Abstract
Under physiological and pathological conditions, extracellular vesicles (EVs) are present in the extracellular compartment simultaneously with soluble mediators. We hypothesized that cytokine effects may be modulated by EVs, the recently recognized conveyors of intercellular messages. In order to test this hypothesis, human monocyte cells were incubated with CCRF acute lymphoblastic leukemia cell line-derived EVs with or without the addition of recombinant human TNF, and global gene expression changes were analyzed. EVs alone regulated the expression of numerous genes related to inflammation and signaling. In combination, the effects of EVs and TNF were additive, antagonistic, or independent. The differential effects of EVs and TNF or their simultaneous presence were also validated by Taqman assays and ELISA, and by testing different populations of purified EVs. In the case of the paramount chemokine IL-8, we were able to demonstrate a synergistic upregulation by purified EVs and TNF. Our data suggest that neglecting the modulating role of EVs on the effects of soluble mediators may skew experimental results. On the other hand, considering the combined effects of cytokines and EVs may prove therapeutically useful by targeting both compartments at the same time.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-014-1618-z) contains supplementary material, which is available to authorized users.
Keywords: Extracellular vesicle, Inflammation, Interleukin-8, Microarray, TNF
Introduction
Extracellular vesicles (EVs) are membrane-surrounded structures. Based on their size and origin, EVs are broadly categorised into the following groups: exosomes, microvesicles (MVs) (also referred to as microparticles, shedding vesicles and ectosomes), and apoptotic bodies [1]. Although there is considerable overlap in the size distribution of the three main groups of EVs, exosomes are roughly 50–100 nm in size, and are generated by the exocytosis of multivesicular bodies. MVs (100–1,000 nm) are produced by budding off the plasma membrane [1]. Apoptotic bodies are generally assumed to be larger, over 1,000 nm in diameter, and are produced via membrane blebbing in the course of apoptosis, although EVs <1,000 nm in diameter are also produced during apoptosis [1]. Several recent reviews have summarized our current knowledge of EV physiology and analytical challenges of the field [1–5].
EVs have long been suspected to transfer messages between cells, and a number of different functions have been associated with EVs, such as antigen presentation [6] and the transfer of mRNA and miRNA (“shuttle” RNA) [7]. EVs may communicate protective messages [8] and promote inflammation [9], and their composition changes depending on the circumstances [10].
The stimulating effect of a T cell-derived supernatant on the human monocyte cell line U937 was observed as early as 1983 [11]. A later study demonstrated that T cell-derived EVs were able to activate human monocytes in a way similar to direct cell-to-cell contact [12].
However, most studies on EVs tend to focus on a single vesicle type, most frequently exosomes, and the effects of different EV populations are rarely studied in combination. Furthermore, EVs (different EV subpopulations and those of multiple cellular origins) and cytokines occur naturally in bodily fluids as a mixture, but no study to date has analyzed the global gene expression effects of a combination of EVs and cytokines on cells.
We hypothesized that EVs modulate the cellular effects of cytokines. Here, we show that the effects of EVs, TNF, or their combination on gene expression of recipient cells are substantially different.
Materials and methods
Cell lines
CCRF-CEM human T cell lymphoma line and U937 human monocyte cell line were obtained from ATCC. The cell lines were cultured in RPMI medium (Sigma, Budapest, Hungary) containing 10 % (v/v) fetal bovine serum (PAA, Budapest, Hungary), 2 mM glutamine, 100 U/mL each penicillin and streptomycin, and 0.5 % ciprofloxacin (Fresenius Kabi, Budapest, Hungary), at 37 °C in 5 % CO2/air. The cell lines were regularly tested for mycoplasma contamination using a DAPI-based mycoplasma detection.
Experimental setup
All experiments were performed in quadruplicate, unless otherwise stated. CCRF cells were grown to confluency. For the experiment, cells were resuspended at 0.5 × 106 cells/mL density in serum-free RPMI medium containing 0.5 % ciprofloxacin, and EV production was allowed to take place for 24 h. To avoid aggregation of EVs, co-sedimentation of protein aggregates with EVs and the potential damage of the membrane of EVs during high-speed centrifugation, we chose a method for studying EVs that we expected to cause the least artefactual effects on EVs. Thus, the supernatant of cells was collected and, in order to produce two different fractions, treated as follows: (1) EV-containing, cell-free supernatant was prepared by centrifuging the supernatant for 10 min at 300g and discarding the pellet; (2) EV-free, cell-free supernatant (control) was prepared by spinning the supernatant at 300g for 10 min, followed by 20,500g for 20 min and 100,000g for 60 min, harvesting the supernatant, and filtering it through a 0.2 µm filter (Millipore, Budapest, Hungary) by hydrostatic pressure.
The two different types of supernatants (total EV-containing and control) were divided in two aliquots each. Next, we added the EV-depleted or EV-containing supernatants (derived from 1 × 106 CCRF cells) to each of the wells of 6-well tissue culture plates. Parallel wells containing EV-depleted or EV-containing supernatants were supplemented with 10 ng/mL TNF (Sigma). U937 cells (5 × 106 cells per well in 5 mL total volume) were added to each well of the plates. The cells were incubated for 24 h at 37 °C in 5 % CO2/air, harvested by spinning at 300g for 5 min and washed once in PBS before RNA extraction. The supernatant was harvested and stored at −80 °C until an ELISA analysis of IL-8 concentration was performed.
Determination of EV concentration and size distribution using qNano
Conditioned tissue culture supernatant of CCRF cells was centrifuged at 300g for 10 min, the pellet was discarded, and the supernatant was submitted to resistive pulse sensing analysis using a qNano instrument (IZON Science, New Zealand) as described previously [13, 14]. Briefly, particle numbers were counted for 3 min using 10 mbar pressure and NP400 or NP200 nanopore membranes. Voltage was set in between 0.1 and 0.25 V in order to achieve a stable 100 nA current. Particle size histograms were recorded when RMS noise was below 12 pA, and particle rate in time was linear. Calibration was performed using known concentration of beads CPC200B (mode diameter: 203 nm) or SKP400D (mode diameter: 335 nm) (both from IZON) diluted 1:1,000 in RPMI. In some experiments we spiked in the EV-depleted CCRF supernatant with known size reference beads (SKP400D at 4.5 × 107 particles/mL concentration) to confirm the removal of EVs of the same size range.
Electron microscopy
In order to characterize the vesicular composition of EV-containing supernatants, and to confirm that EV-free supernatants of CCRF cells did not contain any vesicular structures, we pelleted the conditioned media at 100,000g for 60 min. The supernatants were then carefully removed and the pellets were fixed with 4 % paraformaldehyde in 0.01 M PBS at room temperature for 60 min. Following washing with PBS, the preparations were postfixed in 1 % OsO4 (Taab) for 30 min. After rinsing with distilled water, the pellets were dehydrated in graded ethanol, including block staining with 1 % uranyl-acetate in 50 % ethanol for 30 min, and were embedded in Taab 812 (Taab). After overnight polymerization at 60 °C, the pellets were sectioned and the ultrathin sections were analyzed using a Hitachi 7100 electron microscope equipped with a Megaview II (lower resolution; Soft Imaging System) digital camera.
