Abstract
Drosophila possesses the core gene silencing machinery but, like all insects, lacks the canonical RNA-dependent RNA polymerases (RdRps) that in C. elegans either trigger or enhance two major small RNA-dependent gene silencing pathways. Introduction of two different nematode RdRps into Drosophila showed them to be functional, resulting in differing silencing activities. While RRF-1 enhanced transitive dsRNA-dependent silencing, EGO-1 triggered dsRNA-independent silencing, specifically of transgenes. The strain w; da-Gal4; UAST-ego-1, constitutively expressing ego-1, is capable of silencing transgene including dsRNA hairpin upon a single cross, which created a powerful tool for research in Drosophila. In C. elegans, EGO-1 is involved in transcriptional gene silencing (TGS) of chromosome regions that are unpaired during meiosis. There was no opportunity for meiotic interactions involving EGO-1 in Drosophila that would explain the observed transgene silencing. Transgene DNA is, however, unpaired during the pairing of chromosomes in embryonic mitosis that is an unusual characteristic of Diptera, suggesting that in Drosophila, EGO-1 triggers transcriptional silencing of unpaired DNA during embryonic mitosis.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1218-8) contains supplementary material, which is available to authorized users.
Keywords: Drosophila, RNA-dependent RNA polymerase (RdRp), RNA interference (RNAi), microRNAs (miRNAs)
Introduction
Drosophila melanogaster, along with all insects and the vertebrates, lacks genes encoding members of the canonical RdRp gene family, which are required for the systemic spread of gene silencing by RNA interference (RNAi) in fungi, plants, and some animals [1–4]. Two major forms of RNAi exist in the animal system in which RNAi has been most thoroughly studied, C. elegans. For transitive post-transcription gene silencing (PTGS), RNAi is triggered by double-stranded RNA (dsRNA), which is processed into small interfering RNAs (termed primary siRNAs, ~20–30 nucleotides long) that target a complementary mRNA, leading to its degradation [5]. Transcriptional, dsRNA-independent gene silencing is involved in epigenetic modification and heterochromatin regulation [6]. C. elegans contains multiple RdRp family genes: of these, rrf-1 [7] is required for systemic dsRNA-dependent RNAi in somatic cells, resulting in the unprimed production of secondary siRNAs [8, 9] that are structurally different to the primary siRNAs. These secondary siRNAs enhance silencing of genes targeted by the primary dicer-derived siRNA molecules that are complementary to the target sequence. The other RdRp associated with gene silencing, ego-1, is required for germline transcriptional gene silencing and heterochromatin assembly [10].
Drosophila possesses the core gene silencing machinery [11–13]. Dicer-2 and Ago-2 are able to generate siRNAs from dsRNA, which act to repress transposon transcripts or endogenous mRNAs [14, 15]. The introduction of transgenes producing hairpin dsRNAs allows cell-specific silencing of Drosophila genes [16]. Moreover, a dsRNA-uptake system [17, 18] has recently been shown to allow systemic silencing of viral RNAs in Drosophila [19]. Despite these findings, Drosophila is incapable of systemic RNAi. We therefore asked whether RNAi in Drosophila could be enhanced as observed for systemic RNAi in C. elegans by the introduction of C. elegans RdRp genes.
We have previously shown that GAL4-mediated expression of transgenic C. elegans RdRps in Drosophila does not affect morphological development [20]. However, it remained an open question as to whether these C. elegans RdRps were active in Drosophila at all. We therefore asked whether these RdRps were capable of enhancing RNAi of a specific, known target gene, triggered by a dsRNA corresponding to that gene. In this study we show that C. elegans RdRps RRF-1 and EGO-1 silence Drosophila transgenes by differing mechanisms: RRF-1 was found to enhance transitive, dsRNA-dependent RNAi of target genes, whereas EGO-1 expression resulted in robust silencing specifically of transgenes that was independent of dsRNA.
Results
RRF-1 and EGO-1 could not be observed to enhance silencing of the endogenous gene, pebble
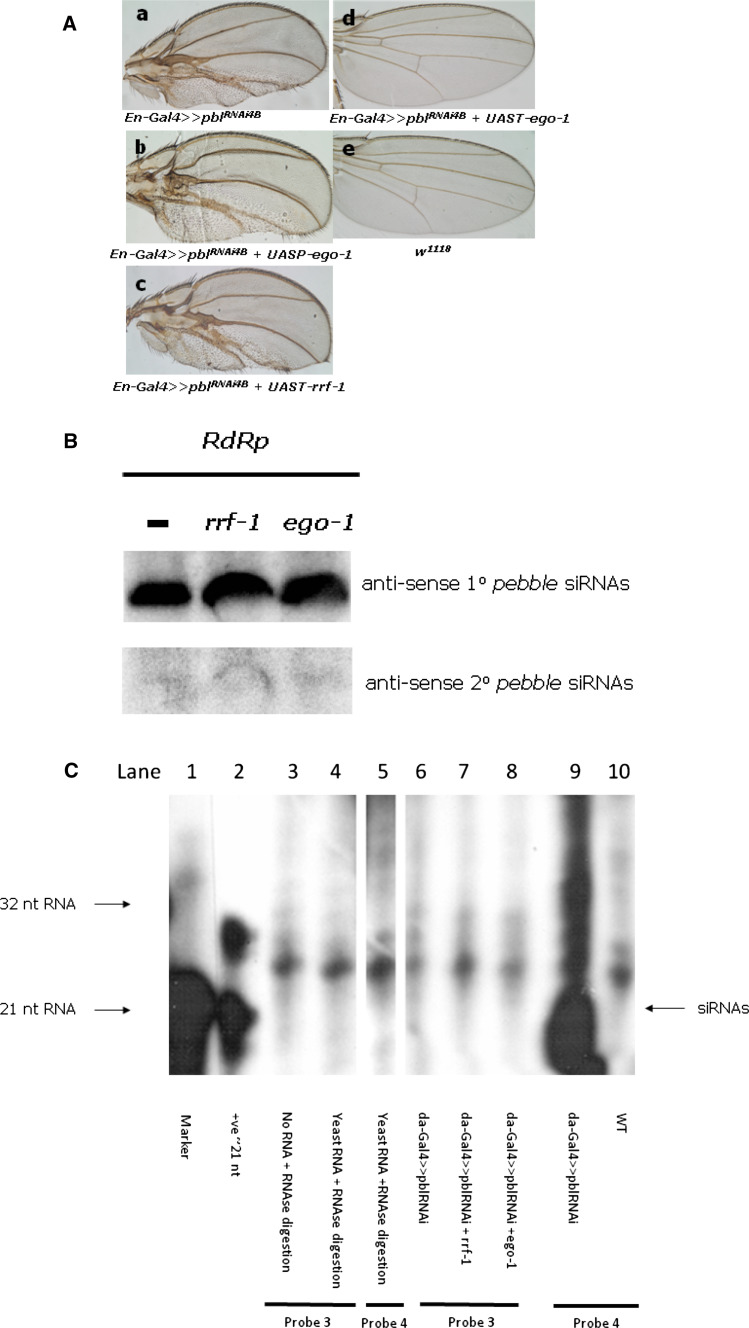
It is known that expression of a dsRNA from a transgenic construct leads to silencing mediated by siRNAs derived solely from dicer-2 activity. In many cases, such silencing is only partial, which provides an assay for enhancement of silencing by the introduction of the C. elegans RdRp genes into transgenic flies. To test this, we selected pebble [21] as the candidate endogenous gene. This gene encodes a guanine nucleotide exchange factor (GEF) that is important for Rho1 activation in dividing cells [22] and therefore required for cytokinesis; knockdown produces multinucleate cells, a cell-autonomous phenotype that is easy to observe. Silencing of pbl specifically in the posterior compartment of the wing disc by combining an en-Gal4 construct with pblRNAi 4B(3.1) [23], a UAS construct that expresses pbl hairpin dsRNA, resulted in disruption of the development of the posterior compartment of the wing. As shown in Fig. 1a, when the RdRp RRF-1 or EGO-1 (in UASP) was co-expressed with the hairpin pbl dsRNA, no change in the wing phenotype was observed.
