Abstract
Neurons using gamma-aminobutyric acid (GABA) as their neurotransmitter are the main inhibitory neurons in the mature central nervous system (CNS) and show great variation in their form and function. GABAergic neurons are produced in all of the main domains of the CNS, where they develop from discrete regions of the neuroepithelium. Here, we review the gene expression and regulatory mechanisms controlling the main steps of GABAergic neuron development: early patterning of the proliferative neuroepithelium, production of postmitotic neural precursors, establishment of their identity and migration. By comparing the molecular regulation of these events across CNS, we broadly identify three regions utilizing distinct molecular toolkits for GABAergic fate determination: telencephalon–anterior diencephalon (DLX2 type), posterior diencephalon–midbrain (GATA2 type) and hindbrain–spinal cord (PTF1A and TAL1 types). Similarities and differences in the molecular regulatory mechanisms reveal the core determinants of a GABAergic neuron as well as provide insights into generation of the vast diversity of these neurons.
Keywords: Central nervous system, GABAergic neuron, Cell differentiation, Neurogenesis, Transcription factor
Introduction: steps of neuronal fate specification during development
Neuronal fate specification during development is a multi-step process through a gradual restriction in cellular competence, culminating in the establishment and maintenance of a defined phenotype (Fig. 1). The earliest step restricting cellular competence during development is patterning of the proliferative neuroepithelial cells: exposure of multipotent progenitor cells to regional patterning molecules secreted by adjacent tissues and local organizers in the developing neural tube [1, 2]. Patterning cues trigger an array of downstream signaling events that are thought to manifest in a combinatorial expression of multitude of downstream homeodomain (HD) transcription factors (TFs), giving cells unique identities depending on their exact spatio-temporal position in the neural tube. Patterning genes establish and maintain domain borders in the CNS, often by mutual cross-repression. Loss of patterning gene function is accompanied with a loss of domain-specific identity, which is often compensated by a respective expansion of adjacent domains.
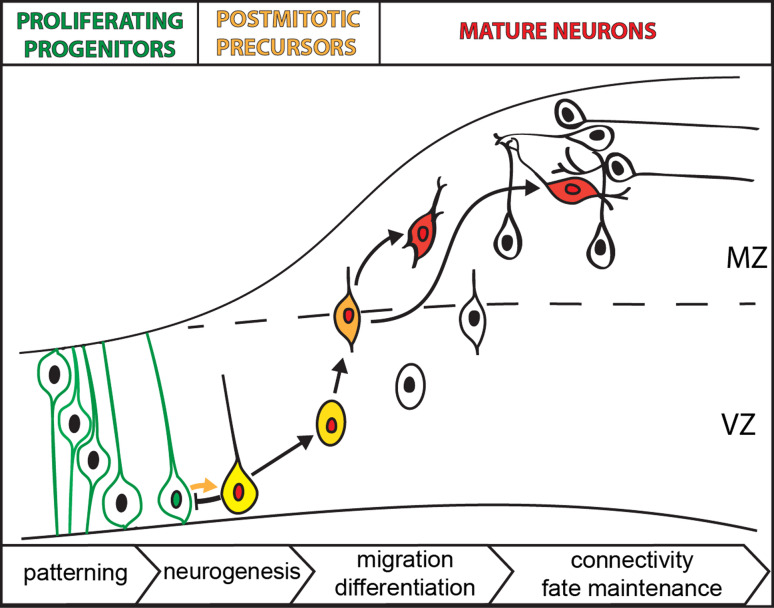
Fig. 1.
Steps of neuronal development. Proliferative, multipotent progenitors reside in the ventricular zone (VZ) where they get a regional identity (patterning) by intercellular signals. Progenitors undergo neurogenic divisions, generating both proliferative progenitor cells (green) and newly born postmitotic precursors (yellow, red nucleus). The lateral inhibition process prevents differentiation of adjacent progenitors. Postmitotic precursors (orange) move out of VZ to mantle zone (MZ) while undergoing terminal differentiation process (red). Once out of the VZ, the precursors may tangentially migrate to their final positions and they establish contacts with their targets. In contrast to this simplified scheme presented here, additional progenitor populations exist in some brain regions such as the forebrain
At the time of neurogenesis—exit of postmitotic precursor cells from the ventricular zone (VZ) neuroepithelium—basic helix-loop-helix (bHLH) genes are central to the timing (and nature) of the execution of differentiation programme in competent progenitors [3] (Fig. 2). bHLH gene activity follows the HD TF expression borders and progenitor cells fated for the same cell type typically express the same bHLH TFs. Such neuronal cell type-specific bHLH factors fall into so-called proneural gene families, showing molecular and functional similarity to Drosophila proneural genes [4]. Antagonism between different bHLH factors is evident in some regions of the developing CNS, but the mechanistic basis of cross-repression between proneural genes has not been demonstrated. The borders of proneural gene expression domains may also be maintained either by patterning genes or by dedicated bHLH or HD TFs that can be widely coexpressed with a particular proneural gene and repress the alternative proneural factors. Recent research shows that HD proteins can act in synergy with proneural proteins in neuronal progenitors to promote a commitment to a particular fate [5, 6]. Fate priming effect could be manifested for example via cross-talk with Notch pathway, chromatin remodeling factors or shifting of proneural gene activity towards cell cycle exit-promoting targets [7, 8]. Direct protein–protein interactions between proneural and HD TF have been shown to facilitate DNA binding and mediate activation of specific target genes [5, 6]. Such interactions provide a direct link between a HD code, proneural genes, and neuronal fate specification in the neural tube.
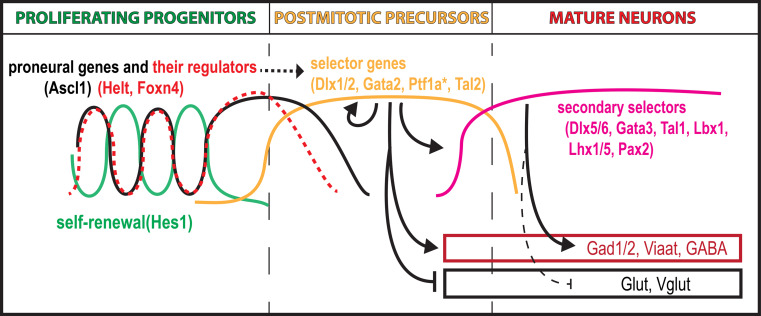
Fig. 2.
Gene regulatory mechanisms underlying GABAergic fate specification. The graphs represent the expression level of key TFs during the course of GABAergic neuron development. In the proliferative progenitor cells, the expression levels of proneural genes and their regulators oscillate with the phase of the cycle opposite to the self-renewal promoting genes (such as Hes1). Cell cycle exit occurs at the high expression phase of proneural genes. At cell cycle exit, proneural gene expression is briefly stabilized and self-renewal genes silenced. At the same time, the expression of primary fate selectors is induced. Primary fate selectors positively autoregulate their own expression and induce subtype-specific genes as well as downstream TFs (secondary selector genes) in the neuronal precursors. As neurons mature, primary fate selector gene expression often ceases while the regulation of subtype-specific genes may be carried out by the secondary selector genes. The asterisk indicates that, unlike the dorsal spinal cord where Ptf1a induction is observed at cell cycle exit, Ptf1a is expressed already at the progenitor stage in the cerebellar ventricular zone
Exposure to an appropriate set of patterning cues and proneural activity, however, does not impose an imperative differentiation regime on neuronal progenitors. Several observations point towards a crucial function of postmitotic selector genes in the control of the identity of neuronal precursors as they move into the mantle zone (MZ). First, although the proliferative neural progenitors already contain specific TF codes, production of definitive neuronal components, including subtype-specific gene products, is activated only at the cell cycle exit. Second, there exist bi- or multipotent progenitor domains that give rise to different types of neurons in parallel or sequentially. Third, inactivation or ectopic expression of certain TFs in newly born neurons can trigger commitment to an alternative fate. For example, GABAergic to glutamatergic fate switch has been observed in striatal precursors upon misexpression of the zinc finger (ZF) TF FEZF2 [9], which normally determines specific subtype identities in cortical glutamatergic neurons and diencephalic identity [10, 11]. Similar examples of TFs driving GABAergic differentiation have also been identified (see below). Thus, the definitive fate specification can occur at the time of or shortly after the neurogenic cell cycle exit. In this step, selector and terminal differentiation genes activate so-called differentiation cassettes—sets of genes allowing the neuron to exhibit its fate-specific function [12] (Fig. 2). Loss or gain of a selector gene function can lead to activation of an alternative differentiation cassette, regardless of the cells’ patterning history. Therefore, this scenario will not affect numbers of postmitotic neural precursors, but alters their cellular subtype composition, function, and connectivity.
The neuronal fate also requires active maintenance [13]. Transcriptional regulatory mechanisms controlling neuronal differentiation often retain activity also in the mature neurons. Recent studies have demonstrated cases of continuous requirement of developmental regulators for the expression of neuron-type-specific genes postnatally [14, 15].
The TFs defining specific progenitor domains, the proneural genes expressed in these domains, and the domain-specific neuronal progeny have been depicted for nearly all brain regions: telencephalon [16, 17], diencephalon [18–20], midbrain [21, 22], rhombomere 1 [23–25], other rhombomeres [26, 27], and spinal cord [28, 29]. In this review, we aim to unite this understanding to provide an overview of both shared and unique mechanisms of GABAergic neurogenesis in the different domains of the developing mammalian CNS (Fig. 3a).
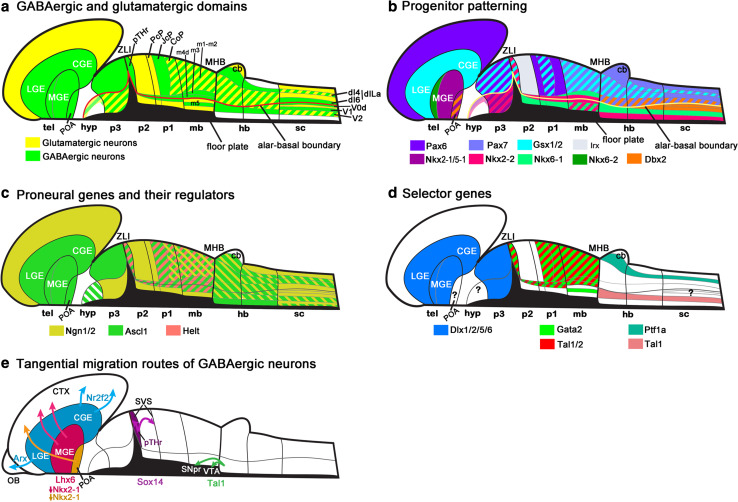
Fig. 3.
GABAergic and glutamatergic domains and their gene-expression in the developing CNS. a Domains producing GABAergic and glutamatergic neurons. b Expression of TFs responsible for patterning of the proliferating progenitors. c Expression of proneural genes and their regulators, which control several aspects of neurogenesis. d Requirements for selector genes in the differentiation of the postmitotic GABAergic precursors. Note that the domain of selector gene expression can be larger than the domain of its function. For example, Gata2 is expressed in the ventral hindbrain, but not required for GABAergic fate selection there. Some of the GABAergic domains have not been associated with a selector gene function to date (labeled with question marks). e Known tangential migration routes and the TFs controlling the migratory behavior in GABAergic cells
Patterning of the neuroepithelial GABAergic progenitor populations
Early brain patterning
The early neural tube compartmentalizes into five large domains along anterior–posterior (AP) axis: secondary prosencephalon, diencephalon, mesencephalon or midbrain, rhombencephalon or hindbrain, and spinal cord. Molecular fingerprints underlying AP patterning of the CNS are established by extracellular signals, such as Wnts, fibroblast growth factors, and their inhibitors, from the organizer tissues both outside and within the neural tube [1, 30, 31]. Early brain domains are further divided into subdomains called neuromeres. A considerable body of fate-mapping studies have now been integrated into a neuromeric model, linking molecular AP coordinates of progenitor cells in a neural tube with their neuronal derivatives in the adult/mature CNS [32–34].
Throughout neuromeres, an array of dorso-ventral (DV) domains can be distinguished. These domains are also established by spatio-temporally graded extracellular signals, such as ventral Sonic hedgehog (SHH) and dorsal bone morphogenetic protein, whose activity is translated into a well-described HD TF code defining distinct progenitor populations [2, 35, 36]. The DV patterning is best understood in the spinal cord, but the same mechanisms appear to operate in the other parts of the CNS.
