Abstract
The prospect of intervening, through the use of a specific molecule, with a cellular alteration responsible for a disease, is a fundamental ambition of biomedical science. Epigenetic-based therapies appear as a remarkable opportunity to impact on several disorders, including cancer. Many efforts have been made to develop small molecules acting as inhibitors of histone deacetylases (HDACs). These enzymes are key targets to reset altered genetic programs and thus to restore normal cellular activities, including drug responsiveness. Several classes of HDAC inhibitors (HDACis) have been generated, characterized and, in certain cases, approved for the use in clinic. A new frontier is the generation of subtype-specific inhibitors, to increase selectivity and to manage general toxicity. Here we will discuss about a set of molecules, which can interfere with the activity of a specific subclass of HDACs: the class IIa.
Keywords: SAHA, HDAC3, HDAC4, HDAC5, HDAC7, HDAC9, MEF2, p21, Therapy, Apoptosis, Cell cycle, Anti-cancer, Neurodegeneration, Inflammation
Introduction
Why to target HDACs?
Every complex cellular adaptation and behavior is supervised by changes in the transcriptional machinery, which align the gene expression profile of a specific cell type to the general requirements of the organism. The harmonic regulation of genes transcribed in a specific instant is the result of an integrated and complex network of signals that controls the activity of different transcriptional players. Transcription factors (TFs), epigenetic regulators and “structural” proteins, constituting the chromatin are the chief protagonists under the tight influence of the environment. Alterations in the signaling networks or in the transcriptional players are responsible for aberrations in tissue homeostasis and triggering events in several different diseases, from neurodegeneration up to cancer [1, 2]. The opportunity to reset the transcriptional subverted context, with the therapeutic perspective of curing/alleviating diseases, straightway attracted scientist’s attention [3, 4].
Perhaps the simplest approach to develop new drugs is the identification of small molecules, acting as inhibitors of an enzymatic activity that is imperative in a specific disease. In the context of gene transcription, post-translational modifications (PTMs) of histones represent realistic targets for the development of epigenetic therapies aimed to amend transcriptional alterations. Acetylation of lysines, placed in histones but also in TFs is an important PTM, exerting both positive (H3K4, 9, 14, 17, 23; H4K5, 8, 12, 16) and negative (in the case of specific TFs) effects on gene expression [5, 6]. Being acetylation reversible and under the scrutiny of different family of enzymes: HATs (histone acetyl transferases) and HDACs (histone deacetylases), it has attracted several interests as a druggable PTM [7]. In particular, during the past decades, many efforts have been made to isolate, synthesize and characterize small molecules targeting HDACs [8]. HDACis are nowadays represented as a considerable fraction of the epigenetic drugs under study and in some circumstances these compounds have been approved for the use in clinic (see below). Importantly, epigenetic drugs in cancer therapy represent an opportunity to revert drug-resistance-associated epigenomes and to prevent or reverse non-responsiveness to anti-cancer drugs [2].
Copious studies on cancer cells’ epigenomes have fully justified the rationale of applying HDACis in anti-cancer therapies. Three major intrinsic features of the neoplastic cells could be subject of specific intervention, thanks to HDACis: (1) cancer cells are characterized by an enhanced degree of heterochromatinization compared to normal cells; which makes cancer genomes inaccessible to DNA-damage response enzymes [9]. The treatment of cancer cells with HDACis relaxes chromatin and allows the activation of the DNA-damage response [9]. (2) Several tumor suppressor genes, including some pro-apoptotic genes, are inactivated in cancer cells because of ipo-acetylated promoters [3, 10, 11]. (3) Alterations of the epigenetic machineries embracing HDACs are frequently observed in tumors [12, 13].
Despite the considerable literature debating the use in epigenetic therapies of pan-HDACi and of class I HDACs specific inhibitors [10, 11, 14–22], reviews specifically discussing of molecules acting as inhibitors of class IIa HDACs, are quite rare. In this manuscript we will discuss specifically of them.
Class IIa HDACs: to be or not to be a lysine deacetylase
In humans there are 18 HDACs grouped into five different classes according to phylogenesis and sequence homology [7]. Class I HDACs (including HDAC1, 2, 3 and 8), class IIb HDACs (including HDAC6 and 10), class III HDACs or Sirtuins (including all Sirtuins from 1 to 7) and class IV (HDAC11) all displaying enzymatic activities [23]. By contrast, when we discuss about class IIa HDACs (HDAC4, 5, 7 and 9) as histone deacetylases, it should be taken into account that these proteins show an extremely low enzymatic activity against acetylated lysines [24, 25] and are rarely associated with histone tails [26].
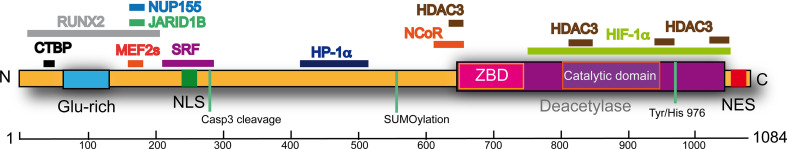
Structurally, class IIa HDACs can be divided into two parts: the N- and the C-terminal regions (Fig. 1). The N-terminal regulates the nuclear import and contains a coiled-coil glutamine-rich domain that is peculiar of the family. This region is highly devoted to protein–protein interactions both in terms of homo- and of heterotypic partners. The C-terminal region contains the catalytic “deacetylase” domain and the nuclear export sequence (Fig. 1). These enzymes are under the control of different signaling pathways, which operate through specific PTMs to influence peculiar aspects of the class IIa biology, including the nuclear/cytoplasmic shuttling (for reviews [7, 12, 27, 28]).
Fig. 1.
