Abstract
Endocytic budding implies the remodeling of a plasma membrane portion from a flat sheet to a closed vesicle. Clathrin- and actin-mediated endocytosis in yeast has proven a very powerful model to study this process, with more than 60 evolutionarily conserved proteins involved in fashioning primary endocytic vesicles. Major progress in the field has been made during the last decades by defining the sequential recruitment of the endocytic machinery at the cell cortex using live-cell fluorescence microscopy. Higher spatial resolution has been recently achieved by developing time-resolved electron microscopy methods, allowing for the first time the visualization of changes in the plasma membrane shape, coupled to the dynamics of the endocytic machinery. Here, we highlight these advances and review recent findings from yeast and mammals that have increased our understanding of where and how endocytic proteins may apply force to remodel the plasma membrane during different stages of the process.
Keywords: Endocytosis, Membrane curvature, Arp2/3 complex, Myosin-I, BAR domain, ENTH domain, Dynamin
Introduction
Clathrin-mediated endocytic budding has been studied comprehensively during the last decades and has therefore become a paradigm for understanding how membranes are deformed and remodeled to produce vesicles for different membrane-trafficking events. The initial molecular models explaining the process came from the interpretation of early electron micrographs showing highly curved clathrin-coated intermediates that bud from the plasma membrane (PM) in association with cargo [1–4]. Subsequent genetic and biochemical studies have identified more than 60 proteins in the pathway, most of them conserved from yeast to mammals ([5–8] and references therein). They include clathrin and its adaptors, the GTPase dynamin, actin, and myosin, proteins bearing membrane-sculpting BAR and ENTH domains and lipid-modifying enzymes. Extensive structural and physicochemical studies have shown that some of these proteins can directly generate and/or stabilize membrane curvature in vitro in a variety of ways; (1) by building curved or helical scaffolds bound to the membrane surface, (2) by asymmetrically inserting hydrophobic protein domains into the lipid bilayer, (3) by creating lipid phase boundaries, and (4) by acting as molecular motors that transduce chemical into mechanical force [9–11]. However, how the budding machineries really exert their function in vivo is still largely unknown.
Remarkable progress into our understanding of how these machineries may cooperate to produce the primary endocytic vesicle within the cell has been made during the last decade, first through the use of live-cell fluorescence microscopy and more recently by the development of time-resolved electron microscopy (TREM) techniques. Yeast has played a pioneering role in all of these advances owing to its simplicity and amenability to genetic manipulation and cell biology experimentation. Although the morphology of endocytic budding in yeast and mammals differs, probably due to the composition of the lipid bilayer, the nature of the cargo, or the turgor pressure [5, 12, 13], numerous studies indicate that the machinery is conserved and that the molecular mechanisms converge [6–8, 14–17], thus making yeast a good model system to study the process. By tagging the yeast endocytic proteins with GFP variants and tracking their relative dynamics using two-color fluorescence microscopy, the seminal work from Drubin’s laboratory and others has defined the precise sequence of molecular events occurring at the sites of endocytosis with a temporal resolution in the order of seconds and a spatial resolution of about 200 nm [18, 19] (see also [6, 7, 20] and references therein) (Fig. 1). This approach demonstrated that, once early components of the endocytic machinery mark the site where an endocytic vesicle will be produced, the rest of the components are recruited in a sequential and highly reproducible manner. Based on the identification of proteins with similar dynamics at endocytic sites, the live-cell imaging defined a number of functional modules that cooperate in vivo to deform the lipid bilayer [19, 21]. The early and the coat modules, including clathrin and its adaptors, first assemble at the cell cortex during a phase of restrained motility that lasts 1–2 min, after which a burst of actin polymerization accompanies a slow inward movement of the coat of around 200 nm. The scission module is finally recruited at the transition from the slow to a fast movement of the endocytic coat components, after which the vesicle uncoats and moves far inside the cell, still accompanied by actin ([6, 7] and references therein). A comprehensive analysis of the dynamics of 34 fluorescently tagged key endocytic proteins in mammalian cells, relative to scission, revealed parallels between yeast and mammals regarding the modular organization of the pathway and the order of recruitment of its components [8].
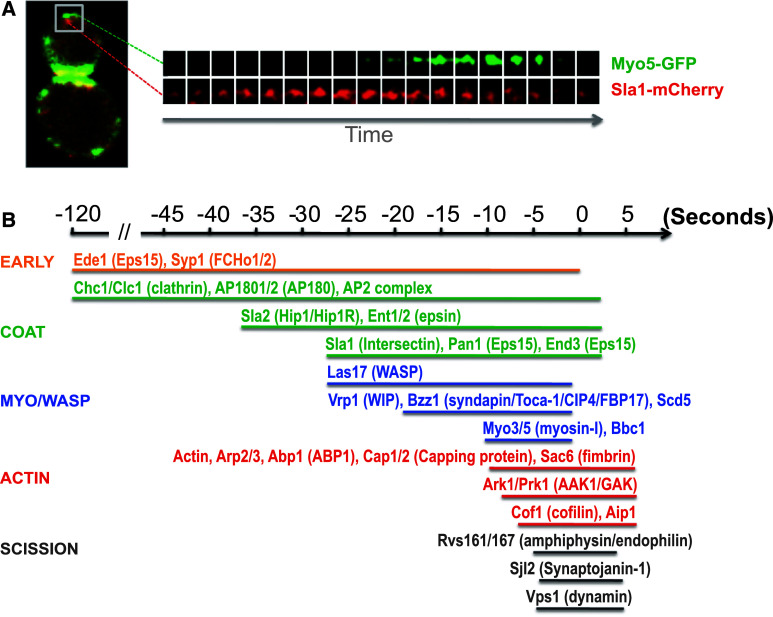
Fig. 1.
Sequential recruitment of endocytic proteins involved in clathrin-mediated endocytosis in yeast, determined by fluorescence microscopy. a Example of a two-color live-cell fluorescence imaging of endocytic proteins in yeast (reproduced from Ref. [119]). Left panel yeast cell expressing an early (Sla1) and a late (Myo5) endocytic protein tagged with mCherry and GFP, respectively, marking the sites of endocytosis. Right panel consecutive fluorescence micrographs from a 2-s time-lapse movie of the endocytic patch in the inset. Frames are approximately 100 × 100 nm. b Sequential recruitment of yeast endocytic proteins during clathrin-mediated endocytosis in yeast based on two-color live-cell fluorescence microscopy studies ([6, 7, 20] and references therein). The color code indicates the functional protein modules based on similar dynamics and functions at endocytic sites. Shown are the most studied components. Yeast nomenclature and the corresponding mammalian homologues or functional counterparts (between parenthesis) are indicated. Time of scission (time = 0 s) was defined by CLEM, relative to Abp1 and Rvs167 [25, 26]
Even though fluorescence live-cell imaging has established the sequence of events leading to the formation of an endocytic vesicle (Fig. 1), its spatial resolution does not allow visualization of how the dynamics of the endocytic components are coupled to changes in the shape of the lipid bilayer. Thus, by only using this technique, it is difficult to answer important questions related to when membrane curvature emerges or when scission occurs relative to the recruitment of the endocytic machinery. In this context, development of time-resolved ultrastructural approaches to study endocytic budding has represented an important breakthrough in the field. In this framework, two approaches have mainly been used for the comprehensive analysis of endocytic budding at the ultrastructural level in the yeast Saccharomyces cerevisiae: quantitative immuno-electron microscopy (QIEM) and correlative light and electron microscopy (CLEM).
The overall architecture of the primary endocytic invaginations in yeast was first defined by QIEM [22, 23] (Fig. 2). In this approach, HA-tagged endocytic proteins in association with PM invaginations are labeled in ultrathin sections from chemically-fixed and resin-embedded yeast cells with gold-conjugated antibodies (Immuno-golds) against the HA tag and localized by immuno-electron microscopy (IEM) (Fig. 2a). The shape of the labeled invaginations and the position of the immuno-golds labeling a particular protein are then described using geometrical parameters, which can be statistically analyzed (Fig. 2b, c). Provided that the amount of data is sufficiently high, the position of the proteins can be described with a resolution down to 7 nm [22, 23]. Previous IEM studies localizing actin, Abp1, and cofilin had already suggested that tubular PM invaginations of about 50 nm in diameter, surrounded by a ribosome-free area, might be the sites of endocytosis in yeast [24]. The QIEM studies unequivocally proved this hypothesis by showing that 18 different endocytic proteins, including clathrin and the clathrin adaptors, specifically associate with these kinds of profiles [22, 23] (Fig. 2e). Further, the statistical analysis of the length of the invaginations labeled for each particular protein, when aligned with their sequential recruitment as assessed by live-cell imaging, demonstrated that membrane curvature during endocytic budding emerges with the onset of actin polymerization; and that once curvature is initiated, the length of the invagination grows linearly up to 180 nm as budding progresses (Fig. 2d). Thus, the length of the invaginations can be used as a temporal marker to define their age. By statistically processing the data and by introducing the time dimension to the study, QIEM can correlate changes in the shape of the membrane with the dynamics of endocytic proteins with unprecedented resolution. These studies confirmed that the components of early and coat modules show similar dynamics at the ultrastructural level and precisely defined their positions as budding progresses. On the other hand, it outlined a much more complex behavior than anticipated for the components of the actin and the Myo/WASP modules (Fig. 2e).
Fig. 2.
