Abstract
DNA damage leads to heritable changes in the genome via DNA replication. However, as the DNA helix is the site of numerous other transactions, notably transcription, DNA damage can have diverse repercussions on cellular physiology. In particular, DNA lesions have distinct effects on the passage of transcribing RNA polymerases, from easy bypass to almost complete block of transcription elongation. The fate of the RNA polymerase positioned at a lesion is largely determined by whether the lesion is structurally subtle and can be accommodated and eventually bypassed, or bulky, structurally distorting and requiring remodeling/complete dissociation of the transcription elongation complex, excision, and repair. Here we review cellular responses to DNA damage that involve RNA polymerases with a focus on bacterial transcription-coupled nucleotide excision repair and lesion bypass via transcriptional mutagenesis. Emphasis is placed on the explosion of new structural information on RNA polymerases and relevant DNA repair factors and the mechanistic models derived from it.
Keywords: X-ray crystallography, Mfd, NusA
Introduction
Given the pervasive nature of DNA-damaging agents, both exogenous and endogenous, DNA lesions and the cellular responses to DNA insults have emerged as key factors in the shaping of genomes throughout evolution and in ensuring normal physiology in both unicellular and multi-cellular organisms. The last three decades have fleshed out, in molecular detail, multiple, overlapping pathways for monitoring and repairing DNA lesions that often interface with other DNA transactions, such as replication, transcription, or chromatin remodeling (Fig. 1). This multitude of DNA repair modalities results in a heterogeneity of DNA damage and repair across genomes, and given its interplay with replication and transcription, also partially accounts for the substitutional strand asymmetry noticed in both bacterial and eukaryotic genomes. Replication can produce substitution rate asymmetries in both the transcribed and untranscribed regions (albeit at different rates on the leading and lagging strands) [1, 2], while transcription results in a lower rate of mutation in the non-coding (transcribed, template) DNA strand as demonstrated for enterobacterial species [3] as well as mammals [4, 5]. On the non-transcribed strand, several studies have shown that common substitutions occur at a higher rate [3, 4, 6, 7]. How can this be explained at a molecular, mechanistic level? The answer may partially lie in the overall, evolutionary conserved architecture of the elongating transcription machinery itself. As numerous structural studies have shown (reviewed in [8]), the overall architecture of RNAP resembles a crab claw, with two pincers formed by the two large subunits (β and β’ in bacteria), which both contribute residues to the centrally located catalytic site of the enzyme (Fig. 3). The overall asymmetry of the geometry of the transcription bubble is to be noted: the template (non-coding) and non-template (coding) strands follow different routes within the body of the elongating RNAP. The template strand passes though channels and narrow grooves within the enzyme, while the non-template strand is well exposed and more susceptible to DNA damage. Indeed, ssDNA is more easily deaminated or depurinated than dsDNA [7, 9, 10]. While the template strand is protected somewhat within the body of RNAP by base-pairing with the nascent RNA, the non-template strand is not, and is subject to ssDNA mutagenesis, also known as transcription-associated mutagenesis (TAM). The rate of TAM is directly proportional to the transcription rate. Mechanistically, TAM relies heavily on chemical modifications such as deamination (reviewed in [11]). Not surprisingly, transcription-induced deamination of the non-template strand appears to be a significant force in genome evolution, as was shown for example for the genomes of the T7 phage and Escherichia coli [7, 10].
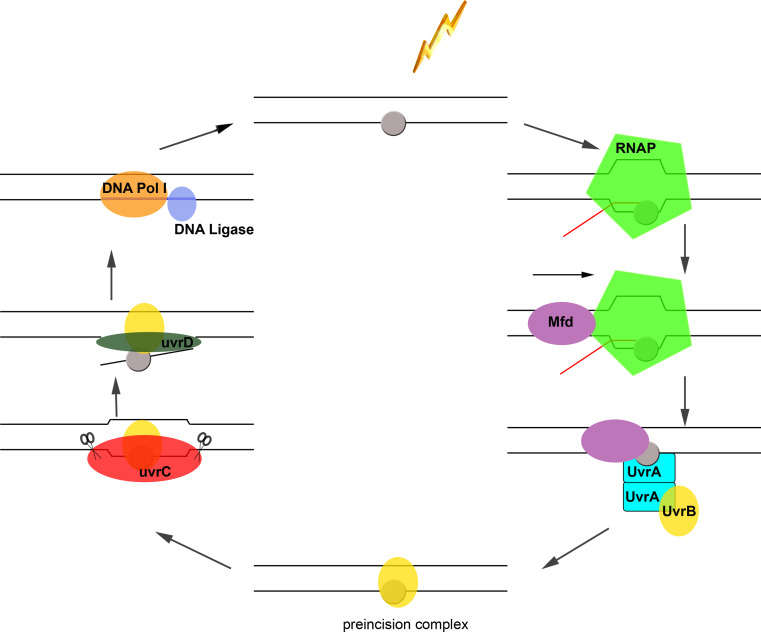
Fig. 1.
Mfd-mediated transcription-coupled DNA repair in bacteria. UV radiation (lightning bolt) induces DNA lesions (grey oval) in the template strand, which induces stalling, and potentially backtracking of elongating RNAP, bound to template DNA (black lines) and nascent transcript (red line). The stalled RNAP complex is recognized by Mfd, which binds the complex from the rear end [67] and subsequently dissociates it after several rounds of ATP hydrolysis [72], during which Mfd tracks and advances on dsDNA [67]. Mfd may then remain bound in proximity to the lesion to recruit the UvrAB complex via direct binding to UvrA. The exact timing of RNAP release within the pathway remains to be established. While the composition of the UvrAB complex recruited during both GG-NER and TC-NER has been a matter of some debate, several recent studies have reported a 2:2 stoichiometry [129, 130]. The presence of two UvrB subunits in the UvrAB complex has been suggested to be required for successive probing of damage of both DNA strands during GG-NER [131]. Due to the high structural similarity and overlapping nature of UvrB/Mfd binding to UvrA [65], it appears likely that Mfd binding may cause dissociation of one UvrB subunit. It is still unknown whether the Mfd-displaced UvrB subunit will eventually form a tight preincision complex with the DNA lesion, or whether this leaves the site so that the second copy of UvrA-bound UvrB can load onto the damaged DNA, as pictured here. UvrC then binds to UvrB to incise the DNA both 3′ and 5′ to the damage, which is followed by removal of the oligonucleotide by UvrD, gap filling and ligation by DNA polymerase I and DNA ligase [62]. The relative sizes of the nucleic acids and proteins depicted schematically do not reflect relative sizes or conformations
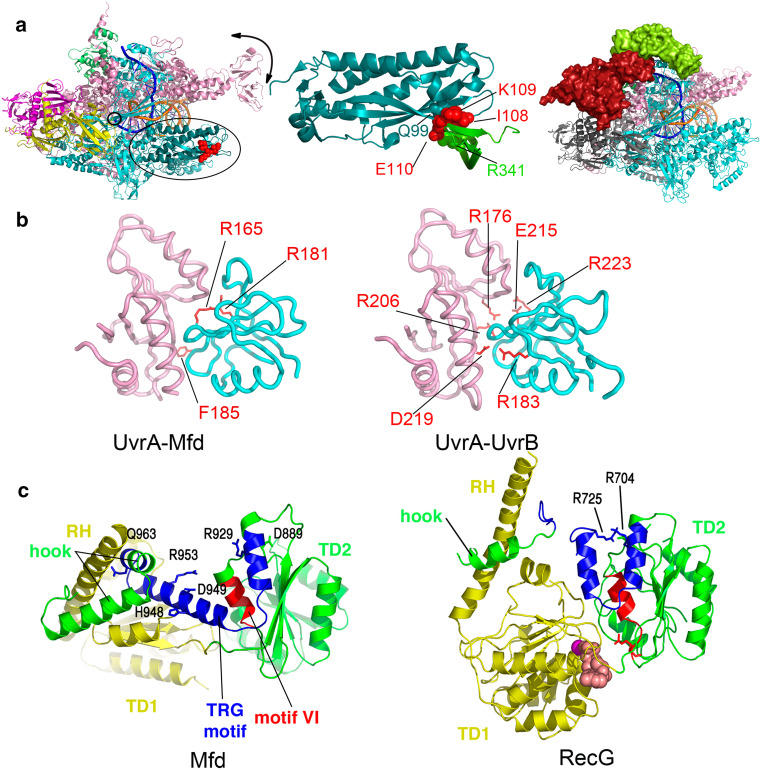
Fig. 3.
