyzed using the dual-luciferase reporter assay system. b HEK293 cells were transfected for 24 h with Flag-tagged FBXO7 isoform-1 or -2 together with the NF-κB luciferase reporter vector and control Renilla luciferase reporter vector. Cells were either unstimulated or treated with 10 ng/ml BMP4 for 24 h. The relative luciferase activity of each cell lysate was measured and normalized to Renilla luciferase activity. Data are expressed as the mean ± SD (n = 3; *p < 0.05, **p < 0.01)
FBXO7 does not affect NRAGE-mediated P19 cell apoptosis in response to BMP4
Based on the report that NRAGE also plays a pivotal role in mediating BMP4-induced apoptosis via the alternative pathway [29, 30], we tested whether FBXO7 also influences the P19 cell apoptosis by accelerating TAK1–TAB1–NRAGE complex formation in response to BMP4. P19 embryonic carcinoma cells are a pluripotent cell line derived from mouse teratomas. To test this possibility, P19 cells were transiently transfected with NRAGE-siRNAs, followed by BMP4 treatment. Cell viability was measured using trypan blue exclusion analyses. As reported, the relative cell death ratio was greatly enhanced with BMP4 stimulation plus treatment with control siRNAs (Supplementary Fig. S3). This effect was not observed in cells with knocked down NRAGE expression (Supplementary Fig. S3). Moreover, NRAGE overexpression caused cell death in P19 cells, and this effect was noted in the absence of BMP4. To further investigate whether FBXO7 regulates NRAGE-mediated apoptosis, P19 cells were transfected with Myc-NRAGE plus wild-type Flag-FBXO7 or the Flag-FBXO7-ΔU mutant and cell viability was evaluated. Overexpression of wild-type FBXO7 or the Flag-FBXO7-ΔU mutant did not affect NRAGE-mediated apoptosis compared to cells transfected with NRAGE alone. These data demonstrate that NRAGE plays a crucial role during the BMP4-initiated cell death process in P19 cells, as previously reported. However, unlike the effect on TAK1–TAB1–NRAGE complex formation and NF-κB activation in HEK293 cells, FBXO7 does not appear to play a regulatory role in BMP4-initiated and NRAGE-mediated P19 cell death.
Discussion
FBXO7 is the causative gene of early onset autosomal recessive Parkinsonian-pyramidal disease (PPD; [4, 5, 29]). PPD differs from typical PD principally by its juvenile onset and the presence of spasticity. Although PPD is a relatively rare form of PD, understanding its genetic and pathological mechanisms may enable new insight into PD pathogenesis and the development of novel therapeutic strategies for PPD as well as for PD [31]. FBXO7 is a member of the SCF ubiquitin E3 ligases, which plays important roles in targeting proteins for ubiquitination. Although SCFFBXO7 is one of the most ubiquitously present SCF ligases in cultured cells [15], not much is known about its substrates for ubiquitination and its role in diseases, including PD. In this study, we identified NRAGE as a new FBXO7-binding protein using a yeast two-hybrid assay. NRAGE is also a substrate for poly-ubiquitination by the SCFFBXO7 E3 ligase complex. In addition to the previously reported three substrates (HURP, cIAP1, and TRAF2; [12–14]), we propose that NRAGE could be a fourth substrate of SCFFBXO7.
Among these substrates, poly-ubiquitination of NRAGE, cIAP1, and TRAF2 do not lead to proteolysis, but may change their biochemical properties. For example, cIAP1 ubiquitination changes its cellular location from the nucleus to the cytoplasm [13]. Consequently, these three targets affect NF-κB signaling after ubiquitination. Although poly-ubiquitination of cIAP1 and TRAF2 suppresses NF-κB signaling [14], our data show that NRAGE enhances NF-κB signaling upon stimulation with BMP. Unlike these targets, HURP ubiquitination results in its proteolysis through the 26S proteasome complex, which consequently regulates Cdk1-cyclin B-mediated cell cycle progression [12].
