Abstract
Understanding how thousands of different neuronal types are generated in the CNS constitutes a major challenge for developmental neurobiologists and is a prerequisite before considering cell or gene therapies of nervous lesions or pathologies. During embryonic development, spinal motor neurons (MNs) segregate into distinct subpopulations that display specific characteristics and properties including molecular identity, migration pattern, allocation to specific motor columns, and innervation of defined target. Because of the facility to correlate these different characteristics, the diversification of spinal MNs has become the model of choice for studying the molecular and cellular mechanisms underlying the generation of multiple neuronal populations in the developing CNS. Therefore, how spinal motor neuron subpopulations are produced during development has been extensively studied during the last two decades. In this review article, we will provide a comprehensive overview of the genetic and molecular mechanisms that contribute to the diversification of spinal MNs.
Keywords: Motor neurons, Spinal cord, Neuronal diversification, Cell differentiation, Embryonic development, Central nervous system
Introduction
“The neurons that constitute the motor cranial nerves and the spinal motor neurons (MNs) are the most important cells of the body” (anonymous). Although this provocative statement may at first glance sound severely overstated, a deeper analysis may eventually lead to reconsider this initial impression. Indeed, one may reasonably estimate that MNs constitute the single output of the CNS. Accordingly, whatever you think, dream, imagine, or want, and however smart or creative you are or particularly well connected your neuronal circuitries are, as long as you do not produce any action or any body movement, one may consider that the activity of your CNS will neither support your own survival nor impact on your environment and is therefore totally useless. Hence, MNs constitute a cell population of critical importance for animal and human life.
How these cells of paramount importance are generated during development has been extensively studied during the last two decades. In particular, the differentiation of spinal MN has become the model of choice for studying the mechanisms that promote the generation of different neuronal populations throughout the CNS. Indeed, a single well-defined progenitor domain generates in a restricted amount of time a collection of a few dozen MN subsets, each characterized by specific molecular markers, a defined location within a specific MN column along the anteroposterior axis of the developing spinal cord and, most importantly, a connectivity pattern with specific target muscles that has been exquisitely, although incompletely, deciphered. Recent articles have provided excellent overviews of MN diversity, survival, positioning, axon pathfinding, and connectivity [1–7]. The goal of the present review is to comprehensively summarize past and recent advances in the understanding of genetic and molecular mechanisms that contribute to the diversification of spinal MNs.
Spinal MN diversity
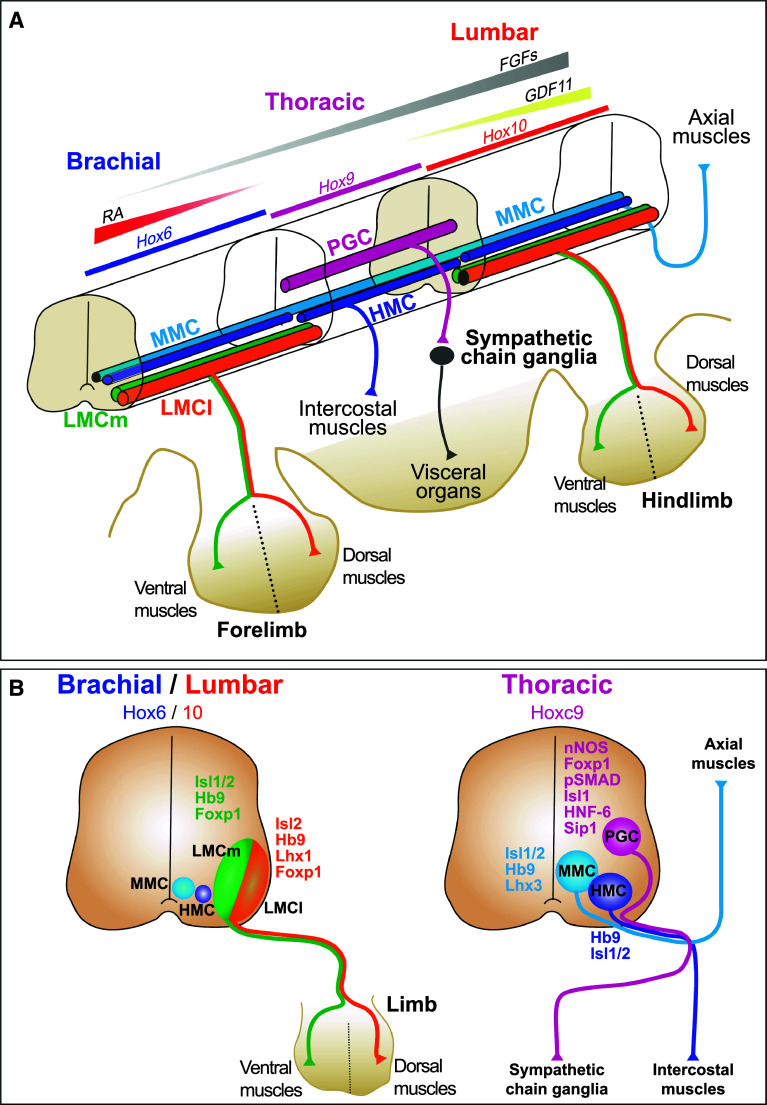
Motor neurons are efferent neurons, i.e., characterized by axonal projection leaving the CNS to reach specific targets in the periphery. All the vertebrate spinal MNs share many common features including large cell body size, neurotransmitter phenotypes, exit route from the CNS through ventral exit points, etc. However, they differentiate early in development into diverse subsets with specific characteristics. Multiple criteria can be used to underline spinal MN diversity. The broadest division segregates MNs that innervate the skeletal muscles of the body, namely somatic MNs, from those that regulate the contraction of the smooth muscles of the visceral organs, called visceral MNs. Neurons from each of these divisions gather at different locations within the spinal cord (Fig. 1). Indeed, visceral MNs are found exclusively at thoracic levels of the spinal cord in a medio-lateral column called the pre-ganglionic column (PGC). The neurons of this column innervate the sympathetic ventral chain wherein they make synapses with the noradrenergic neurons of the paravertebral ganglia, hence its name, and regulate cardiac or smooth muscle contractions and exocrine or endocrine gland activity. In contrast, somatic MNs locate in different columns showing variable extent along the anteroposterior axis of the spinal cord according to the set of skeletal muscle they innervate. MNs that control the axial muscles of the back are found in the medial motor column (MMC), which extends along the entire spinal cord. Neurons that innervate the body wall muscles are located in the hypaxial motor column (HMC), which is present at thoracic levels. In addition, evidence for the presence of a small complement of neurons that display a HMC phenotype at brachial and at lumbar levels has been provided [8, 9], although the target muscles innervated by these MNs remain unknown. Finally, MNs that regulate the activity of the locomotive muscles, including limb muscles and some back muscles that contribute to the movements of the pectoral and of the pelvic girdle, are restricted to the lateral motor columns (LMC), which are exclusively found at the levels of the limbs in the brachial or lumbar regions of the spinal cord. Among these, the medial complement of the LMC neurons (LMCm) innervates the muscles generated in the ventral portion of the developing limb buds, mainly flexor muscles, while the lateral complement (LMCl) connects to muscles generated in the dorsal portion of the limb buds, mainly extensor muscles (Fig. 1).
Fig. 1.
Topographic organization and molecular markers of motor columns along the anteroposterior axis of the developing spinal cord. a Schematic diagram illustrating the three-dimensional distribution of the motor columns. Graded levels of the morphogens FGFs, RA, and GDF11 determine spatial expression of Hox paralog groups that shape the spinal cord into a brachial, a thoracic, and a lumbar portion. At thoracic levels, MNs are distributed in a medial motor column (MMC) that innervates the axial muscles, a hypaxial motor column (HMC) connected to the intercostal muscles and a column of preganglionic MNs (PGC) that project to the sympathetic ganglia chain, which innervates visceral organs. At brachial and lumbar levels, MNs gather in an MMC column, a restricted HMC column, and a lateral motor column (LMC) that innervates locomotor muscles. MNs of the LMC are divided into a medial (LMCm) and a lateral (LMCl) complement that innervate ventral or dorsal limb muscles, respectively. b Location of each motor column and their molecular markers on schematic transverse sections in embryonic spinal cord at brachial/lumbar or at thoracic levels. Hox6 are determinants of the brachial MNs and Hox10 are required for lumbar MN fate, while Hoxc9 establishes thoracic MN identity. FGFs fibroblast growth factors, RA retinoic acid, GDF11 growth differentiation factor 11
Additional levels of diversity include the existence of alpha, beta, and gamma MNs based on the type of muscle fibers (fusal, extrafusal, or both) they innervate, and of different subtypes of alpha MNs that can be classified according to the contractile properties of the motor units that they form with target muscle fibers (fast-twitch fatigable, fast-twitch fatigue-resistant, or slow-twitch fatigue-resistant) (reviewed in more detail in [2]). Finally, it has been demonstrated in the brachial and in the lumbar LMC that MNs that innervate the same muscle gather in so-called motor pools and share identical molecular properties including transcription factor expression profile [10–13], axon guidance and adhesion molecules [14], and neurotransmitter receptors [15]. Whether similar motor pools also exist at thoracic levels and innervate single axial or body wall muscles remains to be determined. Hence, MN is a generic term that actually designates many different neuronal types, each able to modulate the contraction of its specific target muscles in a specific manner. This, as well as the tremendous advances in the understanding of the genetic and molecular mechanisms that drive the diversity of this particular neuronal population, made MNs the model of choice for studying neuronal diversification in the developing CNS. Indeed, all the spinal MNs derive from a single progenitor domain (pMN) located in the ventral portion of the ventricular zone along the anteroposterior axis of the spinal cord. In the next sections, we will briefly summarize the mechanisms that drive the generation of newly born MNs before elaborating on the factors that promote the diversification of this neuronal population during embryonic development.
From pMN progenitors to newly born MNs
Extrinsic cues initiate the spatial patterning of the neuroepithelium in the developing spinal cord. Along the ventro-dorsal axis, Sonic Hedgehog (Shh) initially secreted by the notochord and subsequently produced by the cells of the floor plate provide a positional information that is required for the generation of ventral neuronal populations and triggers the subdivision of the neural progenitors into distinct ordered domains [16, 17]. Increasing concentrations of Shh and progressively longer exposures to Shh signaling impose gradual specification as more ventral progenitor populations [18, 19]. Indeed, high and prolonged Shh levels promote the expression of class II homeobox genes coding for Nkx2.2, Nkx6.1, Nkx6.2, or Pax6 in ventral regions of the neural tube while low levels and brief activation of the Shh pathway is permissive for the expression of class I homeobox genes including those coding for Dbx1, Dbx2, or Irx3, which are therefore restricted to more dorsal regions [18–20]. Molecular interplay between these transcriptional repressors ensures the restriction of Nkx2.2 to the ventral-most progenitor cells that will give rise to V3 interneurons, and of Irx3 to more caudal domains that will produce other ventral interneurons, resulting in the creation of a domain expressing Pax6 and Nkx6 genes that is permissive for MN differentiation [21, 22]. In this domain, retinoic acid (RA) triggers the expression of Olig2, the hallmark of MN and oligodendrocyte progenitors. Indeed, spinal oligodendrocytes are generated from the same progenitor domain at later developmental stages [23–25].
