Abstract
SRC-like adaptor protein (SLAP) is an adaptor protein structurally similar to the SRC family protein kinases. Like SRC, SLAP contains an SH3 domain followed by an SH2 domain but the kinase domain has been replaced by a unique C-terminal region. SLAP is expressed in a variety of cell types. Current studies suggest that it regulates signaling of various cell surface receptors including the B cell receptor, the T cell receptor, cytokine receptors and receptor tyrosine kinases which are important regulator of immune and cancer cell signaling. SLAP targets receptors, or its associated components, by recruiting the ubiquitin machinery and thereby destabilizing signaling. SLAP directs receptors to ubiquitination-mediated degradation and controls receptors turnover as well as signaling. Thus, SLAP appears to be an important component in regulating signal transduction required for immune and malignant cells.
Keywords: SLAP-2, FLT3, c-KIT, PDGFR, EPHA2, EPOR, CSF1R, SYK
Introduction
A majority of cellular processes are controlled by extracellular factors, which bind to cell surface receptors and transduce signals to intracellular signaling molecules. Adaptor proteins play, by linking signaling molecules to the receptors, an important role in signal transduction. This class of proteins contains multiple functional domains and thus can create multi-protein complexes that facilitate signal transduction. Adaptor proteins can regulate cell surface receptor signaling by amplifying the signal or diminishing the signal. In the first case, it recruits signaling molecules that further transduce the signal through the downstream signaling cascades. In the other cases, it recruits negative regulators such as ubiquitin ligases or phosphatases that shut down the signal either by destabilizing the receptor and/or downstream signaling molecule or by de-phosphorylating the receptor, which leads to inactivation of the receptors. SRC-like adaptor protein (SLAP) is an adaptor protein that displays considerable structural homology with SRC [1]. SLAP is expressed in a variety of tissues and known to regulate the signaling of multiple receptors in different systems.
SLAP-deficient mice have been generated. Mice lacking SLAP appear to be healthy, and do not display any obvious physical abnormalities suggesting that SLAP is not essential for embryonic development [2]. However, in adult mice, SLAP negatively regulates differentiation of osteoclasts and proliferation of their precursors [3]. Furthermore, SLAP-deficient mice have higher TCR signaling in comparison to wild-type mice [4]. SLAP2 knockout and SLAP/SLAP2 double-knockout mice have also been described [5]. SLAP2 knockout mice did not show any considerable effect on ZAP70 or TCR signaling. Furthermore, double-knockout mice displayed only the phenotype that SLAP knockout mice show in TCR signaling suggesting that SLAP, but not SLAP2, is required for maintaining TCR signaling. Therefore, it is more likely that SLAP is involved in controlling normal cellular signaling and that loss of function results in aberrant activation of survival pathways.
The SLAP gene and protein
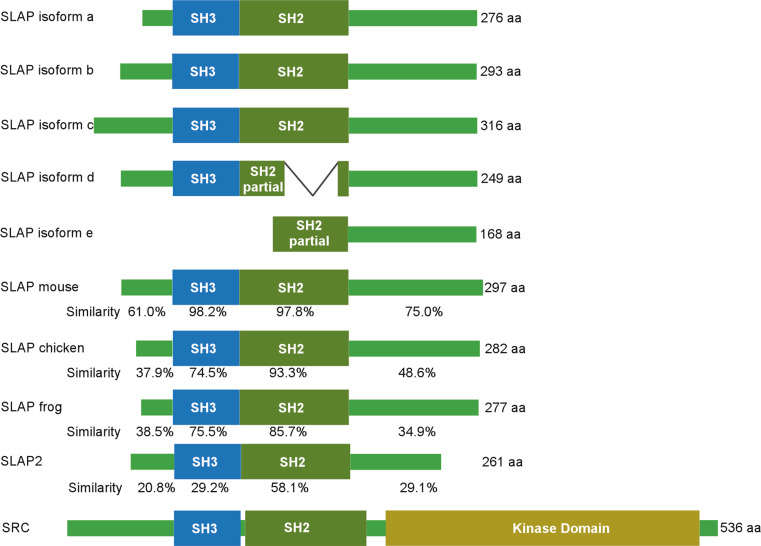
SLAP was initially identified in mice using a yeast two-hybrid screen as a 34 kDa protein [6]. In the same year, SLAP gene was mapped in humans to chromosome 8 (8q22.3) and in mice to chromosome 15 (15 D2) [7]. Both human and mouse SLAP genes contain seven exons and six introns [7–9]. SLAP is also known as SLA and SLA1. The closest homolog of SLAP, SLAP2 was described after identification of SLAP [10, 11]. The SLAP2 gene is located on human chromosome 20q11.23 consisting of seven exons [12]. According to the Gene database (http://www.ncbi.nlm.nih.gov/gene/) the human SLAP gene encodes 5 different isoforms due to the alternative splicing (Fig. 1). The alternative splicing human SLAP gene displays differences in tissue abundance [9]. While the human SLAP2 gene encodes a 261 amino acid protein, an alternative spliced isoform of 210 amino acids (SLAP-2-v) has also been described [12]. SLAP a, b and c isoforms have a short N-terminal uncharacterized region followed by an SH3 and an SH2 domain. All isoforms have a similar C-terminal uncharacterized region. Isoform d lacks a functional SH2 domain and isoform e lacks both SH3 and SH2 domains. The N-terminus of the b, c and d isoforms contains a PAAP motif, indicating the possibility of interaction with SH3 domain-containing proteins. Although the tissue specific roles of different splice variants have not yet been defined, it is likely that due to the difference in the arrangements of functional domains, different splice variants of SLAP play different roles in different tissues.
Fig. 1.
