Abstract
Polycomb group proteins (PcGs) are transcriptional repressors involved in physiological processes whereas PcG deregulation might result in oncogenesis. MYC oncogene is able to regulate gene transcription, proliferation, apoptosis, and malignant transformation. MYC deregulation might result in tumorigenesis with tumor maintenance properties in both solid and blood cancers. Although the interaction of PcG and MYC in cancer was described years ago, new findings are reported every day to explain the exact mechanisms and results of such interactions. In this review, we summarize recent data on the PcG and MYC interactions in cancer, and the putative involvement of microRNAs in the equation.
Keywords: Polycomb group proteins, MYC, EZH2, BMI1, MicroRNA, Cancer
Polycomb group proteins
Polycomb group proteins (PcGs) represent transcriptional repressors of protein-coding gene promoters required for genomic imprinting, chromosome X inactivation (XCI), stem cell plasticity, cell fate determination, and maintenance of the correct cell identity at different developmental stages. Gene repression, which seems to be a dynamic process during differentiation, is achieved via PcG recruitment to target genes. That process involves several different transcription factors (TFs), long non-coding RNAs (lncRNAs), and fusion oncoproteins like PML/RARα and PLZF/RARα. PcGs are closely associated with DNA methylation by recruiting DNA methyltransferases (DNMTs) to target genes [1–6]. Recent findings suggest that XCI spreading seems to be governed by a hierarchy of two types of PcG target sites; the ‘canonical’ sites, which typically contain CpG islands and the ‘non-canonical’ sites that lack H3K4me3 or CpG islands. XCI requires a network involving the lncRNA XIST linked to the proteins HBiX and SMCHD1 [7, 8]. However, previously published data supported that PcG are not required for initiating or maintaining random XCI in mouse embryonic cells [9]. PcG have important regulatory functions during the cell cycle phases repressing cyclins, cyclin-dependent kinase (CDK) inhibitors, the pRB–E2F complex, while they can control DNA synthesis during S phase. PcG affect DNA damage pathways and repair mechanisms. They also regulate apoptosis and prevent the onset of senescence as PcG are downregulated during replicative senescence [10]. For instance, PcG proteins confer the hematopoietic stem cell (HSCs) the ability to act as progenitors and protects them from apoptosis through regulation of the Ink4α/Arf locus. Compositional changes of PcG proteins induces differentiation blockade, leading to malignant hematopoietic phenotypes [11, 12].
PcGs have the ability to re-establish the histone code on newly assembled unmethylated histones, at least in Drosophila [13]. They remain associated with mitotic chromatin in Drosophila S2 cells in order to ensure equal segregation to both new cells, or to keep local concentration of PcG proteins near the DNA at high levels, facilitating rebinding after mitosis [14].
PcG form multiprotein complexes called polycomb repressive complex 1 and 2 (PRC1/2) responsible for gene silencing through post-translational histone modifications. They are involved in monoubiquitylation of lysine 119 of histone H2A (H2AK119ub) and di- and tri-methylation of lysine 27 of histone H3 (H3K27me3), respectively. Multiple forms of the PRC1 exist containing combinations of different PcG proteins (CBX2/4/6/7/8), posterior sex combs proteins PCGF1 (NSPC1), PCGF2 (MEL18), PCGF3, PCGF4 (BMI1), PCGF5, PCGF6 (MBLR), the RING1 and RNF2 (RING2) proteins, three PH proteins (PHC1/2/3 or HPH1/2/3), two Sex combs on midleg (SMCL1/2), and RYBP/YAF2 [1, 15]. PRC2 consists of the core subunits enhancer of zeste 1/2 (EZH1/2), embryonic ectoderm development (EED), the RBBP7/4 (RbAp46/48) protein, and the suppressor of zeste 12 (SUZ12) [16]. Although EZH2 and EZH1 seem to be the catalytic subunit of PRC2, EED specifically recognizes and binds to the repressive trimethylated lysine marks contributing to the affinity of PRC2 for the nucleosomes [17]. PRC2 contains other proteins like AEBP2, PHF1 (PCL1), MTF2 (PCL2), PHF19 (PCL3), and JARID2, which regulate the enzymatic activity of PRC2 and co-localize with PRC2 on target genes. PRC2 is inhibited by H3K4me3 and H3K36me2/3, preventing H3K27me3 occupancy on transcriptionally active genes [18]. On the other hand, PRC1 might affect the ability of TF-IID to remain attached to gene promoters, suggesting a putative transcriptional silencing mechanism. In this way, target genes stay in a poised state to be expressed later during the developmental process [19]. Mounting evidence suggests that both PRC1 and PRC2 complexes can be recruited by lncRNAs on target genes promoting gene silencing [20].
The role of other histone modifications in PRC2 regulation is only now becoming apparent, and very recent data made evident that the member of the PCL family Phf19 regulates PRC2 occupancy. Phf19 recruits specific demethylases to mouse embryonic stem cells (mESCs) during differentiation leading to H3K27me3 and transcriptional silencing [21, 22]. PRC2 subunits might act either as a tumor suppressor (TS) or as an oncogene, suggesting that H3K27me3 may possess dual functions in different cell types [23]. Recent findings demonstrated that the PRC2 complex is allosterically activated by neighboring nucleosomes. Active genes are resistant to PRC2 activity, not only because of active epigenetic marks that antagonize PRC2 activity but also because the chromatin of transcribing genes is less compact, with lower nucleosome density. Therefore, the ability of PRC2 to distinguish active chromatin and dense inactive chromatin highlights the efficiency of PRC2 in maintaining the inactive states of target genes [24]. PHF1 has also been recently found to protect p53 from MDM2 degradation n vitro and in vivo [25].
BMI11 is probably the most studied subunit of PRC1, and is involved in normal stem cell proliferation affecting self-renewal and maintenance of HSCs and neuronal stem cells. It also has an important role in the malignant transformation of normal stem or differentiated cells in cancer stem cells. It is associated with metastasis and chemoresistence to agents routinely used in the battle against cancer. On the contrary, BMI1 silencing enhances the antitumor activity of chemotherapeutic agents [26, 27].
In physiological conditions, Ezh2 is essential for fetal hematopoiesis affecting erythroid differentiation in the fetal liver, while it does not seem to affect self-renewal in adult bone marrow. It might also epigenetically regulate transcriptional programs controlling migration and connectivity of neurons in the cortico–ponto–cerebellar pathway in mice [28, 29]. Ezh2 is closely associated with DNMTs as it is capable of recruiting DNMTs to gene promoters promoting gene silencing through DNA methylation [30].
Deregulated expression of PcG proteins has been identified in several types of hematologic malignancies and solid cancers via modulation of Notch, Hedgehog, and Wnt pathways [31–36]. Intriguingly, EZH2 oncogenic function has been demonstrated not to depend on gene silencing but rather on the co-activation of the transcriptional induction of its target genes in castration-resistant prostate cancer due to EZH2-phosphorylation in a PRC2-independent pattern [37].
An MYC overview
MYC proto-oncogene belongs to a family that includes the c-MYC, the MYCL, and the MYCN homologues, which share general topography. In normal cells, MYC expression is regulated at the transcriptional, post-transcriptional, translational, and post-translational levels. In order to activate gene transcription, MYC forms heterodimers and acts in conjunction with the MYC-associated protein X (MAX), required for regulation of gene transcription, proliferation, apoptosis, and malignant transformation. MYC deregulation has tumor initiating and tumor maintenance properties in both solid and blood cancers. The oncogenic activity is attributed to gene amplification, translocations, and to deregulation of its cofactors such as MAX. MYC acts downstream of receptor signaling pathways, including Wnt, Notch, phosphoinositide 3-kinase (PI3K), and Ras, and interacts with microRNAs (miRNAs), regulating transcription of genes involved in cell growth and proliferation [38–40].
