Abstract
Congenital heart defects represent the most common human birth defects and are often life-threatening. Frequently, they are caused by abnormalities of the outflow tract whose formation results from coordinated development of cells from mesodermal and neural crest origin and depends on the activity of many different transcription factors. However, place, time, and mode of action have only been analyzed for a few of them. Here we assess the contribution of the closely related high-mobility-group transcription factors Sox4 and Sox11 to outflow tract development and determine their function. Using cell-type-specific deletion in the mouse, we show that Sox11 is required for proper development in both mesodermal cells and neural crest cells. Deletion in either mesoderm or neural crest, or both, leads to outflow tract defects ranging from double outlet right ventricle to common arterial trunk. Sox4 supports Sox11 in its function, but has additional roles with relevance for outflow tract formation in other cell types. The two Sox proteins are dispensable during early phases of cardiac neural crest development including neural tube emigration, proliferation, and migration through the pharyngeal arches. They become essential after arrival of the neural crest cells in the outflow tract for their proper differentiation and interaction with each other as well as with the environment through regulation of cytoskeletal, cell adhesion, and extracellular matrix molecules. Our results demonstrate that Sox4 and Sox11 have multiple functions in several cell types during outflow tract formation and may thus help to understand the basis of congenital heart defects in humans.
Keywords: Sox factors, Double outlet right ventricle, Common arterial trunk, Neural crest, Cardiac gene regulatory network
Introduction
Heart development in vertebrates involves extensive morphogenetic processes and the contribution of several different cell types, and is thus highly complex. This developmental complexity partly explains the prevalence of congenital heart defects among birth defects in humans [1]. Congenital heart defects involve defects of ventricular septum, semilunar valves, or outflow tract, and frequently combinations thereof.
Initially, the outflow tract is a single vessel with a myocardial outer layer and an endocardial lining that arises at the anterior pole of the heart from a still unseptated ventricle. As heart development proceeds, this vessel undergoes extensive remodeling, including septation and rotation events. With formation of the aortopulmonary septum, the outflow tract is septated into ascending aorta and pulmonary trunk, which eventually have to join with the two different ventricles [2]. The two main cellular contributions to the outflow tract of the vertebrate heart stem from the second heart field and the cardiac neural crest. The second heart field contributes to both myocardial and endocardial lineages, while cardiac neural crest cells migrate into the conotruncal endocardial cushions and add to the existing mesenchyme, which is endocardially derived [3]. During subsequent formation of the aortopulmonary septum and outflow tract septation both cell populations interact and influence each other’s development reciprocally [4]. Developmental defects in both cell populations can lead to outflow tract malformations and misalignments with the ventricles, including persistent common arterial trunk (CT), double outlet right ventricle (DORV), and transposition of the great arteries.
Among the genes whose mutation or deletion cause outflow tract defects are a number of transcription factors, including Pax3, AP2α, Mef2c, Insm1, SIP1, the two Hand proteins, as well as several Gata, Tbx, Nkx, Fox, and Sox proteins [5]. Several of these have been studied in the mouse and represent excellent candidates for constituents of the transcriptional regulatory network in cardiac neural crest or second heart field-derived cells. Constitutive deletion of Sox4 in the mouse, for instance, impairs proper development of the endocardial ridges into semilunar valves and the outlet portion of the muscular ventricular septum as a consequence of which embryos develop CT and succumb to circulatory failure at embryonic day 14 [6]. Constitutive deletion of Sox11, on the other hand, leads to a wide spectrum of outflow tract phenotypes ranging from DORV to CT, usually in association with a membranous ventricular septum defect [7]. Sox11-deficient mice die shortly after birth. Disease-associated mutations in human SOX4 or SOX11 have not yet been identified.
Both Sox4 and Sox11 belong to the same SoxC subgroup of the Sox transcription factor family. They are closely related with each other and Sox12 as the third member of this subgroup, and believed to exhibit at least partial functional redundancy when co-expressed [8–10]. Nevertheless, deletion of Sox4 and Sox11 alone leads to outflow tract defects, arguing that in this context SoxC factors are functionally more diverse than expected or act in different cell types or at different times of heart development.
To investigate the role of Sox4 and Sox11 in heart development in greater detail, we deleted either one or both genes in developing mouse embryos in different cell types that contribute to the outflow tract region, and studied the consequences for cardiac development. Interestingly, we found that the two related SoxC proteins indeed do not function fully redundantly in this developmental context.
Materials and methods
Mouse husbandry, genotyping, and dissections
Mice used in this study carried the following alleles on a mixed 129SvJ/C57Bl6J background: Sox4 loxP [11], Sox11 loxP [10], and Rosa26 stopflox-EYFP [12]. For conditional deletion of the Sox4 loxP, Sox11 loxP, and Rosa26 stopflox-EYFP alleles, Wnt1::Cre, Nkx2-5 IRES::Cre (Nkx2.5::Cre), AP2a::Cre, Tie2::Cre, Sox2::Cre transgenes were used [13–17]. Genotyping was performed by PCR. Primer sequences are available upon request. Embryos were obtained at 10.5, 13.5, and 17.5 dpc from staged pregnancies.
Histological staining procedures, immunohistochemistry, and TUNEL
For hematoxylin-eosin staining, embryos were incubated overnight in 4 % paraformaldehyde, dehydrated, embedded in paraffin, sectioned on a microtome at 3-μm thickness, dewaxed, rehydrated, and stained in Mayer’s hematoxylin and eosin according to standard protocols.
For immunohistochemistry and TUNEL, embryos underwent fixation in 4 % paraformaldehyde, were frozen at −80 °C in Jung Tissue Freezing Medium (Leica, Nussloch, Germany) after cryoprotection, and then sectioned transversally on a cryotome at 10-μm thickness. The following primary antibodies were used in various combinations: rat antiserum against EYFP (1:1,000 dilution; GERBU Biotechnik, Heidelberg, Germany), guinea pig antisera against Sox4 (1:1,000 dilution; [18]) and Sox11 (1:1,000 dilution; [18]), and rabbit antisera against EYFP (1:2,000 dilution; Molecular Probes, Eugene, OR, USA), NG2 (1:200 dilution; Chemicon, Temecula, CA, USA), and Ki67 (1:500 dilution; Lab Vision, Fremont, CA, USA). Secondary antibodies conjugated to Alexa488 and Cy3 immunofluorescent dyes (Molecular Probes, Eugene, OR, USA, and Dianova, Hamburg, Germany) were used for detection. Phalloidin (1:5,000 dilution; Sigma-Aldrich, St. Louis, MO, USA) staining and TUNEL (Chemicon, Temecula, CA, USA) were performed according to the manufacturer’s protocol.
Quantifications
Numbers of immunoreactive cells and DAPI-positive nuclei per section were counted in relevant areas. Data were obtained from four sections of at least three different embryos for each genotype and embryonic age. Diagrams show mean values ± SEM of biological replicates. Statistical significance was determined by unpaired two-tailed Student’s t test using GraphPad Prism6 software and is mentioned in the corresponding figure legends.
Transfections and luciferase assays
HEK293 cells and Neuro-2a cells were kept on 100-mm dishes in Dulbecco’s modified Eagle’s medium (DMEM) containing 10 % FCS in the presence of penicillin and streptomycin. For luciferase assays, Neuro-2a cells were seeded on 35-mm dishes and transfected with Superfect (Qiagen, Hilden, Germany) using 400 ng of the luciferase reporter plasmid and 250 ng of the effector plasmids per dish [19]. Reporter plasmids contained the luciferase open reading frame under control of the human E-cadherin promoter (Ecad l, corresponding to positions −1,502 to +98 relative to the transcriptional start site, Ecad s, corresponding to positions −220 to +18, gift of J. Behrens, Erlangen) or the human Adam19 promoter (Adam19 l, positions −2,009 to +31, Adam19 s, positions −243 to +31, gift of M. Kreutz, Regensburg) [20]. The Ecad s and Adam19 s promoter fragments were also inserted as a version with mutated Sox binding sites (Ecad sm and Adam19 sm). pCMV5-based effector plasmids for full-length Sox11 and Sox4 were as described [19]. The total amount of plasmid was kept constant using empty pCMV5 vector where needed. Cells were harvested 48 h after transfection, and extracts were assayed for luciferase activity [19]. For production of recombinant proteins, polyethylenimine-transfected HEK293 cells or Superfect-transfected Neuro-2a cells were used, in both cases applying 6–10 μg pCMV-based expression plasmids per 100-mm dish.
Protein extracts and electrophoretic mobility shift assay
Whole cell extracts were prepared from transfected HEK293 and Neuro-2a cells and used for electrophoretic mobility shift assays using 32P-labeled oligonucleotides as probe and poly-dGdC as non-specific competitor under standard conditions [21]. Double-stranded oligonucleotides contained site B of the Mpz promoter [21] or predicted Sox binding sites of the human E-cadherin or Adam19 promoters in wild-type version or after mutation of the Sox binding site. All oligonucleotides had a 5′-GGG overhang for labeling. Sequences were: SiteB CAGAGTATACAATGCCCCTTA, Ecad1 CCAGGCTAGAGGGTCAACGCGT, Ecad1mut CCAGGCTAGAGGCCGCGCTCGT, Ecad2 CACGCACCCCCTCTCAGTGGCG, Ecad2mut CACGCACCCCCTCTGCGTGGCG, Adam19 CGTCCACCACAAATCCCTCCTC, Adam19mut CGTCCACCAGCGATCCCTCCTC.
