Abstract
Tubulin cofactors (TBCs) participate in the folding, dimerization, and dissociation pathways of the tubulin dimer. Among them, TBCB and TBCE are two CAP-Gly domain-containing proteins that together efficiently interact with and dissociate the tubulin dimer. In the study reported here we showed that TBCB localizes at spindle and midzone microtubules during mitosis. Furthermore, the motif DEI/M-COO− present in TBCB, which is similar to the EEY/F-COO− element characteristic of EB proteins, CLIP-170, and α-tubulin, is required for TBCE–TBCB heterodimer formation and thus for tubulin dimer dissociation. This motif is responsible for TBCB autoinhibition, and our analysis suggests that TBCB is a monomer in solution. Mutants of TBCB lacking this motif are derepressed and induce microtubule depolymerization through an interaction with EB1 associated with microtubule tips. TBCB is also able to bind to the chaperonin complex CCT containing α-tubulin, suggesting that it could escort tubulin to facilitate its folding and dimerization, recycling or degradation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1114-2) contains supplementary material, which is available to authorized users.
Keywords: CCT, Microtubule dynamics, +TIPs, Tubulin folding cofactors
Introduction
The cytoskeleton of eukaryotic cells is required for many essential cell processes such as motility, intracellular trafficking, chromosome segregation, and cytokinesis [1]. Microtubules are complex polar polymers of the cytoskeleton that assemble from αβ-tubulin heterodimers. The heterodimers polymerize, forming protofilaments that associate laterally, forming the wall of a hollow cylinder, the microtubule [2, 3]. Therefore, within the microtubule lattice, each single α-tubulin or β-tubulin subunit interacts with four other neighboring tubulin subunits. Each α-tubulin subunit interacts with its β-tubulin partner inside of the heterodimer, with a second β-tubulin subunit from the preceding heterodimer in the protofilament, and laterally with two α-subunits from the two side protofilaments. Thus, the assembly of a microtubule, while preventing unwanted interactions, is a highly complex task that must be properly controlled to avoid critical errors.
Tubulin folding cofactors (TBCs) are a set of different proteins discovered a decade ago in the so-called “postchaperonin” tubulin folding pathway. TBCs are responsible for the achievement of the quaternary conformation of the αβ-heterodimer after tubulin monomers have reached their tertiary structure [4, 5]. More recent studies have shown that in vivo, these proteins are implicated in microtubule dynamics through their ability to dissociate the tubulin heterodimer, and probably by controlling tubulin monomer quality and exchange (shuffling mechanism) [6–8].
TBCB and TBCE are two well-conserved α-tubulin interacting proteins that collaborate in the regulation of microtubule dynamics [6–9]. Both cofactors participate in the α-tubulin folding pathway and are required for cell survival [5, 10], playing important roles in vivo as revealed by the plethora of human disorders in which they are implicated. TBCE mutations cause a syndrome called hypoparathyroidism-retardation-dysmorphism, also known as the Sanjad-Sakati syndrome [11] in humans, and a progressive motor neuropathy in the mouse [12]. TBCB, on the other hand, has been implicated in human cancer [13], neurodevelopmental malformations [14], schizophrenia [15] and neurodegenerative processes [16].
TBCB shares with TBCE two similar domains, a CAP-Gly and a UBL domain, but the cytoskeleton-associated protein glycine-rich (CAP-Gly) domains are localized at the C-terminal position in TBCB and at the N-terminal position in TBCE. While TBCB it is not able to interact with or dissociate the tubulin heterodimer by itself, TBCE is, per se, effective in promoting this dissociation. Nonetheless, TBCE interacts with TBCB, originating the TBCE–TBCB complex, which displays a more efficient stoichiometric tubulin dissociation activity than TBCE alone. Upon dissociation, TBCB, TBCE, and α-tubulin form a stable ternary complex. The disassembly of this ternary complex results in either TBCB and α-tubulin and free TBCE, or TBCE and α-tubulin and free TBCB. Free β-tubulin subunits might be recyclable in the presence of TBCA or TBCD [9].
The function of the CAP-Gly domains of both cofactors is still unknown. This domain is a protein-interaction module that typically plays a role controlling microtubule end dynamics in end-binding proteins (EBs), which can track along microtubule ends [17–19]. In addition to EBs, an increasing number of proteins that control microtubule organization and dynamics, known as microtubule plus-end-tracking proteins (+TIPs), have been identified. These proteins connect to the microtubule plus ends through an interaction with members of the EB family [17–19], the only known protein family that can track microtubule ends autonomously. Recently, a long list of +TIP candidates has been provided by Yu et al. [20], but neither TBCB nor TBCE was included.
In this work, we used a multidisciplinary approach to study the molecular mechanism of TBCB regulation of microtubule dynamics. For this purpose, we cloned the human Tbcb gene and characterized mutant versions of its product, having established that the last three amino acid residues of this protein are crucial for TBCE interaction and efficient tubulin dimer dissociation. The overexpression of the mutated form of TBCB lacking the DEI/M-COO− motif, similar to the EEY/F-COO− element in EB1 and related proteins, produces a massive microtubule destruction in vivo suggesting its role in autoinhibition. Using extensive biophysical and biochemical approaches, we unmasked the molecular mechanism by which TBCB controls microtubule depolymerization by means of EB1. In addition, we showed for the first time that TBCB interacts directly with cytosolic chaperonin containing TCP-1 (CCT) during the folding process of α-tubulin. All the results obtained from this work led us to propose three different models to explain the mechanism by which the C-terminal tail of TBCB protects the microtubule from depolymerization, its role in tubulin folding as a CCT cofactor, and the mechanism by which the deregulation of TBCB activity induces the microtubule catastrophe in living cells.
Materials and methods
Human TBCB gene cloning
The human Tbcb coding sequence was amplified by PCR from a testis cDNA sample (BD Biosciences, USA) using a pair of primers designed with the appropriate restriction enzyme recognition sites at their ends: forward primer 5′ GTG AAG CTT CAT ATG GAG GTG ACG GGG GTG 3′; reverse primer 5′ CGC GGA TCC TCA TAT CTC GTC CAA CCC 3′. The amplified coding sequence was then inserted in the HindIII and BamHI sites of the mammalian expression vector pcDNA3.1 (Invitrogen, Life Technologies, USA) to generate the pcDNA3.1-TBCB recombinant plasmid. Human TBCB was cloned into the pEYFP vector from Clontech (Clontech Laboratories, USA). TBCB∆3 and TBCB∆9 cDNA fragments were produced by PCR. The resulting fragments were cloned into pET29c and sequenced (EMD Millipore Bioscience Novagen, USA). Overexpression and purification of TBCB wild-type, TBCB∆3 and TBCB∆9 are detailed in the Supplementary Material.
TBCA and TBCE protein purification and characterization
Human TBCE cDNA wild-type (accession number U61232) and human TBCA ([21], His-tagged at the C-terminus, see Supplementary Material) were cloned into the pRJ-pFastBac vector [8] for recombinant baculovirus production using the Bac-to-Bac Baculovirus Expression System (Invitrogen, Life Technologies, USA). These were then used to infect commercially obtained Sf9 insect cells to produce recombinant TBCE, which was purified following protocols already described elsewhere with minor modifications [8, 9].
Tubulin dimer dissociating assay and nondenaturing electrophoresis
Aliquots of purified brain tubulin were mixed with different amounts of purified TBCE in a 15-μL reaction mixture containing 50 mM MES (pH 6.7), 1 mM MgCl2, and 1 mM GTP in the absence and presence of a stoichiometric excess of TBCB or TBCB∆3, and incubated for 30 min at 30 °C. The reaction mixtures were diluted with a sucrose-containing native loading buffer and loaded onto a 6 % nondenaturing polyacrylamide gel [22, 23]. Native gels were stained directly with Coomassie Brilliant Blue.
Nonclassical two-dimensional electrophoresis
In the first dimension, the protein complexes were fractionated by charge and shape, and then were denatured and their molecular composition determined in the second dimension. The samples were loaded onto a native 0.75 mm thick minigel (7 × 8 cm) as described previously [22, 23]. After 2 h of electrophoresis, a single running lane containing the native electrophoresed sample was excised with a blade on glass, loaded onto a preparative 1.5 mm thick SDS minigel (7 × 8 cm), and fixed to the gel with 0.5 % agarose prepared in 1×SDS loading buffer. Denaturing electrophoresis was performed for 3 h at 10 mA constant current, after which the gel was stained with Coomassie Blue G-250. In a similar manner, bands of interest were excised with a blade on glass, dried in a Speed-Vac concentrator (Thermo Fisher Scientific, USA) and rehydrated with 1×SDS loading buffer, heated at 90 °C for 2 min, and loaded onto a regular 8.5 % SDS minigel. Electrophoresis was performed as described above.
Crosslinking assays
Different concentrations of TBCB, TBCB∆3 or EB1 were incubated at different molar ratios of glutaraldehyde (EM grade; Sigma Aldrich, Spain) at room temperature and 37 °C and at different times. Finally, TBCB (1 μM), TBCB∆3 (1 μM) or EB1 (1.5 μM) were crosslinked by the addition of 0.05 % glutaraldehyde at 37 °C and the reactions were quenched after 5 min by incubation in 100 mM Tris-HCl (pH 8) for 10 min at the relevant assay temperature. Samples were analyzed by SDS-PAGE (12 % gel) and Coomassie Brilliant Blue G-250 staining.
