Abstract
The past decade has seen the emergence of next-generation sequencing (NGS) technologies, which have revolutionized the field of human molecular genetics. With NGS, significant portions of the human genome can now be assessed by direct sequence analysis, highlighting normal and pathological variants of our DNA. Recent advances have also allowed the sequencing of complete genomes, by a method referred to as whole genome sequencing (WGS). In this work, we review the use of WGS in medical genetics, with specific emphasis on the benefits and the disadvantages of this technique for detecting genomic alterations leading to Mendelian human diseases and to cancer.
Keywords: Exome, Hereditary disease, Mutation, Polymorphism
Introduction
Identification of pathogenic DNA variants causing diseases is one of the main aims of medical genetic investigations. In the past, when direct DNA sequencing possibilities were limited, this goal was achieved only in cases for which the region of the genome harboring a given mutation could be reduced to a manageable size by other procedures, such as family-based linkage or haplotype analyses. In the absence of large pedigrees or of other favorable factors that could help this localization process, disease-causing variants could remain undetected for years. The recent commercialization of next-generation sequencing (NGS) platforms has introduced a substantial methodological shift in mutation detection procedures. Specifically, it has allowed the querying of megabases of DNA at once, through computer-based alignment of millions of short sequence reads [1]. Parallel sequencing of panels of candidate disease genes and whole exome sequencing (WES), investigating all of our protein-coding DNA (~2 % of the human genome), have now become routine procedures in most laboratories.
As the NGS technique develops, the price per sequenced base decreases, to the point that sequencing entire individual genomes is not a prohibitive effort any more. Compared to WES, the use of whole genome sequencing (WGS) in human genetics, and especially in medical genetics, is still in its infancy. The reasons for this delay are mainly two: WGS involves higher costs compared to WES and requires more complex analyses at the computational level. Unlike WES, however, WGS allows the identification of complex DNA variants that are not limited to the coding sequences of the genome and the detection of non-conventional events involving large stretches of DNA (Table 1). Moreover, WGS displays an increased sensitivity with respect to WES in relationship to coding sequences as well, as it analyzes contiguous DNA and allows better sequencing and mapping approaches. More specifically, since it is not limited by constraints originating from discontinuous DNA templates (captured exons), WGS can take advantage of information deriving from a “regional” context. For instance, WGS can identify gene fusions, duplications of exons, and other genetic defects that would likely be missed in the absence of information from surrounding, non-coding DNA, which is seldom targeted by pre-WES purification procedures. Coverage (number of times a given nucleotide is sequenced) in WGS is also in general more uniform, since genomic DNA is provided to the sequencer “as is”, without undergoing selection procedures that may artificially create an uneven representation of the template material to be sequenced.
Table 1.
Features of whole genome sequencing (WGS) vs. whole exome sequencing (WES)
| Feature | WGS | WES |
|---|---|---|
| Exonic variants | Yes | Yes |
| Intronic variants | Yes | No |
| Intergenic variants | Yes | No |
| Indels | Yes | Yes |
| Copy number variations | Yes | Not directly/imprecise |
| Large insertions and deletions | Yes | Not directly/imprecise |
| Transposable elements | Yes | Not directly/imprecise |
Detection of copy number variations, large insertion and deletions, as well as of transposable elements are imprecise in WES since data are available for coding regions only, and these events can originate elsewhere
Unfortunately, the wealth of information produced by WGS, despite being preferable from a theoretical standpoint, may as well represent a burden for the identification of DNA variants meaningful to medical genetics. Such variants typically consist of one or a few mutations that have to be distinguished from thousands of benign DNA changes, and their identification has often been compared to the detection of a needle in a haystack. To follow the same analogy, WGS provides better chances of identifying pathological targets than WES, but at the same time it increases the size of the haystack, to the point that innocuous DNA changes may no longer be recognized as such. The advantages of WGS procedures can therefore be fully achieved only when analytical approaches can efficiently differentiate abnormal DNA changes from the multitude of benign variants that determine normal human heterogeneity.
To better illustrate all of these concepts, this review will focus on the use of WGS as a tool to detect rare DNA variants with a high phenotypic effect, such as germline mutations in Mendelian hereditary disorders and somatic mutations in cancer.
