Abstract
Circadian clocks orchestrate behavioral and physiological processes in a time-of-day dependent manner. The network of clock-controlled genes is intimately interconnected with metabolic regulatory circuits. Circadian clocks rhythmically regulate the expression and activity of key metabolic players, which in turn feed back on the circadian machinery on the transcriptional and post-transcriptional level. Mutations of clock genes are often associated with metabolic defects, especially in lipid and glucose metabolism. Accumulating data suggest that the reciprocal coordination of circadian and metabolic pathways is crucial for cellular homeostasis and the health of the organism.
Keywords: Circadian clock, Energy metabolism, Metabolic syndrome
Introduction
Life on earth evolved in a rhythmic environment where the light, temperature and availability of nutrients cycle in a 24-h period, i.e., the period of the earth’s rotation around its axis. Circadian clocks are timekeeping mechanisms that have evolved to anticipate these daily changes allowing organisms to efficiently establish and maintain cellular homeostasis in a rhythmic environment. They are found in many organisms including cyanobacteria, plants, fungi and animals.
Circadian clocks are characterized by three fundamental properties. The circadian oscillations persist under constant conditions with a period of ~24 h (self-sustained). The period length is stable over a wide range of physiological temperatures (temperature-compensated). Circadian clocks can synchronize with rhythmic environmental cues such as light, temperature and feeding (entrainable). Circadian clocks are cell-autonomous, but systemic cues contribute crucially to the robustness of circadian clocks in animals.
Circadian clocks regulate many metabolic and physiological processes in rhythmic fashion. In cyanobacteria, the circadian clock regulates global gene expression on the level of transcription [1, 2]. The majority of genes are expressed during the light phase when photosynthesis takes place, while oxygen-sensitive reactions, such as nitrogen fixation [3] and purine biosynthesis [4], are confined to the dark phase. Cyanobacterial strains with a functional clock outcompete clock-mutant strains when grown together in a rhythmic environment [5]. Similarly, when strains of the plant Arabidopsis thaliana were grown in a rhythmic environment matching their internal clock, they grew faster, fixed more carbon and survived better than plants with clocks that had endogenous periods deviating from the environment cycles [6]. In mammals, many metabolic pathways, including glycolysis/gluconeogenesis, fatty acid synthesis/fatty acid oxidation and xenobiotic detoxification, are rhythmically coordinated by the circadian clock [7–14]. Reciprocally, various metabolic pathways feed back to the circadian clock. Mounting evidence suggests that a disruption of the circadian clock correlates with an increased prevalence for metabolic disorders. In this review, we discuss recent advances emphasizing the importance of the interlocked relationship of circadian clocks and metabolism for health and disease.
Molecular architecture of the mammalian circadian clock
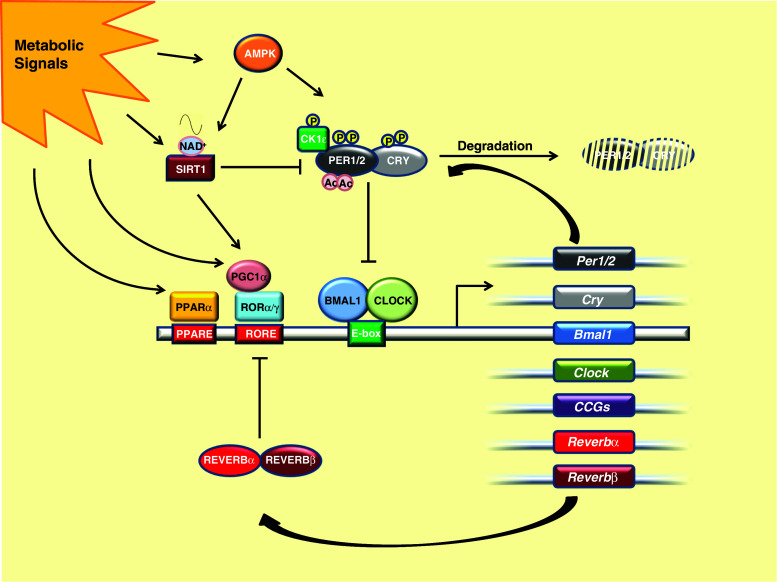
In mammals, the core circadian machinery is based on interconnected transcription feedback loops (Fig. 1). In the positive limb, the transcription activators BMAL1 and CLOCK (or NPAS2 in the brain) form heterodimers, which bind to E-box motifs and activate transcription of target genes, including the core clock genes Period (Per1, Per2 and Per3), Cryptochrome (Cry1 and Cry2), Rev-erbα/ß and Bmal1 itself [15–20]. The PER and CRY proteins form a complex and inhibit BMAL1/CLOCK-driven transcription in a negative feedback loop. This main feedback loop is important since CRY1/2 double knockout (KO) or PER1/2 double KO mice are arrhythmic in constant conditions [21–24]. The transcription repressor REV-ERBα is a major regulator of rhythmic Bmal1 transcription, but its role in circadian rhythm generation was not clear [25, 26]. Recently, it has been shown that the circadian expression of core clock genes and of clock controlled genes (CCGs) is disrupted in liver-specific Rev-erbα and Rev-erbß double-knockout mice. Double knockout of both Rev-erbα and Rev-erbß in the whole body resulted in an altered circadian wheel-running behavior and deregulated lipid metabolism, suggesting REV-ERBα and REV-ERBß are core elements of the circadian clock rather than just elements of a stabilizing loop [13, 27]. The repressive function of REV-ERBs on Bmal1 is counterbalanced by the nuclear receptors RORα/γ and PPARα, whose expression is also regulated by the circadian clock [28–31]. Genome-wide identification of RORα/γ and PPARα binding sites suggests that the cooperation of the REV-ERBs with RORα/γ and PPARα is not restricted to the Bmal1 gene but found in other clock genes and many CCGs [32, 33]. In addition, the transcriptional co-activator PGC1α, a major metabolic regulator, stimulates transcription of Bmal1 and Rev-erbα by promoting the activity of RORα/γ [34].
Fig. 1.
The mammalian molecular clock with major metabolic regulatory points. The main positive limb of mammalian molecular clock is formed by the transcription activator BMAL1/CLOCK, which binds to E-boxes and drives circadian gene expression. The activity of BMAL1/CLOCK is inhibited by PERs and CRYs in a negative feedback loop. Metabolic signals that are conveyed through AMPK and SIRT1 regulate the levels of PER/CRY complexes. SIRT1 deacetylates PER2 and thereby promotes its degradation, whereas AMPK enhances CK1ε activity, which leads to phosphorylation and subsequent degradation of PER2. In addition, phosphorylation of CRY1 by AMPK enhances its degradation. AMPK also activates SIRT1 by increasing cellular NAD+ levels, adding a further critical metabolic feedback regulation [184]. In addition, transcription factors such as RORs, REV-ERBs and PPARs affect the phase and amplitude of the oscillation of Bmal1 expression. Genome-wide ChIPseq analyses suggest that REV-ERBs, RORs and PPARs regulate in addition to Bmal1 also other CCGs
Circadian regulation of the key players in energy metabolism
The metabolic pathways that maintain energy homeostasis are regulated by collaboration of acute signaling systems that instantly respond to changes in metabolites and circadian clocks that anticipate the changes and prepare the molecular environment in a proactive manner. In all organisms analyzed so far, circadian clocks drive rhythmic expression of metabolic genes [9, 35–37] and production of metabolites [38, 39]. Core elements of the circadian clock can either directly regulate rhythmic transcription of metabolic genes or drive rhythmic expression of transcription factors that regulate expression of metabolic genes on a second hierarchical level.
