Abstract
Ablation of tetraspanin protein TSPAN12 from human MDA-MB-231 cells significantly decreased primary tumor xenograft growth, while increasing tumor apoptosis. Furthermore, TSPAN12 removal markedly enhanced tumor-endothelial interactions and increased metastasis to mouse lungs. TSPAN12 removal from human MDA-MB-231 cells also caused diminished association between FZD4 (a key canonical Wnt pathway receptor) and its co-receptor LRP5. The result likely explains substantially enhanced proteosomal degradation of β-catenin, a key effecter of canonical Wnt signaling. Consistent with disrupted canonical Wnt signaling, TSPAN12 ablation altered expression of LRP5, Naked 1 and 2, DVL2, DVL3, Axin 1, and GSKβ3 proteins. TSPAN12 ablation also altered expression of several genes regulated by β-catenin (e.g. CCNA1, CCNE2, WISP1, ID4, SFN, ME1) that may help to explain altered tumor growth and metastasis. In conclusion, these results provide the first evidence for TSPAN12 playing a role in supporting primary tumor growth and suppressing metastasis. TSPAN12 appears to function by stabilizing FZD4–LRP5 association, in support of canonical Wnt-pathway signaling, leading to enhanced β-catenin expression and function.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1444-8) contains supplementary material, which is available to authorized users.
Keywords: TSPAN12, Tetraspanin, β-catenin, Tumor growth, Tumor metastasis
Introduction
Tetraspanin protein TSPAN12 is widely expressed in most human tissues and organs, with the highest levels found in digestive, urogenital, and cardiovascular systems. With its atypically long C-terminal tail, TSPAN12 is phylogenetically one of the earliest tetraspanins (pre-chordate ancestry) and is somewhat distinct from the others except for a distant relationship to Uroplakin-type tetraspanins [1]. Human TSPAN12 mutations are linked to impaired vascularization of the eye within a disease spectrum called FEVR (familial exudative vitreo-retinopathy) [2, 3]. Other genes linked to the FEVR disease in humans include Norrin (NDP), Frizzled 4 (FZD4), and LRP5. Products of these genes collaborate with TSPAN12 to activate the Norrin/β-catenin signaling pathway [2].
TSPAN12 knockout mice display FEVR-type retinal vascular defects and impaired Norrin/β-catenin signaling [4], consistent with disruption of a canonical Wnt signaling pathway. TSPAN12 appears to function by associating with FZD4 (receptor for Norrin) to drive FZD4 oligomerization and signaling, leading to enhanced β-catenin protein expression [4]. The β-catenin pathway is a central regulator of normal cell function, and perturbed β-catenin signaling can affect tumor cell behavior [5, 6]. However, a role for TSPAN12 in cancer pathogenesis had not been demonstrated.
For several reasons, we hypothesized that TSPAN12 may affect cancer cell behavior. First, TSPAN12 can affect β-catenin signaling [4], which may regulate both primary tumor growth and metastasis [6]. Second, the pro-tumorigenic sheddase ADAM10 [7] associates with TSPAN12, leading to ADAM10 maturation [8]. Third, TSPAN12 has been indirectly suggested to have anti-metastatic properties [9]. Fourth, significantly altered TSPAN12 expression has been observed in microarrays of lung, breast, and prostate cancers (see GEO datasets associated with [10–13]). Here, we use basal-type mammary carcinoma MDA-MB-231 cells to show that TSPAN12 plays a critical role in supporting primary tumor growth while inhibiting early stages of metastasis. Also, we provide evidence that TSPAN12 supports molecular association between LRP5 and FZD4, which likely explains promotion of β-catenin signaling. Finally, we demonstrate that TSPAN12 affects expression of over 400 genes, including several key genes regulated by β-catenin.
Materials and methods
Cells, antibodies, other reagents
Cell lines were from American Type Culture Collection (ATCC, USA) and were maintained (for <6 months) in DMEM with addition of 10 % FBS. HUVEC cells (Lonza, USA) were maintained in EBM-2 media (Lonza). Anti-GAPDH (mAb 6C5) was from Millipore (USA). Anti-CD11b (mAb EP1345Y) was from Abcam (USA). Antibodies to LRP5 (mAb D23F7), GSK3β (mAb 27C10), cleaved Caspase-3 (mAb D175), PCNA (mAb PC10), Dvl2 (mAb 30D2), Naked2 (mAb C67C4), Naked1 (mAb C30F10), Axin1 (mAb 2C76H11), LRP6 (mAb C5C7), β-catenin (mAb D10A8), and to Dvl3 (pAb # 3218) were from Cell Signaling Technology (USA). Antibody to FZD4 (pAb C-18) was from Santa Cruz (USA) and to β-catenin (mAb 5H10) was from Millipore. Alexa Fluor 647-conjugated phalloidin and ProLong® Gold antifade media with DAPI (Invitrogen, USA) were used. Alexa Fluor 488 and Alexa Fluor 647-conjugated secondary antibodies were from Invitrogen. Primers for qPCR TSPAN12 (Hs.PT.49a23249; Mm.PT.49a.16486724), GAPDH (Hs.PT.49a.2918858.g; Mm.PT.39a.1) ITGA2 (Hs.PT.49a.15354497), ITGB1 (Hs.PT.49a.19538428), and CD151 (Hs.PT.49a.19132657.g) were from integrated DNA.
Transient and stable gene silencing
For transient TSPAN12 silencing, siRNA duplexes (Dharmacon, USA; M-012466-00 for NM_012338) were transfected using Lipofactamine RNAiMAX (Invitrogen). Control siRNA was from Qiagen (USA). Experiments were performed 3 days post-transfection. Stable TSPAN12 shRNA silencing was as described [8]. Briefly, Lipofectamine 2000 (Invitrogen), together with dR8.91 packaging plasmid and VSVG envelope plasmid, were mixed and transfected into HEK293 cells to generate lentiviral particles. Viral particles were then used to infect cancer cells with shRNA for human TSPAN12 (OpenBiosystems), with selection using Puromycin or FACS sorting.