The medical genome: generalities and common procedures
The human reference genome
Because of the complexity of the human genome, NGS reads from WGS projects cannot be efficiently assembled via de novo procedures, but have to be mapped to a standard template sequence, the human “reference sequence”. This human reference genome is a pooled sequence data of 13 healthy individuals with European ancestry [2], and has gradually evolved with the improvement of sequencing methods. It provides a common and unambiguous system of relationships between genomic coordinates and corresponding DNA bases.
Mapping of sequence reads and identification of variants
Following the generation of the raw DNA sequence reads by an NGS platform, the process of obtaining the full genome sequence of an individual (or, better, a reliable approximation of it), consists of a two-step, computer-based procedure. First, the short NGS reads are mapped to the reference genome by assigning to them specific genomic coordinates. This procedure is in general computer intensive and is achieved by the use of various algorithms (e.g., BWA [3], AGILE [4], NovoAlign [novocraft.com], or FastHASH [5]). Then, mismatches between the reference genome and the individual genome are assessed by a bioinformatic process referred to as “variant calling” (e.g., via software such as GATK [6] or VCMM [7]).
Both mapping and variant calling procedures can be highly parameterized and are susceptible to producing different outputs as a function of such parameters. Therefore, although for a given individual there is only one physical genome, made of DNA, at the present time we can only obtain one or more imperfect representations of it, made of bits and bytes. As a general rule, each step of any genome analysis produces both false positives, i.e., variants that are called but are not physically present in the genome, and false negatives, i.e., variants that are not called but are present in the physical genome. It is therefore important to minimize errors at these initial mapping and variant calling steps, since all of downstream analyses will be made on the assumption that these data are a faithful representation of the physical genome.
General filtering procedures
Since every WGS project produces on average ~4,000,000 called variants [8, 9], identification of mutations relies on a series of filtering procedures that have as goal to recognize rare DNA changes with a pathogenic effect and discard the multitude of variants that are unrelated to the disease studied. Comparison with databases reporting information from the unaffected population such as dbSNP [10], the ESP database (evs.gs.washington.edu), the Exome Aggregation Consortium (ExAC) (exac.broadinstitute.org), etc. represents the most consistent filtering step, under the assumptions that such public databases report (a) reliable information and (b) include polymorphic variants having no direct relationship with genetic diseases. However, these databases have limitations such as the presence of very rare and pathogenic mutations [11] and artifacts [12].
The frequency of the detected variants in the general population could be taken into consideration during filtering procedures, since alleles from some (mostly recessive) diseases may very well be present in the general, unaffected population [13, 14]. Furthermore, most of these entries contain information about genotype and allele frequency in different human populations, allowing as well other important analyses. In addition to comparisons with data providing information on biological variability, filtering from technical errors should also be put in place. NGS platforms as well as mapping and variant calling pipelines tend to produce technical noise (false positives) that is luckily rather constant and sequence specific. Comparison with a small set of control samples sequenced by the same NGS platform and processed by the same informatics pipeline would help to remove errors from the genomes.
Since a considerable amount of variants still survive general filtering, it may be useful to incorporate in the analysis a predictive tool that scores the impact of coding DNA changes on the corresponding protein sequence and, possibly, function. There are currently many software packages that can perform these tasks and compute whether a given variant potentially affects protein formation, expression, and/or interaction with other proteins. Among those that are used most often, we can cite SIFT [15], PROVEAN [16], PolyPhen-2 [17], and GERP++ [18]. Since prediction tools are not always concordant and their output based on different parameters, most studies use a combination of two or more tools to infer the putative pathogenicity of the variants [19–21]. However, it is important to stress that all these packages provide information of predictive nature, and that filtering procedures based on them will have in the end only a relative value.
Databases of disease-associated variants
Many public databases reporting the direct relationship between DNA changes and specific traits exist and are publicly available. Some of them contain information on variants that underlie or are associated with diseases, such as the Human Gene Mutation Database [13] or the Online Mendelian Inheritance in Man database (OMIM) [22]. For structural variations, the Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER) [23] lists copy number variations present in the control and affected populations. For cancer studies, the Catalogue of Somatic Mutations in Cancer (COSMIC) [24] stores bona fide somatic mutations related to human cancers. Some other databases collect the results from pharmacogenetic studies to contribute to the development of individualized treatments (PharmGKB [25], Pharmaco-miR [26]). All these databases have increased substantially in size in recent years, due to NGS and larger and larger genetic studies. If integrated in WGS endeavors, they can be of great help in highlighting genetic variants associated with pathological traits.