Direct regulation of energy metabolism by core clock elements
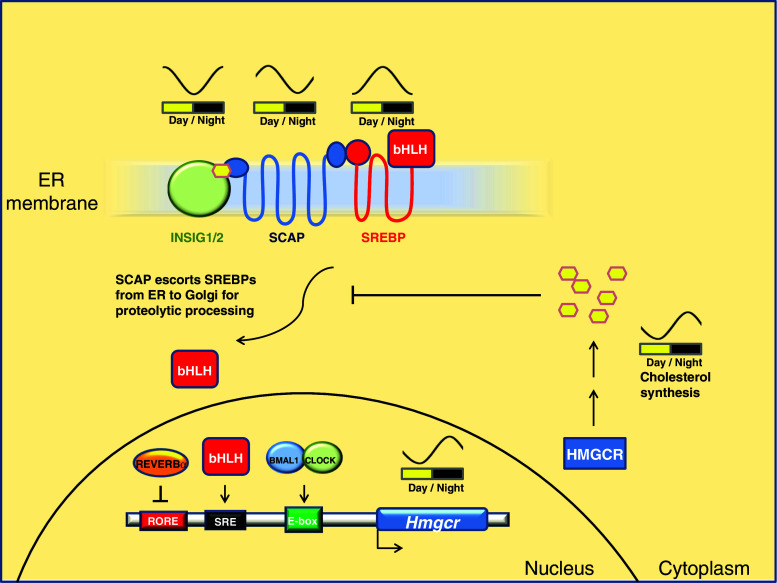
Genome-wide binding analyses of the core clock components BMAL1, CLOCK, NPAS2, PER1/2, CRY1/2 and REV-ERBα/ß suggest that the clock machinery is highly enriched at promoters of genes involved in metabolism, in particular carbohydrate and lipid metabolism [13, 27, 35, 40, 41]. In rats, cholesterol biosynthesis and the activity of the rate-limiting enzyme HMGCoA reductase (HMGCR) follow a circadian rhythm with a peak during the night [42, 43]. BMAL1 and REV-ERBα rhythmically bind to the Hmgcr promoter, and Hmgcr mRNA is circadian with an evening peak [27, 35]. The regulation of cholesterol synthesis by the circadian clock at different steps is an astonishing example showing how strictly the circadian clock can intervene in a biological pathway (Fig. 2). Cellular cholesterol levels are sensed by the SREBP-SCAP-INSIG complex, which is bound to the endoplasmic reticulum (ER) [44]. SREBPs are sterol regulatory element (SRE) binding transcription factors that regulate lipogenesis and cholesterol biosynthesis [45–48]. They activate expression of multiple enzymes in the cholesterol biosynthesis pathway, including HMGCR and HMGCoA synthase [48]. Both SREBP1 and SREBP2 directly bind to Hmgcr promoter [49, 50]. In mouse liver, Srebf1 expression is circadian with an early evening phase [35]. SREBPs are synthesized as inactive, ER membrane-bound proteins, and translocation from the ER to the Golgi is required for their activation. This translocation is mediated by the SREBP cleavage-activating protein (SCAP) [51]. SCAP contains a hexapeptide motif that senses cholesterol levels. High cholesterol concentrations in the cell prevent translocation of the SCAP-SREBP complex from the ER to the Golgi. INSIG1 and INSIG2 are responsible for the sterol-dependent retention of the SCAP-SREBP complex in the ER [52, 53]. In addition to Hmgcr, Insig2 and Scap are also regulated by the circadian clock. Insig2 is regulated by BMAL1 and REV-ERBα, and its expression peaks late at night after cholesterol biosynthesis [11, 35], presumably to sense the cellular cholesterol levels as a quality-control system. In addition, Scap is directly bound by BMAL1 [35, 40], and its mRNA levels peak during the morning before cholesterol synthesis [54]. This is an exquisite example of how the circadian clock coordinates temporal regulation of a metabolic pathway at multiple steps.
Fig. 2.
Regulation of cholesterol biosynthesis by the circadian clock. Expression of HMGCR, a rate-limiting enzyme in cholesterol synthesis, is regulated by the circadian clock. Transcription of Hmgcr is facilitated by SREBPs, which are expressed as inactive precursors. Proteolytic processing is required for the activation of SREBPs. This process is regulated by INSIG1, INSIG2 and SCAP. Srebf1, Insig2 and Scap transcriptions are circadian with different phases. In addition to regulation by SREBPs, REV-ERBα and BMAL1 binding sites are also found in the Hmgcr. The regulation results in evening-specific expression of Hmgcr and cholesterol synthesis. In a negative feedback, cholesterol inhibits Hmgcr expression by promoting retention of the SCAP/SREBP complex in the ER. Sine curves indicate the circadian phases of the corresponding components
In addition to direct transcriptional regulation of metabolic genes by BMAL1/CLOCK and REV-ERBα/ß, the circadian clock impinges on the metabolism by interaction of PER2 and CRY1/2 with key regulators of metabolic pathways [8, 12, 55, 56]. The temporal regulation of gluconeogenesis involves both mechanisms. Although the circadian regulation of gluconeogenesis was observed long ago, the underlying mechanisms were not known [57]. In mammals, feeding and fasting cycles are controlled by the circadian clock. During the fasting period, gluconeogenesis is increased to maintain blood glucose levels. Low energy uptake stimulates release of the hormone glucagon that activates a heterotrimeric G protein and induces cAMP-dependent signaling. This results in protein kinase A (PKA)-dependent phosphorylation of the cAMP regulatory element binding transcription factor (CREB). Transcriptionally active pCREB induces the expression of key gluconeogenic genes, including Pck1 (encoding PEP- carboxykinase), G6pc (encoding G6P-phosphatase) and Pcx (encoding pyruvate carboxylase) [58]. CREB phosphorylation and hence its activity oscillate in a circadian manner [59]. In addition, during fasting, CRY1/2 modulate CREB activity by preventing the glucagon-mediated increase of intracellular cAMP levels by directly interacting with the stimulatory G protein α (Gsα) [8]. Furthermore, BMAL1/CLOCK directly activates gluconeogenic genes in a rhythmic fashion. The promoters of Fbp1 (encoding fructose-1,6-bisphosphatase), Pck1, G6pc and Pcx are bound by BMAL1/CLOCK with a peak during the day when food intake of mice is low and CREB is active [35, 40].
A further example that involves the direct interaction of CRY1/2 with the metabolic pathway is the regulation of glucocorticoid receptor (GR) activity. Glucocorticoids have broad roles in regulating body homeostasis including response to environmental stress and glucose metabolism [60]. CRY1 and CRY2 interact with the GR in a hormone-dependent fashion [56]. This interaction opposes GR-dependent transcription activation and augments the repressive function of the GR. Furthermore, the transcription induction of the gluconeogenic gene Pck1 by the synthetic glucocorticoid dexamethasone is enhanced in CRY-deficient livers, suggesting a general role of CRYs in regulation of gluconeogenesis [56].
Another negative regulator of the circadian clock, PER2, interacts with nuclear receptors including PPARα, PPARγ, REV-ERBα, RORα, HNF4α, TRα and NURR1 [12, 55]. Nuclear receptors are ligand-regulated transcription factors that sense the metabolic state through endocrine and dietary signals and regulate many aspects of metabolism [61, 62]. More than half of the 49 mouse nuclear receptors show rhythmic mRNA expression patterns, adding another layer of circadian control onto the regulation of energy, lipid and glucose metabolism [63]. The interaction of PER2 with the nuclear receptor PPARγ inhibits its transcriptional activity by preventing recruitment of PPARγ to target promoters [12]. In accord with the critical role of PPARγ in adipogenesis and lipid metabolism, PER2 knockout mice show altered lipid metabolism with reduced total triacylglycerol, non-esterified fatty acids and increased fatty acid oxidation [12].