GABAergic progenitor domains in the telencephalon
In the rodent telencephalon, the alar subdivision of the secondary prosencephalon, a sharp boundary (pallial–subpallial boundary) separates mutually exclusive domains of GABAergic and glutamatergic neurogenesis [37]. GABAergic neurons are generated from the subpallial neuroepithelium that comprises morphologically and molecularly distinct domains, the lateral, medial, and caudal ganglionic eminences (LGE, MGE, CGE) and the preoptic area (POA) [38] (Fig. 3a). Furthermore, at least 18 molecularly distinct subdomains have been identified in the subpallium [16]. Subpallial positions, and thus the respective HD TF-expression profiles of GABAergic neuron progenitors, have been associated with co-neurotransmitter and calcium-binding protein expression in the respective progeny, their target position in cortical layers, striatum, or hippocampus, and electrophysiological properties [31, 39–41].
While the TF code downstream of SHH is useful for determining the spatial identity of the domains of interest, there is little evidence supporting their direct involvement in the GABAergic fate determination in telencephalon. An exception to this is Nkx2-1, which is a patterning gene and seems to be involved in tangentially migrating interneuron subtype specification in the MGE and POA of the subpallium [42]. In Nkx2-1 mutant mice, MGE territory transforms into LGE-like neuroepithelium [42] and fails to produce parvalbumin and somatostatin (SST)-positive cortical interneurons [39], as well as SST and neuropeptide Y (NPY)-positive GABAergic neurons of the hippocampus [43]. Recently, POA was shown to contribute SST, PV, reelin, nitric oxide synthase, NPY, and vasoactive intestinal peptide (VIP)-producing GABAergic interneurons in deep layers of cortex [44, 45]. POA progenitors are characterized by Dbx1, Nkx5-1, and Nkx2-1 coexpression. Genetic fate mapping with Dbx1-cre mice showed that migrating POA-derived cells produce NKX2-1 [44], which regulates their tangential migration pattern (see below). In contrast, it has not been demonstrated that NKX2-1 directly regulates genes associated with GABAergic neurotransmission.
Other TFs are also expressed in telencephalic GABAergic progenitors and are more clearly associated with fate-restriction functions (Fig. 3b). This seems to be achieved via suppression of factors promoting alternative fates. HD TFs Gsx1/2 are expressed in all subpallial domains, POA and prosomere 3 (p3, prethalamus) of the diencephalon [16, 46, 47]. GSX2 is involved in defining the pallial–subpallial boundary by inhibiting expression of pallial TFs and specifying the LGE identity [48]. In the Gsx2 mutant, a dual identity switch is observed in the LGE territory. The dorsal LGE acquires a pallium-like phenotype, accompanied by the ventral shift of the pallial–subpallial boundary [49]. As a result, expression of GABAergic markers (Ascl1, Dlx1/2, and Gad1) is lost and pallial glutamatergic markers (Pax6, Ngn2 and Tbr2) upregulated in the dorsal LGE [17, 48, 50]. In contrast, ventral LGE of Gsx1/2 mutants acquires properties similar to the MGE, upregulating Lhx6 and Nkx2-1 expression [17]. Overexpression of either Gsx1 or Gsx2 in dorsal telencephalon induces ectopic expression of GABAergic fate determinants (Ascl1, Dlx1/2) at the expense of glutamatergic fate (Pax6, Tbr1 expression). Interestingly, GSX1 and GSX2 had an opposite impact on neurogenesis, while GSX1 promoted differentiation and GSX2 overexpression suppressed neurogenesis [51, 52]. Thus, GSX1/2 may control both GABAergic progenitor identity and contribute to timing of neurogenesis. In conclusion, it seems that NKX2-1 generally specifies tangentially migrating cortical interneuron progenitor identities in the MGE, CGE, and POA, while NKX2-1-negative cells in LGE remain flexible in their migration patterns. In the LGE (and likely CGE) GSX1/2 promotes commitment to GABAergic fate by suppressing pallial glutamatergic development but also regulates development of regionally specified subpopulations of GABAergic neurons.
In rodents, as discussed above, the regions producing glutamatergic and GABAergic neurons are largely segregated to dorsal (pallial) and ventral (subpallial) regions. In contrast, in primates, the pallial region also produces GABAergic neurons [53, 54]. These neurons appear to have characteristics, such as localization and co-neurotransmitter expression, which are distinct from the GABAergic neurons migrating from the ventral subpallial domains. In humans, up to two-thirds of the cortical GABAergic neurons were estimated to be derived from the dorsal neuroepithelium. In other regions of the brain, such as the midbrain, hindbrain, and spinal cord, the dorsal neuroepithelium gives rise to mixed populations of GABAergic and glutamatergic neurons. Therefore, by the production of GABAergic neurons in the dorsal telencephalon, primate species may realize an existing binary competence in the pallium. Furthermore, restriction to glutamatergic fate does not appear absolute in the rodent pallium either, as dorsal pallial progenitors also generate a specific subset of GABAergic interneurons migrating to the olfactory bulb (OB) [55, 56].
GABAergic progenitor domains in the diencephalon and midbrain
The developing diencephalon is divided into three main domains, prosomeres p3 (prethalamus), p2 (thalamus), and p1 (pretectum), which are further subdivided into smaller units. In the alar region, p1 gives rise to three (precommissural, juxtacommissural, and commissural pretectum; PcP, JcP, and CoP) and p2 to two (rostral and caudal thalamus; pTHr and pTHc) subdomains differing in their gene expression and neural derivatives [18, 57]. The patterning of the developing diencephalon is refined by Zona limitans intrathalamica (ZLI), a signaling center between p3 and p2, which secretes SHH, as well as other signaling molecules. Several diencephalic regions give rise to GABAergic neurons (Fig. 3a). The neuroepithelium of p3 molecularly resembles the GABAergic domains of anterior forebrain expressing Gsx, Dlx, and Arx genes. In addition, FEZF1 and FEZF2 are required for the patterning of p3 by counteracting thalamic (p2) identity, and for proper formation of the organizer ZLI between p3 and p2 [58]. In the alar p2 and p1, GABAergic neurons are derived from the pTHr, JcP, and CoP regions. These express SHH regulated HD TFs, such as Nkx2-2, as well as Pax6 and Pax7, which define regional identities (Fig. 3b) [59]. Gsx1 is expressed in the pTHr, but not required for development of this domain. In addition, throughout the basal p1 and p2, a small parabasal region gives rise to GABAergic neurons ([60], Allen Developing Mouse Brain Atlas, http://developingmouse.brain-map.org).
Several molecularly distinct GABAergic progenitor domains have been identified at different DV levels of the midbrain, as defined by expression of HD TFs, such as Nkx6-1 and Nkx2-2 [21, 22, 24]. The most dorsal regions of the midbrain and the CoP of the diencephalic p1 give rise to intermingled populations of GABAergic and glutamatergic neurons.
GABAergic progenitor domains in the hindbrain and spinal cord
In the hindbrain and spinal cord there are many subgroups of GABAergic progenitors expressing different TFs responsible for their patterning (Fig. 3b). DV patterning divides the spinal cord into 11 progenitor populations (dorsal dP1–dP6 and ventral vP0–vP2, vMN and vP3), which produce distinct types of neurons. In addition to the spatial patterning, the progenitors may change over time as the timing of postmitotic differentiation from the progenitor domain also affects the phenotype of the neurons [61]. However, in the GABAergic neuron lineages, the mechanisms of this temporal patterning are not known.
In the ventral spinal cord, vP0, vP1, and vP2 give rise to molecularly distinct groups of GABAergic neurons. vP2 domain appears multipotent, generating GABAergic V2b (characterized by the expression of Gata2, Gata3, and Tal1), related V2c (Sox1-expressing) and glutamatergic V2a (Chx10-expressing) neurons [62–65]. Forkhead TF Foxn4 is coexpressed with Ascl1 in vP2 [66]. FOXN4 is required for GABAergic V2b and V2c, but not glutamatergic V2a neurogenesis from vP2 progenitors [66]. Overexpression of Foxn4 results in ectopic GABAergic neurogenesis, but only in the presence of ASCL1 [66, 67]. vP0 and vP1 progenitor domains lie within the Dbx2, Ngn1/2-expressing territory [68]. Borders of dP6, vP0, and vP1 domains are defined by cross-repressive interactions between HD TFs NKX6-1/DBX2 and NKX6-2/DBX1 [69]. vP0 and vP1 give rise to a variety of cell types [70–72], but the mechanisms underlying this heterogeneity are poorly understood. Interestingly, the GABAergic nature of vP1 and vP0-derived interneurons is transient—they switch from GABAergic to glycinergic neurotransmission postnatally. vP0 progenitors are bipotent, giving rise to both GABAergic V0d and glutamatergic, EVX1+, V0v commissural interneurons [68, 72]. Dbx1 is expressed in the progenitors of both EVX1+ and EVX1− progeny [68], and its function is required for the vP0 domain identity in both mouse [72, 73] and zebrafish [74]. Inhibitory commissural neurotransmission is affected in Dbx1 mutant mice [72]. However, the specific function of DBX1 in the determination of the GABAergic V0v fate is impossible to assess directly without specific markers for this population. vP1 progenitors are patterned by DBX2 and NKX6-2, and they are fated to EN1+ GABAergic interneurons [69, 75]. In Nkx6-2 mutant spinal cord, ventral expansion of Dbx1 is observed in the VZ concomitant with loss of EN1+ and ectopic EVX1+ cells in MZ [69].
In the dorsal spinal cord (PAX7+), dP4 and dP6 domains generate Lbx1, Pax2 expressing GABAergic neurons of spinal cord dorsal horn [76]. However, dP6 expresses Dbx genes similar to vP0 and vP1, and therefore its patterning resembles the more ventral domains. Early dP4 progenitors express Gsx1/2 and Ascl1 and generate dI4 interneurons. In dI4 cells GSX1/2 specify GABAergic competence by the repression of Ngn2 in the progenitors, demonstrated by ectopic Ngn1/2 expression in the dorsal spinal cord in Gsx1/2 mutant mice, as well as the loss of Ngn1/2 transcripts after overexpression of Gsx2 [77]. In a later wave of neurogenesis, GABAergic dILa interneurons and glutamatergic excitatory dILb neurons are generated from a common progenitor pool in the dorsal spinal cord [78].
In the hindbrain, the progenitor domains appear similar to the spinal cord. Ventrally, GABAergic neurons are derived from Nkx6-1-expressing progenitors [24]. Some of these contribute to GABAergic neurons found in the dopaminergic nuclei of the midbrain [24, 60]. Although PTF1A marks dorsal proliferative GABAergic progenitor populations and is required for GABAergic neuron groups in the hindbrain [79], it is unclear whether it operates as a regional patterning gene. As PTF1A functions as a regulator of differentiation in the dorsal spinal cord, we discuss its function in this context below.
Comparison of the molecular identity of neuroepithelial domains giving rise to GABAergic neurons
In the early neuroepithelium, competence of GABAergic neurogenesis is centralized around the alar–basal boundary region and alar plate (Fig. 3a). The progenitor domains are patterned by multiple TFs, often belonging to the HD family and regulated by extracellular signals (Fig. 3b). Although some of these TFs, such as GSX1, are shared by several GABAergic domains from the telencephalon to spinal cord, none of them is present in all of the GABAergic progenitor domains or appears specific for the GABAergic phenotype (Fig. 3b).
While domains close to alar–basal boundary produce GABAergic neurons either exclusively or in spatially coherent groups, dorsal domains seem predisposed to bimodal competence and typically generate intermixed populations of GABA- and glutamatergic cells. Simultaneous generation of multiple cell types correlates with the overlapping expression of the proneural genes Ascl1 and Ngn1/2 in these regions (see below) (Fig. 3c). However, the production of distinct cell types in such populations is not necessarily temporally synchronized.
Neurogenesis: execution of competence
Neurogenesis and proneural genes
Molecular mechanisms driving neurogenesis are manifold, involving extrinsic signals, cell biological properties of the progenitors, and cell-intrinsic gene regulatory networks [4, 80]. Proneural factors are key transcriptional regulators of neurogenesis and comprise conserved bHLH TFs, such as ASCL1 and NGN1/2. Although first implicated in the neurogenic cell cycle exit and generic neuronal identity, these TFs also appear to directly control other aspects of neuronal development, including progenitor proliferation, precursor fate selection and migration [3, 81]. In general, expression of the proneural genes shows a cyclic pattern in the neural progenitors, is briefly stabilized at the neurogenic cell cycle exit and down-regulated in the differentiating/maturing neurons [82] (Fig. 2). Traditionally, proneural TFs are thought to cell autonomously drive the cell cycle exit and expression of neuron-specific genes and, at the same time, through direct activation of the NOTCH ligand Delta, non-cell-autonomously stimulate NOTCH signaling and maintain stem/progenitor cell properties in the adjacent progenitor cells. The cell cycle exit function of proneural genes has been suggested to involve transcriptional regulation of cell cycle inhibitors as well as antagonism of SOXB1 transcription factors, which support the proliferative neural stem cell identity [83, 84]. However, the cell cycle regulatory function of the proneural genes appears more complex. For example, a recent study suggested that the direct transcriptional targets of ASCL1 include genes both inhibiting and promoting cell cycle progression [81]. The effect of the proneural TF gene on cell cycle may depend on different transcriptional cofactors, mode of expression (cyclic vs. stabilized expression) or post-translational modifications.
