Abstract
Neurodegenerative diseases and other proteinopathies constitute a class of several dozen illnesses etiologically linked to pathological protein misfolding and aggregation. Because of this strong association with disease pathology, cell death, and aging, accumulation of proteins in aggregates or aggregation-associated structures (inclusions) has come to be regarded by many as a deleterious process, to be avoided if possible. Recent work has led us to see inclusion structures and disordered aggregate-like protein mixtures (which we call dynamic droplets) in a new light: not necessarily as a result of a pathological breakdown of cellular order, but as an elaborate cellular architecture regulating function and stress response. In this review, we discuss what is currently known about the role of inclusion structures in cellular homeostasis, stress response, toxicity, and disease. We will focus on possible mechanisms of aggregate toxicity, in contrast to the homeostatic function of several inclusion structures.
Keywords: Chaperone, Ubiquitin, Proteasome, Misfolded protein, Aggregation, PhoC, Inclusion, Inclusion body, Aggresome, JUNQ, IPOD, Stress foci, Stress granules, P-bodies, Dynamic droplets
Introduction
Protein inclusions or aggregate structures have often carried a negative connotation, in part because researchers initially encountered protein aggregation and inclusion body (IB) formation either as biochemists or as clinicians. For a biochemist trying to purify a functional protein, for example from E. coli, aggregation in IBs is usually bad news as it portends a dysfunctional state of the protein that can, at best, be salvaged after prolonged attempts at solubilization and refolding. For a physician, aggregation and IB-like structures are most readily associated with a family of devastating diseases, such as amyotrophic lateral sclerosis (ALS), Alzheimer’s (AD), Parkinson’s (PD), Huntington’s (HD), dementias, ataxias, several additional CAG-repeat diseases, and many other conformational disorders [1, 2].
Although prokaryotic IBs have properties that are largely distinct from eukaryotic aggregate structures, the term “inclusion body” or just “inclusion” has been widely coopted to refer to insoluble or weakly soluble protein deposits in eukaryotes, as well as the classic bacterial IB. When inclusions were first examined as a cell biological phenomenon, much of the focus was still on their role as hallmarks of disease. In several seminal studies [3–19], disease-associated proteins such as polyglutamine (polyQ) Huntingtin, ALS-associated SOD1 mutants (e.g. G93A), the α-synuclein protein and mutants linked to PD and other synucleinopathies, Cystic Fibrosis-causing mutant CFTRΔF508, and AD-associated Aβ-peptides were all shown to accumulate in inclusion structures (sometimes called aggresomes) when expressed in cultured cells or heterologously in other model systems. These inclusion structures exhibited many of the same features as those seen in pathology samples from disease tissue (including co-immunostaining with ubiquitin, chaperones, and staining with amyloid dyes such as Congo Red and Thiophlavin T) [20–24]. Indeed, when these pathogenic proteins were expressed in cultured cells or model organisms they often caused toxicity and cell death [25–35]. It was initially unclear whether inclusions were a non-physiological response to extreme conditions (wide-spread protein misfolding or over-expression of an aggregation-prone protein) or whether they were basic functional units of the cellular response to stress and misfolding. Less clear still was whether the formation of inclusions was part of a protective mechanism utilized by the cell to prevent protein misfolding-induced toxicity [36, 37], a product of dysregulation that may instead cause toxicity, or a peripheral by-product of the quality control machinery having been depleted or having otherwise gone awry [11, 38].
Recent developments, including the discovery of inclusion formation pathways in the single cell eukaryote, Saccharomyces cerevisiae, have the potential to change the way inclusion-like structures are viewed. Whereas initially in the “aggresome model” inclusions were hypothesized to have a single “zip code,” much like a classic organelle, as well as a fixed (juxtanuclear) localization and function, it is now becoming apparent that in both yeast and higher eukaryotes inclusion-like structures exhibit a dynamic ensemble of functions, properties, and resident factors. The goal of this review was to outline recent conceptual advances in the field of cytoplasmic inclusion-like structures, suggesting that they are part of a broader class of membrane-less nano-scale macromolecular assemblies, or Dynamic Droplets, which provide the functional architecture for essential cellular mechanisms including protein and RNA quality control, stress response, and rejuvenation.
Misfolding and aggregation as a consequence of phase transitions
Protein translation and the folding quality control mechanisms that oversee its fidelity take place, for the most part, in the cytoplasm of cells. Far from being a static water-like homogenous entity that it is often made out to be, the cytoplasm is a highly dynamic complex fluid mixture, whose properties are highly prone to stress- and age-dependent changes. One of the most significant factors behind the changes in cytoplasmic properties is the availability of energy in the form of ATP. An emerging consensus is that ATP-hydrolysis is required for the critical activity of maintaining the dynamic fluidity of the cytoplasm [39–42]. Energy production undergoes age-related decline, and stressful events encountered by the cell sap its ATP resources as energy is diverted to combatting damage. When energy resources decline, the cytoplasm drifts away from a liquid-like state and acquires solid-phase properties [43]. Nowhere is the impact of this transition more acute than in the maintenance of protein folding homeostasis [44]. Protein folding is an energetically costly activity, and misfolding due to limited ATP resources only increases demands for chaperones and other energy-dependent factors [45]. This potential for a deadly positive feedback loop is one explanation for the essential role of inclusions in managing quality control by spatially confining misfolded proteins away from bulk cytoplasm. Perhaps not surprisingly, constituent proteins of inclusions like structures (stress granules, p-bodies, RNP granules, and quality control compartments) are prime targets for mutations that cause aging-related disorders accompanied by insoluble protein deposits.
Inclusion structures
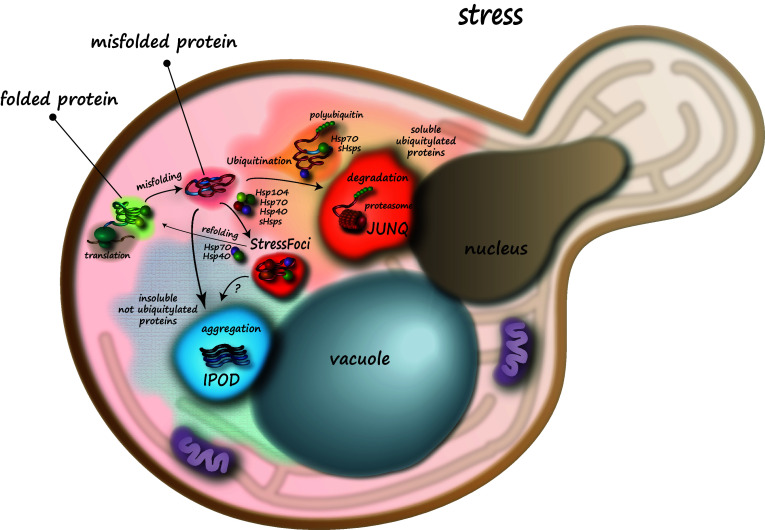
In yeast the cellular architecture of inclusion formation reflects the complexity of the dynamic cellular response to protein misfolding and aggregation (Fig. 1). Severe stress in the form of pH changes (e.g. starvation), oxidative stress, heat or cold shock, or changes in osmolarity, invariably leads to the misfolding of a large subset of the proteome. When a cell is initially presented with an abundance of misfolded proteins, the protein folding quality control machinery must decide between three different fates for the aberrant polypeptides. The misfolded proteins can be refolded, degraded by the ubiquitin–proteasome machinery or autophagy, or converted into a protective insoluble (or amyloid-like) deposit. Refolding, degradation, and active aggregation may happen simultaneously to different subpopulations of the protein misfolding load, or may function in a hierarchical manner, with refolding being the preferred option and active aggregation as a last resort. With elegance and efficiency that is typical of yeast, each of those quality control-associated activities is compartmentalized in a discrete inclusion [46]. It is not yet clear how cells triage between these options at different times and under different conditions, but it is increasingly apparent that inclusion structures play a key role in the decision of what to do with damaged and misfolded proteins.
Fig. 1.