Binding modules within the bacterial TRCFs, Mfd, and Nus. a Binding of the bacterial transcription-repair coupling factors Mfd and NusA to RNAP. Overall architecture of Thermus thermophilus RNAP elongation complex (PDB ID 2O5I) is shown on the left (with RNAP subunits colored as follows: α1 and α2 in magenta and yellow, β in cyan, β’ in pink, ω in green) with the N-terminal region of the β subunit interacting with Mfd-RID colored in deep teal and indicated by an ellipse. Downstream DNA is shown as going into the page, the catalytic center is indicated by a small circle, and movement of the RNAP clamp is indicated schematically by a black arrow. The crystallographic model of Thermus sp. core Mfd-RID/RNAP-β complex (PDB ID 3MLQ) [82] is shown in the middle, while the pseudoatomic model of Bacillus subtilis NusA bound to a TEC, derived from single-particle electron microscopy studies and homology modeling [108], is shown on the right, with the NusA-NTD colored in lime and NusA-CTD, positioned lower down the β flap towards the α-subunit, colored in red. b Mfd and UvrB share a similar mode of binding to UvrA. Shown are core E. coli Mfd-UvrA (left, PDB ID 4DFC) and Geobacillus stearothermophilus UvrA-UvrB (right, PDB ID 3FPN) complexes, with residues important for the respective interaction [79, 132] highlighted as red sticks. Mfd-D2 domain (and the homologous UvrB domain) is colored cyan, and UvrA is shown in pink. c dsDNA translocation and the TRG motif in the Mfd and RecG proteins. Side-by-side views of the translocation modules of nucleotide-free E. coli Mfd (left, PDB ID 2EYQ) and ADP-bound Thermotoga maritima RecG (right, PDB ID 1GM5), illustrating the conformational changes in the relay helix, helicase motif VI (red), TRG motif (blue), and the hook. The two views are aligned such that translocase domain 2 (TD2 or D6 in Mfd, colored green throughout) of each molecule is in identical orientation. Side chains of key residues are shown. Reprinted with permission from [61]
Due to the geometry of the transcription bubble, DNA lesions in the non-template strand can generally be bypassed during transcription elongation. However, DNA lesions in the template strand have a distinct consequence: they impede progression of transcribing RNAPs to various degrees [12, 13] and give rise to asymmetry in DNA repair through preferential targeting of DNA repair machinery to the template strand via transcription-coupled nucleotide excision repair [TC-NER] [4]. Evidence in both enterobacterial [3] and mammalian genomes [4, 5] points to TC-NER as one of the transcription-associated mechanisms resulting in overall reduction of common substitutions in the template strand, such as C → T in bacteria and mammals [3], and, additionally, A → G and G → T substitutions that are frequent in the human genome [5].
That certain parts of the genome are preferentially repaired was suggested very early on by Evelyn Witkin based on her studies of radiation-induced mutagenesis in bacteria [14], as well as by Bockrath and Palmer based on their studies on repair of lesions in the gluV and gluU tRNA genes [15]. Clear experimental evidence for this phenomenon accumulated in the mid 1980s, when several studies from the Hanawalt group demonstrated that: (1) repair is faster in active transcriptional units, as for example in the DHFR gene where the rate of repair increases tenfold compared to that of the genome overall [16, 17], and (2) that within active genes, the transcribed strand is preferentially targeted for repair [18, 19]—hence the distinction between global genome nucleotide excision repair [GG-NER] and TC-NER. This newly identified form of DNA repair was soon found to be associated with human disease—certain mutations in the genes encoding the mammalian transcription-repair coupling factors [TRCFs] CSA and CSB abolished TC-NER, and gave rise to a rare segmental progeroid disorder called Cockayne syndrome [20, 21]. The etiology of the disease—characterized by cachectic dwarfism, neurological impairment, diverse developmental abnormalities, UV sensitivity, but no increased incidence of cancer—is still poorly understood. Studies have suggested that a defect in transcription rather than DNA repair may explain the diverse and systemic effects of the CSA/CSB genes on human physiology [22–24]. After all, the related disease, xeroderma pigmentosum, in which both TC- and GG-NER are abolished due to mutations in the XP genes, has surprisingly distinct manifestations—an incidence of skin cancer that is roughly three orders of magnitude higher, but no neurological dysfunction [25]. TC-NER is also impaired in the somewhat less known UV-sensitive, De Sanctis-Cacchione and cerebro-oculo-facial-skeletal syndromes [26]. The absence of detectable CSB protein or mRNA has been reported to occur in UV-sensitive syndrome, characterized by photosensitivity and mild freckling, and no neurological, developmental abnormalities or skin tumors [27]. More recently, TC-NER has been proposed to have a broader role in disease, and to shape the mutational landscape of human cancers [28].
Despite intense efforts, a clear understanding of the molecular mechanisms underlying TC-NER-associated syndromes is lacking, and the reader is referred to several recent reviews on eukaryotic TC-NER and its connection with disease [29–32], which is beyond the scope of this review. Instead, here, I focus on recent advances in understanding the molecular mechanisms underlying TC-NER and the bypass of DNA lesions by bacterial RNAP, as mediated by the two known bacterial TRCFs, Mfd and NusA. In this context, the two primary roles of these factors are to (1) recognize/remodel or dissociate the damage-stalled RNAP and (2) recruit NER machinery to the lesion via direct binding to the UvrA subunit, which, together with UvrB, senses and verifies the lesion. Once the UvrAB machinery has been recruited, TC-NER proceeds as GG-NER, with excision of the oligonucleotide containing the damage, gap filling, and ligation, as shown in Fig. 1. In addition, this review will also address the role of TRCFs in adaptive mutagenesis and in the bypass of DNA damage through transcriptional mutagenesis [TM], which is not to be confused with TAM.
DNA damage recognition “by proxy” during TC-NER
The decision for DNA damage to be channeled into the TC-NER pathway, as opposed to GG-NER is mediated by RNAP itself, which participates in a detection scheme termed “by proxy” [33]. During this process, the initial damage sensing is not mediated by dedicated DNA repair enzymes as in GG-NER such as by the bacterial UvrAB complex, but rather by RNAP itself, which acts as a proxy (Fig. 1). The lesion-stalled RNAP is inhibitory to repair as it buries the damage under its footprint and prevents the access of repair enzymes [34]. Thus, transcription-repair coupling has been proposed to be directly correlated with the extent to which transcription elongation is stalled or slowed down at DNA damage [13, 34–37].
UV-induced lesions (such as psoralen adducts and cyclobutane pyrimidine dimers) are known to be potent blocks to elongating RNAP [12]. In contrast, oxidative lesions are subtler, and many of them, such as 8-oxoguanines, do not induce transcriptional arrest [38]. One might then conclude that oxidative lesions do not elicit TC-NER. While mutations in the mfd gene (encoding the main E. coli TRCF shown in Fig. 2) reduce the rate of transcription recovery following UV-induced damage, the mfd − mutant does not exhibit increased sensitivity to hydrogen peroxide and it removes oxidative lesions from the genome to levels and with kinetics comparable to actively growing wild-type cells [39]. Thus, in vivo in actively growing E. coli cells, TC-NER does not seem to have a significant contribution to the repair of oxidative damage, although in vitro Mfd was shown to act on ternary elongation complexes [TECs] at abasic sites [40], a frequent intermediate in the repair of oxidative damage. Work from the Doetsch lab has further demonstrated that in non-dividing cells, Mfd processes also occur at 8-oxoguanines as well as uracils [41, 42], highlighting possible effects of growth phase, and the ability of RNAP to bypass lesions through transcriptional mutagenesis, described in “Lesion bypass through transcriptional mutagenesis”.