Schematic representation of class IIa HDACs highlighting the principal domains. As prototype of class IIa we selected HDAC4. Certain interaction partners, as well as the relative HDAC4 sequences involved, are illustrated
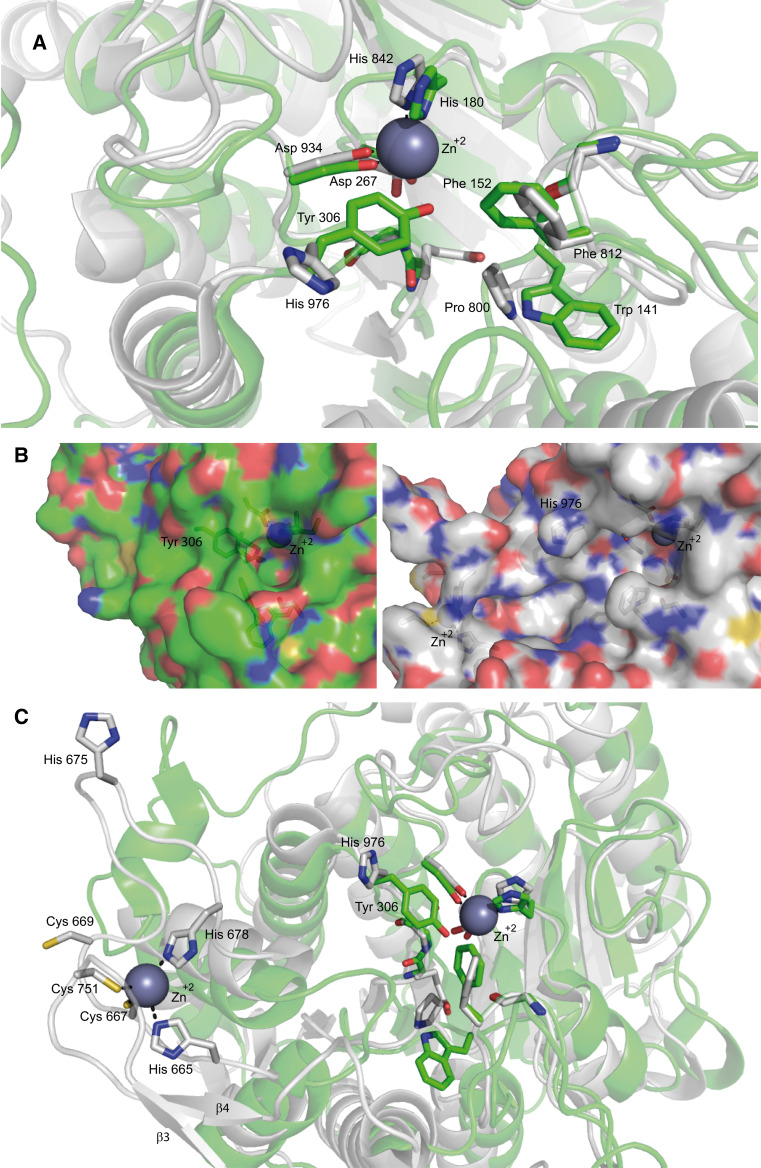
The deacetylase domain is made up of approximately 400 residues (aa) arranged into 21 α-helices and 10 β-strands organized in a single domain, structured around a central “catalytic” Zn2+ ion [29]. Likewise to class I HDACs, 2 aspartates and an histidine coordinate this Zn2+ while 2 other aspartates (Fig. 2a), another histidine, a serine and a leucine coordinate two potassium ions [29–31]. Despite this high similarity, in vertebrates class IIa possess a bigger active site than class I HDACs (Fig. 2b), which impacts on their druggability [26, 29]. The evolution-related event responsible for this structural peculiarity is the mutation of a tyrosine into a histidine, Y967H in HDAC4 [25]. Histidine is sterically less cumbersome and induces the relaxation of the structure. As a consequence, this histidine is far from the central Zn2+ and not able to form hydrogen bonds with the intermediate of the enzymatic reaction (Fig. 2a). The intermediate is, therefore, very unstable, thus resulting in an ineffective reaction. Nevertheless, class IIa can efficiently process alternative substrates such as trifluoroacetyl-lysine. Mechanistically, the presence of the trifluoro group should destabilize the amide bond, hence favoring the reaction even in the absence of transition-state stabilization [25].
Fig. 2.
Representation of class I and class IIa catalytic sites (a, b) and the zinc binding domain (c). a Superimposition of the inhibitor (TFMK)-bound ribbon structure of class I HDAC8 (green) and of class IIa HDAC4 (white) catalytic sites. As mentioned in the text the His 976 is rotated away from the active site differently from Tyr 308 in HDAC8. b Surface representation of class I HDAC8 (green) and class IIa HDAC4 (white) catalytic sites. The figure shows the hydrophilic tunnel necessary for the release of the reaction product in HDAC8 (green), while in HDAC4 (white) the His/Tyr substitution prevents tunnel formation. c Superimposition of the inhibitor (TFMK)-bound ribbon structure of class I HDAC8 (green) and of class IIa HDAC4 (white) catalytic site (right) and zinc binding domain of HDAC4 (left). β3 and β4 are the two antiparallel β-strands involved in the formation of the pocket-like structure in the zinc binding domain. Importantly, His 665 and His 678 in this inhibitor-bound structure are replaced by Cys 669 and His 675 in the coordination of the zinc ion in the Apo-structure. Unfortunately the crystallization of Apo-HDAC4 was unsuccessful and these differences are deduced from crystallographic studies of the mutant GOF (H976Y) of HDAC4 [31]. The coordinates of the protein structures were retrieved from the protein data bank. Amino acids discussed in the text are labeled and shown in stick representation. The accession codes for the protein structures are: 2VQJ (HDAC4) and 1T69 (HDAC8). Figures are edited using PyMOL Molecular graphics system, Schrödinger, LLC
Importantly, replacing back the His with Tyr generates class IIa HDACs with a catalytic efficiency 1,000-fold higher compared to the wild-type (wt) form [25, 31]. Nonetheless, this mutant does not show enhanced repression respect to the wt, at least in the instance of MEF2-dependent transcription, a well-known class IIa partner [25].
Another distinctive feature of class IIa HDAC catalytic site is the existence of a Zinc Binding Domain (ZBD). This ZBD consists in a β-hairpin surrounded by two antiparallel β-strands, forming a pocket-like structure that accommodates a second “structural” zinc ion [29]. In the case of HDAC4 three cysteines (667, 669, 751) and one histidine (675), conserved only among class IIa HDACs, coordinate this Zn2+ and made the so-called “core” of the domain [31] (Fig. 2c). Importantly, the inhibitor-bound structure is shown in this figure, where, respect to the Apo-structure, Cys 669 and His 675 replace His 665 and His 678 in the coordination of the Zn2+.
This domain is extremely flexible and the oxidation of the cysteines involved in Zn2+ coordination (667 and 669 in HDAC4) is sufficient to free the metal, with the consequent opening and deconstruction of the ZBD [31]. Because this domain is head-to-head to the active site (Fig. 2c), it contributes to make the class IIa HDACs’ catalytic site more accessible than that of class I HDACs (Fig. 2b) and does not allow the formation of an efficient hydrophilic tunnel necessary for the release of the acetate reaction product [30, 31].
Old structures and new functions
The enzymatic ineptitude of vertebrates’ class IIa deacetylase domain raises several questions and opens the door to different hypothesis. First, they are not completely silenced enzymes. Because class IIa is capable of processing trifluoroacetyl-lysine with high efficiency, still undiscovered new natural substrates could exist [25]. Alternatively, the described enzymatic activity could simply mark a lab finding, without biological implications. Second, as anticipated above, the absence of improved repressive influence in the case of the gain of function His/Tyr substitution in HDAC4, further demonstrates that class IIa HDACs can repress transcription independently from the deacetylase domain [25]. The relevance of the deacetylase-independent repression is testified by MITR, a splice variant of HDAC9 lacking the deacetylase domain [32]. The existence of MITR supports the possibility that the HDAC domain is of little relevance for the functions of class IIa HDACs and may lead to believe that it is an evolutionary heritage intended to being missed. However, since class IIa deacetylase domain has been preserved behind two duplication events occurred during evolution of vertebrates, evolutionists deny the hypothesis that this domain would be subjected to a negative purifying selection [33].
Although there are evidences pointing to deacetylase-independent activities of class IIa, generation of a mouse model in which, mutated versions of this domain can be analyzed in a physiological context will help our understanding. This point is of crucial relevance for the design and development of class IIa inhibitors.