Structure of SLAP. The SLAP gene is expressed as 5 transcript variants. Variants a, b and c contain intact SH2 and SH3 domains, while the d variant lacks functional SH2 domain and the e variant lacks both domains. The functional domains of SLAP are highly conserved between species including human, mouse, chicken and frog. The uncharacterized C-terminal region displays comparatively poor conservation. SLAP shares considerable sequence similarity with its homolog SLAP2. SLAP has a similar domain structure as SRC except for the presence of a functional kinase domain in SRC
The expression of SLAP
SLAP is expressed in a variety of human and murine tissues (Table 1). Although data from Northern blotting suggest that spleen, lung, thymus and lymph nodes express relatively higher levels of SLAP mRNA [6, 9, 13], expression data from RNAseq and microarray analysis display discrepancies in lungs and other non-hematopoietic organs (Table 1). B cells and T cells from spleen, and thymocytes express moderate levels of SLAP mRNA [13]. Therefore, it has been suggested that SLAP may play an important role in the B cell and T cell lineages. SLAP expression has also been described in Jurkat T cells [14]. Differences in SLAP expression for the same tissue in the Table 1, for example, mouse heart, liver, skeletal muscle and kidney, are not surprising as the experimental settings were different in different studies. Therefore, mRNA levels depicted in Table 1 reflect the relative expression of SLAP within each experimental setting. Besides abundant expression of SLAP in B cells and T cells, several factors induce SLAP expression in hematopoietic cell lines. For example, all-trans retinoic acid (ATRA) induces SLAP expression in U937, HL60 and NB-4 cells [15]. Since, ATRA induces differentiation in these cell lines, it is possible that SLAP expression is required for the cell maturation process [15]. However, the role of SLAP in cell differentiation is still largely unknown. Like ATRA, dexamethasone exposure also markedly increases expression of SLAP in RBL-2H3 mast cells [16]. This regulation is mediated through the transcriptional activation of the glucocorticoid response element (GRE) [17] and thus it has been suggested that SLAP is a regulator of glucocorticoid-induced cell death [18].
Table 1.
SLAP mRNA expression in human and murine tissues using Northern blotting (NB) comparing with RNAseq and microarray data
| Tissue/cell line | Species | NB | References | FPKMa | RPKMb | Expressionc |
|---|---|---|---|---|---|---|
| Heart | Human | + | [9] | 2 | 2.1 | 13 |
| Mouse | + | [6] | ||||
| − | [13] | |||||
| Brain | Human | + | [9] | NF | 3.7 | NF |
| Mouse | ++ | [6] | ||||
| + | [13] | |||||
| Spleen | Mouse | ++++ | [6] | 50 | 38.6 | NF |
| +++ | [13] | |||||
| Lung | Human | +++ | [9] | 16 | 13.3 | 35 |
| Mouse | +++ | [6] | ||||
| + | [13] | |||||
| Liver | Human | + | [9] | 5 | 1.3 | 13 |
| Mouse | + | [6] | ||||
| − | [13] | |||||
| Skeletal muscle | Human | + | [9] | 0 | 0.4 | 17 |
| Mouse | + | [6] | ||||
| − | [13] | |||||
| Kidney | Mouse | + | [6] | 3 | 2 | 8 |
| − | [13] | |||||
| Human | + | [9] | ||||
| Testis | Mouse | − | [6] | 2 | 0.5 | 10 |
| − | [13] | |||||
| Placenta | Human | + | [9] | 8 | NF | 12 |
| Pancreas | Human | + | [9] | 3 | 0.9 | 9 |
| Lymph nodes | Mouse | ++ | [13] | 72 | NF | 91 |
| Thymus | Mouse | ++++ | [13] | NF | NF | 460 |
− absent, + lower, ++ moderate, +++ higher, ++++ very high, NF not found
aData from the Human Protein Atlas
bData from GTExPortal
cData from BioGPS
SLAP2 expression overlaps in many cases with SLAP expression. For example SLAP2 expression is maintained in thymocytes development in a similar fashion to SLAP expression [5]. SLAP2 is highly expressed in bone marrow macrophages, hematopoietic cells such as platelets, monocytes, T cells, B cells, spleen, leukocytes, lung and thymus [3, 10–12, 19, 20]. Therefore, it is more likely that SLAP and SLAP2 share similar functions in many cases.
SLAP in intracellular signaling
So far 18 different proteins have been described to associate with SLAP including cell surface receptors, membrane-bound non-receptor proteins and cytosolic proteins, although in several cases it remains unknown whether the interaction was direct between SLAP and the partner protein (Table 2). Therefore, it has been suggested that SLAP regulates multiple signaling pathways by interacting with various proteins. Since SLAP remains myristoylated in G2 and localizes to the cell membrane [21], it is expected that SLAP associates with membrane-bound proteins. Several receptor tyrosine kinases including EPHA2 [6], FLT3 [22], KIT [23], PDGFR [1] and the cytokine receptor EPOR [24] interact with SLAP in a tyrosine phosphorylation-dependent manner. Receptor tyrosine kinases transduce signal partially through adaptor proteins, which recruits downstream signaling components or negative regulators [25–28]. Thus, association of SLAP with receptor tyrosine kinases may have a role in receptor downstream signaling. SLAP also interacts with multiple intracellular tyrosine kinases, such as SYK [14], LCK [21] and ZAP70 [14] which are involved in T cell receptor (TCR) or B cell receptor (BCR) signaling. SLAP interacts with the TCR and BCR component CD3ζ [14], Igα [29] and LAT [14]. In contrast, SLAP2 interacts with ZAP70 and CD3ζ but not with LAT [11, 19]. There is also evidence that both SLAP and SLAP2 associate with the ubiquitin E3 ligase CBL [11, 14, 19]. Furthermore, SLAP associates with the guanine nucleotide exchange factor VAV1 [13] and SH2 domain-containing leukocyte protein SLP76 [13]. Most of the SLAP-binding proteins are important regulators of T cell and B cell signaling suggesting that SLAP is involved in the regulation of normal immunity.
Table 2.