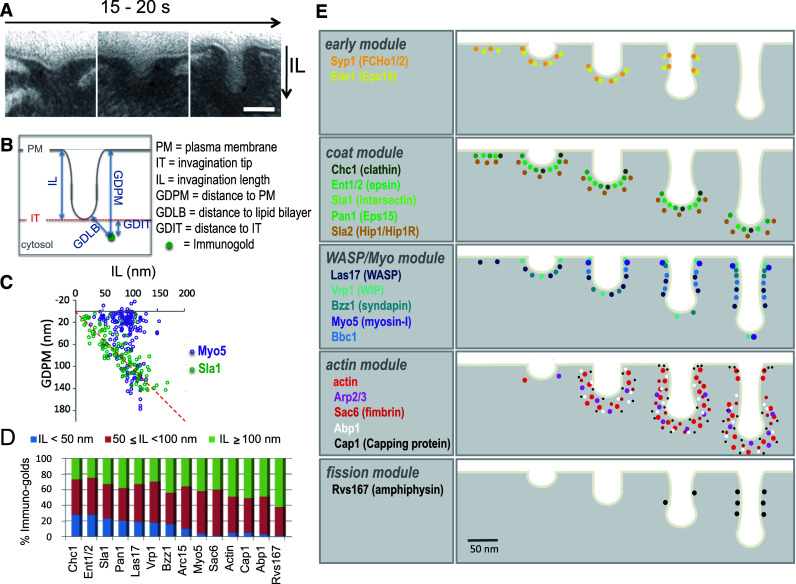
Ultrastructural analysis of endocytic budding by quantitative immuno-electron microscopy (QIEM) a Electron micrographs showing plasma membrane invaginations labeled with immuno-golds against an endocytic coat protein (Sla1) on ultrathin sections of yeast chemically fixed and resin-embedded. Scale bar 100 nm (reproduced from [23]). b Scheme indicating the parameters used to describe the position of the immuno-golds labeling endocytic proteins relative to the invagination (reproduced from [23]). c Dot plot representing the distance of immuno-golds to the basal plasma membrane (PM), versus the length of the associated invagination, for the coat component Sla1 and the myosin-I, Myo5 (data are from [22]). d The average length of the invaginations labeled for different endocytic proteins significantly increases according to their sequential recruitment, only for proteins recruited concomitant with or after Vrp1 and Bzz1. The data indicate that the emergence of membrane curvature coincides with the arrival of these proteins, and that once curvature is generated, the length of the invaginations can be used as a parameter to describe their age. Events occurring before membrane curvature or after scission are not studied by QIEM (data are from [23]). The X-axis represents the level of the basal PM and the red dotted line indicates the position of the invagination tip. In this kind of graph, the spatiotemporal organization of the endocytic protein along the invagination can be monitored. In the examples shown, the position of the immuno-golds indicates that Sla1 remains associated with the tip of the invagination as it grows inside the cytosol. Myo5 is recruited later and appears associated with the invagination base most of the time. In longer invaginations though, just before fission occurs, a pool of Myo5 appears associated with the invagination tip. e The spatio-temporal distribution of 18 endocytic yeast proteins based on QIEM studies [22, 23]. The mammalian homologues or functional counterparts are shown between parentheses
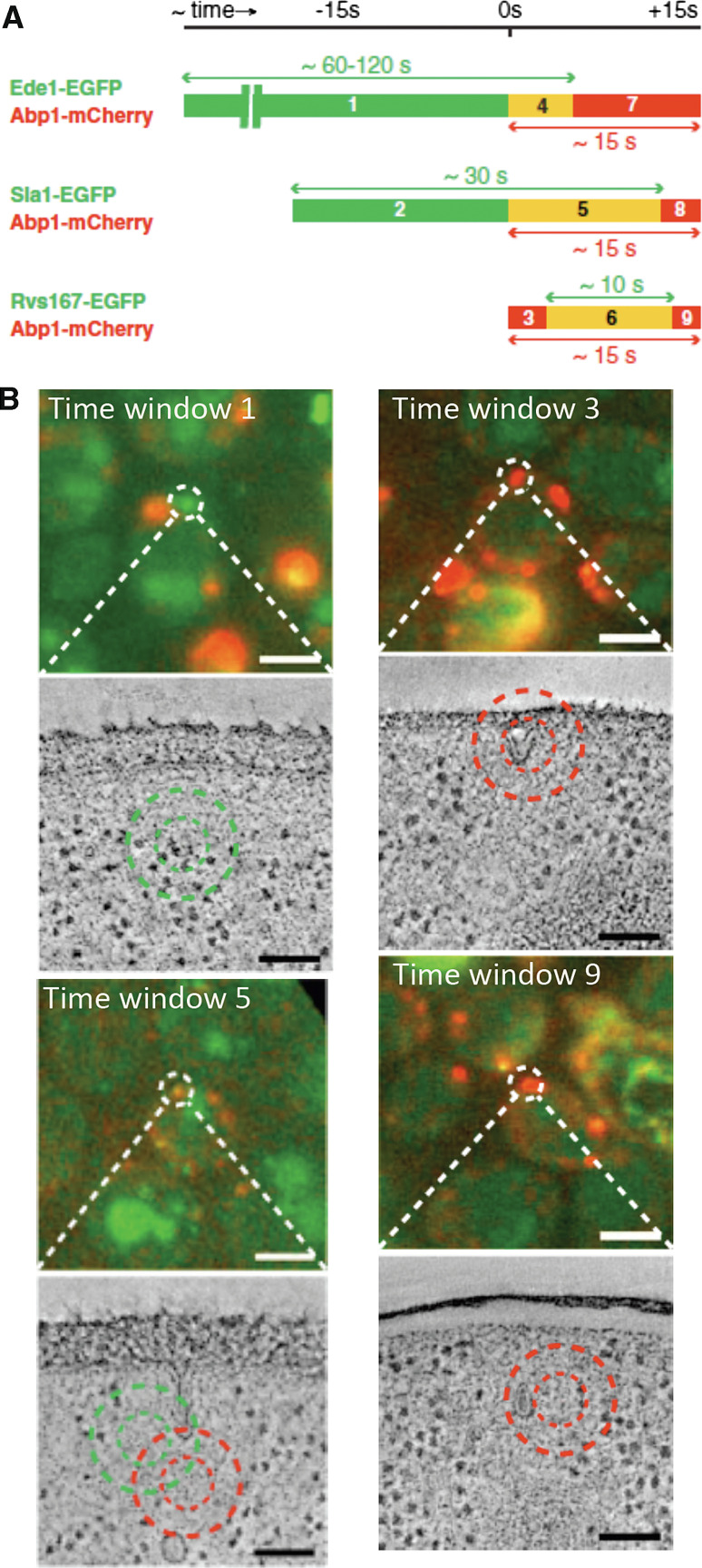
CLEM confirmed the tubular nature of the endocytic profiles in cryofixed, freeze-substituted, and resin-embedded yeast cells [25, 26]. Although the invaginations defined by these studies appear slightly shorter and thinner, as compared to those described by QIEM, the overall morphology of the profiles and the timing of events defined by the two approaches are remarkably consistent (Figs. 2a, 3b). Differences in the fixation, embedding, and/or staining protocols may account for the observed variations. Kukulski et al. [25, 26] use an elegant approach to precisely correlate the dynamics of the proteins at the endocytic sites with three-dimensional topological changes of the PM. Pairs of key fluorescently-tagged endocytic proteins sequentially recruited at endocytic sites were used to label nine time windows during the process (Fig. 3a). By using fluorescent fiducial markers, the endocytic fluorescent patches can then be ascribed to PM profiles on electron tomograms with a resolution of about 100 nm [26] (Fig. 3b). In contrast to conventional transmission electron microscopy, the electron tomography allows the visualization and analysis of the three-dimensional morphology of the complete endocytic invagination. The three-dimensionality is acquired by imaging thin sections of the embedded cells at varying angles with respect to the electron beam. The resulting tilt series of the 2-dimensional images are aligned and merged using special algorithms that synthesize a 3D density map or tomogram [27]. As compared to QIEM, which infers the temporal dimension from the statistical processing of the data, CLEM can unequivocally assign a maturation stage to a particular topology of the PM and can directly analyze the endocytic sites before membrane curvature emergence and after vesicle scission. However, QIEM provides much more detailed information on the architecture of the endocytic machinery relative to the deforming lipid bilayer. Similarly to QIEM, CLEM also estimated that membrane curvature emerges concomitant with massive actin polymerization [23, 26]. Importantly, it also revealed the exact point of vesicle scission and the intriguing morphodynamics of the primary endocytic vesicle [26].
Fig. 3.
Ultrastructure of the yeast endocytic profiles at different stages by correlative light and electron microscopy (CLEM). a Pairs of fluorescently tagged endocytic proteins label a sequence of 9 overlapping time windows along the process (1–9). By using fluorescent fiducial markers, visible in both fluorescence and electron microscopes, each fluorescent patch within a defined time window can be correlated to a plasma membrane profile in electron tomography, with a spatial resolution of about 100 nm. b Overlays of GFP and RFP signals in 300-nm sections of high pressure-frozen resin-embedded yeast cells. Dashed circles mark fluorescent patches corresponding to the indicated time windows. The associated-flat, invaginated and vesicular-profiles are shown below in virtual slices through electron tomograms. Small and big circles mark 50 and 80 % prediction accuracy for mCherry (red) and EGFP (green) positions, respectively. Scale bars in fluorescent micrographs and electron tomographic slices are 2 μm and 100 nm, respectively. Figure is adapted from Ref. [26] with permission from the authors
Here, we review how the outcome of these studies has changed our view on the mechanisms by which membrane-sculpting machineries shape the PM within the cell. In particular, live-cell imaging combined with TREM studies in yeast has contributed significantly to these advances by providing the most detailed dynamic view of a membrane-budding event occurring inside a cell to date. The accumulated data has allowed the dissection of clathrin- and actin-mediated endocytic budding into discrete stages and a significant refinement of the molecular models explaining the process, which will be discussed in the following sections.