Total RNA isolation
Total RNA was isolated from U937 cells using a PureLink RNA Mini Kit (Ambion Life Sciences, Budapest, Hungary) according to the manufacturer’s instructions. The purity and integrity of the RNA was analyzed using Bioanalyzer RNA chips (Agilent, Kromat, Budapest, Hungary), and the RNA concentration was measured using a NanoDrop spectrophotometer.
Microarray analysis
Labeled cRNA was prepared according to the Agilent Low Input QuickAmp Kit protocol and the 4 × 44 k Whole Human Genom oligo microarray (Agilent) was performed according to the protocol. Microarray data were analyzed as outlined below.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [15] and are accessible through GEO Series accession number GSE47897 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47897).
Gene expression data generated by the microarray experiment were first analyzed by Agilent’s GeneSpring GX software. Three replicates of the following four types of samples were studied by the microarray: control, EV, EV + TNF, and TNF, corresponding to treatments of U937 cells. Probes with less than twofold mean change in expression value compared to the control, and probes with compromised flags, were excluded from further analysis. A one-way ANOVA test was performed on the remaining probes in order to detect significant alterations in gene expression, utilizing Tukey’s post hoc test and a Benjamini–Hochberg correction for multiple hypotheses. Corrected p values of <0.05 were considered statistically significant, unless otherwise stated. In order to generate a heatmap of gene expression changes, we used hierarchical clustering on gene entities with Pearson’s centered similarity measure and centroid linkage rule. Individual clusters of genes showing similar changes in expression pattern upon treatment were generated using K-means clustering. GeneMania plugin [16–18] for Cytoscape was used to find and visualize functional clusters of significantly altered genes. Biological function was also derived from gene ontology (GO) terms linked to genes with an altered expression.
Beyond single-gene analysis, gene set enrichment analysis (GSEA) was also used. Using the standard implementation of the method by Subramanian et al. [19], currently called GSEA v.3.1, available from the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp). Given the limited number of conditions, we did all analyses with 1,000 permutations based on the genotype. Gene sets were obtained from the c2, c3, c4, and c5 branches of the Molecular Signatures Database (MSigDB v.3.1) [20], excluding gene sets that contain <15 or more than 500 entities. As a screening test, we investigated all datasets in the corresponding branches of MSigDB and counted the number of gene sets showing a significant enrichment at a false detection rate (FDR) <25 % at nominal p value <5 %. GSEA results of GO terms (c5) were visualized using Cytoscape and its enrichment map [21]. For network generation in enrichment map, we used the standard settings (p value cut-off 0.005; FDR cut-off 0.1; overlap coefficient 0.5; combined constant 0.5). The network of individual words in enriched GO terms was plotted by the wordcloud plugin [22].
Reverse transcription and Taqman assays
In order to validate the results of the microarray, a selected range of genes were tested using Taqman assays. Following extraction, the RNA was reverse transcribed: each 40 µL reaction contained 2 µg RNA, 5 mM MgCl2, 10 mM dNTP (Promega, Budapest, Hungary), random primers (Promega) and 2 µLs of 50 U/µL MuLV reverse transcriptase (Roche, Budaörs, Hungary). The reaction was preformed in a Perkin Elmer DNA Thermal Cycler 480 under the following conditions: 42 °C for 40 min, 99 °C for 5 min, and 20 °C for 5 min.
Quantitative real-time PCR reactions of selected genes including HPRT (endogenous control), CD36, CD82, CNR2, IL8, CCL2, CXCL2, NPC1, ICAM1, MMP9, and SMPD3 were performed using Taqman probes and the reactions were run in an Applied Biosystems 7900HT Fast Real-Time PCR system using the thermal cycling conditions of 20 min at 95 °C, followed by 40 cycles of a 1 s denaturation step at 95 °C and a 20 s annealing and extension step at 60 °C.
Testing the effects of different EV populations on the IL-8 mRNA expression of U937 cells
Two different purified EV fractions were isolated: pure MVs and a mixture of MVs and exosomes (MV + Exo), using the following workflow: 300g 10 min → filtration by gravity through a 5 µm filter → 2,000g for 20 min → filtration by gravity through an 800 nm filter → 12,500g for 10 min (MP) or 20,000g for 40 min (MV + Exo). The MV and MV + Exo pellets were resuspended in EV-depleted conditioned supernatant rather than RPMI (EV-free supernatant was produced using the following workflow: 300g for 10 min → filtration by gravity through a 5 µm filter → 2,000g for 20 min → filtration by gravity through an 800 nm filter → 100,000g for 60 min → filtration through a 0.2 µm filter). MV and MV + Exo pellets, resuspended in EV-free supernatant, were used to treat U937 cells. Total EV-containing supernatant (produced by pelleting cells at 300g for 10 min and filtering the supernatant through a 5 µm filter by gravity) was used as a positive control, and EV-free supernatant (produced as described above) was used as a negative control. The donor to recipient cell ratio in the MV and MV + Exo treatments was 5:1 (i.e. MV or MV + Exo produced by 5 × 106 CCRF cells was used to treat 1 × 106 U937 cells). MVs were used at a concentration of 20 µg/mL, MV + Exo were used at a concentration of 25 µg/mL. U937 cells were incubated for 24 h at 37 °C in 5 % CO2/air, harvested by spinning at 300g for 5 min and washed once in PBS before RNA extraction, and the gene expression levels of IL-8, CCL2, CNR2, and CD36 were measured in the treated cells, using Taqman gene expression assays. Results were expressed as gene expression relative to HPRT (mean ± SEM, n = 4).
Testing the effects of a combination of purified MVs and TNF on the IL-8 mRNA expression of U937 cells
In order to validate the results of the somewhat unorthodox experimental setup used for the microarray experiment, we used purified CCRF-derived MVs (20 µg/mL) with or without TNF (10 ng/mL), resuspended in serum-free RPMI. MVs were purified using the following workflow: 300g for 10 min → filtration by gravity through a 5 µm filter → 2,000g for 20 min → filtration via an 800 nm filter by gravity → 12,500g for 10 min. As a negative control, serum-free RPMI was used, with or without TNF (10 ng/mL). U937 cells were incubated for 24 h at 37 °C in 5 % CO2/air, harvested by spinning at 300g for 5 min and washed once in PBS before RNA extraction, and IL-8 Taqman assays were carried out as described above.
Cytokine ELISAs
In order to confirm the detected gene expression changes on the protein level, IL-8 and CCL2 concentrations were measured in undiluted supernatants of stimulated U937 cells. IL-8 DuoSet ELISA development kit (R&D Biosystems, Biomedica, Budapest, Hungary) and Human MCP-1/CCL2 ELISA kit (Sigma) were used, both according to the manufacturer’s instructions. The TNF concentrations of conditioned media or isolated EVs were measured using a TNF DuoSet ELISA development kit (R&D Biosystems).