SLAP-interacting proteins
| Name | Method and domain involved | Functions | References |
|---|---|---|---|
| CBL | In vitro GST pull-down, co-transfection in COS7 cells, phosphorylation-independent interaction, associates through C-terminal hydrophobic region | positive regulation of NFAT-AP1 activity | [14] |
| Co-immunoprecipitation in transfected Jurkat T cells | Negative regulation | [33] | |
| Co-immunoprecipitation, independent of PDGF stimulation | Negative regulation | [45] | |
| CD247/CD3ζ/TCRζ | Co-immunoprecipitation, TCR activation, indirect interaction, associates through SH2 domain | Positive regulation of NFAT-AP1 activity | [14] |
| Co-immunoprecipitation in Jurkat T, associates through SH2 domain | Down-regulates TCRζ | [32] | |
| CD79A/Igα | Co-immunoprecipitation, TCR activation, associates through SH2 domain | Negative regulation | [29] |
| EPHA2/ECK | In vitro GST pull-down, ligand stimulation-dependent associates through SH2 domain | Unknown | [6] |
| Tumor suppressor functions | [44] | ||
| FLT3 | Co-immunoprecipitation in COS1 cells, ligand stimulation | Positive regulation of AKT, ERK, p38 signaling | [22] |
| KIT | Co-immunoprecipitation in transfected COS1 cells, endogenous in P815 cells, ligand stimulation, associates through SH2 domain | Negative regulation | [23] |
| PDGFRB | Overexpression in COS7 cells, endogenous in NIH3T3 cell line, ligand stimulation-dependent, associates through SH2 domain | SLAP tyrosine phosphorylation, negatively regulates mitogenesis | [1] |
| Binding was not been determined | Strongly inhibits PDGF response | [21] | |
| Co-immunoprecipitation, ligand dependent | Negative regulation | [45] | |
| SYK | Co-immunoprecipitation in transfected COS7 cells, associates through SH2 domain | SLAP dimerization is independent of SYK or LCK activity, required for positive regulation of NFAT-AP1 activity | [14] |
| ZAP70 | |||
| LAT/pp36 | |||
| SLAP | Forms homodimers through C-terminal region | ||
| VAV1 | GST pull-down and immunoprecipitation in SLAP-transfected Jurkat TAg cells | Inhibited TCR-induced NFAT activation | [13] |
| LCK | |||
| SLP76 | |||
| EPOR | Co-immunoprecipitation in overexpressed 293T cells and EI-11 erythroblasts | Negative regulation | [24] |
| UB4EA | Through the SH3 domain | Tumor suppressor functions | [44] |
| GM-CSFRα | GST pull-down, both SH3 and SH2 domains are involved | Regulates monocytic dendritic cell maturation | [41] |
| CSF1R | GST pull-down, CSF1-dependent | Unknown | [47] |
SLAP-mediated regulation of TCR signaling
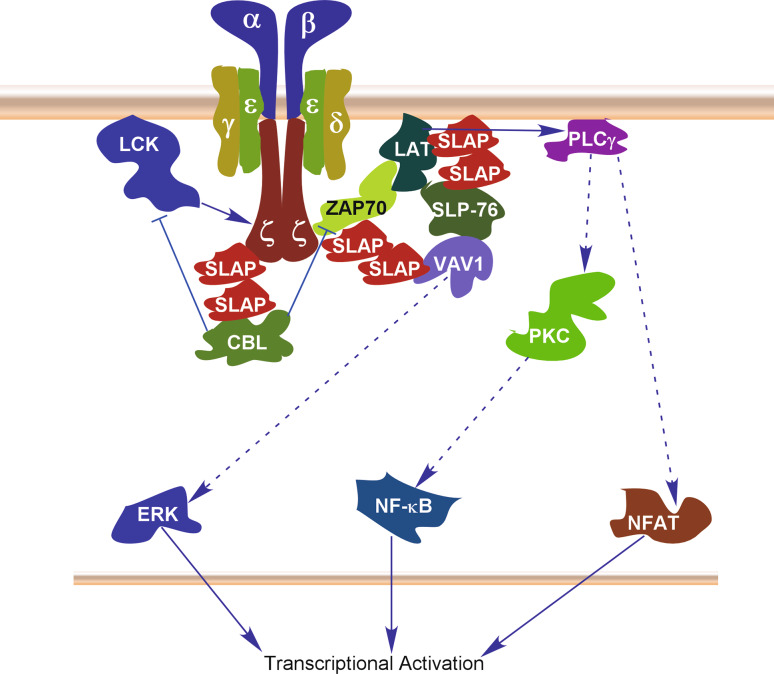
The T cell receptor (TCR) activation is a critical event for T cell development in the thymus. TCR activation is also required for initiating immune responses in mature T cells. Activation of the TCR is initiated by SRC family kinases (such as LCK) resulting in activation of multiple protein tyrosine kinases which in turn phosphorylate various substrates and thereby initiating transcriptional activation of pro-survival genes [30]. Our current understanding suggests that SLAP plays important roles in the control of TCR signaling by regulating at least three different signaling pathways (Fig. 2). SLAP associates with multiple TCR signaling components including CBL, ZAP70, SYK, LAT, CD3ζ, SLP76 and VAV1 upon TCR activation in Jurkat cells [13, 14]. The SH2 domain of SLAP is involved in interaction with those tyrosine-phosphorylated proteins, except CBL that associates through the C-terminal hydrophobic region (Table 2). This probably allows SLAP to recruit the ubiquitin machinery to the signaling proteins which subsequently leads to their degradation. Such regulation has been reported where SLAP binds with CD3ζ-chain of TCR and directs it for proteasomal degradation [31]. In addition, cells lacking SLAP express higher levels of CD3ζ which further suggests that SLAP is required to maintain basal CD3ζ levels [32]. This regulation is mediated through the recruitment of the ubiquitin ligase CBL to SLAP through LCK-dependent phosphorylation [33]. SLAP dimerizes through its C-terminal region which is also required for SLAP-mediated down-regulation of AP1-activity in T cells [14]. This dimerization property allows SLAP to create multi-protein complexes.
Fig. 2.
SLAP in regulation of TCR signaling. Activation of the TCR results in activation of ERK1/2, NF-κB and NFAT signaling through different mechanisms. Activation of TCR signaling initiates tyrosine phosphorylation mediated by tyrosine kinases such as LCK. Tyrosine phosphorylation creates docking site for SLAP that in turn recruits the ubiquitin ligase CBL. SLAP associates with TCRζ, ZAP70 and LAT through its SH2 domain, while CBL associates with SLAP through its C-terminal hydrophobic region. The C-terminal region is also involved in the dimerization of SLAP. Recruitment of CBL directs TCR components to proteasomal degradation resulting in negative regulation of all three pathways
LAT is an upstream activator of PLCγ which in turn activates NFAT and AP-1 in a calcium- and protein kinase C (PKC)-dependent manner. Overexpression of SLAP inhibits the activity of NFAT and AP-1 which can be restored by treatment with ionomycin and phorbol myristate acetate (PMA) [13]. Ionomycin boosts the cellular calcium ion concentration and PMA is a well-known activator of the PKC family proteins [34, 35]. Thus, SLAP controls LAT signaling by deregulating its activity through association. It is also evident that SLAP deficiency leads to up-regulation of TCRβ, CD3, CD4, CD5, CD8 and CD69 levels [2] suggesting that SLAP destabilizes multiple proteins in T cells. Furthermore, SLAP deficiency rescues T cell development in ZAP70 null mice [2]. Therefore, it is suggested that SLAP is an important adaptor protein which is required for the control of normal TCR signaling.