Nude mouse xenograft assays
For ectopic analysis, nude mice were injected s.c. at two sites each with MDA-MB-231 cells (1.5 × 106 per site). For orthotopic analysis, MDA-MB-231 cells were injected into mammary fat pads (1 × 106 per site). Each group contained four mice and experiments were repeated twice. After 32–34 days, mice were sacrificed and tumor weight was determined. Tumor dimensions were measured with calipers and tumor volume = length × width2 × 0.5. Tumors extracted from mice were fixed and embedded in paraffin, and sectioned at 7 μm for H&E staining and immunohistochemistry.
Tumor metastasis
Tail veins of SCID beige mice were injected with 2 × 106 MDA-MB-231 cells. Each group contained four mice. After 5 weeks, the mice were sacrificed and lungs were perfused with India ink, excised, and fixed with Fekete’s solution (680 ml of 95 % ethanol, 200 ml H2O, 80 ml 37 % formaldehyde solution, 40 ml glacial acetic acid). Total tumor colonies on lung surfaces were counted using a stereomicroscope. To assess lung colonization at an early time point, SCID beige mouse tail veins were injected with 2 × 106 MDA-MB-231 cells, that had been labelled with cell-permeable fluorescent dye CFSE (5 μM) prior to injection. After 24 h mice were sacrificed and lungs were extracted and washed in PBS. From five representative fluorescent microscope image fields, the percent of total area positive for CFSE was calculated using ImageJ software. Each experimental group had four mice and experiments were repeated twice.
Cell attachment assays
HUVEC cells (1.5 × 104/well, in triplicate, in 96-well plates) were incubated overnight at 37 °C. MDA-MB-231 cells (5 × 105/ml) were incubated with calcein AM (5 μM) for 30 min, then washed and 100 μl of cell suspension was added to each well of HUVEC cells and incubated at 37 °C for 1 h. After washing three times with PBS, fluorescence was measured using a fluorescein filter set at 495 nm. Fluorescence measured prior to washing = total fluorescence. Measurement of HUVEC cells alone = background fluorescence.
Subcellular localization of β-catenin
MDA-MB-231 cells with control or TSPAN12 shRNA (2 × 104/chamber) were incubated for ~18 h at 37 °C on chamber slides. In some samples, proteosomal inhibitor MG132 (10 μM) was added for the final 6 h. Cells on slides were permeabilized and stained for β-catenin for 30 min. Cells were then washed and mounting media with DAPI (4′,6-diamidino-2-phenylindole) was added. All fluorescent images were recorded using a Leica SP5 confocal microscope. Fluorescence distribution was analyzed using the ImageJ program.
Immunoprecipitation and Western blotting
Cultured MDA-MB-231 cells were lysed at 4 °C (25 mM HEPES, pH 7.5, 150 mM NaCl, and 2.5 mM EDTA) with 1 % Brij 58 (Sigma) plus protease inhibitor cocktail (Roche Applied Science). Immunoprecipitation and western blotting were as described previously [39]. Relative band densities were quantitated using ImageJ software.
Gene array analyses
Mammary fat pad tumors were lysed and RNA was isolated and purified using RNeasy kit (Qiagen). To control for size differences, we chose one of two TSPAN12 null tumors that was comparable in size to one of the two control tumors. Isolated RNA was analyzed using the U133A 2.0 Affymetrix gene chip array (at the Dana-Farber Cancer Institute Microarray Core Facility). Note that there is only ~5 % background binding of mouse genes (from the tumor microenvironment) to human gene array probes [14]. Raw data are publicly available through GEO accession number GSE41892. Raw data were imported into GenePattern [15], normalized and log2-transformed in ExpressionFileCreator and then exported to the Multiplot module to visualize in volcano plot format. Data from GenePattern were then subjected to the “Core analysis” function within Ingenuity Pathway Analysis software (http://www.ingenuity.com).
Results