Assembly of the endocytic machinery on a flat PM
Among the proteins arriving early to the endocytic sites are the components of the early and the coat modules (Fig. 1b). These include clathrin, the yeast epsins Ent1 and Ent2, and the FCHo1/2 yeast homologue, Syp1 [19–21, 28–31] (Fig. 1b). CLEM and QIEM studies have shown that these proteins stay at the PM for a relatively long period before it starts to bend [23, 26] (Fig. 4, Stage 1). Therefore, it is very likely that the clathrin coat remains flat on the PM for most of the time, whilst waiting for cargo to concentrate. Although the localization of cargo at the endocytic profiles has not been assessed by TREM, live-cell imaging analyses demonstrated its recruitment after the arrival of Ede1 and before Sla1 (Fig. 1b), inferring that it accumulates on a flat PM together with the coat components during the early stages [32].
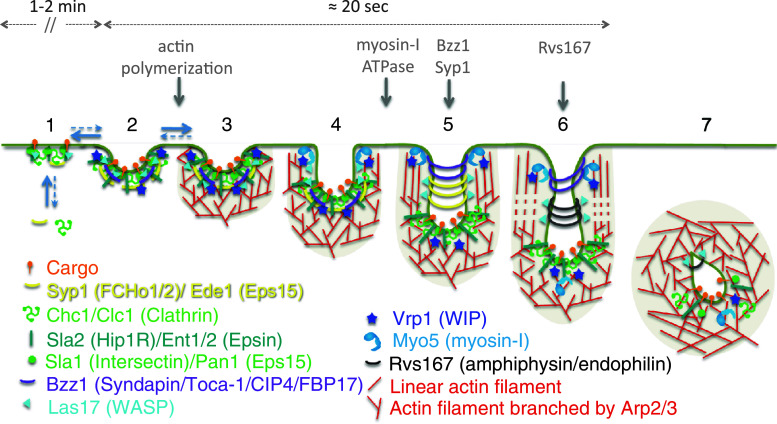
Fig. 4.
Molecular model for clathrin-mediated endocytic budding in yeast based on TREM studies. 1 The early and the coat modules assemble on a flat PM. Clathrin, the clathrin adaptors, and Syp1 presumably remain in an unassembled uncurved conformation and may be involved in regulating cargo selection and concentration. Las17 is recruited during this stage, but its NPF activity remains silenced by the clathrin adaptor Sla1. 2, 3 Membrane invagination and actin polymerization are initiated, most likely in response to a certain threshold of cargo and cargo adaptors. Protein crowding might promote initial membrane curvature. Recruitment of Bzz1 and Vrp1, concomitant with emergence of membrane curvature, could release the inhibition on Las17 by Sla1, which together with Pan1 initiate the assembly of an actin cap, branched by the Arp2/3 complex and cross-linked by Sac6. This actin cap is firmly attached to a curved coat via Sla2 and Ent1. The generated actin cap could stabilize the emerging coated dome-shape invagination. 3, 4 Growth of the branched actin cap promotes the initial elongation of the profile and its transformation from a dome- to a U-shape. At this stage, Las17 moves progressively from the tip to the tubular area of the profile where its NPF activity is probably silenced by Bbc1. Concentration of myosin-I/Vrp1 at the base of the emerging bud promotes appearance of barbed ends facing the PM. Biased Growth of these filaments will push the actin cap and the linked actin coat inward and promote the directed elongation of the profile. 4–6 Further growth of the invaginations from 70 nm onwards requires the motor domain of myosin-I. Constriction requires the concerted action of the myosin-I ATPase and the assembly of the BAR domain containing proteins around the neck. At this stage, additional elongation of the actin filaments with barbed ends facing the PM does not involve branching by the Arp2/3 complex. The motor domain of myosin-I pushes inwards the growing actin filaments firmly attached to the coat causing the elongation and the narrowing of the profile. By sensing the increased membrane curvature or tension of the tubules, the F-BAR proteins Syp1 and Bzz1 move from the tip to the tubular area of the invagination and constrict it. Bzz1 might form a stiff platform necessary for the myosin to generate productive forces. The progressive recruitment of N-BAR proteins Rvs161 and Rvs167 might then promote further elongation and/or constriction of the endocytic tubule. 6 Transient depolymerization of actin at the invagination neck might be required for vesicle scission. The fission reaction probably needs the tubulating activity of Rvs167 and Bzz1, the segregation of lipid domains induced by Sjl2, actin polymerization induced by myosin-I and Las17 and the mechanochemical activity of the yeast dynamin Vps1. 7 The released vesicle continues to move into the cell interior. The movement might be powered by the different NPFs that remain in association with the vesicle including, Pan1, Las17, and Myo5. The localization of the endocytic proteins around the released vesicle is hypothetical. The localizations of Sjl2 and Vps1 on the endocytic profiles have not been described and are therefore not represented in this model. Gray arrows show the critical requirement of different endocytic machineries at different stages of the process (see text for further comments). Equilibrium arrows denote possible reversibility between the indicated stages. Dashed blue arrows points to a possibly less favorable progression from one stage to the other. Yeast nomenclature and the corresponding mammalian homologues or functional counterparts (between parenthesis) are indicated
The observation that the assembly of clathrin, the epsins, and Syp1/FCHo1/2 at the endocytic sites are not immediately coupled to the generation of membrane curvature in yeast contradicts the models for clathrin-mediated endocytic budding in which epsin and FCHo1/2 might induce PM deformation early during the process, through their amphipathic helix and BAR domain, respectively [5, 33, 34]. This generated curvature might then be stabilized by the progressive assembly of clathrin triskelia into hexamers and pentamers forming a basket-like scaffold around the emerging bud [35]. In contrast, the TREM in yeast suggests that clathrin is associated with a flat PM, and only subsequently adopts a hemispherical shape [22, 23, 26, 36]. Thus, either a flat polymerized clathrin network is remodeled to a curved coat [37], or free clathrin triskelia initially establish reversible interactions with the cargo-bound adaptors [38], and only once the membrane bends, can they form a firm curved scaffold that stabilizes the bud (Fig. 4, stages 1, 2).
The F-BAR protein Syp1 is also unlikely to be in a stable curved conformation at early stages of the process. Therefore, its capacity to form helical structures [31, 39, 40] might be initially inhibited, as suggested for other membrane-sculpting proteins. Rao et al. [41] demonstrated that the F-BAR-mediated membrane bending activity of syndapin is subject to autoinhibition by its SH3 domain. Association of the Syndapin-1 SH3 domain with the proline-rich domain of dynamin-1 releases the auto-inhibition and allows polymerization of syndapin into a spiral-like coat [41]. Similarly, the capacity of the GTPase dynamin-2 to switch from the unassembled to the oligomerized helical state is negatively regulated by its GED domain [42, 43]. QIEM shows that, as budding progresses, Syp1 moves from the tip to the neck of the endocytic invaginations and it becomes significantly closer to the lipid bilayer ([23] and our unpublished data) (Fig. 4, stages 3–5). This behavior might reflect a molecular switch that promotes Syp1 polymerization into a helical array and/or direct binding of the F-BAR domain to the lipid bilayer.
There is increasing evidence to suggest that endocytic proteins with membrane deformation capacities might be regulated to fulfill other functions during the early stages of the endocytic process, independently of their ability to bend membranes. One of these functions is to organize cargo loading prior to the initiation of membrane curvature. Syp1 and its mammalian homologues FCHo1/2 have a mu-homology domain that interacts with cargo molecules and adaptors [31, 44, 45], suggesting a role for these proteins in cargo organization during early stages of the process, rather than functioning as initiators of endocytic invaginations. Dynamin-2 is also involved in controlling early rate-limiting steps of the endocytic process in mammalian cells by regulating coat assembly and cargo capture, presumably in its unassembled state [42, 46]. Flat clathrin lattices or free clathrin triskelia and clathrin light chains may regulate similar early functions during endocytosis. In yeast, the clathrin light chain facilitates the transition from the intermediate to late coat stages, probably by regulating actin polymerization at the endocytic sites [20, 47, 48]. Because this transition is also regulated by cargo [20], clathrin may sense the arrival of cargo and adaptors and prevent actin polymerization from occurring before sufficient cargo is present at the sites of endocytosis.
Initiation of membrane curvature and actin polymerization at the endocytic sites
By defining the time at which PM curvature emerges with respect to the arrival of endocytic factors, it is now possible to elucidate which proteins might actually be involved in initial bending of the flat lipid bilayer.
Coupling the emergence of membrane curvature with actin polymerization
An intriguing finding based on the TREM studies in yeast is that initiation of membrane curvature is tightly coupled to the initiation of actin polymerization at the sites of clathrin-coated pit (CCP) formation [23, 26]. CLEM could not detect membrane curvature before the actin binding protein Abp1 is recruited to the endocytic site [26]. On the other hand, the QIEM studies suggest that initial curvature occurs concomitantly with the recruitment of Bzz1 and Vrp1, which marks the onset of the actin-dependent inward movement of the endocytic coat [23, 49]. While these findings agree that actin polymerization and initial membrane curvature are coordinated, it is still not clear whether polymerized actin is the main trigger of PM bending or if membrane curvature initiated by other mechanisms prompts actin polymerization. In agreement with the first hypothesis, Kukulski et al. [26] could not observe endocytic invaginations upon cell treatment with Latrunculin A, a drug that inhibits actin polymerization by sequestering actin monomers. On the contrary, dome-like invaginations specifically decorated with coat proteins could be observed in the QIEM studies in the absence of actin dynamics [23], suggesting that actin polymerization is not essential to initiate membrane curvature. Interestingly, the QIEM studies show that the overall number of dome-like coated invaginations per cell does not increase when actin polymerization is inhibited (our unpublished data), suggesting that the flat versus curved PM profiles might be in a reversible equilibrium in the absence of a dynamic actin cytoskeleton (Fig. 4, stages 1, 2). The construction of an actin cap firmly attached to the endocytic coat could shift this equilibrium toward the dome shape (Fig. 4, stages 2, 3) (see below). This interpretation could reconcile the conflicting results of the CLEM and QIEM studies on this point, since a small fraction of the endocytic coats in the curved conformation might be missed in the CLEM studies unless a significantly higher number of endocytic spots are analyzed. Differences in the fixation and embedding protocols could also provide an alternative explanation for the observed results.