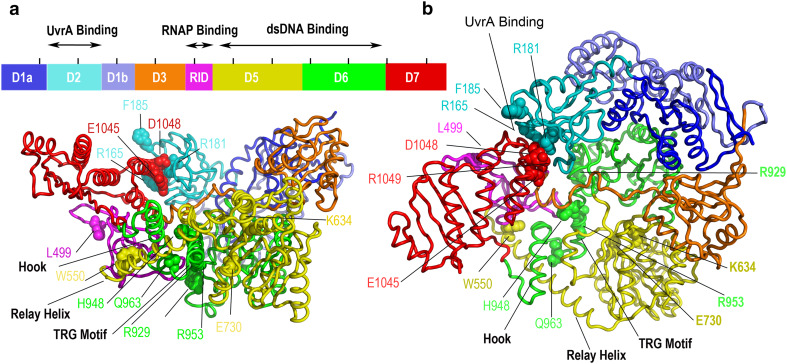
Fig. 2.
Overall architecture of the bacterial Mfd TC-NER machinery (PDB ID 2EYQ) as determined by X-ray crystallography. Nucleotide-free Escherichia coli Mfd is modular and composed of eight domains (D1a–D7) connected by flexible linkers (side view on left, top view on right). Shown as spheres are Mfd residues important for UvrA binding and its inhibition (R165, R181, F185, E1045, E1048, R1049) [65, 132], RNAP binding (L499) [61], dsDNA translocation (H948, Q963, R929, R953) [74], ATP hydrolysis (K634 within the Walker A motif, E730 within the Walker B motif) [65, 80], and the specificity of Mfd for transcription elongation complexes (W550) [75]. Black lines on top of the schematic representation of the Mfd sequence are 100-residue positional marks
Recent evidence points to the possibility that TC-NER may be initiated at RNAP stall sites even in the absence of damage. For example, in eukaryotes, triplet repeat slip-outs in the transcribed strand and R-loops stall RNAPII, and have been proposed to recruit TRCFs to initiate gratuitous TC-NER and lead to triplet instability [43–48]. Protein–DNA complexes also serve as potent RNAP elongation roadblocks and may therefore trigger TC-NER. This was first demonstrated in vitro for CPD-photolyase complexes that were found to stall (and backtrack) the transcription machinery [49], then, to various extents, for base excision repair (BER) intermediates [40, 41], and most recently for alkyltransferase-like (ATL) proteins. Unlike bona fide alkyltransferases, ATL proteins bind DNA damage, but cannot transfer alkyl groups on DNA bases due to the absence of a conserved receptor cysteine, present in alkyltransferases [50]. The yeast ATL protein Atl1 has been studied the best, including at the structural level, and was found to have differential affinity for bulky alkylating damage versus smaller O6-methylguanine [51]. These findings led to a model, according to which the high affinity for bulky damage (as opposed to smaller substrates) is such that long-lived protein-DNA intermediates formed at bulky lesions inhibit GG-NER and at the same time effectively stall TECs for lesions to be channeled into TC-NER rather than GG-NER [51]. Thus, these accessory proteins can regulate the choice between TC-NER and GG-NER. In this context, it is interesting to note that bacterial ATL proteins have also been shown to bind directly to UvrA [52], although DNA excision post UvrA recruitment has not been demonstrated yet.
While RNAP is not normally viewed as a DNA damage sensor, it nevertheless shares important features with DNA damage recognition proteins. First, elongating RNAP complexes scan the genome by virtue of their primary function in RNA polymerization. Secondly, a TEC stalled by a TT dimer has a very long in vitro half-life of ~20 h and a slow off-rate of ~1 × 10−5 s−1 [35]. As noticed before [53], this half-life exceeds even that of a high selectivity damage sensor, such as the UV-damaged-DNA-binding protein, UV-DDB [54], which promotes recognition and directs protein assembly at UV photolesions in eukaryotic NER [55]. Consequently, the arrest/prolonged pausing or “slowing down” of a TEC at a lesion provides the necessary timeframe for the recruitment of repair enzymes and assembly of TC-NER complexes at the site of damage. The assembly of TC-NER intermediates is highly complex, and at its earliest steps relies on: (1) remodeling/repositioning of the TEC, or (2) complete dissociation of the TEC. In essence, both of these mechanisms accomplish the same goal—they render the lesion more accessible to NER enzymes, although promoting accessibility through TEC remodeling appears to be more important in bacteria, as RNAPII, unlike RNAP, was not reported to be inhibitory to repair in vitro [35].
In eukaryotes, which have much longer genes requiring up to 16 h to be transcribed [56], a dissociation-based mechanism would be highly uneconomical as it would require transcription reinitiation at the promoter. As described below, while bacteria employ both types of mechanisms, in eukaryotes, TC-NER factors do not release TECs off the template (at least in vitro), but are thought to reposition them [57]. TC-NER can even occur in the presence of a stalled TEC at a DNA adduct site [58]. The order of assembly and composition of eukaryotic TC-NER intermediates is of significant complexity and still a matter of debate. By comparison, the main bacterial TC-NER pathway mediated by the Mfd protein has been fully reconstituted in vitro [59, 60] and has been described in much clearer terms, including crystallographically and at the single-molecule level.
Mfd-mediated TC-NER
Mfd (also known as TRCF) was initially described as the only factor required for reconstituting TC-NER in vitro [59]. Recent studies have reported that in fact a second bacterial TRCF, NusA, may be part of a separate pathway operating on a subclass of DNA lesions as described in “The NusA protein”.
Mfd is a large (~130-kDa) ATPase with similarity to the Swi/Snf2 chromatin remodelers and a modular architecture (Fig. 2) that integrates three activities: (1) ATP-dependent DNA translocation (2) RNAP binding, and (3) recruitment of UvrAB damage-sensing NER complex through direct binding to the UvrA protein [61–64]. E. coli Mfd remains monomeric irrespective of nucleotide status [65], although recent studies have reported that the Mycobacterium tuberculosis ortholog hexamerizes [66]. While Mfd has no helicase activity [64], it translocates on dsDNA upstream of the transcription bubble, and binds to the rear end of the stalled RNAP, where it requires about 26 bp of dsDNA [67]. In fact, TC-NER was only observed in the lac operon under conditions of low levels of transcription and not under full induction of the operon [68]. Presumably, under high transcription activity, the tightly packed arrays of elongating RNAPs do not allow Mfd to effectively bind upstream of RNAPs within the arrays.
Insightful experiments by the Roberts lab have revealed that by virtue of its dsDNA translocase activity upstream of the TEC, Mfd pushes the RNAP in the downstream direction, and that, at least in vitro, Mfd can rescue backtracked RNAP into productive elongation in such a way that release competes ineffectively with elongation. In the absence of nucleoside triphosphates [NTPs], Mfd was able to dissociate the backtracked and arrested +27 RNAP complex of the T7A1 promoter, but in the presence of both Mfd and NTPs, RNAP was able to escape release at position +27 and arrive at the terminator present in the transcription template [67]. In cases in which RNAP cannot longer advance due to structurally-distorting DNA lesions in the template, NTP starvation or even protein roadblocks ahead of the RNAP, the continued action of the Mfd translocase reanneals DNA at the upstream edge of the transcription bubble and ultimately induces bubble collapse with complete release of the nascent transcript and, ultimately, RNAP [67, 69]. This forward-translocation model is also supported by the finding that elongation complexes formed on heteroduplex templates (carrying substitutions that prevent base-pairing in the transcription bubble or in the duplex region upstream of the bubble) cannot be dissociated by Mfd [69]. Similar effects were also observed for Rho-mediated termination, suggesting that both factors may operate, albeit in different manners, by forward translocation of RNAP [69]. Unlike Mfd, Rho has helicase activity and translocates on the nascent RNA to effectively “peel” it from under the RNAP. There is currently no experimental evidence that Mfd may interact with the RNA transcript, highlighting the differences between Rho and Mfd in effecting forward translocation. Notably, full-length Mfd only possesses dsDNA translocase activity when bound to RNAP [70], although activation for dsDNA translocation, required for termination, can also be achieved in the absence of TECs by deletion of the N-terminal or C-terminal regions of Mfd [70, 71].