RNA preparation from heart outflow tracts for quantitative RT-PCR
Outflow tracts of the heart were dissected from mice at embryonic day 13.5, immediately frozen on dry ice, and stored at −80 °C. Five outflow tracts of each genotype were pooled and processed for RNA isolation using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed in 25-μl reactions using 2 μg RNA, oligo-dT primers, and Moloney murine leukemia virus reverse transcriptase (New England Biolabs, Ipswich, MA, USA). Quantitative PCR using 3 μl of the obtained cDNA was performed on a Bio-Rad Lightcycler according to the manufacturer’s instructions using the ABsolute QPCR SYBR Green Mix (Thermo Scientific, Waltham, MA, USA) with an annealing temperature of 60 °C. Rpl8 transcripts were used for normalization [22]. The following primers were used for detection: 5′-GCCTCTCTGCGACTAAGGTG-3′ and 5′-CAGCAAGCCTCTGAGTCCAA-3′ for Adam9, 5′-CCCCCTAAGAGTGTGGGTCCCG-3′ and 5′-TGGCGATTCAACGCGCAGGT-3′ for Adam19, 5′-GGAAAGGAAGCAGAAGGCTCGGC-3′ and 5′-TGGGAGATGGGGAAGGCTTGGT-3′ for Cx40, 5′-AGTGAAAGAGAGGTGCCCAGACAT-3′ and 5′-TCGGTCTGCGCCACTTTGAGC-3′ for Cx43, 5′-AGCTTTTCCGCGCTCCTGCT-3′ and 5′-AGAGGCAGGGTCGCGGTGGT-3′ for E-cadherin, 5′-TGCCCAACTGTTACTGCCAA-3′ and 5′-GGGCAGAAAAACAACACGGG-3′ for Foxc2, 5′-AAGGAGCGGCGCAGGACTCA-3′ and 5′-CAGCCTGTCCGGCCTTTGGT-3′ for Hand2, 5′-TGCGTCCGGCCTGCTAGAGT-3′ and 5′-GCTCCACCGAAGCGAAGCGA-3′ for Insm1, 5′-GCCTTCACGATCACCATCCT-3′ and 5′-CAGGGCTCCTCTGTGTGAGA-3′ for NG2, 5′-TCCGCACCCACACTGGTGAGA-3′ and 5′-GGAGGCTCTGGGCGGGTACA-3′ for Snail1, 5′-ACTGGACACACACACAGTTAT-3′ and 5′-ATGGCATGGGGGTCTGAAAG-3′ for Snail2, 5′-GCTGCTCCAGCTATGTGTGA-3′ and 5′-AGTGGTGCCAGATCTTTTCCA-3′ for SMA, 5′-TGGACCACCGGCACCCAGAA-3′ and 5′-CGTGGGCAGAGCCACACCTG-3′ for Sox10, 5′-CAGTCTGGGGACAGAGTTCC-3′ and 5′-GACCAGTGGAGCAGACTTGG-3′ for Sox12, 5′-GCAGTCGCTGAACGAGGCGT-3′ and 5′-ACAATGACATCTAGGTCTCCGGCCT-3′ for Twist1, 5′-GTTCGTGTACTGCGGCAAGA-3′ and 5′-ACAGGATTCATGGCCACACC-3′ for Rpl8.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed on HEK293 or Neuro-2a cells after transfection with Sox11 expression plasmid as described previously [23]. Additionally, 16 whole hearts with attached outflow tract region or 16 dissected anterior heart poles from 13.5 dpc-old mouse embryos were used per sample as chromatin source after dissociation with a gentleMACS Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany). After crosslinking proteins to DNA in the presence of 1 % formaldehyde, chromatin was prepared and sheared to an average fragment length of 200–500 bp using a Sonoplus HD2070 homogenizer (Bandelin, Berlin, Germany). Immunoprecipitations were performed overnight at 4 °C using guinea pig antiserum against Sox11 (1:500 dilution) [18] or guinea pig preimmune serum as control, coupled to protein A Sepharose beads. Quantitative PCR was performed on input and precipitated chromatin after crosslink reversal and purification. Primer sequences are available upon request.
Results
SoxC proteins are required for proper development of cardiac neural crest cells
To analyze the role of SoxC proteins in cardiac neural crest cells, we deleted Sox4 or Sox11 using floxed alleles of these genes and a Wnt1::Cre transgene that allows early and efficient gene deletion throughout the cranial, cardiac, and trunk neural crest. The resulting Sox4 fl/fl Wnt1::Cre and Sox11 fl/fl Wnt1::Cre mice are henceforth referred to as Sox4 ΔWnt1 and Sox11 ΔWnt, respectively.
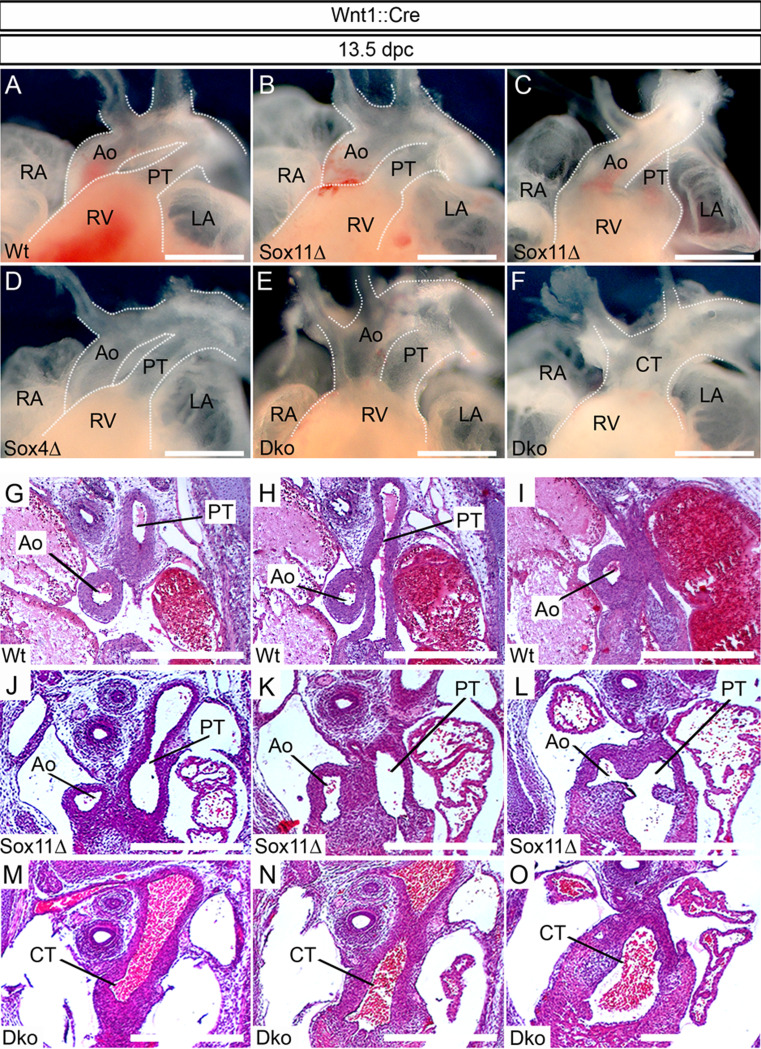
Sox4 ΔWnt1 and Sox11 ΔWnt1 embryos were first analyzed at 13.5 dpc (Fig. 1). All controls (i.e., embryos that were homozygous for the floxed alleles in the absence of Cre recombinase or embryos that carried the Cre transgene in combination with at least one wild-type allele) had a normally developed heart and outflow tract. The same observations were made for all other control genotypes with additional or alternative Cre alleles used during this study. Considering the severe outflow tract defects in Sox4 −/− and Sox11 −/− mice [6, 7] we were surprised to see that most Sox4 ΔWnt1 and Sox11 ΔWnt1 embryos exhibited a macroscopically normal outflow tract region (Figs. 1a, b, d and 3a). Even a more detailed examination of hematoxylin-eosin sections of the anterior heart pole region revealed no major conspicuous aberrations in the majority of mutant embryos in comparison to the wild type (Fig. 1g–i and data not shown). Those defects that were observed in a small minority of embryos all occurred in Sox11 ΔWnt1 and not in Sox4 ΔWnt1 embryos (Fig. 3a). They corresponded to a DORV phenotype (Fig. 1c, j, k, l) and were found in approximately 8 % of Sox11 ΔWnt1 embryos (Fig. 3a). Phenotypic analysis at 17.5 dpc confirmed the results but failed to reveal any additional defects (Fig. 2a–d, g–i). Despite the low percentage of affected Sox11 ΔWnt1 embryos the results pointed to a role of Sox11 in cardiac neural crest cells. They also showed that the outflow tract defects in Sox11 −/− mice cannot be solely attributed to Sox11 functions in neural crest cells.