Although ubiquitination was initially recognized as the reaction that labels intracellular proteins for degradation by the multi-enzymatic proteasome complex, it was later noted that variations in ubiquitin attachment could influence protein function without proteolysis [32]. Ubiquitination is classified as mono-ubiquitination, poly-ubiquitination, and linear ubiquitination. The first mono-ubiquitination is the addition of a single ubiquitin molecule to the substrate protein, which affects the biochemical properties of protein and various cellular processes such as membrane trafficking, endocytosis, and viral budding [28]. The second poly-ubiquitination is the addition of the ubiquitin chain to a single lysine residue on the substrate protein. Following the addition of a single ubiquitin moiety to the substrate, ubiquitin molecules are further added to the first one, yielding a poly-ubiquitin chain [32, 33]. To make the ubiquitin chains, a glycine residue of the ubiquitin molecule is linked to the lysine residue of the next ubiquitin on a substrate. The ubiquitin molecule has seven lysine residues that serve as sites for ubiquitination: Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, and Lys-63. Among these, K48- and K63-linked poly-ubiquitination are well characterized, whereas the function of other lysine chains remains unclear [33, 34].
Lys-48-linked poly-ubiquitin chains usually determine the fate of target proteins for degradation through the proteasome [35]. In contrast, Lys-63-linked chains are not associated with proteasomal degradation of the substrates. Instead, they regulated biochemical and functional properties of the target proteins and a number of their associated cellular processes, such as endocytic trafficking, inflammation, translation, and DNA repair [28]. Finally, linear ubiquitination is quite different from other internal Lys-linked ubiquitination, and is linked on its amino-terminal methionine, which is also referred to as M1 linkage [36]. M1 linkages are created by the linear ubiquitin chain assembly complex (LUBAC), which is a component of the TNF-RSC. LUBAC functions to place M1 linkages on key signaling hubs and modifies the output of many different signal pathways [36]. Among these types of ubiquitination, FBXO7 promotes NRAGE poly-ubiquitination via Lys-63-linkage.
The Ubl domain of FBXO7 is structurally similar to ubiquitin and shares 30 % sequence identity [31]. Several proteins containing Ubl domain have been found to bind the proteasome [37–39]. Moreover, N-terminally located Ubl domains typically directly bind the proteasome [37, 40]. SCF E3 ligase complex also interacts with the proteasome in budding yeast [41]. Therefore, it is thought that Ubl domain of FBXO7 is important for interaction with targets and/or modifying enzymes with the proteasome. It could also play a role in recruiting and transporting substrates to the proteasome for efficient degradation. The F-box is a protein motif with ~50 amino acids that functions as a protein–protein interaction domain. Like FBXO7, many ubiquitin E3 ligases have Ubl or F-box domains, or both. Mutant constructs deleting the Ubl domain or F-box have been widely used as dominant negative controls of protein ubiquitination. In the SCFFBXO7 complex and SCFFBXO7-mediated ubiquitination, the F-box motif links the FBP to other components of the SCF complex by binding to Skp1 [42, 43]. For example, FBXO7 efficiently mediates poly-ubiquitination of cIAP1 and TRAF2, but the FBXO7-ΔF mutant lacks this activity [14]. The present study demonstrates that FBXO7-ΔU is defective in NRAGE poly-ubiquitination.
As it is well known that the F-box domain of F-box protein is required for its binding to Skp1, which in turn associate with cullin1 to make a functional SCF complex [44], our data support the current paradigm for the SCF complex structure and function. In addition, FBXO7-ΔF-box mutant is expected to abolish the SCF-dependent activity of FBXO7. However, compared with wild-type FBXO7, FBXO7-ΔF mutant has decreased, but still substantial activity toward NRAGE. As shown in Fig. 3c, a decent level of endogenous Fbxo7 was detected, which could mediate significant NRAGE poly-ubiquitination. In view of the fact that FBP dimerization is emerging as a major theme for SCF dependent and has independent functions, it is possible that a FBXO7-ΔF mutant forms a dimer with an endogenous FBXO7 via the FP domain [26].