Recent single-molecule studies with magnetic tweezers have revealed that Mfd-mediated release of the TECs involves two distinct ATP-dependent transitions: an initial fast step in which Mfd docks onto the stalled TEC, followed by a slow step resulting in a long-lived intermediate. It has been proposed that the second intermediate is associated with profound and slow conformational changes within Mfd, and that its resolution may involve the binding of downstream factors, specifically UvrA [72]. Given this wealth of information, what is the structural mechanism utilized by Mfd in destabilizing TECs and recruiting UvrA? Although there is currently no structural information on Mfd bound to a TEC to enable a detailed dissection of RNAP recognition and destabilization, crystallographic studies of both nucleotide-free full-length E. coli Mfd [61, 73] and of a core Mfd-UvrA complex [65], SAXS analyses of Mfd in alternative nucleotide states [65], single-molecule [72] and functional studies [40, 65, 67, 70, 74–77] have revealed Mfd architecture and its conformational dynamics as described below.
UvrA recruitment and the UvrB-homology module of Mfd
Proteins of the Mfd family are modular, with domains connected by flexible linkers and interlocked mobile structural elements (Fig. 2). In E. coli, the three N-terminal domains of Mfd (D1a-D2-D1b), named after the homonymous domains of UvrB, share structural similarity with the UvrB protein, although significant sequence identity can only be found in D2 [61]. The UvrB homology module of Mfd does not bind DNA, or turn over ATP, while the homologous region of UvrB binds both DNA and nucleotide [78]. Both in Mfd and UvrB, the D2 domain was shown to bind directly to UvrA [79, 80]. A recent crystallographic study of a core Mfd-UvrA complex revealed that UvrA binds at the D2 intramolecular interface that packs against the C-terminal domain D7 in apo Mfd (Fig. 2a) [65]. Mfd and UvrB share a very similar mode of binding to UvrA, with residues that map to the UvrA-binding surface conserved both within the Mfd and between the Mfd and UvrB families [65]. Of particular importance appear to be Mfd residues R165, R181, and F185 (highlighted in Figs. 2 and 3b), which when mutated, lead to compromise of TC-NER in vivo, and a defect in UvrA binding in vitro [79]. Other currently unknown contact points between Mfd and UvrA cannot be excluded at this time, but the functional defect observed with the R165A R181A F185A triple mutant as well as earlier binding studies with Mfd truncations [80] suggest that D2 is the main UvrA interaction determinant within Mfd. Notably, two of the residues essential for the Mfd-UvrA interaction (R165 and R181) are completely buried by the D2-D7 interaction (Fig. 2), a crystallographic observation, which is also consistent with SAXS studies of Mfd in solution, under close-to-physiological conditions [65]. Unlike UvrB, in which the D2 domain is fully solvent-exposed, full-length Mfd is auto inhibited for UvrA binding. This D2-D7 autoinhibitory interaction provides an effective mechanism for preventing the sequestration of the cellular pool of UvrA, which occurs upon deleting the D7 domain of Mfd and leads to hypersensitivity to UV radiation [79, 80]. The cellular pool of UvrA is low during conditions of normal growth, but is upregulated as part of the SOS response, when levels of UvrAB rise, and therefore a faster GG-NER diminishes the importance of TC-NER [81].
Domain D3 of unknown function
The poorly conserved domain D3, located at the C-terminal end of the UvrB-homology module of Mfd has yet to be been assigned a function, and appears to be mobile during cycles of ATP turnover as shown by SAXS [65]. D3 is connected to D4 or the RNAP interaction domain (RID) via a long and flexible 25-residue linker that spans about 40 Å from one side of the molecule to the other, and whose cleavage via an engineered internal protease recognition site leads to enhanced ATPase activity, presumably due to dissolution of the D2–D7 interaction, which is inhibitory to ATP hydrolysis [71].
RNAP binding via the Tudor-like RID domain
The RID is a Tudor-like domain that binds to the N-terminal region of the β subunit of RNAP (also known as β1) via the so-called IKE motif. In E. coli, the IKE motif is composed of residues I117, K118, and E119 located in the lobe region of the β subunit (Fig. 4a). Mutations within the IKE motif as well as of L499 in Mfd (Fig. 2a) were reported to impair the RNAP–Mfd interaction in two-hybrid assays and affect the ability of Mfd to act on stalled TECs [61, 76]. As shown in Fig. 3a, the RID-β1 interaction is bipartite comprising a central interaction involving an intermolecular β-sheet, whose sequence is conserved across phyla, and a phylum-specific interaction involving residue R341 in Thermus thermophilus Mfd and Q99 in the bound Thermus aquaticus RNAP fragment [82]. Intriguingly, the Mfd-RID-RNAP-β crystal structure features a small register shift of residues 103–111 (region that harbors the key Mfd interaction motif in the T. aquaticus fragment crystallized) [82]. Because this shift has not been observed in any of the many RNAP structures available, it has been suggested that the β region might exist in a dynamic equilibrium of multiple states with Mfd trapping and stabilizing the shifted state [82]. Nevertheless, a cyclobutane pyrimidine dimer-stalled RNAPII (no such structure of bacterial RNAP is available) displays no large characteristic conformational changes [83], and Mfd can bind both TECs and core RNAP [65, 67, 80]. Moreover, a W550A Mfd mutant (Fig. 2) is able to release both transcription initiation complexes and TECs [75]. In pull-down assays, wild-type Mfd still binds a RNAP derivative in which the IKE motif was mutated, but cannot displace it either in vitro or in vivo [76]. Altogether, these data suggest that while the RID provides contacts to ensure polarity and processivity of translocation upstream of the bubble, other contacts with RNAP may exist, and these may be key to explaining TEC susceptibility to Mfd attacks.
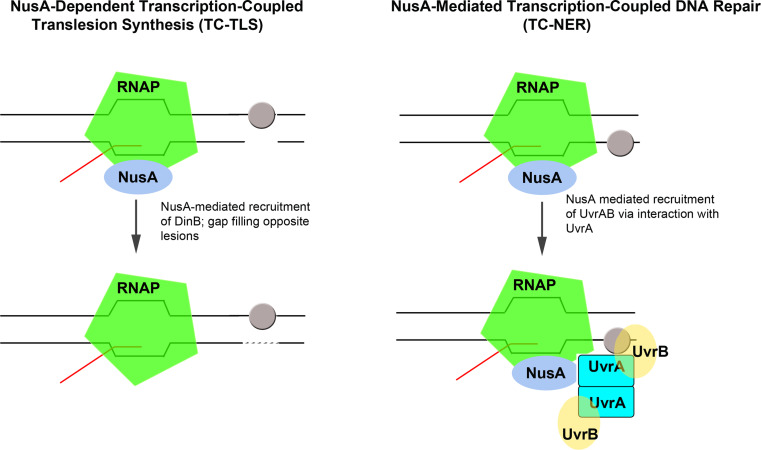
Fig. 4.
NusA-mediated transcription-coupled translesion DNA synthesis (TC-TLS, left) and transcription-coupled DNA repair (TC-NER, right). During TC-TLS, NusA, bound to RNAP recruits the Y-family DNA polymerase DinB to gaps in the DNA opposite from lesions (grey oval) in the non-transcribed strand [104]. This results in filling of the gap (interrupted line) via the action of DinB. During TC-NER, RNAP-bound NusA recruits the UvrAB complex to sites of DNA damage (grey oval), such as nitrofurazone adducts, which do not enter the active site of RNAP. The recruitment of NER machinery via the NusA-UvrA interaction has been proposed to result in excision of the damage and preferential repair of the transcribed strand [100]
Although an Mfd signature domain, the RID is not unique to Mfd. It appears to define a new structural module for binding RNAP also found in the CarD family of transcriptional regulators, which are important for mycobacterial viability and virulence [84, 85]. Both Mfd-RID and CarD-RID interact with the β lobe through similarly located, and partially overlapping set of residues, including M. tuberculosis CarD-R25 (corresponding to Thermus sp. Mfd-RID residue R341 or E. coli Mfd-RID residue L499) and RNAP-β residue E138 (corresponding to Thermus sp. RNAP-β residue Q99, shown in Fig. 3a).