In plants, the alignment of the internal clock with the environmental zeitgebers increases productivity and overall fitness [6]. This could be due to better light harvesting and CO2 fixation since both are under circadian control [64–66]. In A. thaliana, transcriptome analyses suggested that the genes involved in photosynthesis and starch metabolism as well as sugar transporters follow circadian expression [37, 67, 68]. Recently, it has been shown that a nuclear encoded rhythmic transcriptional regulator drives circadian gene expression in the chloroplast, thereby transmitting the circadian timing information between distinct genetic systems [69].
Cyclic activity of metabolic sensors and transcriptional regulators connects the circadian clock and metabolism
NAD/NADH ratio
About a century ago, NAD+ and NADH were identified as essential coenzymes for oxidoreductases. However, the molecules received new attention in the last decade after the identification of NAD+-consuming proteins, such as sirtuins and poly(ADP-ribose) polymerases. The NAD(P)+/NAD(P)H ratio reflects the energy state and reductive power of the cell, so enzymes that depend on NAD+ could act as molecular sensors of the metabolic state. Sirtuins are class III histone deacetylases that require NAD+ in the deacetylation reaction. In addition to histones, sirtuins deacetylate nonhistone proteins, such as transcription factors, thereby controlling their activity [70]. The research investigating their roles as sensors of the metabolic state and executors of downstream signaling revealed tremendously diverse functions of sirtuins [71]. Sirtuins are involved in regulating mitochondrial function, glucose and lipid metabolism, oxidative stress and even lifespan extension [72–79]. Cellular NAD+ levels and levels/activity of the sirtuin SIRT1 follow a circadian rhythm in mice [80–82]. NAD+ can be generated by de novo synthesis starting from tryptophan or by the NAD+ salvage pathway starting from nicotinamide (NAM) [83]. The rate-limiting step of the NAD+ salvage pathway is controlled by the enzyme nicotinamide phosphoribosyltransferase (NAMPT), which catalyzes the synthesis of nicotinamide mononucleotide from NAM [84]. In mice, BMAL1/CLOCK directly regulates NAMPT expression in a circadian manner. Accordingly, cellular NAD+ levels and NAMPT expression are low in Bmal1 KO and Clock ∆19 mice [82, 85]. Since NAD+ has a central role as a cofactor in many metabolic pathways, rhythmic NAD+ levels could transmit the circadian signal to downstream pathways. Analysis of further NAD+-dependent pathways might reveal additional connections of metabolism with the circadian clock.
AMP/ATP ratio
The AMP/ATP ratio is another reflection of the cellular energy state. During starvation, cellular AMP levels increase, and signaling cascades are initiated to provide energy to the cell. For example, AMP-activated protein kinase A (AMPK) activates the catabolic process and inhibits energy-consuming processes, such as the biosynthesis of lipids, carbohydrates and proteins [86, 87]. AMPKs are heterotrimeric protein kinases formed by catalytic α-subunit and regulatory β- and γ-subunits. Phosphorylation of Thr 172 of the α-subunit, primarily by the protein kinase LKB1, tremendously increases AMPK activity [88, 89]. The γ-subunit acts as a sensor of ATP/ADP/AMP concentrations and activates phosphorylation on Thr 172 by binding to AMP [90]. The β subunits regulate the intracellular localization of the AMPK. β1 favors the cytoplasmic localization, and β2 favors the nuclear localization [91]. Lamia et al. [92] observed that expression of β2 mRNA is circadian with a peak during the day in mice. In accordance, they observed that rhythmic nuclear localization of AMPKα1 peaks with the maximal expression of ampkβ2. In addition, rhythmic phosphorylation of Ser792 of Raptor, a well-known substrate of AMPK [93], by AMPK kinase indicates that the kinase activity is circadian [92]. AMPK inhibits lipid biosynthesis by inhibiting processing and the nuclear import of SREBPs [94]. As mentioned earlier, the activity of SREBPs is controlled by the circadian clock by multiple pathways. Additional circadian regulation by AMPK could add another layer of control to determine the amplitude of the SREBP transcriptional activity. As noted in Sect. “Direct regulation of energy metabolism by core clock elements,” the SREBP target genes HMG-CoA reductase and acetyl-CoA carboxylase are the rate-limiting enzymes of fatty acid and sterol synthesis. Their activities are also inhibited by AMPK-dependent phosphorylation, providing a last control step in lipid biosynthesis by the cellular energy state [95–97].
Rhythmic transcription factors as tools to regulate metabolism at a second hierarchical level
Genome-wide circadian transcriptome studies of different organisms suggest that the peak expression of the circadian-controlled genes falls into various phases [7, 9, 35, 36, 68, 98–101]. Analyses of polymerase-II profiles and pre-mRNA levels over a circadian day suggest that rhythmic transcription and post-transcriptional mechanisms contribute to the rhythmic accumulation of mRNAs with different circadian phases [35, 98, 99]. Rhythmic transcription with various phases can be achieved by the circadian control of genes in a second hierarchical level by rhythmic transcription factors. In murine liver, BMAL1/CLOCK directly binds to the promoters of the PARbZip transcription factors DBP, TEF and HLF and drives their rhythmic transcription with a peak during the day [35, 40, 102, 103]. These factors form homo- and heterodimers and activate downstream targets that are mainly involved in the metabolism of xenobiotics. Accordingly, PARbZip triple knockout mice are sensitive to xenobiotic stress and show premature death [104, 105]. In mice, DBP, TEF and HLF regulate the genes involved in detoxification and constitutive androstane receptor (CAR), a key nuclear receptor that functions as a xenobiotic sensor [104, 106]. The expression of Car mRNA peaks at early evening, presumably to prepare the organism for the feeding-related detoxification [63, 104]. This is supported by the observation that oscillation of Dbp mRNA in liver can be regulated by restricted feeding [107].
Nuclear receptors are the sensors of fat-soluble hormones, dietary lipids and vitamins, and they regulate the transcription of genes involved in every aspect of the physiology, including development, reproduction, toxin clearance, immune response, tumor formation, carbohydrate metabolism and lipid metabolism [108]. Since their activity is defined by dietary and endocrine signals, they form the main interface between the cellular environment and gene expression [109]. A comprehensive expression analyses of 49 mouse nuclear receptors in white adipose tissue, brown adipose tissue, muscle and liver over a circadian cycle revealed that 45 nuclear receptors are expressed at least in one tissue, and 25 nuclear receptors display circadian expression profiles [63]. The peak expression of most (18) of the receptors is confined in a short time window (ZT4–ZT8), suggesting a common mechanism regulating their circadian expression. Expression could be directly regulated by BMAL1/CLOCK, since BMAL1 rhythmically binds between ZT4 and ZT8 to eight of these nuclear receptor genes in the liver (i.e., NGFI-B, NOR1, NURR1, PPARα, REV-ERBα, REV-ERBβ, RARα and SHP) [35]. Nevertheless, the phase of expression of GCNF and NURR1 is tissue specific. This observation is particularly interesting since both receptors have critical roles in development [110, 111].