It was previously reported that the ubiquitin ligase Praja1 inhibits neuronal outgrowth in PC12 cells through Lys-48-linked ubiquitination of NRAGE and proteasomal degradation [45]. Praja1 has been implicated in the plasticity of the adult central nervous system and NGF-induced neuronal differentiation. Although Praja1 ubiquitinates NRAGE and subsequently causes its proteasomal degradation [45], our results indicate that FBXO7 mediates NRAGE poly-ubiquitination without its proteolysis during resting conditions and upon BMP4 stimulation. As expected, FBXO7-mediated poly-ubiquitination of NRAGE goes through the Lys-63-linkage rather than through Lys-48.
The present work demonstrates that wild-type FBXO7 and the FBXO7-ΔU mutant display different capacities in NRAGE ubiquitination. Interestingly, there are two isoforms of FBXO7 present in mammalian cells [25]. Isoform-1 is the full-length form of FBXO7, whereas isoform-2 lacks the N-terminal Ubl domain. These two proteins are expressed via alternative splicing of the FBXO7 gene in normal human cells, and isoform-1 is more abundant. In addition, isoform-1 is imperceptible and isoform-2 is severely defective in patients from a Dutch PARK15 family [25]. Like other proteins having Ubl domains, the N-terminal Ubl domain of FBXO7 is thought to interact with ubiquitin receptor proteins [25]. The present work found that FBXO7-isoform-2 could act as a negative regulator of isoform-1 for protein ubiquitination, including for NRAGE. It would be interesting to examine the physiological roles of two isoforms and determine how they are differentially expressed and modulated in neuronal cells. Furthermore, additional experiments are required to investigate whether the mutations in the FBXO7 gene observed in PPD patients affect the differential expression of the two isoforms and consequently, neuronal cell death.
NRAGE is a member of the melanoma antigen family, which has a unique function from other MAGE family proteins. NRAGE plays a critical role in the transmission of pro-apoptotic signals and NF-κB activation in HEK293 cells via the BMP alternative pathway [23]. In the BMP-induced alternative pathway, activation of BMPRs leads to formation of the NRAGE–TAK1–TAB1–XIAP signaling complex, where NRAGE facilitates formation of the complex. Through the signaling cascade, it has also been reported that the TAK1–TAB1–XIAP complex induces IKK-α/β phosphorylation and subsequent activation of NF-κB [23, 29]. Herein, we report that FBXO7 positively regulates NF-κB activity by promoting formation of the TAK1–TAB1–NRAGE complex, which is upstream of the BMP4 alternative signaling pathway. However, the ability of FBXO7 to accelerate formation of the TAK1–TAB1–NRAGE complex does not influence NRAGE-induced apoptosis in P19 cells.
Currently, NRAGE is known to participate only in the “alternative” pathway of BMP signaling. NRAGE facilitates TAK1–TAB1–XIAP complex formation and drives activation of downstream p38 [20]. The BMP-regulated “canonical” pathway would be critical for neurogenesis in developing CNS tissues, such as the cortex [46]. It involves Smad1, Smad5, and Smad4 proteins to activate downstream signaling cascades. It would also be interesting to test whether FBXO7 affects the canonical pathway in response to BMP in neuronal cells, and if so, to determine probable targets of FBXO7-mediated ubiquitination during BMP-initiated downstream signaling.