Retinoic acid produced by the paraxial mesoderm is required, independently from Shh signaling [26], for neurogenesis in the developing spinal cord [27–29]. Indeed, RA contributes to high expression levels of homeobox genes, including Irx3 and Pax6, and thereby enhances the patterning activity of Shh and contributes to define the pMN domain [28]. In more dorsal regions, it also promotes the generation of V0 and V1 interneuron progenitors and the production of differentiated V1 neurons [26]. Subsequently, RA activates its cognate receptors in the pMN and initiates the expression of Olig2 [28]. Although initially coexpressed with Nkx2.2 or Irx3 in cells that will eventually locate in the adjacent progenitor domains, the expression of Olig2 is rapidly refined through mutual cross-repression with Nkx2.2 or Irx3 to become strictly restricted to the MN progenitor domain [25, 30]. Additional factors, including Wnt signaling, which interacts with the Shh pathway to modulate Nkx2.2 expression [31–33], and the Groucho/transducin-like Enhancer of split proteins [34] also contribute to define the ventral boundary of the pMN domain. In turn, recent evidence demonstrates that the interpretation of the Shh gradient, which is continuously at play during this whole process, relies on a gene regulatory network that involves Pax6, Olig2, and Nkx2.2 [35].
In the pMN domain, Olig2 combined to Shh signaling rapidly stimulates the expression of the pro-neural gene Neurogenin2 (Neurog2) (Fig. 2) [21, 25, 36]. This increase in Neurog2 protein levels will determine the onset of MN differentiation. Indeed, the Olig2/Neurog2 ratio serves as a gate for timing proper gene expression during the development of MN progenitors. High levels of Olig2 maintain these cells in a progenitor state, whereas high levels of Neurog2 favor their conversion into post-mitotic MN [21, 37, 38]. Except for the MN determinant Hb9 (see below), the expression of which is directly repressed by Olig2 [37], the physiological targets of Olig2 and Neurog2 in MN progenitors remain unknown. However, the LIM-HD proteins Lhx3 might be downstream of Olig2 [21]. Consistently, Lhx3 and its homolog Lhx4 are jointly required for the generation of spinal MNs (Fig. 2) [39]. The transition from MN progenitors to differentiating neurons is further promoted by the activation of Foxp2 and Foxp4 that cooperatively repress the expression of N-cadherin and thereby promotes the detachment of differentiating MN from the neuroepithelium [40].
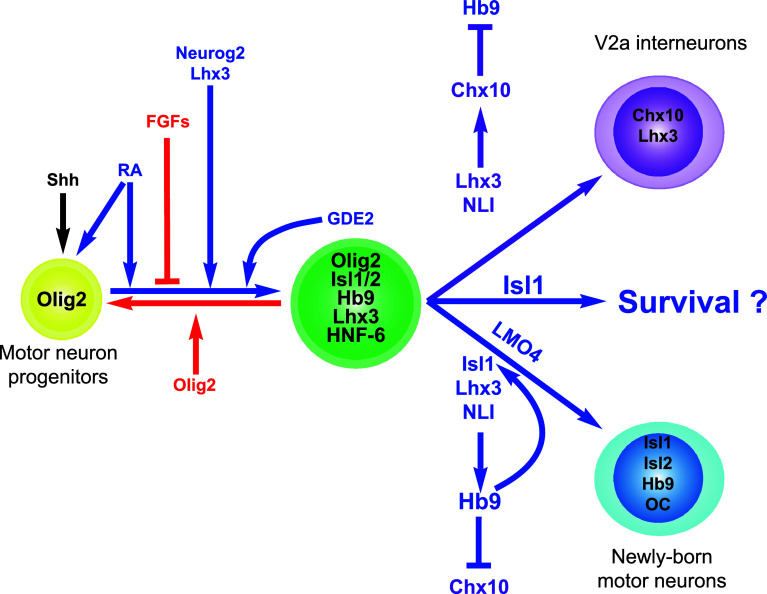
Fig. 2.
MN fate specification and consolidation. A combination of extrinsic signals including Shh, RA, and FGFs cooperate with intrinsic mechanisms that involve Olig2, Neurogenin-2 (Neurog2), and Lhx3 to promote the specification of newly born MN. GDE2 produced by the newly born MN acts in an on-cell autonomous manner to promote MN progenitor differentiation. Lhx3, Isl1, NLI, LMO4, and Hb9 act together to consolidate MN identity. Isl1 may additionally contribute to the survival of newly born MNs. FGFs fibroblast growth factors, RA retinoic acid, Shh Sonic Hedgehog, NLI nuclear Lim-domain interactor, GDE2 glycerophosphodiester phosphodiesterase 2
Epigenetic mechanisms also contribute to the onset of MN differentiation. Indeed, Neurog2 forms with non-activated RA receptors a complex that is recruited on E boxes of MN genes targeted by Neurog2. Upon stimulation by RA, this complex promotes neurogenesis via the recruitment of CBP, a transcription factor with histone acetyltransferase activity, which in turn induces chromatin alterations in MN genes and activation of their transcription. Accordingly, tissue-specific inactivation of CBP, either alone or in combination with that of its paralog gene p300, results in alterations in MN generation [41]. Noticeably, these mutant embryos also showed perturbations in MN migration and axonal projections, suggesting that epigenetic modifications also regulate later aspects of MN development. It should be noted that CBP and p300 are, in liver cells, cofactors of other transcriptional regulators that participate in MN diversification (see below), including Onecut factors [42].
Besides these cell-autonomous processes initially triggered by extrinsic cues, recent observations suggest that non-cell autonomous interactions between MN progenitors and post-mitotic neurons also participate in MN generation. Indeed, in parallel to multiple roles in spinal progenitors (see above), RA stimulates in post-mitotic MNs the expression of the glycerophosphodiester phosphodiesterase GDE2, which turned out to be necessary and sufficient for MN production [43–45]. GDE2 utilizes its extracellular glycerophosphodiester phosphodiesterase activity to induce MN generation by inhibiting, in a non-cell autonomous manner, Notch signaling in neighboring MN progenitors (Fig. 2) [45]. GDE2 activity requires the reduction by the antioxidant scavenger peroxiredoxin1 of an intramolecular disulfide bond that bridges its intracellular N- and C-terminal domains [46] and the formation of a complex with Gαi-GDP, which acts in this context independently from its roles in transducing GPCR signals [44]. Thus, complex genetic and epigenetic processes initiated by extrinsic signals and that eventually give rise to non-cell autonomous crosstalks between differentiating neurons and progenitor cells direct the generation of MNs from the pMN domain.
Anteroposterior patterning of MNs by Hox factors and cofactors
The earlier step of MN diversification involves the activation of different genetic programs along the anteroposterior of the neural tube that eventually results in the formation of distinct motor columns at brachial, thoracic, or lumbar levels of the spinal cord. Indeed, the positional identity of neurons along the anteroposterior axis is established before the first differentiating neurons can be identified in the spinal cord [47], indicating that these characteristics are acquired and maintained by MN progenitors and transmitted to post-mitotic cells. Similar to the ventro-dorsal axis, the spinal cord is patterned along its antero-posterior axis through morphogen gradients and differential distribution of transcription factors that in this case mainly belong to the homeodomain factor family, namely Hox proteins. Although recent and extensive review articles have summarized most of the current knowledge on the action of Hox factors in this process [5, 48], we will briefly overview the main observations and underline the versatile, although somewhat obscure, roles of the Hox cofactors, including Foxp1, in spinal MN diversification. Combinatorial FGF, Wnt, RA, and Gdf11 signals originating from the Hensen’s node and paraxial mesoderm differentially profile the expression of Hox-c and Cdx genes along the anteroposterior axis (Fig. 1a) [47, 49–51]. This indicates that morphogens shape MN diversification from the earlier steps of neural development. Progressive delivery of these extrinsic signals from more anterior to more posterior regions of the neural tube induces a delay in neuronal differentiation between rostral and caudal neurons, including MNs, which remains visible for several days. The regionalization of the expression of Hox genes in response to these extrinsic signals determines the identity of the motor columns along the anteroposterior axis. Indeed, anteriorized expression of Hox-c genes upon induction by FGF8 in progenitor cells results in the conversion of brachial MNs into thoracic MNs [52]. Conversely, progressive inactivation of Hox6 alleles results in gradual erosion of the brachial LMC identity [53]. Similarly, combined inactivation of Hox10 paralog genes results in a posterior shift of the boundary between thoracic and lumbar MNs [54–56] or in an erosion of lumbar LMC identity [8], while ectopic production of Hoxd10 in thoracic segments of chick spinal cord imposes a molecular identity and a connectivity pattern of lumbar LMC neurons [57]. Even more strikingly, inactivation of the single Hoxc9 gene results in the transformation of most of the thoracic MNs (HMC and PGC) into neurons with the molecular identity of limb-innervating MN (LMC), and Hoxc9 is sufficient to repress brachial Hox genes and LMC identity [58]. Oppositely, ectopic expression of these brachial Hox genes at thoracic levels promotes LMC neuron differentiation at the expense of PGC cells [53]. This further indicates that the activity of Hox proteins in patterning the spinal motor columns also relies on mutual cross-repressive interactions [52, 56, 58]. Recent data provide evidence that these interactions establish the caudal boundary of Hox expression domains, whereas Polycomb repressive complex 1 determines their anterior boundary through chromatin modifications [59]. Thus, the constellation of Hox genes and their cross-regulations establish the diversity of MNs in the different segments of the spinal cord. Whether this activity also extends to other spinal populations wherein Hox proteins are present, namely ventral or dorsal interneurons, remain to be investigated. The mechanisms that coordinate the MN anteroposterior patterning activity of the Hox proteins and the action of the proneural factors that stimulate the cell-cycle exit of the MN progenitors and the initiation of MN differentiation remains currently unknown. Of note, Hox and proneural factors synergize in Drosophila to stimulate the differentiation of sensory organs [60].