Along with the enzymatic activity, the deacetylase domain can operate as a scaffold for the recruitment of multi-protein complexes containing class I HDAC3 and other co-repressors [31]. HDAC4 interacts with the RD3 domain of N-CoR [24, 34], while HDAC3 binds the SAINT domain [35] and, as a matter of fact, HDAC4 binds N-CoR/SMRT regardless of HDAC3 and only in a second time the deacetylase is recruited [36]. However, the precise order of the sequential molecular interactions driving the assembly of the multi-protein complex is still waiting for a final verification.
When class IIa HDACs are isolated under native conditions, a lysine deacetylase activity can be measured. This activity is due to class I HDACs co-purified with class IIa [24, 37, 38]. The existence of a heterogeneous repressive complex complicates the assessment of effectiveness and specificity of HDACis, when tested on proteins purified from cells or tissues.
A final consideration refers to a fascinating hypothesis, which attributes to class IIa deacetylase domain the function of acetylated lysine reader [26]. In this view, class IIa could act as readers and interpreters of the histone code, thus orchestrating the epigenetic status thanks to their capability of recruiting additional enzymes, such as methylases [39] or deacetylases [24, 36]. A scenario where class IIa HDACs, acting as molecular scaffolds supervise the introduction of different epigenetic markers, onto specific regions of chromatin or in proximity of different acetylated cellular protein. In this context inhibitors of the deacetylase domain could in principle both interfere with the reading activity or, by promoting structural changes, with the possibility of recruiting additional co-repressors.
Unresolved issues
Biochemically, the enzymatic activity associated to class IIa HDACs could be explained by the recruitment of class I enzymes [24]. Moreover, all the point mutants of the HDAC4 deacetylase domain which, accordingly to Finnin model [40], abrogate its enzymatic activity (H803A, G811A, D838A, D840A, H842A, N845D, D934 N, E973G) demonstrate a perfect correlation between enzymatic activity and the ability to recruit HDAC3 [24]. Classic deacetylase activity is not associated with a cytoplasmic HDAC7 or HDAC4 immunoprecipitated from HEK293 cells and therefore, weakly associated to the mainly nuclear HDAC3 [24, 36]. Similarly, HDAC4 mutants that have lost the ability of binding to N-CoR/SMRT drop the deacetylase activity [24]. Despite in vitro binding experiments prove that the fraction of HDAC3 in complex with HDAC4 is relevant, in vivo HDAC3 preferentially forms homodimers, rather than heterodimers with HDAC4 [41]. Furthermore, the fraction of HDAC4 co-purified with HDAC3 in mammalian cells is extremely low [24, 35, 37].
As aforementioned, another peculiar feature of class IIa deacetylase domain is its sensitivity to redox conditions [31, 42]. Particularly, in HDAC4 the oxidation of cysteines 667 and 669 induces the formation of a disulphide bond that causes the exposition of the NES, the export in the cytoplasm and also the detachment of HDAC3 [31, 42, 43]. This oxidation causes the de-structuration of the HDAC domain because Cys 667 and Cys 669 are directly involved in the “structural” Zn2+ coordination and substrate binding [29, 31] (Fig. 2c). These findings show that researchers should be extremely cautious in verifying the redox status when studying class IIa deacetylase domain.
In addition to nuclear roles of class IIa HDACs, recently, a cytoplasmic enzymatic activity has been reported towards non-histone substrates [reviewed in 44]. During muscle denervation HDAC4, which plays a pro-atrophic role in this context [45, 46] can deacetylate and activate MEKK2 [47]. Kinase engagement culminates in AP-1 activation and cytokines production that stimulate muscle remodeling [47]. Interestingly only the wild-type form, capable of shuttling between the nucleus and the cytoplasm and not a nuclear resident mutant of HDAC4 deacetylated MEKK2. Importantly, this activity is independent from HDAC3 and is not shared with HDAC5 [47]. Paradoxically, MEKK2 activation should activate ERK5 and therefore MEF2s, thus pointing to a positive rather than repressive influence of HDAC4 versus MEF2s [48, 49]. A similar cytoplasmic KDAC (lysine deacetylase) activity of class IIa HDACs was reported towards HIF-1α and STAT-1. Also in these circumstances class IIa deacetylase activity seems to be independent from class I HDACs [44].
Another unresolved issue is the requirement of additional factors to exert the full enzymatic activity. Class I HDACs require particular cofactors both for histone and non-histone substrates [35, 41, 50]. For the enzymatic activity of class IIa HDACs towards the synthetic trifluoroacetyl-lysine or against these cytoplasmic partners, any cofactor seems to be dispensable [25].
The rationale for developing class IIa HDACs inhibitors
HDACis have entered multiple clinical trials principally in virtue of their anti-neoplastic properties [10]. Much more emphasis has been pushed on the identification, synthesis and characterization of class I HDACis. Commonly HDACis show a selective cytotoxicity against tumor cells and weak effects on normal ones [11, 51, 52]. These molecules display cytostatic effects, especially through the induction of p21 and blockage of the cell cycle [53, 54] or by triggering apoptosis via multiple mechanisms [11, 53, 55, 56]. Some HDACis in vivo stimulate also the clearance of tumor cells from the immune system [57, 58] or block angiogenesis [59, 60]. Despite these promising anti-neoplastic properties, entering of HDACis in clinic is slower than expected, principally due to some side effects and toxicity displayed during early-phase clinical trials [14, 61]. In fact, up to now only two HDACis have been approved for the treatment of cutaneous T cell lymphoma: SAHA (Zolinza) in 2006 and Romidepsin/FK-228 in 2009. In 2011 the depsipeptide FK-228 has been further approved for the treatment of peripheral T-cell lymphoma [15]. Considering the recent evidences about a pro-oncogenic potential of class IIa HDACs [12, 37, 38, 62–64] and their impact on epigenetics [65], a stratagem to circumvent the side effects of class I HDACs inhibitors might consist in targeting class IIa HDACs.
Theoretically, targeting class IIa HDACs with specific inhibitors has three major drawbacks:
The high similarity of the catalytic site of these proteins to class I HDACs, which makes selective targeting rather difficult to achieve;
The formal question about the legitimacy of hitting the catalytic site of proteins that are almost enzymatically inactive against acetylated lysines. About this consideration the work of Bottomley et al. [31] explains how targeting of the catalytic site of class IIa HDACs and in particular the Zn2+ atom could impact on the structure of the C-terminus of the proteins, thus compromising their capability to interact with the super complex HDAC3/N-CoR/SMRT. Therefore, targeting class IIa HDAC domain could be an indirect strategy to impact on class I HDACs. By releasing only class IIa driven deacetylation, a more selected transcriptional re-setting can be achieved, which could favor a drop in toxicity.
The methodological approach to measure class IIa HDAC inhibition. Up to now the best-characterized substrate for probing the elusive catalytic activity of vertebrate class IIa histone deacetylases is trifluoroacetyl-lysine [25, 66]. The activity of class I HDACs towards this molecule is indiscernible. Its use as a substrate for the validation of an inhibitor efficiency could exclude all class I HDACs as off-targets. Class IIa HDAC enzymatic activity measured with other methods or with classical substrates (e.g., acetylated H3) or commercial assays, generally based on acetyl-Lys, is extremely low when recombinant proteins are used [24]. Instead, when class IIa are purified from vertebrates the enzymatic activity can be provided by associated class I or IIb enzymes [24, 25, 31, 67]. Therefore, a double check approach should be used to test the potency and specificity of a class IIa HDACis. The potency of the compound should be evaluated by employing trifluoroacetyl-lysine, as a class IIa specific substrate, while its inhibitory activity against other HDAC classes should be excluded using “classical” substrates, such as acetylated lysines. A simplified screening could take advantage from the recently developed trifluoroacetyl-lysine derivative, a trifluoro acetyl-lysine tripeptide named substrate 6, which can be processed by all HDACs, with the exclusion of HDAC10 and 11. This molecule looks like a promising tool for single-run screening aimed to isolate/characterize subtype specific HDACis [68].