Antisera production, immunocytochemistry, and cell cultures
Affinity-purified primary antibodies were produced against purified human TBCB recombinant protein. Rabbit sera were affinity-purified as described previously [24]. For immunocytochemistry, the antibodies used were anti-α-tubulin and anti-β-tubulin (B512 and Tub2.1, respectively) and anti-acetylated tubulin from Sigma-Aldrich. The anti-glutamylated tubulin antibody (GT335) was a gift from Dr. Janke (CNRS, Montpellier, France). Secondary antibodies were Alexa-Fluor-488-conjugated goat anti-rabbit IgG and goat anti-mouse IgG, Alexa-Fluor-647-conjugated goat anti-mouse IgG (Molecular Probes, Invitrogen), Cy3-conjugated goat anti-mouse IgG and goat anti-mouse IgG1, and Cy5-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories). For some experiments, microtubules were depolymerized with 2 μM nocodazole and cold (4 °C) treatment for 30 min.
Microtubule depolymerization experiments
Bovine brain tubulin was purified as described previously [25]. Purified tubulin (20 μM) was incubated at 35 °C for 20 min, and TBCB∆3 (25 μM) or ovalbumin (20 μM) was added for another 20 min in buffer A (MES 100 mM, pH 6.7, EGTA 1 mM and MgCl2 1 mM) with 2 mM GTP and 30 % glycerol. The pellet and supernatant were separated after centrifugation at 45,000×g for 1 h at 30 °C through a 50 % sucrose cushion containing 1 mM GTP.
Fractionation by gel filtration of complexes formed between cofactors TBCB or TBCB∆3 and TBCE
Purified TBCs and complexes formed in reactions conducted at 30 °C for 30 min were fractionated in a Superdex 200 PC 3.2/30 gel-filtration precision column using an Ettan LC system (GE Healthcare) at room temperature. The elution buffer contained 0.1 M MES (pH 6.7), 1 mM MgCl2, 1 mM EGTA, and 25 mM KCl. Fractions of 25 μL were eluted at 40 μL/min and were analyzed by SDS-PAGE.
Confocal microscopy, cell counts, and statistical analysis
Transitory transfection experiments were performed using Lipofectamine Plus reagent (Life Technologies) or the FuGene 6 reagent (Roche) following the manufacturers’ instructions. The GFP:EB1 construct was kindly supplied by Dr. Akhmanova (Utrecht University, The Netherlands). Cell counts shown in Fig. 1c were performed at 30 h after transfection using a 63× Zeiss oil immersion objective starting from a random field and scanning horizontally from that point. Values presented in Fig. 1c were obtained by double immunofluorescence with anti-α-tubulin/TBCB. Values shown in Fig. 6b were obtained by double immunofluorescence with anti-α-tubulin/TBCB combined with Hoechst 33258 and GFP labeling in cotransfection experiments. Cell counts were performed on confocal microscopy projection images using a Nikon A1R confocal microscope. Only cells with no microtubules and healthy-looking nuclei, as assessed by Hoechst staining, were considered. In colocalization experiments, images were scanned sequentially to avoid fluorescent channel emission crosstalk/bleed-through. Data obtained from two different coverslips of at least three different experiments were analyzed using a t test. Statistical analysis and graphing were performed using SigmaPlot 8.0 software (Systat Software, Richmond, CA). Histograms represent mean values and standard error bars.
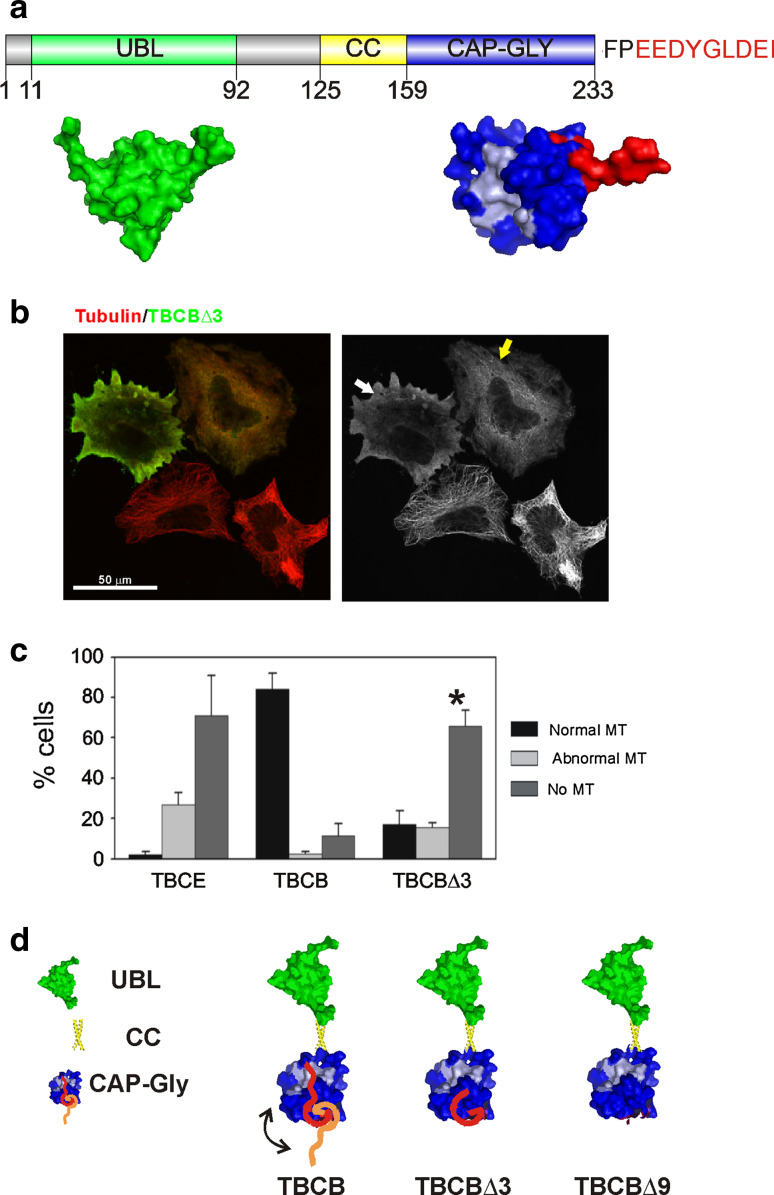
Fig. 1.
TBCB is an autoinhibitory protein. a Schematic drawing of human TBCB depicting the three characterized domains (UBL, coiled-coil and CAP-Gly). The UBL domain (green) corresponds to PDB ID, 1V6E (UBL of murine TBCB), and the CAP-Gly domain (blue) to PDB ID, 1TOV (CAP-Gly domain of F53f4.3) [26]. In light blue are the corresponding residues that form the conserved groove in p150Glued, interacting with the C-terminal peptide of α-tubulin [30, 31]. The last nine residues, which are present in the solved domain of F53f4.3, are shown in red. All structures were drawn using Pymol software (http://www.pymol.org). b Confocal microscopy projection image of TBCBΔ3 overexpression on HeLa cells. TBCBΔ3 (green) produces conspicuous microtubule destruction (white arrow). Moderate TBCBΔ3 levels also severely affect the microtubule cytoskeleton. A high cytoplasmic tubulin background is observed in these two cells. c Statistical analysis of the percentages of cells containing normal, abnormal or absent microtubules in TBCE, TBCB, and TBCBΔ3-overexpressing HeLa cells. A highly significant increase in cells containing a completely destroyed microtubular cytoskeleton is observed when the TBCBΔ3 mutant (asterisk) is overexpressed compared with wild-type TBCB (see also Fig. S1). d The C-terminal region of TBCB functions as an autoinhibitory peptide when bound to the CAP-Gly domain of the protein. The three domains of TBCB are depicted. The N-terminus contains the UBL and the coiled-coil domain, and the C-terminus contains the CAP-Gly domain. The acidic tail of the CAP-Gly domain is shown in red and orange. We propose that the C-terminal tail of TBCB is responsible for the autoinhibition of the protein (red peptide) through interaction with the CAP-Gly domain (blue), specifically with the highly conserved hydrophobic cavity present in the CAP-Gly domain (light blue). In contrast, if the C-terminus region does not interact with the CAP-Gly domain (orange peptide), the protein is derepressed. The hypothetical models of TBCBΔ3 and TBCBΔ9 showing the structure of the protein lacking the last three or nine amino acids are shown
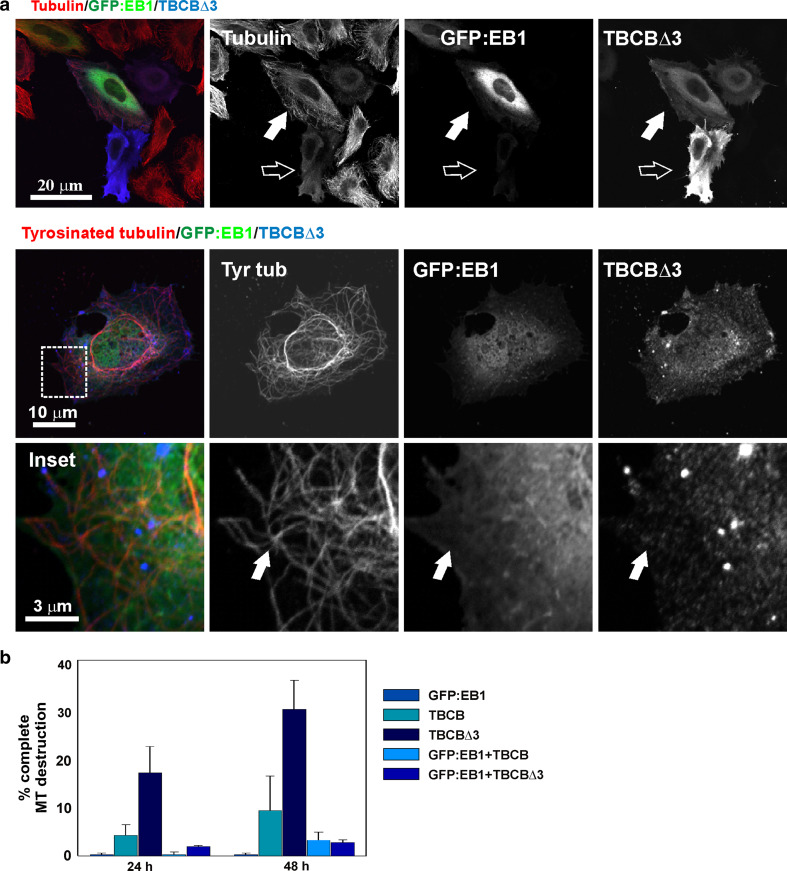
Fig. 6.