TSPAN12 contributes to primary tumor growth in vivo
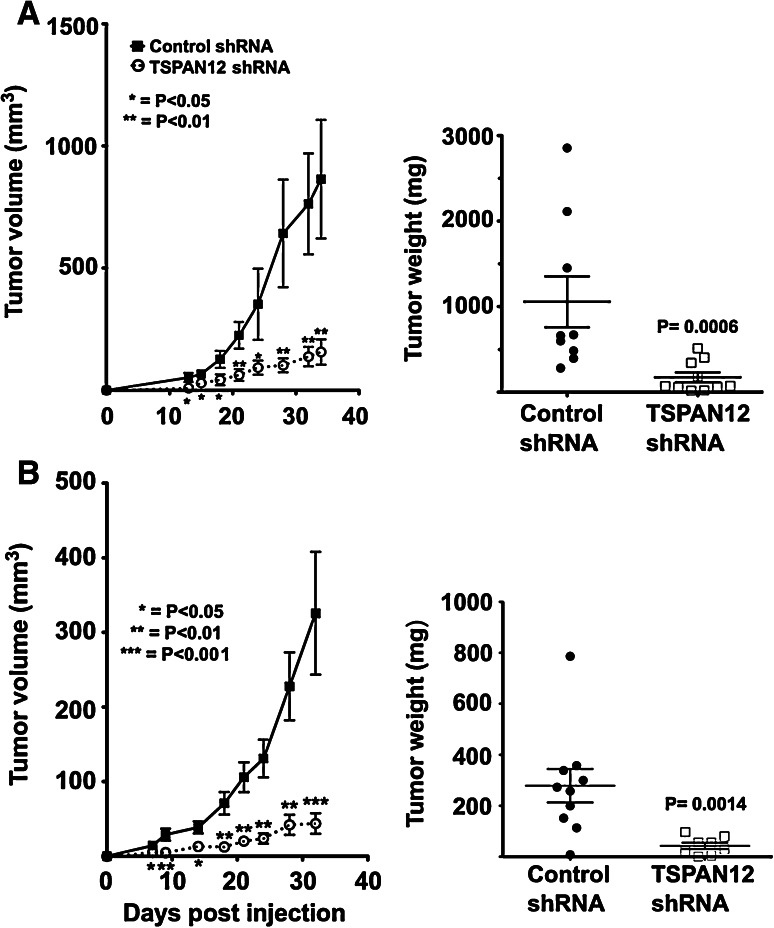
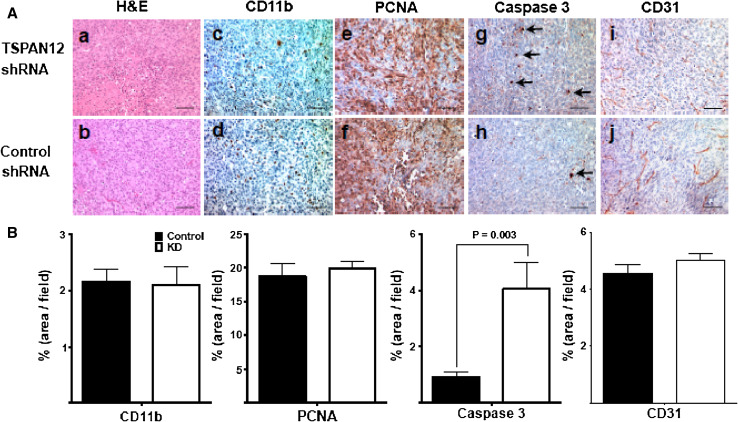
To test whether TSPAN12 affects tumor growth in vivo, human MDA-MB-231 breast cancer cells, with and without TSPAN12 shRNA ablation, were injected ectopically (subcutaneous) or orthotopically (mammary fat pad) into nude mice. Tumor growth was significantly reduced in mice receiving TSPAN12-ablated cells, at both the ectopic and orthotopic sites (Fig. 1a, b). As seen from tumor sections, TSPAN12 ablation did not affect tumor necrosis (Fig. 2a; a, b), cell proliferation (proliferating cell nuclear antigen; PCNA; Fig. 2a; c, d), infiltration of myeloid cells (stained for CD11b; Fig. 2a; e, f), or tumor angiogenesis (blood vessels stained for CD31; Fig. 2a; i, j). However, staining for cleaved caspase-3, a marker for apoptosis, was significantly elevated in sections from TSPAN12-ablated mammary fat pad tumors (Fig. 2a; g, h). TSPAN12 effects on cell death are consistent with gene array results (described below). Quantitation of tumor sections from at least three separate mice (Fig. 2b) confirmed the results shown in Fig. 2a. TSPAN12 was effectively silenced, by >90 %, in MDA-MB-231 cells expressing appropriate shRNA (Suppl. Fig. S1a, S1b) or siRNA (Suppl. Fig. S1c). Upon TSPAN12 knockdown (KD) in MDA-MB-231 cells, and in two other cell lines with high (MDA-MB-175) or low (MCF7) constitutive TSPAN12 expression levels, in vitro cell proliferation was essentially unchanged, as determined by cell counting (Suppl. Fig. S2a). Also, TSPAN12 ablation did not induce apoptosis in MDA-MB-231 cells cultured in vitro (Suppl. Fig. S2b).
Fig. 1.
TSPAN12 supports primary tumor growth. a Human MDA-MB-231 cells, ± stable TSPAN12 ablation, were injected into nude mouse mammary fat pads, and tumor volume was determined (left panel). At day 34, tumor weights were measured (right panel). b MDA-MB-231 cells were injected subcutaneously into nude mice and tumor volumes were determined (left panel) and tumor weight at day 32 was measured (right panel). Error bars ±SEM, P values are from unpaired t tests
Fig. 2.
TSPAN12 effects on tumor physiology. A Tumor sections from control and TSPAN12 ablated tumors were analyzed by H&E staining (a, b). Immunohistochemistry was used to analyze presence of apoptotic marker cleaved caspase-3 (c, d), proliferation marker PCNA (e, f), myeloid marker CD11b (g, h) and blood vessel/angiogenesis marker CD31 (i, j). B In representative tumor sections staining was quantitated as percent stained area/total area. Error bars ±SEM, P values are from unpaired t tests. Bars 100 μM. Note that similar caspase 3 staining results were obtained by counting numbers of stained cells/area (10.3 ± 6.4 SD vs. 44.7 ± 13.6 SD cells/area; P = 0.017)
TSPAN12 affects tumor cell metastasis in vivo
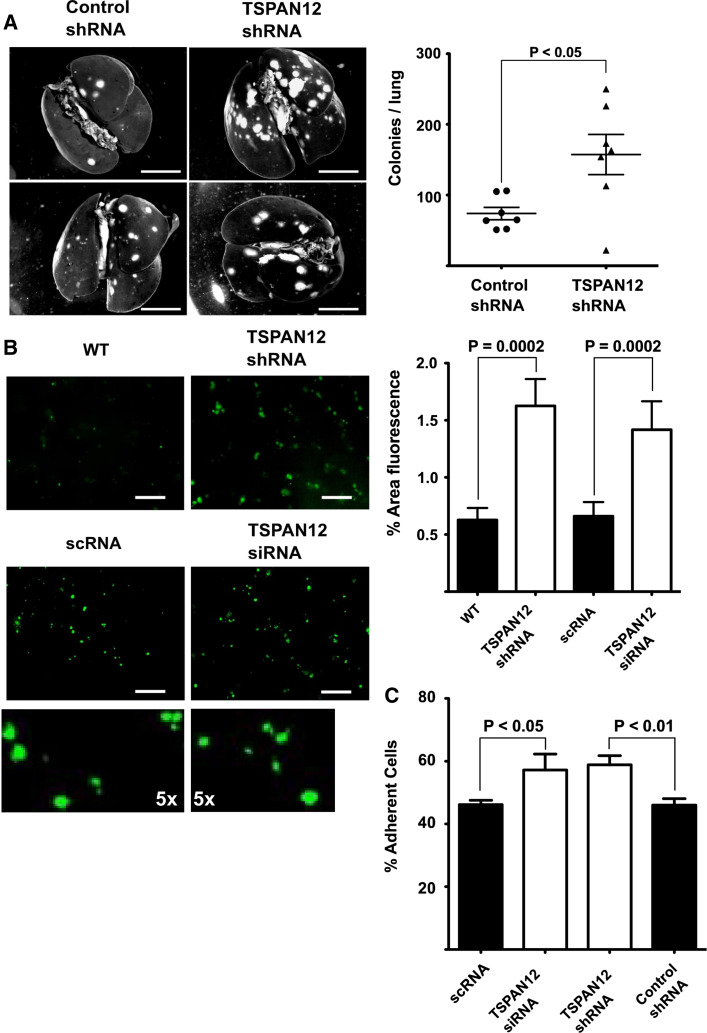
To investigate the role of TSPAN12 in metastasis we injected MDA-MB-231 cells, with and without TSPAN12 ablation, into the tail veins of SCID mice. After 5 weeks, recipients of TSPAN12-ablated cells showed increased numbers of metastatic lung colonies (Fig. 3a). The effect of TSPAN12 ablation was evident early in the metastatic process. Cells lacking TSPAN12 showed significantly more efficient in vivo localization to mouse lungs as early as 24 h post-injection (Fig. 3b). Sizes of 24-h cell deposits were not appreciably different (see higher magnifications in Fig. 3b), thus ruling out increased cell clumping as an explanation for increased metastasis. In accordance with this in vivo data, TSPAN12-ablated cells also showed increased in vitro attachment to HUVEC cells (Fig. 3c). Similar cell attachment results were seen upon targeting of different TSPAN12 mRNA sequences either transiently (siRNA) or stably (shRNA) (Fig. 3b, c). Thus, RNAi off-target effects are unlikely to explain these results.