Fig. 1.
Silencing of the endogenous gene, pebble, in Drosophila and detection of the primary as well as secondary siRNAs. a Expression of canonical C. elegans RdRps does not noticeably enhance RNAi-mediated silencing of the Drosophila endogenous gene, pebble. For activation of the dsRNA expression, flies harboring the pebble gene dsRNA construct pblRNAi 4B(3.1) were crossed with the tissue-specific driver en-Gal4 in the presence (b–d) or absence (a) of one of the RdRps, RRF-1 or EGO-1. Compared with the control (a), noticeable changes in pebble gene silencing in either b or c were not observed; however, expression of higher levels of EGO-1 from the UAST vector gave a wild-type phenotype (d) through comparison with WT (e). b Northern hybridization did not detect any production of secondary siRNAs. Probe 1, designed based on the pebble targeted sequence (Fig. S1), successfully detected 1° siRNAs, while probe 2, designed based on the sequence upstream of the targeted sequence, did not detect any 2° siRNA production regardless of the presence of either of the RdRp genes. We used the RNA samples from UASP-ego-1 rather than UAST-ego-1 as da-gal4-activated UAST-ego-1 automatically silenced pblRNAi 4B(3.1) and gave WT phenotype, i.e., could not suspend the development at pupae stage as occurred in UAST-rrf-1 and UASP-ego-1. c Secondary siRNAs detection with RNAse protection assays. RNAs was prepared from pupae. Lane 1: RNA marker, lane 2: positive control RNA; lanes 3 and 4 are controls for probe 3 with yeast or no RNA; lane 5 is control for probe 4 with RNAse digestion. Lanes 6–8 are the results for detection of 2° siRNAs upstream of the targeted pebble sequence (Fig. S1 and S2) with probe 3; da-Gal4-activated rrf-1 (lane 7) or ego-1 (lane 8) did not generate detectable 2° siRNAs. Lane 9: detection of the primary siRNAs with probe 4, lane 10: no detection in w 1118
The silencing observed normally for the pebble gene, i.e., in the presence of only the dsRNA transgene, is quite pronounced, and the resulting morphologies are variable, ranging up to strongly deformed wings [23]. This means it may not be possible to rely solely on observation of morphological changes upon silencing an endogenous gene to assess enhancement of RNAi. Indeed, RRF-1 had no observable impact on silencing of pebble. In contrast, co-expression of EGO-1 at the high levels enabled by the UAST vector did have an observable impact on silencing, but curiously this was to abolish it, since all progeny with this genotype had a wild-type phenotype (Fig. 1a–d and legend). This was the first observation suggesting that EGO-1 expression directly silenced a transgene, that producing pebble dsRNA.
We next tested whether the expression of RdRps affected the production of siRNAs in these RNAi flies. Although RdRp can enhance silencing by producing extra 2° siRNAs upstream of the dsRNA targeting sequence in C. elegans [8, 9], neither Northern hybridization, nor RNase protection assays, detected production of 2° siRNAs in these flies, while expression of 1° siRNAs was not affected by the expression of rrf-1 or of the UASP-ego-1 transgene (Fig. 1b, c). Since the pebble gene is only expressed in a specific tissue for a limited time, it is possible that the level of any 2° siRNAs produced may be too low for detection.
Silencing by RRF-1 and EGO-1 of an exogenous reporter gene, egfp
The above work showed modulation of pebble silencing to be unsuitable as an experimental system for testing activity of the RdRp transgenes. We therefore asked whether these RdRps could enhance silencing of an exogenous reporter gene, egfp, placed under the control of an ubiquitin/GAL4 construct to ensure expression throughout all tissues and stages. The egfp hairpin dsRNA required to generate primary siRNAs targets almost the complete egfp sequence, leaving only ~28 nt uncovered at the amino-terminal portion of the EGFP coding sequence [16]. In order to provide additional sequences upstream on the same mRNA to allow testing for generation of secondary siRNAs as a consequence of RdRp action, the EGFP coding sequence was translationally fused 3′ to the coding sequence of a Drosophila gene, synaptotagmin (hereafter syt) (syt.EGFP) [24], or of a bacterial gene, NaChBac (NaChBac.EGFP), encoding a Na-channel protein [25].
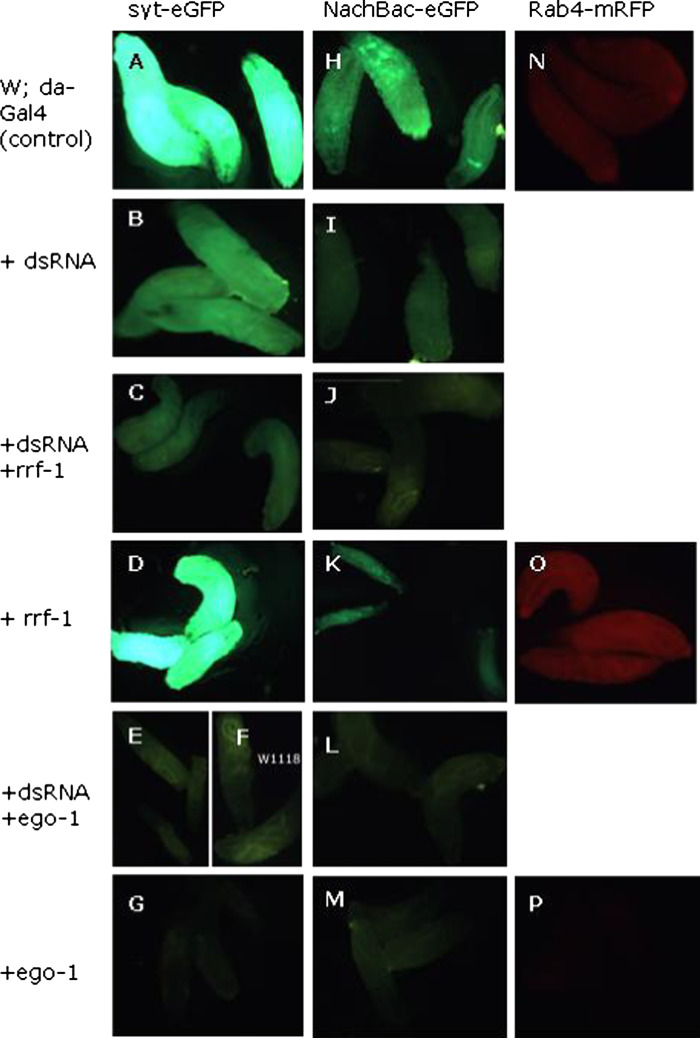
In contrast to the observation with the endogenous pebble gene, we observed that dsRNA-dependent silencing (Fig. 2b, i) of the egfp gene was significantly enhanced in the presence of either of the RdRps (Fig. 2c, e, j, l). To investigate if the corresponding EGFP dsRNA was required for the silencing, we set up parallel crosses omitting only the dsRNA, while keeping all other genes and drivers (i.e., EGFP, RdRp, da-Gal4) unchanged. Surprisingly, EGO-1 still totally silenced the egfp gene (Fig. 2g, m) while RRF-1 did not (Fig. 2d, k), in the absence of the dsRNA; this observation was confirmed using different RdRp lines and a line in which syt.EGFP and the EGFP dsRNA are present on different chromosomes (Supplementary Figure S6). This result was further confirmed by another reporter gene, rfp, encoding the red fluorescence protein (RFP) [26] silencing by EGO-1 without the corresponding rfp dsRNA was observed (Fig. 2p), while no silencing was found by RRF-1 alone (Fig. 2o). qPCR experiments confirmed the RdRp transgenes were expressed (Supplementary Figure S11A and B), and Northern blot and hybridization confirmed the egfp expression (Supplementary Figure 10). Taken together, these results indicate that EGO-1 is an independent silencer that can autonomously silence transgenes; on the other hand, RRF-1 is only capable of enhancing dsRNA-dependent silencing.