All three PPARs (PPARα/δ/γ) regulate lipid metabolism and energy homeostasis; hence, they are molecular targets for treating metabolic disorders [112]. PPARs show circadian expression profiles with dispersed phases in different tissues [63]. PPARs have similar modular structures, and they are all involved in regulating lipid metabolism. Yet, the PPAR isoforms have unique functions in vivo, probably because of their specific response to ligands, distinct expression profiles in different tissues and different biochemical properties [113, 114]. PPARα is the major activator of fatty acid oxidation and is expressed predominantly in liver, heart, brown adipose tissue and kidney [114, 115]. Prolonged fasting induces the hydrolysis of triacylglycerols in adipose tissue and the release of free fatty acids (FFAs) into the bloodstream. The FFAs are taken up by the liver where the expression and activity of PPARα are augmented [116, 117]. PPARα-induced fatty acid oxidation in response to fasting is critical, since the knockout of PPARα results in hypoglycemia, hyperlipidemia, hypoketonemia and fatty liver disease in mice [116, 118]. Accumulating evidence suggests that fatty acid metabolism is regulated by the circadian clock in animals [38, 119]. The PPARα targets Cyp4As and Acox2 are induced by starvation in a PPARα-dependent manner in mice and rats [118, 120]. The Cyp4A family encodes cytochrome P450 enzymes that facilitate the degrading long-chain fatty acids by ω-hydroxylation of fatty acids and related compounds [121]. Acox2 encodes a branched-chain acyl-CoA oxidase, which is involved in the degradation of long-branched fatty acids and bile acid intermediates in peroxisomes. Both Acox2 and Cyp4A10 are rhythmically expressed with a late evening phase in mice [35].
In the filamentous fungus Neurospora crassa, white collar complex (WCC) is the core transcription factor driving light- and circadian-regulated transcription [122–124]. The circadian transcriptional network comprises about 20 % of the genes of Neurospora producing daily rhythms and light responses [125–127]. The direct WCC targets are morning specific and include ~25 transcription factors that could transmit time information to downstream targets [125]. Among these transcription factors is the repressor CSP1. It represses its targets during the day when CSP1 levels are high, and repression is relieved during the evening when CSP1 levels are low [128]. CSP1 binds to promoters of genes that are mainly involved in energy and lipid metabolism. CSP1 represses all six lipid desaturase genes of Neurospora, and at least one of them is rhythmically expressed with an evening-specific phase. Accordingly, lipid saturation and desaturation are modulated by CSP1 [128].
Feedback of metabolic cues to the circadian clock
In addition to the regulation of metabolism by the circadian clock, metabolic cues in turn feed back to the circadian system. A major example is restricted feeding in animals. Mice that are kept in light/dark cycles consume the majority of their food during the night when they are active. In Drosophila, food consumption is also controlled by the clock, being high during the late night to early morning. In both organisms, by changing the time of food availability, the phase of the circadian gene expression can be changed in peripheral organs without altering the phase of cyclic gene expression in the master clock in the brain [59, 107, 129, 130]. Although little is known about the molecular mechanism, the tight coupling of the peripheral clocks to the metabolism suggests a reciprocal regulation of the circadian clock by feeding [131].
Metabolic cues exert their effect on the circadian systems either by triggering post-translational changes of the clock proteins that regulate their activity or by regulating the expression of clock proteins on the level of transcription. In this section, we will discuss examples of such mechanisms in different organisms.
Metabolic sensors modifying the clock components
The cellular metabolic state is reflected by the AMP/ATP, ADP/ATP and NAD(P)+/NAD(P)/H ratios. High AMP/ATP and ADP/ATP ratios promote the activity of AMPK, which then increases the activity or expression of proteins involved in catabolic processes [86]. Since AMPK acts as a central energy sensor, considerable effort has been expended to identify its targets [132]. One target is the negative clock component CRY. AMPK phosphorylates and promotes the degradation of CRY in mice [92]. Interestingly, glucose limitation in the medium or activation of AMPK by the AMP analog AICAR lengthened the period and decreased the amplitude of the circadian oscillations in mouse embryonic fibroblasts. In addition, activation of AMPK by AICAR injection shifts the phase of entrainment in mouse liver [92]. The AMPK phosphorylation site on CRY1 is conserved among organisms in which CRYs act as transcriptional repressors and absent in organisms in which CRYs act as blue light receptors [92, 133]. In addition, increased body size correlates with evolutional conservation of AMPK phosphorylation sites. Hence, it is proposed that the light-induced degradation signals in CRYs may have been evolutionarily replaced by AMPK-dependent degradation to entrain the clock in such organisms where CRYs cannot directly sense light signals anymore [134]. Another negative clock component, PER2, is indirectly regulated by AMPK in mice. Phosphorylation of PER proteins by CK1ɛ and CK1δ facilitates their degradation by the proteasome [135, 136]. AMPK promotes degradation of PER2 by activating CKIɛ, resulting in lower PER2 levels and shortening of the circadian period length [137]. Activation of AMPK by the drug metformin, which is one of the most commonly used drugs for type II diabetes, changed the phase of circadian gene expression in wild-type but not Ampkα2 KO mice [137]. Regulation of the circadian clock by AMPK at multiple steps could act as a resetting signal to coordinate the circadian clock with metabolic changes.
NAD+ is another small molecule that feeds back to the circadian clock according to the energy status of the cell. NAD+ levels oscillate in a circadian fashion [82, 85] and regulate the acetylation of clock components by the NAD+-activated deacetylase SIRT1. On one hand, SIRT1-dependent PER2 deacetylation determines PER2 stability, and SIRT1 downregulation results in higher PER2 levels. On the other hand, interaction of SIRT1 with BMAL1/CLOCK counterbalances CLOCK-mediated acetylation of histone H3 and BMAL1 [80, 81]. Activation of SIRT1 by synthetic molecules alters BMAL1/CLOCK-driven transcription, mainly by decreasing the amplitude of rhythmic transcription [138]. Pharmacological manipulation of SIRT1 has beneficial effects on prevention of obesity and aging [71, 139, 140]. In addition, SIRT1 could be a useful target for the treatment of disorders related with the circadian physiology.
Poly (ADP-ribose) polymerases (PARPs) synthesize ADP polymers using the ADP-ribose group of NAD+. Historically, PARP1 was defined as a DNA repair enzyme. However, recent studies suggest a broader role, including aging, inflammation and energy metabolism [141–143]. Upon activation by DNA damage, PARP1 activity consumes most (80–90 %) of the cellular NAD+ [83]. To reconstitute the NAD+ level, NAD+ must be synthesized by the salvage pathway, which is energy consuming. Since PARP1 is involved in a number of energy-related process and since NAD+ levels are rhythmic, Asher et al. [131] analyzed a possible role of PARP1 in regulating the circadian clock. They found that PARP1 activity is circadian, and its phase is regulated by feeding. PARP1 interacts with BMAL1/CLOCK in a circadian fashion and poly(ADP-ribosyl)ates CLOCK, changing its affinity for DNA. Moreover, the interaction of BMAL1/CLOCK with PER2 and CRYs is phase shifted in the PARP1 knockout mice. The resetting of the circadian phase of the liver clock induced by restricted feeding is delayed PARP1 KO mice [131]. Hence, PARP1 might be involved in food entrainment of peripheral organs by transmitting the feeding signal to circadian clock.
Poly (ADP-ribose) modifications synthesized by PARPs are degraded by poly (ADP-ribose) glycohydrolases. In A. thaliana, a mutation in the tej gene encoding a poly (ADP-ribose) glycohydrolase lengthens the period of the circadian clock [144]. The long-period phenotype can be rescued by a PARP inhibitor, suggesting a role of the poly(ADP-ribosyl)ation in the plant circadian clock although the target protein(s) have not yet been identified.