Hox proteins are known to require interactions with several factors to exert their activity. Different Hox cofactors have been identified in vertebrates, including members of the PBC and MEIS classes of TALE (Three Amino acid Loop Extension) homeodomain proteins and the Forkhead box p1 protein (Foxp1). The PBC class includes vertebrate Pbx homeoproteins, whereas MEIS factors include Meis and Prep homeoproteins. Pbx and Meis/Prep cofactors are able to form trimeric complexes with Hox proteins. Although initially thought to interact through the conserved hexapeptide motif of Hox factors [61–64], multiple evidence suggests that PBC and MEIS factors bind to different regions of the Hox proteins [65, 66]. Interactions with Pbx and Meis/Prep are required for Hox transcriptional activity. Nevertheless, Pbx appears to act not only as Hox cofactors but also independently from Hox proteins to directly regulate distinct target genes and even some HoxA/D genes (reviewed in [63, 64]). Although they are broadly present in the developing spinal cord, the distribution of these Hox cofactors in differentiating MNs has not yet been characterized in detail [67, 68]. Whereas Pbx and Prep proteins regulate different aspects of hindbrain MN development [69, 70] and Hox/Pbx binding sites are present and required for the transcriptional activation of a 125-bp enhancer of the MN determinant gene Hb9 [71], the roles of TALE cofactors in spinal MN development remain elusive [68] and deserve further investigation. Noticeably, recent data demonstrate that Hoxc6-Pbx interaction is required for Hoxc6 to promote brachial LMC characteristics but not for the repression of the thoracic determinant Hoxc9 gene [53], suggesting that only part of the Hox activity in MN development may rely on TALE proteins.
In contrast, the function of Foxp1 in MNs has been deeply characterized [8, 72, 73]. Distinct expression levels of Foxp1 are measured in PGC and in LMC columns, and Hox proteins are required for this expression [72]. Loss- and gain-of function experiments for Foxp1 in mouse and chick embryos suggest that these different levels in Foxp1 are in turn necessary for proper specification of the corresponding motor columns (see below). However, these data do not implicate Foxp1 per se in the anteroposterior patterning of the MN columns [8, 72]. Similarly, sustained Hox5 activity is required for proper development of the phrenic MNs, respiratory neurons that are generated in the rostral portion of the brachial region and provide the sole source of innervation to the diaphragm, without contribution of Foxp1, which is not present in these cells [74]. Thus, the exact requirement for cofactors by each Hox protein during MN development remains to be fully elucidated.
Consolidation of MN identity
Following anteroposterior patterning by the Hox proteins and activation of the proneural factors, MN progenitors exit the cell cycle and initiate neuronal differentiation, as evidenced by activation of the expression of early MN markers including Isl1, Hb9, and Hnf-6. However, the acquisition of this early MN identity is not persistent. Indeed, at variance to what is known for the other neuronal populations of the spinal cord, postmitotic MNs require early consolidation of their identity to maintain their fate. Defective consolidation of MN identity results in a partial or complete transformation of the MN population into dorsally adjacent V2a interneurons [75–78]. This is likely due to the fact that MNs and V2a interneurons share some of their developmental determinants, including Nkx6 and Lhx3 factors [79–83].
The molecular mechanisms involved in the consolidation of MN identity have been exquisitely deciphered (Fig. 2) [77, 79, 81, 83]. In the V2 domain, Lhx3 forms a heteromeric binary complex with the nuclear LIM interactor protein NLI (Ldb, Clim), and the activity of this complex promotes V2a identity and directly stimulates the expression of V2a markers including Chx10 [81]. This complex also prevents the activation of the MN fate-consolidating factor Hb9 [79]. In contrast, the presence of Isl1 and Lhx3 in newly born MNs allows the formation of a heteromeric ternary complex with NLI [81, 83]. This homeodomain complex synergizes with the proneural bHLH factors present in MNs, including the phosphorylated form of Neurog2 [36], to stimulate the expression of genes such as Hb9 [36, 79, 81]. In turn, Hb9 prevents the activation of Chx10 while LMO4 inhibits the formation of the V2a-specific Lhx3-NLI complex [77, 79]. Accordingly, inactivation of Hb9 results in defective consolidation of MN identity and partial conversion into V2a interneurons [75, 78, 84]. Although expressed later in the course of MN differentiation, Runx1 seems to participate in MN fate consolidation by repressing the V2 interneuron differentiation program [85], suggesting a continuous requirement for fate consolidation to maintain MN identity.
In the early phases of MN differentiation, Hb9 indirectly stimulates Isl1 expression [75, 78, 86, 87]. In turn, Isl1 supports Hb9 production, establishing a regulatory loop wherein Isl1 and Hb9 mutually support their expression [77, 79]. Proper control of the levels of Isl proteins is absolutely crucial to ensure consolidation of MN fate. Indeed, analyses of mice carrying a hypomorphic allele or a conditional inactivation of Isl1 demonstrated that progressively reducing the levels of Isl1 favors the differentiation of V2a interneurons at the expense of MN production [76, 77]. Consistently, constitutive inactivation of Isl1 resulted in the complete absence of MNs at very early stages of spinal cord development [88]. This observation was attributed to increased MN cell death, pointing to a possible requirement in Isl1 for newly born MN survival (Fig. 2). However, the generation of V2a interneurons was not investigated in these mice, hindering to formally exclude that defective consolidation of MN identity also contributed to the loss of MNs in these embryos. Although the onset of its production starts slightly later than that of Isl1, it seems that its paralog protein Isl2 also participates in MN fate consolidation process. Indeed, experiments in zebrafish suggest that Isl1 and Isl2 have equivalent abilities to promote MN formation [89]. Furthermore, constitutive inactivation of Isl2 in mouse increases the deficit in MN production that results from decreased Isl1 expression [77]. Therefore, it has been proposed that the global level of Isl proteins, rather than a specific function attributed to each Isl factor, is critical for the consolidation of MN identity as well as for other Isl-dependent processes involved in MN development [89–91]. Tbx20 may also contribute to MN consolidation by supporting in newly born MN the expression of Isl2 and of Hb9 [92].
Early loss of MNs in constitutive Isl1 mutant mice and the lack of a suitable model to efficiently inactivate Isl1 at different stages of MN differentiation prevented the identification of the time window, wherein this factor is required for the consolidation of MN identity and of later roles of Isl1 in MN development. However, recent work describing the consequences of the loss-of-function of the transcription factors of the Onecut family on MN development shed light on these questions. Onecut factors, namely Hepatocyte Nuclear Factor-6 (HNF-6), Onecut-2 (OC-2) and OC-3, are transcriptional activators present in liver, pancreas, and in the CNS during embryonic development [93–95]. These factors have been shown to regulate cell differentiation in the pancreas [96–99] and in the liver [100–102], and to control various aspects of neuronal development [103–107]. In MNs, the three OC factors are dynamically and differentially expressed in each motor column in the course of neuronal differentiation [108]. In particular, the production of HNF-6 is initiated in an “intermediate” cell population wherein Olig2 can still be detected but the early MN markers including Isl1 and Hb9 are already present, therefore corresponding to a transition state between MN progenitors and newly born MNs (Fig. 2) [9, 108]. Recently, it has been shown that OC factors directly contribute to stimulate the expression of Isl1 during MN differentiation [9] by binding the CREST2 enhancer, which drives Isl1 production in spinal MNs [109]. In the combined absence of OC factors, the expression of Isl1 is properly initiated in the “intermediate” population but sharply decreases in newly born and more differentiated MNs, indicating that OC factors are required to maintain Isl1 expression in differentiating MNs. However, the amount of MNs produced in these mutant embryos was normal, as well as the amount of V2a interneurons, and no evidence of defective consolidation of the MN identity could be found. This suggests that the requirement in Isl1 for the consolidation of MN fate is restricted to the time window, wherein Isl1 is present in the “intermediate” MN population [9].
Timing of MN diversification
After initial subdivision upon the action of the Hox factors according to the segmental level they occupy along the anteroposterior axis and consolidation of their MN fate, newly born MNs undergo a diversification process that results in the production of cells displaying distinct molecular identities, migration pathways, axonal trajectories, and target connectivity located in the different motor columns and pools. Although multiple intrinsic and extrinsic mechanisms that tightly control this process have been characterized as discussed below, the timing and the chronology of the events that segregate different MN subsets at thoracic levels of the spinal cord remains unclear.
It was initially anticipated that the segregation between somatic and visceral MN occurs before the separation between MMC and HMC neurons [75, 91]. However, BrdU incorporation experiments [110] and manipulations of the expression of motor column determinants [86] rather suggest that each subdivision is produced at the same developmental stage. Consistent with this second hypothesis, conversion of PGC neurons into HMC cells in the absence of Foxp1 [8, 72] suggests that the somatic and visceral lineages do not diverge earlier than MMC and HMC cells. In contrast, increased production of visceral MNs at the expense of somatic MNs in OC compound mutant embryos without either reduction in the total amount of MNs or modification of the MMC/HMC ratio [9], and the conversion of MMC into HMC neurons in Wnt4/5a/5b compound mutants without alteration of PGC cell number [111], rather favors the hypothesis of an initial segregation of visceral vs. somatic MN lineages followed by a subsequent separation of the MMC or HMC fates. Taking all these observations together, one may propose that the diversification of MNs in the thoracic portion of the spinal cord rely on intrinsic or extrinsic signals delivered to postmitotic cells that retain, at least until embryonic day 10.5, enough plasticity to change their fate according to these signals. However, GDE2 modulates in a non-cell autonomous manner the differentiation potential of MN progenitors to regulate the amount of neurons in specific motor columns, namely HMC and LMC, but not in others [45]. This suggests that part of the program that determines the columnar fate of MNs, including the segregation between somatic or visceral identity, is defined at very early stages of MN development, possibly in MN progenitors. Epigenetic mechanisms activated in progenitor cells in response to the combination of intrinsic and extrinsic signals may transmit such information from MN progenitors to differentiated cells [41]. Indeed, histone H3K4 methylation is associated with active genes and, along with H3K27 methylation, is part of a bivalent chromatin mark that typifies poised developmental genes. Histone methyltransferases and histone demethylases contribute to maintain this poised state, and their inactivation alters the differentiation of progenitor cells along the neural lineage [112, 113]. Hence, these epigenetic signatures could combine to intrinsic and extrinsic cues targeting newly born MNs to eventually and permanently define the molecular identity and the columnar fate of the mature cells. Migration and axonal projection defects of differentiated MNs due to single or combined inactivation of the histone acetyltransferases CBP and p300 are consistent with an involvement of epigenetic mechanisms in the regulation of MN diversification [41].
Segregation of visceral vs. somatic MN fate
One of the main divisions among spinal MNs separates somatic MNs that innervate skeletal muscles from visceral MNs that regulate the contraction of smooth muscle cells through a synaptic relay with the sympathetic neurons of the paravertebral ganglia. Different intrinsic and extrinsic factors have been involved in the separation between these two MN subsets. However, how the action of these different actors is coordinated remained elusive for a long time. The recent description of the roles of the OC factors in this process enabled to propose a comprehensive model that integrates the described functions of all these interveners (Fig. 3) [9].