Spatial architecture of protein folding quality control in yeast. Misfolded proteins are partitioned to distinct protein folding quality control compartments. This process is regulated by the ubiquitination status of the misfolded proteins, their solubility and concentration, and the availability of chaperones. Under acute stress misfolded proteins are initially trapped in stress foci, which are rapidly reversible structures. Over a longer time-scale, proteins that can be ubiquitinated and degraded are sent to the JUNQ for proteasomal degradation. Proteins that are not candidates for proteasomal degradation or are not ubiquitinated are targeted to the IPOD for active aggregation in an amyloid structure. It is unclear whether stress foci can merge with the IPOD
Stress foci
In yeast, proteins that are targeted for refolding are collected in stress foci (SFs): highly dynamic structures which concentrate holdases (sHsp26 and 42), chaperones (Hsp70 and 110), co-chaperones (Sis1 and Ydj1), and disaggregases (Hsp104) together with misfolded proteins [47–50]. SFs form rapidly in response to stress (within minutes), and are disassembled within 30–60 min after return to normal conditions [51, 52]. The fate of proteins that are trapped in SFs but cannot be refolded is not yet clear. It is possible that they eventually merge with the IPOD. SFs have also been observed in mammalian cells, where they exhibit similar properties to yeast, though no refolding from SFs has yet been seen in mammalian cells. Additionally, in yeast and mammals it is not yet clear what governs the motility of SFs (Hsp104, actin, microtubules, or other modes of transport).
JUNQ
Misfolded substrates that are targeted for proteasomal degradation are collected and degraded in JUNQ compartments [46, 52, 53]. From studies that have been carried out to date, it appears that the JUNQ is a degradation domain formed by expansions of the outer nuclear membrane, with misfolded protein substrates binding to the cytosolic surface of the nucleus [46]. Our current model is that by concentrating proteasomes, chaperones (Hsp70/Ssa2, sHsp26, Btn2) [54], and possibly ubiquitination machinery [46, 52, 53] together with degradation substrates in a single central location, the cell gains efficiency while avoiding the undesirable consequences of allowing misfolded proteasomal substrates free migration around the cytoplasm [55, 56]. Recent studies have conclusively demonstrated that the JUNQ is a dynamic functional compartment, rather than a deposition or storage site [52, 53]. In mammalian cells, the JUNQ interacts with intermediate filaments at distinct domains and contains active proteasomes in others [52]. A recent study was able to directly show turnover of misfolded proteins inside the JUNQ [52]. It is becoming clear that the JUNQ goes through several stages of assembly and constitution, starting out as a very dynamic rapidly degrading compartment, and transitioning to somewhat more of an “aggresome”-like (e.g. less mobile, less soluble) state [52]. The composition of proteasomes, chaperones, and cytoskeletal elements in the JUNQ changes in accordance with this.
IPOD
Proteins that are designated for insoluble aggregate deposition are sent to the IPOD. This inclusion is formed on the surface of the vacuole and houses quality control factors which direct the active aggregation (or amyloid formation) of misfolded and aggregation-prone proteins [57]. Several studies point to a protective role of the IPOD [46, 52, 57–61]. There is evidence suggesting that when proteins with a severe tendency to self-associate accumulate in the JUNQ/SFs instead of the IPOD, they cause toxicity [53]. The IPOD colocalizes with several chaperones (sHsp26/42, Hsp104) and cofactors (Btn2), albeit transiently depending on conditions [54, 60]. Hsp42 is a marker for the IPOD, but under stress it can also be observed in SFs. Hsp104, on the other hand, initially localizes to SFs immediately following stress induction, but with time re-distributes to the IPOD, and eventually, following prolonged high levels of stress, can even be observed in the JUNQ [46, 51].
Triage
One of the early triage mechanisms for targeting misfolded proteins to JUNQ or SFs versus the IPOD is the process of ubiquitination [46, 51]. Tagging with ubiquitin is necessary for localization of a misfolded protein to JUNQ and SFs, and when ubiquitination is blocked there is no alternative than targeting to the IPOD, even for proteins that are not highly aggregation-prone [46]. As evidence for the existence of an active aggregation mechanism in the IPOD, soluble misfolded proteins that are forced to go there by blocking ubiquitination alter their biochemical properties and become less soluble than their counterparts localizing to the JUNQ or SFs. De-ubiquitination also plays a role in targeting to JUNQ and SFs, since deletion of a quality control-associated de-ubiquitinating enzyme Ubp3 leads to enhanced targeting of misfolded substrates to the JUNQ for degradation [17, 62, 63].
There is early evidence that the triage decision described above (refolding, degradation, or aggregation) is influenced by the conditions and stress levels of the cell. Under stress, starvation, and upon aging, sequestration in the IPOD becomes the dominant pathway over the more costly and dangerous alternatives of refolding and degrading proteins that are highly likely to misfold or aggregate in a toxic manner [54, 58, 62, 64, 65].
Dynamic droplets: functional inclusion-like structures
It is commonplace to refer to any granular structure in the cytoplasm as an “aggregate.” However, this nomenclature is sometimes misleading, since many of these granular structures are highly dynamic and their formation is reversible. An updated terminology, taking into account liquid-phase and solid-phase structures in the cell is important to distinguish between amyloid-like aggregates (solid phase) and dynamic liquid-phase structures such as p-granules [45]. Hence, we refer to disordered dynamic protein mixtures including Ribonucleoprotein (RNP) granules and disordered inclusions of misfolded proteins as dynamic droplets, to emphasize this distinction. These dynamic droplets include stress granules, p-bodies, starvation-induced fibrils [66], JUNQ, IPOD, stress foci, and aggresomes [46, 51, 66, 67]. The molecules forming liquid phase dynamic droplets are thought to be held together in the absence of membranes by multivalent promiscuous molecular interactions, often generated by low-complexity regions in the constituent proteins [45, 68].
Dynamic droplets consist of a variable assembly of factors, with the constituent factors and their proportions in the droplets itself being a dynamic property that is highly dependent on conditions. For example, SGs and p-bodies share many markers, including Fas-activated serine/threonine phosphoprotein, TIA-1, XRN1, eIF4E, and tristetraprolin [69, 70], and JUNQ, SFs, and IPODs also localize the same set of chaperones at different times and under different conditions [46, 51, 54]. The dynamic association of chaperones with inclusions is an interesting example of spatial organization regulating quality control activity to suit the needs of the cell. During optimal non-stress conditions sHsps are in the IPOD and Hsp104 is diffuse in the cytoplasm. Under acute stress, Hsp104 rapidly enters SFs where is colocalizes with Sis1, sHsps, and Hsp70 [71, 72]. These chaperones appear to mediate re-folding of proteins which may transiently misfold during acute stress, but which may still have the potential to return to a functional state. Under prolonged stress, as refolding becomes less of a priority and toxicity avoidance becomes more important, Hsp104 redistributes to the IPOD where it mediates protective amyloid formation to sequester misfolded substrates from the rest of the cytoplasm [50]. At the same time, Hsp104 can also be observed in the JUNQ, presumably to prevent proteins there from aggregating and blocking proteasomal processing of misfolded proteins. The re-distribution of Hsp104 in response to stress conditions may explain its pleiotropic activities characterized in vitro, while putting them in their in vivo context. Hsp 104 has been shown to both disaggregate amyloids and disordered aggregates [73], and well as to promote amyloid formation [74] and prion seeding [75, 76]. Since it is highly unlikely that a single chaperone is continuously engaged in pleiotropic and often mutually opposing activities, it is possible that its different activities are regulated by dynamic compartmentalization of the Hsp104 and its cofactors in response to different conditions.
Mechanism of inclusion formation
How do misfolded proteins and quality control components get from the cytoplasm to an inclusion? Several mechanisms have been proposed, though the question is far from settled. The classical model, at least in mammalian cells, posits that misfolded proteins and stress foci-like micro-aggregates are delivered to inclusions in a microtubule-dependent manner [77, 78]. This line of reasoning is supported by data showing that disruption of microtubule polymerization leads to cytoplasmic dispersal of multiple stress foci-like structures and prevents their accumulation in a juxtanuclear location or in an inclusion [79]. Disruption of dynein motors has a similar effect [80]. An adaptor protein, HDAC6, is thought to mediate the interaction between microtubules and misfolded proteins, through its ubiquitin-interacting domain, together with Ataxin-1, Ataxin-3, PLIC-1, and p62 [75, 81].