EB1 prevents TBCBΔ3 microtubule destruction. a Triple-labeled confocal microscopy projection images of TBCBΔ3 overexpression on HeLa cells. TBCBΔ3 (green) produces conspicuous microtubule destruction (filled arrows). Moderate TBCBΔ3 levels also severely affect the microtubule cytoskeleton (empty arrows). Cotransfection of TBC∆3 removes GFP:EB1 from tyrosinated microtubule tips. GFP:EB1 comets are no longer observable (arrows). b Statistical analysis of the proportions of cells containing normal, abnormal, or absent microtubules in HeLa cells overexpressing TBCE, TBCB, and TBCBΔ3 24 and 48 h after transfection. A highly significant increase in cells containing a completely destroyed microtubular cytoskeleton is observed when the TBCBΔ3 mutant is overexpressed compared with the wild-type construct
Affinity chromatography
Purified TBCBΔ3 was coupled specifically at its amino terminus using EDC-NHS coupling chemistry with a Hi-trap NHS-activated HP column (GE Healthcare). This column gave complete control over the experimental conditions (extract preparation, column loading, bound partner elution, time, and temperature). Thus, HEK293 cell extracts were prepared, sonicated to fragment microtubules, and loaded into the column at 4 °C to avoid protein degradation. We used a slow loading rate to allow binding of the interactors to the column. HEK293 (300 mg) was resuspended and subjected to a hypotonic shock in Tris buffer 20 mM (pH 7.3) and PMSF 0.5 mM (buffer H). Subsequently, cells were sonicated three times for 30 s at 130 W at 4 °C. Protein extract (1 mL at 18 mg/mL) was applied to the NHS column equilibrated in buffer H. The column was washed with 10 mL of the same buffer, and specifically interacting proteins were eluted with a NaCl gradient.
Results
Autoinhibition of TBCB
TBCB is encoded by a unique gene in the human genome. This protein is composed of two functional structural domains connected by a coiled-coil segment (Fig 1a). At the N-terminus, TBCB contains a ubiquitin-like domain. This domain is spherical (PDB ID, 1V6E), behaves as a monomer of about 14 kDa and is a ubiquitous protein interaction domain present in many unrelated proteins. The C-terminal domain is a CAP-Gly characteristic of +TIP proteins. This domain is also globular, three antiparallel β-sheets and one α-helix, as represented by the Caenorhabditis elegans F53f4.3 protein CAP-Gly domain (Fig 1a, PDB: 1TOV, [26]). The unique α-helix is preceded by a disordered stretch of 17 residues, and the last six or seven amino acid residues protrude from the globular domain. CAP-Gly domains serve as recognition domains for EEY/F-COO− peptides [27]. This sequence assumes an extended conformation, and the side-chain of the terminal tyrosine/phenylalanine packs with several hydrophobic amino acid residues in the CAP-Gly domain [28]. The crystal structure of the CAP-Gly of TBCB (C. elegans) reveals that this domain consists of 84 amino acid residues, and although it does not form a dimer in vitro, the conserved groove, involved in the interaction with EEY/F-COO− elements characteristic of EB, CLIP-170, and α-tubulin, holds the C-terminal peptide of the neighboring molecule in the asymmetric unit of the crystal [26].
The structural prediction [29] for the last nine amino acid residues of human TBCB (Fig 1a) is that of a disordered peptide protruding from the globular domain and thus being able to interact with a CAP-Gly domain groove. Indeed, theoretical models [28, 30] have shown putative interactions between the p150Glued CAP-Gly domain and the C-terminal peptide of EB1 and TBCB.
Taking into account the structural features of the TBCB C-terminal domain, we decided to go further in elucidating the TBCB and TBCB C-terminus interaction and to determine whether the C-terminal region would affect the tubulin binding ability of TBCB. These ideas led us to propose the hypothesis of an autoinhibitory mechanism where the TBCB C-terminal extension folds over the globular part of its own CAP-Gly domain and structurally blocks the conserved groove involved in the interaction with EEY/F-COO− elements characteristic of EBs, CLIP-170, and α-tubulin. For this purpose, we cloned the human Tbcb gene and constructed two Tbcb mutants lacking the last three (TBCBΔ3) and last nine (TBCBΔ9) amino acid residues, predicted to be unstructured. These truncated proteins were transiently overexpressed in HeLa cells and visualized using new polyclonal anti-TBCB antibodies (see Materials and methods).
Previously, we have shown that the overexpression of either TBCE or TBCD in human cell lines leads to the sequestration of free α-tubulin and β-tubulin, respectively, leading to massive microtubule depolymerization [6–8]. On the other hand, murine TBCB overexpression only leads to a moderate microtubule depolymerization effect, probably because of the limiting concentrations under TBCB overexpression conditions, of endogenous TBCE required for the binding and dissociation of the tubulin heterodimer [9].
Unpredictably, overexpression of the TBCBΔ3 mutant in HeLa cells induced massive microtubule destruction, comparable only to that observed upon TBCE overexpression (Fig 1b) [9]. Quantification of the microtubule destruction effect revealed that at 30 h after transfection, over 60 % of the TBCBΔ3-positive cells exhibited no detectable microtubules, while only less than 20 % of the overexpressing cells had an apparently unaffected microtubular cytoskeleton (Fig 1c). Similar results were obtained for the TBCBΔ9 mutant. These findings strongly support the proposed idea that TBCB is self-inhibited by its C-terminus and that the removal of only the last three amino acid residues from this domain is sufficient to activate TBCB to induce microtubule depolymerization (Fig 1d).
These results prompted us to investigate whether TBCBΔ3 was able to depolymerize microtubules assembled in vitro. For this purpose, we purified brain tubulin and the TBCBΔ9 and TBCBΔ3 proteins (Fig. S1a). Stoichiometric amounts of purified TBCBΔ3 or ovalbumin (negative control) were incubated with GTP and polymerized purified tubulin. The incubation mix was then centrifuged, and both the soluble and insoluble fractions were analyzed by SDS-PAGE to determine the amounts of tubulin and TBCBΔ3 proteins in the two fractions (Fig. S1b, see Supplementary Fig. 3 in reference [9]). These experiments revealed that TBCBΔ3 was essentially present in the supernatant fractions. Moreover, similar amounts of tubulin were found in the supernatant and pellet fractions in the presence or absence of TBCBΔ3, which were also similar to the amounts found when ovalbumin was used (Fig. S1b). These results lead to the conclusion that TBCBΔ3 is not able to depolymerize microtubules in vitro, presumably because of the lack of a factor mediating its in vivo effect.
Based on these data and on the structural features of TBCB, we propose that TBCB autoinhibition implies the interaction of the C-terminal tail of this cofactor with its own CAP-Gly domain (Fig 1d). This is also supported by the analysis of a TBCB CAP-Gly domain crystal where two molecules of TBCB were found to interact in this way [26] and by the theoretical model of the interaction with p150Glued CAP-Gly domain. [28, 30]. To test this hypothesis, we performed quantitative binding assays using fluorescence polarization of fluorescein-labeled peptides (Table 1; see also Supplementary Material and Supplementary Table 1). The equilibrium dissociation constants showed that the C-terminal nonapeptide of TBCB (peptide 1, EEDYGLDEI) shows a higher affinity for TBCB (12 μM) than that displayed by the same peptide lacking the last three residues (DEI, peptide 2, 71 μM). Similarly, the truncated mutants TBCB∆3 and TBCB∆9, lacking the last three residues (DEI) and six residues (YGLDEI) of TBCB, showed a comparable reduction (74 and 79 μM) in their affinity for the complete C-terminal nonapeptide of TBCB (peptide 1). In addition, the binding affinities exhibited by peptide 2 for the two truncated TBCB forms were significantly lower than for the full-length protein (178 and 249 μM). Together these results strongly support the idea that the last three amino acid residues of the TBCB C-terminus are involved in the binding to TBCB, thus reinforcing our model of TBCB autoinhibition.