C-MYC expression is high during early embryonic development and persists in the cell cycle phases, whereas it is low or even undetectable in differentiated tissues [41]. A rather unusual feature of MYC is that it can regulate all three RNA polymerases (RNA pol) for a subset of their respective targets. Thus, besides protein-coding genes and many non-coding RNAs, including miRNA and lncRNA genes that are controlled by Pol II, MYC regulates rRNA and tRNA genes that are transcribed by Pol I and Pol III, respectively [42]. MYC exhibit antisenescence functions through its interaction with CDK2 or by repressing cell-cycle inhibitors like p15 ink4b, p21 cip1, activates genes involved in cell cycle progression (CDK4, cyclin D1), and possesses immunomodulatory potential by preventing T-cells from recognizing tumor cells [43, 44].
Lin et al. [45] have recently demonstrated that, in different tumor cells bearing high c-MYC levels, it occupies sites at both promoters and enhancers of the actively transcribed genes across the cancer cell genome. The increase in c-MYC occupancy leads to increased transcription elongation by RNA Pol II and increased levels of transcripts per cell. In that way, c-MYC causes transcriptional amplification, producing elevated levels of transcripts from the existing gene-expression program of tumor cells potentiating the already deregulated transcriptional program of cancer cells. Another intriguing MYC characteristic is that aside from inducing cell proliferation, it can also enhance programmed cell death and paradoxically might induce carcinogenesis by enhancing compensatory proliferation [46]. Myc causes malignant transformation of the cell but it can also induce apoptotic cell death through still unclear mechanisms. It seems that the TS Arf mediates such a switch via inhibition of the ubiquitylation of the c-Myc transcriptional domain. In turn, such inhibition induces the Egr1 gene essential for the c-Myc-induced p53-independent apoptosis observed in double knock out experiments in mouse embryonic fibroblasts (MEFs). That interaction is deregulated in cancer, through the overexpression of Skp2, which inhibits the recruitment of Arf to Egr1 [47].
Myc regulates global chromatin structure contributing to oncogenesis through histone modifications targeting the epigenetic machinery [48]. In mouse naïve cells, Myc binding sites are characterized by active chromatin marks while they are excluded from regions with repressive marks [49]. Target gene repression may be indirect since transcriptional repressors activated by Myc are recruited to Myc target genes as in the case of Ezh2. Ezh2 upregulation during B-cell activation mediates transcriptional repression across the genome via H3K27me3. Overall, the findings of that elegant study determine that Myc is a universal amplifier of gene activation affecting multiple alternative pathways [50].
Here, we summarize data relating to the ongoing progress of the interaction between PcG proteins and MYC oncogene in cancer, based on recent evidence. We report the associations with upstream and downstream effectors, and the networks that arise. Moreover, we describe the putative involvement of miRNAs as an intermediate link between the two afore-mentioned factors.
PcG protein interaction with MYC
PRC1 and MYC
Several years ago, Bmi1 was found to be a partner of Myc within the cell nucleus, leading to lymphomagenesis in a transgenic mice model, suggesting at that time that both factors are members of a transcription regulation complex [51, 52]. Sometime later, it was established that Bmi1 overexpression was able to inhibit Myc-induced apoptosis in MEFs through the negative regulation of Ink4a–Arf in transgenic mice [53]. Since then, several studies have confirmed the positive association between Bmi1 and Myc overexpression. Their interaction led to the formation of circuitries, and repression of TS genes in different subtypes of cancer, and it was also suggested that they may be involved in the generation of cancer stem cells (CSCs) [54–57].
BMI1 gene was found to be a direct transcriptional target of c-MYC in human diploid fibroblasts, and their co-regulation negatively regulated p16 mRNA and protein levels. However, physiological c-MYC levels did not affect p16, while hypoactive c-MYC altered p16 via BMI1. On the other hand, hyperactive c-MYC had a direct effect on CDKN2A (p16) promoter by direct binding to its promoter E-box [58]. These findings suggested that, in part, the combined action of BMI1 and MYC is necessary to modulate apoptosis through modification of cell cycle regulator expression.
Duss and colleagues [59] studied an estrogen-dependent transformation model of human mammary epithelial cells (HMECs) through lentiviral transduction of HMECs with estrogen receptor α (ERα), Myc, Bmi1, and Tert in mice. However, the ERα/Bmi1/Myc/Tert-transduced HMECs failed to survive in normal estrogen level conditions, while estrogen administration promoted tumorigenesis. Cells expressing Bmi1/ERα were biologically active and proliferating when exposed to estrogens. The addition of Myc accelerated tumor growth compared withthe Bmi1/ERα only cells. Given that only Myc cells failed to proliferate and that Bmi1/ERα proliferated slower than the Bmi1/ERα/Myc cells, it can be deduced that these factors are interconnected and increase each other’s activity. In breast cancer cell lines, it was shown that the overexpressed BMI1 positively correlated with Wnt1 and c-MYC expression. On the other hand, it negatively regulated the expression of Wnt inhibitors such as Dickkof (DKK), with the final result being the upregulation of Wnt target such as c-MYC. The specific TF participates in a positive feedback loop activating the transcription of BMI1. In turn, BMI1 itself is a target of Wnt pathway, and DKK1 expression downregulates both c-MYC and BMI1. DKK1 and BMI1 regulate the expression of each other in a negative feedback loop. Thus, the oncogenic activity of Wnt signaling pathway and BMI1 is interconnected in a positive feedback loop via c-MYC [60].
Bmi1 is overexpressed in the granule cell lineage, which can give origin to medulloblastomas as observed in transgenic and knock out mouse models. Upregulated Bmi1 corresponded to the expression of Myc and its upstream regulator Lef1, and a Bmi1–Lef1–Myc axis has been proposed [61]. BMI1 was overexpressed in primary glioma samples and correlated with adverse prognosis and resistance to radio-chemotherapy. Interestingly, BMI1 enhanced the transcriptional activity of the nuclear factor (NF)-κB TF by promoting its nuclear translocation and, most interestingly, MYC expression was induced among other NF-κB targets, suggesting a BMI1–NF-κB–MYC axis in glioma [62]. MYCN and c-MYC overexpression induced proliferation and tumor growth in human neuroblastoma samples and xenografts in a Bmi1-dependent pattern, exhibiting a positive correlation. Knockdown of MycN/c-Myc and BMI1 led to decreased mitosis and karyorrhexis [63]. No correlation between BMI1 and MYCN/c-MYC in gliomas or medulloblastomas was observed, in contrast to the previous study [62], suggesting that the interaction is cell-type specific. Interestingly, in cases of MYCN amplification there was a positive correlation between BMI1 expression and MYCN. Nevertheless, in the non-amplificated MYCN cases, BMI1 expression correlated with c-MYC expression [63]. MycN was found able to protect neuroblastoma precursor cells from death stimuli through p53 repression, whereas Bmi1 overexpression induced the polyubiquitination and proteasomal degradation of p53 in a Ring1a/b-mediated p53 polyubiquitination as observed in a transgenic mice model of neuroblastoma [64].
PcG proteins promote ESCs and adult leukemic stem cell self-renewal maintenance by blocking cell fate decisions, contributing to oncogenesis. Akt phosphorylates Bmi1 at Ser316, both in vitro and in vivo, impairing its chromatin-modifying function. Moreover, phosphorylated Bmi1 exhibits a suppressed growth-promoting potential, and effects on senescence and cellular transformation. Akt-mediated phosphorylation also promotes the dissociation from chromatin and the derepression of the Ink4a–Arf locus, while phosphorylated Bmi1 loses its ability to cooperate with MYC in cellular transformation. Such interaction inhibits self-renewal of hematopoetic progenitor cells and inhibits ubiquitination of H2A. Thus, the PI3K–Akt pathway indirectly fine tunes cell growth by inhibiting Bmi1 through Ser316 phosphorylation [65].