In addition to affecting the cell cycle, proneural genes regulate the identity of the differentiating neuron both directly, by activating neuron-specific genes, and indirectly by activating downstream TF cascades (see below). The expression of distinct proneural genes correlates with the type of neurons produced. For example, Ascl1 is associated with GABAergic and Ngn1/2 glutamatergic neurogenesis [3]. Although likely indirectly, ASCL1 and NGN1/2 have been shown to cross-repress each other in some brain regions, which can contribute to their distinct expression patterns.
Proneural gene function in the telencephalon
In contrast to the diversity of TFs that define and pattern the GABAergic neuron progenitor domains, almost all of the developing GABAergic neurons share the expression of the proneural gene Ascl1 (Fig. 3c). However, although the GABAergic progenitor domains commonly express Ascl1, its role revealed by loss-of-function studies appears to be region-specific. In the ventral telencephalon, especially in MGE, ASCL1 is required in the VZ progenitors to generate sufficient subventricular zone (SVZ) basal progenitors and GABAergic neuron precursors [85]. In turn, increased Ascl1 expression in the dorsal telencephalon can drive ectopic GABAergic neurogenesis [86]. Thus, in addition to the regulation of cell cycle exit, ASCL1 promotes development of GABAergic identity, both directly [81] and through activation of TF genes such as Dlx important for GABAergic identity (see below). Antagonistic interactions between proneural genes were also demonstrated in the dorsal telencephalon, where the loss of Ngn1/2 leads to upregulation of Ascl1 and production of GABAergic neurons [87].
Proneural gene function in the diencephalon and midbrain
The complex roles of proneural factors are demonstrated in the diencephalon and midbrain, where ASCL1 has subdomain-specific functions. In the diencephalon, it promotes cell cycle exit in the p3 domain, inhibits cell cycle exit in the rostral p2 domain (pTHr) and suppresses Ngn2 expression and glutamatergic neurogenesis in the p1 domain [19]. In the dorsal midbrain, ASCL1 is required for GABAergic neurogenesis. In contrast, without ASCL1 GABAergic neurogenesis is only delayed in the ventral midbrain, where Ascl1 controls cell cycle exit in a subdomain-specific fashion [88].
In the midbrain, the proneural gene function appears to be regulated by a bHLH-Orange domain TF HELT. Helt is expressed in a mosaic (likely cyclic) pattern in pretectal (p1), thalamic (pTHr) and midbrain proliferative GABAergic progenitors [19, 21, 89, 90]. Helt mutant mice display a defect in the production of midbrain GABAergic neurons, especially in superior and inferior colliculi, dorsal periaqueductal gray and midbrain reticular formation [90]. Despite these deficiencies, Helt mutant mice display no gross anatomical defects. Instead, except for the ventro-lateral embryonic midbrain (m5 domain), prospective GABAergic progenitors re-specify into glutamatergic phenotype expressing Pou4f1 and Vglut2 [22, 90]. As Ngn1/2 are ectopically expressed in the Helt-deficient VZ, while Helt overexpression results in the loss of Ngn1/2 expression, a model emerges where HELT directs the selection of GABAergic over glutamatergic fate via repression of Ngn1/2, which in turn promote glutamatergic neurogenesis [22]. On the other hand, increase in the numbers of LHX1+, and GAD1+ cells were observed upon Helt overexpression in the presence of ASCL1 [22, 91]. Also in cultured cells, cotransfection of Helt and Ascl1 potentiated GABAergic differentiation [91]. These results indicate that HELT may also act as a priming factor promoting GABAergic neurogenesis upon Ascl1 expression.
In addition to the midbrain, HELT also supports GABAergic progenitor identity of the pretectal progenitors in the diencephalic p1 [92, 93]. However, despite a fate switch to an excitatory phenotype, Ngn1/2 were not upregulated in the p1 of Helt mutants [93].
Proneural gene function in the hindbrain and spinal cord
In the hindbrain and spinal cord, Ascl1 and Ngn1/2 expression patterns less clearly distinguish between GABAergic and other phenotypes. Ascl1 expression is linked to GABAergic neuron production in some ventral (NKX6-1+) and dorsal (GSX1+, PTF1A+) domains. However, the medial (DBX2+) GABAergic populations appear ASCL1 negative. In the developing cerebellum and dorsal spinal cord, ASCL1 is required for development of a subset of late born GABAergic interneurons [94–97]. However, during GABAergic neurogenesis from the cerebellar VZ, also Ngn1/2 are expressed and may guide differentiation of specific subsets of GABAergic neurons [98, 99]. In particular Ngn2, a direct target of PTF1A, has been shown to be important for the cell cycle progression of progenitors of GABAergic Purkinje cells and the maturation of these neurons [99, 100].
Comparison of proneural gene function in brain regions giving rise to GABAergic neurons
In conclusion, a major regulator of GABAergic neurogenesis is the proneural TF ASCL1, which has multiple functions at different stages and regions undergoing production of postmitotic GABAergic precursors. In some domains, the proneural factors ASCL1 and NGN1/2 antagonize each other to control GABAergic vs. glutamatergic neurogenesis. HELT is a regulator of proneural gene activity and the neuronal differentiation balance specifically between ZLI and the midbrain–hindbrain boundary (MHB). In other regions, especially in the hindbrain and spinal cord, both ASCL1 and NGN1/2 may promote generic GABAergic development, but they also regulate differentiation of distinct GABAergic neuron subsets.
Subtype specification in postmitotic neurons
Selector genes and fate determinants
Although the expression of proneural genes can be spatially restricted, proneural activity mostly activates pan-neuronal genes and further regulation of neuronal fate involves additional subtype specification factors. Subtype specification factors are TFs, which act as promoters or inhibitors of a certain cell fate, driving neuronal precursors towards a specific subtype, simultaneously preventing expression of genes characteristic to other cell types. Such bimodal regulators are called selector genes and a few of them have been characterized in the choice between GABAergic vs. glutamatergic neuron phenotype in the mammalian CNS [9, 101–104] (Fig. 2). Terminal selector genes are defined as TFs that are activated at the cell cycle exit and can maintain their own expression by autoregulation during differentiation, retaining activity up to the mature neuron stage. Terminal selector activity leads to both activation of batteries of functionally related genes contributing to particular aspect(s) of neuronal phenotype, and suppression of the determinants for alternative fate(s). The loss of a selector gene does not affect production of postmitotic neuronal precursors or their survival but leads to loss of a specific neuronal identity accompanied by a transformation into an alternative fate.
Currently, a GABAergic over glutamatergic selector function has been unambiguously demonstrated for PTF1A in the dorsal spinal cord, for GATA2/TAL1 complex in the ventral spinal cord, and GATA2 and TAL2 in the midbrain and p1 of diencephalon. GABAergic subtype selector function has been shown for GATA2 in rostral p2 (pTHr) and suggested for DLX TFs in the telencephalon (Fig. 3d).
DLX family TFs and forebrain GABAergic neurons
The expression of Dlx family HD TFs is a uniting feature of the GABAergic neurogenesis domains in the anterior forebrain: telencephalon and diencephalon up to the ZLI [105, 106]. Dlx2 is also expressed in the migratory OB GABAergic neurons in the embryonic and adult brain [107]. Dlx1/2 expression is activated in the VZ and the loss of Dlx1/2 affects gene expression in VZ, SVZ and MZ [108, 109]. However, the targets of DLX1/2 include region-specific progenitor markers, but not general determinants of progenitor identity such as Hes1 [108], supporting the link between DLX function and subtype identity. Dlx1/2 expression in the GEs is regulated by ASCL1 binding on its intergenic enhancers [110]. Conserved DLX/MSX/NKX binding motifs are also found in the Dlx1/2 enhancer, near the ASCL1 binding site [110]. Thus, ASCL1 may interact with HD TFs to induce Dlx1/2 expression. Dlx1/2 enhancers also contain elements through which they positively autoregulate their own expression [111]. During the progression of differentiation, DLX1/2 are gradually replaced with DLX5/6, as DLX2 directly activates Dlx5/6 transcription [105, 109, 112].
DLX1/2 are required for the generation of postmitotic GABAergic interneurons from the SVZ progenitors [113]. As Dlx1 is activated at a slightly later stage than Dlx2, the dose effect in this function should be considered. Dlx1/2 double mutant mice lack all telencephalic GABAergic neurons: from striatum, cortex, OB, and hippocampus. Both defects of migration and failure to maintain GABAergic identity appear to contribute to this phenotype [43, 113, 114].
Dlx genes appear to regulate GABAergic neuron phenotype both directly and by activation of a next layer of transcription factors. Ectopic overexpression of Dlx2 and Dlx5 in forebrain slice cultures was shown to activate the expression of Gad1 and Gad2 [106]. Notably, Dlx2 overexpression could induce similar GABAergic differentiation in the midbrain slices as well [115]. Differential activity of Dlx enhancers seems to correlate with a tendency towards particular subtype of progeny, suggesting that the basis for subtype diversification may be established very early, by cis-regulatory control over Dlx transcription [116]. Indeed, developmental differences in Dlx gene expression have been shown to contribute to interneuron subtype segregation and/or maintenance. For example, the SST+ and NPY+ subtypes were affected in Dlx1 mutant cortex and hippocampus, while CALB2 (CR)+ GABAergic neurons were lost in hippocampus but not in the cortex [117]. Some of the DLX targets are involved in differentiation of forebrain GABAergic neuron subtypes. For example, a zinc-finger homeobox TF ZFHX1B was found to be directly regulated by DLX2 and to drive cortical GABAergic interneuron differentiation as well as to suppress striatal GABAergic neuron differentiation [118].
Microarray studies have listed a multitude of additional subtype-specific and migration-related genes and as targets of DLX1/2, including Calb1, Sst, Arx, and Lmo3/4 [119]. Enriched Gene Ontology categories within de-regulated genes in Arx mutant clearly point to a function in axon guidance and neuronal migration [119]. Cross-comparisons of microarray data from Dlx1/2 and Arx mutants with wild-type samples, as well as DLX5/6-positive and -negative cells revealed that despite a major overlap between the targets of these TFs, there are also significant sets of genes independently regulated either by the DLX family members or ARX. Although an extensive list of putative target genes is now available, the understanding whether a certain TF regulates the transcription of the listed targets directly or indirectly is still mostly lacking. For example, direct binding of ARX was demonstrated only in the enhancers of three ARX-dependent genes [119].
In addition to the telencephalon, DLX TFs regulate GABAergic differentiation also in the p3 region of the anterior diencephalon. Interestingly, DLX2 appears to regulate GABAergic subtype identity in this region [93] (see also discussion below).
GATA2 and the posterior diencephalon and midbrain
Gata2 and Tal2 expression is a hallmark of all GABAergic precursors in the area spanning from ZLI to MHB [19, 21] (Fig. 3d). In the midbrain, Gata2 is activated after Ascl1 and Helt in GABAergic precursors soon after their cell cycle exit [21]. Midbrain GABAergic neuron precursors also express Tal2 in a pattern similar to Gata2 [120, 121]. However, Gata2 and Tal2 are activated independent of each other [121]. Both genes are required for GABAergic neuron development at a postmitotic stage. Mutants of Gata2 show a transformation of all midbrain GABAergic precursors into glutamatergic fates, while it does not affect patterning or proneural gene function in the VZ progenitors [21]. Terminal selector function of GATA2 is strongly suggested by this fate transformation phenotype together with the induction of GABAergic neuron differentiation in ectopic locations by Gata2 overexpression [21]. Similar GABAergic-to-glutamatergic fate transformation was also observed in the midbrain of Tal2 mutants, although the most ventral midbrain is less sensitive to Tal2 inactivation and the two mutants show subregion-specific differences [121]. GATA and TAL factors have been suggested to operate as components of a common transcription factor complex [122]. Similar phenotypes of Gata2 and Tal2 mutants could reflect a partnership in binary terminal selector complex, where either one missing protein would render the complex inactive.
In addition to the GABAergic neurotransmission-related genes, GATA2 and TAL2 are required for the activation of downstream TF genes, including Tal1, Gata3, Six3, and Sox14 [19, 21, 121]. These may regulate the maintenance of the GABAergic phenotype or specific aspects of GABAergic neuron development (see below). For example, SOX14 regulates migration of GABAergic neuron precursors during development of distinct diencephalic nuclei [93].