The requirement of clathrin, epsin and BAR domain-containing proteins in membrane curvature initiation
Other candidates to promote initial membrane curvature are proteins with in vitro membrane sculpting capacity present in the endocytic patch previous to or at initiation of actin polymerization. Those include clathrin, the epsins, and the F-BAR-domain containing proteins Syp1/FCHo1/2 and Bzz1/Syndapin-like [31, 33, 34, 39, 40, 50, 51] (Fig. 1b). In mammalian cells, electron microscopy studies demonstrated that depletion of clathrin precludes the invagination of PM underlying assembled endocytic structures [52]. Conversely, studies in yeast indicate that clathrin is not essential for the formation of the incipient bud at the PM. Coat and cargo can still be internalized in yeast when clathrin is depleted, albeit less efficiently [19, 20, 47, 53], and the endocytic invaginations formed are indistinguishable from those observed in wild-type strains, with regard to their morphology and the localization of the coat adaptor around the tip of the invagination [23]. Thus, clathrin may share redundant functions with other coat components or may not be directly involved in PM invagination or scaffolding of the coat, but might rather have a regulatory role as mentioned above. The differential requirement for clathrin in yeast and mammals might be due to differences in the nature of the internalized cargo and the size and/or the geometry of the endocytic bud, rather than in the capacity to bend the lipid bilayer [5].
Epsins can induce membrane curvature in vitro by inserting the amphipatic helix of their ENTH domains into the lipid bilayer [33]. This insertion has been predicted by a physical model to generate large membrane curvatures [54]. However, recent data show that epsins might not be sufficient to produce significant changes in the topology of the PM by themselves at physiological concentrations [55]. Supporting this view, EM studies have shown that depletion of all epsin isoforms in mammalian cells does not prevent the formation of CCPs [56].
Likewise, the studies in yeast indicate that the BAR-domain proteins Syp1/FCHo1/2 and Bzz1/Syndapin-like are dispensable to generate initial membrane curvature. The mammalian homologues of Syp1, FCHo1/2, have been proposed as nucleators of CCP, which initiate PM curvature [34]. In yeast, however, deletion of Syp1 does not alter the initiation or progression of the coat internalization or the initiation of membrane invagination [21, 23]. Consistent with these observations, recent analysis in mammalian cells and Zebrafish have shown that clathrin structures can still assemble at the PM in the absence of FCHo1/2 [45, 57]. The other yeast F-BAR domain containing protein Bzz1 arrives at the endocytic sites coincident with the emergence of the coated pit [49]; however, deleting Bzz1 alone or together with Syp1 does not abolish endocytic budding ([23, 51] and our unpublished results), indicating that the two yeast F-BAR domain proteins known to localize at the endocytic sites are not essential to produce initial PM bending for endocytic budding.
Initial membrane curvature by molecular crowding
All these data seem to discard the possibility of an exclusive role for one of the classical membrane-sculpting mechanisms in triggering initial membrane curvature at the endocytic sites. Either a high degree of functional redundancy exists or an alternative mechanism is actually responsible for initiating membrane curvature in a cellular context. Interestingly, Stachowiak et al. [55] have recently suggested a novel mechanism through which endocytic molecules could bend membranes by protein–protein crowding. By correlating membrane tubulation with protein densities measured by FRET on the surface of giant unilamellar vesicles, the authors demonstrated that lateral pressure generated by collisions between bound proteins can drive bending. In this mechanism, curvature generation depends on the protein coverage regardless of the mode used to bind the lipid bilayer or the bending capacities of the proteins [55]. In line with these findings, Dannhauser and Ungewickell [50] demonstrated that clathrin is able to form highly curved membrane structures in vitro in the absence of ENTH domains and amphipathic helices, when recruited to membranes via adaptor proteins. It would be interesting to analyze in the same experimental conditions whether any other proteins with the same membrane coverage as clathrin in this assay could induce curvature with similar efficiency. Fluorescence live-cell imaging in yeast has shown that the intensities of the endocytic proteins fused to GFP variants increase progressively during the process and reach their maxima around the time of Vrp1 and Bzz1 arrival, which, according to the QIEM studies, coincides with emergence of curvature [18, 23, 49]. An attractive possibility would be that recruitment of sufficient cargo at the endocytic sites is sensed by the system as the degree of membrane protein coverage (about 20 %, according to Stachowiak et al.) necessary to induce curvature. Curvature could then promote recruitment of the BAR domain protein Bzz1/syndapin-like to trigger actin polymerization, which will in turn stabilize the endocytic invagination and promote progression of budding (see below). Physical models, which integrate parameters such as the number of proteins at each time point of the process and the area of the PM patch occupied by the coat, would be useful to test whether the coverage reached by peripheral membrane proteins at this time point is sufficient to induce membrane curvature. Such parameters could be estimated from live-cell imaging and electron microscopy studies. The exact mechanisms by which endocytic components collaborate to overcome the resistance of the PM to initial bending still need to be identified.
How is actin polymerization initiated at the endocytic sites?
Actin assembly during endocytic budding is essential to elongate the endocytic invaginations and probably also to excise the endocytic vesicles. The Arp2/3 complex and its activators are recruited to the endocytic sites where they are thought to initiate actin polymerization (reviewed in [12, 15, 58]). The Arp2/3 complex nucleates growth of new actin filaments on the side of preexisting ones or “mother filaments”, at a fixed angle of 70 °, thereby forming a branched network. Its nucleating capacity is locally activated by actin nucleating promoting factors (NPF) such as WASP [59, 60]. The endocytic NPFs in yeast include the homologue of WASP Las17, the Eps15–like protein Pan1, and the myosin-Is Myo3/5 [49, 61, 62] (Fig. 4, stage 1). These factors join the endocytic site with different timings and have different dynamics at the ultrastructural level [18, 22, 49] (Fig. 1b). Therefore, they were initially suspected to perform different functions in shaping the nascent endocytic vesicle. Las17 seems to share an early function during the process with Pan1 [49, 63, 64], whereas Myo5 has a prevailing role during elongation of the endocytic invaginations, which can be taken over by Las17 in a myosin-I mutant background [49].
Las17 and Pan1 arrive to the sites of endocytosis and stay for about 20 s on a flat membrane, well before actin binding proteins arrive at the endocytic sites and also before any actin-dependent movement of the coat can be detected [18, 19, 23, 26, 63, 65] (Figs. 1b, 4). During this time, their NPF activities probably remain inhibited by the coat components Sla1 and the Hip1R homologue Sla2, respectively [49, 66–68]. The physico-chemical parameters that initiate actin polymerization are uncertain, but QIEM and biochemical data indicate that the incipient membrane curvature could be the trigger. First, emergence of membrane curvature occurs concomitant with the arrival of Vrp1 and the F-BAR domain containing protein Bzz1 [23, 49]. Second, Bzz1 was shown to release the Sla1 inhibition of Las17 in pyrene-actin polymerization assays [49] and third, the Bzz1-related mammalian proteins Toca1 and FBP17 are able to recruit the N-WASP/WIP complex to liposomes and stimulate its NPF activity in a curvature-dependent manner [69]. Thus, membrane curvature sensing by Bzz1 might provide a molecular switch that elicits actin polymerization. Once again, however, redundant mechanisms might work to couple emergence of membrane curvature to actin polymerization since deletion of Bzz1 alone does not cause a significant delay in endocytic internalization and has little influence in the morphology of the endocytic invaginations [23, 49].
Although the Arp2/3 complex nucleation activity may be involved in initiating actin polymerization at the sites of endocytosis, recent data indicate that the Arp2/3 complex is probably not essential for this function. First, the nucleation activity of the Arp2/3 complex requires a preexisting mother filament [59]. Second, although the Arp2/3 complex is critical for efficient endocytosis in yeast, mutations in the Arp2/3 complex or in the NPFs, which dramatically reduce actin nucleation in vitro, do not significantly affect actin polymerization at the endocytic sites [70, 71]. Recent work suggests that Las17, Pan1, and the WIP homologue Vrp1 could nucleate actin filaments in an Arp2/3-independent manner. Urbanek et al. [64] demonstrated that Las17 has the capacity to nucleate actin polymerization by itself in vitro, via a novel actin binding poly-proline motif. Pan1 and Vrp1 contain similar motifs and thereby they may also nucleate linear actin filaments. These filaments may both recruit the Arp2/3 complex and serve as a mother filament for nucleation of a branched network at the sites of endocytosis. Alternatively, the mother filament may be captured from the cytosol through a contact with another actin-coated endocytic vesicle [72] or by randomly touching surrounding actin networks as suggested from electron micrographs of actin structures associated with CCPs in mammalian cells [73].
Elongation and constriction of the endocytic tubule
Once membrane curvature emerges, maturation of the endocytic invaginations implies their coordinated elongation and reshaping. TREM has demonstrated that invaginations are initially dome-shaped and progressively acquire a U-contour. Subsequent constriction of the invagination neck at approximately one-third of its depth results in omega-shaped profiles, which probably correspond to pre-scission stages [23, 26] (Fig. 4, stages 3–6). The precise localization of the endocytic machinery relative to the endocytic invaginations by QIEM and the ultrastructural analysis of the morphology of the endocytic sites after interfering with key pieces of the endocytic machinery have started to define the molecular mechanisms involved in each particular membrane deformation step [23]. The data indicates that: (1) an actin cap, branched by the Arp2/3 complex and strongly linked to the endocytic coat, is built during the transition from the dome to the U-shape; (2) linear elongation of actin filaments with barbed ends facing the PM probably follows to drive further elongation of the endocytic tubes; (3) the myosin-I motor activity is required to promote growth of the endocytic invagination; and (4) the myosin-I ATPase and the F-BAR-domain proteins cooperate in the constriction of the invagination neck (Fig. 4, stages 3–6).