Germline mutations
WGS in hereditary diseases
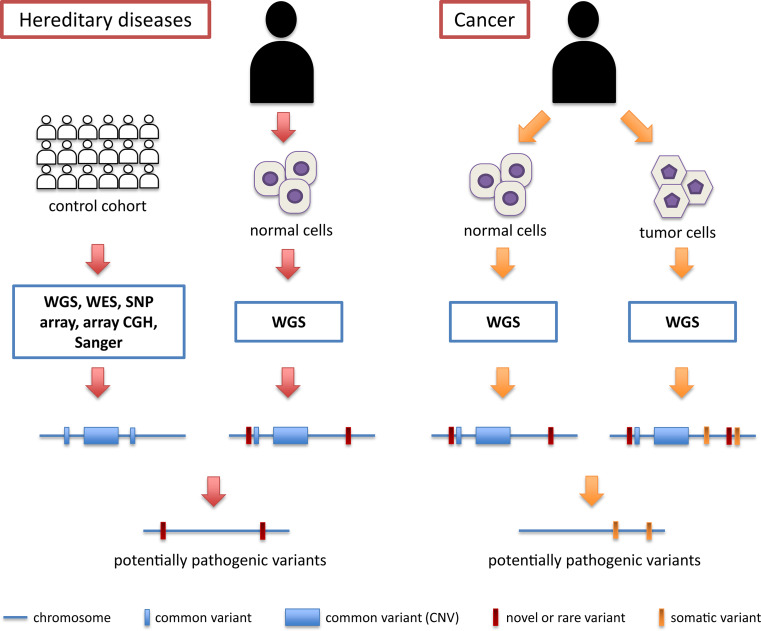
Pathogenic mutations with a high phenotypic effect can either be inherited from a person’s parents (germline mutations) or be acquired throughout life (somatic mutations). Pathologies resulting from germline mutations, which can be transmitted to the following generations, are commonly referred to as hereditary diseases, while somatic DNA injuries are usually not transmittable to the offspring and lead in general to tumors. Both germline and somatic mutations can be efficiently identified by WGS; however, technical and analytical approaches to detect these pathogenic variants are rather different (Fig. 1). A review of the recent literature shows that hereditary complex disorders, for which a combination of common variants in different genes and environmental factors contribute to the pathology, are still mostly investigated via non-NGS techniques. Conversely, WGS is beginning to be systematically used as a tool to understand the causes of Mendelian inherited diseases, resulting from germline mutations in one gene (e.g., [20, 27, 28]).
Fig. 1.
Schematic workflow for the detection of potentially pathogenic DNA variants in hereditary diseases and in cancer. In hereditary diseases, the information from several genomes from a control cohort (white individuals) is assembled to produce a “metagenome” that includes all possible variants (both small events and copy number variations, or CNVs) that are allegedly not causing disease in the general population (blue bars and boxes). Potentially pathogenic variants are then deduced by comparing the WGS information from a patient (black individual) with that of the metagenome. In cancer, there is no need to query a control cohort, since the control information is provided by the genome of normal cells from the same patient. Regardless of their frequency in the general population, all these variants are then subtracted from the pool of DNA changes obtained from the tumor genome, making the detection process of pathogenic variants a more efficient and straightforward procedure
The initial approach for the detection of Mendelian mutations by WGS is virtually the same as that used for WES-based studies. It consists of focusing first on the coding region of genes, more specifically on variants leading to a change in the amino acid sequence of future proteins. However, the real power of WGS emerges when events involving non-coding regions are investigated. Compared to other techniques, WGS allows us to specifically extract information from parts of the genome that are usually neglected, and at a base-pair resolution. Recent WGS studies have indeed shown that a number of unsolved cases from Mendelian disease can be explained by mutations in non-coding regions and, at various degrees, involving coding parts of disease genes (e.g., [8, 29]). Similar examples include the direct identification of gene disruption by the insertion of mobile elements, which are already known to play a significant role in the molecular etiology of hereditary diseases [30], but that are difficult to identify by other NGS techniques than WGS (own unpublished results).