Curiously, mature midbrain contains an exceptional population of GABAergic neurons: development of GABAergic neurons associated with the dopaminergic nuclei [substantia nigra pars reticulata (SNpr), and GABAergic component of ventral tegmental area (VTA)] is independent of GATA2 function. The reason for this is that these GABAergic neurons have their developmental origin in the ventral hindbrain, where GABAergic fate is independent of GATA2, but dependent on TAL1 function [24, 60], (see below).
The selector function of GATA2 appears to be context dependent. In the diencephalic p1, GATA2 acts as a selector for GABAergic over glutamatergic fate as in midbrain: Gata2 mutant GABAergic precursors acquire a glutamatergic phenotype. In pTHr however, the loss of GATA2 imposes a GABAergic subtype respecification: mutant precursors switch to a p3-like differentiation programme, activating Dlx1/2 and Arx expression [19]. On the contrary, inactivation of Dlx2 results in ectopic upregulation of GATA2 targets Tal1 and Sox14 characteristic to the pTHr-like program in the p3 [93]. Thus, DLX2 and GATA2 may act as opposing switches regulating distinct subtypes of GABAergic neurons on the anterior and posterior side of the ZLI, respectively.
TAL1 and PTF1A in the hindbrain and spinal cord
TAL1: ventral hindbrain and spinal cord
In the ventral spinal cord, the V2b population of GABAergic neurons is derived from the most dorsal Nkx6-1-expressing progenitors and is also characterized by Gata2 and Tal1 expression [62, 64, 103]. The early V2 postmitotic precursors are multipotent, giving rise to both GABAergic V2b and glutamatergic V2a neurons, and initially ubiquitously express Gata2 [62, 64]. However, at cell cycle exit, V2 precursors are segregated into two distinct groups by FOXN4-induced NOTCH1–DELTA4 signaling, which initiates the expression of V2b fate determinant Tal1 [67, 101]. The segregation of V2a/b identities is further refined by the LIM-HD transcriptional cofactor LMO4 that bridges GATA2 and TAL1 into a transcriptional complex. This GATA2–TAL1–LMO4 complex binds DNA consensus of specific topology, found for example in intergenic enhancers of Gata2/3 or Gad1 genes [123]. GATA2 as well as TAL1 have been shown to be sufficient for the commitment to V2b phenotype, at the expense of V2a fate [62, 64, 103]. At the same time, the loss of TAL1 results in specific deficiency of V2b neurons [103], while in the Gata2 mutant, the whole V2 precursor population fails to develop [64].
Similar to the ventral spinal cord, the NKX6-1+ progenitors in the ventral hindbrain also produce a population of GABAergic precursors in a TAL1-dependent fashion [60]. However, in contrast to the spinal cord, these GABAergic precursors are not affected by the loss of GATA2 alone. Correlating with the distinct requirements for GATA2, the gene regulatory mechanisms driving Gata2 expression are also different in the GABAergic precursors in the ventral hindbrain and spinal cord compared to the precursors in the midbrain and diencephalon [64].
PTF1A: dorsal hindbrain and spinal cord
Cell lineage tracing has demonstrated that the expression of bHLH TF Ptf1a is restricted to the GABAergic lineage in the cerebellum. Both PAX2+ (c2v) and CORL+ (c2d) GABAergic populations express Ptf1a at the progenitor stage [124]. In accordance with this expression pattern, PTF1A function is central for the GABAergic neurogenesis in the dorsal r1 as Ptf1a mutant mice lack all GABAergic neurons of the cerebellum and cerebellar nuclei [125]. Ectopic PTF1A can induce the differentiation of GABAergic neuron-like cells in dorsal telencephalon, confirming the master regulator properties of this gene [125]. In the absence of PTF1A, the presumptive GABAergic precursors adopt a glutamatergic granule cell phenotype [126]. Although these findings are consistent with a selector function, it is also possible that PTF1A regulates regional patterning rather than neuronal differentiation and activation of neuron-type-specific gene expression in the dorsal hindbrain. Genetic fate mapping and loss-of-function studies have shown that similar to cerebellum, Ptf1a expression is restricted to the GABA- and glycinergic precursors in the cochlear nuclei and this expression is essential for their development [79]. In contrast to its function in GABAergic neurogenesis in the anterior and medial hindbrain, PTF1A is required for the development of both inhibitory neurons and excitatory glutamatergic climbing fiber neurons in the caudal hindbrain (rhombomeres 6–8; [127]). Thus, in the hindbrain, PTF1A has properties of both a patterning factor and a fate selector.
In contrast to the hindbrain, where it is expressed in the proliferative progenitors, Ptf1a is mostly activated after the cell cycle exit in the postmitotic neural precursors of the dorsal spinal cord. PTF1A marks the early born dI4 as well as the late-born dILa GABAergic neuron precursors and instructs their differentiation by allowing the expression of Lhx1/5 and Lbx1, while repressing Tlx3 that acts as a selector gene for the glutamatergic fate [102, 104, 128]. In the absence of PTF1A, spinal cord dorsal horn sensory interneurons fail to activate the GABAergic differentiation markers Pax2, Lhx1/5, and Gad1, and instead are trans-specified into a glutamatergic phenotype (TLX3+, VGLUT2+) [102]. Similarly, LBX1 is both sufficient and necessary for the GABAergic neuron differentiation in dorsal spinal cord [104]. In addition to dI4, Lbx1 is also expressed in dI6 progeny, in a PTF1A-independent manner [76]. The fate switch phenotypes demonstrate dual properties of both PTF1A and LBX1: supporting the GABAergic and suppressing glutamatergic differentiation cassettes. However, while PTF1A is required in all GABAergic neuron subtypes (characterized by Sst, Npy, Pnoc, Penk, and/or galanin coexpression), LBX1 loss-of-function does not affect SST+ or PENK+ subtypes [129].
In summary, bHLH TF PTF1A controls GABAergic as opposed to glutamatergic fate selection in both mammalian cerebellum and dorsal spinal cord. The targets activated by PTF1A include Lhx1/5, Lbx1, Pax2, and Gad1, while glutamatergic determinants are suppressed. Recent ChIP-seq study showed PTF1A binding to enhancers associated with both cell proliferation and cell-fate specification genes [130], supporting the observation that PTF1A can control various aspects during neurogenesis. Indeed, a mechanism where PTF1A outcompetes NOTCH intracellular domain for RBPJ protein binding, causing a switch from Hes expression towards differentiation genes, explains how PTF1A could promote neurogenesis and interact with NOTCH signaling [100, 131].
GABAergic fate in the neural retina
The vertebrate retina is a derivative of anterior neural plate. Retinal field identity is specified by CHX10, RX, and MITF TFs [132]. Retina consists of six cell types, among which amacrine and horizontal cells are inhibitory interneurons (GABA- and glycinergic). Precursors of both amacrine and horizontal cells express Neurod4, Foxn4, and Ptf1a. FOXN4 lies upstream of Ptf1a and also controls the expression of Neurod1, which is required for the cell migration. Ptf1a a does not control Foxn4 or Neurod1 expression or cell migration, but selects GABAergic over glutamatergic cell fate [133, 134]. The downstream targets of PTF1A in retina include Barhl2, Prox1, and Gad1 [135, 136].
Considering the efforts in tracking the evolutionary origins and mechanisms of diversification in retinal cell types [137, 138], it is interesting to note that the GABAergic fate specification mechanisms utilized in amacrine and horizontal cell lineages are more similar to the hindbrain and spinal cord compared to the telencephalic or diencephalic regions.
Maintenance of fate: regulators of differentiation batteries
Fate maintenance involves continued expression of neuron-specific terminal differentiation genes (neurotransmitter synthesizing enzymes, receptors, etc.). Genes allocated to a particular function can be coregulated in transcriptional modules, allowing for a small number of TFs to define very complex functionalities [12, 139]. Many TFs regulating early neural differentiation are also expressed in mature neurons [140]. Recently, temporally controlled mutagenesis experiments have revealed late functions of these TFs in the maintenance of the neuronal identity. For example, inactivation of Nurr1 and Pet1 results in reduction of cell type-specific gene expression in dopaminergic and serotonergic neurons, respectively [14, 15]. Whether the developmental regulators of GABAergic neurons also control their maintenance is currently unknown. The GABAergic fate determining TFs such as DLX2, GATA2, and PTF1A are down-regulated in maturing neural precursors, but some of these TFs can sequentially switch on their paralogs as differentiation progresses. For example, DLX2 binds a DLX/MSX/NKX motif in Dlx5/6 enhancer in telencephalon [109, 141], whereas GATA2 and TAL2 activate Gata3 and Tal1 expression in the midbrain, perhaps via binding to GATA–Ebox motifs similar to the ones involved in the autoregulation of these genes in spinal interneurons [123]. Thus, while the selector genes Gata2 and Tal2 are down-regulated during the final maturation of GABAergic neurons, Gata3 and Tal1 expression continues. Therefore, the latter two factors may be involved in the maintenance of the GABAergic neuron-specific gene expression.
Lim HD TFs have been associated with subtype-specific identities [142], and are often activated by selector factors (Fig. 2): Lhx1/5 are regulated by PTF1A in spinal cord [102] and Lhx1/5 expression is required for maintenance of GABAergic identity in the spinal cord dorsal horn. LHX1/5 loss-of-function causes late down-regulation of Pax2, Gad1, and Viaat expression [143]. Lhx1/5 are also expressed in the PTF1A+ lineage in the cerebellum, where their expression precedes the expression of further subtype-divergent genes Pax2 and Corl2 [124]. Loss of LHX1/5 function results in late loss of CALB1+ Purkinje cell population (CORL2+), leaving other cerebellar cell populations relatively normal [144]. It is likely that while LHX1/5 control general GABA neurotransmission battery, further diversification of cell types can be achieved by additional TFs. Lhx1/5 are also expressed in the GABAergic precursors in midbrain [22], pretectum, and rostral thalamus (pTHr) [145]. Lhx1 expression in midbrain GABAergic, but not glutamatergic populations, requires GATA2 function [21]. However, demonstration of maintenance functions for the selector TFs and their downstream factors would require their inactivation only in the mature GABAergic neurons.
In addition to TFs, microRNAs also regulate neuronal fate, differentiation, and survival [146–148]. Specific microRNA profiles have been linked to differentiated neuron types in mature brain. Comparing microRNA profiles in cortical and cerebellar GABAergic vs. glutamatergic neurons, sets of microRNAs were found enriched in all types of analyzed GABAergic cells. In addition, a number of microRNAs were specifically and differentially expressed in distinct GABAergic neuron subtypes [149]. It will be of great interest to study how microRNAs, likely in collaboration with TFs, establish and maintain subtype-specific gene expression patterns in the GABAergic neurons.
Migration of GABAergic neuron precursors
After the exit from cell cycle and onset of differentiation, neuronal precursors migrate to their final locations in the CNS and form functional synaptic contacts with their targets. In general, neuronal migration occurs radially along radial glial fibers or tangentially along the rostro-caudal or medio-lateral axis [150, 151]. The regulation of neuronal migration involves attractive and repulsive guidance molecules as well as their receptors, cytoskeletal components, cell adhesion, and extracellular matrix molecules [152, 153]. Relatively little is known of the transcriptional regulation of this complex process. In particular, the difficulty in pinpointing a later migratory function for certain transcriptional regulators lies in the fact that they often also affect neuronal fate specification at an earlier phase. In addition, it is hard to determine whether the TFs act cell-autonomously in migrating neurons or in patterning their environment [154]. Here we review the current knowledge on the TFs that regulate especially the tangential migration of GABAergic precursors into different parts of the rodent CNS.
Migration of GABAergic neurons originating from subpallium
Migration of GABAergic cells into the cortex
Cortical GABAergic interneurons comprise a highly heterogenous population that targets both excitatory glutamatergic (pyramidal) neurons as well as other interneurons [155, 156]. Pyramidal neurons are generated locally in the dorsal telencephalon VZ and migrate radially into their final locations in different cortical layers. In contrast, as discussed earlier, the majority of GABAergic interneurons in rodents are born in the subpallium, in the MGE, CGE, and POA (Fig. 3e) [44]. Subpallial GABAergic precursors first migrate along spatially distinct tangential routes into the developing cortex, followed by the final radial distribution into distinct cortical layers. The migrating cells respond to cues in the environment by generating branches in the leading processes [157]. These cues include membrane-bound or diffusible molecules from several families such as semaphorins, neuregulins, neurotrophic factors, chemokines (CXCL12), and neurotransmitters (GABA, dopamine). The set of cues to which each neuron type responds is defined by the set of receptors that the cells express [158–160]. Concerning the migration from the MGE through the deep parts of the septum, a few chemorepulsive factors (SEMA3A and SEMA3F) in the septum and chemoattractive factors (neuregulin 1, NRG1) have been genetically proven [158, 159]. CGE and POA cells take a distinct route to the cortex, suggesting different guidance cues [161–163].