Elongation of the endocytic tubule by two modes of actin polymerization
Initial elongation of the endocytic invaginations and reshaping from a dome to a U-contour is coupled to the assembly of an actin cap around the invagination tip, branched by the Arp2/3 complex and firmly attached to the endocytic coat via Sla2 and Ent1 (Fig. 4, stages 3, 4). QIEM studies have shown that actin initially localizes around the coated endocytic invagination in a ribosome-free area of about 50 nm wide (Fig. 2a), highly enriched in the Arp2/3 complex and the yeast actin bundling protein Sac6/Fimbrin [22, 23, 74]. This observation is consistent with previous rapid-freeze deep-etch electron microscopy studies in yeast cells showing that the Arp2/3 complex accumulates at the tip of conical actin structures with a Y-branched organization, likely corresponding to endocytic actin structures [75, 76]. Thus, at this stage, actin forms a branched and cross-linked structure that caps the emerging bud (Fig. 4, stage 3). Mutations in the Arp2/3 complex and deletion of Sac6 severely affect coat internalization [19, 71, 76], suggesting that crosslinking of the actin network is required for productive force generation. The localization of the NPFs that probably initiate actin polymerization at this stage, Pan1 and Las17, predicts the formation an actin structure with growing ends facing the endocytic coat and pushing it in different directions [22, 49]. Assembly of such a structure could explain the undirected, actin-dependent corralled movement observed by live-cell imaging [18, 19, 65] (Fig. 4, stages 3, 4). Supporting this hypothesis, localization by QIEM of capping proteins (Fig. 2e), which bind the barbed ends of actin filaments, provides evidence for a complex polarity of the actin network around the coated area of the invagination [23].
After the initial burst of actin polymerization, myosin-I arrives to the endocytic sites concomitant with the initiation of massive actin polymerization and with the elongation of the tubular profiles [22, 23, 26, 49]. At this stage, the myosin-I concentrates at the base of the emerging bud together with its co-activator Vrp1/WIP, whereas Las17 starts moving from the tip to the tubular area, where it co-localizes with two of its inhibitors Bbc1 and Syp1 [22, 23, 49] (Fig. 4, stages 4–6). This pattern predicts a switch of the predominant NPF activity from the tip to the base of the invagination, and therefore the appearance of barbed ends facing the PM. Consistent with this view, capping protein appears at the base of the endocytic invaginations, together with Myo5 [23] (Fig. 2e). Biased elongation of these filaments could then power the inward movement of the branched actin network attached to the coat, thereby promoting a directed movement of the coat inside the cytosol [12, 23] (Fig. 4, stages 4–6).
An efficient force transduction by actin polymerization in the model described above requires the presence of a linker that firmly connects the actin network to the membrane (Fig. 4, stages 3–6) [77]. In a recent study, Skruzny et al. [78] have shown that the yeast Hip1R Sla2 and the epsin Ent1 form a complex that stably links the coat to actin. This link is established, on one side, by the cooperative binding of Sla2 and Ent1 to the PM via their lipid binding domains, and on the other side, by their interaction with the actin network via redundant actin binding domains [78]. Consistent with the role of Sla2 and Ent1 as linkers, mutations that disrupt this connection strongly inhibit the internalization of the coat [18, 78]. Also in agreement with a role of Sla2 as a linker between the endocytic coat and actin, QIEM shows that the C-terminus of Sla2 bearing the actin binding domain precisely localizes at the interface between the coat and the actin network, and that SLA2 deletion prevents elongation of the invaginations but not actin polymerization on the endocytic invaginations [18, 23, 79]. The mammalian homologue of Sla2, Hip1R, has also previously been shown to link actin filaments to the clathrin-coated pits [80]. Accordingly, depletion of Hip1R in mammalian cells phenocopies the sla2∆ mutant [18, 81]. However, a branched actin network covering the dome of the CCPs in mammalian cells was not observed in the EM studies on unroofed cells [73], suggesting that either such a network was torn away by sonication during unroofing or such an actin cap does not form in mammalian cells.
Although initial elongation of the tubular profile is promoted by the growth of an actin network branched by the Arp2/3 complex (Fig. 4, stages 3, 4), further elongation of the endocytic invagination is probably accompanied by the extension of linear actin filaments, with barbed ends facing the PM (Fig. 4, stages 5, 6). For invaginations longer than 70 nm, QIEM shows that labeling for the Arp2/3 complex is absent from the base of the profiles. However, actin and other actin binding proteins still accumulate there [22, 23]. Given that the Arp2/3 complex moves inward with the endocytic coat and the addition of actin monomers seems to occur at the base of the endocytic invaginations [18, 19, 23], these observations strongly indicate a change in the mode by which actin polymerization progresses at this stage. Actin monomer addition might progress in an autocatalytic fashion or be powered by the Vrp1 G-actin binding sites. What regulates this change and its functional implications are currently unknown.
Elongation and constriction of the endocytic tubule by myosin-I and BAR domains
The mechano-chemical activity of the myosin-I is required both for elongation and for the constriction of the endocytic invaginations. QIEM shows that mutation of the myosin-I motor domain results in the accumulation of 70-nm-long unconstricted U-shaped invaginations [23]. The motor activity of the myosins-I, located at the base of the invaginations, will push away actin filaments with barbed ends facing the PM [22, 23, 49]. Provided that these actin filaments are firmly attached to the branched actin network linked to the coat, the myosin-I will push the endocytic coat away into the cytosol and, thus, power elongation of the invaginations (Fig. 4, stages 4–6). An important requirement in this model is a stiff platform that immobilizes the endocytic invagination and allows the myosin-I to push the actin filament away from the PM. Recent evidence suggests that the BAR domain protein Bzz1, which co-localizes with Myo5 at this stage, could play this role in yeast [22, 23, 51].
How could the myosin-I also strangle the invagination neck at this stage? Consistent with the localization of Myo5 and actin, the mechano-chemical activity of the myosin could directly constrict a ring with actin filaments arranged into antiparallel arrays [12, 22, 49]. However, it is difficult to envision a simple myosin organization at the base of the invaginations that could simultaneously push away actin filaments with barbed ends facing the PM and constrict a ring of actin filaments with a perpendicular orientation. Alternatively, the elongation of the tubular profile powered by the myosin motor head might generate tension along the invagination and promote assembly of BAR domain-containing proteins at the invagination neck, which in turn constrict the profiles. Consistent with this possibility, QIEM demonstrates that Syp1 and Bzz1 translocate from the tip to the neck of invaginations longer than 70 nm, and that this translocation is precluded when the myosin-I ATPase is mutated [23] (stages 4, 5). Further, deletion of Syp1 or Bzz1, or a mutation in Bzz1 that prevents binding to lipids, increase the number of unconstricted, U-shaped invaginations [23, 51]. In contrast to the myosin-I mutations, though, depletion of the F-BAR-domain proteins does not prevent elongation of the endocytic invaginations [23]. Similar to the yeast F-BAR proteins, the mammalian homologue of Syp1 FCHo2 initially associates with the tip of shallow CCPs but appears at their neck when they acquire more curvature [34]. Likewise, the mammalian counterpart of Bzz1 FBP17 is detected on the tubular necks of pre-scission clathrin-coated pits produced in a cell-free system [82]. Also similar to the F-BAR proteins, IEM in mammalian cells shows that dynamin translocates from the coat area to the neck of clathrin-coated pits when they are sufficiently narrow [83], and dynamin preferentially binds to tubular membranes when they are under tension [84]. Increasing tension and/or curvature resulting from the elongation of the endocytic invaginations might ultimately be the parameter that triggers recruitment of BAR-domain proteins to the invagination neck.
The role of the yeast amphiphysin/endophilin-related proteins Rvs161 and Rvs167 in elongation and constriction of the tubular invaginations remains less defined. These proteins bear an N-BAR domain with a concave structure of smaller diameter, as compared to the F-BAR domains [85]. Consistently, N-BAR domains produce narrower membrane tubes in vitro than the F-BARs [31, 39, 40, 86]. In line with these properties, QIEM studies demonstrate that Rvs167 substitutes for the F-BAR protein, Syp1, at the invagination neck, coincident with the progressive narrowing of the tubule [23]. However, the localization of Rvs167 at this point does not actually coincide with the position of the constriction [23] (Fig. 4, stage 6). Further, the phenotypes reported for deletion of RVS167 are controversial. In one ultrastructural study, deletion of the N-BAR protein causes widening and elongation of the tubule, suggesting a defect in constriction and/or scission, but not in elongation [51]. Conversely, CLEM demonstrates the accumulation of very short endocytic invaginations in an rvs167∆ strain [26], suggesting a defect in elongation. In agreement with this finding, assembly of BAR domain proteins has previously been reported to be required for the stabilization of clathrin-capped elongated tubules produced in a cell-free system [82]. The discrepancy between the two results remains unsolved for the moment. In the studies by Kishimoto et al., the endocytic nature of the invaginations studied could not be unequivocally assessed and, therefore, some long invaginations of another origin could be overrepresented in the rvs167 mutant. Alternatively, if the endocytic coat would disassemble prematurely but the invaginations continue to grow in the amphiphysin mutant, the CLEM will overlook the longest structures. More work is required to dissect the role of amphiphysins at this stage.