Fig. 1.
Early developmental defects of the cardiac outflow tract in mice with neural crest-specific SoxC gene ablations. Macroscopic appearance of the anterior heart pole (a–f) and hematoxylin-eosin stainings of corresponding consecutive serial sections (horizontal plane) (g–o) from wild-type embryos (Wt) (a, g, h, i) at 13.5 dpc or age-matched embryos with Wnt1::Cre-mediated, neural crest-specific SoxC gene ablations. Analyzed genotypes included: Sox11 ΔWnt1 (Sox11Δ) (b, c, j, k, l), Sox4 ΔWnt1 (Sox4Δ) (d), and Sox4 ΔWnt1 Sox11 ΔWnt1 (Dko) (e, f, m, n, o). Observed outflow tract abnormalities included DORV in Sox11 ΔWnt1 and Sox4 ΔWnt1 Sox11 ΔWnt1 embryos (c, e, j, k, l) as well as CT in Sox4 ΔWnt1 Sox11 ΔWnt1 embryos (f, m, n, o). Ao aorta, CT common arterial trunk, LA left atrium, RA right atrium, RV right ventricle, PT pulmonary trunk. Scale bars 500 μm
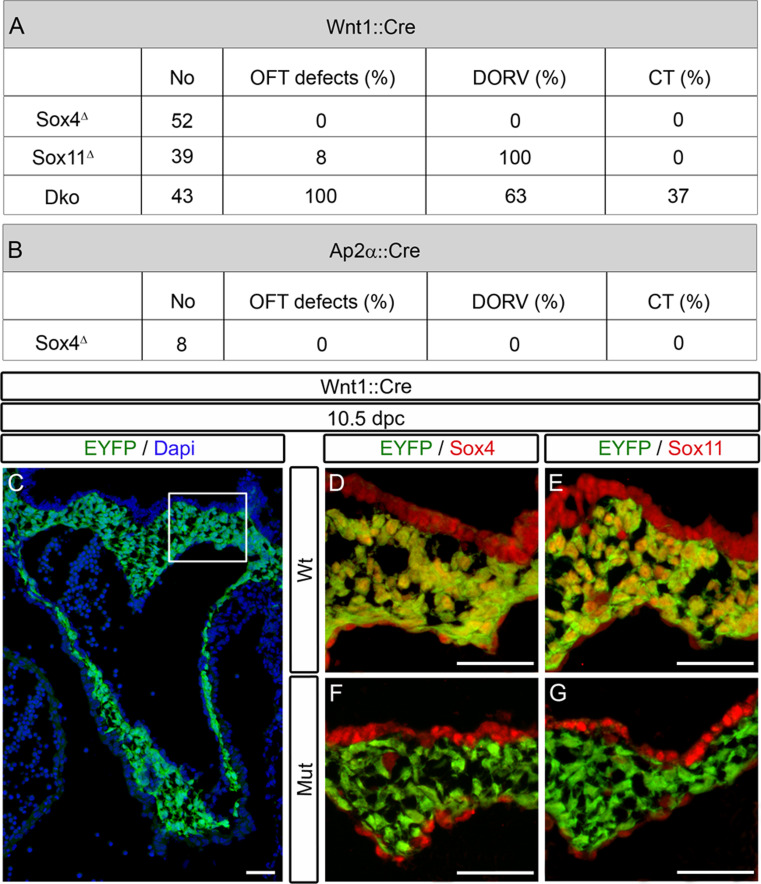
Fig. 3.
Summary of cardiac outflow tract defects and deletion efficiency of SoxC genes in cardiac neural crest. a, b Overview of the number of animals obtained with Wnt1::Cre (a) and AP2α::Cre (b) mediated SoxC deletions (No), and percentage of outflow tract defects (OFT defects) per genotype in total, and subdivided into DORV and CT. c–g Immunohistochemistry was performed at 10.5 dpc with antibodies directed against Sox4 (red in d, f) and Sox11 (red in e, g) on embryos carrying a combination of Rosa26 stopflox-EYFP and Wnt1::Cre alleles (Wt) (c, d, e) or carrying the Rosa26 stopflox-EYFP on a Sox4 ΔWnt1 (f) or Sox11 ΔWnt1 (g) background (Mut). Wnt1::Cre-dependent activation of EYFP (green) from the Rosa26 stopflox-EYFP allele was used to label cells of the neural crest lineage. Nuclei are counterstained with DAPI (blue in c). Pictures in panels d–g were taken from the region boxed in c. Scale bars 50 μm
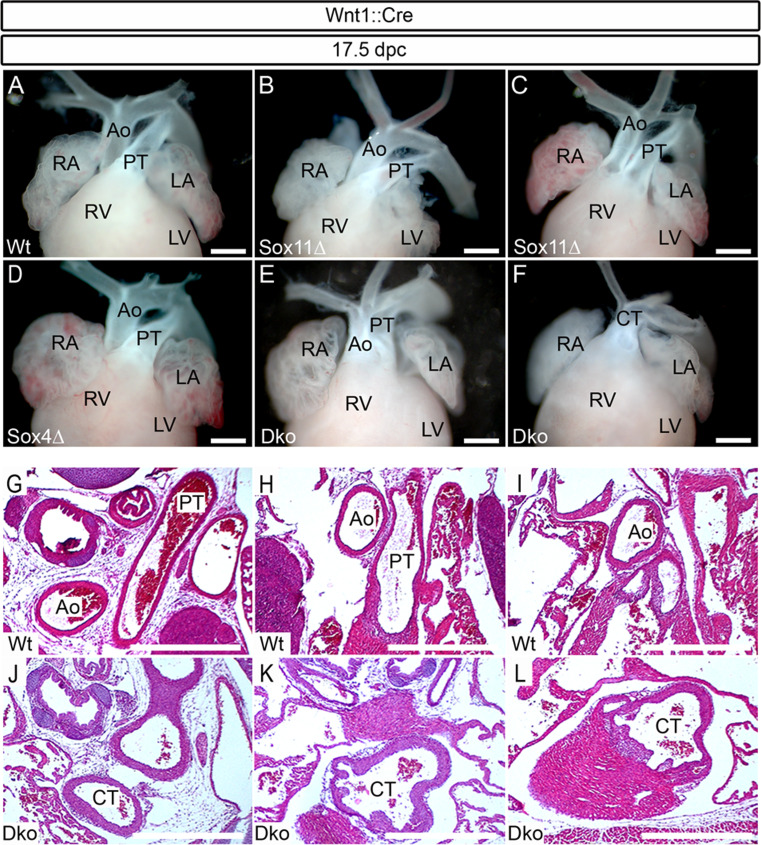
Fig. 2.
Late developmental defects of the cardiac outflow tract in mice with neural crest-specific SoxC gene ablations. Macroscopic appearance of the anterior heart pole (a–f) and hematoxylin-eosin stainings of corresponding consecutive serial sections (horizontal plane) (g–l) from wild-type embryos (Wt) (a, g, h, i) at 17.5 dpc or age-matched embryos with Wnt1::Cre-mediated, neural crest-specific SoxC gene ablations. Analyzed genotypes included: Sox11 ΔWnt1 (Sox11Δ) (b, c), Sox4 ΔWnt1 (Sox4Δ) (d), and Sox4 ΔWnt1 Sox11 ΔWnt1 (Dko) (e, f, j, k, l). Observed outflow tract abnormalities included DORV in Sox11 ΔWnt1 and Sox4 ΔWnt1 Sox11 ΔWnt1 embryos (c, e), whereas CT only occurred in Sox4 ΔWnt1 Sox11 ΔWnt1 embryos (f, j, k, l). Ao aorta, CT common arterial trunk, LA left atrium, RA right atrium, RV right ventricle, PT pulmonary trunk. Scale bars 500 μm
At first sight, the complete absence of outflow tract defects in Sox4 ΔWnt1 embryos argues against a role of Sox4 in the cardiac neural crest. In general, gene deletion by the Wnt1::Cre transgene in cardiac neural crest is very efficient [3]. To confirm the efficiency in case of Sox4 and Sox11 deletion, we genetically labeled cardiac neural crest cells by combining Rosa26 stopflox−EYFP with Wnt1::Cre (Fig. 3c) on an otherwise wild-type background or in mice additionally carrying Sox4 loxP and Sox11 loxP alleles. Whereas EYFP-expressing cardiac neural crest cells were positive for both Sox4 and Sox11 in the heart region of control embryos at 10.5 dpc (Fig. 3d, e), they were almost all negative in the Sox4 ΔWnt1 or Sox11 ΔWnt1 mutant (Fig. 3f, g). Such high Sox4 and Sox11 deletion rates at early times after onset of Cre expression were observed throughout the study (data not shown).