Although the present study demonstrates that NRAGE is a novel substrate of the SCFFBXO7 ubiquitin E3 ligase and examines how FBXO7 regulates NRAGE-mediated BMP signaling, we do not know much regarding the role of FBXO7 in NRAGE-mediated neuronal cell death [20], as well as its implication in PARK15-associated PPD pathogenesis. To address this issue, the physiological effects of FBXO7 on NRAGE-mediated signaling pathway that lead to neuronal cell death should be explored. Among NRAGE-interacting proteins, several putative candidates linked to neuronal cell death or neurodegenerative diseases have already been proposed and their functional link to FBXO7 should be assessed. These candidates include anti-apoptotic factor Che-1 [47], PD- and mitophagy-linked parkin [48], and a cellular prion protein PrP (C) [49]. In addition to the reported FBXO7 substrates (i.e., HURP, cIAP1, TRAF2, and NRAGE), further studies that identify additional binding targets and substrates would help determine how alterations in FBXO7 function and regulation are associated with PD pathogenesis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank H.J. Kuiken, C.Y. You, J.M. Verdi, and G. Takausu for providing plasmids and J.H. Lee for providing P19 cells. The authors also thank Y.J. Oh and Y.K. Jang for their helpful discussion and comments. This work was supported by the National Research Foundation of Korea (NRF) grants (2012R1A1A2021749 and 2007-0056092 to K.C.C.) funded by the Ministry of Science, ICT & Future Planning (MSIP), Republic of Korea. This work was also partially supported by grants from the Korea Healthcare Technology R&D Project, Ministry for Health and Welfare (A092004 and A111653 to K.C.C).
Abbreviations
- BMP
Bone morphogenetic protein
- BMPR
BMP receptor
- DAPI
4,6-Diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle’s medium
- ECL
Enhanced chemiluminescence
- FBP
F-box protein
- FBS
Fetal bovine serum
- FBXO7
F-box only protein 7
- HEK293
Human embryonic kidney 293
- MAGE
Melanoma-associated antigens
- NRAGE
Neurotrophin receptor-interacting MAGE protein
- P19
P19 embryonic carcinoma
- PBS
Phosphate-buffered saline
- PD
Parkinson’s disease
- PI31
Proteasome inhibitor subunit 1
- SCF
Skp1–cullin1–F box protein
- Ubl
Ubiquitin-like
References
- 1.Olanow CW, Tatton WG. Etiology and pathogenesis of Parkinson’s disease. Annu Rev Neurosci. 1999;22:123–144. doi: 10.1146/annurev.neuro.22.1.123. [DOI] [PubMed] [Google Scholar]
- 2.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 3.Thomas B, Beal MF. Parkinson’s disease. Hum Mol Genet. 2007;16:183–194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 4.Di Fonzo A, Dekker MC, Montagna P, Baruzzi A, Yonova EH, Correia Guedes L, Szczerbinska A, Zhao T, Dubbel-Hulsman LO, Wouters CH, de Graaff E, Oyen WJ, Simons EJ, Breedveld GJ, Oostra BA, Horstink MW, Bonifati V. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 5.Paisán-Ruiz C, Guevara R, Federoff M, Hanagasi H, Sina F, Elahi E, Schneider SA, Schwingenschuh P, Bajaj N, Emre M, Singleton AB, Hardy J, Bhatia KP, Brandner S, Lees AJ, Houlden H. Early-onset l-dopa-responsive Parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Mov Disord. 2010;15:1791–1800. doi: 10.1002/mds.23221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shojaee S, Sina F, Banihosseini SS, Kazemi MH, Kalhor R, et al. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500 K SNP arrays. Am J Hum Genet. 2008;82:1375–1384. doi: 10.1016/j.ajhg.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;20:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 8.Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biol. 1999;20:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 9.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature. 2000;6810:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 10.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;6:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 12.Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–32602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- 13.Chang YF, Cheng CM, Chang LK, Jong YJ, Yuo CY. The F-box protein Fbxo7 interacts with human inhibitor of apoptosis protein cIAP1 and promotes cIAP1 ubiquitination. Biochem Biophys Res Commun. 2006;342:1022–1026. doi: 10.1016/j.bbrc.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 14.Kuiken HJ, Egan DA, Laman H, Bernards R, Beijersbergen RL, Dirac AM. Identification of F-box only protein 7 as a negative regulator of NF-κB signaling. J Cell Mol Med. 2012;16:2140–2149. doi: 10.1111/j.1582-4934.2012.01524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JE, Sweredoski MJ, Graham RL, Kolawa NJ, Smith GT, Hess S, Deshaies RJ. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol Cell Proteomics. 2011;10(M110):006460. doi: 10.1074/mcp.M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sang M, Wang L, Ding C, Zhou X, Wang B, Wang L, Lian Y, Shan B. Melanoma-associated antigen genes—an update. Cancer Lett. 2011;302:85–90. doi: 10.1016/j.canlet.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 17.Jordan BW, Dinev D, LeMellay V, Troppmair J, Gotz R, Wixler L, Sendtner M, Ludwig S, Rapp UR. Neurotrophin receptor-interacting mage homologue is an inducible inhibitor of apoptosis protein-interacting protein that augments cell death. J Biol Chem. 2001;276:39985–39989. doi: 10.1074/jbc.C100171200. [DOI] [PubMed] [Google Scholar]
- 18.Kendall SE, Goldhawk DE, Kubu C, Barker PA, Verdi JM. Expression analysis of a novel p75 (NTR) signaling protein, which regulates cell cycle progression and apoptosis. Mech Dev. 2002;117:187–200. doi: 10.1016/S0925-4773(02)00204-6. [DOI] [PubMed] [Google Scholar]
- 19.Bertrand MJ, Kenchappa RS, Andrieu D, Leclercq-Smekens M, Nguyen HN, Carter BD, Muscatelli F, Barker PA, De Backer O. NRAGE, a p75NTR adaptor protein, is required for developmental apoptosis in vivo. Cell Death Differ. 2008;15:1921–1929. doi: 10.1038/cdd.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendall SE, Battelli C, Irwin S, Mitchell JG, Glackin CA, Verdi JM. NRAGE mediates p38 activation and neural progenitor apoptosis via the bone morphogenetic protein signaling cascade. Mol Cell Biol. 2005;25:7711–7724. doi: 10.1128/MCB.25.17.7711-7724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL, Verdi JM, Barker PA. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron. 2000;27:279–288. doi: 10.1016/S0896-6273(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 22.Chalazonitis A, Kessler JA. Pleiotropic effects of the bone morphogenetic proteins on development of the enteric nervous system. Dev Neurobiol. 2012;72:843–856. doi: 10.1002/dneu.22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matluk N, Rochira JA, Karaczyn A, Adams T, Verdi JM. A role for NRAGE in NF-κB activation through the non-canonical BMP pathway. BMC Biol. 2010;8:7. doi: 10.1186/1741-7007-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang EJ, Ahn YS, Chung KC. Protein kinase Dyrk1 activates CREB during the neuronal differentiation in CNS hippocampal progenitor cells. J Biol Chem. 2001;276:39819–39824. doi: 10.1074/jbc.M104091200. [DOI] [PubMed] [Google Scholar]
- 25.Zhao T, De Graaff E, Breedveld GJ, Loda A, Severijnen LA, Wouters CH, Verheijen FW, Dekker MC, Montagna P, Willemsen R, Oostra BA, Bonifati V. Loss of nuclear activity of the FBXO7 protein in patients with parkinsonian-pyramidal syndrome (PARK15) PLoS One. 2011;6:e16983. doi: 10.1371/journal.pone.0016983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirk R, Laman H, Knowles PP, Murray-Rust J, Lomonosov M, el Meziane K, McDonald NQ. Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. J Biol Chem. 2008;283:22325–22335. doi: 10.1074/jbc.M709900200. [DOI] [PubMed] [Google Scholar]
- 27.Ye Y, Blaser G, Horrocks MH, Ruedas-Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492:266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranda M, Sorkin A. Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol Interv. 2007;7:157–167. doi: 10.1124/mi.7.3.7. [DOI] [PubMed] [Google Scholar]