After they cross the pallial–subpallial boundary, GABAergic precursors migrate via a specific set of streams either through the MZ, SVZ, or the subplate [164], staying away from the cortical plate where the pyramidal neurons form the cortical layers [165, 166]. The mechanisms driving intracortical migration are different from the subpallium-to-pallium migration. CXCL12 chemokine signaling mediates this migration by long-range attraction [167–171]. While the MGE-, CGE-, or POA-derived GABAergic interneuron precursors assume different routes during subpallial–pallial migration, the region of origin does not seem to influence their choice of migratory stream in the cortical plate. After the tangential migration into the cortex, interneurons exit from the streams, lose responsiveness to CXCL12 [167], and migrate radially along the radial glia into the cortical plate [163, 168, 172]. Finally, during the first postnatal days in rodents, the interneurons are distributed into specific layers. The organization of interneurons into layers mostly follows inside-out sequence in a manner similar to the pyramidal cells: early born cells occupy deep layers and later-born neurons occupy upper layers [172–174]. It has also been proposed that specific classes of pyramidal cells in different layers may release factors that instruct specific interneurons types to their final destination [172, 175, 176].
The migratory cues and receptors are in turn regulated by region-specific transcription factors [154]. As discussed above, subpallial GABAergic progenitor domains can be distinguished by their differential expression of specific sets of TFs and signaling molecules [38, 163]. Both patterning genes and fate selectors have been linked to the regulation of interneuron migration in the telencephalon, either directly or via downstream TFs. The loss of subpallial interneuron markers DLX1 and DLX2 severely blocks interneuron migration from all subdomains; this is associated with changes in cell shape, survival, and deregulation of cytoskeletal regulators [114]. The reduced expression of cytokine CXCL12 receptor genes Cxcr4 and Cxcr7 may contribute to migration defects observed. CXCL12 attracts Cxcr4 and Cxcr7-expressing interneurons to the cortex in tangential streams and Cxcr7 mutants have defects in leading process morphology during migration [167–169, 177]. Another target of DLX1/2 is the HD TF gene Arx, which is expressed in the subpallium and in migrating interneurons. Arx mutant mice have smaller cortex, hippocampus, and OB [115, 178, 179] and in humans ARX mutations have been implicated in severe cortical migration defects causing lissencephaly [180]. Gene expression profile analyses have identified a number of migration-related genes as ARX targets, including Cxcr7 [119]. In addition, the DLX-dependent switch from tangential to radial migration may be mediated by ARX [181].
CGE cells express a nuclear receptor TF Nr2f2 (Coup-tf2) that has been shown to promote the tangential migration of the CGE-derived interneurons to the cortex [182]. However, targets for this TF have not been identified yet. MGE- and POA-derived GABAergic precursors first express Nkx2-1 and its down-regulation in postmitotic cells is necessary for their migration into the cortex [183]. Down-regulation of Nkx2-1 leads to the expression of neuropilin 2 (Nrp2) in cortical interneurons, which are then repelled away from striatum expressing NRP2 ligands Sema3A/3F. The cells where the Nkx2-1 down-regulation does not occur, migrate into the striatum [183]. In addition to NKX2-1, DLX proteins also regulate Nrp2 expression [184].
MGE-derived cells also express Lhx6 once they exit the cell cycle and this expression is maintained in mature inhibitory neurons in the cortex. Lhx6 expression is lost in NKX2-1 mutant mice [42, 185, 186]. Deletion of Lhx6 in mice results in delayed tangential migration and abnormal distribution of MGE-derived interneurons that accumulate in the cortico-striatal boundary [185, 187].
Migration of GABAergic precursors into the olfactory bulb
The subpallium also generates neurons to other regions such as the septum, basal ganglia, and amygdala. In addition, the dorsal part of the LGE produces precursors that migrate rostrally into the OB along the rostral migratory stream (Fig. 3e). This migration also continues in adult rodents [188–190]. A number of guidance molecules and their receptors have been implicated in precursor migration into the OB such as SLIT1, SLIT2, PSA-NCAM, integrins, EPH/ephrin, and Netrin1 [151, 153, 191].
Very little is known of the TFs regulating precursor migration into the OB. As in cortex, DLX1/2 promote GABAergic fate and tangential migration of interneurons into the OB, potentially regulating ErbB4, Robo2, Slit1, and Prok2 expression [107, 192]. In contrast, interneuron development in the OB is normal in Nkx2-1 mutants, consistent with the expression of Nkx2-1 in the MGE [42]. In addition to TFs involved more broadly in GABAergic development, the serum response factor (SRF) has also been implicated in the migration of interneurons into the OB. SRF regulates a variety of immediate early genes as well as genes encoding for cytoskeletal components. In SRF mutants actin cytoskeleton was severely altered [193].
Migration of GABAergic cells in the diencephalon and midbrain
Much less is known of the transcriptional control of tangential migration of GABAergic interneurons into other regions of the CNS. SOX14 that acts downstream of HELT and DLX1/2, is a marker for GABAergic neurons that migrate from the rostral thalamus and pretectum tangentially to form the subcortical visual shell (SVS) nuclei in diencephalon. In Sox14-deficient mice, the GABAergic neurons were missing from one part of the SVS, the ventral lateral geniculate nucleus [93].
As mentioned above, GABAergic cells in SNpr and VTA originate in the ventral hindbrain and the SNpr and VTA GABAergic neurons require TAL1 for their development [60]. Recent analyses with Pitx3 mutant (Aphakia) mice suggested that midbrain dopaminergic neurons could guide the migration of SNpr and VTA neurons [194]. This is consistent with the observations that the rhombomere 1-derived, TAL1-dependent, and GATA2-independent midbrain GABAergic neurons are found only in a tight association with the dopaminergic neurons in the ventral midbrain [21, 60]. Interestingly, in the forebrain, dopamine receptors D1 and D2 guide the tangential migration of GABAergic neurons [195]. However, the molecular cues guiding the developing SNpr and VTA GABAergic neurons, as well as their exact migratory patterns, timing, and the transcriptional regulation of this process, remain to be elucidated.
Conclusions and open questions
Diverse mechanisms regulate GABAergic neuron differentiation
No single TF involved in patterning, neurogenesis, or differentiation of GABAergic neurons is shared by all the GABAergic neuron populations (Table 1). On the other hand, most of the TFs important for GABAergic neuron development also have functions in other neuronal lineages or cell types. Based on the molecular mechanisms controlling GABAergic neuron development, CNS can be divided into three main subdomains (Fig. 3
Table 1.
Molecular players in key steps of GABAergic neurogenesis
| Function | ||||
|---|---|---|---|---|
| Brain region | Spatial patterning and progenitor specification HD or bHLH proteins | Proneural genes and their regulators bHLH proteins | Postmitotic subtype selection HD, bHLH, ZF | Maturation, migration, maintenance HLH, HD, ZF |
| Telencephalon | MGE: NKX2-1 [42] | ASCL1 [85, 86, 196] | DLX2 [113–115] | ARX [115, 178, 179] |
| DLX5/6 [106, 119] | DLX5/6 [119, 192] | |||
| LGE, CGE: GSX1/2 [48, 50, 77] | ||||
| NR2F2 [182] | ||||
| LHX6 [185] | ||||
| POA: NKX5-1 [45] | ||||
| POA: DBX1 [44] | ||||
| DLX1/2 [106, 108] | ||||
| Diencephalon | NKX2-2 [59, 89] | ASCL1 [91] | p3: DLX2 [93] | p1-2: SOX14 [93] |
| aTAL1 [19, 59] | ||||
| FEZF1/2 [58] | p1-2: HELT [91, 93] | p1-2: GATA2 [19] | ||
| p1-2: aTAL2 | ||||
| aGATA3 [199] | ||||
| GBX2 [197] | ||||
| PAX6 [198] | ||||
| Midbrain | NKX2-2 | ASCL1 [88, 91] | GATA2 [21, 121] | PITX2 [203] |
| NKX6-1 | ||||
| PAX3/7 [200–202] | ||||
| HELT [22, 90] | TAL2 [121] | aGATA3 [60, 199] | ||
| aTAL1 [21, 121] | ||||
| aLHX1/5 [21, 22] | ||||
| Ventral R1 | NKX6-1 [26, 204] | ASCL1 [91] | TAL1 [60] | TAL1 [60] |
| aNGN1/2 | PITX2 [205] | |||
| Dorsal R1 (cerebellum) | EN2 [206–208] | ASCL1 [95, 97, 210] | PTF1A [125, 126] | LHX1/5 [144] |
| NGN1/2 [99] | ||||
| NGN1/2 [25, 99] | ||||
| aCORL, PAX2 [124, 211] | ||||
| PTF1A [125, 209] | ||||
| Dorsal spinal cord | dP4: GSX1/2 [94] | ASCL1 [94, 96] | dI4, dIL, dI6: LBX1 [72, 76, 104] | LBX1 [72, 76, 104] |
| LHX1/5 [143] | ||||
| dI4, dIL: PTF1A [102] | ||||
| NGN1/2 [212] | ||||
| dP6: DBX2 [69, 73] | ||||
| PAX2 [128] | ||||
| Ventral spinal cord | NKX6-1 [36, 69] | ASCL1 [66] | V2b: TAL1 [101] | V2b: TAL1 [63, 103] |
| V0-1: DBX1/2 [36, 69, 72] | ||||
| aNGN1/2 | ||||
| V2: FOXN4 [66, 67] | ||||
| GATA2 [64, 103] | ||||
| LMO4 [123] | ||||
| V0d: aEVX1- [213] | ||||
| GATA3 [62, 63] | ||||
| LMO4 [123] | ||||
| V1: EN1 [71] | ||||
| Mutant phenotype (mouse) | GABAergic progenitors mispatterned or missing | GABAergic neurogenesis delayed and/or diminished | GABAergic precursors respecified to alternative fate | Incomplete differentiation/impaired functionality |
aMarked genes are expressed in GABAergic neurons but their role in fate specification or mechanism of action is unclear
Direct and indirect targets of the precursor patterning and GABAergic fate selector genes
Although TFs driving GABAergic fate selection in the developing mammalian brain have been identified, the mechanistic understanding of the regulatory circuitry is still missing. It is not known how the selector TFs operate with next layer TFs to activate neuron-specific genes, possibly in a relay or feed-forward fashion. It is unclear whether the region-specific targets share enhancer motifs or epigenetic marks and how the patterning, proneural and terminal selector TFs collaborate. If subtype diversity is primed by HD code in progenitors, then we may expect the selector genes to control the GABA/glutamate neurotransmission module alone, but subtype-specific modules such as coneurotransmitters or Ca2+-binding proteins in cooperation with other TFs. This cooperation can manifest by direct physical interactions of selector gene and other TFs (downstream of patterning genes) or alternatively, the regulatory regions of subtype-specific genes may be pre-modulated already in the progenitor stage.
Nature and relative ease of switch between GABAergic and glutamatergic fate
Failure of GABAergic fate specification often leads to adoption of glutamatergic phenotype. The two neurotransmitters are similar in their biochemical synthesis pathways. There are two sources of glutamate in presynaptic terminals. The most prevalent source in functional brain is glutamate recycling, where glutamate is taken up from synaptic clefts by glial cells and presynaptic neurons. In glial cells, glutamate is converted to glutamine and transported to presynaptic neurons where it is converted back to glutamate by glutaminase. Neuronal mitochondria can also produce glutamate as a glucose metabolism intermediate. Presynaptic glutamate is packaged into vesicles by vesicular glutamate transporters (VGLUT, SLC17). Biosynthesis of GABA requires adding one step on top of glutamate biosynthesis pathway, a conversion of glutamate into GABA by glutamic acid decarboxylase (GAD). Thus, the gene batteries for GABA and glutamate biosynthesis naturally largely overlap. In essence, merely switching on Gad and inhibiting Vglut is enough for a transformation from glutamatergic to GABAergic neurotransmitter identity. However, for changed functionality, neurons would have to package and release GABA from synaptic vesicles. Thus, it would make sense to coregulate Gad with vesicular (SLC32) and membrane (SLC6) GABA transporters.
Acknowledgments
We thank Laura Lahti and Maarja Haugas for comments on this manuscript. Our work was supported by the Academy of Finland, Sigrid Juselius Foundation, Finnish Parkinson’s Foundation and the University of Helsinki.