The lack of an outflow tract defect in Sox4 ΔWnt1 mice is thus not attributable to inefficient gene deletion. However, a previous study had shown that gene deletion by the Wnt1::Cre transgene in cardiac neural crest may only be achieved upon, or even after, emigration from the dorsal neural tube [24]. Deletion may then occur too late in Sox4 ΔWnt1 embryos if Sox4 is required in cardiac neural crest cells at the premigratory stage. To address this issue, we combined the Sox4 loxP allele with an AP2α::Cre transgene to generate Sox4 fl/fl AP2α::Cre embryos as this Cre transgene is already active in premigratory neural crest cells [15, 24]. These are referred to as Sox4 ΔAP2α embryos. However, heart and outflow tract region of Sox4 ΔAP2α embryos were phenotypically indistinguishable from those of Sox4 ΔWnt1 embryos and wild-type littermates (Fig. 3b). Corresponding Sox11 ΔAP2α embryos were not generated in this study.
Considering the fact that Sox4 and Sox11 are co-expressed during development in many tissues and cell types [10, 25] and share so many structural properties [19] that functional redundancy has been observed frequently [8, 10, 26, 27], we also analyzed the consequences of joint deletion of both SoxC factors in Sox4 ΔWnt1 Sox11 ΔWnt1 embryos. Intriguingly, all Sox4 ΔWnt1 Sox11 ΔWnt1 embryos that we obtained at 13.5 dpc (Fig. 1e, f, m, n, o) and at 17.5 dpc (Fig. 2e, f, j, k, l) exhibited outflow tract malformations (Fig. 3a). In addition to DORV, which we observed in 63 % of the mutant embryos (Figs. 1e, 2e), we also detected CT as the more severe phenotypic manifestation in 37 % of double-deficient embryos (Figs. 1f, m, n, o and 2f, j, k, l). The fact that additional deletion of Sox4 aggravates the phenotype obtained after deletion of Sox11 provides additional evidence that the floxed Sox4 allele is efficiently deleted by the Wnt1::Cre transgene. Our results show that Sox4 and Sox11 are required for cardiac neural crest development, and that they function in a largely redundant manner. Of the two SoxC proteins, Sox11 appears to be more important for cardiac neural crest development since Sox11 ΔWnt1 embryos but not Sox4 ΔWnt1 embryos show outflow tract malformations.
SoxC protein function is necessary for development of mesodermal cells in the outflow tract region
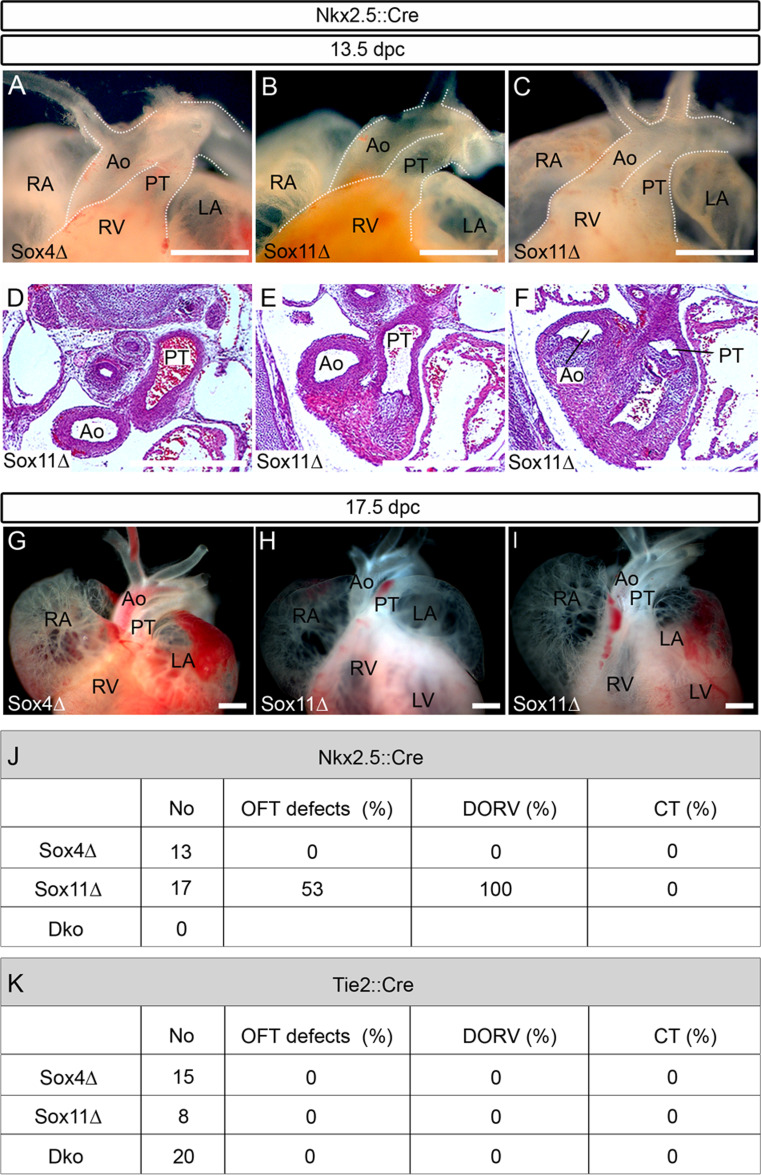
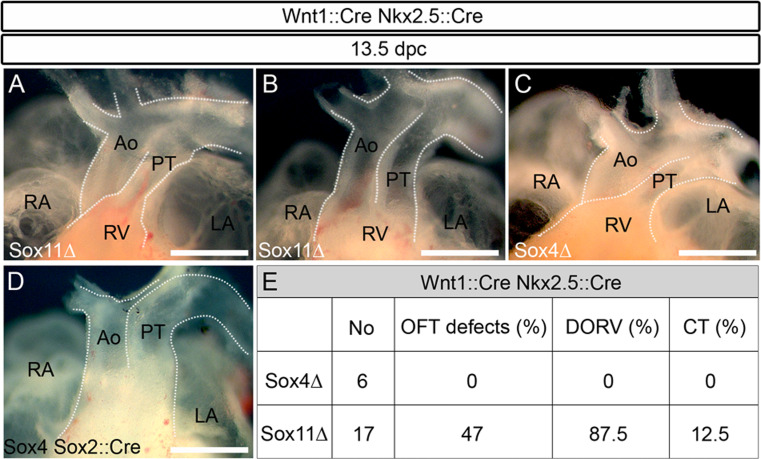
Phenotypic comparison of neural crest-specific and constitutive SoxC mutants argues that the cause of outflow tract malformations in constitutive SoxC mutants cannot be solely attributed to disturbances of neural crest development, and that SoxC proteins are likely active in other contributing cell types as well. To assess the requirement for Sox4 and Sox11 in mesodermal cells of the heart (first and second heart field), we deleted Sox4 or Sox11 using a Nkx2.5::Cre transgene (Fig. 4). The Cre transgene is inserted with preceding IRES into the 3′ UTR of the Nkx2.5 gene so that a bicistronic allele is created that allows simultaneous Nkx2.5 and Cre expression [14]. Consequently, heterozygosity for this allele does not interfere with heart and outflow tract development. Any potential phenotype in Sox4 fl/fl Nkx2.5::Cre and Sox11 fl/fl Nkx2.5::Cre embryos (referred to as Sox4 ΔNkx2.5 and Sox11 ΔNkx2.5 embryos) is thus caused by loss of the Sox protein. At 13.5 and 17.5 dpc, the outflow tracts of Sox4 ΔNkx2.5 embryos were again phenotypically inconspicuous both on a macroscopic and histological level (Fig. 4a, g, j and data not shown). Among Sox11 ΔNkx2.5 embryos, roughly half had a normally appearing outflow tract (Fig. 4b, h, j), whereas the other half exhibited outflow tract defects that all corresponded to DORV (Fig. 4c–f, i, j). We therefore conclude that Sox11 is not only active during outflow tract formation in the cardiac neural crest but also in mesodermal cells. Whether Sox11 is supported in this function by Sox4 could not be analyzed as Sox4 ΔNkx2.5 Sox11 ΔNkx2.5 double-deficient embryos were not obtained because of early developmental defects in multiple other mesodermal lineages [10].
Fig. 4.
Developmental cardiac outflow tract defects in mice with mesodermal or endothelial cell-specific SoxC gene ablations. Macroscopic appearance of the anterior heart pole (a–c, g–i) and hematoxylin-eosin stainings of corresponding consecutive serial sections (horizontal plane) (d–f) from embryos with Nkx2.5::Cre-mediated, mesodermal SoxC gene ablations at 13.5 dpc (a–f) and 17.5 dpc (g–i). Analyzed genotypes included: Sox4 ΔNkx2.5 (Sox4Δ) (a, g) and Sox11 ΔNkx2.5 (Sox11Δ) (b–f, h, i). For age-matched wild-type controls, see Figs. 1 and 2. DORV was observed in a fraction of Sox11 ΔNkx2.5 embryos (c–f, i). Ao aorta, LA left atrium, RA right atrium, RV right ventricle, PT pulmonary trunk. Scale bars 500 μm. j Overview of the number of animals obtained with mesodermal Nkx2.5::Cre-mediated SoxC deletions, and the percentage of outflow tract defects (OFT defects) per genotype in total and subdivided into DORV and CT. k Summary of the number of animals obtained with Tie2::Cre-mediated, endothelial SoxC deletions, and the percentage of outflow tract defects per genotype in total, and subdivided into DORV and CT
Extending our analysis to endothelial cell development we next deleted Sox4 or Sox11 using a Tie2::Cre transgene. In this case, however, neither the resulting Sox4 fl/fl Tie2::Cre nor the Sox11 fl/fl Tie2::Cre embryos (referred to as Sox4 ΔTie2 and Sox11 ΔTie2 embryos) exhibited any outflow tract anomalies at 13.5 and 17.5 dpc (Fig. 4k). Considering that even the double-deficient Sox4 ΔTie2 Sox11 ΔTie2 embryos were phenotypically inconspicuous (Fig. 4k), we conclude that the two SoxC factors have little cell-intrinsic impact on endothelial development.