DNA binding and translocation with a RecA-type motor
The RID connects to the RecA-type ATP-dependent dsDNA translocase (domains D5 and D6, harboring the seven canonical SF2 ATPase motifs) via a long α-helix termed the relay helix [RH]. This is ideally positioned to relay information from the ATP-binding site to the rest of the molecule, and couple ATP hydrolysis to translocation on dsDNA (Fig. 2). There are currently no high-resolution structures of alternative nucleotide states of Mfd, but comparison with the homologous Holliday branch migration RecG motor in its ADP-bound form has revealed a distinct conformation of the RH—straight rather than bent [61]. Other structural elements within the DNA-binding translocase appear to be also important for the mechanochemical cycle. These include the translocation in RecG [TRG] helices [61, 74, 86] as well as the hook helices that wrap around the RH and connect the translocase module to the C-terminal autoinhibitory domain D7 (Fig. 2). A W550A mutation within the RH (Fig. 2) alters the interaction with the hook helices, and presumably the interaction with upstream DNA promoting processivity, and enables Mfd to release not only stalled TECs but also transcription initiation complexes that would not otherwise support Mfd-mediated release [75]. Mutations within the TRG motif, which forms a helical hairpin C-terminal to ATPase motif VI (Figs. 2 and 3c), do not impair DNA binding per se, but are crucial for RNAP displacement, leading to a model in which this motif is essential for transducing the energy of ATP hydrolysis into mechanical work. Conserved residues R929 and R953, located at the base of the helical hairpin, both lead to impairment of RNAP displacement in vitro (with R953A mutation having a more pronounced effect), but leave the ATP hydrolysis and DNA binding functions intact; so do non-conserved residue H948 and conserved residue Q963 [74]. The functional consequences of these mutations within the TRG motif closely recapitulate the hierarchy of defects observed with similar mutants of RecG [86], further pointing to a common mechanism for dsDNA translocation by members of both the Mfd and RecG protein families. The TRG motif likely serves as a spring-loaded structural element during the ATPase cycle and is controlled by an electrostatic “switch” provided by a close interaction between R929 and R953, and a γ-phosphate sensor in ATPase motif VI. The TRG hairpin indirectly senses nucleotide status, and likely cycles between open and closed conformations to modulate binding of dsDNA across the translocase [61]. In both RecG and Mfd, the loop containing Q963 is thought to drive translocation as a swinging arm, directly contacting the DNA, although no DNA binding defect was observed for an Mfd variant carrying a substitution of Q963, located in this loop [74]. Given the complexity of Mfd structure, modeling efforts attempting to define the DNA binding regions of Mfd in detail have remained unfruitful, and therefore, more complete understanding of DNA binding and translocation will require experimental, high-resolution structural models of DNA/TEC-bound Mfd.
The autoinhibitory D7 domain
The translocation module connects through the hook and a flexible linker to the C-terminal domain D7. This domain has an autoinhibitory role with respect to UvrA recruitment due to highly conserved interactions of residues E1045, D1048, and R1049 with D2 [70, 79], as further described in “UvrA recruitment and the UvrB-homology module of Mfd”, but also with respect to the ATPase and dsDNA translocation activities. Several studies have reported that impairing the D2–D7 interaction with domain deletions or amino acid substitutions at the D2–D7 interface breaking the D2–D7 clamp lead to Mfd ATPase hyperactivity as well as the ability to translocate on naked dsDNA [70, 71]. Interestingly, a disulfide bond engineered between the D2 and D7 domains of Mfd, which prevents binding of UvrA, does not compromise transcript release from nucleotide-starved TECs [65], suggesting that simple docking of Mfd to RNAP (prior the transcript release) is not sufficient for unmasking the D2 surface. Recently, Strick and colleagues have suggested, based on single-molecule kinetic data, that several ATP turnover events and remodeling of the TEC are required for Mfd to be activated for UvrA binding [72]. Yet, the precise moment when RNAP dissociates off the template and when UvrA is recruited remain unknown. Simple energetic considerations based on previous biochemical findings suggest that the DNA-RNA hybrid plays a major role in stabilizing the TEC [87], and thus one would expect that once the transcription bubble begins to collapse and the transcript is unwound, RNAP would rapidly dissociate from the template. However, in vitro a second Mfd-TEC intermediate along the pathway, formed after the initial docking of Mfd and ATP turnover, has been observed, and in this intermediate approximately two-thirds of the transcription bubble is rewound. Surprisingly, this intermediate is unusually stable and long-lived in vitro, with a half-life of about 5 min [72]. However, given that covalently tethering D2 to D7 (to prevent UvrA binding) allows for transcript release from the Mfd-TEC complex, and presumably at least partial resolution of this intermediate, it has been suggested that during TEC destabilization, the DNA lesion may become exposed and Mfd may translocate towards it, only then to become fully activated for UvrA recruitment [65].
One major question that remains to be elucidated is how Mfd achieves specificity for stalled TECs. The available data converge on a model in which Mfd may initially not specifically recognize the stalled conformation of RNAP, but rather bind to RNAP irrespective of being stalled or not. Mfd can bind core RNAP, and when mutated, can even dislodge transcription initiation complexes in vitro [75, 80]. This model would parallel reports that other transcription termination factors, such as Rho [88], as well some transcription regulators that act through the RNAP secondary channel (GreA/B, DksA), may associate with RNAP throughout the transcription cycle [89–92]. For example, transcript cleavage factor GreB rescues arrested TECs by promoting the endonucleolytic cleavage of the nascent RNA [93], but is also implicated in regulating transcription initiation by inhibiting the production of abortive transcripts and facilitating promoter escape [94]. The recruitment of Mfd to TECs is incompletely understood, but what has been established is that Mfd appears to be activated by and can only dissociate stalled TECs, as opposed to elongating TECs. This may explain why Mfd overexpression in E. coli can be achieved with no apparent toxicity [73]. This specificity of action has been proposed to be due to a kinetic break imposed by the relative rates of translocation by Mfd and RNAP itself [70]. How forward motion of RNAP eventually results in dissociation of the TEC remains to be determined, but has been speculated to involve an opening of the RNAP clamp (Fig. 3a) [76, 77], which has been observed before, and which modulates processivity [95, 96].
The NusA protein
Despite the prominent role of the Mfd protein in TC-NER, defects in mfd only lead to a modest increase in sensitivity to UV radiation [62], and certain bacterial genomes appear to lack Mfd orthologs altogether [97]. This mild phenotype can be rationalized by the facts that Mfd may act on TEC arrays only under conditions of low transcription [68], that it may become unmasked only when UvrAB levels are low (under conditions of no SOS response induction) [81], and that alternative, partially overlapping modalities of dealing with paused/stalled RNAPs may exist. Indeed, multiple such mechanisms have been uncovered, and these utilize other factors (such as DksA, GreA) reported to be important for UV resistance and the resolving of conflicts between replication and transcription [98], but also employ cooperation between RNAP molecules, which increases transcription efficiency [99]. Moreover, a second bacterial TRCF called NusA has recently been identified based on the increased sensitivity of a NusA (nusA11) mutant to the DNA-damaging agent nitrofurazone [100]. NusA appears to operate in an Mfd-independent TC-NER pathway.
NusA is a well-known transcriptional regulator that associates with RNAP both during elongation and termination, and has specific roles in termination and anti-termination, some of which require its association with other RNAP accessory factors such as λ N and Rho [101]. Several lines of evidence, including a chronic partial SOS induction and increased number of RecA-GFP foci particularly in the stationary phase obtained with the nusA11 strain suggests that this mutant cannot cope with endogenous DNA damage [100]. NusA has been proposed to link the DNA repair and damage tolerance pathways through two different activities. On one hand, NusA recruits DinB, a Y-family polymerase to mediate transcription-coupled translesion synthesis (TC-TLS), presumably in those cases where a DNA lesion in the non-transcribed strand may lead to a large gap in the transcribed strand (Fig. 4). Such events may be attributed to the inability of replicative polymerases to bypass lesions and to replication resuming at the site of the next Okazaki fragment on the lagging strand or, alternatively, to replication restart on the leading strand [102, 103]. Such large gaps could also occur as a result of UvrA-dependent incomplete processing of closely spaced lesions on opposing strands across which RNAP cannot transcribe and stalls [104]. On the other hand, NusA also appears to recruit NER enzymes (UvrA) to sites of nitrofurazone-induced (or similar) adducts [100]. However, no direct measurements of NusA-dependent repair in the template/non-template strand have been reported yet, meaning that equating UvrA binding to actual strand-specific lesion excision needs to be done cautiously.