Flow cytometry of extracellular vesicles
Purified CCRF MV samples were analyzed using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Instrument settings and MV gate were set as described earlier [23–25] using both Megamix beads (BioCytex, Marseille, France) and Silica beads (Kisker Biotech, Steinfurt, Germany). Anti-CD63 PE and anti-CD9 FITC antibodies were purchased from Sigma-Aldrich, and Annexin V-FITC, anti-CD5 FITC, anti-CD71 FITC, and anti-CD33 PE were purchased from BD Biosciences. Annexin V or fluorochrome-conjugated antibodies were added to MV preparations (isolated from 5 mL of tissue culture media/sample) and were incubated for 20 min at RT in the dark. Event numbers of equal sample volumes were counted for 60 s at medium flow rate. EDTA-containing (5 mM) annexin-binding buffer solution or PBS was used to determine the background fluorescence. To verify the vesicular nature of MVs, and to exclude the presence of antibody aggregates, we added 0.1 % Triton X-100 to the samples, as we described previously [24]. This step resulted in prompt disappearance of fluorescent event counts suggesting the presence of membranous structures within the MV gate.
Chemotaxis assay
Chemotactic responsiveness of the THP-1 human monocyte cell line was measured by NeuroProbe® MBB 96 chamber (NeuroProbe, Gaithersburg, MD, USA), which is a two-chamber chemotactic technique. The conditioned media of stimulated or unstimulated U937 cells were placed into wells of 96-well microtitration plate which served as the lower chamber of the system. The THP-1 cells were seeded into the upper chamber which was separated from the lower one by a 5 μm pore size polycarbonate filter (NeuroProbe). After an incubation period of 3 h at 37 °C in a humidified 5 % CO2 atmosphere, the number of positive chemotactic responder cells was determined by alamarBlue® assay (Invitrogen, Carlsbad, CA, USA) by incubating the cells with the dye for 8 h, and measuring the fluorescence signal at 570 nm and 585 nm fluorescence excitation and emission wavelengths using an LS-50B Luminescence Spectrometer (PerkinElmer, Waltham, MA, USA).
Statistical analysis
Beyond the statistical analysis of the microarray results, one-way ANOVA assuming non-normal distribution (Kruskal–Wallis test) with Dunn’s post test was used to analyze the data of the ELISA assays, the chemotaxis assay, and the Taqman assays by GraphPad Prism v.4.
Results
Identification and characterization of EVs in CCRF supernatant
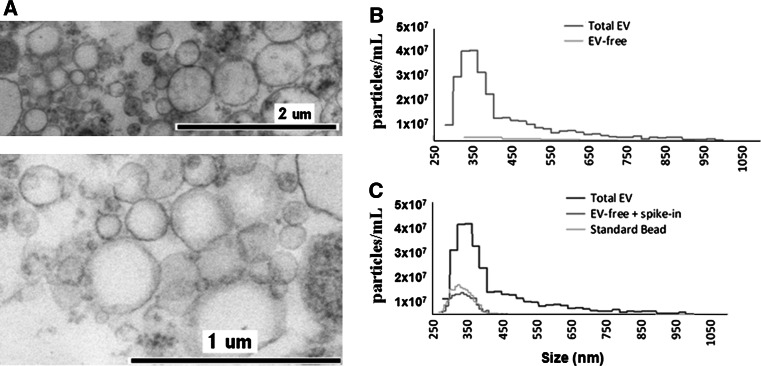
In this study, we used 24 h conditioned medium of CCRF cells as a source of EVs. To confirm the presence of EVs and to characterize the EV composition, we first analyzed the 100,000g pellets of cell-depleted CCRF supernatants by TEM. The pellet contained intact vesicles (Fig. 1a) surrounded by continuous membrane mainly in the size range of MVs. Sporadically, some cup-shaped structures with morphological features of exosomes, at around 100 nm in diameter were also visible.
Fig. 1.
Determination of EV quality, composition and size distribution. a Transmission electron micrograph of CCRF-derived EVs. Transmission electron micrograph of extracellular vesicles pelleted by 100,000g for 60 min from cell-free supernatants of CCRF cells. A Hitachi 7100 electron microscope equipped with a Megaview II (lower resolution; Soft Imaging System) digital camera was used to acquire the images. b IZON qNano analysis of the size distribution and concentration of extracellular vesicles. Cell-depleted CCRF supernatants, containing EVs, were analysed using an NP400 membrane of IZON qNano, at a strech value of 47 mm, 120 nA current, and 10 cm water pressure; at least 500 particles were detected. c Confirmation of EV removal by a known size bead spike. We spiked in the EV-depleted CCRF supernatant with known size reference beads (SKP400D at 4.5 × 107 particles/mL concentration) to confirm that the detected absence of positive signals in the EV-free supernatant was not due to a blockade of the nanopore
Next, an aliquot of the CCRF supernatant containing EVs was analyzed using an NP200 and an NP400 membrane of IZON qNano, which detect particles with roughly over 150 nm and 250 nm diameter, respectively (Fig. 1b). The cell-free, EV-free medium contained virtually no particles in the detected size range (both when using NP200 and NP400 membranes). In order to confirm that this fraction was free of microvesicles, we spiked in the EV-depleted CCRF supernatant with known size reference beads. As shown in Fig 1c, we confirmed that the measured low concentration of EVs was indeed due to the removal of vesicles rather than being a technical artefact (e.g., blockade of the nanopore).
The vesicles used in this study were found to be positive for Annexin V, CD63, CD71, and CD33, and weakly positive for CD5, while they were negative for CD9. Positive staining diminished after adding 0.1 % Triton-X, demonstrating that the positive events were indeed associated with membrane surrounded vesicles (Supplementary Fig. 1).
Effects of EVs, human recombinant TNF or their combination on the gene expression of U937 cells
We next incubated U937 cells with EV-containing or EV-free CCRF human T cell supernatant in the presence or absence of recombinant human TNF. The effects on gene expression were compared using an Agilent gene expression microarray, as shown in details below.