SLAP-mediated regulations of BCR signaling
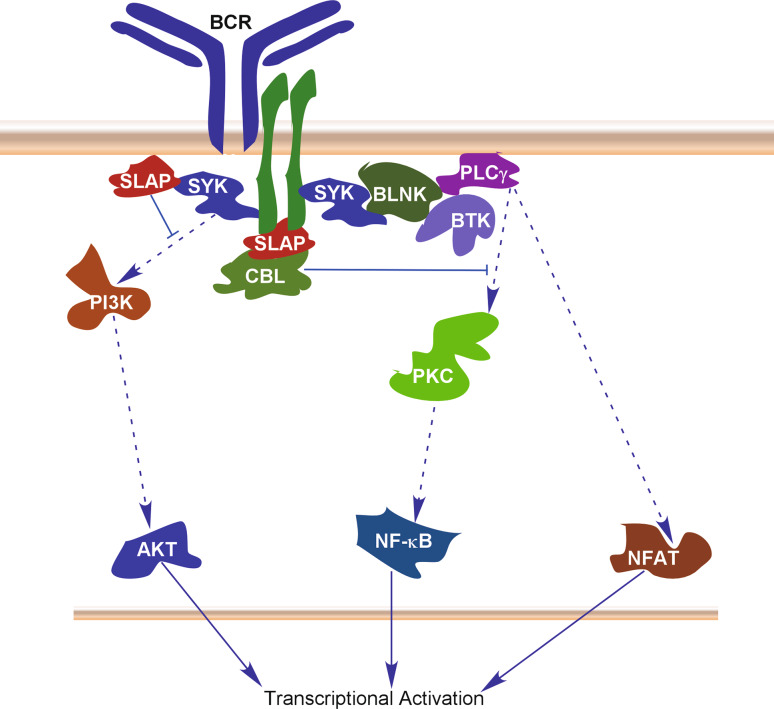
B cell receptor (BCR) signaling is essential for the development and function of B cell. BCR signaling contributes to the antigen-triggered differentiation of memory B cells and antibody-secreting plasma cells [36]. Activation of BCR induces tyrosine phosphorylation on Ig chains, thereby creating docking sites for the SH2 domain-containing tyrosine kinase SYK. These events in turn activate AKT, NF-κB and NFAT signaling leading to transcriptional activation of effector genes. SLAP expression controls the levels of B cell receptor expression and it has been demonstrated that SLAP expression is required to maintain normal levels of BCR expression [37]. SLAP also controls signaling through the BCR complex and B cell development. SLAP has been shown to be directly associated with tyrosine-phosphorylated components of the BCR complex and down-regulates the surface levels and total BCR levels in a CBL-dependent manner [29]. The C-terminal region of SLAP is required for CBL-mediated down-regulation of BCR, suggesting that SLAP recruits the ubiquitin machinery to the signaling proteins through its multiple functional units. SLAP deficiency leads to a decreased number of autoreactive B cells, which is maintained by an inefficient receptor regulation [38]. Since SLAP is capable of binding to SYK as well [14], it is likely that SLAP binds to multiple components of the BCR complex and negatively regulates signaling by directing proteins for CBL-mediated degradation (Fig. 3). SLAP associates with tyrosine-phosphorylated SYK and BCR Igα through its SH2 domain, and destabilizes these two intermediate components of BCR signaling by linking to CBL. Therefore, the SLAP activity is important for maintaining normal BCR signaling.
Fig. 3.
SLAP in regulation of BCR signaling. Activation of BCR results in the activation of AKT, NF-κB and NFAT signaling through different mechanisms. SLAP binds to the BCR components Igα and SYK through its SH2 domain and then recruits CBL resulting in negative regulation of all three pathways. SLAP destabilizes SYK and BCR Igα which are intermediate signaling components of BCR signaling
SLAP-mediated regulations of cytokine signaling
EPOR
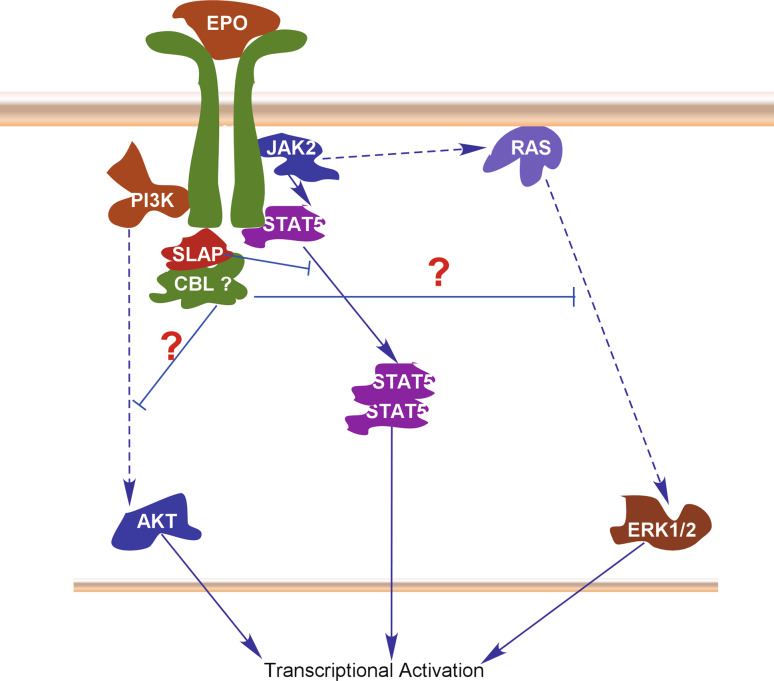
The cytokine erythropoietin (EPO) and its receptor (EPOR) are important regulators of erythroid progenitor cells. Activation of EPOR blocks apoptosis, and promotes terminal differentiation and proliferation of erythroid progenitor cells [39]. These processes are mediated through activation of several signaling cascades including the PI3-K/AKT, JAK2/ERK1/2 and STAT5 pathways [40]. SLAP counteracts EPO-induced anti-apoptotic properties by inhibiting EPO-induced STAT5 activation in erythroblasts [24]. SLAP directly associates with EPO receptor, EPOR and thus controls EPO signaling probably through CBL-dependent degradation of the receptor, as CBL [33] has been shown to be associated with SLAP (Fig. 4). SLAP probably inhibits EPO-induced AKT, STAT5 and ERK1/2 activation by CBL-mediated degradation of EPOR; however, it yet has to be determined whether SLAP down-regulates EPOR signaling through CBL-dependent degradation.