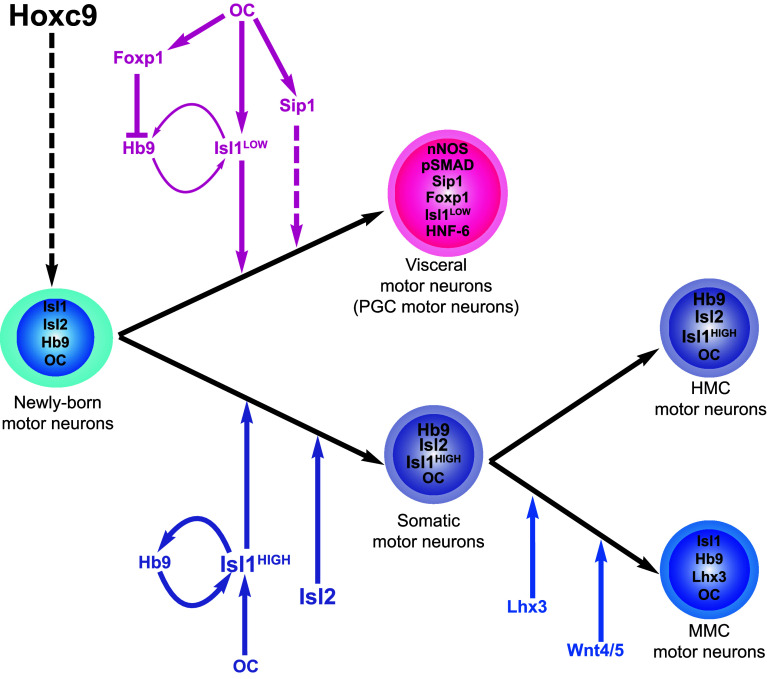
Fig. 3.
MN diversification at thoracic levels of the spinal cord. Hoxc9 establishes thoracic MN identity. The differentiation into somatic or visceral MN is finely tuned by the combined action of OC, Foxp1, Hb9, and Isl proteins. In some newly born MNs, OC factors and a mutual stimulatory loop between Hb9 and Isl1 triggers high Isl protein levels and somatic MN differentiation. In other newly born MNs, OC stimulate Foxp1 expression, which represses Hb9 and thereby weakens the mutual stimulatory loop between Hb9 and Isl1 and reduces Isl levels, which favors visceral MN differentiation. OC also promote Sip1 expression, which supports the generation of PGC cells. Among somatic MNs, Lhx3 and activation of Wnt signaling pathway promote the differentiation of MMC neurons. The HMC fate may constitute the default differentiation fate. nNOS neuronal nitric oxide synthase
Besides their early role in MN identity consolidation, Isl factors participate in the generation of visceral MNs. Indeed, constitutive inactivation of Isl2 results in an expansion of the somatic MNs at the expense of visceral MNs at the thoracic levels of the spinal cord [91], suggesting that Isl2 promotes the differentiation of PGC neurons. As Isl1 and Isl2 seem to share equivalent abilities to promote MN formation [77, 89], the role of Isl1 in this process was also investigated. However, given that Isl1 constitutive mutant embryos die before the time when visceral MN can be detected, the authors relied on the observation that Isl1 expression is severely downregulated in the absence of Hb9 [75, 78] to evaluate the contribution of Isl1 for visceral MN production. The visceral MN differentiation defects observed in the absence of Isl2 were indeed enhanced when Isl1 levels were additionally reduced [91], leading to the conclusion that both Isl factors promote the production of visceral MN. However, this interpretation did not integrate a possible role for Hb9 in this process. Given that it was demonstrated later that Hb9 and Isl proteins contribute to the consolidation of MN identity (see above), alternative interpretations have been proposed for these observations [8, 9]. In addition, the Hox cofactor Foxp1 was also demonstrated to be essential for visceral MN production. Indeed, PGC neurons contain intermediate levels of Foxp1 while this factor is absent from the somatic MN of the MMC and HMC [72]. Ectopic production of intermediate levels of Foxp1, either by overexpression of by inhibition of its negative regulator miR-9, results in the expansion of the PGC neurons at the expense of their somatic counterparts, whereas Foxp1 inactivation or miR-9 overexpression leads to a reduction in visceral MN associated with a specific expansion of the HMC neurons [8, 72, 114]. Although Isl factors and Foxp1 very obviously participate in the segregation between somatic and visceral MN, the relationship between their respective actions remained unclear.
The recent publication of the phenotypic analysis of OC compound mutant embryos enabled to propose a model that fills this gap (Fig. 3) [9]. As previously mentioned, the absence of OC factors results in a severe reduction in the Isl1 expression levels in newly born MNs. Although the expression of Isl2 is not affected, this results in a strong reduction in the global levels of Isl proteins in differentiating MNs. Surprisingly, these perturbations correlate with an increased production of visceral MNs at the expense of their somatic counterpart, arguing against the hypothesis that Isl factors promote visceral MN differentiation. In addition, Isl1 protein levels in wild-type spinal cord are twice as high in prospective somatic MN than in their visceral complement, suggesting that high content in Isl proteins rather favors somatic MN fate whereas low levels would promote visceral MN production (Fig. 3). Consistently, Isl2 overexpression (Fig. S2 in [91]) or stimulation of Isl1 expression by overexpressing Hb9 or the OC factor Hnf6 in chick embryonic spinal cord decrease the amount of visceral MNs in favor of somatic MNs [9, 86, 87, 91]. Therefore, the reduction in visceral MNs observed in the absence of Hb9 and/or Isl factors likely results from defective consolidation of MN identity rather than from a specific contribution of these factors to MN diversification [75, 77–79, 81, 91]. Indeed, MN consolidation defects result in increased production of Lhx3 [75–78], which represses the expression of Foxp1 [8, 72] and visceral MN differentiation [86, 91]. Hence, high contents in Hb9 and Isl proteins would first consolidate MN identity and later promote somatic MN differentiation. In prospective visceral MNs, the presence of Foxp1, which is supported in these cells by OC factors [9], represses the expression of Hb9 [8, 72]. This likely contributes to weaken the mutual stimulatory loop between Hb9 and Isl1, tuning global Isl protein content to intermediate levels that are sufficient for visceral MN differentiation. Therefore, Hb9, Foxp1, Isl, and OC factors cooperate in newly born thoracic MNs to finely tune the differentiation balance between somatic and visceral MNs.
Intriguingly, the ligand BMP5 [86] and the activated form of the BMP signaling intermediates Smad1/5/8 [72] are specifically detected in the PGC neurons from the onset of their differentiation. Moreover, Smad-interacting protein 1 (Sip1, also called Zfhx1b or Zeb2) has recently been found in visceral MNs [9]. Sip1 is able to inhibit transcriptional activity of activated Smad proteins [115] but also to cooperate with the BMP-activated Smad8 to stimulate transcription [115, 116]. Conditional inactivation of Sip1 in differentiating MNs results in a reduction in the amount of PGC neurons, suggesting that Sip1 promotes visceral MN differentiation [9]. Whether Sip1 activity during MN differentiation relies on modulating BMP signaling and whether BMP5 or activation of the BMP pathway also promote PGC neuron development, require further investigations.
Recent data indicate that neonate visceral MNs can be subdivided according to their membrane properties and that these subdivisions may correspond to functionally distinct neuronal subsets [117]. Whether Foxp1, activated Smad1/5/8, Sip1, OC, or Isl factors, all of which are present in subsets of PGC neurons, contribute to the generation of PGC cell diversity during development remains to be establish. Similarly, Reelin [118–120] and Cdk5 [121] control some aspects of the migration and of the final location of the visceral MNs, but whether this additionally contributes to PGC neuron diversification is currently unknown.
Segregation of MMC vs. HMC neurons
Besides high levels of Isl proteins, Lhx3 is a crucial determinant of somatic MN identity (Fig. 3). When overexpressed in the caudal hindbrain, it is able to convert MN with axonal projection that exit the hindbrain dorsally into MN with characteristics of spinal somatic MN including ventral axonal projections [39]. More specifically, Lhx3 is sufficient to determine the identity of MMC neurons. Indeed, maintained expression of Lhx3 in all the differentiating MN suppresses the production of HMC, PGC, and LMC neurons and gates MN differentiation exclusively to an MMC fate [86, 122, 123]. Surprisingly, although Lhx3 and Foxp1 mutually cross-repress their expression [8, 72], the loss of Lhx3 did not result in the expansion of a Foxp1-dependant motor column (PGC or LMC) [39]. Oppositely, the absence of Foxp1 at thoracic levels does not result in an increase in MMC neurons but in expansion of the HMC column at the expense of PGC cells [8, 72]. This raises the possibility that the HMC neuron identity may constitute a default pathway intrinsic to newly born MN that only requires high levels of Isl proteins to be achieved [8].
This also raises the question of the instructive factors upstream of Lhx3 that promote MMC cell fate. Although Lhx3 is transiently produced in all the MN at the transition between progenitors and differentiating cell, it is exclusively maintained in MMC cells at later developmental stages [39]. Wnt signaling seems to participate in this process [111]. Indeed, Wnt4, Wnt5a, and Wnt5b are present in the developing ventral spinal cord and cumulative analysis of their expression pattern suggests the existence of a ventralHIGH to dorsalLOW gradient of Wnt signaling at the time of MN differentiation. Wnt4 and Wnt5 overexpression favors the production of MMC neurons at the expense of their HMC counterpart, whereas progressive accumulation of Wnt4 and Wnt5 mutant alleles results in a progressive decreased production of MMC neurons in favor of HMC cells, without any change in PGC neurons or in the total amount of MN. Accordingly, analyzing Nkx2.2 mutants, where the V3 interneuron progenitor domain is converted into pMN, or chick embryonic spinal cord electroporated with a mutant isoform of the Shh receptor Smoothened that induces the conversion of the V2 interneuron progenitor domain into pMN, it was elegantly shown that ectopic ventral MN differentiate into MMC cells whereas ectopic dorsal MN rather correspond to HMC and PGC cells (Fig. 3). The role of Wnt in MN diversification does not seem to rely on the activation of the Wnt planar cell polarity pathway. Indeed, loop tail mice, which carry a null mutation in the Vangl2/Ltap gene that is essential for the transduction of this pathway, display a normal amount of MN and proportional allocation of MMC neurons [111]. Alternatively, the MMC-promoting role of the Wnt pathway might rely on a direct stimulation of Lhx3 expression by Wnt ligands. Indeed, Wnt is able to activate Lhx3 in neuroendocrine cells of Ciona savignyi embryos [124], although similar regulation in mammalian neural cells has not been demonstrated yet. Interestingly, Wnt signaling is also implicated in somatic MN fate decision in C. elegans [125].