Fig. 3.
TSPAN12 effects on metastasis and cell attachment. a MDA-MB-231 cells, ± TSPAN12 ablation, were injected into tail veins of SCID beige mice (Taconic, USA). After 5 weeks, lung colonies were visualized using India ink (left panels) and total colonies were counted (right panels). Bar 5 mm. b MDA-MB-231 cells were labeled with CFSE, injected into tail veins of SCID beige mice, and then 24-h post-injection lungs were extracted and washed with PBS before representative pictures were taken under a fluorescent microscope (left panels); bar 100 μM. The bottom two left panels show ×5 magnifications of regions from panels directly above. Staining of the four upper panels is quantitated using ImageJ (right panels). c MDA-MB-231 cells, with TSPAN12 ablated either transiently or stably, were incubated with confluent monolayers of HUVEC cells and adhesion was quantitated after 1 h. Error bars ±SEM, P values are from unpaired t tests
TSPAN12 and β-catenin
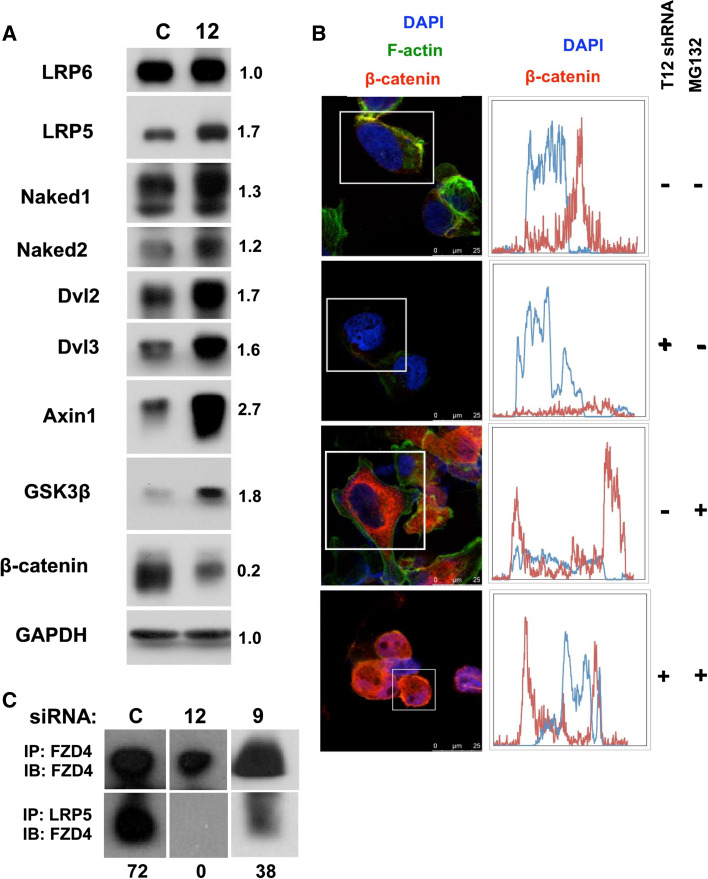
Genetic evidence previously linked TSPAN12 to a Norrin-FZD4–LRP5 signaling pathway, upstream of β-catenin (see introduction). The β-catenin protein and associated signaling pathways can play a prominent role in primary tumor growth as well as metastasis [6]. Hence, to gain mechanistic insight into TSPAN12’s role in MDA-MB-231 cells, we probed TSPAN12 influence on β-catenin. In the absence of TSPAN12, levels of β-catenin were diminished by more than 80 % (Fig 4a), consistent with disrupted FZD4–LRP5 signaling. At the same time, levels of Axin1 and GSK3β were elevated (Fig. 4a, b), consistent with enhanced β-catenin proteosomal degradation, such as occurs when the Wnt signaling pathway is disrupted [16]. Elevated levels of Nkd1 and Nkd2, negative regulators of the Wnt pathway [17, 18], are again consistent with enhanced β-catenin degradation. Altered protein levels for LRP5, Dvl1, and Dvl2, but not LRP6 (Fig. 4a), are also suggestive of dysregulated FZD4–LRP5 signaling.
Fig. 4.