It is important to note that, regardless of the type of mutation, in all Mendelian disorders and within single pedigrees, pathogenic variants always co-segregate with the disease in affected individuals. Therefore, all patients within a family should necessarily share the same mutation(s) but not necessarily the same innocuous DNA variants. This elementary concept of human genetics is one of the most powerful elements of investigation in NGS studies, including WGS, since it allows us to discard benign variants that cannot be immediately recognized as such. One of the first WGS projects that exploited this paradigm is the one performed by Roach et al., who, following the comparison of individual WGS output from two healthy parents and two affected children, could reduce the number of candidates genes, genomewide, from thousands to only four [31].
For monogenic disorders with no genetic heterogeneity, a similar strategy could be extended from a single pedigree to a group of unrelated patients. In these cases, merging genomic data from different patients and different pedigrees represents a much more powerful approach, because unrelated affected individuals would all tend to have rare variants (mutations) only in the disease gene [27]. Conversely, in Mendelian disorders displaying genetic heterogeneity, this approach may lead to false positive results, highlighting as pathogenic benign variants that may be coincidentally shared by a group of patients, and therefore it should not be used.
Identification of recessive, dominant, or X-linked mutations
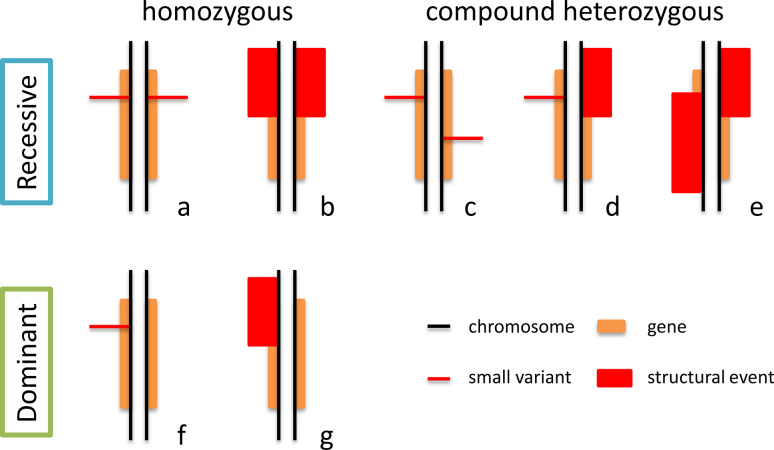
Since heterozygous recessive mutations do not cause disease, they can be present, even at non-negligible frequencies, in the general population [13, 14]. Patients would conversely be either homozygotes for a mutation or compound heterozygotes for two different mutations in the same gene (Fig. 2). This simple genetic concept has tremendous consequences in WGS-based searching for mutations, as only about a dozen genes will harbor rare, non-synonymous variants in the homozygous or compound heterozygous state genomewide. Other methods could be used to identify pathogenic variants, such as the elevated granularity of WGS data, which allows precise haplotype phasing in trios or quartets, to the point that meiotic recombination events in the parents of an index case could be detected. In other words, it is possible to identify all the regions of the genome identical by descent in affected individuals of a kindred, which by definition should harbor both the mutation transmitted from the father and from the mother [31, 32]. An extension of the same concept is autozygosity mapping, which reaches its highest possible precision when WGS information is used. This technique scores stretches of homozygous alleles (usually SNPs) in consanguineous families segregating a recessive disease to detect the single homozygous recessive mutation originating from a heterogeneous mutation in a common ancestor. Since alleles are transmitted from one generation to the other in large genomic “blocks” by meiotic recombination, the genomic region surrounding this homozygous mutation would also be completely homozygous for benign variants, which would act as a beacon for the presence of pathogenic recessive mutations [33].
Fig. 2.