As a potential TRCF, NusA appear to have an activity in common with Mfd—it interacts physically with UvrA as demonstrated using proteomic methods and far-Western blotting [100, 105]. It nevertheless lacks RNAP-release activity. At this point it cannot be ruled out that this activity might require a yet unknown accessory factor. However, it may well be possible that in the case of NusA, RNAP may not need to be dissociated. Mimics of the major adduct induced by nitrofurazone are predicted to stall transcription at position −4 (ahead of the RNAP), and so do not enter the active site of RNAP [100]. This raises the question of whether and how RNAP may sense damage that lies ahead, downstream. To resolve these situations, RNAP might backtrack rather than dissociate to allow for UvrA-dependent repair by NusA [100]. The known association of NusA with RNAP α subunit-CTD and RNA exit channel (Fig. 3c) [106–108] via the NusA acidic repeat domains and its N-terminal domain [NTD], respectively, suggests that NusA may recruit UvrA at the upstream region of RNAP. However, RNAP sites of nitrofurazone resistance and sensitivity map to the leading part of RNAP, including the lineage-specific insertion βi4 (also known as dispensable region 1) [100, 109], which is located ~150 Å away from the RNA exit channel and ~125 Å away from the α-CTD. To reconcile the large distance between the known NusA contact sites in RNAP and sites of nitrofurazone resistance, it has been suggested that UvrA may be recruited via the known upstream RNAP-NusA tethers, but possibly with large accompanying structural rearrangements [100]. The UvrA interaction determinants within NusA remain unknown at this time.
Lesion bypass through transcriptional mutagenesis
At a site of DNA damage on the transcribed strand, an alternative scenario to repair via TC-NER is TM or lesion bypass via ribonucleotide misincorporation. TM provides a mechanism that modifies our largely replication-centric view of mutagenesis, and has largely been studied under non-proliferative, non-dividing conditions, which enabled separation of the effects of transcription from those of replication. Such conditions have been associated with reduced capacities for DNA repair, which therefore competes to a lower extent with TM [110]. Unlike other modes of transcription-mediated mutagenesis (such as for example damage to the transcript itself or inherent lapses in RNAP fidelity), TM is a targeted mechanism—during every bypass event, the same ribonucleotide will be inserted, resulting in a population of identical nascent transcripts, and a consequent change in the entire translational output derived from one particular transcript. Thus, TM is predicted to have a more pronounced contribution to the mutant protein level in the cell compared to the other modes of mutagenesis involving transcription, in which the mutant transcripts generated are random, and thus the level of any mutant translation product is also kept low.
Evidence for TM in vitro is now abundant, and extends to a variety of DNA lesions such as uracil (a product of spontaneous deamination of cytosine), abasic sites, dihydrouracil, 8-oxoguanines, thymine glycols, O6-methylguanine, other bulky damage such as N 6-benzo[a]pyrene diol epoxide, and even single-stranded breaks and gaps [111]. Even in the case of TT dimers, which are a potent block to elongation in vitro [12, 83, 112] and have been viewed as the principal lesion type to be repaired by TC-NER, RNAPs can actually slowly transcribe past damage to bypass it, but in an error-free, non-mutagenic manner following a base pairing rule reminiscent of the A-rule established for DNA polymerases [113]. In the case of alkylation damage and “bulky” UV-induced damage, it has generally been surmised that translocation past the lesion is much slower than usual and bypass is of low efficiency, and therefore these lesions are channeled into TC-NER. Not all bypass is mutagenic—in addition to the TT dimer case described above, RNAPII for example can insert guanine opposite uracil (in addition to adenine) [114] and adenines opposite thymine glycols [115], thus maintaining the original sequence. It must be also underscored that different RNAPs (phage, prokaryotic versus eukaryotic) respond in certain cases differently during TM by inserting distinct miscoding ribonucleotides opposite the same lesion [116]. In some cases (such as gaps and large adducts), read-through leads to small deletions [116].
In the case of more subtle DNA lesions, such as oxidative lesions, bypass by RNAP is, compared to the TT dimer, much more frequent as revealed in vitro by transcription assays, and in vivo by reporter assays based on DNA templates carrying lesions at defined positions [111]. In a luciferase-reporting system in E. coli under non-dividing conditions of novobiocin-induced inhibition of replication, RNAP was reported to bypass uracil and 8-oxoguanine in a mutagenic manner [41, 42]. Evidence for transcriptional mutagenesis in vivo in a variety of organisms has been growing [41, 116], and a recent crystallographic study has even revealed the structural basis for TM through 8-oxoguanine by S. cerevisiae RNAPII. This relies on Hoogsteen rather than Watson–Crick base-pairing between 8-oxoguanine and the inserted adenine [117].
In the E. coli bacterial system and in the case of 8-oxoguanines, TM was found to be dependent of Mfd. In an mfd − mutant, TM was elevated [41], presumably due to the absence of Mfd-mediated release of TECs arrived at the DNA lesion, and consequent bypass. In the case of uracil, the mfd − strain had wild-type levels of TM, but a double ung − mfd − strain (lacking both Mfd and the main uracil DNA glycosylase Ung) had significantly higher TM compared to an ung − strain [42]. One may conclude that Mfd does not recognize abasic intermediates that form during base excision repair [BER], as previously proposed [40, 41], but uracil, otherwise TM should be unchanged or reduced in the ung − mfd − strain because the lack of the uracil glycosylase results in a slower production of abasic sites to direct the potential recruitment of Mfd. Notably, an uvrA − mutM −strain [deficient thus in NER, but also in BER due to absence of the 8-oxoguanine glycosylase MutM] displayed a synergistic increase in TM compared to a mutM − strain, implying that 8-oxoguanines are targeted not only by BER, but also NER [42]. Moreover, mfd was not epistatic to uvrA [42], suggesting that some other repair mechanism in addition to NER, such as BER, may participate in Mfd-dependent processes. These data imply that, to some extent, there may be some flexibility in the type of machinery used through Mfd. NER may intervene in the repair of oxidative lesions in those cases when the preferred BER pathway may be overwhelmed. In the context of BER, Mfd function may be to act as a placeholder at the lesion to block incoming RNAP from bypassing the damage and allowing factor assembly [42]. Validation of this model will, however, require further study.
In toto, these new findings suggest that TM provides a way for cells to complete transcription of a gene and escape from stress, all at the expense of generating pools of aberrant transcripts with possibly altered functionalities. Under certain circumstances, these could confer a temporary growth phenotype, and could potentially allow replication, which would fix the mutation. Such mechanisms have been proposed to give rise to genetic diversity during adaptive mutagenesis and to underlie the accrual of antibiotic resistance. In Bacillus subtilis, mfd inactivation lead to a reduced number of prototrophic revertants to Met+,His+, and Leu+ during starvation [118], and in the same organism, the mutagenic effect of Mfd was reported to be restricted to the stationary phase and not to occur during exponential growth [119]. In Campylobacter jejuni, treatment with the fluoroquinolone ciprofloxacin upregulated mfd as assessed using DNA microarrays, and overexpression of Mfd elevated spontaneous mutation rates, while in contrast, in an mfd − mutant, the rate of spontaneous mutation to ciprofloxacin resistance was reduced by two orders of magnitude [120]. Altogether, these data may point to different roles of Mfd in growing and non-dividing cells. While in growing cells, Mfd is important for reducing mutagenesis and genomic instability via TC-NER and for its role in alleviating head-on collisions between DNA polymerase and RNAP [121], in non-dividing cells, Mfd may actually promote the accrual of mutations via processes that involve transcription of genes under selection [122]. Notably, unlike E. coli, Campylobacter does not possess error-prone DNA polymerases (such as Pol II, Pol IV, Pol V) to be upregulated during the SOS response elicited by DNA damage caused by ciprofloxacin [123]. While the precise mechanism behind these processes remains to be ascertained and may display differences between Gram-positive and Gram-negative bacteria, it has been speculated that TM, or alternatively, a low-fidelity form of DNA repair (employing an error-prone DNA polymerase rather than DNA Pol I) may play a role [119, 120].