Single gene-based analysis by ANOVA, using GeneSpring
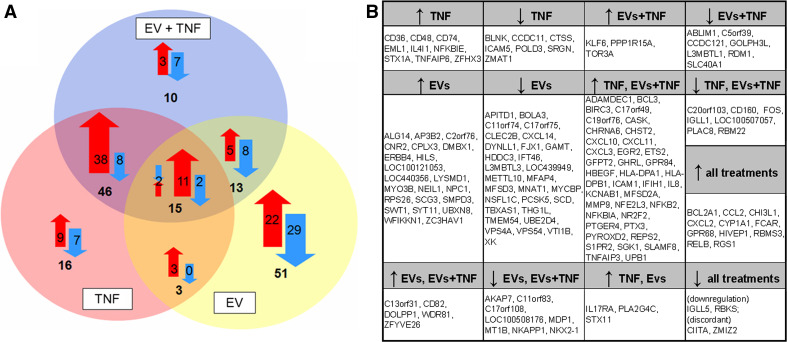
After excluding compromised flags and filtering for at least twofold gene expression changes using ANOVA analysis, a statistically significant (p < 0.05) difference in gene expression was found in the case of 202 probes, which could be mapped to 154 unique annotated genes or loci (Fig. 2b). The mean expression values of these 154 genes were used in all downstream analysis apart from gene set enrichment analysis (GSEA) (where only compromised flags were excluded). The nature of gene expression changes of these 154 genes (i.e. up- or downregulation) following the different treatments were plotted on a Venn diagram (Fig. 2a). TNF treatment, EV treatment and combined treatment induced statistically significant changes in the case of 80, 82, and 84 genes, respectively.
Fig. 2.
Significant gene expression changes based on ANOVA analysis of the microarray data, using GeneSpring. a Venn diagram demonstrating the number of significantly up- or downregulated genes (ANOVA, p < 0.05) after each treatment. Upregulation is indicated by red arrows, downregulation by blue arrows. b A list of genes showing the names of significantly up- or downregulated genes (ANOVA, p < 0.05) after each treatment
Among the genes significantly regulated by EVs there were some genes associated with vesicular trafficking. The multivalent protease inhibitor WFIKKN1 was one of the genes significantly upregulated by EVs. The protein product of this gene has been proposed to act as an inhibitor of serine proteases and metalloproteases [26]. A few genes involved in promoting inflammation, such as CXCL14, a potent neutrophil chemoattractant, as well as TBXAS1 were downregulated by treatment with EVs.
A relatively large group of genes (51 genes) were up- or downregulated following treatment with EVs only. In comparison, the paramount cytokine TNF up- or downregulated the expression levels of as few as 16 genes (exclusive TNF effect) that did not change upon EV or combined EV + TNF treatment.
A large number of genes (38) were upregulated following treatment with both TNF and TNF in combination with EVs. Among these were mainly inflammatory and anti-apoptotic molecules such as IL-8, ICAM-1, and BCL3, and the chemokines CXCL10, CXCL11, and CXCL3. Similarly, all treatments (EVs only, TNF only, combined EVs + TNF treatment) induced the upregulation of a number of inflammatory genes such as CCL2, CXCL2, RELB, and FCAR.
Cluster analysis of interaction patterns based on microarray results
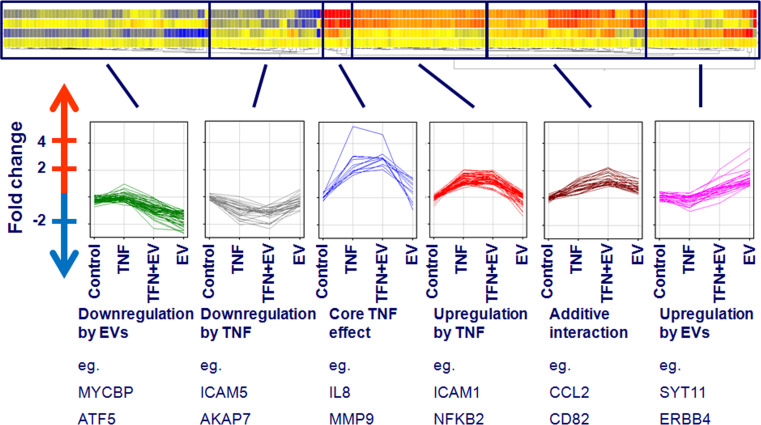
The pattern of interaction between cytokine and EV-related effects was further studied by K-means clustering, and we identified six clusters of significantly altered probes (ANOVA, p < 0.05) (Fig. 3).
Fig. 3.
Heatmap of gene expression changes and clusters of gene interaction patterns. In order to generate a heatmap of gene expression changes, we used hierarchical clustering on gene entities with Pearson’s centered similarity measure and centroid linkage rule. Individual clusters of genes showing similar changes in expression pattern upon treatment were generated using K-means clustering. Here, genes differentially expressed after any one of the treatments (ANOVA, p < 0.05), are hierarchically clustered based on the pattern of effects. Upregulation is indicated by red, downregulation by blue, whereas yellow represents no change compared to control
Clusters #1 and #6: here, the effect could be accounted for by EVs, and the effect of TNF alone was negligible. These clusters contained genes strongly upregulated (cluster 6, e.g., CNR2) or downregulated (cluster 1, e.g., MT1B, CXCL14, TBXAS1, SOCS2, CMYC) by EVs.
Cluster #2: in this cluster, genes were downregulated by both TNF and combined EV + TNF treatment, but unaffected by EVs only.
Cluster #3 and 5: these clusters contained genes where combined EV + TNF treatment resulted in an additive effect compared to single treatments (TNF or EV). Cluster #3 contained genes strongly upregulated by TNF, and the EVs contributed to this effect to a small extent (e.g., CXCL2, CXCL8, CXCL10, and MMP9). Cluster #5 contained genes that were upregulated by both EVs and TNF to a small degree, but the combined effect was more pronounced (e.g., CCL2, CXCL3, CD36, CD82, and CHI3L1).
Cluster #4: here, the effect of EVs on expression of these genes was negligible.
GeneMania analysis and gene set based analysis
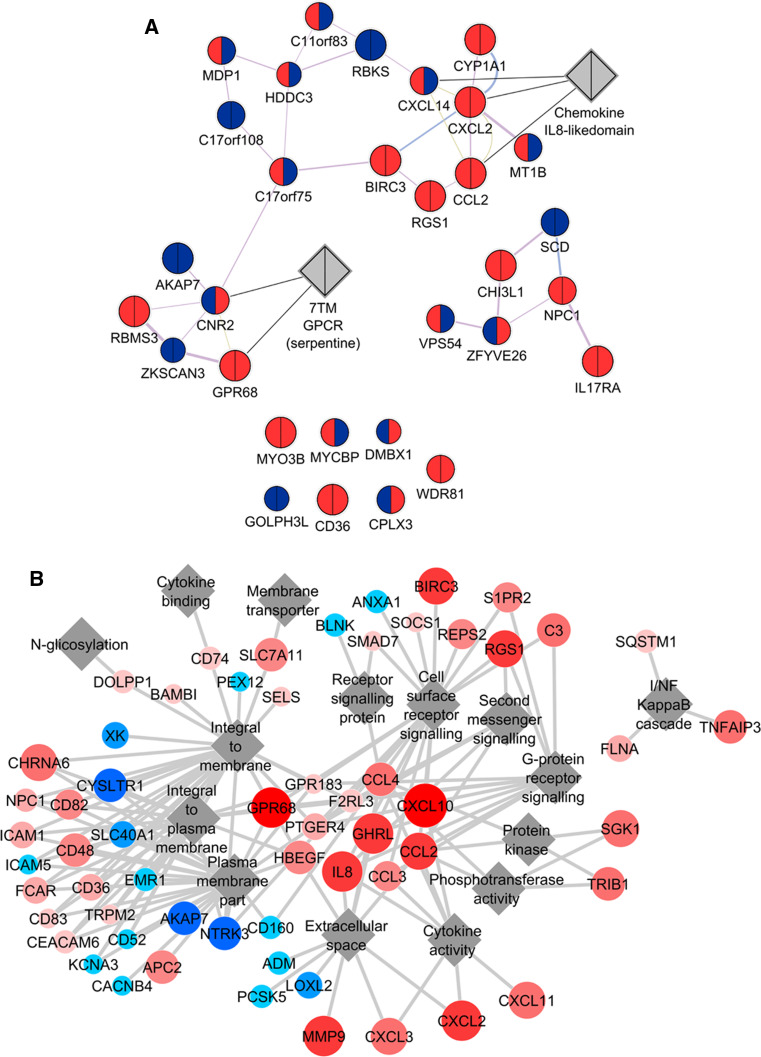
Next, in order to analyze interactions between genes, we used GeneMania plugin [33–35] for Cytoscape to visualize co-expression, co-localization, genetic or physical interactions, and shared functions or domains. Following treatment with EVs only, the expression of IL-8-like domain, Cytochrome P450 enzymes, and proteins with T-SNARE domain or Furin repeat were changed (Supplementary Fig. 2). Genes, significantly differentially regulated after combined EV + TNF treatment, included transmembrane-G-protein-coupled receptors and chemokines with IL-8-like domain (Fig. 4a).