Fig. 2.
C. elegans RdRp RRF-1 enhances dsRNA-triggered silencing of exogenous target genes, whereas silencing of exogenous genes by EGO-1 under the control of UAST is dsRNA-independent. Crosses for obtaining the larvae illustrated above are shown in Supplementary Methods S2, online supplement. Each cross was set up in at least for four replicates, and about 12 1° instar larvae from each cross were randomly selected and checked for consistency of the fluorescence signal; three of these were randomly selected for photography. a, h, and n are positive controls for fluorescence from NaChbac.eGFP, syt.eGFP, or Rab4-mRFP, respectively; b and i show a reduction of green fluorescence signals, indicating that the EGFP transgenes were partially silenced with the introduction of the corresponding EGFP dsRNA; c and j show a dramatic further reduction in the green fluorescence signal after combination with the RdRp gene, rrf-1, indicating a dramatic enhancement of silencing of the EGFP gene. Addition of the rrf-1 gene resulted in no silencing enhancement in the absence of the dsRNA trigger for either GFP [d, k or for the RFP line (o)]. In contrast, total silencing of fluorescent protein gene expression was observed in the presence of ego-1, irrespective of whether the EGFP dsRNA hairpin was present (e, l) or not (g, m); the RFP gene was also silenced totally in the absence of any dsRNA trigger (p). Expression of the EGFP-linked NaChBac gene resulted in suspended larval development at the 1° instar stage (k). f is the negative control from w1118 under a GFP filter (using a Leica MZ11 fluorescence microscope); for the RFP gene a TXR filter was used
Enhancement of silencing by RdRp releases suspension of development due to an exogenous gene, NaChBac, or a dsRNA hairpin
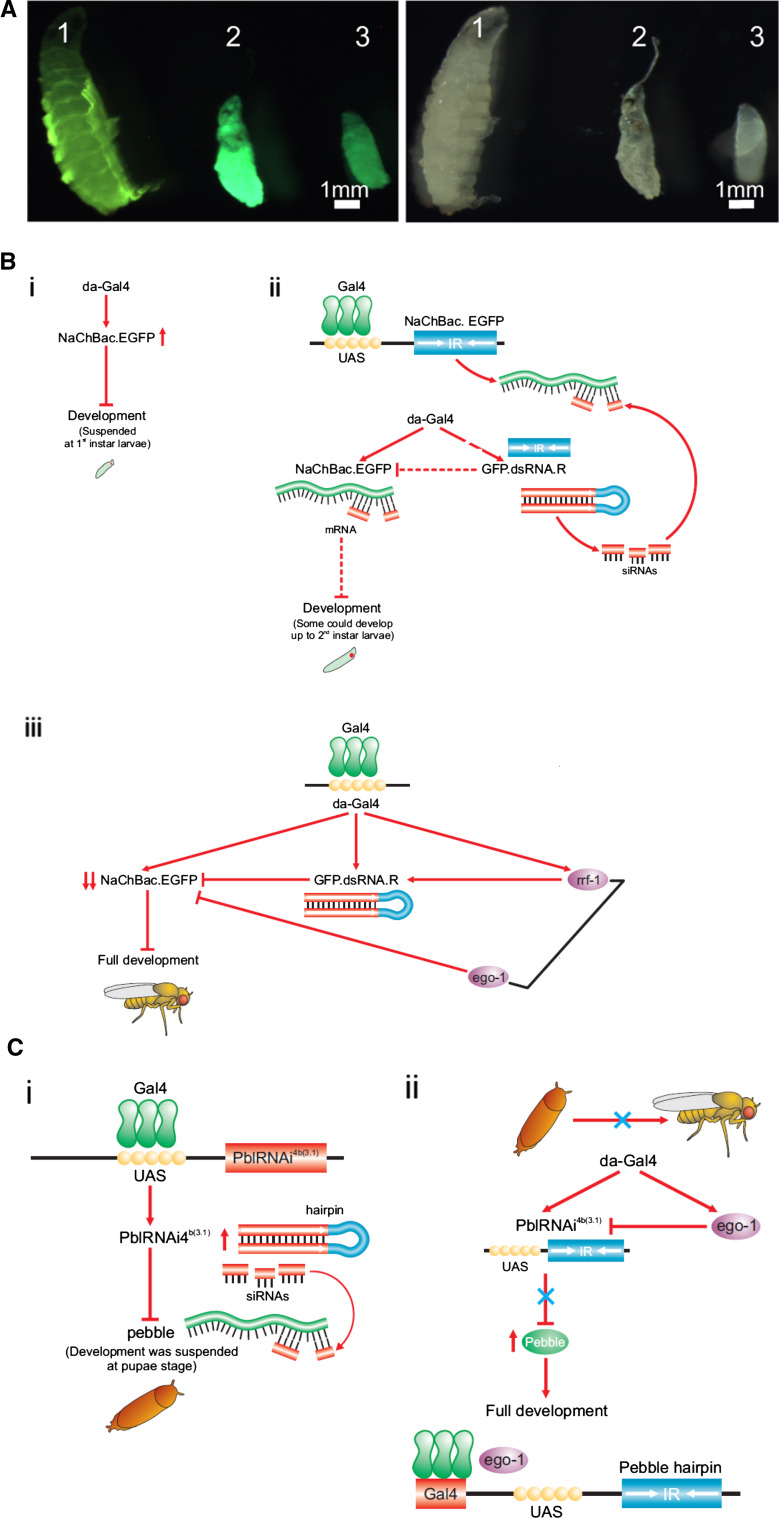
We next asked whether a sensitive test could be developed using a biologically active gene expressed in all tissues and stages, as was the case for the reporter gene egfp. For this purpose, we used the bacterial voltage-gated sodium channel, NaChBac, which as described above, is available as a translation fusion to EGFP. Expression of this fusion transgene in Drosophila has previously been restricted to specific neuron tissues [27]. However, general activation of the NaChBac gene with da-Gal4, which activated EGFP in first instar larvae as shown in Fig. 2h, was also observed to suspend Drosophila development at that stage. This finding established a novel experimental system for testing silencing of a biologically active transgene.