The FoxM1 TF, with known tumor-promoting properties in diverse cancer subtypes, is required for proper mitosis in MEFs, and its overexpression protects cells from oxidative stress-induced senescence. That action is achieved via upregulation of c-Myc, subsequent activation of its downstream target Bmi1, and final suppression of the p19Arf–p53 pathway, resulting in MEFs protection from senescence [66]. These findings shed light into the molecular mechanisms of senescence and oncogenesis at the early transforming stages.
ID1 is an oncogene affecting cell proliferation, cell cycle progression, apoptosis, differentiation, invasion, and angiogenesis. ID1 exclusively affects PRC1 and regulates the expression of MEL-18 and BMI1 in human breast cancer cell lines. It enhances the E3 ligase activity of RING1b through the PI3K/AKT pathway, accumulating H2Aub and proteosomal degradation of geminin, which is a PRC1 target. ID1 induces downregulation of MEL-18 via AKT ser473 phosphorylation and activation of the AKT signaling pathway, and BMI1 activation. ID1 also enhances c-MYC transcription through inhibition of MEL-18. Small interfering RNA (siRNA)-mediated MYC downregulation abolishes ID1-mediated BMI1 upregulation, while ID1 regulated BMI1 transcrition through c-MYC. It can also be suggested that the oncogenic function of c-MYC is attributed/enhanced by the oncogenic power of ID1. It can also be assumed that it affects CSCs biology via BMI1, c-MYC, and geminin, which regulates cell proliferation, differentiation, and genomic stability [67].
Other than Bmi1, PRC1 subunits were also observed to interact with Myc. In fact, Cbx7 is overexpressed, and exhibits tumor-initiating and disease-accelerating properties in contrast to other PcG proteins in transgenic mice. Cbx7 acts independently of Bmi1, repressing Ink4a/Arf TS locus in cooperation with Myc, promoting follicular lymphoma pathogenesis [68]. Members of the posterior sex combs proteins interact and regulate each other at the messenger RNA (mRNA) level. MEL-18 downregulates BMI1 in fibroblasts and accelerates the entry of cells into senescence by upregulating p16 and increasing the growth inhibitory form of phosphorylated Rb. C-MYC binds to BMI1 while MEL-18 is not able to bind directly on BMI1. MEL-18 regulation on BMI1 is achieved by repressing c-MYC and by downregulating AKT. Interestingly, c-MYC overexpression rescued MEL-18 mediated repression of BMI1 expression. MELl-18 expression was reduced in prostate cancer samples and breast cancer, and inversely correlated with BMI1 and c-MYC levels. MEL-18 forced expression also attenuated cancer cell growth through G1 arrest via modulation of AKT signaling [69–73]. Therefore, the existence of a MEL-18–c-MYC–BMI1–p16–pRb pathway regulates cell senescence.
PRC2 and MYC
Less work has been done regarding PRC2 members and MYCc interaction, but the findings are most interesting. All three PRC2 core components were upregulated concomitantly to MYC in cell lines bearing 20q amplification, a chromosomal modification that occurs early in the malignant transformation process [74]. The interaction between EZH2 and c-MYC has been detected in the tumorigenic process as early as the CSC level. EZH2 was overexpressed in primary glioblastoma CSC, promoting aberrant self-renewal by modulating the expression of direct downstream genes such as c-MYC. C-MYC was also capable of rescuing in part the effects of the EZH2 inhibitor 3-deazaneplanocin A (DZNep) treatment on CSC [75]. EZH2 was found to activate c-Myc in breast cancer cells through the ERα and the Wnt pathways, in a Wnt/b-catenin–EZH2–ERα–MYC axis [76].
In MLL-AF9 acute myeloid leukemia (AML), the genes of the MYC module were negatively enriched in EZH2-inactivated leukemic cells. These data suggest a functional link between gene expression programs that are under the control of PRC2 and MYC. These findings could also have therapeutic implications, since modulation of EZH2 might decrease the expression of MYC transcriptional targets at least in specific subtypes of AML [77].
H3K27me3 levels were decreased in prostatic intraepithelial neoplasia and prostatic adenocarcinoma and were independent from the high EZH2 levels. An Myc-expressing mouse model was studied in order to obtain better insight into decreased H3K27me3 levels. The findings supported the hypothesis that in vivo Myc overexpression results in a global decrease of H3K27me3 levels. These data suggested that Myc has the ability to influence epigenetic marks chromatin structure in cancer in a histone methyltransferase-independent pattern [78].
HOXB3 increases expression of DNMT3B, which is recruited and directly bound to RASSF1A promoter, repressing its expression via hypermethylation. DNMT3B recruitment is achieved through interactions with EZH2 and MYC, which is also bound to RASSF1A promoter. In fact, MYC knockdown results in decreased EZH2 expression and DNMT3B recruitment. Epigenetic silencing of RASSF1A through HOXB3 induction of DNMT3B expression is commonly observed in lung adenocarcinomas and several other human cancer cell lines. Thus, a putative repressive mechanism involving the MYC association with EZH2 and DNMT3B on RASSF1A has been suggested [79]. Although this report shows that Myc is important for PRC2 recruitment, it is also true that Myc is not sufficient to recruit PcG proteins, as nearly 5 % of mouse ESC promoters bound by Myc are also bound by PcG proteins [80].
Recently, it has been demonstrated that Myc suppresses the PI3K/Akt pathway through transcriptional upregulation of its negative regulator the PTEN TS, initiating and maintaining gene repression. Furthermore, Ezh2 is activated by Myc-mediated suppression of Akt kinase activity, which in turn leads to Ezh2-mediated gene repression. Myc represses genes via Ezh2-induced H3K27me3, including Myc itself in rat fibroblasts, supporting a role of Ezh2 in Myc-mediated gene repression and autoregulation [81].
MYCN represses clusterin TS through direct interaction with a non-canonical E-box inducing bivalent epigenetic marks and recruitment of repressive enzymes such as histone deacetylases (HDACs) and PcG proteins. MYC recruits EZH2 to clusterin promoter inducing transcriptional silencing both in vivo and in vitro in neuroblastomas. Notably, although binding of MYCN was associated with active chromatin marks such as H3ac and H3K4me2, negative marks such as H3K9me3 and H3k27me3 were also observed immediately downstream of the E-box. In agreement with this observation, several chromatin remodeling factors associated with transcriptional repression such as HDAC 1/2, BMI1, EZH2, and SUZ12 were detected around the E-box or downstream of the E-box sequence in the presence of MYCN. This ‘bivalent’ configuration is typical of repressed, developmentally regulated genes, which are poised to be activated by physiological stimuli [82].
Myc is also involved in stem cell biology, as both c-MYC and MYCN expression is required for ESCs and induced pluripotent stem cell (iPSC) self-renewal that could not be compensated by MYCL, highlighting the different biologic properties of each homolog in ESC biology. This regulatory function of MYC is achieved through the regulation of miRNAs involved in iPSC biology, modulation of cell cycle regulator expression, control of the euchromatin organization, and consequently the epigenetic state of iPSCs [48]. Accumulating evidence shows that PcG proteins and Myc are involved in stem cell biology, and induced pluripotency. Most recent findings establish that c-MYC, together with KLF4, are responsible for initiating the first transcriptional wave during somatic cell reprogramming [83]. Moreover, there is a Myc-centered network that acts independently of the other core TFs (Oct4, Sox2, Nanog) composing sub-signatures represented by the core TF network, the Myc network, and the Polycomb cluster [84]. Intriguingly, recent data revealed that inactivation of Ezh2 in MEFs treated with Oct4, c-Myc, Klf4, and Sox2 TFs resulted in successful reprogramming, probably via Ezh1 recruitment on target genes and H3K27me retention [85].