Table 1.
Equilibrium dissociation constants for the binding of the fluorescein-labeled TBCB and α-tubulin peptides to TBCB, TBCBΔ3, and TBCBΔ9 by fluorescence polarization. Fluorescence polarization binding assays of TBCB with fluorescence-labeled peptides were repeated up to a protein concentration of 100 μM (see also Fig. S5)
| Peptide | Protein | Kd (μM) |
|---|---|---|
| EEDYGLDEI (TBCB) | TBCB | 12 ± 1 |
| TBCB∆3 | 74 ± 3 | |
| TBCB∆9 | 79 ± 4 | |
| EEDYGL (TBCB∆3) | TBCB | 71 ± 3 |
| TBCB∆3 | 178 ± 15 | |
| TBCB∆9 | 249 ± 30 | |
| GEGEEEGEEY (α1/2-tub) | TBCB | 43 ± 2 |
| TBCB∆3 | 113 ± 7 | |
| TBCB∆9 | 145 ± 9 | |
| GEGEEEGEE (α1/2-tub∆Y) | TBCB | 140 ± 4 |
| TBCB∆3 | 312 ± 20 | |
| TBCB∆9 | 472 ± 69 |
Finally, some CAP-Gly domains have been reported to display high affinity for the C-terminal tail of α-tubulin (i.e., CLIP-170 CAP-Gly domain 2) [31]. We have also quantified the binding affinity of TBCB to two different α-tubulin C-terminal peptides (peptide 3 and peptide 4, Tables 1 and S1). Peptide 3 (GEGEEEGEEY) corresponds to the C-terminus of α-tubulin isotypes 1 and 2, and contains the last tyrosine residue, known to be critical for binding to CAP-Gly domains [31]. As expected, all three TBCB proteins (TBCB, TBCB∆3, and TBCB∆9) displayed Kd values for the tubulin peptide 3 up to fourfold higher (43, 113, and 145 μM) than those exhibited for the TBCB C-terminal peptide 1 (12, 74, and 79 μM). The C-terminal tubulin peptide lacking the last tyrosine (peptide 4, GEGEEEGEE) resulted in a threefold reduced affinity for all three TBCB proteins (140, 312, and 472 μM), consistent with the findings of previous studies [31]. Altogether, the results indicate that TBCB interacts with tubulin with a significantly reduced affinity with respect to its autoinhibitory interaction.
The C-terminal acidic tail of TBCB is responsible for TBCE interaction
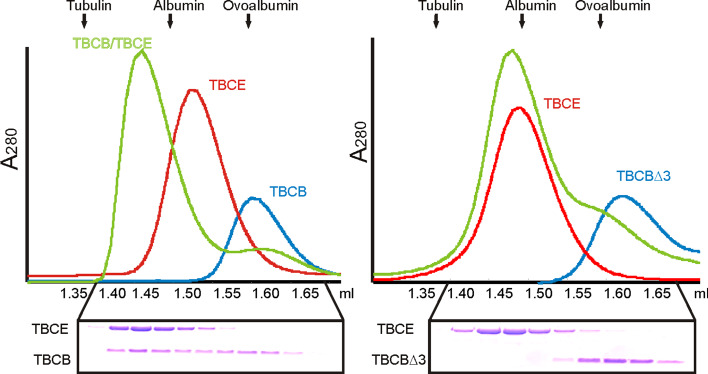
Previous work from our group has shown that when incubated together, purified mammalian TBCB and TBCE produce a new peak on gel filtration analysis chromatograms that corresponds approximately to the sum of the molecular masses of individual TBCB and TBCE [9]. Further analysis of this peak revealed the presence of both cofactors suggesting that purified TBCB forms a binary complex with TBCE (Fig. 2, left panel) [9]. Bearing in mind these results and that the last three amino acid residues of the TBCB C-terminus are critical for its activity, we investigated whether the TBCBΔ3 or TBCBΔ9 truncated proteins are also able to associate with TBCE. Interestingly, when purified TBCE was incubated with purified TBCBΔ3 (Fig. 2, right panel), no additional peaks were detected on gel filtration chromatograms. Indeed, chromatograms only revealed peaks corresponding to the species present when the single purified proteins were analyzed. This observation was confirmed by SDS-PAGE analysis of the corresponding fractions (Fig. 2). These results strongly suggest that none of the TBCB C-terminus deleted mutants is able to interact with TBCE and that the last three amino acid residues of TBCB are essential for TBCE recognition.
Fig. 2.
Biochemical studies of TBCBΔ3. TBCBΔ3, in contrast to complete TBCB, does not form a binary complex with TBCE. Plots of A280 absorbance against elution volume from the size-exclusion chromatography experiments are shown. The elution profiles of TBCB, TBCBΔ3 and TBCE, and the interaction of these proteins were analyzed by gel filtration through a Superdex 200 PC 3.2/30 column (GE-Healthcare). Three curves are shown: TBCB and TBCBΔ3 alone (blue), TBCE alone (red), and the combination of TBCB and TBCE or TBCBΔ3 and TBCE (green). Fractions were subjected to SDS-PAGE. The final concentrations used were 18 μM for TBCE and 15 μM for TBCB and TBCBΔ3
Because the interaction of TBCE and TBCB is required for efficient tubulin heterodimer dissociation activity, the results above predict that the last three amino acid residues of TBCB are also critical for this process. Therefore, we investigated the tubulin heterodimer dissociation activity of TBCE in the presence of either full-length TBCB or TBCBΔ3, and quantified the different molecular species produced by native PAGE (Fig. S2). When tubulin heterodimers were incubated with TBCE in the presence of TBCB, an extra band, which corresponded to the ternary complex composed of TBCB, TBCE and α-tubulin, was detected in native gels. Interestingly, in similar incubations where TBCB was replaced by TBCBΔ3, the ternary complex was absent, and instead an extra band that was slightly retarded in relation to that corresponding to tubulin heterodimers was found. To determine the molecular composition of this new band, we performed an analysis in the second dimension (Fig. S2). This analysis showed that this band corresponded to the binary complex containing α-tubulin and TBCBΔ3, showing that TBCBΔ3 is able to interact with α-tubulin but not with TBCE. Therefore, the complex TBCBΔ3/α-tubulin is formed after dissociation of the tubulin dimer by TBCE.
TBCE and TBCD were characterized as the TBCs that are able to dissociate the tubulin heterodimer [6–8]. To understand better the role of these cofactors in tubulin heterodimer disruption, we decided to investigate the time course of the dissociation activities of TBCE alone and in the presence of stoichiometric amounts of TBCB or TBCBΔ3 (Fig. S2). While TBCE alone dissociated 60 % of the tubulin heterodimers in 30 min, TBCE in the presence of TBCB was able to dissociate about 90 % in less than 30 s, the minimal time required to mix the incubation components and load it into the gel. In contrast, in the same period, but in the presence of TBCBΔ3, only 25 % of tubulin heterodimers were dissociated by TBCE (Fig. S2e, f). Thus, in the presence of TBCE there is a clear difference between the dissociating activities of the TBCB and the TBCBΔ3. Together our data clearly show that the last three residues in TBCB are not required for the formation of the binary complex with α-tubulin. However, they are not only implicated in TBCB autoinhibition but also essential for the interaction of TBCB with TBCE, and are therefore required for the assembly of a more efficient tubulin heterodimer dissociation machine. Moreover, based on these results, we put forward the hypothesis that TBCE would interact with the C-terminus of TBCB, which would lead to the derepression of this cofactor triggering a microtubule catastrophe. However, the observation that neither TBCBΔ3 nor TBCBΔ9 (results not shown) was able to interact with TBCE is still puzzling because their overexpression resulted in massive microtubule depolymerization in HeLa cells (see Fig 1b and 1c).
In solution, TBCB is a monomer as revealed by biophysical studies and crosslinking experiments
The possible TBCB autoinhibition is supported by the interaction observed between the C-terminal peptide molecules in the crystal structure of the CAP-Gly domain of C. elegans TBCB [26]. To obtain clues regarding the behavior of native TBCB that could help us to elucidate the molecular mechanism underlying its autorepressed activity, we used different experimental approaches to investigate whether TBCB is a monomer or a dimer in solution. For this purpose, we first determined the circular dichroism (CD) spectra of the TBCB, TBCBΔ3, and TBCBΔ9 proteins (Supplementary Material). The CD spectra of the three proteins (Fig. S3a, left panels) were characterized by the presence of two minima at 208 and 217 nm, which are indicative of a mixed population of α-helical (20 %, [θ]222/208 = 0.87) and β-strand (30 %) conformations. The absence of a minimum value at 222 nm (typical of all α proteins) and a zero crossing (typical of a α-helices and β-strands) suggests the additional presence of disordered structures including random coils and turns (50 %).