Scission of the endocytic tubule
Even though the involvement of dynamin in the scission of clathrin-coated vesicles in mammalian cells is fairly well established, its endocytic function in yeast is rather limited and, therefore, it was initially postulated that a different fission mechanism might predominate in this organism [12, 87]. The demonstration that fission occurs half-way through the transient recruitment (only 8 s) of Rvs161 and Rvs167 and the observation that Rvs167 concentrates at the invagination neck immediately before fission, suggested a leading role for these proteins in vesicle scission [19, 22, 25, 26]. Accordingly, endocytic coats, possibly failing to pinch off from the PM, can be observed by live-cell imaging retracting towards the cell surface in rvs161 and rvs167 mutants [19, 51]. However, the endocytic defects installed upon depletion of these proteins are only partial, indicating that other components of the endocytic machinery cooperate in vesicles scission. Subsequent works have demonstrated that treatment of cells with low doses of Latrunculin A, alteration of the NPF activity of Las17, and the myosins-I, depletion of the synaptojanin Sjl2 or mutation of the yeast dynamin Vps1 exacerbate the retraction phenotype of the rvs167∆ strain [51, 88]. Thus, the data point to redundant roles of actin polymerization, the PI(4,5)P2 turn over, the tubulating or membrane curvature sensing activity of N-BAR proteins, and the GTPase of dynamin in vesicle scission. However, the molecular mechanism driving vesicle scission in vivo is far from being understood.
Liu et al. [89, 90] have proposed a molecular model to integrate the roles of actin, amphiphysin, and synaptojanin in vesicle scission. According to this model, the endocytic bud first elongates and gains curvature through the driving force of actin and the tubulating activity of BAR proteins. Later recruitment of the synaptojanins then creates a lipid phase boundary between the tubular area of the invagination, where PI(4,5)P2 is protected by the BAR scaffold, and the invagination tip, where the lipids are more exposed to the PI(4,5)P2 5-phosphatase activity of the synaptojanin. The line tension generated at the interface between the two lipid domains can provide force to drive membrane scission [89]. This model is supported by the genetic interactions described in yeast between Rvs167, Sjl2, and the endocytic NPFs [51], by in vivo and in vitro studies suggesting that lipid phase boundaries can drive vesicle scission in the absence of dynamin [91], and by the observation that the enzymatic activity of the mammalian synaptojanin-I is enhanced with increasing curvature [92].
Recent data by CLEM, though, have recently challenged this model. The molecular model proposed by Liu et al. [89] predicts that vesicle scission will occur between the endocytic coat and Rvs167 assembled at the invagination neck. However, the data obtained by Kukulski et al. suggest that scission occurs closer to the basal PM, possibly at the interface between Rvs167 and the F-BAR protein Bzz1, which is assembled at the base of the invaginations together with Myo5 and Vrp1 [22, 23, 26] (Fig. 4, stage 6). The careful morphometric analysis performed by these authors on the released vesicles estimates that the “future scission site” leaves the PM when the invaginations are 60–80 nm in length and moves inwards as the invagination depth increases. According to their calculations, the scission site coincides with the constriction on the invagination neck, which both CLEM and QIEM have positioned at 1/3 of the invagination depth [23, 26]. This constriction appears precisely positioned by the QIEM studies at the interface between Bzz1 and Rvs167, and not between Rvs167 and the endocytic coat [22, 23]. Given that the diameter between the tubules produced by N and F-BAR domains are significantly different [39, 40], the elastic energy at the interphase between Bzz1 and Rvs167 might increase, thereby lowering the energy barrier to fission, similar to what has recently been suggested for dynamin [93]. Future insight into the fission mechanism will have to wait for the exact position of Sjl2, Vps1, and endocytic relevant lipid species along endocytic invaginations.
How actin polymerization can assist in vesicle scission is also not well understood. A model for actin-dependent vesicle scission regulated by BAR proteins has been proposed based on studies in fission yeast [94]. According to this model, the F-BAR proteins Cdc15 and Bzz1 stimulate the NPF activities of myosin-I and WASP at the base and the tip of the endocytic invaginations, respectively, leading to the formation of two actin structures with opposing polarities. Expansion of these actin networks pushes the tip of the invaginating tubule away from the cell surface and generates membrane tension [94]. An alternative model has been proposed based on ultrastructural studies of clathrin-coated pits in mammalian cells. Using platinum replica electron microscopy in combination with electron tomography, Collins et al. [73] demonstrated the presence of branched actin networks with barbed ends facing the lipid bilayer, oriented perpendicular to the invagination neck. Based on these observations, the authors proposed that a constricting force generated by addition of actin monomers on the surface of the invagination neck might lead to vesicle scission. In agreement with this hypothesis, the yeast WASP appears on the surface of the invagination neck at late stages during endocytic budding, where it co-localizes with Rvs167 [22] (Fig. 4, stage 6). Interestingly, the mammalian amphiphysin interacts with N-WASP and stimulates its NPF activity in vitro [95]. Therefore, the NPF activity of Las17 could be reactivated by the disassembly of its inhibitor Syp1 and by the recruitment Rvs167 at this point [19, 26, 30] (Fig. 4, stage 6).
The model that really applies for the role of actin in vesicle scission in yeast is presently unclear. Strikingly, and in apparent contradiction with both models, the QIEM studies demonstrate that, when the endocytic invaginations are at their maximal length, just before scission occurs, actin labeling at the invagination neck is scarce, regardless of the anti-actin antibody used ([22], and our unpublished data). This phenomenon might reflect a need for the transient disruption of the thick actin endocytic network to allow detachment of the vesicles into the cytosol (Fig. 4, stage 6). However, what might trigger complete or partial disassembly of filamentous actin at the invagination neck and its exact functional significance is presently unclear.
Although the evidence for a role for dynamin in endocytic budding in yeast has been conflicting, probably due to differences in the yeast strain backgrounds or in the experimental conditions [7, 51, 87], the recruitment of the yeast dynamin-like protein Vps1 to at least a subset of endocytic sites was recently shown [96]. A recent study suggests that binding of Rvs167 to assembled Vps1 induces a conformational change in dynamin, which stimulates its mechanochemical activity and thereby its disassembly [88]. Recent biochemical studies with the mammalian dynamin, endophilin, and amphiphysin further support this possibility [97]. Vps1 disassembly may then drive membrane scission as suggested for the mammalian dynamin [98].
Unlike Vps1, dynamin is critical for scission of clathrin-coated vesicles in mammalian cells. Knock-down of dynamin in mouse fibroblasts results in accumulation of long clathrin-capped tubules that fail to undergo scission [14]. Nevertheless, while mammalian dynamin can induce membrane tubulation and constriction on its own in vitro, increasing evidence indicates that it may require the aid of some of the mechanisms proposed in yeast to complete scission in vivo, including generation of tension, actin polymerization, and the formation of lipid phase boundaries [89, 92, 99–102].
In a recent study, Boucrot et al. have proposed a novel mechanism for scission in the absence of dynamin. They observe that membrane insertion of amphipathic helices can drive complete membrane scission, which is antoganized by BAR domains, in a cell-free system [56]. Consistent with this observation, depletion of the epsins, which bear amphipathic helices and not BAR domains, in mammalian cells, causes the formation of multi-headed clathrin-coated invaginations that fail to pinch off in spite of dynamin being recruited to these structures. Furthermore, overexpression of the epsins can compensate for the absence of dynamin for endocytic budding [56]. The morphology of the endocytic profiles was not assessed by electron microscopy in yeast lacking Ent1 and Ent2. However, since these proteins are excluded from the area where scission is expected to occur [23], it is unlikely that they have a direct role in vesicle pinching in this organism.
The first seconds in the life of the released vesicle
Early electron microscopy studies have previously identified primary endocytic vesicles in yeast spheroplasts by labeling the endocytic pathway with positively charged nanogolds [103]. CLEM has now provided for the first time informative details on the topology of primary endocytic vesicles and their initial dynamics. The endocytic vesicles appear similar in size but remarkably non-uniform in shape, indicating that they are not enclosed in a spherical coat. They can appear as elongated rugby balls, flattened spheres, or tear-drop-shaped with an average volume of 43,000 ± 24,000 nm3 [26]. Whether the heterogeneity in shape comes from different stages after scission, deformation caused by asymmetrical actin polymerization, differences in the cargo being loaded, or heterogeneity in the fission mechanism is not currently known.
In addition to defining the shape and the size of the endocytic vesicles, CLEM showed that the coat component Sla1 and the amphiphysin remain associated with the newly born vesicle, indicating that complete uncoating generally occurs after scission has occurred [26]. CLEM has also shown that the ribosome-free area surrounding the vesicle continues to expand upon vesicle scission, probably reflecting the progressive growth of an actin network [26]. At this stage, actin filaments may continue to grow by the autocatalytic activity of actin or by NPF activities that still remain associated with the detached vesicle. The disassembly of Sla2 and the epsins by the Sjl2/Synaptojanin-dependent hydrolysis of PI(4,5)P2 may relieve the inhibition on Pan1 still attached to the endocytic coat, [66, 78, 104]. In addition, Las17 and Myo5 could reactivate Arp2/3-dependent actin polymerization on the released vesicle. Even though Las17 and Myo5 could not initially be observed moving with the vesicle into the cytosol in live-cell imaging experiments, QIEM demonstrates that a significant fraction of these proteins is associated with the portion of the membrane that will be released into the cytosol [22]. The actin tails produced by these activities may aid in the detachment of the endocytic vesicle from the PM and/or promote its movement inside the cytosol [18, 19, 26, 63, 65, 78]. Accordingly, actin tails were visualized at the tip of closed clathrin-coated vesicles at the PM by electron tomography in mammalian unroofed cells [73]. CLEM did not visualize actin-labeled vesicles further than 200 nm from the yeast cell surface [26], most likely because the vesicle movement is very fast after coat disassembly [18, 19, 63, 65, 105], and therefore the time window before actin disassembly occurs is very narrow. Alternatively, vesicles might run parallel to the cortex until they lose their actin cap [18, 19].