Fig. 4.
SLAP in regulation of EPOR signaling. Activation of the EPOR results in the activation of AKT, STAT5 and ERK1/2 signaling. SLAP counteracts EPOR signaling probably by recruiting CBL to the EPOR. CBL-dependent degradation of EPOR results in weaker activation of AKT, STAT5 and ERK1/2. Therefore, SLAP negatively regulates all three pathways
GM-CSFR
The cytokine receptor GM-CSFR plays an important role in maturation of dendritic cells. Bone marrow-derived dendritic cells derived from mice deficient of SLAP and its close homolog SLAP2 display increased GM-CSFRβ stability resulting in elevated activation of JAK2, AKT and ERK1/2 signaling in response to GM-CSF stimulation [41]. Although SLAP and SLAP2 regulated stability of GM-CSFRβ subunit, neither SLAP nor SLAP2 associated with GM-CSFRβ. However, only SLAP but not SLAP2 interacted with GM-CSFRα through SH2 and SH3 domains. These dendritic cells display poor response to lipopolysaccharide in IL-12 and TNFα production and therefore fail to stimulate T cells. SLAP function is required in maintaining baseline GM-CSFR signaling which is important for the generation of functionally mature monocytic dendritic cells [41].
SLAP-mediated regulation of RTK signaling
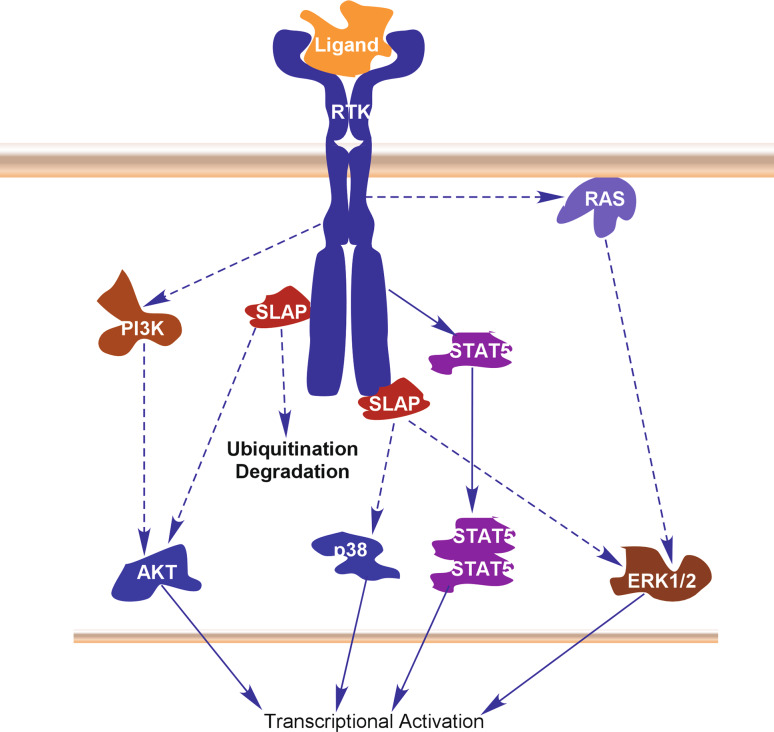
Receptor tyrosine kinases (RTKs) are cell surface receptors that activate diverse signaling cascades upon ligand stimulation. RTK signaling is tightly controlled by regulatory proteins [42]. In general, activation of RTKs initiates various signaling cascades through activation of AKT, p38, ERK1/2 and in some cases STAT5. Like other proteins SLAP associates with RTKs and induces ubiquitination-dependent degradation. Association of SLAP with RTKs is mediated by the SLAP SH2 domain and phospho-tyrosine residue on the RTK. Therefore, SLAP predominantly acts as a negative regulator of RTK signaling. However, it is currently evident that SLAP also cooperates with RTK to potentiate RTK signaling (Fig. 5).
Fig. 5.
SLAP in regulation of RTK signaling. Activation of RTK signaling results in the activation of AKT, p38, STAT5 and ERK1/2 signaling through different mechanisms. SLAP binds to RTKs and display different functions. SLAP associates with phospho-tyrosine residues of the activated receptor through its SH2 domain. While SLAP association increases receptor ubiquitination and accelerates the degradation, it both negatively and positively regulates receptor signaling depending on the receptor type
EPHA2
The adaptor protein SLAP was initially identified as an EPHA2-(also known as ECK) binding protein [6]. EPHA2 is a member of EPH receptors family of the RTKs that binds to membrane-bound ligands denoted ephrins. SLAP binding to EPHA2 leads to recruitment of the N-methyl-d-aspartate (NMDA) receptor as well, and down-regulates this receptor as well as downstream signaling [43]. Furthermore, a SLAP-mediated tumor suppressor functions in colorectal cancer have been found to be mediated through destabilization of the EPHA2 receptor [44]. EPHA2 is a well-known substrate of SRC and by destabilizing EPHA2, SLAP controls SRC-EPHA2-AKT signaling. SLAP recruits the ubiquitin ligase UBE4A to the EPHA2 through the SLAP-SH3 domain and the SLAP-SH2 domain interacts with EPHA2 through pY594 which is a SRC phosphorylation site. Therefore, SLAP function is required for controlling EPHA2 receptor activity which is partially mediated through destabilization of the receptor by ubiquitination.
PDGFR
SLAP negatively regulates PDGF signaling by associating with PDGFRB in response to PDGF stimulation in NIH3T3 cells [1]. The binding sites of SLAP were mapped in PDGFRB. The SLAP-binding sites are completely overlapping with the SRC-binding sites (Y579 and Y581) in PDGFRB, which are located in the juxtamembrane domain. Overexpression of SLAP in NIH3T3 cells inhibits PDGF-induced DNA synthesis and the mitogenic response to both serum and PDGF. An intact SH2 domain was essential for this effect but the SH3 domain was dispensable, suggesting that inhibition is partially due to the competition for binding of SRC to tyrosine-phosphorylated residues in PDGFRB [1, 21]. Later studies also demonstrated that SLAP controls PDGFR signaling by competing with SRC, but it recruits CBL to the receptor as well [45]. Thus, SLAP controls PDGF signaling by two different mechanisms: by limiting access of SRC and by destabilizing the receptor through CBL-mediated ubiquitination.