Possible configurations of pathogenic mutations for autosomal recessive and autosomal dominant conditions. Structural events are usually better or exclusively detected via WGS procedures and therefore genotypes b, d, e, and g may be easily missed by other sequencing techniques
In contrast, in autosomal dominant conditions, only one variant in a specific disease gene gives rise to the pathological phenotype. Compared to recessive cases, it is more difficult to infer pathogenicity of a given DNA variant since, in absence of other information, in principle all of the rare DNA changes detected genomewide can be the mutation causing disease (Fig. 2). Filtering steps as well as the careful use of clinical and public databases and pedigree-based co-segregation analyses become therefore essential. In case the condition is known to display no genetic heterogeneity, then the most powerful tool to infer pathogenicity becomes data merging across different unrelated patients, for the reasons described above.
Recent literature has shown that a substantial proportion of seemingly dominant cases may also result from the presence of de novo mutations [34–37]. In such cases, trio analyses would be the best strategy to choose, since appearance of de novo mutations would be easily scored by subtracting the list of genomic variants of the patient from those of their parents, without in principle the need to filter data from common variation databases.
Finally, procedures for X-linked cases would be substantially the same as those for dominant ones, with the exception that the genomic region to be considered would be limited only to the X chromosome.
Somatic mutations
WGS in cancer
As mentioned, DNA errors can also be acquired somatically through life. Because of age, environment, diet, etc., these mutations are usually not transmitted to the offspring but can accumulate and lead to disease. This is the case of most cancers, where somatic defects lead to a dysregulated cell growth and eventually to tumor and metastasis.
Detection of pathogenic somatic variants via WGS procedures is a much simpler effort, compared to that involving germline mutations in hereditary diseases. Indeed, the cancer genome of a given patient can be directly compared with that from tumor-free tissues from the same individual (usually blood leukocytes). This process eliminates the need for constructing an imprecise reference “metagenome” resulting from cohorts of unrelated patients. In this context, the fact that a given individual’s germline genome carries polymorphic variants, rare DNA changes, or even large structural variations with respect to control genomes is completely irrelevant, since the mutations that count are those present in the cancer genome only (Fig. 1). In other words, the germline genome represents a baseline dataset used as a subtracting factor to obtain an unbiased count of all the acquired somatic mutations. Ley et al. were among the first to apply this method on an acute myeloid leukemia, identifying in the end two known mutations for cancer progression and eight novel mutations that could be used for possible targeted therapy [38].
Cancer appearance, progression, relapse
Since cancer is an evolving disorder, WGS can be used to score tumor progression, relapse, and remission by analyzing its genomic content at different time points. Concerning tumor progression, a study by Ding et al. [39] investigated basal-like breast cancer via four parallel WGS procedures: on the peripheral blood of the patient to obtain a baseline genome, on the primary tumor to detect the somatic mutations, on a brain metastasis to understand metastatic transformation, and on the genome of a human-to-mouse primary tumor xenograph to understand the mechanisms of tumor changes following transplantation.
Tumor evolution in the context of therapeutic treatments can also be studied by WGS, as shown by a report on clonal evolution in acute myeloid leukemia cells, a cancer characterized by frequent relapses following chemotherapy treatment [40]. In this work, the authors noted two distinct patterns of tumor genome evolution: in the first one, the primary tumor clone gained mutations that made it to evolve into the relapse clone and therefore survive treatment; in the second one, chemotherapy applied a selective pressure enabling a specific sub-clone of the initial tumor to expand, and again survive treatment.
Tumorigenic pathways
Although every cancer has a unique landscape of somatic events, in some instances mutations tend to affect common genes, highlighting dysregulation of shared, important pathways for tumor progression. Analyses aimed at identifying such pathways can be done by considering multiple cases of the same tumor, to increase the signal represented by driver mutations (DNA changes providing selective advantage to a cancer cell clone) and minimize the noise deriving from passenger mutations (DNA changes that do not contribute to cancer etiology but accumulate in rapidly expanding clones). In a way, such analyses are very similar to those outlined above for hereditary diseases, for which multiplication of the patients’ or controls’ genomes to be analyzed helps to eliminate DNA changes which are not relevant to the disease. This approach has been applied to a relatively large number of different tumors, for a total of ~150 genomes analyzed [41–49]. In addition to providing new insights into mutation-based differential prognosis, tumor molecular classification, progression mechanisms, etc., comparative WGS on multiple tumor samples helps identifying tumor signatures and mutational spectra across different types of cancer [50] or within the same tumor type, such as smoker and non-smoker lung cancer genomes [43].