TSPAN12 ablation affects FZD4/LRP5/β-catenin pathway. a Lysates from MDA-MB-231 cells, with or without stable TSPAN12 ablation, were fractionated by SDS-PAGE under reducing conditions and then blotted for the indicated proteins. Numbers indicate protein band intensities (TSPAN12 ablation relative to control ablation). Results are representative of multiple experiments. b MDA-MB-231 cells, with and without TSPAN12 ablation, were allowed to adhere to glass slides for ~18 h. In some cases, 10 μM proteosomal inhibitor MG132 was included for the last 6 h. Cells were then fixed with 4 % formaldehyde in PBS for 10 min, permeabilized with 0.2 % Triton X-100 in PBS for 10 min and stained for F-actin and β-catenin before adding mounting media with DAPI. Histograms in right panels indicate fluorescent staining determined linearly across representative cell diameters drawn (but not shown) within white boxes. Red staining represents β-catenin and blue represents DAPI staining of the nucleus. Bar 25 μM. c Control, TSPAN12-ablated and CD9-ablated MDA-MB-231 cells were lysed and then FZD4 and LRP5 were immunoprecipitated (in each case from lysates containing 400 μM total protein). FZD4 present in immunoprecipitated protein complexes was determined by immunoblotting. Note that the amount of FZD4 recovered in lane 2 is 67 % of that in lane 1 (upper panels). Numbers below lower panels represent protein band intensities in lower panels relative to upper panels
When TSPAN12 was present, there was an abundance of non-nuclear β-catenin staining within MDA-MB-231 cells, not overlapping with DAPI-stained nuclei (Fig. 4b, first panels). However, in absence of TSPAN12, minimal β-catenin was present (Fig. 4b, second panels). Treatment with proteosomal inhibitor MG132 (for 6 h) reversed effects of TSPAN12 ablation, thus restoring β-catenin (Fig. 4b, fourth panels) to a level comparable to that in control cells (Fig. 4b, third panels). These results are consistent with TSPAN12 ablation causing β-catenin to be proteosomally degraded.
TSPAN12 did not co-immunoprecipitate with LRP5 or FZD4. Nonetheless, in the presence of TSPAN12, we observed co-immunoprecipitation of FZD4 with LRP5 (Fig. 4c). However, in the absence of TSPAN12, FZD4–LRP5 co-immunoprecipitation was essentially eliminated. Removal of CD9, another tetraspanin protein, did not eliminate FZD4–LRP5 co-immunoprecipitation Fig. 4c). In other control experiments, levels of LRP5 detected by immunoblotting (Fig. 4a) or recovered upon immunoprecipitation with anti-LRP5 antibody (not shown) were not diminished when TSPAN12 was absent. These results suggest that TSPAN12 supports FZD4–LRP5 receptor-coreceptor interaction, thereby stabilizing β-catenin through a canonical Wnt pathway.
To gain unbiased insight into the effects of TSPAN12 on tumor cells, we performed microarray analysis of gene expression. Because tumor growth effects were seen in vivo (e.g., Fig. 1) but not in vivo (Suppl. Fig. S2), human gene expression analysis was done on material from primary mammary fat pad tumors. In TSPAN12 KD tumors, gene expression levels were slightly increased for LRP5 (1.35-fold) and FZD4 (1.27-fold), but essentially unaltered for LRP6, Dvl3, Dvl2, Axin, GSK3b, and β-catenin. Further data processing using GenePattern [9] showed 473 genes either up- or downregulated with a >1.5-fold-change cut-off and a P value of <0.05 (Suppl. Fig. S3; Suppl. Table S1). Data were then subjected to the “Core Analysis” function within the IPA (Ingenuity Pathway Analysis; http://www.ingenuity.com) program, to predict which upstream central node genes might be most significantly associated with the regulation of TSPAN12-dependent genes. Listed among the top potential upstream effectors of TSPAN12-dependent gene regulation was CTNNB1 (β-catenin) (Suppl. Table S2). Although the β-catenin gene itself was not perturbed in TSPAN12 KD tumors, there are 25 TSPAN12-regulated genes known to lie downstream of β-catenin. These include CCNA1 (cyclin A1) and CCNE2 (cyclin E2), downregulated 3.5- and 1.7-fold, respectively, consistent with reduced cell growth [19], and WISP-1, downregulated 1.65-fold, also consistent with reduced tumor growth [20]. Other TSPAN12-dependent, β-catenin-dependent downregulated genes were SFN (down 1.5-fold), ID4 (down 1.6-fold) and ME1 (down 1.6-fold). The SFN (14-3-3σ) and ID4 genes each support tumor development in multiple cancer types [21, 22], and downregulation of ME1 can lead to activation of the p53 tumor suppressor [23]. These results further support a link between TSPAN12 ablation and gene regulation by the β-catenin transcription factor.
Other effects of TSPAN12
Additional gene array results indicate that gene regulation due to TSPAN12 ablation may not solely involve β-catenin. For example, TGFβ1 also appears among potential upstream regulators potentially responsible for altered gene expression due to TSPAN12 ablation (Suppl. Table S2).
IPA “core analysis” also revealed that top networks perturbed by TSPAN12 ablation involve functions such as cell growth and proliferation, cell cycle, movement, death, and survival (Table 1a). Furthermore, the top ‘disease/disorder’ ranked according to significance, was ‘cancer’ (Table 1b), and the top ‘molecular and cellular functions’ included growth, proliferation, death, survival, and cell cycle (Table 1c). Notably, despite reported TSPAN12 effects on ADAM10 maturation in another cell line [8], we did not observe TSPAN12 ablation having any effect on ADAM10 maturation in MDA-MB-231 cells (not shown). TSPAN12 also did not affect ADAM10 gene expression (not shown).
Table 1.
Results of ingenuity pathway analyses of genes affected by TSPAN12 ablation
| (a) Associated network functions | Score |
|---|---|
| Cellular growth and proliferation, embryonic development, tissue morphology | 45 |
| Cell cycle, cellular assembly and organization, DNA replication, recombination, and repair | 41 |
| Cellular movement, cell death and survival, nucleic acid metabolism | 36 |
| Organ morphology, reproductive system development and function, cell morphology | 34 |
| Endocrine system disorders, gastrointestinal disease, hereditary disorder | 30 |
| Name | P value | No. molecules |
|---|---|---|
| (b) Diseases and disorders | ||
| Cancer | 1.84E-18–1.27E-02 | 198 |
| Reproductive system disease | 1.47E-11–1.20E-02 | 111 |
| Gastrointestinal disease | 2.16E-10–1.11E-02 | 90 |
| Endocrine system disorders | 3.92E-07–2.82E-03 | 39 |
| Renal and urological disease | 3.52E-05–1.12E-02 | 44 |
| (c) Molecular and cellular functions | ||
| Cellular growth and proliferation | 7.16E-13–1.18E-02 | 158 |
| Cellular movement | 6.83E-11–1.29E-02 | 107 |
| Cell death and survival | 4.11E-08–1.20E-02 | 140 |
| Cell cycle | 3.95E-07–1.20E-02 | 72 |
| Cellular development | 2.08E-05–1.23E-02 | 138 |
Discussion
Studies of tetraspanin protein TSPAN12 have mostly involved its role in FEVR (familial exudative vitreo-retinopathy) [2–4], with no focus on cancer. Here, we show that TSPAN12 can support tumor growth, by a mechanism involving enhanced cell survival, and associated with FZD4/LRP5/β-catenin signaling. We saw no evidence for TSPAN12 affecting tumor growth by other mechanisms, involving myeloid cell recruitment, tumor cell proliferation, or angiogenesis. Also, TSPAN12 appears to suppress tumor metastasis, likely by modulating tumor cell interaction with endothelial cell layers. Consistent with these results, the top-ranked disease/disorder associated with TSPAN12-regulated gene array results was ‘cancer’.