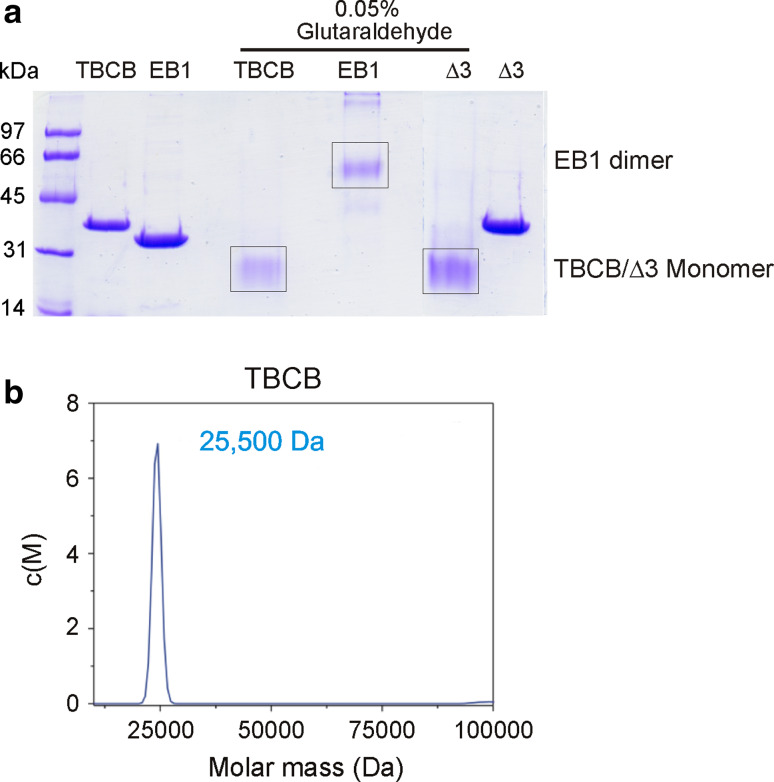
Subsequently, we decided to study the state of aggregation of these proteins by dynamic light scattering (DLS), but obtained inconclusive results (see Supplementary Material and Fig. S3b) [32]. Using gel filtration analysis, we have previously characterized the molecular components of the different complexes formed between TBCB, TBCE, and tubulin [9]. Based on the elution volumes, the estimated molecular mass of TBCB was 30–40 % larger (40 kDa) than that predicted from its amino acid sequence (27 kDa). Curiously, this value (40 kDa) for TBCB coincided with the molecular mass estimated from its mobility on SDS-PAGE. However, the apparent molecular mass of the complexes formed between TBCB and TBCE, and between TBCB and TBCE and α-tubulin, suggested that they were the result of the sum of the molecular masses of the monomeric subunits. These apparent discrepancies in the values of TBCB molecular masses were maintained in the DLS estimations of the molecular mass of TBCB (69 kDa), TBCBΔ3 (53 kDa), and TBCBΔ9 (34 kDa; Fig. S3b). In order to clarify these observations, we decided to investigate the state of oligomerization of the three proteins by crosslinking experiments (Fig. 3a and Fig. S4) and analytical ultracentrifugation (Fig 3b). We used glutaraldehyde as a general protein crosslinker and a protein (EB1) with a very low Kd value for dimer formation as a positive control. Interestingly, purified TBCB, as well as TBCBΔ3, migrated in SDS-PAGE as a single band with an apparent molecular mass of about 38–40 kDa. Nevertheless, when these two proteins were incubated with glutaraldehyde (0.05 %), the band corresponding to 40 kDa, although still visible in trace amounts, was mostly substituted by a new band corresponding to s species with a molecular mass of about 27 kDa (Fig. 3a) in addition to higher forms (Fig. S4). This molecular mass is in agreement with the theoretical molecular mass of these two proteins. On the other hand, the EB1 control protein, which normally migrates as a single band of 32–34 kDa after glutaraldehyde treatment, migrated as a band of 64 kDa, which is consistent with its size in gel filtration experiments (Fig. S3d). TBCB and TBCB∆3 behave unusually in SDS gels and in gel permeation columns and have a strange behavior in DLS as shown in Fig. S3b. The fact that after glutaraldehyde treatment they moved at their expected molecular size position in SDS gels suggests this might have been due to stabilization by intramolecular crosslinking.
Fig. 3.
Biophysical studies of TBCB and TBCBΔ3. a Analysis of quaternary structures of TBCB and TBCB∆3 at 37 °C by crosslinking. Purified proteins at the concentrations indicated were crosslinked using 0.05 % glutaraldehyde and separated by SDS-12 % PAGE. Equal amounts of protein were loaded in each lane. Control lanes with proteins with no treatment are indicated. Crosslinked EB1 at 1.5 μM as a control is indicated (EB1 dimer). b Analytical ultracentrifugation of TBCB. Sedimentation velocity experiments performed at 42,000 rpm and 20 °C yield an estimated molecular mass of 25.5 kDa, in agreement with the molecular mass of the theoretical monomer (27 kDa). See also Fig. S3
These results were confirmed by analytical ultracentrifugation analysis performed at 20 °C at 42,000 rpm (Fig 3b). This analysis showed a molecular mass of TBCB of about 25.5 kDa, corresponding to the size of the monomer (27 kDa). Taken together, all these results led us to conclude that TBCB behaves as a monomer and to suggest that its self-inhibition occurs within the same molecule and not between two or more TBCB molecules.
TBCB localizes to the centrosome and mitotic spindle microtubules
TBCB has been shown to colocalize with Pak1 protein on newly polymerized microtubules [13]. Because the overexpression of TBCB and TBCBΔ3 leads to microtubule depolymerization, we decided to investigate the subcellular distribution of the overexpressed YFP:TBCB protein in HeLa cells throughout the cell cycle.
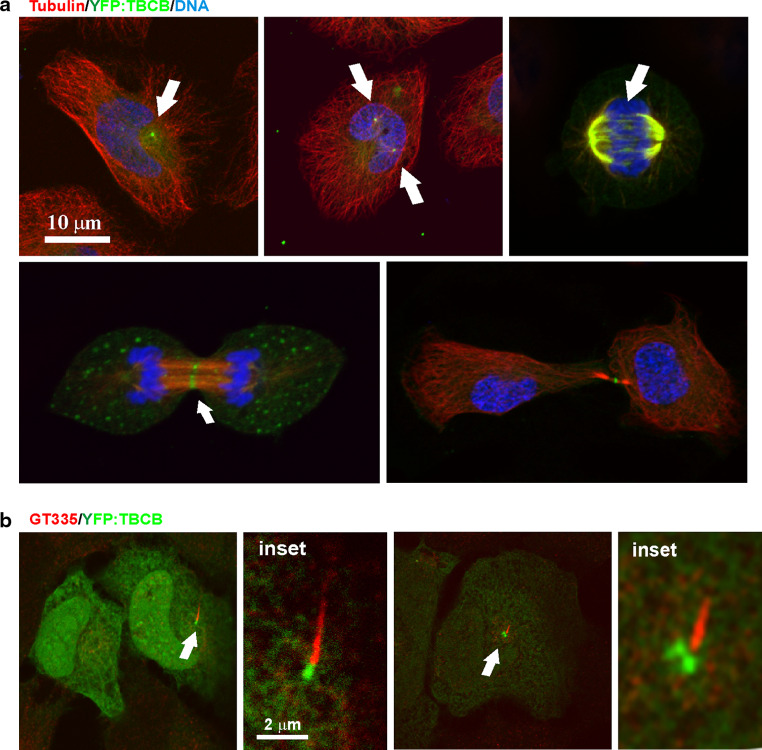
As observed for the wild-type endogenous protein [9], YFP:TBCB is mostly a soluble cytoplasmic protein in interphase cells (Fig 4a). A prominent spot of YFP:TBCB is often localized at the centrosome (double spot) and at the base of the primary cilium (Fig 4b) [33]. Indeed, the distribution of YFP:TBCB during mitosis revealed two clear YFP:TBCB spots during prophase (Fig 4a, top center). Interestingly, as mitosis progressed toward metaphase, TBCB was also localized to spindle microtubules (Fig 4a, top right). This localization was also observed for overexpressed wild-type untagged TBCB by immunostaining (data not shown) and is in accordance with previous analyzes performed for endogenous TBCB in human and mouse cells [9, 13, 35]. Furthermore, with this new analysis, we observed that during anaphase A, YFP:TBCB becomes more visible as thin filaments bridging the midzone, and by anaphase B most of this cofactor had progressively disappeared from the centrosome and was concentrated on the midbody microtubules (Fig 4a, bottom). At the end of telophase, TBCB was apparently absent from the centrosome, concentrating in a unique spot at the midbody. These localization results show that TBCB can bind to microtubules. However, we know from the results described above that TBCB cannot recognize tubulin heterodimers, suggesting this binding to be indirect, occurring through the interaction of TBCB with a microtubule binding protein.