We then asked whether dsRNA-induced partial silencing of the NaChBac.EGFP gene, could overcome this developmental block. Indeed, silencing of the fusion gene, as shown by the reduction in EGFP fluorescence (Fig. 2i), allowed larvae to develop up to the second instar, (Fig. 3a-1), at which point further development was again suspended, consistent with the expected partial silencing of the NaChBac transgene. However, when combined with either one of the RdRps, total silencing was achieved (Fig. 3b, c); this, in turn, released the development suspension conferred by NaChBac, and the larvae were observed to complete full development to adults. While EGO-1 expression allowed completion of development irrespective of the presence of dsRNA, RRF-1 had a similar effect only in the presence of the dsRNA transgene (Fig. 3a-3). These observations confirmed the utility of this novel system for RNAi investigation in Drosophila.
Fig. 3.
Partial silencing of NaChBac:eGFP fusion gene releases suspension of larval development at the 1° instar larvae stage. a Left panel fluorescence images; right panel bright light images of the same larvae. Partial silencing of the EGFP-linked NaChBac gene by introduction of the EGFP dsRNA hairpin gene (1), allows larval development beyond the 1° instar stage whereas development is suspended in larvae expressing EGFP-linked NaChBac (2). Introduction of rrf-1 in the absence of the dsRNA hairpin had no effect on larval development (3). However, the partial silencing achieved by introduction of the EGFP dsRNA is insufficient for full development to adult stage. The green fluorescence signal is decreased (1) compared with no such silencing in 2 and 3. b Schematic model proposed for the genetic interactions observed for EGFP silencing. (i) The general driver da-Gal4 activates the NaChBac.eGFP fusion gene, resulting in the suspension of Drosophila development at the 1° instar stage. (ii) Activation by the general driver da-Gal4 of the EGFP dsRNA hairpin gene, as well as of the NaChBac.eGFP fusion gene, results in partial silencing of the latter and a reduction of the NaChBac.eGFP protein product, partially releasing the developmental arrest, and allowing some larvae to develop further, to the 2° instar stage. (iii) When RRF-1 was introduced, EGFP dsRNA-dependent silencing of the NaChBac.eGFP was dramatically enhanced, allowing larvae to go through full development towards the adult fly stage. In contrast, EGO-1 directly and completely silences the NaChBac.eGFP transgene, independent of the corresponding dsRNA, allowing full development of Drosophila to occur. c Schematic model proposed for the genetic interactions observed for pebble silencing. (i) The general driver da-Gal4 activates the pblRNAi 4B(3.1)dsRNA construct, whose expression silences the pebble gene, resulting in development being suspended at the pupal stage. (ii) General activation of pblRNAi 4B(3.1)as well as EGO-1 (in UAST), resulted in pblRNAi 4B(3.1) expression being repressed by EGO-1, which in turn released pebble silencing imposed by pblRNAi 4B(3.1) and subsequently led to full development
Although the limited extent of pebble expression had earlier been a factor restricting its utility for studies of RNAi enhancement, we unexpectedly found that general activation of the pblRNAi 4B(3.1) dsRNA hairpin construct with da-Gal4 suspended fly development at the pupal stage (Fig. 3c-i, ii). This allowed experiments to ask whether EGO-1 could block expression of this dsRNA hairpin completely to overcome this biological effect. Silencing of the pebble hairpin by en-Gal4-activated EGO-1 had earlier been found to release its silencing of the pebble gene in the wing disc (Fig. 1a–d), co-expression of EGO-1 was indeed observed to release pebble repression in all tissues, therefore rescuing full development (Fig. 3c-i, ii) to the adult stage.
RRF-1, but not EGO-1, enhances transitive RNAi pathway by producing extra secondary siRNAs
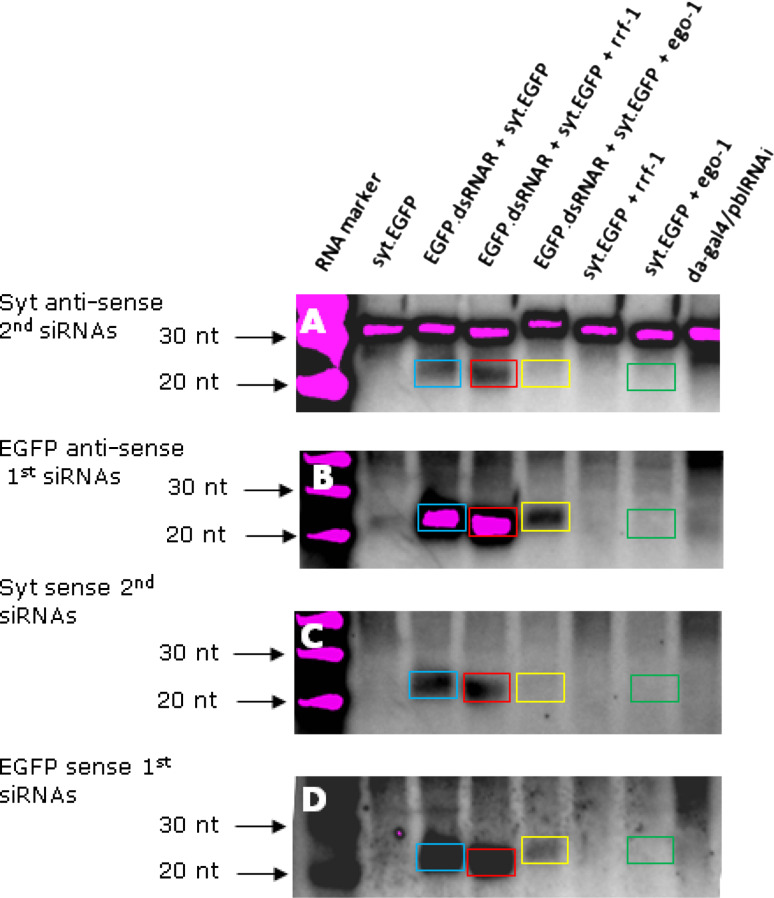
We next asked whether the RdRps produced 2° siRNAs [7–9]. Production of 2° siRNAs is dependent upon presence of 1° siRNAs, as detected in all flies expressing the EGFP dsRNA hairpin (Fig. 4b, d). Both sense and anti-sense 2° siRNAs were detected in flies expressing syt-EGFP and the EGFP dsRNA hairpin (Fig. 4a, c, blue boxes, respectively) in the absence of either rrf-1 or ego-1; their origin, however, remains unknown in the absence of a proven endogenous RdRp. In contrast to observations with the sense 2° siRNAs (Fig. 4c), expression of RRF-1 enhanced anti-sense 2° siRNA levels in flies (Fig. 4a, red box), confirming that RRF-1 enhances RNAi in Drosophila by the transitive pathway.