Fig. 4.
Network analysis of genes modulated by a combined EV + TNF treatment. a Interaction network of genes modulated by EV + TNF treatment compared with TNF treatment alone. Based on the results of the microarray, GeneMania plugin [16–18] for Cytoscape was used to find and visualize functional clusters of significantly altered genes. Gene sets showing significant alteration in any one of the three treatments compared to control (pairwise ANOVA) were queried for co-expression, protein–protein interactions, shared domains, and co-localization. The program was set to find at maximum 20 relevant attributes. This interaction network shows genes significantly differentially expressed after a combined EV + TNF treatment compared to treatment with TNF alone. Upregulation is indicated by red, downregulation by blue, the left side of the nodes is coloured according to gene expression after TNF treatment, the right side according to gene expression after the combined EV + TNF treatment. b Network of GO terms only enriched after a combined EV + TNF treatment, but not TNF treatment alone. Beyond single-gene analysis, gene set enrichment analysis (GSEA) was also applied, using the standard implementation of the method by Subramanian et al. [19], currently called GSEA v3.1, available from the Broad Institute via the web (http://www.broadinstitute.org/gsea/index.jsp). Given the limited number of conditions, we did all analyses with 1,000 permutations based on the genotype. Gene sets were obtained from the c2, c3, c4, and c5 branches of the molecular signatures database (MSigDB v.3.1) [20], excluding gene sets that contain <15 or more than 500 entities. As a screening test, we investigated all datasets in the corresponding branches of MSigDB and counted the number of gene sets showing a significant enrichment at a false detection rate (FDR) <25 % at nominal p value <5 %. GSEA results of gene ontology (GO) terms (c5) were visualized using Cytoscape and its enrichment map [21]. For network generation in the enrichment map, we used the standard settings (p value cut-off 0.005; FDR cut-off 0.1; overlap coefficient 0.5; combined constant 0.5). Here, the top 60 genes of those sets only enriched after the combined EV + TNF treatment, but not after TNF treatment alone, were plotted against related, enriched GO terms. In the figure, genes are connected to the relevant terms. Colour and node size represent expression after a combined EV + TNF treatment. Upregulation is indicated by red, downregulation by blue
For a less restricted analysis of biological significance of microarray results, gene set enrichment was studied on all non-compromised probes. After screening for the number of enriched gene sets by the GSEA method, datasets based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, reactome, chemical and genetic perturbations database, and gene ontologies were found to yield enriched gene sets. Regarding KEGG pathways, innate-like signaling pathways were most enriched in the presence of EVs. The top 60 genes of those sets only enriched after the combined EV + TNF treatment, but not after TNF alone, were plotted against related, enriched GO terms (Fig. 4b). Many plasma membrane proteins, in particular cell surface receptors, were differentially expressed after a combined EV + TNF treatment.
Experimental validation of the microarray results
Validation of gene expression results using Taqman assays
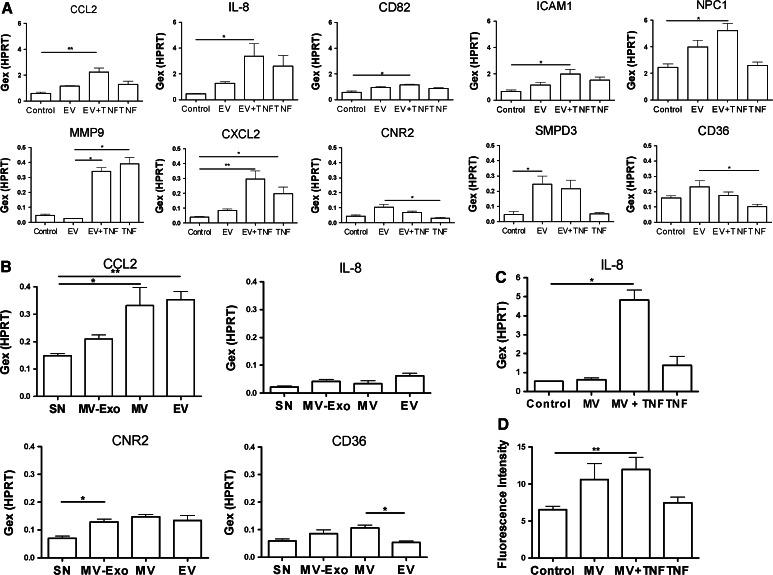
First, we carried out Taqman assays to confirm the microarray data on the expression changes of a select group of genes (Fig. 5a). Both the TNF and the combined EV + TNF treatment upregulated the expression of CCL2. An additive effect of EVs and TNF was observed in the cases of IL-8, CCL2, CD82, ICAM1, and NPC1; a core TNF effect was found in the case of MMP9 expression. A significant effect of both TNF and EV + TNF was observed in the case of CXCL2.
Fig. 5.