Abbreviations
- AP
Anterior-posterior
- bHLH
Basic helix-loop-helix
- cb
Cerebellum
- CGE
Caudal ganglionic eminence
- CNS
Central nervous system
- CoP
Commissural pretectum (mixed GABAergic and glutamatergic)
- dI4, dI6, dILa
Subpopulations of GABAergic neurons in dorsal spinal cord
- DV
Dorso-ventral
- GABA
Gamma-aminobutyric acid
- hb
Hindbrain
- HD
Homeodomain
- hyp
Hypothalamus
- JcP
Juxtacommissural pretectum (GABAergic)
- LGE
Lateral ganglionic eminence
- mb
Midbrain
- m1–m5
Progenitor domains producing GABAergic neurons in midbrain
- MGE
Medial ganglionic eminence
- MHB
Midbrain–hindbrain boundary
- MZ
Mantle zone
- OB
Olfactory bulb
- p1
Prosomere 1, pretectum
- p2
Prosomere 2, thalamus
- p3
Prosomere 3, prethalamus
- PcP
Precommissural pretectum (glutamatergic)
- POA
Preoptic area
- pTHr
Rostral thalamus (GABAergic)
- pTHc
Caudal thalamus (glutamatergic)
- sc
Spinal cord
- SNpr
Substantia nigra pars reticulata
- SVS
Subcortical visual shell nuclei
- SVZ
Subventricular zone
- tel
Telencephalon
- TF
Transcription factor
- V0–V2
Subpopulations of GABAergic neurons in ventral spinal cord
- VTA
Ventral tegmental area
- VZ
Ventricular zone
- ZF
Zinc finger
- ZLI
Zona limitans intrathalamica
Contributor Information
Kaia Achim, Email: kaia.achim@embl.de.
Juha Partanen, Email: juha.m.partanen@helsinki.fi.
References
- 1.Kiecker C, Lumsden A. The role of organizers in patterning the nervous system. Annu Rev Neurosci. 2012;35:347–367. doi: 10.1146/annurev-neuro-062111-150543. [DOI] [PubMed] [Google Scholar]
- 2.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135(15):2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 3.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134(21):3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 4.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135(9):1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 5.Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38(5):731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- 6.Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11(6):831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhou JX, Huang S. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends Genet TIG. 2011;27(2):55–62. doi: 10.1016/j.tig.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouaux C, Arlotta P. Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nat Neurosci. 2010;13(11):1345–1347. doi: 10.1038/nn.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47(6):817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Jeong JY, Einhorn Z, Mathur P, Chen L, Lee S, Kawakami K, Guo S. Patterning the zebrafish diencephalon by the conserved zinc-finger protein Fezl. Development. 2007;134(1):127–136. doi: 10.1242/dev.02705. [DOI] [PubMed] [Google Scholar]
- 12.Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010;33(10):435–445. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmberg J, Hansson E, Malewicz M, Sandberg M, Perlmann T, Lendahl U, Muhr J. SoxB1 transcription factors and Notch signaling use distinct mechanisms to regulate proneural gene function and neural progenitor differentiation. Development. 2008;135(10):1843–1851. doi: 10.1242/dev.020180. [DOI] [PubMed] [Google Scholar]
- 14.Kadkhodaei B, Ito T, Joodmardi E, Mattsson B, Rouillard C, Carta M, Muramatsu S, Sumi-Ichinose C, Nomura T, Metzger D, Chambon P, Lindqvist E, Larsson NG, Olson L, Bjorklund A, Ichinose H, Perlmann T. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci Off J Soc Neurosci. 2009;29(50):15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, Deneris ES. Pet-1 is required across different stages of life to regulate serotonergic function. Nat Neurosci. 2010;13(10):1190–1198. doi: 10.1038/nn.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci Off J Soc Neurosci. 2007;27(36):9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128(2):193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- 18.Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 2010;33(8):373–380. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virolainen SM, Achim K, Peltopuro P, Salminen M, Partanen J. Transcriptional regulatory mechanisms underlying the GABAergic neuron fate in different diencephalic prosomeres. Development. 2012;139(20):3795–3805. doi: 10.1242/dev.075192. [DOI] [PubMed] [Google Scholar]
- 20.Vue TY, Bluske K, Alishahi A, Yang LL, Koyano-Nakagawa N, Novitch B, Nakagawa Y. Sonic hedgehog signaling controls thalamic progenitor identity and nuclei specification in mice. J Neurosci Off J Soc Neurosci. 2009;29(14):4484–4497. doi: 10.1523/JNEUROSCI.0656-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kala K, Haugas M, Lillevali K, Guimera J, Wurst W, Salminen M, Partanen J. Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development. 2009;136(2):253–262. doi: 10.1242/dev.029900. [DOI] [PubMed] [Google Scholar]
- 22.Nakatani T, Minaki Y, Kumai M, Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134(15):2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto M, Hibi M. Development and evolution of cerebellar neural circuits. Dev Growth Differ. 2012;54(3):373–389. doi: 10.1111/j.1440-169X.2012.01348.x. [DOI] [PubMed] [Google Scholar]
- 24.Lahti L, Achim K, Partanen J. Molecular regulation of GABAergic neuron differentiation and diversity in the developing midbrain. Acta Physiol (Oxf) 2013;207(4):616–627. doi: 10.1111/apha.12062. [DOI] [PubMed] [Google Scholar]
- 25.Zordan P, Croci L, Hawkes R, Consalez GG. Comparative analysis of proneural gene expression in the embryonic cerebellum. Dev Dyn Off Publ Am Assoc Anat. 2008;237(6):1726–1735. doi: 10.1002/dvdy.21571. [DOI] [PubMed] [Google Scholar]
- 26.Lebel M, Mo R, Shimamura K, Hui CC. Gli2 and Gli3 play distinct roles in the dorsoventral patterning of the mouse hindbrain. Dev Biol. 2007;302(1):345–355. doi: 10.1016/j.ydbio.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Cordes SP. Molecular genetics of cranial nerve development in mouse. Nat Rev Neurosci. 2001;2(9):611–623. doi: 10.1038/35090039. [DOI] [PubMed] [Google Scholar]
- 28.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101(4):435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 29.Lewis KE. How do genes regulate simple behaviours? Understanding how different neurons in the vertebrate spinal cord are genetically specified. Philos Trans R Soc Lond B Biol Sci. 2006;361(1465):45–66. doi: 10.1098/rstb.2005.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira C, Pombero A, Garcia-Lopez R, Gimeno L, Echevarria D, Martinez S. Molecular mechanisms controlling brain development: an overview of neuroepithelial secondary organizers. Int J Dev Biol. 2010;54(1):7–20. doi: 10.1387/ijdb.092853cv. [DOI] [PubMed] [Google Scholar]
- 31.Beccari L, Marco-Ferreres R, Bovolenta P. The logic of gene regulatory networks in early vertebrate forebrain patterning. Mech Dev. 2013;130(2–3):95–111. doi: 10.1016/j.mod.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Puelles L, Rubenstein JL. Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 1993;16(11):472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 33.Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26(9):469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 34.Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121(12):3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- 35.Briscoe J, Novitch BG. Regulatory pathways linking progenitor patterning, cell fates and neurogenesis in the ventral neural tube. Philos Trans R Soc Lond B Biol Sci. 2008;363(1489):57–70. doi: 10.1098/rstb.2006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briscoe J. Making a grade: Sonic hedgehog signalling and the control of neural cell fate. EMBO J. 2009;28(5):457–465. doi: 10.1038/emboj.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol. 2005;17(6):639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Gelman DM, Marín O, Rubenstein JLR (2012) The Generation of cortical interneurons. In: Noebels JL, Avoli M, Rogawski MA et al (eds) Jasper's basic mechanisms of the epilepsies [Internet], 4th edn. National Center for Biotechnology Information (USA), Bethesda, MD. http://www.ncbi.nlm.nih.gov/books/NBK98190/ [PubMed]
- 39.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci Off J Soc Neurosci. 2004;24(11):2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci Off J Soc Neurosci. 2007;27(41):10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi G, Fishell G. GABAergic interneuron lineages selectively sort into specific cortical layers during early postnatal development. Cereb Cortex. 2011;21(4):845–852. doi: 10.1093/cercor/bhq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126(15):3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 43.Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28(3):727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- 44.Gelman D, Griveau A, Dehorter N, Teissier A, Varela C, Pla R, Pierani A, Marin O. A wide diversity of cortical GABAergic interneurons derives from the embryonic preoptic area. J Neurosci Off J Soc Neurosci. 2011;31(46):16570–16580. doi: 10.1523/JNEUROSCI.4068-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelman DM, Martini FJ, Nobrega-Pereira S, Pierani A, Kessaris N, Marin O. The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J Neurosci Off J Soc Neurosci. 2009;29(29):9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B, Long JE, Flandin P, Pla R, Waclaw RR, Campbell K, Rubenstein JL. Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants. J Comp Neurol. 2013;521(7):1561–1584. doi: 10.1002/cne.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13(6):767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127(20):4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- 49.Carney RS, Cocas LA, Hirata T, Mansfield K, Corbin JG. Differential regulation of telencephalic pallial–subpallial boundary patterning by Pax6 and Gsh2. Cereb Cortex. 2009;19(4):745–759. doi: 10.1093/cercor/bhn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbin JG, Rutlin M, Gaiano N, Fishell G. Combinatorial function of the homeodomain proteins Nkx2.1 and Gsh2 in ventral telencephalic patterning. Development. 2003;130(20):4895–4906. doi: 10.1242/dev.00717. [DOI] [PubMed] [Google Scholar]
- 51.Pei Z, Wang B, Chen G, Nagao M, Nakafuku M, Campbell K. Homeobox genes Gsx1 and Gsx2 differentially regulate telencephalic progenitor maturation. Proc Natl Acad Sci USA. 2011;108(4):1675–1680. doi: 10.1073/pnas.1008824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waclaw RR, Wang B, Pei Z, Ehrman LA, Campbell K. Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron. 2009;63(4):451–465. doi: 10.1016/j.neuron.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417(6889):645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- 54.Petanjek Z, Kostovic I, Esclapez M. Primate-specific origins and migration of cortical GABAergic neurons. Front Neuroanat. 2009;3:26. doi: 10.3389/neuro.05.026.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai Y, Zhang Y, Shen Q, Rubenstein JL, Yang Z. A subpopulation of individual neural progenitors in the mammalian dorsal pallium generates both projection neurons and interneurons in vitro. Stem Cells. 2013;31(6):1193–1201. doi: 10.1002/stem.1363. [DOI] [PubMed] [Google Scholar]
- 56.Kohwi M, Petryniak MA, Long JE, Ekker M, Obata K, Yanagawa Y, Rubenstein JL, Alvarez-Buylla A. A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J Neurosci Off J Soc Neurosci. 2007;27(26):6878–6891. doi: 10.1523/JNEUROSCI.0254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakagawa Y, Shimogori T. Diversity of thalamic progenitor cells and postmitotic neurons. Eur J Neurosci. 2012;35(10):1554–1562. doi: 10.1111/j.1460-9568.2012.08089.x. [DOI] [PubMed] [Google Scholar]
- 58.Hirata T, Nakazawa M, Muraoka O, Nakayama R, Suda Y, Hibi M. Zinc-finger genes Fez and Fez-like function in the establishment of diencephalon subdivisions. Development. 2006;133(20):3993–4004. doi: 10.1242/dev.02585. [DOI] [PubMed] [Google Scholar]
- 59.Jeong Y, Dolson DK, Waclaw RR, Matise MP, Sussel L, Campbell K, Kaestner KH, Epstein DJ. Spatial and temporal requirements for sonic hedgehog in the regulation of thalamic interneuron identity. Development. 2011;138(3):531–541. doi: 10.1242/dev.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Achim K, Peltopuro P, Lahti L, Li J, Salminen M, Partanen J. Distinct developmental origins and regulatory mechanisms for GABAergic neurons associated with dopaminergic nuclei in the ventral mesodiencephalic region. Development. 2012;139(13):2360–2370. doi: 10.1242/dev.076380. [DOI] [PubMed] [Google Scholar]
- 61.Arber S. Motor circuits in action: specification, connectivity, and function. Neuron. 2012;74(6):975–989. doi: 10.1016/j.neuron.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 62.Karunaratne A, Hargrave M, Poh A, Yamada T. GATA proteins identify a novel ventral interneuron subclass in the developing chick spinal cord. Dev Biol. 2002;249(1):30–43. doi: 10.1006/dbio.2002.0754. [DOI] [PubMed] [Google Scholar]
- 63.Smith E, Hargrave M, Yamada T, Begley CG, Little MH. Coexpression of SCL and GATA3 in the V2 interneurons of the developing mouse spinal cord. Dev Dyn Off Publ Am Assoc Anat. 2002;224(2):231–237. doi: 10.1002/dvdy.10093. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Y, Yamamoto M, Engel JD. GATA2 is required for the generation of V2 interneurons. Development. 2000;127(17):3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
- 65.Panayi H, Panayiotou E, Orford M, Genethliou N, Mean R, Lapathitis G, Li S, Xiang M, Kessaris N, Richardson WD, Malas S. Sox1 is required for the specification of a novel p2-derived interneuron subtype in the mouse ventral spinal cord. J Neurosci Off J Soc Neurosci. 2010;30(37):12274–12280. doi: 10.1523/JNEUROSCI.2402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S, Misra K, Matise MP, Xiang M. Foxn4 acts synergistically with Mash1 to specify subtype identity of V2 interneurons in the spinal cord. Proc Natl Acad Sci USA. 2005;102(30):10688–10693. doi: 10.1073/pnas.0504799102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Del Barrio MG, Taveira-Marques R, Muroyama Y, Yuk DI, Li S, Wines-Samuelson M, Shen J, Smith HK, Xiang M, Rowitch D, Richardson WD. A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development. 2007;134(19):3427–3436. doi: 10.1242/dev.005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pierani A, Brenner-Morton S, Chiang C, Jessell TM. A sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell. 1999;97(7):903–915. doi: 10.1016/s0092-8674(00)80802-8. [DOI] [PubMed] [Google Scholar]