Mesodermal and neural crest-specific defects contribute to outflow tract malformation in the absence of Sox11
With Sox11 loss in neural crest and mesodermal cells both leading to outflow tract defects, we next asked what would happen if Sox11 was simultaneously deleted in both cell types. For that purpose, Sox11 ΔWnt1ΔNkx2.5 embryos were generated and analyzed. As previously observed for embryos with mesodermal Sox11 loss, approximately half exhibited outflow tract defects (Fig. 5a, b, e). Thus, the frequency was not increased upon additional Sox11 deletion in the neural crest relative to deletion in mesodermal cells alone. However, the severity of the phenotype increased: a small percentage of Sox11 ΔWnt1ΔNkx2.5 embryos exhibited CT, whereas Sox11 ΔNkx2.5 (and Sox11 ΔWnt1) embryos only presented with DORV.
Fig. 5.
Developmental cardiac outflow tract defects in mice with combined mesodermal and neural crest cell-specific SoxC gene ablations. a–d Macroscopic appearance of the anterior heart pole from embryos with combined neural crest and mesoderm-specific (using Wnt1::Cre and Nkx2.5::Cre), or generalized (using Sox2::Cre) SoxC gene ablations at 13.5 dpc. Analyzed genotypes included: Sox11 ΔWnt1ΔNkx2.5 (Sox11Δ) (a, b), Sox4 ΔWnt1ΔNkx2.5 (Sox4Δ) (c), and Sox4 ΔSox2 (d). DORV was observed in Sox11 ΔWnt1ΔNkx2.5 and Sox4 ΔSox2 embryos (b, d). Ao aorta, LA left atrium, RA right atrium, RV right ventricle, PT pulmonary trunk. Scale bars 500 μm. e Summary of the number of animals obtained with combined Wnt1::Cre and Nkx2.5::Cre-mediated SoxC deletions, and the percentage of outflow tract defects (OFT defects) per genotype in total, and subdivided into DORV and CT
To our surprise, the corresponding Sox4 ΔWnt1ΔNkx2.5 embryos still did not show obvious outflow tract malformations (Fig. 5c, e), thus raising the concern that the deleted Sox4 loxP allele may behave differently than the constitutive null allele. However, Sox2::Cre-dependent deletion of the Sox4 loxP allele throughout the epiblast fully recapitulated the outflow tract defect described for Sox4 −/− embryos (Fig. 5d). We thus conclude that the Sox4 requirement for outflow tract formation cannot even be explained by its combined action in neural crest and mesodermal cells.
Generation, migration, proliferation, and apoptosis of cardiac neural crest cells are not affected by the absence of SoxC proteins
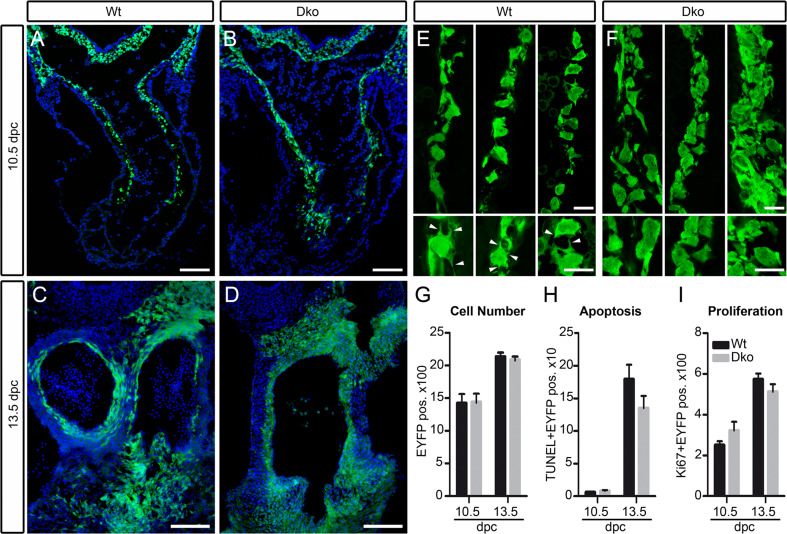
To analyze the role of SoxC proteins in cardiac neural crest cells, we labeled these cells by Wnt1::Cre-dependent activation of EYFP expression from a Rosa26 stopflox-EYFP allele on an otherwise wild-type background or in mice carrying additionally Sox4 loxP and Sox11 loxP alleles. When the cardiac neural crest was followed via its EYFP expression and compared between both genotypes at 10.5 and 13.5 dpc, we failed to detect dramatic differences. Migration of cardiac neural crest through the pharyngeal arches occurred at the right time and in normal numbers (Fig. 6a, b, g and data not shown). Cardiac neural crest cells furthermore reached the anterior heart pole during the same time window and in similar numbers in the Sox4, Sox11 double-deficient mutants as compared to the wild type (Fig. 6c, d, g and data not shown). Rates of proliferation and apoptosis were indistinguishable between the two genotypes both when analyzed on the migratory route or in the target tissue (Fig. 6h, i). We therefore conclude that SoxC proteins do not influence any of these parameters.
Fig. 6.
Analysis of general properties and morphology of cardiac neural crest in mice with neural crest-specific SoxC gene ablations. a–d Immunohistochemistry for EYFP (green) was performed at 10.5 dpc (a, b) and 13.5 dpc (c, d) on embryos carrying a combination of Rosa26 stopflox-EYFP and Wnt1::Cre alleles (Wt) (a, c) or carrying the Rosa26 stopflox-EYFP on a Sox4 ΔWnt1 Sox11 ΔWnt1 background (Dko) (b, d). Nuclei are counterstained with DAPI (blue). e, f High-magnification confocal images of cardiac neural crest cells (green) at 10.5 dpc taken from the wild-type (e) or age-matched Sox4 ΔWnt1 Sox11 ΔWnt1 (f) embryos. Wnt1::Cre-dependent activation of EYFP from the Rosa26 stopflox-EYFP allele was used to label cells of the neural crest lineage. Lower panels show even higher magnification of single cells shown in e, f. Arrowheads mark filopodia between neural crest cells. Scale bars 100 μm (a–d), 15 μm (e, f). g–i Quantification of the total number of cardiac neural crest cells per section (g), the TUNEL-positive apoptotic fraction (h), and the Ki67-positive proliferating fraction (i) at 10.5 and 13.5 dpc in outflow tract regions of embryos carrying a combination of Rosa26 stopflox-EYFP and Wnt1::Cre alleles (Wt, black bars) or carrying the Rosa26 stopflox-EYFP on a Sox4 ΔWnt1 Sox11 ΔWnt1 background (Dko, grey bars). At least four separate sections from three independent embryos were counted for each genotype. Data are presented as mean ± SEM. No statistically significant differences were observed between the two genotypes
Loss of SoxC proteins alters marker gene expression and properties of cardiac neural crest cells
As previously reported [28, 29], neural crest cells often migrate in chain-like formations, with cells contacting each other through filopodia-like processes. Such chains of neural crest cells were regularly observed in the wild-type anterior heart pole at 10.5 dpc, and multiple filopodia-like contacts were usually observed between the cells (Fig. 6e). In age-matched double-deficient Sox4 ΔWnt1 Sox11 ΔWnt1 embryos, cardiac neural crest cells exhibited striking morphological alterations. The cells were no longer arranged in single cell chains. As a consequence, their orientation relative to each other appeared more random. Cells also exhibited an altered and often less compact morphology, and filopodial contacts were much less frequently observed (Fig. 6f).