- 29.Deng H, Liang H, Jankovic J. F-box only protein 7 gene in parkinsonian-pyramidal disease. JAMA Neurol. 2013;70:20–24. doi: 10.1001/jamaneurol.2013.572. [DOI] [PubMed] [Google Scholar]
- 30.Rochira JA, Matluk NN, Adams TL, Karaczyn AA, Oxburgh L, Hess ST, Verdi JM. A small peptide modeled after the NRAGE repeat domain inhibits XIAP-TAB1-TAK1 signaling for NF-κB activation and apoptosis in P19 cells. PLoS One. 2011;6:e20659. doi: 10.1371/journal.pone.0020659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 33.Xu P, Peng J. Characterization of polyubiquitin chain structure by middle-down mass spectrometry. Anal Chem. 2008;80:3438–3444. doi: 10.1021/ac800016w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 35.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 36.Rieser E, Cordier SM, Walczak H. Linear ubiquitination: a newly discovered regulator of cell signaling. Trends Biochem Sci. 2013;38:94–100. doi: 10.1016/j.tibs.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Saeki Y, Sone T, Toh-e A, Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem Biophys Res Commun. 2002;296:813–819. doi: 10.1016/S0006-291X(02)02002-8. [DOI] [PubMed] [Google Scholar]
- 38.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 39.Kleijnen MF, Alarcon RM, Howley PM. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol Biol Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GAG, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 41.Seeger M, Hartmann-Petersen R, Wilkinson CR, Wallace M, Samejima I, Taylor MS, Gordon C. Interaction of the anaphase-promoting complex/cyclosome and proteasome protein complexes with multiubiquitin chain-binding proteins. J Biol Chem. 2003;278:16791–16796. doi: 10.1074/jbc.M208281200. [DOI] [PubMed] [Google Scholar]
- 42.Chen BB, Mallampalli RK. F-box protein substrate recognition: a new insight. Cell Cycle. 2013;12:1009–1010. doi: 10.4161/cc.23071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson DE, Laman H. A competitive binding mechanism between Skp1 and exportin 1 (CRM1) controls the localization of a subset of F-box proteins. J Biol Chem. 2011;286:19804–19815. doi: 10.1074/jbc.M111.220079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crajg KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;3:299–328. doi: 10.1016/S0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 45.Teuber J, Mueller B, Fukabori R, Lang D, Albrecht A, Stork O. The ubiquitin ligase Praja1 reduces NRAGE expression and inhibits neuronal differentiation of PC12 cells. PLoS One. 2013;8:e63067. doi: 10.1371/journal.pone.0063067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morikawa Y, Zehir A, Maska E, Deng C, Schneider MD, Mishina Y, Cserjesi P. BMP signaling regulates sympathetic nervous system development through Smad4-dependent and -independent pathways. Development. 2009;136:3575–3584. doi: 10.1242/dev.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Certo MG, Corbi N, Bruno T, Iezzi S, De Nicola F, Desantis A, Ciotti MT, Mattei E, Floridi A, Fanciulli M, Passananti C. NRAGE associates with the anti-apoptotic factor Che-1 and regulates its degradation to induce cell death. J Cell Sci. 2007;11:1852–1858. doi: 10.1242/jcs.03454. [DOI] [PubMed] [Google Scholar]
- 48.Burchell VS, Nelson DE, Sanchez-Martinez A, Delgado-Camprubi M, Ivatt RM, Pogson JH, Randle SJ, Wray S, Lewis PA, Houlden H, Abramov AY, Hardy J, Wood NW, Whitworth AJ, Laman H, Plun-Favreau H. The Parkinson’s disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat Neurosci. 2013;10:1038. doi: 10.1038/nn.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bragason BT, Palsdottir A. Interaction of PrP with NRAGE, a protein involved in neuronal apoptosis. Mol Cell Neurosci. 2005;2:232–244. doi: 10.1016/j.mcn.2005.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.