Class IIa inhibitors
Three different peculiarities of class IIa HDACs have been exploited to design specific inhibitors:
The catalytic site, and in particular the Zn2+ atoms.
The nuclear/cytoplasmic shuttling.
The N-terminal region and the binding to specific partners, such as the MEF2 family of TFs.
Targeting the Zn2+ binding domain
In accordance to the connecting unit (CU) linker chelator pharmacophore model [16, 69], a classical HDACi is composed of three parts [17]:
The MBG (metal binding group or zinc binding group ZBG), which is a group capable of chelating the Zn2+ in the catalytic site of HDACs (with the exception of sirtuins).
The connecting unit (CU), generally a linker hydrophobic region of five or more carbons, that mimics the acetyl-lysine. It could be linear or aromatic and it perfectly fits to the hydrophobic catalytic site of the targeted HDAC.
The CAP hydrophobic domain (usually aromatic) that interacts with aminoacids delimiting the border of the deacetylase catalytic site.
Slight modifications of the described structure impact both on the specificity and potency of the inhibitor.
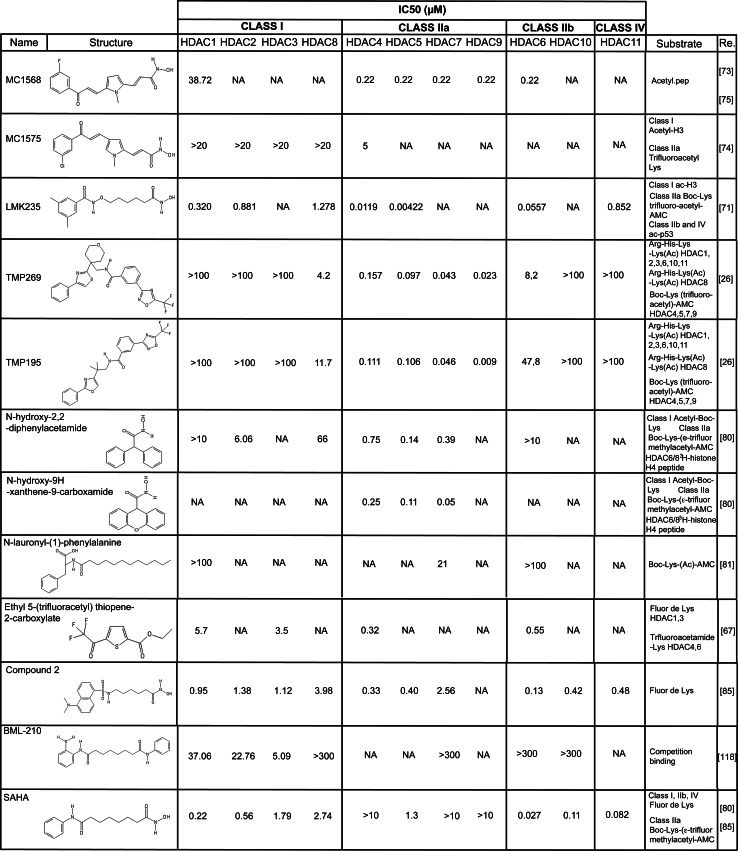
The availability of the crystal structure of the class IIa deacetylase domain [29, 31] has encouraged the development and synthesis of many hydroxamates stemmed from SAHA, with the purpose of selectively influencing class IIa HDACs. In particular to improve specificity, many efforts have been spent in the modification of the CAP and of the ZBG of SAHA. In principle, the selective targeting of class IIa HDACs would require only some changes in the linker region, to better fit the peculiar catalytic site of class IIa HDACs. A recent study effectively demonstrated that slight modifications only in the linker region of SAHA increase the selectivity towards class IIa and class IIb HDACs [70]. However, the achieved results were not as promising as those obtained after modification of both the CAP and the linker region of SAHA [71]. This double tuning seems to be the better strategy to produce SAHA derivatives specific for class IIa HDACs. In a next future, new generation class IIa HDACis could stem from Tasquinimod (described below) that selectively targets the “structural” and not the “catalytic” Zn2+. This peculiarity should increase the specificity because, as discussed above, this “structural zinc” is unique of class IIa HDACs. A summary of the literature data is shown in Fig. 3.
Fig. 3.
Structures and summary of the available literature data on the IC50 for the proposed class IIa inhibitors
The most characterized of these hydroxamate-like drugs, are:
MC1568 and MC1575 (Fig. 3, please note that in Fig. 3 we provide for MC1568 the recently reassigned structure [72]) are two class II HDACs inhibitors specific for HDAC4 and HDAC6 [73–76]. They are derivatives of classical class I HDACs inhibitors aroyl-pyrrolyl–hydroxyamides (APHAs), showing selectivity towards class IIa HDACs. The modified linker region provides this selectivity. Compared to the original class I inhibitors, they exhibit a decreased cytotoxic effect [73]. Despite this fact, MC1568 and MC1575 show some cytostatic effects in melanoma cells [76] and in ER + breast cancer cells [74]. The anti-proliferative effect is provoked by a block in the G1 phase of the cell cycle, through the induction of the Cdk inhibitor p21/Cip1/Waf1 [74]. MC1568 efficacy in cancer cells finds rationality in the capability of up-regulating the tumor suppressor Brahma, repressed by HDAC9 [77]. Curiously, MC1568 has been reported stabilizing the HDAC4-MEF2D complex in differentiated C2C12 myoblasts, thus impairing instead of favoring myogenesis [78].
LMK235 (N-((6-(hydroxyamino)-6-oxoh exyl)oxy)-3,5-dimethylbenzamide) is a hybrid between two classes of class I HDACis: the hydroxamic acids and the benzamides (Fig. 3) [71]. The specificity towards HDAC4 and HDAC5 is conveyed by the hydrophobic dimethyl substituted phenyl ring, which acts as a CAP group, matching class IIa active site better than class I [71]. This modification makes the molecule less toxic and more suitable for the treatment of some malignances, when compared to class I HDACis. Furthermore, LMK235 is able to re-sensitize cancer cells to cisplatin, better than SAHA [71].