Fig. 4.
Northern blot and hybridization for detection of the primary and secondary siRNAs in flies with the transgene egfp RNAi. A diagram illustrating the probes and target sequence is provided in Fig. S3 and S4. a Detection of the syt anti-sense 2° siRNAs with syt sense probe, compared with control (EGFP.dsRNA + syt.eGFP) without either rrf-1 or ego-1, in which the 2° siRNAs (blue) were produced by endogenous non-canonical RdRp activity, the 2° siRNA production in the presence of RRF-1 (red) was dramatically increased, while siRNA production in the presence of EGO-1 was dramatically reduced and difficult to see with naked eye (yellow), indicating an enhanced transitive RNAi pathway in the presence of RRF-1 but not EGO-1. The detection of syt sense 2° siRNAs in c is consistent with the results shown in a. In comparison, in b, EGFP anti-sense 1° siRNAs without RdRp (blue) and with RRF-1 (red) were detected at a similar signal strength, but production of these siRNAs was dramatically decreased (yellow) when EGO-1 was present, indicating an independent, direct silencing of the EGFP dsRNA hairpin transgene by EGO-1, which is consistent with the EGFP sense 1° siRNAs detection in d. In addition, independent silencing by EGO-1 was achieved not through producing 1° or 2° sense or anti-sense siRNAs (green boxes in all panels)
In contrast, 1° siRNAs were reduced and 2° siRNAs abolished in the flies with EGO-1 and the EGFP dsRNA (Fig. 4, yellow boxes). The abolition of even the 1° EGFP siRNAs in the presence of EGO-1 (Fig. 4b, yellow box), suggested that EGO-1 simultaneously silenced the egfp hairpin dsRNA transgene as well as the actual EGFP-transcript (since no dsRNA was required for silencing of this transgene). Silencing of a dsRNA-producing transgene would explain the original observation (Fig. 1, above) that EGO-1 abolished silencing of the endogenous pebble gene.
Transgene silencing by EGO-1
The dsRNA-independent silencing of transgenes, including those producing dsRNA hairpins, by EGO-1 appears to be transcriptional. Evidence that the silencing of the eGFP transgene is transcriptional is that no mRNA was detected (Supplementary Figure S10), no siRNA was produced (Fig. 4a), and the silencing was irreversible (Supplementary Figure S7). Nuclear run-on experiments to confirm transcriptional silencing failed, due to the difficulty of obtaining RNA from the development-suspended larvae and the limited expression level of transgenes. This targeted silencing of transgenes, but not endogenous genes, is consistent with the absence of morphological or other obvious phenotypic changes evident in Drosophila expressing this gene [20]. Silencing of transgenes by EGO-1 cannot be attributed to titration of the GAL4 transactivator required for transgene expression. The ego-1 and rrf-1 genes, and the other genes dependent on GAL4, were expressed from exactly the same expression cassettes, carrying the same complement of UAS sequences. The two RdRp genes nonetheless showed totally different effects.
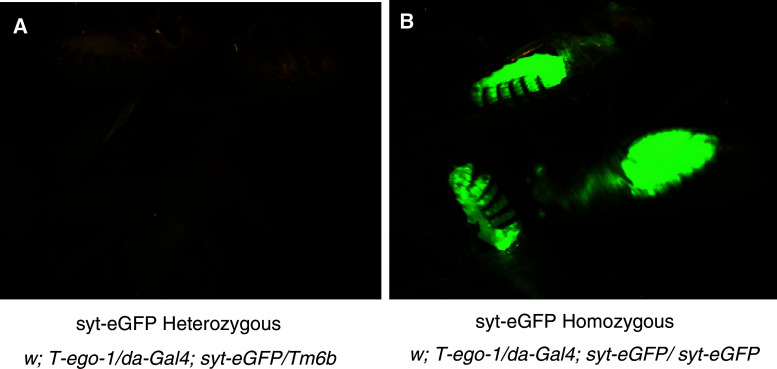
EGO-1 is involved in silencing of unpaired DNA in C. elegans [10]. Since the Drosophila transgenes are actually unpaired in the progeny flies described above, we hypothesized that this is what has made them subject to EGO-1 silencing. For example, the progeny of the cross in Fig. 2g (in which EGO-1 was observed to cause silencing of EGFP) are heterozygous: w; da-gal4/+; T-ego-1/UAST-syt-eGFP. To test this hypothesis, we asked whether the transgene was silenced when paired, by crossing a line homozygous for syt-eGFP and ego-1 with a line homozygous for da-Gal4 and heterozygous for syt-eGFP (Supplementary methods S3). The EGFP was indeed silenced in the progeny flies heterozygous for syt-eGFP (Fig. 5a) but not in those homozygous for this gene (Fig. 5b), supporting the hypothesis of unpaired DNA silencing.
Fig. 5.
Silencing by unpaired DNA mechanism. The crosses used to generate the flies are provided in the Supplementary Methods S3. The flies were checked for genotype and the consistency of EGFP expression/non-expression. Three representative flies were then selected for photography under the same conditions and the same fluorescence microscope parameter. EGFP was silenced in the heterozygous flies (a) but not in the homozygous ones (b)
Silencing by EGO-1 may involve methylation and/or rearrangements of the promoter region of the transgenes
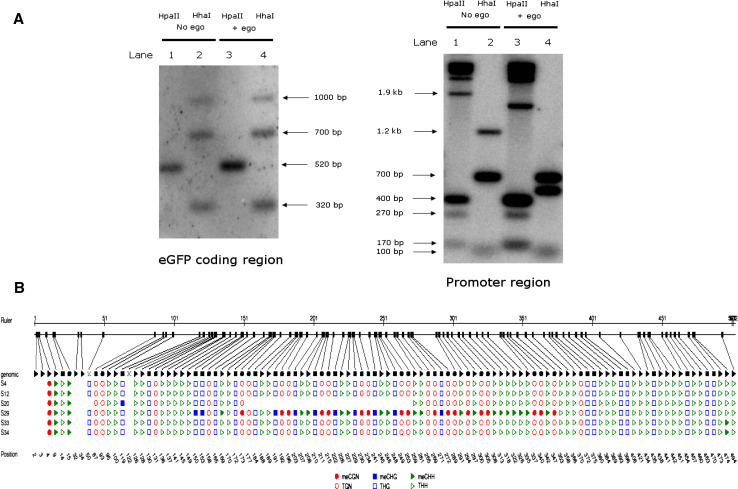
There is ongoing interest in the extent to which DNA methylation occurs in Drosophila, with little evidence for methylation after early embryonic stages [28]. Since the silencing of transgenes by EGO-1 appeared not to be reversible, we next asked whether expression of EGO-1 had altered the chromatin status by stimulating any methylation of DNA. To address this, we first performed Southern blot and hybridization in the EGFP coding region as well as its promoter region. The sizes of fragments detected in the EGFP coding region were consistent with the expected sizes indicated in the diagrams (Supplementary Figure S12) except that the small fragments <100 bp were not detected (Fig. 6a). However, in the promoter region of transgenes, the HpaII and HhaI cutting pattern between the samples with or without EGO-1 obviously altered (Fig. 6a), a possible explanation is that the expression of EGO-1 altered the methylation or demethylation status in the promoter region.