CSF1R
Colony-stimulating factor 1 receptor (CSF1R) regulates proliferation, differentiation and function of macrophages when stimulated with its ligand colony-stimulating factor 1 (CSF1). Activation of CSF1R results in activation of multiple downstream signaling cascades including PI3 K/AKT and RAS/ERK pathways through SH2 domain-containing adaptors or kinases. Since SLAP and SLAP2 genes are constitutively expressed in bone marrow macrophages [3, 46], it is expected that these genes will contribute to the regulation of CSF1R signaling. SLAP2 remains constitutively associated with the CSF1R independent of ligand stimulation [46, 47]. However, how SLAP2 associates with the CSF1R remains debated. While overexpression studies in HEK293 cells suggest that SLAP2 associates with the CSF1R independent of both the N-terminal region, the SH2 domain, the SH3 domain and the C-terminal region [46], another study suggests that association was dependent on the SLAP2-SH2 domain but not on the C-terminal region, as a point mutation in the phosphotyrosine-binding pocket of the SH2 domain completely abolished association in GST pull-down assay [47]. Since SLAP2 constitutively associates with CSF1R, it would be more likely that SLAP2 uses other binding sites than the SH2 domain. However, association with SLAP was completely dependent on CSF1 stimulation [47]. Therefore, it is suggested that the mechanism of interaction between CSF1R and SLAP or between CSF1R and SLAP2 is different. Interaction between SLAP2 and CSF1R results in down-regulation of the CSF1R in response to ligand stimulation of the CSF1R. Furthermore, SLAP2 has been shown to be associated with CBL in primary bone marrow-derived macrophages and expression of a SLAP2 SH2 domain mutant stabilizes CSF1R [10, 47] suggesting that SLAP2 down-regulates CSF1R through CBL-dependent degradation.
FLT3
FLT3 and its ligand (FL) play vital roles in the immune system and also in certain types of cancers. SLAP associates with FLT3 in response to FL stimulation [22]. Although association with SLAP induces accelerated ubiquitination and degradation of FLT3, it also enhances receptor signaling by an unknown mechanism [22]. Probably, the SLAP SH3 domain is involved in the amplification of signals from FLT3. Since SLAP associates with FLT3 through its SH2 domain, the SH3 domain is available to interact with SH3 domain-binding proteins resulting in activation of downstream signaling cascades.
KIT
Another receptor tyrosine kinase KIT and its oncogenic mutants are involved in several cancers including mastocytoma, gastrointestinal stromal tumor (GIST), melanoma and certain type of acute myeloid leukemia. Both wild-type and oncogenic mutants of KIT associate with SLAP. While the interaction between wild-type KIT and SLAP is dependent of stem cell factor (SCF) stimulation, oncogenic KIT-D816V mutant displays constitutive binding [23]. SLAP can block KIT signaling by inducing ubiquitination followed by degradation, but it was unable to block oncogenic KIT-D816V-mediated mitogenic signaling [23]. KIT-D816V can selectively phosphorylate SLAP on three tyrosine residues of which two are located in the C-terminal and involved in dimerization. Probably, KIT-D816V-mediated tyrosine phosphorylation blocks dimerization of SLAP and subsequently makes SLAP inactive, which can be restored by a triple (Y-F) SLAP mutant [23].
SLAP regulates SYK signaling
Besides cell surface receptor tyrosine kinases, SLAP associates with the intracellular tyrosine kinase SYK and partially blocks SYK tyrosine phosphorylation as well as phosphorylation of PLCγ2 and ERK suggesting that SLAP acts as a negative regulator of SYK signaling [16, 48]. Therefore, the role of SLAP in receptor tyrosine kinase signaling is fairly complicated. SLAP can both inhibit and activate signaling downstream of receptors.
Conclusions
Current studies suggest that SLAP mainly acts as an adaptor to recruit the ubiquitination machinery resulting in negative regulation of receptor signaling. Although a majority of studies support this idea, there is sufficient evidence that SLAP can contribute to positive regulation of receptor signaling. The presence of multiple domains, proline-rich motif and the ability to dimerize leads to a multitude of levels of regulation of receptor signaling. Our current knowledge is still mainly limited to the regulation of mitogenesis by SLAP and future studies should define the role of SLAP in the regulation of other cellular processes such as metabolism, cell cycle regulation, intracellular trafficking, etc. The role of SLAP has been defined in the regulation of limited number of receptors, and its expression in a variety of tissues suggests that SLAP might be important in the regulation of diverse signaling pathways. Therefore, identification of novel SLAP-binding proteins will enhance our knowledge about the mechanisms of cellular regulatory mechanisms. Although SLAP is abundantly expressed in the healthy intestinal epithelium, it is strongly down-regulated in 50 % of colorectal cancer patients. Silencing of SLAP enhances tumorigenicity and invasiveness in colorectal cancer cell lines, while overexpression of SLAP suppresses these transforming properties. Regulation of receptor tyrosine kinases frequently mutated or involved in cancers also provide evidence that SLAP is an important regulator for maintaining normal cellular functions.
Acknowledgements
This research was funded by Kungliga Fysiografiska Sällskapet i Lund (JUK), Ollie and Elof Ericssons Stiftelse (JUK), Åke-Wiberg Stiftelse (JUK), Lars Hiertas Minne Stiftelse (JUK), Stiftelsen Olle Engkvist Byggmästare (JUK), Harald Jeanssons Stiftelse and Harald och Greta Jeanssons Stiftelse (JUK), Swedish Childhood Cancer Foundation (JUK), Gunnar Nilsson Cancer Foundation (LR), Swedish Cancer Society (LR), BioCARE cancer network (LR and JUK) and the Swedish Research Council (LR). JUK is a recipient of Assistant Professorship (forskarassistenttjänst) grant from the Swedish Childhood Cancer Foundation.