β-catenin and canonical Wnt signaling
There is considerable evidence for β-catenin being oncogenic [6]. TSPAN12 was previously shown to support canonical β-catenin signaling through a FZD4–LRP5 pathway [4]. Hence, we suspected that TSPAN12 support of primary tumor growth could be explained by TSPAN12 facilitation of β-catenin signaling. Indeed, TSPAN12 supported FZD4–LRP5 signaling, resulting in β-catenin protection from proteosomal degradation. Previous results suggested that TSPAN12 co-localizes with and supports FZD4 oligomerization [4]. We now suggest that TSPAN12 can also promote the association of FZD4 with its co-receptor LRP5. Another abundant tetraspanin protein, CD9, was markedly less effective in supporting FZD4–LRP5 association, thus emphasizing the specific contribution of TSPAN12. Because absence of TSPAN12 was previously shown to have no effect on ligand binding to FZD4 [4], we did not focus on FZD4 ligands. It remains to be determined whether Norrin [4] or another ligand, such as WNT7a [24], is binding to FZD4 in MDA-MB-231 cells.
Consistent with diminished canonical Wnt pathway signaling, TSPAN12 ablation increased the levels of proteins, such as Axin1 and GSK3β, which are involved in β-catenin degradation [16]. In addition, TSPAN12 ablation upregulated negative regulatory proteins Naked1 and Naked2 [17, 18], consistent with diminished signaling through β-catenin. At present, it is not clear why TSPAN12 ablation would cause increased expression of LRP5, Dvl2, and Dvl3, since these proteins typically support canonical Wnt pathway signaling, leading to enhanced β-catenin expression and signaling. One possibility is that TSPAN12 ablation may trigger a regulatory feedback loop, attempting to compensate for diminished function. TSPAN12 ablation perturbed expression of LRP5 but not LRP6. This result is consistent with mouse and human TSPAN12 being linked to FZD4–LRP5 functions [2–4], while not affecting FZD4–LRP6 functions.
DNA array results provide further evidence consistent with TSPAN12 acting through β-catenin. Our unbiased microarray data analysis from mouse xenograft tumors showed that β-catenin was among the top 10 upstream regulators potentially responsible for changes in gene expression due to TSPAN12 ablation. Among these, CCNA1 (cyclin A1), CCNE2 (cyclin E2), WISP-1, SFN (14-3-3σ), and ME1 were downregulated, as expected, in concert with downregulated β-catenin signaling. Furthermore, downregulation of each of these genes is expected to contribute to diminished tumor growth [19–23] and/or decreased cell survival [25–27]. Our results are consistent with another study, in which canonical WNT pathway signaling was essential for MDA-MB-231 xenograft growth [28], Interestingly, in that study [28], some of the most critical WNT-dependent gene expression events were observed in vivo, but not in vitro, thus emphasizing the value of in vivo gene expression experiments.
Effects on tumor metastasis
A correlation between TSPAN12 upregulation and loss of metastasis was noted previously, but TSPAN12 functions were not specifically investigated [9]. Here, we show enhanced metastasis upon TSPAN12 ablation, suggesting that TSPAN12 has a metastasis suppressor function. TSPAN12 ablation did not increase MDA-MB-231 invasion or migration (not shown). However, TSPAN12-ablated cells showed an increase in lung localization as early as 24 h post-tail vein injection, suggestive of increased interaction with tumor vasculature. Consistent with this, we also observed a significant increase in adhesion to endothelial cell monolayers in vitro. Although genes for a few integrin-type adhesion receptor subunits were elevated according to gene array results (Suppl. Table S1), there was no significant increase in protein expression of integrin α2, α3, α6, β1, or β4 subunits at the cell surface, as seen by flow cytometry (not shown).
Metastasis suppression by TSPAN12 could also involve canonical Wnt/β-catenin signaling, because enhanced metastasis is sometimes associated with β-catenin degradation. For example, low β-catenin, coupled with elevated p53 (such as seen upon TSPAN12 ablation), was associated with increased metastasis in colorectal cancer [29], and β-catenin degradation was also linked to increased metastasis in oral squamous cell carcinoma [13]. Furthermore, downregulation of key β-catenin-dependent genes, due to TSPAN12 ablation, may contribute to increased metastasis. For example, downregulation of Wisp-1 [30], SFN (4751), and ID4 [31] may all contribute to increased tumor metastasis, due to elimination of metastasis suppression.
Other TSPAN12 effects
It is possible that TSPAN12 effects on primary tumor growth and metastasis are not entirely through β-catenin. For example, TGFβ1 (listed in Table S2 as a potential mediator of TSPAN12 effects on gene expression) is well known to inhibit primary tumor growth, while stimulating tumor metastasis [32]. Curiously, we did not observe TSPAN12 ablation to affect either ADAM10 or ADAM10 ligands at either genomic or protein levels in MDA-MB-231 cells. Thus, TSPAN12 effects on tumor growth and metastasis appear not to involve ADAM10. TSPAN12 ablation also caused markedly diminished expressions of multiple SPANX family genes. These genes are typically found in testis, and on multiple tumor cell types [33]. It remains to be studied whether diminished SPANX gene expression is critical for altered MDA-MB-231 growth and metastasis observed here, or for the pronounced loss of male fertility that we observed in TSPAN12 knockout mice (not shown).