Fig. 4.
YFP:TBCB is associated with the centrosome and mitotic microtubules. a Confocal microscopy images of YFP:TBCB localization in interphase (top left) and mitotic HeLa cells. YFP:TBCB is mostly cytoplasmic and concentrates at the centrosomes in interphase and prophase HeLa cells (top center, arrows). In anaphase A, YFP:TBCB is clearly associated with spindle microtubules, also decorating microtubules bridging the midzone (bottom left, arrow). A midbody localization pattern is more obvious during anaphase B and telophase (bottom images, arrow), where there is no longer a centrosomal signal. b Relationship of the YFP:TBCB signal to the primary cilium in G1 (left) and G2 HeLa cells (right). Glutamylated tubulin labeling the primary cilium is recognized by the GT335 antibody (red)
TBCBΔ3 interacts with EB1 and the cytosolic chaperonin CCT
That in vivo TBCB is able to promote microtubule destabilization, whereas in vitro it is not able to depolymerize or even to interact with microtubules, strongly suggests that the functions of TBCB in vivo in relation to microtubules should be mediated by an interactor (or interactors). This prompted us to search for TBCB molecular interactors. In a first approach, we performed different experiments such as immunoprecipitation techniques, but were unsuccessful in the identification of any TBCB interactor. This could be ascribed to the fact that we did not use specific conditions required to avoid disruption of weak interactions. To overcome these problems, we constructed an affinity column with bound recombinant untagged TBCBΔ3, the derepressed version of TBCB. Bound proteins or complexes were specifically eluted with a salt gradient that produced a double peak between 100 and 200 mM NaCl, and the corresponding fractions were analyzed by SDS-PAGE (Fig. 5).
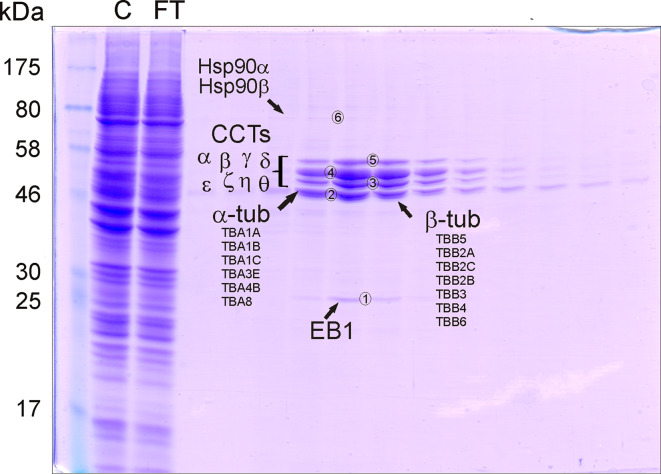
Fig. 5.
Search for TBCB partners using affinity chromatography. Microtubule-bound EB1, Hsp90, and CCT are interactors of TBCB. Purified TBCBΔ3 was purified and coupled specifically to a Hi-Trap NHS-activated HP column (GE Healthcare). Human HEK293 protein extract (1 mL at 18 mg/ml) was applied to the column. Bound proteins were eluted using an NaCl gradient. The SDS-PAGE analysis of the fractions eluted from the NHS column are shown: lane 1 molecular mass marker; lane 2 aliquot of the cell extract (control C); lane 3 unbound proteins eluted in the void volume (flow-through FT); lanes 4–15 fractions eluted with 100–200 mM of NaCl
The different bands (nos. 1–6, Fig. 5) detected in SDS-PAGE gels were subjected to trypsin digestion followed by mass fingerprinting analysis. Band no. 1 was found to correspond to human EB1. The sequence coverage was 83 % corresponding to MARE 1 (UniProt accession C1BKD9, Fig. S5a). Band nos. 3–5 were found to correspond to the eight distinct human CCT subunits required to assemble CCT completely [34]. All the CCT subunits were identified with a sequence coverage higher than 45 %, and for most of the cases, the coverage was about 70 % (Fig. S6a). The presence of complete CCT heterooligomeric particles in the eluted fractions was also confirmed by conventional electron microscopy. CCT is a group II chaperonin mostly committed to the folding of actins and monomeric α- and β-tubulin [34]. Therefore, this is the first evidence that a TBC is able to bind directly to CCT, an interaction that may be relevant for the tubulin folding process. The finding that CCT is one of the interactors of TBCB, as revealed by the use of the affinity column, adds new data to the model of how tubulin folding and dimerization may occur in vivo.
In addition to these interactors, band no. 2 was found to correspond to α-tubulin and β-tubulin (Fig 5). In fact, five α-tubulin isotypes and seven β-tubulin isotypes (Fig. S6b) were unequivocally identified with a sequence coverage for all isotypes higher than 20 % and generally with a value of about 50–60 % (Fig. S6b).These results also show that the HEK293 cell line expresses all known tubulin isotypes. Because TBCBΔ3 does not bind tubulin heterodimers or microtubules in vitro and only binds to α-tubulin, but not to β-tubulin monomers, these results strongly suggest that TBCBΔ3 would bind microtubules through a partner and that this partner could be EB1.
EB1 overexpression prevents TBCBΔ3 microtubule destruction
The finding that EB1 is a TBCB interactor makes this protein the most attractive candidate for explaining how TBCB is able to regulate microtubule dynamics. Because EB1 is known to stabilize the plus ends of microtubules, its interaction with TBCB would explain how TBCB is able to promote a microtubule catastrophe when overexpressed. In this context, we may expect that TBCB has the ability to sequester EB1 from microtubule plus ends. If these hypotheses are true, the overexpression of EB1 would be sufficient to rescue the observed phenotype of microtubule depolymerization when TBCB and TBCBΔ3 are overexpressed. This would also provide evidence of the interaction of EB1 with TBCB in vivo. To examine this model, we cotransfected HeLa cells with wild-type TBCB and GFP:EB1 or TBCBΔ3 and GFP:EB1. As predicted, microtubule destruction resulting from simultaneous overexpression of TBCBΔ3 and GFP:EB1 or TBCB and GFP:EB1 was substantially less accentuated than that observed in cells overexpressing only TBCBΔ3 or TBCB, respectively (Fig. 6a). Furthermore, the typical GFP:EB1 comets, resulting in the localization of this protein at growing microtubule plus ends, were no longer observable (Fig. 6a, bottom), suggesting that excess TBCBΔ3 was interacting with EB1 modifying its intracellular distribution.
To confirm whether TBCB∆3 interacts with EB1 in this system, we next quantified and compared the microtubule destruction effect at specific time points against the background of overexpressing GFP:EB1 + TBCB∆3 versus overexpressing GFP:EB1 + TBCB. Therefore, cotransfected cells and controls were fixed at different time points. Triple labeling experiments revealed that ten times as many cells showed preservation of their microtubule cytoskeleton when cotransfected with both genes (Fig 6b). Hence, this system shows that the TBCB∆3 depolymerization effect is virtually blocked by overexpressed EB1 and thus strongly suggests that TBCB∆3 and EB1 interact in vivo.
Discussion
In this work, we found that the TBCB CAP-Gly domain is autoinhibited by interaction with the last three residues of its C-terminus. Our data also show that these last three residues of TBCB are required for TBCE recognition, interaction and tubulin heterodimer dissociation. Therefore, we have proposed a molecular mechanism explaining how TBCB and TBCE form a binary complex that efficiently recognizes and dissociates the tubulin heterodimer.
Biophysical studies revealed that a TBCB protein lacking the last three amino acid residues (TBCB∆3) behaved as the wild-type protein with a similar CD spectrum and response to unfolding by heat. Crosslinking experiments and analytical ultracentrifugation, in contrast to DLS, showed that TBCB behaved as a monomer of 25.5 kDa. The TBCB∆9 protein, which lacks the last nine amino acid residues, showed a similar CD spectrum and the same unfolding temperature as TBCB and TBCBΔ3. This truncated version of the TBCB protein showed a completely different behavior under heat denaturing conditions, but the DLS analysis also suggested that it is a monomer (Fig. S3). When overexpressed in human cells, TBCB is able to induce microtubule loss [9]. Although initially this could be ascribable to its interaction with endogenous TBCE, we found that the mutant lacking the last three amino acid residues was unable to interact with TBCE and depolymerize microtubules in vivo with a higher efficiency. This suggests that the C-terminal region is an autoinhibitory sequence and that the mechanism of microtubule depolymerization by TBCBΔ3 is TBCE-independent (Fig. 1d). Microtubule destruction was accompanied by an intense tubulin background in the cells when detected with both anti-α-tubulin and anti-β-tubulin antibodies, suggesting the presence of soluble tubulin heterodimers in the cytosol of the cells. This cytoplasmic background is very unusual in cells overexpressing TBCE or TBCD where these two cofactors sequester either α-tubulin or β-tubulin, respectively, upon tubulin heterodimer dissociation, which leads to microtubule depolymerization and microtubule network collapse.