As in bacteria, TM-associated phenotypic changes in eukaryotes are more pronounced in cells deficient in DNA repair, and are attenuated by repair pathways such as such as BER. This is likely due to the fact that under conditions of reduced BER, the half-life of the damage is longer, and therefore more likely to lead to a misincorporation event. In mouse embryonic fibroblasts lacking the OGG1 DNA glycosylase, TM within the HRAS reporter gene led to activation of the MAPK pathway and increased ERK phosphorylation compared to TM in cells containing OGG1, where TM within HRAS occurred, but the downstream effects of MAPK activation were not detected [124]. In addition, elimination of mismatch repair through hMsh2/hMsh6 also led to an increase in TM of 8-oxoguanine [124, 125].
Such phenotypic changes brought about by TM in multicellular organisms may be implicated in tumor development, as individual expressing hypomorphic alleles of BER genes have a higher risk of developing cancer [126] and TM has been proposed to be the mechanism through which driver mutations within the p53 genes appear and lead to tumor development [116]. In addition, TM has also been implicated in neurodegenerative diseases through the generations of prions and protein aggregates [127, 128].
Concluding remarks
Despite recent progress, much work remains to be done on the mechanisms underlying eukaryotic TC-NER. In contrast, bacterial Mfd-dependent TC-NER is currently well understood, and yet, even in this simpler system, important outstanding questions remain to be answered. What kind of conformational changes does RNAP binding trigger in Mfd to tune its various activities and enable dissociation, and vice versa? What are the precise steps leading to formation of the preincision complex during TC-NER? What is the precise role of Mfd in adaptive mutagenesis? What is the nature of the connection between Mfd and BER? In addition, the targeting of the NER enzymes, not only to sites of RNAP stalling, but generally to distinct classes of lesions, appears to be complex, with multiple and structurally diverse factors recruiting the same UvrA subunit of the NER machinery. The targeting of DNA repair enzymes and DNA damage detection will thus continue to be a fertile area of investigation.
Acknowledgments
The author would like to thank Dr. N. Grigorieff and the Damon Runyon Cancer Research Foundation (DRG-1966-08) for support, Dr. Peter Lewis for the coordinates of the NusA-TEC model, and Dr. R. Edayathumangalam for critical reading of the manuscript.
Abbreviations
- TC-NER
Transcription-coupled nucleotide excision repair
- GG-NER
Global genome nucleotide excision repair
- RNAP
RNA polymerase
- TM
Transcriptional mutagenesis
- TEC
Transcription elongation complex
- SAXS
Small-angle X-ray scattering
- TRCF
Transcription-repair coupling factor
- TRG
Translocation in RecG
- RH
Relay helix
- TAM
Transcription-associated mutagenesis
- NTD
N-terminal domain
- CTD
C-terminal domain
References
- 1.Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 2.Marsolier-Kergoat MC, Goldar A. DNA replication induces compositional biases in yeast. Mol Biol Evol. 2012;29:893–904. doi: 10.1093/molbev/msr240. [DOI] [PubMed] [Google Scholar]
- 3.Francino MP, et al. Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science. 1996;272:107–109. doi: 10.1126/science.272.5258.107. [DOI] [PubMed] [Google Scholar]
- 4.Green P, et al. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 5.Mugal CF, et al. Transcription-induced mutational strand bias and its effect on substitution rates in human genes. Mol Biol Evol. 2009;26:131–142. doi: 10.1093/molbev/msn245. [DOI] [PubMed] [Google Scholar]
- 6.Huvet M, et al. Human gene organization driven by the coordination of replication and transcription. Genome Res. 2007;17:1278–1285. doi: 10.1101/gr.6533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beletskii A, Bhagwat AS. Transcription-induced mutations: increase in C to T mutations in the non-transcribed strand during transcription in Escherichia coli . Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekine S, et al. Structural basis of transcription by bacterial and eukaryotic RNA polymerases. Curr Opin Struct Biol. 2012;22:110–118. doi: 10.1016/j.sbi.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Francino MP, Ochman H. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol Biol Evol. 2001;18:1147–1150. doi: 10.1093/oxfordjournals.molbev.a003888. [DOI] [PubMed] [Google Scholar]
- 10.Beletskii A, et al. Mutations induced by bacteriophage T7 RNA polymerase and their effects on the composition of the T7 genome. J Mol Biol. 2000;300:1057–1065. doi: 10.1006/jmbi.2000.3944. [DOI] [PubMed] [Google Scholar]
- 11.Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tornaletti S, Hanawalt PC. Effect of DNA lesions on transcription elongation. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 13.Scicchitano DA, et al. Transcription and DNA adducts: what happens when the message gets cut off? DNA Repair (Amst) 2004;3:1537–1548. doi: 10.1016/j.dnarep.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Witkin EM. Radiation-induced mutations and their repair. Science. 1966;152:1345–1353. doi: 10.1126/science.152.3727.1345. [DOI] [PubMed] [Google Scholar]
- 15.Bockrath RC, Palmer JE. Differential repair of premutational UV-lesions at tRNA genes in E. coli . Mol Gen Genet. 1977;156:133–140. doi: 10.1007/BF00283485. [DOI] [PubMed] [Google Scholar]
- 16.Mullenders LH, et al. Preferential repair of nuclear matrix associated DNA in xeroderma pigmentosum complementation group C. Mutat Res. 1984;141:75–82. doi: 10.1016/0165-7992(84)90014-9. [DOI] [PubMed] [Google Scholar]
- 17.Bohr VA, et al. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 18.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 19.Mellon I, et al. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 20.van Hoffen A, et al. Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orren DK, et al. The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res. 1996;24:3317–3322. doi: 10.1093/nar/24.17.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proietti-De-Santis L, et al. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25:1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balajee AS, et al. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet. 2012;44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 27.Horibata K, et al. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci USA. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 30.Frontini M, Proietti-De-Santis L. Interaction between the Cockayne syndrome B and p53 proteins: implications for aging. Aging. 2012;4:89–97. doi: 10.18632/aging.100439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nouspikel T. DNA repair in mammalian cells : nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;6:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nouspikel T. Nucleotide excision repair and neurological diseases. DNA Repair (Amst) 2008;7:1155–1167. doi: 10.1016/j.dnarep.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Sancar A, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 34.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 35.Selby CP, et al. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 37.Tornaletti S, et al. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem Res Toxicol. 2006;19:1215–1220. doi: 10.1021/tx060103g. [DOI] [PubMed] [Google Scholar]
- 38.Tornaletti S, et al. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Schalow BJ, et al. Mfd is required for rapid recovery of transcription following UV-induced DNA damage but not oxidative DNA damage in Escherichia coli . J Bacteriol. 2012;194:2637–2645. doi: 10.1128/JB.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith AJ, Savery NJ. Effects of the bacterial transcription-repair coupling factor during transcription of DNA containing non-bulky lesions. DNA Repair (Amst) 2008;7:1670–1679. doi: 10.1016/j.dnarep.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Bregeon D, et al. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli . Mol Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 42.Clauson CL, et al. Dynamic flexibility of DNA repair pathways in growth-arrested Escherichia coli . DNA Repair (Amst) 2010;9:842–847. doi: 10.1016/j.dnarep.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salinas-Rios V, et al. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011;39:7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belotserkovskii BP, Hanawalt PC. Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys J. 2011;100:675–684. doi: 10.1016/j.bpj.2010.12.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bentin T, et al. Transcription arrest caused by long nascent RNA chains. Biochim Biophys Acta. 2005;1727:97–105. doi: 10.1016/j.bbaexp.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Belotserkovskii BP, et al. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci USA. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krasilnikova MM, et al. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tornaletti S, et al. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J Biol Chem. 1999;274:24124–24130. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margison GP, et al. Alkyltransferase-like proteins. DNA Repair (Amst) 2007;6:1222–1228. doi: 10.1016/j.dnarep.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 51.Latypov VF, et al. Atl1 regulates choice between global genome and transcription-coupled repair of O(6)-alkylguanines. Mol Cell. 2012;47:50–60. doi: 10.1016/j.molcel.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazon G, et al. The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O(6)-alkylguanine adducts in E. coli . DNA Repair (Amst) 2009;8:697–703. doi: 10.1016/j.dnarep.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 53.Lindsey-Boltz LA, Sancar A. RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Proc Natl Acad Sci USA. 2007;104:13213–13214. doi: 10.1073/pnas.0706316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reardon JT, et al. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: t[c, s]T, T[t, s]T, T[6-4]T, and T[Dewar]T. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 55.Sugasawa K. UV-DDB: a molecular machine linking DNA repair with ubiquitination. DNA Repair (Amst) 2009;8:969–972. doi: 10.1016/j.dnarep.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 56.Tennyson CN, et al. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 57.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 58.Tremeau-Bravard A, et al. Fate of RNA polymerase II stalled at a cisplatin lesion. J Biol Chem. 2004;279:7751–7759. doi: 10.1074/jbc.M309853200. [DOI] [PubMed] [Google Scholar]