QIEM has not been used to describe the position of the endocytic machinery after vesicle release. The observation that most primary endocytic vesicles present a certain degree of asymmetry opens the possibility to apply this technique to define the relative position of the endocytic coat, the actin binding proteins, and the disassembly machinery on the released vesicles. This information could provide interesting insights into the mechanism of vesicle release, uncoating, and subsequent movement.
Conclusions and perspectives
The information provided by the TREM studies in yeast has zoomed out by an order of magnitude our view of endocytic budding in a cellular context, and has allowed the observation for the first time of changes in the lipid bilayer coupled to the dynamics of the endocytic machinery with unprecedented spatio-temporal resolution. By learning when and where the membrane-sculpting proteins exert their function in the cell, we can now refine the molecular models explaining the process (Fig. 4). On the other hand, our magnified view of endocytic budding has helped to define novel steps in the process and has opened new questions and fields for experimentation. For instance, how physical parameters such as the membrane curvature or tension influence the dynamics of molecular assemblies in vivo and vice versa is still not well understood; the exact mechanisms triggering initial membrane bending or scission are still controversial; how lipid domains are organized within the endocytic bud at the ultrastructural level is completely unknown; and how cargo may control progression of budding is still under investigation. Definitive answers to most of these questions will require in vitro assays that recapitulate the budding process in the presence of cargo and purified endocytic complexes, physical modeling, and ultrastructural approaches of increasing resolution, which, in the long term, allow visualization of the actual endocytic complexes at atomic resolution in a cellular context. In this framework, the use of electron tomography on vitrified samples, combined with docking of the atomic structures of the purified endocytic complexes [106], could be the next breakthrough in the field to provide precise three-dimensional information on how these molecular machines exert force to shape the endocytic membranes in situ and at atomic resolution. Electron tomography has been used to determine the topology of the PM during clathrin-mediated endocytic budding in high-pressure frozen, freeze-substituted, and resin-embedded yeast cells [26]. The branched architecture of actin filaments associated with clathrin-coated pits has also been revealed by these techniques on platinum replica of unroofed mammalian cells [73]. The next step now is to directly image these structures in cryo-immobilized frozen hydrated cells in which molecular resolution is preserved due to the lack of stains and chemical fixatives, which generally damage and/or mask real molecular densities [107]. The development of electron microscopy instrumentation and software for correlative light and cryo-electron tomography, as well as addressing the technical difficulties associated with handling and imaging thinned frozen hydrated cells, are key challenges which promise to provide further exciting insight in the future [108–111]. More advances in the field are also expected from the emerging live-cell fluorescence nanoscopy technologies such as STORM (Stochastic Optical Reconstruction Microscopy [112] or STED (Stimulated Emitted Depletion) [113], which circumvent the diffraction limit of light microscopy. Although these technologies do not yet offer sufficient spatiotemporal resolution to analyze the process due to the fast dynamics of the endocytic machinery, the incessant improvement of probes, lasers, detectors, and algorithms is, however, expected to challenge these issues in the next future (for a review, see [114, 115]). Real-time imaging of PM deformation associated with the dynamics of the endocytic components could be potentially resolved by combining the nanoscopy fluorescence techniques with approaches that detect the PM topology in living cells such as IRM (Interference Reflection Microscopy) [116], pTIRFM (polarized-Total Internal Reflection Fluorescence Microscopy) [117] or SICM (Scanning Ion Conductance Microscopy) [118]. Because the basic molecular mechanisms of the process are evolutionarily highly conserved, yeast cells will certainly continue to provide an essential powerful genetic tool for unraveling the molecular mechanisms of endocytic budding by most of these approaches.
Acknowledgments
We thank Felix Campelo, Jon Giblin, and members of the Geli Lab for critical comments on the review, and Marko Kaksonen and Jon Briggs for authorizing the reproduction of part of Figs. 1 and 3 from Ref. [26]. This work was supported by BFU2011-30185 and CSD2009-00016 grants from the Spanish Government to M.I.G.
Abbreviations
- CCP
Clathrin coated pit
- CLEM
Correlative light and electron microscopy
- IEM
Immuno-electron microscopy
- NPF
Nucleation promoting factor
- PM
Plasma membrane
- QIEM
Quantitative immuno-electron microscopy
- TREM
Time-resolved electron microscopy
References
- 1.Heuser J. Three-dimensional visualization of coated vesicle formation in fibroblasts. J Cell Biol. 1980;84:560–583. doi: 10.1083/jcb.84.3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth TF, Porter KR. Yolk protein uptake in the oocyte of the mosquito Aedes aegypti. L. J Cell Biol. 1964;20:313–332. doi: 10.1083/jcb.20.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson RG, Brown MS, Goldstein JL. Role of the coated endocytic vesicle in the uptake of receptor-bound low density lipoprotein in human fibroblasts. Cell. 1977;10:351–364. doi: 10.1016/0092-8674(77)90022-8. [DOI] [PubMed] [Google Scholar]
- 4.Roth MG. Clathrin-mediated endocytosis before fluorescent proteins. Nat Rev Mol Cell Biol. 2006;7:63–68. doi: 10.1038/nrm1783. [DOI] [PubMed] [Google Scholar]
- 5.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 6.Boettner DR, Chi RJ, Lemmon SK. Lessons from yeast for clathrin-mediated endocytosis. Nat Cell Biol. 2012;14:2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 10.Farsad K, De Camilli P. Mechanisms of membrane deformation. Curr Opin Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 11.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 12.Girao H, Geli MI, Idrissi FZ. Actin in the endocytic pathway: from yeast to mammals. FEBS Lett. 2008;582:2112–2119. doi: 10.1016/j.febslet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson SM, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galletta BJ, Mooren OL, Cooper JA. Actin dynamics and endocytosis in yeast and mammals. Curr Opin Biotechnol. 2010;21:604–610. doi: 10.1016/j.copbio.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conibear E. Converging views of endocytosis in yeast and mammals. Curr Opin Cell Biol. 2010;22:513–518. doi: 10.1016/j.ceb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 19.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Carroll SY, Stimpson HE, Weinberg J, Toret CP, Sun Y, Drubin DG. Analysis of yeast endocytic site formation and maturation through a regulatory transition point. Mol Biol Cell. 2012;23:657–668. doi: 10.1091/mbc.E11-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Idrissi FZ, Grotsch H, Fernandez-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol. 2008;180:1219–1232. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idrissi FZ, Blasco A, Espinal A, Geli MI. Ultrastructural dynamics of proteins involved in endocytic budding. Proc Natl Acad Sci USA. 2012;109:E2587–E2594. doi: 10.1073/pnas.1202789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kukulski W, Schorb M, Kaksonen M, Briggs JA. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 27.Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 28.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Toret CP, Lee L, Sekiya-Kawasaki M, Drubin DG. Multiple pathways regulate endocytic coat disassembly in Saccharomyces cerevisiae for optimal downstream trafficking. Traffic. 2008;9:848–859. doi: 10.1111/j.1600-0854.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 30.Boettner DR, D’Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 34.Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heuser J, Kirchhausen T. Deep-etch views of clathrin assemblies. J Ultrastruct Res. 1985;92:1–27. doi: 10.1016/0889-1605(85)90123-5. [DOI] [PubMed] [Google Scholar]
- 36.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.den Otter WK, Briels WJ. The generation of curved clathrin coats from flat plaques. Traffic. 2011;12:1407–1416. doi: 10.1111/j.1600-0854.2011.01241.x. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Zhao X, Baylor L, Kaushal S, Eisenberg E, Greene LE. Clathrin exchange during clathrin-mediated endocytosis. J Cell Biol. 2001;155:291–300. doi: 10.1083/jcb.200104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 40.Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci USA. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sever S, Muhlberg AB, Schmid SL. Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- 44.Burston HE, Maldonado-Baez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol. 2009;185:1097–1110. doi: 10.1083/jcb.200811116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Umasankar PK, Sanker S, Thieman JR, Chakraborty S, Wendland B, Tsang M, Traub LM. Distinct and separable activities of the endocytic clathrin-coat components Fcho1/2 and AP-2 in developmental patterning. Nat Cell Biol. 2012;14:488–501. doi: 10.1038/ncb2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newpher TM, Lemmon SK. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7:574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol Biol Cell. 2011;22:3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Dannhauser PN, Ungewickell EJ. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat Cell Biol. 2012;14:634–639. doi: 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- 51.Kishimoto T, Sun Y, Buser C, Liu J, Michelot A, Drubin DG. Determinants of endocytic membrane geometry, stability, and scission. Proc Natl Acad Sci USA. 2011;108:E979–E988. doi: 10.1073/pnas.1113413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinrichsen L, Meyerholz A, Groos S, Ungewickell EJ. Bending a membrane: how clathrin affects budding. Proc Natl Acad Sci USA. 2006;103:8715–8720. doi: 10.1073/pnas.0600312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Payne GS, Baker D, van Tuinen E, Schekman R. Protein transport to the vacuole and receptor-mediated endocytosis by clathrin heavy chain-deficient yeast. J Cell Biol. 1988;106:1453–1461. doi: 10.1083/jcb.106.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein–protein crowding. Nat Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 56.Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon HT, Kozlov MM. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cocucci E, Aguet F, Boulant S, Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell. 2012;150:495–507. doi: 10.1016/j.cell.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 59.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 60.Cameron LA, Svitkina TM, Vignjevic D, Theriot JA, Borisy GG. Dendritic organization of actin comet tails. Curr Biol. 2001;11:130–135. doi: 10.1016/s0960-9822(01)00022-7. [DOI] [PubMed] [Google Scholar]