To gain an insight into the mechanism by which TBCB causes microtubule depolymerization, we studied whether TBCB was able to depolymerize microtubules in vitro. We demonstrated that TBCB was not a microtubule depolymerizing enzyme in itself (Fig. S1b), which led us to propose the hypothesis of the existence of a TBCB partner that would be implicated in the TBCB microtubule depolymerization mechanism. Specifically, we proposed that TBCB∆3 was derepressed on its presumed ability to bind to a partner through which it would promote microtubule depolymerization. For this reason, we decided to construct an affinity column containing the derepressed version of TBCB. The reasoning behind the use of the derepressed TBCB mutant was to increase the possibility of identifying partners that would not be easy to discover with the wild-type repressed protein.
Therefore, we performed TBCB∆3 affinity binding studies by constructing an affinity column with this polypeptide bound to a matrix and incubating it with a soluble human cell protein extract. This derepressed TBCB protein was able to interact with Hsp90, CCT, and EB1. Indeed, the amount of Hsp90 that appeared to be bound to the affinity column containing TBCB∆3 was low compared with the amounts of EB1 and CCT. Consequently, and although this interaction has a putative role, we did not continue studying this interaction. The same was true for CCT, although we can envisage an important role for the CCT–TBCB interaction in the process of CCT-mediated α-tubulin folding.
Model of TBCB-mediated α-tubulin folding bound to CCT
This is the first time that a TBC was found to associate with a chaperonin containing TCP-1 (CCT, [34]). The possibility of TBCB being a CCT substrate was excluded because TBCB interacts with CCT while binding α-tubulin [9]. This strongly suggests that the interaction of TBCB with CCT might contribute to proper tubulin folding and dimerization. The interaction of TBCB with CCT is also supported by the fact that after incubation of different amounts of TBCB with purified bovine CCT, an extra band with a higher molecular mass than those corresponding to CCT or TBCB was recognized by anti-TBCB antiserum (our unpublished results). This band migrated at the same position as the extra band containing CCT, α-tubulin, and TBCB previously observed to occur in in vitro translation assays [9]. Together, these results led us to propose a model in which TBCB would recognize α-tubulin bound to CCT. It has been shown in vitro and in vivo that free α-tubulin or β-tubulin will aggregate in the absence of a partner (the other tubulin partner or TBCs [4, 5]). Therefore, our proposed mechanism predicts that α-tubulin would be released from CCT bound to TBCB insuring that the α-tubulin monomer would never aggregate. Later, the monomeric tubulin subunit would be transferred to TBCE for dimer assembly and incorporation into growing microtubules or would be transferred to the degradative pathway involving the proteasome if not properly folded [36].
Model of TBCB-mediated microtubule depolymerization
Previous studies have shown that TBCB is regulated by phosphorylation being a substrate of Pak1 as revealed in a yeast two-hybrid screen. Pak1 directly phosphorylates TBCB, and both proteins colocalize on newly polymerized microtubules [13]. We have also shown that the YFP:TBCB protein colocalizes with microtubules of the mitotic spindle, and as mitosis progresses, the staining gradually increases in microtubules of the spindle midzone where active polymerizing microtubules are present. Moreover, TBCB depletion in neuronal cells by siRNA induces axonal extension and growth cone detachment, suggesting a role for TBCB at microtubule tips [35]. Recently, TBCB was identified as a target for nitrogen-containing bisphosphonates (N-BP), drugs extensively used in the treatment of bone diseases. In fact, TBCB is upregulated in mammalian cells after N-BP treatment inducing the loss of microtubule architecture at sites of active microtubule assembly, such as neuronal protrusions [37]. This effect might be explained by the observation that overexpression of TBCB in growth cones leads to microtubule depolymerization [35].
EB proteins are dimeric proteins formed from two functional domains [38]. The N-terminal domain mediates microtubule binding, while the C-terminal domain consists of a coiled coil responsible for dimerization and an unstructured tail. Several models have been proposed to explain EB binding to the plus end of microtubules. Crystal structures of the C-terminal domain of EB1 and the CAP-Gly domains of the dynactin subunit p150Glued led Hayashi and coworkers [39] to postulate that EB1 is autoinhibited and that this conformation is unhampered by binding of a CAP-Gly-containing protein (p150Glued). Although TBCB and TBCE contain a CAP-Gly domain, these proteins were not described as localizing at microtubule tips as are other +TIP proteins. TBCE was also a good candidate for such a role, but we could not see an interaction with EB1 in vitro. While the model proposed by Hayashi and coworkers [39] is well supported by the different structures solved, the mechanism by which EBs bind to microtubules remains unclear. Although as predicted, the removal of the EB tail blocked binding to partners [40], it had no effect in vivo on microtubule plus-end accumulation. Also, removal of the C-terminal tail of EBs does not alter the global conformation of the protein [40], which does not support the model proposed by Hayashi and coworkers [39]. In addition, Buey and colleagues [41] suggested that the negative charge of the domain is responsible for the specificity of the EBs to the microtubule tip. Despite the different models that try to explain the preference of EB proteins for the growing microtubule plus end, it seems that EBs recognize an inaccessible region of tubulin in the GTP-bound form [42]. It is thought that the binding of EBs to microtubule tips is dynamic, being characterized by rounds of binding and unbinding [19]. Although we detected an interaction of TBCB∆3 with EB1, in vivo and using our polyclonal antibodies against human or murine TBCB, we never found typical EB comets. TBCB is a CAP-Gly-containing protein, but when we performed double immunolabeling with GFP-EB1 and TBCB we could not colocalize TBCB to the microtubule tips that were clearly seen for EB1. For this reason, it was surprising to find that EB1 was one of the major interactors of TBCB. Thus, we decided to study the in vitro interaction of TBCB with EB1 using purified untagged proteins. Unexpectedly, we found no interaction under the conditions tested (Fig. S3d). We could not rule out the possibility that specific posttranslational modifications in the EB1 protein were required for TBCB binding. This would not be the case for TBCB because the protein used in the affinity column was purified from E. coli and could not have posttranslational modifications. The detailed mass spectrometric analysis of the EB1 polypeptide showed acetylation at alanine 2 (Fig. S5a). This cotranslational acetylation occurrence takes place only in eukaryotes. This usual modification probably does not add functional diversity to the EB1 polypeptide, supporting the notion that TBCB and EB1 do not interact unless EB1 is derepressed. We isolated microtubule fragments containing different α-tubulin and β-tubulin polypeptides (Fig. 5 and Fig. S6b) bound to TBCB∆3. In our experiments (this work; [9]), TBCB did not interact along the microtubule, although we cannot rule out an interaction with microtubule ends. Isolation of microtubule fragments with bound EB1 provides strong evidence that TBCB∆3 binds and sequesters EB1 from microtubule ends leading to microtubule depolymerization [43].
We have established that the C-terminal peptide of TBCB is required for binding to TBCE and for efficient tubulin heterodimer dissociation (Fig. 2 and Fig. S2). TBCB also recognizes EB1 at the plus end of the microtubule, and our results suggest that this interaction takes place when EB1 is also derepressed, probably after interaction with another +TIP protein or after binding to microtubules. Finally, we also demonstrated that cotransfection with EB1 prevents TBCB∆3 microtubule destruction and that cells recover their normal phenotype, confirming that TBCB∆3 and EB1 interact in vivo. This is more than sufficient to justify microtubule destabilization, but, as TBCB forms an active heterodimer with TBCE in tubulin dissociation, we suggest that this is also the mechanism by which these proteins regulate microtubule dynamics. In this way, TBCB participates in microtubule dynamics, and as shown here, the deregulation of TBCB activity induces a microtubule catastrophe in living cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was funded by BFU2007-64882, BFU2008-01344 and BFU2010-18948, the Consolider-Ingenio Spanish Ministry of Education and Science Centrosome-3D and the Instituto de Formación e Investigacion Marqués de Valdecilla. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Laura Alvarez and Begoña Ubilla for excellent technical assistance. Mass spectrometry analysis was performed in the Proteomics Core Facility-SGIKER, a member of ProteoRed, at the University of the Basque Country.
Conflict of interest
None of the authors have a financial interest related to this work.