References
- 1.Roche S, Alonso G, Kazlauskas A, Dixit VM, Courtneidge SA, Pandey A. Src-like adaptor protein (Slap) is a negative regulator of mitogenesis. Curr Biol. 1998;8:975–978. doi: 10.1016/S0960-9822(98)70400-2. [DOI] [PubMed] [Google Scholar]
- 2.Sosinowski T, Killeen N, Weiss A. The Src-like adaptor protein downregulates the T cell receptor on CD4+ CD8+ thymocytes and regulates positive selection. Immunity. 2001;15:457–466. doi: 10.1016/S1074-7613(01)00195-9. [DOI] [PubMed] [Google Scholar]
- 3.Kim HJ, Zou W, Ito Y, et al. Src-like adaptor protein regulates osteoclast generation and survival. J Cell Biochem. 2010;110:201–209. doi: 10.1002/jcb.22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friend SF, Peterson LK, Kedl RM, Dragone LL. SLAP deficiency increases TCR avidity leading to altered repertoire and negative selection of cognate antigen-specific CD8+ T cells. Immunol Res. 2013;55:116–124. doi: 10.1007/s12026-012-8354-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dragone LL, Shaw LA, Myers MD, Weiss A. SLAP, a regulator of immunoreceptor ubiquitination, signaling, and trafficking. Immunol Rev. 2009;232:218–228. doi: 10.1111/j.1600-065X.2009.00827.x. [DOI] [PubMed] [Google Scholar]
- 6.Pandey A, Duan H, Dixit VM. Characterization of a novel Src-like adapter protein that associates with the Eck receptor tyrosine kinase. J Biol Chem. 1995;270:19201–19204. doi: 10.1074/jbc.270.33.19201. [DOI] [PubMed] [Google Scholar]
- 7.Angrist M, Wells DE, Chakravarti A, Pandey A. Chromosomal localization of the mouse Src-like adapter protein (Slap) gene and its putative human homolog SLA. Genomics. 1995;30:623–625. doi: 10.1006/geno.1995.1289. [DOI] [PubMed] [Google Scholar]
- 8.Kratchmarova I, Sosinowski T, Weiss A, Witter K, Vincenz C, Pandey A. Characterization of promoter region and genomic structure of the murine and human genes encoding Src like adapter protein. Gene. 2001;262:267–273. doi: 10.1016/S0378-1119(00)00516-3. [DOI] [PubMed] [Google Scholar]
- 9.Meijerink PH, Yanakiev P, Zorn I, et al. The gene for the human Src-like adaptor protein (hSLAP) is located within the 64-kb intron of the thyroglobulin gene. Eur J Biochem. 1998;254:297–303. doi: 10.1046/j.1432-1327.1998.2540297.x. [DOI] [PubMed] [Google Scholar]
- 10.Holland SJ, Liao XC, Mendenhall MK, et al. Functional cloning of Src-like adapter protein-2 (SLAP-2), a novel inhibitor of antigen receptor signaling. J Exp Med. 2001;194:1263–1276. doi: 10.1084/jem.194.9.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey A, Ibarrola N, Kratchmarova I, et al. A novel Src homology 2 domain-containing molecule, Src-like adapter protein-2 (SLAP-2), which negatively regulates T cell receptor signaling. J Biol Chem. 2002;277:19131–19138. doi: 10.1074/jbc.M110318200. [DOI] [PubMed] [Google Scholar]
- 12.Loreto MP, McGlade CJ. Cloning and characterization of human Src-like adaptor protein 2 and a novel splice isoform, SLAP-2-v. Oncogene. 2003;22:266–273. doi: 10.1038/sj.onc.1206114. [DOI] [PubMed] [Google Scholar]
- 13.Sosinowski T, Pandey A, Dixit VM, Weiss A. Src-like adaptor protein (SLAP) is a negative regulator of T cell receptor signaling. J Exp Med. 2000;191:463–474. doi: 10.1084/jem.191.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang J, Sawasdikosol S, Chang JH, Burakoff SJ. SLAP, a dimeric adapter protein, plays a functional role in T cell receptor signaling. Proc Natl Acad Sci USA. 1999;96:9775–9780. doi: 10.1073/pnas.96.17.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohtsuki T, Hatake K, Ikeda M, et al. Expression of Src-like adapter protein mRNA is induced by all-trans retinoic acid. Biochem Biophys Res Commun. 1997;230:81–84. doi: 10.1006/bbrc.1996.5887. [DOI] [PubMed] [Google Scholar]
- 16.Hiragun T, Peng Z, Beaven MA. Cutting edge: dexamethasone negatively regulates Syk in mast cells by up-regulating SRC-like adaptor protein. J Immunol. 2006;177:2047–2050. doi: 10.4049/jimmunol.177.4.2047. [DOI] [PubMed] [Google Scholar]
- 17.Park SK, Beaven MA. Mechanism of upregulation of the inhibitory regulator, src-like adaptor protein (SLAP), by glucocorticoids in mast cells. Mol Immunol. 2009;46:492–497. doi: 10.1016/j.molimm.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansha M, Carlet M, Ploner C, et al. Functional analyses of Src-like adaptor (SLA), a glucocorticoid-regulated gene in acute lymphoblastic leukemia. Leukoc Res. 2010;34:529–534. doi: 10.1016/j.leukres.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 19.Loreto MP, Berry DM, McGlade CJ. Functional cooperation between c-Cbl and Src-like adaptor protein 2 in the negative regulation of T-cell receptor signaling. Mol Cell Biol. 2002;22:4241–4255. doi: 10.1128/MCB.22.12.4241-4255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugihara S, Katsutani S, Deckmyn H, Fujimura K, Kimura A. Roles of Src-like adaptor protein 2 (SLAP-2) in GPVI-mediated platelet activation SLAP-2 and GPVI signaling. Thromb Res. 2010;126:e276–e285. doi: 10.1016/j.thromres.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Manes G, Bello P, Roche S. Slap negatively regulates Src mitogenic function but does not revert Src-induced cell morphology changes. Mol Cell Biol. 2000;20:3396–3406. doi: 10.1128/MCB.20.10.3396-3406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazi JU, Rönnstrand L. Src-Like adaptor protein (SLAP) binds to the receptor tyrosine kinase Flt3 and modulates receptor stability and downstream signaling. PLoS ONE. 2012;7:e53509. doi: 10.1371/journal.pone.0053509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazi JU, Agarwal S, Sun J, Bracco E, Rönnstrand L. Src-like-adaptor protein (SLAP) differentially regulates normal and oncogenic c-Kit signaling. J Cell Sci. 2014;127:653–662. doi: 10.1242/jcs.140590. [DOI] [PubMed] [Google Scholar]