ESCs can undergo rapid self-renewal and can differentiate into any cell type. That feature depends on TFs, including Oct4, Sox2, and Nanog, that form the core pluripotency network. This ESC-specific network interacts with both the Myc-based transcription network and a chromatin-modifying complex network including PRC2. Together these three networks occupy and regulate a large number of target genes essential for the self-renewal and differentiation of ESCs. Myc and PRC2 seem to be under the control of the Utf1 TF, which prevents PRC2 binding, while it blocks the Myc–Arf feedback loop, ensuring rapid proliferation of ESCs [86]. Thus, the three modules do not act separately but rather in conjunction, with Utf1 being the intermediate link.
Myc is involved in the transcriptional regulation of ESCs pluripotency network and in histone methylation mediated by PcG. Transcriptional activation of the entire PRC2, and not of individual core members, contributes to high H3K27me3 levels, therefore keeping bivalent genes silent. The final result of the Myc–PRC2 interaction was a maintained ESCs undifferentiated state, as established in double knockdown experiments, and the cell cycle progression maintaining self-renewal of ESCs [87].
Ben-Porath et al. [88] sought to determine whether the regulatory networks that characterize ESCs are also active in cancer. Four groups were defined, with 13 partially overlapping gene sets; the Polycomb targets and the Myc targets were among them. Their data suggested that poorly differentiated tumors display a molecular pattern similar to ESCs, and that cancer cells in such tumors are biologically closer to normal undifferentiated stem cells than are cells in well differentiated tumors.
C-MYC overexpression in human umbilical cord blood-marrow stroma cells (hUCB-MSCs) induces the expression of PcG complex genes, and most PcG genes are down-regulated after HDAC2 inhibition. However, the expression level of PHC1, PHC2, RING1, and EZH2 are not down-regulated after HDAC2 inhibition. This might indicate that these genes are not under the control of HDAC2 or c-MYC. A c-MYC regulatory feature might also be the regulation of PcG gene expression via HDAC2 control. As a result, cell proliferation and differentiation of adult stem cells is affected [89]. These studies on adult stem cells and iPSCs could act as guides to get further insight in the first steps of carcinogenesis.
Poly-MYCroRNAs: “ménage a trois”
miRNAs are a class of non-coding RNAs with regulatory function of gene expression. They are involved in the regulation of physiological processes, in pluripotency, in reprogramming and in several different diseases including cancer [90–92]. Their function is achieved through modulation of cell signaling, differentiation, proliferation, organogenesis, development, and apoptosis. miRNA genes are distributed in all human chromosomes except for the Y chromosome. Half of them are found in clusters and they are transcribed as polycystronic primary transcripts [93, 94].
More than 2,000 human mature miRNA sequences are included in miRBase release 19 accounting for the 1–2 % of the human genome, with the ability to control the activity of nearly 50 % of all coding genes [95].
miRNA biogenesis is a complex multistep process involving several proteins. Canonical and alternative miRNA biogenesis pathways have been described, and our knowledge in their biogenesis is expanding every day. In the canonical pathway, the pri-miRNAs generated by RNA Pol II are cleaved in the nucleus by the RNase III DROSHA in conjunction with the DGCR8 protein. These new pre-miRNAs are exported to the cytoplasm by Exportin-5 protein, where they are further cleaved by DICER, reaching their final ~22 nt length. The resulting double-stranded small RNA is loaded onto the Argonaute (Ago) proteins, forming the effector complex RNA-induced silencing complex (RISC). One RNA strand remains attached to the Ago as a mature miRNA while the other strand is degraded [94, 96]. Alternative miRNA biogenesis pathways that bypass DICER and/or DROSHA/DGCR8 have also been identified [97, 98].
miRNAs are able to regulate their target gene expression by base pairing with the 3′ untranslated region (UTR) of the target mRNA, although there is evidence supporting that targets can be located in the 5′UTR or in coding regions of genes. miRNA target sites exhibit perfect matching between the nucleotides 2–7 of a single miRNA and the mRNA, whereas mutations in the target genes might lead to novel target sites [99]. Through that interaction, a single miRNA is capable of repressing tens to hundreds of targets. Several mechanisms of target regulation have been proposed, including stimulation of translation, endonucleolytic cleavage, deadenylation and degradation of the mRNA, inhibition of translation initiation, and inhibition after translational initiation. All these mechanisms finally consolidate gene silencing [100].
miRNAs possess regulatory functions of fundamental signaling pathways such as Wnt, NOTCH, Hedgehog, RAS, and MAPK/PI3K/AKT [101, 102]. MiRNAs have the ability to act either as TS or as oncogenes depending on the type of genetic or epigenetic abnormality present. They have multiple roles in tumor genesis and progression, as they are capable of modulating oncogenic, TS pathways, and metastasis pathways, including c-MYC, p53, RAS, and BCR/ABL, the TWIST1–miR10b–HOXD10 pathway. Nevertheless, their expression can be regulated by other oncogenes or TS [102, 103]. They are involved in almost all types of human cancer, and they can classify human cancer according to the differentiation state and developmental lineage of the cancer [104]. Their expression is significantly associated with major cancer outcomes, while they can be used as biomarkers for disease progression and response to treatment [104–108]. A more detailed description of miRNA biogenesis, function, and regulation is beyond the scope of the present manuscript.
miRNAs act at the transcriptional and post-transcriptional level and they are regulated by TFs, forming complex regulatory networks affecting each other’s expression. Both TFs and miRNAs form feedback and feedforward loops through which their target gene expression is regulated [109]. Thus, miRNAs are closely associated with MYC. For example miR-150 is under the negative control of MLL-fusion/Myc/Lin28 axis in MLL-rearranged AML [110]. Moreover, several other TS miRNAs are downregulated by MYC in lymphoma cells by direct binding to their promoters, promoting lymphomagenesis [111]. MiR-22 and MYC form feedforward loops in cells that exit a quiescent state and enter a proliferative state characteristic of malignant transformation [112]. MYC has the ability to suppress proline oxidase and proline metabolism in cancer cells through mir-23b* upregulation, affecting in that way another hallmark of cancer [113].
However, miRNAs also interact with the PcG members. EZH2 is under the control of several different miRNAs affecting H3K27me3 levels of target genes, whereas EZH2 itself activates or represses other miRNAs [114]. Specific miRNAs have the ability to promote myogenesis and terminal differentiation in mouse myoblast cell lines. Among them, the upregulated expression of miR-26a was required during terminal differentiation in order to induce rapid and efficient silence of Ezh2, which is a negative regulator of myogenesis Ezh2 [115].
The interaction between MYC-miRNAs and PcG proteins is quite complex, as these factors compose sophisticated modulatory networks. MiR-29 has dual oncogenic or TS function depending on the cellular context and it seems that acts as TS in MYC-associated lymphomas. MiR-29 is MYC-repressed with the cooperation of HDAC3 and EZH2 in MYC-associated lymphomas cell lines and in primary samples. Interestingly, MYC recruits EZH2 and SUZ12 at miR-29 promoter, promoting its epigenetic silencing. MYC depletion leads to decreased recruitment of RNA pol II, HDAC3 levels, and EZH2, promoting increased histone acetylation and decreased H3K27me3, respectively. An important finding of this study is the identification of the MYC–miR-26a–EZH2–miR-494 positive feedback loop that sustains MYC activity and consequent miR-29 repression. MYC leads to EZH2 upregulation through miR-26a repression and in turn EZH2 suppresses miR-494, which targets MYC. That loop confers the MYC oncogene persistent high protein levels and further repression of miR-29. This study sheds light on a putative mechanism of EZH2 activation and contribution to tumor aggressive transformation [116].