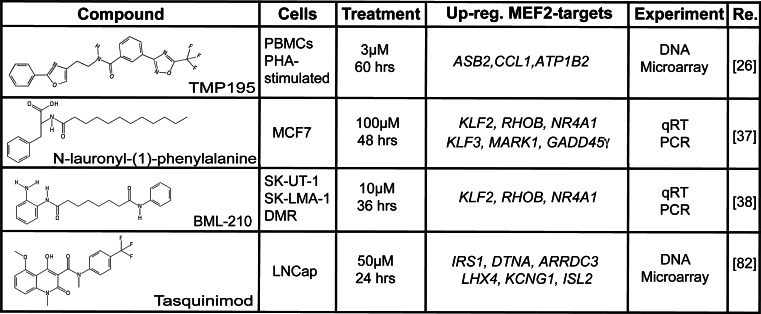
TMP269 and 195 (Figs. 3, 5) are two recently developed class IIa HDACis in which the classical hydroxamic Zn2+ binding domain is substituted by a trifluoromethyloxadiazolyl group (TFMO) [26] that highly resembles the trifluoromethylketone (TFMK) adopted by Bottomley and colleagues in their biochemical study of the ZBD [31]. The ring structure of the TFMO group increases its stability with respect to the highly unstable TFMK series of compounds [79]. Moreover, this TFMO moiety, differently from hydroxamate, acts as a non-chelating metal binding group, which interacts with the “catalytic” Zn2+, through weak electrostatic interactions. As a consequence, the TFMO series has fewer off-targets compared to hydroxamates. Augmented selectivity is indirectly proved by gene expression profile studies in (PHA)-activated human peripheral blood mononuclear cells (PBMC) (Fig. 5). In these cells SAHA modulates the expression of 4,556 genes, whereas TMP195 regulates only 76 genes [26]. Curiously this finding is in accordance to what was observed in fibroblasts, where HDAC4 directly modulate only 76 genes [38]. To better characterize the transcriptome profile induced by their TFMO series of compounds, Lobera and colleagues purified T cells (CD3+), B cells (CD19+) and monocytes (CD14+) from the PHA-stimulated PBMC population and separately treated the three sub-populations with TMP195. T and B cells turned out to be very low sensitive to TMP195 (17 and 36 genes regulated, respectively); on the contrary the effect of the compound on monocytes was impressive (587 genes) and was not due to an increase in the expression of class IIa HDACs in these cells compared to the other two cell types. In particular the inhibitor interfered with monocytes to macrophages M-CSF (macrophage colony-stimulating factor)-induced differentiation. These findings candidate class IIa HDACs as druggable targets for immunological diseases [18, 71].
N-hydroxy-2,2-diphenylacetamide and N-hydroxy-9H-xanthene-9-carboxamide (respectively, compound 6 and 13 in the original manuscript) are two diphenylmethylene hydroxamic acids characterized by Besterman group as class IIa HDACs specific inhibitors active in the μM range [80]. Both molecules exhibit a certain degree of symmetry and the second compound could be considered as the rigidification of the diphenyl moiety of the first (Fig. 3). This modification increases the specificity of the molecule towards HDAC7 [80].
N-lauroyl-(l)-phenylalanine is a class IIa HDACi active in the μM range (Fig. 3) [81]. It was identified during a screening of a commercial available library of compounds. The specificity was scored not merely by classical measurements of HDAC activity but also through a fluorescence assay, which exploits the competition between a fluorescent substrate and the putative inhibitor for each purified HDAC [81]. This molecule shows anti-tumoral properties against ER+ breast cancer cells and can influence the expression of some MEF2-target genes (Fig. 5) [37].
Ethyl 5-(trifluoroacetyl)thiophene-2-carboxylate [67] is the founder of a class of compounds, the trifluoroacetylthiophenes, that targets class II HDACs (class IIa and HDAC6) with some specificity. It was identified during a screening of a commercially available library of compounds using both the wt and the GOF mutant of HDAC4 as targets. It is a tripartite molecule characterized by: (i) a trifluoromethyl ketone group that chelates the active site zinc in a bidentate manner, (ii) the central thiophene ring that fits perfectly to class IIa active site and (iii) the amide group that interacts with the surrounding residues. The chemistry and the tri-functional nature of this compound justify its specificity.
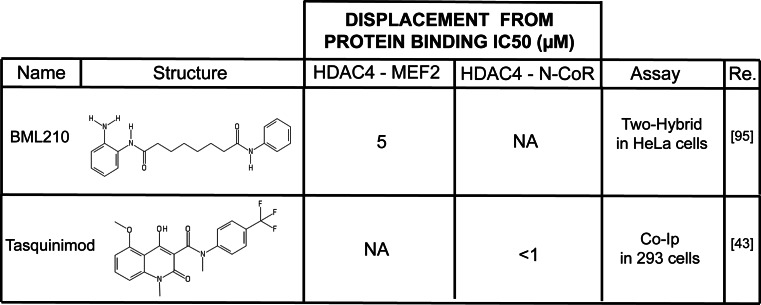
Tasquinimod (Fig. 4) is a promising drug for the treatment of advanced castration resistant prostate cancers [82, 83]. It acts by perturbing the tumor microenvironment. Differently from the aforementioned molecules it was not rationally designed or screened to target HDACs. Nevertheless, this carboxamide is able to enter the ZBD of HDAC4, keeping it in the inactive form and thus reorganizing the HDAC4 catalytic site. Tasquinimod-induced structural changes are causative of N-CoR/SMRT/HDAC3 displacement [43]. This finding is surprisingly considering the pronounced steric hindrance of the molecule, which is profoundly different from all SAHA derivatives. However, by virtue of its selective targeting of the “structural” Zn2+”, Tasquinimod molecular backbone could substitute SAHA as starting model for the development of specific inhibitors. From a molecular point of view the inactivation of HDAC4 prevents HIF-1α deacetylation, thus inducing its destabilization. Clinically, in hypoxic conditions the activation of HIF-1α transcriptional program stimulates the differentiation of tumor infiltrating myeloid derived suppressor cells into tumor-associated macrophage, which secrete pro-angiogenic factors [84]. Authors, therefore, proposed Tasquinimod as an anti-angiogenetic drug, which anti-cancer efficacy is being evaluated in pre-clinical models [43].
Fig. 5.
Summary of the available literature data on the effect of class IIa HDACs inhibitors on MEF2s-dependent transcription. MEF2s are the foremost characterized transcriptional partners of class IIa HDACs. Hence, an effect of these inhibitors on the expression of MEF2s target genes is an important read-out of their activity
Fig. 4.
Structure and binding interference properties of BML210 and Tasquinimod, two compounds capable of altering interaction of class IIa HDACs with their partners
These last three molecules are considered unconventional inhibitors because, even though characterized by a tripartite motif, they are not SAHA derivatives.
Targeting the nuclear-cytoplasmic shuttling
In 2011, Brown group made the first attempt of blocking class II HDACs in the cytoplasm [85]. Starting from the structure of SAHA, they generated a couple of molecules by substituting the amino-phenyl group with a fluorescent dansyl group. This modification increases the specificity for class II HDACs in spite of a loss of reactivity against class I HDACs. If used in the µM range, the most effective molecule of the series, named compound 2 (Fig. 3), increases the fraction of cytoplasmic HDAC4 in prostate cancer cells PC3. The authors suggested that since the inhibitor accumulates in the cytoplasm, it binds HDAC4, thus impeding the interaction with importin-1α. As a consequence, the inhibitor increases the fraction of cells in the G1 phase of the cell cycle, the levels of p21/Cip1/Waf1, of acetylated H3 and tubulin. The increase of tubulin acetylation is probably due to the inhibition of HDAC6 [86] and seems to be unrelated to the suppression of class IIa [85].