Validation of the microarray results using Taqman assays, testing for involvement of different EV populations including purified MVs and functional significance of the gene expression changes (chemotaxis assay). Changes in mRNA and protein levels of selected genes upon different treatments were detected by Taqman real-time PCR and ELISA assays. a The mRNA expression of CCL2, IL-8, CD82, ICAM1, NPC1, MMP9, CXCL2, CNR2, SMPD3, and CD36 genes were detected by Taqman real-time PCR assays (mean ± SEM, n = 4). Results are expressed as relative gene expression referred to HPRT (Gex). Data were analyzed by one-way ANOVA assuming non-normal distribution (Kruskal–Wallis test) with Dunn’s post test. *p < 0.05, **p < 0.01. b In order to identify the type of EVs responsible for the observed effects, we also tested purified MVs and a preparation containing both MVs and exosomes. Relative gene expressions of CCL2, IL-8, CNR2 and CD36 were referred to HPRT. Data were analyzed by one-way ANOVA assuming non-normal distribution (Kruskal–Wallis test) with Dunn’s post test. *p < 0.05, **p < 0.01. c In order to prove the existence of a cross-talk between MVs and cytokines, we used purified MV populations, resuspended in RPMI, to treat U937 cells. The gene expression levels of IL-8 were monitored in the treated cells, using Taqman gene expression assays (mean ± SEM, n = 3). Data were analyzed by one-way ANOVA assuming non-normal distribution (Kruskal–Wallis test) with Dunn’s post test. *p < 0.05. d To assess the functional significance of the upregulated chemokines, we carried out a chemotaxis assay monitoring the migration of THP1 human monocytic cell line towards the conditioned medium of differentially stimulated U937 cells. Prior to the chemotaxis assay, U937 cells were incubated with isolated MVs alone or in combination with TNF. Our data suggest that only the simultaneous presence of MVs and TNF induced a significant increase in the chemotactic activity of U937 cell supernatant [**p < 0.01, one-way ANOVA assuming non-normal distribution (Kruskal–Wallis test) with Dunn’s post test]
EV-containing supernatant caused a non-significant upregulation of CNR2 and a significant increase in SMPD3 gene expression. Similar tendencies were observed in the cases of CNR2 and CD36: the difference between TNF and the combined EV + TNF treatment was significant. As TNF alone caused a tendency to decrease mRNA levels of CNR2 and CD36, we propose that EVs and TNF had an antagonistic effect on the gene expression levels of CD36 and CNR2.
Validation of microarray data at the protein level
Next, we wanted to confirm changes of CCL2 and IL-8 expression at the protein level. Therefore, we measured CCL2 and interleukin-8 in the supernatant of U937 cells following treatment with EV-containing or EV-free supernatant ± TNF (n = 6). In the case of CCL2, we found slightly, but not significantly, elevated levels after all three treatments of U937 cells. In contrast, the additive effect of the EV-containing supernatant and TNF on IL-8 protein production was statistically significant (Supplementary Fig. 3).
Validation using purified EV populations
In order to elucidate which EV population was responsible for the regulatory effects, we assessed the effect of purified MVs and MV + Exo secreted by CCRF cells in comparison with the effects of the total EV content of CCRF’s conditioned media. As shown in Fig. 5b, purified MVs upregulated the expression of CCL2, CNR2, and CD36 in a statistically significant manner. The MV + Exo treatment significantly upregulated CNR2, and there was also a tendency towards an increase in the case of IL-8, CCL2, and CD36. Therefore, these results support the role of MVs rather than exosomes in the induction of the analyzed genes.
Finally, we wanted to demonstrate that our observations could also be reproduced using purified EVs. To this end, we purified CCRF-derived MVs which we subsequently resuspended in RPMI medium. These MVs were used, with or without the addition of TNF (10 ng/mL), to treat U937 cells. Compared to the treatment with MVs or TNF only, the combined MV + TNF treatment resulted in a statistically significant upregulation of IL-8 mRNA levels (Fig. 5c) that was substantially higher than those observed in the presence of either MVs or TNF alone. Further experimental validation is described and shown in Supplementary Figs. 3 and 4.
Chemotaxis assay to demonstrate the functional significance of cytokine modulating effects of EVs
To assess the functional significance of the upregulated chemokines, we carried out a chemotaxis assay monitoring the migration of THP1 human monocytic cell line towards the conditioned medium of differentially stimulated U937 cells. Prior to the chemotaxis assay, U937 cells were incubated with isolated MVs alone or in combination with TNF. After incubation, U937 cells were washed 3×, and the conditioned medium of U937 cells was used in subsequent chemotaxis assays. Our data suggest that only the simultaneous presence of MVs and TNF induced a significant increase in the chemotactic activity of U937 cell supernatant (p < 0.01, one-way ANOVA) (Fig. 5d).
Discussion
Over the past half century, numerous studies investigated the effects of soluble mediators in different in vitro settings to gain an insight to the functions of cytokines/chemokines. Approximately 15 years ago, a novel field evolved demonstrating the presence and physiological/pathological significance of purified EV preparations in vitro for cellular effects. Even though in vivo soluble mediators and extracellular vesicles are present simultaneously in the extracellular space, up until now, their combined presence has not been taken into consideration. Thus, the present study was carried out to fill this gap.
In order to test cellular effects of EVs, it is widely accepted to use purified EVs. However, this approach requires caution because of the lack of quality control tests for EV integrity. A common problem with vesicle pellets is aggregation, which makes it difficult to ensure even EV load in different treatments. EVs, in particular MVs, may also get damaged in the course of serial ultracentrifugation (similar to necrosis of cells). A third common confounding factor is co-pelleting of protein aggregates with EVs [24]. Furthermore, using pellets results in testing concentrated vesicles. For these reasons, we treated recipient cells with cell-depleted supernatant of EV-donor cells. As a negative control, we used supernatant depleted both in cells and EVs. This way, we were able to dissect the effects of cytokines present in the supernatant from the effects of EVs. We proved the validity of this experimental design by reproducing the interaction of EVs and TNF using purified MVs. Intriguingly, treatment with a combination of purified MVs and TNF induced significantly higher IL-8 mRNA levels than treatment with MVs or TNF alone, producing a synergistic rather than additive effect. One possible explanation for this synergistic effect could have been the binding of TNF on the surface of MVs, since TNF has been shown to associate with EVs [27]. However, we were able to experimentally rule out this possibility (Supplementary Fig. 4). Therefore, we speculated that convergent signaling pathways rather than TNF accumulation on the surface of MVs were responsible for the observed additive/synergistic effect. There might be several possible explanations for the synergism between the effect of TNF and EVs. In the case of IL-8, one possible mechanism suggested by GSEA analysis involves the upregulation of nuclear factor NF-kappaB, which acts both upstream of IL-8 and downstream of TNF. In another set of GSEA analyses (focusing on transcription factors that might induce the gene expression changes observed in the presence of EVs), sterol regulatory element-1 (SREBF1) appeared the most likely transcription factor responsible for the EV-induced effects. SREBF1 does not directly influence the gene expression of IL-8. However, according to GeneMania, it physically interacts with CREB1, which is involved in the regulation of IL-8. This parallel signaling pathway could also potentially aid the upregulation of IL-8 in the presence of TNF. Yet another possibility for the modulating effect of EVs would be a mechanism involving their RNA cargo, mainly miRNAs. Although our bioinformatics approach using miRwalk [28] did not result in any clue on a specific set of miRNAs that might be involved in the effect, this possibility cannot be excluded.