Ezh2 can epigenetically repress miRNAs, enhancing the expression of Bmi1 and Ring2, promoting H2AK119ub in advanced prostate cancer. Therefore, miRNAs also act as the intermediate link for the coordinated function of PRC1 and PRC2 in cancer [117]. The interaction between miRNAs and PcG proteins could indirectly affect the fate of the hematopoietic progenitor. In fact, an miR-223/PcG axis regulates the NFI-A gene affecting hematopoietic cell lineage determination [118]. MiR-18a and miR-19a were upregulated and transctivated by MycN in neuroblastoma and they negatively regulated ERα1 expression [119].
MYC and EZH2 were overexpressed and positively correlated in primary samples and in a mouse model of prostatic intraepithelial neoplasia. However, as MYC regulates the expression of miR-26a/b, and miR-26a/b targets EZH2 in prostate cancer cells only and not in breast cancer or AML, it could be suggested that the MYC–miR-26a/b–EZH2 interaction presents a tissue-specific pattern. Final result of the deregulated axis is the maintained proliferative capacity of cancer cells. Therefore, EZH2 can also represent an early prostate cancer contributor and a driver of disease progression, and its expression could be enhanced by MYC via two mechanisms (directly or through miR-26a/b) [120].
MYC has the ability to alter PRC2 by interacting with the TS miR-26a or by targeting E2F1 proapoptotic protein [121]. miR-26a is downregulated in MYC-induced lymphoma, whereas its direct negative target EZH2 is overexpressed [122]. That interaction might result in MYC-induced EZH2 expression through downregulation of its target miRNA. In AML samples, MYC directly enhanced EZH2 transcription, while it also repressed miR-26a transcription [123].
miR-26a was also downregulated in nasopharyngeal (NPC) primary samples and cell lines promoting tumor growth, while negatively correlated with EZH2 levels. miR-26a directly targeted EZH2 in NPC cells and it was demonstrated that ectopic EZH2 expression rescued miR-26a cell growth inhibition and cell cycle arrest. The effect of forced miR-26a expression on tumor growth inhibition was in part mediated by downregulation of c-MYC together with other cell-cycle regulators [124].
Treatment options
MYC seemed to represent an attractive therapeutic target as it fulfilled the required criteria for optimal therapeutic efficacy [125]. However, the enthusiasm has switched to skepticism, and a search for alternative approaches for several reasons [126]. Drugs able to inhibit c-MYC/MAX dimerization, and to decrease global H3K9ac and increase H3K9me2 levels, such as Omomyc, have been developed [127, 128]. Other alternative approaches for inhibiting MYC expression have been developed, such as inhibition of the bromodomain and extraterminal (BET) subfamily of human bromodomain proteins, or the use of antogomirs and miRNA mimics to inhibit or activate oncogenic and TS miRNAs, respectively, that are associated with Myc function [129–131].
Similarly, pharmacologic inhibition of EZH2-activating mutations in lymphoma has been reported. DZNep is the most studied EZH2 inhibitor in different cancer subtypes with a targeting potential that reaches the CSC compartment [132–135].
However, there is no evidence of how inhibition of MYC or PRC could affect each other’s expression. Moreover, none of the currently known drugs have been reported to affect both MYC and PcG and modulate their interaction. Recently, it was described that genistein, a botanical isoflavone enriched in soybean products, induces the expression of p21waf1 and p16ink4a and downregulates both Bmi1 and c-Myc [136]. C-Myc itself was determined to be an HDAC inhibitor target, which also possesses the ability to indirectly suppress Bmi1 and EZH2 transcription in breast cancer cell lines, leading to reactivation of PcG target genes [137]. The effect of HDAC inhibitors and of genistein on Myc in cancer biology should be further explored.
Conclusions
It is widely accepted that PcG proteins and MYC are involved in several physiogical processes, induced pluripotency, and cancer, acting independently or in conjunction. PcG proteins regulate chromatin organization, whereas MYC might also control global chromatin organization through regulation of target gene transcription. PcG proteins also exhibit gene-expression regulator characteristics and might act as fine tuners of MYC-induced changes in cell biology. Although MYC binding to its targets correlates with specific epigenetic changes, it is unclear whether MYC establishes these marks or is recruited to target promoters as a consequence of chromatin modifications. MYC and PcG protein interactions are involved in the regulation of most important signaling pathways (Fig. 1). MYC might promote the transcription of TS, which in turn suppress signaling transduction and finally enhances its oncogenic potential via PcG target gene repression. Moreover, MYC, PcG proteins, and miRNAs form complex networks, feedforward and feedback loops regulating gene expression and controlling each other’s expression (Fig. 2). PcG proteins have been shown to be able to substitute MYC in induced pluripotency. However, given that the function of these genes is not completely clear and understood, we must be prudent before using them widely. Accumulating data continuously describe new complex circuitries in which PcG proteins and MYC are deregulated in cancer, with prospective therapeutic implications against both blood and solid cancers.
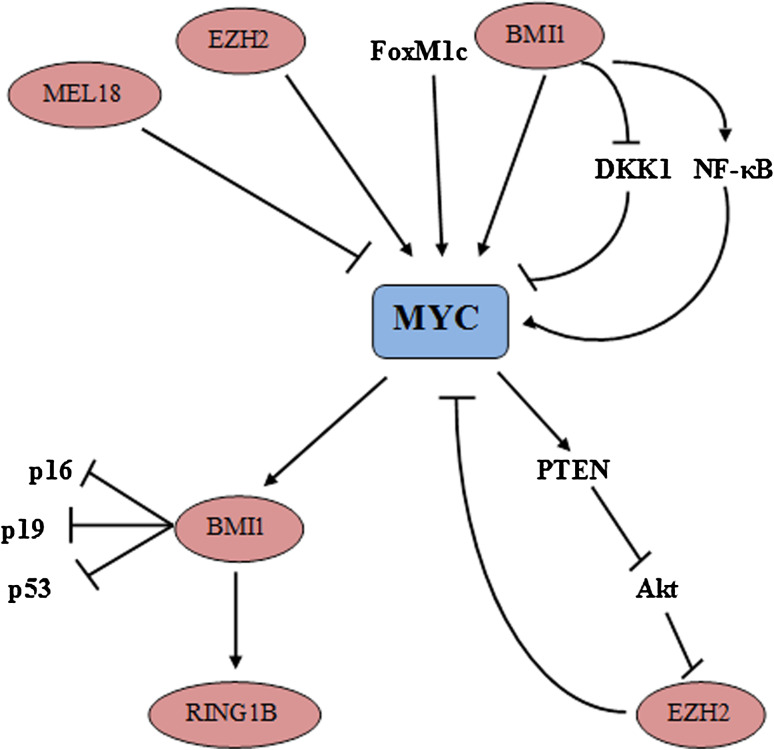
Fig. 1.