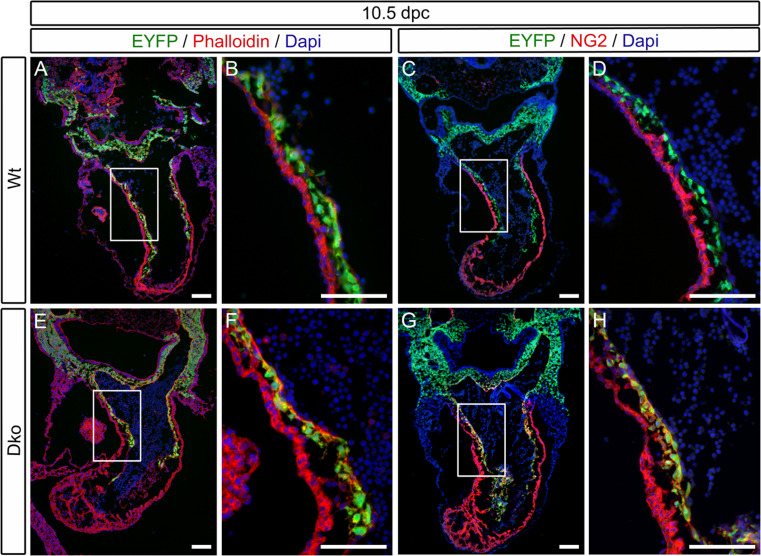
In line with this altered morphology, cardiac neural crest cells in the double mutant also exhibited abnormalities in their cytoskeletal microfilament network as visualized by phalloidin staining. In wild-type cardiac neural crest cells, overall phalloidin staining was relatively low at 10.5 dpc, whereas it was prominent in Sox4 ΔWnt1 Sox11 ΔWnt1 cardiac neural crest cells (compare Fig. 7a, b to e, f). Another abnormality concerned expression of the extracellular matrix glycoprotein component NG2, which is turned on by a fraction of neural crest-derived cells in the outflow tract region as they develop into pericytes. This normally occurs well after the onset of outflow tract remodeling so that NG2 is not yet expressed in wild-type cardiac neural crest cells at 10.5 dpc as evident from the fact that Wnt1::Cre-induced EYFP expression does not co-localize with NG2 expression in the wild-type outflow tract at this time (Fig. 7c, d). In contrast, NG2 is detected in substantial amounts in EYFP-labeled cardiac neural crest cells of age-matched Sox4 ΔWnt1 Sox11 ΔWnt1 embryos at 10.5 dpc (Fig. 7g, h). By 13.5 dpc, phalloidin and NG2 staining in wild-type cardiac neural crest had caught up with mutant ones so that differences were no longer visible (data not shown; see also results from RT-PCR in Fig. 8a). We conclude from these data that cardiac neural crest cells prematurely lose characteristics of migratory cells in the absence of Sox4 and Sox11 and turn on expression of pericyte markers in an untimely manner. It is conceivable that such misregulation during a critical time window may then prevent cardiac neural crest cells from functioning properly in outflow tract remodeling and formation of the aortopulmonary septum.
Fig. 7.
Analysis of marker gene expression of cardiac neural crest in mice with neural crest-specific SoxC gene ablations. Co-immunohistochemistry was performed at 10.5 dpc on embryos carrying a combination of Rosa26 stopflox-EYFP and Wnt1::Cre alleles (Wt) (a–d) or carrying the Rosa26 stopflox-EYFP on a Sox4 ΔWnt1 Sox11 ΔWnt1 background (Dko) (e–h). Antibodies directed against EYFP were used to detect cells of neural crest origin (green). Actin microfilaments were detected by fluorescently labeled phalloidin (a, b, e, f) and NG2 expression by specific antibodies (c, d, g, h) (both in red). Nuclei were counterstained with DAPI (blue). The area shown at higher magnification in b, d, f, h is boxed in a, c, e, g. Scale bars 100 μm
Fig. 8.
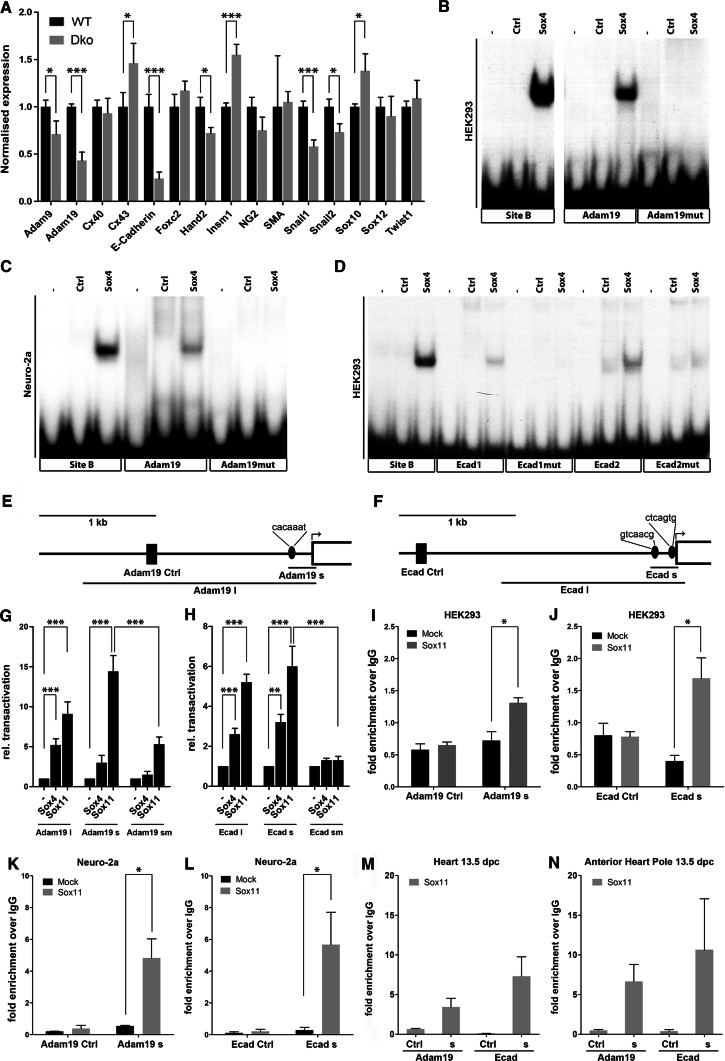
Analysis of SoxC target genes in neural crest cells. a Expression levels were determined by quantitative RT-PCR on RNA prepared from the anterior heart pole of wild-type (Wt, black bars) and Sox4 ΔWnt1 Sox11 ΔWnt1 (Dko, grey bars) embryos at 13.5 dpc for several genes as indicated below the bars. Transcript levels were normalized to Rpl8 levels in the respective samples. For each RNA source, RT-PCRs were performed on at least three independent samples. b–d Electrophoretic mobility shift assays with oligonucleotides encompassing potential Sox binding sites from the Adam19 (b, c) and E-cadherin (d) promoters and the known Sox binding site B from the Mpz promoter. For each confirmed binding site, a wild-type (Adam19, Ecad1, Ecad2) and a mutant (Adam19mut, Ecad1mut, Ecad2mut) version were used. Oligonucleotides were incubated in the absence (−) or presence of protein extracts from mock-transfected cells (Ctrl) or cells expressing a carboxyterminally truncated Sox4 protein (Sox4) as indicated above the lanes. Both HEK293 (b, d) and Neuro-2A cells (c) were used. e, f Schematic representation of Adam19 and E-cadherin upstream regions. Location of the short (Adam19 s, Ecad s) and long (Adam19 l, Ecad l) promoter regions used for luciferase assays as well as position of Sox binding sites (ellipses), transcriptional start sites (arrows) and control regions (Adam19 Ctrl, Ecad Ctrl, black boxes) for ChIP are indicated. The exact binding site sequences are listed above the ellipses. Scale bar, 1 kb. g, h Transient transfections were performed in Neuro-2a cells with luciferase reporters under control of the Adam19 (g) or E-cadherin (h) promoters either in a short version encompassing the proximal promoter (274 bp for Adam19 and 238 bp for E-cadherin) or a long version including additional upstream sequences (2,040 bp for Adam19 and 1,600 bp for E-cadherin). Both proximal promoters were furthermore employed in the wild-type version and as a variant with inactivated Sox binding sites (Adam19 sm, Ecad sm). Empty pCMV5 expression plasmids (−) or expression plasmids for Sox4 and Sox11 were co-transfected as indicated below the bars. Luciferase activities were determined in ten experiments, each performed in triplicates. The luciferase activity obtained for a promoter in the absence of ectopic transcription factor was arbitrarily set to 1. Fold inductions in the presence of transcription factors are presented as mean ± SEM. i–n ChIP was performed on chromatin from HEK293 cells (i, j), Neuro-2a cells (k, l), whole hearts (m), and anterior heart pole tissue (n) from mouse embryos at 13.5 dpc. In case of HEK293 and Neuro-2a cells, chromatin was prepared from both untransfected (mock, black bars) and Sox11-transfected (Sox11, grey bars) cells. Precipitation was with antibodies directed against Sox11 and control IgGs. Quantitative PCRs were carried out on immunoprecipitated chromatin to determine the relative enrichment of proximal promoters (Adam19 s, Ecad s) and upstream control regions (Adam Ctrl, Ecad Ctrl) from Adam19 (i, k, m, n) and E-cadherin (j, l, m, n) gene loci in the Sox11 precipitate over control. Experiments were performed for five biological replicates in i–l, for three in m, and for two in n with each PCR in triplicate. Fold enrichment of PCR products from Sox11-precipitated chromatin over IgG-precipitated chromatin is presented as mean ± SEM. Differences between samples in a, g, h, i, j, k, l were statistically significant as indicated according to Student’s t test (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). No statistical analysis was performed for m and n because of the low sample number
Further evidence for such a model came from the observation that several markers that are typically expressed in cardiac neural crest cells at this stage and are known to be relevant for their development exhibited strikingly altered expression levels (Fig. 8a). This was most conspicuously the case for E-cadherin and Adam19. Cadherins are not only involved in epithelial-to-mesenchymal transition at the premigratory stage and in interactions between migratory cells, they also influence the mesenchymal-to-epithelial transition post-migration [30, 31]. Expression of the metalloprotease Adam19 in cardiac neural crest cells is essential for proper outflow tract remodeling [32]. This altered expression of genes whose products influence extracellular matrix, cytoskeletal organization, or mesenchymal-to-epithelial transition, has the potential to interfere with proper outflow tract remodeling.