Our study is the first to compare the effects of EVs in the presence or absence of a cytokine on the global gene expression profile of recipient cells in a hypothesis-free system. Here, we show that the presence of T cell-derived EVs or TNF, alone or in combination, causes differential gene expression patterns of U937 monocyte cells. The expression of 51 genes was statistically significantly altered following treatment with EVs compared to control. Thus, EVs are sufficient to regulate the expression of a large number of genes.
As demonstrated by a network of GO terms, a combined EV + TNF treatment not only enhanced a cluster of inflammatory cytokines and members of the NF-κB pathway but also caused a prominent effect on genes associated with cell surface receptor signaling, cytokine binding, membrane transport, and genes associated with the plasma membrane (Fig. 4b; Supplementary Fig. 5).
Interestingly, we found that the expression of only 16 genes was significantly altered due to the effects of TNF alone, compared to the control group. Most of the genes modulated by TNF (80 in total) in this study were also affected by a combined treatment with TNF and EVs, and by EV treatment alone. This may suggest that EVs fine-tune the effects of TNF, and that a cross-talk exists between cytokines and EVs. Importantly, several anti-inflammatory genes were significantly upregulated by treatment with EVs. Thus, EVs, besides promoting the induction of inflammation, may also potentially contribute to the resolution of inflammation. A similar pattern of regulation was observed by Wahlgren et al. [29] who looked at the combined effects of IL-2 and autologous exosomes on the cytokine profile of T lymphocytes. Conspicuously, we found that the genes repressed by treatment with EVs in this study outnumbered the genes enhanced by EV treatment. Furthermore, the number of repressed genes was also large (29) compared with the number of genes downregulated in the other treatments in this study. Exosomes transfer mRNA and microRNA between cells (termed exosomal shuttle RNA) [7], and a similar role of MVs has also been shown [30]. The extensive regulatory function of microRNAs in the immune system via repressing target genes is well known [31], and so microRNAs in EVs are potentially responsible for repressing target genes in recipient cells.
IL-8, a potent chemoattractant for neutrophils, basophils, and T cells, and an activator of neutrophils, appeared to be a central player in the effects of all treatments compared with control. Thus, we also validated its expression at the mRNA and protein level. The induction of IL-8 expression at the mRNA and protein levels in monocytes by EVs is in keeping with data by Baj-Krzyworzeka et al. [32]. These authors observed that tumor-derived MVs not only serve as a storage pool of IL-8 mRNA and protein but also induce the de novo production of IL-8 and several CC cytokines (CCL2-5) by monocytes [32]. IL-8 production by synoviocytes was also induced by platelet-derived MVs in a seminal study on the role of platelet-derived MVs in arthritis [9].
GSEA results showed a strong association of the EV-regulated gene modules with those of a previous study on the effects of oxidized phospholipids on endothelial cells [33]. An interesting connection with the above study on oxidized phospholipids is our finding that treatment with EVs induced strong upregulation of CD36 (Fig. 5a). The scavenger receptor CD36 plays a variety of essential roles in the body. It recognises oxidized phospholipids [34], binds apoptotic cells via interacting with oxidised phosphatidylserine [35], and also appears to be important in the uptake of vesicles. At least some of the binding of phosphatidylserine-containing vesicles to monocyte cells could be attributed to CD36 [36]. Therefore, it is not surprising that treatment with EVs caused an upregulation of one of their putative receptors.
The G-protein-coupled cannabinoid receptor 2 (CNR2, CB2) was strongly induced by treatment with EVs, and this effect was diminished by the additional presence of TNF. CNR2 is the “peripheral” cannabinoid receptor, mostly expressed by cells of the immune system and responsible for mediating the non-psychoactive effects of cannabinoids such as analgesia and immunosuppression [37]. Although relatively little is known about the cannabinoid receptors, CNR2 has evoked medical interest as an inducer of apoptosis, an inhibitor of angiogenesis and skin tumor growth [38], and a potent anti-inflammatory and immunosuppressive agent [39]. CNR2 is also a target receptor of the dietary cannabinoid beta-caryophyllene, which, via the stimulation of CNR2, was able to achieve potent anti-inflammatory effects in wild-type, but not in Cnr2-/- mice [40]. The potent upregulation of CNR2 by EVs may also reflect the anti-inflammatory effects of EVs.
This study proved that the combined effects of EVs and cytokines differ from their independent effects and points out that EVs likely modify the effects of inflammatory cytokines. The data presented here give a new insight into inflammatory processes. We propose that testing the combined effects of soluble mediators and extracellular vesicles may model the in vivo effects of these mediators on cells more accurately than testing them separately. Our data may provide an explanation why targeting of certain soluble mediators does not always lead to the desired therapeutic effects. Furthermore, uncoupling the interaction of EVs and soluble mediators may open a novel avenue for a more effective therapeutic intervention.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful for Ms. Andrea Orbán’s and Mrs. Rita Antónia Fekete Heszné’s excellent technical assistance and Eszter Tóth for her contribution to some of the experiments. This work was funded by OTKA NK 84043, OTKA PD104369, Baross Gábor (REG-KM-09-1-2009-0010), and Marie Curie Networks for Initial Training-ITN-FP7-PEOPLE-2011-ITN, PITN-GA-2011-289033. Furthermore, this research was supported by TÁMOP 4.2.4. A/1-11-1-2012-0001 “National Excellence Program––Elaborating and operating an inland student and researcher personal support system” and subsidized by the European Union and co-financed by the European Social Fund.
Conflict of interest
The authors declare no conflict of interest.
Abbreviations
- CNR2
Cannabinoid receptor 2
- EV
Extracellular vesicle
- Gex (HPRT)
Gene expression (relative to HPRT)