- 69.Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell TM, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31(5):743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- 70.Alvarez FJ, Jonas PC, Sapir T, Hartley R, Berrocal MC, Geiman EJ, Todd AJ, Goulding M. Postnatal phenotype and localization of spinal cord V1-derived interneurons. J Comp Neurol. 2005;493(2):177–192. doi: 10.1002/cne.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sapir T, Geiman EJ, Wang Z, Velasquez T, Mitsui S, Yoshihara Y, Frank E, Alvarez FJ, Goulding M. Pax6 and engrailed 1 regulate two distinct aspects of Renshaw cell development. J Neurosci Off J Soc Neurosci. 2004;24(5):1255–1264. doi: 10.1523/JNEUROSCI.3187-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M. Genetic identification of spinal interneurons that coordinate left–right locomotor activity necessary for walking movements. Neuron. 2004;42(3):375–386. doi: 10.1016/s0896-6273(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 73.Pierani A, Moran-Rivard L, Sunshine MJ, Littman DR, Goulding M, Jessell TM. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron. 2001;29(2):367–384. doi: 10.1016/s0896-6273(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 74.Gribble SL, Nikolaus OB, Dorsky RI. Regulation and function of Dbx genes in the zebrafish spinal cord. Dev Dyn Off Publ Am Assoc Anat. 2007;236(12):3472–3483. doi: 10.1002/dvdy.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matise MP, Joyner AL. Expression patterns of developmental control genes in normal and Engrailed-1 mutant mouse spinal cord reveal early diversity in developing interneurons. J Neurosci Off J Soc Neurosci. 1997;17(20):7805–7816. doi: 10.1523/JNEUROSCI.17-20-07805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34(4):535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 77.Kriks S, Lanuza GM, Mizuguchi R, Nakafuku M, Goulding M. Gsh2 is required for the repression of Ngn1 and specification of dorsal interneuron fate in the spinal cord. Development. 2005;132(13):2991–3002. doi: 10.1242/dev.01878. [DOI] [PubMed] [Google Scholar]
- 78.Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr Opin Neurobiol. 2003;13(1):42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 79.Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y, Kawaguchi Y, Nabeshima Y, Hoshino M. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136(12):2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- 80.Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20(6):707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 81.Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, Dolle D, Bithell A, Ettwiller L, Buckley N, Guillemot F. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25(9):930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(Suppl 1):S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 83.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by So1–3 activity. Nat Neurosci. 2003;6(11):1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 84.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 85.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 86.Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16(3):324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14(1):67–80. [PMC free article] [PubMed] [Google Scholar]
- 88.Peltopuro P, Kala K, Partanen J. Distinct requirements for Ascl1 in subpopulations of midbrain GABAergic neurons. Dev Biol. 2010;343(1–2):63–70. doi: 10.1016/j.ydbio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 89.Kataoka A, Shimogori T. Fgf8 controls regional identity in the developing thalamus. Development. 2008;135(17):2873–2881. doi: 10.1242/dev.021618. [DOI] [PubMed] [Google Scholar]
- 90.Guimera J, Weisenhorn DV, Wurst W. Megane/Heslike is required for normal GABAergic differentiation in the mouse superior colliculus. Development. 2006;133(19):3847–3857. doi: 10.1242/dev.02557. [DOI] [PubMed] [Google Scholar]
- 91.Miyoshi G, Bessho Y, Yamada S, Kageyama R. Identification of a novel basic helix-loop-helix gene, Heslike, and its role in GABAergic neurogenesis. J Neurosci Off J Soc Neurosci. 2004;24(14):3672–3682. doi: 10.1523/JNEUROSCI.5327-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guimera J, Vogt Weisenhorn D, Echevarria D, Martinez S, Wurst W. Molecular characterization, structure and developmental expression of Megane bHLH factor. Gene. 2006;377:65–76. doi: 10.1016/j.gene.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 93.Delogu A, Sellers K, Zagoraiou L, Bocianowska-Zbrog A, Mandal S, Guimera J, Rubenstein JL, Sugden D, Jessell T, Lumsden A. Subcortical visual shell nuclei targeted by ipRGCs develop from a Sox14+-GABAergic progenitor and require Sox14 to regulate daily activity rhythms. Neuron. 2012;75(4):648–662. doi: 10.1016/j.neuron.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 94.Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9(6):770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- 95.Sudarov A, Turnbull RK, Kim EJ, Lebel-Potter M, Guillemot F, Joyner AL. Ascl1 genetics reveals insights into cerebellum local circuit assembly. J Neurosci Off J Soc Neurosci. 2011;31(30):11055–11069. doi: 10.1523/JNEUROSCI.0479-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wildner H, Muller T, Cho SH, Brohl D, Cepko CL, Guillemot F, Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133(11):2105–2113. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]
- 97.Grimaldi P, Parras C, Guillemot F, Rossi F, Wassef M. Origins and control of the differentiation of inhibitory interneurons and glia in the cerebellum. Dev Biol. 2009;328(2):422–433. doi: 10.1016/j.ydbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 98.Lundell TG, Zhou Q, Doughty ML. Neurogenin1 expression in cell lineages of the cerebellar cortex in embryonic and postnatal mice. Dev Dyn Off Publ Am Assoc Anat. 2009;238(12):3310–3325. doi: 10.1002/dvdy.22165. [DOI] [PubMed] [Google Scholar]
- 99.Florio M, Leto K, Muzio L, Tinterri A, Badaloni A, Croci L, Zordan P, Barili V, Albieri I, Guillemot F, Rossi F, Consalez GG. Neurogenin 2 regulates progenitor cell-cycle progression and Purkinje cell dendritogenesis in cerebellar development. Development. 2012;139(13):2308–2320. doi: 10.1242/dev.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Henke RM, Savage TK, Meredith DM, Glasgow SM, Hori K, Dumas J, MacDonald RJ, Johnson JE. Neurog2 is a direct downstream target of the Ptf1a–Rbpj transcription complex in dorsal spinal cord. Development. 2009;136(17):2945–2954. doi: 10.1242/dev.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng CY, Yajima H, Burns CE, Zon LI, Sisodia SS, Pfaff SL, Sharma K. Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron. 2007;53(6):813–827. doi: 10.1016/j.neuron.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132(24):5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- 103.Muroyama Y, Fujiwara Y, Orkin SH, Rowitch DH. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005;438(7066):360–363. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- 104.Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8(11):1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- 105.Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JL. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414(2):217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 106.Stuhmer T, Puelles L, Ekker M, Rubenstein JL. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb Cortex. 2002;12(1):75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- 107.Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci Off J Soc Neurosci. 2007;27(12):3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512(4):556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci Off J Soc Neurosci. 2000;20(2):709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Poitras L, Ghanem N, Hatch G, Ekker M. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development. 2007;134(9):1755–1765. doi: 10.1242/dev.02845. [DOI] [PubMed] [Google Scholar]
- 111.Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JL. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci. 2009;40(2):167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Panganiban G, Rubenstein JL. Developmental functions of the distal-less/Dlx homeobox genes. Development. 2002;129(19):4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 113.Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19(1):27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 114.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278(5337):474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 115.Colasante G, Collombat P, Raimondi V, Bonanomi D, Ferrai C, Maira M, Yoshikawa K, Mansouri A, Valtorta F, Rubenstein JL, Broccoli V. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci Off J Soc Neurosci. 2008;28(42):10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ghanem N, Yu M, Long J, Hatch G, Rubenstein JL, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci Off J Soc Neurosci. 2007;27(19):5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8(8):1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- 118.McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio LA, Huylebroeck D, Higashi Y, Rubenstein JL. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77(1):83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fulp CT, Cho G, Marsh ED, Nasrallah IM, Labosky PA, Golden JA. Identification of Arx transcriptional targets in the developing basal forebrain. Hum Mol Genet. 2008;17(23):3740–3760. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Herberth B, Minko K, Csillag A, Jaffredo T, Madarasz E. SCL, GATA-2 and Lmo2 expression in neurogenesis. Int J Dev Neurosci Off J Int Soc Dev Neurosci. 2005;23(5):449–463. doi: 10.1016/j.ijdevneu.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 121.Achim K, Peltopuro P, Lahti L, Tsai H, Zachariah A, Åstrand M, Salminen M, Rowitch D, Partanen J. The role of Tal2 and Tal1 in the differentiation of midbrain GABAergic neuron precursors. Biol Open. 2013 doi: 10.1242/bio.20135041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ogilvy S, Ferreira R, Piltz SG, Bowen JM, Gottgens B, Green AR. The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins. Mol Cell Biol. 2007;27(20):7206–7219. doi: 10.1128/MCB.00931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Joshi K, Lee S, Lee B, Lee JW, Lee SK. LMO4 controls the balance between excitatory and inhibitory spinal V2 interneurons. Neuron. 2009;61(6):839–851. doi: 10.1016/j.neuron.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Minaki Y, Nakatani T, Mizuhara E, Inoue T, Ono Y. Identification of a novel transcriptional corepressor, Corl2, as a cerebellar Purkinje cell-selective marker. Gene Expr Patterns GEP. 2008;8(6):418–423. doi: 10.1016/j.gep.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 125.Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47(2):201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 126.Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci USA. 2007;104(12):5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yamada M, Terao M, Terashima T, Fujiyama T, Kawaguchi Y, Nabeshima Y, Hoshino M. Origin of climbing fiber neurons and their developmental dependence on Ptf1a. J Neurosci Off J Soc Neurosci. 2007;27(41):10924–10934. doi: 10.1523/JNEUROSCI.1423-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7(5):510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- 129.Huang M, Huang T, Xiang Y, Xie Z, Chen Y, Yan R, Xu J, Cheng L. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev Biol. 2008;322(2):394–405. doi: 10.1016/j.ydbio.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 130.Meredith DM, Borromeo MD, Deering TG, Casey B, Savage TK, Mayer PR, Hoang C, Tung KC, Kumar M, Shen C, Swift GH, Macdonald RJ, Johnson JE. Program specificity for Ptf1a in pancreas versus neural tube development correlates with distinct collaborating cofactors and chromatin accessibility. Mol Cell Biol. 2013;33(16):3166–3179. doi: 10.1128/MCB.00364-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hori K, Cholewa-Waclaw J, Nakada Y, Glasgow SM, Masui T, Henke RM, Wildner H, Martarelli B, Beres TM, Epstein JA, Magnuson MA, Macdonald RJ, Birchmeier C, Johnson JE. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22(2):166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Graw J. Eye development. Curr Top Dev Biol. 2010;90:343–386. doi: 10.1016/S0070-2153(10)90010-0. [DOI] [PubMed] [Google Scholar]
- 133.Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133(22):4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- 134.Jusuf PR, Almeida AD, Randlett O, Joubin K, Poggi L, Harris WA. Origin and determination of inhibitory cell lineages in the vertebrate retina. J Neurosci Off J Soc Neurosci. 2011;31(7):2549–2562. doi: 10.1523/JNEUROSCI.4713-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134(6):1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- 136.Dullin JP, Locker M, Robach M, Henningfeld KA, Parain K, Afelik S, Pieler T, Perron M. Ptf1a triggers GABAergic neuronal cell fates in the retina. BMC Dev Biol. 2007;7:110. doi: 10.1186/1471-213X-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lamb TD, Collin SP, Pugh EN., Jr Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat Rev Neurosci. 2007;8(12):960–976. doi: 10.1038/nrn2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2(2):109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 139.Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, Rubenstein JL, Horvath S, Geschwind DH. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Holmberg J, Perlmann T. Maintaining differentiated cellular identity. Nat Rev Genet. 2012;13(6):429–439. doi: 10.1038/nrg3209. [DOI] [PubMed] [Google Scholar]