It must be underlined that the IC50 values of these new inhibitors have been estimated by measuring the enzymatic activities of HDACs purified from mammalian cells, using the Fluor–de-Lys substrate [85]. Therefore, in the case of class IIa HDACs, it must be intended as indirect, deriving principally from the associated class I HDACs.
The strategy of interfering with class IIa HDACs nuclear accumulation could be attractive in oncology, as increasing evidences demonstrate that nuclear resident class IIa can display oncogenic functions [37, 38], but it might also present some drawbacks. First of all, class IIa HDACs possess also cytoplasmic functions [reviewed in 44], which could be amplified after inhibition of their nuclear import. Moreover, the cytoplasmic accumulation of class IIa HDACs is sometimes an indirect still uncertain effect of class I inhibition. For example the class I/II inhibitor LBH589, which is a SAHA derivate, confines HDAC4 in the cytoplasm in irradiated non-small cell lung cancer cells [87]. Considering all these drawbacks, the nucleus/cytoplasmic shuttling of class IIa HDACs seems to be the less druggable feature of these proteins.
Class IIa HDACs N-terminus, which allows their interaction with some partners, such as MEF2 family of TFs
As discussed above, class IIa HDACs’ N-terminal region (Fig. 1) mediates the interaction with multiple partners and contains a glutamine-rich domain (with the exception of HDAC7) that allows homo- and heterodimerization among the different class IIa members [12, 88]. The best-characterized class IIa transcriptional partners are the MEF2s proteins [49, 89]. Several of the biological functions attributed to class IIa HDACs are the results of the MEF2s transcriptional repression [27, 37, 38]. The phenotype of the single knock-out of class IIa HDACs could be explained as the effect of MEF2 over-activation in bone (HDAC4), heart (HDAC5/9) and cardiovascular system (HDAC7), in relation to the district in which the single HDACs are more abundant [90–92]. Hence, the design of an inhibitor that displaces class IIa HDACs from MEF2s could be a good approach to selectively interfere with this specific repressive exploit. A limitation to this strategy concerns the promiscuity of the class IIa HDACs sequence required for this interaction (aa 166-184 in HDAC4). In fact, this stretch of amino acids is also involved in the interaction with additional partners, among which, the nucleoporin Nup155 [93] and the demethylase JARID1B [94] (Fig. 1). An alternative plan to influence the MEF2-HDAC axis could be targeting the region of MEF2s that interacts with class IIa HDACs. Using this approach, BML-210 (Figs. 3, 4, 5), a weak class I HDAC benzamide inhibitor, was found to interact through its aminophenyl-group with the hydrophobic residues of MEF2s (aa 66-69) thus displacing class IIa HDACs [95]. Using the crystal structure of the HDAC9–MEF2B complex as a guide [96], authors generated a panel of more powerful BML-210 derivatives. In the next future it will be important to further improve the specificity of these compounds to exclude residual targeting of class I HDACs.
Conclusions and perspectives
The identification of molecules that could reset the transcriptional profile in neoplastic cells has raised many hopes for new anti-cancer therapies [97]. Unfortunately today this goal has been only partially reached. Nevertheless an epigenetic therapy against cancer is still subject of intense research. A new impetus in this field was given by the discovery of the demethylases [98, 99] and the synthesis of their specific inhibitors [100]. A more niche-research concerns class IIa HDACs and their selective inhibitors, which are hypothesized to be less powerful than pan-HDACis but more specific. However, these studies are still in their infancy and the applicability of class IIa HDACis in clinic requires still intense laboratory characterization. Additional experiments and data are mandatory to characterize and understand the contribution of these molecules to epigenetic changes in vivo. Up to now, information about the impact of class IIa HDACis on RNA non-coding world and the role of class IIa HDACs in stemness maintenance are very limited [101]. In parallel the efforts trying to design, isolate and characterize new compounds, acting as epigenetic regulators must persist. In addition, a robust in vitro pre-clinical characterization of molecules already available is needed to define: their molecular mechanism of action, their ideal context of utilization and off-targets effects. All these efforts are justified by the benefits that drug-induced genetic reprogramming could exert on different diseases.
Certainly anti-cancer therapy is the first and most important scope. Nevertheless, the involvement of class IIa HDACs in the regulation of Glut4 [102–105], of the NF-kB pathway [106, 107] and of many neuronal activities [108–111] could stimulate studies about the employment of class IIa HDACis for the treatment of diseases other than cancer, such as diabetes [112], neurodegenerative disorders [113, 114] and inflammatory diseases [26, 115–118]. There are opportunities out there; we just have to find out what is the best compound for each specific application.
Acknowledgments
Research activities were supported by FIRB (Progetto RBAP11S8C3_002), PRIN (Progetto 2010W4J4RM_002) and AIRC to CB.
References
- 1.Gräff J, Tsai L-H. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 2.Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 3.Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011;68:1681–1702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mihaylova MM, Vasquez DS, Ravnskjaer K, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X-J, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delcuve G, Khan D, Davie J. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Micco R, Sulli G, Dobreva M, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 11.Henderson C, Brancolini C. Apoptotic pathways activated by histone deacetylase inhibitors: implications for the drug-resistant phenotype. Drug Resist Updat. 2003;6:247–256. doi: 10.1016/s1368-7646(03)00067-0. [DOI] [PubMed] [Google Scholar]
- 12.Clocchiatti A, Florean C, Brancolini C. Class IIa HDACs: from important roles in differentiation to possible implications in tumourigenesis. J Cell Mol Med. 2011;15:1833–1846. doi: 10.1111/j.1582-4934.2011.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aygün O, Mehta S, Grewal SIS. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol. 2013;20:547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prince HM, Bishton MJ, Harrison SJ. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. 2009;15:3958–3969. doi: 10.1158/1078-0432.CCR-08-2785. [DOI] [PubMed] [Google Scholar]
- 15.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009;280:211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand P. Inside HDAC with HDAC inhibitors. Eur J Med Chem. 2010;45:2095–2116. doi: 10.1016/j.ejmech.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 17.Giannini G, Cabri W, Fattorusso C, Rodriquez M. Histone deacetylase inhibitors in the treatment of cancer: overview and perspectives. Future Med Chem. 2012;4:1439–1460. doi: 10.4155/fmc.12.80. [DOI] [PubMed] [Google Scholar]
- 18.Thaler F, Mercurio C. Towards selective inhibition of histone deacetylase isoforms: what has been achieved, where we are and what will be next. Chem Med Chem. 2014;9:523–536. doi: 10.1002/cmdc.201300413. [DOI] [PubMed] [Google Scholar]