Control of LMC neuron generation and diversification
The production and diversification of the LMC neurons have been investigated in great detail (Fig. 4). Indeed, the in-depth characterization of numerous motor pools and the exquisite selectivity of each of them for a specific appendicular muscle enabled to address the whole process of neuronal development, from specification to synaptogenesis, in this single well-defined model. In the framework of this review article, we will only briefly emphasize the mechanisms that contribute to the diversity of the LMC neurons and of the motor pools. We refer to more specific reviews for a complete overview of LMC cell axonal growth and connectivity [2, 4–6].
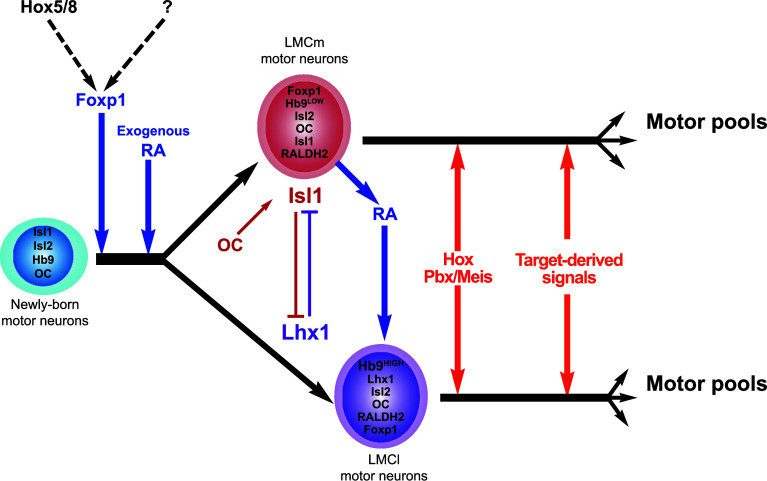
Fig. 4.
MN diversification at limb levels. Exogenous RA from the paraxial mesoderm induces the generation of LMC neurons with LMCm identity. These cells rapidly initiate Raldh2 expression and RA synthesis, which promotes the generation of a later LMC population with LMCl identity. Hox proteins, possibly generic Hox5-8 paralogs, are necessary for high Foxp1 levels in the brachial LMC. Cross-repressive interactions between Isl1 and Lhx1, respectively, expressed by LMCm and LMCl neurons, stabilize this segregation and determine settling position and axonal projection of MNs in each division. Upon the influence of intrinsic factors, including Hox proteins and their cofactors, and of target-derived signals, subdivision of LMC neurons proceeds further and results in the formation of motor pools that individually innervate single target muscles. RALDH2 retinaldehyde dehydrogenase 2, RA retinoic acid
As they specifically innervate limb and some back muscles involved in locomotion, including for example the cutaneous maximus and the latissimus dorsi [13], LMC neurons are generated in register with the embryonic limb fields. RA is the major extrinsic determinant that promotes LMC cell production at brachial level of the developing spinal cord. During embryonic development, RA is mainly synthesized by retinaldehyde dehydrogenase 2 (RALDH2) [126–129]. RALDH2 is dynamically expressed within the paraxial mesoderm directly adjacent to the spinal cord. Initially, it is found at all axial levels, but at later stages, higher levels are observed in forelimb and in hindlimb regions [127, 128, 130, 131]. Blockade of RA signaling in newly born MNs prevents brachial LMC differentiation and rather favors the production of cells displaying the characteristics of thoracic MN. Noticeably, lumbar MN do not show a similar dependence to RA, suggesting that other extrinsic factors control their production [132].
At both levels, a first wave of LMC neuron differentiation generates the prospective LMCm cells that will eventually innervate the ventral complement of the appendicular musculature [132, 133]. Interestingly, these MNs rapidly activate the expression of Raldh2 and become themselves a source of RA [134]. Subsequently, the combination of RA produced by early LMC cells [131–134] and by the nearby paraxial mesoderm [133] is necessary for a second wave of LMC neuron production that will eventually constitute the LMCl subdivision, which innervates the dorsal complement of appendicular muscles [131–134]. Interestingly, the firstly generated LMC cells do not need to acquire the specific identity of LMCm neurons for initiating Raldh2 expression and RA production. Indeed, OC compound mutant embryos lack LMCm neurons but generate a normal amount of RALDH2-producing LMC cells that all display LMCl characteristics, including molecular identity and axonal projections [9]. This indicates that the ability to express Raldh2 and to produce RA is a general property of LMC neurons and does not require proper columnar subdivision. Finally, RA produced by differentiated LMCm and LMCl neurons is necessary for maintenance of these cells, as MN-specific inactivation of Raldh2 results in a global loss of LMC neurons at brachial and at lumbar levels of the spinal cord [133]. Hox factors are also involved in this maintenance process, as the expression of the RARβ receptors in the LMC requires the presence of Hoxc8 [131].
Hence, RA signaling from the paraxial mesoderm and from the LMC neurons contributes to successively generate distinct medial and lateral complements. However, additional mechanisms are required to consolidate this initial columnar division and to provide each of these complements with their own characteristics. Soon after their generation upon induction by RA, the early LMC cells initiate Isl1 expression [135]. In these cells, Isl1 represses the expression of Lhx1 and determines their LMCm identity by directing their settling position in the medial portion of the LMC [136] and stimulating the expression of EphB1, which orients their axons towards the ventral mesenchyme of the limb buds [137]. In contrast, LMC neurons that are generated subsequently initiate the expression of Lhx1 [135]. In these cells, Lhx1 represses the expression of Isl1 and imposes specific characteristics of LMCl cells including their settling position into the lateral portion of the LMC and the expression of EphA4 and of the GDNF receptor Ret, which orient the axons towards the dorsal mesenchyme of the limb buds [136, 138–140]. Thus, the cross-repressive interaction between Isl1 and Lhx1 consolidates the columnar subdivision of the LMC initiated by RA. OC factors are required in the LMCm for the production of Isl1. In OC mutant embryos, LMC cells amount normally but lack Isl1 expression and therefore adopt an LMCl identity [9]. As mentioned above, Isl1 and Lhx1 coordinate multiple aspects of LMC neurons identity, including settling position and axonal projections. However, it seems that distinct downstream mechanisms control these two characteristics. Indeed, Foxp1 and Lhx1 cooperate in LMCl cells to maintain high levels of the Reelin signaling intermediate Dab1, which contributes to the lateral position of these cells within the LMC but does not impact on their axonal projection [73]. Surprisingly, expansion of the LMCl observed in OC mutant embryos did not result in a lateral displacement of the ectopic LMCl neurons [9]. This suggests that unknown effectors may intervene downstream of OC factors in this process.
Further diversification of LMC neurons divides this population into motor pools [141, 142] where each innervates a single appendicular muscle [11]. Different mechanisms contribute to the specification and to the maintenance of the motor pools, including cell-autonomous processes and target-derived extrinsic signals. Hox proteins specify the identity of distinct motor pools along the anteroposterior axis of the LMC and additionally provide an intrasegmental coding for motor pool diversity. Alterations of the Hox code result in conversions of motor pool identity and coordinated redirection of their axons towards alternative muscle target [10, 53]. Downstream of Hox factors [52], a post-mitotic period of Nkx6.1 activity is required for the specification of the motor pools that innervate the gracilis posterior and the adductor muscles independently from target-derived signals [11]. Similarly, PEA3 is necessary for the development of the motor pools that target the cutaneous maximus, the latissimus dorsi and the pectoralis minor muscles [13, 143]. However, activation of PEA3 in these motor pools and proper specification of their neuronal identity require in MNs the activation of GDNF signaling by muscle-derived GDNF [144]. Subsequently, HGF signaling through its receptor Met is necessary in a non-cell autonomous manner for the expansion of Pea3 expression to more anterior LMC populations [145]. Hence, the diversification of LMC neurons into motor pools is ensured by the integration of cell-autonomous and non-cell autonomous mechanisms. Although the mechanisms whereby subsets of LMC neurons that display the same molecular identity eventually gather into motor pools remain partly elusive, type II cadherins and semaphorins are specifically expressed in defined LMC pools and participate in their segregation [13, 14, 146].
Conclusions and perspectives
The quest of spinal MNs for their proper molecular identity requires passing numerous hurdles and integrating at each step intrinsic determinants with extrinsic cues produced by the nearby or more distant environment. As MNs constitute the model of choice for studying the mechanisms that promote neuronal diversification during embryonic development, numerous molecular and cellular mechanisms involved in their differentiation have been identified. However, several aspects related to the diversification of this neuronal population into multiple subsets, the control of their migration and of their final location within the ventral spinal cord, and the exquisite manner their axon is able to find its way toward its specific muscle or sympathetic ganglion target request further investigations. In particular, the non-cell autonomous modulation of each of these processes by secreted molecules known to be expressed in the neural tube during MN development but whose influence on MN fate remains elusive would deserve additional research. Similarly, whether MNs establish crosstalks to coordinate their differentiation, their migration, or their sorting into different columns or different pools should be further investigated.
Beyond improving our basic knowledge of the mechanisms that regulate neuronal diversification, understanding the roles of the MN developmental determinants also constitutes a prerequisite for the development of cell replacement or gene therapies of MN disorders. Indeed, grafting either naive stem/induced pluripotent stem (iPS) cells or pre-differentiated cells, or modifying the genome of altered MNs or surrounding cells, can provide a therapeutic benefit in animal models of MN alterations (reviewed in [147–150]). These include models for amyotrophic lateral sclerosis [148, 150–152], spinal muscular atrophy [153, 154], spinal muscular atrophy with respiratory distress type 1 [155, 156] and traumatic spinal cord lesions (reviewed in [148]). The most critical issue regarding replacement cell therapy for MNs consists of the challenge for graft-derived or neo-generated MNs to establish innervation of peripheral targets located centimeters away from their cell body. However, experiments in mouse models established the proof-of-principle of this approach by demonstrating eventual reinnervation of skeletal muscles after MN grafting into the spinal cord [156, 157]. A deeper knowledge of the mechanisms that ensure MN diversification would benefit the three distinct aspects of these therapeutic approaches. First, it provides markers to precisely evaluate the developmental status of MN-like neurons generated from naive or predifferentiated cells grafted into an injured spinal cord. Second, it constitutes a read-out of the developmental status of stem/iPS cells undergoing in vitro differentiation into MNs [158]. Third, it generates tools to orient in vitro the differentiation of stem/iPS cells toward specific MN subsets [159, 160]. Hence, increasing our understanding of the mechanisms that regulate MN diversification should contribute to further improvement of the therapeutic approaches for MN disorders.