- 61.Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- 62.Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- 63.Galletta BJ, Chuang DY, Cooper JA. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urbanek AN, Smith AP, Allwood EG, Booth WI, Ayscough KR. A novel actin-binding motif in Las17/WASP nucleates actin filaments independently of Arp2/3. Curr Biol. 2013;23:196–203. doi: 10.1016/j.cub.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 65.Kim K, Galletta BJ, Schmidt KO, Chang FS, Blumer KJ, Cooper JA. Actin-based motility during endocytosis in budding yeast. Mol Biol Cell. 2006;17:1354–1363. doi: 10.1091/mbc.E05-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toshima J, Toshima JY, Duncan MC, Cope MJ, Sun Y, Martin AC, Anderson S, Yates III, Mizuno K, Drubin DG. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feliciano D, Di Pietro SM. SLAC, a complex between Sla1 and Las17, regulates actin polymerization during clathrin-mediated endocytosis. Mol Biol Cell. 2012;23:4256–4272. doi: 10.1091/mbc.E11-12-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- 69.Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27:2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galletta BJ, Carlsson AE, Cooper JA. Molecular analysis of Arp2/3 complex activation in cells. Biophys J. 2012;103:2145–2156. doi: 10.1016/j.bpj.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch M, Drubin DG. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Basu R, Chang F. Characterization of dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr Biol. 2011;21:905–916. doi: 10.1016/j.cub.2011.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collins A, Warrington A, Taylor KA, Svitkina T. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol. 2011;21:1167–1175. doi: 10.1016/j.cub.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skau CT, Courson DS, Bestul AJ, Winkelman JD, Rock RS, Sirotkin V, Kovar DR. Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J Biol Chem. 2011;286:26964–26977. doi: 10.1074/jbc.M111.239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodal AA, Kozubowski L, Goode BL, Drubin DG, Hartwig JH. Actin and septin ultrastructures at the budding yeast cell cortex. Mol Biol Cell. 2005;16:372–384. doi: 10.1091/mbc.E04-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young ME, Cooper JA, Bridgman PC. Yeast actin patches are networks of branched actin filaments. J Cell Biol. 2004;166:629–635. doi: 10.1083/jcb.200404159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 78.Skruzny M, Brach T, Ciuffa R, Rybina S, Wachsmuth M, Kaksonen M. Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc Natl Acad Sci USA. 2012;109:E2533–E2542. doi: 10.1073/pnas.1207011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buser C, Drubin DG. Ultrastructural imaging of endocytic sites in Saccharomyces cerevisiae by transmission electron microscopy and immunolabeling. Microsc Microanal. 2013;19:381–392. doi: 10.1017/S1431927612014304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Engqvist-Goldstein AE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol. 2001;154:1209–1223. doi: 10.1083/jcb.200106089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Engqvist-Goldstein AE, Zhang CX, Carreno S, Barroso C, Heuser JE, Drubin DG. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell. 2004;15:1666–1679. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu M, Huang B, Graham M, Raimondi A, Heuser JE, Zhuang X, De Camilli P. Coupling between clathrin-dependent endocytic budding and F-BAR-dependent tubulation in a cell-free system. Nat Cell Biol. 2010;12:902–908. doi: 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Evergren E, Tomilin N, Vasylieva E, Sergeeva V, Bloom O, Gad H, Capani F, Shupliakov O. A pre-embedding immunogold approach for detection of synaptic endocytic proteins in situ. J Neurosci Methods. 2004;135:169–174. doi: 10.1016/j.jneumeth.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 84.Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107:4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 86.Youn JY, Friesen H, Kishimoto T, Henne WM, Kurat CF, Ye W, Ceccarelli DF, Sicheri F, Kohlwein SD, McMahon HT, et al. Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis. Mol Biol Cell. 2010;21:3054–3069. doi: 10.1091/mbc.E10-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geli MI, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111(Pt 8):1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- 88.Smaczynska-de II R, Allwood EG, Mishra R, Booth WI, Aghamohammadzadeh S, Goldberg MW, Ayscough KR (2012) Yeast dynamin Vps1 and amphiphysin Rvs167 function together during endocytosis. Traffic 13:317–328 [DOI] [PubMed]
- 89.Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J, Sun Y, Oster GF, Drubin DG. Mechanochemical crosstalk during endocytic vesicle formation. Curr Opin Cell Biol. 2010;22:36–43. doi: 10.1016/j.ceb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Romer W, Pontani LL, Sorre B, Rentero C, Berland L, Chambon V, Lamaze C, Bassereau P, Sykes C, Gaus K, et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell. 2010;140:540–553. doi: 10.1016/j.cell.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 92.Chang-Ileto B, Frere SG, Chan RB, Voronov SV, Roux A, Di Paolo G. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev Cell. 2011;20:206–218. doi: 10.1016/j.devcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morlot S, Galli V, Klein M, Chiaruttini N, Manzi J, Humbert F, Dinis L, Lenz M, Cappello G, Roux A. Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell. 2012;151:619–629. doi: 10.1016/j.cell.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamada H, Padilla-Parra S, Park SJ, Itoh T, Chaineau M, Monaldi I, Cremona O, Benfenati F, De Camilli P, Coppey-Moisan M, et al. Dynamic interaction of amphiphysin with N-WASP regulates actin assembly. J Biol Chem. 2009;284:34244–34256. doi: 10.1074/jbc.M109.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smaczynska-de II R, Allwood EG, Aghamohammadzadeh S, Hettema EH, Goldberg MW, Ayscough KR (2010) A role for the dynamin-like protein Vps1 during endocytosis in yeast. J Cell Sci 123:3496–3506 [DOI] [PMC free article] [PubMed]
- 97.Meinecke M, Boucrot E, Camdere G, Hon WC, Mittal R, McMahon HT. Cooperative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. J Biol Chem. 2013;288:6651–6661. doi: 10.1074/jbc.M112.444869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 100.Taylor MJ, Lampe M, Merrifield CJ. A feedback loop between dynamin and actin recruitment during clathrin-mediated endocytosis. PLoS Biol. 2012;10:e1001302. doi: 10.1371/journal.pbio.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramachandran R. Vesicle scission: dynamin. Semin Cell Dev Biol. 2011;22:10–17. doi: 10.1016/j.semcdb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 102.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prescianotto-Baschong C, Riezman H. Morphology of the yeast endocytic pathway. Mol Biol Cell. 1998;9:173–189. doi: 10.1091/mbc.9.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huckaba TM, Gay AC, Pantalena LF, Yang HC, Pon LA. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 2004;167:519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yahav T, Maimon T, Grossman E, Dahan I, Medalia O. Cryo-electron tomography: gaining insight into cellular processes by structural approaches. Curr Opin Struct Biol. 2011;21:670–677. doi: 10.1016/j.sbi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 107.Milne JL, Borgnia MJ, Bartesaghi A, Tran EE, Earl LA, Schauder DM, Lengyel J, Pierson J, Patwardhan A, Subramaniam S. Cryo-electron microscopy–a primer for the non-microscopist. FEBS J. 2013;280:28–45. doi: 10.1111/febs.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pierson J, Vos M, McIntosh JR, Peters PJ. Perspectives on electron cryo-tomography of vitreous cryo-sections. J Electron Microsc (Tokyo) 2011;60(Suppl 1):S93–100. doi: 10.1093/jmicro/dfr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pierson J, Fernandez JJ, Bos E, Amini S, Gnaegi H, Vos M, Bel B, Adolfsen F, Carrascosa JL, Peters PJ. Improving the technique of vitreous cryo-sectioning for cryo-electron tomography: electrostatic charging for section attachment and implementation of an anti-contamination glove box. J Struct Biol. 2010;169:219–225. doi: 10.1016/j.jsb.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 110.Rigort A, Bauerlein FJ, Villa E, Eibauer M, Laugks T, Baumeister W, Plitzko JM. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci USA. 2012;109:4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rigort A, Bauerlein FJ, Leis A, Gruska M, Hoffmann C, Laugks T, Bohm U, Eibauer M, Gnaegi H, Baumeister W, et al. Micromachining tools and correlative approaches for cellular cryo-electron tomography. J Struct Biol. 2010;172:169–179. doi: 10.1016/j.jsb.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 112.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hein B, Willig KI, Wurm CA, Westphal V, Jakobs S, Hell SW. Stimulated emission depletion nanoscopy of living cells using SNAP-tag fusion proteins. Biophys J. 2010;98:158–163. doi: 10.1016/j.bpj.2009.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Agrawal U, Reilly DT, Schroeder CM. Zooming in on biological processes with fluorescence nanoscopy. Curr Opin Biotechnol. 2013;24:646–653. doi: 10.1016/j.copbio.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 115.van de Linde S, Heilemann M, Sauer M. Live-cell super-resolution imaging with synthetic fluorophores. Annu Rev Phys Chem. 2012;63:519–540. doi: 10.1146/annurev-physchem-032811-112012. [DOI] [PubMed] [Google Scholar]
- 116.Llobet A, Beaumont V, Lagnado L. Real-time measurement of exocytosis and endocytosis using interference of light. Neuron. 2003;40:1075–1086. doi: 10.1016/s0896-6273(03)00765-7. [DOI] [PubMed] [Google Scholar]
- 117.Anantharam A, Onoa B, Edwards RH, Holz RW, Axelrod D. Localized topological changes of the plasma membrane upon exocytosis visualized by polarized TIRFM. J Cell Biol. 2010;188:415–428. doi: 10.1083/jcb.200908010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shevchuk AI, Novak P, Taylor M, Diakonov IA, Ziyadeh-Isleem A, Bitoun M, Guicheney P, Lab MJ, Gorelik J, Merrifield CJ, et al. An alternative mechanism of clathrin-coated pit closure revealed by ion conductance microscopy. J Cell Biol. 2012;197:499–508. doi: 10.1083/jcb.201109130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Grotsch H, Giblin JP, Idrissi FZ, Fernandez-Golbano IM, Collette JR, Newpher TM, Robles V, Lemmon SK, Geli MI. Calmodulin dissociation regulates Myo5 recruitment and function at endocytic sites. EMBO J. 2010;29:2899–2914. doi: 10.1038/emboj.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]