TSPAN12 comparisons to other tetraspanins
A supporting role for TSPAN12 during FZD4–LRP5 association and function fits a paradigm seen for other tetraspanin proteins. Examples include CD9 indirectly facilitating diphtheria toxin binding to its receptor [34], CD81 enhancing α4β1 integrin interaction with VCAM-1 [35], and C. elegans TSP-15 aiding dual oxidase functions needed for exoskeletal development [36].
Among other tetraspanin proteins, CD151 can also support primary tumor growth [37–40]. However, CD151 appears to function largely by modulating functions of closely associated integrins [41], whereas TSPAN12 does not associate closely with integrins (not shown). Other tetraspanins, such as CD82 and CD9, can also suppress metastasis [42, 43] by mechanisms that may involve β-catenin. However, CD82 and CD9 appear to inhibit rather than stimulate canonical Wnt signaling and the mechanisms do not specifically involve FZD4 [44, 45].
Summary and implications
Our investigation of TSPAN12 in cancer models provides new insights into TSPAN12 functions, and demonstrates that TSPAN12 may be useful as a marker during tumor progression at primary and secondary sites. Our experiments were carried out using MDA-MB-231 cells. It remains to be seen to what extent TSPAN12 contributes to basal type breast cancers in general, and to other types of cancer. In this regard, TSPAN12 gene expression levels are significantly altered in highly metastatic versus poorly metastatic prostate cancer cells and in normal versus squamous cell carcinomas present in lung biopsies (see GEO datasets associated with [10, 46]).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by National Institutes of Health Grant CA42368 (to M.E.H.).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Garcia-Espana A, Chung PJ, Sarkar IN, Stiner E, Sun TT, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 2008;91:326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Poulter JA, Ali M, Gilmour DF, Rice A, Kondo H, Hayashi K, Mackey DA, Kearns LS, Ruddle JB, Craig JE, Pierce EA, Downey LM, Mohamed MD, Markham AF, Inglehearn CF, Toomes C. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:248–253. doi: 10.1016/j.ajhg.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EA, Arts P, Wieskamp N, Strom TM, Ayuso C, Tilanus MA, Bouwhuis S, Mukhopadhyay A, Scheffer H, Hoefsloot LH, Veltman JA, Cremers FP, Collin RW. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:240–247. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Junge HJ, Yang S, Burton JB, Paes K, Shu X, French DM, Costa M, Rice DS, Ye W. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139:299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 5.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des. 2009;15:2288–2299. doi: 10.2174/138161209788682442. [DOI] [PubMed] [Google Scholar]
- 8.Xu D, Sharma C, Hemler ME. Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. FASEB J. 2009;23:3674–3681. doi: 10.1096/fj.09-133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lara H, Wang Y, Beltran AS, Juarez-Moreno K, Yuan X, Kato S, Leisewitz AV, Cuello FM, Licea AF, Connolly DC, Huang L, Blancafort P. Targeting serous epithelial ovarian cancer with designer zinc finger transcription factors. J Biol Chem. 2012;287:29873–29886. doi: 10.1074/jbc.M112.360768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 12.Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, Eneman J, Crocker A, White J, Tessitore J, Stanley M, Harlow S, Weaver D, Muss H, Plaut K. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat. 2009;114:47–62. doi: 10.1007/s10549-008-9982-8. [DOI] [PubMed] [Google Scholar]
- 13.Kudo Y, Kitajima S, Ogawa I, Hiraoka M, Sargolzaei S, Keikhaee MR, Sato S, Miyauchi M, Takata T. Invasion and metastasis of oral cancer cells require methylation of E-cadherin and/or degradation of membranous beta-catenin. Clin Cancer Res. 2004;10:5455–5463. doi: 10.1158/1078-0432.CCR-04-0372. [DOI] [PubMed] [Google Scholar]
- 14.Creighton CJ, Bromberg-White JL, Misek DE, Monsma DJ, Brichory F, Kuick R, Giordano TJ, Gao W, Omenn GS, Webb CP, Hanash SM. Analysis of tumor-host interactions by gene expression profiling of lung adenocarcinoma xenografts identifies genes involved in tumor formation. Mol Cancer Res. 2005;3:119–129. doi: 10.1158/1541-7786.MCR-04-0189. [DOI] [PubMed] [Google Scholar]
- 15.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 16.Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T, Clevers H. Wnt signaling through inhibition of beta-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Rousset R, Mack JA, Wharton KA, Jr, Axelrod JD, Cadigan KM, Fish MP, Nusse R, Scott MP. Naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev. 2001;15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wharton KA, Jr, Zimmermann G, Rousset R, Scott MP. Vertebrate proteins related to Drosophila Naked Cuticle bind Dishevelled and antagonize Wnt signaling. Dev Biol. 2001;234:93–106. doi: 10.1006/dbio.2001.0238. [DOI] [PubMed] [Google Scholar]
- 19.Yasmeen A, Berdel WE, Serve H, Muller-Tidow C. E- and A-type cyclins as markers for cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2003;3:617–633. doi: 10.1586/14737159.3.5.617. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Corcoran RB, Welsh JW, Pennica D, Levine AJ. WISP-1 is a Wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]
- 21.Perathoner A, Pirkebner D, Brandacher G, Spizzo G, Stadlmann S, Obrist P, Margreiter R, Amberger A. 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin Cancer Res. 2005;11:3274–3279. doi: 10.1158/1078-0432.CCR-04-2207. [DOI] [PubMed] [Google Scholar]