Interestingly, one of the genes with unaltered expression after combined deletion of Sox4 and Sox11 in cardiac neural crest cells was Sox12, the third member of the SoxC subgroup (Fig. 8a). It thus seems that there is no substantial upregulation of the remaining SoxC member in an attempt to compensate the loss of Sox4 and Sox11.
SoxC proteins directly influence expression of E-cadherin and Adam19
To analyze whether E-cadherin and Adam19 were direct target genes of SoxC proteins, we searched for the presence of potential Sox binding sites in the respective promoter regions and detected one site in the proximal Adam19 promoter, whereas the proximal E-cadherin promoter contained two such sites (Fig. 8e, f). Electrophoretic mobility shift analysis revealed that Sox4 as well as Sox11 recognized these sites (Fig. 8b–d and data not shown). Considering that all oligonucleotide probes were labeled with similar efficiencies and that similar results were obtained with extracts from transfected HEK293 and Neuro-2a neuroblastoma cells as protein source, binding of both proteins to the Adam19 site appeared stronger than to the two E-cadherin sites (compare Fig. 8b, c to d). HEK293 cells were chosen because of their widespread use for protein production, while Neuro-2a cells were chosen because of their ontogenetic relationship and similarity to neural crest cells. Sox4 and Sox11 also activated both promoters in luciferase assays upon co-transfection in Neuro-2a cells (Fig. 8g, h). For both genes, the proximal promoter containing the binding sites was at least as efficiently activated as a longer promoter fragment that additionally included more distal sequences indicating that the SoxC effect is indeed mediated by the proximal promoter region. In line with observations on other target gene promoters [25, 33], Sox11 activated luciferase expression more efficiently than Sox4. Whether this higher activity in vitro is causally related to the higher impact of Sox11 on cardiac neural crest cell development remains to be studied in future experiments.
To further prove that promoter activation is a direct consequence of binding to these regions, we first introduced mutations in both Adam19 promoter and E-cadherin promoter that prevented SoxC proteins from binding to the identified sites (Fig. 8b, c, d). As a consequence, activation of both promoters by either Sox4 or Sox11 was strongly diminished (Fig. 8g, h). Additionally, we studied by ChIP whether transfected Sox11 would associate with the endogenous E-cadherin and Adam19 promoters in HEK293 and Neuro-2a cells. Both promoters were significantly enriched in chromatin precipitated with antibodies directed against Sox11 as compared to precipitates with control immunoglobulins in HEK293 cells (Fig. 8i, j) as well as in Neuro-2a cells (Fig. 8k, l). Enrichment was furthermore only observed for the promoter regions when cells were transfected with Sox11 before ChIP, and could not be obtained for other more distally located regions from the respective gene loci (Fig. 8i–l). Intriguingly, both promoters were also specifically precipitated from chromatin that was prepared from whole hearts or anterior heart poles of 13.5-dpc-old embryos as tissues that included the outflow tract region (Fig. 8m, n). Thus it appears very likely that the influence of SoxC proteins on both E-cadherin and Adam19 is direct and occurs in cardiac neural crest cells.
Discussion
Congenital heart defects represent the most common type of birth defect in humans and occur with a frequency of 10 in 1,000 with three of them requiring early postnatal intervention [1]. Many of these defects represent or include malformations of the outflow tract. This region is formed by mesodermal cells from the second heart field, cardiac neural crest cells, and endothelial cells that cooperate and reciprocally influence each others’ development [4].
Over the years, a number of transcription factors have been identified that are required for outflow tract formation [5]. How these factors work has only been addressed for some, and turns out to be complicated, as transcription factors may be active in any of the cell types that participate in outflow tract formation and may affect any of the numerous processes that these cells have to undergo before or while becoming part of the outflow tract [34].
Since constitutive deletion of Sox4 or Sox11 leads to outflow tract defects [6, 7], our first aim was the identification of the affected cell type. Our analysis clearly shows that Sox11 is required in both cardiac neural crest and mesodermal cells. The observed phenotype corresponded in all cases to DORV. Interestingly, simultaneous deletion of Sox11 in neural crest as well as mesodermal cells led to increased phenotypic severity as a substantial number of embryos now manifested with CT instead of DORV. These findings raise the intriguing possibility that Sox11 may be involved in the reciprocal signaling that occurs between these cell types during outflow tract formation.
At least in the neural crest, Sox11 is furthermore supported in its function by the related Sox4 as only the combined deletion of both SoxC factors led to a fully penetrant outflow tract defect. Whereas neural crest-specific deletion of Sox11 led to an incidence of outflow tract defects of 8 %, the additional deletion of Sox4 increased rates to 100 %. Both factors thus function redundantly in the cardiac neural crest as previously seen in central nervous system and other organs throughout the developing embryo [8, 10]. From our results, we furthermore conclude that the impact of Sox11 on cardiac neural crest and mesodermal development is stronger than the one of Sox4.
Considering that the outflow tract defect in constitutive Sox4-deficient mice is more severe than the one in constitutive Sox11-deficient mice [6, 7], we were surprised that even the combined deletion of Sox4 in cardiac neural crest and mesoderm remained phenotypically inapparent. We do not think that this is due to trivial reasons, as Cre-dependent deletion of the floxed Sox4 allele was rapid and near complete in targeted cell types and as combination of the same allele with a pan-epiblast Cre deleter yielded the same phenotype previously observed in constitutive Sox4 null mutants. However, it is conceivable that the Sox4 requirement for outflow tract formation is based on its action in a cell type that was not hit by our deletion strategy, or at a time at which the applied Cre recombinases were not yet active. However, considering that Wnt1::Cre is active already very early throughout the neural crest [13], and that we obtained the same phenotype when the earlier deleting AP2α::Cre [24] was applied, it appears unlikely that Sox4 deletion in the neural crest occurred too late. The same can be argued for the heart mesoderm, as Nkx2.5::Cre expression sets in very early and efficiently [14]. One option therefore is that the outflow tract defect observed in constitutive Sox4-deficient mice results from effects on neuroectodermal or mesodermal development that precede neural crest or heart field formation. Alternatively, Sox4 effects on neural crest or second heart field mesoderm may be indirect and not cell-autonomous. It could for instance be envisaged that Sox4 acts in cells that provide essential cues for the second heart field or the cardiac neural crest. Pharyngeal endoderm cells are an interesting candidate as they have been shown in the past to influence outflow tract development and septation through multiple mechanisms [35, 36]. Future experiments will have to clarify this issue.
The second aim of our study was to understand how SoxC transcription factors act molecularly in the cardiac neural crest. Our studies revealed that cardiac neural crest cells deficient for both Sox4 and Sox11 emigrated normally from the neural tube. We furthermore failed to detect any gross abnormalities during the migratory phase. Rates of proliferation and apoptosis were comparable to the wild type so that normal numbers of cardiac neural crest cells reached the outflow tract region at appropriate times. We therefore conclude that SoxC factors are not required for the generation, multiplication, maintenance, or migration of the cardiac neural crest.
The changes we observed occurred after the cardiac neural crest cells had entered the outflow tract region. SoxC proteins thus affect later stages of cardiac neural crest development. Alterations included different morphology and arrangement of cardiac neural crest cells relative to each other. Less filopodial contacts were observed between cells and their chain-like arrangement was lost so that they no longer appeared as a single cell layer along the outflow tract. This went along with alterations in cytoskeletal organization as evident from phalloidin stainings. Similar alterations in packing and morphologic appearance of cardiac neural crest cells with associated cytoskeletal alterations have been observed in neural crest-specific Disheveled mutants arguing that SoxC function in the cardiac neural crest may be related to non-canonical Wnt signaling and the planar cell polarity pathway [37]. Further abnormalities included a precocious expression of the pericyte marker and proteoglycan NG2 in these cells as well as reduced E-cadherin and Adam19 expression. For E-cadherin and Adam19, the SoxC effect appeared to be direct. A previous study had identified Cx43 as another direct target for SoxC proteins in the heart [38]. Although we were unable to detect a reduction of Cx43 expression in mice with neural crest-specific SoxC deletion (likely because Cx43 is expressed in mesodermal cells as well), it supports the notion that SoxC proteins influence a number of genes that are upregulated during differentiation and determine the interaction of cardiac neural crest cells among each other and with their environment. In the absence of SoxC proteins the differentiation program is not properly implemented and interactions are dramatically altered.