O-Linked ß-N-acetylglucosamine (O-GlcNAc) is a post-translation modification regulated by metabolism, particularly via glucose levels [145]. Reciprocally, it is involved in regulating key metabolic events, such as insulin signaling, gluconeogenesis and lipogenesis [146–148]. Recently, O-GlcNAcylation gained attention in the regulation of the circadian clock. O-GlcNAcylation is circadian in mice and Drosophila [149, 150]. In return, both the positive and negative components of the circadian clock are rhythmically O-GlcNAcylated in mice. BMAL1/CLOCK O-GlcNAcylation results in stabilization of proteins by preventing phosphorylation-dependent ubiquitination [151]. Overexpression of O-GlcNAc transferase in mice resulted in the perturbations of Bmal1 oscillation and aberrant circadian rhythms of glucose homeostasis [151]. Changes in the O-GlcNAcylation capacity affect the circadian period in mice and Drosophila [150]. In mice, this is accomplished in part by blocking the CK1-dependent PER2 phosphorylation site S662 by O-GlcNAcylation, a position that corresponds to a phoshorylation site in human PER2 implicated in regulation of the sleep phase [152]. Finally, rhythmic O-GlcNAcylation of dPER in Drosophila regulates its subcellular localization [153]. Hence, O-GlcNAcylation has the potential to affect the circadian clock at multiple levels to coordinate its phase with glucose metabolism.
Regulation of core clock gene expression by metabolic signals
A further way of transmitting metabolic signals to the circadian clock is by transcriptional regulation of the clock components. By changing the expression levels of the core clock components, the phase, amplitude and period of circadian rhythms can be modulated. In the mammalian circadian clock, Bmal1 transcription is regulated by different transcription regulators that are coupled to metabolism. The nuclear receptors REV-ERBα and REV-ERBβ bind to ROR elements (RORE) in the Bmal1 promoter and repress Bmal1 transcription, whereas RORα, RORβ and RORγ act as transcriptional activators of Bmal1 by also binding to RORE [25–28, 154]. The competition between REV-ERBs and RORs determines Bmal1 expression levels. Bmal1 mRNA is anti-phasic to REV-ERB expression, and its rhythm is blunted in Rev-Erbα/β double-KO mice, suggesting REV-ERBs are the major components regulating the amplitude of the rhythmic Bmal1 transcript levels. In addition to REV-ERBs and RORs, the nuclear hormone receptor PPARα regulates the expression of Bmal1 by directly binding to its promoter [29]. In turn, PPARα expression is controlled by the circadian clock in rodents, suggesting a reciprocal regulation [29, 30, 63]. Although Pparα knockout mice do not show altered rhythmic locomotor activity [29], activation of PPARα by bezafibrate results in a phase shift of locomotor activity [155]. Moreover, Bmal1 expression is lower in the liver of Pparα KO mice, and fenofibrate, another PPARα ligand, activates Bmal1 expression rat fibroblasts in a PPARα-dependent manner [29]. PPARα regulates genes involved in fatty acid oxidation, lipid transport, ketogenesis, gluconeogenesis, glycogen metabolism and inflammation [156]. Pparα mRNA is induced during fasting, and PPARα plays an important role in the adaptive response to fasting [116]. Moreover, fatty acids and derivatives are endogenous ligands of PPARα [157]. The role of PPARα in energy metabolism and its connection to the circadian clock represent a further example of how the circadian clock and metabolism are coordinated.
PGC1α, a key transcriptional co-activator that regulates energy metabolism in mammals, is expressed in a circadian manner [34]. PGC1α exerts its effects by regulating the activity of transcription factors, such as PPARγ, GR, HNF4α, ERRα and FOXO1 [158]. Liu et al. [34] showed that, by activating RORα, PGC1α activates the transcription of Bmal1 and integrates energy metabolism with the circadian clock. Pgc1α KO mice have a lengthened period of circadian locomotor activity, and Bmal1 expression is altered.
As opposed to the activating roles of RORα, PPARα and PGC1α, the rhythmic transcription repressor TIEG1/KLF10/mGIF inhibits Bmal1 transcription by directly binding to a GC box in the Bmal1 promoter [159]. This repression is additive to the repression by REVERBα. Feeding starved mice or adding glucose to cultured rat fibroblasts in vitro induces TIEG1/KLF10/mGIF expression, connecting energy input to the cells to the regulation of Bmal1 expression [159]. A corresponding regulation by glucose has been reported for the Neurospora circadian clock. Expression of WC1, a subunit of the circadian transcription activator WCC, is inhibited in a glucose-dependent manner by the rhythmic transcription repressor CSP1 [160]. Adding glucose to the medium induces expression of CSP1, which in turn represses wc1 transcription. The absence of CSP1 results in period shorting in high- but not low-glucose conditions, suggesting a CSP1-dependent metabolic compensation of the period of the circadian clock of Neurospora [160].
Circadian clock dysfunctions implicated in metabolic disorders
The vital role of the circadian players in regulating metabolism is self-evident in genetically modified circadian mutant animals. In mice, liver-specific deletion of Bmal1 results in fasting-induced hypoglycemia, whereas deletion in the pancreas leads to diabetes [161–163]. This emphasizes the local role of BMAL1 in these peripheral tissues since the general circadian locomotor activity is not altered, and the master clock in the brain is intact. Whole-body Bmal1 KO mice show an arrhythmic clock phenotype in constant darkness [16], accelerated aging [164] and disruptions in the diurnal variation of glucose and triglycerides [165]. Deletion of Clock in the whole body results in metabolic syndrome [166]. These mice have impaired diurnal feeding cycles and show hyperleptinemia, hyperlipidemia, hepatic steatosis, hyperinsulinemia and hyperglycemia. Mice expressing a dominant-negative allele of Clock (Clock Δ19/Δ19) show hyperglycemia and defects in insulin release from the pancreas [163]. When challenged with a high-fat diet, ∆Cry1/∆Cry2 double-knockout mice show hyperinsulinemia and weight gain exceeding the wt controls [167]. REV-ERBα and REV-ERBβ have crucial roles in regulating metabolism and the circadian clock [13, 27, 168]. The modest metabolic phenotype of the Rev-erbα KO [11, 41, 169] was exacerbated in the double KO of Rev-erbα and Rev-erbβ, showing the redundant roles of REV-ERB isoforms. The deficiency of both isoforms resulted in hyperlipidemia, hepatic steatosis and hyperglycemia [13, 27, 168]. Moreover, the administration of REV-ERB agonists to diet-induced obese mice resulted in fat loss and improved dyslipidemia and hyperglycemia by regulating the expression of key metabolic genes [168]. Recently, Woldt et al. [170] showed that REV-ERBα modulates the oxidative capacity of muscle by regulating mitochondrial biogenesis and autophagy. Further metabolic phenotypes associated with mutations of murine clock elements are listed in Table 1.
Table 1.