- 19.Dekker FJ, van den Bosch T, Martin NI. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov Today. 2014;19:654–660. doi: 10.1016/j.drudis.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 21.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Zhong Q. Histone deacetylase inhibitors and cell death. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischle W, Dequiedt F, Hendzel MJ, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 25.Lahm A, Paolini C, Pallaoro M, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobera M, Madauss KP, Pohlhaus DT, et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol. 2013;9:319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 27.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–5467. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014;6:139–150. doi: 10.2217/epi.13.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuetz A, Min J, Allali-Hassani A, et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J Biol Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vannini A, Volpari C, Filocamo G, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci USA. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bottomley MJ, Lo Surdo P, Di Giovine P, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci USA. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Huang EY, Zhang J, Miska EA, et al. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 35.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischle W, Dequiedt F, Fillion M, et al. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 37.Clocchiatti A, Di Giorgio E, Ingrao S, et al. Class IIa HDACs repressive activities on MEF2-depedent transcription are associated with poor prognosis of ER+ breast tumors. FASEB J. 2013;27:942–954. doi: 10.1096/fj.12-209346. [DOI] [PubMed] [Google Scholar]
- 38.Di Giorgio E, Clocchiatti A, Piccinin S, et al. MEF2 is a converging hub for HDAC4 and PI3K/Akt-induced transformation. Mol Cell Biol. 2013;33:4473–4491. doi: 10.1128/MCB.01050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, McKinsey T, Olson E. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finnin MS, Donigian JR, Cohen A, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 41.Yang W-M, Tsai S-C, Wen Y-D, et al. Functional domains of histone deacetylase-3. J Biol Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- 42.Ago T, Liu T, Zhai P, et al. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 43.Isaacs JT, Antony L, Dalrymple SL, et al. Tasquinimod is an allosteric modulator of HDAC4 survival signaling within the compromised cancer microenvironment. Cancer Res. 2013;73:1386–1399. doi: 10.1158/0008-5472.CAN-12-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clocchiatti A, Di Giorgio E, Demarchi F, Brancolini C. Beside the MEF2 axis: unconventional functions of HDAC4. Cell Signal. 2013;25:269–276. doi: 10.1016/j.cellsig.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Cohen TJ, Barrientos T, Hartman ZC, et al. The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J. 2009;23:99–106. doi: 10.1096/fj.08-115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moresi V, Williams AH, Meadows E, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi MC, Cohen TJ, Barrientos T, et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell. 2012;47:122–132. doi: 10.1016/j.molcel.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Cavanaugh JE, Wang Y, et al. ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc Natl Acad Sci USA. 2003;100:8532–8537. doi: 10.1073/pnas.1332804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grégoire S, Tremblay AM, Xiao L, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- 50.Grégoire S, Xiao L, Nie J, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess A, Ruefli A, Beamish H, et al. Histone deacetylase inhibitors specifically kill nonproliferating tumour cells. Oncogene. 2004;23:6693–6701. doi: 10.1038/sj.onc.1207893. [DOI] [PubMed] [Google Scholar]
- 52.Bolden JE, Shi W, Jankowski K, et al. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis. 2013;4:e519. doi: 10.1038/cddis.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vrana JA, Decker RH, Johnson CR, et al. Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene. 1999;18:7016–7025. doi: 10.1038/sj.onc.1203176. [DOI] [PubMed] [Google Scholar]
- 54.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gammoh N, Lam D, Puente C, et al. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci. 2012;109:6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henderson C, Mizzau M, Paroni G, et al. Role of caspases, Bid, and p53 in the apoptotic response triggered by histone deacetylase inhibitors trichostatin-A (TSA) and suberoylanilide hydroxamic acid (SAHA) J Biol Chem. 2003;278:12579–12589. doi: 10.1074/jbc.M213093200. [DOI] [PubMed] [Google Scholar]
- 57.Maeda T, Towatari M, Kosugi H, Saito H. Up-regulation of costimulatory/adhesion molecules by histone deacetylase inhibitors in acute myeloid leukemia cells. Blood. 2000;96:3847–3856. [PubMed] [Google Scholar]
- 58.Magner WJ, Kazim AL, Stewart C, et al. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol. 2000;165:7017–7024. doi: 10.4049/jimmunol.165.12.7017. [DOI] [PubMed] [Google Scholar]
- 59.Rossig L, Li H, Fisslthaler B, et al. Inhibitors of histone deacetylation downregulate the expression of endothelial nitric oxide synthase and compromise endothelial cell function in vasorelaxation and angiogenesis. Circ Res. 2002;91:837–844. doi: 10.1161/01.res.0000037983.07158.b1. [DOI] [PubMed] [Google Scholar]
- 60.Deroanne CF, Bonjean K, Servotte S, et al. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002;21:427–436. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- 61.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 62.Wilson AJ, Byun D-S, Nasser S, et al. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell. 2008;19:4062–4075. doi: 10.1091/mbc.E08-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mottet D, Pirotte S, Lamour V, et al. HDAC4 represses p21(WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene. 2009;28:243–256. doi: 10.1038/onc.2008.371. [DOI] [PubMed] [Google Scholar]
- 64.Rad R, Rad L, Wang W, et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hohl M, Wagner M, Reil J, et al. HDAC4 controls histone methylation in response to elevated cardiac load. J Clin Invest. 2013;123:1359–1370. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones P, Altamura S, De Francesco R, et al. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008;18:1814–1819. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 67.Jones P, Bottomley MJ, Carfí A, et al. 2-Trifluoroacetylthiophenes, a novel series of potent and selective class II histone deacetylase inhibitors. Bioorg Med Chem Lett. 2008;18:3456–3461. doi: 10.1016/j.bmcl.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 68.Bradner JE, West N, Grachan ML, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sternson SM, Wong JC, Grozinger CM, Schreiber SL. Synthesis of 7200 small molecules based on a substructural analysis of the histone deacetylase inhibitors trichostatin and trapoxin. Org Lett. 2001;3:4239–4242. doi: 10.1021/ol016915f. [DOI] [PubMed] [Google Scholar]
- 70.Henkes LM, Haus P, Jäger F, et al. Synthesis and biochemical analysis of 2,2,3,3,4,4,5,5,6,6,7,7-dodecafluoro-N-hydroxy-octanediamides as inhibitors of human histone deacetylases. Bioorg Med Chem. 2012;20:985–995. doi: 10.1016/j.bmc.2011.11.041. [DOI] [PubMed] [Google Scholar]
- 71.Marek L, Hamacher A, Hansen FK, et al. Histone deacetylase (HDAC) inhibitors with a novel connecting unit linker region reveal a selectivity profile for HDAC4 and HDAC5 with improved activity against chemoresistant cancer cells. J Med Chem. 2013;56:427–436. doi: 10.1021/jm301254q. [DOI] [PubMed] [Google Scholar]
- 72.Fleming CL, Ashton TD, Gaur V, et al. Improved synthesis and structural reassignment of MC1568: a class IIa selective HDAC inhibitor. J Med Chem. 2014;57:1132–1135. doi: 10.1021/jm401945k. [DOI] [PubMed] [Google Scholar]
- 73.Mai A, Massa S, Pezzi R, et al. Class II (IIa)-selective histone deacetylase inhibitors. 1. Synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J Med Chem. 2005;48:3344–3353. doi: 10.1021/jm049002a. [DOI] [PubMed] [Google Scholar]
- 74.Duong V, Bret C, Altucci L, et al. Specific activity of class II histone deacetylases in human breast cancer cells. Mol Cancer Res. 2008;6:1908–1919. doi: 10.1158/1541-7786.MCR-08-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nebbioso A, Dell’Aversana C, Bugge A, et al. HDACs class II-selective inhibition alters nuclear receptor-dependent differentiation. J Mol Endocrinol. 2010;45:219–228. doi: 10.1677/JME-10-0043. [DOI] [PubMed] [Google Scholar]