Other potential models for delivery exist as well. The dispersal of misfolded proteins as a result of microtubule disruption could be an indirect effect—the structure of the cytoplasm and intermediate filaments depend on the microtubule and actin cytoskeleton. Disrupting actin or tubulin could result in pleiotropic effects on cytoplasmic architecture and may not necessarily indicate that misfolded proteins travel directly on microtubules. Stress foci-like particles and other aggregate foci, for their part, do not move in a unidirectional fashion, at least in data acquired so far [51]. If delivery is not based on direct transport on microtubules, what mechanisms could account for the accumulation of misfolded proteins in juxtanuclear inclusions like the JUNQ? Several options exist. For example, it is not clear that foci are actually transported and merged with an inclusion. Influx and egress of misfolded proteins in stress foci and other foci could be transient, and the inclusion could form diffuse misfolded proteins as opposed to a hierarchical pathway of diffuse protein to foci to inclusion, as is often thought. The diffuse misfolded proteins may simply have a higher affinity for the JUNQ, either because of specific chaperone receptors stationed therein, or because of a “sticky” cytoskeletal platform such as a high VIF concentration [82]. Fluorescence loss in photobleaching (FLIP) and PhotoConversion (PhoC) studies (utilizing these techniques to alter the fluorescence state of a sub-population of cytoplasmic proteins to then monitor their movement) have shown that the cytoplasm is churned with relatively rapid kinetics; hence a given misfolded protein “sees” the inclusion many times per minute, and has, therefore, the option of retention within it [83, 84].
Another possibility is that the cytoskeleton functions as a “net” rather than a “railroad” to deliver foci and proteins to the inclusion. In mammalian cells, for example, Inclusion formation coincides with VIF collapse—it is, therefore, possible that rather than dynein-based delivery directly on microtubules, “sticky” VIF filaments collect stress foci and misfolded proteins and transport them to the inclusion by interacting both with tubulin and actin. Direct time-resolved observation of foci dynamics in live cells will be essential for determining the precise method of delivery from cytoplasm to inclusion.
It is important to note that misfolded proteins or stress foci are not the only particles that must be delivered to inclusions. Recent studies provide novel observations of lysosomal trafficking towards the inclusion as well. The proposed mechanism is similar to the one described above—rather than directed microtubule transport, the study suggests that lack of transport around the inclusion site leads to the accumulation of lysosomes in what the report terms an “entrapment zone” [78].
Inclusions: function vs toxicity
Although inclusions, stress foci, stress granules, and other dynamic droplets appear to occupy a liquid state in their native functional constitution, under stressful or disease-associated conditions they can “mature” into solid-state aggregates. For example, mutations in genes coding for RNA quality control proteins TAR DNA-binding protein 43 (TDP-43) and fused in sarcoma (FUS), and in the gene chromosome 9 open reading frame 72 (C9orf72), cause ALS pathology [85–89]. There are many ongoing investigations aimed at deciphering how these two RNA-binding proteins and the dipeptide products of C9orf72 cause disease. So far, it appears that accumulation of FUS, TDP43, and dipeptides in stress granules, results in a disruption of RNA metabolism and possibly translation [90–95]. Indeed, many ALS pathologies exhibit a TDP43 positive inclusion, even when the underlying mutation is in SOD1, or in another protein [90, 96]. Since many of the proteins comprising stress granules have glutamine-rich or prion-like domains and since they associate in the droplet through many multivalent interactions, the presence of a protein with high aggregation propensity may facilitate a transition from a dynamic liquid phase structure to a more immobile, insoluble, “aggregate” like structure.
As noted above, JUNQ inclusions exhibit a similar tendency to transition from a dynamic droplet to a significantly less mobile inclusion containing aggregated protein. In yeast, prolonged exposure to high levels of stress causes the JUNQ to increase in size, become less mobile, and accumulate Hsp104 (suggesting the presence of aggregated species that are Hsp104 substrates) [46]. This phenomenon has been studied at higher resolution in mammalian cells [52, 53]. As the amount or aggregation propensity of misfolded proteins increases in the JUNQ, its mobility steadily declines. In conjunction with this, the degradative capacity of the JUNQ declines as well, with JUNQ-localized proteasomes becoming inactive. In these stages of initial aggregation, Hsp70 appears to be the limiting reagent, and over-expression of the stress-induced version of Hsp70 restores both mobility and degradative function, while also rescuing toxicity. A protective alternative is furnished by the IPOD—when toxic aggregating species (such as ALS-linked mutant SOD1) are rerouted from the JUNQ to the IPOD, toxicity is rescued as well [53].
From JUNQ to aggresome
We suggest that these changes in JUNQ properties brought on by accumulation of large amounts of substrate or the accumulation of aggregation-prone substrate corresponds to changes in JUNQ function. When the JUNQ becomes immobile, large, and more reminiscent of an “aggresome,” actin filaments redistribute around it [52]. It is possible that this coincides with the recruitment of lysosomes to the JUNQ, transforming its function from proteasomal degradation to autophagic degradation, as a last resort.
The term aggresome was initially used to describe all inclusions observed in mammalian cells, generated by expressing disease-associated proteins such as polyglutamine huntingtin, mutant SOD1, CFTRΔF508, synphillin, PrP, and other pathologically linked proteins [77, 97–101]. According to the aggresome model, all of these proteins formed juxtanuclear inclusions that interacted with the MTOC, were surrounded by (VIF), and contained aggregated forms of the misfolded protein that was being expressed. During the last decade, however, it has been well documented that many misfolded proteins which earlier thought to be aggresome substrates form inclusions that do not necessarily colocalize with the MTOC [52], nor always exhibit a VIF cage [52]. Moreover, juxtanuclear inclusions are far from uniformly aggregated, with recent studies reporting a range of mobility properties, from extremely dynamic, soluble, and mobile, to insolubly aggregated [17, 52, 53]. In light of this, what we suggest, is that this range of observations in mammalian cells represent different inclusion types (JUNQ-like and IPOD-like), as well as a time- and stress- dependent maturation of certain dynamic JUNQs to immobile inclusion. The term aggresome has been useful so far as a general concept of a central location for collecting aggregated proteins, but it will now have to be refined as our understanding of dynamic droplets and different inclusion structures expands.
One important question that remains unanswered so far is whether inclusions and other dynamic droplets are ad hoc compartments that form in response to stress, or whether these are constitutive functional units that become more pronounced and, therefore, easier to detect upon stress induction. Given standard imaging approaches, it is often easy to conclude that a given cellular structure is formed when the accumulation of a marker used to visualize it in a distinct location. However, it is equally true that the underlying structure may be present constitutively below detection limits, until acute stress makes markers more pronounced. Stress Granules, for example, are hypothesized by some to exist constitutively, as translation factories, until severe stress causes translation to stall and for Stress Granule factors to accumulate to above the detection limit [102].
Functional amyloids
From recent studies in the field it is clear that many dynamic droplets are functional compartments rather than “aggregates,” as they are often called. What about actual amyloid aggregates in the cell? Can they also have function, are they inert protective structures, or a harmful and undesirable byproduct of the propensity of proteins to aggregate? Turning to yeast which, much unlike human neuronal cells, have a high tolerance for amyloidogenic proteins, amyloid-containing IPODs appear to serve simultaneously as a defense mechanism preventing the accumulation of aggregated and misfolded proteins in the cytoplasm, and as a functional compartment [46, 58, 59]. A number of studies have observed a protective role for amyloid IPODs in yeast and mammals [38, 46, 53]. It seems that highly amyloidogenic proteins do decidedly less damage to cellular homeostasis when they can be efficiently stored in an IPOD inclusion. However, another report highlighted a potentially important function for IPODs, in the creation of amyloids for transition into a yeast prion state [57, 59]. The inheritance of the yeast prion state [PSI+] requires the aggregation of the translation terminator Sup35, through its prion domain (PrD). The prionogenesis of Sup35 requires a maturation process, from diffuse cytosolic localization, to perivacuolar ribbons, to compaction in an IPOD inclusion. Surprisingly, only cells that managed to pack Sup35 PrD into the IPOD were able to transmit [PSI+] prion state to progeny, whereas cells in various intermediary states such as ribbons failed to do so. Critically, Sup35 PrD fibrils that were packed into IPODs were highly fragmented, whereas ring-localized PrD was organized into long un-interrupted fibrils. This study suggests that Sup35 PrD is actively converted into prionogenic fibrils in the IPOD by Hsp104’s fragmentation activity. This is particularly interesting in light of other data showing that Hsp104 re-distributes to the IPOD only under certain conditions (such as under heat stress of oxidative stress [46, 51] helping to explain why these conditions are favorable for de novo prion induction. A key question is whether this dual functionality (sequestration of aggregates and prion seeding) of the IPOD has been evolutionarily conserved. Yeast are a relatively aggregation-tolerant organisms, and have been suggested to exploit the multitude of amyloidogenic prion-like proteins encoded in its genome [103] as the basis for phenotypic switches, helping sub-populations gain fitness advantages under diverse and stressful conditions [104]. It is interesting to speculate that in multicellular eukaryotes the IPOD serves only a protective role and that amyloid fragmentation for the sake of prion induction is an intolerable risk for multicellular organisms; hence the Hsp104 amyloid seed-generating activity is not conserved from yeast to mammals [105].