The mutations of clock elements are associated with metabolic phenotypes
| Gene | Function | Metabolic phenotype |
|---|---|---|
| Clock | Transcription factor, main regulator of the circadian clock together with BMAL1 | Metabolic syndrome in clock KO mice |
| Bmal1 | Transcription factor, main regulator of the circadian clock together with CLOCK | Hypoglycemia in liver-specific KO. Reduced insulin secretion in pancreas-specific KO. Whole body Bmall KO mice show accelerated aging together with disrupted diurnal regulation of glucose and triglycerides levels |
| Per2 | Inhibits BMAL1/CLOCK driven transcription | Impaired lipid homeostasis |
| Cry1, Cry2 | Inhibit BMAL1/CLOCK driven transcription | Depletion of Cry1 and Cry2 causes hyperglycemia [8]. High-fat diet results in hyperinsulinemia and excess weight gain in double-knockout mice |
| Reverbα, Reverbβ | Nuclear receptors involved in rhythmic regulation of genes by binding to ROR elements. Repress Bmall expression | Reverb α/β double-KO mice show hyperlipidemia, hepatic steatosis and hyperglycemia |
| Rom Rory | Nuclear receptors, activate the expression of genes by binding to ROR elements | sg/sg (rorα mutant) mice show lower cholesterol levels and resistance to diet-induced obesity [185, 186] |
| PGC1a | Transcriptional coactivator, targets include RORs and PPARs | Pgc1α KO mice show reduced mitochondrial function, resistance to diet-induced obesity and altered thermogenic response [187, 188] |
In humans, mutations or polymorphisms in the clock genes seem to be associated with metabolic phenotypes, such as type 2 diabetes or obesity [171–175]. Moreover, accumulating data suggest that the disruptions of the circadian clock by shift work or social jet-lag are also associated with obesity and diabetes [176–179]. These epidemiological data were supported by experimental data showing that circadian misalignment of the circadian clock and metabolism results in reduced glucose tolerance and insulin insensitivity [180]. Presumably, feeding and sleeping at abnormal circadian times cause misalignment of the peripheral clocks with the environmental day/night cycle that result in metabolic problems (see “BOX1”). In a rat model of “shift work” based on forced activity during the inactive phase, temporal patterns of food intake and metabolic oscillations were altered [181]. Such rats gained more weight and acquired glucose intolerance and microvesicular steatosis [182]. Hatori et al. [183] showed that limiting the feeding time only to the active phase of mice prevented the development of metabolic disorders when they were fed with a high-fat diet. Mice fed a normal diet on a time-restricted basis also showed healthier metabolic markers than mice fed ad libitum [183]. The data suggested that without changing the caloric intake, feeding at the correct circadian time could protect against obesity, hyperinsulinemia, hepaticsteatosis and inflammation.
Concluding remarks
Circadian clocks are involved in regulating many aspects of metabolism, and they coordinate energy intake and expenditure. The feedback loops of the circadian clock and the regulatory networks of metabolic genes are interconnected in a complex manner and at various levels. Perturbing the circadian clock leads to metabolic anomalies especially in glucose and lipid metabolism. The occurrence of metabolic disorders increases in developed societies at an accelerating rate. A detailed understanding of the reciprocal interaction of circadian clocks and metabolic pathways may open new perspectives toward the treatment of such disorders.
Appendix
Box 1
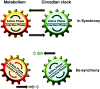
Metabolic events and the circadian clock work in synchrony. Consecutive events in the “active phase” and “inactive phase” are both drivers and outputs of the circadian clock and of metabolism. The active phase is associated mainly with locomotion, feeding and catabolic reactions, whereas the inactive phase is associated with sleep, fasting and anabolic reactions. At night, circadian clocks set the metabolism to a state where energy expenditure is expected to be low for diurnal animals including humans. Challenges, such as eating or exercise at nighttime, are not anticipated by the circadian clock. Hence, the unprepared metabolism has to respond instantly to such perturbations, which desynchronizes metabolism and the circadian clock.
References
- 1.Liu Y, Tsinoremas NF, Johnson CH, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9(12):1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 2.Woelfle MA, Johnson CH. No promoter left behind: global circadian gene expression in cyanobacteria. J Biol Rhythms. 2006;21(6):419–431. doi: 10.1177/0748730406294418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitsui AKS, Takahashi A, Ikemoto H, Arai T. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature. 1986;323:720–722. [Google Scholar]
- 4.Liu Y, Tsinoremas NF, Golden SS, Kondo T, Johnson CH. Circadian expression of genes involved in the purine biosynthetic pathway of the cyanobacterium Synechococcus sp. strain PCC 7942. Mol Microbiol. 1996;20(5):1071–1081. doi: 10.1111/j.1365-2958.1996.tb02547.x. [DOI] [PubMed] [Google Scholar]
- 5.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14(16):1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Dodd AN, Salathia N, Hall A, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 7.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 8.Zhang EE, Liu Y, Dentin R, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16(10):1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 10.Claudel T, Cretenet G, Saumet A, Gachon F. Crosstalk between xenobiotics metabolism and circadian clock. FEBS Lett. 2007;581(19):3626–3633. doi: 10.1016/j.febslet.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Le Martelot G, Claudel T, Gatfield D, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimaldi B, Bellet MM, Katada S, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12(5):509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugge A, Feng D, Everett LJ, et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masri S, Patel VR, Eckel-Mahan KL, et al. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci USA. 2013;110(9):3339–3344. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293(5529):506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 16.Bunger MK, Wilsbacher LD, Moran SM, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gekakis N, Staknis D, Nguyen HB, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 18.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA. 1998;95(10):5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King DP, Zhao Y, Sangoram AM, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17(14):R538–R539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 21.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286(5440):768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 22.Kume K, Zylka MJ, Sriram S, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 23.van der Horst GT, Muijtjens M, Kobayashi K, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 24.Zheng B, Albrecht U, Kaasik K, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 25.Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 26.Ueda HR, Hayashi S, Chen W, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 27.Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato TK, Panda S, Miraglia LJ, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Canaple L, Rambaud J, Dkhissi-Benyahya O, et al. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20(8):1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- 30.Lemberger T, Saladin R, Vazquez M, et al. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271(3):1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- 31.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20(5):391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 32.Takeda Y, Jothi R, Birault V, Jetten AM. RORgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012;40(17):8519–8535. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boergesen M, Pedersen TA, Gross B, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol. 2012;32(4):852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447(7143):477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 35.Koike N, Yoo SH, Huang HC, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correa A, Lewis ZA, Greene AV, March IJ, Gomer RH, Bell-Pedersen D. Multiple oscillators regulate circadian gene expression in Neurospora. Proc Natl Acad Sci USA. 2003;100(23):13597–13602. doi: 10.1073/pnas.2233734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harmer SL, Hogenesch JB, Straume M, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290(5499):2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 38.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9(2):e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards PA, Muroya H, Gould RG. In vivo demonstration of the circadian thythm of cholesterol biosynthesis in the liver and intestine of the rat. J Lipid Res. 1972;13(3):396–401. [PubMed] [Google Scholar]
- 43.Shapiro DJ, Rodwell VW. Diurnal variation and cholesterol regulation of hepatic HMG-CoA reductase activity. Biochem Biophys Res Commun. 1969;37(5):867–872. doi: 10.1016/0006-291x(69)90972-3. [DOI] [PubMed] [Google Scholar]
- 44.Dong XY, Tang SQ, Chen JD. Dual functions of Insig proteins in cholesterol homeostasis. Lipids Health Dis. 2012;11:173. doi: 10.1186/1476-511X-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magana MM, Osborne TF. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J Biol Chem. 1996;271(51):32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 46.Vallett SM, Sanchez HB, Rosenfeld JM, Osborne TF. A direct role for sterol regulatory element binding protein in activation of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene. J Biol Chem. 1996;271(21):12247–12253. doi: 10.1074/jbc.271.21.12247. [DOI] [PubMed] [Google Scholar]