Direct upstream regulators and downstream effectors of MYC oncogene are shown. MYC expression is directly regulated by other transcription factors (FoxM1c) and by different polycomb proteins (EZH2, BMI1, Mel-18). BMI1 also regulates MYC via other transcription factors (nuclear factor [NF]-κB) or via Wnt pathway inhibitors such as Dickkof (DKK1). EZH2 is able to activate c-MYC in a Wnt/b-catenin–EZH2–ERα–MYC axis. MYC represses genes via EZH2, including MYC itself, forming autoregulatory loops with EZH2. MYC controls the polycomb repressive complex (PRC)-1 member RING1B through regulation of its direct downstream regulator BMI1. In turn, BMI1 affects the expression of cell cycle regulators. For a more detailed description see the text
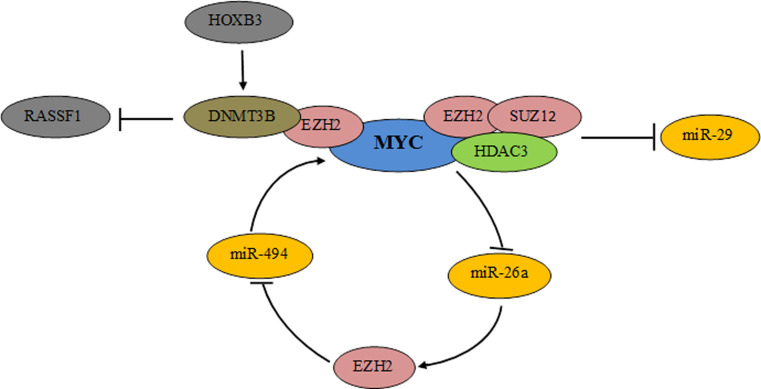
Fig. 2.
MYC interacts with the histone methyltransferase EZH2 and recruits the DNA methyltransferase DNMT3B, expression of which is enhanced by HOXB3, on the promoter of RASSF1 tumor suppressor repressing its expression. MYC also binds to EZH2/SUZ12 and in cooperation with the histone deacetylase HDAC3 promote silencing of miR-29. MYC is a part of a MYC/miR-26a/EZH2/miR-494 regulatory loop enhancing MYC expression
Acknowledgments
The authors would like to thank Nikolaos Benetatos MD for critical review of the manuscript. The authors apologize to those authors whose work has not been cited.
Conflict of interest
None.
References
- 1.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 2.Bantignies F, Cavalli G. Polycomb group proteins: repression in 3D. Trends Genet. 2011;27:454–464. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the polycomb perspective. Dev Cell. 2010;19:651–661. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Viré E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, Fuks F, Helin K, Di Croce L. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Boukarabila H, Saurin AJ, Batsché E, Mossadegh N, van Lohuizen M, Otte AP, Pradel J, Muchardt C, Sieweke M, Duprez E. The PRC1 polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23:1195–1206. doi: 10.1101/gad.512009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. Spreading of X chromosome inactivation via a hierarchy of defined polycomb stations. Genome Res. 2012;22:1864–1876. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, Obuse C. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1–HBiX1 pathway. Nat Struct Mol Biol. 2013;20:566–573. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- 9.Kalantry S, Magnuson T. The polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Perez D, Piris MA, Sanchez-Beato M. Polycomb proteins in hematologic malignancies. Blood. 2010;116:5465–5475. doi: 10.1182/blood-2010-05-267096. [DOI] [PubMed] [Google Scholar]
- 12.Radulović V, Haan G, Klauke K. Polycomb-group proteins in hematopoietic stem cell regulation and hematopoietic neoplasms. Leukemia. 2013;27(3):523–33. doi: 10.1038/leu.2012.368. [DOI] [PubMed] [Google Scholar]
- 13.Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Follmer NE, Wani AH, Francis NJ. A polycomb group protein is retained at specific sites on chromatin in mitosis. PLoS Genet. 2012;8:e1003135. doi: 10.1371/journal.pgen.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luis NM, Morey L, Di Croce L, Benitah SA. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell. 2012;11:16–21. doi: 10.1016/j.stem.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Müller J, Thomä NH. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann L, Ferrari R, Vashisht AA, Wohlschlegel JA, Kurdistani SK, Carey M. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J Biol Chem. 2012;287:35784–35794. doi: 10.1074/jbc.M112.397430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockdorff N. Noncoding RNA and polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballaré C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, Carlomagno T, Benitah SA, Di Croce L. Phf19 links methylated Lys36 of histone H3 to regulation of polycomb activity. Nat Struct Mol Biol. 2012;19:1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brien GL, Gambero G, O’Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, Lohan AJ, Ferguson N, Shi X, Sinha KM, Loftus BJ, Cagney G, Bracken AP. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol. 2012;19:1273–1281. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 23.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, Han Z, Chai J, Zhou XJ, Gao S, Zhu B. Dense chromatin activates polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337(6097):971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Wang C, Zhang P, Gao K, Wang D, Yu H, Zhang T, Jiang S, Hexige S, Hong Z, Yasui A, Liu JO, Huang H, Yu L. Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis. J Biol Chem. 2013;288:529–539. doi: 10.1074/jbc.M111.338996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crea F, Duhagon Serrat MA, Hurt EM, Thomas SB, Danesi R, Farrar WL. BMI1 silencing enhances docetaxel activity and impairs antioxidant response in prostate cancer. Int J Cancer. 2011;128:1946–1954. doi: 10.1002/ijc.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30:372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, Shinga J, Koseki H, Iwama A. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118:6553–6561. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- 29.Di Meglio T, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, Hrycaj SM, Roska B, Peters AH, Eichmann A, Wellik D, Ducret S, Rijli FM. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science. 2013;339:204–207. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: time for clinical application? Crit Rev Oncol Hematol. 2012;83:184–193. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, Asp P, Hadler M, Rigo I, De Keersmaecker K, Patel J, Huynh T, Utro F, Poglio S, Samon JB, Paietta E, Racevskis J, Rowe JM, Rabadan R, Levine RL, Brown S, Pflumio F, Dominguez M, Ferrando A, Aifantis I. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hock H. A complex polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26:751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richly H, Aloia L, Di Croce L. Roles of the polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tönnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 35.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 36.Ueda T, Sanada M, Matsui H, Yamasaki N, Honda ZI, Shih LY, Mori H, Inaba T, Ogawa S, Honda H. EED mutants impair polycomb repressive complex 2 in myelodysplastic syndrome and related neoplasms. Leukemia. 2012;26:2557–2560. doi: 10.1038/leu.2012.146. [DOI] [PubMed] [Google Scholar]
- 37.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, Xu H, Cato L, Thornton JE, Gregory RI, Morrissey C, Vessella RL, Montironi R, Magi-Galluzzi C, Kantoff PW, Balk SP, Liu XS, Brown M. EZH2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albihn A, Johnsen JI, Henriksson MA. MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res. 2010;107:163–224. doi: 10.1016/S0065-230X(10)07006-5. [DOI] [PubMed] [Google Scholar]
- 39.Buendia MA, Bourre L, Cairo S. Myc target miRs and liver cancer: small molecules to get Myc sick. Gastroenterology. 2012;142:214–218. doi: 10.1053/j.gastro.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 40.Uribesalgo I, Benitah SA, Di Croce L. From oncogene to tumor suppressor: the dual role of Myc in leukemia. Cell Cycle. 2012;11:1757–1764. doi: 10.4161/cc.19883. [DOI] [PubMed] [Google Scholar]
- 41.Cascón A, Robledo M. MAX and MYC: a heritable breakup. Cancer Res. 2012;72:3119–3124. doi: 10.1158/0008-5472.CAN-11-3891. [DOI] [PubMed] [Google Scholar]