References
- 1.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Amos LA. Focusing-in on microtubules. Curr Opin Struct Biol. 2000;10:236–241. doi: 10.1016/S0959-440X(00)00070-1. [DOI] [PubMed] [Google Scholar]
- 3.Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH. Microtubule structure at 8 A resolution. Structure. 2002;10:1317–1328. doi: 10.1016/S0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 4.Lewis SA, Tian G, Cowan NJ. The alpha- and beta-tubulin folding pathways. Trends Cell Biol. 1997;12:479–484. doi: 10.1016/S0962-8924(97)01168-9. [DOI] [PubMed] [Google Scholar]
- 5.López-Fanarraga M, Avila J, Guasch A, Coll M, Zabala JC. Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J Struct Biol. 2001;135:219–229. doi: 10.1006/jsbi.2001.4386. [DOI] [PubMed] [Google Scholar]
- 6.Bhamidipati A, Lewis SA, Cowan NJ. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martín L, Fanarraga ML, Aloria K, Zabala JC. Tubulin folding cofactor D is a microtubule destabilizing protein. FEBS Lett. 2000;470:93–95. doi: 10.1016/S0014-5793(00)01293-X. [DOI] [PubMed] [Google Scholar]
- 8.Kortazar D, Carranza G, Bellido J, Villegas JC, Fanarraga ML, Zabala JC. Native tubulin-folding cofactor E purified from baculovirus-infected Sf9 cells dissociates tubulin dimers. Protein Expr Purif. 2006;49:196–202. doi: 10.1016/j.pep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Kortazar D, Fanarraga ML, Carranza G, Bellido J, Villegas JC, Avila J, Zabala JC. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp Cell Res. 2007;313:425–436. doi: 10.1016/j.yexcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Feierbach B, Nogales E, Downing KH, Stearns T. Alf1p, a CLIP-170 domain-containing protein, is functionally and physically associated with alpha-tubulin. J Cell Biol. 1999;144:113–124. doi: 10.1083/jcb.144.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parvari R, Hershkovitz E, Grossman N, Gorodischer R, Loeys B, Zecic A, Mortier G, Gregory S, Sharony R, Kambouris M, Sakati N, Meyer BF, Al Aqeel AI, Al Humaidan AK, Al Zanhrani F, Al Swaid A, Al Othman J, Diaz GA, Weiner R, Khan KT, Gordon R, Gelb BD, HRD/Autosomal Recessive Kenny-Caffey Syndrome Consortium Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat Genet. 2002;32:448–452. doi: 10.1038/ng1012. [DOI] [PubMed] [Google Scholar]
- 12.Martin N, Jaubert J, Gounon P, Salido E, Haase G, Szatanik M, Guénet JL. A missense mutation in Tbce causes progressive motor neuronopathy in mice. Nat Genet. 2002;32:443–447. doi: 10.1038/ng1016. [DOI] [PubMed] [Google Scholar]
- 13.Vadlamudi RK, Barnes CJ, Rayala S, Li F, Balasenthil S, Marcus S, Goodson HV, Sahin AA, Kumar R. p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol Cell Biol. 2005;25:3726–3736. doi: 10.1128/MCB.25.9.3726-3736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian G, Jaglin XH, Keays DA, Francis F, Chelly J, Cowan NJ. Disease-associated mutations in TUBA1A result in a spectrum of defects in the tubulin folding and heterodimer assembly pathway. Hum Mol Genet. 2010;19:3599–3613. doi: 10.1093/hmg/ddq276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martins-de-Souza D, Gattaz WF, Schmitt A, Rewerts C, Maccarrone G, Dias-Neto E, Turck CW. Prefrontal cortex shotgun proteome analysis reveals altered calcium homeostasis and immune system imbalance in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2009;259:151–163. doi: 10.1007/s00406-008-0847-2. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Ding J, Allen E, Zhu P, Zhang L, Vogel H, Yang Y. Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin-proteasome pathway. Curr Biol. 2005;15:2050–2055. doi: 10.1016/j.cub.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 17.Schuyler SC, Pellman D. Microtubule “plus end tracking proteins”: the end is just the beginning. Cell. 2001;105:421–424. doi: 10.1016/S0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 18.Akhmanova A, Hoogenrad CC. Microtubule plus end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Galjart N. Plus-end-tracking proteins and their interactions at microtubule ends. Curr Biol. 2010;20:R528–R537. doi: 10.1016/j.cub.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Yu KL, Keijzer N, Hoogenraad CC, Akhmanova A. Isolation of novel +TIPs and their binding partners using affinity purification techniques. Methods Mol Biol. 2011;777:293–316. doi: 10.1007/978-1-61779-252-6_21. [DOI] [PubMed] [Google Scholar]
- 21.Llosa M, Aloria K, Campo R, Padilla R, Avila J, Sánchez-Pulido L, Zabala JC. The beta-tubulin monomer release factor (p14) has homology with a region of the DnaJ protein. FEBS Lett. 1996;397:283–289. doi: 10.1016/S0014-5793(96)01198-2. [DOI] [PubMed] [Google Scholar]
- 22.Zabala JC, Cowan NJ. Tubulin dimer formation via the release of alpha- and beta-tubulin monomers from multimolecular complexes. Cell Motil Cytoskel. 1992;23:222–230. doi: 10.1002/cm.970230306. [DOI] [PubMed] [Google Scholar]
- 23.Fanarraga ML, Carranza G, Castaño R, Nolasco S, Avila J, Zabala JC. Nondenaturing electrophoresis as a tool to investigate tubulin complexes. Methods Cell Biol. 2010;95:59–75. doi: 10.1016/S0091-679X(10)95005-X. [DOI] [PubMed] [Google Scholar]
- 24.Lajoie-Mazenc I, Tollon Y, Detraves C, Julian M, Moisand A, Gueth-Hallonet C, Debec A, Salles-Passador I, Puget A, Mazarguil H. Recruitment of antigenic gamma-tubulin during mitosis in animal cells: presence of gamma-tubulin in the mitotic spindle. J Cell Sci. 1994;107:2825–2837. doi: 10.1242/jcs.107.10.2825. [DOI] [PubMed] [Google Scholar]
- 25.Avila J, Soares H, Fanarraga ML, Zabala JC. Isolation of microtubules and microtubule proteins. Curr Protoc Cell Biol. 2008;39:3.29.1–3.29.28. doi: 10.1002/0471143030.cb0329s39. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Finley J, Liu ZJ, Qiu SH, Chen H, Luan CH, Carson M, Tsao J, Johnson D, Lin G, Zhao J, Thomas W, Nagy LA, Sha B, DeLucas LJ, Wang BC, Luo M. Crystal structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain. J Biol Chem. 2002;277:48596–48601. doi: 10.1074/jbc.M208512200. [DOI] [PubMed] [Google Scholar]
- 27.Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, Steinmetz MO. Structure-function relationship of CAP-Gly domains. Nat Struct Mol Biol. 2007;14:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 28.Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, Steinmetz MO. Key interaction modes of dynamic +TIP networks. Mol Cell. 2006;23:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Kaur H, Garg A, Raghava GP. PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept Lett. 2007;14:626–630. doi: 10.2174/092986607781483859. [DOI] [PubMed] [Google Scholar]
- 30.Steinmetz MO, Akhmanova A. Capturing protein tails by CAP-Gly domains. Trends Biochem Sci. 2008;33:535–545. doi: 10.1016/j.tibs.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Mishima M, Maesaki R, Kasa M, Watanabe T, Fukata M, Kaibuchi K, Hakoshima T. Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proc Natl Acad Sci U S A. 2007;104:10346–10351. doi: 10.1073/pnas.0703876104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borgstahl GE. How to use dynamic light scattering to improve the likelihood of growing macromolecular crystals. Methods Mol Biol. 2007;363:109–129. doi: 10.1007/978-1-59745-209-0_6. [DOI] [PubMed] [Google Scholar]
- 33.Bloodgood RA. From central to rudimentary to primary: the history of an underappreciated organelle whose time has come. The primary cilium. Methods Cell Biol. 2009;94:3–52. doi: 10.1016/S0091-679X(08)94001-2. [DOI] [PubMed] [Google Scholar]
- 34.Muñoz IG, Yébenes H, Zhou M, Mesa P, Serna M, Park AY, Bragado-Nilsson E, Beloso A, de Cárcer G, Malumbres M, Robinson CV, Valpuesta JM, Montoya G. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol. 2011;18:14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Fanarraga M, Carranza G, Bellido J, Kortazar D, Villegas JC, Zabala JC. Tubulin cofactor B plays a role in the neuronal growth cone. J Neurochem. 2007;100:1680–1687. doi: 10.1111/j.1471-4159.2006.04328.x. [DOI] [PubMed] [Google Scholar]
- 36.Voloshin O, Gocheva Y, Gutnick M, Movshovich N, Bakhrat A, Baranes-Bachar K, Bar-Zvi D, Parvari R, Gheber L, Raveh D. Tubulin chaperone E binds microtubules and proteasomes and protects against misfolded protein stress. Cell Mol Life Sci. 2010;67:2025–2038. doi: 10.1007/s00018-010-0308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bivi N, Romanello M, Harrison R, Clarke I, Hoyle DC, Moro L, Ortolani F, Bonetti A, Quadrifoglio F, Tell G, Delneri D. Identification of secondary targets of N-containing bisphosphonates in mammalian cells via parallel competition analysis of the barcoded yeast deletion collection. Genome Biol. 2009;10:R93. doi: 10.1186/gb-2009-10-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell. 2005;19:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 40.Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, Akhmanova A. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol Biol Cell. 2005;16:5334–5345. doi: 10.1091/mbc.E05-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buey RM, Mohan R, Leslie K, Walzthoeni T, Missimer JH, Menzel A, Bjelic S, Bargsten K, Grigoriev I, Smal I, Meijering E, Aebersold R, Akhmanova A, Steinmetz MO. Insights into EB1 structure and the role of its C-terminal domain for discriminating microtubule tips from the lattice. Mol Biol Cell. 2011;22:2912–2923. doi: 10.1091/mbc.E11-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akhmanova A, Steinmetz MO. Microtubule end binding: EBs sense the guanine nucleotide state. Curr Biol. 2011;21:R283–R285. doi: 10.1016/j.cub.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 43.Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.