Aggregation in disease models
What then is the relationship between dynamic droplets/inclusions observed in cultured cells or simple models like yeast, and the cell physiology or pathology in animals, from organismal models of disease to human pathology? So far studies forging the connection have been few and have relied mostly on fixed samples and immunohistochemistry. Several studies have also examined fixed brain slices from mice expressing disease-associated proteins and human samples from individuals affected by neurodegenerative diseases [106–108]. The main preliminary conclusion that emerges from these reports is that affected brain and spinal cord tissue contains inclusion structures that co-stain with many of the markers that localize to inclusions in cell culture models, but the connection to functional dynamic inclusions observed so far in tissue culture remains unclear.
Despite the fact that it is often difficult to draw connections between tissue culture and phenomena observed in human brains, studying disease-associated proteins in simple cell models like cultured cells and yeast can offer important insight into the cellular pathology caused by expression or abundance of these proteins. Simple animal models have also provided significant insight on disease pathology. The nematode Caenorhabditis elegans offers one of the best models for studying the interface of cell biology and multi-cellular organismal regulation. Several studies have observed inclusions, mostly formed by disease-associated proteins and other simple aggregation prone models, in live animals [95, 109, 110]. These studies would suggest that similar inclusions to JUNQ, IPOD, and others observed in yeast in cultured cells exist in a multicellular animal context, though the question of their biological function has not been fully explored. For example, mutant SOD1 aggregation and the resulting cellular toxicity and organismal pathology are very well recapitulated in many different model systems, from cultured cells, to C. elegans, to mice and rats, showing remarkably similar pathology in all these models [53, 94, 95, 111]. Moreover, the mutant SOD1 pathology observed in simple models seems to closely mirror human pathology. In particular, a number of studies have demonstrated that the accumulation of mutant SOD1 (mutSOD1) in inclusions that show similar characteristics across species, is closely correlated to the onset of toxicity.
One of the most striking features of all neurodegenerative diseases, including ALS, Parkinson’s, Huntington’s, and Alzheimer’s is that the underlying toxic mechanism lies dormant for several decades before triggering relatively rapid degeneration, suggesting that pathology manifests in some cells, some of the time, under certain conditions. Hence, to understand the origins of disease pathology and to validate potential therapeutics, the molecular basis for late onset and cell specificity of disease must be clearly defined. For HD, ALS, spinocerebellar ataxia type 3 (SCA3), and many other neurodegenerative diseases, the extreme cellular and temporal specificity of toxicity onset is one of the least-understood facets of the disease. Due to their tractability and rapid aging, simple animal models that are designed to faithfully recapitulate the cellular and organismal pathology of disease will be extremely valuable in probing the mechanisms of neurodegeneration.
For example, studies of heterologously expressed α-synuclein have shed light on poorly understood facets of its cell-biological function and potential mechanisms of pathology, which have otherwise remained elusive [26, 112, 113]. Using yeast as a model, recent reports have shown that α-synuclein, when over-expressed or carrying a mutation, interferes with proper endo-membrane homeostasis, possibly disrupting endocytic sorting [114]. It remains for further study in neuronal sub-types to determine whether this triggers the dysfunction observed in Parkinson’s pathology, and why it initially effects dopaminergic cells. It is important to note, however, that modeling α-synuclein and other disease-associated proteins in yeast offers an extremely effective platform for screening and designing therapeutic molecules to target specific interactions between the disease protein and cellular machinery that is deemed to be pathogenic [115].
Inclusions and aging
Whether inclusions and dynamic droplets are a response to stress or a constitutive part of cellular architecture, they have recently been attributed an additional unexpected function. Besides maintaining quality control within the cell, inclusions mediate quality control across multiple generations of cells in a population. Several recent reports have demonstrated that inclusions are partitioned asymmetrically during mitosis and that this mechanism mediates the replicative aging process, or replicative rejuvenation, of dividing cells. The process of aging manifests a global decline in a wide range of cellular functions. Although discrete aging factors have been difficult to pinpoint, these factors are encompassed by the accumulation of damage in organelles (e.g. mitochondria, vacuoles, membranes) and the cytoplasm (e.g. accumulation of misfolded and aggregation-prone proteins). A number of important studies have shown that bacteria, yeast, and mammalian cells, all use a complex and multifaceted machinery to prevent the inheritance of damaged materials, and in particular damaged and aggregated proteins, by the new generation of cells [116–120]. By spatially restricting damaged proteins in inclusions and then asymmetrically partitioning inclusions during mitosis, cells are able to withhold misfolded and aggregated proteins from specific lineages (Fig. 2). Since these aberrant protein conformers are key determinants of aging, the ability to control their inheritance is crucial for avoiding aging in specific cells [121, 122].
Fig. 2.
Replicative rejuvenation in yeast. Misfolded and damaged proteins, aggregates, and old or damaged organelles are asymmetrically partitioned during mitosis through various mechanisms including compartmentalization and active transport on actin filaments. Retention of JUNQ and IPOD in mother cell is mediated by their attachment to the membranes of the nucleus and the vacuole, respectively. This process generates a daughter cell free from damage at the level of the proteome and with new “more fit” organelles
The key property enabling inclusions to be inherited asymmetrically is their ability to spatially sequester misfolded proteins and to interact with organelles and cytoskeleton to ensure the polarity of their inheritance after mitosis. In yeast, the JUNQ and IPOD inclusions are uniformly retained in the mother cells during budding. The JUNQ remains attached to the nuclear membrane, and the IPOD to the vacuolar membrane [51]. During cytokinesis the organelles are divided in a way that the regions tethered to the inclusions remain in the mother cell. It is not yet clear how the segregation is ensured at the level of organelles; however, it has been shown that misfolded proteins that are not sequestered in inclusions, such as the proteins in SFs, can be inherited by daughter cells as well [51]. This provides an intriguing possibility that stress foci are inherited as a hedge mechanism: the misfolded proteins in JUNQ and IPOD are terminal so there is no benefit to inheriting them, whereas the proteins in stress foci still have the potential for refolding. Although inheriting them can be detrimental if stress persists (since they are already misfolded), if the stress subsides the proteins in stress foci can easily be put back into circulation without the need to synthesize them from scratch. This is similar to mRNA stored in stress granules, which can be recycled for rapid reinitiation of translation once there is no more stress [123].
In mammalian cells as well, asymmetric inheritance of inclusions has emerged as a key mediator of replicative rejuvenation and aging. An important earlier study suggested that some insoluble amyloid inclusions, similar to the IPOD in yeast, were partitioned asymmetrically in dividing mammalian-derived cell lines [124]. It was left open to interpretation whether this is a deterministic phenomenon (an insoluble amyloid inclusion is not easy to divide in two; hence one cell will have to inherit it whether in a polar division or just randomly), or whether there is an aging polarity to mammalian mitoses similar to what is observed in yeast.
A recent study has taken a second look at the phenomenon of inclusion asymmetry in mammalian cells. Looking at different types of inclusions (JUNQ and IPOD) in several different cell lines, this study showed that, surprisingly, dynamic JUNQ compartments are also inherited in an asymmetric fashion during mammalian cytokinesis [52]. JUNQ and IPOD are usually inherited by the same cell (in CHO, HEK, N2a, and HeLa cells), suggesting that the divisions of these cells might actually be polar with respect to damage (Fig. 3). To support this hypothesis, the study observed a slight fitness advantage for cells that do not inherit the JUNQ and are, therefore, free of its damaged contents. Another study [125] in drosophila similarly showed that intestinal stem cells partition damaged proteins to differentiating progeny, confirming other organism-level drosophila studies showing asymmetric inheritance of damaged and ubiquitinated proteins by specific cell types [124, 126].