- 47.Hua X, Yokoyama C, Wu J, et al. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci USA. 1993;90(24):11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakakura Y, Shimano H, Sone H, et al. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochem Biophys Res Commun. 2001;286(1):176–183. doi: 10.1006/bbrc.2001.5375. [DOI] [PubMed] [Google Scholar]
- 49.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4(7):e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo YK, Jeon TI, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13(4):367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102(3):315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 52.Yang T, Espenshade PJ, Wright ME, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110(4):489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 53.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci USA. 2002;99(20):12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes ME, DiTacchio L, Hayes KR, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24(4):345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamia KA, Papp SJ, Yu RT, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480(7378):552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kida K, Nishio T, Yokozawa T, Nagai K, Matsuda H, Nakagawa H. The circadian change of gluconeogenesis in the liver in vivo in fed rats. J Biochem. 1980;88(4):1009–1013. doi: 10.1093/oxfordjournals.jbchem.a133051. [DOI] [PubMed] [Google Scholar]
- 58.Jitrapakdee S. Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. Int J Biochem Cell Biol. 2011;44(1):33–45. doi: 10.1016/j.biocel.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Witchel SF, DeFranco DB. Mechanisms of disease: regulation of glucocorticoid and receptor levels–impact on the metabolic syndrome. Nat Clin Pract Endocrinol Metab. 2006;2(11):621–631. doi: 10.1038/ncpendmet0323. [DOI] [PubMed] [Google Scholar]
- 61.Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 63.Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 64.Fredeen AL, Hennessey TL, Field CB. Biochemical correlates of the circadian rhythm in photosynthesis in Phaseolus vulgaris . Plant Physiol. 1991;97(1):415–419. doi: 10.1104/pp.97.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gould PD, Diaz P, Hogben C, et al. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 2009;58(5):893–901. doi: 10.1111/j.1365-313X.2009.03819.x. [DOI] [PubMed] [Google Scholar]
- 66.Hartwell J. The co-ordination of central plant metabolism by the circadian clock. Biochem Soc Trans. 2005;33(Pt 5):945–948. doi: 10.1042/BST20050945. [DOI] [PubMed] [Google Scholar]
- 67.Haydon MJ, Bell LJ, Webb AA. Interactions between plant circadian clocks and solute transport. J Exp Bot. 2011;62(7):2333–2348. doi: 10.1093/jxb/err040. [DOI] [PubMed] [Google Scholar]
- 68.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9(8):R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noordally ZB, Ishii K, Atkins KA, et al. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science. 2013;339(6125):1316–1319. doi: 10.1126/science.1230397. [DOI] [PubMed] [Google Scholar]
- 70.Feige JN, Auwerx J. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol. 2008;20(3):303–309. doi: 10.1016/j.ceb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13(4):225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong L, D’Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 74.Hirschey MD, Shimazu T, Goetzman E, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanfi Y, Naiman S, Amir G, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 76.Liu Y, Dentin R, Chen D, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong L, Mostoslavsky R. Fine tuning our cellular factories: sirtuins in mitochondrial biology. Cell Metab. 2011;13(6):621–626. doi: 10.1016/j.cmet.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Someya S, Yu W, Hallows WC, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jing E, Emanuelli B, Hirschey MD, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci USA. 2011;108(35):14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 81.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31(2):194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279(49):50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 85.Ramsey KM, Yoshino J, Brace CS, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hardie DG, Carling D, Gamblin SJ. AMP-activated protein kinase: also regulated by ADP? Trends Biochem Sci. 2011;36(9):470–477. doi: 10.1016/j.tibs.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 88.Woods A, Johnstone SR, Dickerson K, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13(22):2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 89.Shaw RJ, Kosmatka M, Bardeesy N, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA. 2004;101(10):3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 2013;18(4):556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suzuki A, Okamoto S, Lee S, Saito K, Shiuchi T, Minokoshi Y. Leptin stimulates fatty acid oxidation and peroxisome proliferator-activated receptor alpha gene expression in mouse C2C12 myoblasts by changing the subcellular localization of the alpha2 form of AMP-activated protein kinase. Mol Cell Biol. 2007;27(12):4317–4327. doi: 10.1128/MCB.02222-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lamia KA, Sachdeva UM, DiTacchio L, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Y, Xu S, Mihaylova MM, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13(4):376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clarke PR, Hardie DG. Regulation of HMG-CoA reductase: identification of the site phosphorylated by the AMP-activated protein kinase in vitro and in intact rat liver. EMBO J. 1990;9(8):2439–2446. doi: 10.1002/j.1460-2075.1990.tb07420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sato R, Goldstein JL, Brown MS. Replacement of serine-871 of hamster 3-hydroxy-3-methylglutaryl-CoA reductase prevents phosphorylation by AMP-activated kinase and blocks inhibition of sterol synthesis induced by ATP depletion. Proc Natl Acad Sci USA. 1993;90(20):9261–9265. doi: 10.1073/pnas.90.20.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carlson CA, Kim KH. Regulation of hepatic acetyl coenzyme A carboxylase by phosphorylation and dephosphorylation. J Biol Chem. 1973;248(1):378–380. [PubMed] [Google Scholar]
- 98.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Le Martelot G, Canella D, Symul L, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10(11):e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16(6):833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gachon F. Physiological function of PARbZip circadian clock-controlled transcription factors. Ann Med. 2007;39(8):562–571. doi: 10.1080/07853890701491034. [DOI] [PubMed] [Google Scholar]
- 103.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38(3):369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 104.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4(1):25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 105.Gachon F, Fonjallaz P, Damiola F, et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18(12):1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qatanani M, Moore DD. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab. 2005;6(4):329–339. doi: 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- 107.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McKenna NJ, Cooney AJ, DeMayo FJ, et al. Minireview: evolution of NURSA, the nuclear receptor signaling atlas. Mol Endocrinol. 2009;23(6):740–746. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582(1):2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276(5310):248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 111.Zechel C. The germ cell nuclear factor (GCNF) Mol Reprod Dev. 2005;72(4):550–556. doi: 10.1002/mrd.20377. [DOI] [PubMed] [Google Scholar]
- 112.Ahmadian M, Suh JM, Hah N, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19(5):557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10(4):355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 114.Poulsen L, Siersbaek M, Mandrup S. PPARs: fatty acid sensors controlling metabolism. Semin Cell Dev Biol. 2012;23(6):631–639. doi: 10.1016/j.semcdb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 115.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142(10):4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 116.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103(11):1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc Natl Acad Sci USA. 1993;90(6):2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96(13):7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bray MS, Young ME. Regulation of fatty acid metabolism by cell autonomous circadian clocks: time to fatten up on information? J Biol Chem. 2011;286(14):11883–11889. doi: 10.1074/jbc.R110.214643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kroetz DL, Yook P, Costet P, Bianchi P, Pineau T. Peroxisome proliferator-activated receptor alpha controls the hepatic CYP4A induction adaptive response to starvation and diabetes. J Biol Chem. 1998;273(47):31581–31589. doi: 10.1074/jbc.273.47.31581. [DOI] [PubMed] [Google Scholar]