Acknowledgments
Research at the NEDI laboratory was supported by the “Fonds spéciaux de recherche” (FSR) of the UCL, by the Fund for Scientific Medical Research (Grant #3.4.538.10.F) of the F.R.S.-FNRS, by the “Actions de Recherche Concertées (ARC)” #10/15-026 of the “Direction générale de l’Enseignement non obligatoire et de la Recherche scientifique—Direction de la Recherche scientifique—Communauté française de Belgique” and granted by the “Académie universitaire ‘Louvain’”, and by the “Association Belge contre les Maladies neuro-Musculaires asbl” (ABMM). C.F. was supported by the FSR (UCL). F.C. is a Research Associate of the F.R.S.-FNRS.
Abbreviations
- Foxp
Forkhead box p
- HMC
Hypaxial motor column
- HNF-6
Hepatocyte nuclear factor-6
- iPS
Induced pluripotent stem
- LMC
Lateral motor column
- MMC
Medial motor column
- MN
Motor neuron
- Neurog2
Neurogenin 2
- OC
Onecut
- PGC
Preganglionic column
- pMN
Motor neuron progenitor
References
- 1.Jessell TM, Surmeli G, Kelly JS. Motor neurons and the sense of place. Neuron. 2011;72(3):419–424. doi: 10.1016/j.neuron.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- 3.Alaynick WA, Jessell TM, Pfaff SL. SnapShot: spinal cord development. Cell. 2011;146(1):178–178. doi: 10.1016/j.cell.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonanomi D, Pfaff SL. Motor axon pathfinding. Cold Spring Harb Perspect Biol. 2010;2(3):a001735. doi: 10.1101/cshperspect.a001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- 6.di Dalla Torre Sanguinetto SA, Dasen JS, Arber S. Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr Opin Neurobiol. 2008;18(1):36–43. doi: 10.1016/j.conb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Levine AJ, Lewallen KA, Pfaff SL. Spatial organization of cortical and spinal neurons controlling motor behavior. Curr Opin Neurobiol. 2012;22(5):812–821. doi: 10.1016/j.conb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59(2):226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy A, Francius C, Rousso DL, Seuntjens E, Debruyn J, Luxenhofer G, Huber AB, Huylebroeck D, Novitch BG, Clotman F. Onecut transcription factors act upstream of Isl1 to regulate spinal motoneuron diversification. Development. 2012;139(17):3109–3119. doi: 10.1242/dev.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123(3):477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 11.De Marco Garcia NV, Jessell TM. Early motor neuron pool identity and muscle nerve trajectory defined by postmitotic restrictions in Nkx6.1 activity. Neuron. 2008;57(2):217–231. doi: 10.1016/j.neuron.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95(3):393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- 13.Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, Arber S. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35(5):877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 14.Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109(2):205–216. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- 15.Rekling JC, Funk GD, Bayliss DA, Dong XW, Feldman JL. Synaptic control of motoneuronal excitability. Physiol Rev. 2000;80(2):767–852. doi: 10.1152/physrev.2000.80.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101(4):435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 17.Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87(4):661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- 18.Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, Novitch BG, Briscoe J, Sasai N. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8(6):e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450(7170):717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 20.Oosterveen T, Kurdija S, Alekseenko Z, Uhde CW, Bergsland M, Sandberg M, Andersson E, Dias JM, Muhr J, Ericson J. Mechanistic differences in the transcriptional interpretation of local and long-range Shh morphogen signaling. Dev Cell. 2012;23(5):1006–1019. doi: 10.1016/j.devcel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31(5):773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 22.Oh S, Huang X, Liu J, Litingtung Y, Chiang C. Shh and Gli3 activities are required for timely generation of motor neuron progenitors. Dev Biol. 2009;331(2):261–269. doi: 10.1016/j.ydbio.2009.05.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109(1):75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 24.Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12(13):1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 26.Pierani A, Brenner-Morton S, Chiang C, Jessell TM. A sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell. 1999;97(7):903–915. doi: 10.1016/s0092-8674(00)80802-8. [DOI] [PubMed] [Google Scholar]
- 27.Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40(1):65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- 28.Novitch BG, Wichterle H, Jessell TM, Sockanathan S. A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron. 2003;40(1):81–95. doi: 10.1016/j.neuron.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Wilson L, Gale E, Chambers D, Maden M. Retinoic acid and the control of dorsoventral patterning in the avian spinal cord. Dev Biol. 2004;269(2):433–446. doi: 10.1016/j.ydbio.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 30.Chen JA, Huang YP, Mazzoni EO, Tan GC, Zavadil J, Wichterle H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69(4):721–735. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei Q, Jeong Y, Misra K, Li S, Zelman AK, Epstein DJ, Matise MP. Wnt signaling inhibitors regulate the transcriptional response to morphogenetic Shh-Gli signaling in the neural tube. Dev Cell. 2006;11(3):325–337. doi: 10.1016/j.devcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135(2):237–247. doi: 10.1242/dev.012054. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, McDonnell K, Taketo MM, Bai CB. Wnt signaling determines ventral spinal cord cell fates in a time-dependent manner. Development. 2008;135(22):3687–3696. doi: 10.1242/dev.021899. [DOI] [PubMed] [Google Scholar]
- 34.Todd KJ, Lan-Chow-Wing N, Salin-Cantegrel A, Cotter A, Zagami CJ, Lo R, Stifani S. Establishment of motor neuron-V3 interneuron progenitor domain boundary in ventral spinal cord requires Groucho-mediated transcriptional corepression. PLoS ONE. 2012;7(2):e31176. doi: 10.1371/journal.pone.0031176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148(1–2):273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma YC, Song MR, Park JP, Henry Ho HY, Hu L, Kurtev MV, Zieg J, Ma Q, Pfaff SL, Greenberg ME. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron. 2008;58(1):65–77. doi: 10.1016/j.neuron.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19(2):282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31(5):757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- 39.Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95(6):817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- 40.Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, Morrisey EE, Novitch BG. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74(2):314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S, Lee B, Lee JW, Lee SK. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron. 2009;62(5):641–654. doi: 10.1016/j.neuron.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lannoy VJ, Rodolosse A, Pierreux CE, Rousseau GG, Lemaigre FP. Transcriptional stimulation by hepatocyte nuclear factor-6. Target-specific recruitment of either CREB-binding protein (CBP) or p300/CBP-associated factor (p/CAF) J Biol Chem. 2000;275(29):22098–22103. doi: 10.1074/jbc.M000855200. [DOI] [PubMed] [Google Scholar]
- 43.Rao M, Sockanathan S. Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science. 2005;309(5744):2212–2215. doi: 10.1126/science.1117156. [DOI] [PubMed] [Google Scholar]
- 44.Periz G, Yan Y, Bitzer ZT, Sockanathan S. GDP-bound Galphai2 regulates spinal motor neuron differentiation through interaction with GDE2. Dev Biol. 2010;341(1):213–221. doi: 10.1016/j.ydbio.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabharwal P, Lee C, Park S, Rao M, Sockanathan S. GDE2 regulates subtype-specific motor neuron generation through inhibition of Notch signaling. Neuron. 2011;71(6):1058–1070. doi: 10.1016/j.neuron.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Y, Sabharwal P, Rao M, Sockanathan S. The antioxidant enzyme Prdx1 controls neuronal differentiation by thiol-redox-dependent activation of GDE2. Cell. 2009;138(6):1209–1221. doi: 10.1016/j.cell.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordstrom U, Maier E, Jessell TM, Edlund T. An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 2006;4(8):e252. doi: 10.1371/journal.pbio.0040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakami Y, Tanaka M. Evolution of motor innervation to vertebrate fins and limbs. Dev Biol. 2011;355(1):164–172. doi: 10.1016/j.ydbio.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Liu JP, Laufer E, Jessell TM. Assigning the positional identity of spinal motor neurons: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron. 2001;32(6):997–1012. doi: 10.1016/s0896-6273(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 50.Liu JP. The function of growth/differentiation factor 11 (Gdf11) in rostrocaudal patterning of the developing spinal cord. Development. 2006;133(15):2865–2874. doi: 10.1242/dev.02478. [DOI] [PubMed] [Google Scholar]
- 51.Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129(22):5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- 52.Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425(6961):926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- 53.Lacombe J, Hanley O, Jung H, Philippidou P, Surmeli G, Grinstein J, Dasen JS. Genetic and functional modularity of hox activities in the specification of limb-innervating motor neurons. PLoS Genet. 2013;9(1):e1003184. doi: 10.1371/journal.pgen.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin AW, Carpenter EM. Hoxa10 and Hoxd10 coordinately regulate lumbar motor neuron patterning. J Neurobiol. 2003;56(4):328–337. doi: 10.1002/neu.10239. [DOI] [PubMed] [Google Scholar]
- 55.Wu Y, Wang G, Scott SA, Capecchi MR. Hoxc10 and Hoxd10 regulate mouse columnar, divisional and motor pool identity of lumbar motoneurons. Development. 2008;135(1):171–182. doi: 10.1242/dev.009225. [DOI] [PubMed] [Google Scholar]
- 56.Misra M, Shah V, Carpenter E, McCaffery P, Lance-Jones C. Restricted patterns of Hoxd10 and Hoxd11 set segmental differences in motoneuron subtype complement in the lumbosacral spinal cord. Dev Biol. 2009;330(1):54–72. doi: 10.1016/j.ydbio.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah V, Drill E, Lance-Jones C. Ectopic expression of Hoxd10 in thoracic spinal segments induces motoneurons with a lumbosacral molecular profile and axon projections to the limb. Dev Dyn. 2004;231(1):43–56. doi: 10.1002/dvdy.20103. [DOI] [PubMed] [Google Scholar]