Fig. 3.
Replicative rejuvenation in mammalian cells. Inclusions are asymmetrically inherited in mammalian cells during division. Daughter cells lacking JUNQ and IPOD may have survival advantage compared to daughter cell that retains the inclusions. Question marks imply mechanisms for asymmetry that are not yet clear
Mechanism of replicative rejuvenation
What is the mechanism that enables asymmetric partitioning? There are several pathways known to be involved in mitotic partitioning. (1) The actin cytoskeleton can use myosin motors to selectively import healthy mitochondria and other organelles into daughter cells (this has been shown in yeast). (2) Attachment to the centrosome via the microtubule cytoskeleton can result in asymmetric inheritance. (3) Sequestration in compartments (e.g. RNP granules or inclusions) combined with an association with organelles or cytoskeleton can also be used to partition materials during mitosis; and finally, (4) diffusion barriers in the endomembrane system can ensure asymmetry in polar divisions [51, 117–119, 127].
In the case of misfolded and damaged proteins, evidence points to a diverse set of mechanisms. Several studies have shown a clear dependence of asymmetric inheritance of misfolded proteins in yeast on an intact actin cytoskeleton. When actin polymerization or anchoring at the bud is impaired, misinheritance and premature aging ensue [65, 128–130]. It is not yet completely clear whether the role of actin is primarily in the maintenance of cytoplasmic organization, enabling proper sequestration of misfolded proteins in inclusions, or whether it has a more direct role in unidirectional (retrograde) transport of misfolded proteins out of the bud and back into the mother cell [130]. Although it has been clearly shown that stress foci containing misfolded proteins can easily pass into the bud and be inherited by daughter and even granddaughter cells, retrograde transport remains a possibility for their return into the mother cell. On the other hand, stress foci do appear to colocalize with actin cables, at least in certain situations, and do not appear to move very much in the cytoplasm, certainly not diffusively enough to rule out a level of regulation with respect to which of them enter the bud and which do not [65].
Once misfolded proteins are sequestered in inclusions, a non-actin-based mechanism ensures their retention. Yeast mother cells retain JUNQ and IPOD inclusions by attaching them to membranes [51]. In future it will be important to determine whether this mechanism of attachment intersects with endomembrane diffusion barriers.
In mammalian cells, the asymmetric mitotic partitioning of JUNQ inclusions depends on the interaction of the inclusion with the VIF, the microtubule cytoskeleton, and the microtubule organizing center (MTOC) [52]. JUNQs are associated with VIF and physically linked to the MTOC. When the MTOC is displaced towards the edge of the cell via the expression of Nesprin 4, a protein mediating cell polarity, the JUNQ moves with the MTOC away from the nucleus. The JUNQ and associated VIF stay intact during mitosis (despite being quite dynamic and mobile); hence the interesting model that emerges is one in which the JUNQ is tethered by microtubules to one of the centrosomes—perhaps the same one in each division. Another report in drosophila suggested as well that asymmetric segregation of ubiquitinated proteins is centrosome-driven [126]. It is important to mention that, unlike mammalian and Drosophila cells, yeast shows no association between inclusions (JUNQ or IPOD) and the spindle pole body (SBP), or the yeast functional homologue of the MTOC.
Surprisingly, VIF itself was shown to be inherited asymmetrically in dividing mammalian cell lines; hence it could play the role of establishing polarity in a variety of mitoses [52]. However, the IPOD does not always co-segregate with the JUNQ and has no perceptible association with VIF, suggesting that, although there is still a strong bias towards polarity, IPODs containing insoluble proteins (e.g. polyglutamine aggregates) are sometimes misinherited during division.
Frontiers
While much work has been done to further our understanding of the effect of disease-associated protein aggregation and inclusion formation on cell viability and animal physiology, we have almost no models for examining their molecular pathology in functional neurons in their physiological context (in a live animal model of disease). Moreover, we do not yet have a clear understanding of what is special about certain neurons, making them extraordinarily susceptible to the toxicity of accumulating damaged and misfolded proteins. Are neurons metabolically hyperactive and hence generate more oxidatively damaged proteins than other cells? Do highly functional neurons over-burden their cellular trafficking machinery making it susceptible to dysfunction? Do neurons accumulate more protein damage because they are the longest-living post-mitotic cells in the organism and, therefore, cannot partition damage to other lineages during division? Are neurons slow to up-regulate important chaperones? Do certain neurons require precisely timed mRNA processing and delivery to the synapse, making RNA metabolism their “Achilles’ heel”? All of these models for neuronal specificity of many neurodegenerative diseases are compelling, but unfortunately, save for a few pilot studies, so far they are preliminary [94, 95, 131–133]. In the future, it will be essential to apply reductionist cell biological approaches to functional neural circuits in live animals, thanks to a proliferation of optogenetic techniques and molecular probes enabling real-time reporting on key cellular functions [134–136]. This will help determine what makes neuronal function cell-biologically unique in their sensitivity to different types of stress, at the single-neuron level and in individual circuits. Discovering more “bottleneck” factors that predispose neurons to proteinopathy would have a profound impact on our understanding of the disease and on translational initiatives.
Concluding remarks
The past decade has seen a conceptual transformation in our understanding of the role of compartmentalization and spatial architecture in the basic functions of the cytoplasm, especially in protein quality control and homeostasis. The boundaries between a single molecule diffusely localized in the cytoplasm, a molecular complex, membrane-less dynamic droplets, and larger immobile inclusion structures, are much more fluid than we previously thought. New tools in both concept and methodology are urgently needed to forge ahead in our understanding of how cells regulate folding and stress response over time, in different cell-type contexts, during aging, and at different conditions.
Conceptually, it is important to advance our cell biological characterization of granular compartments beyond diffuse versus aggregated. This also requires methodological advances, especially in terms of the incorporation of imaging approaches with higher resolution and over time.
This is especially critical for understanding the onset and progression of diseases linked to protein misfolding, which are kept at bay for years by proteostasis machinery, onset after years of exposure to the underlying etiological cause, and are exacerbated by stress and aging.
Acknowledgments
We tried to cite all primary literature pertaining to inclusions and inclusion-like structures. We apologize to any colleagues if we unintentionally missed their studies. We thank Mark Kaganovich, Jeremy England, Richard Gardner, Ehud Cohen, and members of the Kaganovich lab for discussion and feedback on the manuscript. This work was supported by the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC-StG2013 337713 DarkSide starting grant, as well as an Israel Science Foundation Grant ISF 843/11; a German Israel Foundation Grant GIFI-1201-242.13/2012; an Israel Health Ministry grant under the framework of E-Rare-2, a Niedersachsen-Israel Research Program grant, and a grant from the American Federation for Aging Research.