- 121.Muller DN, Schmidt C, Barbosa-Sicard E, et al. Mouse Cyp4a isoforms: enzymatic properties, gender- and strain-specific expression, and role in renal 20-hydroxyeicosatetraenoic acid formation. Biochem J. 2007;403(1):109–118. doi: 10.1042/BJ20061328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276(5313):763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 123.Baker CL, Loros JJ, Dunlap JC. The circadian clock of Neurospora crassa . FEMS Microbiol Rev. 2012;36(1):95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297(5582):815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 125.Smith KM, Sancar G, Dekhang R, et al. Transcription factors in light and circadian clock signaling networks revealed by genome-wide mapping of direct targets for Neurospora WHITE COLLAR COMPLEX. Eukaryot Cell. 2010;9(10):1549–1556. doi: 10.1128/EC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28(8):1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vitalini MW, de Paula RM, Park WD, Bell-Pedersen D. The rhythms of life: circadian output pathways in Neurospora. J Biol Rhythms. 2006;21(6):432–444. doi: 10.1177/0748730406294396. [DOI] [PubMed] [Google Scholar]
- 128.Sancar G, Sancar C, Brugger B, et al. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol Cell. 2011;44(5):687–697. doi: 10.1016/j.molcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 129.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 130.Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13(6):639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142(6):943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 132.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24(4):948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 134.Jordan SD, Lamia KA. AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol. 2012;366(2):163–169. doi: 10.1016/j.mce.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eide EJ, Woolf MF, Kang H, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25(7):2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci USA. 2011;108(39):16451–16456. doi: 10.1073/pnas.1107178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Um JH, Yang S, Yamazaki S, et al. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282(29):20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- 138.Bellet MM, Nakahata Y, Boudjelal M, et al. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc Natl Acad Sci USA. 2013;110(9):3333–3338. doi: 10.1073/pnas.1214266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nogueiras R, Habegger KM, Chaudhary N, et al. Sirtuin 1 and sirtuin 3: physiological modulators of metabolism. Physiol Rev. 2012;92(3):1479–1514. doi: 10.1152/physrev.00022.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Villalba JM, Alcain FJ. Sirtuin activators and inhibitors. Biofactors. 2012;38(5):349–359. doi: 10.1002/biof.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84(2):137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 142.Mangerich A, Burkle A. Pleiotropic cellular functions of PARP1 in longevity and aging: genome maintenance meets inflammation. Oxid Med Cell Longev. 2012;2012:321653. doi: 10.1155/2012/321653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26(5):417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Panda S, Poirier GG, Kay SA. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the arabidopsis circadian oscillator. Dev Cell. 2002;3(1):51–61. doi: 10.1016/s1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 145.Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13(5):312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 146.Yang X, Ongusaha PP, Miles PD, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451(7181):964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 147.Ruan HB, Han X, Li MD, et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab. 2012;16(2):226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Anthonisen EH, Berven L, Holm S, Nygard M, Nebb HI, Gronning-Wang LM. Nuclear receptor liver X receptor is O-GlcNAc-modified in response to glucose. J Biol Chem. 2010;285(3):1607–1615. doi: 10.1074/jbc.M109.082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Durgan DJ, Pat BM, Laczy B, et al. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem. 2011;286(52):44606–44619. doi: 10.1074/jbc.M111.278903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kaasik K, Kivimae S, Allen JJ, et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17(2):291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li MD, Ruan HB, Hughes ME, et al. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013;17(2):303–310. doi: 10.1016/j.cmet.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291(5506):1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 153.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26(5):490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ueda HR, Chen W, Adachi A, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 155.Shirai H, Oishi K, Kudo T, Shibata S, Ishida N. PPARalpha is a potential therapeutic target of drugs to treat circadian rhythm sleep disorders. Biochem Biophys Res Commun. 2007;357(3):679–682. doi: 10.1016/j.bbrc.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 156.Schupp M, Lazar MA. Endogenous ligands for nuclear receptors: digging deeper. J Biol Chem. 2010;285(52):40409–40415. doi: 10.1074/jbc.R110.182451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab. 2012;23(7):351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 158.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582(1):46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hirota T, Kon N, Itagaki T, Hoshina N, Okano T, Fukada Y. Transcriptional repressor TIEG1 regulates Bmal1 gene through GC box and controls circadian clockwork. Genes Cells. 2010;15(2):111–121. doi: 10.1111/j.1365-2443.2009.01371.x. [DOI] [PubMed] [Google Scholar]
- 160.Sancar G, Sancar C, Brunner M. Metabolic compensation of the Neurospora clock by a glucose-dependent feedback of the circadian repressor CSP1 on the core oscillator. Genes Dev. 2012;26(21):2435–2442. doi: 10.1101/gad.199547.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105(39):15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54(1):120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rudic RD, McNamara P, Curtis AM, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Barclay JL, Shostak A, Leliavski A, et al. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab. 2013;304(10):E1053–E1063. doi: 10.1152/ajpendo.00512.2012. [DOI] [PubMed] [Google Scholar]
- 168.Solt LA, Wang Y, Banerjee S, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485(7396):62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Raspe E, Duez H, Mansen A, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43(12):2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 170.Woldt E, Sebti Y, Solt LA, et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19(8):1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Woon PY, Kaisaki PJ, Braganca J, et al. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci USA. 2007;104(36):14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gomez-Abellan P, Hernandez-Morante JJ, Lujan JA, Madrid JA, Garaulet M. Clock genes are implicated in the human metabolic syndrome. Int J Obes (Lond) 2008;32(1):121–128. doi: 10.1038/sj.ijo.0803689. [DOI] [PubMed] [Google Scholar]
- 173.Garcia-Rios A, Perez-Martinez P, Delgado-Lista J, et al. A Period 2 genetic variant interacts with plasma SFA to modify plasma lipid concentrations in adults with metabolic syndrome. J Nutr. 2012;142(7):1213–1218. doi: 10.3945/jn.111.156968. [DOI] [PubMed] [Google Scholar]
- 174.Englund A, Kovanen L, Saarikoski ST, et al. NPAS2 and PER2 are linked to risk factors of the metabolic syndrome. J Circadian Rhythms. 2009;7:5. doi: 10.1186/1740-3391-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2008;32(4):658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 176.Roenneberg T, Allebrandt KV, Merrow M, Vetter C. Social jetlag and obesity. Curr Biol. 2012;22(10):939–943. doi: 10.1016/j.cub.2012.03.038. [DOI] [PubMed] [Google Scholar]
- 177.Suwazono Y, Dochi M, Sakata K, et al. A longitudinal study on the effect of shift work on weight gain in male Japanese workers. Obesity (Silver Spring) 2008;16(8):1887–1893. doi: 10.1038/oby.2008.298. [DOI] [PubMed] [Google Scholar]
- 178.Di Lorenzo L, De Pergola G, Zocchetti C, et al. Effect of shift work on body mass index: results of a study performed in 319 glucose-tolerant men working in a Southern Italian industry. Int J Obes Relat Metab Disord. 2003;27(11):1353–1358. doi: 10.1038/sj.ijo.0802419. [DOI] [PubMed] [Google Scholar]
- 179.Pan A, Schernhammer ES, Sun Q, Hu FB. Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS Med. 2011;8(12):e1001141. doi: 10.1371/journal.pmed.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Salgado-Delgado R, Angeles-Castellanos M, Buijs MR, Escobar C. Internal desynchronization in a model of night-work by forced activity in rats. Neuroscience. 2008;154(3):922–931. doi: 10.1016/j.neuroscience.2008.03.066. [DOI] [PubMed] [Google Scholar]
- 182.Salgado-Delgado RC, Saderi N, Basualdo Mdel C, Guerrero-Vargas NN, Escobar C, Buijs RM. Shift work or food intake during the rest phase promotes metabolic disruption and desynchrony of liver genes in male rats. PLoS ONE. 2013;8(4):e60052. doi: 10.1371/journal.pone.0060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hatori M, Vollmers C, Zarrinpar A, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Canto C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mamontova A, Seguret-Mace S, Esposito B, et al. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98(24):2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- 186.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283(26):18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 187.Lin J, Wu PH, Tarr PT, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 188.Leone TC, Lehman JJ, Finck BN, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3(4):e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]