- 59.Selby CP, et al. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selby CP, Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci USA. 1991;88:8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 62.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 63.Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 65.Deaconescu AM, et al. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc Natl Acad Sci USA. 2012;109:3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prabha S, et al. Distinct properties of hexameric but functionally conserved Mycobacterium tuberculosis transcription-repair coupling factor. PLoS One. 2011;6:e19131. doi: 10.1371/journal.pone.0019131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park J-S, et al. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 68.Kunala S, Brash DE. Intragenic domains of strand-specific repair in Escherichia coli . J Mol Biol. 1995;246:264–272. doi: 10.1006/jmbi.1994.0082. [DOI] [PubMed] [Google Scholar]
- 69.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith AJ, et al. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy MN, et al. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res. 2009;37:6042–6053. doi: 10.1093/nar/gkp680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Howan K, et al. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490:431–434. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deaconescu AM, Darst SA. Crystallization and preliminary structure determination of Escherichia coli Mfd, the transcription-repair coupling factor. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:1062–1064. doi: 10.1107/S1744309105035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chambers AL, et al. A DNA translocation motif in the bacterial transcription–repair coupling factor, Mfd. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith AJ, et al. Multipartite control of the DNA translocase, Mfd. Nucleic Acids Res. 2012;40:10408–10416. doi: 10.1093/nar/gks775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–764. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deaconescu AM, et al. Interplay of DNA repair with transcription: from structures to mechanisms. Trends Biochem Sci. 2012;37:543–552. doi: 10.1016/j.tibs.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Assenmacher N, et al. Structural basis for transcription-coupled repair: the N terminus of Mfd resembles UvrB with degenerate ATPase motifs. J Mol Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 79.Manelyte L, et al. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40:714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 81.Crowley DJ, Hanawalt PC. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6–4 photoproducts, in UV-irradiated Escherichia coli . J Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Westblade LF, et al. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brueckner F, et al. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 84.Stallings CL, et al. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weiss LA, et al. Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J Bacteriol. 2012;194:5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mahdi AA, et al. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sidorenkov I, et al. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 88.Epshtein V, et al. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nickels BE, Hochschild A. Regulation of RNA polymerase through the secondary channel. Cell. 2004;118:281–284. doi: 10.1016/j.cell.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 90.Furman R, et al. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012;40:3392–3402. doi: 10.1093/nar/gkr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rutherford ST, et al. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rutherford ST, et al. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sosunova E, et al. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hsu LH, et al. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sevostyanova A, et al. The beta subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weixlbaumer A, et al. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roberts J, Park JS. Mfd, the bacterial transcription repair coupling factor: translocation, repair and termination. Curr Opin Microbiol. 2004;7:120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 98.Trautinger BW, et al. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 99.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 100.Cohen SE, et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli . Proc Natl Acad Sci USA. 2010;107:15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang X, Lewis PJ. The interaction between RNA polymerase and the elongation factor NusA. RNA Biol. 2010;7:272–275. doi: 10.4161/rna.7.3.12021. [DOI] [PubMed] [Google Scholar]
- 102.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 103.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 104.Cohen SE, et al. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli . J Bacteriol. 2009;191:665–672. doi: 10.1128/JB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli . Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 106.Mah TF, et al. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–2675. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shankar S, et al. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol Cell. 2007;27:914–927. doi: 10.1016/j.molcel.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang X, et al. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 2009;10:997–1002. doi: 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Opalka N, et al. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8:e1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu G, et al. Base excision repair, aging and health span. Mech Ageing Dev. 2008;129:366–382. doi: 10.1016/j.mad.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Doetsch PW. Translesion synthesis by RNA polymerases: occurrence and biological implications for transcriptional mutagenesis. Mutat Res. 2002;510:131–140. doi: 10.1016/s0027-5107(02)00258-0. [DOI] [PubMed] [Google Scholar]
- 112.Donahue BA, et al. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Walmacq C, et al. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol Cell. 2012;46:18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuraoka I, et al. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J Biol Chem. 2003;278:7294–7299. doi: 10.1074/jbc.M208102200. [DOI] [PubMed] [Google Scholar]
- 115.Charlet-Berguerand N, et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bregeon D, Doetsch PW. Transcriptional mutagenesis: causes and involvement in tumour development. Nat Rev Cancer. 2011;11:218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Damsma GE, Cramer P. Molecular basis of transcriptional mutagenesis at 8-oxoguanine. J Biol Chem. 2009;284:31658–31663. doi: 10.1074/jbc.M109.022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ross C, et al. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis . J Bacteriol. 2006;188:7512–7520. doi: 10.1128/JB.00980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Robleto EA, et al. Mfd and transcriptional derepression cause genetic diversity in Bacillus subtilis . Front Biosci (Elite Ed) 2012;4:1246–1254. doi: 10.2741/455. [DOI] [PubMed] [Google Scholar]
- 120.Han J, et al. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni . PLoS Pathog. 2008;4:e1000083. doi: 10.1371/journal.ppat.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martin HA, et al. Transcriptional de-repression and Mfd are mutagenic in stressed Bacillus subtilis cells. J Mol Microbiol Biotechnol. 2011;21:45–58. doi: 10.1159/000332751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parkhill J, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 124.Saxowsky TT, et al. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci USA. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bregeon D, et al. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5:e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maynard S, et al. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.El-Agnaf OM, et al. Aggregates from mutant and wild-type alpha-synuclein proteins and NAC peptide induce apoptotic cell death in human neuroblastoma cells by formation of beta-sheet and amyloid-like filaments. FEBS Lett. 1998;440:71–75. doi: 10.1016/s0014-5793(98)01418-5. [DOI] [PubMed] [Google Scholar]
- 128.van Leeuwen FW, Hol EM. Molecular misreading of genes in Down syndrome as a model for the Alzheimer type of neurodegeneration. J Neural Transm Suppl. 1999;57:137–159. doi: 10.1007/978-3-7091-6380-1_9. [DOI] [PubMed] [Google Scholar]
- 129.Pakotiprapha D, et al. Structure and mechanism of the UvrA-UvrB DNA damage sensor. Nat Struct Mol Biol. 2012;19:291–298. doi: 10.1038/nsmb.2240. [DOI] [PubMed] [Google Scholar]
- 130.Pakotiprapha D, Jeruzalmi D. Small-angle X-ray scattering reveals architecture and A(2)B(2) stoichiometry of the UvrA-UvrB DNA damage sensor. Proteins. 2013;81:132–139. doi: 10.1002/prot.24170. [DOI] [PubMed] [Google Scholar]
- 131.Verhoeven EE, et al. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J. 2002;21:4196–4205. doi: 10.1093/emboj/cdf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pakotiprapha D, et al. A structural model for the damage-sensing complex in bacterial nucleotide excision repair. J Biol Chem. 2009;284:12837–12844. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]