Fig. 6.
Methylation detection and rearrangement of the promoter region of the transgenes. a Southern hybridization detected the cutting pattern unchanged in the EGFP coding region, and altered in the transgenes’ promoter regions, respectively. DNA samples in lane 1: w; da-Gal4/+; UAST-syt.eGFP/+ cut with HpaII; lane 2: w; da-Gal4/+; UAST-syt.eGFP/+ cut with HhaI; lane 3: w; da-Gal4/+; UAST-syt.eGFP/T-ego-1 cut with HpaII; lane 4: w; da-Gal4/+; UAST-syt.eGFP/T-ego-1 cut with HhaI. The cutting pattern in EGFP coding region was not affected by expression of the EGO-1 gene (left); in contrast; the pattern in transgenes’ promoter regions was altered by the co-expression of EGO-1 (right). b Bisulfite sequencing analysis in the promoter regions of the transgenes in the presence or absence of EGO-1. Sample S4: syt.eGFP only without ego-1, S12, S20: Syt.eGFP with ego-1, S29, S33, S34: ego-1 only. “N stands for any nucleotide and “H” is either A, C, or T
In light of the lack of resolution obtained by the Southern experiment, we next performed bisulfite sequencing to analyze individual cytosine residues in the promoter regions of the transgenes (Fig. 6b). The promoter regions from the transgenes syt.eGFP, in the absence of EGO-1 (S4), in the presence of EGO-1 (S12 and S20), or only that from EGO-1 (S29, S33, and S34), were compared using VectorNTI (Invitrogen) and CYMATE (GMI) [29]. In sample S29, a fragment between ~151 and 375 bp was found to be heavily methylated in all three patterns CGN, CHG, and CHH, sequence analysis showed that it was comprised of 5 × Gal4 UAS and the first half of HS promoter (Supplementary Figure S13). This indicated that the effects of expression of EGO-1 were not universal in all cells. Moreover, it was striking that a fragment between ~151 and 260 bp in sample S20 was deleted, possibly as a consequence of EGO-1 modulation of the transgene’s expression. Further sequence analysis confirmed that this deleted fragment contained 5 × Gal4 UAS, which is critical in the GAL4-UAS system for successful transgene expression. It should be noted that the observed incidence was relatively low, with only one sample in ten being affected. Deletion of a portion of the transgene promoter was observed as a possible consequence of EGO-1 being co-expressed.
Discussion
In this study, we show that C. elegans RNA-directed RNA polymerases RRF-1 and EGO-1, are capable of activity when introduced into Drosophila as transgenes. By taking advantage of the partial silencing induced by dsRNA, we investigated whether these RdRps were capable of enhancing RNAi. In accordance with their different roles in C. elegans, we confirmed their ability to function by mechanisms dependent and independent, respectively, of dsRNA.
Novel insights from RdRp-enhancement of endogenous and trans-gene silencing
The use of transgenes encoding reporter proteins allowed testing for enhancement of dsRNA-dependent silencing by RRF-1. Beyond the experiments using only the reporter genes, work with two biologically active target genes was required to assess how effective the enhancement of RNAi was. The endogenous target gene used initially, pbl [21], was silenced quite effectively by dsRNA. The pronounced, if variable, deformation caused by this silencing meant it was not possible to confirm any enhancement when the en-Gal4 construct was used to drive the dsRNA and RRF-1 transgenes. Use of the da-Gal4-activator, on the other hand, to drive the dsRNA transgene enabled general silencing of pbl at a level sufficient to kill all progeny at the pupa stage. While this is of interest as the first demonstration that general silencing of pbl is lethal, it was not possible to assess any enhancement by RRF-1.
To demonstrate that RRF-1 could enhance RNAi of a biologically active gene required establishment of a novel system for RNAi investigation in Drosophila, involving modulation of NaChBac.eGFP silencing. The voltage-gated Na-channel protein [25] encoded by NaChBac has previously been used to study neuron excitation in Drosophila [27, 30] but only with its expression restricted to specific tissues and neurons., Our finding that general activation of the sodium channel NaChBac (as a translational fusion protein with EGFP) suspends development at the first instar larval stage generates a valuable research tool. Previously, over-expression of a potassium channel protein (Kir2.1) in Drosophila has also been shown to prove lethal [30]. The RRF-1-enhanced silencing of the NaChBac.eGFP gene allowed full completion of development, whereas dsRNA-induced partial silencing resulted in larvae developing only up to the 2° instar.
Does ego-1 induce mitotic silencing of unpaired DNA?
During meiosis, homologous chromosomes are paired to allow recombination and segregation. DNA sequences that are unpaired during meiosis (e.g. as a result of chromosomal rearrangements) are silenced in C. elegans germline cells by a process in which the RdRp EGO-1 has been implicated [10, 31]; in Neurospora crassa its orthologue SAD-1 displays similar activity [32]. The observed silencing of transgenes elicited by EGO-1 in Drosophila occurs in the absence of meiosis, e.g. in the direct progeny of the w; da-Gal4; UAST - ego-1 × w; +; UAS-syt.eGFP crosses. The silencing of a transgene like UAS-syt.eGFP by EGO-1 may however be explained by an unusual aspect of mitosis in Drosophila: the mitotic pairing of homologous chromosomes in somatic cells that has long been known to occur in Diptera [33]. Since the transgene (UAS-syt.eGFP) is unpaired in the progeny of this cross, it would be susceptible to detection by an EGO-1-induced process during mitotic pairing of chromosomes that would then result in its silencing.
Mitotic pairing underlies genetic phenomena such as transvection [34], which affects gene function [35, 36]. Work in Drosophila has shown transvection may be evident as silencing of paired genes (pairing-sensitive silencing, PSS) or of unpaired genes (un-paired silencing, UPS) [37–39]. Transvectional PSS affects genes carrying Polycomb group (PcG) response elements (PRE), which are involved in homeotic gene regulation [40–43]. While these studies, interestingly, suggested a role for RNAi pathway genes in pairing-sensitive transcriptional gene silencing (TGS), more recent work has found long ncRNAs to be involved in PRE silencing, so that the role of the RNAi pathway genes and short RNAs in pairing-sensitive TGS remains largely unclear [44, 45]. Interestingly, Kavi and Birchler [46] have recently suggested that the RNA polymerase polII may be associated with RNAi during heterochromatin silencing. Such suggestions are consistent with the observation that polII in the yeast Schizosaccharomyces pombe has RdRp activity [47].
The EGO-1-dependent silencing observed for unpaired genes in the present work is unlikely to be related to UPS. This specific form of transvection was observed under quite different experimental circumstances, in a study using a transgene driven by a copia LTR promoter which showed silencing was dependent on the histone acetyl transferase mof [48]. This was therefore a specific finding for a particular type of transgene, and required a promoter and a histone acetylation activity that cannot be linked to what is known about EGO-1 and its role in meiotic silencing. Overall, the differences between transvection, and the TGS observed in the present study, make it very unlikely that our observations result from transvection. This leads to the possibility that ego-1 may therefore trigger mitotic silencing of these unpaired transgenes by a mechanism similar to that used in meiotic silencing of unpaired DNA in C. elegans.