- 76.Venza I, Visalli M, Oteri R, et al. Class II-specific histone deacetylase inhibitors MC1568 and MC1575 suppress IL-8 expression in human melanoma cells. Pigment Cell Melanoma Res. 2013;26:193–204. doi: 10.1111/pcmr.12049. [DOI] [PubMed] [Google Scholar]
- 77.Kahali B, Gramling SJ, Marquez SB, et al. Identifying targets for the restoration and reactivation of BRM. Oncogene. 2014;33:653–664. doi: 10.1038/onc.2012.613. [DOI] [PubMed] [Google Scholar]
- 78.Nebbioso A, Manzo F, Miceli M, et al. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009;10:776–782. doi: 10.1038/embor.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ontoria JM, Altamura S, Di Marco A, Ferrigno F, Laufer R, Muraglia E, Palumbi MC, Rowley M, Scarpelli R, Schultz-Fademrecht C, Serafini S, Steinkühler CJP. Identification of novel, selective, and stable inhibitors of class II histone deacetylases. Validation studies of the inhibition of the enzymatic activity of HDAC4 by small molecules as a novel approach for cancer therapy. J Med Chem. 2009;52:6782–6789. doi: 10.1021/jm900555u. [DOI] [PubMed] [Google Scholar]
- 80.Tessier P, Smil DV, Wahhab A, et al. Diphenylmethylene hydroxamic acids as selective class IIa histone deacetylase inhibitors. Bioorg Med Chem Lett. 2009;19:5684–5688. doi: 10.1016/j.bmcl.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 81.Haus P, Korbus M, Schröder M, Meyer-Almes F-J. Identification of selective class II histone deacetylase inhibitors using a novel dual-parameter binding assay based on fluorescence anisotropy and lifetime. J Biomol Screen. 2011;16:1206–1216. doi: 10.1177/1087057111424605. [DOI] [PubMed] [Google Scholar]
- 82.Olsson A, Björk A, Vallon-Christersson J, et al. Tasquinimod (ABR-215050), a quinoline-3-carboxamide anti-angiogenic agent, modulates the expression of thrombospondin-1 in human prostate tumors. Mol Cancer. 2010;9:107. doi: 10.1186/1476-4598-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dalrymple SL, Becker RE, Zhou H, et al. Tasquinimod prevents the angiogenic rebound induced by fractionated radiation resulting in an enhanced therapeutic response of prostate cancer xenografts. Prostate. 2012;72:638–648. doi: 10.1002/pros.21467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Corzo CA, Condamine T, Lu L, et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207:2439–2453. doi: 10.1084/jem.20100587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kong Y, Jung M, Wang K, et al. Histone deacetylase cytoplasmic trapping by a novel fluorescent HDAC inhibitor. Mol Cancer Ther. 2011;10:1591–1599. doi: 10.1158/1535-7163.MCT-10-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Li N, Caron C, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geng L, Cuneo KC, Fu A, et al. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of gamma-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer Res. 2006;66:11298–11304. doi: 10.1158/0008-5472.CAN-06-0049. [DOI] [PubMed] [Google Scholar]
- 88.Guo L, Han A, Bates DL, et al. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc Natl Acad Sci USA. 2007;104:4297–4302. doi: 10.1073/pnas.0608041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miska EA, Karlsson C, Langley E, et al. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vega RB, Matsuda K, Oh J, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 91.Chang S, McKinsey TA, Zhang CL, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang S, Young BD, Li S, et al. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 93.Kehat I, Accornero F, Aronow BJ, Molkentin JD. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol. 2011;193:21–29. doi: 10.1083/jcb.201101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barrett A, Santangelo S, Tan K, et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer. 2007;121:265–275. doi: 10.1002/ijc.22673. [DOI] [PubMed] [Google Scholar]
- 95.Jayathilaka N, Han A, Gaffney KJ, et al. Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2. Nucleic Acids Res. 2012;40:5378–5388. doi: 10.1093/nar/gks189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Han A, He J, Wu Y, et al. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J Mol Biol. 2005;345:91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 97.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 98.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 99.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 100.Hoffmann I, Roatsch M, Schmitt ML, et al. The role of histone demethylases in cancer therapy. Mol Oncol. 2012;6:683–703. doi: 10.1016/j.molonc.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhuang Q, Qing X, Ying Y, et al. Class IIa histone deacetylases and myocyte enhancer factor 2 proteins regulate the mesenchymal-to-epithelial transition of somatic cell reprogramming. J Biol Chem. 2013;288:12022–12031. doi: 10.1074/jbc.M113.460766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McGee SL, van Denderen BJW, Howlett KF, et al. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 103.Sparling DP, Griesel BA, Weems J, Olson AL. GLUT4 enhancer factor (GEF) interacts with MEF2A and HDAC5 to regulate the GLUT4 promoter in adipocytes. J Biol Chem. 2008;283:7429–7437. doi: 10.1074/jbc.M800481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weems J, Olson AL. Class II histone deacetylases limit GLUT4 gene expression during adipocyte differentiation. J Biol Chem. 2011;286:460–468. doi: 10.1074/jbc.M110.157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Raichur S, Hooi Teh S, Ohwaki K, et al. Histone deacetylase 5 regulates glucose uptake and insulin action in muscle cells. J Mol Endocrinol. 2012;49:203–211. doi: 10.1530/JME-12-0095. [DOI] [PubMed] [Google Scholar]
- 106.Kumar A, Lin Z, SenBanerjee S, Jain MK. Tumor necrosis factor alpha-mediated reduction of KLF2 is due to inhibition of MEF2 by NF-kappaB and histone deacetylases. Mol Cell Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baek Y-S, Haas S, Hackstein H, et al. Identification of novel transcriptional regulators involved in macrophage differentiation and activation in U937 cells. BMC Immunol. 2009;10:18. doi: 10.1186/1471-2172-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bolger TA, Zhao X, Cohen TJ, et al. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J Biol Chem. 2007;282:29186–29192. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 109.Morrison BE, Majdzadeh N, D’Mello SR. Histone deacetylases: focus on the nervous system. Cell Mol Life Sci. 2007;64:2258–2269. doi: 10.1007/s00018-007-7035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.West AE. Regulated shuttling of the histone deacetylase HDAC5 to the nucleus may put a brake on cocaine addiction. Neuron. 2012;73:1–3. doi: 10.1016/j.neuron.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 111.Li J, Chen J, Ricupero CL, et al. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med. 2012;18:783–790. doi: 10.1038/nm.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hara N, Alkanani AK, Dinarello CA, Zipris D. Histone deacetylase inhibitor suppresses virus-induced proinflammatory responses and type 1 diabetes. J Mol Med. 2014;92:93–102. doi: 10.1007/s00109-013-1078-1. [DOI] [PubMed] [Google Scholar]
- 113.Herman D, Jenssen K, Burnett R, et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 114.Burli RW, Luckhurst CA, Aziz O, et al. Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington’s disease. J Med Chem. 2013;56:9934–9954. doi: 10.1021/jm4011884. [DOI] [PubMed] [Google Scholar]
- 115.Tao R, de Zoeten EF, Ozkaynak E, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 116.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de Zoeten EF, Wang L, Sai H, et al. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010;138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bantscheff M, Hopf C, Savitski MM, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]