Conclusions
From a genetic standpoint, there is nothing more exhaustive than the full sequence of a genome. It is therefore easy to predict that, when costs associated with WGS substantially decrease and better analytical tools are available, this procedure will become the technique of choice for most medical genetics investigations. WGS can in fact detect features of the human genome, such as copy number variations and intronic mutations, that other techniques cannot or struggle to identify, and that are becoming increasingly relevant to human genetic pathology. Furthermore, it is conceivable that many different genetic tests, which are currently performed as individual analyses (array CGH, sequence-specific mutation detection, gene panel screening, etc.) could be soon replaced by a single WGS run, which in fact can provide all of this information at once.
However, for WGS to become a popular tool in research and a routine test for DNA diagnosis, a few improvements still have to be made. From a clinical standpoint, diagnosis of the disease has to be very accurate, especially in terms of inheritance, because all downstream analyses would depend on it. Also, since a person’s whole genome is unveiled, the risk of incidental findings is very high, revealing the need for integrating ethical policies adapted to this specific test. On the technical side, sequencing errors and noise have to be better estimated and eliminated, since false positive findings or long processing times are not compatible with diagnostic needs. This could be done by optimizing the reference genome, databases of common variants, prediction software, and also pre-WGS experimental design (e.g., by including specific information of a patient’s family). In a more distant future, complex diseases will probably also be approached by WGS, to fully exploit the wealth of information that this technique produces in the context of variants that are not pathogenic per se, but that can cause disease via additive or multiplicative effects.
Acknowledgments
We would like to acknowledge the Swiss National Science Foundation (Grant 310030_138346) and Fond’action contre le cancer Lausanne for their financial support. We are also indebted to Dr. Aaron F. McDaid for his critical revision of this text.
References
- 1.Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011;52:413–435. doi: 10.1007/s13353-011-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(2010) E pluribus unum. Nat Methods 7:331 [DOI] [PubMed]
- 3.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misra S, Agrawal A, Liao WK, Choudhary A. Anatomy of a hash-based long read sequence mapping algorithm for next generation DNA sequencing. Bioinformatics. 2011;27:189–195. doi: 10.1093/bioinformatics/btq648. [DOI] [PubMed] [Google Scholar]
- 5.Xin H, Lee D, Hormozdiari F, Yedkar S, Mutlu O, Alkan C. Accelerating read mapping with FastHASH. BMC Genom. 2013;14(Suppl 1):S13. doi: 10.1186/1471-2164-14-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigemizu D, Fujimoto A, Akiyama S, Abe T, Nakano K, Boroevich KA, Yamamoto Y, Furuta M, Kubo M, Nakagawa H, Tsunoda T. A practical method to detect SNVs and indels from whole genome and exome sequencing data. Sci Rep. 2013;3:2161. doi: 10.1038/srep02161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiguchi KM, Tearle RG, Liu YP, Oh EC, Miyake N, Benaglio P, Harper S, Koskiniemi-Kuendig H, Venturini G, Sharon D, Koenekoop RK, Nakamura M, Kondo M, Ueno S, Yasuma TR, Beckmann JS, Ikegawa S, Matsumoto N, Terasaki H, Berson EL, Katsanis N, Rivolta C. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc Natl Acad Sci USA. 2013;110:16139–16144. doi: 10.1073/pnas.1308243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen L, Chernikoff D, Cheung A, Chirita R, Curson B, Ebert JC, Hacker CR, Hartlage R, Hauser B, Huang S, Jiang Y, Karpinchyk V, Koenig M, Kong C, Landers T, Le C, Liu J, McBride CE, Morenzoni M, Morey RE, Mutch K, Perazich H, Perry K, Peters BA, Peterson J, Pethiyagoda CL, Pothuraju K, Richter C, Rosenbaum AM, Roy S, Shafto J, Sharanhovich U, Shannon KW, Sheppy CG, Sun M, Thakuria JV, Tran A, Vu D, Zaranek AW, Wu X, Drmanac S, Oliphant AR, Banyai WC, Martin B, Ballinger DG, Church GM, Reid CA. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 10.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenna KP, McLaughlin RL, Hardiman O, Bradley DG. Using reference databases of genetic variation to evaluate the potential pathogenicity of candidate disease variants. Hum Mutat. 2013;34:836–841. doi: 10.1002/humu.22303. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Sun S. Comparing a few SNP calling algorithms using low-coverage sequencing data. BMC Bioinformatics. 2013;14:274. doi: 10.1186/1471-2105-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiguchi KM, Rivolta C. Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS One. 2012;7:e41902. doi: 10.1371/journal.pone.0041902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 16.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter. 2013;7(Unit7):20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li MX, Kwan JS, Bao SY, Yang W, Ho SL, Song YQ, Sham PC. Predicting mendelian disease-causing non-synonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 2013;9:e1003143. doi: 10.1371/journal.pgen.1003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, Klein TE. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rukov JL, Wilentzik R, Jaffe I, Vinther J, Shomron N (2013) Pharmaco-miR: linking microRNAs and drug effects. Brief Bioinform [DOI] [PMC free article] [PubMed]
- 27.Gonzaga-Jauregui C, Lotze T, Jamal L, Penney S, Campbell IM, Pehlivan D, Hunter JV, Woodbury SL, Raymond G, Adesina AM, Jhangiani SN, Reid JG, Muzny DM, Boerwinkle E, Lupski JR, Gibbs RA, Wiszniewski W. Mutations in VRK1 associated with complex motor and sensory axonal neuropathy plus microcephaly. JAMA Neurol. 2013;70:1491–1498. doi: 10.1001/jamaneurol.2013.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bainbridge MN, Wiszniewski W, Murdock, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, Muzny DM, Hoang LD, Yousaf S, Lupski JR, Gibbs RA. Whole-genome sequencing for optimized patient management. Sci Trans Med. 2011;3:87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BB, Kleefstra T, Brunner HG, Vissers LE, Veltman JA (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature [DOI] [PubMed]
- 30.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 31.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dewey FE, Chen R, Cordero SP, Ormond KE, Caleshu C, Karczewski KJ, Whirl-Carrillo M, Wheeler MT, Dudley JT, Byrnes JK, Cornejo OE, Knowles JW, Woon M, Sangkuhl K, Gong L, Thorn CF, Hebert JM, Capriotti E, David SP, Pavlovic A, West A, Thakuria JV, Ball MP, Zaranek AW, Rehm HL, Church GM, West JS, Bustamante CD, Snyder M, Altman RB, Klein TE, Butte AJ, Ashley EA. Phased whole-genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet. 2011;7:e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alkuraya FS. Homozygosity mapping: one more tool in the clinical geneticist’s toolbox. Genet Med. 2010;12:236–239. doi: 10.1097/GIM.0b013e3181ceb95d. [DOI] [PubMed] [Google Scholar]
- 34.Kariminejad A, Barzegar M, Abdollahimajd F, Pramanik R, McGrath JA. Olmsted syndrome in an Iranian boy with a new de novo mutation in TRPV3. Clin Exp Dermatol. 2014;39:492–495. doi: 10.1111/ced.12318. [DOI] [PubMed] [Google Scholar]