Conservation of pathways required for ego-1 induced silencing of unpaired DNA
The small RNA based gene silencing mechanism controlled by EGO-1 in C. elegans [31, 49] involves histone H3K9 dimethylation, which requires a specific histone methyltransferase (HMTase). Similarly in Drosophila, the HMTase encoded by Su(Var)3-9 plays a central role in heterochromatin gene silencing [50]. Indeed, the heterochromatin Su(Var)3-9 HMTases are highly conserved in eukaryotes [51], and introduction of the human gene into Drosophila allows partial rescue of silencing defects due to Su(Var)3-9 mutation [52].
The H3K9 HMTases have furthermore been shown to direct DNA methylation in fungi, plants and mammals [53, 54]. However methylation in Drosophila DNA remains controversial [55], having previously only been reported restricted to the early stages of embryonic development [56]. Our observation of possible methylation associated with EGO-1-dependent silencing is a further example of how the introduction of heterologous transgenes can be used in the study of this question. Previously, introduction of a mammalian Dnmt3a showed it to be active in Drosophila [57]; the biological effects evident upon DNA hypermethylation required the H3-K9 HMTase Su(Var)3-9, suggesting that further insights into possible methylation associated with EGO-1-dependent silencing could come from use of mutants in this Drosophila gene. In Drosophila, H3K9me2 marks are associated with the GREEN type of chromatin identified by [58]. A link between H3K9 trimethylation and postembryonic DNA methylation in Drosophila, mediated by the methyl cytosine binding domain protein dSETDB1, was recently identified by [59]. Finally, it will also be of interest to extend this study to ask whether the introduction of RdRps provides Drosophila with a novel genome-defense or RNA-based immunity capability against DNA or RNA viral genomes.
Materials and methods
Constructs and fly strains
Constructs for RdRp expression were as described in Duan et al. [20]. Fly strains used in this study in Supplementary Table 1 are from Ozdros, Australia (http://www.ozdros.com), Strains in Supplementary Table 2 are from Bloomington stock centre, USA, and strains in Supplementary Table 3 were created in this study using strains from Supplementary Table 1, Supplementary Table 2, or strains as described in Duan et al. [20].
Total RNA isolation
Adult flies, pupae, or 1° instar larvae were collected for total RNA extraction with TRIzol reagent (Invitrogen), RNA quality and quantity were checked with a Nano-Drop® Spectrophotometer ND-1000 (ThermoFisher Scientific) and an Agilent 2100 Bioanalyser (Agilent Technologies).
Quantitative real-time-PCR and result normalization
Initially, RT-PCR was performed to obtain the first strand cDNA with SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen), qPCR with the cDNA then was performed using iTaq™ SYBR® Green Supermix with ROX (BIO-RAD) on a machine 7900 HT Fast Real-Time PCR system (Applied Biosystems). Four biological replicates were included for each sample. A normalized value for absolute expression in each line was obtained by calculating the mean from the four replicates followed by a deduction of the mean values from no RT controls before divided by the actin mean value.
Small RNAs detection
Small RNAs was detected based on small RNA Northern blotting and hybridization [60, 61]. In general, ~20 μg total RNA for each sample was run on a 15 % polyacrylamide gel, and electroblotted onto Amersham Hybond™-N+ membrane (GE Healthcare), RNA was fixed on the membrane by UV crosslinking with 1200J using a UVP CL-1000 UV crosslinker. The membrane was hybridized overnight with isotope-labeled probes before exposure in a phosphor screen and detected with a FLA-5000 PhosphorImager (Fujifilm). Extended exposure time was required for weak signals. For re-probing, the membrane was washed with 0.1 % boiling SDS to remove the hybridized probe.
Small RNA probe preparation
Probes for small RNA hybridization were labeled with T7 RNA polymerase (Promega) and P32-UTP (PerkinElmer) based on template DNA containing a T7 RNA polymerase promoter. The labeled RNA was treated with RNAse-free DNAse (Promega), and purified using a GE Healthcare Radiolabeled Probe Purification Kit G-50 (GE Healthcare) according to the manufacturer’s instruction. For siRNA detection, probes were fragmented to ~50 nt with 200 mM carbonate solution before adding for hybridization.
RNAse protection assay
The RNAse protection assay was performed according to the instructions in the mirVana™ miRNA detection kit (Ambion). The hybridization reaction solutions were run on a 45 ml 15 % acrylamide gel followed by exposure with X-ray film in a cassette at −80 °C for 3–5 days (depending on the strength of the signals) before development on a AGFA CP1000.
Southern blotting and hybridization
Restriction fragment was detected based on Southern blotting [62]. In brief, genomic DNA was extracted, and followed by purification with phenol/chloroform extraction, ~15 μg gDNA for each sample was used for restriction digestion with HapII or HhaI, extended digestions (up to 3 h) were required until a pre-check on the gel showed that a smear was formed in each lane indicating the majority of gDNA was digested, DNA was run on a 1 % agarose gel. The gel was then depurinated in 250 mM HCl and denatured for 2 × 15 min, followed by neutralization at least 30 min. The gel was blotted overnight onto an Amersham Hybond™-N+ membrane (GE Healthcare) and UV-cross linked as Northern membrane preparation. The probes were radioactively labeled with ++P32-CTP (PerkinElmer) using a Ready-to-go bead (Amersham Biosciences), and cleaned on an Illustra™ probe Quant™ G50 Micro column (GE Healthcare) according to the manufacturer’s instructions. The membrane was hybridized overnight with the probe, and signal was detected with a FLA-5000 PhosphorImager (Fujifilm).
mRNA Northern blotting and hybridization
mRNA Northern hybridization was similar as Southern blotting and hybridization except that total RNA and 1.2 % formaldehyde gel were used. The membrane was hybridized with DNA probes labeled with Ready-to-go DNA labeling Beads (-dCTP) (GE Healthcare).
Bisulphite PCR and sequencing
The method for bisulphite PCR was based on Wang et al. [63]. Initially each DNA sample (2–5 μg) was treated with bisulphite using MethylEasy™ Xceed (Human Genetic Signature) according to the manufacturer‘s instruction. A hot-start PCR was performed using AmpliTaq Gold® 360 DNA polymerase (Applied Biosystems). Nested PCR product was checked and purified from the gel, followed by cloning into the pGEM-T easy vector (Promega) and transformed into DH10BT (Invitrogen). Plasmids were prepared from each individual colony and sequenced with M13 primer through Micromon (Monash University, Australia).
Sequence analysis
The sequencing results were initially analyzed by aligned to Drosophila pUAST expression vector using VectorNTI (Invitrogen), and individual Cytosine methylation was confirmed by non-“C to T” conversion, methylation pattern were analyzed by CYMATE (GMI) [29].