References
- 1.Aguzzi A, O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9(3):237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 2.Renner M, Melki R. Protein aggregation and prionopathies. Pathol Biol (Paris) 2014;62(3):162–168. doi: 10.1016/j.patbio.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Rajan RS, Illing ME, Bence NF, Kopito RR. Specificity in intracellular protein aggregation and inclusion body formation. Proc Natl Acad Sci USA. 2001;98(23):13060–13065. doi: 10.1073/pnas.181479798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 5.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143(7):1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotto J, Fox S, Morimoto R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110(Pt 23):2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4(10):826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto G, Kim S, Morimoto RI. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J Biol Chem. 2006;281(7):4477–4485. doi: 10.1074/jbc.M509201200. [DOI] [PubMed] [Google Scholar]
- 9.Perez MK, Paulson HL, Pendse SJ, Saionz SJ, Bonini NM, Pittman RN. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol. 1998;143(6):1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 1998;21(12):516–520. doi: 10.1016/s0166-2236(98)01276-4. [DOI] [PubMed] [Google Scholar]
- 11.Treusch S, Cyr DM, Lindquist S. Amyloid deposits: protection against toxic protein species? Cell Cycle. 2009;8(11):1668–1674. doi: 10.4161/cc.8.11.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97(4):1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA. 1997;94(24):12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281(5384):1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 15.Cleveland DW, Liu J. Oxidation versus aggregation—how do SOD1 mutants cause ALS? Nat Med. 2000;6(12):1320–1321. doi: 10.1038/82122. [DOI] [PubMed] [Google Scholar]
- 16.Outeiro TF, Putcha P, Tetzlaff JE, Spoelgen R, Koker M, Carvalho F, Hyman BT, McLean PJ. Formation of toxic oligomeric alpha-synuclein species in living cells. PLoS ONE. 2008;3(4):e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Gedalya T, Cohen E. Quality control compartments coming of age. Traffic. 2012;13(5):635–642. doi: 10.1111/j.1600-0854.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- 18.Dillin A, Cohen E. Ageing and protein aggregation-mediated disorders: from invertebrates to mammals. Philos Trans R Soc Lond B Biol Sci. 2011;366(1561):94–98. doi: 10.1098/rstb.2010.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.England JL, Kaganovich D. Polyglutamine shows a urea-like affinity for unfolded cytosolic protein. FEBS Lett. 2011;585(2):381–384. doi: 10.1016/j.febslet.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Lin WL, Dickson DW. Ultrastructure of ubiquitin-positive, TDP-43-negative neuronal inclusions in cerebral cortex of C9ORF72-linked frontotemporal lobar degeneration/amyotrophic lateral sclerosis. Neuropathology. 2012;32(6):679–681. doi: 10.1111/j.1440-1789.2012.01305.x. [DOI] [PubMed] [Google Scholar]
- 21.Waxman EA, Giasson BI. Induction of intracellular tau aggregation is promoted by alpha-synuclein seeds and provides novel insights into the hyperphosphorylation of tau. J Neurosci. 2011;31(21):7604–7618. doi: 10.1523/JNEUROSCI.0297-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forman MS, Farmer J, Johnson JK, Clark CM, Arnold SE, Coslett HB, Chatterjee A, Hurtig HI, Karlawish JH, Rosen HJ, Van Deerlin V, Lee VM, Miller BL, Trojanowski JQ, Grossman M. Frontotemporal dementia: clinicopathological correlations. Ann Neurol. 2006;59(6):952–962. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frid P, Anisimov SV, Popovic N. Congo red and protein aggregation in neurodegenerative diseases. Brain Res Rev. 2007;53(1):135–160. doi: 10.1016/j.brainresrev.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Bhat KP, Yan S, Wang CE, Li S, Li XJ. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc Natl Acad Sci USA. 2014;111(15):5706–5711. doi: 10.1073/pnas.1402215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues M, Tenreiro S, Franssens V, Reichert AS, Outeiro TF, Winderickx J, Ludovico P. SNCA (alpha-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy. 2012;8(10):1494–1509. doi: 10.4161/auto.21275. [DOI] [PubMed] [Google Scholar]
- 26.Zondler L, Miller-Fleming L, Repici M, Goncalves S, Tenreiro S, Rosado-Ramos R, Betzer C, Straatman KR, Jensen PH, Giorgini F, Outeiro TF. DJ-1 interactions with alpha-synuclein attenuate aggregation and cellular toxicity in models of Parkinson’s disease. Cell Death Dis. 2014;5:e1350. doi: 10.1038/cddis.2014.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 28.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans . Proc Natl Acad Sci USA. 2002;99(16):10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volovik Y, Marques FC, Cohen E. The nematode Caenorhabditis elegans: a versatile model for the study of proteotoxicity and aging. Methods. 2014;68(3):458–464. doi: 10.1016/j.ymeth.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 30.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 31.Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105(20):7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2008;105(17):6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Shinobu LA, Ward CM, Young D, Cleveland DW. Elevation of the Hsp70 chaperone does not effect toxicity in mouse models of familial amyotrophic lateral sclerosis. J Neurochem. 2005;93(4):875–882. doi: 10.1111/j.1471-4159.2005.03054.x. [DOI] [PubMed] [Google Scholar]
- 34.Parone PA, Da Cruz S, Han JS, McAlonis-Downes M, Vetto AP, Lee SK, Tseng E, Cleveland DW. Enhancing mitochondrial calcium buffering capacity reduces aggregation of misfolded SOD1 and motor neuron cell death without extending survival in mouse models of inherited amyotrophic lateral sclerosis. J Neurosci. 2013;33(11):4657–4671. doi: 10.1523/JNEUROSCI.1119-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Angelo F, Vignaud H, Di Martino J, Salin B, Devin A, Cullin C, Marchal C. A yeast model for amyloid-beta aggregation exemplifies the role of membrane trafficking and PICALM in cytotoxicity. Dis Model Mech. 2013;6(1):206–216. doi: 10.1242/dmm.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J, Arrasate M, Shaby BA, Mitra S, Masliah E, Finkbeiner S. Quantitative relationships BETWEEN huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into Huntington’s disease molecular pathogenesis. J Neurosci. 2010;30(31):10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nucifora LG, Burke KA, Feng X, Arbez N, Zhu S, Miller J, Yang G, Ratovitski T, Delannoy M, Muchowski PJ, Finkbeiner S, Legleiter J, Ross CA, Poirier MA. Identification of novel potentially toxic oligomers formed in vitro from mammalian-derived expanded huntingtin exon-1 protein. J Biol Chem. 2012;287(19):16017–16028. doi: 10.1074/jbc.M111.252577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kryndushkin D, Ihrke G, Piermartiri TC, Shewmaker F. A yeast model of optineurin proteinopathy reveals a unique aggregation pattern associated with cellular toxicity. Mol Microbiol. 2012;86(6):1531–1547. doi: 10.1111/mmi.12075. [DOI] [PubMed] [Google Scholar]
- 39.Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA. Cytoplasmic diffusion: molecular motors mix it up. J Cell Biol. 2008;183(4):583–587. doi: 10.1083/jcb.200806149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyman AA, Brangwynne CP. Beyond stereospecificity: liquids and mesoscale organization of cytoplasm. Dev Cell. 2011;21(1):14–16. doi: 10.1016/j.devcel.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, Lippincott-Schwartz J, Mackintosh FC, Weitz DA. Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell. 2014;158(4):822–832. doi: 10.1016/j.cell.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. P Natl Acad Sci USA. 2012;109(19):7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parry BR, Surovtsev IV, Cabeen MT, O’Hem CS, Dufresne ER, Jacobs-Wagner C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156(1–2):183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CF, Brangwynne CP, Gharakhani J, Hyman AA, Julicher F. Spatial organization of the cell cytoplasm by position-dependent phase separation. Phys Rev Lett. 2013;111(26):088101. doi: 10.1103/PhysRevLett.111.088101. [DOI] [PubMed] [Google Scholar]
- 45.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149(6):1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):U1036–U1088. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter GM, Smith MC, Wisen S, Basrur V, Elenitoba-Johnson KSJ, Duennwald ML, Kumar A, Gestwicki JE. Ordered assembly of heat shock proteins, Hsp26, Hsp70 Hsp90, and Hsp104, on expanded polyglutamine fragments revealed by chemical probes. J Biol Chem. 2011;286(47):40486–40493. doi: 10.1074/jbc.M111.284448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. Disassembling protein aggregates in the yeast cytosol: the cooperation of Hsp26 with SSA1 and Hsp104. J Biol Chem. 2005;280(25):23861–23868. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- 49.Alberti S (2012) Molecular mechanisms of spatial protein quality control. Prion 6(5):2–4. doi:10.4161/Pri.22470 [DOI] [PMC free article] [PubMed]