- 58.Jung H, Lacombe J, Mazzoni EO, Liem KF, Jr, Grinstein J, Mahony S, Mukhopadhyay D, Gifford DK, Young RA, Anderson KV, Wichterle H, Dasen JS. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron. 2010;67(5):781–796. doi: 10.1016/j.neuron.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golden MG, Dasen JS. Polycomb repressive complex 1 activities determine the columnar organization of motor neurons. Genes Dev. 2012;26(19):2236–2250. doi: 10.1101/gad.199133.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gutzwiller LM, Witt LM, Gresser AL, Burns KA, Cook TA, Gebelein B. Proneural and abdominal Hox inputs synergize to promote sensory organ formation in the Drosophila abdomen. Dev Biol. 2010;348(2):231–243. doi: 10.1016/j.ydbio.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delval S, Taminiau A, Lamy J, Lallemand C, Gilles C, Noel A, Rezsohazy R. The Pbx interaction motif of Hoxa1 is essential for its oncogenic activity. PLoS ONE. 2011;6(9):e25247. doi: 10.1371/journal.pone.0025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Remacle S, Abbas L, De Backer O, Pacico N, Gavalas A, Gofflot F, Picard JJ, Rezsohazy R. Loss of function but no gain of function caused by amino acid substitutions in the hexapeptide of Hoxa1 in vivo. Mol Cell Biol. 2004;24(19):8567–8575. doi: 10.1128/MCB.24.19.8567-8575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capellini TD, Zappavigna V, Selleri L. Pbx homeodomain proteins: TALEnted regulators of limb patterning and outgrowth. Dev Dyn. 2011;240(5):1063–1086. doi: 10.1002/dvdy.22605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291(2):193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 65.Hudry B, Remacle S, Delfini MC, Rezsohazy R, Graba Y, Merabet S. Hox proteins display a common and ancestral ability to diversify their interaction mode with the PBC class cofactors. PLoS Biol. 2012;10(6):e1001351. doi: 10.1371/journal.pbio.1001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams TM, Williams ME, Innis JW. Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction. Dev Biol. 2005;277(2):457–471. doi: 10.1016/j.ydbio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 67.Roberts VJ, van Dijk MA, Murre C. Localization of Pbx1 transcripts in developing rat embryos. Mech Dev. 1995;51(2–3):193–198. doi: 10.1016/0925-4773(95)00364-9. [DOI] [PubMed] [Google Scholar]
- 68.Rottkamp CA, Lobur KJ, Wladyka CL, Lucky AK, O’Gorman S. Pbx3 is required for normal locomotion and dorsal horn development. Dev Biol. 2008;314(1):23–39. doi: 10.1016/j.ydbio.2007.10.046. [DOI] [PubMed] [Google Scholar]
- 69.Cooper KL, Leisenring WM, Moens CB. Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Dev Biol. 2003;253(2):200–213. doi: 10.1016/s0012-1606(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 70.Deflorian G, Tiso N, Ferretti E, Meyer D, Blasi F, Bortolussi M, Argenton F. Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development. 2004;131(3):613–627. doi: 10.1242/dev.00948. [DOI] [PubMed] [Google Scholar]
- 71.Nakano T, Windrem M, Zappavigna V, Goldman SA. Identification of a conserved 125 base-pair Hb9 enhancer that specifies gene expression to spinal motor neurons. Dev Biol. 2005;283(2):474–485. doi: 10.1016/j.ydbio.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 72.Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134(2):304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 73.Palmesino E, Rousso DL, Kao TJ, Klar A, Laufer E, Uemura O, Okamoto H, Novitch BG, Kania A. Foxp1 and lhx1 coordinate motor neuron migration with axon trajectory choice by gating Reelin signalling. PLoS Biol. 2010;8(8):e1000446. doi: 10.1371/journal.pbio.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Philippidou P, Walsh CM, Aubin J, Jeannotte L, Dasen JS. Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat Neurosci. 2012;15(12):1636–1644. doi: 10.1038/nn.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23(4):659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 76.Liang X, Song MR, Xu Z, Lanuza GM, Liu Y, Zhuang T, Chen Y, Pfaff SL, Evans SM, Sun Y. Isl1 is required for multiple aspects of motor neuron development. Mol Cell Neurosci. 2011;47(3):215–222. doi: 10.1016/j.mcn.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Song MR, Sun Y, Bryson A, Gill GN, Evans SM, Pfaff SL. Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development. 2009;136(17):2923–2932. doi: 10.1242/dev.037986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thaler J, Harrison K, Sharma K, Lettieri K, Kehrl J, Pfaff SL. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23(4):675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- 79.Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14(6):877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sander M, Paydar S, Ericson J, Briscoe J, Berber E, German M, Jessell TM, Rubenstein JL. Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 2000;14(17):2134–2139. doi: 10.1101/gad.820400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein–protein interactions. Cell. 2002;110(2):237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- 82.Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell TM, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31(5):743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- 83.Bhati M, Lee C, Nancarrow AL, Lee M, Craig VJ, Bach I, Guss JM, Mackay JP, Matthews JM. Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J. 2008;27(14):2018–2029. doi: 10.1038/emboj.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38(5):731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- 85.Stifani N, Freitas AR, Liakhovitskaia A, Medvinsky A, Kania A, Stifani S. Suppression of interneuron programs and maintenance of selected spinal motor neuron fates by the transcription factor AML1/Runx1. Proc Natl Acad Sci USA. 2008;105(17):6451–6456. doi: 10.1073/pnas.0711299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.William CM, Tanabe Y, Jessell TM. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130(8):1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
- 87.Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95(1):67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- 88.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84(2):309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 89.Hutchinson SA, Eisen JS. Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development. 2006;133(11):2137–2147. doi: 10.1242/dev.02355. [DOI] [PubMed] [Google Scholar]
- 90.Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30(2):423–436. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- 91.Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41(3):337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- 92.Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, Hui CC, Henkelman RM, Nemer M, Black BL, Nagy A, Bruneau BG. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132(10):2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 93.Jacquemin P, Pierreux CE, Fierens S, van Eyll JM, Lemaigre FP, Rousseau GG. Cloning and embryonic expression pattern of the mouse Onecut transcription factor OC-2. Gene Expr Patterns. 2003;3(5):639–644. doi: 10.1016/s1567-133x(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 94.Landry C, Clotman F, Hioki T, Oda H, Picard JJ, Lemaigre FP, Rousseau GG. HNF-6 is expressed in endoderm derivatives and nervous system of the mouse embryo and participates to the cross-regulatory network of liver-enriched transcription factors. Dev Biol. 1997;192(2):247–257. doi: 10.1006/dbio.1997.8757. [DOI] [PubMed] [Google Scholar]
- 95.Vanhorenbeeck V, Jacquemin P, Lemaigre FP, Rousseau GG. OC-3, a novel mammalian member of the ONECUT class of transcription factors. Biochem Biophys Res Commun. 2002;292(4):848–854. doi: 10.1006/bbrc.2002.6760. [DOI] [PubMed] [Google Scholar]
- 96.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, Rousseau GG, Lemaigre FP. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20(12):4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jacquemin P, Lemaigre FP, Rousseau GG. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev Biol. 2003;258(1):105–116. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 98.Pierreux CE, Poll AV, Kemp CR, Clotman F, Maestro MA, Cordi S, Ferrer J, Leyns L, Rousseau GG, Lemaigre FP. The transcription factor hepatocyte nuclear factor-6 controls the development of pancreatic ducts in the mouse. Gastroenterology. 2006;130(2):532–541. doi: 10.1053/j.gastro.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 99.Vanhorenbeeck V, Jenny M, Cornut JF, Gradwohl G, Lemaigre FP, Rousseau GG, Jacquemin P. Role of the Onecut transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev Biol. 2007;305(2):685–694. doi: 10.1016/j.ydbio.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 100.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19(16):1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, Rousseau GG, Lemaigre FP. The Onecut transcription factor HNF6 is required for normal development of the biliary tract. Development. 2002;129(8):1819–1828. doi: 10.1242/dev.129.8.1819. [DOI] [PubMed] [Google Scholar]
- 102.Margagliotti S, Clotman F, Pierreux CE, Beaudry JB, Jacquemin P, Rousseau GG, Lemaigre FP. The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration. Dev Biol. 2007;311(2):579–589. doi: 10.1016/j.ydbio.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 103.Audouard E, Schakman O, Rene F, Huettl RE, Huber AB, Loeffler JP, Gailly P, Clotman F. The Onecut transcription factor HNF-6 regulates in motor neurons the formation of the neuromuscular junctions. PLoS ONE. 2012;7(12):e50509. doi: 10.1371/journal.pone.0050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chakrabarty K, Von Oerthel L, Hellemons A, Clotman F, Espana A, Groot Koerkamp M, Holstege FC, Pasterkamp RJ, Smidt MP. Genome wide expression profiling of the mesodiencephalic region identifies novel factors involved in early and late dopaminergic development. Biol Open. 2012;1(8):693–704. doi: 10.1242/bio.20121230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Espana A, Clotman F. Onecut factors control development of the Locus Coeruleus and of the mesencephalic trigeminal nucleus. Mol Cell Neurosci. 2012;50(1):93–102. doi: 10.1016/j.mcn.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 106.Espana A, Clotman F. Onecut transcription factors are required for the second phase of development of the A13 dopaminergic nucleus in the mouse. J Comp Neurol. 2012;520(7):1424–1441. doi: 10.1002/cne.22803. [DOI] [PubMed] [Google Scholar]
- 107.Hodge LK, Klassen MP, Han BX, Yiu G, Hurrell J, Howell A, Rousseau G, Lemaigre F, Tessier-Lavigne M, Wang F. Retrograde BMP signaling regulates trigeminal sensory neuron identities and the formation of precise face maps. Neuron. 2007;55(4):572–586. doi: 10.1016/j.neuron.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 108.Francius C, Clotman F. Dynamic expression of the Onecut transcription factors HNF-6, OC-2 and OC-3 during spinal motor neuron development. Neuroscience. 2010;165(1):116–129. doi: 10.1016/j.neuroscience.2009.09.076. [DOI] [PubMed] [Google Scholar]
- 109.Uemura O, Okada Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol. 2005;278(2):587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 110.Yip YP, Zhou G, Capriotti C, Yip JW. Location of preganglionic neurons is independent of birthdate but is correlated to reelin-producing cells in the spinal cord. J Comp Neurol. 2004;475(4):564–574. doi: 10.1002/cne.20212. [DOI] [PubMed] [Google Scholar]