Fluorescence microscopy
The EGFP- or RFP-expressing larvae were collected from vials using 20 % sucrose, a pre-check (~100) with a UV fluorescence microscope (Leica MZ11 fluorescence microscope) was performed to show the fluorescence signals were consistent in flies with the same genotype, and about 12 larvae were randomly selected for photography using the program QCapture. For the EGFP gene, a EGFP2 filter was used at the excitation wavelength of 480 nm (excitation filter) and emission wavelength of 510 nm (barrier filter), a TXR filter was used for the RFP gene images.
Transgenic flies accession: The RdRp transgenic flies were deposited in Ozdros, Australia (http://www.ozdros.com).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Eleanor Maine, Syracuse University, USA, and Darryl Conte and Craig Mello, University of Massachusetts Medical School, USA, respectively, for generously providing the cDNAs of C. elegans ego-1 and rrf-1. We thank Joanne Milverton for microinjection and JianBin Wang for valuable assistance with the Drosophila crosses. We also thank Peter Grewe, Darren Obbard and some anonymous reviewers for valuable discussions and comments on the manuscript. Work in the authors’ laboratories on miRNAs is supported by the CSIRO Emerging Science Initiative on Cellular Reprogramming and the Special Research Centre of the Australian Research Council for the Molecular Genetics for Development.
Conflict of interest
The authors declare no competing financial interests.
References
- 1.Agrawal N, et al. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cogoni C, Macino G. Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10:638–643. doi: 10.1016/S0959-437X(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 3.May RC, Plasterk RH. RNA interference spreading in C. elegans . Methods Enzymol. 2005;392:308–315. doi: 10.1016/S0076-6879(04)92018-6. [DOI] [PubMed] [Google Scholar]
- 4.Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;39:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Obbard DJ, Gordon KH, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans . EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 9.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 10.Maine EM, et al. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 12.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 13.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 14.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 15.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roignant JY, et al. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila . RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saleh MC, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncb1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulvila J, et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- 19.Saleh MC, et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan G, et al. Expression of Caenorhabditis elegans RNA-directed RNA polymerase in transgenic Drosophila melanogaster does not affect morphological development. Transgenic Res. 2010;19:1121–1128. doi: 10.1007/s11248-010-9372-y. [DOI] [PubMed] [Google Scholar]
- 21.Prokopenko SN, et al. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila . Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Impel A, et al. Regulation of the Rac GTPase pathway by the multifunctional Rho GEF Pebble is essential for mesoderm migration in the Drosophila gastrula . Development. 2009;136:813–822. doi: 10.1242/dev.026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shandala T, Gregory SL, Dalton HE, Smallhorn M, Saint R. Citron kinase is an essential effector of the Pbl-activated Rho signalling pathway in Drosophila melanogaster . Development. 2004;131:5053–5063. doi: 10.1242/dev.01382. [DOI] [PubMed] [Google Scholar]
- 24.Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 25.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 26.Chang H (2004) Chang constructs and insertions. Personal communication FlyBase
- 27.Hodge JJ. Ion channels to inactivate neurons in Drosophila . Front Mol Neurosci. 2009;2:13. doi: 10.3389/neuro.02.013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet. 2011;27:127–131. doi: 10.1016/j.tig.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- 30.Luan H, et al. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila . J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maine EM. Meiotic silencing in Caenorhabditis elegans . Int Rev Cell Mol Biol. 2010;282:91–134. doi: 10.1016/S1937-6448(10)82002-7. [DOI] [PubMed] [Google Scholar]
- 32.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/S0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 33.McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta. 2004;1677(1–3):165–180. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Lewis EB. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster . Am Nat. 1954;88:225–239. doi: 10.1086/281833. [DOI] [Google Scholar]
- 35.Golic MM, Golic KG. A quantitative measure of the mitotic pairing of alleles in Drosophila melanogaster and the influence of structural heterozygosity. Genetics. 1996;143:385–400. doi: 10.1093/genetics/143.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tartof KD, Henikoff S. Trans-sensing effects from Drosophila to humans. Cell. 1991;65:201–203. doi: 10.1016/0092-8674(91)90153-P. [DOI] [PubMed] [Google Scholar]
- 37.Duncan IW. Transvection effects in Drosophila . Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 38.Kennison JA, Southworth JW. Transvection in Drosophila . Adv Genet. 2002;46:399–420. doi: 10.1016/S0065-2660(02)46014-2. [DOI] [PubMed] [Google Scholar]
- 39.Wu CT, Morris JR. Transvection and other homology effects. Curr Opin Genet Dev. 1999;9:237–246. doi: 10.1016/S0959-437X(99)80035-5. [DOI] [PubMed] [Google Scholar]
- 40.Grimaud C, et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 41.Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression in Drosophila: gene silencing of alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/S0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- 42.Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression of nonhomologous transgenes in Drosophila involves mutually related endogenous sequences. Cell. 1999;99:35–46. doi: 10.1016/S0092-8674(00)80060-4. [DOI] [PubMed] [Google Scholar]
- 43.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila . Mol Cell. 2002;9:315–327. doi: 10.1016/S1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 44.Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6:129–137. doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- 45.Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 46.Kavi HH, Birchler JA. Interaction of RNA polymerase II and the small RNA machinery affects heterochromatic silencing in Drosophila . Epigenetics Chromatin. 2009;2:15. doi: 10.1186/1756-8935-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–449. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
- 48.Matyunina LV, Bowen NJ, McDonald JF. LTR retrotransposons and the evolution of dosage compensation in Drosophila . BMC Mol Biol. 2008;9:55. doi: 10.1186/1471-2199-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smardon A, et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans . Curr Biol. 2000;10:169–178. doi: 10.1016/S0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 50.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila . Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 51.Krauss V. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica. 2008;133:93–106. doi: 10.1007/s10709-007-9184-z. [DOI] [PubMed] [Google Scholar]
- 52.Schotta G, et al. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 54.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/S0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 55.Schaefer M, Lyko F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat Genet. 2010;42:920–921. doi: 10.1038/ng1110-920. [DOI] [PubMed] [Google Scholar]
- 56.Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster . Nature. 2000;408(6812):538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 57.Weissmann F, et al. DNA hypermethylation in Drosophila melanogaster causes irregular chromosome condensation and dysregulation of epigenetic histone modifications. Mol Cell Biol. 2003;23:2577–2586. doi: 10.1128/MCB.23.7.2577-2586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filion GJ, Steensel B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat Genet. 2010;42(4):5–6. doi: 10.1038/ng0110-4. [DOI] [PubMed] [Google Scholar]
- 59.Gou D, et al. SETDB1 is involved in postembryonic DNA methylation and gene silencing in Drosophila . PLoS One. 2010;5:e10581. doi: 10.1371/journal.pone.0010581. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Brosnan CA, et al. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis . Proc Natl Acad Sci USA. 2007;104:14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitter N, Sulistyowati E, Dietzgen RG. Cucumber mosaic virus infection transiently breaks dsRNA-induced transgenic immunity to Potato virus Y in tobacco. Mol Plant Microbe Interact. 2003;16:936–944. doi: 10.1094/MPMI.2003.16.10.936. [DOI] [PubMed] [Google Scholar]
- 62.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/S0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 63.Wang MB, Wesley SV, Finnegan EJ, Smith NA, Waterhouse PM. Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA. 2001;7:16–28. doi: 10.1017/S1355838201001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.