- GSEA
Gene set enrichment analysis
- HPRT
Hypoxanthine-guanine phosphoribosyltransferase
- ICAM1
Intracellular adhesion molecule 1
- IL-8
Interleukin 8
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NPC1
Niemann–Pick disease C1
- MMP9
Matrix metalloproteinase 9
- SMPD3
Sphingomyelin phosphodiesterase 3
- TEM
Transmission electron microscopy
- TNF
Tumor necrosis factor
Footnotes
K. Szabó-Taylor and E. I. Buzás contributed equally.
References
- 1.György B, Szabó T, Pásztói M, Pál Z, Misják P, Aradi B, László V, Pállinger É, Pap E, Kittel Á, Nagy G, Falus A, Buzás E. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Théry C, Wauben MH, Hochberg F (2013) Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles 27(2) [DOI] [PMC free article] [PubMed]
- 3.Gutiérrez-Vázquez C, Villarroya-Beltri C, Mittelbrunn M, Sánchez-Madrid F. Transfer of extracellular vesicles during immune cell–cell interactions. Immunol Rev. 2013;251(1):125–142. doi: 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 8.Eldh M, Ekström K, Valadi H, Sjöstrand M, Olsson B, Jernös M, Lötvall J. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS ONE. 2010;5(12):e15353. doi: 10.1371/journal.pone.0015353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boilard E, Nigrovic PA, Larabee K, Watts GFM, Coblyn JS, Weinblatt ME, Massarotti EM, Remold-O’Donnell E, Farndale RW, Ware J, Lee DM. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science. 2010;327(5965):580–583. doi: 10.1126/science.1181928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong OG, Verhaar MC, Chen Y, Vader P, Gremmels H, Posthuma G, Schiffelers RM, Gucek M, van Balkom BWM. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1:18396. doi: 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moscicki RA, Amento EP, Krane SM, Kurnick JT, Colvin RB. Modulation of surface antigens of a human monocyte cell line, U937, during incubation with T lymphocyte-conditioned medium: detection of T4 antigen and its presence on normal blood monocytes. J Immunol. 1983;131(2):743–748. [PubMed] [Google Scholar]
- 12.Scanu A, Molnarfi N, Brandt KJ, Gruaz L, Dayer J-M, Burger D. Stimulated T cells generate microparticles, which mimic cellular contact activation of human monocytes: differential regulation of pro- and anti-inflammatory cytokine production by high-density lipoproteins. J Leukoc Biol. 2008;83(4):921–927. doi: 10.1189/jlb.0807551. [DOI] [PubMed] [Google Scholar]
- 13.Patkó D, György B, Németh A, Szabó-Taylor K, Kittel Á, Buzás E, Horváth R. Label-free optical monitoring of surface adhesion of extracellular vesicles. Sens Actuators B-chem. 2013;188:697–701. doi: 10.1016/j.snb.2013.07.035. [DOI] [Google Scholar]
- 14.de Vrij J, Maas SL, van Nispen M, Sena-Esteves M, Limpens RW, Koster AJ, Leenstra S, Lamfers ML, Broekman ML. Quantification of nanosized extracellular membrane vesicles with scanning ion occlusion sensing. Nanomedicine. 2013;8(9):1443–1458. doi: 10.2217/nnm.12.173. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: nCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26(22):2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(suppl 2):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oesper L, Merico D, Isserlin R, Bader GD (2011) WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol Med 6(7). doi:10.1186/1751-0473-6-7 [DOI] [PMC free article] [PubMed]
- 23.Lacroix R, Robert S, Poncelet P, Kasthuri RS, Key NS, Dignat-George F. ISTH SSC workshop. Standardization of platelet-derived microparticle enumeration by flow cytometry using calibrated beads: results of ISTH SSC collaborative workshop. J Thromb Haemost. 2010;8(2010):2571–2574. doi: 10.1111/j.1538-7836.2010.04047.x. [DOI] [PubMed] [Google Scholar]
- 24.György B, Módos K, Pállinger É, Pálóczi K, Pásztói M, Misják P, Deli MA, Sipos Á, Szalai A, Voszka I, Polgár A, Tóth K, Csete M, Nagy G, Gay S, Falus A, Kittel Á, Buzás EI. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117(4):e39–e48. doi: 10.1182/blood-2010-09-307595. [DOI] [PubMed] [Google Scholar]
- 25.György B, Szabó TG, Turiák L, Wright M, Herczeg P, Lédeczi Z, Kittel A, Polgár A, Tóth K, Dérfalvi B, Zelenák G, Böröcz I, Carr B, Nagy G, Vékey K, Gay S, Falus A, Buzás EI. Improved flow cytometric assessment reveals distinct microvesicle (cell-derived microparticle) signatures in joint diseases. PLoS ONE. 2012;7:e49726. doi: 10.1371/journal.pone.0049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trexler M, Bányai L, Patthy L. A human protein containing multiple types of protease-inhibitory modules. Proc Natl Acad Sci USA. 2001;98(7):3705–3709. doi: 10.1073/pnas.061028398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolowos W, Gaipl US, Sheriff A, Voll RE, Heyder P, Kern P, Kalden JR, Herrmann M. Microparticles shed from different antigen-presenting cells display an individual pattern of Surface molecules and a distinct potential of allogeneic T-cell activation. Scand J Immunol. 2005;61(3):226–233. doi: 10.1111/j.1365-3083.2005.01551.x. [DOI] [PubMed] [Google Scholar]
- 28.Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk––Database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44 (5):839–847. 10.1016/j.jbi.2011.05.002 [DOI] [PubMed]
- 29.Wahlgren J, Karlson DLT, Glader P, Telemo E, Valadi H. Activated human T cells secrete exosomes that participate in IL-2-mediated immune response signaling. PLoS ONE. 2012 doi: 10.1371/journal.pone.0049723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridandapani S, Paulaitis ME, Piper MG, Marsh CB. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121(6):984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 32.Baj-Krzyworzeka M, Weglarczyk K, Mytar B, Szatanek R, Baran J, Zembala M. Tumour-derived microvesicles contain interleukin-8 and modulate production of chemokines by human monocytes. Anticancer Res. 2011;31(4):1329–1335. [PubMed] [Google Scholar]
- 33.Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, Yang W-P, He A, Truong A, Patel S, Nelson SF, Horvath S, Berliner JA, Kirchgessner TG, Lusis AJ. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci USA. 2006;103(34):12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller YI, Choi S-H, Wiesner P, Fang L, Harkewicz R, Hartvigsen K, Boullier AS, Gonen A, Diehl CJ, Que X, Montano E, Shaw PX, Tsimikas S, Binder CJ, Witztum JL. Oxidation-specific epitopes are danger-associated molecular patterns recognized by pattern recognition receptors of innate immunity. Circ Res. 2011;108(2):235–248. doi: 10.1161/CIRCRESAHA.110.223875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenberg ME, Sun M, Zhang R, Febbraio M, Silverstein R, Hazen SL. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med. 2006;203(12):2613–2625. doi: 10.1084/jem.20060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tait JF, Smith C. Phosphatidylserine receptors: role of CD36 in binding of anionic phospholipid vesicles to monocytic cells. J Biol Chem. 1999;274(5):3048–3054. doi: 10.1074/jbc.274.5.3048. [DOI] [PubMed] [Google Scholar]
- 37.Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365(6441):61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- 38.Casanova M, Blázquez C, Martinez-Palacio J, Villanueva C, Fernández-Acenero M, Huffman J, Jorcano J, Guzmán M. Inhibition of skin tumor growth and angiogenesis in vivo by activation of cannabinoid receptors. J Clin Investig. 2003;111(1):43–50. doi: 10.1172/JCI200316116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitzcharles M-A, McDougall J, Ste-Marie PA, Padjen I. Clinical implications for cannabinoid use in the rheumatic diseases: potential for help or harm? Arthritis Rheum. 2012;64(8):2417–2425. doi: 10.1002/art.34522. [DOI] [PubMed] [Google Scholar]
- 40.Gertsch J. Anti-inflammatory cannabinoids in diet. Commun Integr Biol. 2008;1(1):26–28. doi: 10.4161/cib.1.1.6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.