- 111.Agalliu D, Takada S, Agalliu I, McMahon AP, Jessell TM. Motor neurons with axial muscle projections specified by Wnt4/5 signaling. Neuron. 2009;61(5):708–720. doi: 10.1016/j.neuron.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144(4):513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 2011;30(22):4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31(3):809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22(10):2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoshimoto A, Saigou Y, Higashi Y, Kondoh H. Regulation of ocular lens development by Smad-interacting protein 1 involving Foxe3 activation. Development. 2005;132(20):4437–4448. doi: 10.1242/dev.02022. [DOI] [PubMed] [Google Scholar]
- 117.Zimmerman A, Hochman S. Heterogeneity of membrane properties in sympathetic preganglionic neurons of neonatal mice: evidence of four subpopulations in the intermediolateral nucleus. J Neurophysiol. 2010;103(1):490–498. doi: 10.1152/jn.00622.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Phelps PE, Rich R, Dupuy-Davies S, Rios Y, Wong T. Evidence for a cell-specific action of Reelin in the spinal cord. Dev Biol. 2002;244(1):180–198. doi: 10.1006/dbio.2002.0580. [DOI] [PubMed] [Google Scholar]
- 119.Yip YP, Capriotti C, Yip JW. Migratory pathway of sympathetic preganglionic neurons in normal and reeler mutant mice. J Comp Neurol. 2003;460(1):94–105. doi: 10.1002/cne.10634. [DOI] [PubMed] [Google Scholar]
- 120.Yip YP, Kronstadt-O’Brien P, Capriotti C, Cooper JA, Yip JW. Migration of sympathetic preganglionic neurons in the spinal cord is regulated by Reelin-dependent Dab1 tyrosine phosphorylation and CrkL. J Comp Neurol. 2007;502(4):635–643. doi: 10.1002/cne.21318. [DOI] [PubMed] [Google Scholar]
- 121.Yip YP, Capriotti C, Drill E, Tsai LH, Yip JW. Cdk5 selectively affects the migration of different populations of neurons in the developing spinal cord. J Comp Neurol. 2007;503(2):297–307. doi: 10.1002/cne.21377. [DOI] [PubMed] [Google Scholar]
- 122.Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406(6795):515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- 123.Shirasaki R, Lewcock JW, Lettieri K, Pfaff SL. FGF as a target-derived chemoattractant for developing motor axons genetically programmed by the LIM code. Neuron. 2006;50(6):841–853. doi: 10.1016/j.neuron.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 124.Satou Y, Imai KS, Satoh N. Early embryonic expression of a LIM-homeobox gene Cs-lhx3 is downstream of beta-catenin and responsible for the endoderm differentiation in Ciona savignyi embryos. Development. 2001;128(18):3559–3570. doi: 10.1242/dev.128.18.3559. [DOI] [PubMed] [Google Scholar]
- 125.Schneider J, Skelton RL, Von Stetina SE, Middelkoop TC, van Oudenaarden A, Korswagen HC, Miller DM., 3rd UNC-4 antagonizes Wnt signaling to regulate synaptic choice in the C. elegans motor circuit. Development. 2012;139(12):2234–2245. doi: 10.1242/dev.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao D, McCaffery P, Ivins KJ, Neve RL, Hogan P, Chin WW, Drager UC. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur J Biochem. 1996;240(1):15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]
- 127.Niederreither K, McCaffery P, Drager UC, Chambon P, Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62(1):67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 128.Swindell EC, Thaller C, Sockanathan S, Petkovich M, Jessell TM, Eichele G. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev Biol. 1999;216(1):282–296. doi: 10.1006/dbio.1999.9487. [DOI] [PubMed] [Google Scholar]
- 129.Mic FA, Haselbeck RJ, Cuenca AE, Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129(9):2271–2282. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Blentic A, Gale E, Maden M. Retinoic acid signalling centres in the avian embryo identified by sites of expression of synthesising and catabolising enzymes. Dev Dyn. 2003;227(1):114–127. doi: 10.1002/dvdy.10292. [DOI] [PubMed] [Google Scholar]
- 131.Vermot J, Schuhbaur B, Le Mouellic H, McCaffery P, Garnier JM, Hentsch D, Brulet P, Niederreither K, Chambon P, Dolle P, Le Roux I. Retinaldehyde dehydrogenase 2 and Hoxc8 are required in the murine brachial spinal cord for the specification of Lim1+ motoneurons and the correct distribution of Islet1+ motoneurons. Development. 2005;132(7):1611–1621. doi: 10.1242/dev.01718. [DOI] [PubMed] [Google Scholar]
- 132.Sockanathan S, Perlmann T, Jessell TM. Retinoid receptor signaling in postmitotic motor neurons regulates rostrocaudal positional identity and axonal projection pattern. Neuron. 2003;40(1):97–111. doi: 10.1016/s0896-6273(03)00532-4. [DOI] [PubMed] [Google Scholar]
- 133.Ji SJ, Zhuang B, Falco C, Schneider A, Schuster-Gossler K, Gossler A, Sockanathan S. Mesodermal and neuronal retinoids regulate the induction and maintenance of limb innervating spinal motor neurons. Dev Biol. 2006;297(1):249–261. doi: 10.1016/j.ydbio.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 134.Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94(4):503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- 135.Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79(6):957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 136.Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38(4):581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- 137.Luria V, Krawchuk D, Jessell TM, Laufer E, Kania A. Specification of motor axon trajectory by ephrin-B:EphB signaling: symmetrical control of axonal patterning in the developing limb. Neuron. 2008;60(6):1039–1053. doi: 10.1016/j.neuron.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 138.Helmbacher F, Schneider-Maunoury S, Topilko P, Tiret L, Charnay P. Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Development. 2000;127(15):3313–3324. doi: 10.1242/dev.127.15.3313. [DOI] [PubMed] [Google Scholar]
- 139.Kramer ER, Knott L, Su F, Dessaud E, Krull CE, Helmbacher F, Klein R. Cooperation between GDNF/Ret and ephrinA/EphA4 signals for motor-axon pathway selection in the limb. Neuron. 2006;50(1):35–47. doi: 10.1016/j.neuron.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 140.Dudanova I, Gatto G, Klein R. GDNF acts as a chemoattractant to support ephrinA-induced repulsion of limb motor axons. Curr Biol. 2010;20(23):2150–2156. doi: 10.1016/j.cub.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 141.Landmesser L. The development of motor projection patterns in the chick hind limb. J Physiol. 1978;284:391–414. doi: 10.1113/jphysiol.1978.sp012546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Landmesser L. The distribution of motoneurones supplying chick hind limb muscles. J Physiol. 1978;284:371–389. doi: 10.1113/jphysiol.1978.sp012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lamballe F, Genestine M, Caruso N, Arce V, Richelme S, Helmbacher F, Maina F. Pool-specific regulation of motor neuron survival by neurotrophic support. J Neurosci. 2011;31(31):11144–11158. doi: 10.1523/JNEUROSCI.2198-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Haase G, Dessaud E, Garces A, de Bovis B, Birling M, Filippi P, Schmalbruch H, Arber S, deLapeyriere O. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35(5):893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 145.Helmbacher F, Dessaud E, Arber S, deLapeyriere O, Henderson CE, Klein R, Maina F. Met signaling is required for recruitment of motor neurons to PEA3-positive motor pools. Neuron. 2003;39(5):767–777. doi: 10.1016/s0896-6273(03)00493-8. [DOI] [PubMed] [Google Scholar]
- 146.Cohen S, Funkelstein L, Livet J, Rougon G, Henderson CE, Castellani V, Mann F. A semaphorin code defines subpopulations of spinal motor neurons during mouse development. Eur J Neurosci. 2005;21(7):1767–1776. doi: 10.1111/j.1460-9568.2005.04021.x. [DOI] [PubMed] [Google Scholar]
- 147.Nizzardo M, Simone C, Falcone M, Locatelli F, Riboldi G, Comi GP, Corti S. Human motor neuron generation from embryonic stem cells and induced pluripotent stem cells. Cell Mol Life Sci. 2010;67(22):3837–3847. doi: 10.1007/s00018-010-0463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Suter DM, Krause KH. Neural commitment of embryonic stem cells: molecules, pathways and potential for cell therapy. J Pathol. 2008;215(4):355–368. doi: 10.1002/path.2380. [DOI] [PubMed] [Google Scholar]
- 149.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137(1):13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 150.Nizzardo M, Simone C, Falcone M, Riboldi G, Rizzo F, Magri F, Bresolin N, Comi GP, Corti S. Research advances in gene therapy approaches for the treatment of amyotrophic lateral sclerosis. Cell Mol Life Sci. 2012;69(10):1641–1650. doi: 10.1007/s00018-011-0881-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Corti S, Locatelli F, Papadimitriou D, Del Bo R, Nizzardo M, Nardini M, Donadoni C, Salani S, Fortunato F, Strazzer S, Bresolin N, Comi GP. Neural stem cells LewisX+ CXCR4+ modify disease progression in an amyotrophic lateral sclerosis model. Brain. 2007;130(Pt 5):1289–1305. doi: 10.1093/brain/awm043. [DOI] [PubMed] [Google Scholar]
- 152.Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Simone C, Falcone M, Riboldi G, Govoni A, Bresolin N, Comi GP. Systemic transplantation of c-kit+ cells exerts a therapeutic effect in a model of amyotrophic lateral sclerosis. Hum Mol Genet. 2010;19(19):3782–3796. doi: 10.1093/hmg/ddq293. [DOI] [PubMed] [Google Scholar]
- 153.Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Ronchi D, Simone C, Falcone M, Papadimitriou D, Locatelli F, Mezzina N, Gianni F, Bresolin N, Comi GP. Embryonic stem cell-derived neural stem cells improve spinal muscular atrophy phenotype in mice. Brain. 2010;133(Pt 2):465–481. doi: 10.1093/brain/awp318. [DOI] [PubMed] [Google Scholar]
- 154.Corti S, Nizzardo M, Simone C, Falcone M, Nardini M, Ronchi D, Donadoni C, Salani S, Riboldi G, Magri F, Menozzi G, Bonaglia C, Rizzo F, Bresolin N, Comi GP. Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med. 2012;4(165):165ra162. doi: 10.1126/scitranslmed.3004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Corti S, Locatelli F, Papadimitriou D, Donadoni C, Del Bo R, Crimi M, Bordoni A, Fortunato F, Strazzer S, Menozzi G, Salani S, Bresolin N, Comi GP. Transplanted ALDHhiSSClo neural stem cells generate motor neurons and delay disease progression of nmd mice, an animal model of SMARD1. Hum Mol Genet. 2006;15(2):167–187. doi: 10.1093/hmg/ddi446. [DOI] [PubMed] [Google Scholar]
- 156.Corti S, Nizzardo M, Nardini M, Donadoni C, Salani S, Del Bo R, Papadimitriou D, Locatelli F, Mezzina N, Gianni F, Bresolin N, Comi GP. Motoneuron transplantation rescues the phenotype of SMARD1 (spinal muscular atrophy with respiratory distress type 1) J Neurosci. 2009;29(38):11761–11771. doi: 10.1523/JNEUROSCI.2734-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nogradi A, Pajer K, Marton G. The role of embryonic motoneuron transplants to restore the lost motor function of the injured spinal cord. Ann Anat. 2011;193(4):362–370. doi: 10.1016/j.aanat.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 158.Hashimoto T, Jaakkola T, Sherwood R, Mazzoni EO, Wichterle H, Gifford D. Lineage-based identification of cellular states and expression programs. Bioinformatics. 2012;28(12):i250–i257. doi: 10.1093/bioinformatics/bts204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wichterle H, Peljto M (2008) Differentiation of mouse embryonic stem cells to spinal motor neurons. Curr Protoc Stem Cell Biol Chap. 1:Unit 1H.1.1–1H.1.9 [DOI] [PubMed]
- 160.Peljto M, Wichterle H. Programming embryonic stem cells to neuronal subtypes. Curr Opin Neurobiol. 2011;21(1):43–51. doi: 10.1016/j.conb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]