- 42.Lüscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494:145–160. doi: 10.1016/j.gene.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 43.Lee S, Schmitt CA, Reimann M. The Myc/macrophage tango: oncogene-induced senescence Myc style. Semin Cancer Biol. 2011;21:377–384. doi: 10.1016/j.semcancer.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjö S, Murga M, Fernandez-Capetillo O, Barbacid M, Larsson LG, Amati B. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–59. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- 45.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Tai Y, Lisanti MP, Liao DJ. c-Myc induction of programmed cell death may contribute to carcinogenesis: a perspective inspired by several concepts of chemical carcinogenesis. Cancer Biol Ther. 2011;1:615–626. doi: 10.4161/cbt.11.7.14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Spears E, Boone DN, Li Z, Gregory MA, Hann SR. Domain-specific c-Myc ubiquitylation controls c-Myc transcriptional and apoptotic activity. Proc Natl Acad Sci USA. 2013;110:978–983. doi: 10.1073/pnas.1208334110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith K, Dalton S. Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5:947–959. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 52.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 53.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guney I, Sedivy JM. Cellular senescence, epigenetic switches and c-Myc. Cell Cycle. 2006;5:2319–2323. doi: 10.4161/cc.5.20.3348. [DOI] [PubMed] [Google Scholar]
- 55.Cenci T, Martini M, Montano N, D’Alessandris QG, Falchetti ML, Annibali D, Savino M, Bianchi F, Pierconti F, Nasi S, Pallini R, Larocca LM. Prognostic relevance of c-Myc and BMI1 expression in patients with glioblastoma. Am J Clin Pathol. 2012;138:390–396. doi: 10.1309/AJCPRXHNJQLO09QA. [DOI] [PubMed] [Google Scholar]
- 56.Joensuu K, Hagström J, Leidenius M, Haglund C, Andersson LC, Sariola H, Heikkilä P. Bmi-1, c-myc, and Snail expression in primary breast cancers and their metastases—elevated Bmi-1 expression in late breast cancer relapses. Virchows Arch. 2011;459:31–39. doi: 10.1007/s00428-011-1096-8. [DOI] [PubMed] [Google Scholar]
- 57.Ochiai H, Takenobu H, Nakagawa A, Yamaguchi Y, Kimura M, Ohira M, Okimoto Y, Fujimura Y, Koseki H, Kohno Y, Nakagawara A, Kamijo T. Bmi1 is a MYCN target gene that regulates tumorigenesis through repression of KIF1Bbeta and TSLC1 in neuroblastoma. Oncogene. 2010;29:2681–2690. doi: 10.1038/onc.2010.22. [DOI] [PubMed] [Google Scholar]
- 58.Guney I, Wu S, Sedivy JM. Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16 (INK4a) Proc Natl Acad Sci USA. 2006;103:3645–3650. doi: 10.1073/pnas.0600069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duss S, André S, Nicoulaz AL, Fiche M, Bonnefoi H, Brisken C, Iggo RD. An oestrogen-dependent model of breast cancer created by transformation of normal human mammary epithelial cells. Breast Cancer Res. 2007;9:R38. doi: 10.1186/bcr1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho JH, Dimri M, Dimri GP. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J Biol Chem. 2013;288:3406–3418. doi: 10.1074/jbc.M112.422931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Behesti H, Bhagat H, Dubuc AM, Taylor MD, Marino S. Bmi1 overexpression in the cerebellar granule cell lineage of mice affects cell proliferation and survival without initiating medulloblastoma formation. Dis Model Mech. 2013;6:49–63. doi: 10.1242/dmm.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, Gong LY, Song LB, Jiang LL, Liu LP, Wu J, Yuan J, Cai JC, He M, Wang L, Zeng M, Cheng SY, Li M. Oncoprotein Bmi-1 renders apoptotic resistance to glioma cells through activation of the IKK-nuclear factor-kappaB pathway. Am J Pathol. 2010;176:699–709. doi: 10.2353/ajpath.2010.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang R, Cheung NK, Vider J, Cheung IY, Gerald WL, Tickoo SK, Holland EC, Blasberg RG. MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas. FASEB J. 2011;25:4138–4149. doi: 10.1096/fj.11-185033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calao M, Sekyere EO, Cui HJ, Cheung BB, Thomas WD, Keating J, Chen JB, Raif A, Jankowski K, Davies NP, Bekkum MV, Chen B, Tan O, Ellis T, Norris MD, Haber M, Kim ES, Shohet JM, Trahair TN, Liu T, Wainwright BJ, Ding HF, Marshall GM. Direct effects of Bmi1 on p53 protein stability inactivates oncoprotein stress responses in embryonal cancer precursor cells at tumor initiation. Oncogene. 2012 doi: 10.1038/onc.2012.368. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y, Liu F, Yu H, Zhao X, Sashida G, Deblasio A, Harr M, She QB, Chen Z, Lin HK, Di Giandomenico S, Elf SE, Yang Y, Miyata Y, Huang G, Menendez S, Mellinghoff IK, Rosen N, Pandolfi PP, Hedvat CV, Nimer SD. Akt phosphorylates the transcriptional repressor bmi1 to block its effects on the tumor-suppressing ink4a-arf locus. Sci Signal. 2012;5:ra77. doi: 10.1126/scisignal.2003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li SK, Smith DK, Leung WY, Cheung AM, Lam EW, Dimri GP, Yao KM. FoxM1c counteracts oxidative stress-induced senescence and stimulates Bmi-1 expression. J Biol Chem. 2008;283:16545–16553. doi: 10.1074/jbc.M709604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian T, Lee JY, Park JH, Kim HJ, Kong G. Id1 enhances RING1b E3 ubiquitin ligase activity through the Mel-18/Bmi-1 polycomb group complex. Oncogene. 2010;29:5818–5827. doi: 10.1038/onc.2010.317. [DOI] [PubMed] [Google Scholar]
- 68.Scott CL, Gil J, Hernando E, Teruya-Feldstein J, Narita M, Martínez D, Visakorpi T, Mu D, Cordon-Cardo C, Peters G, Beach D, Lowe SW. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci USA. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JY, Jang KS, Shin DH, Oh MY, Kim HJ, Kim Y, Kong G. Mel-18 negatively regulates INK4a/ARF-independent cell cycle progression via Akt inactivation in breast cancer. Cancer Res. 2008;68:4201–4209. doi: 10.1158/0008-5472.CAN-07-2570. [DOI] [PubMed] [Google Scholar]
- 70.Guo WJ, Zeng MS, Yadav A, Song LB, Guo BH, Band V, Dimri GP. Mel-18 acts as a tumor suppressor by repressing Bmi-1 expression and down-regulating Akt activity in breastcancer cells. Cancer Res. 2007;67:5083–5089. doi: 10.1158/0008-5472.CAN-06-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang W, Lin T, Huang J, Hu W, Xu K, Liu J. Analysis of Mel-18 expression in prostate cancer tissues and correlation with clinicopathologic features. Urol Oncol. 2011;29:244–251. doi: 10.1016/j.urolonc.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 72.Guo WJ, Datta S, Band V, Dimri GP. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol Biol Cell. 2007;18:536–546. doi: 10.1091/mbc.E06-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Häyry V, Mäkinen LK, Atula T, Sariola H, Mäkitie A, Leivo I, Keski-Säntti H, Lundin J, Haglund C, Hagström J. Bmi-1 expression predicts prognosis in squamous cell carcinoma of the tongue. Br J Cancer. 2010;102:892–897. doi: 10.1038/sj.bjc.6605544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tabach Y, Kogan-Sakin I, Buganim Y, Solomon H, Goldfinger N, Hovland R, Ke XS, Oyan AM, Kalland KH, Rotter V, Domany E. Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLoS One. 2011;6:e14632. doi: 10.1371/journal.pone.0014632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suvà ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, Marquez VE, Clément V, Stamenkovic I. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 76.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, Sun L, Zhang Y, Chen Y, Li R, Zhang Y, Hong M, Shang Y. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, Xie H, Orkin SH, Armstrong SA. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci USA. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pellakuru LG, Iwata T, Gurel B, Schultz D, Hicks J, Bethel C, Yegnasubramanian S, De Marzo AM. Global levels of H3K27me3 track with differentiation in vivo and are deregulated by MYC in prostate cancer. Am J Pathol. 2012;181:560–569. doi: 10.1016/j.ajpath.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palakurthy RK, Wajapeyee N, Santra MK, Gazin C, Lin L, Gobeil S, Green MR. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol Cell. 2009;36:219–230. doi: 10.1016/j.molcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaur M, Cole MD. MYC acts via the PTEN tumor suppressor to elicit auto regulation and genome-wide gene repression by activation of the Ezh2 methyltransferase. Cancer Res. 2013;73:695–705. doi: 10.1158/0008-5472.CAN-12-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corvetta D, Chayka O, Gherardi S, et al. Physical interaction between MYCN and polycomb repressive complex 2 (PRC2) in neuroblastoma: functional and therapeutic implications. J Biol Chem. 2013;288(12):8332–41. doi: 10.1074/jbc.M113.454280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, Bar-Nur O, Cheloufi S, Stadtfeld M, Figueroa ME, Robinton D, Natesan S, Melnick A, Zhu J, Ramaswamy S, Hochedlinger K. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fragola G, Germain PL, Laise P, Cuomo A, Blasimme A, Gross F, Signaroldi E, Bucci G, Sommer C, Pruneri G, Mazzarol G, Bonaldi T, Mostoslavsky G, Casola S, Testa G. Cell reprogramming requires silencing of a core subset of polycomb targets. PLoS Genet. 2013;9:e1003292. doi: 10.1371/journal.pgen.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jia J, Zheng X, Hu G, Cui K, Zhang J, Zhang A, Jiang H, Lu B, Yates J, 3rd, Liu C, Zhao K, Zheng Y. Regulation of pluripotency and self-renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell. 2012;151:576–589. doi: 10.1016/j.cell.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neri F, Zippo A, Krepelova A, Cherubini A, Rocchigiani M, Oliviero S. Myc regulates the transcription of the PRC2 gene to control the expression of developmental genes in embryonic stem cells. Mol Cell Biol. 2012;32:840–851. doi: 10.1128/MCB.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhandari DR, Seo KW, Jung JW, Kim HS, Yang SR, Kang KS. The regulatory role of c-MYC on HDAC2 and PcG expression in human multipotent stem cells. J Cell Mol Med. 2011;15:1603–1614. doi: 10.1111/j.1582-4934.2010.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 91.Benetatos L, Hatzimichael E, Londin E, Vartholomatos G, Loher P, Rigoutsos I, Briasoulis E. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814. doi: 10.1007/s00018-012-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs in pluripotency and reprogramming. Nat Cell Biol. 2012;14:1114–1121. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 94.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;2006(22):165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 95.Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10:103. doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and argonaute. Nature. 2012;486:541–544. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xhemalce B, Robson SC, Kouzarides T. Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell. 2012;151:278–288. doi: 10.1016/j.cell.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:8468–8470. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 100.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 101.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 102.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. MicroRNAs in cancer management. Lancet Oncol. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 104.Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst. 2012;104:528–540. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen PS, Su JL, Hung MC. Dysregulation of microRNAs in cancer. J Biomed Sci. 2012;19:90. doi: 10.1186/1423-0127-19-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S. A microRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 108.Zhao H, Wang D, Du W, Gu D, Yang R. MicroRNA and leukemia: tiny molecule, great function. Crit Rev Oncol Hematol. 2010;74:149–155. doi: 10.1016/j.critrevonc.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 109.Kunej T, Godnic I, Horvat S, Zorc M, Calin GA. Cross talk between microRNA and coding cancer genes. Cancer J. 2012;18:223–231. doi: 10.1097/PPO.0b013e318258b771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, Wang J, Arnovitz S, Elkahloun A, Price C, Hong GM, Ren H, Kunjamma RB, Neilly MB, Matthews JM, Xu M, Larson RA, Le Beau MM, Slany RK, Liu PP, Lu J, Zhang J, He C, Chen J. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Polioudakis D, Bhinge AA, Killion PJ, Lee BK, Abell NS, Iyer VR. A Myc–microRNA network promotes exit from quiescence by suppressing the interferon response and cell-cycle arrest genes. Nucleic Acids Res. 2013;41(4):2239–54. doi: 10.1093/nar/gks1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA. 2012;109:8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Benetatos L, Voulgaris E, Vartholomatos G, Hatzimichael E. Non-coding RNAs and EZH2 interactions in cancer: long and short tales from the transcriptome. Int J Cancer. 2012 doi: 10.1002/ijc.27859. [DOI] [PubMed] [Google Scholar]
- 115.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase enhancer of zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 116.Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, Zhang Y, Chan JC, Fu K, Marquez VE, Chen-Kiang S, Moscinski LC, Seto E, Dalton WS, Wright KL, Sotomayor E, Bhalla K, Tao J. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell. 2012;22:506–523. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 117.Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X, Wang R, Li Y, Dahiya A, Wang L, Pandhi M, Lonigro RJ, Wu YM, Tomlins SA, Palanisamy N, Qin Z, Yu J, Maher CA, Varambally S, Chinnaiyan AM. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20:187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N, Mancini M, Nanni M, Cimino G, Lo-Coco F, Grignani F, Nervi C. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119:4034–4046. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 119.Lovén J, Zinin N, Wahlström T, Müller I, Brodin P, Fredlund E, Ribacke U, Pivarcsi A, Påhlman S, Henriksson M. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci USA. 2010;107:1553–1558. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–683. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sander S, Bullinger L, Wirth T. Repressing the repressor: a new mode of MYC action in lymphomagenesis. Cell Cycle. 2009;8:556–559. doi: 10.4161/cc.8.4.7599. [DOI] [PubMed] [Google Scholar]
- 122.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Möller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 123.Salvatori B, Iosue I, Djodji Damas N, Mangiavacchi A, Chiaretti S, Messina M, Padula F, Guarini A, Bozzoni I, Fazi F, Fatica A. Critical role of c-Myc in acute myeloid leukemia involving direct regulation of miR-26a and histone methyltransferase EZH2. Genes Cancer. 2011;2:585–592. doi: 10.1177/1947601911416357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, Li XP. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 125.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sodir NM, Evan GI. Finding cancer’s weakest link. Oncotarget. 2011;2:1307–1313. doi: 10.18632/oncotarget.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Whitfield JR, Soucek L. Tumor microenvironment: becoming sick of Myc. Cell Mol Life Sci. 2012;69:931–934. doi: 10.1007/s00018-011-0860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, Soucek L, Nasi S. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS One. 2011;6:e22284. doi: 10.1371/journal.pone.0022284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Frenzel A, Lovén J, Henriksson MA. Targeting MYC-regulated miRNAs to combat cancer. Genes Cancer. 2010;1:660–667. doi: 10.1177/1947601910377488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Crea F, Fornaro L, Bocci G, Sun L, Farrar WL, Falcone A, Danesi R. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 2012;31:753–761. doi: 10.1007/s10555-012-9387-3. [DOI] [PubMed] [Google Scholar]
- 133.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, Scott MP, Jin L, Smith JJ, Olhava EJ, Chesworth R, Moyer MP, Richon VM, Copeland RA, Keilhack H, Pollock RM, Kuntz KW. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 134.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 135.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, Gu J, Ardayfio O, Zhang JH, Yan X, Fang J, Mi Y, Zhang M, Zhou T, Feng G, Chen Z, Li G, Yang T, Zhao K, Liu X, Yu Z, Lu CX, Atadja P, Li E. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci USA. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li Y, Chen H, Hardy TM, Tollefsbol TO. Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein. PLoS One. 2013;8:e54369. doi: 10.1371/journal.pone.0054369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bommi PV, Dimri M, Sahasrabuddhe AA, Khandekar J, Dimri GP. The polycomb group protein BMI1 is a transcriptional target of HDAC inhibitors. Cell Cycle. 2010;9:2663–2673. doi: 10.4161/cc.9.13.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]