- 35.Grozeva D, Carss K, Spasic-Boskovic O, Parker MJ, Archer H, Firth HV, Park SM, Canham N, Holder SE, Wilson M, Hackett A, Field M, Floyd JA, Hurles M, Raymond FL. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet. 2014;94:618–624. doi: 10.1016/j.ajhg.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 37.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Ropke A, Moog U, Riess A, Thiel CT, Tzschach A, Wiesener A, Wohlleber E, Zweier C, Ekici AB, Zink AM, Rump A, Meisinger C, Grallert H, Sticht H, Schenck A, Engels H, Rappold G, Schrock E, Wieacker P, Riess O, Meitinger T, Reis A, Strom TM. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 38.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A, Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC, Graubert TA, DiPersio JF, Wilson RK. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM, Jr, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, McMichael JF, Wallis JW, Lu C, Shen D, Harris CC, Dooling DJ, Fulton RS, Fulton LL, Chen K, Schmidt H, Kalicki-Veizer J, Magrini VJ, Cook L, McGrath SD, Vickery TL, Wendl MC, Heath S, Watson MA, Link DC, Tomasson MH, Shannon WD, Payton JE, Kulkarni S, Westervelt P, Walter MJ, Graubert TA, Mardis ER, Wilson RK, DiPersio JF. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, Bassaganyas L, Baumann T, Juan M, Lopez-Guerra M, Colomer D, Tubio JM, Lopez C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernandez JM, Puente DA, Freije JM, Velasco G, Gutierrez-Fernandez A, Costa D, Carrio A, Guijarro S, Enjuanes A, Hernandez L, Yague J, Nicolas P, Romeo-Casabona CM, Himmelbauer H, Castillo E, Dohm JC, de Sanjose S, Piris MA, de Alava E, San Miguel J, Royo R, Gelpi JL, Torrents D, Orozco M, Pisano DG, Valencia A, Guigo R, Bayes M, Heath S, Gut M, Klatt P, Marshall J, Raine K, Stebbings LA, Futreal PA, Stratton MR, Campbell PJ, Gut I, Lopez-Guillermo A, Estivill X, Montserrat E, Lopez-Otin C, Campo E. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J, Klijn C, Guillory J, Bhatt D, Vartanian S, Walter K, Chan J, Holcomb T, Dijkgraaf P, Johnson S, Koeman J, Minna JD, Gazdar AF, Stern HM, Hoeflich KP, Wu TD, Settleman J, de Sauvage FJ, Gentleman RC, Neve RM, Stokoe D, Modrusan Z, Seshagiri S, Shames DS, Zhang Z. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 2012;22:2315–2327. doi: 10.1101/gr.140988.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 45.Kiel MJ, Velusamy T, Betz BL, Zhao L, Weigelin HG, Chiang MY, Huebner-Chan DR, Bailey NG, Yang DT, Bhagat G, Miranda RN, Bahler DW, Medeiros LJ, Lim MS, Elenitoba-Johnson KS. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209:1553–1565. doi: 10.1084/jem.20120910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss GJ, Liang WS, Demeure MJ, Kiefer JA, Hostetter G, Izatt T, Sinari S, Christoforides A, Aldrich J, Kurdoglu A, Phillips L, Benson H, Reiman R, Baker A, Marsh V, Von Hoff DD, Carpten JD, Craig DW. A pilot study using next-generation sequencing in advanced cancers: feasibility and challenges. PLoS ONE. 2013;8:e76438. doi: 10.1371/journal.pone.0076438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Craig DW, O’Shaughnessy JA, Kiefer JA, Aldrich J, Sinari S, Moses TM, Wong S, Dinh J, Christoforides A, Blum JL, Aitelli CL, Osborne CR, Izatt T, Kurdoglu A, Baker A, Koeman J, Barbacioru C, Sakarya O, De La Vega FM, Siddiqui A, Hoang L, Billings PR, Salhia B, Tolcher AW, Trent JM, Mousses S, Von Hoff D, Carpten JD. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol Cancer Ther. 2013;12:104–116. doi: 10.1158/1535-7163.MCT-12-0781. [DOI] [PubMed] [Google Scholar]
- 49.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, Romero-Cordoba S, Trevino V, Vazquez-Santillan K, Guadarrama AS, Wright AA, Rosenberg MW, Duke F, Kaplan B, Wang R, Nickerson E, Walline HM, Lawrence MS, Stewart C, Carter SL, McKenna A, Rodriguez-Sanchez IP, Espinosa-Castilla M, Woie K, Bjorge L, Wik E, Halle MK, Hoivik EA, Krakstad C, Gabino NB, Gomez-Macias GS, Valdez-Chapa LD, Garza-Rodriguez ML, Maytorena G, Vazquez J, Rodea C, Cravioto A, Cortes ML, Greulich H, Crum CP, Neuberg DS, Hidalgo-Miranda A, Escareno CR, Akslen LA, Carey TE, Vintermyr OK, Gabriel SB, Barrera-Saldana HA, Melendez-Zajgla J, Getz G, Salvesen HB, Meyerson M. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]