- 22.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 23.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobov IB, Rao S, Carroll TJ, Vallance JE, Ito M, Ondr JK, Kurup S, Glass DA, Patel MS, Shu W, Morrisey EE, McMahon AP, Karsenty G, Lang RA. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature. 2005;437:417–421. doi: 10.1038/nature03928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji P, Baumer N, Yin T, Diederichs S, Zhang F, Beger C, Welte K, Fulda S, Berdel WE, Serve H, Muller-Tidow C. DNA damage response involves modulation of Ku70 and Rb functions by cyclin A1 in leukemia cells. Int J Cancer. 2007;121:706–713. doi: 10.1002/ijc.22634. [DOI] [PubMed] [Google Scholar]
- 26.Wegiel B, Bjartell A, Culig Z, Persson JL. Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. Int J Cancer. 2008;122:1521–1529. doi: 10.1002/ijc.23261. [DOI] [PubMed] [Google Scholar]
- 27.Maiese K, Chong ZZ, Shang YC, Wang S. Targeting disease through novel pathways of apoptosis and autophagy. Expert Opin Ther Targets. 2012;16:1203–1214. doi: 10.1517/14728222.2012.719499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res. 2009;11:R32. doi: 10.1186/bcr2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pancione M, Forte N, Fucci A, Sabatino L, Febbraro A, Di BA, Daniele B, Parente D, Colantuoni V. Prognostic role of beta-catenin and p53 expression in the metastatic progression of sporadic colorectal cancer. Hum Pathol. 2010;41:867–876. doi: 10.1016/j.humpath.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Soon LL, Yie TA, Shvarts A, Levine AJ, Su F, Tchou-Wong KM. Overexpression of WISP-1 down-regulated motility and invasion of lung cancer cells through inhibition of Rac activation. J Biol Chem. 2003;278:11465–11470. doi: 10.1074/jbc.M210945200. [DOI] [PubMed] [Google Scholar]
- 31.Umetani N, Mori T, Koyanagi K, Shinozaki M, Kim J, Giuliano AE, Hoon DS. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene. 2005;24:4721–4727. doi: 10.1038/sj.onc.1208538. [DOI] [PubMed] [Google Scholar]
- 32.Ikushima H, Miyazono K. TGFbeta signaling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 33.Kouprina N, Mullokandov M, Rogozin IB, Collins NK, Solomon G, Otstot J, Risinger JI, Koonin EV, Barrett JC, Larionov V. The SPANX gene family of cancer/testis-specific antigens: rapid evolution and amplification in African great apes and hominids. Proc Natl Acad Sci USA. 2004;101:3077–3082. doi: 10.1073/pnas.0308532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitamura T, Iwamoto R, Umata T, Yomo T, Urabe I, Tsuneoka M, Mekada E. The 27-kD diphtheria toxin receptor-associated protein (DRAP27) from Vero Cells is the monkey homologue of human CD9 antigen: expression of DRAP27 elevates the number of diphtheria toxin receptors on toxin-sensitive cells. J Cell Biol. 1992;118:1389–1399. doi: 10.1083/jcb.118.6.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feigelson SW, Grabovsky V, Shamri R, Levy S, Alon R. The CD81 tetraspanin facilitates instantaneous leukocyte VLA-4 adhesion strengthening to vascular cell adhesion molecule 1 (VCAM-1) under shear flow. J Biol Chem. 2003;278:51203–51212. doi: 10.1074/jbc.M303601200. [DOI] [PubMed] [Google Scholar]
- 36.Moribe H, Konakawa R, Koga D, Ushiki T, Nakamura K, Mekada E. Tetraspanin is required for generation of reactive oxygen species by the dual oxidase system in Caenorhabditis elegans . PLoS Genet. 2012;8:e1002957. doi: 10.1371/journal.pgen.1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadej R, Romanska H, Baldwin G, Gkirtzimanaki K, Novitskaya V, Filer AD, Krcova Z, Kusinska R, Ehrmann J, Buckley CD, Kordek R, Potemski P, Eliopoulos AG, Lalani E, Berditchevski F. CD151 regulates tumorigenesis by modulating the communication between tumor cells and endothelium. Mol Cancer Res. 2009;7:787–798. doi: 10.1158/1541-7786.MCR-08-0574. [DOI] [PubMed] [Google Scholar]
- 38.Yang XH, Richardson AL, Torres-Arzayus MI, Zhou P, Sharma C, Kazarov AR, Andzelm MM, Strominger JL, Brown M, Hemler ME. CD151 accelerates breast cancer by regulating a6 integrin functions, signaling, and molecular organization. Cancer Res. 2008;68:3204–3213. doi: 10.1158/0008-5472.CAN-07-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Yang XH, Xu F, Sharma C, Wang HX, Knoblich K, Rabinovitz I, Granter SR, Hemler ME. Tetraspanin CD151 plays a key role in skin squamous cell carcinoma. Oncogene. 2013;32:1772–1783. doi: 10.1038/onc.2012.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng X, Li Q, Hoff J, Novak M, Yang H, Jin H, Erfani SF, Sharma C, Zhou P, Rabinovitz I, Sonnenberg A, Yi A, Zhou P, Stipp CS, Kaetzel DM, Hemler ME, Yang HY. Integrin-associated CD151 drives ErbB2-evoked mammary tumor onset and metastasis. Neoplasia. 2012;14:678–689. doi: 10.1593/neo.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kazarov AR, Yang X, Stipp CS, Sehgal B, Hemler ME. An extracellular site on tetraspanin CD151 determines a3 and a6 integrin-dependent cellular morphology. J Cell Biol. 2002;158:1299–1309. doi: 10.1083/jcb.200204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miranti CK. Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cell Signal. 2009;21:196–211. doi: 10.1016/j.cellsig.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 43.Miyake M, Inufusa H, Adachi M, Ishida H, Hashida H, Tokuhara T, Kakehi Y. Suppression of pulmonary metastasis using adenovirally motility related protein-1 (MRP-1/CD9) gene delivery. Oncogene. 2000;19:5221–5226. doi: 10.1038/sj.onc.1203919. [DOI] [PubMed] [Google Scholar]
- 44.Chigita S, Sugiura T, Abe M, Kobayashi Y, Shimoda M, Onoda M, Shirasuna K. CD82 inhibits canonical Wnt signaling by controlling the cellular distribution of beta-catenin in carcinoma cells. Int J Oncol. 2012;41:2021–2028. doi: 10.3892/ijo.2012.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang CL, Liu D, Masuya D, Kameyama K, Nakashima T, Yokomise H, Ueno M, Miyake M. MRP-1/CD9 gene transduction downregulates Wnt signal pathways. Oncogene. 2004;23:7475–7483. doi: 10.1038/sj.onc.1208063. [DOI] [PubMed] [Google Scholar]
- 46.Wong SY, Haack H, Kissil JL, Barry M, Bronson RT, Shen SS, Whittaker CA, Crowley D, Hynes RO. Protein 4.1B suppresses prostate cancer progression and metastasis. Proc Natl Acad Sci USA. 2007;104:12784–12789. doi: 10.1073/pnas.0705499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.