During neuronal differentiation, SoxC proteins are required for the timely establishment of neuronal properties and maturation [26, 39]. During this process, SoxC proteins affect neuronal morphology [40, 41] and influence expression of several cytoskeletal proteins including β3-Tubulin and Doublecortin [26, 27, 42]. In analogy, SoxC proteins may coordinate the timely differentiation of cardiac neural crest cells in the outflow tract region. The role of Sox4 and Sox11 in the cardiac neural crest appears to occur later than the action of most other transcription factors that have been identified so far. These are either already active at the premigratory stage and required for neural crest cell emigration or expansion of the precursor cell pool such as SIP1 and Pax3 [43, 44], or they support survival and proper interaction with surrounding tissues during the migratory phase such as Foxc1, Foxc2, and AP2α [45, 46]. They may also direct early specification decisions in the postmigratory phase such as Twist1 [47]. While this argues that SoxC proteins may fulfill the function of downstream effectors, future experiments will have to define the exact ways in which Sox4 and Sox11 are activated by and interact with other transcription factors known to be relevant for outflow tract formation.
Acknowledgments
Jürgen Behrens and Marina Kreutz are acknowledged for the gift of Ecadherin and Adam19 plasmids, respectively. This work was supported by a grant from the DFG to E.S. (So251/3-1) and from a National Health and Medical Research Council Australia Fellowship to RPH (573705).
References
- 1.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/S0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 2.Webb S, Qayyum SR, Anderson RH, Lamers WH, Richardson MK. Septation and separation within the outflow tract of the developing heart. J Anat. 2003;202:327–342. doi: 10.1046/j.1469-7580.2003.00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snider P, Olaopa M, Firulli AB, Conway SJ. Cardiovascular development and the colonizing cardiac neural crest lineage. Sci World J. 2007;7:1090–1113. doi: 10.1100/tsw.2007.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Kodo K, Yamagishi H. A decade of advances in the molecular embryology and genetics underlying congenital heart defects. Circ J. 2011;75:2296–2304. doi: 10.1253/circj.CJ-11-0636. [DOI] [PubMed] [Google Scholar]
- 6.Schilham MW, Oosterwegel MA, Moerer P, Ya J, Deboer PAJ, Vandewetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 7.Sock E, Rettig SD, Enderich J, Bösl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thein DC, Thalhammer JM, Hartwig AC, Crenshaw EB, 3rd, Lefebvre V, Wegner M, Sock E. The closely related transcription factors Sox4 and Sox11 function as survival factors during spinal cord development. J Neurochem. 2010;115:131–141. doi: 10.1111/j.1471-4159.2010.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattaram P, Penzo-Méndez A, Sock E, Colmenares C, Kaneko KJ, DePamphilis ML, Wegner M, Lefebvre V. Organogenesis relies on Sox4, Sox11, and Sox12 for survival of neural and mesenchymal progenitor cells. Nat Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penzo-Mendez A, Dy P, Pallavi B, Lefebvre V. Generation of mice harboring a Sox4 conditional null allele. Genesis. 2007;45:776–780. doi: 10.1002/dvg.20358. [DOI] [PubMed] [Google Scholar]
- 12.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 14.Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nk2–5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 15.Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Gene Expr Patterns. 2002;2:93–97. doi: 10.1016/S0925-4773(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 18.Hoser M, Baader SL, Bosl MR, Ihmer A, Wegner M, Sock E. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J Neurosci. 2007;27:5495–5505. doi: 10.1523/JNEUROSCI.1384-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and Sox proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 20.Ehrnsperger A, Rehli M, Thu-Hang P, Kreutz M. Epigenetic regulation of the dendritic cell-marker gene ADAM19. Biochem Biophys Res Commun. 2005;332:456–464. doi: 10.1016/j.bbrc.2005.04.149. [DOI] [PubMed] [Google Scholar]
- 21.Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero expression is regulated by the glial transcription factor Sox10. Mol Cell Biol. 2000;20:3198–3209. doi: 10.1128/MCB.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellerer S, Schreiner S, Stolt CC, Bösl MR, Wegner M. Replacement of the Sox10 transcription factor by Sox8 reveals incomplete functional equivalence. Development. 2006;133:2875–2886. doi: 10.1242/dev.02477. [DOI] [PubMed] [Google Scholar]
- 23.Schlierf B, Werner T, Glaser G, Wegner M. Expression of Connexin47 in oligodendrocytes is regulated by the Sox10 transcription factor. J Mol Biol. 2006;361:11–21. doi: 10.1016/j.jmb.2006.05.072. [DOI] [PubMed] [Google Scholar]
- 24.Olaopa M, Zhou HM, Snider P, Wang J, Schwartz RJ, Moon AM, Conway SJ. Pax3 is essential for normal cardiac neural crest morphogenesis but is not required during migration nor outflow tract septation. Dev Biol. 2011;356:308–322. doi: 10.1016/j.ydbio.2011.05.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoser M, Potzner MR, Koch JMC, Bösl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals non-reciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28:4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu L, Berti L, Masserdotti G, Covic M, Michaelidis TM, Doberauer K, Merz K, Rehfeld F, Haslinger A, Wegner M, Sock E, Lefebvre V, Couillard-Despres S, Aigner L, Berninger B, Lie DC. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J Neurosci. 2012;32:3067–3080. doi: 10.1523/JNEUROSCI.4679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teddy JM, Kulesa PM. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development. 2004;131:6141–6151. doi: 10.1242/dev.01534. [DOI] [PubMed] [Google Scholar]
- 29.Young HM, Bergner AJ, Anderson RB, Enomoto H, Milbrandt J, Newgreen DF, Whitington PM. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev Biol. 2004;270:455–473. doi: 10.1016/j.ydbio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Koyanagi M, Urbich C, Chavakis E, Hoffmann J, Rupp S, Badorff C, Zeiher AM, Starzinski-Powitz A, Haendeler J, Dimmeler S. Differentiation of circulating endothelial progenitor cells to a cardiomyogenic phenotype depends on E-cadherin. FEBS Lett. 2005;579:6060–6066. doi: 10.1016/j.febslet.2005.09.071. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, High FA, Epstein JA, Radice GL. N-cadherin is required for neural crest remodeling of the cardiac outflow tract. Dev Biol. 2006;299:517–528. doi: 10.1016/j.ydbio.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komatsu K, Wakatsuki S, Yamada S, Yamamura K, Miyazaki J, Sehara-Fujisawa A. Meltrin beta expressed in cardiac neural crest cells is required for ventricular septum formation of the heart. Dev Biol. 2007;303:82–92. doi: 10.1016/j.ydbio.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 33.Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104:933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- 35.Goddeeris MM, Schwartz R, Klingensmith J, Meyers EN. Independent requirements for Hedgehog signaling by both the anterior heart field and neural crest cells for outflow tract development. Development. 2007;134:1593–1604. doi: 10.1242/dev.02824. [DOI] [PubMed] [Google Scholar]
- 36.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha T, Wang B, Evans S, Wynshaw-Boris A, Wang J. Disheveled mediated planar cell polarity signaling is required in the second heart field lineage for outflow tract morphogenesis. Dev Biol. 2012;370:135–144. doi: 10.1016/j.ydbio.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boogerd CJ, Wong LY, van den Boogaard M, Bakker ML, Tessadori F, Bakkers J, t Hoen PA, Moorman AF, Christoffels VM, Barnett P. Sox4 mediates Tbx3 transcriptional regulation of the gap junction protein Cx43. Cell Mol Life Sci. 2011;68:3949–3961. doi: 10.1007/s00018-011-0693-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usui A, Mochizuki Y, Iida A, Miyauchi E, Satoh S, Sock E, Nakauchi H, Aburatani H, Murakami A, Wegner M, Watanabe S. The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development. 2013;140:740–750. doi: 10.1242/dev.090274. [DOI] [PubMed] [Google Scholar]
- 40.Jankowski MP, Cornuet PK, McIlwrath S, Koerber HR, Albers KM. SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience. 2006;143:501–514. doi: 10.1016/j.neuroscience.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin L, Lee VM, Wang Y, Lin JS, Sock E, Wegner M, Lei L. Sox11 regulates survival and axonal growth of embryonic sensory neurons. Dev Dyn. 2011;240:52–64. doi: 10.1002/dvdy.22489. [DOI] [PubMed] [Google Scholar]
- 42.Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conway SJ, Bundy J, Chen J, Dickman E, Rogers R, Will BM. Decreased neural crest stem cell expansion is responsible for the conotruncal heart defects within the splotch (Sp(2H))/Pax3 mouse mutant. Cardiovasc Res. 2000;47:314–328. doi: 10.1016/S0008-6363(00)00098-5. [DOI] [PubMed] [Google Scholar]
- 44.Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am J Hum Genet. 2003;72:465–470. doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brewer S, Jiang X, Donaldson S, Williams T, Sucov HM. Requirement for AP-2α in cardiac outflow tract morphogenesis. Mech Dev. 2002;110:139–149. doi: 10.1016/S0925-4773(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 46.Seo S, Kume T. Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Dev Biol. 2006;296:421–436. doi: 10.1016/j.ydbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Vincentz JW, Firulli BA, Lin A, Spicer DB, Howard MJ, Firulli AB. Twist1 controls a cell-specification switch governing cell fate decisions within the cardiac neural crest. PLoS Genet. 2013;9:e1003405. doi: 10.1371/journal.pgen.1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]