- 24.Lebigot I, Gardellin P, Lefebvre L, Beug H, Ghysdael J, Quang CT. Up-regulation of SLAP in FLI-1-transformed erythroblasts interferes with EpoR signaling. Blood. 2003;102:4555–4562. doi: 10.1182/blood-2003-06-2077. [DOI] [PubMed] [Google Scholar]
- 25.Kazi JU, Kabir NN, Flores-Morales A, Rönnstrand L. SOCS proteins in regulation of receptor tyrosine kinase signaling. Cell Mol Life Sci. 2014;71:3297–3310. doi: 10.1007/s00018-014-1619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabir NN, Sun J, Rönnstrand L, Kazi JU. SOCS6 is a selective suppressor of receptor tyrosine kinase signaling. Tumour Biol. 2014 doi: 10.1007/s13277-014-2542-4. [DOI] [PubMed] [Google Scholar]
- 27.Kabir NN, Kazi JU. Grb10 is a dual regulator of receptor tyrosine kinase signaling. Mol Biol Rep. 2014;41:1985–1992. doi: 10.1007/s11033-014-3046-4. [DOI] [PubMed] [Google Scholar]
- 28.Lennartsson J, Rönnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 29.Dragone LL, Myers MD, White C, et al. Src-like adaptor protein (SLAP) regulates B cell receptor levels in a c-Cbl-dependent manner. Proc Natl Acad Sci USA. 2006;103:18202–18207. doi: 10.1073/pnas.0608965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu G, Rybakin V, Brzostek J, Paster W, Acuto O, Gascoigne NR. Fine-tuning T cell receptor signaling to control T cell development. Trends Immunol. 2014;35:311–318. doi: 10.1016/j.it.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ersek B, Molnar V, Balogh A, et al. CD3zeta-chain expression of human T lymphocytes is regulated by TNF via Src-like adaptor protein-dependent proteasomal degradation. J Immunol. 2012;189:1602–1610. doi: 10.4049/jimmunol.1102365. [DOI] [PubMed] [Google Scholar]
- 32.Myers MD, Dragone LL, Weiss A. Src-like adaptor protein down-regulates T cell receptor (TCR)-CD3 expression by targeting TCRzeta for degradation. J Cell Biol. 2005;170:285–294. doi: 10.1083/jcb.200501164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers MD, Sosinowski T, Dragone LL, et al. Src-like adaptor protein regulates TCR expression on thymocytes by linking the ubiquitin ligase c-Cbl to the TCR complex. Nat Immunol. 2006;7:57–66. doi: 10.1038/ni1291. [DOI] [PubMed] [Google Scholar]
- 34.Kazi JU, Kabir NN, Rönnstrand L. Protein kinase C (PKC) as a drug target in chronic lymphocytic leukemia. Med Oncol. 2013;30:757. doi: 10.1007/s12032-013-0757-7. [DOI] [PubMed] [Google Scholar]
- 35.Kazi JU. The mechanism of protein kinase C regulation. Front Biol. 2011;6:328–336. [Google Scholar]
- 36.Kurosaki T. Regulation of BCR signaling. Mol Immunol. 2011;48:1287–1291. doi: 10.1016/j.molimm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Dragone LL, Myers MD, White C, Sosinowski T, Weiss A. SRC-like adaptor protein regulates B cell development and function. J Immunol. 2006;176:335–345. doi: 10.4049/jimmunol.176.1.335. [DOI] [PubMed] [Google Scholar]
- 38.Peterson LK, Pennington LF, Shaw LA, et al. SLAP deficiency decreases dsDNA autoantibody production. Clin Immunol. 2014;150:201–209. doi: 10.1016/j.clim.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kagaya Y, Asaumi Y, Wang W, et al. Current perspectives on protective roles of erythropoietin in cardiovascular system: erythropoietin receptor as a novel therapeutic target. Tohoku J Exp Med. 2012;227:83–91. doi: 10.1620/tjem.227.83. [DOI] [PubMed] [Google Scholar]
- 40.Munugalavadla V, Kapur R. Role of c-Kit and erythropoietin receptor in erythropoiesis. Crit Rev Oncol Hematol. 2005;54:63–75. doi: 10.1016/j.critrevonc.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Liontos LM, Dissanayake D, Ohashi PS, Weiss A, Dragone LL, McGlade CJ. The Src-like adaptor protein regulates GM-CSFR signaling and monocytic dendritic cell maturation. J Immunol. 2011;186:1923–1933. doi: 10.4049/jimmunol.0903292. [DOI] [PubMed] [Google Scholar]
- 42.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 43.Semerdjieva S, Abdul-Razak HH, Salim SS, et al. Activation of EphA receptors mediates the recruitment of the adaptor protein Slap, contributing to the downregulation of N-methyl-d-aspartate receptors. Mol Cell Biol. 2013;33:1442–1455. doi: 10.1128/MCB.01618-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naudin C, Sirvent A, Leroy C, et al. SLAP displays tumour suppressor functions in colorectal cancer via destabilization of the SRC substrate EPHA2. Nat Commun. 2014;5:3159. doi: 10.1038/ncomms4159. [DOI] [PubMed] [Google Scholar]
- 45.Sirvent A, Leroy C, Boureux A, Simon V, Roche S. The Src-like adaptor protein regulates PDGF-induced actin dorsal ruffles in a c-Cbl-dependent manner. Oncogene. 2008;27:3494–3500. doi: 10.1038/sj.onc.1211011. [DOI] [PubMed] [Google Scholar]
- 46.Manes GA, Masendycz P, Nguyen T, et al. A potential role for the Src-like adapter protein SLAP-2 in signaling by the colony stimulating factor-1 receptor. FEBS J. 2006;273:1791–1804. doi: 10.1111/j.1742-4658.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 47.Pakuts B, Debonneville C, Liontos LM, Loreto MP, McGlade CJ. The Src-like adaptor protein 2 regulates colony-stimulating factor-1 receptor signaling and down-regulation. J Biol Chem. 2007;282:17953–17963. doi: 10.1074/jbc.M701182200. [DOI] [PubMed] [Google Scholar]
- 48.Park SK, Qiao H, Beaven MA. Src-like adaptor protein (SLAP) is upregulated in antigen-stimulated mast cells and acts as a negative regulator. Mol Immunol. 2009;46:2133–2139. doi: 10.1016/j.molimm.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]