- 141.Zhou QP, Le TN, Qiu X, Spencer V, de Melo J, Du G, Plews M, Fonseca M, Sun JM, Davie JR, Eisenstat DD. Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res. 2004;32(3):884–892. doi: 10.1093/nar/gkh233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet TIG. 2000;16(2):75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 143.Pillai A, Mansouri A, Behringer R, Westphal H, Goulding M. Lhx1 and Lhx5 maintain the inhibitory-neurotransmitter status of interneurons in the dorsal spinal cord. Development. 2007;134(2):357–366. doi: 10.1242/dev.02717. [DOI] [PubMed] [Google Scholar]
- 144.Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA. 2007;104(32):13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moreno N, Bachy I, Retaux S, Gonzalez A. LIM-homeodomain genes as developmental and adult genetic markers of Xenopus forebrain functional subdivisions. J Comp Neurol. 2004;472(1):52–72. doi: 10.1002/cne.20046. [DOI] [PubMed] [Google Scholar]
- 146.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci Off J Soc Neurosci. 2011;31(9):3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204(7):1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol. 2013;25(2):215–221. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73(1):35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 151.Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 152.de Castro F, Bribian A. The molecular orchestra of the migration of oligodendrocyte precursors during development. Brain Res Brain Res Rev. 2005;49(2):227–241. doi: 10.1016/j.brainresrev.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 153.Ghashghaei HT, Lai C, Anton ES. Neuronal migration in the adult brain: are we there yet? Nat Rev Neurosci. 2007;8(2):141–151. doi: 10.1038/nrn2074. [DOI] [PubMed] [Google Scholar]
- 154.Chedotal A, Rijli FM. Transcriptional regulation of tangential neuronal migration in the developing forebrain. Curr Opin Neurobiol. 2009;19(2):139–145. doi: 10.1016/j.conb.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 155.Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5(10):793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 156.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321(5885):53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Baudoin JP, Viou L, Launay PS, Luccardini C, Espeso Gil S, Kiyasova V, Irinopoulou T, Alvarez C, Rio JP, Boudier T, Lechaire JP, Kessaris N, Spassky N, Metin C. Tangentially migrating neurons assemble a primary cilium that promotes their reorientation to the cortical plate. Neuron. 2012;76(6):1108–1122. doi: 10.1016/j.neuron.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 158.Marin O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JL. Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science. 2001;293(5531):872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- 159.Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marin O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44(2):251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 160.Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, Hollt V, Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci Off J Soc Neurosci. 2003;23(12):5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yozu M, Tabata H, Nakajima K. The caudal migratory stream: a novel migratory stream of interneurons derived from the caudal ganglionic eminence in the developing mouse forebrain. J Neurosci Off J Soc Neurosci. 2005;25(31):7268–7277. doi: 10.1523/JNEUROSCI.2072-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zimmer G, Rudolph J, Landmann J, Gerstmann K, Steinecke A, Gampe C, Bolz J. Bidirectional ephrinB3/EphA4 signaling mediates the segregation of medial ganglionic eminence- and preoptic area-derived interneurons in the deep and superficial migratory stream. J Neurosci Off J Soc Neurosci. 2011;31(50):18364–18380. doi: 10.1523/JNEUROSCI.4690-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Marin O. Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur J Neurosci. 2013;38(1):2019–2029. doi: 10.1111/ejn.12225. [DOI] [PubMed] [Google Scholar]
- 164.Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2(11):780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 165.Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci Off J Soc Neurosci. 1999;19(18):7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat Neurosci. 1999;2(5):461–466. doi: 10.1038/8131. [DOI] [PubMed] [Google Scholar]
- 167.Li G, Adesnik H, Li J, Long J, Nicoll RA, Rubenstein JL, Pleasure SJ. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J Neurosci Off J Soc Neurosci. 2008;28(5):1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lopez-Bendito G, Sanchez-Alcaniz JA, Pla R, Borrell V, Pico E, Valdeolmillos M, Marin O. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J Neurosci Off J Soc Neurosci. 2008;28(7):1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tiveron MC, Rossel M, Moepps B, Zhang YL, Seidenfaden R, Favor J, Konig N, Cremer H. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J Neurosci Off J Soc Neurosci. 2006;26(51):13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sessa A, Mao CA, Colasante G, Nini A, Klein WH, Broccoli V. Tbr2-positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev. 2010;24(16):1816–1826. doi: 10.1101/gad.575410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zarbalis K, Choe Y, Siegenthaler JA, Orosco LA, Pleasure SJ. Meningeal defects alter the tangential migration of cortical interneurons in Foxc1hith/hith mice. Neural Dev. 2012;7:2. doi: 10.1186/1749-8104-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pla R, Borrell V, Flames N, Marin O. Layer acquisition by cortical GABAergic interneurons is independent of Reelin signaling. J Neurosci Off J Soc Neurosci. 2006;26(26):6924–6934. doi: 10.1523/JNEUROSCI.0245-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fairen A, Cobas A, Fonseca M. Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. J Comp Neurol. 1986;251(1):67–83. doi: 10.1002/cne.902510105. [DOI] [PubMed] [Google Scholar]
- 174.Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci Off J Soc Neurosci. 2003;23(12):5113–5122. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hevner RF, Daza RA, Englund C, Kohtz J, Fink A. Postnatal shifts of interneuron position in the neocortex of normal and reeler mice: evidence for inward radial migration. Neuroscience. 2004;124(3):605–618. doi: 10.1016/j.neuroscience.2003.11.033. [DOI] [PubMed] [Google Scholar]
- 176.Lodato S, Rouaux C, Quast KB, Jantrachotechatchawan C, Studer M, Hensch TK, Arlotta P. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69(4):763–779. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JL. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69(1):61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cobos I, Broccoli V, Rubenstein JL. The vertebrate ortholog of Aristaless is regulated by Dlx genes in the developing forebrain. J Comp Neurol. 2005;483(3):292–303. doi: 10.1002/cne.20405. [DOI] [PubMed] [Google Scholar]
- 179.Colombo E, Collombat P, Colasante G, Bianchi M, Long J, Mansouri A, Rubenstein JL, Broccoli V. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J Neurosci Off J Soc Neurosci. 2007;27(17):4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, Matsuo M, Kamijo S, Kasahara M, Yoshioka H, Ogata T, Fukuda T, Kondo I, Kato M, Dobyns WB, Yokoyama M, Morohashi K. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32(3):359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 181.Friocourt G, Parnavelas JG (2011) Identification of Arx targets unveils new candidates for controlling cortical interneuron migration and differentiation. Front Cell Neurosci. doi:10.3389/fncel.2011.00028 [DOI] [PMC free article] [PubMed]
- 182.Kanatani S, Yozu M, Tabata H, Nakajima K. COUP-TFII is preferentially expressed in the caudal ganglionic eminence and is involved in the caudal migratory stream. J Neurosci Off J Soc Neurosci. 2008;28(50):13582–13591. doi: 10.1523/JNEUROSCI.2132-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nobrega-Pereira S, Kessaris N, Du T, Kimura S, Anderson SA, Marin O. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59(5):733–745. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Le TN, Du G, Fonseca M, Zhou QP, Wigle JT, Eisenstat DD. Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J Biol Chem. 2007;282(26):19071–19081. doi: 10.1074/jbc.M607486200. [DOI] [PubMed] [Google Scholar]
- 185.Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci Off J Soc Neurosci. 2007;27(12):3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Du T, Xu Q, Ocbina PJ, Anderson SA. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 2008;135(8):1559–1567. doi: 10.1242/dev.015123. [DOI] [PubMed] [Google Scholar]
- 187.Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510(1):79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11(1):173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 189.Luskin MB, Boone MS. Rate and pattern of migration of lineally-related olfactory bulb interneurons generated postnatally in the subventricular zone of the rat. Chem Senses. 1994;19(6):695–714. doi: 10.1093/chemse/19.6.695. [DOI] [PubMed] [Google Scholar]
- 190.Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264(5162):1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 191.Hakanen J, Duprat S, Salminen M. Netrin1 is required for neural and glial precursor migrations into the olfactory bulb. Dev Biol. 2011;355(1):101–114. doi: 10.1016/j.ydbio.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 192.Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54(6):873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Alberti S, Krause SM, Kretz O, Philippar U, Lemberger T, Casanova E, Wiebel FF, Schwarz H, Frotscher M, Schutz G, Nordheim A. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc Natl Acad Sci USA. 2005;102(17):6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vasudevan A, Won C, Li S, Erdelyi F, Szabo G, Kim KS. Dopaminergic neurons modulate GABA neuron migration in the embryonic midbrain. Development. 2012;139(17):3136–3141. doi: 10.1242/dev.078394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Crandall JE, McCarthy DM, Araki KY, Sims JR, Ren JQ, Bhide PG. Dopamine receptor activation modulates GABA neuron migration from the basal forebrain to the cerebral cortex. J Neurosci Off J Soc Neurosci. 2007;27(14):3813–3822. doi: 10.1523/JNEUROSCI.5124-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14(4–5):355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- 197.Chen L, Guo Q, Li JY. Transcription factor Gbx2 acts cell-nonautonomously to regulate the formation of lineage-restriction boundaries of the thalamus. Development. 2009;136(8):1317–1326. doi: 10.1242/dev.030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mastick GS, Andrews GL. Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci. 2001;17(1):190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- 199.Zhao GY, Li ZY, Zou HL, Hu ZL, Song NN, Zheng MH, Su CJ, Ding YQ. Expression of the transcription factor GATA3 in the postnatal mouse central nervous system. Neurosci Res. 2008;61(4):420–428. doi: 10.1016/j.neures.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 200.Agarwala S, Sanders TA, Ragsdale CW. Sonic hedgehog control of size and shape in midbrain pattern formation. Science. 2001;291(5511):2147–2150. doi: 10.1126/science.1058624. [DOI] [PubMed] [Google Scholar]
- 201.Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133(9):1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- 202.Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, Czerny T, Brodski C, Ang SL, Wurst W, Simeone A. Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development. 2004;131(9):2037–2048. doi: 10.1242/dev.01107. [DOI] [PubMed] [Google Scholar]
- 203.Waite MR, Skidmore JM, Billi AC, Martin JF, Martin DM. GABAergic and glutamatergic identities of developing midbrain Pitx2 neurons. Dev Dyn Off Publ Am Assoc Anat. 2011;240(2):333–346. doi: 10.1002/dvdy.22532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lorente-Canovas B, Marin F, Corral-San-Miguel R, Hidalgo-Sanchez M, Ferran JL, Puelles L, Aroca P. Multiple origins, migratory paths and molecular profiles of cells populating the avian interpeduncular nucleus. Dev Biol. 2012;361(1):12–26. doi: 10.1016/j.ydbio.2011.09.032. [DOI] [PubMed] [Google Scholar]
- 205.Waite MR, Skaggs K, Kaviany P, Skidmore JM, Causeret F, Martin JF, Martin DM. Distinct populations of GABAergic neurons in mouse rhombomere 1 express but do not require the homeodomain transcription factor PITX2. Mol Cell Neurosci. 2012;49(1):32–43. doi: 10.1016/j.mcn.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sgaier SK, Lao Z, Villanueva MP, Berenshteyn F, Stephen D, Turnbull RK, Joyner AL. Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development. 2007;134(12):2325–2335. doi: 10.1242/dev.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sillitoe RV, Stephen D, Lao Z, Joyner AL. Engrailed homeobox genes determine the organization of Purkinje cell sagittal stripe gene expression in the adult cerebellum. J Neurosci Off J Soc Neurosci. 2008;28(47):12150–12162. doi: 10.1523/JNEUROSCI.2059-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wilson SL, Kalinovsky A, Orvis GD, Joyner AL. Spatially restricted and developmentally dynamic expression of engrailed genes in multiple cerebellar cell types. Cerebellum. 2011;10(3):356–372. doi: 10.1007/s12311-011-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, Millen KJ. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133(15):2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- 210.Kim EJ, Battiste J, Nakagawa Y, Johnson JE. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Mol Cell Neurosci. 2008;38(4):595–606. doi: 10.1016/j.mcn.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Maricich SM, Herrup K. Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. J Neurobiol. 1999;41(2):281–294. doi: 10.1002/(sici)1097-4695(19991105)41:2<281::aid-neu10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 212.Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132(12):2709–2719. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Moran-Rivard L, Kagawa T, Saueressig H, Gross MK, Burrill J, Goulding M. Evx1 is a postmitotic determinant of v0 interneuron identity in the spinal cord. Neuron. 2001;29(2):385–399. doi: 10.1016/s0896-6273(01)00213-6. [DOI] [PubMed] [Google Scholar]