- 50.Shiber A, Breuer W, Brandeis M, Ravid T. Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol Biol Cell. 2013;24(13):2076–2087. doi: 10.1091/mbc.E13-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2(4):738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 52.Ogrodnik M, Salmonowicz H, Brown R, Turkowska J, Sredniawa W, Pattabiraman S, Amen T, Abraham AC, Eichler N, Lyakhovetsky R, Kaganovich D. Dynamic JUNQ inclusion bodies are asymmetrically inherited in mammalian cell lines through the asymmetric partitioning of vimentin. Proc Natl Acad Sci USA. 2014;111(22):8049–8054. doi: 10.1073/pnas.1324035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weisberg SJ, Lyakhovetsky R, Werdiger AC, Gitler AD, Soen Y, Kaganovich D. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc Natl Acad Sci USA. 2012;109(39):15811–15816. doi: 10.1073/pnas.1205829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell. 2012;23(16):3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gallagher PS, Oeser ML, Abraham AC, Kaganovich D, Gardner RG. Cellular maintenance of nuclear protein homeostasis. Cell Mol Life Sci. 2014;71(10):1865–1879. doi: 10.1007/s00018-013-1530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fredrickson EK, Gallagher PS, Candadai SVC, Gardner RG. Substrate recognition in nuclear protein quality control degradation is governed by exposed hydrophobicity that correlates with aggregation and insolubility. J Biol Chem. 2013;288(9):6130–6139. doi: 10.1074/jbc.M112.406710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyedmers J. Patterns of [PSI+] aggregation allow insights into cellular organization of yeast prion aggregates. Prion. 2012;6(3):191–200. doi: 10.4161/pri.18986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treusch S, Lindquist S. An intrinsically disordered yeast prion arrests the cell cycle by sequestering a spindle pole body component. J Cell Biol. 2012;197(3):369–379. doi: 10.1083/jcb.201108146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. P Natl Acad Sci USA. 2010;107(19):8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanneganti V, Kama R, Gerst JE. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol Biol Cell. 2011;22(10):1648–1663. doi: 10.1091/mbc.E10-11-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polling S, Mok YF, Ramdzan YM, Turner BJ, Yerbury JJ, Hill AF, Hatters DM. Misfolded polyglutamine, polyalanine, and superoxide dismutase 1 aggregate via distinct pathways in the cell. J Biol Chem. 2014;289(10):6669–6680. doi: 10.1074/jbc.M113.520189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oling D, Eisele F, Kvint K, Nystrom T. Opposing roles of Ubp3-dependent deubiquitination regulate replicative life span and heat resistance. EMBO J. 2014;33(7):747–761. doi: 10.1002/embj.201386822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. Mol Biol Cell. 1999;10:86a. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hill SM, Hao X, Liu B, Nystrom T. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae . Science. 2014;344(6190):1389–1392. doi: 10.1126/science.1252634. [DOI] [PubMed] [Google Scholar]
- 65.Song J, Yang Q, Yang J, Larsson L, Hao X, Zhu X, Malmgren-Hill S, Cvijovic M, Fernandez-Rodriguez J, Grantham J, Gustafsson CM, Liu B, Nystrom T. Essential genetic interactors of SIR2 required for spatial sequestration and asymmetrical inheritance of protein aggregates. PLoS Genet. 2014;10(7):e1004539. doi: 10.1371/journal.pgen.1004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrovska I, Nuske E, Munder MC, Kulasegaran G, Malinovska L, Kroschwald S, Richter D, Fahmy K, Gibson K, Verbavatz JM, Alberti S (2014) Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. Elife 3:5–11. doi:10.7554/Elife.02409 [DOI] [PMC free article] [PubMed]
- 67.Ben-Gedalya T, Lyakhovetsky R, Yedidia Y, Bejerano-Sagie M, Kogan NM, Karpuj MV, Kaganovich D, Cohen E. Cyclosporin-A-induced prion protein aggresomes are dynamic quality-control cellular compartments. J Cell Sci. 2011;124(Pt 11):1891–1902. doi: 10.1242/jcs.077693. [DOI] [PubMed] [Google Scholar]
- 68.Thirumalai D, Reddy G, Straub JE. Role of water in protein aggregation and amyloid polymorphism. Accounts Chem Res. 2012;45(1):83–92. doi: 10.1021/ar2000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;170(5):847. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoecklin G, Kedersha N. Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol. 2013;768:197–211. doi: 10.1007/978-1-4614-5107-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu BD, Larsson L, Franssens V, Hao XX, Hill SM, Andersson V, Hoglund D, Song J, Yang XX, Oling D, Grantham J, Winderickx J, Nystrom T. Segregation of protein aggregates involves actin and the polarity machinery. Cell. 2011;147(5):959–961. doi: 10.1016/j.cell.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 72.Summers DW, Wolfe KJ, Ren HY, Cyr DM. The type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS ONE. 2013;8(1):e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304(5678):1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 74.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27(20):2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, Shorter J. Operational plasticity enables Hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151(4):778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5(2):251–262. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zaarur N, Meriin AB, Gabai VL, Sherman MY. Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin 1. J Biol Chem. 2008;283(41):27575–27584. doi: 10.1074/jbc.M802216200. [DOI] [PubMed] [Google Scholar]
- 78.Zaarur N, Meriin AB, Bejarano E, Xu XB, Gabai VL, Cuervo AM, Sherman MY. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol. 2014;34(7):1336–1348. doi: 10.1128/MCB.00103-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muchowski PJ, Ning K, D’Souza-Schorey C, Fields S. Requirement of an intact microtubule cytoskeleton for aggregation and inclusion body formation by a mutant huntingtin fragment. P Natl Acad Sci USA. 2002;99(2):727–732. doi: 10.1073/pnas.022628699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strom AL, Shi P, Zhang FJ, Gal J, Kilty R, Hayward LJ, Zhu HN. Interaction of amyotrophic lateral sclerosis (ALS)-related mutant copper-zinc superoxide dismutase with the dynein-dynactin complex contributes to inclusion formation. J Biol Chem. 2008;283(33):22795–22805. doi: 10.1074/jbc.M800276200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yao TP. The role of ubiquitin in autophagy-dependent protein aggregate processing. Genes Cancer. 2010;1(7):779–786. doi: 10.1177/1947601910383277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478(7368):260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wustner D, Solanko LM, Lund FW, Sage D, Schroll HJ, Lomholt MA. Quantitative fluorescence loss in photobleaching for analysis of protein transport and aggregation. BMC Bioinformatics. 2012;13:296. doi: 10.1186/1471-2105-13-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fossati M, Borgese N, Colombo SF, Francolini M. Visualization of endoplasmic reticulum subdomains in cultured cells. J Vis Exp. 2014;84:e50985. doi: 10.3791/50985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, Kim CE, Frackelton EC, Solski JA, Williams KL, Clay-Falcone D, Elman L, McCluskey L, Greene R, Hakonarson H, Kalb RG, Lee VM, Trojanowski JQ, Nicholson GA, Blair IP, Bonini NM, Van Deerlin VM, Mourelatos Z, Shorter J, Gitler AD. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21(13):2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gitler AD, Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5(3):179–187. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.West JP, 3rd, Gitler AD. Cell biology. Clogging information flow in ALS. Science. 2014;345(6201):1118–1119. doi: 10.1126/science.1259461. [DOI] [PubMed] [Google Scholar]
- 88.Hart MP, Brettschneider J, Lee VMY, Trojanowski JQ, Gitler AD. Distinct TDP-43 pathology in ALS patients with ataxin 2 intermediate-length polyQ expansions. Acta Neuropathol. 2012;124(2):221–230. doi: 10.1007/s00401-012-0985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bonini NM, Gitler AD. Model organisms reveal insight into human neurodegenerative disease: ataxin-2 intermediate-length polyglutamine expansions are a risk factor for ALS. J Mol Neurosci. 2011;45(3):676–683. doi: 10.1007/s12031-011-9548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201(3):361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gitler AD. TDP-43 and FUS/TLS yield a target-rich haul in ALS. Nat Neurosci. 2012;15(11):1467–1469. doi: 10.1038/nn.3243. [DOI] [PubMed] [Google Scholar]
- 92.Liu-Yesucevitz L, Lin AY, Ebata A, Boon JY, Reid W, Xu YF, Kobrin K, Murphy GJ, Petrucelli L, Wolozin B. ALS-linked mutations enlarge TDP-43-enriched neuronal RNA granules in the dendritic arbor. J Neurosci. 2014;34(12):4167–4174. doi: 10.1523/JNEUROSCI.2350-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM, Finkbeiner S, Gitler AD, Bonini NM. Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet. 2014;46(2):152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang J, Farr GW, Zeiss CJ, Rodriguez-Gil DJ, Wilson JH, Furtak K, Rutkowski DT, Kaufman RJ, Ruse CI, Yates JR, Perrin S, Feany MB, Horwich AL. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. P Natl Acad Sci USA. 2009;106(5):1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang J, Farr GW, Hall DH, Li F, Furtak K, Dreier L, Horwich AL (2009) An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. Plos Genet 5(1):2–6. doi:10.1371/journal.pgen.1000350 [DOI] [PMC free article] [PubMed]
- 96.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J. Gitler AD (2009) TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284(37):25459. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mishra RS, Bose S, Gu Y, Li R, Singh N. Aggresome formation by mutant prion proteins: the unfolding role of proteasomes in familial prion disorders. J Alzheimers Dis. 2003;5(1):15–23. doi: 10.3233/jad-2003-5103. [DOI] [PubMed] [Google Scholar]
- 98.Swinnen E, Buttner S, Outeiro TF, Galas MC, Madeo F, Winderickx J, Franssens V. Aggresome formation and segregation of inclusions influence toxicity of alpha-synuclein and synphilin-1 in yeast. Biochem Soc Trans. 2011;39(5):1476–1481. doi: 10.1042/BST0391476. [DOI] [PubMed] [Google Scholar]
- 99.Wang H, Ying Z, Wang G. Ataxin-3 regulates aggresome formation of copper-zinc superoxide dismutase (SOD1) by editing K63-linked polyubiquitin chains. J Biol Chem. 2012;287(34):28576–28585. doi: 10.1074/jbc.M111.299990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adachi H, Kurooka T, Otsu W, Inaba M. The forced aggresome formation of a bovine anion exchanger 1 (AE1) mutant through association with deltaF508-cystic fibrosis transmembrane conductance regulator upon proteasome inhibition in HEK293 cells. Jpn J Vet Res. 2010;58(2):101–110. [PubMed] [Google Scholar]
- 101.Cohen E, Taraboulos A. Scrapie-like prion protein accumulates in aggresomes of cyclosporin A-treated cells. EMBO J. 2003;22(3):404–417. doi: 10.1093/emboj/cdg045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Pietrement O, Pastre D. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 2014;42(13):8678–8691. doi: 10.1093/nar/gku582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20(3):125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 106.Prudencio M, Borchelt DR. Superoxide dismutase 1 encoding mutations linked to ALS adopts a spectrum of misfolded states. Mol Neurodegener. 2011;6:77. doi: 10.1186/1750-1326-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sacino AN, Thomas MA, Ceballos-Diaz C, Cruz PE, Rosario AM, Lewis J, Giasson BI, Golde TE. Conformational templating of alpha-synuclein aggregates in neuronal-glial cultures. Mol Neurodegener. 2013;8:17. doi: 10.1186/1750-1326-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Young D, Mayer F, Vidotto N, Schweizer T, Berth R, Abramowski D, Shimshek DR, van der Putten PH, Schmid P. Mutant huntingtin gene-dose impacts on aggregate deposition, DARPP32 expression and neuroinflammation in HdhQ150 mice. PLoS ONE. 2013;8(9):e75108. doi: 10.1371/journal.pone.0075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. P Natl Acad Sci USA. 2009;106(35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nussbaum-Krammer CI, Morimoto RI. Caenorhabditis elegans as a model system for studying non-cell-autonomous mechanisms in protein-misfolding diseases. Dis Models Mech. 2014;7(1):31–39. doi: 10.1242/dmm.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gitler AD, Lehmann R. Modeling human disease. Science. 2012;337(6092):269. doi: 10.1126/science.1227179. [DOI] [PubMed] [Google Scholar]
- 112.Auluck PK, Caraveo G, Lindquist S. Alpha-synuclein: membrane interactions and toxicity in Parkinson’s disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 113.Eisbach SE, Outeiro TF. Alpha-synuclein and intracellular trafficking: impact on the spreading of Parkinson’s disease pathology. J Mol Med (Berl) 2013;91(6):693–703. doi: 10.1007/s00109-013-1038-9. [DOI] [PubMed] [Google Scholar]
- 114.Tenreiro S, Reimao-Pinto MM, Antas P, Rino J, Wawrzycka D, Macedo D, Rosado-Ramos R, Amen T, Waiss M, Magalhaes F, Gomes A, Santos CN, Kaganovich D, Outeiro TF (2014) Phosphorylation modulates clearance of alpha-synuclein inclusions in a yeast model of Parkinson’s disease. PLoS Genet 10(5):11–14. doi:10.1371/journal.pgen.1004302 [DOI] [PMC free article] [PubMed]
- 115.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302(5651):1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nystrom T, Liu BD. Protein quality control in time and space—links to cellular aging. FEMS Yeast Res. 2014;14(1):40–48. doi: 10.1111/1567-1364.12095. [DOI] [PubMed] [Google Scholar]
- 117.Nystrom T. Aging: filtering out bad mitochondria. Curr Biol. 2013;23(23):R1037–R1039. doi: 10.1016/j.cub.2013.10.049. [DOI] [PubMed] [Google Scholar]
- 118.Nystrom T, Liu BD. The mystery of aging and rejuvenation—a budding topic. Curr Opin Microbiol. 2014;18:61–67. doi: 10.1016/j.mib.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 119.Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Frei SB, Snapp EL, Barral Y (2014) A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. Elife 3:3–11. doi:10.7554/eLife.01883 [DOI] [PMC free article] [PubMed]
- 120.Lippuner AD, Julou T, Barral Y. Budding yeast as a model organism to study the effects of age. FEMS Microbiol Rev. 2014;38(2):300–325. doi: 10.1111/1574-6976.12060. [DOI] [PubMed] [Google Scholar]
- 121.Lindner AB, Madden R, Dernarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. P Natl Acad Sci USA. 2008;105(8):3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coelho M, Dereli A, Haese A, Kuhn S, Malinovska L, DeSantis ME, Shorter J, Alberti S, Gross T, Tolic-Norrelykke IM. Fission yeast does not age under favorable conditions, but does so after stress. Curr Biol. 2013;23(19):1844–1852. doi: 10.1016/j.cub.2013.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anderson P, Kedersha N. RNA granules. Journal of Cell Biology. 2006;172(6):803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MAWH, van der Want JJL, de Vos RAI, Brunt ER, Sibon OCM, Kampinga HH. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4(12):2325–2335. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bufalino MR, DeVeale B, van der Kooy D. The asymmetric segregation of damaged proteins is stem cell-type dependent. J Cell Biol. 2013;201(4):523–530. doi: 10.1083/jcb.201207052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: unequal segregation of proteins destined for degradation. P Natl Acad Sci USA. 2008;105(22):7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sinsimer KS, Lee JJ, Thiberge SY, Gavis ER. Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Rep. 2013;5(5):1169–1177. doi: 10.1016/j.celrep.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae . P Natl Acad Sci USA. 2007;104(26):10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Erjavec N, Cvijovic M, Klipp E, Nystrom T. Selective benefits of damage partitioning in unicellular systems and its effects on aging. P Natl Acad Sci USA. 2008;105(48):18764–18769. doi: 10.1073/pnas.0804550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu B, Larsson L, Franssens V, Hao X, Hill SM, Andersson V, Hoglund D, Song J, Yang X, Oling D, Grantham J, Winderickx J, Nystrom T. Segregation of protein aggregates involves actin and the polarity machinery. Cell. 2011;147(5):959–961. doi: 10.1016/j.cell.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 131.Li J, Li T, Zhang X, Tang Y, Yang J, Le W. Human superoxide dismutase 1 overexpression in motor neurons of Caenorhabditis elegans causes axon guidance defect and neurodegeneration. Neurobiol Aging. 2014;35(4):837–846. doi: 10.1016/j.neurobiolaging.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 132.Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31(45):16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9(4):e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spokoini R, Shamir M, Keness A, Kaganovich D (2013) 4D imaging of protein aggregation in live cells. J Vis Exp 74:4–6. doi:10.3791/50083 [DOI] [PMC free article] [PubMed]
- 135.Shipley FB, Clark CM, Alkema MJ, Leifer AM. Simultaneous optogenetic manipulation and calcium imaging in freely moving C. elegans . Front Neural Circuits. 2014;8:28. doi: 10.3389/fncir.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baker SM, Buckheit RW, 3rd, Falk MM. Green-to-red photoconvertible fluorescent proteins: tracking cell and protein dynamics on standard wide-field mercury arc-based microscopes. BMC Cell Biol. 2010;11:15. doi: 10.1186/1471-2121-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]